Abstract
The aim of this study was to investigate the prevalence and transferability of resistance in tetracycline-resistant Escherichia coli isolates recovered from beef cattle in South Korea. A total of 155 E. coli isolates were collected from feces in South Korea, and 146 were confirmed to be resistant to tetracycline. The tetracycline resistance gene tet(A) (46.5%) was the most prevalent, followed by tet(B) (45.1%) and tet(C) (5.8%). Strains carrying tet(A) plus tet(B) and tet(B) plus tet(C) were detected in two isolates each. In terms of phylogenetic grouping, 101 (65.2%) isolates were classified as phylogenetic group B1, followed in decreasing order by D (17.4%), A (14.2%), and B2 (3.2%). Ninety-one (62.3%) isolates were determined to be multidrug resistant by the disk diffusion method. MIC testing using the principal tetracyclines, namely, tetracycline, chlortetracycline, oxytetracycline, doxycycline, and minocycline, revealed that isolates carrying tet(B) had higher MIC values than isolates carrying tet(A). Conjugation assays showed that 121 (82.9%) isolates could transfer a tetracycline resistance gene to a recipient via the IncFIB replicon (65.1%). This study suggests that the high prevalence of tetracycline-resistant E. coli isolates in beef cattle is due to the transferability of tetracycline resistance genes between E. coli populations which have survived the selective pressure caused by the use of antimicrobial agents.
INTRODUCTION
Antimicrobial resistance in humans and animals is considered a problem worldwide. Resistance to antimicrobial agents impedes the effective prevention and treatment of infectious disease, and thus, many governments have planned and implemented national programs for monitoring resistance in humans and animals (1–4). Surveillance data show that the inadequate selection and extensive use of antimicrobials result in the emergence and spread of resistant bacteria, particularly multidrug-resistant bacteria, and increase resistance to newer compounds, such as tetracycline-class antimicrobials (5).
The tetracyclines are one of the most widely used classes of antimicrobial agents in human and veterinary medicine because they have several advantages, which include a broad spectrum of activity, low cost, oral administration, and few side effects (6). After chlortetracycline was introduced into clinical medicine in 1948, many derivatives, such as tetracycline, oxytetracycline, doxycycline, and minocycline, were developed, and today, these derivatives are widely used to treat disease and as growth promoters in the food animal industry. However, the widespread and indiscriminate use of tetracyclines has subjected bacterial populations to selection pressure and increased the prevalence of tetracycline resistance (6, 7).
Tetracycline resistance is generally caused by the acquisition of a tetracycline resistance (tet) gene, as these genes are associated with primary resistance mechanisms, which involve active efflux pumps, ribosomal protection, and enzyme inactivation (8). To date, more than 40 different resistance genes have been identified (7). In Gram-negative bacteria, the most important mechanism involves the efflux pump system, which is encoded by tetracycline resistance genes tet(A), tet(B), tet(C), tet(D), and tet(G) (6).
Although most Escherichia coli strains are considered harmless commensal bacteria of the gastrointestinal tracts of humans and animals, pathogenic strains that can cause several intestinal and extraintestinal infections exist. Surveillance of E. coli isolates is also considered to provide an excellent means of monitoring antimicrobial resistance in food and the environment because of the wide range of hosts of E. coli and because it easily acquires resistance (9). Thus, the degrees of resistance in commensal and pathogenic E. coli strains provide indicators of antimicrobial selection in their environment, and tetracycline-resistant E. coli strains could be used for surveillance for tetracycline resistance in humans and animals. Studies have reported tetracycline-resistant E. coli strains in various environments (8, 10–13), but only a small number of studies have been conducted in animals.
The aim of this study was to determine the prevalence of tetracycline-resistant E. coli isolates in South Korean beef cattle and determine the phenotypes and genotypes of these isolates with a view toward investigating the transferabilities of tetracycline resistance determinants between E. coli isolates.
MATERIALS AND METHODS
Bacterial strains.
In total, 290 E. coli strains were isolated from feces collected from clinically healthy beef cattle during 2011 and 2012 (14). E. coli isolates that showed resistance and intermediate resistance to tetracycline were obtained by culture on MacConkey agar plates containing tetracycline at a concentration of 8 μg/ml (the MIC of tetracycline for E. coli indicating tetracycline resistance is ≥16 μg/ml) (15). As a result, 155 E. coli isolates were selected for analysis. E. coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used as quality control organisms in antimicrobial susceptibility tests and MIC tests.
Antimicrobial susceptibility testing.
The E. coli isolates were tested for susceptibility by the disk diffusion method in accordance with the guidelines issued by the Clinical and Laboratory Standards Institute (CLSI) (15). The antimicrobial disks (Oxoid, Basingstoke, United Kingdom) used in this study included ampicillin (10 μg), streptomycin (25 μg), gentamicin (10 μg), chloramphenicol (C, 30 μg), nalidixic acid (30 μg), ciprofloxacin (5 μg), trimethoprim-sulfamethoxazole (1.25/23.75 μg), and tetracycline (30 μg) disks.
Detection of tetracycline resistance genes.
All 155 tetracycline-resistant isolates were tested by multiplex PCR for the presence of the tet(A), tet(B), tet(C), tet(D), and tet(G) genes, as described previously (16). Bacterial DNA for PCR was obtained by suspending colonies of bacteria grown on tryptic soy broth (TSB) in 500 μl of ultrapure water and boiling at 100°C for 10 min. The oligonucleotide primers used in this study are shown in Table 1. The PCRs included a negative and a positive control, and reactions were run in duplicate to confirm the results. Sequence alignments were performed by use of a search of the GenBank database via the National Center for Biotechnology Information website with the BLAST program (http://www.ncbi.nlm.nih.gov/BLAST).
