Abstract
Methane (CH4) emission by carbon-rich cryosols at the high latitudes in Northern Hemisphere has been studied extensively. In contrast, data on the CH4 emission potential of carbon-poor cryosols is limited, despite their spatial predominance. This work employs CH4 flux measurements in the field and under laboratory conditions to show that the mineral cryosols at Axel Heiberg Island in the Canadian high Arctic consistently consume atmospheric CH4. Omics analyses present the first molecular evidence of active atmospheric CH4-oxidizing bacteria (atmMOB) in permafrost-affected cryosols, with the prevalent atmMOB genotype in our acidic mineral cryosols being closely related to Upland Soil Cluster α. The atmospheric (atm) CH4 uptake at the study site increases with ground temperature between 0 °C and 18 °C. Consequently, the atm CH4 sink strength is predicted to increase by a factor of 5–30 as the Arctic warms by 5–15 °C over a century. We demonstrate that acidic mineral cryosols are a previously unrecognized potential of CH4 sink that requires further investigation to determine its potential impact on larger scales. This study also calls attention to the poleward distribution of atmMOB, as well as to the potential influence of microbial atm CH4 oxidation, in the context of regional CH4 flux models and global warming.
Introduction
After 7 years of a steady-state concentration between 1999 and 2006, atmospheric (atm) CH4 has been increasing steadily at 5 p.p.b. year−1 (Dlugokencky et al., 2009). At 1.80 p.p.m.v. in 2011, the atm CH4 concentration is 217 times lower than that of carbon dioxide (CO2); however, it accounts for 18% of the total radiative forcing by long-lived greenhouse gases (IPCC, 2013) and has a global warming potential 75 times greater than CO2 over a 20-year timescale (Shindell et al., 2009). Although cost-effective strategies could be applied to mitigate CH4 production due to anthropogenic activities (~64% of current global total CH4 emissions), future global CH4 emissions will depend greatly on the uncertain responses of natural ecosystems to climate change, especially in wetlands and permafrost-affected areas in the Arctic and sub-Arctic regions (Dlugokencky et al., 2011; Graham et al., 2012).
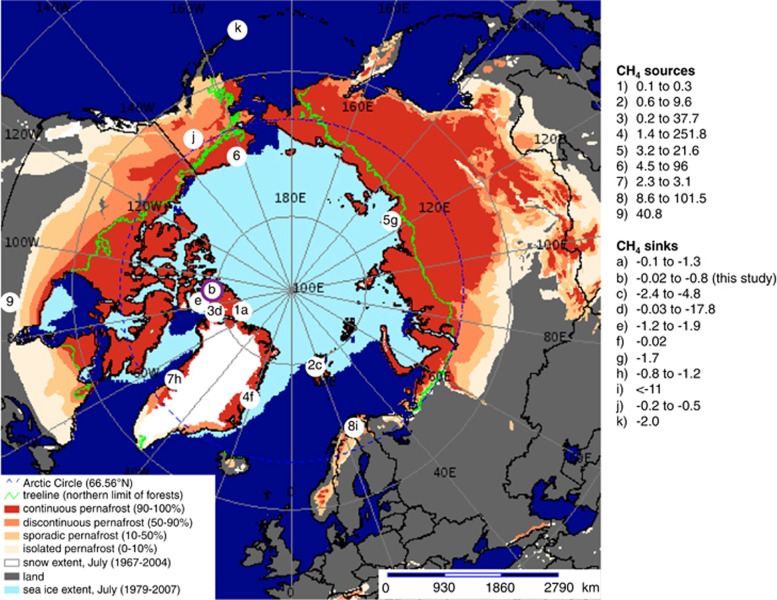
Permafrost, defined as ‘ground that remains ⩽0 °C for at least 2 consecutive years' (Van Everdingen, 1998), is overlain by an active layer that thaws seasonally. By 2100, it is projected that the mean annual air temperature in the Arctic will have increased by up to 10 °C (IPCC, 2013), which would result in warming of the 17.8–22.8 × 106 km2 of permafrost (Zhang et al., 2008; Hugelius et al., 2014) that contains 1034±183 or 1104±133 Pg of soil organic carbon in the top 3 m (Hugelius et al., 2014). If 20–59% of the permafrost has thawed down to 0.5–1 m by 2200 as predicted (Schaefer et al., 2011), this large carbon pool would become available for microbial mineralization into greenhouse gases and may amplify warming. As a result of the concern about this positive feedback response that further intensifies warming, the transition of carbon-rich permafrost into CH4-emitting wetlands has been the focus of considerable research (1–9 in Figure 1; Supplementary Table S1), even though the majority (87%) of Arctic permafrost is comprised of mineral (that is, carbon-poor) cryosols (Hugelius et al., 2014). Observations of atm CH4 uptake were first reported in Alaskan permafrost sites in early 1990 s (Whalen and Reeburgh, 1990). Since then, atm CH4 uptake has occasionally been reported in other permafrost-affected sites (a–k in Figure 1; Supplementary Table S1), including cryosols of high organic carbon and water saturation levels where CH4 emission was expected. Recently reported CH4 fluxes of polar desert mineral cryosols at Ellesmere Island, Canada over five consecutive summers also indicate consumption of atm CH4 (a in Figure 1; Emmerton et al., 2014). In the present study, we investigated the CH4 feedback response of mineral cryosols on Axel Heiberg Island (AHI) in the Canadian high Arctic by both in situ flux measurements and laboratory experiments, identified the active functional groups responsible for the feedback process and evaluated how these cryosols would respond to changing climate.
Figure 1.
CH4 field fluxes in the Northern Circumpolar permafrost region. Sites that showed net CH4 release are sources (numbers) whereas those showed net atm CH4 consumption are sinks (letters). The CH4 fluxes are rounded up to the first decimal place except for values <0.1 mg CH4-C m−2 day−1. Purple outline indicates that the presence of atmMOB is supported by microbial data. Information of soil characteristics is provided in Supplementary Table S1. Background map was generated using the interactive tool ‘The Atlas of the Cryosphere' (Maurer, 2007) available at the National Snow and Ice Data Center website (http://nsidc.org/data/atlas/).
Materials and methods
Site description
The study site, characterized by high-centered ice-wedge polygons of acidic tundra, is located at an upland polygonal terrain in proximity to the McGill Arctic Research Station at Expedition Fjord (79°24'57"N, 90°45'46"W), AHI, Nunavut, Canada. The average depth of the active layer varied between 60 and 73 cm (mid-summer 2009–2011; Allan et al., 2014). The mean soil temperatures in mid-summer (14 July 2013) at both the polygon interior and the ice wedge trough were 9±0.8 °C at 5-cm depth and decreased to 3±1.4 °C at 20-cm depth. Soil temperatures were measured by LiCOR thermistor (Maxim Integrated Products, San Jose, CA, USA). The polygon (16 × 16 m2) from which cryosols were collected for incubation and molecular studies was sparsely vegetated but root materials were present to 15-cm depth (Stackhouse et al., submitted). Soil characteristics and pore water chemistry has been analyzed by Stackhouse et al. (submitted). Briefly, the cryosols were slightly acidic (pH 5.5–6). The top 10 cm of the active layer had up to 6 wt% of organic carbon but much less (~1 wt%) from 10 cm down to the permafrost table during summer. Total nitrogen did not vary with depth (0.1 wt%).
