ABSTRACT
Highly invasive, community-acquired Klebsiella pneumoniae infections have recently emerged, resulting in pyogenic liver abscesses. These infections are caused by hypervirulent K. pneumoniae (hvKP) isolates primarily of capsule serotype K1 or K2. Hypervirulent K1 isolates belong to clonal complex 23 (CC23), indicating that this clonal lineage has a specific genetic background conferring hypervirulence. Here, we apply whole-genome sequencing to a collection of K. pneumoniae isolates to characterize the phylogenetic background of hvKP isolates with an emphasis on CC23. Most of the hvKP isolates belonged to CC23 and grouped into a distinct monophyletic clade, revealing that CC23 is a unique clonal lineage, clearly distinct from nonhypervirulent strains. Separate phylogenetic analyses of the CC23 isolates indicated that the CC23 lineage evolved recently by clonal expansion from a single common ancestor. Limited grouping according to geographical origin was observed, suggesting that CC23 has spread globally through multiple international transmissions. Conversely, hypervirulent K2 strains clustered in genetically unrelated groups. Strikingly, homologues of a large virulence plasmid were detected in all hvKP clonal lineages, indicating a key role in K. pneumoniae hypervirulence. The plasmid encodes two siderophores, aerobactin and salmochelin, and RmpA (regulator of the mucoid phenotype); all these factors were found to be restricted to hvKP isolates. Genomic comparisons revealed additional factors specifically associated with CC23. These included a distinct variant of a genomic island encoding yersiniabactin, colibactin, and microcin E492. Furthermore, additional novel genomic regions unique to CC23 were revealed which may also be involved in the increased virulence of this important clonal lineage.
IMPORTANCE
During the last 3 decades, hypervirulent Klebsiella pneumoniae (hvKP) isolates have emerged, causing severe community-acquired infections primarily in the form of pyogenic liver abscesses. This syndrome has so far primarily been found in Southeast Asia, but increasing numbers of cases are being reported worldwide, indicating that the syndrome is turning into a globally emerging disease. We applied whole-genome sequencing to a collection of K. pneumoniae clinical isolates to reveal the phylogenetic background of hvKP and to identify genetic factors associated with the increased virulence. The hvKP isolates primarily belonged to clonal complex 23 (CC23), and this clonal lineage was revealed to be clearly distinct from nonhypervirulent strains. A specific virulence plasmid was found to be associated with hypervirulence, and novel genetic determinants uniquely associated with CC23 were identified. Our findings extend the understanding of the genetic background of the emergence of hvKP clones.
INTRODUCTION
Klebsiella pneumoniae has traditionally been considered an opportunistic pathogen and is a common cause of nosocomial infections (1). However, starting in the mid-1980s, a distinctive syndrome of community-acquired invasive K. pneumoniae infections, primarily in the form of pyogenic liver abscesses, has emerged (2–5). These infections are often complicated by devastating metastatic infections, including endophthalmitis and meningitis. Remarkably, in contrast to most other K. pneumoniae infections, approximately half the cases occur in younger, otherwise healthy individuals. The invasive syndrome has mostly been reported in Taiwan and South Korea, where K. pneumoniae has become the most common etiologic agent of liver abscess over the last decades. Thus, K. pneumoniae liver abscess is now considered an endemic disease in Taiwan, where an almost 60% rise in the annual incidence from 1996 to 2004 has been observed (6). In South Korea, 78.2% of liver abscess cases in 2004 and 2005 were caused by K. pneumoniae compared to only 3.3% in the period from 1970 to 1979 (7). Although the K. pneumoniae liver abscess syndrome has been primarily reported within Southeast Asia, an increasing number of cases reported from other geographic regions, including North America and Europe, indicates that the syndrome is turning into a globally emerging disease (5, 8). Indeed, studies from U.S. institutions have reported that K. pneumoniae recently has surpassed Escherichia coli as the most common cause of liver abscess (9, 10).
The K. pneumoniae strains causing these invasive infections are termed hypervirulent and characteristically express a distinct hypermucoviscous phenotype when grown on agar plates (11–13). This may be related to overexpression of capsule polysaccharides. The capsule is recognized as an important virulence factor in K. pneumoniae that protects the bacteria from phagocytosis and the bactericidal effect of serum (1). Of the 78 capsular serotypes described, the hypervirulent K. pneumoniae (hvKP) isolates primarily belong to serotype K1 and, to a lesser degree, K2 (7, 12–15). Notably, it has been established that the invasive isolates exhibit significantly increased virulence in animal models compared to K. pneumoniae isolates from other infection types, supporting that these isolates indeed are hypervirulent (12, 16).
A number of putative virulence factors have been associated with hvKP. These include RmpA (regulator of the mucoid phenotype) and the aerobactin siderophore, which has been found in some strains to be encoded by a large virulence plasmid (17, 18). Furthermore, additional iron acquisition systems, such as yersiniabactin encoded by an integrative and conjugative element (ICE) (ICEKp1) and the kfu operon, as well as a region associated with allantoin metabolism and a fimbrial gene cluster, kpc, have been associated with specific hvKP strains (19–22).
