Abstract
Embryonic stem cells (ESCs) are characterized by their ability to self-renew and to differentiate into all cell types of a given organism. Understanding the molecular mechanisms that govern the ESC state is of great interest not only for basic research—for instance, ESCs represent a perfect system to study cellular differentiation in vitro—but also for their potential implications in human health, as these mechanisms are likewise involved in cancer progression and could be exploited in regenerative medicine. In this minireview, we focus on the latest insights into the molecular mechanisms mediated by the pluripotency factors as well as their roles during differentiation. We also discuss recent advances in understanding the function of the epigenetic regulators, Polycomb and MLL complexes, in ESC biology.
INTRODUCTION
Within 2 days after fertilization, a mouse oocyte has undergone a series of cellular divisions and has developed into the morula embryo. The totipotent cells within the morula then divide and further specialize to form the hollow blastocyst sphere. The outer layer of the blastocyst contains the trophectoderm cells, while the inner cell mass (ICM) contains the pluripotent embryonic stem cells (ESCs) that will give rise in the developing embryo to all cell types of the three germ layers—ectoderm, mesoderm, and endoderm. Mouse ESCs were first isolated by Evans and Kaufman in 1981 (1) and have since been extensively studied. Under proper cell culture conditions, ESCs can divide and self-renew indefinitely, yet under differentiation stimuli, ESCs can also differentiate into virtually all cell types of the organism.
Which molecular mechanisms control the decision of ESCs to self-renew or to differentiate? During the last decades, several transcription factors have been identified to be essential for ESC pluripotency. These transcription factors regulate pluripotency by a so-called “pluripotency network” that regulates their own expression and coregulates the expression of other key transcription factors through multiple mechanisms. Interestingly, pluripotency is controlled at the transcriptional levels of genes through specific signaling pathways and epigenetic factors.
Epigenetics is the study of heritable changes in gene expression that are not caused by changes at the DNA sequence level. The Polycomb and MLL (myeloid-lineage leukemia) complexes are two of the best-characterized epigenetic machineries implicated in ESC pluripotency and differentiation. Although pluripotency factors do not physically interact with Polycomb and MLL complexes, they coregulate lineage-specific genes important for ESC differentiation.
In this review, we discuss the most recent advances in understanding mouse ESC pluripotency and differentiation, paying particular attention to Oct4, Nanog, and Sox2, as well as to factors involved in the exit from pluripotency. Finally, we discuss the function and the molecular mechanisms of the Polycomb and MLL complexes in mouse ESC pluripotency.
MASTER REGULATORS OF ESC IDENTITY: THE Oct4, Sox2, AND Nanog TRIUMVIRATE
Pluripotency and self-renewal are considered unwavering features of ESCs, yet the ICM of the developing embryo has a half-life of only about a day. The pluripotent state is therefore an ephemeral biological state that is tightly regulated by transcription factors, signaling pathways, and epigenetic machineries (Fig. 1). Several transcription factors have been shown to be indispensable in regulating the pluripotent state of ESCs in vivo and in vitro. Among these factors, we will discuss the biological function of the triumvirate of Oct4, Sox2, and Nanog.
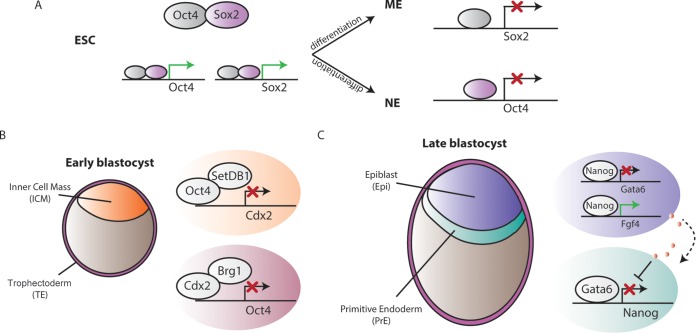
FIG 1.
Transcriptional regulation of lineage-specifying decisions by pluripotent factors. (A) In ESCs, Sox2 and Oct4 (as a heterodimer) activate their own transcription, but during differentiation to mesoendoderm (ME) and neuroectoderm (NE) lineages, Sox2 and Oct4 negatively regulate each other's expression. (B) Reciprocal inhibition between Oct4 and Cdx2 determines inner cell mass (ICM) and trophectoderm (TE) specification in the early blastocyst. (C) Nanog and Gata6 orchestrate differentiation to epiblast (Epi) and primitive endoderm (PrE) in the late blastocyst. Moreover, Nanog-expressing cells in the epiblast terminally differentiate PrE cells in a non-cell-autonomous manner by secreting Fgf4 to the extracellular medium.
Oct4, Sox2, and Nanog expression and functions.
The transcription factor Oct4 (octamer-binding transcription factor 4) was one of the first factors identified as a master regulator of ESC pluripotency (2–4). It is highly expressed in the blastocyst ICM (5), and its expression rapidly decreases during embryo development; indeed, Oct4 is widely used as a reporter marker to assess for differentiation (6). Accordingly, Oct4 null embryos fail to form a pluripotent ICM and do not develop beyond the blastocyst stage (5). Oct4 expression is tightly regulated, and reduction of only 50% causes spontaneous differentiation of ESCs toward the trophectoderm (TE) lineage (7).
Another master regulator of ESC pluripotency is Sox2 (Sry box-containing gene 2) (8). Similar to Oct4, Sox2 is also highly expressed in the ICM, where it is essential to retain the maximum pluripotency capacity of the ESCs (9). Changes of Sox2 expression in ESCs also trigger differentiation: overexpression induces differentiation toward neuroectodermal cells (10), whereas Sox2 deletion results in TE differentiation. Furthermore, targeted disruption of Sox2 in vivo results in peri-implantation lethality due to a strong impairment of the ICM (8). Importantly, Sox2 expression is not restricted to ESCs: in adult mice, Sox2 expression is maintained in many adult stem/progenitor stem cells (8, 11).
Nanog is another transcription factor involved in the ESC pluripotency network (12). Nanog is highly expressed not only in the ICM but also in the epiblast cells of the embryo (13). Similar to Oct4- and Sox2-null mutants, Nanog-null embryos fail to develop. However, in contrast to those from Oct4- and Sox2-null mutants, ESCs from Nanog mutant embryos can be derived and maintained in culture, indicating that Nanog is dispensable for ESC self-renewal (14).
Oct4, Sox2, and Nanog are implicated in a regulatory network.
The Oct4, Sox, and Nanog (abbreviated OSN) triumvirate does not work independently but rather is involved in an intricate regulatory circuitry in which other transcription factors are also implicated (15–19). Chromatin immunoprecipitation sequencing (ChIP-seq) experiments have revealed that pluripotency factors cooccupy gene regulatory elements in a large spectrum of genes, in what is called multiple transcription factor-binding loci (MTL) (18). Chen and colleagues resolved 3,583 MTLs, 43.4% of which were cooccupied by Oct4, Sox2, and Nanog, indicating a functional cooperation in gene regulation (17). Moreover, OSN triumvirate regulate their expression positively by binding to their own promoters, cooccupy and activate expression of other genes essential for maintaining ESC pluripotency, and cooperate to repress lineage-specific transcription factors, the silencing of which is essential to prevent exit from pluripotency and spontaneous differentiation.
The ability of the OSN triumvirate to positively or negatively regulate gene expression relies on their ability to interact with specific transcription factors and epigenetic machineries, and great efforts have been made recently to characterize the interactome of these three factors (6, 20–22). Despite a few differences between these reports, all of them converged in identifying numerous associated proteins, including nucleosome-remodeling complexes (such as SWI/SNF [23] and NuRD [21]), histone methyltransferases (i.e., SetDB1 [24] and Wdr5 [24]), enhancer-associated factors (i.e., Mediator [25]), and pluripotency factors (20, 21, 26). For instance, it has been shown that Oct4 and Nanog associate with proteins of the NuRD complex, Mta1/2 and HDAC1/2, to compose a unique complex (termed NODE) that has a deacetylation activity comparable to that of the NuRD complex, connecting Oct4 and Nanog to repressor functions (21). In contrast, the OSN proteins have also been reported to strongly participate in actively transcribed regions in the genome. Indeed, Young and colleagues demonstrated that the OSN proteins recruit Mediator, and therefore RNA polymerase II, and activate transcription of many genes that ultimately characterize ESC biology (25).
Interestingly, Oct4 and Sox2 have been related to a group of proteins with apparently divergent functions from the heterotrimeric XPC-nucleotide excision repair (NER) complex, which has been implicated in DNA damage repair. Fong and colleagues reported that the XPC complex functions as a coactivator that is required to enhance Nanog transcriptional activity. Combined loss-of-function studies of these proteins compromised mouse ESC pluripotency as well as reprogramming efficiency in mouse embryonic fibroblasts (MEFs) (27).
PLURIPOTENCY FACTORS IN CELL FATE COMMITMENT
In addition to their role in maintaining ESC pluripotency and self-renewal, the OSN proteins have also been proposed to be involved in early cell fate decisions (16, 28). These factors endow ESCs with plasticity, preparing the cells to rapidly respond to any differentiation stimuli. In fact, OSN are indispensable for proper differentiation. Loh and Lim recently reviewed this idea of pluripotent factors also being prodifferentiation genes (28), yet we would like to pursue this hypothesis.
Several studies have shown that Oct4 has explicit lineage-specifying functions (16, 29–32). Initial reports revealed that ESCs expressing higher Oct4 levels induce differentiation toward the mesoendoderm lineage (33). Indeed, Thompson and colleagues showed a specific cross-regulation between Oct4 and Sox2 in early stages of germ layer differentiation (16). In ESCs, expression of these two factors decreases once the cells enter the differentiation program; however, Oct4 and Sox2 are required in these cells to specify the following mesoendoderm (ME) and neuroectoderm (NE) lineages, respectively. In ME cells, Oct4 occupies the enhancer regulatory region of Sox2, inhibiting its expression. In contrast, Oct4 is not expressed in NE cells, so that Sox2 can bind to its enhancer region to activate its own transcription. Thus, when they are coexpressed, Oct4 and Sox2 repress differentiation, but when they are asymmetrically expressed, they can activate specific differentiation programs while inhibiting others (16).
