Abstract
Invasion of the intestinal epithelium is a critical step in Salmonella enterica infection and requires functions encoded in the gene cluster known as Salmonella Pathogenicity Island 1 (SPI-1). Expression of SPI-1 genes is repressed by l-arabinose, and not by other pentoses. Transport of l-arabinose is necessary to repress SPI-1; however, repression is independent of l-arabinose metabolism and of the l-arabinose-responsive regulator AraC. SPI-1 repression by l-arabinose is exerted at a single target, HilD, and the mechanism appears to be post-translational. As a consequence of SPI-1 repression, l-arabinose reduces translocation of SPI-1 effectors to epithelial cells and decreases Salmonella invasion in vitro. These observations reveal a hitherto unknown role of l-arabinose in gene expression control and raise the possibility that Salmonella may use L-arabinose as an environmental signal.
Keywords: Salmonella pathogenicity island 1, Salmonella invasion, l-arabinose, HilD
INVASION of epithelial cells by Salmonella enterica requires the expression of genes located in a 40-kb region known as Salmonella pathogenicity island 1 (SPI-1) (Lostroh and Lee 2001; Altier 2005; Jones 2005). SPI-1 is a cluster of ∼40 genes organized in several transcriptional units that encodes a Type 3 Secretion System (TTSS) and effector proteins that are translocated into epithelial cells (Lostroh and Lee 2001). Inside the epithelial cell, the effector proteins trigger cytoskeleton rearrangements necessary for Salmonella invasion (Darwin and Miller 1999a). SPI-1 also encodes transcriptional activators responsible for its own expression: HilA, HilC, HilD, and InvF (Lostroh and Lee 2001; Ellermeier and Slauch 2007). HilA is a transcriptional activator of the OmpR/ToxR family (Lee et al. 1992; Bajaj et al. 1995) and activates transcription of genes encoding components of the TTSS and the transcriptional activator InvF (Bajaj et al. 1996). In association with the SicA chaperone, InvF boosts transcription of the sicA/sip operon, which encodes effector proteins (Darwin and Miller 1999b; Eichelberg and Galan 1999). Transcription of hilA is directly activated by HilC and HilD, both members of the AraC/XylS family of transcriptional regulators (Schechter and Lee 2001). HilC and HilD relieve repression of the hilA promoter by the nucleoid proteins H-NS and Hha (Olekhnovich and Kadner 2006) and can activate expression of the invF and sicA/sip transcriptional units independently of HilA (Rakeman et al. 1999; Akbar et al. 2003). Furthermore, HilD activates transcription of hilC and of hilD itself (Ellermeier et al. 2005) by direct binding to both promoters (Olekhnovich and Kadner 2002). Together with a transcriptional activator encoded outside SPI-1, RtsA (Ellermeier and Slauch 2003), SPI-1 transcription factors form a regulatory network that governs SPI-1 expression in response to environmental and physiological factors (Supporting Information, Figure S1) (Jones 2005; Ellermeier and Slauch 2007).
During passage through the digestive tract, Salmonella encounters environmental conditions that affect SPI-1 expression. In the stomach, SPI-1 is repressed at acidic pH (Behlau and Miller 1993; Bajaj et al. 1996), thus preventing invasion. The proximal part of the small intestine is under the influence of digestive fluids coming from the stomach, and the pH remains slightly acidic. In the duodenum, SPI-1 is repressed by bile, thereby inhibiting invasion of the proximal section of the small intestine (Prouty and Gunn 2000). In the large intestine, SPI-1 is repressed by the short chain fatty acids propionate and butyrate, synthesized by the intestinal microbiota (Lawhon et al. 2002). The gradients that repress SPI-1 along the digestive tract, however, leave a region, the ileum, that permits SPI-1 expression. As a consequence, the ileum is the portion of the small intestine that Salmonella preferentially invades (Jones et al. 1994).
In this study, we describe that the pentose l-arabinose represses SPI-1 expression. l-Arabinose is the second most abundant pentose in nature (Schadel et al. 2010; Seiboth and Metz 2011) and is found in hemicellulose and pectin in plant cell walls (Abedon et al. 2006; Schadel et al. 2010). Salmonella can use l-arabinose as the sole carbon source (Gutnick et al. 1969). l-Arabinose is transported into Salmonella cells by a permease encoded by the araE gene (Lee et al. 1981, 1982). Inside the cell, l-arabinose is sequentially transformed into l-ribulose, l-ribulose-5-phosphate, and d-xylulose-5-phosphate by the action of l-arabinose isomerase, ribulokinase, and l-ribulose-5-phosphate-4-epimerase, respectively (Englesberg 1961; Englesberg et al. 1962). L-Ribulose-5-phosphate and d-xylulose-5-phosphate are substrates for the pentose phosphate pathway, which produces glycolytic intermediates (Figure S2). l-Arabinose isomerase, ribulokinase, and l-ribulose-5-phosphate-4-epimerase are encoded by the araA, araB, and araD genes, respectively (Englesberg 1961; Englesberg et al. 1962), arranged in a single transcriptional unit, the araBAD operon (Gross and Englesberg 1959; Englesberg 1961; Englesberg et al. 1962; Lee et al. 1984). Expression of the araBAD operon and the araE gene is induced by l-arabinose (Lee et al. 1980, 1982) and requires the transcriptional regulator AraC (Englesberg et al. 1965; Lee et al. 1981). The araC gene is located upstream of the araBAD operon, but it is transcribed in divergent orientation (Lee et al. 1984). AraC bound to l-arabinose activates transcription from the araBAD and araE promoters. However, in the absence of l-arabinose, AraC acts as a repressor (Schleif 2010).
The phenomenon described in this study, SPI-1 repression by l-arabinose, is independent of l-arabinose catabolism and of the regulatory protein AraC. We describe epistasis analysis indicating that the target of l-arabinose within SPI-1 is the transcription factor HilD and present evidence that the inhibitory mechanism may be post-translational. HilD inhibition causes SPI-1 repression, which results in impaired secretion of SPI-1 effectors and reduced invasion of epithelial cells.
Materials and Methods
Bacterial strains, plasmids, bacteriophages, and strain construction
All the S. enterica strains listed in Table 1 belong to serovar Typhimurium and derive from the mouse-virulent strain American Type Culture Collection (ATCC) 14028. For simplicity, S. enterica serovar Typhimurium is often abbreviated as S. enterica. Targeted gene disruption was achieved using pKD13 or pKD3 (Datsenko and Wanner 2000). Antibiotic resistance cassettes introduced during strain construction were excised by recombination with plasmid pCP20 (Datsenko and Wanner 2000). The oligonucleotides used for disruption (labeled “UP” and “DO”) are listed in Table S1, together with the oligonucleotides (labeled “E”) used for allele verification by the polymerase chain reaction. For the construction of transcriptional and translational lac fusions in the Salmonella chromosome, FRT sites generated by excision of Kmr cassettes (Datsenko and Wanner 2000) were used to integrate either pCE37 or pCE40 (Ellermeier et al. 2002). An exception was the hilD::lac477 translational fusion, constructed using the method described by M. Hensel and co-workers (Gerlach et al. 2007). Unless specified otherwise, all lac fusions used in this study are translational. Addition of a 3xFLAG epitope tag to protein-coding DNA sequences was carried out using plasmid pSUB11 (Kmr, 3xFLAG) as template (Uzzau et al. 2001). Plasmid pIZ1902 (pXG10-hilD) was constructed by cloning a DNA fragment encompassing the hilD transcription start point and the hilD transcriptional terminator (Lopez-Garrido et al. 2014) on BrfBI-NheI restriction sites in pXG10 (Urban and Vogel 2007). Transductional crosses using phage P22 HT105/1 int201 (Schmieger 1972 and G. Roberts, unpublished results) were used for strain construction operations involving chromosomal markers. The transduction protocol was described elsewhere (Garzon et al. 1995). To obtain phage-free isolates, transductants were purified by streaking on green plates. Phage sensitivity was tested by cross-streaking with the clear-plaque mutant P22 H5.
Table 1. Strains used in this study.
