Abstract
Given the fundamental roles of microRNAs (miRNAs) in physiological, developmental and pathological processes, we hypothesized that genes involved in miRNA biogenesis contribute to human complex traits. For thirteen such genes, we evaluated the relationship between transcription and two classes of complex traits, namely cellular growth and sensitivity to various chemotherapeutic agents in a set of lymphoblastoid cell lines. We found a highly significant correlation between protein argonaute-2 (AGO2) expression and cellular growth rate (Bonferroni-adjusted p < 0.05), and report additional miRNA biogenesis genes with suggestive associations with either cellular growth rate or chemotherapeutic sensitivity. AGO2 expression was found to be correlated with multiple drug sensitivity phenotypes. Furthermore, small interfering RNA (siRNA) knockdown of AGO2 resulted in cellular growth inhibition in an ovarian cancer cell line (OVCAR3), supporting the role of this miRNA biogenesis gene in cell proliferation in cancer cells. Expression quantitative trait loci mapping indicated that genetic variation (in the form of both single nucleotide polymorphisms (SNPs) and copy number variations (CNVs)) that may regulate the expression of AGO2 can have downstream effects on cellular-growth-dependent complex phenotypes.
Keywords: microRNA biogenesis genes, cellular proliferation, chemotherapeutic sensitivity
Introduction
MicroRNAs (miRNAs), the non-coding small RNA molecules that have been shown to play important roles in post-transcriptional gene expression, are predicted to target a third of all human mRNAs (1,2). Studies have demonstrated the role of miRNAs in diverse cellular, developmental and pathological processes (3) as well as in drug sensitivity (4,5). Given the important role of miRNAs, we hypothesized that genes affecting miRNA biogenesis and function may broadly influence complex phenotypes, and altering the expression and/or the function of these genes may have substantial downstream phenotypic effects.
Many gene products have been found to be involved in miRNA biogenesis and function (6–9). miRNA biogenesis is a highly complex and finely tuned series of biological processes, including transcription, nuclear processing, export from nucleus, and cytoplasmic processing. We selected thirteen genes that have been shown to be directly involved in converting primary miRNA to pre-miRNA (DGCR8 and DROSHA (RNASEN)), exporting pre-miRNA from nucleus to cytosol (RAN and XPO5), and regulating post-transcriptional and translational processes (DICER1, TARBP2, PRKRA, AGO1, AGO2, AGO3, AGO4, DDX20 and GEMIN4). Furthermore, to explore the functional significance of these genes as well as to evaluate the effect of genetic variation on their expression, we conducted our study in a collection of cell lines from the International HapMap project (10). The primary reason for the choice of such a discovery model is the readily available genomic, transcriptomic and miRNA expression data along with a wide range of phenotypic information (e.g. intrinsic cellular growth rate, cellular sensitivity to different drugs) in these cell lines (11–14).
In this study, we evaluated the expression correlation between each of the thirteen miRNA biogenesis/function related genes and cell growth rate and sensitivity to various drugs. Expression quantitative trait loci (eQTL) mapping was performed to quantify the effect of genetic variation (in the form of SNPs and CNVs) on the expression of these miRNA biogenesis genes. Functional validation experiment was carried out in cancer cell lines.
Materials and Methods
Cell lines
EBV-transformed B-lymphoblastoid cell lines (LCLs) from the International HapMap consortium were purchased from the Coriell Institute for Medical Research (Camden, NJ). Fifty-three unrelated CEU (Utah residents with northern and western European ancestry) and 54 unrelated YRI (Yoruba people from Ibadan, Nigeria) samples were used for this study. These LCLs were maintained as suspension cultures in RPMI 1640 with supplements described previously (15). For functional studies, OVCAR-3, an ovarian cancer cell line, was procured from ATCC (Manassas, VA) and grown as an adherent culture in RPMI-1640 medium with 20% fetal bovine serum (Atlanta Biologicals, GA) and 0.01 mg/mL bovine insulin. OVCAR-3 cells were passaged every three days at a ratio of 1:3.
Genomic, transcriptomic, microRNAomic expression information and intrinsic cellular growth rate, drug sensitivity data
We included in our study a total of 13 genes that have been shown to influence miRNA biogenesis and function (6–9). The expression levels of these genes in the HapMap samples were obtained from Gene Expression Omnibus (GEO, GSE7761) which was quantified using microarray-based gene expression profiling (Affymetrix GeneChip® Human Exon 1.0 ST array).
Genome-wide miRNA expression in these HapMap samples were characterized using the Exiqon miRCURY LNA arrays v.10.0 (Exiqon array), obtained from GEO (GSE34406)(4,16). SNP genotypes were downloaded from International HapMap database (www.hapmap.org, release 27). CNV data were obtained based on the Conrad et al 2010 publication (17).