Oct4 is also involved in the first cell fate decision in the developing embryo that occurs at the 16-cell stage, in which the outermost cells segregate from the inner cells to form the extraembryonic TE compartment. Oct4 and Cdx2 direct the divergence between the TE and the ICM in the embryo (31). A reciprocal inhibition of the transcriptional activities between both factors results in a mutually antagonistic expression pattern: Cdx2 represses Oct4 expression in the TE compartment, while Oct4 represses Cdx2 in the ICM. Additional studies resolved the epigenetic mechanisms by which this mutual silencing is achieved. In the TE cells, the chromatin remodeling protein Brg1 cooperates with Cdx2 to ensure Oct4 repression (34), while in the ICM, the H3K9 histone methyltransferase Setdb1 interacts with Oct4 to silence Cdx2 expression and other TE-associated genes (24).
Nanog is also involved in regulating cell fate commitment at early stages of development. Soon after the ICM and TE lineages have been specified, a second differentiation process results in the formation of two new layers of specialized cells: the primitive endoderm (PrE), which will form the extraembryonic endoderm tissues of the placenta, and the epiblast cells, which will give rise to the three germ layers of the embryo. Before implantation, Nanog and Gata6 are expressed in a mutually exclusive manner, which has been described as a “salt-and-pepper” expression pattern, as they inhibit each other's expression (35). Moreover, Nanog depletion in ESCs induces expression of PrE markers, suggesting that only repressing Nanog is sufficient to induce PrE differentiation and that forcing expression of Gata6 causes Nanog downregulation, concomitantly with aberrant differentiation of the ICM toward PrE cells (36). It has been shown that the Fgf/RTK signaling pathway plays an essential role in arbitrating this battle between Nanog and Gata6, as Grb2 mutants (a protein essential for the transduction in this signaling pathway) fail to generate PrE cells, as all the ICM cells express Nanog. Furthermore, Nanog-expressing cells induce full PrE differentiation in a non-cell-autonomous manner by activating the expression and secretion of Fgf4 to the extracellular medium, which in turn triggers the expression of later markers of PrE in Gata6-expressing cells, thereby reinforcing PrE cell identity (36, 37).
NONPLURIPOTENT FACTORS INVOLVED IN CELL FATE COMMITMENT
During the last decades, the mechanisms that govern ESC pluripotency and self-renewal have been thoroughly dissected. Notwithstanding, the mechanisms by which ESCs exit the pluripotent state and embark to differentiation are poorly understood.
Tcf3 (transcription factor 3) is one of the best-characterized transcription factors involved in the progression from ground-state pluripotency (38, 39) to cell fate commitment. Depletion of Tcf3 enhances ESC self-renewal (38). Tcf3 is part of the TCF/LEF family of transcription factors, which usually activates transcription following Wnt pathway activation (40, 41). However, in ESCs, Tcf3 mainly acts as a repressor (38). Unexpectedly, while Tcf3 also binds to the Oct4, Sox2, and Nanog promoters, it is not capable per se of repressing their expression, which only occur under differentiation stimuli (i.e., in the absence of Wnt signal). In the presence of Wnt signal, β-catenin is free to enter the nucleus and abrogate Tcf3 function (39). Therefore, a continuous battle between the forces that activate/repress the Wnt signaling pathway is necessary to keep ESCs in the pluripotency state.
Recent studies have revealed that there are additional factors involved in destabilizing the ground state of ESCs and thus directing the progression from self-renewal to cell fate commitment. An efficient functional assay to search for new potential genes implicated in the extinction of pluripotency entails detection of persistent self-renewal features under differentiation-tolerant culture conditions. Betschinger and coworkers applied a large-scale small interfering RNA screen to interrogate which factors are involved in the exit from pluripotency (42). In this assay, cells that are depleted for Flcn (follicullin) and Tsc2 (tuberous sclerosis 2) retain Oct4 expression and self-renew when they are cultured in a permissive differentiation media. Further characterization revealed that both Flcn and Tsc2 are involved in the subcellular localization of the Tfe3 (transcription factor binding to IGHM enhancer 3) protein, as knockdown of these proteins increased the nuclear Tfe3 concentration. Equally, enforced nuclear Tfe3 expression enables ESCs to avoid cell fate commitment through the transcriptional upregulation of the pluripotency factor Esrrb. Altogether, these data revealed novel players that thrust ESCs to cell fate commitment by constraining Tfe3 in the cytoplasmic compartment (42).
A similar experimental approach using haploid ESCs identified additional factors not previously connected to regulating ESC fate commitment: the zinc finger protein Zfp706 and the RNA-binding protein Pum1. A differentiation-permissive medium was not enough to trigger differentiation after either Zfp706 or Pum1 had been depleted, and pluripotent colonies could be isolated 10 days after culturing the ESCs in this medium. Mechanistically, Zfp706 regulates Klf4, whose downregulation is known to be required to exit pluripotency (43). Pum1 regulates mRNA stability by binding a highly conserved motif at the 3′ untranslated region of several mRNAs. Potential Pum1 mRNA targets were identified by using a bioinformatic prediction analysis and also gene expression profiles after Pum1 depletion. Interestingly, mRNAs of pluripotency-related transcription factors, such as Klf4, Tfcp2l1, Tbx3, and Esrrb, were identified and validated as Pum1 targets. Thus, Pum1 contributes to the exit of self-renewal by directly regulating mRNA transcripts of several pluripotency factors (44).
In the past few years, long noncoding RNAs (lncRNAs) have emerged as key regulators of pluripotency as well as of the transition to the differentiated state. Indeed, increasing evidence proves that these molecules are essential to ensure pluripotency (45, 46). In this direction, Guttman and colleagues showed that the effects of knockdown of large intergenic noncoding RNAs (lincRNA) in ESCs are comparable to those of knockdown of well-known pluripotent factors (47). Better understood is the role of the lncRNA X-inactive specific transcript (Xist) in X chromosome inactivation (XCI) during female ESC differentiation and its connection to the pluripotency network (48). Future investigations will undoubtedly uncover novel functions of lncRNAs in ESCs.
These results suggest that disassembly of the pluripotency network takes place at multiple levels, which need to be concomitantly regulated to allow ESCs to determine their cell fates.
EXTRINSIC SIGNALING IN CELL FATE COMMITMENT
A central question in stem cell biology is how extrinsic signaling pathways modulate ESC pluripotency and differentiation. Since ESCs were first derived in 1981 (1), great efforts have been made to develop cell culture conditions in the laboratory that resemble ICM conditions. This did not occur until 1988, when the contribution of the cytokine leukemia inhibitory factor (LI) was discovered. LIF is secreted by fibroblasts that are cocultured with the ESCs (the so-called feeder cells) and is indispensable for the proliferation of ESCs cultured in media supplemented with fetal bovine serum (49, 50). LIF binds to the LIFR/gp130 receptor, which drives activation of the JAK-STAT pathway and expression of STAT3 (51). STAT3 forms a dimer and regulates the expression of pluripotency factors, like Klf4, Nanog, and Tfcp2l1 (52–54). Although LIF is essential to derive and maintain ESCs, LIF−/− embryos are viable and fertile (55).
LIF can only sustain self-renewal when ESCs are cultured with fetal bovine serum, suggesting that the serum contains essential components to prevent exit from pluripotency. Indeed, Smith and colleagues discovered that the BMP4-mediated signaling pathway supports pluripotency in the presence of LIF (56). BMP4 has been extensively studied during development and found to induce expression of Id proteins in pluripotent ESCs, which in turn indirectly inhibits the expression of the bHLH factors involved in differentiation. In agreement with the functional link between Id proteins and BMP4, overexpression of Id proteins is sufficient to maintain pluripotency in cells cultured with only LIF but no BMP4 or serum (56).
The fibroblast growth factor (FGF) signaling pathway additionally regulates lineage specification in the embryo. FGF4 is the main FGF expressed in the early stages of the embryo and in ESCs. This stem cell-specific expression pattern relies on the presence of a distant enhancer in the FGF4 promoter that is under the control of the pluripotency factors Oct4 and Sox2, which positively activate its transcription (57). FGF4 plays a role not only in the ICM but also in the TE lineage. Accordingly, trophoblast stem (TS) cells are highly enriched with the FGF4 receptor, FGF2r (58). In fact, FGF4- and FGF2r-null embryos display a similar phenotype, as both are lethal at the peri-implantation stage (59, 60). Tanaka and coworkers were able to derive TS cells from blastocysts by treating the desegregated blastocysts with medium containing FGF4. Therefore, the FGF4-mediated signal contributes to maintain trophectoderm cells in a proliferating and undifferentiated state (61).
The function of FGF4 in ESCs has also been extensively studied. FGF4, through the activation of the Erk1/2 signaling cascade, acts as an autoinductive stimulus for ESCs to exit self-renewal and to enter into the differentiation program. FGF4-null cells can be cultured and do not show any apparent defects, but they do not differentiate when they are cultured in medium that induces neuronal or mesodermal differentiation, suggesting that FGF4 signaling, through the Erk1/2 pathway, may act as one of the primary signals directing cell fate commitment (62). Accordingly, Erk1/2 inhibitors have been extensively used to improve the efficiency of ESC derivation (63).
ESCs maintained in serum and LIF are often morphologically heterogeneous and express pluripotency factors in a heterogeneous manner, indicating that the pluripotent state is unstable. Ying and colleagues defined specific culture conditions in which ESCs are more protected from differentiation stimuli, thereby resembling the transcriptional status of the ESCs in vivo (64). This serum-free medium contains LIF and inhibitors of two kinases, PD0325901 (Mek kinase inhibitor) and CHIR99021 (GSK3 kinase inhibitor). This combination of molecules helps to maintain a naive ground state of the ESCs (64).
