Abstract
Essential bacterial genes located within operons are particularly challenging to study independently because of coordinated gene expression and the nonviability of knockout mutants. Essentiality scores for many operon genes remain uncertain. Antisense RNA (asRNA) silencing or in-frame gene disruption of genes may help establish essentiality but can lead to polar effects on genes downstream or upstream of the target gene. Here, the Escherichia coli ribF-ileS-lspA-fkpB-ispH operon was used to evaluate the possibility of independently studying an essential gene using expressed asRNA and target gene overexpression to deregulate coupled expression. The gene requirement for growth in conditional silencing strains was determined by the relationship of target mRNA reduction with growth inhibition as the minimum transcript level required for 50% growth (MTL50). Mupirocin and globomycin, the protein inhibitors of IleS and LspA, respectively, were used in sensitization assays of strains containing both asRNA-expressing and open reading frame-expressing plasmids to examine deregulation of the overlapping ileS-lspA genes. We found upstream and downstream polar silencing effects when either ileS or lspA was silenced, indicating coupled expression. Weighted MTL50 values (means and standard deviations) of ribF, ileS, and lspA were 0.65 ± 0.18, 0.64 ± 0.06, and 0.76 ± 0.10, respectively. However, they were not significantly different (P = 0.71 by weighted one-way analysis of variance). The gene requirement for ispH could not be determined due to insufficient growth reduction. Mupirocin and globomycin sensitization experiments indicated that ileS-lspA expression could not be decoupled. The results highlight the inherent challenges associated with genetic analyses of operons; however, coupling of essential genes may provide opportunities to improve RNA-silencing antimicrobials.
INTRODUCTION
Many essential genes are located within operons, which can cause difficulties when studying gene functions for individual open reading frames (ORFs). Escherichia coli has 302 essential genes (1); 218 are in operons, of which 112 potentially are problematic due to their location (e.g., located upstream of other essential genes). Unsurprisingly, a shotgun method for creating silencers in E. coli found multiple essential gene silencing events arising from single expressed antisense sequences (2). In another study, certain operon genes (e.g., eno, fbaA, and pgk), categorized as essential for growth by knockout (negative) evidence, were not essential for growth by knockdown evidence (3). One reason for these discrepancies may be that essential gene knockouts have downstream polar effects on the expression of an intact nonessential gene. This outcome is possible for eno, as it is located downstream of pyrG, an essential gene (4). A polar effect from knockout was observed in the ackA-pta operon, where the disruption of ackA reduced Pta activity by 31% and the disruption of pta reduced AckA activity by 38% compared to that of the wild type (WT) (5).
Antisense silencing is a useful tool for controlling gene expression without genetic modification of the target gene. It is particularly useful for silencing essential genes because a knockdown of expression can be achieved, maintaining cell viability so long as sufficient levels of the silenced mRNA remain available for translation. Plasmids designed to express a short antisense sequence, called expressed antisense RNA (asRNA), are inexpensive to produce, do not suffer from delivery problems, can be conditionally controlled by different promoters (6, 7), and can be expanded easily to target a large number of genes (2, 8). Due to the complexities of operon gene silencing, the use of expressed asRNA has been limited largely to monocistronic genes. When expressed asRNA is applied to an operon, there is often no way to ensure that only the intended target ORF is affected. One study on the ackA-pta operon of E. coli showed downstream and upstream polar effects when either gene was silenced (5), while another study on the sol operon of Clostridium acetobutylicum showed upstream polarity effects by reduced gene product levels (9). The use of antisense peptide nucleic acids (PNA) for silencing of genes in operons suffers similar problems; the lac operon in E. coli and the cmeABC operon in Campylobacter jejuni showed downstream polarity through reduced transcript or protein levels (10, 11).
To evaluate the possibility of independently silencing operon genes by previously established methods (12), we used the E. coli ribF-ileS-lspA-fkpB-ispH operon (NC_000913.3). This operon was chosen because it contains four genes (ribF, ileS, lspA, and ispH) thought to be essential for growth by knockout evidence, and each is involved in a different biological/biochemical pathway; hence, the four genes seem unlikely to regulate one another. The gene ribF encodes an enzyme needed for the synthesis of essential metabolites flavin mononucleotide (FMN)/flavin adenine dinucleotide (FAD), ileS encodes isoleucyl-tRNA synthetase, required for translation, lspA encodes prolipoprotein signal peptidase, required for cell wall maturation, and ispH encodes an enzyme needed for isoprenoid biosynthesis (13). In addition, transcriptional units for these genes are known (14–16), there is transposon-mediated knockout (negative) evidence of the essentiality for the four genes (1, 17), and ileS-lspA overlap by one nucleotide. Finally, there are inhibitors for IleS (18, 19) and LspA (20), enabling us to assay for sensitization after silencing.
MATERIALS AND METHODS
Construction of conditional silencing strains.
Antisense RNA sequences were expressed from pHN1257, an expression vector containing a kanamycin resistance gene, an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter, Ptrc, and a multiple cloning site (MCS) flanked by inverted repeats, termed paired termini, for stabilizing an inserted antisense sequence (5). Antisense sequences of 100 to 160 bases around the ribosome-binding site (RBS) and coding regions of target genes that have minimal predicted secondary structures by M-fold were chosen and amplified with appropriate primers using MG1655 genomic DNA (gDNA) as the template (Table 1). MG1655 gDNA was extracted with a GenElute bacterial genomic kit (Sigma). PCR was carried out using Phusion high-fidelity PCR master mix with HF buffer (New England BioLabs). Amplicons of the expected sizes were column purified using a MinElute PCR purification kit (Qiagen) and digested with NcoI and XhoI (Fermentas), followed by ligation to similarly digested pHN1257. Ligation reactions were transformed to DH5α cells (New England BioLabs), plated on LB-Miller broth supplemented with kanamycin (50 μg/ml; Sigma), and screened by colony PCR using a plasmid-specific forward primer, pHN1257_3751F, and an insert-specific reverse primer (e.g., AS_ribF-R4) (Table 1). Colony PCR was carried out using Crimson Taq DNA polymerase (New England BioLabs). Positive DH5α transformants were used for plasmid preparations (plasmid minikit; Qiagen) of asRNA-expressing plasmids pHNRF4, pHNIS, pHNLA2, pHNLA3, pHNLA4, pHNLA5, and pHNIH (Table 2), which were sequenced to verify the cloning of inserts and transformed into MG1655 to obtain conditional silencing strains.
TABLE 1.
