Abstract
Novel processing strategies for hydrolysis and fermentation of lignocellulosic biomass in a single reactor offer large potential cost savings for production of biocommodities and biofuels. One critical challenge is retaining high enzyme production in the presence of elevated product titers. Toward this goal, the cellulolytic, ethanol-producing bacterium Clostridium phytofermentans was adapted to increased ethanol concentrations. The resulting ethanol-tolerant (ET) strain has nearly doubled ethanol tolerance relative to the wild-type level but also reduced ethanol yield and growth at low ethanol concentrations. The genome of the ET strain has coding changes in proteins involved in membrane biosynthesis, the Rnf complex, cation homeostasis, gene regulation, and ethanol production. In particular, purification of the mutant bifunctional acetaldehyde coenzyme A (CoA)/alcohol dehydrogenase showed that a G609D variant abolished its activities, including ethanol formation. Heterologous expression of Zymomonas mobilis pyruvate decarboxylase and alcohol dehydrogenase in the ET strain increased cellulose consumption and restored ethanol production, demonstrating how metabolic engineering can be used to overcome disadvantageous mutations incurred during adaptation to ethanol. We discuss how genetic changes in the ET strain reveal novel potential strategies for improving microbial solvent tolerance.
INTRODUCTION
The conversion of lignocellulosic biomass to fuels and commodities represents a large-scale, renewable alternative to petroleum. This multistep bioconversion is traditionally performed in a series of independent processes, but consolidated bioprocessing (CBP) is an alternative paradigm with potential economic advantages (1). In CBP, enzyme production, hydrolysis, and fermentation occur in a single reactor, leading to savings in capital and operating costs as well as increased efficiencies due to system synergies (2). Here we studied Clostridium phytofermentans, a promising CBP candidate that ferments plant biomass primarily to ethanol (3, 4). C. phytofermentans hydrolyzes pretreated corn stover (both glucans and xylans) with efficiencies similar to those seen with simultaneous saccharification and cofermentation (SSCF) using commercial enzymes and xylose-fermenting yeast (Saccharomyces cerevisiae) (5). Fermentation of pretreated corn stover by C. phytofermentans reaches a titer of 7 g liter−1 ethanol (6), and stable cocultures of C. phytofermentans and S. cerevisiae cdt-1 ferment ∼70 g liter−1 cellulose to 22 g liter−1 ethanol (7), which is an ethanol concentration that reduces C. phytofermentans growth. Thus, application of CBP bacteria such as C. phytofermentans will likely require improving their solvent tolerances without compromising enzyme production or fermentation of soluble carbohydrates to ethanol.
Considerable effort has focused on adapting clostridia to increased solvent levels and investigating the genetic and physiological changes associated with adaptation to solvents (8–13). Other studies have shown increased ethanol production in clostridia that primarily produce fermentation products other than ethanol. C. cellulolyticum expressing pyruvate decarboxylase and alcohol dehydrogenase (ADH) overcame pyruvate accumulation and shifted fermentation products from lactate to acetate and ethanol (14). In C. thermocellum, redirection of carbon flow through pyruvate kinase (15), inactivation of lactate dehydrogenase and phosphotransacetylase (16), and deletion of hydrogenases (17) all improve ethanol production. These results demonstrate that, although the genetic tools are being developed only now, engineering improved ethanol production in cellulolytic clostridia is possible. However, development of strains that are ethanol tolerant (ET) and that also produce ethanol in high titers remains a significant challenge.
Here we sought to develop a strain of C. phytofermentans with both improved resistance and production of ethanol, particularly from cellulose. We isolated an ethanol-tolerant (ET) C. phytofermentans strain by serial transfer into increasing ethanol levels and characterized its growth and fermentation properties. We sequenced the ET strain genome to reveal genomic mutations that arose during adaptation and overcame reduced ethanol yield in the ET strain by heterologous expression of an alternative ethanol formation pathway. We discuss how the findings from this study improve our understanding of how microbes adapt to elevated concentrations of solvents such as ethanol.
MATERIALS AND METHODS
Culturing.
C. phytofermentans ISDg (ATCC 700394) was grown anaerobically by preparing cultures in a Coy anaerobic chamber with a 1.5% H2/98.5% N2 atmosphere. Cultures were incubated without shaking at 30°C in GS2 medium (18) adjusted to a pH of 7 and supplemented with carbon sources as described elsewhere in the text. Growth kinetics were monitored by optical density at 600 nm (OD600) in sealed 100-well microtiter plates (Bioscreen 9502550) as previously described (19); cultures were briefly shaken to resuspend cells before each optical density measurement. Cellulose, cellobiose, and glucose cultures for substrate consumption and fermentation product analysis were grown in 100-ml serum bottles, which were sealed with butyl rubber stoppers after degassing.
The C. phytofermentans ethanol-tolerant (ET) strain was selected by serial transfer (1:50 dilution) into culture tubes containing 10 ml of GS2 medium supplemented with increasing ethanol concentrations. Starting with cultures in 4% (vol/vol) (31.5 g liter−1) ethanol, cultures were transferred weekly to fresh medium containing the same ethanol concentration and to medium with a 1%-higher ethanol concentration. If no growth was observed at the higher concentration after 1 week, cultures were retransferred to the same ethanol concentrations. If a culture grew at a higher ethanol concentration, this culture was transferred again to that ethanol concentration and to a 1%-greater ethanol concentration. Growth was observed at 5% ethanol after 7 weekly transfers, 6% after 13 transfers, and 7% after 19 transfers. Each time the ethanol tolerance improved, cells were plated, individual colonies were picked, and liquid cultures were reinoculated to ensure that selection was based on a specific strain with increased ethanol tolerance and not on a consortium of strains that collectively survived the increased ethanol concentration. The ET strain is thus a colony-purified isolate from a mother culture that grew in GS2 medium supplemented with 7% (vol/vol) ethanol. After colony purification, it was confirmed the ET strain has ethanol resistance similar to that of the mother culture.
Cellulose and fermentation analysis.
The level of cellulose remaining in the culture was measured by taking a 1-ml sample from a 10-ml culture tube with a sterile syringe, placing it in a preweighed 1.7-ml microcentrifuge tube, and centrifuging at 13,000 × g for 10 min. The supernatant was removed, and the cellulose pellet was washed and centrifuged again at 13,000 × g for 10 min. The rinsed pellet was placed at 70°C to dry until a constant mass was reached. The contribution of cellular biomass to total cellulose weight was not accounted for and was assumed to be minimal due to low anaerobic biomass yields.