We have previously generated an intrinsic cellular growth (iGrowth) phenotype in over 500 HapMap LCLs (11). The iGrowth for the CEU and YRI samples were used in this study. Data on sensitivity to chemotherapeutic drugs carboplatin, cisplatin, daunorubicin and etoposide were queried using a publicly available pharmacogenomics resource we developed (www.PACdb.org) (14).
Integrative analysis of miRNA biogenesis genes, miRNA expression, genomic and other cellular phenotypes
We combined the CEU and YRI samples and performed linear regression of gene expression against iGrowth or IC50 for each drug independently with the ancestral group as covariate. Linear regression analysis was also performed for each of the cellular phenotypes in the HapMap CEU and YRI samples separately using the R Statistical Software (http://www.r-project.org/) (18). For this study, Bonferroni-adjusted p < 0.05 was considered significant, but we report all suggestive associations (p<0.05).
SNP and CNV associations with miRNA biogenesis gene expression were identified using SCANdb (www.scandb.org), an online gene expression regulation database we developed (19). SNP and CNV associations with cellular growth rate and drug sensitivity were evaluated by generalized linear regression assuming an additive genetic model. The presence of a negative correlation between expression of a miRNA biogenesis gene and genome-wide miRNA expression was tested through linear regression.
Functional validation
Functional evaluation of the biological role of AGO2 was subsequently conducted in OVCAR-3 cell line, an ovarian cancer cell line. The rationale for selecting OVCAR-3 cells as a model was the observed common over-expression of AGO2 in primary ovarian cancers (data obtained through The Cancer Genome Atlas [TCGA] data query (Supplemental Fig 1)). Gene knockdown was conducted through small interfering RNA (siRNA). Specifically, siAGO2 (Cat. No. 1027416, 25nM) and scrambled control (AllStars negative control siRNA, Cat No. 1027292), were purchased from Qiagen. Transfection experiments were conducted using DharmaFECT 1 (Dharmacon™). The effect of transfection was confirmed by measuring AGO2 expression at 0, 24 and 48 hours post transfection using quantitative PCR (qPCR). The cellular growth rate was measured using CellTiter-Glo luminescent cell viability assay (Promega) at 0, 24, 48 and 72 hours post transfection. Two-way ANOVA was performed to compare cellular growth rate obtained after siAGO2 and that from scramble control. P<0.05 was considered statistically significant for validation.
Results
miRNA biogenesis/function related genes in human complex traits
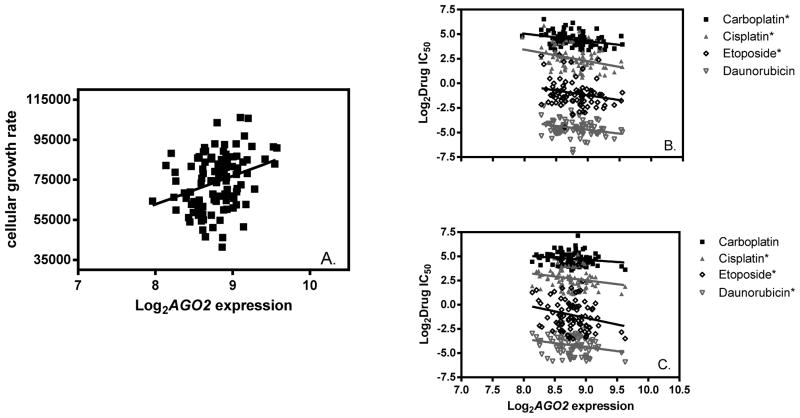
The expression levels of 13 genes directly involved in miRNA biogenesis and function were compared with iGrowth and sensitivity to each of 4 chemotherapeutic agents (carboplatin, cisplatin, daunorubicin and etoposide) independently. In the pooled CEU and YRI samples, AGO2 (p=4×10−6) showed a highly significant correlation (Bonferroni-adjusted p < 0.05) with iGrowth, and several additional miRNA biogenesis genes showed suggestive associations: DGCR8 (p=0.0002), DROSHA (p=0.075), PRKRA (p=0.033) and TARBP2 (p=0.066). Higher AGO2 expression was correlated with faster cellular growth in the combined CEU and YRI LCLs (Figure 1A). In each ancestral group (CEU or YRI), 3 genes had expression levels that were correlated with at least one of the four drug IC50s (Table 1 for all nominal associations, p<0.05). Notably, AGO2 expression was correlated with almost all drugs evaluated in both populations with increasing expression level resulting in lower IC50, suggesting greater sensitivity to these agents (Figure 1B and 1C).