| Strain designation | Genotype or description | Source |
|---|---|---|
| ATCC 14028 | Wild type | ATCC |
| SV5999 | ΔaraA | This study |
| SV5297 | Φ(invF′-lacZ+)(Hyb) | Lopez-Garrido and Casadesus (2010) |
| SV6000 | ΔaraA Φ(invF′-lacZ+)(Hyb) | This study |
| SV6205 | ΔaraA Φ(hilA′-lacZ+)(Hyb) | This study |
| SV6206 | ΔaraA Φ(sipB′-lacZ+)(Hyb) | This study |
| SV6207 | ΔaraA Φ(prgH′-lacZ+)(Hyb) | This study |
| SV6209 | ΔaraA invF::3xFLAG | This study |
| SV6208 | ΔaraA hilA::3xFLAG | This study |
| SV6210 | ΔaraA sipB::3xFLAG | This study |
| SV6211 | ΔaraA prgH::3xFLAG | This study |
| SV5284 | Φ(hilA′-lacZ+)(Hyb) | Lopez-Garrido and Casadesus (2010) |
| SV6218 | ΔxylA Φ(hilA′-lacZ+)(Hyb) | This study |
| SV6213 | ΔaraA ΔaraE Φ(hilA′-lacZ+)(Hyb) | This study |
| SV6212 | ΔaraBAD ΔaraC Φ(hilA′-lacZ+)(Hyb) | This study |
| SV7138 | ΔaraE ΔaraC Φ(hilA′-lacZ+)(Hyb) | This study |
| SV6244 | ΔaraA PLtetO-araE Φ(hilA′-lacZ+)(Hyb) | This study |
| SV6245 | ΔaraBAD ΔaraC PLtetO-araE Φ(hilA′-lacZ+)(Hyb) | This study |
| SV6219 | ΔaraA Φ(hilC′-lacZ+)(Hyb) | This study |
| SV6222 | ΔaraA ΔhilD Φ(hilC′-lacZ+)(Hyb) | This study |
| SV6220 | ΔaraA Φ(rtsA′-lacZ+)(Hyb) | This study |
| SV6223 | ΔaraA ΔhilD Φ(rtsA′-lacZ+)(Hyb) | This study |
| SV6221 | ΔaraA Φ(invH′-lacZ+)(Hyb) | This study |
| SV6224 | ΔaraA ΔhilD Φ(invH′-lacZ+)(Hyb) | This study |
| SV6417 | ΔaraA Φ(hilD-lacZ1) | This study |
| SV6419 | ΔaraA Φ(hilD′-lacZ477)(Hyb) | This study |
| SV6421 | ΔaraA Φ(hilD-lacZ930) | This study |
| SV7139 | ΔaraA PLtetO-hilD Φ(hilD-lacZ930) | This study |
| SV7140 | ΔaraA PLtetO-hilD | This study |
| SV6657 | ΔaraA PLtetO-hilD Φ(hilA′-lacZ+)(Hyb) | This study |
| SV6658 | ΔaraA PLtetO-hilD Φ(invF′-lacZ+)(Hyb) | This study |
| SV6659 | ΔaraA Φ(rtsA′-lacZ+)(Hyb)/pXG10 | This study |
| SV6660 | ΔaraA Δspi-1 Φ(rtsA′-lacZ+)(Hyb)/pXG10 | This study |
| SV6661 | ΔaraA Δspi-1 Φ(rtsA′-lacZ+)(Hyb)/pXG10-hilD | This study |
| SV6199 | ΔaraE | This study |
| SV6197 | ΔaraC | This study |
| SV6243 | PLtetO-araE | This study |
| SV6201 | ΔxylA | This study |
| SV6423 | ΔaraA Φ(sipA′-cya+)(Hyb) | This study |
| SV6424 | ΔaraA ΔaraE Φ(sipA′-cya+)(Hyb) | This study |
| SV6425 | ΔaraA ΔprgH Φ(sipA′-cya+)(Hyb) | This study |
Expression of araE from a heterologous promoter was achieved by replacing its native promoter with the PLtetO promoter (Lutz and Bujard 1997). A fragment containing the cat gene and the PLtetO promoter was amplified by PCR using pXG1 as template (Urban and Vogel 2007) and primers PLtetOUP and PLtetODO (Table S1). The PCR product was treated with DpnI to remove template traces. The construction was inserted in the chromosome by λ Red recombinase-mediated recombination (Datsenko and Wanner 2000), and Cmr colonies were selected. Insertion of the construction was verified by PCR using a pair of primers specific for the cat gene and the target gene (Table S1).
Growth conditions
Luria–Bertani (LB) broth was used as standard liquid medium. Solid media were prepared by the addition of 1.5% agar. l-Arabinose, d-arabinose, d-xylose, or sucrose was added from 20% stocks prepared in distilled water. Batch cultures were performed in LB or LB supplemented with the appropriate sugar and incubated at 37° with shaking at 220 × g. Samples were taken when the cultures had reached stationary phase (OD600 2–2.5). For translocation and invasion assays, an overnight bacterial culture of the appropriate strain was diluted 1:50 in LB and LB + 0.2% l-arabinose and was incubated at 37° until stationary phase (OD600 ∼2). Carbon-free medium (NCE) (Maloy and Roth 1983) supplemented with the appropriate carbon source was used as minimal medium. Green plates were prepared according to Chan et al. (1972), except that methyl blue (Sigma Chemical, St. Louis) was substituted for aniline blue.
pH curves
An overnight culture of Salmonella was diluted 1:50 in LB and in LB containing 0.01, 0.02, 0.05, 0.1, 0.2, 0.5, and 1% l-arabinose. The cultures were incubated at 37° until OD600 ∼2.5. The cultures were then centrifuged for 20 min at 2600 × g, and the pH of the supernatant was determined using a pH meter Basic 20 (Crison Instruments, Barcelona, Spain).
Protein extracts and Western blot analysis
Total protein extracts were prepared from bacterial cultures grown in LB or LB + l-arabinose at 37° until stationary phase (final OD600 ∼2.5). Bacterial cells were collected by centrifugation (16,000 × g, 2 min) and suspended in 100 μl of Laemmli sample buffer [1.3% SDS, 10% (v/v) glycerol, 50 mM Tris–HCl, 1.8% β-mercaptoethanol, 0.02% bromophenol blue, pH 6.8]. Proteins were resolved by Tris–Tricine–PAGE using 12% gels. Conditions for protein transfer have been described elsewhere (Jakomin et al. 2008). Optimal dilutions of primary antibodies were as follows: anti-Flag M2 monoclonal antibody (1:5,000, Sigma Chemical) and anti-GroEL polyclonal antibody (1:20,000, Sigma). Goat anti-mouse horseradish peroxidase-conjugated antibody (1:5000, Bio-Rad, Hercules, CA) or goat anti-rabbit horseradish-peroxidase-conjugated antibody (1:20,000, Santa Cruz Biotechnology, Heidelberg, Germany) were used as secondary antibodies. Proteins recognized by the antibodies were visualized by chemoluminescence using the luciferin–luminol reagents in a LAS3000 mini imaging system (Fujifilm, Tokyo). For quantification, the intensity of the bands was determined using the MultiGauge software (Fujifilm). GroEL was used as loading control.
RNA extraction and Northern blot analysis
A 2-ml aliquot from a stationary culture (OD600 ∼2) was centrifuged at 16,000 × g, 4°, for 5 min. The pellet was resuspended in 100 µl of a solution of lysozyme (3 mg/ml, Sigma Chemical). Cell lysis was facilitated by three consecutive freeze–thaw cycles. After lysis, RNA was extracted using 1 ml of Trizol reagent (Invitrogen, Carlsbad, CA), according to manufacturer instructions. Finally, total RNA was resuspended in 30 µl of RNase-free water for subsequent uses. The quality and quantity of the RNA was determined using a ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). For Northern blot analysis, 10 µg of total RNA was loaded per well and electrophoresed in denaturing 1% agarose formaldehyde gels. Transfer and fixation to Hybond-N+ membranes (GE Healthcare, Little Chalfont, UK) were performed by vacuum using 0.05 M NaOH. Filters were then hybridized using an internally labeled [(α-32P)UTP] riboprobe specific for the upstream (5′) 300 nucleotides of the hilD coding sequence, generated by in vitro transcription using the MEGAscript T7 Transcription Kit (Invitrogen). Hybridization was carried out at 65°. As a control of RNA loading and transfer efficiency, the filters were hybridized with a riboprobe for the RNase P messenger RNA (mRNA) gene (rnpB). Images of radioactive filters were obtained with a FLA-5100 imaging system (Fujifilm), and quantification was performed using MultiGauge software (Fujifilm).
β-Galactosidase assays
Levels of β-galactosidase activity were assayed using the CHCl3–sodium dodecyl sulfate permeabilization procedure (Miller 1972). Data are averages and standard deviations of three or more independent experiments.
Preparation of HeLa cells for infection assays
HeLa cells (ATCC CCL2) were cultured in tissue culture medium (Dulbecco’s modified essential medium supplemented with 10% fetal calf serum and 2 mM l-glutamine). For routine cultivation, 60 µg/ml penicillin and 100 µg/ml streptomycin were added to the culture medium. The day before infection, ∼1.5 × 105 HeLa cells were seeded, using 24-well plates (Costar, Corning, NY). Each well contained 1 ml of tissue culture medium without antibiotics. Cells were grown at 37°, 5% CO2, to obtain 80% confluency. One hour before infection, the culture medium was removed and replaced by 0.5 ml fresh tissue culture medium without antibiotics.
Translocation assays
Aliquots of bacterial suspensions were added to reach a multiplicity of infection of 15:1 bacteria/HeLa cell. Plates were centrifuged at 200 × g for 5 min and incubated at 37°, 5% CO2, for 10 min. Infected cells were then washed three times with PBS and lysed with a 0.1 M HCl solution containing 0.1% Triton X-100. Quantification of cAMP was performed using the Direct Cyclic AMP Enzyme Immunoassay Kit (Array Designs, Ann Arbor, MI) following the manufacturer’s instructions.
Invasion assays
Bacterial cultures were grown overnight at 37° in LB, diluted into fresh medium (1:50), and incubated at 37° until stationary phase (OD600 ∼2). Bacterial suspensions were added to reach a multiplicity of infection of 50:1 bacteria:HeLa cell. HeLa cells were infected for 30 min, washed three times with PBS, incubated in fresh tissue culture medium containing 100 µg/ml gentamicin for 1.5 hr, and washed three times with PBS. Numbers of viable intracellular bacteria were obtained by lysing infected cells with 1% Triton X-100 (prepared in PBS) and subsequent plating. Invasion rates were calculated as the ratio between viable intracellular bacteria and viable bacteria added to infect the HeLa cells.
Results
SPI-1 expression is downregulated in the presence of l-arabinose
pBAD vectors are a set of plasmids that contain the PBAD promoter of the l-arabinose operon and the regulatory gene araC (Guzman et al. 1995). In the presence of l-arabinose, transcription from the PBAD promoter is turned on, while in its absence transcription is extremely low. Use of pBAD vectors thus allows conditional, l-arabinose-dependent expression of cloned genes (Guzman et al. 1995). We considered the possibility of using pBAD vectors to study SPI-1 expression. As a preliminary control, we examined whether SPI-1 expression was affected by the presence of l-arabinose (in a strain without pBAD). For this purpose, we measured the β-galactosidase activity of an invF::lac fusion in LB and in LB + 0.2% l-arabinose. As shown in Figure 1A, invF::lac expression was reduced around fourfold in LB + l-arabinose. This observation suggested that the presence of l-arabinose in the culture medium might repress expression of SPI-1 genes.