Interestingly, multilocus sequence typing (MLST) has revealed that the vast majority of hvKP strains of serotype K1 belong to the same clonal complex, CC23 (23–29). In contrast, hvKP strains of serotype K2 have been found to belong to different, unrelated MLST types (27, 29, 30). A recent study that applied a 694-gene core genome MLST scheme to a diverse K. pneumoniae collection revealed that K1 CC23 isolates form a distinct clonal group and are associated with specific virulence factors whereas hypervirulent K2 isolates are genetically more diverse (31). The prominence of the hypervirulent K1 CC23 clone is intriguing and indicates that this clonal lineage has a specific genetic background conferring hypervirulence and also possibly increased environmental fitness.
Here, we sequenced and analyzed the genomes of a collection of clinically and geographically diverse K. pneumoniae isolates to characterize the phylogenetic background and evolution of hvKP strains with emphasis on the CC23 clonal lineage. Furthermore, by comparative genomics analyses we identify factors which are specifically associated with hvKP strains and which may play a role in their increased virulence.
RESULTS
Strain collection.
A collection of 30 hvKP strains, isolated from cases of liver abscess or community-acquired pneumonia, submitted to the WHO Collaborating Centre for Reference and Research on Escherichia and Klebsiella at Statens Serum Institut, Denmark, was included in the study. The strains were isolated in the period from 1996 to 2012 and originated from seven different countries in Africa, Asia, Europe, and North America (see Table S1 in the supplemental material).
The vast majority (28/30) of the hvKP isolates were found to belong to capsule serotype K1. The remaining two strains belonged to serotype K2. All the strains exhibited the characteristic hypermucoviscous phenotype shown by the formation of a mucoviscous string when a loop was passed through a colony.
Remarkably, MLST analysis revealed that 27 of the 28 hvKP K1 isolates belonged to CC23. The last K1 isolate belonged to ST260, which differs from ST23 at two alleles. In contrast to the K1 isolates, the two K2 isolates belonged to two unrelated ST types, ST25 and ST86. To investigate whether the observed high degree of clonality of the K1 hvKP isolates reflected a general clonality of strains belonging to the K1 serotype, two K1 reference strains and two additional K1 blood isolates were included. The K1 reference strains belonged to ST82, differing from ST23 at four alleles, and the blood isolates belonged to ST249, differing at three alleles.
Whole-genome phylogenetic analysis.
To understand the clonal diversity and phylogenetic relatedness of the hvKP isolates, the strain collection described above (including the K1 reference strains and blood isolates) and 35 clinical but nonhypervirulent strains, including nine strains belonging to the carbapenem-resistant ST258 clone, were subjected to whole-genome sequencing (WGS) (see Table S1 in the supplemental material). Furthermore, 11 previously published K. pneumoniae genomes (see Table S2) were included in the phylogenetic analysis, including the closed genome of the Taiwanese CC23 strain NTUH-K2044 isolated from a patient with liver abscess and meningitis (17). Thus, a total of 80 strains were included in the final phylogenetic analysis. A total of 72,371 single nucleotide polymorphisms (SNPs) were identified in the conserved core genome of the strains and were used to construct a maximum-likelihood phylogenetic tree (Fig. 1). All the hvKP CC23 isolates were found to cluster in a single clade, clearly separated from the other isolates, revealing a high degree of clonality of the CC23 group. Although the CC23 isolates were found to be more closely related to the K1 reference strains and the K1 blood isolates than the isolates of other capsule serotypes, the CC23 cluster was clearly distinct from the other K1 strains. Notably, CAS686, the only K1 hvKP isolate not belonging to CC23 but to the ST23 double-locus variant ST260, formed a distinct branch and was clearly separated from the CC23 clade (Fig. 1).
FIG 1 .
Rooted maximum-likelihood phylogeny of 80 K. pneumoniae isolates based on 72,371 total SNPs, including 41,171 parsimony-informative SNPs. Hypervirulent clonal lineages are marked with shadings. Columns 1 and 2 show the presence of the virulence plasmid pLVPK and the allS region associated with allantoin metabolism, respectively. Columns 3, 4, and 5 show the presence of colibactin (pks), yersiniabactin (fyuA), and microcin E492 (mE492) in the KPHPI208 genomic island, respectively. The presence and absence of genetic elements are indicated by black and white, respectively. The asterisks indicate bootstrap support values of ≥90%.
Whereas the hvKP K1 isolates were found to exhibit a high degree of clonality, the two hvKP K2 strains were found to be genetically unrelated and clustered in two distinct clades. Interestingly, the ST86 isolate, CAS688, clustered closely together with two previously described hvKP K2 strains, CG43 and hvKP1, also belonging to ST86, revealing a high degree of clonality of hypervirulent ST86 K2 isolates.
Recent studies have reported high levels of recombination in K. pneumoniae (32, 33). Thus, recombinant regions were identified and removed using Gubbins (34); however, both the clonality of the CC23 lineage and the distinctness of the different hypervirulent K2 lineages were maintained (see Fig. S1 in the supplemental material). Interestingly, after recombination filtering, the ST260 isolate CAS686 clustered closely with the CC23 isolates, indicating that this hybrid strain is the result of recombination between an ST23 strain and another K. pneumoniae isolate from a novel clonal lineage (see below).