CROSS TALK BETWEEN PLURIPOTENCY AND EPIGENETIC FACTORS
As mentioned above, the OSN factors not only positively regulate their expression by binding to their own promoters but also repress lineage-specific transcription factors to prevent exit from pluripotency and spontaneous differentiation. Transcription of genes encoding epigenetic factors such as Polycomb proteins (i.e., Eed, Jarid2, and Cbx7), Polycomb-associated proteins (Max), and members of the MLL complexes (Wdr5 and Ash2l) appears to be under the control of the pluripotency network (22, 25, 65), and their expression is positively regulated by the OSN proteins.
Interestingly, Polycomb and MLL complexes are in turn required during the pluripotent state to maintain lineage-specific transcription factors in a silenced state, to avoid exiting pluripotency (66, 67). Therefore, a cross talk between OSN proteins and several epigenetic factors is required for maintaining ESC pluripotency as well as for ensuring proper ESC differentiation. Other epigenetic factors involved in DNA hydroxymethylation and methylation, such as Tet (ten-eleven translocation) and DNA methyltransferase (Dnmt) enzymes, and also other histone modifications, have also been strongly associated with ESC pluripotency and differentiation (68). However, in the next sections, we focus on the architecture of the Polycomb and MLL complexes, their recruitment to chromatin mechanisms, and their biological functions in pluripotent and differentiating ESCs.
THE Polycomb GROUP OF PROTEINS IN PLURIPOTENCY AND DIFFERENTIATION
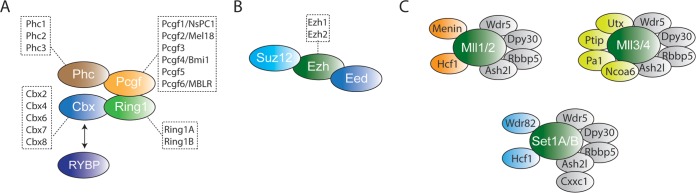
Polycomb group proteins (PcG) are essential epigenetic regulators of stem cell identity and development (69, 70). The two major classes of PcG complexes are the Polycomb repressive complex 1 (PRC1) and the Polycomb repressive complex 2 (PRC2) (66, 71, 72) (Fig. 2). The core PRC2 complex contains Suz12, Eed, and the histone methyltransferase enzymes Ezh1/2, which catalyze di- and trimethylation on lysine 27 of histone H3 (H3K27me2/3) (73). PRC1 complexes can be further divided into two main subcomplexes, namely, the canonical and noncanonical PRC1 (cPRC1 and ncPRC1, respectively). cPRC1 consists of Pcgf2/4, Polyhomeiotic 1/2/3 (Phc1/2/3), the Cbx proteins (Cbx2/4/6/7/8), and the E3-ligase subunit Ring1A/B, which monoubiquitinates histone H2A at lysine 119 (H2AK119ub). In contrast, ncPRC1 does not contain a Cbx protein but instead contains RYBP/YAF2, Pcgf1/3/5, and Ring1A/B (74–77) (Fig. 3).
FIG 2.
Polycomb and MLL complex architecture. (A) PRC1 core subunits. The PRC1 complex contains the Ring1A/B, Cbx, Pcgf, and Phc subunits. Ring1A/B directly interact with the Pcgf and Cbx subunits, and Phc proteins directly interact with Pcgf proteins. (B) PRC2 core subunits. The PRC2 complex contains either Ezh1 or Ezh2. Suz12 and Eed directly interact with Ezh proteins. (C) Set1A/B and MLL complex architecture. These complexes contain Wdr5, Dpy30, Ash2l, and Rbbp5 as well as specific cofactors, and the different complexes are classified by the enzyme that they contain.
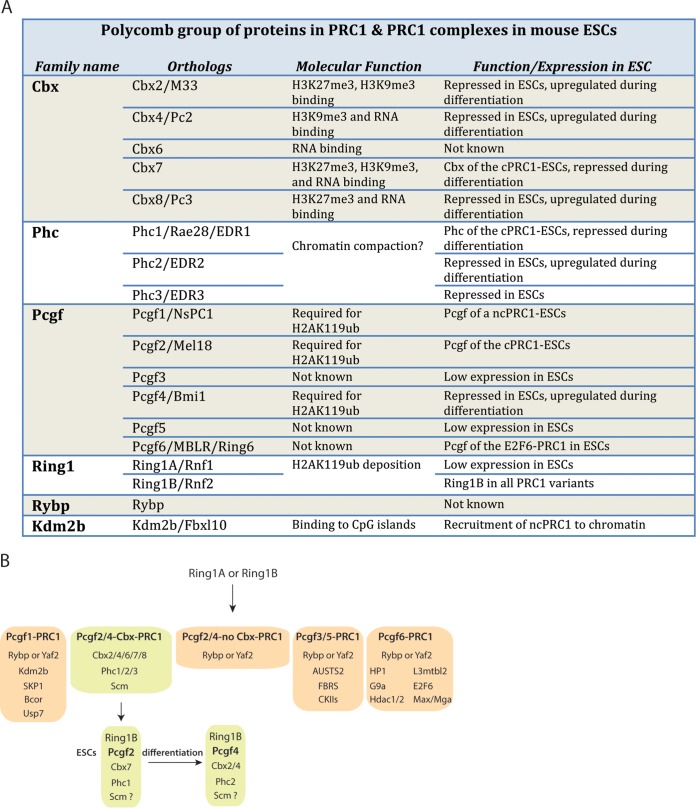
FIG 3.
PRC1 subunits and variants. (A) Table of the different PRC1 variants in mouse ESCs, classified by their genes, orthologues, molecular functions, and expression or biological functions. (B) Classification of PRC1 variants based on data from reference 76. Different cPRC1 complexes are assembled in ESCs compared to differentiating ESCs.
Polycomb-mediated gene repression mechanisms and target genes in ESCs.
In pluripotent ESCs, Polycomb complexes are recruited to lineage-specific gene promoters and are required to maintain gene repression (78–80). The molecular mechanisms of Polycomb-mediated gene repression are constantly under examination and debate. The classical mode of action of Polycomb complexes is the hierarchical model in which the PRC2 complex is recruited to chromatin by cofactors and/or RNA to deposit H3K27me3 (69). Once H3K27me3 is established, the PRC1 complex is recruited through the chromodomain of the Cbx protein, and Ring1A/B deposits H2AK119ub. While this model is widely established, other Polycomb recruitment mechanisms must exist, as the hierarchical model does not clarify, for instance, why a functional PRC1 complex is still recruited to the inactivated X chromosome in differentiating female ESCs even in the absence of PRC2 (81), or why H2AK119ub levels remain unaffected in ESCs that lack a functional PRC2 complex (82). Recently, it has been proposed that an ncPRC1 complex, containing RYBP but no Cbx proteins, deposits H2AK119ub independently on the PRC2 complex (75).
Recently, using a de novo targeting system in ESCs, the PRC2 complex was shown to be recruited to chromatin in a PRC1-dependent manner (83). PRC1 complexes containing Pcgf1, Pcgf3, or Pcgf5 are first recruited to DNA, where they monoubiquitate H2AK119, which then drives PRC2 recruitment. Using the same targeting system, those authors also suggested that canonical PRC1 complexes fail to deposit H2AK119ub (83).
The biological function of H2AK119ub is still poorly understood. Elegant studies by Koseki and colleagues indicated that the enzymatic activity of Ring1B in ESCs is necessary for gene repression and chromatin compaction (84). Moreover, H2AK119ub serves as a recruitment platform for Zrf1 (zuotin-related factor 1). Upon differentiation of NT2 cells (human teratocarcinoma cells) and ESCs, Zrf1 binds to H2AK119ub and displaces the PRC1 complex from chromatin, resulting in upregulation of lineage genes with differentiation (85, 86).
Molecular functions of the canonical and noncanonical PRC1 complexes in ESCs.
Recently, variations of PRC1 complexes have been identified in 293T cells and ESCs. In mouse ESCs, cPRC1 mainly contains Cbx7, Phc1, Mel18/Pcgf2, and Ring1B (87, 88) (Fig. 3). Cbx7 depletion strongly reduces chromatin binding of Ring1B and Mel18. In turn, Cbx7 recruitment is fully dependent on H3K27me3, suggesting that the cPRC1 recruitment follows the hierarchical model in ESCs (87). ncPRC1 has now been characterized in ESCs and shown to contain Kdm2b/Fbxl10 (H3K36me3 lysine demethylase), RYBP, Pcgf1/NsPC1, and Ring1B (77, 89) (Fig. 3). Interestingly, Fbxl10 is a H3K36me3 demethylase and contains a CXXC domain (90). Chromatin recruitment of this ncPRC1 complex depends on the CXXC domain of Kdm2b, which drives this ncPRC1 to CpG islands. Importantly, neither ncPRC1 recruitment nor its activity depends on Kdm2b histone demethylase activity (77, 89).
Do these complexes have redundant functions in ESCs?
H2AK119ub global levels are not affected upon Cbx7 depletion and are only slightly reduced at target genes, yet depletion of either Kdm2b or Pcgf1 strongly reduces H2AK119ub. While Ring1B recruitment to chromatin is strongly reduced upon Cbx7 or Fbxl10 depletion, Fbxl10 is not required for Cbx7 recruitment (77, 87), suggesting that cPRC1 is important for efficient Ring1B recruitment while ncPRC1 is required for the enzymatic activity of Ring1B. RYBP appears to be also important for global and local H2AK119ub deposition (75–77, 87, 89), and its depletion seems to induce Ring1B instability (75). However, as this regulation might not be due to Ring1B degradation (74, 76, 77, 91), the mechanism by which RYBP induces H2AK119ub deposition by Ring1B remains elusive.
