Abstract
In this study, the metabolic and physiological potential evaluator system based on Kyoto Encyclopedia of Genes and Genomes (KEGG) functional modules was employed to establish a functional classification of archaeal species and to determine the comprehensive functions (functionome) of the previously uncultivated thermophile “Candidatus Caldiarchaeum subterraneum” (Ca. C. subterraneum). A phylogenetic analysis based on the concatenated sequences of proteins common among 142 archaea and 2 bacteria, and among 137 archaea and 13 unicellular eukaryotes suggested that Ca. C. subterraneum is closely related to thaumarchaeotic species. Consistent with the results of the phylogenetic analysis, clustering and principal component analyses based on the completion ratio patterns for all KEGG modules in 79 archaeal species suggested that the overall metabolic and physiological potential of Ca. C. subterraneum is similar to that of thaumarchaeotic species. However, Ca. C. subterraneum possessed almost no genes in the modules required for nitrification and the hydroxypropionate–hydroxybutyrate cycle for carbon fixation, unlike thaumarchaeotic species. However, it possessed all genes in the modules required for central carbohydrate metabolism, such as glycolysis, pyruvate oxidation, the tricarboxylic acid (TCA) cycle, and the glyoxylate cycle, as well as multiple sets of sugar and branched chain amino acid ABC transporters. These metabolic and physiological features appear to support the predominantly aerobic character of Ca. C. subterraneum, which lives in a subsurface thermophilic microbial mat community with a heterotrophic lifestyle.
Introduction
Since the existence of undescribed physiological types of archaea in addition to extremophiles and methanogens in marine plankton was demonstrated in 1992 [1, 2], subsequent studies have characterized the archaeal abundance and diversity in the overall biosphere, suggesting that archaeal species have significant impacts on carbon and nitrogen cycling [3]. In addition to traditional cultivation methods, culture-independent metagenomic analyses, particularly those based on high-throughput sequencing, are now feasible methods to obtain genomic sequences of uncultivated archaea that thrive in various environments [4, 5]. In fact, the complete genomes of uncultivated archaea, including those within candidate phyla, have been recovered solely from metagenomic sequences [6–8]. Another culture-independent approach to identify genomes from uncultivated microbes is single-cell genomics, which involves the amplification and sequencing of DNA from single cells collected directly from terrestrial and marine environmental samples [9–12].
We obtained the complete genome of an uncultivated archaeal species in the candidate phylum, Hot Water Crenarchaeotic Group I, from the metagenomes of a microbial mat community that flourishes along a subsurface geothermal stream [13–15]. The phylogenetic position of the microbe based on this composite genome was similar to that of thaumarchaeotic species such as Nitrosopumilus maritimus and Cenarchaeum symbiosum. However, we proposed that this crenarchaeotic group should be considered a novel phylum, Aigarchaeota, owing to its unique genomic traits that were distinct from those of known phyla and candidate divisions such as Crenarchaeota, Euryarchaeota, “Korarchaeota”, Nanoarchaeota, and Thaumarchaeota; accordingly, we named the microbe Candidatus Caldiarchaeum subterraneum (Ca. C. subterraneum) [14].
A primary objective of genomic analyses of uncultivated Ca. C. subterraneum is to deduce its lifestyle on the basis of potential comprehensive functions (functionome) of genes harbored in its genome. We performed a detailed genomic analysis of its metabolic and physiological features, but it was difficult to capture its overall functional potential and to compare it with that of other archaea. The functional categories defined by the Kyoto Encyclopedia of Genes and Genomes (KEGG) [16] and SEED [17] databases are extremely broad with respect to metabolic and physiological features. Additionally, it is difficult to automatically annotate each function solely by assigning ortholog identifiers (IDs). However, KEGG provides an easy method for the computational annotation and characterization of more detailed individual functions using four types of functional modules: pathway, molecular complex, functional set, and signature modules [18]. These modules comprise small units of subpathways and multiple molecules such as the subunits of transporters and receptors, which encompass broad functional categories, including energy metabolism, carbohydrate and lipid metabolism, and nucleotide and amino acid metabolism. Previously, we developed a method to evaluate the potential functionome by calculating the module completion ratios (MCRs) for KEGG modules [19]. We also developed a metabolic and physiological potential evaluator (MAPLE) to automate a series of steps used in this method [20]. In the present study, we applied the MAPLE system (http://www.genome.jp/) to the functional classification of archaea and characterized the potential functionome of the Ca. C. subterraneum genome.
In this study, we performed clustering and statistical analyses to determine functional classifications based on the MCR patterns of 79 archaeal species, including the uncultivated Ca. C. subterraneum. We also determined that the metabolic potential of Ca. C. sunterraneum based on overall MCR patters was similar to that of thaumarchaeotic species.
Materials and Methods
Phylogenetic analysis based on common orthologous genes
The orthologous relationships between each Ca. C. subterraneum gene and the genes of other prokaryotic species were identified using the DomClust program [21] based on the all-against-all similarities between Ca. C. subterraneum and 143 prokaryotic species (141 archaeal species and two bacterial species, as described in S1 Table) from the MBGD database [22]. Similarly, the orthologous relationships between Ca. C. subterraneum and 149 other organisms (136 archaeal species and 13 unicellular eukaryotic species, as listed in S1 Table) were identified. The resulting ortholog table was visualized and analyzed using the RECOG system (http://mbgd.genome.ad.jp/RECOG/). To determine the phylogenetic position of Ca. C. subterraneum among archaeal species, phylogenetic analyses were performed using a concatenated alignment of single-copy protein-coding genes without domain splitting, which were selected using the MBGD database. Twenty-two orthologous families conserved in 142 archaea and two bacteria, or 12 orthologous families conserved in 137 archaea and 13 unicellular eukaryotes were used for each phylogenetic analysis. The sequences of genes in these families were aligned using MUSCLE [23] with the parameter "—maxiterate 1000" and the resulting alignments were processed using the Gblocks program [24] with the default settings to eliminate poorly aligned positions. The processed alignments were concatenated and subjected to a phylogenetic analysis. The phylogenetic position of Ca. C. subterraneum was inferred by MEGA6 using the maximum-likelihood method [25]. The substitution model LG+ had the lowest Akaike information criterion (or, AIC) value and was selected as the best-fit model by ProtTest 2.4 [26]. According to this result, the LG+G model was employed to produce the maximum-likelihood tree (1,000 bootstrap replicates using the LG model with gamma distributed rates, partial deletion of gaps, and a site coverage cutoff of 95%).
Calculation of the MCR and pathway analysis
MCRs of all KEGG functional modules (228 pathways, 271 molecular complexes, 86 functional sets, and 8 signature modules) in each type of archaea were calculated based on a Boolean algebra-like equation [19] using the MAPLE system [20]. MAPLE is an automatic system that is used to map genes in an individual genome and metagenome to functional modules defined by KEGG. It calculates the MCR, the percentage of a module component filled with input genes, as follows: it assigns a KEGG orthology (KO) ID to the query genes using the KEGG Automatic Annotation Server (KAAS), maps the KO-assigned genes to the KEGG functional modules, and finally calculates an MCR for each functional module. If all genes are assigned to all KO IDs in each reaction according to the Boolean algebra-like equation, the MCR is 100%. For this analysis, one genome was selected from each archaeal genus (a total of 79 genomes, including Ca. C. subterraneum) listed in S2 Table and a reference genome set was constructed to cover all of the completely sequenced prokaryotes (as of August 2, 2014). When the type species for each genus was registered in the KEGG GENOME database, it was selected as a representative in this study; if not, the first registered species for each genus was selected as a representative. If there were multiple genomic sequences for a single species, the earliest registered sequence was selected. The KEGG pathway [27] and module databases were used for pathway construction [18].