Primer sequences and qPCR efficiencies
| Primer | Sequencea | Target gene | Purpose and/or detailsb |
|---|---|---|---|
| Q/rpoA_F1 | CCGAGGTTGAGATTGATGGT | rpoA | qPCR; 500 nM F1, 300 nM R1, E = 104% |
| Q/rpoA_R1 | CTTCCTGAACGCCTTCTTTG | ||
| Q/ribF_F1 | GCAGAGTGTGGCGTTGATTA | ribF | qPCR; 500 nM F1, 400 nM R1, E = 102% |
| Q/ribF_R1 | CACCAGAAGATCGCTGATGA | ||
| Q/ileS_F | ACAAAACGCCGATCATCTTC | ileS | qPCR; 400 nM F, 500 nM R, E = 102% |
| Q/ileS_R | ACTGCACGCCTTTGATCTCT | ||
| Q/lspA_F | GCGGCGTTTAGTTTCCTTG | lspA | qPCR; 400 nM F, 500 nM R, E = 102% |
| Q/lspA_R | AATCGCAATACCGGCAAAG | ||
| Q/ispH_F1 | CATCTGCTACGCCACGACTA | ispH | qPCR; 400 nM F1, 500 nM R1, E = 101% |
| Q/ispH_R1 | AACACAACTTCCGCCTGTTC | ||
| Q/16S_F2 | TGCATCTGATACTGGCAAGC | rrsA | qPCR; 400 nM F2, 500 nM R2, E = 97% |
| Q/16S_R2 | ACCTGAGCGTCAGTCTTCGT | ||
| Q/plsC_F | CCGTTCAAGACTGGAGCATT | plsC | qPCR; 500 nM F, 500 nM R, E = 100% |
| Q/plsC_R | GAGACGCACACGGGAATAAT | ||
| Q/polA_F | CGGCAACGGTGATTTCTTAT | polA | qPCR; 400 nM F, 500 nM R, E = 99% |
| Q/polA_R | CTTTTTCCAGCTTCGCAATC | ||
| Q/mreD_F | AATGTGGGCACAGGTTTTGT | mreD | qPCR; 500 nM F, 300 nM R, E = 104% |
| Q/mreD_R | GCCACCAGGTAAGCAATGAT | ||
| Q/zipA_F | ATAAACCGAAGCGCAAAGAA | zipA | qPCR; 400 nM F, 300 nM R, E = 100% |
| Q/zipA_R | CCGCTTGTTGAATGCTGTTA | ||
| Q/fldA_F | GACATTGCAAAAAGCAGCAA | fldA | qPCR; 400 nM F, 300 nM R, E = 100.6% |
| Q_fldA_R | ACCAGTTTGCCGTTGAAATC | ||
| AS_ribF-F4 | ACACCATGGGCCTTCTTGCGGGGCCTG | ribF (−50 to +50) | Antisense construct |
| AS_ribF-R4 | AACTCGAGGACCGCTGTACAAGGTATACTCGGA | ||
| AS_ileS-F1 | GGACACCATGGGATCGCCACGCATCG | ileS (−59 to +61) | |
| AS_ileS-R1 | TGGAACTCGAGTAACAAAACCGGCTTAAGC | ||
| AS_lspA-F2 | GGACACCATGGCGACTACCACCAGCCAC | lspA (−45 to +55) | |
| AS_lspA-R2 | TGGAACTCGAGTGTCAGCAACGTCGC | ||
| AS_ispH-F1 | GGACACCATGGCCGTAAATGGCCAGCG | ispH (−49 to +89) | Antisense construct |
| AS_ispH-R1 | TGGAACTCGAGCATTTTGATATTGAAGTGCTGG | ||
| pHN1257_3751F | GCATAATTCGTGTCGCTCAA | pHN1257 | Screening transformants |
| pBAD_F | ATGCCATAGCATTTTTATCC | pBAD | |
| pBAD-R | TCTGATTTAATCTGTATCAGG | ||
| pBAD18SacI_R | GTAGAGCTCTTCCTCCTGCTAGCCCAA | p1G2 | Removal of gfp, insertion of SacI and XhoI in p1G2 for cloning of ORFs |
| pBAD18XhoI_F | TGTCTCGAGGATGATGAGTCGACCTGCA | ||
| ribF_FSacI | TAAGAGCTCATGAAGCTGATACGCG | ribF | Construction of p1G2RF |
| ribF_RXhoI | CTACTCGAGTTAAGCCGGTTTTGTTAG | ||
| ileS_FSacI | TAAGAGCTCATGAGTGACTATAAATCAACC | ileS | Construction of p1G2IS |
| ileS_RXhoI | CTACTCGAGTCAGGCAAACTTACGT | ||
| lspA_FsacI | TAAGAGCTCATGAGTCAATCGATCTGTTCA | lspA | Construction of p1G2LA |
| lspA_RXhoI | CTACTCGAGTTATTGTTTTTTCGCTCTAGAAG | ||
| ispH_FSacI | TAAGAGCTCATGCAGATCCTGTTGGC | ispH | Construction of p1G2IH |
| ispH_RXhoI | CTACTCGAGTTAATCGACTTCACGAATATCG |
Underlined sequences indicate restriction endonuclease sites.
E is the percent amplification efficiency.
TABLE 2.
Plasmids and bacteria in this study
| Strain/plasmid | Relevant feature(s) | Function | Reference or source |
|---|---|---|---|
| E. coli strains | |||
| MG1655 | F−, λ−, rph-1 | Expression, PCR | CGSC, Yale University |
| DH5α | fhuA2Δ(argF-lacZ)U169 phoA glnV44 Φ80 Δ(lacZ)M15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17 | Subcloning | New England Biolabs |
| Plasmids | |||
| pHN1257 | pSC101H ori, Kanr, lacIq, Ptrc, laco-PT-MCS | asRNA expression vector | 5 |
| pHNRF4 | pHN1257 carrying ribF asRNA targeting −50 to +50 of ribF | IPTG-inducible asRNA expression | This study |
| pHNIS | pHN1257 carrying ileS asRNA targeting −59 to +61 of ileS | ||
| pHNLA2 | pHN1257 carrying lspA asRNA targeting −45 to +55 of lspA | ||
| pHNLA3 | pHN1257 carrying lspA asRNA targeting +1 to +100 of lspA | ||
| pHNLA4 | pHN1257 carrying lspA asRNA targeting +21 to +138 of lspA | ||
| pHNLA5 | pHN1257 carrying lspA asRNA targeting +51 to +151 of lspA | ||
| pHNIH | pHN1257 carrying ispH asRNA targeting −49 to +89 of ispH | ||
| p1G2 | pBR322/ColE ori, Cmr, PBAD, AraC N6I, V65G, L133M, E165G, E169V, C280* | ORF expression vector | 23 |
| p1G2RF | p1G2 carrying ribF | l-Arabinose-inducible ORF expression | This study |
| p1G2IS | p1G2 carrying ileS | ||
| p1G2LA | p1G2 carrying lspA | ||
| p1G2IH | p1G2 carrying ispH |
Construction of ORF-expressing plasmids.
Target gene overexpression was driven by the arabinose-inducible promoter PBAD. An arabinose-inducible plasmid was considered, because overexpression of some essential genes is toxic to cells unless they are tightly controlled and expressed at low levels (21, 22). However, IPTG prevents efficient expression from the arabinose-inducible promoter PBAD. To improve compatibility, we used a plasmid derived from pBAD with a mutated AraC (N6I, V65G, L133M, E165G, E169V, and C280* [where * represents a stop codon]), kindly provided by Jay Keasling (Table 2), shown to improve expression in the presence of IPTG (23). The gfp in this plasmid was removed by inverse PCR using primers pBAD18SacI_R and pBAD18XhoI_F (Table 1) and Phusion high-fidelity PCR master mix. Inserts of ribF, lspA, ileS, and ispH were amplified from MG1655 gDNA with gene-specific forward and reverse primers containing SacI and XhoI 5′ ends, respectively (Table 1). Amplicons of p1G2 and ORFs were column purified (MinElute PCR purification kit; Qiagen), digested with SacI and XhoI, ligated with T4 DNA ligase, transformed to chemically competent DH5α, and selected on LB-Miller agar supplemented with chloramphenicol (30 μg/ml). Recombinant plasmids were sequenced to verify cloning of inserts.
Construction of rescue strains.
Rescue strains each contain a pair of asRNA-expressing plasmid (pHNRF4, pHNIS, pHNLA2, and pHNIH in Table 2) and ORF-expressing plasmid (p1G2RF, p1G2IS, p1G2LA, p1G2IH in Table 2). Conditional silencing strains were made competent by CaCl2 treatment (24) for transformation of ORF-expressing plasmids and selected on LB-Miller agar supplemented with kanamycin (50 μg/ml; Sigma) and chloramphenicol (30 μg/ml; Sigma). Positive transformants were confirmed by PCR using two primer pairs: pHN1257_3751F and either ASribF-R4, ASileS-R1, ASlspA-R2, or ASispH-R1 to detect asRNA and pBAD_F/pBAD_R (Table 1) to detect ORFs.