Are the cPRC1 and ncPRC1 complexes recruited only to lineage-specific genes? Or, in contrast, is there a certain degree of gene target selectivity by these PRC1 complexes? Lineage-specific genes are cooccupied by either both cPRC1 and ncPRC1 or only the cPRC1 complex (Fig. 4A and B). These genes also contain high levels of PRC2 and are strongly repressed. Interestingly, a subset of ncPRC1 target genes (classified for the presence of RYBP and Ring1B, but without Cbx7), have low levels of H2AK119ub and PRC2 binding and are in turn less repressed; these genes are involved in germ cell fate and cellular metabolism (74) (Fig. 4B).
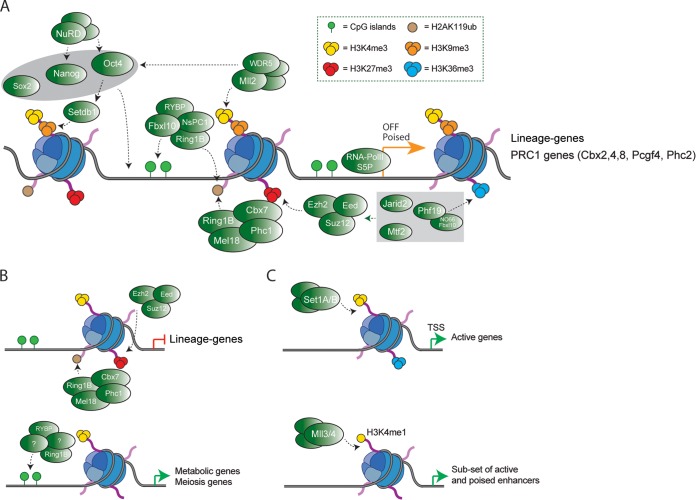
FIG 4.
Target genes of Polycomb and MLL complexes in ESCs. (A) Although lineage-specific genes are classified as bivalent genes because of the presence of both H3K27me3 and H4K4me3, they are also decorated with two other repressive marks: H3K9me3 (deposited by Setdb1) and H2AK119ub (deposited by the PRC1 complex). Pluripotency factors, Polycomb, MLL complexes, and Setdb1 are required to maintain low levels of expression of lineage-specific genes. (B) cPRC1 is also recruited to lineage genes that also contain PRC2 but not the Rybp-PRC1 complex. Additionally, the Rybp-PRC1 complex is recruited to metabolic and cell cycle-related genes that are active. (C) Set1A/B complexes deposit H3K4me3 at TSS of active genes in ESCs. The MLL3/4 complexes deposit H3K4me1 at both poised and active enhancers in ESCs.
Molecular functions of PRC2 complexes in ESCs.
The PRC2 complex architecture is not as heterogeneous as the PRC1 complex (Fig. 2), yet several cofactors have recently been identified that associate it with PRC2 members in ESCs (92). In the past years, the Jumanji protein Jarid2 was identified as the first protein to interact directly, via its interaction with Suz12, with the core PRC2 complex in ESCs. Jarid2 cooccupies a large set of PRC2 target genes, and its depletion greatly reduces PRC2 occupancy at promoters. Whether Jarid2 positively or negatively regulates PRC2 activity in ESCs is under debate (93–97). Jarid2 contains an AT-rich interaction domain and was therefore postulated to be a recruiter to PRC2, yet to date it has not been demonstrated that Jarid2 can directly bind to DNA. Thus, it seems that Jarid2 is a component but not a recruitment factor, of a specific PRC2 complex in ESCs.
In Drosophila melanogaster, the Polycomb protein PCL interacts with Su(z) (97), and PCL mutant flies have reduced levels of H3K27me3. In mammals, there are three PCL orthologues: Pcl1, Pcl2/Mtf2, and Pcl3/Phf19 (66). Consistent with the fly phenotype, Phf19 facilitates PRC2 recruitment to chromatin and therefore H3K27me3 deposition (98–100). Surprisingly, although Mtf2 knockdown ESCs contain increased global levels of H3K27me3, PRC2 recruitment and H3K27me3 levels are reduced at some PRC2 target genes (101). Pcl proteins contain a Tudor domain, which recognizes H3K36me3 (98–100). Mechanistically, Phf19 recruits the H3K36me3 demethylases NO66 and Kdm2b to active genes through its binding to H3K36me3, thus favoring PRC2 recruitment and resulting in gene repression (98–100). More recently, C17orf96 was identified as a new PRC2-associated protein in a complex also containing Jarid2 and Pcl2. Although C17orf96 function in ESCs has not yet been interrogated on a genome-wide basis, C17orf96 might be required for PRC2 stability and enzymatic activity (102).
In sum, four polypeptides, Jarid2, Phf19, Mtf2, and C17orf96, have been recently identified as new cofactors that substoichiometrically associate with different PRC2 complexes in ESCs (Fig. 4A). This raises numerous questions regarding the biological and molecular functions of these PRC2 subcomplexes. Are they functionally redundant in gene regulation? Do they coregulate the same target genes? Why does Mtf2 depletion enhance global PRC2 activity but reduce H3K27me3 and PRC2 binding at target genes? A comprehensive analysis of Jarid2, Phf19, Mtf2, and C17orf96 target genes and gene expression profiles in ESCs depleted by these cofactors will shed light on the functional regulation of these PRC2 subcomplexes.
The exact mechanisms that drive PRC2 recruitment remain largely elusive. Recently, it was postulated that the PRC2 complex binds via Ezh2 to nascent RNAs (ezRNAs) at genes that lack H3K27me3, have intermediate or low levels of PRC2, and are transcriptionally active, suggesting that ezRNAs inhibit PRC2 activity (103). While another report corroborated the interaction of ezRNAs with the PRC2 complex, it also suggested that a lack of H3K27me3 is not a prerequisite for the ezRNA-PRC2 interaction (104).
Polycomb-MEDIATED PHENOTYPES IN ESCs AND MICE
PRC1 subunits.
Among all PRC1 members, mice with a mutant Ring1B display the most severe phenotype, and mice lacking Ring1B are embryonic lethal due to gastrulation defects. Mutant mice for other PRC1 subunits are viable (i.e., Ring1A, Mel18, Bmi1, Cbx2, and Phc1), yet they show homeotic transformations and other abnormalities of the axial skeleton (69). Cbx7 mutant mice are viable, with increased body length, and develop lung and liver tumors (105).
In ESCs, Ring1A-null or Ring1B-null cells proliferate and self-renew normally (82, 106). While a single Ring1A depletion does not have a major impact on gene regulation, Ring1B is required to maintain gene repression of lineage-specific genes (78, 82). In contrast to PRC2-null ESCs, complete impairment of all of the PRC1 complexes (in Ring1A/B double knockout ESCs) strongly affects self-renewal (107), suggesting that the PRC2 and PRC1 complexes are not completely functionally redundant. Depleting Cbx7, RYBP, or Fbxl10 in ESCs does not compromise self-renewal of the E14Tg2a ESC strain, but depleting RYBP in TT2 ESCs decreases their proliferation (76, 77, 87, 91).
Cbx7 is dispensable for ESC proliferation, although it is required to maintain gene repression of a large amount of PRC1 target genes. The aberrant upregulation of lineage-specific genes results in defects during differentiation of the ESCs into embryoid bodies (EBs), and more specifically, toward the neuroectoderm lineage (87, 88). While Cbx7 is strongly downregulated during differentiation, Cbx2/4/8 are assembled into new cPRC1 complexes during EB differentiation. Interestingly, depletion of Cbx2 and Cbx4 resulted in lineage-specific phenotypes, indicating not only that Cbx proteins have nonoverlapping functions in pluripotent and differentiating ESCs but also that the expression of Cbx2 and Cbx4 is important to drive the newly assembled cPRC1 complexes to specific and de novo loci during differentiation (87, 88).
RYBP-null or knockdown ESCs also deregulate PRC1 target genes, and unlike Cbx7, RYBP is necessary during mesoderm differentiation (74, 76, 91). RYBP is not functionally restricted to the ncPRC1 complex but is probably present in other chromatin-associated complexes; therefore, the precise contribution of RYBP in PRC1-mediated phenotypes is unclear. Although Fbxl10 depletion strongly impairs Ring1B activity and reduces global levels of H2AK119ub, changes in gene expression are very subtle (77, 89). The lack of gene activation is probably due to the persistent binding of Cbx7 and PRC2 in cells depleted for Fbxl10. ESCs depleted for both Cbx7 and Fbxl10 have not yet been reported, but double Cbx7- and RYBP-depleted cells strongly deregulate genes bound by both cPRC1 and ncPRC1 (74), indicating a functional cooperation in gene regulation of different PRC1 complexes in ESCs.
PRC2.
Mice with mutations in core PRC2 components display embryonic lethality at different developmental stages: Ezh2-null mice are embryonic lethal at day embryonic day 8.5 (E8.5), Eed-null mice at E9.5, and Suz12 mice between E8.5 and E10.5 (69).
ESCs with mutant Eed, Suz12, or Ezh2 ESCs self-renew and proliferate normally but have a strongly compromised differentiation capacity (108–113). Surprisingly, the precise contribution of each of the PRC2 subunits during ESC lineage specification is still poorly understood. It has been shown that Eed-null ESCs lose their cellular identity and are not able differentiate toward any specific cell lineage, yet another report indicated that Eed−/− ESCs are capable of developing teratomas that contain all three cell lineages but also have an overrepresentation of ectoderm and mesoderm tissues (114). Mutant Suz12 ESCs display defects during neuroectodermal and EB differentiation, and ESCs lacking Ezh2 fail to differentiate toward the mesoendoderm lineage (115).
Regarding the PRC2-associated proteins in ESCs, Jarid2 is also essential during embryogenesis (116). While Jarid2 and C17orf96 are also dispensable for ESC self-renewal, Mtf2-depleted ESCs self-renew faster (101), and Phf19-depleted cells display a certain degree of spontaneous differentiation (100). Jarid2, Mtf2, and Phf19 are essential regulators of ESC differentiation, although they are rapidly downregulated during differentiation (93, 95, 96, 100, 102, 117).