Figure 1. Relationships among AGO2 expression, cellular growth rate and drug sensitivity in the HapMap LCLs.
A) Correlation between AGO2 expression and cellular growth rate in HapMap CEU and YRI samples (n=107); B) Correlation between AGO2 expression and each of four drug sensitivity phenotypes in HapMap CEU samples (n=53); and C) Correlation between AGO2 expression and each of four drug sensitivity phenotypes in HapMap YRI samples (n=54). A “*” next to the drug name represents suggestively significant correlation between AGO2 expression and drug IC50 (p<0.05). All drug IC50 values are in μmol unit.
Table 1.
miRNA biogenesis genes whose expression levels correlated with a drug IC50 (P<0.05).
| Genes | Drugs | Population | Beta value | P-value |
|---|---|---|---|---|
| DGCR8 | daunorubicin | YRI | 0.079935 | 0.0027 |
| DGCR8 | etoposide | YRI | 0.055369 | 0.0003 |
| AGO2 | carboplatin | CEU | −0.13163 | 0.0023 |
| AGO2 | cisplatin | CEU | −0.08967 | 0.0003 |
| AGO2 | daunorubicin | CEU | −0.086 | 0.0043 |
| AGO2 | etoposide | CEU | −0.03606 | 0.0123 |
| AGO2 | cisplatin | YRI | −0.06142 | 0.0244 |
| AGO2 | daunorubicin | YRI | −0.05969 | 0.0181 |
| AGO2 | etoposide | YRI | −0.03971 | 0.0259 |
| AGO3 | daunorubicin | CEU | −0.0772 | 0.0337 |
| GEMIN4 | cisplatin | CEU | −0.05419 | 0.0407 |
| GEMIN4 | daunorubicin | CEU | −0.09949 | 0.0030 |
| GEMIN4 | etoposide | CEU | −0.044 | 0.0301 |
| PRKRA | etoposide | YRI | −0.04417 | 0.0366 |
Functional validation of AGO2 in a cancer cell line
To explore the role of miRNA biogenesis genes in cancers, we analyzed The Cancer Genome Atlas (TCGA) dataset, in which a large number of tumors representing over 20 different types of cancers have undergone genomic profiling (http://www.cbioportal.org/public-portal/), for the miRNA biogenesis genes. We found that genetic mutations and altered gene expression are common for AGO2 in various types of cancers (including ovarian, breast, liver, prostate, uterine, head and neck cancers). More importantly, over 30% of the primary ovarian cancer samples evaluated by TCGA showed AGO2 over-expression relative to normal, making ovarian cancer a good candidate in evaluating the role of AGO2 through gene knockdown (Supplemental Figure 1).
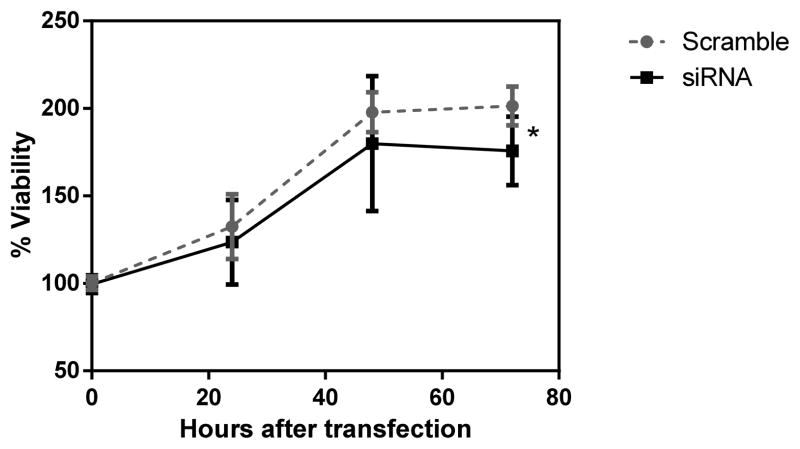
We conducted AGO2 inhibition experiment in an ovarian cancer cell line (OVCAR3) using siRNA. The transfection of siAGO2 resulted in significantly decreased expression of AGO2 compared to scramble control (quantified through qPCR. Supplemental Figure 2). Subsequently, we observed a significant cellular growth inhibition after siAGO2 transfection when compared to that of control (two-way ANOVA p=0.036, Figure 2). This growth inhibition effect is most pronounced at 72 hours post transfection (t-test p= 0.002).
Figure 2. The effect of AGO2 inhibition on OVCAR-3 cellular growth.