Bacterial growth for induction of asRNA and/or ORF expression.
Overnight cultures were grown at 37°C, with shaking at 180 rpm for 16 to 18 h. Cultures were standardized to 5 × 105 CFU/ml by the measurement of the optical density at 600 nm (OD600) of a 1:10 dilution and adjustment to an OD600 value of 0.003 in LB-Miller broth supplemented with appropriate antibiotics (25). Standardized cultures were added in 180-μl volumes to each well of a 96-well plate. For induction of asRNA expression, conditional silencing strains were treated with different IPTG concentrations in 20 μl per well and incubated in a microplate reader (SpectraMax; Molecular Diagnostics) at 37°C for 5 h. Growth was monitored every 5 min after shaking for 15 s. For induction of asRNA expression and ORF expression, rescue strains were treated with IPTG and l-arabinose in 20 μl per well. The IPTG and l-arabinose combination resulting in growth rescue was determined for each rescue strain by titration of IPTG causing growth inhibition against 0.008, 0.016, 0.032, 0.064, 0.125, 0.25, and 0.5 mM l-arabinose. Plates were incubated in the microplate reader at 37°C for 18 h, and growth was monitored every 15 min after shaking for 10 s.
Bacterial growth for RNA extraction, cDNA synthesis, and qPCR.
Conditional silencing strains were induced with IPTG as described above. The growth of cultures was monitored by OD550 readings every 5 min after shaking for 5 s in a microplate reader (SpectraMax 340PC; Molecular Diagnostics), and all cultures were harvested when the untreated culture increased to an OD550 of ≈0.1. OD550 readings of cultures in the time period with the same IPTG treatment were used to calculate an average representative growth rate (ΔOD/Δt). Cultures with the same IPTG treatment but grown in different wells of a 96-well plate were pooled for total RNA extraction and DNase I treatment (RiboPure bacterium kit; Life Technologies). RNA was checked for gDNA contamination by qPCR using 16S primers (Table 1) and SsoFast EvaGreen supermix (Bio-Rad) before cDNA synthesis of 100 ng RNA (iScript reverse transcription supermix; Bio-Rad). Samples of cDNA were diluted 10-fold, and 4 μl was used in each qPCR. Nontemplate controls were included for each primer pair, and technical duplicates for each sample were done in each run.
Microarray data analysis for reference genes in qPCR.
Genes with the least variable expression under IPTG induction were sought in the microarray data obtained from the GEO database (26) under accession number GSE17505 (27). In addition to using the MAS5-analyzed data set, microarray data also were normalized under RMA and MAS5 methods within the affy package (28). The probe sets were ranked in order of increasing standard deviations (SD) across the whole data set for each data subset. The probe sets with the lowest average ranking had the least variation and were considered the most invariant. Seven candidate reference genes were chosen from the two lists for validation of expression stability under experimental conditions.
Validation of primers for qPCR.
Primer pairs for qPCR were designed by Primer3 with a melting temperature (Tm) of 60°C and product sizes of 100 to 200 bp (Table 1). Primer concentrations (300 to 500 nM) were optimized for use in SsoFast EvaGreen supermix (Bio-Rad) in a CFX96 real-time PCR detection system (CFX Manager v2; Bio-Rad) with 10 ng MG1655 gDNA per reaction mixture. Primer efficiency percentages were determined using 10-fold serial dilutions of MG1655 gDNA (10 pg to 1 μg), and only primers with efficiencies within 5% of one another were selected for use (Table 1). Melting-curve analyses were carried out for all reactions.
Validation of reference genes and analysis of mRNA levels by RT-qPCR.
The following samples were tested for candidate reference gene stability with the indicated IPTG concentrations: MG1655 containing pHN1257 induced with IPTG (0, 0.03, 0.1, 0.4, and 0.8 mM), MG1655 containing pHNIS (0 and 0.03 mM), MG1655 containing pHNLA2 (0 and 0.06 mM), and MG1655 containing pHNIH (0 and 0.8 mM). Total RNA and cDNA were prepared as described above. Raw quantification cycle (Cq) values were analyzed in qbase+ (Biogazelle, Belgium), and the expression stability (M value) of each candidate reference gene, rrs, mreD, fldA, plsC, zipA, polA, and ispB, was calculated as the average pairwise variation for a gene with all other tested genes (29). The lower the M value, the more stable the expression. Stepwise exclusion of the gene with the highest M value allows ranking of the tested genes according to their expression stability, and from this the optimal number of reference genes may be calculated (29). To determine the expression of genes conditionally silenced by asRNA, Cq values were normalized against the two most stably expressed references genes with user-defined PCR efficiencies (Table 1) in qbase+ (Biogazelle). Relative mRNA levels then were calculated as induced normalized Cq divided by uninduced normalized Cq.
Determination of MTL50 values and statistical analysis.
Relative reductions in growth rate were plotted against relative reductions in mRNA over the same range of IPTG concentrations for each experimental repeat in Prism 6 and fitted to a four-parameter dose-response curve used for determining effective concentration inhibiting growth by 50% (EC50). In this instance, the minimum transcript level required for 50% growth (MTL50) was determined. Both the x and y values were variable in each experiment, and a weighted mean and standard error method was used to account for variability. The weighted MTL50 means, standard deviations, and standard errors were calculated using the formulae shown below, where Xw is weighted mean, SDw is weighted SD, SEw is weighted standard error, A is the MTL50 of one experiment, a is the standard error of A, and n is the number of experimental repeats.
| (1) |
| (2) |
| (3) |
Weighted one-way analysis of variance (ANOVA) was carried out using weighted means and weighted standard errors.
MIC and growth sensitization assays.
Stock solutions of synthetic globomycin (30) were dissolved in dimethyl sulfoxide (DMSO) (5.3 mg/ml), and mupirocin (AppliChem) was dissolved in ethanol (50 mg/ml). To determine MIC values, 2-fold serial dilutions of antibiotic stocks (2 to 256 μg/ml) were made in LB-Miller broth and tested for bacterial growth inhibition. The solvents DMSO and ethanol also were included in the assay to ensure they were not inhibiting growth. Bacterial cultures were prepared as described above, and 180 μl was added to 20 μl of antibiotics per well. Plates were incubated at 37°C for 24 h, and growth was visually scored.
Downregulated expression of a drug target gene results in bacterial cell sensitization to the protein inhibitor (2). This mechanism has been used to validate the mechanism of action of antibacterials (2, 31) and was applied here to validate gene specificity of asRNA silencers. Antibiotic sensitization of conditional silencing strains was carried out by titration of either globomycin or mupirocin (0, 0.01, 0.1, 1, 2, 4, 8, 16, and 32 μg/ml) against IPTG concentrations effective for growth inhibition in a checkerboard format before the addition of 180 μl bacterial culture to a final volume of 200 μl. Plates were incubated at 37°C for 24 h, and growth was visually scored. Where there was sensitization in the presence of IPTG, the assay was repeated twice more, with the lowest IPTG concentration resulting in this phenotype. Antibiotic sensitization of rescue strains pHNIS/p1G2LA and pHNLA2/p1G2IS was carried out at 0.03 mM and 0.1 mM IPTG, respectively, with or without 0.03 mM l-arabinose, in the presence of globomycin and mupirocin as described above. The combination of IPTG and l-arabinose was determined in rescue assays as described above.
RESULTS
Silencing of ribF, ileS, lspA, and ispH was gene specific and revealed different requirement stringencies.
