Abstract
Chemoreceptors are at the beginnings of chemosensory signaling cascades that mediate chemotaxis. Most bacterial chemoreceptors are functionally unannotated and are characterized by a diversity in the structure of their ligand binding domains (LBDs). The data available indicate that there are two major chemoreceptor families at the functional level, namely, those that respond to amino acids or to Krebs cycle intermediates. Since pseudomonads show chemotaxis to many different compounds and possess different types of chemoreceptors, they are model organisms to establish relationships between chemoreceptor structure and function. Here, we identify PP2861 (termed McpP) of Pseudomonas putida KT2440 as a chemoreceptor with a novel ligand profile. We show that the recombinant McpP LBD recognizes acetate, pyruvate, propionate, and l-lactate, with KD (equilibrium dissociation constant) values ranging from 34 to 107 μM. Deletion of the mcpP gene resulted in a dramatic reduction in chemotaxis toward these ligands, and complementation restored a native-like phenotype, indicating that McpP is the major chemoreceptor for these compounds. McpP has a CACHE-type LBD, and we present data indicating that CACHE-containing chemoreceptors of other species also mediate taxis to C2 and C3 carboxylic acids. In addition, the LBD of NbaY of Pseudomonas fluorescens, an McpP homologue mediating chemotaxis to 2-nitrobenzoate, bound neither nitrobenzoates nor the McpP ligands. This work provides further insight into receptor structure-function relationships and will be helpful to annotate chemoreceptors of other bacteria.
INTRODUCTION
Many microorganisms have developed chemotactic mechanisms that allow them to rapidly respond to environmental changes by actively moving toward more favorable environments. Data suggest that the access to compounds that are required for growth is a major reason for chemotaxis, and chemotactic movement was observed, for instance, toward sugars, amino acids, organic acids, or inorganic phosphate (1–5). In addition, chemotaxis has been observed for other classes of compounds, such as neurotransmitters, plant hormones, or quorum-sensing signals (6–10). The specificity of a chemotactic response is determined by the chemoreceptor that is at the beginning of the chemosensory signaling cascade.
Chemoreceptors are typically composed of a cytosolic methyl-accepting signaling domain and a ligand binding domain (LBD) that is frequently located in the extracytoplasmic space. Ligand recognition at the LBD generates a molecular stimulus that is transmitted to the other extension of the chemoreceptor, where it modulates CheA autokinase activity. Phosphoryl groups are then transferred to the CheY response regulator, which in turn permits its interaction with the flagellar motor to ultimately mediate chemotaxis (1).
In contrast to the conserved methyl-accepting signaling domain, chemoreceptor LBDs show a high degree of diversity. The very large majority of chemoreceptor LBDs are unannotated in the SMART database (11). However, LBDs can be classified according to their size into cluster I domains (120 to 210 amino acids) and cluster II domains (220 to 300 amino acids) (11). Cluster I domains include the 4-helix bundle, GAF, PAS, CHASE, and CACHE domains (12–16). So far, two cluster II domain types have been identified, which are the helical bimodular (HBM) (17) and the double PDC (PhoQ/DcuS/CitA) domains (18, 19).
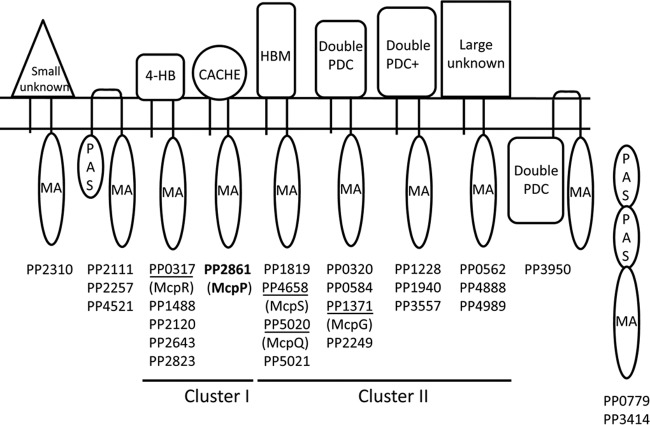
Pseudomonads are ubiquitously present in different ecological niches, and many strains are characterized by a metabolic diversity, as evidenced by the fact that some Pseudomonas putida strains can use more than 100 different compounds for growth (20). This metabolic versatility appears to be reflected in a chemotactic versatility, and so far 140 compounds have been identified to induce chemotaxis in Pseudomonas strains (21), including organic acids, amino acids, differently substituted aromatic hydrocarbons, biphenols, nucleotide bases, sugars, inorganic phosphate, metal ions, or peptides. P. putida KT2440, the model organism of this study, has 27 chemoreceptors that differ in their topologies and LBD types (Fig. 1). So far only the McpS, McpQ, McpR, and McpG receptors have been annotated with a function, and they mediate chemotaxis toward different organic and amino acids (3, 10, 19, 22). The diversity in the type of chemoreceptors and chemoattractants makes pseudomonads ideal model organisms to address a central question in the field (2), namely, to establish chemoreceptor structure-function relationships.
FIG 1.
Predicted topologies and ligand binding domains of P. putida KT2440 chemoreceptors. Annotation is based on the fold recognition by the Phyre2 algorithm (46) and consensus secondary structure predictions (47). Topologies are based on the prediction of transmembrane regions using the DAS algorithm (48). MA, methyl-accepting domain; 4-HB, 4-helix bundle domain; HBM, helical bimodular domain; double PDC, repeat of PhoQ/DcuS/CitA domain (18); double PDC+, double PDC domain containing a 40- to 50-amino-acid insert into double PDC domains. Functionally annotated receptors are underlined. HAMP domains have been omitted, and classification is based on receptor topology and LBD type.
As shown in Fig. 1, most chemoreceptors of P. putida KT2440 form paralogous groups. One of the exceptions is the receptor encoded by the open reading frame PP2861, which is predicted to have a CACHE-type LBD. CACHE domains are ubiquitously present in bacterial signal transduction systems and also form parts of one- and two-component systems (23). PP2861 is paralogous (58% sequence identity) to the CACHE domain-containing NbaY receptor from Pseudomonas fluorescens, which was shown to mediate chemotaxis toward 2-nitrobenzoate (24). Since P. putida shows chemotaxis to benzoate (25, 26) and substituted benzoates, including nitro derivatives (27), we hypothesized that PP2861 may be the corresponding chemoreceptor. Here we report the functional annotation of PP2861, which we term McpP. We show that it recognizes and responds specifically to some C2 and C3 carboxylic acids, thus representing a novel chemoreceptor type.
MATERIALS AND METHODS
Strains and plasmids.
