Abstract
Trace amine-associated receptors (TAARs) are G Protein-Coupled Receptors that function as vertebrate olfactory receptors. Like odorant receptors, TAARs constitute an ever-evolving sensory subsystem, with individual TAARs recognizing particular chemicals and some evoking stereotyped behaviors. Several TAARs mediate aversion or attraction towards volatile amines that include the mouse odor trimethylamine, the predator odor 2-phenylethylamine, and the death-associated odor cadaverine. TAAR-expressing sensory neurons achieve monoallelic receptor expression, use canonical olfactory signaling molecules, and target a dedicated olfactory bulb region. In mouse, TAAR4 and TAAR5 are encoded by adjacent genes and localize to adjacent glomeruli, yet mediate opposing behaviors. Future studies are needed to understand how TAAR-expressing sensory neurons engage higher-order neural circuits to encode odor valence.
Introduction
Trace amine-associated receptors (TAARs) were discovered in 2001 [1,2], and initially proposed to recognize low abundance neurotransmitters termed trace amines [3]. This early model of TAAR function, for which these receptors were named, was largely based on studies involving TAAR1. Subsequent work revealed a functional dichotomy in the TAAR family, with all other TAARs instead serving predominantly or exclusively as chemosensory receptors in the main olfactory system [4]. Much progress has since been made to understand TAAR signaling- from detection of odors to production of behaviors.
The TAAR family
TAARs are distantly related to biogenic amine receptors and not phylogenetically related to canonical odorant receptors (ORs) or other chemosensory receptors. The TAAR repertoire varies between species, with 15 full-length TAARs in mouse (mTAARs), 17 in rat (rTAARs), 6 in macaque (macTAARs), and 6 in human (hTAARs) [5–8]. In these mammals, Taar genes are found in a single genomic cluster and, except for Taar2, contain one translated exon. Mammalian TAAR orthologs generally display sequence conservation across species, unlike vomeronasal receptors that are rapidly diverging [5]. In teleost fish, the TAAR family is larger, more variable, and encoded by genes on multiple chromosomes [6,9,10]. There are 48 full-length TAARs in stickleback, 27 in salmon, 25 in medaka, 18 in pufferfish, and 112 in zebrafish (zTAARs), with most fish TAARs part of a unique clade that arose in teleosts [9–11]. While the precise timing is debated, TAARs likely originated early in vertebrates, around the emergence of jawed fish [10–12].
TAAR Expression
All TAARs except TAAR1 function as olfactory receptors, based on studies in rodent, primate, and fish [4,7,10]. TAAR expression is highly enriched in the olfactory system by quantitative PCR (qPCR) analysis, with little or no expression in other tissues examined [4]. In the nose, TAARs display characteristic hallmarks of OR expression: confinement to sparse sensory neurons (~0.1%) and distribution within a spatial zone (Figure 1) [4]. TAARs are also expressed in the neonatal Grueneberg ganglion [13], a distinct olfactory substructure, but not the vomeronasal organ [4].
Figure 1. TAAR expression in the olfactory system.
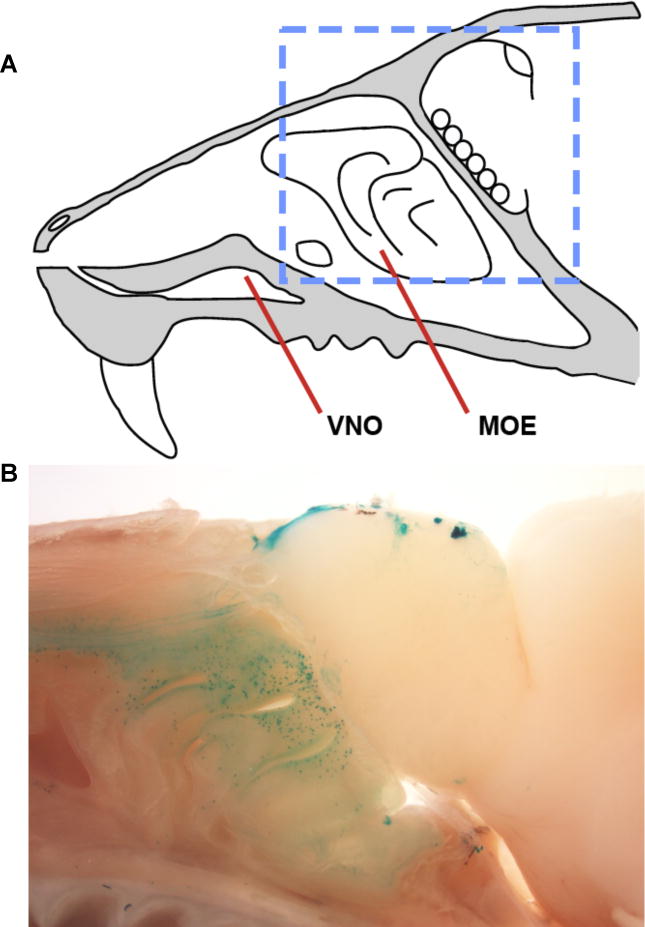
(A) Anatomy of the mouse nose in sagittal view, with major chemosensory structures displayed (VNO- vomeronasal organ; MOE-main olfactory epithelium. (B) A sagittal section of olfactory tissue with TAAR-expressing neurons genetically labeled. Images are derived from TAAR5 knockout mice, in which β-galactosidase is expressed from the endogenous Taar5 locus. Labeled neurons include a subset of all TAAR neuron types (see discussion of receptor re-selection following Taar pseudogene expression), and are visualized by wholemount X-gal staining. The imaged region is indicated in panel A by the blue dashed box. Multiple TAAR-innervating glomeruli are observed in the olfactory bulb, as shown in Figure 4A. Image in panel A is adapted from [50].
