Abstract
Culturing before DNA extraction represents a major time-consuming step in whole-genome sequencing of slow-growing bacteria, such as Mycobacterium tuberculosis. We report a workflow to extract DNA from frozen isolates without reculturing. Prepared libraries and sequence data were comparable with results from recultured aliquots of the same stocks.
TEXT
In recent years, studies employing whole-genome sequencing (WGS) of Mycobacterium tuberculosis isolates have demonstrated its value in understanding transmission patterns, recurrent tuberculosis (TB), development of drug resistance, and bacterial evolution (1–9). In parallel, improvements of next-generation sequencing platforms and library preparation workflows make it possible to determine the genome sequence from bacterial DNA samples in the time span of 1 week. Reculturing isolates for DNA isolation has been reported as a necessary step in published WGS studies (2, 4, 7, 10). As culturing of slow-growing bacteria, such as M. tuberculosis, takes from 1 week to several weeks, it constitutes the main time-consuming process in WGS projects (11–14). While initially large amounts of DNA were required for reliable WGS library preparation, newly developed library preparation protocols for bacterial samples typically require about 1 to 10 ng of DNA (e.g., Illumina Nextera XT [1 ng], New England BioLabs NEBNext Ultra [5 to 1,000 ng], and Bioo Scientific NEXTflex ChIP-Seq [1 to 10 ng]). Therefore, faster and reliable methods for DNA isolation would enable a considerable decrease in the time needed to perform WGS analyses. In this regard, a recent study proposed a WGS workflow starting from early positive liquid cultures of the MGIT system (15), potentially enhancing the speed of WGS procedures as part of routine diagnostics.
Furthermore, reculturing of isolates from even well-maintained frozen stocks can fail entirely, usually excluding the respective isolate from any further analysis. A recent publication reported failure rates of up to 50% for M. tuberculosis glycerol stocks (16). New methods enabling WGS analysis directly from frozen stocks without reculturing can rescue genotype information, especially from historic collections of isolates.
In this study, we investigated and tested a protocol for performing WGS of DNA extracted directly from frozen glycerol stocks, which were all historic isolates from patients diagnosed with fully susceptible TB between 1992 and 2012, circumventing the step of reculturing altogether.
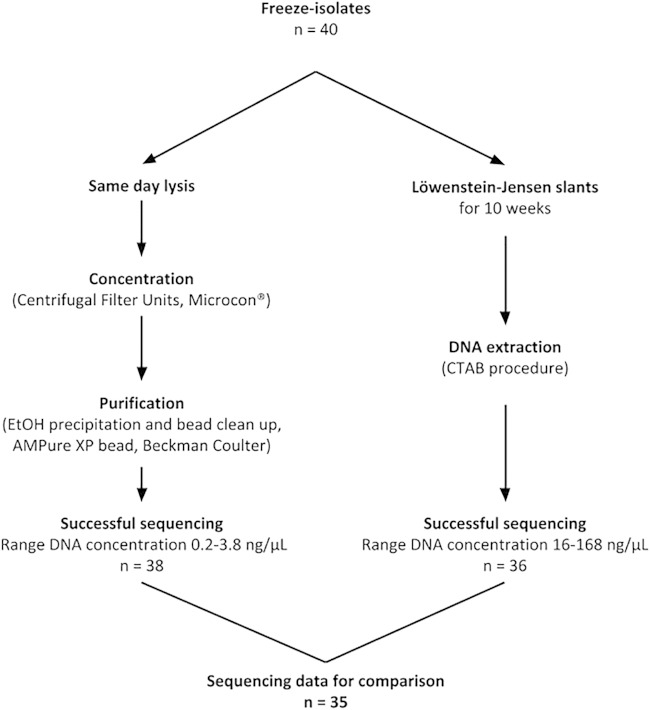
For validation, we sequenced DNA from cultured aliquots of the same frozen stocks in parallel. In total, we included 40 frozen glycerol stocks (1992 to 2012) stored at −80°C at the International Reference Laboratory of Mycobacteriology at the Statens Serum Institut (Copenhagen). All were processed according to a standard lysis protocol with heat inactivation and sonication as is usually used for PCR (17, 18). Lysates were concentrated with Microcon filters (Merck KGaA, Darmstadt, Germany), followed by a purification with