TABLE 1.
Primers used in this study
| Target gene | Primer | Sequence | Amplicon size (bp) | GenBank accession no. | Reference |
|---|---|---|---|---|---|
| tet(A) | TetA-F | GCTACATCCTGCTTGCCTTC | 210 | X61367 | 16 |
| TetA-R | CATAGATCGCCGTGAAGAGG | ||||
| tet(B) | TetB-F | TTGGTTAGGGGCAAGTTTTG | 659 | J01830 | 16 |
| TetB-R | GTAATGGGCCAATAACACCG | ||||
| tet(C) | TetC-F | CTTGAGAGCCTTCAACCCAG | 418 | J01749 | 16 |
| TetC-R | ATGGTCGTCATCTACCTGCC | ||||
| tet(D) | TetD-F | AAACCATTACGGCATTCTGC | 787 | L06798 | 16 |
| TetD-R | GACCGGATACACCATCCATC | ||||
| tet(G) | TetG-F | GCTCGGTGGTATCTCTGCTC | 468 | S52437 | 16 |
| TetG-R | AGCAACAGAATCGGGAACAC | ||||
| chuA | ChuA-F | GACGAACCAACGGTCAGGAT | 279 | HQ284193 | 17 |
| ChuA-R | TGCCGCCAGTACCAAAGACA | ||||
| yjaA | Yja-F | TGAAGTGTCAGGAGACGCTG | 211 | HQ284194 | 17 |
| Yja-R | ATGGAGAATGCGTTCCTCAAC | ||||
| TspE4C2 | TspE4C2-F | GAGTAATGTCGGGGCATTCA | 152 | HQ284195 | 17 |
| TspE4C2-R | CGCGCCAACAAAGTATTACG |
Phylogenetic grouping.
The phylogenetic tree described by Clermont et al. was used to classify all E. coli isolates into one of four phylogenetic groups, that is, groups A, B1, B2, and D (17). Triplex PCR was used to determine the phylogenetic groupings by targeting two genes (chuA and yjaA) and an anonymous DNA fragment (TspE4.C2) (17). The result of phylogenetic typing was used to compare the pattern of antimicrobial resistance and the tet gene distributions among the E. coli isolates tested in this study.
Determination of MICs of principal tetracyclines.
To investigate the phenotypic characteristics of tetracycline-resistant isolates, the MIC values of the principal tetracycline antibiotics, tetracycline, chlortetracycline, oxytetracycline, doxycycline, and minocycline, were determined using the broth dilution method (15). All antimicrobials used in this study were tested in 2-fold dilutions from 1 to 2,048 μg/ml. MIC tests were conducted in triplicate for each sample.
Conjugation assay and plasmid replicon typing.
To determine the transferability of tetracycline resistance, conjugation assays were conducted on tetracycline-resistant isolates using the broth mating method. E. coli J53 Azr was used as the recipient strain, and tetracycline-resistant isolates served as the donors (18). Eight-hour cultures of recipient and donor cells grown in Luria-Bertani (LB) broth at 37°C were mixed with each other at a ratio of 1:1, and the mixture was incubated for 20 h. To identify resistance carried by plasmids, 100-μl aliquots of these mixtures were spread onto tryptic soy agar (TSA) plates containing tetracycline (8 μg/ml) and sodium azide (200 μg/ml) and incubated at 37°C for 20 h. PCR was used to confirm that the transconjugants carried the tet gene of their donors. Multiplex PCR was conducted on all donors and transconjugants to type the plasmid replicons, as described previously (19).
Statistical analysis.
Data were analyzed using IBM SPSS Statistics, version 21, software (SPSS Inc., Chicago, IL). The distributions of the tet genes were analyzed using the chi-square test. To compare the different tet genes and MIC values, survival analysis was carried out using the Kaplan-Meier method, and the curves so obtained were compared using the log-rank test. P values of <0.05 were considered statistically significant.
RESULTS
Antimicrobial resistance profile.
Among 155 E. coli isolates, 146 (94.2%) isolates were resistant to tetracycline, as determined using the disk diffusion method. The tetracycline-resistant isolates detected in this study showed concurrent resistance to streptomycin (82.2%), ampicillin (45.3%), nalidixic acid (32.8%), chloramphenicol (28.8%), trimethoprim-sulfamethoxazole (25.3%), ciprofloxacin (10.3%), and gentamicin (5.5%) (Table 2). Of these 146 tetracycline-resistant E. coli isolates, 91 (62.3%) were multidrug resistant. The most frequent combination of multidrug resistance was tetracycline-streptomycin-ampicillin, which was detected in 20 (13.7%) isolates. Five (3.4%) isolates in phylogenetic group B2 showed resistance to streptomycin; resistance to no other antimicrobial was found (Table 2).
TABLE 2.
Resistances of 146 tetracycline-resistant E. coli isolates in different phylogenetic groups to other antimicrobials
| Phylogenetic group | No. (%) of strains showing antimicrobial resistancea |
||||||
|---|---|---|---|---|---|---|---|
| AMP | GN | STR | C | SXT | NA | CIP | |
| Total | 66 (45.3) | 8 (5.5) | 120 (82.2) | 42 (28.8) | 37 (25.3) | 48 (32.8) | 15 (10.3) |
| A | 14 (9.6) | 3 (2.1) | 15 (10.3) | 7 (4.8) | 7 (4.8) | 6 (4.1) | 5 (3.4) |
| B1 | 43 (29.5) | 4 (2.7) | 75 (51.4) | 33 (22.6) | 26 (17.8) | 24 (16.4) | 10 (6.8) |
| B2 | 5 (3.4) | ||||||
| D | 9 (6.2) | 1 (0.7) | 25 (17.1) | 2 (1.4) | 4 (2.7) | 18 (12.3) | |
AMP, ampicillin; GN, gentamicin; STR, streptomycin; C, chloramphenicol; SXT, sulfamethoxazole-trimethoprim; NA, nalidixic acid; CIP, ciprofloxacin.