Each Taar allele defines a unique sensory neuron population that does not express other Taars or Ors [4,14,15]. The epigenetic signature observed on silenced Or genes is absent from Taar genes, suggesting a different gene choice mechanism [14]. Neurons selecting a Taar pseudogene express a second receptor with bias towards re-selection of a second Taar gene [14,15], raising the possibility that TAAR neurons undergo committed cell fate determination prior to receptor choice. Furthermore, the second Taar allele is usually chosen from the other chromosome [15], suggesting blockade of the originally chosen cluster as occurs during vomeronasal receptor re-selection [16]. Cell fate markers, transcription factors, locus control regions, and epigenetic modifications that contribute to TAAR neuron specification are unknown.
Within sensory neurons, TAAR proteins are localized to olfactory cilia, the site of odor detection, and to axons, where they may participate in axon guidance [14]. TAARs and ORs likely engage similar intracellular pathways, as TAAR neurons express Gαolf and other canonical olfactory signaling molecules [4,17]. Furthermore, activated TAARs couple to cAMP pathways in heterologous cells [4,18], and pharmacological inhibition of adenylyl cyclase blocks odor responses in TAAR neurons [17].
Uniquely, TAAR1 does not function as an olfactory receptor in mice, macaque, human, or zebrafish [4,7,10]. TAAR1 expression is reported in many cell types [1–3], although levels are generally quite low as qPCR analysis across various tissues only detects expression in pancreas [19]. Knock-in mice containing a β-galactosidase reporter at the endogenous Taar1 locus revealed reporter expression in rare neurons, including midbrain dopamine neurons and brainstem serotonin neurons [20]. The TAAR family displays a functional dichotomy reminiscent of the mouse formyl peptide receptor family, which includes different receptors for innate immunity and vomeronasal chemoreception [21,22].
TAAR ligands and molecular recognition properties
TAARs share homology with biogenic amine receptors, for which the biochemical and structural basis of ligand recognition is well understood [23,24]. Biogenic amine receptors recognize amines through a key salt bridge involving a conserved transmembrane three aspartic acid (Asp3.32; Ballesteros-Weinstein indexing) [24]. Asp3.32 is retained in all hTAARs (6/6), most mTAARs (13/15), and some zTAARs (27/112), suggesting that many TAARs would retain amine recognition capacity. High throughput chemical screens and medicinal chemistry approaches revealed ligands for TAAR1, TAAR3, TAAR4, TAAR5, TAAR7s, TAAR8s, TAAR9, and TAAR13c [1,2,4,18,25,26]. Each of these TAARs detects amines, with TAAR1 detecting various biogenic amines and olfactory TAARs detecting volatile odors, some also natural products (Figure 2).
Figure 2. TAAR ligands, expression patterns, and behavioral roles.
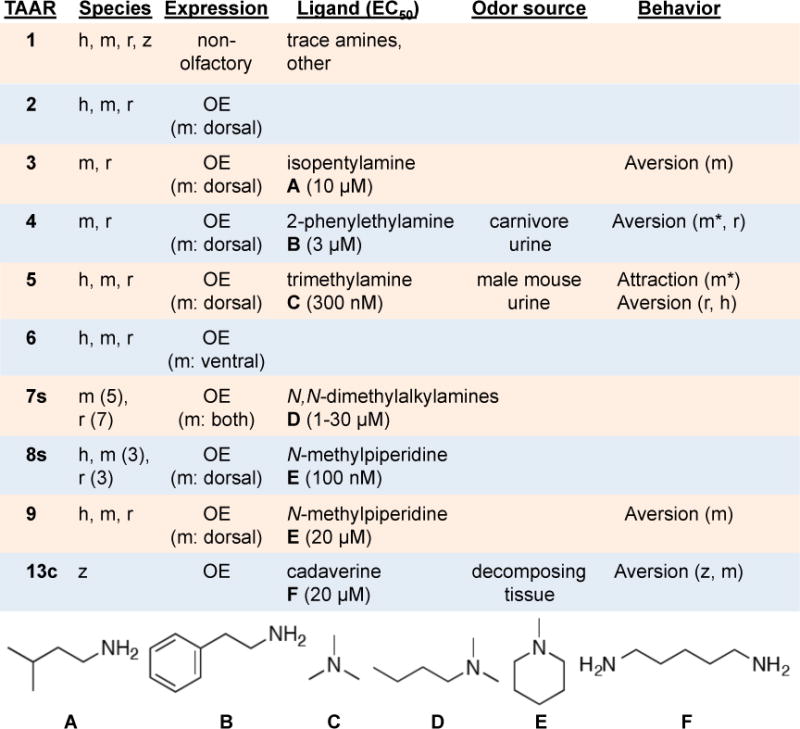
Human (h), mouse (m), rat (r), and/or zebrafish (z) genomes contain genes from indicated TAAR subfamilies. Mice and rats have multiple TAAR7s and TAAR8s, as indicated in parenthesis. All mammalian subfamilies, but not all teleost subfamilies, are depicted. All TAARs except TAAR1 are expressed in olfactory epithelium (OE), and in mouse, all TAARs are located dorsally except TAAR6, TAAR7a, and TAAR7b. The identities, ecological sources, and evoked behavioral responses of TAAR ligands are shown (*behavioral response lost in TAAR knockout mice, EC50 determined in HEK-293 cells).