The strains and plasmids used are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Relevant genotype and/or descriptiona | Source or reference |
|---|---|---|
| Escherichia coli | ||
| BL21(DE3) | F− ompI hsdSB (rB− mB−) gal dam met | 49 |
| DH5α | F− endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR nupG ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 hsdR17 (rK− mK+) phoA supE44 λ−; host for DNA manipulations | 50 |
| TK1132 | Apr; E. coli DH5α derivative carrying plasmid pJRG1 | This work |
| CC118 λpir | Rifr; Δ(ara-leu) araD ΔlacX74 galE galK phoA20 thi-1 rpsE rpoB argE(Am) recA1 Tn7 λpir | 51 |
| TK1133 | Rifr Smr; E. coli CC118 λpir derivative carrying pJRG2 | This work |
| HB101 | Smr; K-12/B hybrid; recA thi pro leu hsdRM with pRK600 | 53, 54 |
| Pseudomonas putida | ||
| KT2440R | Rifr derivative of P. putida KT2440, wild type | 52 |
| TK1134 | Rifr Apr; P. putida KT2440R Δpp2861 | This work |
| P. fluorescens KU-7 | Wild type, grows on 2-nitrobenzoate, chemotactic to 2-nitrobenzoate | 55 |
| Plasmids | ||
| pGEMT | Apr; cloning vector | Promega |
| pUC18NotI | Apr; similar to pUC18 but with NotI sites flanking the multiple-cloning site | 51 |
| pJRG1 | pUC18NotI derivative containing a 1-kb HindIII-EcoRI fragment from the P. putida KT2440R genome (containing upstream and downstream regions of pp2861) | This work |
| pRK600 | Cmr; ori of ColE1; RK2-mob+ RK2-tra+; conjugational helper plasmid | 54 |
| pKNG101 | Smr; suicide vector; mob sac | 56 |
| pJRG2 | Smr; pKNG101 derivative containing a 1.1-kb NotI fragment from pJRG1 cloned into pKNG101 | This work |
| pET28b | Kmr; protein expression plasmid | Novagen |
| pET28b-McpP-LBD | Kmr; pET28b containing McpP LBD | This work |
| pET28b-NbaY-LBD | Kmr; pET28b containing NbaY LBD | This work |
| pBBR1MCS2_START | Kmr; oriRK2 mobRK2 | 57 |
| pMAMV240 | Kmr; a 2.1-kb PCR fragment containing the mcpP gene and its promoter region was cloned into the HindIII/XbaI sites of pBBR1MCS2-START | This work |
Ap, ampicillin; Cm, chloramphenicol; Km, kanamycin; Rif, rifampin; Sm, streptomycin.
Construction of expression plasmids for McpP LBD and NbaY LBD.
The DNA fragment of mcpP encoding amino acids Arg33 to Ser194 was amplified with the primers 5′-TGCATATGCGGCAGATCCATGG-3′ and 5′-TACGGATCCCTACGAAGCGTCG-3′ using genomic DNA of P. putida KT2440R. Similarly, the DNA fragment of nbaY encoding amino acids Met31 to Ala193 was amplified using the primers 5′-GGAATTCCATATGATGCTCAACCAAATCCGC AACGAT-3′ and 5′-AAGAATTCTCACGCACTGATAACTTTTTCGCGGAACT-3′ using genomic DNA of Pseudomonas fluorescens KU-7 as the template. Both sets of primers contained restriction sites for NdeI and BamHI. The resulting PCR products were digested with these enzymes and then cloned into the expression plasmid pET28b(+) linearized with the same enzymes. The resulting plasmids, termed pET28-McpP-LBD and pET28-NbaY-LBD, were verified by DNA sequencing of the insert and flanking regions.
Overexpression and purification of McpP LBD and NbaY LBD.
Escherichia coli BL21(DE3) containing pET28-McpP-LBD was grown in 2-liter Erlenmeyer flasks containing 500 ml LB medium supplemented with 50 μg/ml kanamycin at 30°C to an optical density at 660 nm (OD660) of 0.6. At this point, protein production was induced by adding 0.1 mM isopropyl-thiogalactopyranoside (IPTG), and the cultures were shifted to 18°C. After overnight incubation, cells were harvested by centrifugation at 10,000 × g for 30 min. Cell pellets were resuspended in buffer A (30 mM Tris-HCl, 300 mM NaCl, 10 mM imidazole and 5% [vol/vol] glycerol, pH 8.0) and broken with a French press at 1,000 lb/in2. After centrifugation at 20,000 × g for 1 h, the supernatant was loaded onto a 5-ml HisTrap column (Amersham Bioscience), washed with five column volumes of buffer A, and eluted with an imidazole gradient of 45 to 500 mM in buffer A.
A slightly modified protocol was used to purify the NbaY LBD. E. coli BL21(DE3) containing pET28b-NbaY-LBD was grown in 2-liter Erlenmeyer flasks containing 400 ml of 2× YT medium supplemented with 50 μg/ml kanamycin at 30°C to an OD660 of 0.6, at which time protein expression was induced by adding 0.1 mM IPTG. Growth was continued at 18°C overnight before cells were harvested by centrifugation at 6,000 × g for 30 min. Cell pellets were resuspended in buffer B (20 mM Tris-HCl, 500 mM NaCl, 10 mM imidazole, 0.1 mM EDTA, 5% [vol/vol] glycerol, pH 7.8) and broken with a French press at 1,000 lb/in2. After centrifugation at 20 000 × g for 1 h, the supernatant was loaded onto a 5-ml HisTrap column (Amersham Bioscience), washed with five column volumes of buffer B, and eluted with an imidazole gradient of 45 to 500 mM in buffer B.
Isothermal titration calorimetry (ITC).
Experiments were conducted on a VP microcalorimeter (Microcal, Amherst, MA) at 20°C. Proteins were dialyzed overnight against polybuffer [5 mM Tris-HCl, 5 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), 5 mM morpholineethanesulfonic acid (MES), 10% (vol/vol) glycerol, 150 mM NaCl, pH 8.0] and placed into the sample cell. Typically, 10 μM protein was introduced into the sample cell and titrated with 0.5 to 5 mM ligand solutions that were prepared in dialysis buffer immediately before use. The mean enthalpies measured from the injection of ligands into the buffer were subtracted from raw titration data prior to data analysis with the MicroCal version of ORIGIN. Data were fitted with the “one binding site model” of ORIGIN.
CD spectroscopy.
Circular dichroism (CD) experiments were performed using a Jasco J-715 (Tokyo, Japan) spectropolarimeter equipped with a thermostatically controlled cell holder. Far-UV CD spectra were recorded with a 1-mm-path-length quartz cuvette using a bandwidth of 2 nm, a scan rate of 100 nm/min, a response time of 4 s, and an average of 5 scans. The NbaY LBD was at 16.4 μM in 5 mM Tris-HCl–5 mM PIPES–5 mM MES, pH 8.0. Spectra were corrected for buffer contributions to the signal. Analyses were performed at 25°C, 50°C, and 85°C.
Intrinsic tryptophan fluorescence spectroscopy.
Fluorescence measurements were performed on a PTI spectrofluorimeter (Photon Technology International) equipped with the photomultiplier detection system 814. The NbaY LBD was dialyzed into 5 mM Tris-HCl–5 mM PIPES–5 mM MES (pH 8.0), adjusted to 4 μM, loaded into a 3-mm quartz cuvette, and placed into the cell holder thermostated at 25°C. An excitation wavelength of 290 nm was used, and emission spectra were recorded between 300 and 500 nm in 2-nm steps. Slit widths of 4 nm were used for both excitation and emission.
Growth experiments.