Fermentation product concentrations were measured in 0.22-μm-pore-size-filtered culture supernatant using an Agilent 1100 high-performance liquid chromatograph (HPLC) with a Jasco RI-1531 refractive index detector (RID) and an Aminex HPX-87H cation exchange column (Bio-Rad). The HPLC was run using a 0.01 M sulfuric acid mobile phase, 65°C column temperature, 30°C RID temperature, 25 μl sample volume, and 0.6 ml/min operating flow rate. Product formation is reported relative to the concentration in the medium at the point of inoculation. Gas phase measurements were made by removing 1 ml of headspace and injecting 100 μl into a gas chromatograph (Model 8610C multiple-gas analyzer; SRI Instruments). Argon was used as a carrier gas and was adjusted to 30 lb/in2 gauge pressure. A stainless steel molecular sieve (13×) and silica gel-packed columns were used for sample separation, and the components were detected using a thermal conductivity detector (TCD). The column compartment temperature was held initially at 40°C for 3.5 min and then ramped to 160°C for 2 min and to 300°C for 10 min, after which the column was allowed to cool to 40°C for the remainder of the sample run.
Genome sequencing and variant analysis of the ET strain.
A total of 12 μg of genomic DNA was extracted from a 4-ml ET strain culture using a Sigma GenElute bacterial genomic DNA kit (NA2110). DNA was sequenced on an Illumina MiSeq instrument with an insertion size of 795 bp and 300-bp paired-end reads. A total of 5,077,282 reads passed quality filtering and mapping by Picard Tools (https://github.com/broadinstitute/picard), yielding approximately 250-fold genome coverage. Sequence variants (single nucleotide polymorphisms [SNPs] and indels) in the ET genome relative to the reference strain genome (NCBI accession no. NC_010001.1) were identified using the Genome Analysis Toolkit (GATK) (20) (see the supplemental material for detailed descriptions of the filtering and variant-calling methods).
ADH purification and activity measurements.
The Cphy3925-encoding genes from the wild-type (WT) and ET strains were cloned by ligation-independent cloning (21) into pET-22B(+) as previously described (19). Genes were cloned with C-terminal His tags using primer pair cphy3925F/cphy3925R (Table 1) and confirmed by sequencing. Plasmids were transformed into Escherichia coli BL21(DE3) (Novagen 70235) and grown in 50 ml TB medium (12 g liter−1 tryptone, 24 g liter−1 yeast extract, 4 ml liter−1 glycerol) to an OD600 of 1, and expression was induced by adding 500 μM IPTG (isopropyl-β-d-thiogalactopyranoside) and incubating overnight at 20°C. Cells were pelleted, resuspended in lysis buffer (50 mM phosphate buffer [pH 8], 0.5 M NaCl, 10 mM imidazole, 15% glycerol, 1 mM Pefabloc [Sigma 76307]), and lysed by sonication (Cole-Parmer Vibracell CV33) with lysozyme (Novagen 71230). His-tagged proteins were purified from 50 ml culture on nickel-nitrilotriacetic acid (Ni-NTA) spin columns (Qiagen 31014) and visualized on 12% SDS-PAGE gels (Novex 12% Bis-Tris gel NP0342BOX). Enzyme activities were measured as described in reference 22 in 100 μl of 100 mM Tris-HCl (pH 8) containing cofactor [0.2 mM NADH(P)H or 2 mM NAD(P)+] and substrate (300 μM acetyl coenzyme A [acetyl-CoA] or CoA, 18 mM acetaldehyde, 2 M ethanol) as appropriate. NAD(P)H was measured as 340-nm absorbance (extinction coefficient, 6.22 mM−1 · cm−1) using a SAFAS UVmc2 spectrophotometer at room temperature.
TABLE 1.
Bacterial strains, primers, and plasmids used in this study
| Strain name | Genotype or descriptiona | Source or sequence |
|---|---|---|
| Strains | ||
| Clostridium phytofermentans ISDg | ATCC type strain 700394 | Susan Leschine Laboratory, University of Massachusetts, Amherst, Amherst, MA, USA |
| E. coli Top 10 | hsdR mcrA endA1 recA1 rpsL (Strr) (cloning strain) | Invitrogen Corporation |
| E. coli S17-1 | RP4-2 (Km::Tn7 Tc::Mu-1) recA1 endA1 (conjugal strain) | Yale E. coli Stock Center |
| Primers | ||
| pdcAdhB_F | Forward primer for amplification of pdc and adhB from pES120 | 5′-TTTTTCGAATTCACCGGATCCCTGCAGTAGGAGGAATTAACC-3′ |
| pdcAdhB_R | Reverse primer for amplification of pdc and adhB from pES120 | 5′-ATATTTCGATCGATTGCATGCTTAGAAAGCGCTCAGGAAGAG-3′ |
| pQexp_F | Forward primer to confirm pdc-adhB insertion in pQexp | 5′-AAACCTAGGTAATTGAGGAAAGTTACAATTA-3′ |
| pQexp_R | Reverse primer to confirm pdc-adhB insertion in pQexp | 5′-GAATGGCGCCTGATGCG-3′ |
| cphy3925F | Forward primer to amplify Cphy3925 coding sequence | 5′-AAAGAAGGAGATAGGATCATGACGAAGAAAGTGGAATTA-3′ |
| cphy3925R | Reverse primer to amplify Cphy3925 coding sequence | 5′-GTGTAATGGATAGTGATCTTAATGGTGATGGTGATGATGTTTACCGTAGTACACTTTAAGATAG-3′ |
| Plasmids | ||
| pES120 | Source of Z. mobilis pdc and adhB genes | Jay Keasling Laboratory, University of California, Berkeley, Berkeley, CA, USA |
| pQexp | Replicating plasmid for C. phytofermentans | Andrew Tolonen Laboratory, Genoscope-CEA, Évry, France |
| pQexpE | pQexp with Z. mobilis pdc and adhB cloned into the unique BamHI and PvuI sites | This study |
Str, streptomycin, Km, kanamycin; Tc, tetracycline.