TAAR1
TAAR1 functions in the brain to regulate neuron excitability and neurotransmitter usage [3,20,27]. TAAR1 agonists include putative trace amines (tyramine, octopamine, tryptamine, 2-phenylethylamine), psychoactive chemicals (amphetamines), thyronamines, and other amines [1,2,4,26,28]. These chemicals exert various physiological effects, presumably through TAAR1-dependent and TAAR1-independent mechanisms; for example, thryonamine-induced hypothermia persists in TAAR1 knockout mice [29]. Synthetic agonists and antagonists with improved specificity for TAAR1 acutely modulate aminergic neuron firing with effects eliminated in TAAR1 knockout mice [30,31]. Furthermore, aminergic neurons fire at elevated rates in TAAR1 knockout mice [20], with TAAR1 inhibitory effects mediated through Gαs and inwardly rectifying potassium channels [30].
Olfactory TAARs
Ligands for olfactory TAARs are small, volatile chemicals. Agonists for 6 mTAARs, 7 rTAARs, 1 zTAAR, 1 macTAAR, and 1 hTAAR were discovered using a heterologous expression system based on cAMP-dependent reporter induction [4,7,18,32]. These 16 olfactory TAARs detect different volatile amines, with affinities (0.1–10 μM) like OR affinities in similar assays. Amines that activate TAAR3 (isopentylamine), TAAR4 (2-phenylethylamine), and TAAR13c (cadaverine) include primary amines derived from natural amino acids by decarboxylation [4,25]. Other TAARs prefer tertiary amines, including TAAR5 (trimethylamine), TAAR7s (N,N-dimethylalkylamines), TAAR8s (N-methylpiperidine), and TAAR9 (N-methylpiperidine) [4,18]. TAARs that detect primary or tertiary amines form different clades on the TAAR phylogenetic tree (Figure 3) [18]. A third TAAR clade arose in teleosts with widespread mutation of amine-contacting Asp3.32 and it is unclear what they detect [10]. TAAR-expressing olfactory sensory neurons recognize the same amines identified in cell culture experiments [17,33], but with enhanced sensitivity [17]. Coupling to downstream signaling components is presumably optimized in olfactory sensory neurons so activation of a small number of receptors is sufficient to cause neuron depolarization.
Figure 3. Mapping odor preferences on the mammalian TAAR phylogeny.
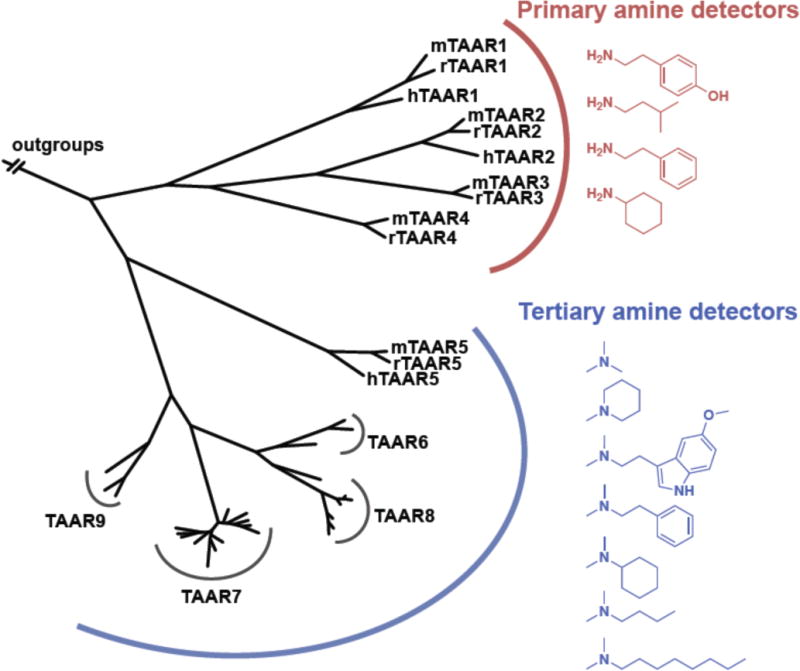
Two branches of the TAAR phylogenetic tree prefer primary amines (TAARs 1–4) and tertiary amines (TAARs 5–9). zTAAR13c is not depicted but clusters with primary amine detectors. A third major TAAR clade evolved in teleosts (not shown), with almost all losing the canonical Asp3.32 amine recognition site. Image adapted from [18].