As expected, the carbapenem-resistant ST258 isolates clustered together, illustrating the close genetic relationship of the ST258 lineage, whereas the remaining nonhypervirulent isolates were generally genetically diverse and formed several clearly separated clades in the phylogenetic tree (Fig. 1).
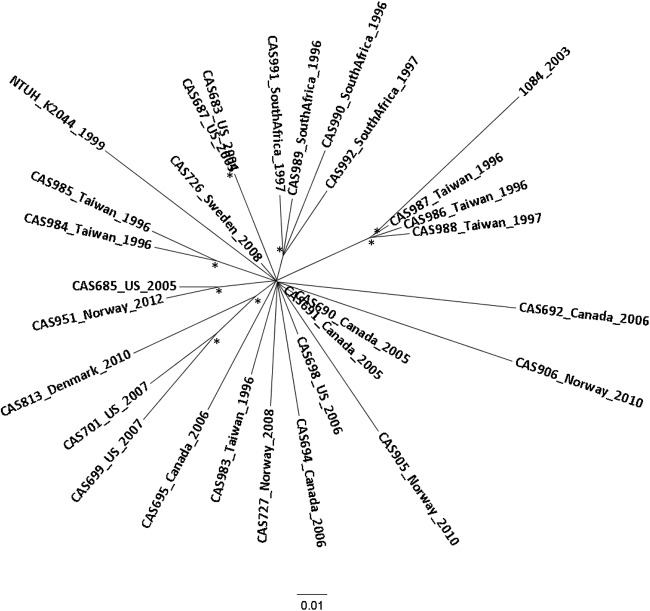
Phylogenetic relationship of CC23.
To further resolve the phylogenetic relationship of the CC23 cluster, we performed a separate analysis including only the CC23 isolates. To increase the size of the conserved core genome from which genetic polymorphisms are called, the CC23 clade was investigated separately to construct a high-resolution phylogenetic tree (Fig. 2). The inferred relationship between the isolates exhibited a star-like topology, suggesting a recent clonal expansion of the CC23 lineage from a common ancestor. Geographical clustering was not observed except for four isolates from South Africa and a cluster of four Taiwanese isolates.
FIG 2 .
High-resolution phylogenetic analysis of 29 CC23 isolates of different geographical origins. The radial maximum-likelihood tree was based on 2,181 total SNPs, including 342 parsimony-informative SNPs after removal of recombinant regions. Isolates are labeled with country of origin and year of sampling. The asterisks indicate bootstrap support values of ≥90%.
The ST260 isolate, CAS686, is a CC23 hybrid strain.
As described above, one hvKP K1 isolate, CAS686, did not belong to CC23 but formed a separate branch in the phylogenetic tree (Fig. 1); however, after recombination filtering the isolate clustered closely with the CC23 isolates (see Fig. S1 in the supplemental material). Indeed, analysis of the composition of the ST260 isolate revealed that a large proportion of the genome of the ST260 isolate showed a remarkably high degree of similarity to CC23 corresponding to approximately 2.9 Mbp (55%) of the NTUH-K2044 genome (see Fig. S2). Thus, according to SNP mapping, most of the divergence was located in regions corresponding to 2.4 Mbp (45%) of the NTUH-K2044 genome, revealing a chimeric composition of CAS686 between a CC23 strain and a strain of unknown clonal origin (see Fig. S2). Indeed, when separate phylogenetic analysis of the non-CC23 segment of the ST260 genome was performed, it was clearly distinct from the CC23 clade (see Fig. S3).
Virulence plasmid.
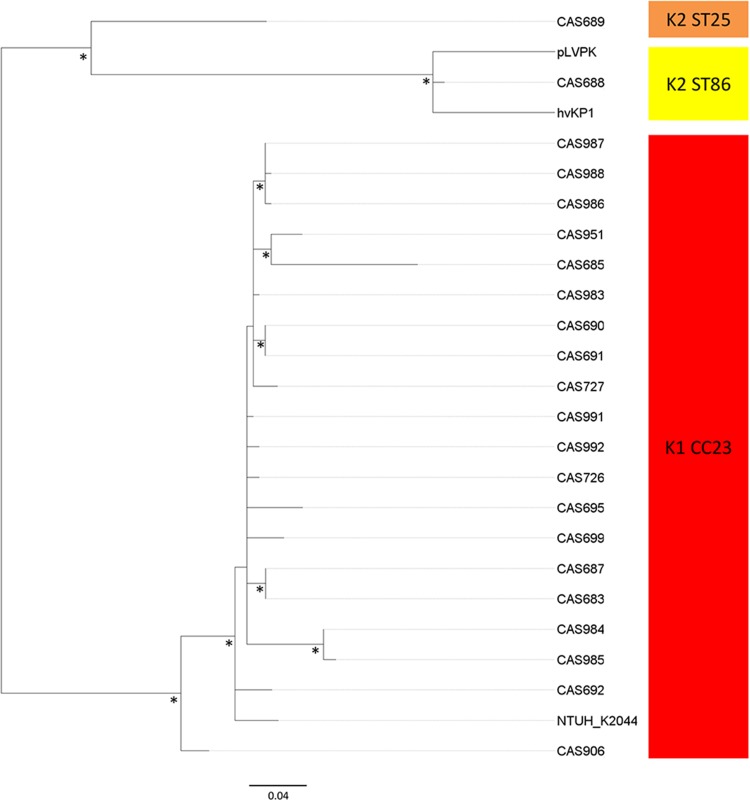
The prototype ST23 liver abscess strain NTUH-K2044 has been shown to carry a 224-kb plasmid, pNTUH-K2044, highly similar to the large virulence plasmid pLVPK of the hvKP K2 strain CG43 (17, 18). The plasmid carries virulence-associated genes, including those for the iron acquisition systems aerobactin (iucABCD-iutA) and salmochelin (iroBCDN) and two different variants of a gene involved in expression of the characteristic hypermucoviscous phenotype of hvKP, rmpA (regulator of the mucoid phenotype) and rmpA2. Interestingly, plasmids closely resembling the large virulence plasmid were detected in all the hvKP isolates examined, including the two K2 