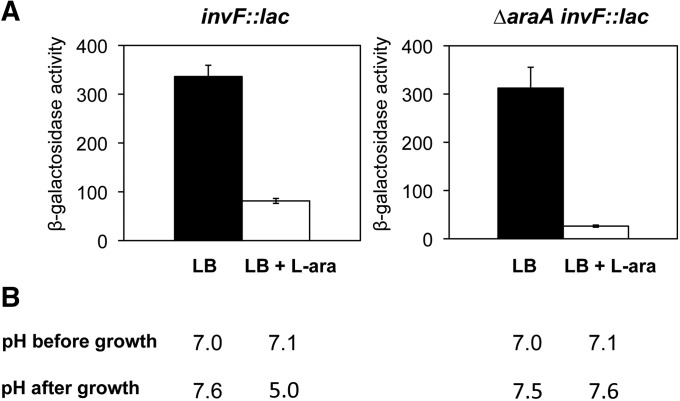
Figure 1.
Regulation of invF by l-arabinose. (A) β-Galactosidase activity of an invF::lac fusion in LB (black bars) and in LB + 0.2% l-arabinose (white bars). (Left) Activities measured in an AraA+ background. (Right) Activities measured in an AraAˉ background. (B) pH of the culture medium measured before bacterial inoculation (before growth) and after the bacterial culture had reached an OD600 of ∼2.5 (after growth).
SPI-1 repression in the presence of l-arabinose is independent of l-arabinose catabolism
The carbon sources for enteric bacteria in LB are catabolizable amino acids, not sugars (Sezonov et al. 2007). Under such conditions, enteric bacteria undergo gluconeogenesis, and the culture medium is slightly alkalinized during growth. However, if a catabolizable sugar is added to LB, Salmonella uses it as a carbon source and undergoes glycolysis. As a consequence, pyruvic acid is produced and the culture medium is acidified (Figure S2). Because S. enterica is able to regulate SPI-1 expression in response to pH (Bajaj et al. 1996), we considered the possibility that repression of SPI-1 by l-arabinose could be an indirect effect of acidification. We measured the pH of LB and LB + 0.2% l-arabinose before inoculation (before growth) and when the bacterial population had reached an OD600 of ∼2.5 (after growth) (Figure 1B, left). Before growth, both LB and LB + l-arabinose showed a pH near neutrality (7.0 and 7.1, respectively). However, after bacterial growth the pH of LB was slightly alkaline (7.6) while the pH of LB + l-arabinose was 5.0. This difference indicated that catabolism of l-arabinose by Salmonella acidifies indeed the culture medium and supported the possibility that SPI-1 repression by l-arabinose might be a consequence of acidification. To test this hypothesis, we investigated the effect of l-arabinose in a mutant lacking l-arabinose isomerase (AraA), the enzyme that catalyzes the first step of l-arabinose catabolism (Englesberg 1961). As expected, an AraA– mutant was unable to grow in minimal medium with l-arabinose as the sole carbon source (Figure S3). More interestingly, growth of the AraA– mutant did not cause acidification of the culture medium upon growth in LB + 0.2% l-arabinose (pH 7.6 compared with 7.5 in LB without l-arabinose) (Figure 1B). Absence of adicification was likewise observed at higher l-arabinose concentrations (Figure S3). Thus, if SPI-1 repression in the presence of l-arabinose was due to culture medium acidification, it should not be observed in an AraA– mutant. However, to our surprise, an invF::lac fusion was found to be repressed >10-fold by 0.2% l-arabinose in an AraA– background (Figure 1A, right), suggesting that SPI-1 repression is exerted by l-arabinose itself, rather than being a consequence of culture medium acidification. To avoid indirect effects caused by medium acidification, further experiments were performed in an AraAˉ background.
The entire SPI-1 is repressed by l-arabinose even at low concentrations
To investigate whether l-arabinose repressed only a subset of SPI-1 genes or the entire island, we analyzed the expression of additional SPI-1 genes (Figure 2). We chose genes encoded in different transcriptional units and representing different functional categories: hilA, which encodes a major transcriptional activator of SPI-1; invF, which encodes a transcriptional activator of a subset of SPI-1 genes; sipB, which encodes an effector protein; and prgH, which encodes a component of the SPI-1 TTSS. SPI-1 gene expression was compared in LB and LB + 0.2% l-arabinose by two independent methods: (1) measuring the β-galactosidase activity of lac fusions (Figure 2A); and (2) determining the protein levels by Western blot (Figure 2B). Both the β-galactosidase activities and the protein levels decreased in the presence of l-arabinose (Figure 2).
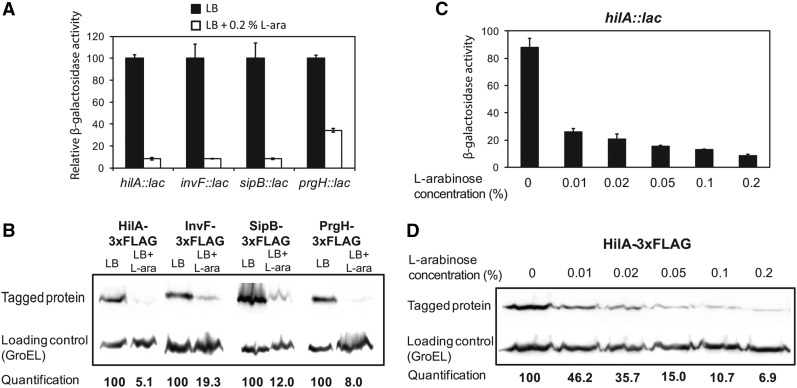
Figure 2.
Dose-dependent regulation of SPI-1 by l-arabinose. (A) β-Galactosidase activities of hilA::lac, invF::lac, sipB::lac, and prgH::lac fusions in the absence (black bars) and in the presence (white bars) of 0.2% l-arabinose, measured in AraAˉ background. Due to disparity between the β-galactosidase activities of the fusions, the β-galactosidase activities of individual lac fusions in LB without l-arabinose have been normalized to 100. The actual β-galactosidase activities in LB were 410, 308, 5384, and 1184 Miller units for hilA::lac, invF::lac, sipB::lac, and prgH::lac, respectively. (B) Levels of HilA, InvF, SipB, and PrgH in protein extracts from AraAˉ strains grown in LB with and without 0.2% l-arabinose. Proteins tagged with the 3xFLAG epitope were detected by Western blotting using anti-FLAG commercial antibodies. The loading control was GroEL in all cases. (C) β-Galactosidase activity of a hilA::lac fusion in an AraAˉ strain grown in the presence of increasing concentrations of l-arabinose. (D) Level of HilA-3xFLAG in protein extracts from an AraAˉ strain grown in increasing concentrations of l-arabinose. The HilA level was normalized to GroEL for quantification.
Next we analyzed the effect of different concentrations of arabinose on SPI expression, using the central SPI-1 regulator HilA as a reporter. The effect of varying the concentration of l-arabinose concentration was tested again by β-galactosidase assays and Western blotting. A gradual decrease in hilA expression as the l-arabinose concentration increased was observed in both the β-galactosidase assays (Figure 2C) and the Western blot analyses (Figure 2D). Note that, at the lower concentration of l-arabinose assayed (0.01%, equivalent to 66.7 µM), hilA expression was still reduced (more than threefold in β-galactosidase assays and more than twofold in Western blots). Dose-dependent repression suggests the ocurrence of an analog response in which l-arabinose is used by Salmonella as a signal.
Effect of pentoses other than l-arabinose on SPI-1 expression
In addition to l-arabinose, Salmonella can metabolize other pentoses (Gutnick et al. 1969). An example is d-xylose, the most abundant pentose in nature (Seiboth and Metz 2011). The first reaction in d-xylose catabolism is catalyzed by d-xylose isomerase, encoded by the xylA gene (Shamanna and Sanderson 1979). To examine the effect of d-xylose on SPI-1 expression, we constructed a strain carrying a xylA deletion. The xylA mutant is unable to use d-xylose as sole carbon source (Figure S4), thus avoiding a change in the pH of the culture medium due to d-xylose catabolism. Expression of a hilA::lac fusion was monitored in a XylAˉ background, growing the strain in LB, LB + 0.2% d-xylose, and LB + 1% d-xylose. As shown in Figure 3 (center), the presence of d-xylose in the culture medium did not alter hilA::lac expression. However, the same fusion was strongly repressed in the presence of either 0.2 or 1% l-arabinose (Figure 3, left).
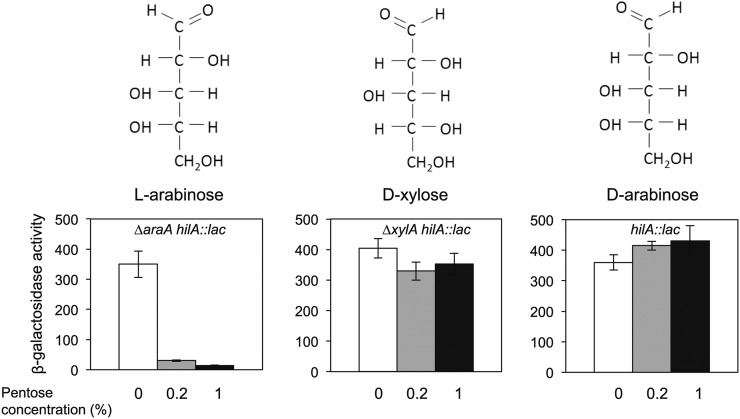
Figure 3.