Phylogenetic classification.
Of the 155 E. coli isolates, 101 (65.2%) isolates were classified as phylogenetic group B1; 27 (17.4%) were classified as group D, which is associated with pathogenic bacteria; 22 (14.2%) were classified as group A; and 5 (3.2%) were classified as group B2, the phylogenetic lineage associated with virulent extraintestinal strains (Table 3).
TABLE 3.
Distributions of tetracycline resistance genes in E. coli isolates in the four identified phylogenetic groups
| Phylogenetic group | No. (%) isolates with the following tetracycline resistance gene(s): |
|||||
|---|---|---|---|---|---|---|
| Total | tet(A) | tet(B) | tet(C) | tet(A) plus tet(B) | tet(B) plus tet(C) | |
| Total | 155 (100) | 72 (46.5) | 70 (45.1) | 9 (5.8) | 2 (1.3) | 2 (1.3) |
| A | 22 (14.2) | 6 (3.9) | 9 (5.8) | 5 (3.2) | 2 (1.3) | |
| B1 | 101 (65.2) | 41 (26.5) | 54 (34.8) | 4 (2.6) | 2 (1.3) | |
| B2 | 5 (3.2) | 5 (3.2) | ||||
| D | 27 (17.4) | 25 (16.1) | 2 (1.3) | |||
Prevalence of tetracycline resistance determinants.
All 155 isolates carried at least one of the tet genes examined. PCR detection of single tet determinants showed that 142 (91.6%) isolates carried tet(A) or tet(B) only: 72 (46.5%) harbored tet(A) only, and 70 (45.1%) isolates harbored tet(B) only. tet(C) was detected in 11 (7.1%) isolates. Four (2.6%) isolates contained two tet genes: tet(A) plus tet(B) in two (1.3%) isolates and tet(B) plus tet(C) in two (1.3%) isolates. tet(D) and tet(G) were not detected. The distributions of tet(A) and tet(B) in the phylogenetic groups were not significantly different (chi-square test, P > 0.05) (Table 3).
MIC values of tetracycline-class antimicrobials.
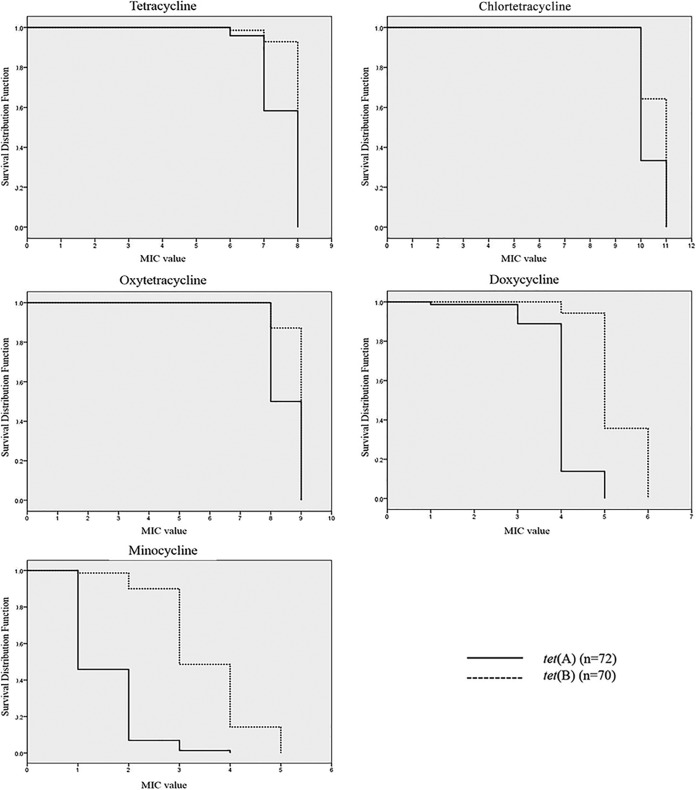
The MIC distributions of tetracycline, chlortetracycline, oxytetracycline, doxycycline, and minocycline for each group of isolates containing the same tet genes are shown in Table 4. The MIC values of all tetracyclines for isolates susceptible by the disk diffusion method were higher than the breakpoint (MIC ≥ 16 μg/ml). The MIC of chlortetracycline (range, 1,024 to 2,048 μg/ml) was much higher than the MICs of the four other tetracyclines. Resistance to minocycline (MIC ≥ 16 μg/ml) was observed for 35 (22.6%) isolates, and the genomes of 34 of these isolates encoded only the tet(B) resistance determinant. In fact, the average MICs for isolates containing the tet(B) gene were higher than those for isolates harboring the tet(A) gene (Fig. 1). Furthermore, the differences in the MICs between isolates containing tet(A) or tet(B) were greater for doxycycline and minocycline than the other three tetracyclines (Fig. 1).
TABLE 4.