Overnight cultures were grown in M9 minimal medium (28) supplemented with 10 mM glucose, which was then used to inoculate 20 ml of fresh M9 minimal medium supplemented with the appropriate carbon source to an initial OD600 of 0.05. The cultures were then put into an orbital platform at 200 rpm and allowed to grow at 30°C. OD600 readings were taken every 30 min using a Lambda 20 UV-visible spectrophotometer (Perkin-Elmer). Cultures were maintained in the exponential phase by periodic dilutions with fresh prewarmed M9 medium supplemented with different carbon sources.
Construction of the P. putida pp2861 (mcpP) mutant KT2440R.
Two 0.5-kb DNA fragments containing the upstream and downstream sequences of the pp2861 gene were amplified using the primer pairs 5′-ATAAAGCTTAATCTTGCTGCGAAACTCCC-3′ and 5′-CTAGTCTAGACTAGCCATCAGCTCCCGCATTGTT-3′ for the upstream region and 5′-CTAGTCTAGACTAGTGTTGCGGCAGTTCCGGGT-3′ and 5′-AAGAATTCTGGGCGTGGCAGACGGGAG-3′ for the downstream region. Primers contained sequences that introduced HindIII and XbaI as well as XbaI and EcoRI restriction sites (underlined) into the upstream and downstream fragments, respectively. Both fragments were cloned into pUC18NotI and transformed into E. coli DH5α, giving rise to strain TK1132. The NotI fragment containing the upstream and downstream sequences was excised from pJRG1 and cloned into pKNG101 to produce pJRG2. The resulting plasmid, pJRG2, was transformed into E. coli CC118λpir to generate E. coli TK1133 (Table 1). Plasmid pJRG2 was mobilized from E. coli TK1133 into P. putida KT2440R by triparental mating using E. coli HB101(pRK600) as the helper strain. In contrast to E. coli, P. putida KT2440R is able to grow on benzoate. Therefore, the selection of plasmid cointegrates of P. putida KT2440R was accomplished using M9 minimal medium (28) supplemented with 10 mM benzoate and 100 μg/ml streptomycin. The Smr colonies were unable to grow on LB medium containing 5% (wt/vol) sucrose, confirming that the plasmid pJRG2 with its sacB gene had integrated into these strains. The transconjugant was grown overnight in streptomycin-free LB medium, diluted 1,000-fold, and, following incubation for 12 h, serially diluted and plated on LB medium plates with or without 15% (wt/vol) sucrose. PCR analysis of one of the Sucr Sms clones confirmed that gene deletion had occurred.
Motility and chemotaxis assays. (i) Swim plate motility assays.
Bacteria from single colonies were grown overnight on LB medium supplemented with 10 μg/ml of rifampin. Two-microliter aliquots of bacterial suspension were transferred to the centers of swim agar plates (10% LB, 0.25% [wt/vol] agar). Plates were incubated at 30°C overnight and motility monitored the following day.
(ii) Plate gradient assays.
Bacteria were grown overnight in minimal saline (MS) medium (29) supplemented with 10 mM succinate and diluted to an OD600 of 0.8 to 1 with fresh MS medium. Cells were then washed twice with MS medium by consecutive resuspension and centrifugation at 3,750 × g for 3 min. Square petri dishes were filled with 80 ml MS agar (0.25% agar, wt/vol) medium containing 25 mM glycerol. Plates were cooled to room temperature for at least 30 min. Along the vertical central line of the plate, 10-μl aliquots of a 10 mM chemoattractant solution, dissolved in MS medium, were placed at regular distances. Plates were incubated at 4°C for 12 to 16 h to allow the formation of the chemoattractant gradient. Two-microliter aliquots of bacterial suspension were then placed horizontally to each of the chemoattractant deposits with a distance of 2.5 cm to the vertical line. Plates were incubated at 30°C for 16 to 20 h prior to the inspection of chemotaxis.
(iii) Quantitative capillary chemotaxis assay.
A modified version of the capillary assay was used (30). MS medium supplemented with 10 mM succinate was inoculated with P. putida KT2440R or KT2440R Δpp2861 and grown to early logarithmic phase. Cultures were then centrifuged at 1,667 × g at 4°C for 5 min and washed twice with 50 mM potassium phosphate–20 μM EDTA–0.05% (wt/vol) glycerol, pH 7.0. Cells were resuspended in the same buffer to an OD600 of 0.08 to 0.1, and 230-μl aliquots of bacterial suspension were then placed into the wells of a 96-well plate. Capillaries (Microcaps; Drummond Scientific) were heat sealed at one end and warmed over fire, and the open end was inserted into the chemoattractant solution for filling. The pH of the chemoattractant solution had been adjusted to that of the bacterial suspension. MS medium lacking chemoattractants was used as a control. After immersion of the open ends of the capillaries in the bacterial suspension (for 30 min), capillaries were removed and the open ends rinsed and placed into a microcentrifuge tube containing 1 ml MS medium. The sealed end was broken, and the content was emptied into the tube by a short centrifugation. One hundred microliters of the resulting cell suspension was plated out and incubated at 30°C. Colonies were counted after growth on M9 medium supplemented with 10 mM succinate at 30°C for 24 h.
Construction of an mcpP-containing plasmid for complementation.
Primers 5′-ATAAAGCTTAATCTTGCTGCGAAACTCCC-3′ and 5′-CTAGTCTAGACTAGTCAGACCCGGAACTGCCGCAACA-3′ were used to amplify the mcpP gene and its promoter region. The resulting 2.1-kb PCR fragment was cloned into the HindIII and XbaI sites of pBBR1MCS2-START. The insert was confirmed by PCR and sequencing. The resulting plasmid, pMAMV240, was transformed into P. putida TK1134 by electroporation.
RESULTS
The PP2861 LBD recognizes C2 and C3 carboxylic acids.
The DNA sequence encoding the fragment flanked by the two transmembrane regions of PP2861, harboring the LBD, was cloned into an expression vector, and the protein was expressed as a His tag fusion protein in E. coli and purified from the soluble fraction of the cell lysate. The purified protein was subjected to microcalorimetric binding studies (31) to identify its cognate ligands. Based on our hypothesis that this NbaY homologue may be involved in recognizing benzoates, the McpP LBD was titrated with benzoate and a number of its derivatives, including 2-nitrobenzoate (listed in Table S1 in the supplemental material). However, none of these compounds was found to bind.
Since the only other characterized chemoreceptor with a CACHE domain is the malate-specific PA2652 of P. aeruginosa (32), we continued microcalorimetric ligand screening by titrations with malate and other dicarboxylates (see Table S1 in the supplemental material), which, however, did not bind. We therefore extended the ligand screen to different monocarboxylic acids. Whereas glyoxylate did not bind, acetate was identified as a PP2681-LBD ligand (Fig. 2), and data analysis revealed a KD (equilibrium dissociation constant) of 34 ± 5 μM (Table 2). Interestingly, the C3 carboxylic acid propionate bound with the same affinity, but the C4 acid butyrate failed to bind (Fig. 2; Table 2). Further experiments showed that C3 acids pyruvate and l-lactate bound to the PP2861 LBD with affinities of 39 ± 3 and 107 ± 11 μM, respectively (Fig. 2; Table 2). In contrast, d-lactate and phosphoenolpyruvate were unable to bind. In order to continue the assessment of the PP2861 LBD ligand spectrum, we tested different compounds, such as propanol, acetone, or amino acids (see Table S1 in the supplemental material), which, however, did not bind. We can therefore conclude that the PP2861 LBD binds acetate, pyruvate, l-lactate, and propionate and renamed the receptor McpP.