Clustering and PCA analyses based on the MCR pattern
To characterize the overall MCR pattern of Ca. C. subterraneum based on comparisons with those of the other 78 archaea, the pairwise Euclidean distances between the overall MCR patterns for each archaea were used and the complete-linkage clustering method was employed for the functional classification of 79 archaea using an R statistical package [28]. KEGG modules with the same MCR values ranging from 0% to 100% in all archaea, including Ca. C. subterraneum, were excluded from this analysis. Similar to the clustering analysis, the overall MCR patterns of Ca. C. subterraneum and the other 78 archaea, except for the modules with no differences in the MCR values among all archaea, were subjected to a principal components analysis (PCA). PCA was performed using the R statistical environment.
Results
Phylogenetic position of Ca. C. subterraneum among archaea
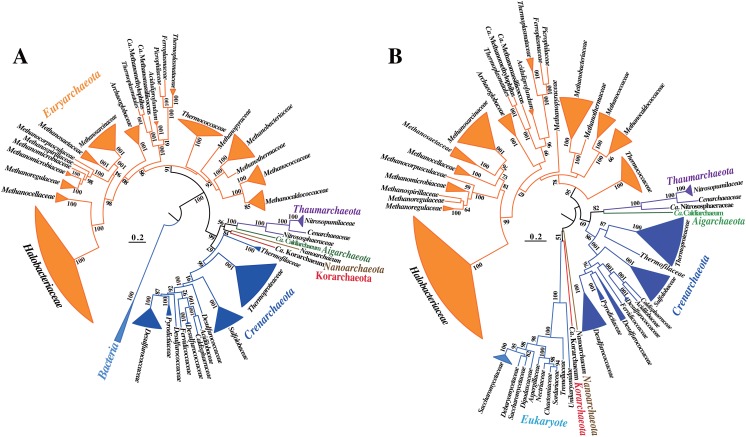
To confirm the phylogenetic position of the thermophile Ca. C. subterraneum among archaea, we conducted a genome-wide phylogenetic analysis using the maximum-likelihood method with the LG+G substitution model, which was not used in the previous study [14] and we obtained a concatenated alignment of 22 broadly conserved proteins among 79 archaea, including Ca. C. subterraneum and two representative bacterial species (Escherichia coli and Bacillus subtilis) as an outgroup (S1 Table). The phylogenetic tree demonstrated that Ca. C. subterraneum formed a cluster with five species within Thaumarchaeota (Fig 1A). The thermophile “Candidatus Nitrososphaera gargensis” (Ca. N. gargensis) [29, 30] was somewhat distantly related to other mesophilic thaumarchaeotic species; species within the families Nitrosopumilaceae and Cenoarchaeaceae comprised a sub-cluster. Thus, Ca. C. subterraneum was closely related to the thermophile Ca. N. gargensis based on this tree (Fig 1A). We constructed another phylogenetic tree using the concatenated alignment of 12 broadly conserved genes among 137 archaea, including Ca. C. subterraneum, and 13 unicellular eukaryotes as the outgroup. The topology was similar to that of the former tree except for the phylogenetic position of the species within “Korarchaeota” and Nanoarchaeota, but there was no difference in the phylogenetic relationships between Ca. C. subterraneum and thaumarchaeotic species (Fig 1B). “Korarchaeota” formed a cluster with the eukaryotic cluster, which comprised several sub-clusters, but Nanoarchaeota was independent in this tree, although these two species formed a cluster in the previous tree.
Fig 1. Phylogenetic position of “Candidatus Caldiarchaeum subterraneum” based on concatenated common protein sequences.
A maximum-likelihood tree was constructed using MEGA [25]. The phylogenetic trees were collapsed at the family level except for Nanoarchaeum, “Ca. Korarchaeum,” and “Ca. Caldiarchaeum.” (A) Concatenated alignment of the sequences of 22 common proteins with no paralogs and domain splitting (hisS, pheS, valS, rpl11p, rpl14p, rpl1P, rpl22p, rpl2p, rpl3p, rpl5p, rpl6p, rplW, rps10p, rps11p, rps19p, rps2P, rps3p, rps4p, rps5p, rps7p, rps8p, and rps9p) among 142 archaea, including “Ca. A. autotrophicum” and two bacteria as an outgroup, as listed in S1 Table. (B) Concatenated alignment of the sequences of 12 common protein sequences without paralogs and domain splitting (eif2G, rfcL, eif6, rpl10e, rpl15p, rpl18p, rpl7ae, rplP0, rps19p, rps3p, rps5p, and rio1) among 137 archaea, including “Ca. C. subterraneum” and 13 unicellular eukaryotes as the outgroup (S1 Table). The numbers indicate the bootstrap support expressed as percentages. Bootstrap values of less than 50% were omitted.
Functional classification of archaea based on the MCR patterns of the KEGG modules
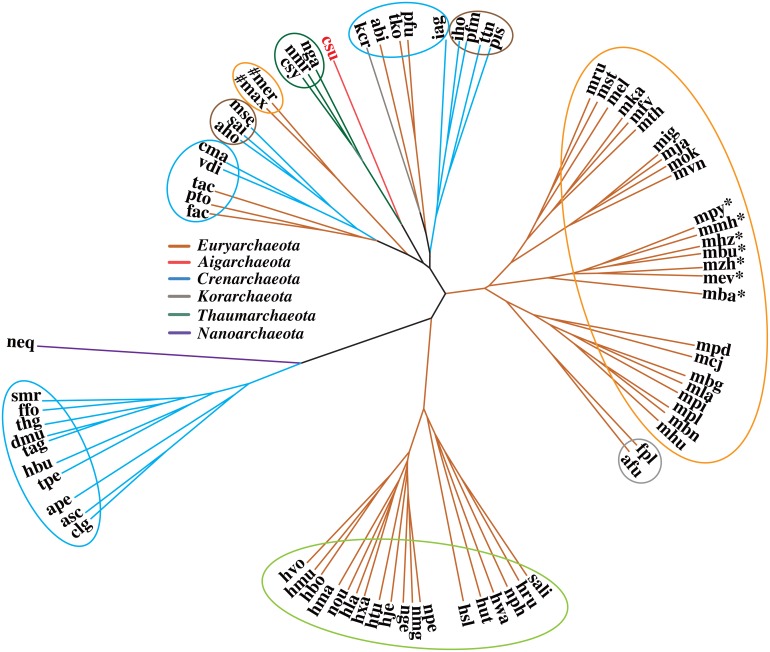
The MCR of all KEGG modules in 79 archaea, including Ca. C. subterraneum, was calculated to evaluate their metabolic and physiological potential using the MAPLE system [20] (S1 Fig and S3 Table). The MCR patterns of 273 modules, except those with identical MCR values, among 79 archaea were subjected to a clustering analysis to obtain functional classifications of archaea. Three thaumarchaeotic species formed a small cluster with Ca. C. subterraneum, suggesting similar overall metabolic and physiological potential (Fig 2). Thaumarchaeotic species have been known as chemolithoautotrophic ammonia oxidizers, but recent marine isolates, Nitrosopumilus maritimus strains PS0 and HCA1, display obligate mixotrophy [31]. Thus, Ca. C. subterraneum is similar to thaumarchaeotic species with respect to not only phylogeny, but also metabolic and physiological potential. Species within Euryarchaeota were divided into two major clusters, i.e., halophiles and methanogens, and small clusters such as hyperthermophilic sulfur reducers and hyperthermophilic acidophiles or neutrophiles. The methanogens isolated from environmental samples such as terrestrial soils and a hydrothermal vent formed a large cluster with three sub-clusters. However, interestingly, two other methanogens, “Ca. Methanomassiliicoccus intestinalis” [32] and “Ca. Methanomethylophilus alvus” [33], isolated from human feces, formed an independent cluster, although the two species were distantly related based on the phylogeny (Fig 2). This result indicates that the difference in habitat clearly reflects the overall difference in metabolic and physiological potential, whereas methanogenesis is a common phenotypic property. Both species are only able to generate methane from methanol, while all other species can generate methane from CO2. In addition, seven species that formed another cluster, Methanosarcina barkeri, Methanococcoides burtonii, Methanohalophilus mahii, Methanohalobium evestigatum, Methanosalsum zhilinae, Methanolobus psychrophilus, and Methanomethylovorans hollandica, have the ability to generate methane from all 4 possible substrates, CO2, acetate, methanol, and methylamine (Fig 2). Species within Crenarchaeota were divided into three clusters and a major cluster comprised of heterotrophic (hyper)-thermophilic acidophiles or neutrophiles. The members that formed this cluster were sulfur compound reducers, except Fervidicoccus fontis [34] and Thermosphaera aggregans [35]. Three other crenarchaeotic species with similar phenotypic properties to the members that formed the major cluster were spread between two different clusters comprising chemolithoautotrophic or mixotrophic (hyper)-thermophilic acidophiles.