Plasmid construction and transformations.
pQexpE is derived from pQexp (23), a plasmid that replicates stably with erythromycin selection in E. coli (200 μg ml−1 erythromycin) and C. phytofermentans (40 μg ml−1erythromycin on plates and 200 μg ml−1 erythromycin in liquid culture). To construct pQexpE, the Z. mobilis pdc and adhB genes were PCR amplified from pES120 (24) using primer pair pdcAdhB_F/dcAdhB_R (Table 1) and cloned into the unique BamHI and PvuI sites of pQexp. The insertion was confirmed by sequencing using primer pair pQexp_F/pQexp_R. pQexpE was conjugally transferred to C. phytofermentans using donor strain E. coli S17-1. Conjugation was performed as described in references 23 and 25, except polyethersulfone membranes were used to support 50-μl culture mixtures and only nalidixic acid without trimethoprim was used to select against E. coli following mating. Positive C. phytofermentans transconjugants containing pQexpE were confirmed by colony PCR using primer pair pQexp_F/pQexp_R to amplify the 2.9-kb pdc-adhB operon.
Nucleotide sequence accession number.
The FASTQ-formatted DNA sequencing files for the ET genome were submitted to the European Nucleotide Archive under primary accession no. PRJEB7255.
RESULTS AND DISCUSSION
Isolation and physiology of an ethanol-tolerant C. phytofermentans strain.
The growth of wild-type (WT) C. phytofermentans was monitored in cultures supplemented with 0, 2, 4, 6, or 7% (vol/vol) ethanol (Fig. 1A), and generation times (hours) and maximum cell densities (OD600) were calculated (see Table S1 in the supplemental material). The WT strain cultures grew similarly at 0% ethanol and 2% ethanol but growth was significantly inhibited at 4% ethanol, and very little growth was observed at 6 and 7% ethanol. In contrast, the ET strain grew at 4, 6, and 7% ethanol, with a maximum cell density (OD600) at 7% ethanol similar to that of the WT strain at 4% ethanol (Fig. 1B; see also Table S1).
FIG 1.
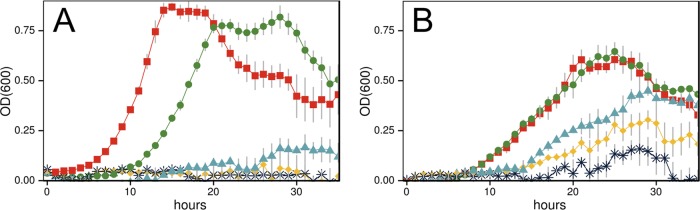
Growth of wild-type (WT) (A) and ethanol-tolerant (ET) (B) C. phytofermentans strains at 30°C in GS2 medium with 3 g liter−1 glucose supplemented with ethanol at the following levels (vol/vol): 0% (red squares), 2% (green circles), 4% (blue triangles), 6% (yellow diamonds), and 7% (black X's). Data points of growth (OD600) represent means of results from triplicate 400-μl cultures in sealed microtiter plates; error bars represent 1 standard deviation.
While the ET strain was more ethanol resistant, it grew more slowly than the WT strain and grew at reduced cell yields in cultures without added ethanol (Fig. 1). Glucose consumption by the ET strain was also slower than glucose consumption by the WT strain, reflecting the reduced growth rate. When grown on 30 g liter−1 glucose, the ET strain consumed only 20 g liter−1 substrate in 200 h, while the WT strain completely exhausted the glucose in less than 150 h (see Fig. S1 in the supplemental material). Thus, the enhanced growth of the ET strain at a high ethanol concentration is accompanied by reduced growth under standard conditions, supporting the idea that the ET strain has altered physiology, with results similar to those seen with C. thermocellum adapted to 5% ethanol (26, 27). However, we observed no morphological differences between ET and WT colonies grown on GS2 agar plates or cells grown in liquid GS2 medium (see Fig. S2).
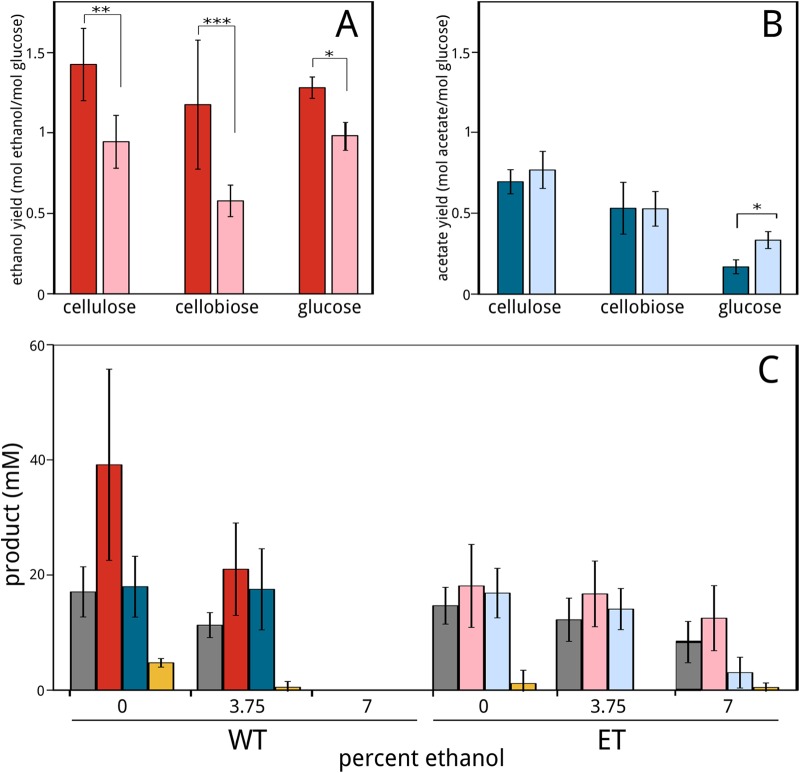
The ET strain also produces less ethanol per unit of sugar consumption than the WT strain. For example, ethanol yield (moles of product per mole of glucose equivalent consumed) by the ET strain decreased 25 to 50% relative to that seen with the WT strain when growing on cellulose, cellobiose, and glucose (Fig. 2A), whereas the acetate yields of the strains were similar (Fig. 2B). We also measured cellobiose consumption and formation of the primary fermentation products ethanol, acetate, and formate in WT and ET cultures grown in cellobiose medium supplemented with 0, 3.75, or 7% ethanol (Fig. 2C). Ethanol production by the WT strain was significantly lower at 3.75% ethanol, and cellobiose consumption and fermentation ceased at 7% ethanol. In contrast, cellobiose consumption and ethanol production by the ET strain decreased only slightly with increased ethanol supplementation, demonstrating the robustness of the ethanol resistance phenotype. In all, these results highlight the phenotypic advantages of the ET strain with respect to metabolizing and producing ethanol at elevated ethanol concentrations but also indicate that those advantages occur at the expense of lower ethanol yields and slower growth at low ambient concentrations of ethanol.