FIG 2.
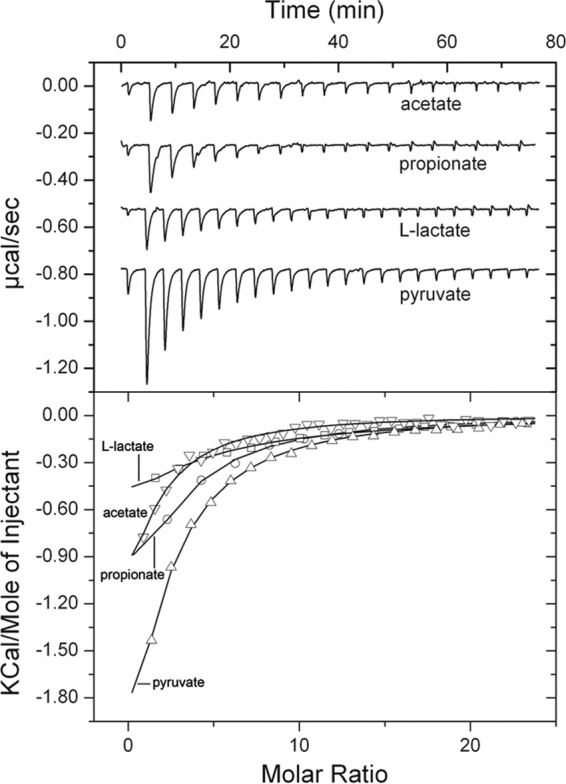
Microcalorimetric titration of McpP-LBD with different C2 and C3 carboxylic acids. Upper panel, raw data for the titration of 10 μM protein with 2 mM ligand solutions. In all cases the first injection was of 1.6 μl, followed by a series of 9.6-μl injections. Lower panel, dilution-corrected and concentration-normalized integrated peak areas of the raw data. Fitting was done using the “one binding site model” of the MicroCal version of ORIGIN. Shown are results of representative experiments, and thermodynamic data (presented in Table 2) are means and standard deviations from three independent experiments.
TABLE 2.
Thermodynamic parameters of ligand binding derived from the microcalorimetric titration of the McpP LBD with different ligandsa
| Ligand | KA (M−1) | KD (μM) | ΔH (kcal/mol) | nb |
|---|---|---|---|---|
| Acetate | (2.9 ± 0.4) × 104 | 34 ± 5 | −5.11 ± 3.1 | 0.77 ± 0.4 |
| Propionate | (2.9 ± 0.3) × 104 | 34 ± 4 | −5.35 ± 3.3 | 1.37 ± 0.6 |
| Pyruvate | (2.6 ± 0.2) × 104 | 39 ± 3 | −9.94 ± 2.6 | 0.89 ± 0.2 |
| l-Lactate | (9.3 ± 1) × 103 | 107 ± 11 | −5.20 ± 4.1 | 1.26 ± 0.8 |
Data are means and standard deviations from three experiments.
Stoichiometry of binding of ligands to the McpP LBD.
The McpP ligands and nitrobenzoates do not bind to NbaY LBD.
Since McpP and NbaY are paralogous, we were also interested in identifying the compounds that are recognized by NbaY. To this end, we followed a similar strategy and subjected the purified NbaY LBD to microcalorimetric titrations with different nitrobenzoates. Since no binding was observed, ITC experiments with the four McpP ligands and further compounds (see Table S1 in the supplemental material) were carried out. However, none of these compounds bound to the NbaY LBD.
Circular dichroism and intrinsic fluorescence spectroscopy experiments were conducted to determine whether the NbaY LBD corresponded to folded protein. As shown in Fig. S1A in the supplemental material, the maximum of the intrinsic tryptophan fluorescence emission spectrum was at 330 nm. This is consistent with a significant protection of the Trp residues from the solvent, which is a typical feature of proteins in its folded state (33). As a control, the spectrum was recorded in the presence of the chaotropic agent guanidine hydrochloride (GdnHCl) (see Fig. S1A in the supplemental material), which causes protein unfolding and exposure of Trp residues to the solvent. Under these conditions, the maximum was shifted to 344 nM, indicting that the presence of GdnHCl had induced protein unfolding. Figure S1B in the supplemental material shows a far-UV circular dichroism spectrum of the NbaY LBD, from which α-helical and β-strand contents of 18% and 30%, respectively, were calculated using the procedure described by Bohm et al. (34). Figure S1B in the supplemental material also shows that the exposure of the protein to heat caused loss of the CD signal, indicative of protein unfolding. The fact that both high concentrations of GdnHCl and heat treatment caused protein unfolding suggests that the NbaY LBD was present as a folded protein.
The four McpP ligands support growth of P. putida KT24440.
Many chemoattractants serve as carbon sources for growth (21). To verify whether P. putida KT2440 can use the identified McpP ligands as sole carbon sources, growth experiments were conducted in minimal medium supplemented with the individual compounds at 10 mM. As shown in Fig. S2 in the supplemental material, all four McpP ligands supported growth of P. putida KT2440R. In contrast to the case for l-lactate, acetate, and pyruvate, a lag phase of more than 4 h was observed for propionate.
Mutation of the mcpP gene largely reduces chemotaxis to McpP ligands.
It has been shown that chemoreceptors feed into signaling pathways that either mediate chemotaxis or carry out alternative cellular functions such as controlling the levels of second messengers (35). In subsequent experiments, we wanted to establish whether McpP is involved in chemotaxis and, if so, determine its contribution to the taxis of P. putida toward the four ligands identified. To this end, we constructed an mcpP deletion mutant by homologous recombination. To assess any potential effect of this mutation on bacterial motility, the wild type (wt) and the mutant strain were analyzed by swim plate assays. As shown in Fig. 3A, the motilities of the wt and mcpP mutant strain are comparable.
FIG 3.