When functional expression is achieved, TAAR orthologs from different species recognize similar ligands, although sensitivities vary [7,18,32,33]. An exception is the rapidly evolving TAAR7 subfamily, where recent mutations changed ligand recognition properties [18]. The mouse TAAR7 lineage provides direct evidence for functional expansion of the olfactory receptor repertoire through a pattern of gene duplication and subsequent mutation.
Modeling TAAR-ligand interactions
Structural models were generated for agonist-bound TAAR1, TAAR7e, and TAAR7f based on crystallographic data for the β-adrenergic receptor [18,28,34]. These modeled receptors contain the canonical seven transmembrane α-helical fold and other conserved features of rhodopsin-like GPCRs. Modeling and mutational analysis predict ligand binding to occur in the membrane plane through an Asp3.32 salt bridge and other ligand contacts largely distributed across transmembrane α-helices. Ligand selectivity differences between TAAR7e and TAAR7f, highly related receptors with distinct response profiles, are determined by two adjacent transmembrane 3 residues (3.37, 3.38) [18]. In TAAR7e, Van der Waals interactions between the ligand and Ser3.37 are a key selectivity determinant, with the bulkier side chain of Tyr3.37 in TAAR7f sterically limiting ligand size [18].
TAARs that recognize ethological odors
Several TAARs detect natural odors derived from urine, microbial metabolism, and other ecological sources. Urine directly activates mTAAR4, mTAAR5, mTAAR7f, rTAAR8c, and rTAAR9 [4,33,35], with mTAAR4 and mTAAR5 detecting species-specific metabolites (see below). Agonists for mTAAR3 (isopentylamine) and mTAAR7e (5-methoxy-N,N-dimethyltryptamine) are also reported in urine [36,37]. Isopentylamine is a biogenic amine produced by leucine decarboxylation and its ecological salience is unclear, as it is innately aversive to rodents [38]. zTAAR13c and TAAR5 detect microbially derived amines emitted in decomposing flesh (see below) [7,25].
TAAR4
TAAR4 detects a carnivore odor that repels rodents [35]. Chemical fractionation of bobcat urine revealed 2-phenylethylamine to be the natural TAAR4 agonist, and quantitative analysis across 38 mammalian species indicated enriched production by carnivores, with some (lion, tiger, serval, jaguar) releasing >1,000-fold more than other animals [35]. Mice and rats innately avoid 2-phenylethylamine upon first exposure, and enzymatic depletion of 2-phenylethylamine from lion urine impairs its repellant properties [35]. Genetically defined TAAR4 olfactory sensory neurons display unusually low threshold 2-phenylethylamine responses [17], and knockout of TAAR4 eliminates avoidance of 2-phenylethylamine and bobcat urine [39]. Together, these findings provide evidence that mTAAR4 is a kairomone receptor. Interestingly, the Taar4 gene is conserved in many carnivores [40], and 2-phenylethylamine is proposed to be a tiger pheromone [41]. Perhaps 2-phenylethylamine and TAAR4 evoke different behaviors in predators and prey.
TAAR5
TAAR5 detects trimethylamine, an abundant and sexually dimorphic mouse odor that evokes species-specific behaviors [4,33]. TAAR5 detects male mouse urine with exquisite sensitivity due to high abundance of trimethylamine (5 mM) [4], and enzymatic depletion of trimethylamine from mouse urine abrogates responses [33]. Mice release >1,000-fold more trimethylamine than rats and some other rodents [33]. Recent evolutionary forces sculpted the Mus trimethylamine biosynthesis pathway, which involves both gut bacteria and host enzymes [33,42].
Trimethylamine is a byproduct of commensal microflora-mediated nutrient metabolism, and, in most species, catabolized by a host enzyme (FMO3) [42]. In mice, FMO3 expression is tightly repressed in kidney and male liver, leading to abundant and sexually dimorphic trimethylamine production [33]. Trimethylamine is also found in other microbial odor sources [7], so evoked perceptions may be context-dependent.
Presented alone, trimethylamine evokes species-specific behaviors. Rats and humans avoid trimethylamine [33,43], but mice display concentration-dependent responses with attraction to lower (urinary) levels and aversion to higher levels [33]. Perhaps predator aversion to trimethylamine provided the selective pressure for enriched production in mouse scent. TAAR5 knockout mice lose behavioral attraction to trimethylamine, and have decreased attraction to mouse scent [33]. However, avoidance of high trimethylamine persists in TAAR5 knockout mice [33], suggesting detection by another low-affinity receptor that mediates dominant aversion responses. Roles for another receptor are supported by TAAR5-independent trimethylamine anosmias in humans [32].
How does trimethylamine evoke species-specific behaviors? One possibility is that aversion-and attraction-mediating olfactory receptors display variable affinities across species. Mouse and rat TAAR5 have ~200-fold improved trimethylamine sensitivity compared with human and macaque orthologs [4,7,32,33], and perhaps an aversion-mediating olfactory receptor also has species-specific affinity. A second possibility, which does not invoke a second receptor, is that mouse and rat TAAR5 differentially couple to neural circuits associated with attraction and aversion. A third possibility is that mouse attraction to trimethylamine in scent marks involves learned override of a developmentally established aversion circuit. Neural systems that control behavioral responses to species-specific odors, such as TAAR4 and TAAR5 ligands, must be sufficiently dynamic across evolution to ensure species-appropriate behaviors.
