Summary
The widespread use of chloroquine to treat Plasmodium falciparum infections has resulted in the selection and dissemination of variant haplotypes of the primary resistance determinant PfCRT. These haplotypes have encountered drug pressure and within-host competition with wild-type drug-sensitive parasites. To examine these selective forces in vitro, we genetically engineered P. falciparum to express geographically diverse PfCRT haplotypes. Variant alleles from the Philippines (PH1 and PH2, which differ solely by the C72S mutation) both conferred a moderate gain of chloroquine resistance and a reduction in growth rates in vitro. Of the two, PH2 showed higher IC50 values, contrasting with reduced growth. Furthermore, a highly mutated pfcrt allele from Cambodia (Cam734) conferred moderate chloroquine resistance and enhanced growth rates, when tested against wild-type pfcrt in co-culture competition assays. These three alleles mediated cross-resistance to amodiaquine, an antimalarial drug widely used in Africa. Each allele, along with the globally prevalent Dd2 and 7G8 alleles, rendered parasites more susceptible to lumefantrine, the partner drug used in the leading first-line artemisinin-based combination therapy. These data reveal ongoing region-specific evolution of PfCRT that impacts drug susceptibility and relative fitness in settings of mixed infections, and raise important considerations about optimal agents to treat chloroquine-resistant malaria.
Keywords: malaria, PfCRT, drug accumulation, chloroquine resistance, fitness, transfection
Introduction
In functionally constrained genes, the rise of non-synonymous mutations may decrease an organism’s fitness by steering it away from a long-optimized machinery of closely interacting components. Without a specific selective pressure, the more fit wild-type usually predominates at the population level and the less-fit variants are either eliminated or persist at low frequencies (Mitchell-Olds et al., 2007). An example of selective pressure is the use of drugs to treat human pathogens. As such, these constitute an ideal group in which to study the balance between surviving drug pressure and remaining competitive with wild-type organisms.
Generally, fitness costs associated with initial resistance-conferring mutations will lead to the attrition of mutants upon the removal of drug pressure, allowing for the reemergence of surviving wild-type organisms (Hastings and Donnelly, 2005). However, prolonged drug exposure can provide pathogens the opportunity to acquire additional mutations, either within the primary resistance determinant or within secondary factors, which compensate for the initial fitness cost (Brown et al., 2010). Competition between mutants carrying those compensatory mutations can then take place, leading to a mutant population with a reduced fitness cost that eventually can successfully compete with wild-type organisms in a drug-free environment (Levin et al., 2000). Fitness costs and the prevalence of initial resistance-conferring mutations can also be influenced by the use of other drugs to replace the failed therapeutic agent, particularly in instances where their efficacy is impacted by the same resistance determinants.
The malarial parasite Plasmodium falciparum, which caused an estimated 198 million clinical cases and 584,000 deaths in 2013 (WHO, 2014), is a prime example of a human pathogen that repeatedly encounters drug pressure and within-host competition among parasite strains. For much of the 20th century, the antimalarial treatment of choice was chloroquine (CQ), a drug characterized by its rapidity of action, safety and low cost. This drug prevents the detoxification of reactive iron-containing heme that is liberated as a result of hemoglobin proteolysis in the acidic digestive vacuole (DV) of intra-erythrocytic parasites (Fitch, 2004). CQ resistance (CQR) emerged slowly, but by the early 1990s had taken hold across virtually all the malaria-endemic world (Wellems and Plowe, 2001). The cellular basis for CQR has been attributed to reduced CQ accumulation in the DV, resulting in diminished access of CQ to its otherwise toxic heme target (Saliba et al., 1998).
At the molecular level, CQR has been traced primarily to amino acid changes in the DV transmembrane protein PfCRT (Fidock et al., 2000). These include K76T, ubiquitous among CQ-resistant strains and a highly sensitive marker of CQ treatment failure, as well as 3–8 additional PfCRT polymorphisms that produce region-specific haplotypes (Ecker et al., 2012). These haplotypes reflect a handful of origins of mutant pfcrt that disseminated under drug pressure in selective sweeps across the world (Nash et al., 2005; Kidgell et al., 2006; Mu et al., 2010; Park et al., 2012). At least 34 different mutant PfCRT haplotypes have been reported, contrasting with a conserved wild-type haplotype in CQ-sensitive parasites (Isozumi et al., 2010; Ecker et al., 2012; Baro et al., 2013). In Malawi, the prevalent African mutant form of PfCRT (CVIET haplotype at positions 72–76, found in strains including Dd2) largely disappeared within several years of CQ withdrawal, presumably due to a fitness cost that rendered this variant less competitive than pfcrt wild-type parasites in the absence of drug pressure (Kublin et al., 2003; Mita et al., 2003; Laufer et al., 2010).
The notion that mutation of PfCRT negatively impacts parasite fitness in field settings is supported by recent in vitro metabolomic and allelic competition investigations, which revealed a defect in hemoglobin catabolism and reduced relative growth rates in vitro, interpreted as a proxy of fitness costs for the CQ-resistant Dd2 and 7G8 PfCRT haplotypes (Lewis et al., 2014). Consistent with these findings, a fitness disadvantage was observed for CQ-resistant parasites during the dry season in The Gambia, when drug pressure is transiently removed and transmission is low (Ord et al., 2007). This fitness disadvantage was recapitulated in southern Zambia at the level of vectorial selection, whereby the wild-type (K76) form of PfCRT was significantly enriched in the infected Anopheles arabiensis mosquitoes compared to its baseline prevalence in the local infected human populations (Mharakurwa et al., 2013). Field studies in South America and Asia, however, have documented no or only modest attrition in mutant PfCRT forms, including 7G8 (SVMNT haplotype at positions 72–76), despite the discontinued use of CQ for the treatment of P. falciparum malaria for over two decades (Wang et al., 2005; Chen et al., 2008; Griffing et al., 2010). Those studies have led to the suggestion that the fitness cost of parasites harboring the 72SVMNT76 haplotype may be less severe than that of parasites carrying the 72CVIET76 signature (Sa and Twu, 2010).
Aside from regional differences in the choice of antimalarial drug regimens that may help sustain variant PfCRT haplotypes (Ecker et al., 2012), studies of human and murine parasites highlight a potential selective advantage of mutant pfcrt alleles in enhancing human to mosquito transmission of parasites following CQ treatment. Among Sudanese parasite isolates bearing the K76T mutation in PfCRT, a higher gametocyte carriage rate was observed as compared to parasites encoding wild-type PfCRT (Osman et al., 2007). Furthermore, P. berghei parasites engineered to express the P. falciparum 7G8 pfcrt variant protected early gametocytes against CQ action and were transmitted at higher levels compared to drug-sensitive parasites (Ecker et al., 2011). These observations underscore the complexity of factors that collectively determine the fitness of Plasmodium parasites, of which relative growth rates in infected erythrocytes is but one component, with others including antimalarial drug susceptibility profiles, the impact of host immunity and transmission dynamics, differences in gametocyte production and infectivity, competition between strains within mosquitoes, and growth differences that could manifest during the liver stages (Walliker et al., 2005; Rosenthal, 2013).
Here, we dissect the specific contribution of geographically distinct PfCRT haplotypes to parasite fitness and antimalarial drug susceptibility. Our study includes novel haplotypes that have not been previously assessed in a controlled genetic background, including two closely-related PfCRT isoforms from the Philippines (Chen et al., 2005), as well as an allele from Cambodia that harbors nine mutations, an exceptionally high number (Durrand et al., 2004). These alleles were assessed alongside the geographically widespread Dd2 and 7G8 alleles in vitro in drug susceptibility assays as well as mixed-infection competition assays. We also investigated how various mutant PfCRT haplotypes impact CQ accumulation and parasite response to other antimalarials in current clinical use. Our results highlight the importance of regional PfCRT haplotypes in contributing to parasite fitness and define a novel allele in Cambodia that appears to have overcome the hurdle of reduced fitness associated with less mutated pfcrt forms, while still maintaining a moderate degree of CQR.
Results
Generation of Isogenic Parasite Lines Expressing Asian pfcrt Alleles From the Endogenous Locus by Allelic Exchange
We engineered the mutant pfcrt alleles PH1 and PH2 (from the Philippines) and Cam734 (from Cambodia) into P. falciparum CQ-sensitive parasites via allelic exchange. The recipient CQ-sensitive strain C1GC03 was genetically modified from the GC03 parasite line, a progeny of the HB3 ×Dd2 genetic cross (Su et al., 1997), in a prior round of transfection. Briefly, the highly interrupted endogenous wild-type pfcrt gene sequence was replaced with a shortened sequence containing all exons and intron 1, rendering this line more amenable to pfcrt allelic exchange (Sidhu et al., 2002). PH1 and PH2 represent two common PfCRT haplotypes in the Philippines (Chen et al., 2003) that differ from one another at position 72 and that are notable for lacking the common A220S mutation but harboring the two novel mutations A144T and L160Y (Table 1). Cam734 comprises ~20% of all pfcrt alleles in Cambodia and is a highly mutated pfcrt allele, differing from the wild-type allele at 9 positions (Durrand et al., 2004).
Table 1.