The general methodology of this study is presented here. A detailed version is provided in the Supplementary Material, which includes the measurements of CH4 uptake in the field and under laboratory conditions, the setup of incubation experiments, various molecular analyses and the characterization of CH4 uptake rates by the mineral cryosols.
Field flux measurements
In situ CH4 fluxes were measured in July 2011–2013 using a Picarro soil CO2-CH4 gas analyzer (Picarro Inc., Santa Clara, CA, USA) or Los Gatos Fast Methane Analyzer (Los Gatos Research Inc., Mountain View, CA, USA) (Allan et al., 2014; Stackhouse et al., submitted and this study). Surface fluxes were measured in replicates using open-circuit dark chambers with continuous gas replacement from the air in 2011–2013 or closed-static chamber in 2013. Soil temperatures at corresponding depths were measured by LiCOR thermistor (Maxim Integrated Products).
Abundance of methanotrophs and methanogens in metagenomic studies
As part of the long-term intact core-warming experiments, 4 g of cryosols were collected and processed as described in Stackhouse et al. (submitted) and Supplementary Material. Total DNA was extracted using the Fast DNA SPIN Kit (MP Biomedical, Irvine, CA, USA) (Vishnivetskaya et al., 2014). The extracted DNA samples were used to prepare metagenome shotgun libraries using the Illumina Nextera DNA Library Preparation Kit (Illumina, Inc., San Diego, CA, USA) and sequenced on an Illumina HiSeq 2000 platform (Chauhan et al., 2014). Raw data from 16 near-surface (at 5-cm depth) cryosol samples representing different time point (T=1 week, 6 and 12 months) were processed and analyzed using MG-RAST (Meyer et al., 2008). All gene features were taxonomically assigned for determining the microbial compositions. The relative abundance of each individual genus was not statistically different at alpha=0.05 (analysis of variance in STatistical Analysis of Metagenomic Profiles (STAMP; Parks and Beiko, 2010)), and therefore means (and s.ds.) of all 16 samples were reported. Only the methanotrophic and methanogenic genera (as reviewed by Nazaries et al., 2013) with relative abundances >0.001% were reported.
Assembly of pmo gene from metagenomes
De novo co-assembly of raw sequences from 10 libraries (five 1-week and five 6-months thawed samples at 5-cm depth) were performed using MetaVelvet (Namiki et al., 2012). Functional classifications were annotated separately via IMG/ER and MG-RAST (ID: 4530050.3). Contigs identified as ‘methane monooxygenase' were searched for protein-coding genes because they may contain fragments of more than one gene. Phylogenetic affiliation of individual gene was queried against the National Center for Biotechnology Information (NCBI) non-redundant protein database using BlastX. Co-assembly of sequences from multiple libraries usually masks the genetic variations within and between populations or libraries, and the resultant contigs likely contain mixed genetic signals from the dominant population. We used the prefix pan- to indicate that the detected genes are not derived from a single clonal population.
De novo assembly is more preferred when compared with reference-based assembly because the latter prevents the discovery of new genotypes or variants by setting an a priori framework for read alignment. Nonetheless, raw reads of five 1-week thawed samples (5-cm depth) were mapped to the representative pmoCAB operon of Upland Soil Cluster α (USCα) recovered by bacterial artificial chromosome cloning (GenBank Acc. No. CT005232) (Ricke et al., 2005) to demonstrate that a complete pmoCAB operon of the USCα genotype was successfully assembled from our data.
Phylogenetic analyses of pmo genes
Phylogenetic trees were constructed from deduced amino-acid (aa) sequences for pmoA, pmoB and pmoC genes that encodes for α, β and γ subunit of particulate methane monooxygenase (pMMO), respectively. All sequences had no frame-shift errors and no curation was applied. Three data sets, one for each gene, were created. Each data set contains the aa sequences of pan-pmo genes from this study and reference sequences downloaded from NCBI (http://www.ncbi.nlm.nih.gov/). For the latter where aa sequences were not available, nucleotide sequences were downloaded and translated. Positions covered by more than half of the sequences were included while unaligned and ambiguous positions were trimmed. RAxML (v7.2.7 alpha) (Stamatakis, 2006; Stamatakis et al., 2008) was used to search for the best-scoring maximum likelihood tree with the aa evolutionary model selected by ProtTest (v3.3) (Darriba et al., 2011) and empirically estimated base frequencies, and to perform a rapid bootstrap analysis of 100 iterations in single run.
Assembly of pmo genes from metatranscriptome
Cryosols for metatranscriptomic analysis were collected on 15 July 2013 from an ice-wedge polygon (79°24'57"N, 90°45'48"W), namely polygon interior and trough. The samples were preserved using LifeGuard Soil Preservation Solution (MO BIO Laboratories, Inc., Carlsbad, CA, USA) and stored at −20 °C. Total RNA was extracted from 15 g of soil using the RNA PowerSoil Total RNA Isolation Kit (MO BIO Laboratories, Inc.). Illumina TruSeq libraries were generated from the total RNA following the manufacturer's protocols (Illumina) and sequenced on MiSeq (1 × 150 nt). Raw reads were quality controlled and assembled (built-in assembler velvet) using CLC Genomics Workbench (version 7.0; CLC Bio, Boston, MA, USA), and annotated by MG-RAST (ID 4548476.3 and 4548477.3 for the polygon interior and trough samples, respectively). Transcript contigs containing pmoB genes were translated and used as template for the alignment of the peptide sequences detected from the proteome experiment (as described below).
Identification of pMMO in metaproteome
Cores showing high CH4 uptake flux in the intact core-thawing experiment (Stackhouse et al., submitted) were selected. Cryosols at 5-cm depth from 1-week cores, with in situ water saturation conditions, were subsampled and kept frozen at −20 °C. Three grams of cryosol was mixed with sodium dodecyl sulfate-based lysis buffer, and the slurry were subjected to protein extraction and trypsin proteolysis described earlier (Chourey et al., 2010). Digested peptides were separated on an in-house packed SCX (Luna)-C18 (Aqua) column and analyzed by an LTQ-Orbitrap (Thermo Fisher Scientific, San Jose, CA, USA) coupled to an Ultimate 3000 HPLC system (Dionex, Thermo Fisher Scientific Inc., Waltham, MA, USA). The raw spectra acquired by 12-step MS/MS runs were searched via SEQUEST v27 (Eng et al., 1994) against a custom pMMO database using parameters described elsewhere (Thompson et al., 2006; Sharma et al., 2012). Sequences of common contaminants such as trypsin and keratin were also concatenated to the database. Identification of at least two peptide sequences per protein was set as the criterion for positive protein identifications. Three technical replicates were analyzed for each protein sample.