Significant inhibition of cell growth observed in OVCAR3 at 72 hours post-transfection (t test p=0.002, two-way ANOVA, p=0.036). Cellular growth was determined using CellTiter Glo reagent. “si” represents siRNA treatment while “scr” represents the control experiment.
Genetic variation, miRNA biogenesis genes and downstream miRNA expression
To identify genetic effect on the miRNA biogenesis genes, we performed eQTL mapping for the 13 miRNA processing genes. We found a number of expression-associated SNPs (eSNPs, at nominal p≤10−4) for each of the 13 genes in either the CEU or YRI samples (Supplementary Table 1). In addition, a number of CNVs were found to be associated with 5 of these 13 genes (p≤10−4, Supplementary Table 1), including CNVR5446_full and DROSHA (CEU, p=4.1E-5), CNVR841.1 and DICER1 (YRI, p=1.4×10−5), CNVR5184_full and AGO2 (YRI, p=5.1×10−6), CNVR81.1 and AGO3 (YRI, p=2.6×10−5) and CNVR6423.1 and GEMIN4 (YRI, p=2.3×10−5).
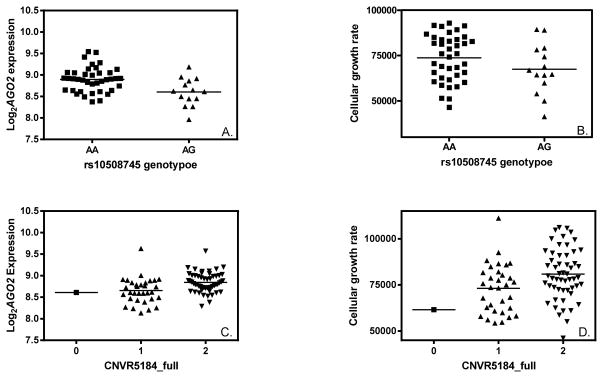
We tested the identified eSNPs for association with cellular growth. Particularly for AGO2, 38 eSNPs were identified in CEU (p≤10−4, MAF>0.1). An example of such an eSNP (rs10508745) for AGO2 is shown in Fig 3A (p=0.0001). Notably, this SNP rs10508745 was also associated with cellular growth rate in CEU (Fig 3B, p=0.0006). A CNV (CNVR5184_full) was significantly associated with AGO2 expression in YRI (Fig 3C, p=5×10−6); CNVR5184_full was also associated with cellular growth rate in this population (Fig 3D, p=0.006).
Figure 3. Genetic variations associated with AGO2 expression are also associated with cellular proliferation.
A) rs10508745 and AGO2 expression association in HapMap CEU samples (p=0.0001); B) rs10508745 and cellular growth rate association in CEU (p=0.0006); C) CNVR5184_full and AGO2 expression association in YRI (p=5×10−6); D) CNVR5184_full and cellular growth rate association in YRI (p=0.006).
For 12 of the 13 miRNA biogenesis genes, we found significant negative correlations between gene expression and miRNA expression in the HapMap samples (p≤10−4, FDR<0.05). The one exception was AGO2, for which we did not observe expression correlation between AGO2 and any of the miRNAs (p>0.05), suggesting that AGO2 may affect miRNA function rather than expression.
Discussion
In this study, we evaluated the effect of key miRNA biogenesis genes on two human complex traits (cellular proliferation and drug sensitivity) with implications for a range of complex phenotypes and implicated AGO2 in cellular proliferation and cellular sensitivity to drug. The effect of AGO2 in cell growth was originally observed in LCLs derived from apparently healthy individuals and subsequently validated in an ovarian cancer cell line.
It has been shown that genes involved in miRNA biogenesis can affect multiple human physiological and pathological functions. For example, DICER was found to play an essential role in thyroid function (20), hepatocyte survival, metabolism, tumor suppression (21) and in tumorigenesis (22). RNASEN (DROSHA) regulates cell proliferation and affects survival in esophageal cancer patients (23). RNASEN, DGCR8, DICER, TARBP2, and PRKRA were found to interact with miRNAs and affect liver regeneration (24). Furthermore, factors affecting miRNA biogenesis and function may also affect an individual’s susceptibility to pollutants (25) and sensitivity to drugs (26). Kovalchuk et al. observed decreased protein expression of DICER and Argonaute 2 (encoded by AGO2) in the doxorubicin resistant MCF-7 breast cancer cell line compared to the parent doxorubicin sensitive MCF-7 cells (26).