Transformation status and PfCRT haplotype of recombinant and wild-type lines.
| Functional PfCRT haplotype |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clone | Parent | Transfection plasmid* | 72 | 74 | 75 | 76 | 144 | 148 | 160 | 194 | 220 | 271 | 326 | 333 | 356 | 371 |
| GC03 | HB3 × Dd2 | - | C | M | N | K | A | L | L | I | A | Q | N | T | I | R |
| C1GC03 | GC03 | phDHFR-crt-GC03Pf3' | C | M | N | K | A | L | L | I | A | Q | N | T | I | R |
| C8PH1-II | C1GC03 | pBSD-crt-PH1Py3' | C | M | N | T | T | L | Y | I | A | Q | D | T | I | R |
| C10PH2-I,II | C1GC03 | pBSD-crt-PH2Py3' | S | M | N | T | T | L | Y | I | A | Q | D | T | I | R |
| C12Cam734-I,II | C1GC03 | pBSD-crt-Cam734Py3' | C | I | D | T | F | I | L | T | S | E | N | S | I | R |
| C2GC03 | C1GC03 | pBSD-crt-Dd2Pf3' | C | M | N | K | A | L | L | I | A | Q | N | T | I | R |
| C4Dd2 | C1GC03 | pBSD-crt-Dd2Py3' | C | I | E | T | A | L | L | I | S | E | S | T | T | I |
| C67G8 | C1GC03 | pBSD-crt-7G8Py3' | S | M | N | T | A | L | L | I | S | Q | D | T | L | R |
| HB3 | - | - | C | M | N | K | A | L | L | I | E | Q | N | T | I | R |
| Dd2 | - | - | C | I | E | T | A | L | L | I | S | E | S | T | T | I |
| 7G8 | - | - | S | M | N | T | A | L | L | I | S | Q | D | T | L | R |
Plasmids harboring different pfcrt allelic sequences were transfected into CQ-sensitive C1GC03 parasites to generate the recombinant mutant and control lines. Grey shading indicates residues that differ from the wild-type sequence.
To generate recombinants, C1GC03 parasites were electroporated with the plasmids pBSD-crt-PH1Py3', pBSD-crt-PH2Py3', and pBSD-crt-Cam734Py3', containing exons 2–13 of the three Asian pfcrt alleles (Fig. 1A). Transfected parasite cultures were obtained following exposure to blasticidin and WR99210 to select for expression of blasticidin S-deaminase (bsd) from the transfection plasmid and human dihydrofolate reductase (dhfr) in the C1GC03 parental line, respectively.
Fig. 1. pfcrt allelic exchange strategy and molecular characterization of clones.
(A) Schematic representation of single-site crossover between a pBSD-based pfcrt transfection plasmid and the functional pfcrt locus of C1GC03, leading to expression of a recombinant allele (PH1, PH2, or Cam734; see Table 1) from the endogenous full-length promoter. The diagram illustrates transfection of the CQ-sensitive C1GC03 clone with pBSD-crt-PH1Py3’. This construct contained a pfcrt sequence with a deletion in exon 1, no introns between exons 2–13 and a downstream 0.7 kb 3' UTR sequence from the P. yoelii ortholog pycrt. Homologous recombination upstream of codon positions 74–76 resulted in generation of a functional pfcrt allele containing all the point mutations from the mutant PH1 allele, under the control of pfcrt 5' UTR and pycrt 3' UTR regulatory elements. Downstream remnant pfcrt fragments were truncated in exon 1, had a 5' in-frame stop codon and lacked a promoter. (B) PCR-based analysis of the recombinant clones and parental lines (primer positions illustrated in Fig. 1A). Parasite strain and primer details are provided in Table 1 and Table S1, respectively. (C) Southern blot hybridization of gDNA digested with SalI and ClaI and subsequently probed with a pfcrt fragment from exon 2. The positions of the restriction sites and exon 2-specific probe are indicated in Fig. 1A. Panels B and C include the clone C8PH1-I, which was subsequently found to have undergone a spontaneous mutation in codon 326 and was removed from further analysis.
PCR was used to identify transfected lines that had undergone homologous recombination and single-site crossover into the pfcrt locus. Recombinant clones were then obtained by limiting dilution. Clones from each successfully integrated transfection were selected for further characterization and were termed C8PH1-I, C8PH1-II, C10PH2-I, C10PH2-II, C12Cam734-I, and C12Cam734-II. Following our earlier reports (Sidhu et al., 2002), the superscript indicates the pfcrt allele, with the Roman numeral indicating the clone. For comparison, we included C1GC03, which was generated using the same allelic exchange strategy (Sidhu et al., 2002) and which expresses the canonical wild-type allele (Table 1).
To confirm the clonality of these lines, we performed PCR with primers P1 and P3 that targeted parasites with integrated plasmid, and primers P1 and P2 that were specific for the original C1GC03 locus (Fig. 1A, 1B; Table S1). Primers P1 and P3 yielded the expected 1.3 kb band from the clones that had undergone two rounds of recombination, but not from the first-round C1GC03 or parental GC03 parasites. PCR with primers P1 and P2 resulted in expected bands of 3.3 kb from the unmodified genomic pfcrt locus in GC03 and 1.7 kb from the first round of recombination present in the C1GC03 line (Fig. 1B). Southern blot analysis of genomic DNA (gDNA) digested with SalI+ClaI revealed band sizes of ~16.3 kb, 8.1 kb, 7.7 kb and 1.2 kb, consistent with plasmid integration into the C1GC03 pfcrt locus (Fig. 1C). GC03 parasites and linearized plasmid DNA showed the predicted 9.4 kb and 7.7 kb bands, respectively.
Sequencing of the functional recombinant pfcrt locus amplified from gDNA, which was performed shortly after limiting dilution cloning, confirmed the expected full-length sequence of pfcrt in the individual lines. Amplification of the pfcrt locus from cDNA and gDNA and subsequent sequencing of the polymorphic region encoding for amino acids 72–76 confirmed the exclusive expression of the integrated allele in the new second-round recombinants (data not shown). Real-time PCR analysis was also performed using two independent preparations of parasite RNA from synchronized ring-stage cultures. These were assayed for pfcrt and the housekeeping gene actin (PFL2215w) on 4–8 independent occasions with each sample tested in triplicate per assay. Kruskal-Wallis tests showed no significant differences in pfcrt transcript levels between any pfcrt-modified lines (Table S2). The same finding of statistically indistinguishable expression levels was observed by quantitative Western blot analysis of protein extracts from these recombinant lines (Table S2).
Ongoing characterization of these recombinant lines, during the lengthy period of propagation required to complete their phenotypic assessment, revealed a highly unusual event in the C8PH1-I clone. Over time, a subpopulation arose that outgrew the original line. This subpopulation was found to have undergone reversion of the N326D mutation back to the wild-type N326 codon in pfcrt, with the other three PH1 mutations being retained (K76T, A144T and L160Y; Table 1). Interestingly, this revertant was found to be CQ sensitive, implicating N326D as an important contributor to CQR, consistent with a previous report (Summers et al., 2014). Nevertheless, the advent of a spontaneous sequence reversion in this line made us cautious about using the C8PH1-I line, and it was excluded from further consideration in this present study. Repeated sequence analysis of other recombinant lines during long-term culture confirmed the genotypes of all other lines under investigation. Analysis of the PH1 haplotype was therefore confined to the C8PH1-II clone, whose genotype was closely monitored and remained stable over time.
Mutant Southeast Asian pfcrt Alleles Influence Susceptibility To Locally Used Drugs
Using these recombinant isogenic lines, we assayed the impact of different PfCRT haplotypes (Table 1) on parasite susceptibility to CQ as well as other antimalarials. For comparison, we included C2GC03, C4Dd2, and C67G8, which were generated using the same genetic strategy (Sidhu et al., 2002). These encode the wild-type GC03 haplotype, the Dd2 haplotype commonly found in Asia and Africa, and the 7G8 haplotype that is widespread in South America and the Pacific region, respectively (Sa et al., 2009). For reference, we also included the non-recombinant lines GC03, Dd2 and 7G8.
We note that our CQ values, both for resistant and sensitive strains, are lower than earlier reports (Sidhu et al., 2002; Lakshmanan et al., 2005; Valderramos et al., 2010). One important technical difference is that we reduced the HEPES concentration from the earlier 50 mM to the current 25 mM. Our detailed studies have since revealed that this decrease in the HEPES concentration leads to a substantial reduction in half-maximal inhibitory concentration (IC50) values for CQ-sensitive and even more so for CQ–resistant parasites, as detailed in the Supporting Information (SI; see text and Figures S2–S4). This is one of the variables that can produce differences in CQ IC50 values (others include genetic differences between strains maintained long-term in separate laboratories and the choice of assay). Relative differences between strains in a given dataset are thus recognized to provide the most informative data (Ekland and Fidock, 2008). Consequently, we focus below primarily on relative differences between parasite lines expressing distinct pfcrt alleles.