Microcosm incubation experiments
Two sets of microcosms were set up to study the effect of water saturation and temperature on atm CH4 oxidation rates. A frozen core collected in April 2011 (Stackhouse et al., submitted) was dissected into sections for every 10 cm. The peripheral rim of 5-cm thick was discarded to remove any potential contaminants from the core liner. The pristine cryosols were put into sterile Whirl-pak bags and homogenized by hand. The 0–10-cm section was used in this experiment. Cryosols were preconditioned to attain the desired water saturation levels of 33, 66 and 100%, respectively, by storing subsamples in a desiccator at 4 °C. Eight-to-10 g (wet weight) of cryosols were then put into vials and sealed with treated butyl rubber stoppers and Al-crimps. Blank vials containing no soils were used to track abiotic gas exchanges and minor instrumental drift. All treatments were run in triplicates. The headspace was flushed for 2–4 min with manufactured air (20% O2, 80% N2, 2.0 p.p.m.v. CH4, 400 p.p.m.v. CO2, 53±21 p.p.b.v. H2 and 95±14 p.p.b.v. CO). Additional manufactured air was injected with a gas-tight glass syringe to overpressurize the vials to 1.5 atm. One set of 12 vials was incubated at 4 °C while another set was incubated at 10 °C. Gas was sampled from the headspace at T=0, twice per week for the first 2 weeks and weekly for another 2 weeks (period of incubation=31 days). Analysis was performed on Picarro iCO2 (Model no. G2101-I) using the G2101-i coordinator (Picarro Inc.). The detection limit for CH4 is 50 p.p.b.v.
Temperature coefficients (Q10)
Q10 is used to measure the rate of change of a chemical or biological reaction as a consequent of temperature increase of 10 °C. Q10 values were computed for methanotrophy using (1) mean CH4 oxidation rates obtained from microcosms for each treatment and corresponding incubation temperatures; and (2) mean CH4 oxidation rates reported in the literature for which temperature data were available. For the latter, only rates collected from a temperature difference of >5 °C were used.
Arrhenius relationship between in situ CH4 uptake flux and surface soil temperature
We are cautious about quantitatively extrapolating the observed effects of temperature and water saturation under laboratory conditions to the real situation and model prediction, therefore only field CH4 fluxes and the corresponding surface soil temperatures measured during 2011–2013 expeditions were used to determine the Arrhenius relationship. Fluxes measured by open-circuit chambers were used to determine the relationship, whereas those measured by closed-static chambers were excluded to eliminate the variations resulted from different collection methods (Whalen et al., 1992). Natural logarithm of CH4 uptake fluxes (y axis) was plotted against 1000/temperature (x axis) to create an Arrhenius plot, which showed a potential change in the slope. The data were then analyzed using the ‘Segmented' R package (http://cran.r-project.org/web/packages/segmented/). For comparative purpose, other data were overlain on the plot (Figure 4), which include: (1) CH4 fluxes estimated from our microcosm experiments at 2.0 p.p.m.v. of CH4; (2) CH4 fluxes estimated from intact core-thawing experiments (Stackhouse et al., submitted); and (3) atm CH4 oxidation sites at lower latitudes.
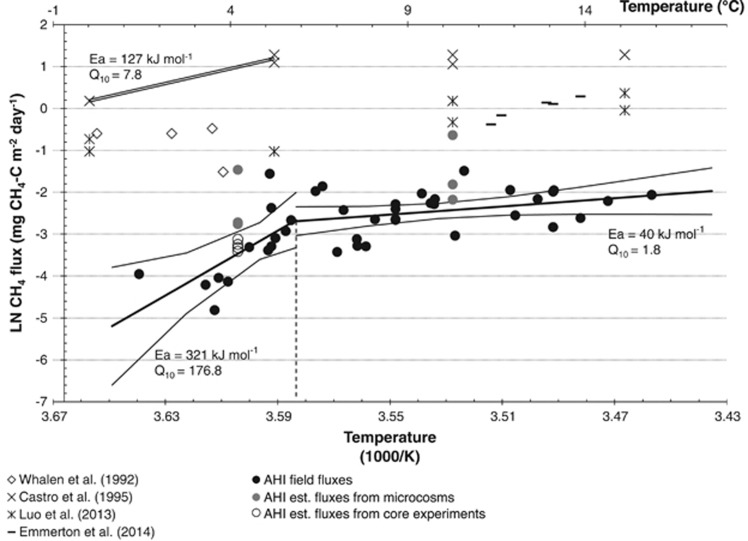
Figure 4.
Arrhenius relationship of CH4 uptake and surface soil temperature at AHI. Expected ln-CH4 fluxes (thick line) and 95% confident intervals (thin line) were estimated from best-fit linear regression analyses. The breakpoint temperature reads 3.588 (eq. 5.6 °C), as indicated by the dashed line. Values of activation energy (Ea) and temperature coefficient (Q10) were derived from respective slopes. Results estimated from forest soils (parallel lines, Castro et al., 1995) are provided as a reference.
Estimation of monthly and annual atm CH4 uptake fluxes
Monthly air temperatures at AHI during 1990 s and 2090 s were simulated through the Climate Model Intercomparison Project (CMIP5) using eight climate models (BCC-CSM1.1, CCSM4, CSIRO-Mk3.6.0, GFDL-ESM2M, GISS-E2-R, HadGEM2-AO, IPSL-CM5A-MR and NorESM1-M) (Taylor et al., 2012). The ‘high emissions scenario' assuming mitigation policies in action (RCP8.5) was used to project the climate change in 2090s. Monthly and annual atm CH4 uptakes were estimated for temperatures from each model, and multi-model means were obtained by taking average across all models.
Air temperatures at Eureka, Ellesmere Island, Nunavut, Canada (N80°00'03', W86°00'25' 112 km NE of AHI) were available for 2010 and 2011 through Total Carbon Column Observing Network (TCCON) (Wunch et al., 2011). Temperature data (T) from late March to August 2011 was used. Missing data was gap-filled by linear interpolation between the two neighboring values. Mean daily temperatures were calculated by averaging multiple measurements on the day, which were then averaged to give monthly temperatures. The monthly and annual uptake fluxes were estimated in a similar manner as that using the CMIP5-simulated temperatures. Eureka is located at higher latitude where the temperature is slightly cooler than that at AHI. CH4 uptake fluxes therefore were also calculated for T+1 °C and T+6 °C which, respectively, are more representative for our study site and also to mimic severe summer warming, which was not projected by climate models.
Results and Discussion
In situ atmospheric CH4 consumption by mineral cryosols
Measurement of in situ surface CH4 flux at various locations at the AHI site during the initial thaw and into mid-summer from 2011 to 2013 consistently showed atm CH4 uptake (b in Figure 1; Supplementary Table S1). The CH4 uptake rates at AHI (−0.1 to −0.8 mg CH4-C m−2 day−−1) are at the lower end of the range (−0.02 to −17.8 mg CH4-C m−2 day−1) observed among other sites in the Arctic and sub-Arctic region (a–k in Figure 1), but are comparable with that of other terrestrial systems at lower latitudes (−0.1 to −1.0 mg CH4-C m−2 day−1) (Luo et al., 2013). The comparison of the scarce data on CH4 uptake at the northern latitudes suggests that mineral cryosols act as a constant active atm CH4 sink (this study and Emmerton et al., 2014) in part because of their low soil organic carbon availability, low vegetation cover and low moisture content (Supplementary Table S1). This field observation was followed up with molecular analyses and incubation experiments to understand this atm CH4 oxidation process that is known to be mediated by microorganisms.