FIG 2.
C. phytofermentans yields (mole of product per mole of glucose equivalent consumed) of ethanol from WT (dark red) and ET (light red) strains (A) and acetate from WT (dark blue) and ET (light blue) strains (B). Statistical differences between WT and ET yield averages are indicated at P values of <0.01 (*), <0.05 (**), and <0.1 (***) using Student's t test. (C) C. phytofermentans WT and ET cellobiose consumption and fermentation products in GS2 cellobiose medium with 0, 3.75, or 7% (vol/vol) added ethanol. Bars show concentrations (in millimoles) of cellobiose consumption (gray) and production of ethanol (red), acetate (blue), and formate (yellow) by WT (dark bars) and ET (light bars) strains. All cultures were grown in serum bottles containing GS2 medium at 30°C with 30 g liter−1 cellulose, 10 g liter−1 cellobiose, or 30 g liter−1 glucose and measured after 14 days. Bars represent averages of the results from 4 cultures; error bars represent 1 standard deviation.
Genome sequence of the ET strain.
The C. phytofermentans ET strain genome contains 12 variants relative to the WT strain (Table 2), many fewer than a C. thermocellum ET genome with similar ethanol resistance that had 200 to 500 changes (9). While ethanol resistance is likely a complex, multigenic trait, the small number of changes in the ET strain genome shed light on DNA variants that could have functional roles in ethanol tolerance. Two of the 12 mutations were in genes encoding transcriptional regulators that could effectuate broad gene expression changes: PolB, the β-subunit of DNA-directed RNA polymerase, and a LysR regulator, Cphy3040. LysR-type regulators often colocalize in the genome with their targets (28) and the gene encoding Cphy3040 is adjacent to a gene encoding a NAD-dependent aldehyde dehydrogenase, suggesting that this regulator is related to alcohol formation.
TABLE 2.
Genomic DNA variants in the ET straina
| Protein | Length (amino acids) | Amino acid variant | Confidence (Q) value | Annotation |
|---|---|---|---|---|
| Energy and metabolism | ||||
| Cphy3925 (AdhE) | 872 | G609D | 8,929 | Fe-dependent bifunctional acetaldehyde-CoA/alcohol dehydrogenase |
| Cphy0215 (RnfA) | 191 | C26S | 5,569 | Ferredoxin:NAD+ oxidoreductase (Fno); couples electron transfer from reduced ferredoxin to NAD+ with cation transport out of the cell to create an electrochemical gradient |
| Cphy3255 | 259 | 20-bp insertion at Q91 | 15,938 | FMN-dependent nitroreductase |
| Transport | ||||
| Cphy0543 (MgtA/MgtB) | 920 | K417N | 8,661 | P-type ATPase for Mg2+ uptake transporter or Ca2+/Mg2+ antiporter |
| Cphy3778 | 258 | 1-bp deletion, Y239 frameshift | 11,687 | Na+ efflux transporter, ABC-2 transmembrane permease (PFAM accession no. PF06182) |
| Membrane and cell wall | ||||
| Cphy0233 (PlsD) | 237 | D80N | 9,246 | Membrane synthesis: glycerol-3-phosphate acyltransferase; transfers a fatty acid to the 1 position of glycerol-3-phosphate |
| Cphy0107 (MurC) | 469 | G115D | 8,841 | Peptidoglycan synthesis: ATP-dependent ligation of l-alanine and UDP-N-acetylmuramic acid to form UDP-N-acetylmuramyl-l-alanine |
| Gene regulation | ||||
| Cphy3040 | 301 | D182N | 8,214 | LysR transcriptional activator/repressor |
| Cphy3687 (PolB) | 1,278 | D189Y | 8,824 | β-Subunit of DNA-directed RNA polymerase |
| Noncoding changes | ||||
| Cphy0267 | 524 | Q65Q (synonymous) | 7,470 | Modification methyltransferase |
| Intergenic G→T transversion | None | 52 | Transversion between 2 convergently transcribed genes, encoding Cphy1313 and Cphy1314 | |
| Cphy3036 (ApbE) | 381 | L244L (synonymous) | 9,011 | Thiamine biosynthesis: membrane-associated lipoprotein |
Data include the gene name, encoded protein length, amino acid (if coding) or DNA variant, confidence value (Q) of the variant call, and annotation of the mutated protein. The probability (P) that a variant exists in the genome is reported as a Phred-scaled probability, i.e., Q = −10 × log10(1 − P), meaning that a Q value of 100 indicates an error probability (1 − P) of 10−10 (see GATK reports in the supplemental material for more information). FMN, flavin mononucleotide.
Ethanol increases the permeability of the cell membrane, resulting in toxic leakage of metabolites out of the cell (29). Ethanol resistance thus often involves membrane modifications such as altered protein content (8) or longer chain fatty acids and more plasmalogen lipids (30) that increase membrane rigidity to mitigate the fluidizing effect of ethanol. The ET strain has a D80N change in the putative acyl-acceptor binding pocket (NCBI accession no. cd07989) of Cphy0233, a homolog of C. butyricum PlsD that transfers a fatty acyl group to the sn-1 position of glycerol-3-phosphate in phospholipid biosynthesis (31). The D80N Cphy0233 mutation may thus enable synthesis of a more rigid, ethanol-resistant cell membrane by altering which fatty acids are incorporated into phospholipids.
The ET strain has a C26S mutation in the RnfA subunit of the membrane-bound Rnf complex that couples efflux of H+ (32) or Na+ (33) with electron transfer from reduced ferredoxin to NAD+ (34). The resulting electrochemical gradient is harnessed by an F0F1 ATPase for ATP synthesis. The C. phytofermentans Rnf complex (Cphy0211 to Cphy0216) and FoF1 ATPase (Cphy3735 to Cphy3742) are highly expressed on all tested carbon sources (19) and may be important for energy conservation, similarly to C. ljungdahlii (32). However, Rnf generates NADH, which may not be tolerated by the ET strain that cannot reoxidize NADH by AdhE-mediated ethanol formation (see below). The C26S RnfA variant may thus cripple the Rnf complex, which sacrifices ATP production, but may benefit the ET strain by balancing cellular NADH/NAD+ ratios.