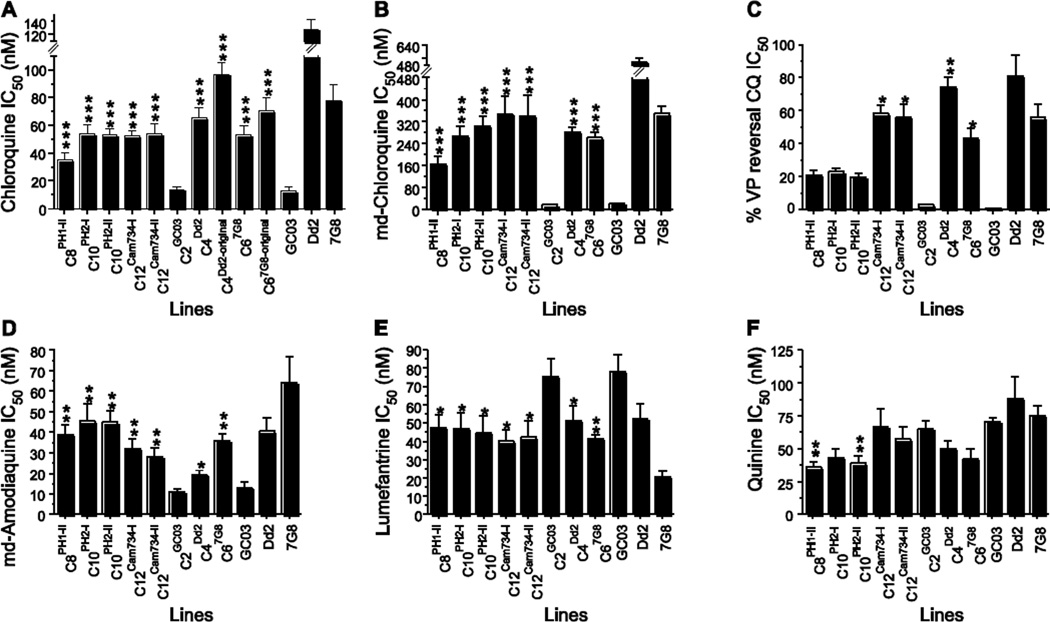
For CQ, all recombinant lines expressing mutant pfcrt alleles (C8PH1, C10PH2, C12Cam734, C4Dd2 and C67G8) had a statistically significant, 2.5 to 4.7–fold increase in mean IC50 value relative to the isogenic recombinant C2GC03 line expressing the wild-type allele (mean IC50 value of 14 nM) (Fig. 2A; Table S3). Of note, C10PH2 and C12Cam734 yielded CQ IC50 values (53–54 nM) that were comparable in these assays to the recombinant C4Dd2 and C67G8 parasites expressing the most globally prevalent mutant pfcrt alleles (Sidhu et al., 2002). We note, however, that C4Dd2 and C67G8 are no longer as CQ-resistant as when they were originally generated (in 2002) and characterized by several groups (Sidhu et al., 2002; Lakshmanan et al., 2005; Gligorijevic et al., 2008). This has also been observed in an independent recent study that employed these lines (Hrycyna et al., 2014). C4Dd2 and C67G8 currently display CQ IC50 values that are now 51% and 68% of the parental Dd2 and 7G8 lines respectively (Table S3), as compared to the initial report that documented corresponding values of 76% and 90%. These relative levels of CQR are illustrated for the original C4Dd2 and C67G8 lines, alongside the now-attenuated lines, in Fig. 2A. Thus, our present data with the more recently generated PH1, PH2 and Cam734 pfcrt-expressing lines identify these as only modestly CQR, with relatively low CQ IC50 values. Strikingly, the two C10PH2 clones demonstrated higher CQ mean IC50 values as compared to C8PH1-II parasites (53–54 nM compared to 35 nM). This finding implicates amino acid 72, which is the only polymorphism that distinguishes the PH2 and PH1 alleles (Table 1), as an important determinant of the degree of CQR.
Fig. 2. Susceptibility of pfcrt-modified clones to selected antimalarial drugs.
Mean IC50 values ± SEM for the indicated parasite strains subjected to clinically significant antimalarials, as measured in vitro using [3H]-hypoxanthine incorporation assays. We note that the CQ values, both for resistant and sensitive strains, are markedly lower than earlier reports (Sidhu et al., 2002; Lakshmanan et al., 2005; Valderramos et al., 2010), and coincide with our reducing the HEPES concentration from the earlier 50 mM to the current 25 mM (see SI). CQ values for the original C4Dd2 and C67G8 lines, presented as a proportion of their IC50 values of the reference Dd2 and 7G8 lines, are included to illustrate the attenuation of the CQR phenotypes of these lines over time. The C4Dd2-original CQ mean±SEM IC50 value of 96.8±8.7 nM is comparable to the values of 91.8±10.7 nM and 100.3±15.5 nM recently reported with two pfcrt-modified GC03 clones engineered to express the Dd2 allele (i.e. analogous to C4Dd2) using customized zinc-finger nucleases (Straimer et al., 2002). VP reversal was calculated as the IC50 of CQ + 0.8 µM VP divided by the IC50 of CQ. VP reversal values for CQ, md-CQ and md-ADQ are provided in Table S4. Mann-Whitney U tests were used to assess for statistically significant differences between a recombinant line expressing mutant pfcrt and the CQ-sensitive line C2GC03 expressing wild-type pfcrt. *P<0.05; **P<0.01; ***P<0.001. IC50 and IC90 values, numbers of assays, and tests for significance are reported in Table S3.
Similar observations were made with the CQ metabolite monodesethyl-chloroquine (md-CQ), which yields much higher IC50 values in CQ-resistant parasites, allowing for greater discrimination between resistant and sensitive lines (Sidhu et al., 2002). C10PH2 and C12Cam734 clones all yielded mean IC50 values (267–346 nM) that were comparable to C4Dd2 and C67G8 (282 and 262 nM, respectively; Fig. 2B; Table S3). In comparison, the CQ-sensitive lines C2GC03 and GC03 showed mean IC50 values of 17–22 nM. Of note, the mean md-CQ IC50 value (164 nM) of the C8PH1 clone was considerably lower than the IC50 values of the two C10PH2 lines (267 and 305 nM), again supporting a direct role for the C72S mutation in augmenting the degree of CQR.
Prior work has shown that CQ-resistant parasites can be chemosensitized to CQ and md-CQ by the resistance-reversing agent verapamil (VP) (Krogstad et al., 1987; Martin et al., 1987). The primary determinant of this reversibility trait has been mapped by quantitative trait loci analysis to mutant pfcrt (Patel et al., 2010) and is more pronounced in the presence of the Dd2 allele as compared to the 7G8 allele (Mehlotra et al., 2001; Sidhu et al., 2002; Sa et al., 2009). To assess VP reversibility in our recombinant lines, we performed drug assays in the presence or absence of 0.8 µM VP, and compared IC50 values. Results showed a high degree of resistance reversal (74–85%) for C4Dd2 and the reference Dd2 line, and an intermediate degree (43–56%) for C67G8 and 7G8 parasites (Fig. 2C). These values were significantly different from the CQ-sensitive C2GC03 line that along with GC03 showed no growth inhibition with this concentration of VP. Intriguingly, both C8PH1 and C10PH2 showed only a slight degree of reversal, which did not attain statistical significance. A similar lack of significant reversal was also observed with the metabolite md-CQ (Table S3). This agrees with an earlier report of Philippine isolates (Chen et al., 2003) and is consistent with a recent study that associated PfCRT N75 (present in both haplotypes; Table 1) with minimal VP reversal (Sa et al., 2009). We note that C12Cam734 parasites, which harbor the novel N75D mutation, were also subject to a significant degree (56–72%) of VP reversal of CQ and md-CQ resistance, at levels intermediate to the Dd2 and 7G8 alleles (Fig. 2C; Table S3).
We extended these studies to monodesethyl-amodiaquine (md-AQ), the clinically relevant metabolite of amodiaquine, a 4-aminoquinoline drug formerly used in monotherapy in many South American, African and Asian countries, including the Philippines (Sa et al., 2009). This drug continues to be clinically important because of its incorporation into the widely used amodiaquine-artesunate combination (Wells et al., 2009). Our studies reveal a substantial impact of both Philippine pfcrt alleles, as well as the Cam734 allele, on md-AQ responses (Fig. 2D), resulting in a 2.5 to 4-fold increase in IC50 values compared to C2GC03. Of all alleles tested, the smallest gain in md-AQ resistance was afforded by expression of the recombinant Dd2 allele that mediates relatively high-level CQR (see C4Dd2 responses in Fig. 2A, 2D). These data implicate an important role for the PfCRT mutations unique to the Philippine and Cambodian alleles in reducing parasite susceptibility to amodiaquine.
Importantly, every mutant pfcrt allele significantly increased susceptibility to the arylaminoalcohol drug lumefantrine (LMF; Fig. 2E), the partner drug comprising the most widely used artemisinin-based combination therapy (ACT), artemether-lumefantrine (Wells et al., 2009). Mean IC50 reductions were 37–47%, as compared to C2GC03 (Table S3). It is worth noting that a similar decrease in LMF IC50 values in parasites expressing mutant pfcrt as compared to isogenic parasites expressing the wild-type allele was earlier found to be associated with a significant reduction in the prevalence of mutant pfcrt in field isolates that recrudesced in patients treated with artemether-lumefantrine (Sisowath et al., 2009). These findings support the therapeutic advantage of using LMF in areas of CQ-resistant malaria.
A similar trend was observed with the arylaminoalcohol mefloquine and the endoperoxide artemisinin. However the differences in IC50 values did not attain statistical significance (Table S3). The PH1 and PH2 pfcrt alleles, but not Cam734, also significantly increased parasite susceptibility to quinine (Fig. 2F), a centuries-old drug used to treat severe malaria. Quinine resistance is known to be multifactorial, with quantitative trait loci analyses implicating mutant pfcrt and pfmdr1 as major determinants (Ferdig et al., 2004; Sanchez et al., 2011; Sanchez et al., 2014). Our data support the hypothesis that the direction and magnitude of the effect of mutant pfcrt on quinine response depends on the genetic background and PfCRT haplotype (Cooper et al., 2002; Sidhu et al., 2002; Lakshmanan et al., 2005; Cooper et al., 2007). Finally, no differences were observed with piperaquine (Table S3), an ACT partner drug that comprises two CQ 4-aminoquinoline rings tethered together with a spacer (Wells et al., 2009), consistent with this drug being equally potent against parasites expressing wild-type or common mutant variants of pfcrt (Pascual et al., 2013).