isolates, although some strains appeared to carry variants in which specific regions had undergone deletions (see Fig. S4 in the supplemental material). Even though some regions of the plasmid (e.g., regions encoding copper and silver resistance) were also detected among the other isolates, the virulence plasmid per se was found to be uniquely associated with hvKP strains (Fig. 1; see also Fig. S4). The only exception was the previously sequenced 2,3-butanediol-producing K2 isolate KCTC 2242 of unknown virulence potential, which carried a 202-kb plasmid with high homology to pLVPK.
To further investigate the phylogenetic relationship of the virulence plasmid, a separate phylogenetic analysis on the diversity of the conserved portions of the 224-kb pNTUH-K2044 plasmid was performed. To maintain as much informative sequence as possible, eight CC23 isolates shown to carry plasmid variants in which regions from 10 to 65 kb were deleted were excluded from the initial analysis, as were regions with evidence of horizontal gene transfer (HGT). The inferred relationship between the pNTUH-K2044 plasmid variants shows a clear separation into two distinct branches, one encompassing all the CC23 K1 isolates and one containing the K2 isolates (Fig. 3). This suggested that the virulence plasmid was acquired only once by each clone, followed by clonal dissemination within the distinct lineages. A single acquisition of the virulence plasmid into the CC23 lineages is further supported by the observed similarities in the subclustering of isolates in the phylogenetic tree of CC23 and the plasmid (Fig. 2 and 3, respectively).
FIG 3 .
Phylogenetic analysis of the virulence plasmid. The maximum-likelihood tree was based on 206 total SNPs, including 86 parsimony-informative SNPs after removal of recombinant regions. CC23 and K2 isolates are marked with red and yellow shadings, respectively. Eight CC23 isolates shown to carry plasmid variants in which regions from 10 to 65 kb were deleted were not included but analyzed individually (see text for details). The asterisks indicate bootstrap support values of ≥90%.
Individual analysis of the plasmid diversity among the eight CC23 isolates carrying major deletion variants revealed that these all cluster within the pNTUH-K2044 CC23 clade (results not shown).
Genomic island associated with an integrative and conjugative element (ICE).
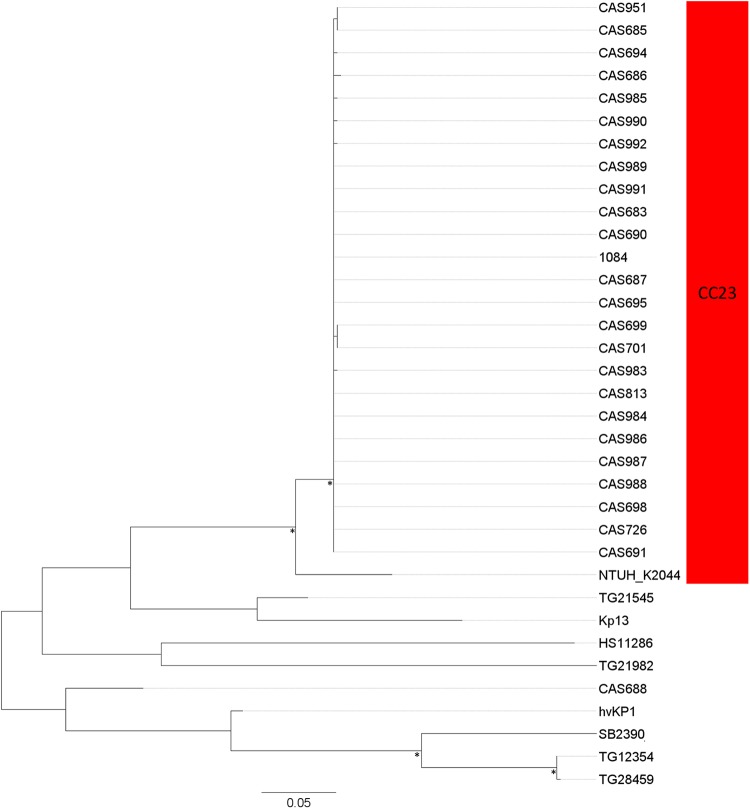
A 76-kb integrative and conjugative element (ICEKp1) integrated at an Asn-tRNA locus in the CC23 hvKP strain NTUH-K2044 has previously been described (19). ICEKp1 is divided into three regions, one similar to the high-pathogenicity island (HPI) of Yersinia species encoding the siderophore yersiniabactin followed by a middle region similar to a part of the large virulence plasmid, including the genes encoding the iron acquisition system salmochelin and a variant of the rmpA gene. The latter region of ICEKp1 contains genes responsible for conjugative transfer of the ICE followed by six open reading frames (ORFs) encoding hypothetical proteins.