To test the specificity of expressed asRNA against the essential genes ribF, ileS, lspA, and ispH, conditional silencing strains were induced with IPTG and growth was monitored. Growth inhibition was observed for expressed asRNA against ileS, lspA, and ribF but not ispH (see Fig. S1 in the supplemental material). We then tested whether the conditional silencing strains could be sensitized to relevant protein inhibitors and whether the growth inhibition due to gene silencing could be rescued by target gene overexpression. Previously we showed that overexpression of a gene targeted by expressed asRNA resulted in rescue from growth inhibition (12); therefore, it may provide a way of compensating for polar silencing effects. Mupirocin inhibits the isoleucyl-tRNA synthetase encoded by ileS (19), and globomycin inhibits the prolipoprotein signal peptidase II encoded by lspA (20). There are no known protein inhibitors of RibF and IspH. Both ileS- and lspA-silencing strains were sensitized to both mupirocin and globomycin with induction of asRNA, while the ribF- and ispH-silencing strains did not change in susceptibility to globomycin or mupirocin with induction of asRNA (Table 3). As ileS and lspA overlap by one nucleotide, it was possible that silencing of either gene resulted in polar silencing of the other, and sensitization to both protein inhibitors of LspA and IleS was observed.
TABLE 3.
Sensitization of conditional silencing strains to mupirocin and globomycin
| Conditional silencer | Target gene | Fold reduction in MICa of: |
|
|---|---|---|---|
| Mupirocin | Globomycin | ||
| pHNRF4 | ribF | 0 | 0 |
| pHNIS | ileS | 8b | 2b |
| pHNLA2 | lspA | 2c | 2c |
| pHNIH | ispH | 0 | 0 |
Fold reduction relative to the level for uninduced cultures from three experiments. The MIC of mupirocin and globomycin was 16 μg/ml in uninduced conditional silencing strains.
At 0.03 mM IPTG.
At 0.1 mM IPTG.
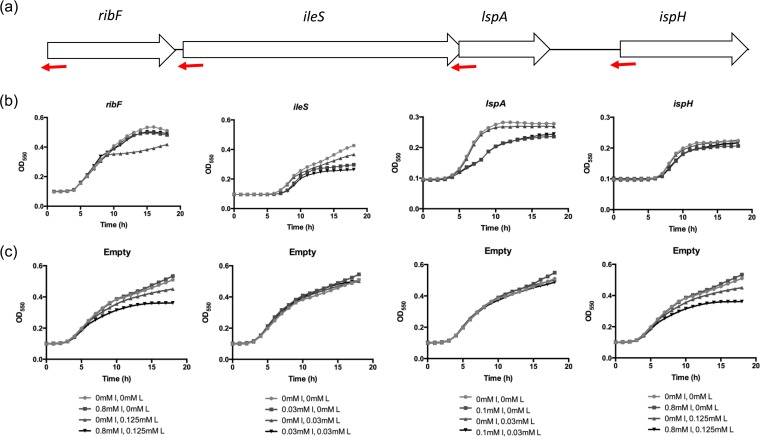
Another method for determining silencer specificity is by overexpressing the gene target to supply functional protein to a level that would counteract the effects of silencing. Logically, the reverse also would indicate the specificity of a silencer in cases where gene overexpression is toxic, since depletion of a chromosomal gene product by silencing could equilibrate levels of the overexpressed gene to that in a normal cell. This method is useful when protein inhibitors are lacking, as is the case for RibF and IspH. Growth inhibition due to overexpression of ribF in rescue strains was greater than growth inhibition due to silencing of either gene (Fig. 1b), and growth was rescued with expression of asRNA at 0.8 mM IPTG and the corresponding ORF at 0.125 mM l-arabinose, indicating that the expressed antisense silencer of ribF was gene specific (Fig. 1b). Although growth of the empty plasmid control strain was reduced with the addition of 0.125 mM l-ara (Fig. 1c), growth of the ribF rescue strains at 0.8 mM IPTG and 0.125 mM l-ara was similar to that of uninduced cultures. Growth inhibition due to silencing of ileS, lspA, or ispH in rescue strains was greater than growth inhibition due to overexpression of the target genes compared to levels for uninduced cultures and control strains (Fig. 1b and c). However, growth was not rescued with expression of asRNA and ORF for the ileS and lspA strains, suggesting either the silencers were not gene specific or the two genes were tightly coupled. As growth inhibition due to ispH silencing was weak, rescue in growth was weak (Fig. 1b and c).
FIG 1.
Validation of asRNA silencing specificity through overexpression of targeted ORF in rescue strains. (a) Gene organization of the operon, with asRNA complementary regions in red. (b) Growth of rescue strains induced with IPTG (I) for asRNA expression and l-arabinose (L) for ORF expression. (c) Growth of control rescue strains containing empty p1G2 plasmid at the same inducer concentrations as those used for panel b. Concentrations of IPTG (I) and l-arabinose (L) are indicated. Mean 18-h growth curves are plotted (n = 3).
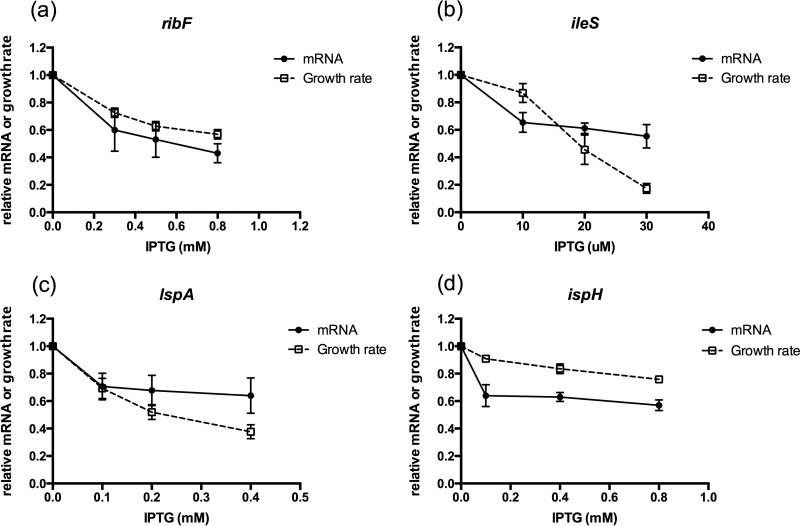
We then analyzed target mRNA reduction along with growth inhibition. In order to accurately normalize gene expression to that of reference genes, we selected seven genes displaying stable expression based on microarray data and determined gene expression stability in 11 samples (see Table S1 in the supplemental material). The optimal combination of stably expressed genes was zipA and rrs, which were used for normalization of qPCR data. Three IPTG concentrations of up to 1 mM, resulting in a reduction of growth rate, were chosen for each silencing strain, and the corresponding mRNA reduction of target genes was determined. Reductions in ribF and ispH mRNA were greater than reductions in growth rates, particularly in ispH, where only a 24% reduction in growth rate was achieved with 43% reduction in mRNA (Fig. 2). In contrast, reductions in ileS and lspA mRNA were less than reductions in growth rates at the two highest inducer concentrations tested (Fig. 2).
FIG 2.
Bacterial growth rate (ΔOD/Δt) and mRNA reductions with IPTG induction of expressed antisense-targeting essential genes. Growth rate and mRNA reductions were calculated relative to those of uninduced cultures (averages ± SD; n = 3).