AGO2, a gene coding Argonaute 2 protein, plays a significant role in the regulation of post-transcriptional and translational processes through miRNA and siRNA (27). It is an important component of RNA induced silencing complex (RISC) catalyzing RNA interference (RNAi) (28) which leads to alteration of many bioprocesses. In our study, the expression of AGO2 was found to be highly correlated with cellular proliferation in LCLs and with cellular sensitivity to multiple chemotherapeutic agents. Given the mechanism of action for these cytotoxic agents, for which cytotoxicity is highly dependent on cellular proliferation rate (29), it is plausible that the observed relationship between AGO2 expression and drug sensitivity may be a consequence of this gene’s effect on cell growth. To examine the correlation between AGO2 and cell growth in cancers, we utilized the comprehensive TCGA data. We found AGO2 is commonly over-expressed in many cancers when compared to the adjacent normal tissue. This observation finds support in the literature. For example, up-regulation of AGO2 mRNA and protein was observed in urothelial carcinoma of bladder compared to paired normal bladder (30). Chang et al. reported AGO2 over-expression in head and neck squamous cell carcinoma (HNSCC) cells (31). Multiple myeloma cell lines showed increased expression of AGO2 through DNA copy number gain (32) while AGO2 was overexpressed in pleural effusions when compared to primary ovarian caricinoma in patients with disease that spread beyond the ovary (33). In addition, knocking down AGO2 was shown to induce apoptosis in HL-60 (myeloid leukemia cells) and was, furthermore, involved in regulation of siRNA mediated RNAi pathways in HEK-293 cells (34). However, to our knowledge, no study to date has reported on the effect of AGO2 in ovarian cancer. Our AGO2 knockdown experiment in OVCAR3 resulted in decreased cell proliferation in ovarian cancer. Taken together, we have shown evidence of the effect of AGO2 on cell proliferation in both normal and cancer cells.
To further examine how AGO2 affects cell proliferation, we performed expression correlation study between this gene and all expressed miRNAs in the genome. Surprisingly, we did not observe any expression correlation between AGO2 and any of the miRNA expression in HapMap samples. AGO2 is one of the 4 argonaute proteins (AGO1-AGO4) that play major roles in guiding siRNAs or miRNAs to perform post-transcriptional gene silencing or activation (35). AGO1, AGO3 and AGO4 are present as tandem copies on chromosome 1 whereas AGO2 is present on its own on chromosome 8. Differing from the other 3 AGO subfamily proteins, AGO2 can carry out both site-specific cleavage and non-cleavage mediated inhibition while the other AGO proteins are restricted to non-cleavage mediated inhibition (36). Based on these, we speculated that the variable AGO2 expression may result in differential miRNA-target gene binding, with downstream regulation of various cellular processes, rather than having a direct effect on miRNA expression.
Lastly, we hypothesized that genetic polymorphisms (in the form of SNPs and CNVs) that regulate the expression of genes in the miRNA biogenesis pathway may influence cellular growth. Indeed, we identified several SNPs/CNVs that were associated with the transcription of miRNA biogenesis genes. Both, the SNP rs10508745 and a CNV (CNVR5184_full) were associated with AGO2 expression and with cellular growth. These findings support a highly complex network of relationships among genetic variants, miRNA biogenesis and biological function.
In summary, through a comprehensive evaluation of 13 miRNA biogenesis genes, we found that AGO2 expression was associated with both cellular growth rate and sensitivity to multiple chemotherapeutic drugs. AGO2 inhibition results in cellular growth inhibition in an ovarian cancer cell line. Finally, eQTLs for AGO2 may provide genetic effects on cellular proliferation.
Supplementary Material
Acknowledgments
We are grateful for the excellent technical support provided by Pharmacogenomics of Anti-cancer Agent Research (PAAR) group cell line core. We would also like to thank Ms. Catherine Park and Stephanie Joseph for their assistant in optimizing functional experimental conditions.
This study was supported by NIH/NCI grant R21 CA139278 and by NIH/NIGMS grant UO1GM61393. RSH also received support from NIH/NIGMS grant K08GM089941, Circle of Service Foundation Early Career Investigator award, University of Chicago Cancer Center Support Grant (#P30 CA14599), Breast Cancer SPORE Career Development Award [CA125183] a Conquer Cancer Foundation of ASCO Translational Research Professorship award In Memory of Merrill J. Egorin, MD, and pilot grant from NIH/NCATS grant UL1RR024999. HKI received support from NIH/NCI K12CA139160 and NIH/NIDDK P30 DK020595 (DRTC).
All authors have read the journal’s authorship agreement and the manuscript has been reviewed and approved by all named authors.