ethanol (EtOH) precipitation and bead clean up (AMPure XP bead; Beckman Coulter, Krefeld, Germany) (15). For culture, aliquots of frozen stocks were put on solid Löwenstein-Jensen (LJ) slants and incubated at 35°C. After approximately 10 weeks of growth, DNA extraction was performed with the traditional cetyltrimethylammonium bromide (CTAB) procedure (19) from visible colonies. For each extraction method, we measured final DNA concentrations with the Qubit 2.0 Fluorometer (Fig. 1; see also the supplemental material for further details). Libraries for WGS were prepared from DNA samples with the Nextera XT kits and run on Illumina next-generation sequencing platforms (MiSeq) as instructed by the manufacturer (Illumina, San Diego, CA, USA). Respective fastq files were submitted to the EMBL EBI ENA short read archive (accession number PRJEB9308). Reads were mapped to the genome of the M. tuberculosis reference strain H37Rv (GenBank accession number NC_000962.3) with the alignment program SARUMAN (20). For variant detection in mapped reads, we employed minimum thresholds of 10× coverage and 75% allele frequency and filtered results for repetitive regions (1, 2). Detected variants were manually curated using the Integrative Genome Viewer software (21) to visualize mapped reads.
FIG 1.
The workflow of DNA extraction from 40 frozen glycerol stocks with same day direct lysis from frozen glycerol stocks or reculturing of aliquots.
Of the 40 selected glycerol stocks, 35 were successfully sequenced from DNA extracted from glycerol stocks and culture. Five isolates were excluded due to failure to grow (n = 4) on any media (LJ, liquid Dubos, blood agar) or continuous failure of library preparation from DNA directly isolated from stock (n = 2). For the remaining 35 isolates, the median DNA concentration from direct isolation was 0.67 ng/μl (range, 0.2 to 3.8 ng/μl). DNA extraction with direct lysis of glycerol stocks, DNA concentration, and purification takes less than a day in total. In comparison, most samples grown on LJ medium are normally visible within 3 to 8 weeks, and DNA extraction with the CTAB procedure takes 3 days.
For the two methods, mapping of the genome to the reference strain revealed a genome-wide coverage in the range of 98.8% to 99.5% and medians of 99.3% for glycerol stocks and 99.4% for cultured aliquots. The median percentages of the reference genome fulfilling thresholds for variant detection was 97.9% (range, 96.6% to 98.9%) for glycerol stocks and 98.5% (range, 97.5% to 98.9%) for cultured aliquots.
Analysis of whole-genome sequencing data revealed only two instances where paired genotypes from glycerol stocks or cultured material differed in detected variants. In these cases, sequence data for cultivated aliquots exhibited one additional single nucleotide polymorphism (SNP) compared to the H37Rv reference genome. We detected one SNP in the rRNA gene Rvnr01 at position 1471870 for sample GE0078 and one nonsynonymous SNP in the Rv3085 gene-coding region at position 3199069 for sample GE0079 (see Table 1).
TABLE 1.