MICs of tetracycline antimicrobials for E. coli isolates with different tetracycline resistance genes
| Antimicrobial | Gene profile | No. of strains | Avg MIC (μg/ml) | No. of isolates for which the MIC (μg/ml) was: |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 | 512 | 1,024 | 2,048 | ||||
| Tetracycline | tet(A) | 72 | 200.0 | 3 | 27 | 42 | |||||||||
| tet(B) | 70 | 245.9 | 1 | 4 | 65 | ||||||||||
| tet(C) | 9 | 23.1 | 5 | 4 | |||||||||||
| tet(A) and tet(B) | 2 | 256.0 | 2 | ||||||||||||
| tet(B) and tet(C) | 2 | 256.0 | 2 | ||||||||||||
| Chlortetracycline | tet(A) | 72 | 1,365.3 | 48 | 24 | ||||||||||
| tet(B) | 70 | 1,682.3 | 25 | 45 | |||||||||||
| tet(C) | 9 | 170.7 | 6 | 3 | |||||||||||
| tet(A) and tet(B) | 2 | 1,536 | 1 | 1 | |||||||||||
| tet(B) and tet(C) | 2 | 1,536 | 1 | 1 | |||||||||||
| Oxytetracycline | tet(A) | 72 | 384.0 | 36 | 36 | ||||||||||
| tet(B) | 70 | 479.1 | 9 | 61 | |||||||||||
| tet(C) | 9 | 49.8 | 4 | 5 | |||||||||||
| tet(A) and tet(B) | 2 | 512.0 | 2 | ||||||||||||
| tet(B) and tet(C) | 2 | 384.0 | 1 | 1 | |||||||||||
| Doxycycline | tet(A) | 72 | 17.3 | 1 | 7 | 54 | 10 | ||||||||
| tet(B) | 70 | 42.5 | 4 | 41 | 25 | ||||||||||
| tet(C) | 9 | 5.8 | 6 | 2 | 1 | ||||||||||
| tet(A) and tet(B) | 2 | 32.0 | 2 | ||||||||||||
| tet(B) and tet(C) | 2 | 32.0 | 2 | ||||||||||||
| Minocycline | tet(A) | 72 | 3.3 | 39 | 28 | 4 | 1 | ||||||||
| tet(B) | 70 | 13.7 | 1 | 6 | 29 | 24 | 10 | ||||||||
| tet(C) | 9 | 1.1 | 8 | 1 | |||||||||||
| tet(A) and tet(B) | 2 | 6.0 | 1 | 1 | |||||||||||
| tet(B) and tet(C) | 2 | 8.0 | 2 | ||||||||||||
FIG 1.
Survival curves (obtained by the Kaplan-Meier method) of E. coli isolates harboring tet(A) or tet(B) for resistance to the tetracycline family of antimicrobials. The survival rates of the E. coli isolates are compared with the MIC values of the five tetracyclines (tetracycline, chlortetracycline, oxytetracycline, doxycycline, and minocycline). Full and dotted lines, survival rates of tet(A)-carrying and tet(B)-carrying strains, respectively. a, the MIC values of the five tetracyclines were log transformed (base 2).
Conjugative transfer of plasmid-mediated tetracycline resistance genes.
Of the 146 tetracycline-resistant isolates, 121 (82.9%) isolates were found to transfer the tet gene to the recipient strain in conjugation assays. Transfer frequencies ranged from 1.26 × 10−8 to 9.26 × 10−6. For 121 isolates possessing tet(A) or tet(B), the transconjugants possessed the same tet gene as their donors. Interestingly, for isolates containing tet(A) plus tet(B) or tet(B) plus tet(C), the transconjugants carried only the tet(B) gene. Plasmid replicon typing revealed that the most frequent replicon in the transconjugants was IncFIB, which was found in 95 (65.1%) isolates, and this was followed by Frep (45.2%), IncI1 (25.3%), IncP (24.7%), IncFIA (19.2%), and IncY (17.1%). The results of the conjugation assay with E. coli isolates included in phylogenetic groups B2 and D are shown in Table 5. The tetracycline resistance gene was successfully transferred for all except two isolates in these phylogenetic groups. IncFIB was the most frequent plasmid replicon detected in transconjugants of these groups (Table 5).
TABLE 5.