Homologues of ICEKp1 were detected in all but three of the 27 CC23 isolates as well as the ST260 CC23 hybrid strain (hereafter termed CC23-related isolates). Whereas the region encoding yersiniabactin was conserved, the middle region encoding salmochelin and RmpA was not detected in the ICE region of any of the CC23-related isolates except NTUH-K2044. Furthermore, in all CC23-related isolates the six ORFs in the third region of ICEKp1 encoding hypothetical proteins were replaced by a 50-kb region (pks) encoding the polyketide genotoxin colibactin. Thus, the ICE region of all the CC23 isolates analyzed in this study resembled a genomic island recently described in the Taiwanese ST23 liver abscess strain 1084 (35). The 208-kb genomic island of strain 1084, KPHPI208, is divided into eight genomic modules (GM1 to GM8) where GM2 and GM3 are homologous to the ICEKp1 region responsible for conjugative transfer and yersiniabactin synthesis, respectively (see Fig. S5 in the supplemental material). Homologues to all genomic modules of KPHPI208 were detected in all 27 yersiniabactin-positive CC23 isolates and the CC23 hybrid strain, except for GM8, which was detected in only three isolates, and GM2, which was absent in three isolates (see Fig. S5). Notably, GM6 encodes the bacteriocin microcin E492, which may play a role in pathogenicity. Only homologues of GM2, GM3, GM5, and GM7 were detected in NTUH-K2044, further emphasizing the divergence of the genomic island in this specific strain.
In contrast to the high prevalence of homologues of KPHPI208 among the CC23 isolates, variants of the genomic island were detected in only nine of 50 non-CC23-related strains included in this study (see Fig. S5 and Tables S1 and S2 in the supplemental material). Notably, three of these were hypervirulent K2 strains. Overall, the structure of the genomic region varied among the non-CC23-related strains (see Fig. S5). The colibactin-encoding module was detected in only two strains, one of which was the hvKP K2 strain 52.145, and the microcin E492-encoding module was not detected in the genomic island of any of the non-CC23-related isolates. Thus, the variant of the genomic island encoding all three putative virulence-related factors, microcin E492, yersiniabactin, and colibactin, was found to be unique to the CC23-related strains (Fig. 1; see also Fig. S5 in the supplemental material).
To further investigate the phylogenetic relationship of the genomic island variants, the relationship between the conserved parts of the island was inferred by a phylogenetic analysis based on polymorphic sites within this region after exclusion of putative HGTs. All CC23-related isolates, except NTUH-K2044, distributed into a monophyletic clade with very low sequence diversity, indicating a recent single acquisition of the genomic island into the CC23 lineage (Fig. 4). In contrast, the genomic islands of the non-CC23-related strains formed several clearly separated clades in the analysis. The genomic island of NTUH-K2044 was clearly divergent from the other CC23 strains, indicating that the island may have undergone a major recombination event in this specific strain.
FIG 4 .
Phylogeny of the conserved parts of the KPHPI208 genomic island. The maximum-likelihood tree was based on 474 total SNPs, including 269 parsimony-informative SNPs after removal of recombinant regions. CC23 isolates are marked with red shading. The asterisks indicate bootstrap support values of ≥90%.
Virulence genes previously associated with hypervirulence.
The virulence plasmid encodes the iron acquisition systems aerobactin and salmochelin and two variants of the regulator of the mucoid phenotype, RmpA and RmpA2. Remarkably, except for the two K1 reference strains A5054 and 8045, which both carried an rmpA variant (8045 also carried aerobactin), these three virulence factors were detected only in the genomes of hvKP strains (see Tables S1 and S2 in the supplemental material). Interestingly, the K2 strain Kp52.145, the only hvKP strain not carrying a pLVPK homologue, also possessed these three virulence factors, further emphasizing their close association with K. pneumoniae hypervirulence.
Due to shifts in a poly(G) and/or a poly(A) tract of the rmpA2 gene observed in all CC23-related strains, the gene was found to encode a truncated product of 99 to 126 amino acids compared to the 212-amino-acid product encoded by the pLVPK rmpA2 gene. This suggests that the RmpA2 protein may be nonfunctional or have a different functionality in the CC23 lineage than that described for the RmpA2 variant in pLVPK. The ST86 K2 isolate CAS688 was found to possess the full 212-amino-acid RmpA2 variant of pLVPK, whereas a 100-amino-acid truncated variant was detected in the other hvKP K2 isolate, CAS689.
Three chromosomal regions associated with increased virulence have been identified in NTUH-K2044. The allS region contains genes associated with allantoin metabolism, the kfu region encodes an iron uptake system, and a 56-kb putative pathogenicity island encodes hypothetical proteins and carries the fimbrial gene cluster kpc.