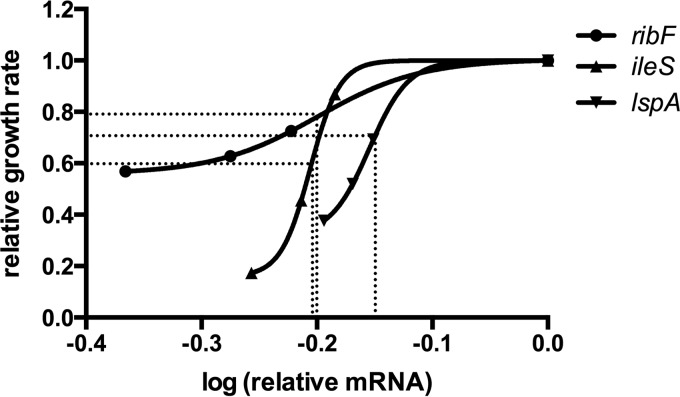
Log10 values (mRNA reduction) then were plotted against values for relative growth reduction as a dose-response curve to enable calculation of differences in gene requirement for growth, indicated by the MTL50 (12). This value measures the minimum level of transcript required to sustain 50% cell viability, so that a value approaching 0 indicates no requirement (e.g., nonessential gene) and a value approaching 1 indicates high requirement for viability. For this purpose, an additional IPTG dose (0.04 mM) for maximum inhibitory response was used for ileS. Similar to determining EC50s from a dose-response curve, the mean weighted MTL50 of ribF, ileS, and lspA were calculated as 0.65 ± 0.18, 0.64 ± 0.06, and 0.76 ± 0.10, respectively (Fig. 3). However, the MTL50 values were not significantly different (P = 0.71 by weighted one-way ANOVA). MTL50 for ispH could not be determined, as growth reductions were not sufficient for good curve fits in two of three experiments.
FIG 3.
Use of mRNA and growth rate covariation to determine the MTL50 of essential genes ribF, ileS, and lspA. Transformed mean values of relative mRNA levels were plotted against mean values of the relative growth rate shown in Fig. 2 (n = 3). Dotted lines indicate the minimum transcript level value of each gene (x axis) required to support 50% of maximal (uninduced) bacterial growth (y axis). Curves and MTL50 were generated in GraphPad Prism 6.
Silencing of ileS or lspA results in polar effects.
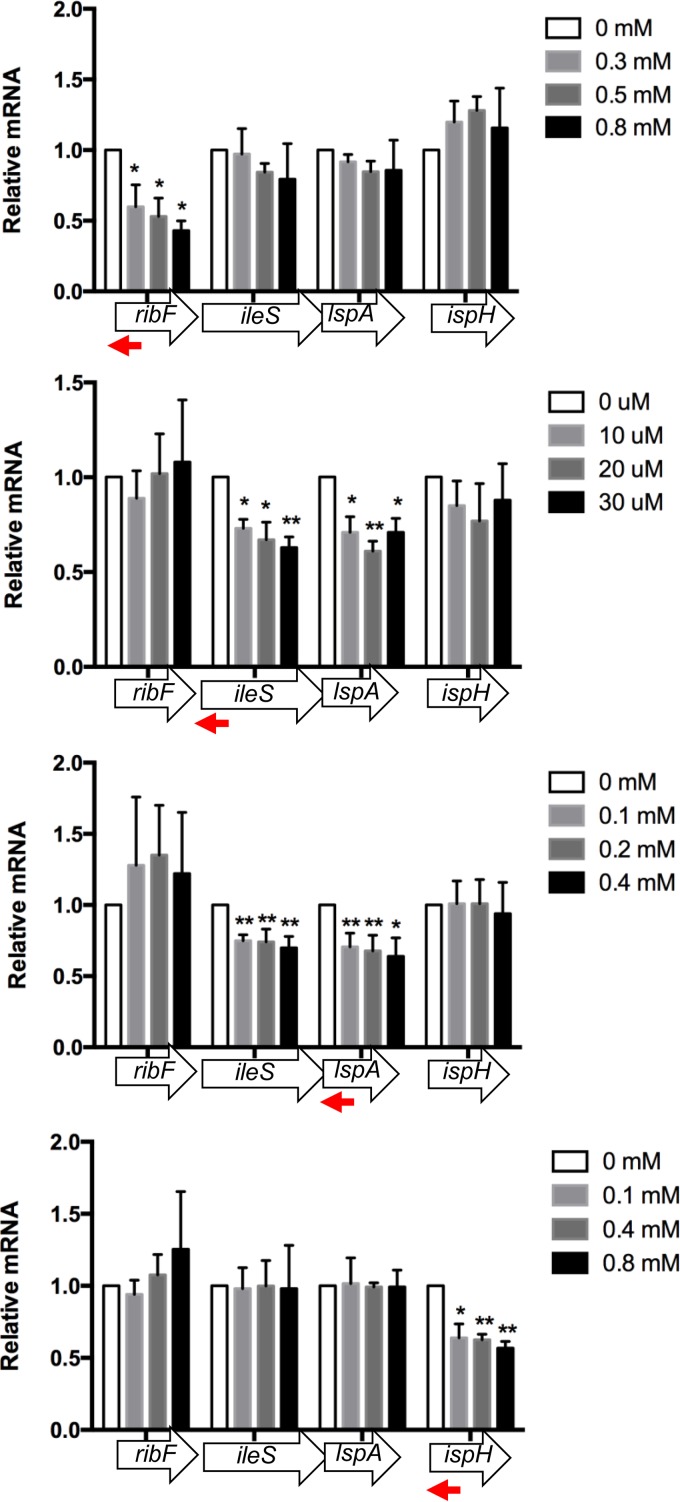
We next examined polar silencing effects for each silencer by measuring the mRNA levels of all genes in the operon upon silencer induction. The ribF and ispH silencers did not result in polar silencing, indicating that independent gene silencing in this operon is possible (Fig. 4). However, ileS asRNA induction significantly reduced downstream transcripts of lspA, and lspA asRNA induction significantly reduced upstream transcripts of ileS (Fig. 4). These results may explain the earlier observation of cross-sensitization of both ileS and lspA silencing strains to mupirocin and globomycin (Table 3) and the lack of growth rescue when asRNA and ORFs were expressed together (Fig. 1b).
FIG 4.
Expression profiles of each essential gene within the ribF-ileS-lspA-fkpB-ispH operon upon silencing one essential gene. Target regions for expressed antisense sequences specific for individual target genes are indicated by red arrows. Averages ± SD of mRNA levels relative to those of the uninduced control are shown (n = 3). *, P < 0.05; **, P < 0.01.
To examine whether we could silence lspA without an upstream effect on ileS, we created three more expressed antisense plasmids, pHNLA3, pHNLA4, and pHNLA5 (Table 2), avoiding the ileS ORF while targeting the coding gene of lspA. The expression levels of all four essential genes in the operon were determined from three experimental repeats, and each silencing strain was tested for sensitization to globomycin (Table 4). Results indicated that silencing of lspA was coordinated with ileS, and silencers targeting regions further into the coding region (i.e., pHNLA4 and pHNLA5) were not effective compared to silencers targeting regions at the start of the coding region (i.e., pHNLA2 and pHNLA3) (32). We were not able to create asRNAs that silenced lspA or ileS; hence, their expression had to be discoordinated by compensating for the effects of the nontargeted gene.
TABLE 4.
Efficacy of asRNA silencers targeting lspA
| Antisense plasmid | Target region of lspA | Growth inhibition with IPTG | Reduction in mRNA levela |
Fold reduction in globomycin MICb | |||
|---|---|---|---|---|---|---|---|
| ribF | ileS | lspA | ispH | ||||
| pHNLA2 | −45 to +55 | Yes | − | + | + | − | 4 |
| pHNLA3 | +1 to +100 | Yes | − | + | + | − | 4 |
| pHNLA4 | +21 to +138 | Yes (slight) | − | − | − | − | 0 |
| pHNLA5 | +51 to +151 | Yes (slight) | − | − | − | − | 0 |
Only mRNA significantly reduced (n = 3; P < 0.05) in a dose-dependent manner are considered. −, no reduction; +, reduction.