Characterization and transferability of resistance in E. coli isolates classified into phylogenetic groups B2 and D
| Strain | Phylogenetic group | Resistance phenotypea | Resistance gene | MICb (μg/ml) |
Plasmid replicon typec | Transconjugants |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TET | OXY | CTC | DOX | MIN | Transferability | tet gene | Plasmid replicon typec | |||||
| 60 | B2 | TE, S | tet(B) | 256 | 512 | 1,024 | 32 | 8 | FIB, Y, I1, Frep | + | tet(B) | FIB, I1, Frep |
| 61 | B2 | TE, S | tet(B) | 256 | 512 | 1,024 | 32 | 16 | FIB, Y, I1, Frep | − | ||
| 62 | B2 | TE, S | tet(B) | 256 | 512 | 1,024 | 32 | 8 | FIB, Y, I1 | + | tet(B) | FIB, I1 |
| 64 | B2 | TE, S | tet(B) | 256 | 512 | 1,024 | 32 | 8 | FIB, Y, I1 | + | tet(B) | FIB, I1 |
| 68 | B2 | TE, S | tet(A) | 256 | 512 | 1,024 | 32 | 8 | FIB, Y, I1, Frep | − | ||
| 90 | D | TE, S, AMP | tet(A) | 256 | 512 | 2,048 | 32 | 4 | P, FIA, FIB, Frep | + | tet(A) | FIA, FIB, Frep |
| 106 | D | TE, S | tet(B) | 256 | 512 | 2,048 | 32 | 16 | Frep | + | tet(B) | Frep |
| 123 | D | TE, NA | tet(A) | 256 | 512 | 1,024 | 16 | 2 | FIB, Frep | + | tet(A) | FIB, Frep |
| 124 | D | TE, GN, SXT, C, S, NA, AMP | tet(B) | 256 | 512 | 2,048 | 64 | 8 | FIA, FIB, Frep | + | tet(B) | FIB, Frep |
| 127 | D | TE, S, AMP | tet(A) | 256 | 512 | 2,048 | 32 | 4 | P, I1 | + | tet(A) | I1 |
| 128 | D | TE, AMP | tet(A) | 256 | 512 | 2,048 | 16 | 4 | FIB, I1 | + | tet(A) | Frep, I1 |
| 133 | D | TE, S, NA | tet(A) | 256 | 512 | 1,024 | 2 | 2 | FIB, Frep | + | tet(A) | Frep |
| 135 | D | TE, S, NA | tet(A) | 256 | 512 | 1,024 | 8 | 2 | FIB | + | tet(A) | FIB |
| 136 | D | TE, S, NA | tet(A) | 256 | 512 | 1,024 | 16 | 2 | FIB, Frep | + | tet(A) | FIB |
| 147 | D | TE, S, NA | tet(A) | 256 | 512 | 1,024 | 16 | 2 | FIB | + | tet(A) | FIB |
| 148 | D | TE, S, NA | tet(A) | 256 | 512 | 1,024 | 8 | 2 | FIB, Frep | + | tet(A) | FIB, Frep |
| 152 | D | TE, S, NA | tet(A) | 256 | 512 | 1,024 | 16 | 2 | FIB, Frep | + | tet(A) | FIB, Frep |
| 153 | D | TE, S, NA | tet(A) | 256 | 512 | 1,024 | 16 | 2 | FIB, Frep | + | tet(A) | FIB, Frep |
| 156 | D | TE, S, NA | tet(A) | 256 | 512 | 1,024 | 16 | 2 | FIB, Frep | + | tet(A) | FIB |
| 162 | D | TE, S, NA | tet(A) | 128 | 256 | 1,024 | 32 | 2 | FIB, Frep | + | tet(A) | FIB, Frep |
| 163 | D | TE, S, NA | tet(A) | 128 | 256 | 1,024 | 16 | 2 | FIB, Frep | + | tet(A) | FIB, Frep |
| 164 | D | TE, S, NA | tet(A) | 128 | 256 | 1,024 | 16 | 2 | FIB, Frep | + | tet(A) | FIB, Frep |
| 167 | D | TE, S, NA | tet(A) | 128 | 256 | 1,024 | 16 | 2 | FIB, Frep | + | tet(A) | FIB |
| 172 | D | TE, S, NA | tet(A) | 128 | 256 | 1,024 | 16 | 2 | FIB, Frep | + | tet(A) | FIB |
| 173 | D | TE, S, NA | tet(A) | 128 | 256 | 1,024 | 16 | 2 | FIB, Frep | + | tet(A) | FIB, Frep |
| 174 | D | TE, S, NA | tet(A) | 128 | 256 | 1,024 | 16 | 2 | FIB, Frep | + | tet(A) | FIB, Frep |
| 175 | D | TE, S, NA | tet(A) | 256 | 512 | 1,024 | 8 | 2 | FIB, Frep | + | tet(A) | FIB |
| 177 | D | TE, S, AMP | tet(A) | 64 | 256 | 2,048 | 16 | 4 | P, FIA, FIB, Frep | + | tet(A) | FIB, Frep |
| 178 | D | TE, S, AMP | tet(A) | 64 | 256 | 2,048 | 16 | 4 | P, FIA, FIB, Frep | + | tet(A) | FIB, Frep |
| 192 | D | TE, SXT, C, S, AMP | tet(A) | 256 | 512 | 1,024 | 16 | 2 | P, FIB, Frep | + | tet(A) | FIB, Frep |
| 194 | D | TE, SXT, S, AMP | tet(A) | 128 | 256 | 1,024 | 16 | 2 | Frep | + | tet(A) | Frep |
| 198 | D | TE, SXT, S, AMP | tet(A) | 128 | 256 | 1,024 | 32 | 2 | Frep | + | tet(A) | Frep |
TE, tetracycline; S, streptomycin; GN, gentamicin; SXT, sulfamethoxazole-trimethoprim; C, chloramphenicol; NA, nalidixic acid; AMP, ampicillin.
TET, tetracycline; OXY, oxytetracycline; CTC, chlortetracycline; DOX, doxycycline; MIN, minocycline.
FIB, IncFIB replicon; I1, IncI1 replicon; P, IncP replicon, FIA, IncFIA replicon; Y, IncY replicon.
DISCUSSION
In the present study, all tetracycline-resistant isolates carried either tet(A) or tet(B), suggesting that these genes are important for the development of tetracycline resistance. Actually, tet(A) and/or tet(B), encoding efflux mechanisms, has been reported to be the most common tetracycline resistance determinant in E. coli isolates from humans and animals in many countries (12, 13, 20–22). Previous studies conducted in cattle disagree: some have reported that the tet(A) determinant is dominant in E. coli isolates recovered from cattle (23–25), whereas others found tet(B) to be dominant (26–28). In the present study, the prevalences of tet(A) and tet(B) were almost equal at 46.5% and 45.1%, respectively, which is consistent with other reports that showed a similar distribution pattern for the tet gene in E. coli isolates recovered from animals (23, 29). The degree of resistance to tetracycline is associated with the presence of tet(B) (10). In the present study, MIC testing showed that E. coli isolates carrying only tet(B) appeared to have higher MIC values for tetracycline, chlortetracycline, oxytetracycline, doxycycline, and minocycline, which concurs with previous reports (10, 13, 30). Furthermore, we found that the MIC values for isolates carrying tet(B) were significantly higher for doxycycline and minocycline. These results are consistent with those of a previous study, in which tet(B) was found to confer resistance to expanded-spectrum tetracyclines, including minocycline and doxycycline (31).