Why do genetic mutations of different PRC2 core components result in different phenotypes during ESC differentiation? A possible explanation is that the protein stability and function of the other PRC2 subunits differentially change when only one is depleted; for instance, ESCs lacking Eed have reduced levels of Ezh2 and normal Suz12 levels (114, 115). In contrast, Ezh2 knockout ESCs have normal Eed and Suz12 protein levels, and Ezh1 functionally compensates for the lack of Ezh2 (115), while Suz12 knockout ESCs are impaired in Ezh2 stability and Ezh2-mediated enzymatic activity (109). Furthermore, Ezh2 has been recently shown to have PRC2-independent functions in cancer (70). Although it seems clear that Eed, Suz12, and Ezh2 are only associated with the PRC2 complex in pluripotent ESCs, it remains unclear whether they exhibit PRC2-independent functions during ESC differentiation or whether other cofactors expressed during differentiation are assembled into new PRC2 complexes that in turn might have specific biological functions. Therefore, identifying further PRC2 subunits that associate during ESC differentiation will shed light on the biological function of PRC2 during differentiation.
SET1A/B AND MLL COMPLEXES IN ESCs
In mammals, the Set1A/B and MLL family of complexes (MLL1 to -4) are responsible for methylation of the lysine 4 at the histone H3 (67, 118, 119). These can be further classified, depending on the methyltransferase enzyme they contain as well as on their specific and common cofactors (Fig. 2 and 4C).
In ESCs, lineage-specific genes marked by H3K4me3 and H3K27me3 are defined as bivalent genes, which, despite the presence of H3K4me3, are expressed only at low levels (80, 120). Upon differentiation stimuli, bivalent genes become either activated, with a concomitant loss of H3K27me3, or fully repressed, with a loss of H3K4me3 (121). These observations led to a model in which ESCs keep lineage-specific genes silent but poised for rapid activation upon differentiation. Recently, by combining ChIP-seq and depletion experiments, specific functions of each of the MLL enzymes in ESC pluripotency and differentiation have been elucidated. While Set1A/B complexes deposit H3K4me3 at the transcription start site (TSS) of active genes in ESCs (122), Mll2 specifically deposits H3K4me3 at the bivalent genes (118). In colon cancer cells, Mll3 and Mll4 monomethylate H3K4 at a subset of enhancers, and in ESCs, Mll4 occupies enhancers (119) (Fig. 4C). The molecular mechanism(s) that confers methylation selectivity to the MLL enzymes remains elusive.
Mll2-knockout ESCs can self-renew yet have proliferation defects due to the reduced expression of the antiapoptotic factor Bcl2 (B-cell CLL/lymphoma 2), whose expression is directly regulated by Mll2 (123). Because Mll2 specifically trimethylates lysine 4 of histone H3 at bivalent genes, H3K4me3 global levels remain unaltered. Analysis of the differentiation potential of Mll2−/− ESCs revealed an Mll2-dependent differentiation delay toward cardiomyocytes (mesoderm) and neuroectodermal cells. In contrast, a lack of Mll2 enhanced ESC differentiation toward the endoderm lineage. Therefore, Mll2−/− ESCs are able to generate cells from all three germ layers, yet with an altered timing of lineage commitment.
Wdr5 and Ash2l are subunits present in all Set1A/B and MLL complexes (Fig. 2). As their expression is rapidly downregulated upon ESC differentiation, they might exert specific functions that maintain the ESC state. Interestingly, WDR5 targets and directly controls the expression of several pluripotency genes, and its depletion results in loss of ESC self-renewal (124). While Mll2 depletion does not alter global H3K4me3 levels, Wdr5 knockdown strongly reduces global H3K4me3 levels and upregulates H3K4me1/2 levels. Wdr5 also interacts with Oct4 (124) and has been found in pulldown experiments by using Ring1B as bait (125), although Ring1B-mediated functions of Wdr5 have not yet been characterized. Additionally, Polycomb and MLL complexes have opposing functions in gene regulation, and it has not yet been determined which subunit of the PRC1 complex directly interacts with Wdr5. Genome-wide assessment of Wdr5 and Oct4 indicated that they cooccupy a subset of gene promoters together with Rbbp5, Sox2, and Nanog, providing a possible molecular mechanism by which Wdr5 is necessary for ESC self-renewal (124). Depletion of Ash2l also downregulates the expression of several pluripotency factors, concomitant with the upregulation of differentiation-associated genes of all three germ layers, resulting in a loss of ESC self-renewal (126).
DPY30 is another core subunit of all Set1A/B and MLL complexes. In contrast to Wdr5, H3K4me3 levels are reduced at both bivalent and ESC-specific genes upon DPY30 depletion. DPY30 is dispensable for ESC self-renewal (127), indicating that DPY30-mediated loss of H3K4me3 at pluripotency genes is not sufficient to counteract gene repression. Indeed, DPY30 expression is maintained during ESC differentiation, and deposition of H3K4me3 to activated lineage-specific genes is DPY30 dependent (127), thus suggesting a dual role of DPY30 in maintaining proper H3K4me3 levels at pluripotency and lineage-specific genes.
The H3K27me3 histone demethylase Utx was recently identified as a specific subunit of the MLL3/4 complexes (128). Utx is encoded on the X chromosome but escapes X chromosome inactivation. While female Utx-null mice are embryonic lethal before E12.5, male Utx-null mice are viable, although they are smaller and have reduced survival rates (129, 130). Female mutant Utx ESCs self-renew normally and can give rise to teratomas, yet EBs derived from mutant Utx ESCs retain high levels of Oct4 and Nanog and do not properly differentiate (131). Surprisingly, upon RA-induced differentiation, the H3K27me3 levels in Utx-null cells are higher than those in wild-type differentiating cells, indicating that active H3K27me3 demethylation occurs in the absence of Utx during RA-mediated differentiation. Moreover, at the HOXB cluster, H3K4me3 levels are increased in cells lacking Utx (131), suggesting a MLL complex-independent function of Utx. In male ESCs, Utx is also not required for self-renewal and proliferation, but it is required for mesoderm and ectoderm differentiation. Interestingly, the contribution of Utx in ESC differentiation appears to be independent of its enzymatic activity (132, 133).
Rbbp5 genome-wide binding in ESCs has been investigated recently. The histone variant H2A.Z colocalizes in a subset of enhancers with low levels of H3K4me3 and Rbbp5 (134), and its depletion results in reduced Oct4 and Rbbp5 occupancy not only at active genes in ESCs but also at enhancers that contain low levels of H3K4me3. As mentioned, Mll4 knockdown does not affect global H3K4me3 levels but impairs the H2A.Z binding and reduces H3K4me3 at enhancers (134).
CONCLUDING REMARKS
While much work remains to be done, it is clear that the roles of epigenetic factors and the regulation of fate of ESCs are strongly connected, maybe even more so than previously believed. In the past years, our understanding of the mechanisms that govern the proliferation of pluripotent stem cells and of the processes that lead to lineage choice has dramatically increased. This knowledge will significantly contribute to identifying more effective conditions for reprogramming differentiated cells into pluripotent conditions, which will facilitate the development of specialized stem cell-based therapies. Moreover, given the multiple similarities between undifferentiated ESCs with initiating cancer cells, the identification of novel drugs targeting chromatin modifiers, together with the optimization of those already available, will surely improve cancer therapies.
ACKNOWLEDGMENTS
We are grateful to V. A. Raker for critical reading of the manuscript. We also thank members of the Di Croce laboratory for helpful discussions.
This work was supported by grants from the Spanish Ministerio de Educación y Ciencia (SAF2013-48926-P), from AGAUR, from La Marato TV3, and from the European Commission's 7th Framework Program 4DCellFate (grant number 277899 to L.D.C.).