Chemical structure of the pentoses l-arabinose, d-xylose, and d-arabinose (Top) and their effect on SPI-1 expression (Bottom). β-Galactosidase activity of a hilA::lac fusion in LB and LB + 0.2% and 1% of the appropriate pentose. Experiments with l-arabinose and d-xylose were performed in AraAˉ and XylAˉ backgrounds, respectively.
d-Arabinose, the d-stereoisomer of arabinose, cannot be used by Salmonella as sole carbon source (Gutnick et al. 1969). However, certain Salmonella mutants can metabolize d-arabinose (Old and Mortlock 1977). In such mutants, d-arabinose induces the synthesis of two enzymes usually involved in l-fucose metabolism: l-fucose isomerase and l-fuculokinase. These enzymes have a bifunctional activity and can convert d-arabinose into d-ribulose-1-phosphate. Figure 3 (right) shows that hilA::lac expression is similar in LB, LB + 0.2% d-arabinose, and LB + 1% d-arabinose, thus ruling out any effect of d-arabinose on SPI-1 expression.
The conclusion from these experiments was that the effect of l-arabinose on SPI-1 expression is specific, rather than a property shared with other pentoses such as d-xylose and d-arabinose.
SPI-1 repression requires l-arabinose transport
Trying to understand better SPI-1 repression by l-arabinose, we investigated whether l-arabinose exerted its repressive action from outside the cell or needed to be transported inside. In Salmonella, l-arabinose is transported by a specific permease encoded by the araE gene (Lee et al. 1981, 1982). Note that Salmonella does not harbor the AraFG transport system found in Escherichia coli (Kolodrubetz and Schleif 1981). A Salmonella AraE– mutant constructed ad hoc was unable to grow in minimal medium containing 0.1% l-arabinose as the sole carbon source grew slowly in the presence of 0.2% l-arabinose and grew like the wild-type strain in 1% l-arabinose (Figure S3). These observations suggest that the AraE permease is essential for transport of l-arabinose when the extracellular concentration is low; higher l-arabinose concentrations, however, may permit transport by AraE-independent routes. This view is supported by the ability of an AraE– mutant to acidify the culture medium when grown on high concentrations of l-arabinose (Figure S3). Note that the wild-type strain acidified the culture medium below pH 5 with 0.2% l-arabinose, while the AraE– mutant needed 1% l-arabinose to produce a similar acidification.
If l-arabinose needs to be inside the cell to repress SPI-1, we reasoned, repression should not be observed in an AraE– mutant grown at low l-arabinose concentrations. At higher concentrations, however, l-arabinose would enter the cell through AraE-independent pathways, repressing SPI-1. If, on the contrary, extracellular l-arabinose was able to repress SPI-1 expression, repression should be observed in AraE+ and AraEˉ backgrounds at both low and high l-arabinose concentrations. To ascertain between these alternative possibilities, we examined the expression of a hilA::lac fusion in isogenic AraA– and AraA– AraE– strains grown on different concentrations of l-arabinose (Figure 4A). The araE mutation nearly suppressed hilA repression in the presence of 0.2% l-arabinose. In the presence of higher l-arabinose concentrations, however, hilA remained significantly repressed. On the other hand, in all the l-arabinose concentrations assayed, repression was higher in the AraE+ background than in the AraEˉ background. Altogether, these observations indicate that l-arabinose needs to be transported inside the cell to repress SPI-1.
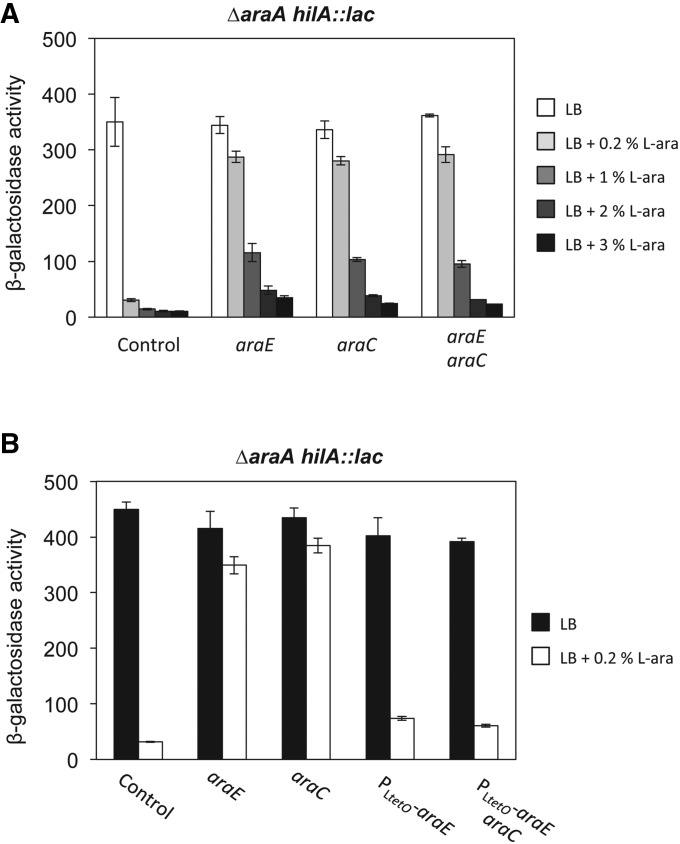
Figure 4.
Roles of AraE and AraC in SPI-1 regulation by l-arabinose. (A) β-Galactosidase activity of a hilA::lac fusion in AraAˉ (control), AraAˉ AraEˉ, AraAˉ AraCˉ, and AraAˉ AraEˉ AraCˉ backgrounds. β-Galactosidase activities were measured in LB and LB + 0.2, 1, 2, and 3% l-arabinose. (B) β-Galactosidase activity of an hilA::lac fusion in AraAˉ (control), AraAˉ AraEˉ, AraAˉ AraCˉ, AraAˉ PLtetO-araE, and AraAˉ AraCˉ PLtetO-araE backgrounds, grown in LB and LB + 0.2% l-arabinose.
To rule out the possibility that repression at high l-arabinose concentrations might be caused by high osmolarity, hilA::lac expression was examined also in the presence of sucrose (up to 3%). Expression of hilA::lac remained unaffected under all the sucrose concentrations assayed (Figure S5).
SPI-1 repression by l-arabinose is independent of AraC
In the bacterial cytoplasm, l-arabinose binds AraC and causes an allosteric change that enables AraC to activate the expression of genes involved in l-arabinose catabolism, including araE (Schleif 2010). The evidence that l-arabinose needs to be inside the cell to repress SPI-1 raised the possibility that activation of AraC by l-arabinose might be directly responsible for SPI-1 repression. To test this hypothesis, we compared the effect of increasing concentrations of l-arabinose on the expression of a hilA::lac fusion in AraA– and AraA– AraC– backgrounds. Lack of AraC suppressed hilA repression by 0.2% l-arabinose (Figure 4A, third column). However, this observation did not necessarily indicate that AraC is involved in l-arabinose repression. As AraC activates araE transcription, suppression in an AraCˉ background might be caused by a decrease of intracellular l-arabinose due to lack of AraE permease. In fact, hilA expression was increasingly repressed in an AraCˉ background as the l-arabinose concentration increased, and the repression pattern was very similar to that observed in an AraE– mutant (Figure 4A, second column). To ascertain whether AraC was directly involved in SPI-1 regulation or merely necessary for AraE permease synthesis, two different approaches were undertaken:
If suppression of SPI-1 regulation by l-arabinose in AraCˉ background was due to lack of AraE permease, an araE mutation should be epistatic over an araC mutation. Thus, a double-mutant AraE– AraC– and an AraE– mutant should display the same phenotype. However, if the effect of the araC mutation was independent of AraE permease, araE and araC mutations should have additive effects. As shown in Figure 4, an AraE– AraC– double mutant and an AraE– mutant displayed the same phenotype.
If AraC is needed for AraE permease synthesis only, constitutive araE expression should prevent AraC-mediated suppression of SPI-1 repression. AraC-independent expression of araE was achieved by replacing the native araE promoter with the PLtetO promoter (Lutz and Bujard 1997). The resulting strain grew as well as the wild type in minimal medium containing 0.1% l-arabinose (Figure S6). In addition, it acidified the culture medium like the wild type when grown in the presence of different concentrations of l-arabinose, suggesting that AraE permease was produced indeed (Figure S6). When araE was expressed constitutively, lack of AraC no longer suppressed hilA repression by l-arabinose (Figure 4B).
Altogether, the above observations suggest that, once inside the cell, l-arabinose represses SPI-1 expression by an AraC-independent mechanism.
l-Arabinose-dependent repression of SPI-1 is transmitted via HilD
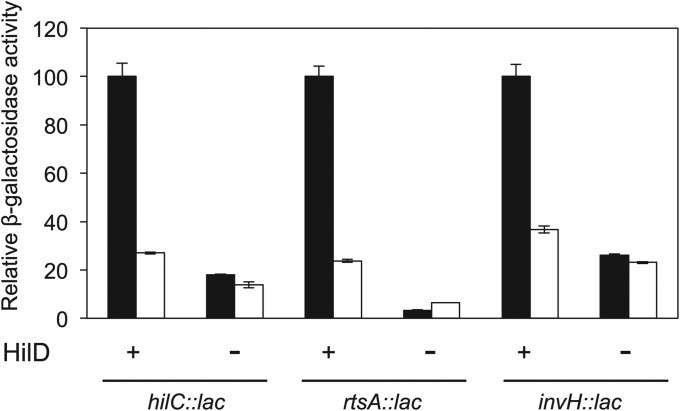
SPI-1 expression is controlled by a regulatory network of SPI-1-encoded transcriptional activators. On top of the network are the transcriptional activators HilA, HilC, and HilD (Figure S1). The fact that hilA expression is regulated by l-arabinose might indicate either that l-arabinose directly regulates hilA or that the regulatory input is transmitted to hilA via HilC and/or HilD. Lack of HilC reduces hilA expression 2- to 3-fold (Lopez-Garrido and Casadesus 2010), while addition of 0.2% l-arabinose to the culture medium represses hilA >10-fold. This discrepancy makes unlikely that HilC is responsible for transmission of l-arabinose regulation. In contrast, lack of HilD reduces hilA expression >100-fold (Lopez-Garrido and Casadesus 2010). Thus, we examined the possibility that l-arabinose might regulate SPI-1 expression via HilD. For this purpose, we analyzed the effect of l-arabinose on the expression of three HilD-activated genes (hilC, rtsA, and invH). Strains carrying hilC::lac, rtsA::lac, or invH::lac fusions in AraAˉ HilD+ and AraAˉ HilDˉ backgrounds were used, and β-galactosidase activities were measured in the presence and in the absence of 0.2% l-arabinose. Expression of all three genes was found to decrease in the presence of l-arabinose in a HilD+ background (Figure 5). In a HilDˉ background, expression of the hilC::lac, rtsA::lac, and invH::lac fusions is reduced but not completely abolished, and similar levels of β-galactosidase activity are detected in LB and LB + l-arabinose (Figure 5). Loss of l-arabinose repression in a hilD background thus provides evidence that l-arabinose-dependent regulation of SPI-1 requires a functional hilD gene.