Fold reduction in MIC at 0.2 mM IPTG was determined relative to the level of uninduced cultures in two repeated assays. Growth was scored visually.
Polar silencing effects of ileS and lspA cannot be decoupled.
The ileS gene is transcribed in two transcriptional units, while lspA is transcribed in three transcriptional units (Fig. 5a). To decouple the silencing polarity of ileS and lspA, overexpression of either gene should result in a phenotype representative of the uncomplemented, silenced gene. That is, previously observed cross-sensitivity to mupirocin or globomycin shown in Table 3 should be reduced or eliminated. For an ileS-specific silencing phenotype, a strain containing ileS asRNA and lspA overexpression (pHNIS/p1G2LA) (Fig. 5b) was made for antibiotic sensitization assays with 0.03 mM IPTG for induction of ileS antisense and either with 0.03 mM l-arabinose or without l-arabinose for induction of lspA expression. The IPTG and l-arabinose concentrations used here were the same as those for conditional silencing strains (Table 3) and rescue strains (Fig. 1). The pHNIS/p1G2LA strain had MICs of 8 μg/ml and 32 μg/ml for mupirocin and globomycin, respectively, whereas the conditional silencing strain had a MIC of 16 μg/ml to both antibiotics under noninduced conditions (Table 3). With ileS asRNA expression under IPTG induction, the range of fold reduction in mupirocin MICs with lspA expression under l-arabinose induction was similar to that of cultures lacking l-arabinose (Table 5). With ileS asRNA expression under IPTG induction, the reduction in globomycin MIC also was the same with and without lspA expression under l-arabinose induction (Table 5). Therefore, simultaneous expression of lspA and antisense sequence against ileS did not improve sensitization to mupirocin over globomycin. However, the pHNIS/p1G2LA strain displayed lower mupirocin MICs and higher globomycin MICs than the pHNIS conditional silencing strain.
FIG 5.
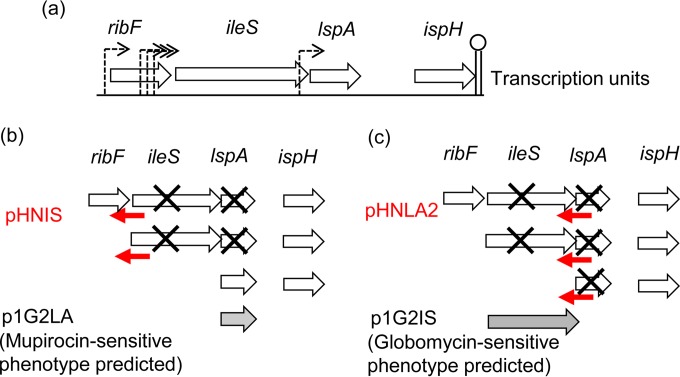
Designs for decoupling iles-lspA expression by asRNA silencing and nontarget gene expression. (a) Known transcriptional units of the ribF-ileS-lspA-fkpB-ispH operon showing only the essential genes (14–16). Dashed arrows indicate promoter sites. (b) The ileS silencer, pHNIS (red arrow), silences ileS and lspA (crosses) on two of three possible transcriptional units. For an ileS-specific response with pHNIS, lspA should be expressed from p1G2LA (gray arrow) to compensate for loss of lspA expression. This is predicted to result in a mupirocin-sensitive phenotype. (c) The lspA silencer, pHNLA2 (red arrow), silences ileS and lspA (crosses) on all possible transcriptional units. For an lspA-specific response from pHNLA2, ileS should be expressed from p1G2IS (gray arrow) to compensate for loss of ileS expression. This is predicted to result in a globomycin-specific phenotype.
TABLE 5.
Sensitization of strains expressing asRNA and nontarget ORF to mupirocin and globomycin
| Conditional silencer | Gene silenced (mM IPTG) | ORF expressed (mM l-ara) | Fold reduction in MIC ofc: |
|||
|---|---|---|---|---|---|---|
| Mupirocin |
Globomycin |
|||||
| 0 mM l-ara | 0.03 mM l-ara | 0 mM l-ara | 0.03 mM l-ara | |||
| pHNIS/p1G2LAa | ileS (0.03) | lspA (0.03) | 8–16 | 8–32 | 4–8 | 4–8 |
| pHNLA2/p1G2ISb | lspA (0.1) | ileS (0.03) | 4 | 4 | 4 | 4 |
MICs of mupirocin and globomycin were 8 μg/ml and 32 μg/ml, respectively, without IPTG and l-arabinose (l-ara) induction.
MIC of mupirocin and globomycin was 16 μg/ml without IPTG and l-ara induction.
Fold reduction relative to the level of uninduced asRNA cultures from three experiments.
To test for a phenotype specific for silencing of lspA, a strain containing lspA asRNA and ileS overexpression (i.e., pHNLA2/p1G2IS) (Fig. 5c) was constructed and used in antibiotic sensitization experiments as described above, except 0.01 mM IPTG was used as the most effective induction concentration. In contrast to the pHNIS/p1G2LA strain, antibiotic MICs of the pHNLA2/p1G2IS strain remained the same as those for the conditional silencing strain of lspA under noninduced conditions (i.e., 16 μg/ml) (Table 3). With lspA asRNA expression induced under IPTG, the fold reductions in mupirocin and globomycin MICs were the same with and without ileS expression induced under l-arabinose (Table 5). There was no reduction in cross-sensitization to mupirocin, indicating uncoupling of ileS from lspA was not achieved with simultaneous expression of ileS and antisense sequence against lspA.
DISCUSSION
For the E. coli ribF-ileS-lspA-fkpB-ispH operon, we determined gene requirement stringencies for bacterial growth and the effect of silencing one gene on the gene expression of the rest of the operon. Although three promoters of ileS were identified within ribF (33, 34), silencing of ribF did not lead to downstream polar effects on the expression of ileS. This suggests an advantage of using asRNA over disruption or deletion of ribF, which could remove ileS promoter sites and result in downstream inhibition of ileS transcription. Two studies were not able to knock out ribF (yaaC) (1, 17), but one study did (35), leaving uncertainty about its requirement. Our results support ribF as an essential gene.
The MTL50 value for lspA was not significantly greater than that for ileS and ribF; however, other studies have scored the gene requirement of lspA as greater than that of ileS in terms of the viability of knockout mutants in rich and minimal media (1). It is possible that growth in minimal media enhances differences in MTL50 values. Indeed, we observed greater growth inhibition by pHNRF4 in M9 minimal media than in LB (unpublished data). Insufficient growth reduction from the ispH silencer may be due to silencer design, although up to a 43% reduction in ispH transcripts was achieved. Alternatively, it may show that 57% of normal levels of transcript and/or enzyme was sufficient to support growth in a rich medium. A study examining the depletion of ispH (lytB) under arabinose induction in a mutated ispH strain found cells were able to grow for about 4 h before cell death (36). It is probable that ispH requirement for growth is the lowest of the four essential genes examined under the tested conditions.
Upstream silencing in an operon through expression of asRNA does not appear to be limited to overlapping genes. In creating a genome-wide library of E. coli silencers, upstream silencing effects were shown indirectly in the rpsF-priB-rpsR-rplI operon, where a silencer against a nonessential gene (rplI) resulted in growth inhibition, presumably due to a polar silencing effect on the upstream, nonoverlapping, and essential gene, rpsR, that was 41 nucleotides away (2). A study by Tummala et al. applied similar tools to two nonoverlapping, nonessential genes in the tricistronic aad-ctfA-ctfB operon in C. acetobutylicum (9). Silencing of ctfB reduced CtfA and CtfB levels (9); ctfA and ctfB are one nucleotide apart. Therefore, proximity and transcription units may be factors in upstream polar silencing, since it did not occur for ribF when ileS was targeted or for lspA when ispH was targeted. Naturally occurring antisense small noncoding RNAs (sRNA) have been reported to repress genes downstream of the targeted ORF. Two examples are sRNAs of the phoPQ operon in E. coli (37) and the YPK_1206-1205 operon in Yersinia pseudotuberculosis (38), where sRNAs silence the target and downstream genes. It will be interesting to see if upstream silencing exists in nature as more sRNAs are identified and characterized.