TAAR13c
zTAAR13c detects rotting flesh odors through the lysine decomposition product cadaverine [25]. Structure-activity analysis reveals a preference for medium-sized, odd-chained diamines, and indicates a binding pocket with two distinct cation recognition sites [25]. Cadaverine activates zTAAR13c in olfactory sensory neurons, and evokes innate avoidance responses in zebrafish [25]. Cadaverine also repels mice, a behavior lost in TAAR cluster knockouts [39]. Identifying an mTAAR that recognizes cadaverine (mice lack a TAAR13 ortholog) would provide a valuable tool for mechanistic analysis of odor aversion behavior.
TAAR-activated neural circuits
Olfactory sensory neurons provide highly organized inputs to the olfactory bulb at specialized structures termed glomeruli [44,45]. TAAR-expressing sensory neurons converge on glomeruli of stereotyped position within a dorsomedial domain (Figure 4A) [14,15]. Interestingly, TAAR4 and TAAR5 neurons, which mediate different behaviors, target adjacent glomeruli (Figure 4B) [14]. Furthermore, neurons that initially express a Taar5 pseudogene can secondarily express Taar4 [14], suggesting that coupling to aversion or attraction circuitry may occur after receptor choice. In vivo calcium imaging revealed that TAAR glomeruli detect amines and natural odors previously identified in cell culture studies [39]. Amine-detecting glomeruli are also found in olfactory bulb regions that lack TAAR input [46], which makes sense since TAARs recognize some but not all amine odors. Interestingly, 2-phenylethylamine responses are sparser in the olfactory bulb [39] than olfactory epithelium [35], and of imaging-accessible glomeruli, are apparently confined to the TAAR4 glomerulus [39], perhaps due to lateral inhibition through intrinsic olfactory bulb circuits.
Figure 4. Projections of TAAR neurons in the brain.
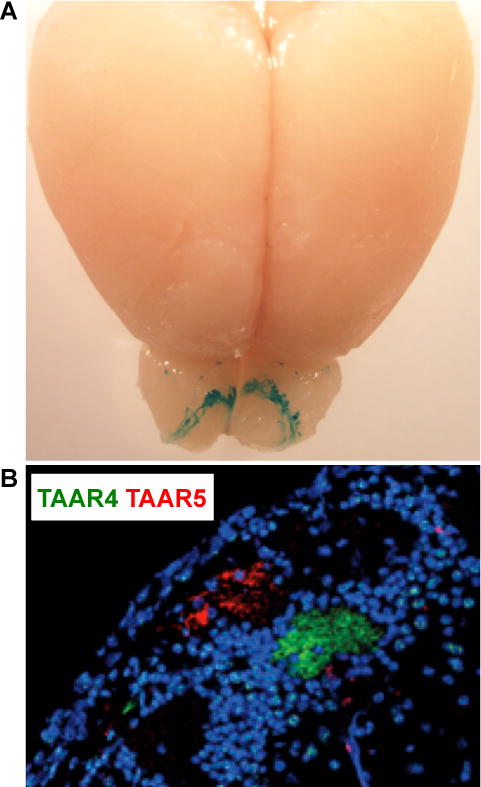
(A) Wholemount view of TAAR inputs in the olfactory bulb. Subsets of all TAAR-expressing neuron types were simultaneously visualized using genetic approaches, as described for Figure 1B. (B) Two-color immunohistochemistry for TAAR4 (green) and TAAR5 (red) labels adjacent glomeruli in the olfactory bulb. Image adapted from [14].
TAAR-activated neural circuits are uncharacterized beyond the olfactory bulb. Perhaps TAAR4 and TAAR5 provide stereotyped inputs to higher-order olfactory nuclei that mediate innate odor aversion and attraction behaviors, such as the cortical amygdala [47]. Future studies are generally needed to understand the structure and function of neural circuits that process odor valence, and how connections from particular classes of sensory neurons are specified [48].
Conclusion
TAARs, like ORs, provide an ever-changing landscape for sensory biology, with the TAAR repertoire expanding, contracting, and mutating across the phylogeny. Differently sized families of gustatory GPCRs mediate fundamentally distinct functions: bitter versus savory/sweet [49]. Several TAARs detect volatile and aversive amines, but the olfactory system is capable of discarding ligand-based or function-based constraints on TAAR evolution. Particular TAARs have mutated to recognize new ligands, with almost an entire teleost clade losing the canonical amine-recognition motif. Furthermore, while some TAARs detect aversive odors, TAAR-mediated behaviors can vary across species. Perhaps the tremendous flexibility inherent to olfactory system development allows for lineage-specific functional changes in the TAAR family. The ability of particular TAARs to mediate aversion and attraction behavior provides an exciting opportunity for mechanistic unraveling of odor valence encoding.
Acknowledgments
Figures 1B and 4A are unpublished images of Dheeraj Roy and SDL. This work was supported by an NIH grant (RO1DC013289).