Fig 2. Functional classification of archaea based on the MCR patterns of the KEGG modules.
csu, Ca. Caldiarchaeum subterraneum; nga, Ca. Nitrososphaera gargensis; nmr, Nitrosopumilus maritimus; csy, Cenarchaeum symbiosum. Full species names for all other abbreviations are listed in S2 Table. Orange circle, methanogens; yellow-green circle, halophiles; gray circle, hyperthermophilic facultative autotrophic sulfur reducers; blue circle, heterotrophic (hyper)-thermophilic acidophiles or neutrophiles; brown circle, chemolithoautotrophic or facultative autotrophic (hyper)-thermophilic acidophiles; green circle, facultative autotrophic ammonia oxidizers. *, methanogens, which can generate methane from all possible substrates (CO2, acetate, methanol, and methylamine); #, methanogens isolated from the human gut, which can generate methane only from methanol.
PCA of the MCR patterns
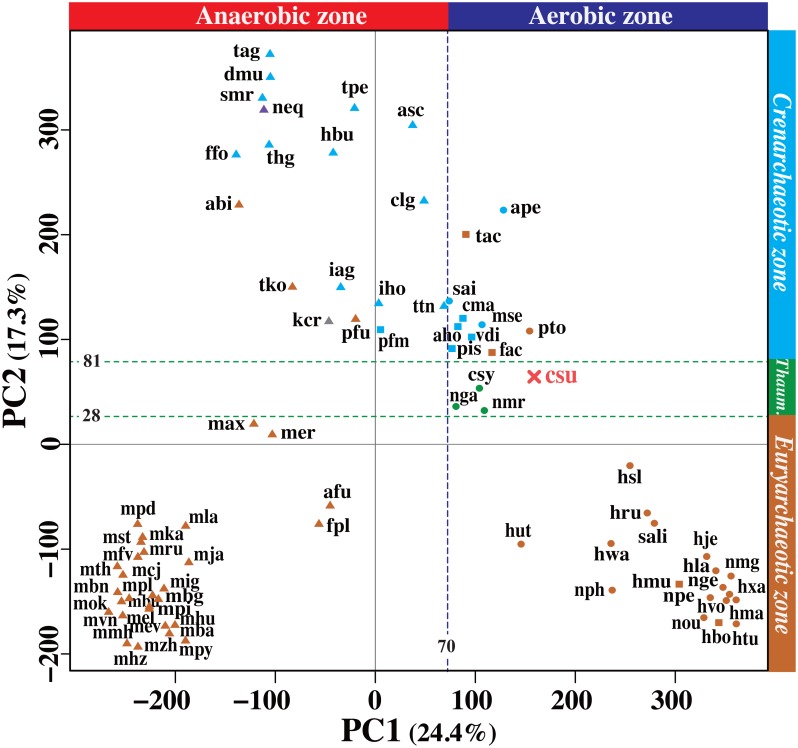
Hierarchical clustering (Fig 2) provides an intuitive means to visualize a classification; however, it cannot explain the dominant factors (i.e., reaction modules, in this study) that determine the clusters owing to limitations of the method. Thus, we performed a PCA, which explains the main factors determining the clusters by avoiding problems that result from multicollinearity (i.e., overlap among modules). The overall physiological features that characterize aerobes and anaerobes were identified according to the preliminary PCA of the MCR patterns of the archaeal species. In this analysis, the first principal component (PC1) was correlated with the oxygen requirement for growth, so that anaerobes and aerobes could be distinguished along the first axis. The second PC (PC2) was correlated with the phylogenetic classification at the phylum level, such as Crenarchaeota, Thaumarchaeota, and Euryarchaeota. Thus, we used the PCA to confirm whether Ca. C. subterraneum possessed a signature similar to that of other aerobes as well as phylogenetically related species. As shown in Fig 3, all aerobic archaeal species, including facultative anaerobes, were located to the right-hand side of the border (ca. 70), which discriminated aerobes from anaerobes, except for Pyrolobus fumarii [36], a facultative microaerophile. Ca. C. subterraneum was positioned at the right-hand side of the border along with the aerobes Picrophilus torridus [37], Aeropyrum pernix [38], and Halorhabdus utahensis [39]. KEGG modules such as methanogenesis (M00356), pyruvate:ferredoxin oxidoreductase (M00310), succinate dehydrogenase (M00149), and cytochrome c oxidase (M00155) contributed to the variance along the first principal axis, although the factor loading of each module was small because the analysis included over 270 modules (S2A Fig).
Fig 3. Distribution of “Ca. C. subterraneum” along the first and second PCA axes based on MCR patterns.
Obligate and facultative anaerobes are denoted by triangles and squares, respectively. Aerobes are denoted by circles. X indicates uncultivated species. Light brown, Euryarchaeota; light blue, Crenarchaeota; green, Thaumarchaeota; gray, “Korarchaeota”; purple, Nanoarchaeota; csu, “Ca. Caldiarchaeum subterraneum”. Full species names for all other abbreviations are listed in S2 Table. MCR patterns of 273 modules (pathway: 161, structural complex: 101, and functional set: 11), except for those with the same MCR values, among 79 archaea were subjected to this analysis.
Most of the euryarchaeotic species were located in a euryarchaeotic zone along the second axis, except for several (hyper)-thermophilic heterotrophs. These exceptional euryarchaeotic species were mixed within the crenarchaeotic zone, together with Nanoarchaeum equitans [40] and “Ca. Korarchaeum cryptofilum” [9]. Consequently, Ca. C. subterraneum was located in the thaumarchaeotic zone (ca. 28–81). In this case, modules, such as the archaeal exosome (M00390), which was identified in archaea, except for the halophiles and half of the methanogens, and DNA polymerase II complex (M00261), were not identified in Crenarchaeota and contributed to the variance in PC2, as shown in S2B Fig. These results demonstrate that Ca. C. subterraneum is probably an aerobe that is similar to thaumarchaeotic species in overall metabolic and physiological potential; however, more careful examinations of these properties are required owing to the low percentages of variance explained (see Discussion).