The allS region was detected in all the CC23-related isolates, whereas it was absent from all but one of the non-CC23 strains, illustrating the close association of this region with the CC23 lineage (Fig. 1; see also Table S1 in the supplemental material). The only allS-positive non-CC23-related isolate was the hvKP K2 isolate CAS689.
Also, kfu was detected in all CC23-related isolates. However, the kfu region was also detected in all four non-CC23 K1 isolates, suggesting a general association of kfu with isolates of the K1 serotype. Furthermore, kfu was also detected in an additional seven of the nonhypervirulent strains, indicating that kfu is relatively widespread in K. pneumoniae.
We found the kpc island to be present in all but one of the CC23 isolates (see Table S1 in the supplemental material). However, only partial regions were present in three isolates and the island was not detected in the ST23 hybrid strain. Among the non-CC23-related isolates, partial islands were detected only in the two K1 blood isolates with conservation of the kpc fimbrial gene cluster.
Identification of unique genes in the CC23 lineage.
Genomic comparisons among the 80 diverse K. pneumoniae isolates were used to identify genes uniquely present in the CC23 clade, potentially involved in its observed hypervirulence. This analysis identified four regions ranging from 2 to ~23 kb (see Table S4 in the supplemental material). Two of these regions were associated with integrases and had significantly lower GC content (34% compared to the overall GC content of the NTUH-K2044 genome at 58%), suggesting horizontal acquisition from other organisms. The 23-kb region was homologous to a chromosomal region in the Klebsiella variicola strain At-22 (98% nucleotide identity). This region encodes putative transcriptional regulators; putative metabolism-associated enzymes, including oxidases, reductases, and hydrolases; and a number of hypothetical proteins. Genes encoding putative membrane proteins and hypothetical proteins were present in the three other unique regions.
Six additional regions with a high degree of association with CC23 were identified (see Table S3 in the supplemental material). Two of these were low-GC-content regions and were associated with integrases, indicating horizontal acquisition. Remarkably, a 5.6-kb region encoding hypothetical proteins (KP1_2357 to KP1_2370) was conserved in all but two CC23-related isolates and otherwise detected in only a highly virulent K2 strain.
Antibiotic resistance genes.
The presence of antibiotic resistance coding genes in the hvKP isolates was investigated by use of the web-based tool ResFinder (36). As expected, all isolates carried a chromosomal SHV beta-lactamase which mediates the intrinsic resistance to ampicillin of K. pneumoniae. Only one hvKP isolate, the Danish liver abscess isolate CAS813, was found to carry additional antibiotic resistance-associated genes. These were the beta-lactamase gene blaTEM-2, the aminoglycoside resistance genes aac(3)-IIa and aadA11, sulfonamide resistance genes sul1 and sul2, and dfrB1, encoding resistance to trimethoprim. In agreement, CAS813 was found to be phenotypically resistant to ampicillin, gentamicin, streptomycin, sulfonamide, and trimethoprim.
DISCUSSION
The remarkable evolution of K. pneumoniae infections, including a recent steep rise in the prevalence of severe community-acquired invasive infections in otherwise healthy individuals, is alarming. While this syndrome has mainly been observed in Southeast Asia, it is increasingly being recognized also in the Western world, indicating that it is developing into a globally emerging disease (5, 8). Traditional typing methods, including capsule serotyping and MLST analysis, have revealed that many of these infections are caused by related strains, suggesting that specific hvKP clones have emerged. This is particularly observed in the CC23 clonal lineage belonging to capsular serotype K1, which has been shown to be by far the most prevalent clone causing invasive infections in several studies (23–29). Liver abscess patients have been found to carry the same strain in their gastrointestinal tract as the one causing infection (37). Furthermore, a high fecal carriage rate of CC23 has been observed among individuals in Southeast Asia (26, 38), which possibly contributes to the high prevalence of invasive infections in this region and indicates that CC23 isolates are highly effective colonizers of the human intestinal tract.
However, analyses of the genetic repertoire of the hvKP CC23 clonal lineage and its position in the population structure of K. pneumoniae in general and in relation to other hypervirulent K. pneumoniae clones have so far been carried out by only one study applying core genome MLST (31). To investigate the evolutionary history of the hvKP clones, we sequenced the genomes of 69 K. pneumoniae isolates, including 30 hvKP isolates of different geographical origins.
The close association of CC23 with hypervirulence was confirmed, as all but three hvKP isolates were found to belong to CC23 even though the strain collection included isolates from four different continents collected over a 16-year period. Notably, this included four community-acquired pneumonia isolates from South Africa. In contrast to the Western world, K. pneumoniae is an important cause of severe community-acquired pneumonia in Africa (3), but this is to our knowledge the first time that the CC23 clone has been directly linked to this phenomenon.
We verified the close clonal relationship of the CC23 isolates, as they all grouped into a distinct monophyletic clade (Fig. 1). This is in agreement with a recent study which showed that CC23 formed a distinct clonal group when applying core genome MLST (31). Importantly, by including K1 isolates not associated with hypervirulence, we could show that the CC23 isolates form a distinct phylogenetic clade clearly separated from the nonhypervirulent K1 isolates. Thus, CC23 does not merely reflect a general clonality of isolates belonging to the K1 serotype but is indeed a unique clonal lineage with an exclusive genetic composition.