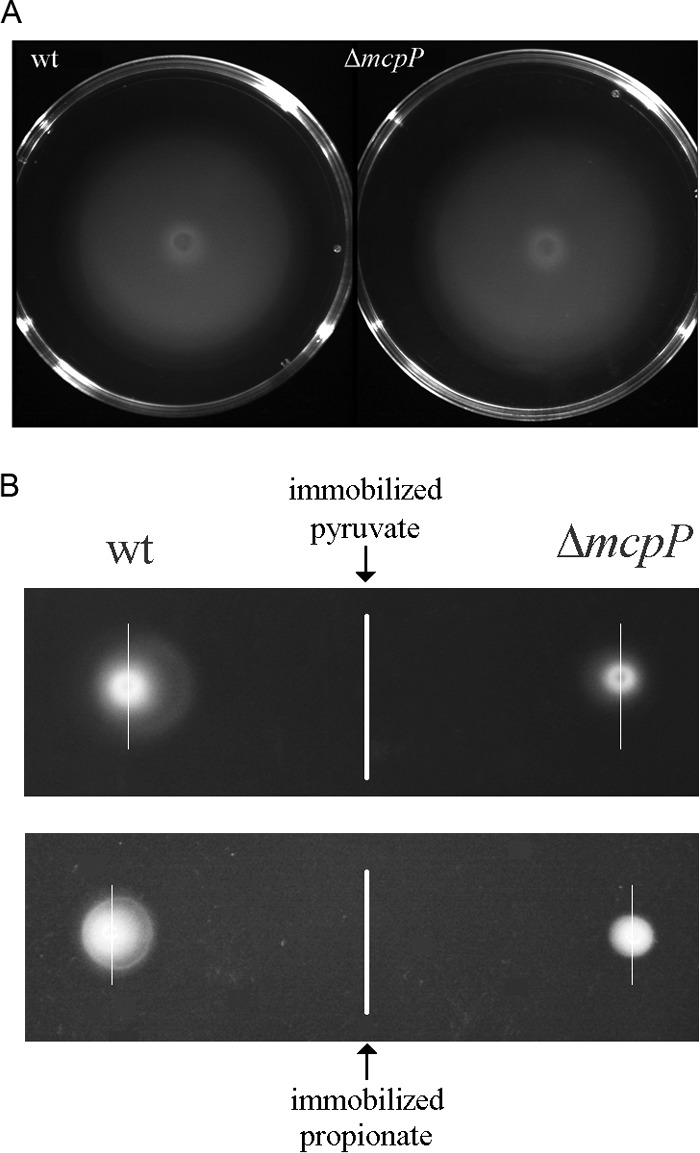
Mutation of the mcpP gene does not alter motility but reduces chemotaxis to pyruvate and propionate. (A) Swim plate assays of P. putida KT2440R and its mcpP-deficient mutant. Shown are representative images. (B) Plate gradient chemotaxis assays in which 10 mM pyruvate or propionate were deposited along a vertical line on MS agar plates. After overnight incubation for gradient formation, suspensions of wt or mcpP mutant strains were deposited at either side. Plates were inspected the following day for chemotaxis. This assay was repeated 3 times, and representative images are shown. To visualize the acentric spread, thin vertical lines that go through the center of bacterial deposition are shown.
We then assessed the contribution of McpP to chemotaxis toward the four ligands identified using plate gradient assays. In this assay, a 10 mM ligand solution is immobilized on agar plates, which are then left overnight for gradient formation. At identical distances from the immobilized compounds, aliquots of the wt or mutant strain are placed. Using pyruvate and propionate as chemoattractants (Fig. 3B), an acentric spread of bacteria toward the immobilized compound was observed for the wt strain. The corresponding chemotaxis indices, as calculated according to Pham and Parkinson (36), were 0.70 ± 0.05 (n = 8) for pyruvate and 0.60 ± 0.02 (n = 8) for propionate, indicative of chemotaxis. The mcpP mutant cells showed a minor spread that was only slightly acentric, and chemotaxis indices of 0.54 ± 0.02 (n = 8) for pyruvate and 0.53 ± 0.05 (n = 8) for propionate were obtained. Both values are close to 0.5 (observed in the absence of taxis) and are indicative of some residual chemotaxis toward these compounds.
Assessment of the concentration dependence of McpP-mediated taxis.
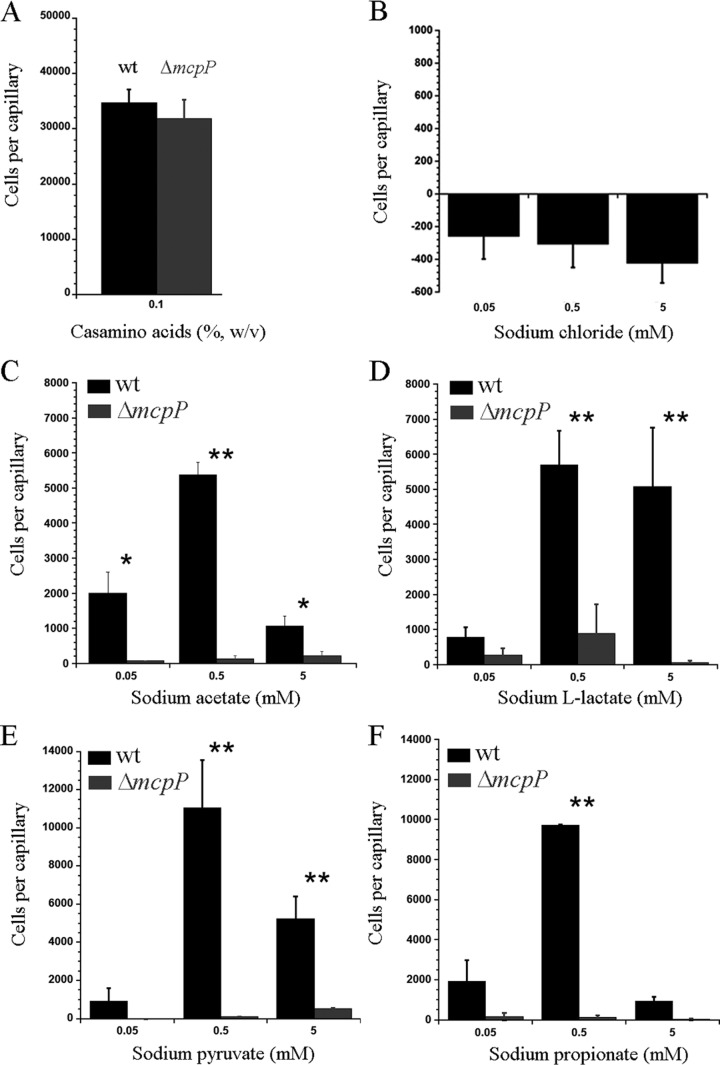
We have then conducted quantitative capillary chemotaxis assays to determine the concentration dependence of the response. Prior to the study of McpP ligands, a number of control experiments were conducted. As a positive control, we studied the chemotaxis to Casamino Acids, which is mediated by a different chemoreceptor. As shown in Fig. 4A, the responses of the wt and mcpP mutant strains were similar, confirming that the mcpP mutation did not alter motility in a nonspecific manner. Since the McpP ligands used were sodium salts, we also measured the response of the wt strain to sodium chloride (Fig. 4B). Very minor repellent responses toward NaCl were measured, which allowed the use of sodium salts for further experiments.
FIG 4.
Quantitative capillary chemotaxis assays of P. putida KT2440R toward Casamino Acids (A) (control), NaCl (B) (control), sodium acetate (C), sodium l-lactate (D), sodium pyruvate (E), and sodium propionate (F). Data were corrected with the number of cells that migrated into buffer-containing capillaries. Data are means and standard deviations from at least three independent experiments. *, P < 0.1; **, P < 0.05 (by Student's t test).
Quantitative capillary assays with the four McpP ligands were conducted over a ligand concentration range of 5 nM to 50 mM. Significant responses were observed only in the concentration range between 50 μM and 5 mM, and replicate measurements at this range were carried out to determine the responses precisely. The wt strain showed, for all four ligands, optimal responses at a concentration of 0.5 mM (Fig. 4C to F). At this concentration, the magnitude of response toward pyruvate and propionate was around twice that toward acetate and l-lactate. In all cases, chemotaxis responses at 50 μM and 5 mM were inferior to that at 0.5 mM. For all four ligands, the mutation of mcpP reduced taxis to very low levels. For complementation assays, we cloned the mcpP gene and its promoter region into the vector pBBR1MCS2 and introduced the resulting plasmid (pMAMV240) into the mcpP mutant. As a control, pBBRMCS2 was introduced into the wt and mcpP mutant strains. Assays of chemotaxis of the resulting three strains toward 0.5 mM solutions of the four McpP ligands were conducted (see Fig. S3 in the supplemental material). Statistical analysis of the resulting data revealed that the response of the complemented mutant was similar to that of the wt for three ligands, whereas chemotaxis of the complemented mutant to l-lactate was superior to that of the wt strain. Taken together, these data show that McpP is the primary chemoreceptor of P. putida KT2440 for acetate, l-lactate, pyruvate, and propionate.