Atmospheric CH4-oxidizing bacteria detected by metagenomics
From the top 5 cm of intact cores in a long-term warming study (Stackhouse et al., submitted), we extracted DNA for metagenomic analyses. All gene features predicted by the MG-RAST M5NR database revealed that known methanotrophic taxa accounted for only 0.7% of all annotated sequences (Supplementary Table S2A) in comparison to the 55% of Proteobacteria and Actinobacteria (Chauhan et al., 2014). Among the methanotrophic community, α-proteobacterial (Type II) methanotrophs dominated over γ-proteobacterial (Type I) or verrucomicrobial methanotrophs (Supplementary Table S2A). The abundance of the methanotrophic community at our study site decreased with depth (Stackhouse et al., submitted).
Atm CH4 oxidation primarily occurs by reaction with hydroxyl radicals (OH ) in the troposphere and stratosphere but is also mediated by aerobic CH4-oxidizing bacteria (MOB) that have high affinity for atm CH4, or atmospheric CH4-oxidizing bacteria (atmMOB) (Dlugokencky et al., 2011). Soils consuming atmospheric CH4, for example, forest soils, show low Michaelis–Menten constants (Km), ranging from 2 to 348 p.p.m.v. (Knief et al., 2003; Bender and Conrad, 1992). As genetic information on atmMOB has been limited to a collection of pmoA genes encoding for the α subunit of pMMO (Degelmann et al., 2010; Martineau et al., 2014; Bárcena et al., 2011) and a 42-kb-long sequence of the genotype USCα (Ricke et al., 2005), the search for pmo genes has been the only way to detect the yet-to-be cultured atmMOB whose phylogenetic identity of 16S rRNA gene has remained elusive.
De novo co-assembly of raw sequences yielded 13 contigs that contained genes affiliated to MMO and homologous enzymes (Supplementary Table S3A). Six of these contigs (528–1301 nt) yielded seven pan-pmo gene fragments. We estimated the abundance of these pan-pmo genes by mapping read sequences against the 13 contigs. The low percentage of pmo genes confirmed that MOB are a minority in the cryosol community (<1%) (Supplementary Table S3A). The phylogeny of our pan-pmoA, pmoB and pmoC genes (Supplementary Figures S1–S3) suggests that the prevalent pMMO-possessing MOB in our cryosol samples are closely related to USCα, which appears to be the dominant MOB in acidic soils where atm CH4 uptake was reported (Knief et al., 2003; Ricke et al., 2005; Martineau et al., 2014), such as our samples (pH 5.5–6).
In acidic forest soil samples where only USCα-like pmoA genes were detected, the 16S rRNA genes from α-proteobacterial families Methylocystaceae and Beijerinckiaceae were detected and were more abundant than γ-proteobacterial MOB (Lau et al., 2007), which is similar to what was found in this study. Although the phylogenetic identity of USCα cannot be resolved down to genus level, it is logical to think that, similar to the 16S rRNA genes, protein-coding genes of USCα are homologous to those of these two families. Hence, the gene features in our metagenomes being assigned to these two families may come from the USCα-like atmMOB. Unfortunately, these annotation results derived from all gene features do not give a reliable basis to deduce the relative abundance of our USCα-like atmMOB.
MOB strains grown under high CH4 concentrations in the laboratory are found unable to oxidize CH4 at low concentrations (<600 p.p.m.v.). However, exceptions have been reported in the literature. For example, Methylobacter albus BG8 oxidized atm CH4 when supplemented with methanol (Benstead et al., 1998), and Methylocystis sp. SC2 oxidized atm CH4 by expressing its high-affinity pMMO (Baani and Liesack, 2008). Genes related to these two genera, albeit at low abundance, were detected in our metagenomic data (Supplementary Table S2A), but the specific strains aforementioned were not detected. Although the contribution from other MOB cannot be excluded, USCα-like atmMOB is likely responsible for most of the observed atm CH4 uptake at our site.
Technical problems such as insufficient sequencing depth in metagenomic analyses and primer bias in PCR-based assays (McDonald et al., 2008) partly explain why atmMOB has not been reported in previous molecular studies carried out in the Arctic region (Supplementary Table S4). To our knowledge, this study presents the first report of USCα-like atmMOB in metagenomic libraries, indicating that our data may represent the largest inventory of USCα genomes available, which will facilitate the search for additional genes belonging to USCα and hence improve the identification of its phylogenetic lineage.
Active atmospheric CH4-oxiding bacteria revealed by metatranscriptomics and metaproteomics
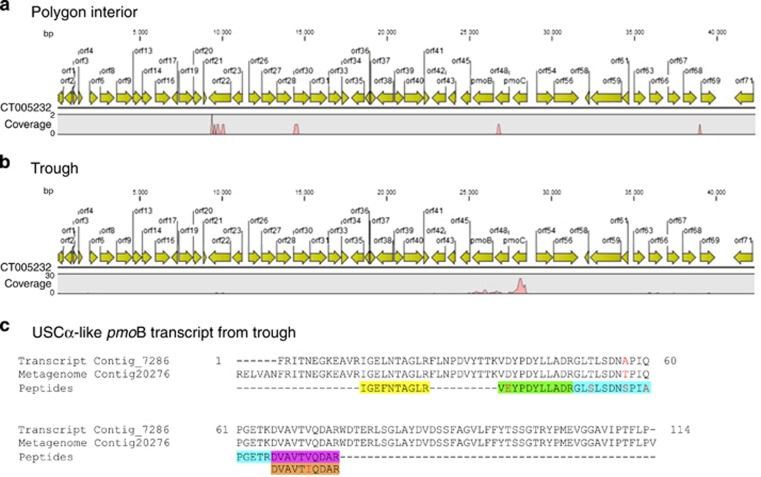
The metatranscriptomes detected transcripts closely related to USCα (Figures 2a and b), which confirms in situ atmMOB activity and its responsibility for the in situ atm CH4 uptake measured at our study site. Shown by the multiple sequence alignment of translated aa sequences of pmoB genes, our USCα-like pan-pmoB genes do contain the conserved residues (H33, H137 and H139) coordinating the di-Cu center, the active site of pMMO (Balasubramanian et al., 2010) (Supplementary Figure S4). The number of raw reads related to USCα pmo genes in the polygon trough sample was about 100-fold higher than that in the polygon interior sample (118 vs 1 reads). Thirty-four percent of 8039 transcript contigs (>200 bp) assembled from the trough sample mapped to our metagenome contigs, including the 13 MMO-containing contigs (Supplementary Table S3B). Surprisingly none of the transcript contigs from the polygon interior sample matched with our MMO-containing metagenome contigs. The higher pmo gene expression at the more vegetated, moist trough suggests that, like other MOB (Liebner et al., 2011), the USCα-like atmMOB has preference for vegetated micro-niches in polygonal terrains.
Figure 2.
Transcripts of methane monooxygenase relating to atmMOB USCα. Mapping of metatranscriptomic reads from (a) the polygon interior and (b) the trough samples to the USCα pmoCAB operon (GenBank Acc. No. CT005232; Ricke et al., 2005). The pmoA gene in CT005232 is annotated as orf48. (c) Alignment of a translated aa sequence of the transcript contig 7286 and five unique peptides (color boxes) identified from a separate proteomic experiment. Mismatches are denoted by red letters.