In a previous study, tet(C) was frequently identified in E. coli isolates recovered from a commercial beef processing plant (32). However, we found tet(C) in only nine strains isolated from beef cattle, and those isolates showed susceptibility, but with low MIC values, to tetracycline, which concurs with the findings of previous studies (8, 33). Interestingly, the prevalences of tet(C) in E. coli isolates recovered from animals was reported to be higher than the prevalences of tet(C) in E. coli isolates recovered from meat and meat products (8), which suggests that some processing stages may reduce tetracycline resistance in E. coli.
Several studies have described E. coli isolates carrying more than two tet genes (11, 34, 35). In South Korea, 40% of E. coli strains isolated from cows and pigs in slaughterhouses were found to have two different tet genes (36), and in the present study, four E. coli isolates were found to carry more than two tet genes. Although the prevalence of isolates containing both tet(A) and tet(B) in the present study was lower than that reported in previous studies (11, 34), we found two isolates harboring tet(B) and tet(C), which is the first report of this combination in E. coli strains isolated from beef cattle in South Korea. However, this conflicts with the findings of a previous study, in which tet(C) was always found with tet(A) (37). Our study also showed that two isolates that carried more than one tet gene did not have higher MIC values than isolates that harbored one tet gene. This phenomenon was described in a previous study, in which it was proposed that the acquisition of more than one tet gene is caused by strong selective pressure rather than a selective advantage (35).
The long-term use of tetracycline confers resistance to other antimicrobial agents by E. coli. This phenomenon, called coselection, could be the result of tet genes being located on the same mobile genetic elements, such as plasmids, transposons, or integrons, as other resistance genes (38). In the present study, many isolates were resistant to tetracycline and other antimicrobials, and 62.3% of tetracycline-resistant isolates exhibited multidrug resistance. Thus, coselection has important implications, as it means that tetracycline resistance has contributed much to the increased prevalence of multidrug resistance in E. coli.
Phylogenetic groups B2 and D are associated with pathogenicity, whereas strains of groups A and B1 are classified as nonpathogenic commensal strains (17, 39). In the present study, most isolates were classified as group B1 (65.2%). This is consistent with the results of other studies that found that bovine E. coli isolates most frequently belong to group A and/or B1 (25, 40). Twenty-seven isolates (17.4%) were classified as group D, even though they were cultured from clinically healthy cattle in this study.
Conjugative transfer is the most common mechanism for the delivery of antimicrobial resistance between Gram-negative isolates because plasmid conjugation can occur at a high frequency and transfer resistance genes (41). In the present study, most tetracycline-resistant isolates (82.9%) exhibited conjugative transfer, which means that most tet genes are carried and transferred by conjugative plasmids. Therefore, we presume that the horizontal transfer of tet genes provides an effective mechanism for the widespread distribution of tetracycline resistance in bacterial populations and explains the high prevalence of tetracycline-resistant E. coli isolates.
In South Korea, although the use of tetracyclines as feed additives was entirely banned in July 2011, in 2013, about 40% of bovine E. coli isolates were found to be resistant to tetracycline (42). Accordingly, we propose that the high prevalence of tetracycline resistance in E. coli is probably due to the horizontal transfer of tet determinants from E. coli isolates carrying tet genes which have survived selective pressure caused by the use of tetracycline derivatives. We hope that these findings can be utilized as basic data for epidemiologic studies and studies to assess the risk of tetracycline resistance.
ACKNOWLEDGMENTS
This study was supported by the Rural Development Administration (PJ008970012012), BK21 PLUS, and the Research Institute for Veterinary Science, Seoul National University, Seoul, South Korea.