The ET strain also has mutations in two transporters putatively involved in cation homeostasis. Cphy0543 is homologous to MgtA, a P-type ATPase upregulated at low ambient Mg2+ concentrations (35) to mediate Mg2+ uptake (36) or Ca2+/Mg2+ antiport (37). Cphy3778, the membrane component of an ABC transporter (PFAM accession no. PF06182), appears to be cotranscribed with Cphy3780, an ABC-type Na+ efflux protein (NCBI accession no. cd03267). In Bacillus subtilis, this Na+ efflux system is induced by ethanol and is proposed to compensate for an influx of extracellular Na+ resulting from a weakened membrane barrier (38). The variants in these cation transporters may increase their activities to alleviate cation leakage due to ethanol stress.
AdhE activities.
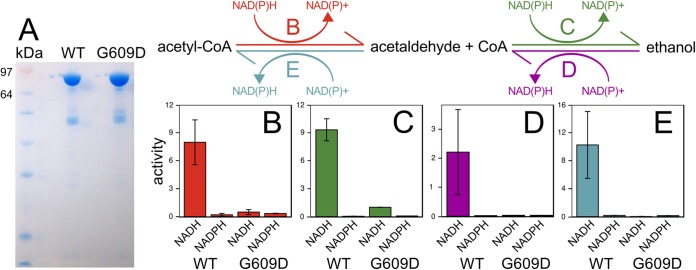
The ET strain has a G609D variant in Cphy3925 AdhE, a putative acetaldehyde-CoA dehydrogenase and alcohol dehydrogenase (ADH). The G609D mutation is in a conserved position in the active site of the C-terminal ADH domain (NCBI accession no. cd08178). A previous study reported an ethanol-tolerant C. thermocellum strain with AdhE mutations (P704L and H735R) that shifted the cofactor specificity from NADH to NADPH, which was proposed to confer ethanol resistance by altering the internal redox balance (9). To determine the effect of the G609D mutation on Cphy3925 enzyme activity, we purified WT and ET versions of the enzyme (Fig. 3A) and tested their in vitro catalysis of the two-step, bidirectional reactions converting acetyl-CoA to ethanol using either NADH or NADPH cofactors.
FIG 3.
Comparison of activities of purified Cphy3925 AdhE from wild-type and ET strains. (A) SDS-PAGE gel of purified Cphy3925 from the wild-type (WT) and ET (G609D) strains showing single bands of the expected 95-kDa molecular mass. (B to E) Reactions for the two-step, bidirectional interconversion of acetyl-CoA acetaldehyde and ethanol: reduction of 300 μM acetyl-CoA to acetaldehyde (red) (B), reduction of 18 mM acetaldehyde to ethanol (green) (C), oxidation of 2 M ethanol to acetaldehyde (purple) (D), and oxidation of 18 mM acetaldehyde to acetyl-CoA (blue) (E). Enzyme activities are shown in millimoles of NAD(P)H per micromole of enzyme per second measured using NADH(P)H or NAD(P)+ cofactors. Bar heights represent averages of duplicate activity measurements, and error bars represent 1 standard deviation.
The mutated Cphy3925 lost NAD(H)-dependent activities, but, unlike the mutated AdhE in C. thermocellum, the G609D mutation did not result in NADPH-dependent ADH activity (Fig. 3B to E). Instead, our results support the notion that the ET strain arrested AdhE-mediated interconversion of acetyl-CoA, acetaldehyde, and ethanol, which helps explain why the C. phytofermentans ET strain had lower ethanol yield. AdhE loss of function could mitigate ethanol stress by reducing intracellular levels of ethanol and its highly toxic precursor, acetaldehyde. C. phytofermentans encodes four Fe-dependent ADHs in addition to Cphy3925 as well as a Zn-dependent ADH. All 6 ADHs are expressed, and Cphy3925 and Cphy1029 are among the most highly expressed proteins on all tested carbon sources (19, 39). C. phytofermentans thus likely produces ethanol by the concerted action of multiple ADHs, and these other ADHs, especially Cphy1029, are responsible for ethanol produced by the ET strain.
Ethanol pathway engineering.
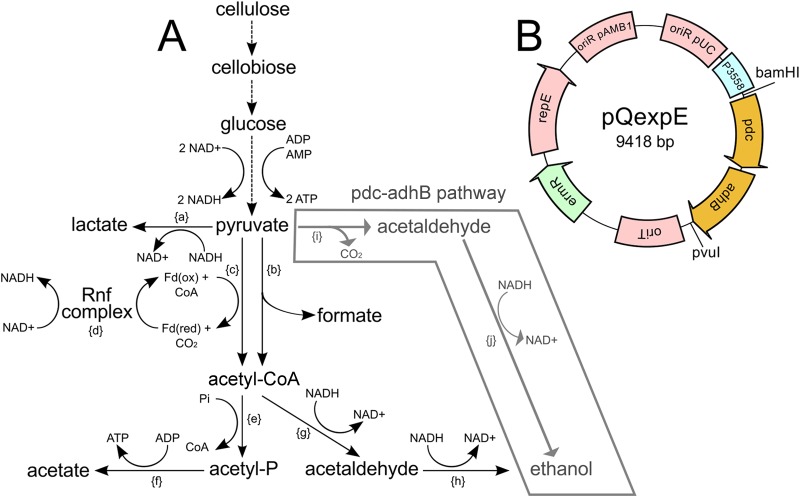
To augment ethanol production by the ET strain, an alternative ethanol production pathway comprised of pyruvate decarboxylase (Pdc) and alcohol dehydrogenase (AdhB) from Zymomonas mobilis (Fig. 4A) was transferred into C. phytofermentans on the replicating pQexpE plasmid (Fig. 4B). Together, these enzymes couple decarboxylation of pyruvate to ethanol with the oxidation of NADH and thus represent an alternative to the AdhE ethanol formation pathway. We chose to express foreign enzymes rather than a WT copy of Cphy3925 because AdhE multimerizes (40) such that the mutant AdhE could have a dominant-negative effect in a merodiploid.
FIG 4.