MCR patterns involved in carbon fixation
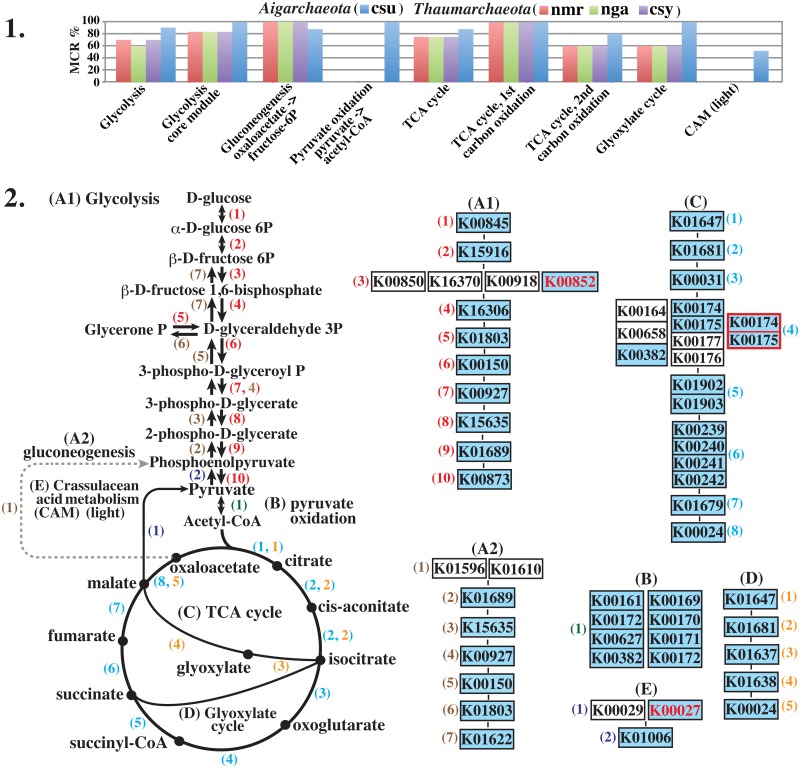
The results of our analyses suggested that Ca. C. subterraneum is similar to thaumarchaeotic species, both phylogenetically and in terms of its overall metabolic and physiological potential. Thus, we focused on the MCR patterns of six types of carbon fixation modules in Ca. C. subterraneum and thaumarchaeotic species, i.e., Nitrosopumilus maritimus [41], Cenarchaeum symbiosum [42], and “Ca. Nitrososphaera gargensis” [29], which fix carbon in the hydroxypropionate–hydroxybutyrate (HP-HB) cycle [43, 44]. We found that Ca. C. subterraneum could not complete any of the carbon fixation modules and the MCR values were very low except for the reductive tricarboxylic acid (TCA) cycle (M00173) (Fig 4-1). Apparently, Ca. C. subterraneum had a high completion ratio of more than 90% for the reductive TCA cycle module, but the most important reaction step (steps 11_1 or 11_2 and 12) catalyzed by ATP-citrate lyase [45] or citryl-CoA synthetase and citryl-CoA lyase, which are the key enzymes in this cycle [46, 47], was not complete (Fig 4-2-A). The MCR of the reductive TCA cycle sometimes has a high value in other organisms because the enzymes used by this cycle are shared among many other pathway modules. For example, the enzymes used for steps 4 to 10 of the reductive TCA cycle (Fig 4-2-A) are shared in all steps of the TCA cycle, except for citrate synthase involved in step 1 (Fig 5-2-C). In fact, Ca. C. subterraneum possesses enzymes related to all reaction steps shared between both cycles. If Ca. C. subterraneum harbors different types of key enzymes for the reductive TCA cycle, it could perform carbon fixation, but it is parsimonious to infer that it has an extremely poor carbon fixation potential. The thaumarchaeotic species were also unable to complete any of the carbon fixation modules, but are known to possess the HP-HB cycle for carbon fixation (Fig 4-2B). The HP-HB cycle is not complete in thaumarchaeotic species because the genes responsible for 5 reaction steps (steps 2, 3, 6, 10, and 11), which are common to both crenarchaeotic and thaumarchaeotic HP-HB cycles, have not yet been identified in their genomes using bioinformatics methods [44]. The K numbers corresponding to the enzymes responsible for steps 10 (K18601: succinyl-CoA reductase) and 11 (K18602: succinate semialdehyde reductase) are assigned to the Nmar_1608 gene, and the paralogous Nmar_1110 and Nmar_161 genes, respectively, in the KEGG database; however, these K numbers were incorrectly assigned by KEGG. Recently, it has been experimentally confirmed that Nmar_1110 is malonic semialdehyde reductase, responsible for step 3 [48]. This misannotation by KEGG should be corrected immediately. The thaumarchaeotic HP-HB cycle, which is thought to have arisen independently in thaumarchaeal and crenarchaeal lineages by convergent evolution, is the most energy-efficient aerobic pathway for CO2 fixation, and the activities of all enzymes employed by the HP-HB cycle, which comprises 15 reaction steps, have been confirmed experimentally [44]. Thus, the remaining 4 unidentified genes that did not show significant homology to crenarchaeotic enzymes should be present in their genomes. Almost no enzymes for this cycle were identified bioinformatically in the Ca. C. subterraneum genome, but the overall MCR patterns for other carbon fixation modules were similar to those of thaumarchaeotic species (S1 Fig and S3 Table).
Fig 4. Comparison of MCR patterns for carbon fixation in “Ca. C. subterraneum” and three thaumarchaeotic species.
1. MCR patterns of six major carbon fixation pathways. 3–HP bicycle, 3–hydroxypropionate bicycle; HP–HB cycle, hydroxypropionate–hydroxybutyrate cycle; dicarboxylate HB cycle, dicarboxylate hydroxybutyrate cycle; 2, Mapping patterns of genes to two carbon fixation modules. Numbers in parentheses show the order of reaction steps in each module. In each “K number” set of components of the module, the vertically connected and horizontally located K numbers indicate complexes and alternatives, respectively [19]. (A) Reductive TCA cycle (M00173). *Specific enzymes for this pathway; §found only in Cenarchaeum symbiosum. (B) HP–HB cycle (M00375). Numbers in red show the reaction steps for which the corresponding genes were not identified in the genomes of the three thaumarchaeotic species, although their enzymatic activities have been detected experimentally [44]. *The gene responsible for step 3 has been experimentally identified in the N. maritimus genome very recently, whereas the K number has not yet been assigned to the gene (Nmar_1110) by KEGG [48]. **Because the K numbers were incorrectly assigned to the genes of thaumarchaeotic species, the genes responsible for steps 10 and 11 have not yet been identified in their genomes.
Fig 5. Major central carbohydrate metabolism in “Ca. C. subterraneum.”.
1. Comparison of the MCR patterns between aigarchaeotic and thaumarchaeotic species. csu, Ca. Caldiarchaeum subterraneum; nmr, Nitrosopumilus maritimus; nga, Ca. Nitrososphaera gargensis; csy, Cenarchaeum symbiosum. 2. Pathway map for central carbohydrate metabolism (left side) and mapping pattern of the modules corresponding to the pathway map in Ca. C. subterraneum (right side). In each “K number” set of components of the module, the vertically connected and horizontally located K numbers indicate complexes and alternatives, respectively [19]. K numbers in blue boxes indicate the orthologous genes identified in Ca. C. subterraneum. Grayish dashed lines and arrows show missing reaction steps in the pathway. Numbers in parentheses on each module component correspond to those on the pathway map. A1, glycolysis (M0001): (1), glucokinase; (2), glucose/mannose-6-phosphate isomerase; (3), 6-phosphofructokinase or ADP-dependent phosphofructokinase/glucokinase; (4), fructose-bisphosphate aldolase; (5), triosephosphate isomerase; (6), glyceraldehyde-3-phosphate dehydrogenase; (7), phosphoglycerate kinase; (8), 2,3-bisphosphoglycerate-independent phosphoglycerate mutase; (9), enolase; (10), pyruvate kinase. A2, gluconeogenesis (M0003): (1), ADP-dependent phosphofructokinase/glucokinase; (2), enolase; (3), 2,3-bisphosphoglycerate- independent phosphoglycerate mutase; (4), phosphoglycerate kinase; (5), glyceraldehyde-3-phosphate dehydrogenase, (6), triosephosphate isomerase; (7), fructose 1,6-bisphosphate aldolase/phosphatase. B, pyruvate oxidation (M00307): (1), pyruvate dehydrogenase. C, TCA (Krebs) cycle (M0009): (1), citrate synthase; (2), aconitate hydratase; (3), isocitrate dehydrogenase; (4), 2-oxoglutarate dehydrogenase; (5), succinyl-CoA synthetase; (6), succinate dehydrogenase; (7), fumarate hydratase, class II; (8), malate dehydrogenase. D, glyoxylate cycle (M00012): (1), citrate synthase; (2), aconitate hydratase; (3), isocitrate lyase; (4), malate synthase; (5), malate dehydrogenase. M number shows identifier of each KEGG module. E, Crassulacean acid metabolism–light (M00169): (1), malate dehydrogenase; (2), pyruvate, orthophosphate dikinase. K numbers shown in red have not been assigned to the KEGG module, but similar enzymatic reactions to the K numbers assigned to the module have been confirmed. The reaction component shown by the red frame in the TCA cycle shows αβ-heterodimeric 2-oxoacid:ferredoxin oxidoreductase [55, 56], which is not yet reflected in the KEGG module.