DISCUSSION
A significant number of chemoreceptors that mediate chemotaxis toward organic acids of the Krebs cycle have been identified, such as McpS of P. putida KT2440 (3, 37), its three homologues in P. putida F1 (22), Mcp2201 of Comamonas testosteroni (38), the citrate-specific Tcp of Salmonella enterica serovar Typhimurium (39, 40), or the malate-specific PA2652 of P. aeruginosa (32). Although chemotaxis to non-Krebs cycle organic acids has been reported for different species (25, 41–44), little information is available on the corresponding chemoreceptors. Here we report that McpP specifically responds to C2 and C3 carboxylic acids, which corresponds to a novel chemoreceptor ligand profile. Within P. putida KT2440, the receptors for Krebs cycle and non-Krebs cycle organic acids belong to different families. Whereas McpS is a cluster II receptor with a helical bimodular (HBM) LBD (17, 19), McpP forms part of cluster I and has a CACHE domain for ligand recognition. Pseudomonads are model organisms for the study of chemotaxis, and a recent review contains a list of all known chemoeffectors of this genus (21). So far there are no reports on the chemotaxis to pyruvate, propionate, and l-lactate, and our data thus expand the list of known chemoattractants for pseudomonads.
We sought to determine whether CACHE domain-containing chemoreceptors in other species may also respond to C2 and C3 carboxylic acids. To this end we conducted a BLAST search in the Protein Data Bank using the McpP LBD sequence. The domains with highest sequence similarity were CACHE-type LBDs of chemoreceptors from Anaeromyxobacter dehalogenans (PDB 4K08) and of Vibrio parahaemolyticus (accession no. Q87T87, PDB 4EXO). These chemoreceptors are uncharacterized and the LBD structures unpublished. As shown in Fig. 5, both structures are similar to the homology model of the McpP LBD. Most interestingly, these structures contain in the ligand binding pocket of the CACHE domain either bound acetate (4K08) or pyruvate (4EXO), suggesting that they may be chemoattractants of the corresponding receptors (Fig. 5). Acetate was part of the crystallization buffer of the 4K08 structure, whereas pyruvate must have been copurified with the protein of structure 4EXO.
FIG 5.
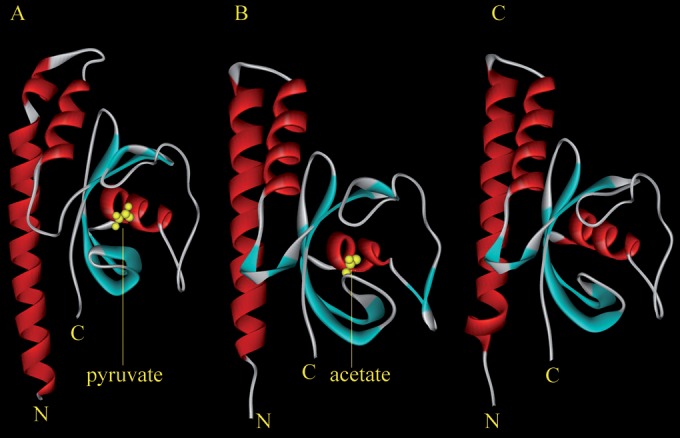
CACHE domains of chemoreceptors that recognize pyruvate and acetate. (A) Structure of a CACHE-type LBD of an uncharacterized chemoreceptor from Vibrio parahaemolyticus (PDB 4EXO) in complex with pyruvate. (B) Structure of a CACHE-type LBD of an uncharacterized chemoreceptor from Anaeromyxobacter dehalogenans in complex with acetate (PDB 4K08). (C) Homology model of the McpP LBD. The model was generated using Phyre2 (46). Ninety-one percent of the McpP LBD residues were modeled at >90% confidence.
Bacterial LDBs are characterized by a high degree of sequence divergence (12, 13, 45), which hampers a functional annotation based on sequence similarities. Although the McpP LBD and 4EX0 bind the same ligand, these domains show only 22% sequence identity. The inspection of the 4EXO structure shows that pyruvate binding is accomplished by nine amino acids, of which four establish hydrogen bonds (see Fig. S4A in the supplemental material). Interestingly, eight of the nine amino acids involved in pyruvate recognition are conserved (5 identical and 3 with strong similarity) in the alignment of the McpP LBD and 4EXOB sequences (see Fig. S4B in the supplemental material), suggesting that pyruvate recognition at the McpP LBD occurs in a manner similar to that in 4EXOB. Taken together, the data suggest that CACHE LBD-containing chemoreceptors function as C2 and C3 carboxylic acid chemoreceptors in different bacterial species.
An interesting feature of the McpP-mediated chemotaxis is that optimal responses for all four ligands are observed at a concentration of 500 μM (Fig. 4). As mentioned above, the McpS chemoreceptor of P. putida KT2440 mediates chemotaxis toward Krebs cycle organic acids and, using quantitative capillary chemotaxis assays, we have previously reported the McpS dose-response ratios (3). For the three primary McpS ligands, malate, succinate, and fumarate, strong responses were obtained at a concentration of 1 μM, a concentration at which no significant responses were measured for McpP ligands. These data indicate a higher sensitivity of the bacterium to Krebs cycle intermediates compared to the C2 and C3 carboxylic acids.
The NbaY chemoreceptor of P. fluorescens was shown to mediate chemotaxis to 2-nitrobenzoate (24). This function is also supported by the fact that the nbaY gene is located next to a gene cluster encoding the proteins for 2-nitrobenzoate degradation. The NbaY LBD could be produced in large amounts as a highly soluble protein, and its analysis by CD and intrinsic fluorescence spectroscopy indicates that the protein is folded. As shown in the Fig. S5 in the supplemental material, the homology model of the NbaY LBD is similar to the structures/models of the other CACHE domains displayed in Fig. 5. However, microcalorimetric titrations of this protein with different benzoate derivatives and the McpP ligands did not reveal any binding. The data thus suggest that ligand recognition at NbaY may not occur directly but may require previous binding to a periplasmic ligand binding protein.
Given the diversity of bacterial chemoreceptors, their functional annotation is a major research need in the field. Here we report a chemoreceptor with a novel ligand spectrum and present data suggesting that CACHE domain-containing chemoreceptors of diverse species may be involved in the chemotaxis to C2 and C3 carboxylic acids, a finding that may facilitate the functional annotation of chemoreceptors.
Supplementary Material
ACKNOWLEDGMENTS
We thank Peter Lau for providing P. fluorescens strain KU-7.