(A) Diagram of C. phytofermentans carbon metabolism showing insertion of the Z. mobilis pdc-adhB alternative ethanol formation pathway. Enzymatic steps: {a}, lactate dehydrogenase (Cphy1117); {b}, pyruvate formate lyase (Cphy1174); {c}, pyruvate ferredoxin oxidoreductase (Cphy3558); {d}, Rnf ferredoxin:NAD+ oxidoreductase complex (Cphy0211 to Cphy0216); {e}, phosphate acetyltransferase (Cphy1326); {f}, acetate kinase (Cphy1327); {g}, acetaldehyde dehydrogenase (Cphy1428 or Cphy3925); {h}, alcohol dehydrogenase (Cphy3925 or Cphy1029); {i}, Zymomonas mobilis pyruvate decarboxylase (pdc); {j}, alcohol dehydrogenase (adhB). Dashed lines represent multienzyme reactions where all enzymes are not listed. (B) Plasmid map of pQexpE for pdc-adhB expression in C. phytofermentans. Plasmid features: Gram-negative pUC origin of replication (oriR pUC), C. phytofermentans pyruvate ferredoxin oxidoreductase promoter (P3558) to express the Z. mobilis pdc-adhB genes, the RP4 conjugal origin of transfer (oriT), the Gram-negative/Gram-positive erythromycin resistance gene from TN1545 (ermR), the Gram-positive pAMB1 origin (oriR pAMB1), and repE-encoded protein.
Expression of pQexpE increased cellulolysis by ∼30% in both the WT and ET strains (Fig. 5A) and boosted ethanol production by 70% relative to the ET strain (P < 0.01), thereby restoring ethanol yields to WT levels (Fig. 5B). CO2 production increased disproportionately relative to H2 production in WT and ET strains expressing pQexpE (Table 3). Elevated CO2 synthesis is likely due to increased pyruvate decarboxylation by the Pdc enzyme. Previous results showed that Pdc/AdhB expression enhanced cellulolysis and ethanol production in WT C. cellulolyticum, which was proposed to result from consumption of excess pyruvate that otherwise leads to metabolic arrest (14). Increased metabolism (cellulolysis and production of CO2 and ethanol) by C. phytofermentans expressing Pdc/AdhB might be due to alleviation of inhibition by excess pyruvate. Alternatively, expression or activity of glycolytic enzymes might be regulated by NADH levels such that NADH reoxidation by Pdc/AdhB stimulates glycolysis, which results in increased substrate utilization.
FIG 5.
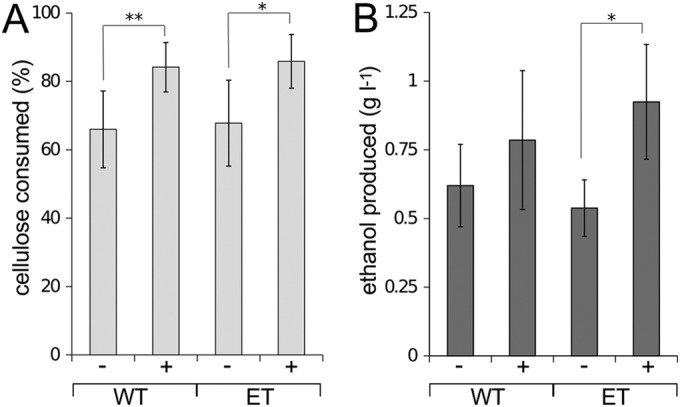
Cellulose consumption (A) and ethanol production (B) by non-plasmid-bearing (−) and pdc-adhB-containing (+) WT and ET cultures at 30°C in GS2 medium with 3 g liter−1 α-cellulose after 30 days. Bars represent averages of the results from duplicate cultures in serum bottles from three independent experiments, and error bars represent 1 standard deviation. Statistical differences between treatments are indicated at P values of <0.01 (*) and <0.05 (**) using Student's t test.
TABLE 3.
Carbon dioxide and hydrogen gas production by C. phytofermentans WT and ET cultures with or without the Z. mobilis pdc-adhB genes after 2 weeks of growth at 30°C in GS2 medium containing 100 g liter−1 α-cellulosea
| Strain | Gas produced (ml) | H2 (mM) | CO2 (mM) | H2/CO2 (mol/mol) |
|---|---|---|---|---|
| WT | 26.3 ± 0.4 | 4.21 ± 0.11 | 6.26 ± 0.08 | 0.67 |
| WT (pdc-adhB) | 40.0 ± 5.4 | 4.62 ± 0.13 | 10.02 ± 0.07 | 0.46 |
| ET | 25.2 ± 4.7 | 3.47 ± 0.12 | 4.46 ± 0.19 | 0.78 |
| ET (pdc-adhB) | 31.0 ± 2.3 | 4.41 ± 0.15 | 7.20 ± 0.13 | 0.61 |
Data represent averages ± standard deviations of the results from four replicates.
Conclusions.
In this study, we investigated the genetic basis and phenotypic consequences of microbial ethanol tolerance by isolating, characterizing, and engineering an ethanol-resistant (ET) strain of Clostridium phytofermentans. The ET strain grows at higher ethanol concentrations than the wild-type strain (Fig. 1) and continues to produce ethanol at a 7% ambient ethanol concentration (Fig. 2C) but has impaired growth (Fig. 1) and ethanol yield (Fig. 2A) relative to the wild type. The genome sequence of the ET strain revealed 12 mutations in genes involved in diverse aspects of metabolism (Table 2), including a G609D variant in the bifunctional acetaldehyde CoA/alcohol dehydrogenase AdhE that abolishes its activity (Fig. 3). We complemented the AdhE mutation in the ET strain by expressing pyruvate decarboxylase (Pdc) and alcohol dehydrogenase B (AdhB) from Zymomonas mobilis on the pQexpE plasmid (Fig. 4), which boosted substrate conversion (Fig. 5A) and restored ethanol production (Fig. 5B).
Additional work is needed to enhance C. phytofermentans' plant biomass degradation and ethanol formation rates and product titers. Recently, improvement of C. phytofermentans' growth on cellobiose, cellulose, and xylan by experimental evolution yielded strains that also produced ethanol more quickly (41). The genome sequence of the ET strain presented here suggests other novel approaches to potentially improve ethanol resistance and production. For example, our results suggest that further studies on ethanol resistance should focus on PlsD-mediated fatty acid incorporation into phospholipids, LysR-regulated gene expression patterns, overexpression of the Rnf complex to stimulate AdhE-mediated ethanol production, and prevention of cation leakage.