Figure 5.
β-Galactosidase activities of hilC::lac, rtsA::lac, and invH::lac fusions in isogenic AraAˉ HilD+ and AraAˉ HilDˉ backgrounds. Black bars represent β-galactosidase activities in LB. White bars represent β-galactosidase activities in LB + 0.2% l-arabinose. Due to disparity between the β-galactosidase activities of the different fusions, the β-galactosidase activities of individual lac fusions in the HilD+ background grown in LB without l-arabinose have been normalized to 100. The actual β-galactosidase activities were 320, 2513, and 1134 Miller units for hilC::lac, rtsA::lac, and invH::lac, respectively.
Regulation of HilD by l-arabinose is post-transcriptional
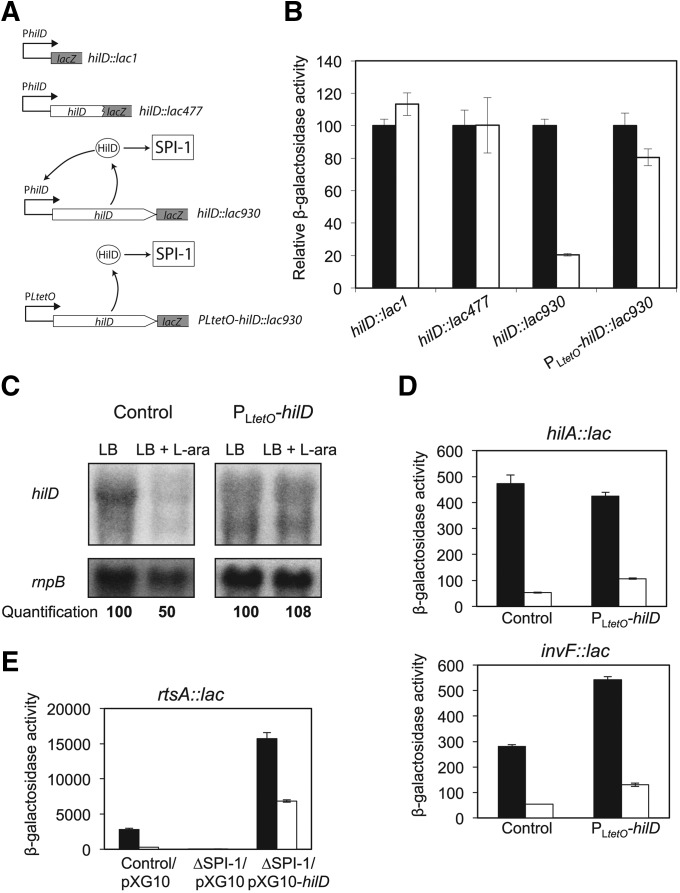
The evidence that l-arabinose regulates SPI-1 expression through HilD suggested that hilD expression itself might be controlled by l-arabinose. To examine this possibility, we monitored the expression of two different hilD::lac fusions in LB and LB + 0.2% l-arabinose: (1) hilD::lac1, a transcriptional fusion in which the lacZ gene is inserted exactly at the transcription start point (Lopez-Garrido and Casadesus 2012); and (2) hilD::lac477, a translational fusion in which the lacZ gene is inserted at position 477 of the hilD-coding sequence (Figure 6A). To our surprise, similar β-galactosidase levels were observed in both fusions in the absence and in the presence of l-arabinose (Figure 6B). This observation suggested that l-arabinose does not regulate hilD transcription or hilD translation initiation. However, when the level of hilD mRNA was analyzed by Northern blotting (Figure 6, panel C), a twofold reduction was detected in the presence of l-arabinose.
Figure 6.
Post-translational regulation of HilD by l-arabinose. (A) Diagram of the hilD::lac fusions used in this study. (B) Relative β-galactosidase activities of hilD::lac1, hilD::lac 477, hilD::lac930, and PLtetO-hilD::lac930 fusions in LB (black bars) and LB + 0.2% l-arabinose (white bars). Measurements were performed in an AraAˉ background. To facilitate visual perception of differences, the β-galactosidase activities of individual lac fusions in LB without l-arabinose have been normalized to 100. The actual β-galactosidase activities in LB were 177, 61, 494, and 220 Miller units for hilD::lac1, hilD::lac 477, hilD::lac930, and PLtetO-hilD::lac930, respectively. (C) Level of hilD mRNA in extracts from AraAˉ (control) and AraAˉ PLtetO-hilD strains, grown in LB and LB + 0.2% l-arabinose. hilD mRNA was detected by Northern blotting using a 32P-labeled riboprobe complementary to the 5′ region of hilD mRNA. rnpB mRNA was used as loading control. (D) β-Galactosidase activities of hilA::lac and invF::lac fusions in LB (black bars) and LB + 0.2% l-arabinose (white bars). Measurements were performed in AraAˉ (control) and AraAˉ PLtetO-hilD strains. (E) β-Galactosidase activity of an rtsA::lac fusion in LB (black bars) and LB + 0.2% l-arabinose (white bars) in the following backgrounds: AraAˉ with pXG10 empty plasmid, AraAˉ ∆SPI-1 with pXG10 empty plasmid, and AraAˉ ∆SPI-1 with pXG10-hilD plasmid.
The discrepancy between hilD::lac fusion expression (not responsive to l-arabinose) and hilD mRNA level (responsive to arabinose) led us consider the following possibility: the hilD::lac fusions used above generate hilD null mutations, while Northern blotting was carried out in a HilD+ background. Transcription of hilD undergoes autogenous activation by the HilD protein (Ellermeier et al. 2005). If l-arabinose impaired the ability of HilD to activate gene transcription, the reduction in the hilD mRNA level observed in the presence of l-arabinose might be due to loss of the HilD positive feedback loop. If such were the case, hilD transcription should be l-arabinose-dependent in a strain carrying a functional HilD protein. To examine this possibility, we used a hilD::lac transcriptional fusion constructed immediately downstream of the hilD stop codon (hilD::lac930). This fusion leaves the hilD-coding sequence intact, thus allowing autogenous activation of hilD transcription (Figure 6A) (Lopez-Garrido and Casadesus 2012). Figure 6B shows that the β-galactosidase activity of the hilD::lac930 fusion is reduced around fivefold in the presence of 0.2% l-arabinose. To determine whether this regulatory pattern reflected the occurrence of HilD autoregulation, we expressed hilD from the heterologous promoter PLtetO (Figure 6A) (Lopez-Garrido et al. 2014). The β-galactosidase activity of the hilD::lac930 fusion transcribed from PLtetO was similar in LB and LB + 0.2% l-arabinose (Figure 6B). Furthermore, the level of hilD mRNA did not decrease in the presence of l-arabinose when hilD was transcribed from PLteto (Figure 6C). Altogether, these results suggest that the target of l-arabinose is the HilD protein. A conceivable model is that l-arabinose might interfere with HilD activity, thus affecting transcription of hilD itself and of the remaining SPI-1 genes. This hypothesis is supported by the following observations:
The effect of l-arabinose on the activity of hilA::lac and invF::lac fusions was monitored upon hilD expression from PLtetO. Expression of both fusions was reduced around fourfold in the presence of 0.2% l-arabinose (Figure 6D).
The effect of l-arabinose on the activity of a rtsA::lac fusion was also monitored. The rtsA gene is located outside SPI-1 but is directly activated by HilD. This trait permits study of rtsA regulation in the absence of SPI-1, expressing hilD ectopically. For this purpose, we cloned hilD on pXG-10 under the control of the PLtetO promoter. Regulation of an rtsA::lac fusion by l-arabinose was examined in three different backgrounds: AraA– with the pXG-10 vector (empty), AraA– ∆SPI-1 with the pXG-10 vector (empty), and AraA– ∆SPI-1 with pIZ1902 (pXG10-hilD). As shown in Figure 6E, the rtsA::lac fusion was regulated by l-arabinose in a strain with SPI-1. Deletion of SPI-1 decreased β-galactosidase activity and suppressed regulation by l-arabinose. However, ectopic expression of hilD from a plasmid restored both the level of rtsA::lac expression and regulation by l-arabinose in the absence of SPI-1.
These observations support the view that l-arabinose may interact with the HilD protein and confirms that HilD alone is sufficient for transmission of l-arabinose-dependent regulation to other Salmonella genes.