MCR patterns involved in central metabolism and transporters
The carbon fixation potential of Ca. C. subterraneum was extremely poor despite previous suggestions that it has an autotrophic lifestyle [3, 13]. Thus, we analyzed the MCR patterns involved in central carbohydrate metabolism as well as the transporters related to sugar and amino acid transport, and compared the patterns with those of thaumarchaeotic species (S3 Fig and Fig 5-1). Ca. C. subterraneum did not complete the KEGG module for the Embden–Meyerhof (EM) pathway comprising 10 reaction steps because two enzymes responsible for step 3, ATP-dependent 6-phosphofructokinase (ATP-PFK) (K00850 or K16370) and ADP-dependent phosphofructokinase/glucokinase (K00918), were not identified in the genome (Fig 5-2-A1). However, another type of ATP-PFK that shows no sequence similarity to classical ATP-PFKs has been purified and characterized in the two crenarchaeotic species Aeropyrum pernix and Desulfurococcus amylolyticus [49–51]. Both enzymes are in the PFK-B family within the ribokinase superfamily based on sequence similarities. In the KEGG database, the orthologous group containing ATP-PFK from A. pernix (APE_0012) belonging to the PFK-B family is defined as ribokinase (EC: 2.7.1.15) and the different K number of K00852 is assigned to this orthologous group. At present, K00852 has not yet been assigned to the module for the EM pathway, but it should be assigned to reaction step 3 in this module together with currently assigned KOs. In the Ca. C. subterraneum genome, there are two K00852-assigned genes, CSUB_C0883 and CSUB_C1035, and APE_0012 and CSUB_C1035 share 74.4% similarity (ca. 30% identity). Thus, the gene product of CSUB_C1035 presumably acts as ATP-PFK, responsible for reaction step 3.
Similar to the enzymes involve in the reductive TCA cycle and TCA cycle, the enzymes used for 4 to 9 reaction steps in the EM pathway modules are shared with those involved in reaction steps 2 to 7 of the gluconeogenesis module (Fig 5-2-A2). Actually, Ca. C. subterraneum completed the gluconeogenesis module with a high MCR of 85.7%, although phosphoenolpyruvate carboxykinase, which is responsible for reaction step 1, was not identified. Ca. C. subterraneum possesses genes encoding malate dehydrogenase: oxaloacetate-decarboxylating (CSUB_C1205) and pyruvate orthophosphate dikinase (CSUB_C1231), which enable the conversion of malate to phosphoenolpyruvate via pyruvate. A series of reactions have been defined as the KEGG module for crassulacean acid metabolism (CAM)-light (M00169), comprising 2 reaction steps, and K00029 and K01006 were assigned to each step as shown in Fig 5E. The gene product of CSUB_C1205 shows ca. 85% similarity (56% identity) to the K00029-assigned gene (SSO_2869) from Sulfolobus solfataricus [52], but a different K number, K00027, is assigned to this gene in KEGG database. Accordingly Ca. C. subterraneum cannot complete the current M00169 module, to which K00027 is not assigned for unclear reasons. One of two specific enzymes in the module for gluconeogenesis, fructose 1,6-bisphosphate aldolase/phosphatase, was identified in both Ca. C. subterraneum and thaumarchaeotic species [53, 54]. Ca. C. subterraneum also completed the pyruvate oxidation module connected to the TCA cycle.
All of the thaumarchaeotic species showed a high MCR value for the EM pathway because they completed the gluconeogenesis modules. Two of three thaumarchaeotic species, N. maritimus and C. symbiosum, completed the EM pathway with an MCR of 70%, but lacked glucokinase (step 1) and pyruvate kinase (step 10) as well as the enzyme (6-phosphofructokinase or ADP-dependent phosphofructokinase/glucokinase) responsible for step 3 (S3 Fig). However, thermophilic “Ca. N. gargensis” possessed an ATP-PFK within the PFK-B family showing 75% similarity (ca. 32% identity) to that of Ca. C. subterraneum, unlike the two other thaumarchaeotic species. Ca. C. subterraneum possesses at least three sets of multiple sugar and simple sugar ABC transporters (M00207 and M00221), while the thaumarchaeotic species possess no sugar transporters as shown in Fig 6. Thus, this organism presumably has the ability to metabolize the sugars taken up by these transporters via the EM pathway.
Fig 6. Sugar and blanched chain amino acid (BCAA) ABC transporters module abundance in Ca. C. subterraneum and thaumarchaeotic species.
K numbers in blue boxes indicate the orthologous genes identified in each species. csu, Ca. Caldiarchaeum subterraneum; nmr, Nitrosopumilus maritimus; nga, Ca. Nitrososphaera gargensis; csy, Cenarchaeum symbiosum. Grayish characters indicate the species with no gene set in the module. M numbers indicate the module IDs defined by KEGG.
Ca. C. subterraneum did not complete the current KEGG module for the TCA cycle, which comprises eight reaction steps; the genes encoding the γ (K00177) and δ (K00176) subunits of 2-oxoglutarate ferredoxin oxidoreductase (step 3) were not identified in the genome, but those for the α (K00174) and β (K00175) subunits were identified. Another enzyme type with a different subunit composition, such as an αβ (heterodimer), α2β2 (dimer of heterodimer), and α2 homodimer, has been identified [55–58]. However, these reaction components are not yet connected to step 4 in the current KEGG module; accordingly, this module should be improved to enable more precise bioinformatic characterization of archaeal metabolism. The gene products of CSUB_C1635 (α subunit) and CSUB_C1634 (β subunit) in the Ca. C. subterraneum genome show sequence similarity of more than 84% to each subunit of the αβ-type enzyme from Sulfolobus tokodaii strain 7 [55]. Thus, the αβ-type heterodimeric enzyme from Ca. C. subterraneum is thought to function in reaction step 4 of the TCA cycle module, similar to in S. tokodaii. In addition, Ca. C. subterraneum possesses the glyoxylate cycle to bypass the decarboxylation steps in the TCA cycle. As mentioned above, because this organism also possesses malate dehydrogenase (CSUB_C1205) and pyruvate orthophosphate dikinase (CSUB_C1231), which enable the conversion of malate to phosphoenolpyruvate, this cycle connected to the gluconeogenesis pathway is thought to be involved in the production of cell components. In contrast to Ca. C. subterraneum, thaumarchaeotic species possess almost none of the key enzymes responsible for the glyoxylate cycle (S3 Fig).
Ca. C. subterraneum possesses five sets of blanched chain amino acids (BCAA), such as leucine, isoleucine, and valine ABC transporters, whereas among the thaumarchaeotic species, C. symbiosum possesses only one set (Fig 6). Except for valine, BCAAs are converted to acetyl-CoA via the degradation pathways identified in many bacteria and eukaryotic species. The degradation pathways of leucine and isoleucine have not yet been identified in archaea, including Ca. C. subterraneum, although genes related to these pathways have been partially identified in their genomes. Thus, this organism may employ an unknown alternative pathway to convert these amino acids to acetyl-CoA. Moreover, a citrate transporter (CSUB_C0827), which is one of the metabolites in the TCA cycle that possibly stimulates growth, was identified in the Ca. C. subterraneum genome.