REFERENCES
- 1.Cizman M. 2003. The use and resistance to antibiotics in the community. Int J Antimicrob Agents 21:297–307. doi: 10.1016/S0924-8579(02)00394-1. [DOI] [PubMed] [Google Scholar]
- 2.Aarestrup FM. 2004. Monitoring of antimicrobial resistance among food animals: principles and limitations. J Vet Med B Infect Dis Vet Public Health 51:380–388. doi: 10.1111/j.1439-0450.2004.00775.x. [DOI] [PubMed] [Google Scholar]
- 3.Tadesse DA, Zhao S, Tong E, Ayers S, Singh A, Bartholomew MJ, McDermott PF. 2012. Antimicrobial drug resistance in Escherichia coli from humans and food animals, United States, 1950–2002. Emerg Infect Dis 18:741–749. doi: 10.3201/eid1805.111153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee SJ, Lee DS, Choe HS, Shim BS, Kim CS, Kim ME, Cho YH. 2011. Antimicrobial resistance in community-acquired urinary tract infections: results from the Korean Antimicrobial Resistance Monitoring System. J Infect Chemother 17:440–446. doi: 10.1007/s10156-010-0178-x. [DOI] [PubMed] [Google Scholar]
- 5.Levy SB, Marshall B. 2004. Antibacterial resistance worldwide: causes, challenges and responses. Nat Med 10:S122–S129. doi: 10.1038/nm1145. [DOI] [PubMed] [Google Scholar]
- 6.Chopra I, Roberts M. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev 65:232–260. doi: 10.1128/MMBR.65.2.232-260.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts MC. 2005. Update on acquired tetracycline resistance genes. FEMS Microbiol Lett 245:195–203. http://dx.doi.org/10.1016/j.femsle.2005.02.034. doi: 10.1016/j.femsle.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 8.Koo HJ, Woo GJ. 2011. Distribution and transferability of tetracycline resistance determinants in Escherichia coli isolated from meat and meat products. Int J Food Microbiol 145:407–413. doi: 10.1016/j.ijfoodmicro.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 9.Erb A, Sturmer T, Marre R, Brenner H. 2007. Prevalence of antibiotic resistance in Escherichia coli: overview of geographical, temporal, and methodological variations. Eur J Clin Microbiol Infect Dis 26:83–90. http://dx.doi.org/10.1007/s10096-006-0248-2. doi: 10.1007/s10096-006-0248-2. [DOI] [PubMed] [Google Scholar]
- 10.Gow SP, Waldner CL, Harel J, Boerlin P. 2008. Associations between antimicrobial resistance genes in fecal generic Escherichia coli isolates from cow-calf herds in western Canada. Appl Environ Microbiol 74:3658–3666. doi: 10.1128/AEM.02505-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sengelov G, Halling-Sorensen B, Aarestrup FM. 2003. Susceptibility of Escherichia coli and Enterococcus faecium isolated from pigs and broiler chickens to tetracycline degradation products and distribution of tetracycline resistance determinants in E. coli from food animals. Vet Microbiol 95:91–101. doi: 10.1016/S0378-1135(03)00123-8. [DOI] [PubMed] [Google Scholar]
- 12.Karami N, Nowrouzian F, Adlerberth I, Wold AE. 2006. Tetracycline resistance in Escherichia coli and persistence in the infantile colonic microbiota. Antimicrob Agents Chemother 50:156–161. doi: 10.1128/AAC.50.1.156-161.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tuckman M, Petersen PJ, Howe AYM, Orlowski M, Mullen S, Chan K, Bradford PA, Jones CH. 2007. Occurrence of tetracycline resistance genes among Escherichia coli isolates from the phase 3 clinical trials for tigecycline. Antimicrob Agents Chemother 51:3205–3211. doi: 10.1128/AAC.00625-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shin SW, Byun J, Jung M, Shin M, Yoo HS. 2014. Antimicrobial resistance, virulence genes and PFGE-profiling of Escherichia coli isolates from South Korean cattle farms. J Microbiol 52:785–793. doi: 10.1007/s12275-014-4166-1. [DOI] [PubMed] [Google Scholar]
- 15.Clinical and Laboratory Standards Institute. 2013. Performance standards for antimicrobial susceptibility testing; twenty-third informational supplement. M100-S23. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 16.Ng LK, Martin I, Alfa M, Mulvey M. 2001. Multiplex PCR for the detection of tetracycline resistant genes. Mol Cell Probes 15:209–215. doi: 10.1006/mcpr.2001.0363. [DOI] [PubMed] [Google Scholar]
- 17.Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol 66:4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang MG, Sahm DF, Jacoby GA, Hooper DC. 2004. Emerging plasmid-mediated quinolone resistance associated with the qnr gene in Klebsiella pneumoniae clinical isolates in the United States. Antimicrob Agents Chemother 48:1295–1299. doi: 10.1128/AAC.48.4.1295-1299.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson TJ, Wannemuehler YM, Johnson SJ, Logue CM, White DG, Doetkott C, Nolan LK. 2007. Plasmid replicon typing of commensal and pathogenic Escherichia coli isolates. Appl Environ Microbiol 73:1976–1983. doi: 10.1128/AEM.02171-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahmed MO, Clegg PD, Williams NJ, Baptiste KE, Bennett M. 2010. Antimicrobial resistance in equine faecal Escherichia coli isolates from North West England. Ann Clin Microbiol Antimicrob 9:12. doi: 10.1186/1476-0711-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwaiger K, Holzel C, Bauer J. 2010. Resistance gene patterns of tetracycline resistant Escherichia coli of human and porcine origin. Vet Microbiol 142:329–336. doi: 10.1016/j.vetmic.2009.09.066. [DOI] [PubMed] [Google Scholar]
- 22.Hu GZ, Pan YS, Wu H, Hu H, Xu R, Yuan L, Liu JH, Feng JK. 2013. Prevalence of tetracycline resistance genes and identification of tet(M) in clinical isolates of Escherichia coli from sick ducks in China. J Med Microbiol 62:851–858. doi: 10.1099/jmm.0.051896-0. [DOI] [PubMed] [Google Scholar]