Abbreviations
- AGO1
Argonaute RISC catalytic component 1
- AGO2
Argonaute RISC catalytic component 2
- AGO3
Argonaute RISC catalytic component 3
- AGO4
Argonaute RISC catalytic component 4
- CEU
Centre d’Etude du Polymorphisme Humain (CEPH) people from Utah, USA
- CNV
Copy Number Variation
- DDX20
DEAD (Asp-Glu-Ala-Asp) box polypeptide 20
- DGCR8
DiGeorge syndrome critical region gene 8
- DICER1
Drosophila, Homolog of, 1
- eQTL
expression Quantitative trait Locus
- FDR
False Discovery Rate
- GEMIN4
Gem (nuclear organelle) associated protein 4
- GEO
Gene Expression Omnibus
- HNSCC
Head and Neck Squamous Cell Carcinoma
- iGrowth
Intrinsic cellular growth
- LCLs
Lymphoblastoid Cell Lines
- miRNA
microRNA
- PRKRA
Protein kinase, interferon-inducible double stranded RNA dependent activator
- RAN
Ras-related nuclear protein
- RNAi
RNA interference
- RNASEN
Ribonuclease type III, nuclear
- siRNA
small interfering RNA
- SNP
Single Nucleotide Polymorphism
- TARBP2
TAR RNA-binding protein 2
- TCGA
The Cancer Genome Atlas
- XPO5
Exportin-5
- YRI
Yoruba people from Ibadan, Nigeria
Footnotes
Conflict of Interest: All authors have read the journal’s policy on disclosure of potential conflicts of interest and they have none to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RHA, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell [Internet] 2005 Jan 14;120(1):21–4. doi: 10.1016/j.cell.2004.12.031. [cited 2014 Sep 4]; Available from: http://www.ncbi.nlm.nih.gov/pubmed/15652478. [DOI] [PubMed] [Google Scholar]
- 2.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell [Internet] 2005 Jan 14;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [cited 2014 Jul 9]; Available from: http://www.ncbi.nlm.nih.gov/pubmed/15652477. [DOI] [PubMed] [Google Scholar]
- 3.Chitwood DH, Timmermans MCP. Small RNAs are on the move. Nature [Internet] 2010 Sep 23;467(7314):415–9. doi: 10.1038/nature09351. [cited 2014 Sep 24]; Available from: http://www.ncbi.nlm.nih.gov/pubmed/20864994. [DOI] [PubMed] [Google Scholar]
- 4.Huang RS, Gamazon ER, Ziliak D, Wen Y, Im HK, Zhang W, et al. Population differences in microRNA expression and biological implications. RNA Biol. 8(4):692–701. doi: 10.4161/rna.8.4.16029. [Internet]. [cited 2013 Jun 27]; Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3225983&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Søkilde R, Kaczkowski B, Podolska A, Cirera S, Gorodkin J, Møller S, et al. Global microRNA analysis of the NCI-60 cancer cell panel. Mol Cancer Ther. 2011 Mar;10(3):375–84. doi: 10.1158/1535-7163.MCT-10-0605. [Internet] [cited 2014 Oct 27] Available from: http://www.ncbi.nlm.nih.gov/pubmed/21252286. [DOI] [PubMed] [Google Scholar]
- 6.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008 Feb;9(2):102–14. doi: 10.1038/nrg2290. [Internet] [cited 2014 Jul 10]; Available from: http://www.ncbi.nlm.nih.gov/pubmed/18197166. [DOI] [PubMed] [Google Scholar]
- 7.Gregory RI, Yan K-P, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004 Nov 11;432(7014):235–40. doi: 10.1038/nature03120. [Internet] [cited 2014 Nov 5]; Available from: http://www.ncbi.nlm.nih.gov/pubmed/15531877. [DOI] [PubMed] [Google Scholar]
- 8.Hiard S, Charlier C, Coppieters W, Georges M, Baurain D. Patrocles: a database of polymorphic miRNA-mediated gene regulation in vertebrates. Nucleic Acids Res. 2010 Jan;38:D640–51. doi: 10.1093/nar/gkp926. [Internet] [cited 2014 Oct 27] Database issue. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2808989&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boominathan L. The tumor suppressors p53, p63, and p73 are regulators of microRNA processing complex. PLoS One. 2010 Jan;5(5):e10615. doi: 10.1371/journal.pone.0010615. [Internet] [cited 2014 Oct 14] Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2868896&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The International HapMap Project. Nature. 2003 Dec 18;426(6968):789–96. doi: 10.1038/nature02168. [Internet] [cited 2014 Nov 28]; Available from: http://www.ncbi.nlm.nih.gov/pubmed/14685227. [DOI] [PubMed] [Google Scholar]