Translocation of SPI-1 effectors and epithelial cell invasion are reduced in the presence of l-arabinose
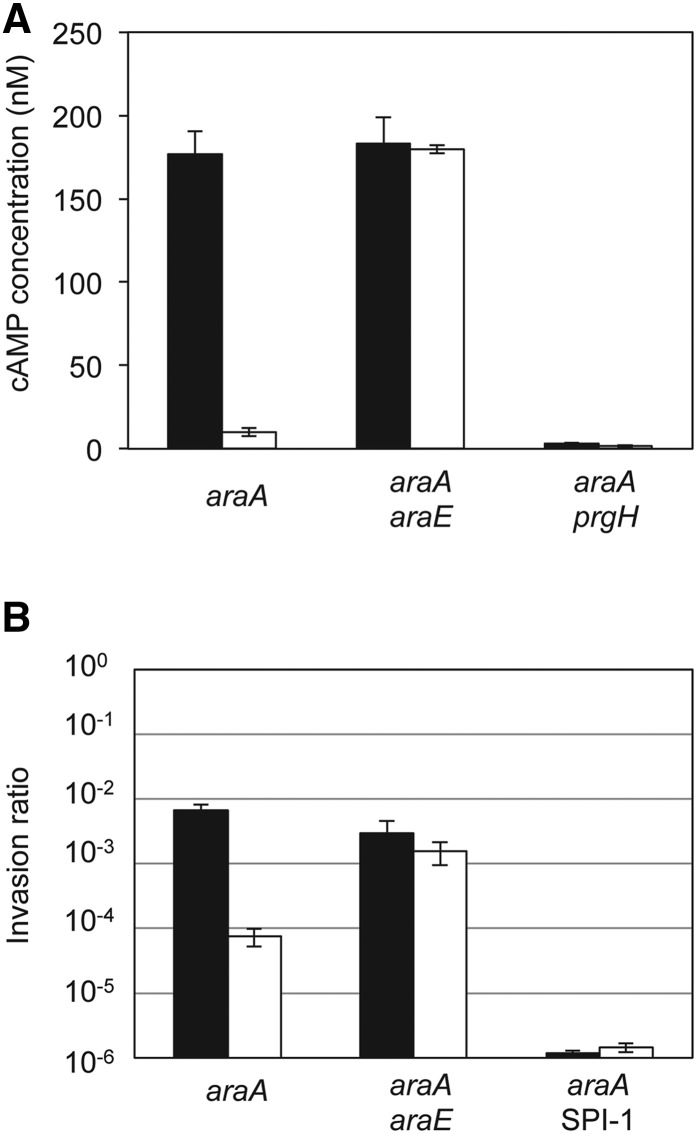
To determine whether SPI-1 repression by l-arabinose affected the interaction between Salmonella and host cells, we analyzed translocation of SPI-1 effectors into eukaryotic HeLa cells (Figure 7A). A fusion of the SPI-1 gene sipA with the cyaA gene of Bordetella pertussis was used for this purpose. Bordetella adenylate cyclase requires calmodulin to synthesize cAMP (Wolff et al. 1980; Greenlee et al. 1982; Glaser et al. 1989). Thus, cAMP will be produced only if the SipA-CyaA fusion protein is translocated into eukaryotic cells. The sipA::cyaA fusion was introduced in isogenic AraA–, AraA– AraE–, and AraA– PrgH– backgrounds, and the strains were grown in LB and LB + 0.2% l-arabinose before mixing with HeLa cells. Translocation was estimated by measuring the amount of cAMP and was strongly reduced in the presence of l-arabinose in the AraA– background (Figure 7A). However, in the AraA– AraE– background, addition of l-arabinose did not reduce translocation, reflecting the need of l-arabinose transport to repress SPI-1 expression (Figure 7A). The AraA– PrgH– strain was used as negative control since prgH encodes an essential component of the SPI-1 T3SS. These results confirmed that SPI-1 repression by l-arabinose impairs the interaction between Salmonella and epithelial host cells.
Figure 7.
(A) Translocation of a SipA-CyaA fusion protein into HeLa cells. Translocation was determined as the amount of cAMP produced upon infection. Black bars represent translocation levels of a bacterial culture in LB, and white bars represent translocation levels of a bacterial culture in LB + 0.2% l-arabinose. (B) Invasion of eukaryotic HeLa cells by Salmonella strains grown in LB (black bars) or LB + 0.2% l-arabinose (white bars). Invasion rates were determined as the ratio of viable intracellular bacteria vs. viable bacteria added to infect the HeLa cells. Note that a logarithmic scale has been used for the y-axis.
We then analyzed if the impaired translocation of SPI-1 effectors caused a reduction in Salmonella invasion. The ability to invade epithelial HeLa cells was determined for isogenic AraA–, AraA– AraE–, and AraA– ∆SPI-1 strains, grown on either LB or LB + 0.2% l-arabinose. Invasion by an AraA– strain decreased almost 100-fold in the presence of arabinose. As expected, inactivation of AraE permease nearly suppressed invasion inhibition in the presence of l-arabinose. In the absence of SPI-1, invasion was reduced >5000-fold, and no further reduction was observed when l-arabinose was added to the culture medium (Figure 7B). These observations suggest that SPI-1 repression in the presence of l-arabinose impairs Salmonella interaction with eukaryotic epithelial cells, causing inhibition of invasion.
Discussion
Regulation of virulence genes by sugars has been previously reported. In the Gram-positive pathogen Listeria monocytogenes, expression of virulence genes regulated by the master regulator PrfA is repressed in the presence of sugars transported by the phosphoenolpyruvate-sugar phosphotransferase system (PTS) (Park and Kroll 1993; Milenbachs et al. 1997; De Las Heras et al. 2011). However, repression is not observed in the presence of non-PTS sugars (Ripio et al. 1997; Joseph et al. 2008; Stoll et al. 2008; De Las Heras et al. 2011). In Streptococcus pyogenes, production of surface M protein, a major virulence determinant, is affected by the sugar source (Pine and Reeves 1978). Transcription of the gene encoding the M protein is indirectly activated by carbon catabolic repression (CCR) through the virulence gene regulator Mga. CCR also controls virulence gene expression in Clostridium perfringens (Varga et al. 2004) and Staphylococcus aureus (Morse et al. 1969). In S. enterica, evidence exists that PTS-dependent sugars may repress expression of invasion genes: Crp– Cya– mutants of S. enterica serovar Choleraesuis are attenuated in pigs (Kennedy et al. 1999), which correlates with the inability of Crp– mutants to secrete SPI-1 TTSS effectors (Chen et al. 2010). In addition, it has been reported that Mlc, a global regulator of carbon metabolism that can be induced by the PTS-sugars glucose and mannose (Plumbridge 2002), activates SPI-1 expression by repressing transcription of the hilE gene, which encodes a negative regulator of SPI-1 (Lim et al. 2007).
The ability of l-arabinose, a non-PTS sugar, to regulate the expression of genes involved in l-arabinose catabolism has been known for half a century (reviewed in Schleif 2010). Like l-arabinose catabolism, SPI-1 repression requires transport of l-arabinose by the AraE permease, a requirement that admits two alternative interpretations: (1) l-arabinose has to be inside the cell to repress SPI-1; or (2) transport of l-arabinose by AraE is necessary for SPI-1 repression. The latter possibility seems unlikely because a signal transduction system associated with AraE has not been described. Furthermore, the view that SPI-1 repression is exerted by l-arabinose itself is supported by two observations: (1) Salmonella AraE– mutants can grow with 1% l-arabinose as sole carbon source, indicating the existence of alternative transport mechanisms; and (2) l-arabinose represses SPI-1 expression in Salmonella AraE– mutants if provided at concentrations of 1% or higher.
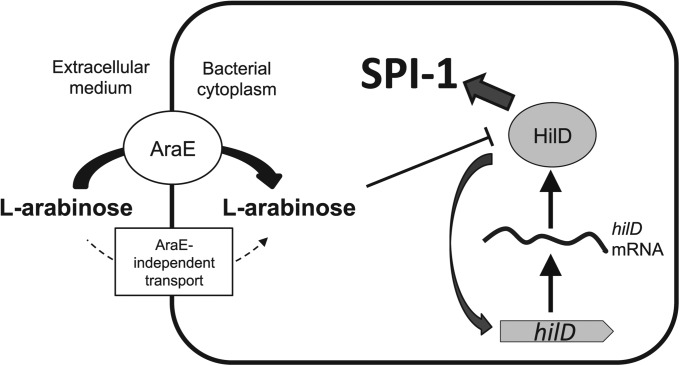
Epistasis analysis indicates that the target of l-arabinose is the transcription factor HilD, and experiments with gene fusions suggest that regulation is post-transcriptional. Because HilD is an AraC-like transcription factor, it is tempting to propose that l-arabinose might regulate HilD activity by directly binding to the protein. A tentative model of SPI-1 regulation by l-arabinose is depicted in Figure 8.
Figure 8.
Model of SPI-1 regulation by l-arabinose. l-Arabinose is transported inside the cell by AraE permease. AraE-independent transport may also occur when l-arabinose is provided at high concentrations. Inside the cell, l-arabinose inhibits the HilD protein. Repression of hilD by l-arabinose is further transmitted to the rest of SPI-1. The regulatory effect is amplified by the feedback loop that activates hilD transcription.
l-Arabinose is a plant-derived sugar, and the possession of a specific system for l-arabinose catabolism may enable Salmonella to use l-arabinose as carbon source in l-arabinose-rich environments. In addition, l-arabinose may serve as a signal for SPI-1 repression under certain circumstances. The observation that Salmonella grown in the presence of l-arabinose fails to invade epithelial cells (Figure 7) suggests that l-arabinose might inhibit invasion in the animal intestine. l-Arabinose is poorly absorbed during digestion by mammals (Cori 1925) and chickens (Wagh and Waibel 1967), and its presence in the intestine after ingestion is supported by experimental evidence: PBAD promoter expression is induced in the intestine of mice fed with plant components (Loessner et al. 2009). Furthermore, l-arabinose catabolism is required for efficient colonization of the large intestine by commensal and pathogenic strains of E. coli (Fabich et al. 2008). l-Arabinose supports efficient growth of Salmonella in vitro and might be a good carbon source in the intestine. Thus, the presence of l-arabinose in the intestine might be detected by Salmonella as a signal for repression of invasion. If such were the case, l-arabinose-rich compounds in the diet could prevent infections by Salmonella. Consistent with this possibility, dietary addition of arabinoxylooligosaccharides, made of l-arabinose and d-xylose, has been shown to protect poultry against oral infection by S. enterica serovar Enteritidis (Eeckhaut et al. 2008).