Reduced Chloroquine Accumulation Alone Does Not Account for Differences in the Degree of Chloroquine Resistance
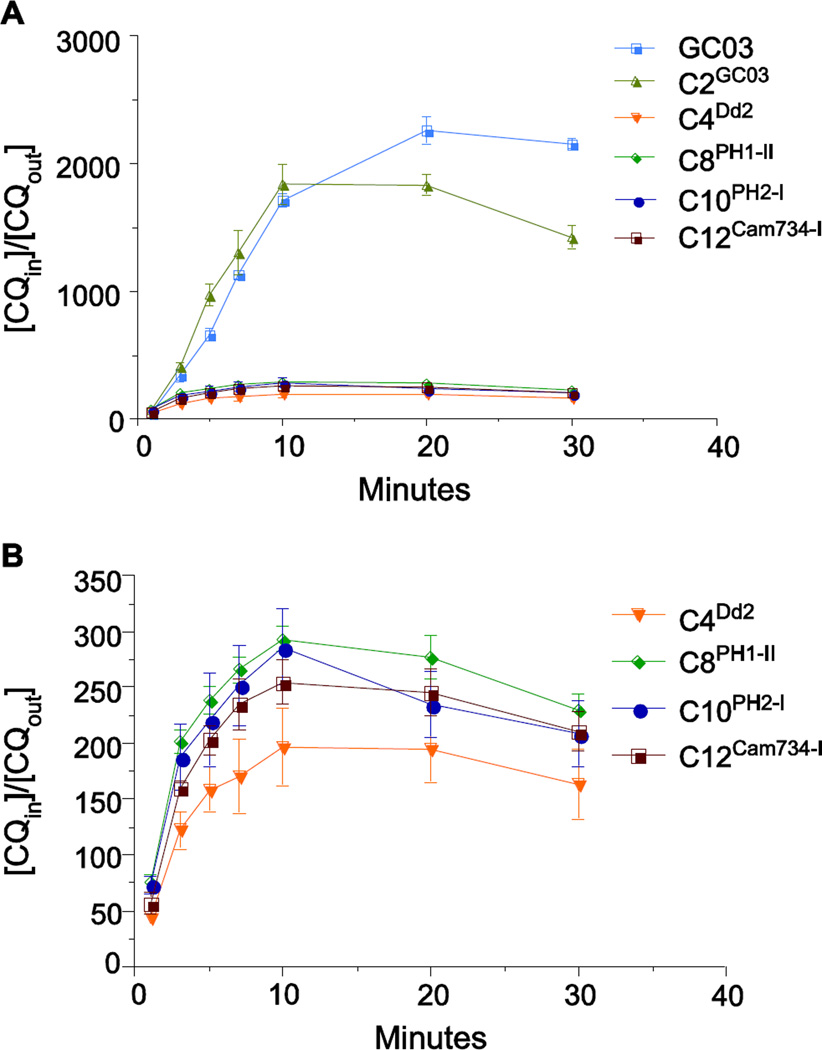
Resistance to CQ has been associated with reduced drug accumulation in the DV and has previously been attributed to mutant PfCRT-mediated efflux of CQ from this acidic organelle (Krogstad et al., 1987; Martin et al., 1987; Valderramos and Fidock, 2006; Martin et al., 2009). To investigate the influence of the Southeast Asian pfcrt alleles on CQ accumulation, we measured the kinetics of [3H]-CQ uptake in cultured parasites. The CQ-sensitive control lines GC03 and C2GC03 showed a rapid increase in CQ accumulation, as measured by a very high ratio (up to 2,300) of total intracellular CQ to extracellular CQ ([CQin]/[CQout]) (Fig. 3A). In contrast, the four pfcrt-variant recombinant lines (C4Dd2, C8PH1-I, C10PH2-I, and C12Cam734-I) accumulated minimal levels of CQ ([CQin]/[CQout] values of ~200–300). Thus, these mutant PfCRT haplotypes, all expressed in the same genetic background, were associated with a very similar reduction of intracellular CQ levels (Fig. 3B), despite marked differences in their CQ IC50 values (Fig. 2). These data lead us to suggest that reduced CQ accumulation might represent only one means by which mutant PfCRT dictates the degree of CQR.
Fig. 3. Chloroquine accumulation of pfcrt-modified parasite lines.
[3H]-CQ accumulation is represented as the ratio of the total intracellular CQ to extracellular CQ ([CQin]/[CQout]). Values represent the mean ± SEM determined from three independent experiments performed in duplicate. Data are shown in (A) for the full set of pfcrt-modified lines and the CQ-sensitive control GC03, and in (B) exclusively for the lines expressing mutant pfcrt (note the reduced Y-axis scale).
PfCRT Haplotypes Influence Parasite Growth Rates in Co-Culture in vitro Competition Assays
To measure the extent to which pfcrt mutations influence the relative growth rates of asexual blood stage parasites, serving as a partial proxy for assessing fitness costs, we performed in vitro co-culture competition assays. In these assays, two lines were mixed in 1:1 ratios and the proportions of the individual pfcrt alleles were quantified by pyrosequencing every 4 days, on average, over a 2–3 month period. Data from these assays were converted using the ratio of the natural logs of the allelic frequencies, based on an assumption of exponential growth, and were subjected to linear regression (Maree et al., 2000; Mita et al., 2004). Linear regression R2 values were generally high (average 0.64, reflecting an acceptable goodness of fit; Table S5). In total, 34 co-culture assays were followed, yielding 518 measurements of pfcrt allelic frequencies over time (Table S5). From these measurements, we computed the mean relative growth rate value for each line, as detailed in the SI.
Results indicated that the isogenic clones C8PH1-II and C10PH2-I, expressing the PH1 and PH2 mutant pfcrt alleles respectively, each displayed reduced in vitro growth when independently co-cultured with C2GC03 parasites (Figs. 4; 5A). Of the two mutant pfcrt parasites, C10PH2-I parasites showed the more substantial loss of relative growth rate in vitro. This finding was consistent with direct competition assays between C10PH2-I and C8PH1-II, which revealed reduced relative growth rates with the former (Table S5). This contrasted with CQ and md-CQ IC50 values that were higher for C10PH2-I (Fig. 2; Table S3). These results suggest a state of balanced polymorphisms whereby the PH1 allele could be predicted to fare better than the more unfit PH2 in mixed infections in the absence of CQ drug pressure, whereas the moderately more CQR PH2 allele could be more competitive in the presence of CQ and thus be retained in the population. Intriguingly, parallel studies with the CQ-resistant C12Cam734-I line documented that these parasites were consistently as fit in terms of growth rates, if not slightly more so, when compared to C2GC03 in vitro. These data suggest that the Cam734 allele, harboring an unusually high number of point mutations (Table 1), has achieved a functional state that might reduce the fitness cost typically observed with mutant pfcrt alleles in endemic settings.
Fig. 4. Relative growth rate plots for mixed competition assays between pfcrt-modified lines.
Parasite cultures were initiated at ~1:1 ratios and allelic proportions were measured over time for up to 45 generations by pyrosequencing. The Y-axis illustrates the proportion of the first listed allele comprising the mixed cultures. Each color represents a separate assay. Data are collectively summarized in Fig. 5A and Table S5.
Fig. 5. Influence of pfcrt alleles on relative growth rates and chloroquine resistance.
(A) Relative growth rate values for the individual pfcrt-modified parasite lines were measured following pyrosequencing-based assessment of changes in pfcrt allele frequencies in mixed cultures maintained for up to 45 generations. For each combination, 1–6 independent competition assays were performed in duplicate (summarized in Table S5; data plotted in Fig. 4). The histogram depicts the relative growth rate of the first line compared to the second, e.g. for C8PH1-II vs. C2GC03 the negative value reflects the reduced relative growth rate of C8PH1-II. (B) Distribution of various pfcrt alleles in the major malaria-endemic regions. (C) Mean relative growth rate differences between wild-type and mutant pfcrt alleles (in the GC03 background) presented as a function of the effect of these alleles on CQ IC50 values (based on data presented in Tables S3 and S5 and depicted in Figs. 2 and 5A). Circle size indicates the estimated worldwide frequency of the pfcrt allele, approximated from literature reports and database summaries on the number of clinical cases and the distribution of pfcrt alleles (see Table S6)
In contrast to Cam734, we observed a substantial reduction in relative growth rates associated with the Dd2 pfcrt allele when comparing C4Dd2 to C2GC03, a finding consistent with reports of reduced fitness associated with this allele in African parasite populations (Kublin et al., 2003; Mita et al., 2003; Ord et al., 2007). A more modest reduction in relative growth rates was observed with the 7G8 allele present in C67G8. The reduced relative growth rates of the Dd2 and 7G8 alleles is consistent with a recent report that also assessed these recombinant parasite lines in mixed culture experiments (Lewis et al., 2014). In our studies, C67G8 consistently outcompeted C4Dd2 in mixed cultures in four independent experiments (Fig. 4; Table S5). Importantly, when testing non-recombinant lines, we found the Dd2 line to have moderately increased growth rates relative to lines expressing wild-type pfcrt (GC03 and HB3), suggesting that Dd2 harbors additional compensatory mutations in its genome that corrected the growth defect associated with expression of its pfcrt allele. A summary of the influence of pfcrt alleles on in vitro growth rates in these co-competition assays is depicted in Fig. 5A. Independent support for these data came from measurements of in vitro growth rates for individual lines, which were monitored daily for seven days in three independent assays per line. Calculated multiplication rates per 48 hr revealed that in comparison to parasites expressing the pfcrt wild-type GC03 allele, parasites expressing the Cam734 allele showed an equivalent growth rate, whereas parasites expressing the PH2, Dd2 and 7G8 alleles displayed slower rates of growth (Fig. S1).