References
- 1.Borowsky B, Adham N, Jones KA, Raddatz R, Artymyshyn R, Ogozalek KL, Durkin MM, Lakhlani PP, Bonini JA, Pathirana S, Boyle N, et al. Trace amines: Identification of a family of mammalian g protein-coupled receptors. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(16):8966–8971. doi: 10.1073/pnas.151105198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bunzow JR, Sonders MS, Arttamangkul S, Harrison LM, Zhang G, Quigley DI, Darland T, Suchland KL, Pasumamula S, Kennedy JL, Olson SB, et al. Amphetamine, 3,4-methylenedioxymethamphetamine, lysergic acid diethylamide, and metabolites of the catecholamine neurotransmitters are agonists of a rat trace amine receptor. Mol Pharmacol. 2001;60(6):1181–1188. doi: 10.1124/mol.60.6.1181. [DOI] [PubMed] [Google Scholar]
- 3.Lindemann L, Hoener MC. A renaissance in trace amines inspired by a novel gpcr family. Trends Pharmacol Sci. 2005;26(5):274–281. doi: 10.1016/j.tips.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 4.Liberles SD, Buck LB. A second class of chemosensory receptors in the olfactory epithelium. Nature. 2006;442(7103):645–650. doi: 10.1038/nature05066. [DOI] [PubMed] [Google Scholar]
- 5.Grus WE, Zhang J. Distinct evolutionary patterns between chemoreceptors of 2 vertebrate olfactory systems and the differential tuning hypothesis. Molecular biology and evolution. 2008;25(8):1593–1601. doi: 10.1093/molbev/msn107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hashiguchi Y, Nishida M. Evolution of trace amine associated receptor (taar) gene family in vertebrates: Lineage-specific expansions and degradations of a second class of vertebrate chemosensory receptors expressed in the olfactory epithelium. Molecular biology and evolution. 2007;24(9):2099–2107. doi: 10.1093/molbev/msm140. [DOI] [PubMed] [Google Scholar]
- 7*.Horowitz LF, Saraiva LR, Kuang D, Yoon KH, Buck LB. Olfactory receptor patterning in a higher primate. J Neurosci. 2014;34(37):12241–12252. doi: 10.1523/JNEUROSCI.1779-14.2014. This study described the macaque TAAR family, and found macTAAR5 to be activated by spoiled fish odor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nei M, Niimura Y, Nozawa M. The evolution of animal chemosensory receptor gene repertoires: Roles of chance and necessity. Nature reviews. 2008;9(12):951–963. doi: 10.1038/nrg2480. [DOI] [PubMed] [Google Scholar]
- 9.Hashiguchi Y, Furuta Y, Nishida M. Evolutionary patterns and selective pressures of odorant/pheromone receptor gene families in teleost fishes. PLoS One. 2008;3(12):e4083. doi: 10.1371/journal.pone.0004083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hussain A, Saraiva LR, Korsching SI. Positive darwinian selection and the birth of an olfactory receptor clade in teleosts. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(11):4313–4318. doi: 10.1073/pnas.0803229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tessarolo JA, Tabesh MJ, Nesbitt M, Davidson WS. Genomic organization and evolution of the trace amine-associated receptor (taar) repertoire in atlantic salmon (salmo salar) G3 (Bethesda) 2014;4(6):1135–1141. doi: 10.1534/g3.114.010660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Libants S, Carr K, Wu H, Teeter JH, Chung-Davidson YW, Zhang Z, Wilkerson C, Li W. The sea lamprey petromyzon marinus genome reveals the early origin of several chemosensory receptor families in the vertebrate lineage. BMC Evol Biol. 2009;9(180) doi: 10.1186/1471-2148-9-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fleischer J, Schwarzenbacher K, Breer H. Expression of trace amine-associated receptors in the grueneberg ganglion. Chemical senses. 2007;32(6):623–631. doi: 10.1093/chemse/bjm032. [DOI] [PubMed] [Google Scholar]
- 14.Johnson MA, Tsai L, Roy DS, Valenzuela DH, Mosley C, Magklara A, Lomvardas S, Liberles SD, Barnea G. Neurons expressing trace amine-associated receptors project to discrete glomeruli and constitute an olfactory subsystem. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(33):13410–13415. doi: 10.1073/pnas.1206724109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pacifico R, Dewan A, Cawley D, Guo C, Bozza T. An olfactory subsystem that mediates high-sensitivity detection of volatile amines. Cell Rep. 2012;2(1):76–88. doi: 10.1016/j.celrep.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roppolo D, Vollery S, Kan CD, Luscher C, Broillet MC, Rodriguez I. Gene cluster lock after pheromone receptor gene choice. EMBO J. 2007;26(14):3423–3430. doi: 10.1038/sj.emboj.7601782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Pacifico R, Cawley D, Feinstein P, Bozza T. Ultrasensitive detection of amines by a trace amine-associated receptor. J Neurosci. 