- 11.Im HK, Gamazon ER, Stark AL, Huang RS, Cox NJ, Dolan ME. Mixed effects modeling of proliferation rates in cell-based models: consequence for pharmacogenomics and cancer. PLoS Genet. 2012;8(2):e1002525. doi: 10.1371/journal.pgen.1002525. [Internet] 2012/02/22 ed. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22346769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.LaCroix B, Gamazon ER, Lenkala D, Im HK, Geeleher P, Ziliak D, et al. Integrative analyses of genetic variation, epigenetic regulation, and the transcriptome to elucidate the biology of platinum sensitivity. BMC Genomics. 2014 Jan;15:292. doi: 10.1186/1471-2164-15-292. [Internet] [cited 2014 Dec 5] Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3996490&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weng L, Ziliak D, Im HK, Gamazon ER, Philips S, Nguyen AT, et al. Genome-wide discovery of genetic variants affecting tamoxifen sensitivity and their clinical and functional validation. Ann Oncol. 2013 Jul;24(7):1867–73. doi: 10.1093/annonc/mdt125. [Internet] [cited 2014 Dec 5]; Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3690911&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gamazon ER, Duan S, Zhang W, Huang RS, Kistner EO, Dolan ME, et al. PACdb: a database for cell-based pharmacogenomics. Pharmacogenet Genomics. 2010 Apr;20(4):269–73. doi: 10.1097/FPC.0b013e328337b8d6. [Internet] [cited 2014 Dec 5]; Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2914089&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang RS, Kistner EO, Bleibel WK, Shukla SJ, Dolan ME. Effect of population and gender on chemotherapeutic agent-induced cytotoxicity. Mol Cancer Ther. 2007;6(1):31–6. doi: 10.1158/1535-7163.MCT-06-0591. [Internet] 2007/01/24 ed. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17237264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gamazon ER, Ziliak D, Im HK, LaCroix B, Park DS, Cox NJ, et al. Genetic architecture of microRNA expression: implications for the transcriptome and complex traits. Am J Hum Genet. 2012;90(6):1046–63. doi: 10.1016/j.ajhg.2012.04.023. [Internet] 2012/06/05 ed. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22658545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010 Apr 1;464(7289):704–12. doi: 10.1038/nature08516. [Internet] [cited 2014 Jul 10]; Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3330748&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: R Foundation for Statistical Computing; Vienna, Austria: 2014. [Internet] Available from: http://www.r-project.org/ [Google Scholar]
- 19.Gamazon ER, Huang RS, Cox NJ. SCAN: a systems biology approach to pharmacogenomic discovery. Methods Mol Biol. 2013 Jan;1015:213–24. doi: 10.1007/978-1-62703-435-7_14. [Internet] [cited 2014 Nov 5] Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4032625&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frezzetti D, Reale C, Calì G, Nitsch L, Fagman H, Nilsson O, et al. The microRNA-processing enzyme Dicer is essential for thyroid function. PLoS One. 2011 Jan;6(11):e27648. doi: 10.1371/journal.pone.0027648. [Internet] [cited 2014 Oct 27] Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3221669&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sekine S, Ogawa R, Ito R, Hiraoka N, McManus MT, Kanai Y, et al. Disruption of Dicer1 induces dysregulated fetal gene expression and promotes hepatocarcinogenesis. Gastroenterology. 2009 Jun;136(7):2304–15. e1–4. doi: 10.1053/j.gastro.2009.02.067. [Internet] [cited 2014 Nov 5]; Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3167383&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007 May;39(5):673–7. doi: 10.1038/ng2003. [Internet] [cited 2014 Oct 18]; Available from: http://www.ncbi.nlm.nih.gov/pubmed/17401365. [DOI] [PubMed] [Google Scholar]
- 23.Sugito N, Ishiguro H, Kuwabara Y, Kimura M, Mitsui A, Kurehara H, et al. RNASEN regulates cell proliferation and affects survival in esophageal cancer patients. Clin Cancer Res. 2006 Dec 15;12(24):7322–8. doi: 10.1158/1078-0432.CCR-06-0515. [Internet] [cited 2014 Oct 27]; Available from: http://www.ncbi.nlm.nih.gov/pubmed/17121874. [DOI] [PubMed] [Google Scholar]