Detecting a specific protein from a low-abundance population (that is, rare proteins) is challenging given the high protein diversity in soils. Nonetheless, metaproteome profiling of the near-surface cryosols from intact cores subjected to warming experiments yielded pmoB peptides. Five of the six unique pmoB peptides aligned to the same USCα-like pmoB-transcript contig (Figure 2c and Supplementary Table S5), which confirms that the active USCα-like atmMOB in situ is also active in our warming experiments performed on intact cores that exhibit net atm CH4 uptake under all studied conditions (Stackhouse et al., submitted). The absence of pmoA and pmoC peptides in our samples may be partly due to the low extractability of these proteins as they are highly hydrophobic and embedded in the membrane lipid layer or partly because of their lower abundance relative to pmoB peptides. The more frequent detection of pmoB has also been reported in other CH4-oxidizing samples (Paszczynski et al., 2011). Among the three subunits of pMMO, β subunits may serve a regulatory role as they do for the soluble form of MMO (Murrell et al., 2000) and therefore show a higher expression level.
The detection of active USCα at AHI indicates that high Arctic mineral cryosols are a previously unidentified habitat of atmMOB (Kolb, 2009) and the biogeographic range of this methanotrophic group extends to 80°N. The search for atmMOB in other Arctic cryosols may help to explain the phenomenon of atm CH4 uptake in the polar region.
Atmospheric CH4 consumption is sensitive to temperature and soil moisture content
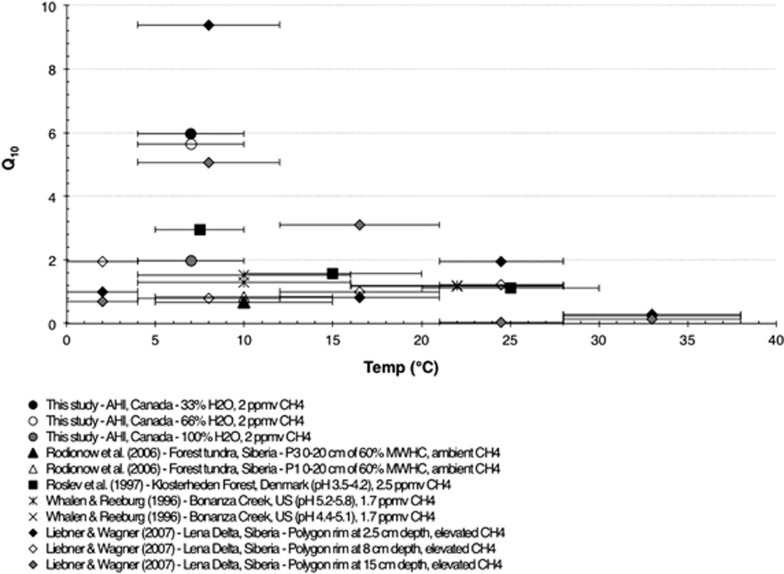
A factorial microcosm experiment simulating natural thaw conditions was designed to study the effects of temperature and water saturation on atm CH4 oxidation. CH4 (initial concentration at 2.0 p.p.m.v.) was consumed under all conditions by the end of a 31-day incubation (Table 1). Assuming a first-order rate law (Whalen and Reeburgh, 1990), estimated CH4 oxidation rates at 33% saturation were threefold to fourfold faster than at 66% and 100% saturations at each temperature and twofold faster at 10 °C than at 4 °C (Table 1). Thermal sensitivity of atm CH4 oxidation was quantitatively assessed using the Q10, which describes the rate of change of a reaction resulting from an increment of 10 °C. At 100% saturations, the Q10 values fell within the range of 1.3–2.9 values previously reported from incubation experiments carried out at atmospheric or elevated CH4 concentrations (Figure 3). In contrast, the Q10 value was 6 at 33% and 66% saturations (Figure 3). In contrast, the Q10 value was 8 at 33% saturation (Figure 3). Thus, lower water saturation not only increased the CH4 oxidation rate, presumably by promoting the availability of O2 and CH4 to atmMOB, but also allowed atmMOB to be more responsive to temperature change. A similar interactive effect of water saturation and temperature on atm CH4 oxidation was observed in boreal forest soils (Whalen and Reeburgh, 1996). High Q10 values were also estimated for CH4 oxidation in Siberian cryosols at elevated CH4 concentrations (Figure 3), suggesting that under some conditions thermal sensitivity of CH4 oxidation is as high as that reported for methanogenesis, which exhibits Q10 values of 0.6–12, in some boreal and temperate wetlands (Whalen, 2005).
Table 1. CH4 oxidation rates (mean±s.e.m.) estimated from microcosm experiments.
|
Incubation condition |
Final [CH4] (ppmv) | Rate constant, k ((g of soil)−1 day−1) | Estimated CH4 oxidation rate at 1.813 ppmv (ng C (g of soil)−1 day−1) | Estimated CH4 flux (mg C m−2 day−1) | |
|---|---|---|---|---|---|
| Temp. (°C) | Water saturation (%) | ||||
| 4 | 33 | 0.6 | −0.012±0.005 (R2=0.940) | −2.76±1.15 | −0.25±0.10 |
| 4 | 66 | 1.4 | −0.002±0.001 (R2=0.691) | −0.45±0.21 | −0.04±0.02 |
| 4 | 100 | 1 | −0.003±0.001 (R2=0.740) | −0.59±0.19 | −0.05±0.02 |
| 10 | 33 | 0.1 | −0.036±0.006 (R2=0.944) | −8.06±1.36 | −0.73±0.12 |
| 10 | 66 | 0.6 | −0.006±0.001 (R2=0.821) | −1.26±0.32 | −0.11±0.03 |
| 10 | 100 | 0.4 | −0.004±0.001 (R2=0.700) | −0.89±0.16 | −0.08±0.01 |
Figure 3.
Temperature coefficients (Q10) estimated from incubation experiments. All symbols mark the mid-point of the temperature range (denoted by the error bars) from which the Q10 was calculated. MWHC, maximum water-holding capacity.
Field fluxes of atm CH4 uptake at AHI displayed different kinetic properties as a function of ground temperature between 0 and 18 °C. The breakpoint in temperature response, however, occurred at a lower temperature (5.6 °C) than previously reported from the temperate region (Castro et al., 1995), and the rate of increase was reduced remarkably at temperatures >5.6 °C (Figure 4). A stronger thermal sensitivity of atm CH4 uptake at cooler temperatures suggests that the USCα at high northern latitudes may represent an ecotype containing cold-adapted pMMO.
Atmospheric CH4 uptake in warming scenarios
Climate models predicted a seasonal difference in warming over the century, with 10–14 °C warming in fall/winter and 3–5 °C warming in spring/summer (Supplementary Figure S5). As a consequence, atm CH4 uptake fluxes would take place over more months each year, which would then result in a fivefold increase in the annual atm CH4 uptake to −1.8 mg CH4-C m−2 year−1 (Table 2). However, the mean summer temperatures simulated for the 2090 s were 5–10 °C cooler than the temperatures measured at nearby Eureka, Ellesmere Island in 2011 (Supplementary Figure S5). This summer warming further increased the annual uptake fluxes by a factor of 4–6 (−8.0 to −11.5 mg CH4-C m−2 year−1) (Table 2). Although 2011 could have been an abnormally warm year (field observations and Sturtevant and Oechel, 2013), this indicates that warming of 10 °C at the Arctic across all months is possible (Supplementary Figure S5).