REFERENCES
- 1.Evans MJ, Kaufman MH. 1981. Establishment in culture of pluripotential cells from mouse embryos. Nature 292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 2.Rosner MH, Vigano MA, Ozato K, Timmons PM, Poirier F, Rigby PW, Staudt LM. 1990. A POU-domain transcription factor in early stem cells and germ cells of the mammalian embryo. Nature 345:686–692. doi: 10.1038/345686a0. [DOI] [PubMed] [Google Scholar]
- 3.Scholer HR, Dressler GR, Balling R, Rohdewohld H, Gruss P. 1990. Oct-4: a germline-specific transcription factor mapping to the mouse t-complex. EMBO J 9:2185–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okamoto K, Okazawa H, Okuda A, Sakai M, Muramatsu M, Hamada H. 1990. A novel octamer binding transcription factor is differentially expressed in mouse embryonic cells. Cell 60:461–472. doi: 10.1016/0092-8674(90)90597-8. [DOI] [PubMed] [Google Scholar]
- 5.Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. 1998. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95:379–391. doi: 10.1016/S0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 6.Brambrink T, Foreman R, Welstead GG, Lengner CJ, Wernig M, Suh H, Jaenisch R. 2008. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell 2:151–159. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niwa H, Miyazaki J, Smith AG. 2000. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet 24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 8.Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. 2003. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev 17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, Takahashi K, Okochi H, Okuda A, Matoba R, Sharov AA, Ko MS, Niwa H. 2007. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol 9:625–635. doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- 10.Kopp JL, Ormsbee BD, Desler M, Rizzino A. 2008. Small increases in the level of Sox2 trigger the differentiation of mouse embryonic stem cells. Stem Cells 26:903–911. doi: 10.1634/stemcells.2007-0951. [DOI] [PubMed] [Google Scholar]
- 11.Arnold K, Sarkar A, Yram MA, Polo JM, Bronson R, Sengupta S, Seandel M, Geijsen N, Hochedlinger K. 2011. Sox2(+) adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell 9:317–329. doi: 10.1016/j.stem.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. 2003. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113:643–655. doi: 10.1016/S0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 13.Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. 2003. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113:631–642. doi: 10.1016/S0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 14.Silva J, Nichols J, Theunissen TW, Guo G, van Oosten AL, Barrandon O, Wray J, Yamanaka S, Chambers I, Smith A. 2009. Nanog is the gateway to the pluripotent ground state. Cell 138:722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, Wong KY, Sung KW, Lee CW, Zhao XD, Chiu KP, Lipovich L, Kuznetsov VA, Robson P, Stanton LW, Wei CL, Ruan Y, Lim B, Ng HH. 2006. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet 38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 16.Thomson M, Liu SJ, Zou LN, Smith Z, Meissner A, Ramanathan S. 2011. Pluripotency factors in embryonic stem cells regulate differentiation into germ layers. Cell 145:875–889. doi: 10.1016/j.cell.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rizzino A. 2013. Concise review. The Sox2-Oct4 connection: critical players in a much larger interdependent network integrated at multiple levels. Stem Cells 31:1033–1039. doi: 10.1002/stem.1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, Loh YH, Yeo HC, Yeo ZX, Narang V, Govindarajan KR, Leong B, Shahab A, Ruan Y, Bourque G, Sung WK, Clarke ND, Wei CL, Ng HH. 2008. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 19.Rodda DJ, Chew JL, Lim LH, Loh YH, Wang B, Ng HH, Robson P. 2005. Transcriptional regulation of nanog by OCT4 and SOX2. J Biol Chem 280:24731–24737. doi: 10.1074/jbc.M502573200. [DOI] [PubMed] [Google Scholar]
- 20.Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, Orkin SH. 2006. A protein interaction network for pluripotency of embryonic stem cells. Nature 444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- 21.Liang J, Wan M, Zhang Y, Gu P, Xin H, Jung SY, Qin J, Wong J, Cooney AJ, Liu D, Songyang Z. 2008. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat Cell Biol 10:731–739. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- 22.van den Berg DL, Snoek T, Mullin NP, Yates A, Bezstarosti K, Demmers J, Chambers I, Poot RA. 2010. An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell 6:369–381. doi: 10.1016/j.stem.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ho L, Crabtree GR. 2010. Chromatin remodelling during development. Nature 463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan P, Han J, Guo G, Orlov YL, Huss M, Loh YH, Yaw LP, Robson P, Lim B, Ng HH. 2009. Eset partners with Oct4 to restrict extraembryonic trophoblast lineage potential in embryonic stem cells. Genes Dev 23:2507–2520. doi: 10.1101/gad.1831909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J, Young RA. 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao Z, Cox JL, Gilmore JM, Ormsbee BD, Mallanna SK, Washburn MP, Rizzino A. 2012. Determination of protein interactome of transcription factor Sox2 in embryonic stem cells engineered for inducible expression of four reprogramming factors. J Biol Chem 287:11384–11397. doi: 10.1074/jbc.M111.320143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fong YW, Inouye C, Yamaguchi T, Cattoglio C, Grubisic I, Tjian R. 2011. A DNA repair complex functions as an Oct4/Sox2 coactivator in embryonic stem cells. Cell 147:120–131. doi: 10.1016/j.cell.2011.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loh KM, Lim B. 2011. A precarious balance: pluripotency factors as lineage specifiers. Cell Stem Cell 8:363–369. doi: 10.1016/j.stem.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 29.Le Bin GC, Munoz-Descalzo S, Kurowski A, Leitch H, Lou X, Mansfield W, Etienne-Dumeau C, Grabole N, Mulas C, Niwa H, Hadjantonakis AK, Nichols J. 2014. Oct4 is required for lineage priming in the developing inner cell mass of the mouse blastocyst. Development 141:1001–1010. doi: 10.1242/dev.096875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeVeale B, Brokhman I, Mohseni P, Babak T, Yoon C, Lin A, Onishi K, Tomilin A, Pevny L, Zandstra PW, Nagy A, van der Kooy D. 2013. Oct4 is required ∼E7.5 for proliferation in the primitive streak. PLoS Genet 9:e1003957. doi: 10.1371/journal.pgen.1003957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, Rossant J. 2005. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell 123:917–929. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- 32.Teo AK, Arnold SJ, Trotter MW, Brown S, Ang LT, Chng Z, Robertson EJ, Dunn NR, Vallier L. 2011. Pluripotency factors regulate definitive endoderm specification through eomesodermin. Genes Dev 25:238–250. doi: 10.1101/gad.607311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeineddine D, Papadimou E, Chebli K, Gineste M, Liu J, Grey C, Thurig S, Behfar A, Wallace VA, Skerjanc IS, Puceat M. 2006. Oct-3/4 dose dependently regulates specification of embryonic stem cells toward a cardiac lineage and early heart development. Dev Cell 11:535–546. doi: 10.1016/j.devcel.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 34.Kidder BL, Palmer S, Knott JG. 2009. SWI/SNF-Brg1 regulates self-renewal and occupies core pluripotency-related genes in embryonic stem cells. Stem Cells 27:317–328. doi: 10.1634/stemcells.2008-0710. [DOI] [PubMed] [Google Scholar]
- 35.Singh AM, Hamazaki T, Hankowski KE, Terada N. 2007. A heterogeneous expression pattern for Nanog in embryonic stem cells. Stem Cells 25:2534–2542. doi: 10.1634/stemcells.2007-0126. [DOI] [PubMed] [Google Scholar]
- 36.Frankenberg S, Gerbe F, Bessonnard S, Belville C, Pouchin P, Bardot O, Chazaud C. 2011. Primitive endoderm differentiates via a three-step mechanism involving Nanog and RTK signaling. Dev Cell 21:1005–1013. doi: 10.1016/j.devcel.2011.10.019. [DOI] [PubMed] [Google Scholar]
- 37.Bessonnard S, De Mot L, Gonze D, Barriol M, Dennis C, Goldbeter A, Dupont G, Chazaud C. 2014. Gata6, Nanog and Erk signaling control cell fate in the inner cell mass through a tristable regulatory network. Development 141:3637–3648. doi: 10.1242/dev.109678. [DOI] [PubMed] [Google Scholar]
- 38.Cole MF, Johnstone SE, Newman JJ, Kagey MH, Young RA. 2008. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev 22:746–755. doi: 10.1101/gad.1642408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wray J, Kalkan T, Gomez-Lopez S, Eckardt D, Cook A, Kemler R, Smith A. 2011. Inhibition of glycogen synthase kinase-3 alleviates Tcf3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nat Cell Biol 13:838–845. doi: 10.1038/ncb2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brantjes H, Roose J, van De Wetering M, Clevers H. 2001. All Tcf HMG box transcription factors interact with Groucho-related co-repressors. Nucleic Acids Res 29:1410–1419. doi: 10.1093/nar/29.7.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rousset R, Mack JA, Wharton KA Jr, Axelrod JD, Cadigan KM, Fish MP, Nusse R, Scott MP. 2001. Naked cuticle targets dishevelled to antagonize Wnt signal transduction. Genes Dev 15:658–671. doi: 10.1101/gad.869201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Betschinger J, Nichols J, Dietmann S, Corrin PD, Paddison PJ, Smith A. 2013. Exit from pluripotency is gated by intracellular redistribution of the bHLH transcription factor Tfe3. Cell 153:335–347. doi: 10.1016/j.cell.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang J, Chan YS, Loh YH, Cai J, Tong GQ, Lim CA, Robson P, Zhong S, Ng HH. 2008. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol 10:353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- 44.Leeb M, Dietmann S, Paramor M, Niwa H, Smith A. 2014. Genetic exploration of the exit from self-renewal using haploid embryonic stem cells. Cell Stem Cell 14:385–393. doi: 10.1016/j.stem.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fatica A, Bozzoni I. 2014. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 15:7–21. doi: 10.1038/nri3777. [DOI] [PubMed] [Google Scholar]
- 46.Flynn RA, Chang HY. 2014. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell 14:752–761. doi: 10.1016/j.stem.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES. 2011. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Navarro P, Chambers I, Karwacki-Neisius V, Chureau C, Morey C, Rougeulle C, Avner P. 2008. Molecular coupling of Xist regulation and pluripotency. Science 321:1693–1695. doi: 10.1126/science.1160952. [DOI] [PubMed] [Google Scholar]