We considered the possible effects of pHNLA2 on ileS due to gene overlap with lspA and created silencers targeting deeper into the lspA coding sequence to avoid nonspecific effects on ileS. However, these silencers were less efficient, possibly because they are not likely to repress translation initiation. Many known mRNA degradation mechanisms are centered on the RBS or initiated by activities occurring around the RBS (39). For instance, bacterial mRNA is vulnerable to degradation by RNases through direct entry (40) when ribosome and RNAP are physically separated or uncoupled by translation repression (41). Another RBS-centered mechanism is activation of mRNA cleavage to occur at a site distal from mRNA-asRNA binding at the RBS (42). Therefore, while a silencer targeting the RBS is more effective, it may result in nonspecific activity in complex overlapping gene targets, such as iles-lspA.
Cross-sensitization of ileS and lspA-silencing strains to globomycin and mupirocin could not be reduced by complementation, indicating the complexity in expression and regulation of the two overlapping essential genes. Systemic physiological effects of translation errors from silencing ileS and cell wall weakening from silencing lspA may add to the difficulty of reducing cross-sensitization. An E. coli mutant with defective IleS was shown to increase antibiotic susceptibility to mupirocin and other antibiotics because of the cumulative global effects of incorrect codon translation (43). Inhibition of LspA by globomycin leads to the accumulation of prolipoprotein within the periplasm and cell death (44, 45), presumably as a result of the loss in cell wall structure and integrity (46). It is possible that cells simultaneously expressing ileS and antisense against lspA suffered from cell wall malformation and increased permeability to mupirocin without depletion of IleS.
In summary, this study shows essential genes in an operon may have different requirements for growth in rich media, and it was not possible to decouple expression of overlapping ileS and lspA genes using a combination of expressed antisense and ORF expression vectors due to inherent complexities in gene regulation. However, this strategy enabled the examination of essential gene associations within an operon and is a powerful tool for prokaryotic genetics. Overlapping putative essential genes in operons identified by knockout techniques may need closer examination prior to accurate scoring of gene function and requirements. Potentially, such gene structures can be used to our advantage as gene targets for antimicrobials.
Supplementary Material
ACKNOWLEDGMENTS
This project was funded by the Wellcome Trust, The Royal Veterinary College, and The Heptagon Fund.
We thank Kenneth Rudd and Nobutaka Nakashima for helpful discussions and advice and Ruby Chang for help on statistics.
Footnotes
This article has been assigned the approval number PPB_01000 by the RVC.
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01444-15.
REFERENCES
- 1.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meng J, Kanzaki G, Meas D, Lam CK, Crummer H, Tain J, Xu HH. 2012. A genome-wide inducible phenotypic screen identifies antisense RNA constructs silencing Escherichia coli essential genes. FEMS Microbiol Lett 329:45–53. doi: 10.1111/j.1574-6968.2012.02503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakashima N, Ohno S, Yoshikawa K, Shimizu H, Tamura T. 2014. A vector library for silencing central carbon metabolism genes with antisense RNAs in Escherichia coli. Appl Environ Microbiol 80:564–573. doi: 10.1128/AEM.02376-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weng M, Makaroff CA, Zalkin H. 1986. Nucleotide sequence of Escherichia coli pyrG encoding CTP synthetase. J Biol Chem 261:5568–5574. [PubMed] [Google Scholar]
- 5.Nakashima N, Tamura T. 2009. Conditional gene silencing of multiple genes with antisense RNAs and generation of a mutator strain of Escherichia coli. Nucleic Acids Res 37:e103. doi: 10.1093/nar/gkp498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan XX, Rose K, Margolin W, Chen Y. 2004. DNA enzyme generated by a novel single-stranded DNA expression vector inhibits expression of the essential bacterial cell division gene ftsZ. Biochemistry 43:1111–1117. doi: 10.1021/bi035164h. [DOI] [PubMed] [Google Scholar]
- 7.Nakashima N, Tamura T. 2013. Gene silencing in Escherichia coli using antisense RNAs expressed from doxycycline-inducible vectors. Lett Appl Microbiol 56:436–442. doi: 10.1111/lam.12066. [DOI] [PubMed] [Google Scholar]
- 8.Ji Y, Zhang B, Van SF, Horn Warren P, Woodnutt G, Burnham MK, Rosenberg M. 2001. Identification of critical staphylococcal genes using conditional phenotypes generated by antisense RNA. Science 293:2266–2269. doi: 10.1126/science.1063566. [DOI] [PubMed] [Google Scholar]
- 9.Tummala SB, Junne SG, Papoutsakis ET. 2003. Antisense RNA downregulation of coenzyme A transferase combined with alcohol-aldehyde dehydrogenase overexpression leads to predominantly alcohologenic Clostridium acetobutylicum fermentations. J Bacteriol 185:3644–3653. doi: 10.1128/JB.185.12.3644-3653.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dryselius R, Nikravesh A, Kulyte A, Goh S, Good L. 2006. Variable coordination of cotranscribed genes in Escherichia coli following antisense repression. BMC Microbiol 6:97. doi: 10.1186/1471-2180-6-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oh E, Zhang Q, Jeon B. 2014. Target optimization for peptide nucleic acid (PNA)-mediated antisense inhibition of the CmeABC multidrug efflux pump in Campylobacter jejuni. J Antimicrob Chemother 69:375–380. doi: 10.1093/jac/dkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goh S, Boberek JM, Nakashima N, Stach J, Good L. 2009. Concurrent growth rate and transcript analyses reveal essential gene stringency in Escherichia coli. PLoS One 4:e6061. doi: 10.1371/journal.pone.0006061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altincicek B, Kollas A, Eberl M, Wiesner J, Sanderbrand S, Hintz M, Beck E, Jomaa H. 2001. LytB, a novel gene of the 2-C-methyl-d-erythritol 4-phosphate pathway of isoprenoid biosynthesis in Escherichia coli. FEBS Lett 499:37–40. doi: 10.1016/S0014-5793(01)02516-9. [DOI] [PubMed] [Google Scholar]
- 14.Kamio Y, Lin CK, Regue M, Wu HC. 1985. Characterization of the ileS-lsp operon in Escherichia coli. Identification of an open reading frame upstream of the ileS gene and potential promoter(s) for the ileS-lsp operon. J Biol Chem 260:5616–5620. [PubMed] [Google Scholar]
- 15.Miller KW, Bouvier J, Stragier P, Wu HC. 1987. Identification of the genes in the Escherichia coli ileS-lsp operon. Analysis of multiple polycistronic mRNAs made in vivo. J Biol Chem 262:7391–7397. [PubMed] [Google Scholar]