DNA concentration and reference genome coverage for DNA extracted from directly lysed glycerol stocks or from recultured aliquotsa
| Year | Sample no. | DNA from directly lysed samplesb |
DNA from recultured samples |
Comparison |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Qubit (HS) before purification (ng/μl) | Qubit (HS) after purification (ng/μl) | Cov any (mean) | Cov any (%) | Cov unamb (%) | Visible growth | Qubit (HS) after 1:2 dilution (ng/μl) | Cov any (mean) | Cov any (%) | Cov unamb (%) | Library from (gly/LJ) | No. of SNPs | ||
| 1992 | GE0005 | 0.08 | 0.07 | 52.1 | 99.4 | 98.3 | No | Yes/no | |||||
| 1992 | GE0006 | 0.22 | 0.16 | 63.6 | 99.4 | 98.2 | Yes | 29 | 92.4 | 99.4 | 98.5 | Yes/yes | 0 |
| 1992 | GE0007 | 0.39 | 0.15 | Yes | 79 | 56.4 | 99.2 | 97.6 | No/yes | ||||
| 1992 | GE0008 | 0.71 | 0.37 | 61.7 | 99.4 | 98.3 | Yes | 16 | 83.7 | 99.5 | 98.7 | Yes/yes | 0 |
| 1992 | GE0009 | 0.71 | 0.23 | 64.7 | 99.3 | 98.3 | No | Yes/no | |||||
| 1992 | GE0011 | 0.47 | 0.99 | No | No/no | ||||||||
| 1992 | GE0012 | 0.18 | 0.18 | 94.3 | 98.9 | 98.0 | No | Yes/no | |||||
| 2002 | GE0022 | Too low | 0.53 | 64.4 | 99.4 | 98.1 | Yes | 63 | 111.3 | 99.4 | 98.6 | Yes/yes | 0 |
| 2003 | GE0028 | 0.37 | 0.37 | 59.9 | 99.2 | 97.7 | Yes | 87 | 90.0 | 99.3 | 98.2 | Yes/yes | 0 |
| 2003 | GE0030 | 0.30 | 0.75 | 92.4 | 99.4 | 98.5 | Yes | 57 | 78.7 | 99.4 | 98.5 | Yes/yes | 0 |
| 2005 | GE0039 | 0.20 | 0.17 | 68.0 | 99.2 | 97.9 | Yes | 74 | 111.6 | 99.4 | 98.6 | Yes/yes | 0 |
| 2005 | GE0040 | 0.70 | 0.22 | 92.3 | 99.5 | 98.9 | Yes | 37 | 118.4 | 99.4 | 98.6 | Yes/yes | 0 |
| 2005 | GE0041 | 0.40 | 0.27 | 70.8 | 99.4 | 98.3 | Yes | 33 | 105.3 | 99.5 | 98.7 | Yes/yes | 0 |
| 2005 | GE0042 | 0.71 | 0.91 | 63.6 | 99.3 | 97.6 | Yes | 14 | 107.1 | 99.4 | 98.7 | Yes/yes | 0 |
| 2005 | GE0044 | 0.48 | 0.30 | 71.0 | 99.3 | 97.9 | Yes | 42 | 102.2 | 99.4 | 98.5 | Yes/yes | 0 |
| 2005 | GE0045 | 0.09 | 1.30 | 73.4 | 99.3 | 98.1 | Yes | 50 | 89.3 | 99.3 | 98.3 | Yes/yes | 0 |
| 2005 | GE0046 | 0.36 | 0.34 | 66.5 | 99.3 | 98.1 | Yes | 90 | 89.5 | 99.4 | 98.6 | Yes/yes | 0 |
| 2005 | GE0047 | 0.76 | 0.89 | 67.7 | 99.3 | 98.2 | Yes | 28 | 77.2 | 99.4 | 98.5 | Yes/yes | 0 |
| 2005 | GE0048 | 0.32 | 0.44 | 54.4 | 99.3 | 98.0 | Yes | 37 | 65.1 | 99.3 | 98.2 | Yes/yes | 0 |