- 24.Shu J, Kren BT, Xia Z, Wong PY-P, Li L, Hanse EA, et al. Genomewide microRNA down-regulation as a negative feedback mechanism in the early phases of liver regeneration. Hepatology. 2011 Aug;54(2):609–19. doi: 10.1002/hep.24421. [Internet] [cited 2014 Nov 5]; Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3145019&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilker EH, Alexeeff SE, Suh H, Vokonas PS, Baccarelli A, Schwartz J. Ambient pollutants, polymorphisms associated with microRNA processing and adhesion molecules: the Normative Aging Study. Environ Health. 2011 Jan; doi: 10.1186/1476-069X-10-45. [Internet] [cited 2014 Nov 5];10:45. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3124411&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed]
- 26.Kovalchuk O, Filkowski J, Meservy J, Ilnytskyy Y, Tryndyak VP, Chekhun VF, et al. Involvement of microRNA-451 in resistance of the MCF-7 breast cancer cells to chemotherapeutic drug doxorubicin. Mol Cancer Ther. 2008 Jul;7(7):2152–9. doi: 10.1158/1535-7163.MCT-08-0021. [Internet] [cited 2014 Oct 6]; Available from: http://www.ncbi.nlm.nih.gov/pubmed/18645025. [DOI] [PubMed] [Google Scholar]
- 27.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004 Jul 23;15(2):185–97. doi: 10.1016/j.molcel.2004.07.007. [Internet] [cited 2014 Oct 29]; Available from: http://www.ncbi.nlm.nih.gov/pubmed/15260970. [DOI] [PubMed] [Google Scholar]
- 28.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song J-J, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004 Sep 3;305(5689):1437–41. doi: 10.1126/science.1102513. [Internet] [cited 2014 Jul 15]; Available from: http://www.ncbi.nlm.nih.gov/pubmed/15284456. [DOI] [PubMed] [Google Scholar]
- 29.Stark AL, Zhang W, Mi S, Duan S, O’Donnell PH, Huang RS, et al. Pharmacogenomics J. 6. Vol. 10. Nature Publishing Group; 2010. Heritable and non-genetic factors as variables of pharmacologic phenotypes in lymphoblastoid cell lines; pp. 505–12. [Internet] Available from: http://www.ncbi.nlm.nih.gov/pubmed/20142840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang F-Q, Huang J-H, Liu M, Yang F-P, Li W, Wang G-C, et al. Argonaute 2 is up-regulated in tissues of urothelial carcinoma of bladder. Int J Clin Exp Pathol. 2014 Jan;7(1):340–7. [Internet] [cited 2014 Oct 29]; Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3885489&tool=pmcentrez&rendertype=abstract. [PMC free article] [PubMed] [Google Scholar]
- 31.Chang SS, Smith I, Glazer C, Hennessey P, Califano JA. EIF2C is overexpressed and amplified in head and neck squamous cell carcinoma. ORL J Otorhinolaryngol Relat Spec. 2010 Jan;72(6):337–43. doi: 10.1159/000320597. [Internet] [cited 2014 Oct 29]; Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2975733&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou Y, Chen L, Barlogie B, Stephens O, Wu X, Williams DR, et al. High-risk myeloma is associated with global elevation of miRNAs and overexpression of EIF2C2/AGO2. Proc Natl Acad Sci U S A. 2010 Apr 27;107(17):7904–9. doi: 10.1073/pnas.0908441107. [Internet] [cited 2014 Oct 29]; Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2867889&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vaksman O, Stavnes HT, Kaern J, Trope CG, Davidson B, Reich R. miRNA profiling along tumour progression in ovarian carcinoma. J Cell Mol Med. 2011 Jul;15(7):1593–602. doi: 10.1111/j.1582-4934.2010.01148.x. [Internet] [cited 2014 Oct 29]; Available from: http://www.ncbi.nlm.nih.gov/pubmed/20716115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Naoghare PK, Tak YK, Kim MJ, Han E, Song JM. Knock-down of argonaute 2 (AGO2) induces apoptosis in myeloid leukaemia cells and inhibits siRNA-mediated silencing of transfected oncogenes in HEK-293 cells. Basic Clin Pharmacol Toxicol. 2011 Oct;109(4):274–82. doi: 10.1111/j.1742-7843.2011.00716.x. [Internet] [cited 2014 Oct 29]; Available from: http://www.ncbi.nlm.nih.gov/pubmed/21535412. [DOI] [PubMed] [Google Scholar]
- 35.Höck J, Meister G. The Argonaute protein family. Genome Biol. 2008 Jan;9(2):210. doi: 10.1186/gb-2008-9-2-210. [Internet] [cited 2015 Jan 20] Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2374724&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valdmanis PN, Gu S, Schüermann N, Sethupathy P, Grimm D, Kay MA. Expression determinants of mammalian argonaute proteins in mediating gene silencing. Nucleic Acids Res. 2012 Apr;40(8):3704–13. doi: 10.1093/nar/gkr1274. [Internet] [cited 2015 Jan 20]; Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3333847&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.