Separate phylogenetic analysis of the CC23 isolates revealed a star-like phylogeny (Fig. 2), further indicating that the CC23 lineage has evolved recently by clonal expansion from a single common ancestor. This could suggest that the CC23 lineage has developed rapidly, perhaps after acquirement of specific genetic elements conferring increased virulence or environmental fitness (e.g., the virulence plasmid or the KPHPI208 genomic island), as has been described elsewhere with other successful K. pneumoniae clones (e.g., CC258).
Interestingly, only minimal geographical grouping of the isolates was observed in the CC23 phylogeny, suggesting that CC23 has spread globally through multiple international transmission events rather than by local expansions. Indeed, infections with hvKP have in some cases been linked to recent travel to Asia. However, in several cases a link to travel or close contact with individuals from areas of endemicity could not be established, suggesting that the CC23 clonal lineage has already established itself outside Asia (29, 39–44). The location where the CC23 lineage first evolved is unknown; however, the fact that the invasive syndrome was first reported from Taiwan, as well as the high prevalence of invasive K. pneumoniae infections in this region, strongly indicates Southeast Asia.
Among the hvKP K1 isolates, only one isolate was found that did not belong strictly to the CC23 clade but instead formed a separate branch in the phylogeny. This isolate was revealed to be a hybrid strain with approximately half of its chromosomal DNA resembling CC23 and the other half originating from a K. pneumoniae isolate belonging to another clonal lineage. Intriguingly, a recent study revealed that the epidemic carbapenem-resistant ST258 clone also is a hybrid strain, presumably as a result of a large recombination event (33). Our finding of a second example of a hybrid strain suggests that, as described for other important pathogens, genomic hybridizations may be a frequent phenomenon in K. pneumoniae that occasionally may lead to the introduction of novel important genotypes as exemplified by ST258.
In contrast to the high degree of clonality of the K1 CC23 isolates, the two hvKP K2 strains were found to be genetically unrelated. However, the existence of specific hypervirulent K2 clonal lineages was supported by the close genetic relationship of the ST86 K2 isolate with two previously described hvKP strains, CG43 (45) and hvKP1 (46), which together formed a distinct ST86 clade (Fig. 1).
Due to the different genetic backgrounds of the hypervirulent K1 and K2 isolates, we hypothesized that their common hypervirulent phenotype could be associated with shared genetic elements. An element found in all hvKP strains in the collection is the large virulence plasmid which mediates the hypermucoviscous phenotype and encodes iron acquisition systems (aerobactin and salmochelin). This plasmid was absent in all of the nonhypervirulent strains except the 2,3-butanediol-producing K2 isolate KCTC 2242 of unknown virulence potential (Fig. 1). A large virulence plasmid encoding these factors was first described in the highly virulent K2 strain Kp52.145 (derived from the K2 serotype reference strain B5055) (47–49). However, in our collection, the virulence plasmid present in the hvKP strains was found to be unrelated to the large virulence plasmid of strain Kp52.145 but instead to be homologous to the large virulence plasmid pLVPK of the hvKP K2 strain CG43. The conservation of three virulence-related factors on different plasmids in hypervirulent strains is intriguing and indicates a key role of these factors, and virulence plasmids, in K. pneumoniae hypervirulence. Indeed, animal studies have revealed significantly reduced virulence of plasmid-cured derivatives of both Kp52.145 and CG43 (47, 50). Also, strain 1084, a CC23 strain that is apparently lacking the virulence plasmid, was found to have significantly attenuated virulence compared to a hypermucoviscous isolate (51, 52). Interestingly, strain 1084 could still cause fatal infections in mice (51), revealing that the high virulence of CC23 is not solely related to the virulence plasmid. It could be speculated that the 2,3-butanediol-producing K2 isolate KCTC 2242 is hypervirulent, as it both possesses the virulence plasmid and phylogenetically clusters in a clade with other hvKP isolates, thus raising concerns about the biosafety of using this isolate as a production strain.
Separate phylogenetic analysis of the plasmids revealed distinct clades according to serotype. This suggests that a plasmid has been acquired independently by each clonal lineage, followed by clonal dissemination. Furthermore, it could be speculated that acquisition of the virulence plasmid by CC23 played a significant role in the clonal expansion of this lineage.
The large virulence plasmid of CG43 contains two variants of rmpA, designated rmpA and rmpA2, which have both been shown to activate capsule production, resulting in a hypermucoviscous phenotype as well as increasing the virulence in mice (53, 54). In agreement with previous studies, we found a close association of rmpA with hypervirulence, as all hvKP strains were rmpA positive whereas no variants of this gene were detected in any of the nonhypervirulent strains, except the two K1 reference strains. Furthermore, we found that all CC23-related strains and one of the K2 strains carried truncated variants of RmpA2. A high prevalence of truncated RmpA2 variants has previously been reported but not been generally related to CC23 (55). It could be speculated that the presence of a truncated variant in this highly successful clone indicates that it confers an advantage perhaps via a more subtle activation of capsule expression in comparison to a strain with two fully functional variants present. Notably, in agreement with the presence of a full-length copy of RmpA, all the CC23 isolates expressed the hypermucoviscous phenotype.