Discussion
A primary objective of genomic analyses is to deduce the potential functionome of individual organisms. Since the genome of uncultured Ferroplasma type II [59] from a natural acidophilic biofilm has been nearly complete for more than a decade, uncultivated archaeal genomes, even within candidate phyla, have been recovered by metagenomics or a hybrid method of metagenomics and single-cell genomics from various environmental samples [9–11, 14, 60]. However, evaluating the potential functionome remains difficult because standard methodology has not yet been established to extract functional features such as those related to metabolism, energy generation, and transportation systems. Thus, we developed a new method to evaluate the potential functionome by calculating the MCRs for KEGG modules [19]. Recently, we launched the MAPLE system to automate a series of steps used in this method [20]. In the present study, we applied this new method to the functional classification of archaeal species and we determined the functional characteristics of the previously uncultured Ca. C. subterraneum in a comparative analysis with other archaea based on MCR patterns.
MCR is an easy-to-understand measure to evaluate functional potential. Generally, there is a correlation between the completeness of a KEGG module and the likelihood that an organism can perform the physiological function corresponding to the module. However, when KOs used for a module are shared with other independent modules, i.e., the reductive TCA cycle (Fig 4) and TCA cycle (Fig 5), the MCR does not necessarily reflect the working probability of each functional module. Thus, the relationship between the MCR of the targeted module and others to which the same KOs are assigned and the contribution of the specific KOs for each module to the MCR should be considered when a module is not complete. In this study, the MCR patterns of 273 modules, regardless of the relationships between modules, were subjected to a PCA to characterize the overall physiological features of archaea. Consequently, 79 archaea were classified into two groups (aerobes and anaerobes) according to PC1 and into 3 major taxonomic groups, Euryarchaeota, Crenarchaeota, and Thaumarchaeota, according to PC2. These interpretations according to the loadings in PC1 and PC2 are biologically plausible (see Results). However, some exceptions were observed. For example, some euryarchaeal species belonged to the Crenarchaeota group. These outliers can be explained by the low percentages of variance along PC1 and PC2 (Fig 3). In general, a PCA provides understandable explanation by reducing high-dimensional features (i.e., the series of reaction modules, in this study) and by avoiding problems associated with multicollinearity, i.e., overlap between modules; however, such a reduction in dimensionality was not effective in this study owing to the data complexity. Thus, we are unable to provide a deeper discussion. The difficulty in interpreting the results is a known limitation of the PCA [61]. Accordingly, higher-level statistical analyses are necessary, such as the sparse PCA [61]. However, we did not use the sparse PCA owing to the higher computational cost. For a more detailed analysis, additional physiological properties (i.e., growth temperature and nutrient requirements) of archaea are also necessary for biologically meaningful interpretations of the principle components. In this study, the relationship between physiological/taxonomic properties and PCs was not comprehensively evaluated owing to the lack of physiological data for each species, excluding oxygen requirement. Thus, it is possible that oxygen requirement and taxonomic groups are not optimal descriptors of PC1 and PC2, respectively. Some data on physiological properties is available [62]; however, the collection of more physiological properties of archaeal species is important for future studies.
The genome of Ca. C. subterraneum was reconstructed from a metagenomic library obtained from a subsurface thermophilic microbial mat community in a 70°C hot water stream in a Japanese epithermal mine [14]. Because this hot water stream contains low levels of organic matter, the microbial mat community at the oxic–anoxic interface is considered to be supported by geological energy and carbon sources such as CO2 and CH4, which are supplied by the geological aquifer [63–65]. The microbial mat comprised at least 16 prokaryotic phylotypes, including phyla such as Chloroflexi, Proteobacteria, Deinococcus-Thermus, and Thaumarchaeota. However, Ca. C. subterraneum was dominant together with “Candidatus Acetothermus autotrophicum” (Ca. A. autotrophicum), the genomic sequence of which has almost been completed [15]. The genomic analysis of Ca. A. autotrophicum demonstrated its chemolithoautotrophic potential based on homo-acetogenesis via H2 and CO2, although the detailed mechanisms for energy generation still remain unclear. In contrast to Ca. A. autotrophicum, Ca. C. subterraneum has an extremely poor carbon fixation potential, but completed the modules responsible for major central carbohydrate metabolism. Ca. C. subterraneum also possesses all enzymes responsible for the glyoxylate cycle, which has been well studied in Escherichia coli [66]. E. coli can grow using the glyoxylate cycle, which is an anaplerotic pathway of the TCA cycle when acetate serves as the sole carbon source for growth. Therefore, Ca. C. subterraneum also may use acetate produced by acetogenic Ca. A. autotrophicum as a carbon source for growth in the microbial mat community. Moreover, Ca. C. subterraneum possesses multiple sets of ABC transporters required for the uptake of sugars and BCAAs, thereby supporting its heterotrophic lifestyle in the microbial mat community. In addition to Ca. A. autotrophicum, another potentially chemolithoautotrophic phylotype that is closely related to “Ca. Nitrosocaldus yellowstonii” (Thaumarchaeota), Hydrogenobacter thermophilus (Aquificae), Hydrogenophilus thermoluteolus (Betaproteobacteria), and a methanotrophic phylotype similar to Methylohalobius crimensis (Gammaproteobacteria), which probably use CO2 and CH4 supplied by the geothermal aquifer, coexist with Ca. C. subterraneum. However, the abundance of these phylotypes was low in the microbial mat community [15]. Given the abundance and diversity of these chemolithoautotrophic and methanotrophic phylotypes, they may serve as the primary energy and carbon source suppliers in the microbial mat community, and contribute to the primary production of the heterotrophic phylotypes, including Ca. C. subterraneum in the anaerobic organic-depleted state.
In conclusion, using the MAPLE system, the archaeal species registered in the KEGG GENOME database were classified into various groups that possess similar phenotypic properties, such as halophiles, methanogens, ammonia oxidizers, heterotrophic (hyper)-thermophilic acidophiles or neutrophiles, and hyperthermophilic facultative autotrophic sulfur reducers. Moreover, methanogens were classified into two major groups with different ecological niches, the human gut and other earth environments. The functional classification of previously uncultivated Ca. C. subterraneum using the MAPLE system and subsequent metabolic analyses based on the MCR patterns of individual KEGG modules, supported the predominantly aerobic nature of Ca. C. subterraneum, which lives in a subsurface thermophilic microbial mat community with a heterotrophic lifestyle. However, it is still challenging for the archaeal science community to elucidate metabolic features using bioinformatics methods because many of the unusual features of archaeal metabolism revealed to date are not reflected in the KEGG modules. Thus, the MAPLE system will be more helpful for characterizing archaeal metabolism, even in uncultured archaea, when the KEGG modules incorporate more archaeal metabolic features. Also, manual data curation and literature mining are still important strategies when implement MAPLE.
Supporting Information
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
We thank Prof. S. Goto of the Bioinformatics Center of Kyoto University, Japan and the staff of KEGG for providing useful information about the KEGG modules. We also thank Dr. H. Huang of Chuo University, Tokyo, Japan for his technical assistance.
Data Availability
All relevant data are within the paper and its Supporting Information files. All sequence data used in this study are available in NCBI database. MAPLE system used in this study is easily accessible from the GenomeNet (http://www.genome.jp/).
Funding Statement
This research was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan to H.T. (Nos. 26550053 and 20310124). This research was also supported in part by the collaborative research programs of the National Institute for Basic Biology (NIBB). Computational resources were provided partly by the Data Integration and Analysis Facility of NIBB. Mitsubishi Research Institute Inc. provided support in the form of salaries for T.T., but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific role of this author (T.T.) is articulated in the ‘author contributions’ section.