Table 2. Estimated monthly and annual CH4 uptake fluxes at Axel Heiberg Island, Nunavut, Canada.
| Multi-model mean 1900–1999 | Multi-model mean 2090–2099 | T 2011 | T+1°C | T+6°C | |
|---|---|---|---|---|---|
| Est. monthly flux (μg CH4-C m−2 month−1) | |||||
| Jan | 0 | 0 | NA | NA | NA |
| Feb | 0 | 0 | NA | NA | NA |
| Mar | 0 | 0 | NA | NA | NA |
| Apr | 0 | 0 | 0 | 0 | 0 |
| May | 0 | 0 | 0 | 0 | 0 |
| Jun | −8 | −25 | −82 | −87 | −117 |
| (UCI: −25; LCI: −2) | (UCI: −41; LCI: −15) | (UCI: −100; LCI: −67) | (UCI: −104; LCI: −73) | (UCI: −172; LCI: −80) | |
| Jul | −19 | −32 | −109 | −115 | −154 |
| (UCI: −35; LCI: −10) | (UCI: −51; LCI: −38) | (UCI: −148; LCI −80) | (UCI: −166; LCI: −80) | (UCI: −301; LCI: −79) | |
| Aug | −5 | −18 | −71 | −76 | −103 |
| (UCI: −21; LCI: −1) | (UCI: −34; LCI: −10) | (UCI: −96; LCI: −53) | (UCI: −97; LCI: −60) | (UCI: −133; LCI: −79) | |
| Sept | 0 | −5 (UCI: -22; LCI: 1) | 0 | 0 | 0 |
| Oct | 0 | 0 | NA | NA | NA |
| Nov | 0 | 0 | NA | NA | NA |
| Dec | 0 | 0 | NA | NA | NA |
| Est. annual flux (mg CH4-C m−2 year−1) | |||||
| −0.4 | −1.8 | −8 | −8.5 | −11.5 | |
| (UCI: −0.9; LCI: −0.2) | (UCI: −3.1; LCI: −1.6) | (UCI: −10.5; LCI: −6.1) | (UCI: −11.3; LCI: −6.5) | (UCI: −18.6; LCI: −7.3) | |
Abbreviations: LCI, lower confidence interval; NA, not available; UCI, upper confidence interval.
In addition to warming, climate change over the next century is also expected to affect the atmospheric circulation, cloudiness and precipitation pattern. Recent climate modeling results have predicted that the loss of sea ice in the Arctic Ocean would result in increased wintertime precipitation over Arctic land, including the Canadian Archipelago (Deser et al., 2010; Higgins and Cassano, 2009). A critical question is whether such hydroclimatic changes would also result in increased soil moisture during summer (June–August) when microbial activities are their greatest (Table 2). This will likely depend on the local landscape and ground water hydrological transport.
Our study site is located on the slope (176-m elevation) within 200 m from a small lake (Colour Lake, 10 ha). Based on our field observations in the past 3 years, snowmelt increased soil moisture to saturated level during initial spring thaw when the temperature was low and when the measured atm CH4 oxidation rates were also low. However, the surface cryosols, where atmMOB was most abundant, became dry (15–20 wt%) in summer when the temperature was warmer and when the measured atm CH4 oxidation rates were higher. In light of the underground hydrological system that facilitates drainage of snowmelt, we therefore expect the acidic mineral surface cryosols at our upland site to stay dry during summer. The temperature dependence of atm CH4 uptake was determined based upon field fluxes measured from initial thaw to mid-summer, that is, ‘initial wet/cold and later dry/warm' conditions. An increase in precipitation during summer would lower the atm CH4 oxidation rates, as suggested by our microcosm experiments. Our predictions of CH4 uptake increasing with future warming are therefore likely to be among the high estimates (that is, dry soils). Nonetheless, substantial uncertainties remain in determining the combined effects of twenty-first century changes in precipitation, evapotranspiration, land surface hydrology and permafrost structure on soil moisture (Hinzman et al., 2013; Avis et al., 2011), hence vegetation cover and ultimately the balance between atm CH4 oxidation and methanogenesis. Additional studies accounting for the links between microbial processes and hydroclimate should be carried out to reduce this uncertainty.
Net CH4 flux in soils is a balance between methanogenesis and CH4 oxidation where both reactions are sensitive to temperature change. It may then be a concern that future warming may convert our study site from a sink of CH4 to a source if methanogenesis is enhanced relative to methanotrophy as a result of increases in soil temperature and moisture. Methanogens were present at the lowest abundance (0.16% of total sequences) in near-surface cryosols (Supplementary Table S2B; Stackhouse et al., submitted) where the oxic condition is inhibitory to methanogenesis. Depth profiling of dissolved CH4 in the intact core-warming experiments detected methanogenesis taking place at depths near the permafrost table at 4 °C; however, pore water CH4 concentrations diminished towards the surface and became undetectable at 5 cm (Stackhouse et al., submitted). In situ depth profiling of pore gas showed a minimum CH4 concentration of <53 p.p.b.v. (at 23 cm) and then progressively increases with depth. Anaerobic microcosm experiments performed on active layer samples from this site yielded methanogenic rates of 0.4–0.9 nmols of CH4-C g−1 day−1 at 4 °C (Allan et al., 2014). Such a production rate extrapolated over the 70-cm depth of the active layer would be equivalent to 1.2–2.8 mg CH4-C m−2 day−1 if the soils were strictly anaerobic and no microbial oxidation were taking place. These results demonstrated that the relatively low amount of CH4 being produced at depth is completely oxidized before reaching the atmosphere (open circles in Figure 4).
As a decreasing temperature gradient as a function of depth will exist naturally down to the permafrost table, (atm) CH4 oxidation in the upper layers is anticipated to be greater than methanogenesis at the lower depths. AHI should continue to be a CH4 sink under this assumption. Nonetheless, further investigations are required to determine: (i) the range of CH4 production rates by subsurface methanogenesis that the aerobic methanotrophic community can accommodate before they cease providing a net atm CH4 sink; (ii) the critical conditions (for example, temperature and water saturation level) under which this offset takes place; and (iii) changes in permafrost dynamics as well as the potential topographical and hydrological modifications as a result of warming.
Under expected patterns of climate change, AHI will continue to be a microbial atm CH4 sink and the sink strength will vary distinctly in accordance with seasonal and inter-annual temperature variability. We recommend this two-kinetics temperature dependence of atm CH4 uptake be implemented in CH4 flux modeling to improve the predictive power. In addition, if atmMOB are ubiquitous in mineral cyrosols (and potentially present in peatlands), permafrost-affected cryosols (particularly mineral cryosols) have likely been modulating the rate of atm CH4 increase. Further studies are required to determine the impact on the regional CH4 fluxes, for example, (i) whether the uptake of atm CH4 during summer months partially accounts for the dip of ~50 p.p.b.v. in atm CH4 concentration and the δ13C enrichment observed seasonally in summer at high latitudes (Dlugokencky et al., 2009; Pickett-Heaps et al., 2011; Houweling et al., 2000) and (ii) whether a CH4 sink at high latitudes attributes to the unexplained bowl-shape variation in the CH4 isotopic record across the Holocene period (Sowers, 2010).
Acknowledgments
We thank the Canadian Polar Continental Shelf Program (PCSP) for their logistical support and McGill University's High Arctic Research Station. The project was supported by US Department of Energy, Office of Science, Office of Biological and Environmental Research (DE-SC0004902) to TCO and SMP; NSF grant (ARC-0909482) to ACL; and grants from Canada Foundation for Innovation (CFI) (206704) and the Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant Program (298520-05) and the Northern Research Supplements Program (305490-05) to LGW. We also thank the reviewers for their valuable comments.