We acknowledge financial support from FEDER funds and Fondo Social Europeo through grants from the Junta de Andalucía (grants P09-RNM-4509 and CVI-7335 to T.K.) and the Spanish Ministry for Economy and Competitiveness (grants BIO2010-16937 and BIO2013-42297 to T.K.). M.A.M was supported by a Spanish Ministry of Economy and Competitiveness Postdoctoral Research Program, Juan de la Cierva (BVA-2009-0200).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01529-15.
REFERENCES
- 1.Sourjik V, Wingreen NS. 2012. Responding to chemical gradients: bacterial chemotaxis. Curr Opin Cell Biol 24:262–268. doi: 10.1016/j.ceb.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kato J, Kim HE, Takiguchi N, Kuroda A, Ohtake H. 2008. Pseudomonas aeruginosa as a model microorganism for investigation of chemotactic behaviors in ecosystem. J Biosci Bioeng 106:1–7. doi: 10.1263/jbb.106.1. [DOI] [PubMed] [Google Scholar]
- 3.Lacal J, Alfonso C, Liu X, Parales RE, Morel B, Conejero-Lara F, Rivas G, Duque E, Ramos JL, Krell T. 2010. Identification of a chemoreceptor for tricarboxylic acid cycle intermediates: differential chemotactic response towards receptor ligands. J Biol Chem 285:23126–23136. doi: 10.1074/jbc.M110.110403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mesibov R, Adler J. 1972. Chemotaxis toward amino acids in Escherichia coli. J Bacteriol 112:315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adler J. 1966. Chemotaxis in bacteria. Science 153:708–716. doi: 10.1126/science.153.3737.708. [DOI] [PubMed] [Google Scholar]
- 6.Antunez-Lamas M, Cabrera E, Lopez-Solanilla E, Solano R, Gonzalez-Melendi P, Chico JM, Toth I, Birch P, Pritchard L, Liu H, Rodriguez-Palenzuela P. 2009. Bacterial chemoattraction towards jasmonate plays a role in the entry of Dickeya dadantii through wounded tissues. Mol Microbiol 74:662–671. doi: 10.1111/j.1365-2958.2009.06888.x. [DOI] [PubMed] [Google Scholar]
- 7.Bansal T, Englert D, Lee J, Hegde M, Wood TK, Jayaraman A. 2007. Differential effects of epinephrine, norepinephrine, and indole on Escherichia coli O157:H7 chemotaxis, colonization, and gene expression. Infect Immun 75:4597–4607. doi: 10.1128/IAI.00630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hegde M, Englert DL, Schrock S, Cohn WB, Vogt C, Wood TK, Manson MD, Jayaraman A. 2011. Chemotaxis to the quorum-sensing signal AI-2 requires the Tsr chemoreceptor and the periplasmic LsrB AI-2-binding protein. J Bacteriol 193:768–773. doi: 10.1128/JB.01196-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rader BA, Wreden C, Hicks KG, Sweeney EG, Ottemann KM, Guillemin K. 2011. Helicobacter pylori perceives the quorum-sensing molecule AI-2 as a chemorepellent via the chemoreceptor TlpB. Microbiology 157:2445–2455. doi: 10.1099/mic.0.049353-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reyes-Darias JA, Garcia V, Rico-Jimenez M, Corral-Lugo A, Lesouhaitier O, Juarez-Hernandez D, Yang Y, Bi S, Feuilloley M, Munoz-Rojas J, Sourjik V, Krell T. 29 April 2015. Specific gamma-aminobutyrate (GABA) chemotaxis in Pseudomonads with different lifestyle. Mol Microbiol doi: 10.1111/mmi.13045. [DOI] [PubMed] [Google Scholar]
- 11.Lacal J, Garcia-Fontana C, Munoz-Martinez F, Ramos JL, Krell T. 2010. Sensing of environmental signals: classification of chemoreceptors according to the size of their ligand binding regions. Environ Microbiol 12:2873–2884. doi: 10.1111/j.1462-2920.2010.02325.x. [DOI] [PubMed] [Google Scholar]
- 12.Ulrich LE, Zhulin IB. 2005. Four-helix bundle: a ubiquitous sensory module in prokaryotic signal transduction. Bioinformatics 21(Suppl 3):45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anantharaman V, Aravind L. 2000. Cache—a signaling domain common to animal Ca(2+)-channel subunits and a class of prokaryotic chemotaxis receptors. Trends Biochem Sci 25:535–537. doi: 10.1016/S0968-0004(00)01672-8. [DOI] [PubMed] [Google Scholar]
- 14.Mougel C, Zhulin IB. 2001. CHASE: an extracellular sensing domain common to transmembrane receptors from prokaryotes, lower eukaryotes and plants. Trends Biochem Sci 26:582–584. doi: 10.1016/S0968-0004(01)01969-7. [DOI] [PubMed] [Google Scholar]
- 15.Moglich A, Ayers RA, Moffat K. 2009. Structure and signaling mechanism of Per-ARNT-Sim domains. Structure 17:1282–1294. doi: 10.1016/j.str.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinez SE, Beavo JA, Hol WG. 2002. GAF domains: two-billion-year-old molecular switches that bind cyclic nucleotides. Mol Interv 2:317–323. doi: 10.1124/mi.2.5.317. [DOI] [PubMed] [Google Scholar]
- 17.Ortega A, Krell T. 2014. The HBM domain: introducing bimodularity to bacterial sensing. Protein Sci 23:332–336. doi: 10.1002/pro.2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Z, Hendrickson WA. 2010. Structural characterization of the predominant family of histidine kinase sensor domains. J Mol Biol 400:335–353. doi: 10.1016/j.jmb.2010.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pineda-Molina E, Reyes-Darias JA, Lacal J, Ramos JL, Garcia-Ruiz JM, Gavira JA, Krell T. 2012. Evidence for chemoreceptors with bimodular ligand-binding regions harboring two signal-binding sites. Proc Natl Acad Sci U S A 109:18926–18931. doi: 10.1073/pnas.1201400109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Timmis KN. 2002. Pseudomonas putida: a cosmopolitan opportunist par excellence. Environ Microbiol 4:779–781. doi: 10.1046/j.1462-2920.2002.00365.x. [DOI] [PubMed] [Google Scholar]
- 21.Sampedro I, Parales RE, Krell T, Hill JE. 2015. Pseudomonas chemotaxis. FEMS Microbiol Rev 39:17–46. doi: 10.1111/1574-6976.12081. [DOI] [PubMed] [Google Scholar]
- 22.Parales RE, Luu RA, Chen GY, Liu X, Wu V, Lin P, Hughes JG, Nesteryuk V, Parales JV, Ditty JL. 2013. Pseudomonas putida F1 has multiple chemoreceptors with overlapping specificity for organic acids. Microbiology 159:1086–1096. doi: 10.1099/mic.0.065698-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ulrich LE, Koonin EV, Zhulin IB. 2005. One-component systems dominate signal transduction in prokaryotes. Trends Microbiol 13:52–56. doi: 10.1016/j.tim.2004.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iwaki H, Muraki T, Ishihara S, Hasegawa Y, Rankin KN, Sulea T, Boyd J, Lau PC. 2007. Characterization of a pseudomonad 2-nitrobenzoate nitroreductase and its catabolic pathway-associated 2-hydroxylaminobenzoate mutase and a chemoreceptor involved in 2-nitrobenzoate chemotaxis. J Bacteriol 189:3502–3514. doi: 10.1128/JB.01098-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harwood CS, Rivelli M, Ornston LN. 1984. Aromatic acids are chemoattractants for Pseudomonas putida. J Bacteriol 160:622–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harwood CS. 1989. A methyl-accepting protein is involved in benzoate taxis in Pseudomonas putida. J Bacteriol 171:4603–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parales RE. 2004. Nitrobenzoates and aminobenzoates are chemoattractants for Pseudomonas strains. Appl Environ Microbiol 70:285–292. doi: 10.1128/AEM.70.1.285-292.