References
- 1. Fuhrman JA, McCallum K, Davis AA. Novel major archaebacterial group from marine plankton. Nature 1992; 356: 148–149. [DOI] [PubMed] [Google Scholar]
- 2. DeLong ED. Archaea in costal marine environments. Proc Natl Acad Sci USA 1992; 89: 5685–5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Offre P, Spang A, Schleper C. Archaea in biogeochemical cycles. Annu Rev Microbiol. 2013; 67: 437–457. 10.1146/annurev-micro-092412-155614 [DOI] [PubMed] [Google Scholar]
- 4. Hendelsman J. Metagenomics. Application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev. 2004; 68: 669–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hugenholtz P, Tyson GW. Metagenomics. Nature 2008; 455: 481–483. 10.1038/455481a [DOI] [PubMed] [Google Scholar]
- 6. Blainey PC. The future is now: single-cell genomics of bacteria and archaea. FEMS Microbial Rev. 2013; 37: 407–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stepanauskas R. Single cell genomics: an individual look at microbes. Curr Opin Microbiol. 2012; 15: 613–620. 10.1016/j.mib.2012.09.001 [DOI] [PubMed] [Google Scholar]
- 8. Baker BJ, Comolli LR, Dick GJ, Hauser LJ, Hyatt D, Dill BD, et al. Enigmatic, ultrasmall, uncultivated Archaea. Proc Natl Acad Sci USA 2010; 107: 8806–8811. 10.1073/pnas.0914470107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Elkins JG, Podar M, Graham DE, Makarova KS, Wolf Y, Randau L, et al. A korarchaeal genome reveals insights into the evolution of the Archaea. Proc Natl Acad Sci USA 2008; 105: 8102–8107. 10.1073/pnas.0801980105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Narasingarao P, Podell S, Ugalde JA, Brochier-Armanet C, Emerson JB, Brocks JJ, et al. De novo metagenomic assembly reveals abundant novel major lineage of Archaea in hypersaline microbial communities. ISME J. 2012; 6: 81–93. 10.1038/ismej.2011.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ghai R, Pašić L, Fernández AB, Martin-Cuadrado AB, Mizuno CM, McMahon KD, et al. New abundant microbial groups in aquatic hypersaline environments. Sci Rep. 2011; 1: 135 10.1038/srep00135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rinke C, Schwientek P, Sczyrba A, Ivanova N, Anderson I, Cheng JF, et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature 2013; 499: 431–437. 10.1038/nature12352 [DOI] [PubMed] [Google Scholar]
- 13. Nunoura T, Hirayama H, Takami H, Oida H, Nishi S, Shimamura S, et al. Genetic and functional properties of uncultivated thermophilic crenarchaeotes from a subsurface gold mine as revealed by analysis of genome fragments. Environ Microbiol. 2005; 7: 1967–1984. [DOI] [PubMed] [Google Scholar]
- 14. Nunoura T, Takaki Y, Kakuta J, Nishi S, Sugahara J, Kazama H, et al. Insights into the evolution of Archaea and eukaryotic protein modifier systems revealed by the genome of a novel archaeal group. Nucleic Acids Res. 2011; 39: 3204–3223. 10.1093/nar/gkq1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Takami H, Noguchi H, Takaki H, Uchiyama I, Toyoda A, Nishi S, et al. A deeply branching thermophilic bacterium with an ancient acetyl-CoA pathway dominates a subsurface ecosystem. PLoS ONE 2012; 7: e30559 10.1371/journal.pone.0030559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000; 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005; 33: 5691–5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, et al. KEGG for linking genomes to life and environment. Nucleic Acids Res. 2008; 36: D480–D484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Takami H, Taniguchi T, Moriya Y, Kuwahara T, Kanehisa M, Goto S. Evaluation method for the potential functionome harbored in the genome and metagenome. BMC Genomics 2012; 13: 699 10.1186/1471-2164-13-699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Takami H. New method for comparative functional genomics and metagenomics using KEGG module In: Nelson K, editor. Encyclopedia of metagenomics. Berlin, Heidelberg: Springer-Verlag; 2014; 525–539. [Google Scholar]
- 21. Uchiyama I. Hierarchical clustering algorithm for comprehensive orthologous-domain classification in multiple genomes. Nucleic Acids Res. 2006; 34: 647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Uchiyama I, Higuchi T, Kawai M. MBGD update 2010: toward a comprehensive resource for exploring microbial genome diversity. Nucleic Acids Res. 2010; 38: D361–D365. 10.1093/nar/gkp948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004; 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007; 56: 564–577. [DOI] [PubMed] [Google Scholar]
- 25. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013; 30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Abascal F, Zardoya R, Posada D. ProTest: selection of best-fit models of protein evolution. Bioinformatics 2005; 21: 2104–2105. [DOI] [PubMed] [Google Scholar]
- 27. Nikitin F, Rance B, Itoh M, Kanehisa M, Lisacek F. Using protein motif combinations to update KEGG pathway maps and orthologue tables. Genome Inform. 2004; 15: 266–275. [PubMed] [Google Scholar]
- 28. R Core Team. R: A language and environment for statistical computing. In: R foundation for statistical computing, Vienna, Austria: (http://www.r-project.org/) 2014. [Google Scholar]
- 29. Torre JR, Walker CB, Ingalls AE, Könneke M, Stahl DA. Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ Microbiol. 2008; 10: 810–818. 10.1111/j.1462-2920.2007.01506.x [DOI] [PubMed] [Google Scholar]
- 30. Spang A, Poehlein A, Offre P, Zumbrägel S, Halder S, Rychlik N, et al. The genome of the ammonia-oxidizing Candidatus Nitrososphaera gargensis: insights into metabolic versatility and environmental adaptations. Environ Microbiol. 2012; 14: 3123–3145. [DOI] [PubMed] [Google Scholar]
- 31. Qin W, Amin SA, Martens-Habbena W, Walker CB, Urakawa H, Devol AH, et al. Marine ammonia-oxidizing archaeal isolates display obligate mixotrophy and wide ecotypic variation. Proc Natl Acad Sci USA 2014; 111: 12504–12509. 10.1073/pnas.1324115111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Borrel G, Harris HMB, Parisot N, Gaci N, Tottey W, Mihajlovski A, et al. Genome sequence of “Candidatus Methanomassiliicoccus intestinalis” Issoire-Mx1, a third Thermoplasmatales-related methanogenic archaeon from human feces. Genome Announc. 2013; 1: e00453–13. 10.1128/genomeA.00453-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Borrel G, Harris HMB, Totteya W, Mihajlovskia A, Parisot N, Peyretaillade E, et al. Genome sequence of “Candidatus Methanomethylophilus alvus” Mx1201, a methanogenic archaeon from the human gut belonging to a seventh order of methanogens. J Bacteriol. 2012; 194: 6944–6945. 10.1128/JB.01867-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Perevalova AA, Bidzhieva SK, Kublanov IV, Hinrichs KU, Liu XL, Mardanov AV, et al. Fervidicoccus fontis gen. nov., sp. nov., an anaerobic, thermophilic crenarchaeote from terrestrial hot springs, and proposal of Fervidicoccaceae fam. nov. and Fervidicoccales ord. nov. Int J Syst Evol Microbiol. 2010; 60: 2082–2088. 10.1099/ijs.0.019042-0 [DOI] [PubMed] [Google Scholar]
- 35. Huber R, Dyba D, Huber H, Burggraf S, Rachel R. Sulfur-inhibited Thermosphaera aggregans sp. nov., a new genus of hyperthermophilic archaea isolated after its prediction from environmentally derived 16S rRNA sequences. Int J Syst Bacteriol. 1998; 48: 31–38. [DOI] [PubMed] [Google Scholar]