| 2005 | GE0049 | 0.29 | 0.24 | 59.3 | 99.3 | 98.0 | Yes | 42 | 58.2 | 99.4 | 98.2 | Yes/yes | 0 |
| 2005 | GE0050 | 0.36 | 0.39 | 64.2 | 99.3 | 97.8 | Yes | 26 | 56.0 | 99.4 | 98.3 | Yes/yes | 0 |
| 2006 | GE0052 | 0.22 | 0.78 | 71.7 | 99.3 | 98.1 | Yes | 37 | 102.5 | 99.5 | 98.8 | Yes/yes | 0 |
| 2005 | GE0054 | 1.93 | 2.32 | 73.0 | 99.3 | 98.0 | Yes | 50 | 87.1 | 99.4 | 98.7 | Yes/yes | 0 |
| 2005 | GE0055 | 1.53 | 3.17 | 75.3 | 99.4 | 98.0 | Yes | 20 | 65.3 | 99.4 | 98.4 | Yes/yes | 0 |
| 2006 | GE0056 | 0.79 | 1.21 | 82.5 | 99.4 | 98.3 | Yes | 59 | 85.9 | 99.4 | 98.6 | Yes/yes | 0 |
| 2006 | GE0057 | 0.69 | 0.90 | 56.3 | 99.4 | 97.9 | Yes | 81 | 93.0 | 99.3 | 98.5 | Yes/yes | 0 |
| 2006 | GE0058 | 0.66 | 0.64 | 67.6 | 99.3 | 97.9 | Yes | 34 | 75.3 | 99.4 | 98.4 | Yes/yes | 0 |
| 2008 | GE0069 | 0.68 | 0.68 | 67.6 | 99.4 | 98.1 | Yes | 65 | 91.6 | 99.4 | 98.6 | Yes/yes | 0 |
| 2008 | GE0070 | 0.30 | 0.37 | 69.8 | 99.4 | 98.1 | Yes | 20 | 67.9 | 99.3 | 98.2 | Yes/yes | 0 |
| 2009 | GE0072 | 1.05 | 1.18 | 55.7 | 99.2 | 97.6 | Yes | 74 | 85.0 | 99.4 | 98.5 | Yes/yes | 0 |
| 2009 | GE0073 | 1.28 | 1.47 | 87.0 | 99.2 | 97.6 | Yes | 31 | 69.3 | 99.4 | 98.5 | Yes/yes | 0 |
| 2009 | GE0074 | 1.56 | 1.61 | 70.2 | 99.2 | 97.5 | Yes | 101 | 111.3 | 99.5 | 98.8 | Yes/yes | 0 |
| 2009 | GE0077 | 2.05 | 3.76 | 75.7 | 99.3 | 98.1 | Yes | 71 | 88.5 | 99.4 | 98.6 | Yes/yes | 0 |
| 2009 | GE0078 | 2.15 | 2.55 | 69.3 | 99.4 | 98.1 | Yes | 107 | 118.4 | 99.5 | 98.8 | Yes/yes | 1 |
| 2009 | GE0079 | 2.01 | 1.45 | 59.4 | 99.3 | 97.9 | Yes | 169 | 117.1 | 99.5 | 98.9 | Yes/yes | 1 |
| 2009 | GE0080 | 0.66 | 0.54 | 60.3 | 99.1 | 96.9 | Yes | 65 | 72.3 | 99.3 | 98.2 | Yes/yes | 0 |
| 2009 | GE0081 | 1.87 | 1.66 | 64.7 | 99.3 | 97.8 | Yes | 50 | 60.9 | 99.3 | 98.1 | Yes/yes | 0 |
| 2009 | GE0082 | 1.44 | 1.43 | 63.0 | 99.2 | 97.6 | Yes | 98 | 75.1 | 99.4 | 98.4 | Yes/yes | 0 |
| 2012 | GE0148 | 0.69 | 0.70 | 70.2 | 98.8 | 97.7 | Yes | 33 | 56.3 | 98.8 | 97.5 | Yes/yes | 0 |
| 2012 | GE0151 | 0.70 | 0.65 | 67.9 | 99.3 | 97.9 | Yes | 91 | 112.2 | 99.5 | 98.8 | Yes/yes | 0 |
In total, analysis of sequencing data from paired DNA samples differed in two cases. An additional SNP was found for the recultured aliquot: for sample GE0078 one SNP at position 1471870 in the rRNA gene Rvnr01 and for sample GE0079 one nonsynonymous SNP at position 3199069 in the Rv3085 gene.