Supplementary Material
ACKNOWLEDGMENTS
C. phytofermentans ISDg was kindly provided by Susan Leschine (University of Massachusetts, Amherst).
A.C.T. was supported by a CNRS Chaire d'Excellence and the Genoscope-CEA. T.R.Z. was supported by a GRFP Fellowship from the National Science foundation (grant no. DGE1255832) and a John and Jeannette McWhirter Fellowship from The Pennsylvania State University. M.R. was supported by the Faculty Abroad Program, SRM University. Anaerobic growth capabilities for this work were financed by the Genoscope-CEA and a grant from the Department of Energy Advanced Research Project Agency—Energy (ARPA-e; no. DE-AR0000092) to W.R.C.
We thank Alex Rajangam for facilitating M.R.'s international exchange, Sergio Florez for useful feedback, Patrick Hillery and Sam Bjork for experimental assistance, and Nymul Khan for supporting GC analysis. We acknowledge the Huck Institute of the Life Sciences Shared Fermentation Facility for providing HPLC analytical capabilities and the Genoscope-CEA Sequencing Platform for sequencing the ET strain. Finally, we thank George Church and Noel Goddard for facilitating a sabbatical leave for W.R.C. at Harvard, where these studies were initiated.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00619-15.
REFERENCES
- 1.Lynd LR, van Zyl WH, McBride JE, Laser M. 2005. Consolidated bioprocessing of cellulosic biomass: an update. Curr Opin Biotechnol 16:577–583. doi: 10.1016/j.copbio.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 2.Lu Y, Zhang Y-HP, Lynd LR. 2006. Enzyme-microbe synergy during cellulose hydrolysis by Clostridium thermocellum. Proc Natl Acad Sci U S A 103:16165–16169. doi: 10.1073/pnas.0605381103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Warnick TA, Methé BA, Leschine SB. 2002. Clostridium phytofermentans sp. nov., a cellulolytic mesophile from forest soil. Int J Syst Evol Microbiol 52:1155–1160. doi: 10.1099/ijs.0.02125-0. [DOI] [PubMed] [Google Scholar]
- 4.Tolonen AC, Petit E, Blanchard JL, Warnick T, Leschine SB. 2013. Technologies to study plant biomass fermentation using the model bacterium Clostridium phytofermentans, p 114–139. In Sun J, Ding SY, Peterson JD (ed), Biological conversion of biomass for fuels and chemicals: explorations from natural utilization systems. Royal Society of Chemistry, Cambridge, United Kingdom. doi: 10.1039/9781849734738-00114. [DOI] [Google Scholar]
- 5.Jin M, Balan V, Gunawan C, Dale BE. 2011. Consolidated bioprocessing (CBP) performance of Clostridium phytofermentans on AFEX-treated corn stover for ethanol production. Biotechnol Bioeng 108:1290–1297. doi: 10.1002/bit.23059. [DOI] [PubMed] [Google Scholar]
- 6.Jin M, Gunawan C, Balan V, Dale BE. 2012. Consolidated bioprocessing (CBP) of AFEXTM-pretreated corn stover for ethanol production using Clostridium phytofermentans at a high solids loading. Biotechnol Bioeng 109:1929–1936. doi: 10.1002/bit.24458. [DOI] [PubMed] [Google Scholar]
- 7.Zuroff TR, Xiques SB, Curtis WR. 2013. Consortia-mediated bioprocessing of cellulose to ethanol with a symbiotic Clostridium phytofermentans/yeast co-culture. Biotechnol Biofuels 6:59. doi: 10.1186/1754-6834-6-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shao X, Raman B, Zhu M, Mielenz JR, Brown SD, Guss AM, Lynd LR. 2011. Mutant selection and phenotypic and genetic characterization of ethanol-tolerant strains of Clostridium thermocellum. Appl Microbiol Biotechnol 92:641–652. doi: 10.1007/s00253-011-3492-z. [DOI] [PubMed] [Google Scholar]
- 9.Brown SD, Guss AM, Karpinets TV, Parks JM, Smolin N, Yang S, Land ML, Klingeman DM, Bhandiwad A, Rodriguez M, Raman B, Shao X, Mielenz JR, Smith JC, Keller M, Lynd LR. 2011. Mutant alcohol dehydrogenase leads to improved ethanol tolerance in Clostridium thermocellum. Proc Natl Acad Sci U S A 108:13752–13757. doi: 10.1073/pnas.1102444108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu X, Cui J, Feng Y, Fa Y, Zhang J, Cui Q. 2013. Metabolic adaption of ethanol-tolerant Clostridium thermocellum. PLoS One 8:e70631. doi: 10.1371/journal.pone.0070631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang S, Giannone RJ, Dice L, Yang ZK, Engle NL, Tschaplinski TJ, Hettich RL, Brown SD. 2012. Clostridium thermocellum ATCC27405 transcriptomic, metabolomic and proteomic profiles after ethanol stress. BMC Genomics 13:336. doi: 10.1186/1471-2164-13-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ezeji T, Milne C, Price ND, Blaschek HP. 2010. Achievements and perspectives to overcome the poor solvent resistance in acetone and butanol-producing microorganisms. Appl Microbiol Biotechnol 85:1697–1712. doi: 10.1007/s00253-009-2390-0. [DOI] [PubMed] [Google Scholar]
- 13.Tomas CA, Beamish J, Papoutsakis ET. 2004. Transcriptional analysis of butanol stress and tolerance in Clostridium acetobutylicum. J Bacteriol 186:2006–2018. doi: 10.1128/JB.186.7.2006-2018.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guedon E, Desvaux M, Petitdemange H. 2002. Improvement of cellulolytic properties of Clostridium cellulolyticum by metabolic engineering. Appl Environ Microbiol 68:53–58. doi: 10.1128/AEM.68.1.53-58.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng Y, Olson DG, Zhou J, Herring CD, Joe Shaw A, Lynd LR. 2013. Redirecting carbon flux through exogenous pyruvate kinase to achieve high ethanol yields in Clostridium thermocellum. Metab Eng 15:151–158. doi: 10.1016/j.ymben.