- 49.Smith AG, Heath JK, Donaldson DD, Wong GG, Moreau J, Stahl M, Rogers D. 1988. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature 336:688–690. doi: 10.1038/336688a0. [DOI] [PubMed] [Google Scholar]
- 50.Williams RL, Hilton DJ, Pease S, Willson TA, Stewart CL, Gearing DP, Wagner EF, Metcalf D, Nicola NA, Gough NM. 1988. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature 336:684–687. doi: 10.1038/336684a0. [DOI] [PubMed] [Google Scholar]
- 51.Niwa H, Burdon T, Chambers I, Smith A. 1998. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev 12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bourillot PY, Aksoy I, Schreiber V, Wianny F, Schulz H, Hummel O, Hubner N, Savatier P. 2009. Novel STAT3 target genes exert distinct roles in the inhibition of mesoderm and endoderm differentiation in cooperation with Nanog. Stem Cells 27:1760–1771. doi: 10.1002/stem.110. [DOI] [PubMed] [Google Scholar]
- 53.Do DV, Ueda J, Messerschmidt DM, Lorthongpanich C, Zhou Y, Feng B, Guo G, Lin PJ, Hossain MZ, Zhang W, Moh A, Wu Q, Robson P, Ng HH, Poellinger L, Knowles BB, Solter D, Fu XY. 2013. A genetic and developmental pathway from STAT3 to the OCT4-NANOG circuit is essential for maintenance of ICM lineages in vivo. Genes Dev 27:1378–1390. doi: 10.1101/gad.221176.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martello G, Bertone P, Smith A. 2013. Identification of the missing pluripotency mediator downstream of leukaemia inhibitory factor. EMBO J 32:2561–2574. doi: 10.1038/emboj.2013.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stewart CL, Kaspar P, Brunet LJ, Bhatt H, Gadi I, Kontgen F, Abbondanzo SJ. 1992. Blastocyst implantation depends on maternal expression of leukaemia inhibitory factor. Nature 359:76–79. doi: 10.1038/359076a0. [DOI] [PubMed] [Google Scholar]
- 56.Ying QL, Nichols J, Chambers I, Smith A. 2003. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 115:281–292. doi: 10.1016/S0092-8674(03)00847-X. [DOI] [PubMed] [Google Scholar]
- 57.Yuan H, Corbi N, Basilico C, Dailey L. 1995. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev 9:2635–2645. doi: 10.1101/gad.9.21.2635. [DOI] [PubMed] [Google Scholar]
- 58.Lanner F, Rossant J. 2010. The role of FGF/Erk signaling in pluripotent cells. Development 137:3351–3360. doi: 10.1242/dev.050146. [DOI] [PubMed] [Google Scholar]
- 59.Arman E, Haffner-Krausz R, Chen Y, Heath JK, Lonai P. 1998. Targeted disruption of fibroblast growth factor (FGF) receptor 2 suggests a role for FGF signaling in pregastrulation mammalian development. Proc Natl Acad Sci U S A 95:5082–5087. doi: 10.1073/pnas.95.9.5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Feldman B, Poueymirou W, Papaioannou VE, DeChiara TM, Goldfarb M. 1995. Requirement of FGF-4 for postimplantation mouse development. Science 267:246–249. doi: 10.1126/science.7809630. [DOI] [PubMed] [Google Scholar]
- 61.Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. 1998. Promotion of trophoblast stem cell proliferation by FGF4. Science 282:2072–2075. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- 62.Kunath T, Saba-El-Leil MK, Almousailleakh M, Wray J, Meloche S, Smith A. 2007. FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development 134:2895–2902. doi: 10.1242/dev.02880. [DOI] [PubMed] [Google Scholar]
- 63.Burdon T, Stracey C, Chambers I, Nichols J, Smith A. 1999. Suppression of SHP-2 and ERK signalling promotes self-renewal of mouse embryonic stem cells. Dev Biol 210:30–43. doi: 10.1006/dbio.1999.9265. [DOI] [PubMed] [Google Scholar]
- 64.Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. 2008. The ground state of embryonic stem cell self-renewal. Nature 453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim J, Chu J, Shen X, Wang J, Orkin SH. 2008. An extended transcriptional network for pluripotency of embryonic stem cells. Cell 132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Di Croce L, Helin K. 2013. Transcriptional regulation by Polycomb group proteins. Nat Struct Mol Biol 20:1147–1155. doi: 10.1038/nsmb.2669. [DOI] [PubMed] [Google Scholar]
- 67.Herz HM, Garruss A, Shilatifard A. 2013. SET for life: biochemical activities and biological functions of SET domain-containing proteins. Trends Biochem Sci 38:621–639. doi: 10.1016/j.tibs.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Young RA. 2011. Control of the embryonic stem cell state. Cell 144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Morey L, Helin K. 2010. Polycomb group protein-mediated repression of transcription. Trends Biochem Sci 35:323–332. doi: 10.1016/j.tibs.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 70.Laugesen A, Helin K. 2014. Chromatin repressive complexes in stem cells, development, and cancer. Cell Stem Cell 14:735–751. doi: 10.1016/j.stem.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 71.Simon JA, Kingston RE. 2009. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol 10:697–708. doi: 10.1038/nrn2731. [DOI] [PubMed] [Google Scholar]
- 72.Scelfo A, Piunti A, Pasini D. 2014. The controversial role of the Polycomb group proteins in transcription and cancer: how much do we not understand Polycomb proteins? FEBS J 282:1703–1722. doi: 10.1111/febs.13112. [DOI] [PubMed] [Google Scholar]
- 73.Cao R, Zhang Y. 2004. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev 14:155–164. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 74.Morey L, Aloia L, Cozzuto L, Benitah SA, Di Croce L. 2013. RYBP and Cbx7 define specific biological functions of polycomb complexes in mouse embryonic stem cells. Cell Rep 3:60–69. doi: 10.1016/j.celrep.2012.11.026. [DOI] [PubMed] [Google Scholar]
- 75.Tavares L, Dimitrova E, Oxley D, Webster J, Poot R, Demmers J, Bezstarosti K, Taylor S, Ura H, Koide H, Wutz A, Vidal M, Elderkin S, Brockdorff N. 2012. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell 148:664–678. doi: 10.1016/j.cell.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gao Z, Zhang J, Bonasio R, Strino F, Sawai A, Parisi F, Kluger Y, Reinberg D. 2012. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell 45:344–356. doi: 10.1016/j.molcel.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu X, Johansen JV, Helin K. 2013. Fbxl10/Kdm2b recruits polycomb repressive complex 1 to CpG islands and regulates H2A ubiquitylation. Mol Cell 49:1134–1146. doi: 10.1016/j.molcel.2013.01.016. [DOI] [PubMed] [Google Scholar]
- 78.Riising EM, Comet I, Leblanc B, Wu X, Johansen JV, Helin K. 2014. Gene silencing triggers polycomb repressive complex 2 recruitment to CpG islands genome wide. Mol Cell 55:347–360. doi: 10.1016/j.molcel.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 79.Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, Adli M, Kasif S, Ptaszek LM, Cowan CA, Lander ES, Koseki H, Bernstein BE. 2008. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 81.Schoeftner S, Sengupta AK, Kubicek S, Mechtler K, Spahn L, Koseki H, Jenuwein T, Wutz A. 2006. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J 25:3110–3122. doi: 10.1038/sj.emboj.7601187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Leeb M, Wutz A. 2007. Ring1B is crucial for the regulation of developmental control genes and PRC1 proteins but not X inactivation in embryonic cells. J Cell Biol 178:219–229. doi: 10.1083/jcb.200612127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Blackledge NP, Farcas AM, Kondo T, King HW, McGouran JF, Hanssen LL, Ito S, Cooper S, Kondo K, Koseki Y, Ishikura T, Long HK, Sheahan TW, Brockdorff N, Kessler BM, Koseki H, Klose RJ. 2014. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell 157:1445–1459. doi: 10.1016/j.cell.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Endoh M, Endo TA, Endoh T, Isono K, Sharif J, Ohara O, Toyoda T, Ito T, Eskeland R, Bickmore WA, Vidal M, Bernstein BE, Koseki H. 2012. Histone H2A mono-ubiquitination is a crucial step to mediate PRC1-dependent repression of developmental genes to maintain ES cell identity. PLoS Genet 8:e1002774. doi: 10.1371/journal.pgen.1002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Aloia L, Di Stefano B, Sessa A, Morey L, Santanach A, Gutierrez A, Cozzuto L, Benitah SA, Graf T, Broccoli V, Di Croce L. 2014. Zrf1 is required to establish and maintain neural progenitor identity. Genes Dev 28:182–197. doi: 10.1101/gad.228510.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Richly H, Rocha-Viegas L, Ribeiro JD, Demajo S, Gundem G, Lopez-Bigas N, Nakagawa T, Rospert S, Ito T, Di Croce L. 2010. Transcriptional activation of polycomb-repressed genes by ZRF1. Nature 468:1124–1128. doi: 10.1038/nature09574. [DOI] [PubMed] [Google Scholar]
- 87.Morey L, Pascual G, Cozzuto L, Roma G, Wutz A, Benitah SA, Di Croce L. 2012. Nonoverlapping functions of the Polycomb group Cbx family of proteins in embryonic stem cells. Cell Stem Cell 10:47–62. doi: 10.1016/j.stem.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 88.O'Loghlen A, Munoz-Cabello AM, Gaspar-Maia A, Wu HA, Banito A, Kunowska N, Racek T, Pemberton HN, Beolchi P, Lavial F, Masui O, Vermeulen M, Carroll T, Graumann J, Heard E, Dillon N, Azuara V, Snijders AP, Peters G, Bernstein E, Gil J. 2012. MicroRNA regulation of Cbx7 mediates a switch of Polycomb orthologs during ESC differentiation. Cell Stem Cell 10:33–46. doi: 10.1016/j.stem.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Farcas AM, Blackledge NP, Sudbery I, Long HK, McGouran JF, Rose NR, Lee S, Sims D, Cerase A, Sheahan TW, Koseki H, Brockdorff N, Ponting CP, Kessler BM, Klose RJ. 2012. KDM2B links the Polycomb repressive complex 1 (PRC1) to recognition of CpG islands. eLife 1:e00205. doi: 10.7554/eLife.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Agger K, Christensen J, Cloos PA, Helin K. 2008. The emerging functions of histone demethylases. Curr Opin Genet Dev 18:159–168. doi: 10.1016/j.gde.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 91.Hisada K, Sanchez C, Endo TA, Endoh M, Roman-Trufero M, Sharif J, Koseki H, Vidal M. 2012. RYBP represses endogenous retroviruses and preimplantation- and germ line-specific genes in mouse embryonic stem cells. Mol Cell Biol 32:1139–1149. doi: 10.1128/MCB.06441-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vizan P, Beringer M, Ballare C, Di Croce L. 2014. Role of PRC2-associated factors in stem cells and disease. FEBS J 282:1723–1735. doi: 10.1111/febs.13083. [DOI] [PubMed] [Google Scholar]