2013;33(7):3228–3239. doi: 10.1523/JNEUROSCI.4299-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrero DM, Wacker D, Roque MA, Baldwin MW, Stevens RC, Liberles SD. Agonists for 13 trace amine-associated receptors provide insight into the molecular basis of odor selectivity. ACS Chem Biol. 2012;7(7):1184–1189. doi: 10.1021/cb300111e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Regard JB, Sato IT, Coughlin SR. Anatomical profiling of g protein-coupled receptor expression. Cell. 2008;135(3):561–571. doi: 10.1016/j.cell.2008.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindemann L, Meyer CA, Jeanneau K, Bradaia A, Ozmen L, Bluethmann H, Bettler B, Wettstein JG, Borroni E, Moreau JL, Hoener MC. Trace amine-associated receptor 1 modulates dopaminergic activity. J Pharmacol Exp Ther. 2008;324(3):948–956. doi: 10.1124/jpet.107.132647. [DOI] [PubMed] [Google Scholar]
- 21.Liberles SD, Horowitz LF, Kuang D, Contos JJ, Wilson KL, Siltberg-Liberles J, Liberles DA, Buck LB. Formyl peptide receptors are candidate chemosensory receptors in the vomeronasal organ. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(24):9842–9847. doi: 10.1073/pnas.0904464106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riviere S, Challet L, Fluegge D, Spehr M, Rodriguez I. Formyl peptide receptor-like proteins are a novel family of vomeronasal chemosensors. Nature. 2009;459(7246):574–577. doi: 10.1038/nature08029. [DOI] [PubMed] [Google Scholar]
- 23.Nygaard R, Zou Y, Dror RO, Mildorf TJ, Arlow DH, Manglik A, Pan AC, Liu CW, Fung JJ, Bokoch MP, Thian FS, et al. The dynamic process of beta(2)-adrenergic receptor activation. Cell. 2013;152(3):532–542. doi: 10.1016/j.cell.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi L, Javitch JA. The binding site of aminergic g protein-coupled receptors: The transmembrane segments and second extracellular loop. Annu Rev Pharmacol Toxicol. 2002;42:437–467. doi: 10.1146/annurev.pharmtox.42.091101.144224. [DOI] [PubMed] [Google Scholar]
- 25*.Hussain A, Saraiva LR, Ferrero DM, Ahuja G, Krishna VS, Liberles SD, Korsching SI. High-affinity olfactory receptor for the death-associated odor cadaverine. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(48):19579–19584. doi: 10.1073/pnas.1318596110. The authors identified TAAR13c as a receptor for cadaverine, an odor of putrefied tissue that is innately aversive to zebrafish. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scanlan TS, Suchland KL, Hart ME, Chiellini G, Huang Y, Kruzich PJ, Frascarelli S, Crossley DA, Bunzow JR, Ronca-Testoni S, Lin ET, et al. 3-iodothyronamine is an endogenous and rapid-acting derivative of thyroid hormone. Nat Med. 2004;10(6):638–642. doi: 10.1038/nm1051. [DOI] [PubMed] [Google Scholar]
- 27.Wolinsky TD, Swanson CJ, Smith KE, Zhong H, Borowsky B, Seeman P, Branchek T, Gerald CP. The trace amine 1 receptor knockout mouse: An animal model with relevance to schizophrenia. Genes Brain Behav. 2007;6(7):628–639. doi: 10.1111/j.1601-183X.2006.00292.x. [DOI] [PubMed] [Google Scholar]
- 28.Reese EA, Norimatsu Y, Grandy MS, Suchland KL, Bunzow JR, Grandy DK. Exploring the determinants of trace amine-associated receptor 1’s functional selectivity for the stereoisomers of amphetamine and methamphetamine. J Med Chem. 2014;57(2):378–390. doi: 10.1021/jm401316v. [DOI] [PubMed] [Google Scholar]
- 29.Panas HN, Lynch LJ, Vallender EJ, Xie Z, Chen GL, Lynn SK, Scanlan TS, Miller GM. Normal thermoregulatory responses to 3-iodothyronamine, trace amines and amphetamine-like psychostimulants in trace amine associated receptor 1 knockout mice. J Neurosci Res. 2010;88(9):1962–1969. doi: 10.1002/jnr.22367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bradaia A, Trube G, Stalder H, Norcross RD, Ozmen L, Wettstein JG, Pinard A, Buchy D, Gassmann M, Hoener MC, Bettler B. The selective antagonist epptb reveals taar1-mediated regulatory mechanisms in dopaminergic neurons of the mesolimbic system. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(47):20081–20086. doi: 10.1073/pnas.0906522106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Revel FG, Moreau JL, Gainetdinov RR, Bradaia A, Sotnikova TD, Mory R, Durkin S, Zbinden KG, Norcross R, Meyer CA, Metzler V, et al. Taar1 activation modulates monoaminergic neurotransmission, preventing hyperdopaminergic and hypoglutamatergic activity. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(20):8485–8490. doi: 10.1073/pnas.1103029108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wallrabenstein I, Kuklan J, Weber L, Zborala S, Werner M, Altmuller J, Becker C, Schmidt A, Hatt H, Hummel T, Gisselmann G. Human trace amine-associated receptor taar5 can be activated by trimethylamine. PLoS One. 2013;8(2):e54950. doi: 10.1371/journal.pone.0054950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33**.Li Q, Korzan WJ, Ferrero DM, Chang RB, Roy DS, Buchi M, Lemon JK, Kaur AW, Stowers L, Fendt M, Liberles SD. Synchronous evolution of an odor biosynthesis pathway and behavioral response. Curr Biol. 2013;23(1):11–20. doi: 10.1016/j.cub.2012.10.047. Knockout mice lacking TAAR5 lose attraction to the sexually dimorphic mouse odor trimethylamine, showing that an evolutionarily conserved olfactory receptor can mediate a species-specific behavior. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan ES, Naylor JC, Groban ES, Bunzow JR, Jacobson MP, Grandy DK, Scanlan TS. The molecular basis of species-specific ligand activation of trace amine-associated receptor 1 (taar(1)) ACS Chem Biol. 2009;4(3):209–220. doi: 10.1021/cb800304d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35**.Ferrero DM, Lemon JK, Fluegge D, Pashkovski SL, Korzan WJ, Datta SR, Spehr M, Fendt M, Liberles SD. Detection and avoidance of a carnivore odor by prey. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(27):11235–11240. doi: 10.1073/pnas.1103317108. This study found that TAAR4 detects 2-phenylethylamine, a carnivore odor that repels rodents. Based on this finding, the authors first proposed a role for some TAARs in innate avoidance behavior. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishimura K, Utsumi K, Yuhara M, Fujitani Y, Iritani A. Identification of puberty-accelerating pheromones in male mouse urine. J Exp Zool. 1989;251(3):300–305. doi: 10.1002/jez.1402510306. [DOI] [PubMed] [Google Scholar]
- 37.Sitaram BR, Lockett L, Blackman GL, McLeod WR. Urinary excretion of 5-methoxy-n,n-dimethyltryptamine, n,n-dimethyltryptamine and their n-oxides in the rat. Biochemical pharmacology. 1987;36(13):2235–2237. doi: 10.1016/0006-2952(87)90159-6. [DOI] [PubMed] [Google Scholar]
- 38.Kobayakawa K, Kobayakawa R, Matsumoto H, Oka Y, Imai T, Ikawa M, Okabe M, Ikeda T, Itohara S, Kikusui T, Mori K, et al. Innate versus learned odour processing in the mouse olfactory bulb. Nature. 2007;450(7169):503–508. doi: 10.1038/nature06281. [DOI] [PubMed] [Google Scholar]
- 39**.Dewan A, Pacifico R, Zhan R, Rinberg D, Bozza T. Non-redundant coding of aversive odours in the main olfactory pathway. Nature. 2013 doi: 10.1038/nature12114. Knockout mice lacking TAAR4 lose aversion to 2-phenylethylamine and bobcat urine, and knockout mice lacking all TAARs lose aversion and glomerular responses to some TAAR ligands. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Staubert C, Boselt I, Bohnekamp J, Rompler H, Enard W, Schoneberg T. Structural and functional evolution of the trace amine-associated receptors taar3, taar4 and taar5 in primates. PLoS One. 2010;5(6):e11133. doi: 10.1371/journal.pone.0011133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poddar-Sarkar M, Brahmachary RL. Pheromones of tiger and other big cats. 2014 [PubMed] [Google Scholar]
- 42.Bain MA, Fornasini G, Evans AM. Trimethylamine: Metabolic, pharmacokinetic and safety aspects. Curr Drug Metab. 2005;6(3):227–240. doi: 10.2174/1389200054021807. [DOI] [PubMed] [Google Scholar]
- 43.Mitchell SC, Smith RL. Trimethylaminuria: The fish malodor syndrome. Drug metabolism and disposition: the biological fate of chemicals. 2001;29(4 Pt 2):517–521. [PubMed] [Google Scholar]
- 44.Ressler KJ, Sullivan SL, Buck LB. Information coding in the olfactory system: Evidence for a stereotyped and highly organized epitope map in the olfactory bulb. Cell. 1994;79(7):1245–1255. doi: 10.1016/0092-8674(94)90015-9. [DOI] [PubMed] [Google Scholar]
- 45.Vassar R, Chao SK, Sitcheran R, Nunez JM, Vosshall LB, Axel R. Topographic organization of sensory projections to the olfactory bulb. Cell. 1994;79(6):981–991. doi: 10.1016/0092-8674(94)90029-9. [DOI] [PubMed] [Google Scholar]
- 46.Takahashi YK, Nagayama S, Mori K. Detection and masking of spoiled food smells by odor maps in the olfactory bulb. J Neurosci. 2004;24(40):8690–8694. doi: 10.1523/JNEUROSCI.2510-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Root CM, Denny CA, Hen R, Axel R. The participation of cortical amygdala in innate, odour-driven behaviour. Nature. 2014;515(7526):269–273. doi: 10.1038/nature13897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Q, Liberles SD. Aversion and attraction through olfaction. Current Biology. doi: 10.1016/j.cub.2014.11.044. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yarmolinsky DA, Zuker CS, Ryba NJ. Common sense about taste: From mammals to insects. Cell. 2009;139(2):234–244. doi: 10.1016/j.cell.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liberles SD. Mammalian pheromones. Annu Rev Physiol. 2014;76:151–175. doi: 10.1146/annurev-physiol-021113-170334. [DOI] [PMC free article] [PubMed] [Google Scholar]