- 36. Blöchl E, Rachel R, Burggraf S, Hafenbradl D, Jannasch HW, Stetter KO. Pyrolobus fumarii, gen. and sp. nov., represents a novel group of archaea, extending the upper temperature limit for life to 113°C. Extremophiles 1997; 1: 14–21. [DOI] [PubMed] [Google Scholar]
- 37. Fütterer O, Angelov A, Liesegang H, Gottschalk G, Schleper C, Schepers B, et al. Genome sequence of Picrophilus torridus and its implications for life around pH 0. Proc Natl Acad Sci USA 2004; 101: 9091–9096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sako Y, Nomura N, Uchida A, Ishida Y, Morii H, Koga Y, et al. Aeropyrum pernix gen. nov., sp. nov., a novel aerobic hyperthermophilic archaeon growing at temperatures up to 100 degrees C. Int J Syst Bacteriol. 1996; 46: 1070–1077. [DOI] [PubMed] [Google Scholar]
- 39. Wainø M, Tindall BJ, Ingvorsen K. Halorhabdus utahensis gen. nov., sp. nov., an aerobic, extremely halophilic member of the Archaea from Great Salt Lake, Utah. Int J Syst Evol Microbiol. 2000; 50: 183–190. [DOI] [PubMed] [Google Scholar]
- 40. Waters E, Hohn MJ, Ahel I, Graham DE, Adams MD, Barnstead M, et al. The genome of Nanoarchaeum equitans: insights into early archaeal evolution and derived parasitism. Proc Natl Acad Sci USA 2003; 100: 12984–12988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Walker CB, de la Torre JR, Klotz MG, Urakawa H, Pinel N, Arp DJ, et al. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci USA 2010; 107: 8818–8823. 10.1073/pnas.0913533107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hallam SJ, Konstantinidis KT, Putnam N, Schleper C, Watanabe Y, Sugahara J, et al. Genomic analysis of the uncultivated marine creanarchaeote Cenarchaeum symbiosum . Proc Natl Acad Sci USA 2006; 103: 18296–18301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Berg IA, Kockelkorn D, Buckel W, Fuchs G. A 3-hydroxypropionate/ 4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in archaea. Science 2007; 318: 1782–1786. [DOI] [PubMed] [Google Scholar]
- 44. Könneke M, Schubert DM, Brown PC, Hügler M, Standfest S, Schwander T, et al. Ammonia-oxidizing archaea use the most energy-efficient aerobic pathway for CO2 fixation. Proc Natl Acad Sci USA 2014; 111: 8239–8244. 10.1073/pnas.1402028111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kanao T, Fukui H, Atomi H, Imanaka T. ATP-citrate lyase from the green sulfur bacterium Chlorobium limcola is a heteromeric enzyme composed of two distinct gene products. Eur J Biochem. 2001; 268: 1670–1678. [PubMed] [Google Scholar]
- 46. Aoshima M, Ishii M, Igarashi Y. A novel enzyme, citryl-CoA synthetase, catalysing the first step of the citrate cleavage reaction in Hydrogenobacter thermophileus TK-6. Mol Microbiol. 2004; 52: 751–761. [DOI] [PubMed] [Google Scholar]
- 47. Aoshima M, Ishii M, Igarashi Y. A novel enzyme, citryl-CoA lyase, catalysing the second step of the citrate cleavage reaction in Hydrogenobacter thermophileus TK-6. Mol Microbiol. 2004; 52: 763–770. [DOI] [PubMed] [Google Scholar]
- 48. Otte J, Mall A, Schubert DM, Könneke M, Berg A. Malonic semialdehyde reductase from the archaeon Nitrosopumilus maritimus is involved in the sutotrophic 3-hydroxypropionate/4-hydroxybutyrate cycle. Appl Environ Microbiol. 2015; 81: 1700–1707. 10.1128/AEM.03390-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hansen T, Schonheit P. Sequence, expression, and characterization of the first archaeal ATP-dependent 6-phosphofructokinase, a nonallosteric enzyme related to the phosphofructokinase-B sugar kinase family, from the hyperthermophilic crenarchaeote Aeropyrum pernix . Arch Microbiol. 2001; 177: 62–69. [DOI] [PubMed] [Google Scholar]
- 50. Hansen T, Schonheit P. Purification and properties of the first identified, archaeal, ATP-dependent 6-phosphofructokinase, an extremely thermophilic non-allosteric enzyme, from the hyperthermophile Desulfurococcus amylolyticus . Arch Microbiol. 2000; 173: 103–109. [DOI] [PubMed] [Google Scholar]
- 51. Ronimus RS, Morgan HW. The biochemical properties and phylogenies of phosphofructokinases from extremophiles. Extremophiles 2001; 5: 357–373. [DOI] [PubMed] [Google Scholar]
- 52. Bartolucci S, Rella R, Guagliardi A, Raia CA, Gambacorta A, Rosa MD, et al. Malic enzyme from archaebacterium Sulfolobus solfataricus . J Biol Chem. 1987; 262: 7725–7731. [PubMed] [Google Scholar]
- 53. Say RF, Fuchs G. Fructose 1,6-bisphosphate aldolase/phosphatase may be an ancestral gluconeogenesis enzyme. Nature 2010; 464: 1077–1081. 10.1038/nature08884 [DOI] [PubMed] [Google Scholar]
- 54. Bräsen C, Esser D, Rauch B, Siebers B. Carbohydrate metabolism in Archaea: Current insights into unusual enzymes and pathways and their regulation. Microbiol Mol Biol Rev. 2014; 78: 89–175. 10.1128/MMBR.00041-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Xhang Q, Iwasaki T, Wakagi T, Oshima T. 2-oxoacid:ferredoxin oxidereductase from the thermoacidphilic archaeon, Sulfolobus sp. strain 7. J Biochem. 1996; 120: 587–599. [DOI] [PubMed] [Google Scholar]
- 56. Yan Xhen, Fushinobu S, Wakagi T. Four Cys residues in heterodimeric 2-oxoacid:ferredoxin oxidereductase are required for CoA-dependent oxidative decarboxylation but not for a non-oxidative decarboxylation. Biochim Biophys Acta 2014; 1844: 736–743. 10.1016/j.bbapap.2014.01.015 [DOI] [PubMed] [Google Scholar]
- 57. Kerscher L, Oesterhelt D. Purification ans properties of two 2-oxoacid:ferredoxin oxidoreductases from Halobacterium halobium . Eur J Biochem. 1981; 116: 587–594. [DOI] [PubMed] [Google Scholar]
- 58. Lindblad A, Jansson J, Brostedt E, Johansson M, Hellman U, Nordlund S. Identification and sequence of a nifJ-like gene in Rhodospirillum rubrum: partial characterization of a mutant unaffected in nitrogen fixation. Mol Microbiol. 1996; 20: 559–568. [DOI] [PubMed] [Google Scholar]
- 59. Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 2004; 428: 37–43. [DOI] [PubMed] [Google Scholar]
- 60. Bakera BJ, Comollib LR, Dicka GJ, Hauserc LJ, Hyattc D, Dilld BD, et al. Enigmatic, ultrasmall, uncultivated Archaea. Proc Natl Acad Sci USA 2010; 107: 8806–8811. 10.1073/pnas.0914470107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zou H, Hastie T, Tibshirani R. Sparse principal component analysis, J Comp Graph Stat. 2006; 15: 265–286. [Google Scholar]
- 62. Takemoto K, Borjigin S. Metabolic network modularity in archaea depends on growth conditions. PLoS ONE 2011; 6: e25874 10.1371/journal.pone.0025874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Takai K, Hirayama H, Sakihama Y, Inagaki F, Yamato Y, Horikoshi K. Isolation and metabolic characteristics of previously uncultured members of the order Aquificales in a subsurface gold mine. Appl Environ Microbiol. 2002; 68: 3046–3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hirayama H, Takai K, Inagaki F, Yamato Y, Suzuki M, Nealson KH, et al. Bacterial community shifts along a subsurface geothermal water stream in a Japanese gold mine. Extremophiles 2005; 9: 169–184. [DOI] [PubMed] [Google Scholar]
- 65. Nishizawa M, Koba K, Makabe A, Yoshida N, Kaneko M, Hiraoe S, et al. Nitrification-driven forms of nitrogen metabolism in microbial mat communities thriving along an ammonium-enriched subsurface geothermal stream. Geochim Cosmochim Acta 2013; 113: 152–173. [Google Scholar]
- 66. Kornberg HL. The role and control of the glyoxylate cycle in Escherichia coli . Biochem J. 1966; 99: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. All sequence data used in this study are available in NCBI database. MAPLE system used in this study is easily accessible from the GenomeNet (http://www.genome.jp/).