- 93.Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J. 2009. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell 139:1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pasini D, Cloos PA, Walfridsson J, Olsson L, Bukowski JP, Johansen JV, Bak M, Tommerup N, Rappsilber J, Helin K. 2010. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature 464:306–310. doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- 95.Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D. 2010. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev 24:368–380. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Landeira D, Sauer S, Poot R, Dvorkina M, Mazzarella L, Jorgensen HF, Pereira CF, Leleu M, Piccolo FM, Spivakov M, Brookes E, Pombo A, Fisher C, Skarnes WC, Snoek T, Bezstarosti K, Demmers J, Klose RJ, Casanova M, Tavares L, Brockdorff N, Merkenschlager M, Fisher AG. 2010. Jarid2 is a PRC2 component in embryonic stem cells required for multi-lineage differentiation and recruitment of PRC1 and RNA polymerase II to developmental regulators. Nat Cell Biol 12:618–624. doi: 10.1038/ncb2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Savla U, Benes J, Zhang J, Jones RS. 2008. Recruitment of Drosophila Polycomb-group proteins by Polycomblike, a component of a novel protein complex in larvae. Development 135:813–817. doi: 10.1242/dev.016006. [DOI] [PubMed] [Google Scholar]
- 98.Qin S, Guo Y, Xu C, Bian C, Fu M, Gong S, Min J. 2013. Tudor domains of the PRC2 components PHF1 and PHF19 selectively bind to histone H3K36me3. Biochem Biophys Res Commun 430:547–553. doi: 10.1016/j.bbrc.2012.11.116. [DOI] [PubMed] [Google Scholar]
- 99.Brien GL, Gambero G, O'Connell DJ, Jerman E, Turner SA, Egan CM, Dunne EJ, Jurgens MC, Wynne K, Piao L, Lohan AJ, Ferguson N, Shi X, Sinha KM, Loftus BJ, Cagney G, Bracken AP. 2012. Polycomb PHF19 binds H3K36me3 and recruits PRC2 and demethylase NO66 to embryonic stem cell genes during differentiation. Nat Struct Mol Biol 19:1273–1281. doi: 10.1038/nsmb.2449. [DOI] [PubMed] [Google Scholar]
- 100.Ballare C, Lange M, Lapinaite A, Martin GM, Morey L, Pascual G, Liefke R, Simon B, Shi Y, Gozani O, Carlomagno T, Benitah SA, Di Croce L. 2012. Phf19 links methylated Lys36 of histone H3 to regulation of Polycomb activity. Nat Struct Mol Biol 19:1257–1265. doi: 10.1038/nsmb.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Walker E, Chang WY, Hunkapiller J, Cagney G, Garcha K, Torchia J, Krogan NJ, Reiter JF, Stanford WL. 2010. Polycomb-like 2 associates with PRC2 and regulates transcriptional networks during mouse embryonic stem cell self-renewal and differentiation. Cell Stem Cell 6:153–166. doi: 10.1016/j.stem.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang Z, Jones A, Sun CW, Li C, Chang CW, Joo HY, Dai Q, Mysliwiec MR, Wu LC, Guo Y, Yang W, Liu K, Pawlik KM, Erdjument-Bromage H, Tempst P, Lee Y, Min J, Townes TM, Wang H. 2011. PRC2 complexes with JARID2, MTF2, and esPRC2p48 in ES cells to modulate ES cell pluripotency and somatic cell reprogramming. Stem Cells 29:229–240. doi: 10.1002/stem.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kaneko S, Son J, Bonasio R, Shen SS, Reinberg D. 2014. Nascent RNA interaction keeps PRC2 activity poised and in check. Genes Dev 28:1983–1988. doi: 10.1101/gad.247940.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Davidovich C, Zheng L, Goodrich KJ, Cech TR. 2013. Promiscuous RNA binding by Polycomb repressive complex 2. Nat Struct Mol Biol 20:1250–1257. doi: 10.1038/nsmb.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Forzati F, Federico A, Pallante P, Abbate A, Esposito F, Malapelle U, Sepe R, Palma G, Troncone G, Scarfo M, Arra C, Fedele M, Fusco A. 2012. CBX7 is a tumor suppressor in mice and humans. J Clin Invest 122:612–623. doi: 10.1172/JCI58620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, Koseki H, Brockdorff N. 2004. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell 7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 107.Endoh M, Endo TA, Endoh T, Fujimura Y, Ohara O, Toyoda T, Otte AP, Okano M, Brockdorff N, Vidal M, Koseki H. 2008. Polycomb group proteins Ring1A/B are functionally linked to the core transcriptional regulatory circuitry to maintain ES cell identity. Development 135:1513–1524. doi: 10.1242/dev.014340. [DOI] [PubMed] [Google Scholar]
- 108.Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. 2004. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J 23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. 2007. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol 27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang J, Mager J, Schnedier E, Magnuson T. 2002. The mouse PcG gene eed is required for Hox gene repression and extraembryonic development. Mamm Genome 13:493–503. doi: 10.1007/s00335-002-2182-7. [DOI] [PubMed] [Google Scholar]
- 111.O'Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T. 2001. The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol 21:4330–4336. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Faust C, Schumacher A, Holdener B, Magnuson T. 1995. The eed mutation disrupts anterior mesoderm production in mice. Development 121:273–285. [DOI] [PubMed] [Google Scholar]
- 113.Faust C, Lawson KA, Schork NJ, Thiel B, Magnuson T. 1998. The Polycomb-group gene eed is required for normal morphogenetic movements during gastrulation in the mouse embryo. Development 125:4495–4506. [DOI] [PubMed] [Google Scholar]
- 114.Leeb M, Pasini D, Novatchkova M, Jaritz M, Helin K, Wutz A. 2010. Polycomb complexes act redundantly to repress genomic repeats and genes. Genes Dev 24:265–276. doi: 10.1101/gad.544410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, Orkin SH. 2008. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell 32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fisher CL, Fisher AG. 2011. Chromatin states in pluripotent, differentiated, and reprogrammed cells. Curr Opin Genet Dev 21:140–146. doi: 10.1016/j.gde.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 117.Pasini D, Cloos PAC, Walfridsson J, Olsson L, Bukowski J-P, Johansen JV, Bak M, Tommerup N, Rappsilber J, Helin K. 2009. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature 464:3006–3610. doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- 118.Hu D, Garruss AS, Gao X, Morgan MA, Cook M, Smith ER, Shilatifard A. 2013. The Mll2 branch of the COMPASS family regulates bivalent promoters in mouse embryonic stem cells. Nat Struct Mol Biol 20:1093–1097. doi: 10.1038/nsmb.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hu D, Gao X, Morgan MA, Herz HM, Smith ER, Shilatifard A. 2013. The MLL3/MLL4 branches of the COMPASS family function as major histone H3K4 monomethylases at enhancers. Mol Cell Biol 33:4745–4754. doi: 10.1128/MCB.01181-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 121.Voigt P, Tee WW, Reinberg D. 2013. A double take on bivalent promoters. Genes Dev 27:1318–1338. doi: 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Clouaire T, Webb S, Skene P, Illingworth R, Kerr A, Andrews R, Lee JH, Skalnik D, Bird A. 2012. Cfp1 integrates both CpG content and gene activity for accurate H3K4me3 deposition in embryonic stem cells. Genes Dev 26:1714–1728. doi: 10.1101/gad.194209.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lubitz S, Glaser S, Schaft J, Stewart AF, Anastassiadis K. 2007. Increased apoptosis and skewed differentiation in mouse embryonic stem cells lacking the histone methyltransferase Mll2. Mol Biol Cell 18:2356–2366. doi: 10.1091/mbc.E06-11-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ang YS, Tsai SY, Lee DF, Monk J, Su J, Ratnakumar K, Ding J, Ge Y, Darr H, Chang B, Wang J, Rendl M, Bernstein E, Schaniel C, Lemischka IR. 2011. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell 145:183–197. doi: 10.1016/j.cell.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sanchez C, Sanchez I, Demmers JA, Rodriguez P, Strouboulis J, Vidal M. 2007. Proteomics analysis of Ring1B/Rnf2 interactors identifies a novel complex with the Fbxl10/Jhdm1B histone demethylase and the Bcl6 interacting corepressor. Mol Cell Proteomics 6:820–834. doi: 10.1074/mcp.M600275-MCP200. [DOI] [PubMed] [Google Scholar]
- 126.Wan M, Liang J, Xiong Y, Shi F, Zhang Y, Lu W, He Q, Yang D, Chen R, Liu D, Barton M, Songyang Z. 2013. The trithorax group protein Ash2l is essential for pluripotency and maintaining open chromatin in embryonic stem cells. J Biol Chem 288:5039–5048. doi: 10.1074/jbc.M112.424515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jiang H, Shukla A, Wang X, Chen WY, Bernstein BE, Roeder RG. 2011. Role for Dpy-30 in ES cell-fate specification by regulation of H3K4 methylation within bivalent domains. Cell 144:513–525. doi: 10.1016/j.cell.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D, Di Croce L, Shiekhattar R. 2007. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science 318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 129.Shpargel KB, Sengoku T, Yokoyama S, Magnuson T. 2012. UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLoS Genet 8:e1002964. doi: 10.1371/journal.pgen.1002964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, Iwase S, Alpatov R, Issaeva I, Canaani E, Roberts TM, Chang HY, Shi Y. 2007. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- 131.Welstead GG, Creyghton MP, Bilodeau S, Cheng AW, Markoulaki S, Young RA, Jaenisch R. 2012. X-linked H3K27me3 demethylase Utx is required for embryonic development in a sex-specific manner. Proc Natl Acad Sci U S A 109:13004–13009. doi: 10.1073/pnas.1210787109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wang C, Lee JE, Cho YW, Xiao Y, Jin Q, Liu C, Ge K. 2012. UTX regulates mesoderm differentiation of embryonic stem cells independent of H3K27 demethylase activity. Proc Natl Acad Sci U S A 109:15324–15329. doi: 10.1073/pnas.1204166109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Morales Torres C, Laugesen A, Helin K. 2013. Utx is required for proper induction of ectoderm and mesoderm during differentiation of embryonic stem cells. PLoS One 8:e60020. doi: 10.1371/journal.pone.0060020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hu G, Cui K, Northrup D, Liu C, Wang C, Tang Q, Ge K, Levens D, Crane-Robinson C, Zhao K. 2013. H2A.Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation. Cell Stem Cell 12:180–192. doi: 10.1016/j.stem.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]