Discussion
Genome-wide studies of P. falciparum populations have demonstrated a remarkable degree of recent evolution in the pfcrt coding sequence, beginning with several independent origins of variant alleles. These alleles have spread across malaria-endemic regions as selective sweeps driven by intense drug pressure (Wootton et al., 2002; Kidgell et al., 2006; Volkman et al., 2007; Mu et al., 2010). Primary origins have been localized to South America and Papua New Guinea (independent sources of the 7G8 variant haplotype), the Philippines (PH1 and PH2), the Thai-Cambodian border (Dd2), and most likely Cambodia (Cam734; Fig. 5B) (Ecker et al., 2012). CQR in Africa resulted from the introduction of variant pfcrt of Southeast Asian origin (Ariey et al., 2006), whose insidious impact on malaria rates was highlighted by reports from Senegal showing nearly a six-fold increase in malarial deaths following the arrival of resistant strains (Snow et al., 2001; Trape, 2001). Our study of geographically distinct PfCRT haplotypes reveals an intricate balance between CQR and parasite growth rates, which provide an in vitro proxy of fitness. Our data also highlight the emergence (in Cambodia) of the Cam734 allele (Durrand et al., 2004), which appears to have succeeded in mediating a moderate degree of resistance while concurrently maintaining in vitro growth rates at least as good if not better than wild-type pfcrt (Fig. 5C). Notably, Cambodia is a known hotbed of multidrug resistance, beginning with CQ and pyrimethamine-sulfadoxine and more recently with emerging resistance to artemisinin derivatives (Dondorp et al., 2011; Ariey et al., 2014; Straimer et al., 2015). We ascribe the lack of an observable growth rate defect in the Cam734 allele to its complex set of mutations (Table 1), which include the L148I, I194T and T333S mutations not present in other parasites studied herein. We posit that these mutations have evolved to compensate for a loss of fitness bestowed by an initial set of mutations that were sufficient to confer CQR, including but not limited to K76T (Lakshmanan et al., 2005). The ability of Plasmodium parasites to acquire a set of mutations simultaneously conferring CQR and enhanced fitness has previously been documented in murine studies, in which CQ-resistant P. chabaudi parasites outgrew their sensitive counterparts, even when mice were inoculated with a nine-fold excess of sensitive parasites (Rosario et al., 1978). Recent metabolomic studies show that the widespread Dd2 and 7G8 alleles cause increased levels of intracellular peptides in asexual blood stage parasites, presumably stemming from impaired hemoglobin digestion that restricts the supply of amino acids required for parasite proliferation (Lewis et al., 2014). These recent findings provide a potential mechanistic explanation for the fitness costs observed with the Dd2 and 7G8 haplotypes. Further studies are required to assess whether the Cam734 allele corrects this abnormal accumulation of hemoglobin-derived peptides.
Our findings also illustrate a singular impact of PfCRT residue 72, which is the sole sequence distinction between the Philippine PH1 allele that mediates marginal CQR and the PH2 allele that is moderately more resistant (Fig. 5C). This residue is also associated with differences in relative growth rates in vitro (Fig. 4; 5A). How this single amino acid difference might impact protein conformation, stability or post-translational modifications remains to be determined. During these experiments, we isolated a separate recombinant C8PH1 parasite, referred to as C8PH1-II, which underwent a spontaneous loss of the N326D mutation during extended culture. This reversion back to the wild-type residue was associated with an increased rate of parasite propagation and a loss of CQR. The reversion to a CQS phenotype by the N326D mutation is consistent with a recent report that showed a lack of CQ transport in Xenopus oocyte-based heterologous expression assays, in contrast to the PH1 PfCRT variant that showed CQ transport behavior (Summers et al., 2014). These data highlight a requirement for multiple PfCRT mutations in producing the CQR phenotype, arguing against the notion that CQR results solely from the K76T mutation and that the other mutations in this protein compensate solely for loss of function. This conclusion is supported by transport studies in an oocyte expression system, which provided evidence that the K76T mutation needed to be accompanied by either the N75E (Southeast Asian PfCRT variants) or the N326D mutation (Latin American and oceanic PfCRT variants) to attain a CQ transport function (Summers et al., 2014). While two mutational changes sufficed for a basal CQ transport activity, additional mutations were required for full activity. The order in which these mutations were added was important to avoid reductions in CQ transport activity (Summers et al., 2014).
Epidemiological studies have found that some parasites harboring the PfCRT K76T mutation have low to moderate CQ IC50 values that do not meet the standard definition of CQR and in some instances are similar to values observed with CQ-sensitive parasites, for example Cambodian isolates harboring the Cam734 allele (Durrand et al., 2004). Studies are ongoing to dissect the role of the PfCRT SNPS that are unique to this haplotype. Furthermore, in vitro selection studies have shown that parasites harboring the Dd2 pfcrt allele acquired a C101F mutation that resulted in a loss of CQR despite the presence of K76T (Eastman et al., 2011). Thus, while K76T continues to be an important molecular marker of CQR, recent evidence suggests that additional PfCRT SNPs can substantially modify the CQ response, in some cases causing an attenuation or loss of the resistance phenotype. We also note that while PfCRT is widely recognized to be the primary mediator of CQR, several studies point to a requirement for secondary determinants to augment CQR, including pfmdr1 (Sidhu et al., 2002; Sa et al., 2009; Patel et al., 2010; Valderramos et al., 2010; Gaviria et al., 2013).
In our mixed infection studies with isogenic pfcrt-modified clones we also observed that parasites displayed the greatest loss of asexual blood stage growth when expressing the Dd2 allele, consistent with its progressive disappearance from high-transmission African settings in the absence of CQ pressure (Mita et al., 2003; Ord et al., 2007; Mwai et al., 2009; Laufer et al., 2010; Frosch et al., 2011). This contrasts with the situation in Southeast Asia where the Dd2 allele remains at high frequencies in the absence of CQ pressure. Fewer mixed infections in Asia compared to Africa likely result in less opportunity for competition with the wild-type allele. The lesser fitness cost observed with the 7G8 allele, which is prevalent in South America and the Pacific region, is concordant with studies from these regions showing the continued presence of mutant pfcrt despite minimal CQ use in recent decades to treat P. falciparum malaria (Mu et al., 2010). We note that modest selective pressure on mutant pfcrt may also have come from the use of CQ to treat P. vivax infections, which are common outside of Africa (Price et al., 2007). Overall, our in vitro mixed competition relative growth rate data would suggest that the idea of reintroducing CQ into regions where prolonged drug removal has led to the near disappearance of resistant strains (Juliano et al., 2007; Laufer et al., 2010) is suitable only in areas harboring mutant pfcrt alleles such as Dd2 that cause reduced fitness, and would be less applicable to regions harboring relatively “fit” alleles such as Cam734.
The loss of CQ efficacy across the globe, followed by a short-lived dependence on the sulfadoxine-pyrimethamine antifolates, has resulted in recent years in the global adoption of ACTs (White, 2008; Eastman and Fidock, 2009). Notably, our study shows that all mutant pfcrt alleles tested herein, including the two that are most prevalent (Dd2 and 7G8), increase parasite susceptibility to lumefantrine. This is particularly significant as this drug, partnered with artemether (CoArtem®), is globally the most widely used antimalarial (Wells et al., 2009). While the fold change in lumefantrine IC50 values is relatively low (≤ 2-fold; Fig. 2E; Table S3), we note that a clinical trial from Tanzania observed significant selection against mutant pfcrt parasites harboring the 72CVIET76 PfCRT haplotype following artemether-lumefantrine treatment (Sisowath et al., 2009).
Amodiaquine-artesunate is another ACT that is often used in Africa (Olliaro and Mussano, 2000). We observed significantly reduced parasite susceptibility to the amodiaquine metabolite md-AQ with every tested mutant pfcrt allele. This includes both Philippine alleles, obtained from a country where amodiaquine has been used as an antimalarial treatment for over 40 years (Sa et al., 2009). We thus posit that amodiaquine could have been a major contributor to the emergence and/or maintenance of the PH1 and PH2 alleles. An important role for amodiaquine in driving the spread of the 72SVMNT76 PfCRT haplotype (present in 7G8) has recently been proposed based on studies of parasites from South America and Asia (Sa et al., 2009; Beshir et al., 2010), and amodiaquine pressure could conceivably account for the apparent recent spread of this haplotype into Africa and in India (Alifrangis et al., 2006; Gama et al., 2010; Mixson-Hayden et al., 2010). In addition, as with many Asian countries, CQ has continued to be used to treat patients infected with P. vivax, thus sustaining local CQ pressure that could influence the course of mixed infections of P. vivax and P. falciparum. Overall, our data support the use of artemether-lumefantrine in preference to amodiaquine-artesunate to treat CQ-resistant malaria. We also found no evident effect of mutant pfcrt on the efficacy of piperaquine, an ACT partner drug with excellent clinical efficacy and post-treatment prophylactic activity (Wells et al., 2009).
Our mechanistic investigations into CQR provide evidence that mutant PfCRT-mediated CQR can be phenotypically distinguished from reduced intracellular CQ accumulation, as also noted by a previous study (Sanchez et al., 2011). This was particularly evident with C8PH1 parasites that, despite having only a nominal degree of CQR, displayed kinetics of intracellular CQ accumulation that paralleled the other, more CQ-resistant parasites. In contrast, CQ-sensitive parasites showed 7–10 fold higher levels of CQ accumulation (Fig. 4). Thus, all mutant PfCRT variants shared an ability to reduce CQ accumulation. This study agrees with recent evidence that reduced CQ accumulation is not the sole cause of CQR (Cabrera et al., 2009; Sanchez et al., 2011; Baro et al., 2013). We posit that mutant PfCRT generally reduces CQ accumulation, and that this is an essential feature of CQR, but that various PfCRT haplotypes differ in a second respect that further contributes to the CQR phenotype. One possibility is that the higher degree of CQR reflects varying degrees to which PfCRT functions to also reduce the cellular toxicity associated with CQ action, possibly by negating the effect of CQ on preventing the buildup of reactive heme-iron or oxygen species liberated following hemoglobin proteolysis. Another possibility is that the drug competes with a yet to be identified physiological substrate for transport via PfCRT and that this competition impacts on the natural function of the transporter. In this context it is interesting to note that PfCRT appears capable of simultaneously accepting different substrates at distinct but antagonistically interacting binding sites (Bellanca et al., 2014). Binding of two different substrates might result in an inactive transporter or one with substantially reduced activity, depending on the nature of the substrates bound (Bellanca et al., 2014). How a geographic PfCRT variant copes with its drug and physiological transport functions is likely determined by its specific amino acid substitutions. Further dissection of the biochemical parameters associated with heme detoxification and CQ action can now be achieved using the series of isogenic pfcrt-modified lines described herein.