Author contribution
MCYL, BTS, ACL, JR, RLH, SMP, LGW and TCO designed research. MCYL, BTS, ACL, AC, TAV, KC, NCSM, PCB, GL-G, NB, JR, WHP, CRO, DMM and TCO performed research. All the authors analyzed data. MCYL wrote the manuscript and all authors commented on the drafts.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Allan J, Ronholm J, Mykytczuk NCS, Greer CW, Onstott TC, Whyte LG. Methanogen community composition and rates of methane production in Canadian high Arctic permafrost soils. Environ Microbiol Rep. 2014;6:136–144. doi: 10.1111/1758-2229.12139. [DOI] [PubMed] [Google Scholar]
- Avis CA, Weaver AJ, Meissner KJ. Reduction in areal extent of high-latitude wetlands in response to permafrost thaw. Nat Geosci. 2011;4:444–448. [Google Scholar]
- Baani M, Liesack W. Two isozymes of particulate methane monooxygenase with different methane oxidation kinetics are found in Methylocystis sp. strain SC2. Proc Natl Acad Sci USA. 2008;105:10203–10208. doi: 10.1073/pnas.0702643105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian R, Smith SM, Rawat S, Yatsunyk LA, Stemmler TL, Rosenzweig AC. Oxidation of methane by a biological dicopper centre. Nature. 2010;465:115–119. doi: 10.1038/nature08992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bárcena TG, Finster KW, Yde JC. Spatial patterns of soil development, methane oxidation, and methanotrophic diversity along a receding glacier forefield, Southeast Greenland. Arctic Antarct Alp Res. 2011;43:178–188. [Google Scholar]
- Bender M, Conrad R. Kinetics of CH4 oxidation in oxic soils exposed to ambient air or high CH4 mixing ratios. FEMS Microbiol Ecol. 1992;101:261–270. [Google Scholar]
- Benstead J, King GM, Williams HG. Methanol promotes atmospheric methane oxidation by methanotrophic cultures and soils. Appl Environ Microbiol. 1998;64:1091–1098. doi: 10.1128/aem.64.3.1091-1098.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro MS, Steudler PA, Melillo JM, Aber JD, Bowden RD. Factors controlling atmospheric methane consumption by temperate forest soils. Global Biogeochem Cycles. 1995;9:1–10. [Google Scholar]
- Chauhan A, Layton AC, Vishnivetskaya TA, Williams D, Pfiffner SM, Rekepalli B, et al. Metagenomes from thawing low-soil-organic-carbon mineral cryosols and permafrost of the Canadian high Arctic. Genome Announc. 2014;2:e01217–14. doi: 10.1128/genomeA.01217-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chourey K, Jansson JK, VerBerkmoes N, Shah M, Chavarria KL, Tom LM, et al. Direct cellular lysis/protein extraction protocol for soil metaproteomics. J Proteome Res. 2010;9:6615–6622. doi: 10.1021/pr100787q. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degelmann DM, Borken W, Drake HL, Kolb S. Different atmospheric methane-oxidizing communities in European beech and Norway spruce soils. Appl Environ Microbiol. 2010;76:3228–3235. doi: 10.1128/AEM.02730-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deser C, Tomas R, Alexander M, Lawrence D. The seasonal atmospheric response to projected Arctic sea ice loss in the late twenty-first century. J Clim. 2010;23:333–351. [Google Scholar]
- Dlugokencky EJ, Bruhwiler L, White JWC, Emmons LK, Novelli PC, Montzka SA, et al. Observational constraints on recent increases in the atmospheric CH4 burden. Geophys Res Lett. 2009;36:L18803. [Google Scholar]
- Dlugokencky EJ, Nisbet EG, Fisher R, Lowry D. Global atmospheric methane: budget, changes and dangers. Philos Trans A Math Phys Eng Sci. 2011;369:2058–2072. doi: 10.1098/rsta.2010.0341. [DOI] [PubMed] [Google Scholar]
- Emmerton CA, St. Louis VL, Lehnherr I, Humphreys ER, Rydz E, Kosolofski HR. The net exchange of methane with high Arctic landscapes during the summer growing season. Biogeosci Discuss. 2014;11:1673–1706. [Google Scholar]
- Eng JK, McCormack AL, Yates JRI. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Van Everdingen RO.(ed) (1998 revised in 2005). Multi-Language Glossary of Permafrost and Related Ground-Ice Terms. National Snow and Ice Data Center/World Data Center for Glaciology: Boulder, CO, USA.
- Graham DE, Wallenstein MD, Vishnivetskaya TA, Waldrop MP, Phelps TJ, Pfiffner SM, et al. Microbes in thawing permafrost: the unknown variable in the climate change equation. ISME J. 2012;6:709–712. doi: 10.1038/ismej.2011.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins ME, Cassano JJ. Impacts of reduced sea ice on winter Arctic atmospheric circulation, precipitation, and temperature. J Geophys Res. 2009;114:D16107. [Google Scholar]
- Hinzman LD, Deal CJ, McGuire AD, Mernild SH, Polyakov IV, Walsh JE. Trajectory of the Arctic as an integrated system. Ecol Appl. 2013;23:1837–1868. doi: 10.1890/11-1498.1. [DOI] [PubMed] [Google Scholar]
- Houweling S, Dentener F, Lelieveld J, Walter B, Dlugokencky E. The modeling of tropospheric methane: how well can point measurements be reproduced by a global model. J Geophys Res. 2000;105:8981–9002. [Google Scholar]
- Hugelius G, Strauss J, Zubrzycki S, Harden JW, Schuur EAG, Ping CL, et al. Improved estimates show large circumpolar stocks of permafrost carbon while quantifying substantial uncertainty ranges and identifying remaining data gaps. Biogeosci Discuss. 2014;11:4771–4822. [Google Scholar]
- IPCC. 2013IPCC. Stocker TF, IPCC. Qin D, IPCC. Plattner G-K, IPCC. Tignor M, IPCC. Allen SK, IPCC. Boschung J.(eds)Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change Cambridge University Press: Cambridge, UK; New York, NY, USA; p1535 [Google Scholar]
- Knief C, Lipski A, Dunfield PF. Diversity and activity of methanotrophic bacteria in different upland soils. Appl Environ Microbiol. 2003;69:6703–6714. doi: 10.1128/AEM.69.11.6703-6714.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb S. The quest for atmospheric methane oxidizers in forest soils. Environ Microbiol Rep. 2009;1:336–346. doi: 10.1111/j.1758-2229.2009.00047.x. [DOI] [PubMed] [Google Scholar]
- Lau E, Ahmad A, Steudler PA, Cavanaugh CM. Molecular characterization of methanotrophic communities in forest soils that consume atmospheric methane. FEMS Microbiol Ecol. 2007;60:490–500. doi: 10.1111/j.1574-6941.2007.00308.x. [DOI] [PubMed] [Google Scholar]
- Liebner S, Wagner D. Abundance, distribution and potential activity of methane oxidizing bacteria in permafrost soils from the Lena Delta, Siberia. Environ Microbiol. 2007;9:107–117. doi: 10.1111/j.1462-2920.2006.01120.x. [DOI] [PubMed] [Google Scholar]