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abril MA, Michan C, Timmis KN, Ramos JL. 1989. Regulator and enzyme specificities of the TOL plasmid-encoded upper pathway for degradation of aromatic hydrocarbons and expansion of the substrate range of the pathway. J Bacteriol 171:6782–6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muraki T, Taki M, Hasegawa Y, Iwaki H, Lau PC. 2003. Prokaryotic homologs of the eukaryotic 3-hydroxyanthranilate 3,4-dioxygenase and 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase in the 2-nitrobenzoate degradation pathway of Pseudomonas fluorescens strain KU-7. Appl Environ Microbiol 69:1564–1572. doi: 10.1128/AEM.69.3.1564-1572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adler J. 1973. A method for measuring chemotaxis and use of the method to determine optimum conditions for chemotaxis by Escherichia coli. J Gen Microbiol 74:77–91. doi: 10.1099/00221287-74-1-77. [DOI] [PubMed] [Google Scholar]
- 31.Krell T. 2008. Microcalorimetry: a response to challenges in modern biotechnology. Microb Biotechnol 1:126–136. doi: 10.1111/j.1751-7915.2007.00013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alvarez-Ortega C, Harwood CS. 2007. Identification of a malate chemoreceptor in Pseudomonas aeruginosa by screening for chemotaxis defects in an energy taxis-deficient mutant. Appl Environl Microbiol 73:7793–7795. doi: 10.1128/AEM.01898-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Righetti PG, Verzola B. 2001. Folding/unfolding/refolding of proteins: present methodologies in comparison with capillary zone electrophoresis. Electrophoresis 22:2359–2374. doi:. [DOI] [PubMed] [Google Scholar]
- 34.Bohm G, Muhr R, Jaenicke R. 1992. Quantitative analysis of protein far UV circular dichroism spectra by neural networks. Protein Eng 5:191–195. doi: 10.1093/protein/5.3.191. [DOI] [PubMed] [Google Scholar]
- 35.Wuichet K, Zhulin IB. 2010. Origins and diversification of a complex signal transduction system in prokaryotes. Sci Signal 3:ra50. doi: 10.1126/scisignal.2000724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pham HT, Parkinson JS. 2011. Phenol sensing by Escherichia coli chemoreceptors: a nonclassical mechanism. J Bacteriol 193:6597–6604. doi: 10.1128/JB.05987-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lacal J, Garcia-Fontana C, Callejo-Garcia C, Ramos JL, Krell T. 2011. Physiologically relevant divalent cations modulate citrate recognition by the McpS chemoreceptor. J Mol Recognit 24:378–385. doi: 10.1002/jmr.1101. [DOI] [PubMed] [Google Scholar]
- 38.Ni B, Huang Z, Fan Z, Jiang CY, Liu SJ. 2013. Comamonas testosteroni uses a chemoreceptor for tricarboxylic acid cycle intermediates to trigger chemotactic responses towards aromatic compounds. Mol Microbiol 90:813–823. doi: 10.1111/mmi.12400. [DOI] [PubMed] [Google Scholar]
- 39.Yamamoto K, Imae Y. 1993. Cloning and characterization of the Salmonella typhimurium-specific chemoreceptor Tcp for taxis to citrate and from phenol. Proc Natl Acad Sci U S A 90:217–221. doi: 10.1073/pnas.90.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iwama T, Ito Y, Aoki H, Sakamoto H, Yamagata S, Kawai K, Kawagishi I. 2006. Differential recognition of citrate and a metal-citrate complex by the bacterial chemoreceptor Tcp. J Biol Chem 281:17727–17735. doi: 10.1074/jbc.M601038200. [DOI] [PubMed] [Google Scholar]
- 41.Lambert A, Takahashi N, Charon NW, Picardeau M. 2012. Chemotactic behavior of pathogenic and nonpathogenic Leptospira species. Appl Environ Microbiol 78:8467–8469. doi: 10.1128/AEM.02288-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cuppels DA. 1988. Chemotaxis by Pseudomonas syringae pv. tomato. Appl Environ Microbiol 54:629–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hugdahl MB, Beery JT, Doyle MP. 1988. Chemotactic behavior of Campylobacter jejuni. Infect Immun 56:1560–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poole PS, Armitage JP. 1988. Motility response of Rhodobacter sphaeroides to chemotactic stimulation. J Bacteriol 170:5673–5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taylor BL, Zhulin IB. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev 63:479–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 47.Deleage G, Blanchet C, Geourjon C. 1997. Protein structure prediction. Implications for the biologist. Biochimie 79:681–686. [DOI] [PubMed] [Google Scholar]
- 48.Cserzo M, Wallin E, Simon I, von Heijne G, Elofsson A. 1997. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng 10:673–676. doi: 10.1093/protein/10.6.673. [DOI] [PubMed] [Google Scholar]
- 49.Studier FW, Moffatt BA. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol 189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 50.Woodcock DM, Crowther PJ, Doherty J, Jefferson S, DeCruz E, Noyer-Weidner M, Smith SS, Michael MZ, Graham MW. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res 17:3469–3478. doi: 10.1093/nar/17.9.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Herrero M, de Lorenzo V, Timmis KN. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol 172:6557–6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Espinosa-Urgel M, Ramos JL. 2004. Cell density-dependent gene contributes to efficient seed colonization by Pseudomonas putida KT2440. Appl Environ Microbiol 70:5190–5198. doi: 10.1128/AEM.70.9.5190-5198.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boyer HW, Roulland-Dussoix D. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol 41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 54.Kessler B, de Lorenzo V, Timmis KN. 1992. A general system to integrate lacZ fusions into the chromosomes of gram-negative eubacteria: regulation of the Pm promoter of the TOL plasmid studied with all controlling elements in monocopy. Mol Gen Genet 233:293–301. doi: 10.1007/BF00587591. [DOI] [PubMed] [Google Scholar]
- 55.Hasegawa Y, Muraki T, Tokuyama T, Iwaki H, Tatsuno M, Lau PC. 2000. A novel degradative pathway of 2-nitrobenzoate via 3-hydroxyanthranilate in Pseudomonas fluorescens strain KU-7. FEMS Microbiol Lett 190:185–190. doi: 10.1111/j.1574-6968.2000.tb09284.x. [DOI] [PubMed] [Google Scholar]
- 56.Kaniga K, Delor I, Cornelis GR. 1991. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109:137–141. doi: 10.1016/0378-1119(91)90599-7. [DOI] [PubMed] [Google Scholar]
- 57.Obranic S, Babic F, Maravic-Vlahovicek G. 2013. Improvement of pBBR1MCS plasmids, a very useful series of broad-host-range cloning vectors. Plasmid 70:263–267. doi: 10.1016/j.plasmid.2013.04.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.