HS, high sensitivity; cov, genome coverage; unamb, unambiguous; gly, glycerol stocks.
Library preparation and whole-genome sequencing succeeded for 38 (95%) of the glycerol stocks with a DNA yield well in the range required for WGS library preparation (range, 0.2 to 3.8 ng/μl). In contrast, culture and DNA extraction succeeded for 36 of the 40 (90.0%) glycerol stocks. In total, DNA was obtained from 39 of the 40 lysed frozen stocks with one or both methods. Reference mapping-based analysis gave excellent results, with at least 96.6% of the reference genome covered at high quality (10× coverage, 75% allele identity) for directly isolated DNA and DNA extracted from cultured stocks.
In conclusion, DNA extraction, library preparation, and sequencing was successful in three samples that exhibited no growth on LJ slants; hence, the proposed method can even be used for samples where reculturing is not possible. As results are comparable between DNA obtained directly from frozen stocks versus DNA extracted from cultured aliquots, this study shows that reculturing of frozen glycerol stocks is not needed in order to perform whole-genome sequencing analysis for epidemiologic research. On the contrary, reculturing might introduce mutations in the genome, which likely happened in two cases for our sample set. In samples with distinct clones, culturing might also change the relative ratio for intrapatient evolving clones (7) and for coinfections (22). This would be even more pronounced for strains carrying fitness-reducing resistance mutations (23). Since direct lysis of frozen stocks, concentration, and purification can be performed in 1 day while reculturing and DNA extraction with the CTAB procedure takes several weeks, this finding can facilitate implementation of WGS as a routine clinical tool.
Nucleotide sequence accession number.
Sequence reads were submitted to the EMBL EBI ENA short read archive under accession number PRJEB9308.
Supplementary Material
ACKNOWLEDGMENTS
We thank Timothy Walker, Viola Schleusener, and Christine Rohde for helpful discussions and Antonina Votintseva and Louise Pankhurst for excellent advice and help with the bead-based DNA purification protocol. We also thank the laboratory staff at the International Reference Laboratory of Mycobacteriology, Statens Serum Institut, Denmark for their help with sample preparation.
All authors contributed to the conception and design of the study, the analyses of data, and writing of the article.
Parts of this work have been supported by the Commission for Scientific Research in Greenland (0605-00062B), Seed Funding Initiative for Collaboration of the Universities of Southern Denmark, Kiel, Hamburg, and Aarhus, Lundbeck Foundation (R151-2013-14628), Novo Nordisk Foundation (7651), the European Union TB-PAN-NET (FP7-223681), Patho-Ngen-Trace (FP7-278864-2) projects, and the German Center for Infection Research (DZIF). The funders had no role in study design, data collection or analysis, the decision to publish, or preparation of the manuscript.
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00662-15.