Experimental Procedures
Parasite Culture, Transfection, and Selection and Characterization of Integrant Clones
Parasites were cultured at 37°C in human red blood cells in Albumax-containing culture medium, as described (Fidock et al., 1998). Isogenic lines expressing variant pfcrt alleles were generated following transfection of the C1GC03 clone (Sidhu et al., 2002). This clone was previously generated from GC03 (a progeny of the HB3×Dd2 genetic cross (Wellems et al., 1990)), and expresses wild-type pfcrt from a recombinant locus lacking introns 2–12 (Fig. 1). C1GC03 parasites were propagated to ~8% ring stage parasitemia and electroporated with 50 µg of plasmid (pBSD-crt-PH1Py3’, pBSD-crt-PH2Py3’ or pBSD-crt-Cam734Py3’; Table 1; Fig. 1). Transformed parasites were selected using 2.5 nM WR99210 (Jacobus Pharmaceuticals; Princeton, NJ) and 2.5 µM Blasticidin HCl (Invitrogen). Successfully transfected parasites were detectable in culture 2–3 weeks post-transfection and cloned by limiting dilution once plasmid integration was detected. Details of plasmid construction, and of the molecular characterization of parasite lines are provided in the SI (primers listed in Table S1). pfcrt-modified transgenic lines will be made available upon request and are being deposited in the MR4 Malaria Reagent Repository.
In vitro Drug Susceptibility Assays
Parasite susceptibilities to antimalarial drugs were assessed in vitro as described (Fidock et al., 1998) using 72 hr [3H]-hypoxanthine assays (see SI). IC50 values were calculated by non-linear extrapolation. Statistical analyses employed Mann-Whitney U tests.
In vitro Mixed Culture Competition Assays, Pyrosequencing and Determination of Relative Growth Rate Values
For growth competition assays, two parasite lines were mixed 1:1 and seeded in duplicate or triplicate at an initial parasitemia of 0.6% ring stage parasites, in drug-free medium. Parasitemias were maintained between 0.3% and 8% to assure optimal growth conditions. Two to six separate competition assays were performed for each drug-free mixture and each assay was monitored for an average of 66 days (range 43–90; Fig. 4; Table S5). To determine the ratio of both strains in the mixture over time, saponin-lysed parasite pellets of the mixed cultures were collected on average every four days (range 2–9) and DNA was extracted using DNeasy Blood & Tissue Kits (Qiagen). The DNA was then used for ratiometric determination of individual allele frequencies in these mixed cultures by pyrosequencing of codon position 72 or 76 (detailed in the SI). To calculate the relative growth rates of individual parasite lines, the relative proportion of the two distinct pfcrt alleles (whose values were always between 0 and 1, inclusive) were natural log-transformed and linear regression was applied to estimate the relative growth rate value, as detailed in the SI. These values, along with the calculated SEM and R2 values, are listed in Table S5.
Chloroquine Accumulation Assays
These were performed, as previously described (Sanchez et al., 2003), using magnet-purified, sorbitol-synchronized trophozoites (detailed in SI). The amount of accumulated intracellular [3H]-CQ was calculated as the ratio of [CQin]/[CQout], normalized to 1×106 infected erythrocytes.
Supplementary Material
Acknowledgements
We are grateful to Drs. Thierry Fandeur (Centre de Recherche Médicale et Sanitaire, Niamey, Niger) and Qin Cheng (Australian Army Malaria Institute, Enoggera, Queensland, Australia) for their kind gifts of Cam734 and PH1 pfcrt cDNA, and Dr. Liyong Deng, Division of Molecular Genetics, Columbia University College of Physicians and Surgeons, New York, for assistance with pyrosequencing. We also thank all members of the Fidock lab for helpful discussions. Funding for this work was provided in part by the NIH (R01 AI50234, to D.A.F.) and an Investigator in Pathogenesis of Infectious Diseases Award from the Burroughs Wellcome Fund (to D.A.F.). Geoffrey Johnston is a recipient of a Graduate Research Fellowship from the National Science Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supporting Information
Supplemental Materials and Methods and Supplemental Text
Table S1. Primers used in this study.
Table S2. Quantitative RT-PCR and Western blot analysis of pfcrt expression in transgenic lines.
Table S3. Mean IC50 values for pfcrt-modified recombinant and control lines and their statistical comparisons.
Table S4. Percent VP reversibility of 4-aminoquinoline IC50 values for pfcrt-modified recombinant and control lines.
Table S5. Relative rate differences from pairwise in vitro competition assays.
Table S6. Estimated prevalence of pfcrt alleles in malaria-endemic regions.
Fig. S1. Growth rates of parental and pfcrt-modified recombinant P. falciparum lines.
Fig. S2. CQ responses of CQ-sensitive and CQ-resistant parasites in the presence of varying [HEPES].
Fig. S3. CQ IC50 values of parasites in varying [HEPES] at a fixed culture medium pH of 7.40.
Fig. S4. The effect of HEPES concentration on the susceptibility of Dd2 parasites at pH 7.40 to various antimalarials.
Author Contributions
Conceived and designed the experiments: IP AML DAF. Performed the experiments: IP AE REL AML MJA JS PHH EP DJJ OCF CS AML. Analyzed the data: IP SJG GLJ AE SKD AML ML DAF. Wrote the paper: IP SJG GLJ AML DAF.
The authors have no conflict of interest to declare.
References
- Alifrangis M, Dalgaard MB, Lusingu JP, Vestergaard LS, Staalsoe T, Jensen AT, et al. Occurrence of the Southeast Asian/South American SVMNT haplotype of the chloroquine-resistance transporter gene in Plasmodium falciparum in Tanzania. J Infect Dis. 2006;193:1738–1741. doi: 10.1086/504269. [DOI] [PubMed] [Google Scholar]
- Ariey F, Fandeur T, Durand R, Randrianarivelojosia M, Jambou R, Legrand E, et al. Invasion of Africa by a single pfcrt allele of South East Asian type. Malar J. 2006;5:34. doi: 10.1186/1475-2875-5-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois AC, Khim N, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baro NK, Callaghan PS, Roepe PD. Function of resistance conferring Plasmodium falciparum chloroquine resistance transporter isoforms. Biochemistry. 2013;52:4242–4249. doi: 10.1021/bi400557x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellanca S, Summers RL, Meyrath M, Dave A, Nash MN, Dittmer M, et al. Multiple drugs compete for transport via the Plasmodium falciparum chloroquine resistance transporter at distinct but interdependent sites. J Biol Chem. 2014;289:36336–36351. doi: 10.1074/jbc.M114.614206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beshir K, Sutherland CJ, Merinopoulos I, Durrani N, Leslie T, Rowland M, Hallett RL. Amodiaquine resistance in Plasmodium falciparum malaria in Afghanistan is associated with the pfcrt SVMNT allele at codons 72 to 76. Antimicrob Agents Chemother. 2010;54:3714–3716. doi: 10.1128/AAC.00358-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KM, Costanzo MS, Xu W, Roy S, Lozovsky ER, Hartl DL. Compensatory mutations restore fitness during the evolution of dihydrofolate reductase. Mol Biol Evol. 2010;27:2682–2690. doi: 10.1093/molbev/msq160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera M, Paguio MF, Xie C, Roepe PD. Reduced digestive vacuolar accumulation of chloroquine is not linked to resistance to chloroquine toxicity. Biochemistry. 2009;48:11152–11154. doi: 10.1021/bi901765v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Gao Q, Wang S, Wang G, Gatton M, Cheng Q. No genetic bottleneck in Plasmodium falciparum wild-type pfcrt alleles reemerging in Hainan Island, China, following high-level chloroquine resistance. Antimicrob Agents Chemother. 2008;52:345–347. doi: 10.1128/AAC.00711-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Kyle DE, Pasay C, Fowler EV, Baker J, Peters JM, Cheng Q. pfcrt allelic types with two novel amino acid mutations in chloroquine-resistant Plasmodium falciparum isolates from the Philippines. Antimicrob Agents Chemother. 2003;47:3500–3505. doi: 10.1128/AAC.47.11.3500-3505.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Wilson DW, Pasay C, Bell D, Martin LB, Kyle D, Cheng Q. Origin and dissemination of chloroquine-resistant Plasmodium falciparum with mutant pfcrt alleles in the Philippines. Antimicrob Agents Chemother. 2005;49:2102–2105. doi: 10.1128/AAC.49.5.2102-2105.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper RA, Ferdig MT, Su XZ, Ursos LM, Mu J, Nomura T, et al. Alternative mutations at position 76 of the vacuolar transmembrane protein PfCRT are associated with chloroquine resistance and unique stereospecific quinine and quinidine responses in Plasmodium falciparum . Mol Pharmacol. 2002;61:35–42. doi: 10.1124/mol.61.1.35. [DOI] [PubMed] [Google Scholar]
- Cooper RA, Lane KD, Deng B, Mu J, Patel JJ, Wellems TE, et al. Mutations in transmembrane domains 1, 4 and 9 of the Plasmodium falciparum chloroquine resistance transporter alter susceptibility to chloroquine, quinine and quinidine. Mol Microbiol. 2007;63:270–282. doi: 10.1111/j.1365-2958.2006.05511.x. [DOI] [PubMed] [Google Scholar]