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 16.Argyros DA, Tripathi SA, Barrett TF, Rogers SR, Feinberg LF, Olson DG, Foden JM, Miller BB, Lynd LR, Hogsett DA, Caiazza NC. 2011. High ethanol titers from cellulose by using metabolically engineered thermophilic, anaerobic microbes. Appl Environ Microbiol 77:8288–8294. doi: 10.1128/AEM.00646-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biswas R, Zheng T, Olson DG, Lynd LR, Guss AM. 2015. Elimination of hydrogenase active site assembly blocks H2 production and increases ethanol yield in Clostridium thermocellum. Biotechnol Biofuels 8:20. doi: 10.1186/s13068-015-0204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cavedon K, Leschine SB, Canale-Parola E. 1990. Cellulase system of a free-living, mesophilic clostridium (strain C7). J Bacteriol 172:4222–4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boutard M, Cerisy T, Nogue P-Y, Alberti A, Weissenbach J, Salanoubat M, Tolonen AC. 2014. Functional diversity of carbohydrate-active enzymes enabling a bacterium to ferment plant biomass. PLoS Genet 10:e1004773. doi: 10.1371/journal.pgen.1004773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aslanidis C, de Jong PJ. 1990. Ligation-independent cloning of PCR products (LIC-PCR). Nucleic Acids Res 18:6069–6074. doi: 10.1093/nar/18.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan RT, Chen JS. 1990. Coenzyme A-acylating aldehyde dehydrogenase from Clostridium beijerinckii NRRL B592. Appl Environ Microbiol 56:2591–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tolonen AC, Chilaka AC, Church GM. 2009. Targeted gene inactivation in Clostridium phytofermentans shows that cellulose degradation requires the family 9 hydrolase Cphy3367. Mol Microbiol 74:1300–1313. doi: 10.1111/j.1365-2958.2009.06890.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bokinsky G, Peralta-Yahya PP, George A, Holmes BM, Steen EJ, Dietrich J, Lee TS, Tullman-Ercek D, Voigt CA, Simmons BA, Keasling JD. 2011. Synthesis of three advanced biofuels from ionic liquid-pretreated switchgrass using engineered Escherichia coli. Proc Natl Acad Sci U S A 108:19949–19954. doi: 10.1073/pnas.1106958108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tolonen AC, Cerisy T, El-Sayyed H, Boutard M, Salanoubat M, Church GM. 25 May 2014, posting date. Fungal lysis by a soil bacterium fermenting cellulose. Environ Microbiol doi: 10.1111/1462-2920.12495. [DOI] [PubMed] [Google Scholar]
- 26.Williams TI, Combs JC, Lynn BC, Strobel HJ. 2007. Proteomic profile changes in membranes of ethanol-tolerant Clostridium thermocellum. Appl Microbiol Biotechnol 74:422–432. doi: 10.1007/s00253-006-0689-7. [DOI] [PubMed] [Google Scholar]
- 27.Biswas R, Prabhu S, Lynd LR, Guss AM. 2014. Increase in ethanol yield via elimination of lactate production in an ethanol-tolerant mutant of Clostridium thermocellum. PLoS One 9:e86389. doi: 10.1371/journal.pone.0086389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maddocks SE, Oyston PCF. 2008. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 154(Pt 12):3609–3623. doi: 10.1099/mic.0.2008/022772-0. [DOI] [PubMed] [Google Scholar]
- 29.Ingram LO. 1990. Ethanol tolerance in bacteria. Crit Rev Biotechnol 9:305–319. [DOI] [PubMed] [Google Scholar]
- 30.Timmons MD, Knutson BL, Nokes SE, Strobel HJ, Lynn BC. 2009. Analysis of composition and structure of Clostridium thermocellum membranes from wild-type and ethanol-adapted strains. Appl Microbiol Biotechnol 82:929–939. doi: 10.1007/s00253-009-1891-1. [DOI] [PubMed] [Google Scholar]
- 31.Heath RJ, Goldfine H, Rock CO. 1997. A gene (plsD) from Clostridium butyricum that functionally substitutes for the sn-glycerol-3-phosphate acyltransferase gene (plsB) of Escherichia coli. J Bacteriol 179:7257–7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tremblay P-L, Zhang T, Dar SA, Leang C, Lovley DR. 2012. The Rnf complex of Clostridium ljungdahlii is a proton-translocating ferredoxin:NAD+ oxidoreductase essential for autotrophic growth. mBio 4:e00406-412. doi: 10.1128/mBio.00406-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Biegel E, Müller V. 2010. Bacterial Na+-translocating ferredoxin:NAD+ oxidoreductase. Proc Natl Acad Sci U S A 107:18138–18142. doi: 10.1073/pnas.1010318107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Biegel E, Schmidt S, Müller V. 2009. Genetic, immunological and biochemical evidence for a Rnf complex in the acetogen Acetobacterium woodii. Environ Microbiol 11:1438–1443. doi: 10.1111/j.1462-2920.2009.01871.x. [DOI] [PubMed] [Google Scholar]
- 35.Snavely MD, Gravina SA, Cheung TT, Miller CG, Maguire ME. 1991. Magnesium transport in Salmonella typhimurium. Regulation of mgtA and mgtB expression J Biol Chem 266:824–829. [PubMed] [Google Scholar]
- 36.Hmiel SP, Snavely MD, Florer JB, Maguire ME, Miller CG. 1989. Magnesium transport in Salmonella typhimurium: genetic characterization and cloning of three magnesium transport loci. J Bacteriol 171:4742–4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neef J, Andisi VF, Kim KS, Kuipers OP, Bijlsma JJE. 2011. Deletion of a cation transporter promotes lysis in Streptococcus pneumoniae. Infect Immun 79:2314–2323. doi: 10.1128/IAI.00677-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng J, Guffanti AA, Krulwich TA. 1997. A two-gene ABC-type transport system that extrudes Na+ in Bacillus subtilis is induced by ethanol or protonophore. Mol Microbiol 23:1107–1120. doi: 10.1046/j.1365-2958.1997.2951656.x. [DOI] [PubMed] [Google Scholar]
- 39.Tolonen AC, Haas W, Chilaka AC, Aach J, Gygi SP, Church GM. 2011. Proteome-wide systems analysis of a cellulosic biofuel-producing microbe. Mol Syst Biol 7:461. doi: 10.1038/msb.2010.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kessler D, Leibrecht I, Knappe J. 1991. Pyruvate-formate-lyase-deactivase and acetyl-CoA reductase activities of Escherichia coli reside on a polymeric protein particle encoded by adhE. FEBS Lett 281:59–63. doi: 10.1016/0014-5793(91)80358-A. [DOI] [PubMed] [Google Scholar]
- 41.Mukherjee S, Thompson LK, Godin S, Schackwitz W, Lipzen A, Martin J, Blanchard JL. 2014. Population level analysis of evolved mutations underlying improvements in plant hemicellulose and cellulose fermentation by Clostridium phytofermentans. PLoS One 9:e86731. doi: 10.1371/journal.pone.0086731. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.