REFERENCES
- 1.Roetzer A, Diel R, Kohl TA, Rückert C, Nübel U, Blom J, Wirth T, Jaenicke S, Schuback S, Rüsch-Gerdes S, Supply P, Kalinowski J, Niemann S. 2013. Whole genome sequencing versus traditional genotyping for investigation of a Mycobacterium tuberculosis outbreak: a longitudinal molecular epidemiological study. PLoS Med 10:e1001387. doi: 10.1371/journal.pmed.1001387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walker TM, Ip CLC, Harrell RH, Evans JT, Kapatai G, Dedicoat MJ, Eyre DW, Wilson DJ, Hawkey PM, Crook DW, Parkhill J, Harris D, Walker AS, Bowden R, Monk P, Smith EG, Peto TEA. 2013. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect Dis 13:137–146. doi: 10.1016/S1473-3099(12)70277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bryant JM, Schürch AC, van Deutekom H, Harris SR, de Beer JL, de Jager V, Kremer K, van Hijum SA, Siezen RJ, Borgdorff M, Bentley SD, Parkhill J, van Soolingen D. 2013. Inferring patient to patient transmission of Mycobacterium tuberculosis from whole genome sequencing data. BMC Infect Dis 13:110. doi: 10.1186/1471-2334-13-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walker TM, Lalor MK, Broda A, Saldana Ortega L, Morgan M, Parker L, Churchill S, Bennett K, Golubchik T, Giess AP, Del Ojo Elias C, Jeffery KJ, Bowler ICJW, Laurenson IF, Barrett A, Drobniewski F, McCarthy ND, Anderson LF, Abubakar I, Thomas HL, Monk P, Smith EG, Walker AS, Crook DW, Peto TEA, Conlon CP. 2014. Assessment of Mycobacterium tuberculosis transmission in Oxfordshire, UK, 2007-12, with whole pathogen genome sequences: an observational study. Lancet Respir Med 2:285–292. doi: 10.1016/S2213-2600(14)70027-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guerra-Assunção JA, Houben RMGJ, Crampin AC, Mzembe T, Mallard K, Coll F, Khan P, Banda L, Chiwaya A, Pereira RPA, McNerney R, Harris D, Parkhill J, Clark TG, Glynn JR. 2015. Recurrence due to relapse or reinfection with Mycobacterium tuberculosis: a whole-genome sequencing approach in a large, population-based cohort with a high HIV infection prevalence and active follow-up. J Infect Dis 211:1154–1163. doi: 10.1093/infdis/jiu574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eldholm V, Norheim G, von der Lippe B, Kinander W, Dahle UR, Caugant DA, Mannsåker T, Mengshoel AT, Dyrhol-Riise AM, Balloux F. 2014. Evolution of extensively drug-resistant Mycobacterium tuberculosis from a susceptible ancestor in a single patient. Genome Biol 15:490. doi: 10.1186/s13059-014-0490-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Merker M, Kohl TA, Roetzer A, Truebe L, Richter E, Rüsch-Gerdes S, Fattorini L, Oggioni MR, Cox H, Varaine F, Niemann S. 2013. Whole genome sequencing reveals complex evolution patterns of multidrug-resistant Mycobacterium tuberculosis Beijing strains in patients. PLoS One 8:e82551. doi: 10.1371/journal.pone.0082551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galagan JE. 2014. Genomic insights into tuberculosis. Nat Rev Genet 15:307–320. doi: 10.1038/nrg3664. [DOI] [PubMed] [Google Scholar]
- 9.Merker M, Blin C, Mona S, Duforet-Frebourg N, Lecher S, Willery E, Blum M, Rüsch-Gerdes S, Mokrousov I, Aleksic E, Allix-Béguec C, Antierens A, Augustynowicz-Kopec E, Ballif M, Barletta F, Beck HP, Barry CE, Bonnet M, Borroni E, Campos-Herrero I, Cirillo D, Cox H, Crowe S, Crudu V, Diel R, Drobniewski F, Fauville-Dufaux M, Gagneux S, Ghebremichael S, Hanekom M, Hoffner S, Jiao W-W, Kalon S, Kohl TA, Kontsevaya I, Lillebæk T, Maeda S, Nikolayevskyy V, Rasmussen M, Rastogi N, Samper S, Sanchez-Padilla E, Savic B, Shamputa IC, Shen A, Sng L-H, Stakenas P, Toit K, Varaine F, Vukovic D, Wahl C, Warren R, Supply P, Niemann S, Wirth T. 2015. Evolutionary history and global spread of the Mycobacterium tuberculosis Beijing lineage. Nat Genet 47:242–249. doi: 10.1038/ng.