- 23.Guerra B, Junker E, Schroeter A, Malorny B, Lehmann S, Helmuth R. 2003. Phenotypic and genotypic characterization of antimicrobial resistance in German Escherichia coli isolates from cattle, swine and poultry. J Antimicrob Chemother 52:489–492. doi: 10.1093/jac/dkg362. [DOI] [PubMed] [Google Scholar]
- 24.Sharma R, Munns K, Alexander T, Entz T, Mirzaagha P, Yanke LJ, Mulvey M, Topp E, McAllister T. 2008. Diversity and distribution of commensal fecal Escherichia coli bacteria in beef cattle administered selected subtherapeutic antimicrobials in a feedlot setting. Appl Environ Microbiol 74:6178–6186. doi: 10.1128/AEM.00704-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karczmarczyk M, Walsh C, Slowey R, Leonard N, Fanning S. 2011. Molecular characterization of multidrug-resistant Escherichia coli isolates from Irish cattle farms. Appl Environ Microbiol 77:7121–7127. doi: 10.1128/AEM.00601-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walk ST, Mladonicky JM, Middleton JA, Heidt AJ, Cunningham JR, Bartlett P, Sato K, Whittam TS. 2007. Influence of antibiotic selection on genetic composition of Escherichia coli populations from conventional and organic dairy farms. Appl Environ Microbiol 73:5982–5989. doi: 10.1128/AEM.00709-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mirzaagha P, Louie M, Sharma R, Yanke LJ, Topp E, McAllister TA. 2011. Distribution and characterization of ampicillin- and tetracycline-resistant Escherichia coli from feedlot cattle fed subtherapeutic antimicrobials. BMC Microbiol 11:78. doi: 10.1186/1471-2180-11-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sawant AA, Hegde NV, Straley BA, Donaldson SC, Love BC, Knabel SJ, Jayarao BM. 2007. Antimicrobial-resistant enteric bacteria from dairy cattle. Appl Environ Microbiol 73:156–163. doi: 10.1128/AEM.01551-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Momtaz H, Rahimi E, Moshkelani S. 2012. Molecular detection of antimicrobial resistance genes in E. coli isolated from slaughtered commercial chickens in Iran. Veterinar Med 57:193–197. [Google Scholar]
- 30.Blake DP, Humphry RW, Scott KP, Hillman K, Fenlon DR, Low JC. 2003. Influence of tetracycline exposure on tetracycline resistance and the carriage of tetracycline resistance genes within commensal Escherichia coli populations. J Appl Microbiol 94:1087–1097. doi: 10.1046/j.1365-2672.2003.01937.x. [DOI] [PubMed] [Google Scholar]
- 31.Huys G, Cnockaert M, Vaneechoutte M, Woodford N, Nemec A, Dijkshoorn L, Swings J. 2005. Distribution of tetracycline resistance genes in genotypically related and unrelated multiresistant Acinetobacter baumannii strains from different European hospitals. Res Microbiol 156:348–355. doi: 10.1016/j.resmic.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 32.Aslam M, Service C. 2006. Antimicrobial resistance and genetic profiling of Escherichia coli from a commercial beef packing plant. J Food Prot 69:1508–1513. [DOI] [PubMed] [Google Scholar]
- 33.Chalmers G, Kozak GK, Hillyer E, Reid-Smith RJ, Boerlin P. 2010. Low minimum inhibitory concentrations associated with the tetracycline-resistance gene tet(C) in Escherichia coli. Can J Vet Res 74:145–148. [PMC free article] [PubMed] [Google Scholar]
- 34.Lanz R, Kuhnert P, Boerlin P. 2003. Antimicrobial resistance and resistance gene determinants in clinical Escherichia coli from different animal species in Switzerland. Vet Microbiol 91:73–84. doi: 10.1016/S0378-1135(02)00263-8. [DOI] [PubMed] [Google Scholar]
- 35.Bryan A, Shapir N, Sadowsky MJ. 2004. Frequency and distribution of tetracycline resistance genes in genetically diverse, nonselected, and nonclinical Escherichia coli strains, isolated from diverse human and animal sources. Appl Environ Microbiol 70:2503–2507. doi: 10.1128/AEM.70.4.2503-2507.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cho JK, Kim KS. 2008. Antimicrobial resistance and distribution of tetracycline resistance genes of Escherichia coli isolated from human and livestock in slaughterhouse. Kor J Vet Public Health 32:213–223. [Google Scholar]
- 37.Maynard C, Fairbrother JM, Bekal S, Sanschagrin F, Levesque RC, Brousseau R, Masson L, Lariviere S, Harel J. 2003. Antimicrobial resistance genes in enterotoxigenic Escherichia coli O149:K91 isolates obtained over a 23-year period from pigs. Antimicrob Agents Chemother 47:3214–3221. doi: 10.1128/AAC.47.10.3214-3221.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gophna U, Parket A, Hacker J, Ron EZ. 2003. A novel ColV plasmid encoding type IV pili. Microbiology 149:177–184. doi: 10.1099/mic.0.25858-0. [DOI] [PubMed] [Google Scholar]
- 39.Cocchi S, Grasselli E, Gutacker M, Benagli C, Convert M, Piffaretti JC. 2007. Distribution and characterization of integrons in Escherichia coli strains of animal and human origin. FEMS Immunol Med Microbiol 50:126–132. doi: 10.1111/j.1574-695X.2007.00242.x. [DOI] [PubMed] [Google Scholar]
- 40.Houser BA, Donaldson SC, Padte R, Sawant AA, DebRoy C, Jayarao BM. 2008. Assessment of phenotypic and genotypic diversity of Escherichia coli shed by healthy lactating dairy cattle. Foodborne Pathog Dis 5:41–51. doi: 10.1089/fpd.2007.0036. [DOI] [PubMed] [Google Scholar]
- 41.Sunde M, Norstrom M. 2006. The prevalence of, associations between and conjugal transfer of antibiotic resistance genes in Escherichia coli isolated from Norwegian meat and meat products. J Antimicrob Chemother 58:741–747. doi: 10.1093/jac/dkl294. [DOI] [PubMed] [Google Scholar]
- 42.Animal and Plant Quarantine Agency. 2013. Antimicrobial use in livestock and monitoring of antimicrobial resistance in animal and carcass, p 26–27. South Korea Food and Drug Administration, Seoul, South Korea. [Google Scholar]