- Dondorp AM, Fairhurst RM, Slutsker L, Macarthur JR, Breman JG, Guerin PJ, et al. The threat of artemisinin-resistant malaria. The New England journal of medicine. 2011;365:1073–1075. doi: 10.1056/NEJMp1108322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrand V, Berry A, Sem R, Glaziou P, Beaudou J, Fandeur T. Variations in the sequence and expression of the Plasmodium falciparum chloroquine resistance transporter (Pfcrt) and their relationship to chloroquine resistance in vitro . Mol Bochem Parasitol. 2004;136:273–285. doi: 10.1016/j.molbiopara.2004.03.016. [DOI] [PubMed] [Google Scholar]
- Eastman RT, Dharia NV, Winzeler EA, Fidock DA. Piperaquine resistance is associated with a copy number variation on chromosome 5 in drug-pressured Plasmodium falciparum parasites. Antimicrob Agents Chemother. 2011;55:3908–3916. doi: 10.1128/AAC.01793-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman RT, Fidock DA. Artemisinin-based combination therapies: a vital tool in efforts to eliminate malaria. Nat Rev Microbiol. 2009;7:864–874. doi: 10.1038/nrmicro2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker A, Lakshmanan V, Sinnis P, Coppens I, Fidock DA. Evidence that mutant PfCRT facilitates the transmission to mosquitoes of chloroquine-treated Plasmodium gametocytes. J Infect Dis. 2011;203:228–236. doi: 10.1093/infdis/jiq036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker A, Lehane AM, Clain J, Fidock DA. PfCRT and its role in antimalarial drug resistance. Trends Parasitol. 2012;28:504–514. doi: 10.1016/j.pt.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekland EH, Fidock DA. In vitro evaluations of antimalarial drugs and their relevance to clinical outcomes. International journal for parasitology. 2008;38:743–747. doi: 10.1016/j.ijpara.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdig MT, Cooper RA, Mu J, Deng B, Joy D, Su XZ, Wellems TE. Dissecting the loci of low-level quinine resistance in malaria parasites. Mol Microbiol. 2004;52:985–997. doi: 10.1111/j.1365-2958.2004.04035.x. [DOI] [PubMed] [Google Scholar]
- Fidock DA, Nomura T, Talley AK, Cooper RA, Dzekunov SM, Ferdig MT, et al. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell. 2000;6:861–871. doi: 10.1016/s1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidock DA, Nomura T, Wellems TE. Cycloguanil and its parent compound proguanil demonstrate distinct activities against Plasmodium falciparum malaria parasites transformed with human dihydrofolate reductase. Mol Pharmacol. 1998;54:1140–1147. doi: 10.1124/mol.54.6.1140. [DOI] [PubMed] [Google Scholar]
- Fitch CD. Ferriprotoporphyrin IX, phospholipids, and the antimalarial actions of quinoline drugs. Life sciences. 2004;74:1957–1972. doi: 10.1016/j.lfs.2003.10.003. [DOI] [PubMed] [Google Scholar]
- Frosch AE, Venkatesan M, Laufer MK. Patterns of chloroquine use and resistance in sub-Saharan Africa: a systematic review of household survey and molecular data. Malar J. 2011;10:116. doi: 10.1186/1475-2875-10-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama BE, Pereira-Carvalho GA, Lutucuta Kosi FJ, Almeida de Oliveira NK, Fortes F, Rosenthal PJ, et al. Plasmodium falciparum isolates from Angola show the StctVMNT haplotype in the pfcrt gene. Malar J. 2010;9:174. doi: 10.1186/1475-2875-9-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaviria D, Paguio MF, Turnbull LB, Tan A, Siriwardana A, Ghosh D, et al. A process similar to autophagy is associated with cytocidal chloroquine resistance in Plasmodium falciparum . PloS one. 2013;8:e79059. doi: 10.1371/journal.pone.0079059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gligorijevic B, Purdy K, Elliott DA, Cooper RA, Roepe PD. Stage independent chloroquine resistance and chloroquine toxicity revealed via spinning disk confocal microscopy. Mol Biochem Parasitol. 2008;159:7–23. doi: 10.1016/j.molbiopara.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffing S, Syphard L, Sridaran S, McCollum AM, Mixson-Hayden T, Vinayak S, et al. pfmdr1 amplification and fixation of pfcrt chloroquine resistance alleles in Plasmodium falciparum in Venezuela. Antimicrob Agents Chemother. 2010;54:1572–1579. doi: 10.1128/AAC.01243-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings IM, Donnelly MJ. The impact of antimalarial drug resistance mutations on parasite fitness, and its implications for the evolution of resistance. Drug Resist Updat. 2005;8:43–50. doi: 10.1016/j.drup.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Hrycyna CA, Summers RL, Lehane AM, Pires MM, Namanja H, Bohn K, et al. Quinine dimers are potent inhibitors of the Plasmodium falciparum chloroquine resistance transporter and are active against quinoline-resistant P. falciparum . ACS chemical biology. 2014;9:722–730. doi: 10.1021/cb4008953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isozumi R, Uemura H, Le DD, Truong VH, Nguyen DG, Ha VV, et al. Longitudinal survey of Plasmodium falciparum infection in Vietnam: characteristics of antimalarial resistance and their associated factors. Journal of clinical microbiology. 2010;48:70–77. doi: 10.1128/JCM.01449-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano JJ, Kwiek JJ, Cappell K, Mwapasa V, Meshnick SR. Minority-variant pfcrt K76T mutations and chloroquine resistance, Malawi. Emerg Infect Dis. 2007;13:872–877. doi: 10.3201/eid1306.061182. [DOI] [PubMed] [Google Scholar]
- Kidgell C, Volkman SK, Daily J, Borevitz JO, Plouffe D, Zhou Y, et al. A systematic map of genetic variation in Plasmodium falciparum . PLoS Pathog. 2006;2:e57. doi: 10.1371/journal.ppat.0020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogstad DJ, Gluzman IY, Kyle DE, Oduola AM, Martin SK, Milhous WK, Schlesinger PH. Efflux of chloroquine from Plasmodium falciparum: mechanism of chloroquine resistance. Science. 1987;238:1283–1285. doi: 10.1126/science.3317830. [DOI] [PubMed] [Google Scholar]
- Kublin JG, Cortese JF, Njunju EM, Mukadam RA, Wirima JJ, Kazembe PN, et al. Reemergence of chloroquine-sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi. J Infect Dis. 2003;187:1870–1875. doi: 10.1086/375419. [DOI] [PubMed] [Google Scholar]
- Lakshmanan V, Bray PG, Verdier-Pinard D, Johnson DJ, Horrocks P, Muhle RA, et al. A critical role for PfCRT K76T in Plasmodium falciparum verapamil-reversible chloroquine resistance. EMBO J. 2005;24:2294–2305. doi: 10.1038/sj.emboj.7600681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufer MK, Takala-Harrison S, Dzinjalamala FK, Stine OC, Taylor TE, Plowe CV. Return of chloroquine-susceptible falciparum malaria in Malawi was a reexpansion of diverse susceptible parasites. J Infect Dis. 2010;202:801–808. doi: 10.1086/655659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin BR, Perrot V, Walker N. Compensatory mutations, antibiotic resistance and the population genetics of adaptive evolution in bacteria. Genetics. 2000;154:985. doi: 10.1093/genetics/154.3.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis IA, Wacker M, Olszewski KL, Cobbold SA, Baska KS, Tan A, et al. Metabolic QTL analysis links chloroquine resistance in Plasmodium falciparum to impaired hemoglobin catabolism. PLoS genetics. 2014;10:e1004085. doi: 10.1371/journal.pgen.1004085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maree AF, Keulen W, Boucher CA, De Boer RJ. Estimating relative fitness in viral competition experiments. J Virol. 2000;74:11067–11072. doi: 10.1128/jvi.74.23.11067-11072.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin RE, Marchetti RV, Cowan AI, Howitt SM, Broer S, Kirk K. Chloroquine transport via the malaria parasite’s chloroquine resistance transporter. Science. 2009;325:1680–1682. doi: 10.1126/science.1175667. [DOI] [PubMed] [Google Scholar]
- Martin SK, Oduola AM, Milhous WK. Reversal of chloroquine resistance in Plasmodium falciparum by verapamil. Science. 1987;235:899–901. doi: 10.1126/science.3544220. [DOI] [PubMed] [Google Scholar]
- Mehlotra RK, Fujioka H, Roepe PD, Janneh O, Ursos LM, Jacobs-Lorena V, et al. Evolution of a unique Plasmodium falciparum chloroquine-resistance phenotype in association with pfcrt polymorphism in Papua New Guinea and South America. Proc Natl Acad Sci USA. 2001;98:12689–12694. doi: 10.1073/pnas.221440898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mharakurwa S, Sialumano M, Liu K, Scott A, Thuma P. Selection for chloroquine-sensitive Plasmodium falciparum by wild Anopheles arabiensis in Southern Zambia. Malar J. 2013;12:453. doi: 10.1186/1475-2875-12-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita T, Kaneko A, Lum J, Zungu I, Tsukahara T, Eto H, et al. Expansion of wild type allele rather than back mutation in pfcrt explains the recent recovery of chloroquine sensitivity of Plasmodium falciparum in Malawi. Mol Biochem Parasitol. 2004;135:159–163. doi: 10.1016/j.molbiopara.2004.01.011. [DOI] [PubMed] [Google Scholar]
- Mita T, Kaneko A, Lum JK, Bwijo B, Takechi M, Zungu I, et al. Recovery of chloroquine sensitivity and low prevalence of the Plasmodium falciparum chloroquine resistance transporter gene mutation K76T following the discontinuance of chloroquine use in Malawi. Am J Trop Med Hyg. 2003;68:413–415. [PubMed] [Google Scholar]
- Mitchell-Olds T, Willis JH, Goldstein DB. Which evolutionary processes influence natural genetic variation for phenotypic traits? Nat Rev Genet. 2007;8:845–856. doi: 10.1038/nrg2207. [DOI] [PubMed] [Google Scholar]