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stucki D, Ballif M, Bodmer T, Coscolla M, Maurer A-M, Droz S, Butz C, Borrell S, Längle C, Feldmann J, Furrer H, Mordasini C, Helbling P, Rieder HL, Egger M, Gagneux S, Fenner L. 2014. Tracking a tuberculosis outbreak over 21 years: strain-specific single-nucleotide polymorphism typing combined with targeted whole-genome sequencing. J Infect Dis 211:1306–1316. doi: 10.1093/infdis/jiu601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crump JA, Morrissey AB, Ramadhani HO, Njau BN, Maro VP, Reller LB. 2011. Controlled comparison of BacT/Alert MB system, manual Myco/F lytic procedure, and isolator 10 system for diagnosis of Mycobacterium tuberculosis bacteremia. J Clin Microbiol 49:3054–3057. doi: 10.1128/JCM.01035-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cook GM, Berney M, Gebhard S, Heinemann M, Cox RA, Danilchanka O, Niederweis M. 2009. Physiology of mycobacteria. Adv Microb Physiol 55:81–182. doi: 10.1016/S0065-2911(09)05502-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naveen G, Peerapur BV. 2012. Comparison of the Lowenstein-Jensen medium, the Middlebrook 7H10 medium and MB/BacT for the isolation of Mycobacterium tuberculosis (MTB) from clinical specimens. J Clin Diagn Res 6:1704–1709. doi: 10.7860/JCDR/2012/4603.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feyzioglu B, Dogan M, Sanli OO, Ozdemir M, Baykan M. 2014. Comparison of the performance of TK system with LJ and MGIT methods in the diagnosis of tuberculosis. Int J Clin Exp Med 7:1084–1088. [PMC free article] [PubMed] [Google Scholar]
- 15.Votintseva AA, Pankhurst LJ, Anson LW, Morgan MR, Gascoyne-Binzi D, Walker TM, Quan TP, Wyllie DH, Del Ojo Elias C, Wilcox M, Walker AS, Peto TEA, Crook DW. 2015. Mycobacterial DNA extraction for whole-genome sequencing from early positive liquid (MGIT) cultures. J Clin Microbiol 53:1137–1143. doi: 10.1128/JCM.03073-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang T-S, Chen Y-S, Lee SS-J, Tu H-Z, Liu Y-C. 2005. Preservation of clinical isolates of Mycobacterium tuberculosis complex directly from MGIT culture tubes. Ann Clin Lab Sci 35:455–458. [PubMed] [Google Scholar]
- 17.Kirschner P, Springer B, Vogel U, Meier A, Wrede A, Kiekenbeck M, Bange FC, Böttger EC. 1993. Genotypic identification of mycobacteria by nucleic acid sequence determination: report of a 2-year experience in a clinical laboratory. J Clin Microbiol 31:2882–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hillemann D, Weizenegger M, Kubica T, Richter E, Niemann S. 2005. Use of the genotype MTBDR assay for rapid detection of rifampin and isoniazid resistance in Mycobacterium tuberculosis complex isolates. J Clin Microbiol 43:3699–3703. doi: 10.1128/JCM.43.8.3699-3703.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Soolingen D, Hermans PW, de Haas PE, Soll DR, van Embden JD. 1991. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol 29:2578–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blom J, Jakobi T, Doppmeier D, Jaenicke S, Kalinowski J, Stoye J, Goesmann A. 2011. Exact and complete short-read alignment to microbial genomes using Graphics Processing Unit programming. Bioinformatics 27:1351–1358. doi: 10.1093/bioinformatics/btr151. [DOI] [PubMed] [Google Scholar]
- 21.Thorvaldsdóttir H, Robinson JT, Mesirov JP. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niemann S, Richter E, Rüsch-Gerdes S, Schlaak M, Greinert U. 2000. Double infection with a resistant and a multidrug-resistant strain of Mycobacterium tuberculosis. Emerg Infect Dis 6:548–551. doi: 10.3201/eid0605.000518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gagneux S, Long CD, Small PM, Van T, Schoolnik GK, Bohannan BJM. 2006. The competitive cost of antibiotic resistance in Mycobacterium tuberculosis. Science 312:1944–1946. doi: 10.1126/science.1124410. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.