- 16.Salgado H, Peralta-Gil M, Gama-Castro S, Santos-Zavaleta A, Muniz-Rascado L, Garcia-Sotelo JS, Weiss V, Solano-Lira H, Martinez-Flores I, Medina-Rivera A, Salgado-Osorio G, Alquicira-Hernandez S, Alquicira-Hernandez K, Lopez-Fuentes A, Porron-Sotelo L, Huerta AM, Bonavides-Martinez C, Balderas-Martinez YI, Pannier L, Olvera M, Labastida A, Jimenez-Jacinto V, Vega-Alvarado L, Del Moral-Chavez V, Hernandez-Alvarez A, Morett E, Collado-Vides J. 2013. RegulonDB v8.0: omics data sets, evolutionary conservation, regulatory phrases, cross-validated gold standards and more. Nucleic Acids Res 41:D203–D213. doi: 10.1093/nar/gks1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerdes SY, Scholle MD, Campbell JW, Balazsi G, Ravasz E, Daugherty MD, Somera AL, Kyrpides NC, Anderson I, Gelfand MS, Bhattacharya A, Kapatral V, D'Souza M, Baev MV, Grechkin Y, Mseeh F, Fonstein MY, Overbeek R, Barabasi AL, Oltvai ZN, Osterman AL. 2003. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J Bacteriol 185:5673–5684. doi: 10.1128/JB.185.19.5673-5684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas CM, Hothersall J, Willis CL, Simpson TJ. 2010. Resistance to and synthesis of the antibiotic mupirocin. Nat Rev Microbiol 8:281–289. doi: 10.1038/nrmicro2278. [DOI] [PubMed] [Google Scholar]
- 19.Yanagisawa T, Lee JT, Wu HC, Kawakami M. 1994. Relationship of protein structure of isoleucyl-tRNA synthetase with pseudomonic acid resistance of Escherichia coli. A proposed mode of action of pseudomonic acid as an inhibitor of isoleucyl-tRNA synthetase. J Biol Chem 269:24304–24309. [PubMed] [Google Scholar]
- 20.Dev IK, Harvey RJ, Ray PH. 1985. Inhibition of prolipoprotein signal peptidase by globomycin. J Biol Chem 260:5891–5894. [PubMed] [Google Scholar]
- 21.Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H. 2005. Complete set of ORF clones of Escherichia coli ASKA library (A Complete Set of E. coli K-12 ORF Archive): unique resources for biological research. DNA Res 12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- 22.Goh S, Good L. 2008. Plasmid selection in Escherichia coli using an endogenous essential gene marker. BMC Biotechnol 8:61. doi: 10.1186/1472-6750-8-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee SK, Chou HH, Pfleger BF, Newman JD, Yoshikuni Y, Keasling JD. 2007. Directed evolution of AraC for improved compatibility of arabinose- and lactose-inducible promoters. Appl Environ Microbiol 73:5711–5715. doi: 10.1128/AEM.00791-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 25.Nakashima N, Goh S, Good L, Tamura T. 2012. Multiple-gene silencing using antisense RNAs in Escherichia coli. Methods Mol Biol 815:307–319. doi: 10.1007/978-1-61779-424-7_23. [DOI] [PubMed] [Google Scholar]
- 26.Edgar R, Domrachev M, Lash AE. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haddadin FT, Harcum SW. 2005. Transcriptome profiles for high-cell-density recombinant and wild-type Escherichia coli. Biotechnol Bioeng 90:127–153. doi: 10.1002/bit.20340. [DOI] [PubMed] [Google Scholar]
- 28.Gautier L, Cope L, Bolstad BM, Irizarry RA. 2004. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 29.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sarabia F, Chammaa S, Garcia-Ruiz C. 2011. Solid phase synthesis of globomycin and SF-1902 A5. J Org Chem 76:2132–2144. doi: 10.1021/jo1025145. [DOI] [PubMed] [Google Scholar]
- 31.Ji Y, Yin D, Fox B, Holmes DJ, Payne D, Rosenberg M. 2004. Validation of antibacterial mechanism of action using regulated antisense RNA expression in Staphylococcus aureus. FEMS Microbiol Lett 231:177–184. doi: 10.1016/S0378-1097(03)00931-5. [DOI] [PubMed] [Google Scholar]
- 32.Dryselius R, Aswasti SK, Rajarao GK, Nielsen PE, Good L. 2003. The translation start codon region is sensitive to antisense PNA inhibition in Escherichia coli. Oligonucleotides 13:427–433. doi: 10.1089/154545703322860753. [DOI] [PubMed] [Google Scholar]
- 33.Tokunaga M, Loranger JM, Chang SY, Regue M, Chang S, Wu HC. 1985. Identification of prolipoprotein signal peptidase and genomic organization of the lsp gene in Escherichia coli. J Biol Chem 260:5610–5615. [PubMed] [Google Scholar]
- 34.Nonaka G, Blankschien M, Herman C, Gross CA, Rhodius VA. 2006. Regulon and promoter analysis of the E. coli heat-shock factor, σ32, reveals a multifaceted cellular response to heat stress. Genes Dev 20:1776–1789. doi: 10.1101/gad.1428206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arigoni F, Talabot F, Peitsch M, Edgerton MD, Meldrum E, Allet E, Fish R, Jamotte T, Curchod ML, Loferer H. 1998. A genome-based approach for the identification of essential bacterial genes. Nat Biotechnol 16:851–856. doi: 10.1038/nbt0998-851. [DOI] [PubMed] [Google Scholar]
- 36.McAteer S, Coulson A, McLennan N, Masters M. 2001. The lytB gene of Escherichia coli is essential and specifies a product needed for isoprenoid biosynthesis. J Bacteriol 183:7403–7407. doi: 10.1128/JB.183.24.7403-7407.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coornaert A, Chiaruttini C, Springer M, Guillier M. 2013. Post-transcriptional control of the Escherichia coli PhoQ-PhoP two-component system by multiple sRNAs involves a novel pairing region of GcvB. PLoS Genet 9:e1003156. doi: 10.1371/journal.pgen.1003156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu P, Zhang Y, Li L, Hu Y, Huang L, Li Y, Rayner S, Chen S. 2012. Small non-coding RNA SraG regulates the operon YPK_1206-1205 in Yersinia pseudotuberculosis. FEMS Microbiol Lett 331:37–43. doi: 10.1111/j.1574-6968.2012.02548.x. [DOI] [PubMed] [Google Scholar]
- 39.Kaberdin VR, Blasi U. 2006. Translation initiation and the fate of bacterial mRNAs. FEMS Microbiol Rev 30:967–979. doi: 10.1111/j.1574-6976.2006.00043.x. [DOI] [PubMed] [Google Scholar]
- 40.Mackie GA. 2013. RNase E: at the interface of bacterial RNA processing and decay. Nat Rev Microbiol 11:45–57. doi: 10.1038/nrmicro2930. [DOI] [PubMed] [Google Scholar]
- 41.Proshkin S, Rahmouni AR, Mironov A, Nudler E. 2010. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science 328:504–508. doi: 10.1126/science.1184939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prevost K, Desnoyers G, Jacques JF, Lavoie F, Masse E. 2011. Small RNA-induced mRNA degradation achieved through both translation block and activated cleavage. Genes Dev 25:385–396. doi: 10.1101/gad.2001711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bacher JM, de Crecy-Lagard V, Schimmel PR. 2005. Inhibited cell growth and protein functional changes from an editing-defective tRNA synthetase. Proc Natl Acad Sci U S A 102:1697–1701. doi: 10.1073/pnas.0409064102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hussain M, Ichihara S, Mizushima S. 1980. Accumulation of glyceride-containing precursor of the outer membrane lipoprotein in the cytoplasmic membrane of Escherichia coli treated with globomycin. J Biol Chem 255:3707–3712. [PubMed] [Google Scholar]
- 45.Yakushi T, Tajima T, Matsuyama S, Tokuda H. 1997. Lethality of the covalent linkage between mislocalized major outer membrane lipoprotein and the peptidoglycan of Escherichia coli. J Bacteriol 179:2857–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lai JS, Philbrick WM, Hayashi S, Inukai M, Arai M, Hirota Y, Wu HC. 1981. Globomycin sensitivity of Escherichia coli and Salmonella typhimurium: effects of mutations affecting structures of murein lipoprotein. J Bacteriol 145:657–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.