- Mixson-Hayden T, Jain V, McCollum AM, Poe A, Nagpal AC, Dash AP, et al. Evidence of selective sweeps in genes conferring resistance to chloroquine and pyrimethamine in Plasmodium falciparum isolates in India. Antimicrob Agents Chemother. 2010;54:997–1006. doi: 10.1128/AAC.00846-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu J, Myers RA, Jiang H, Liu S, Ricklefs S, Waisberg M, et al. Plasmodium falciparum genome-wide scans for positive selection, recombination hot spots and resistance to antimalarial drugs. Nat Genet. 2010;42:268–271. doi: 10.1038/ng.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwai L, Ochong E, Abdirahman A, Kiara SM, Ward S, Kokwaro G, et al. Chloroquine resistance before and after its withdrawal in Kenya. Malar J. 2009;8:106. doi: 10.1186/1475-2875-8-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash D, Nair S, Mayxay M, Newton PN, Guthmann JP, Nosten F, Anderson TJ. Selection strength and hitchhiking around two anti-malarial resistance genes. Proc Biol Sci. 2005;272:1153–1161. doi: 10.1098/rspb.2004.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olliaro P, Mussano P. Amodiaquine for treating malaria. Cochrane Database Syst Rev. 2000 doi: 10.1002/14651858.CD000016. CD000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ord R, Alexander N, Dunyo S, Hallett R, Jawara M, Targett G, et al. Seasonal carriage of pfcrt and pfmdr1 alleles in Gambian Plasmodium falciparum imply reduced fitness of chloroquine-resistant parasites. J Infect Dis. 2007;196:1613–1619. doi: 10.1086/522154. [DOI] [PubMed] [Google Scholar]
- Osman ME, Mockenhaupt FP, Bienzle U, Elbashir MI, Giha HA. Field-based evidence for linkage of mutations associated with chloroquine (pfcrt/pfmdr1) and sulfadoxine-pyrimethamine (pfdhfr/pfdhps) resistance and for the fitness cost of multiple mutations in P. falciparum . Infection, genetics and evolution : journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2007;7:52–59. doi: 10.1016/j.meegid.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Park DJ, Lukens AK, Neafsey DE, Schaffner SF, Chang HH, Valim C, et al. Sequence-based association and selection scans identify drug resistance loci in the Plasmodium falciparum malaria parasite. Proc Natl Acad Sci USA. 2012;109:13052–13057. doi: 10.1073/pnas.1210585109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual A, Madamet M, Bertaux L, Amalvict R, Benoit N, Travers D, et al. In vitro piperaquine susceptibility is not associated with the Plasmodium falciparum chloroquine resistance transporter gene. Malar J. 2013;12:431. doi: 10.1186/1475-2875-12-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel JJ, Thacker D, Tan JC, Pleeter P, Checkley L, Gonzales JM, et al. Chloroquine susceptibility and reversibility in a Plasmodium falciparum genetic cross. Mol Microbiol. 2010;78:770–787. doi: 10.1111/j.1365-2958.2010.07366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price RN, Tjitra E, Guerra CA, Yeung S, White NJ, Anstey NM. Vivax malaria: neglected and not benign. Am J Trop Med Hyg. 2007;77:79–87. [PMC free article] [PubMed] [Google Scholar]
- Rosario VE, Hall R, Walliker D, Beale GH. Persistence of drug-resistant malaria parasites. Lancet. 1978;1:185–187. doi: 10.1016/s0140-6736(78)90616-5. [DOI] [PubMed] [Google Scholar]
- Rosenthal PJ. The interplay between drug resistance and fitness in malaria parasites. Mol Microbiol. 2013;89:1025–1038. doi: 10.1111/mmi.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sa JM, Twu O. Protecting the malaria drug arsenal: halting the rise and spread of amodiaquine resistance by monitoring the PfCRT SVMNT type. Malar J. 2010;9:374. doi: 10.1186/1475-2875-9-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sa JM, Twu O, Hayton K, Reyes S, Fay MP, Ringwald P, Wellems TE. Geographic patterns of Plasmodium falciparum drug resistance distinguished by differential responses to amodiaquine and chloroquine. Proc Natl Acad Sci USA. 2009;106:18883–18889. doi: 10.1073/pnas.0911317106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saliba KJ, Folb PI, Smith PJ. Role for the Plasmodium falciparum digestive vacuole in chloroquine resistance. Biochem Pharmacol. 1998;56:313–320. doi: 10.1016/s0006-2952(98)00140-3. [DOI] [PubMed] [Google Scholar]
- Sanchez CP, Liu CH, Mayer S, Nurhasanah A, Cyrklaff M, Mu J, et al. A HECT ubiquitin-protein ligase as a novel candidate gene for altered quinine and quinidine responses in Plasmodium falciparum . PLoS genetics. 2014;10:e1004382. doi: 10.1371/journal.pgen.1004382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez CP, Mayer S, Nurhasanah A, Stein WD, Lanzer M. Genetic linkage analyses redefine the roles of PfCRT and PfMDR1 in drug accumulation and susceptibility in Plasmodium falciparum . Mol Microbiol. 2011;82:865–878. doi: 10.1111/j.1365-2958.2011.07855.x. [DOI] [PubMed] [Google Scholar]
- Sanchez CP, Stein W, Lanzer M. Trans stimulation provides evidence for a drug efflux carrier as the mechanism of chloroquine resistance in Plasmodium falciparum . Biochemistry. 2003;42:9383–9394. doi: 10.1021/bi034269h. [DOI] [PubMed] [Google Scholar]
- Sidhu AB, Verdier-Pinard D, Fidock DA. Chloroquine resistance in Plasmodium falciparum malaria parasites conferred by pfcrt mutations. Science. 2002;298:210–213. doi: 10.1126/science.1074045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisowath C, Petersen I, Veiga MI, Mortensson A, Premji Z, Bjorkman A, et al. In vivo selection of Plasmodium falciparum parasites carrying the chloroquine-susceptible pfcrt K76 allele after treatment with artemether-lumefantrine in Africa. J Infect Dis. 2009;199:750–757. doi: 10.1086/596738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow RW, Trape JF, Marsh K. The past, present and future of childhood malaria mortality in Africa. Trends Parasitol. 2001;17:593–597. doi: 10.1016/s1471-4922(01)02031-1. [DOI] [PubMed] [Google Scholar]
- Straimer J, Gnadig NF, Witkowski B, Amaratunga C, Duru V, Ramadani AP, et al. Drug resistance. K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science. 2015;347:428–431. doi: 10.1126/science.1260867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Kirkman LA, Fujioka H, Wellems TE. Complex polymorphisms in an approximately 330 kDa protein are linked to chloroquine-resistant P. falciparum in Southeast Asia and Africa. Cell. 1997;91:593–603. doi: 10.1016/s0092-8674(00)80447-x. [DOI] [PubMed] [Google Scholar]
- Summers RL, Dave A, Dolstra TJ, Bellanca S, Marchetti RV, Nash MN, et al. Diverse mutational pathways converge on saturable chloroquine transport via the malaria parasite’s chloroquine resistance transporter. Proc Natl Acad Sci USA. 2014;111:E1759–E1767. doi: 10.1073/pnas.1322965111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trape JF. The public health impact of chloroquine resistance in Africa. The American journal of tropical medicine and hygiene. 2001;64:12–17. doi: 10.4269/ajtmh.2001.64.12. [DOI] [PubMed] [Google Scholar]
- Valderramos SG, Fidock DA. Transporters involved in resistance to antimalarial drugs. Trends Pharmacol Sci. 2006;27:594–601. doi: 10.1016/j.tips.2006.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valderramos SG, Valderramos JC, Musset L, Purcell LA, Mercereau-Puijalon O, Legrand E, Fidock DA. Identification of a mutant PfCRT-mediated chloroquine tolerance phenotype in Plasmodium falciparum . PLoS Pathog. 2010;6:e1000887. doi: 10.1371/journal.ppat.1000887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkman S, Sabeti P, Decaprio D, Neafsey D, Schaffner S, Milner D, et al. A genome-wide map of diversity in Plasmodium falciparum . Nat Genet. 2007;39:113–119. doi: 10.1038/ng1930. [DOI] [PubMed] [Google Scholar]
- Walliker D, Hunt P, Babiker H. Fitness of drug-resistant malaria parasites. Acta Tropica. 2005;94:251–259. doi: 10.1016/j.actatropica.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Wang X, Mu J, Li G, Chen P, Guo X, Fu L, et al. Decreased prevalence of the Plasmodium falciparum chloroquine resistance transporter 76T marker associated with cessation of chloroquine use against P. falciparum malaria in Hainan, People's Republic of China. Am J Trop Med Hyg. 2005;72:410–414. [PubMed] [Google Scholar]
- Wellems TE, Panton LJ, Gluzman IY, do Rosario VE, Gwadz RW, Walker-Jonah A, Krogstad DJ. Chloroquine resistance not linked to mdr-like genes in a Plasmodium falciparum cross. Nature. 1990;345:253–255. doi: 10.1038/345253a0. [DOI] [PubMed] [Google Scholar]
- Wellems TE, Plowe CV. Chloroquine-resistant malaria. J Infect Dis. 2001;184:770–776. doi: 10.1086/322858. [DOI] [PubMed] [Google Scholar]
- Wells TN, Alonso PL, Gutteridge WE. New medicines to improve control and contribute to the eradication of malaria. Nat Rev Drug Discov. 2009;8:879–891. doi: 10.1038/nrd2972. [DOI] [PubMed] [Google Scholar]
- White NJ. Qinghaosu (artemisinin): the price of success. Science. 2008;320:330–334. doi: 10.1126/science.1155165. [DOI] [PubMed] [Google Scholar]
- WHO. World Malaria Report 2014. 2014 http://wwwwhoint/malaria/publications/world_malaria_report_2014/report/en/.
- Wootton JC, Feng X, Ferdig MT, Cooper RA, Mu J, Baruch DI, et al. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum . Nature. 2002;418:320–323. doi: 10.1038/nature00813. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.