SPI-1 repression by l-arabinose could also play a role outside the animal host. As a plant-derived sugar, l-arabinose accumulates in the soil. It has been shown that Salmonella is able to persist in the soil for long periods (Islam et al. 2004). Because synthesis of the SPI-1 TTSS causes growth retardation (Sturm et al. 2011; Ali et al. 2014), SPI-1 repression by l-arabinose may improve the fitness of Salmonella in the soil. On the other hand, Salmonella can colonize plant surfaces (Barak et al. 2002; Brandl and Mandrell 2002) as well as intercellular spaces inside the plants (Franz et al. 2007). Plant colonization by Salmonella might be used as a strategy for recolonizing animal hosts (Tyler and Triplett 2008). Salmonella mutants lacking components of the SPI-1 TTSS colonize plants more efficiently than the wild type (Iniguez et al. 2005), perhaps because the presence of a functional TTSS in the Salmonella surface triggers a defense response by the plant (Iniguez et al. 2005). In such a scenario, detection of l-arabinose during plant colonization might contribute to turning down SPI-1 expression.
An additional implication of this study concerns the use of l-arabinose and other sugars for ectopic induction of gene expression, a tool routinely used in biological research. Metabolizable sugars can produce changes in cell physiology. In addition, as shown above, sugars can have metabolism-independent effects, potentially interfering with the subject of study.
Supplementary Material
Acknowledgments
We thank Dick D’Ari for encouraging the development of this project; David Reyes and Karina Ramijan for performing experiments not included in the manuscript; and the assistance provided by Modesto Carballo, Laura Navarro, and Cristina Reyes of the Servicio de Biología, Centro de Investigación, Tecnología e Innovación de la Universidad de Sevilla. This study was supported by grants BIO2010-15023 and BIO2013-44220-R from the Ministerio de Economía y Competitividad (MINECO) of Spain and the European Regional Fund; grant CSD2008-00013 from the Consolider-Ingenio 2010 Program of MINECO; and grant CVI-5879 from the Consejería de Innovación, Ciencia y Empresa, Junta de Andalucía, Spain. J.L.G. and I.C. were supported by fellowships of the MINECO program “Formación del Profesorado Universitario.”
Footnotes
Communicating editor: A. Hochschild
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.178103/-/DC1.
Literature Cited
- Abedon B. G., Hatfield R. D., Tracy W. F., 2006. Cell wall composition in juvenile and adult leaves of maize (Zea mays L.). J. Agric. Food Chem. 54: 3896–3900. [DOI] [PubMed] [Google Scholar]
- Akbar S., Schechter L. M., Lostroh C. P., Lee C. A., 2003. AraC/XylS family members, HilD and HilC, directly activate virulence gene expression independently of HilA in Salmonella typhimurium. Mol. Microbiol. 47: 715–728. [DOI] [PubMed] [Google Scholar]
- Ali S. S., Soo J., Rao C., Leung A. S., Ngai D. H.-M., et al. , 2014. Silencing by H-NS potentiated the evolution of Salmonella. PLoS Pathog. 10: e1004500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altier C., 2005. Genetic and environmental control of Salmonella invasion. J. Microbiol. 43: 85–92. [PubMed] [Google Scholar]
- Bajaj V., Hwang C., Lee C. A., 1995. HilA is a novel OmpR/ToxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol. Microbiol. 18: 715–727. [DOI] [PubMed] [Google Scholar]
- Bajaj V., Lucas R. L., Hwang C., Lee C. A., 1996. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol. Microbiol. 22: 703–714. [DOI] [PubMed] [Google Scholar]
- Barak J. D., Whitehand L. C., Charkowski A. O., 2002. Differences in attachment of Salmonella enterica serovars and Escherichia coli O157:H7 to alfalfa sprouts. Appl. Environ. Microbiol. 68: 4758–4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behlau I., Miller S. I., 1993. A PhoP-repressed gene promotes Salmonella typhimurium invasion of epithelial cells. J. Bacteriol. 175: 4475–4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandl M. T., Mandrell R. E., 2002. Fitness of Salmonella enterica serovar Thompson in the cilantro phyllosphere. Appl. Environ. Microbiol. 68: 3614–3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan R. K., Botstein D., Watanabe T., Ogata Y., 1972. Specialized transduction of tetracycline resistance by phage P22 in Salmonella typhimurium. II. Properties of a high-frequency-transducing lysate. Virology 50: 883–898. [DOI] [PubMed] [Google Scholar]
- Chen Z. W., Hsuan S. L., Liao J. W., Chen T. H., Wu C. M., et al. , 2010. Mutations in the Salmonella enterica serovar Choleraesuis cAMP-receptor protein gene lead to functional defects in the SPI-1 type III secretion system. Vet. Res. 41: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cori C. F., 1925. The fate of sugar in the animal body. I. The rate of absorption of hexoses and pentoses from the intestinal tract. J. Biol. Chem. 70: 691–715. [Google Scholar]
- Darwin K. H., Miller V. L., 1999a Molecular basis of the interaction of Salmonella with the intestinal mucosa. Clin. Microbiol. Rev. 12: 405–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin K. H., Miller V. L., 1999b InvF is required for expression of genes encoding proteins secreted by the SPI1 type III secretion apparatus in Salmonella typhimurium. J. Bacteriol. 181: 4949–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko K. A., Wanner B. L., 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97: 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de las Heras A., Cain R. J., Bielecka M. K., Vazquez-Boland J. A., 2011. Regulation of Listeria virulence: PrfA master and commander. Curr. Opin. Microbiol. 14: 118–127. [DOI] [PubMed] [Google Scholar]
- Eeckhaut V., Van Immerseel F., Dewulf J., Pasmans F., Haesebrouck F., et al. , 2008. Arabinoxylooligosaccharides from wheat bran inhibit Salmonella colonization in broiler chickens. Poult. Sci. 87: 2329–2334. [DOI] [PubMed] [Google Scholar]
- Eichelberg K., Galan J. E., 1999. Differential regulation of Salmonella typhimurium type III secreted proteins by pathogenicity island 1 (SPI-1)-encoded transcriptional activators InvF and HilA. Infect. Immun. 67: 4099–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellermeier C. D., Slauch J. M., 2003. RtsA and RtsB coordinately regulate expression of the invasion and flagellar genes in Salmonella enterica serovar Typhimurium. J. Bacteriol. 185: 5096–5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellermeier C. D., Janakiraman A., Slauch J. M., 2002. Construction of targeted single copy lac fusions using lambda red and FLP-mediated site-specific recombination in bacteria. Gene 290: 153–161. [DOI] [PubMed] [Google Scholar]
- Ellermeier C. D., Ellermeier J. R., Slauch J. M., 2005. HilD, HilC and RtsA constitute a feed forward loop that controls expression of the SPI1 type three secretion system regulator hilA in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 57: 691–705. [DOI] [PubMed] [Google Scholar]
- Ellermeier J. R., Slauch J. M., 2007. Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr. Opin. Microbiol. 10: 24–29. [DOI] [PubMed] [Google Scholar]
- Englesberg E., 1961. Enzymatic characterization of 17 L-arabinose negative mutants of Escherichia coli. J. Bacteriol. 81: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englesberg E., Anderson R. L., Weinberg R., Lee N., Hoffee P., et al. , 1962. L-arabinose-sensitive, L-ribulose 5-phosphate 4-epimerase-deficient mutants of Escherichia coli. J. Bacteriol. 84: 137–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englesberg E., Irr J., Power J., Lee N., 1965. Positive control of enzyme synthesis by gene C in the L-arabinose system. J. Bacteriol. 90: 946–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabich A. J., Jones S. A., Chowdhury F. Z., Cernosek A., Anderson A., et al. , 2008. Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect. Immun. 76: 1143–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz E., Visser A. A., Van Diepeningen A. D., Klerks M. M., Termorshuizen A. J., et al. , 2007. Quantification of contamination of lettuce by GFP-expressing Escherichia coli O157:H7 and Salmonella enterica serovar Typhimurium. Food Microbiol. 24: 106–112. [DOI] [PubMed] [Google Scholar]
- Garzon A., Cano D. A., Casadesus J., 1995. Role of Erf recombinase in P22-mediated plasmid transduction. Genetics 140: 427–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach R. G., Holzer S. U., Jackel D., Hensel M., 2007. Rapid engineering of bacterial reporter gene fusions by using red recombination. Appl. Environ. Microbiol. 73: 4234–4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser P., Elmaoglou-Lazaridou A., Krin E., Ladant D., Barzu O., et al. , 1989. Identification of residues essential for catalysis and binding of calmodulin in Bordetella pertussis adenylate cyclase by site-directed mutagenesis. EMBO J. 8: 967–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenlee D. V., Andreasen T. J., Storm D. R., 1982. Calcium-independent stimulation of Bordetella pertussis adenylate cyclase by calmodulin. Biochemistry 21: 2759–2764. [DOI] [PubMed] [Google Scholar]
- Gross J., Englesberg E., 1959. Determination of the order of mutational sites governing L-arabinose utilization in Escherichia coli B/r by transduction with phage Plbt. Virology 9: 314–331. [DOI] [PubMed] [Google Scholar]
- Gutnick D., Calvo J. M., Klopotowski T., Ames B. N., 1969. Compounds which serve as the sole source of carbon or nitrogen for Salmonella typhimurium LT-2. J. Bacteriol. 100: 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman L. M., Belin D., Carson M. J., Beckwith J., 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177: 4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iniguez A. L., Dong Y., Carter H. D., Ahmer B. M., Stone J. M., et al. , 2005. Regulation of enteric endophytic bacterial colonization by plant defenses. Mol. Plant Microbe Interact. 18: 169–178. [DOI] [PubMed] [Google Scholar]