- Liebner S, Zeyer J, Wagner D, Schubert C, Pfeiffer E-M, Knoblauch C. Methane oxidation associated with submerged brown mosses reduces methane emissions from Siberian polygonal tundra. J Ecol. 2011;99:914–922. [Google Scholar]
- Luo GJ, Kiese R, Wolf B, Butterbach-Bahl K. Effects of soil temperature and moisture on methane uptake and nitrous oxide emissions across three different ecosystem types. Biogeosciences. 2013;10:3205–3219. [Google Scholar]
- Martineau C, Pan Y, Bodrossy L, Yergeau E, Whyte LG, Greer CW. Atmospheric methane oxidizers are present and active in Canadian high Arctic soils. FEMS Microbiol Ecol. 2014;89:257–269. doi: 10.1111/1574-6941.12287. [DOI] [PubMed] [Google Scholar]
- Maurer J. Atlas of the Cryosphere. National Snow and Ice Data Center. Digital media: Boulder, CO, USA; 2007. [Google Scholar]
- McDonald IR, Bodrossy L, Chen Y, Murrell JC. Molecular ecology techniques for the study of aerobic methanotrophs. Appl Environ Microbiol. 2008;74:1305–1315. doi: 10.1128/AEM.02233-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F, Paarmann D, M DS, Olson R, Glass EM, Kubal M. The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell JC, Gilbert B, McDonald IR. Molecular biology and regulation of methane monooxygenase. Arch Microbiol. 2000;173:325–332. doi: 10.1007/s002030000158. [DOI] [PubMed] [Google Scholar]
- Namiki T, Hachiya T, Tanaka H, Sakakibara Y. MetaVelvet: an extension of Velvet assembler to de novo metagenome assembly from short sequence reads. Nucleic Acids Res. 2012;40:e155. doi: 10.1093/nar/gks678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazaries L, Murrell JC, Millard P, Baggs L, Singh BK. Methane, microbes and models: fundamental understanding of the soil methane cycle for future predictions. Environ Microbiol. 2013;15:2395–2417. doi: 10.1111/1462-2920.12149. [DOI] [PubMed] [Google Scholar]
- Parks DH, Beiko RG. Identifying biologically relevant differences between metagenomic communities. Bioinformatics. 2010;26:715–721. doi: 10.1093/bioinformatics/btq041. [DOI] [PubMed] [Google Scholar]
- Paszczynski AJ, Paidisetti R, Johnson AK, Crawford RL, Colwell FS, Green T, et al. Proteomic and targeted qPCR analyses of subsurface microbial communities for presence of methane monooxygenase. Biodegradation. 2011;22:1045–1059. doi: 10.1007/s10532-011-9462-4. [DOI] [PubMed] [Google Scholar]
- Pickett-Heaps CA, Jacob DJ, Wecht KJ, Kort EA, Wofsy SC, Diskin GS, et al. Magnitude and seasonality of wetland methane emissions from the Hudson Bay Lowlands (Canada) Atmos Chem Phys. 2011;11:3773–3779. [Google Scholar]
- Ricke P, Kube M, Nakagawa S, Erkel C, Reinhardt R, Liesack W. First genome data from uncultured Upland Soil Cluster alpha methanotrophs provide further evidence for a close phylogenetic relationship to Methylocapsa acidiphila B2 and for high-affinity methanotrophy involving particulate methane monooxygenase. Appl Environ Microbiol. 2005;71:7472–7482. doi: 10.1128/AEM.71.11.7472-7482.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionow A, Flessa H, Kazansky O, Guggenberger G. Organic matter composition and potential trace gas production of permafrost soils in the forest tundra in northern Siberia. Geoderma. 2006;135:49–62. [Google Scholar]
- Roslev P, Iversen N, Henriksen K. Oxidation and assimilation of atmospheric methane by soil methane oxidizers. Appl Environ Microbiol. 1997;63:874–880. doi: 10.1128/aem.63.3.874-880.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer K, Zhang T, Bruhwiler L, Barrett AP. Amount and timing of permafrost carbon release in response to climate warming. Tellus B. 2011;63:165–180. [Google Scholar]
- Sharma R, Dill BD, Chourey K, Shah M, VerBerkmoes NC, Hettich RL. Coupling a detergent lysis/cleanup methodology with intact protein fractionation for enhanced proteome characterization. J Proteome Res. 2012;11:6008–6018. doi: 10.1021/pr300709k. [DOI] [PubMed] [Google Scholar]
- Shindell DT, Faluvegi G, Koch DM, Schmidt GA, Unger N, Bauer SE. Improved attribution of climate forcing to emissions. Science. 2009;326:716–718. doi: 10.1126/science.1174760. [DOI] [PubMed] [Google Scholar]
- Sowers T. Atmospheric methane isotope records covering the Holocene period. Quat Sci Rev. 2010;29:213–221. [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Sturtevant CS, Oechel WC. Spatial variation in landscape-level CO2 and CH4 fluxes from arctic coastal tundra: influence from vegetation, wetness, and the thaw lake cycle. Glob Chang Biol. 2013;19:2853–2866. doi: 10.1111/gcb.12247. [DOI] [PubMed] [Google Scholar]
- Taylor KE, Stouffer RJ, Meehl GA. An overview of CMIP5 and the experiment design. Bull Am Meteorol Soc. 2012;93:485–498. [Google Scholar]
- Thompson MR, VerBerkmoes NC, Chourey K, Shah M. Dosage-dependent proteome response of Shewanella oneidensis MR-1 to acute chromate challenge. J Proteome Res. 2006;6:1745–1757. doi: 10.1021/pr060502x. [DOI] [PubMed] [Google Scholar]
- Vishnivetskaya T, Layton A, Lau M, Chauhan A, Meyers A, Murphy J, et al. Commercial DNA extraction kits impact observed microbial community composition in permafrost samples. FEMS Microb Ecol. 2014;87:217–230. doi: 10.1111/1574-6941.12219. [DOI] [PubMed] [Google Scholar]
- Whalen SC. Biogeochemistry of methane exchange between natural wetlands and the atmosphere. Environ Eng Sci. 2005;22:73–94. [Google Scholar]
- Whalen SC, Reeburgh WS. Consumption of atmospheric methane by tundra soils. Nature. 1990;346:160–162. [Google Scholar]
- Whalen SC, Reeburgh WS. Moisture and temperature sensitivity of CH4 oxidation in boreal soils. Soil Biol Biochem. 1996;28:1271–1281. [Google Scholar]
- Whalen SC, Reeburgh WS, Barber VA. Oxidation of methane in boreal forest soils: a comparison of seven measures. Biogeochemistry. 1992;16:181–211. [Google Scholar]
- Wunch D, Toon GC, Blavier J-FL, Washenfelder RA, Notholt J, Connor BJ, et al. The Total Carbon Column Observing Network. Phil Trans R Soc A. 2011;369:2087–2112. doi: 10.1098/rsta.2010.0240. [DOI] [PubMed] [Google Scholar]
- Zhang T, Barry RG, Knowles K, Heginbottom JA, Brown J. Statistics and characteristics of permafrost and ground-ice distribution in the Northern Hemisphere. Polar Geogr. 2008;31:47–68. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.