- Islam M., Morgan J., Doyle M. P., Phatak S. C., Millner P., et al. , 2004. Persistence of Salmonella enterica serovar Typhimurium on lettuce and parsley and in soils on which they were grown in fields treated with contaminated manure composts or irrigation water. Foodborne Pathog. Dis. 1: 27–35. [DOI] [PubMed] [Google Scholar]
- Jakomin M., Chessa D., Baumler A. J., Casadesus J., 2008. Regulation of the Salmonella enterica std fimbrial operon by DNA adenine methylation, SeqA, and HdfR. J. Bacteriol. 190: 7406–7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B. D., 2005. Salmonella invasion gene regulation: a story of environmental awareness. J. Microbiol. 43(Spec. No.): 110–117. [PubMed] [Google Scholar]
- Jones B. D., Ghori N., Falkow S., 1994. Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized M cells of the Peyer’s patches. J. Exp. Med. 180: 15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph B., Mertins S., Stoll R., Schar J., Umesha K. R., et al. , 2008. Glycerol metabolism and PrfA activity in Listeria monocytogenes. J. Bacteriol. 190: 5412–5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy M. J., Yancey R. J., Jr, Sanchez M. S., Rzepkowski R. A., Kelly S. M., et al. , 1999. Attenuation and immunogenicity of Delta-cya Delta-crp derivatives of Salmonella choleraesuis in pigs. Infect. Immun. 67: 4628–4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodrubetz D., Schleif R., 1981. l-Arabinose transport systems in Escherichia coli K-12. J. Bacteriol. 148: 472–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawhon S. D., Maurer R., Suyemoto M., Altier C., 2002. Intestinal short-chain fatty acids alter Salmonella typhimurium invasion gene expression and virulence through BarA/SirA. Mol. Microbiol. 46: 1451–1464. [DOI] [PubMed] [Google Scholar]
- Lee C. A., Jones B. D., Falkow S., 1992. Identification of a Salmonella typhimurium invasion locus by selection for hyperinvasive mutants. Proc. Natl. Acad. Sci. USA 89: 1847–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. H., Heffernan L., Wilcox G., 1980. Isolation of ara-lac gene fusions in Salmonella typhimurium LT2 by using transducing bacteriophage Mud(Ap lac). J. Bacteriol. 143: 1325–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. H., Al-Zarban S., Wilcox G., 1981. Genetic characterization of the araE gene in Salmonella typhimurium LT2. J. Bacteriol. 146: 298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. H., Russo R. J., Heffernan L., Wilcox G., 1982. Regulation of L-arabinose transport in Salmonella typhimurium LT2. Mol. Gen. Genet. 185: 136–141. [DOI] [PubMed] [Google Scholar]
- Lee J. H., Nishitani J., Wilcox G., 1984. Genetic characterization of Salmonella typhimurium LT2 ara mutations. J. Bacteriol. 158: 344–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S., Yun J., Yoon H., Park C., Kim B., et al. , 2007. Mlc regulation of Salmonella pathogenicity island I gene expression via hilE repression. Nucleic Acids Res. 35: 1822–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loessner H., Leschner S., Endmann A., Westphal K., Wolf K., et al. , 2009. Drug-inducible remote control of gene expression by probiotic Escherichia coli Nissle 1917 in intestine, tumor and gall bladder of mice. Microbes Infect. 11: 1097–1105. [DOI] [PubMed] [Google Scholar]
- Lopez-Garrido J., Casadesus J., 2010. Regulation of Salmonella enterica pathogenicity island 1 by DNA adenine methylation. Genetics 184: 637–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Garrido J., Casadesus J., 2012. Crosstalk between virulence loci: regulation of Salmonella enterica pathogenicity island 1 (SPI-1) by products of the std fimbrial operon. PLoS ONE 7: e30499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Garrido J., Puerta-Fernandez E., Casadesus J., 2014. A eukaryotic-like 3′ untranslated region in Salmonella enterica hilD mRNA. Nucleic Acids Res. 42: 5894–5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lostroh C. P., Lee C. A., 2001. The Salmonella pathogenicity island-1 type III secretion system. Microbes Infect. 3: 1281–1291. [DOI] [PubMed] [Google Scholar]
- Lutz R., Bujard H., 1997. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1–I2 regulatory elements. Nucleic Acids Res. 25: 1203–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloy S. R., Roth J. R., 1983. Regulation of proline utilization in Salmonella typhimurium: characterization of put:Mud(Ap, lac) operon fusions. J. Bacteriol. 154: 561–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milenbachs A. A., Brown D. P., Moors M., Youngman P., 1997. Carbon-source regulation of virulence gene expression in Listeria monocytogenes. Mol. Microbiol. 23: 1075–1085. [DOI] [PubMed] [Google Scholar]
- Miller J. H., 1972. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Morse S. A., Mah R. A., Dobrogosz W. J., 1969. Regulation of staphylococcal enterotoxin B. J. Bacteriol. 98: 4–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Old D., Mortlock R. D., 1977. The metabolism of D-arabinose by Salmonella typhimurium. J. Gen. Microbiol. 101: 341–344. [DOI] [PubMed] [Google Scholar]
- Olekhnovich I. N., Kadner R. J., 2002. DNA-binding activities of the HilC and HilD virulence regulatory proteins of Salmonella enterica serovar Typhimurium. J. Bacteriol. 184: 4148–4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olekhnovich I. N., Kadner R. J., 2006. Crucial roles of both flanking sequences in silencing of the hilA promoter in Salmonella enterica. J. Mol. Biol. 357: 373–386. [DOI] [PubMed] [Google Scholar]
- Park S. F., Kroll R. G., 1993. Expression of listeriolysin and phosphatidylinositol-specific phospholipase C is repressed by the plant-derived molecule cellobiose in Listeria monocytogenes. Mol. Microbiol. 8: 653–661. [DOI] [PubMed] [Google Scholar]
- Pine L., Reeves M. W., 1978. Regulation of the synthesis of M protein by sugars, Todd Hewitt broth, and horse serum, in growing cells of Streptococcus pyogenes. Microbios 21: 185–212. [PubMed] [Google Scholar]
- Plumbridge J., 2002. Regulation of gene expression in the PTS in Escherichia coli: the role and interactions of Mlc. Curr. Opin. Microbiol. 5: 187–193. [DOI] [PubMed] [Google Scholar]
- Prouty A. M., Gunn J. S., 2000. Salmonella enterica serovar Typhimurium invasion is repressed in the presence of bile. Infect. Immun. 68: 6763–6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakeman J. L., Bonifield H. R., Miller S. I., 1999. A HilA-independent pathway to Salmonella typhimurium invasion gene transcription. J. Bacteriol. 181: 3096–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripio M. T., Brehm K., Lara M., Suarez M., Vazquez-Boland J. A., 1997. Glucose-1-phosphate utilization by Listeria monocytogenes is PrfA dependent and coordinately expressed with virulence factors. J. Bacteriol. 179: 7174–7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadel C., Blochl A., Richter A., Hoch G., 2010. Quantification and monosaccharide composition of hemicelluloses from different plant functional types. Plant Physiol. Biochem. 48: 1–8. [DOI] [PubMed] [Google Scholar]
- Schechter L. M., Lee C. A., 2001. AraC/XylS family members, HilC and HilD, directly bind and derepress the Salmonella typhimurium hilA promoter. Mol. Microbiol. 40: 1289–1299. [DOI] [PubMed] [Google Scholar]
- Schleif R., 2010. AraC protein, regulation of the L-arabinose operon in Escherichia coli, and the light switch mechanism of AraC action. FEMS Microbiol. Rev. 34: 779–796. [DOI] [PubMed] [Google Scholar]
- Schmieger H., 1972. Phage P22-mutants with increased or decreased transduction abilities. Mol. Gen. Genet. 119: 75–88. [DOI] [PubMed] [Google Scholar]
- Seiboth B., Metz B., 2011. Fungal arabinan and L-arabinose metabolism. Appl. Microbiol. Biotechnol. 89: 1665–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sezonov G., Joseleau-Petit D., D’Ari R., 2007. Escherichia coli physiology in Luria-Bertani broth. J. Bacteriol. 189: 8746–8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamanna D. K., Sanderson K. E., 1979. Uptake and catabolism of D-xylose in Salmonella typhimurium LT2. J. Bacteriol. 139: 64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoll R., Mertins S., Joseph B., Muller-Altrock S., Goebel W., 2008. Modulation of PrfA activity in Listeria monocytogenes upon growth in different culture media. Microbiology 154: 3856–3876. [DOI] [PubMed] [Google Scholar]
- Sturm A., Heinemann M., Arnoldini M., Benecke A., Ackermann M., et al. , 2011. The cost of virulence: retarded growth of Salmonella typhimurium cells expressing type III secretion system 1. PLoS Pathog. 7: e1002143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler H. L., Triplett E. W., 2008. Plants as a habitat for beneficial and/or human pathogenic bacteria. Annu. Rev. Phytopathol. 46: 53–73. [DOI] [PubMed] [Google Scholar]
- Urban J. H., Vogel J., 2007. Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res. 35: 1018–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzzau S., Figueroa-Bossi N., Rubino S., Bossi L., 2001. Epitope tagging of chromosomal genes in Salmonella. Proc. Natl. Acad. Sci. USA 98: 15264–15269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga J., Stirewalt V. L., Melville S. B., 2004. The CcpA protein is necessary for efficient sporulation and enterotoxin gene (cpe) regulation in Clostridium perfringens. J. Bacteriol. 186: 5221–5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagh P. V., Waibel P. E., 1967. Alimentary absorption of L-arabinose and D-xylose in chicks. Proc. Soc. Exp. Biol. Med. 124: 421–424. [DOI] [PubMed] [Google Scholar]
- Wolff J., Cook G. H., Goldhammer A. R., Berkowitz S. A., 1980. Calmodulin activates prokaryotic adenylate cyclase. Proc. Natl. Acad. Sci. USA 77: 3841–3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.