An integrative and conjugative element, ICEKp1, containing virulence genes, including those for the siderophores yersiniabactin and salmochelin and a third copy of rmpA, has been described in the CC23 strain NTUH-K2044 (19). Except for the conserved presence of yersiniabactin, we found that the composition of the ICE-containing region was remarkably different from NTUH-K2044 in all CC23 isolates and resembled the genomic island, KPHPI208, previously described in isolate 1084 (35). Thus, the rmpA and the salmochelin region appear to be specific for the ICE region in the NTUH-K2044 strain, whereas these regions are generally restricted to the virulence plasmid in all other CC23 isolates. Intriguingly, we found gene clusters encoding colibactin and microcin E492 in the genomic island of all CC23 isolates except NTUH-K2044. Colibactin is a genotoxin shown to induce DNA damage in host cells in vivo and in vitro and has been suggested to play a role in the pathogenesis of inflammatory bowel disease and to promote colorectal cancer (56–58). Colibactin has primarily been associated with the E. coli B2 phylogenetic lineage, where it has been associated with long-term gastrointestinal colonization and bacteremia (59, 60). Notably, colibactin expression in the 1084 CC23 isolate was recently found to induce DNA damage in liver parenchymal cells of infected mice (35). Microcin E492 is a bacteriocin active against members of the Enterobacteriaceae family but has also been shown to induce apoptosis in human cell lines (61, 62). Thus, it may provide a competitive advantage during gastrointestinal colonization or play a direct role in virulence.
Intriguingly, we found that the KPHPI208 variant containing colibactin, yersiniabactin, and microcin E492 was restricted to CC23 (Fig. 1). It could be speculated that this region may play a key role in the success of this clone by promoting virulence and/or environmental fitness. Worryingly, a significantly increased risk of subsequent colorectal cancer has been observed in patients with K. pneumoniae liver abscess compared to patients with liver abscess caused by other microbes (63). In this respect, the finding that colibactin is generally present in the hypervirulent CC23 lineage is concerning and should be further investigated. As for the virulence plasmid, the phylogenetic analysis indicated that the genomic island was recently introduced into the CC23 lineages by a single acquisition of the element. Remarkably, the region involved in mobilization of the genomic island was absent in three isolates, suggesting that the island may be stably integrated in the genome of some CC23 isolates.
The ability to acquire iron in the host environment plays a key role in K. pneumoniae pathogenicity. Indeed, a remarkable number of different iron acquisition systems were detected in CC23. Thus, in addition to the virulence plasmid-encoded siderophores aerobactin and salmochelin, and the KPHPI208-encoded yersiniabactin, the chromosomally carried kfu operon was also conserved in all CC23 isolates. Indeed, recent studies have established that hvKP isolates produce more siderophores than do other isolates and that this trait enhances virulence (64). The enhanced siderophore production in hvKP isolates was found to be mainly related to aerobactin production (65). Our finding of aerobactin being strongly associated with hvKP strains and being encoded by a virulence plasmid further indicates a key role of aerobactin and the virulence plasmid in K. pneumoniae hypervirulence. The presence of multiple iron acquisition systems in CC23 isolates may ensure optimal iron acquisition in different host environments. For instance, kfu was shown to be important for virulence in mice after gastrointestinal inoculation but not when the bacteria were inoculated intraperitoneally, suggesting that kfu primarily promotes gastrointestinal colonization (22).
Another factor that may promote the ability of CC23 to colonize the intestinal tract is the allS region, associated with allantoin metabolism, which also has been shown to contribute to virulence in mice after gastrointestinal inoculation but not when the bacteria were inoculated intraperitoneally (21). The previously reported close association of the allS region with the CC23 genetic background is supported by this study, as it was detected in all CC23-related isolates but otherwise only in one of the hvKP K2 isolates (Fig. 1). Similarly, a high prevalence of the kpc fimbrial gene cluster both among the CC23 isolates and in two nonhypervirulent K1 strains suggests a possible general association of this fimbria with the K1 serotype. In contrast to a previous study (23), we did not find any association of the nonfimbrial adhesin CF29K and CC23, as this adhesin was not detected in any of the isolates analyzed.
Using genome-wide comparisons, we identified four novel regions unique for the CC23 isolates as well as six additional regions with a high degree of association with CC23 (see Table S3 in the supplemental material). Several of these regions were associated with transposases and/or integrases and had a significantly lower GC content, suggesting horizontal acquisition from other organisms. It could be speculated that the regions play a role in the hypervirulence of CC23 or confer increased fitness (e.g., increased gastrointestinal colonization ability). However, as most of the genes encoded hypothetical proteins, additional studies are needed to clarify this. Even if the CC23 unique genes are not associated with CC23 pathogenicity, they can serve as novel markers for easy identification of this important emerging clonal lineage.
Most hvKP isolates have so far been reported to be generally susceptible to antibiotics (5). In agreement, except for blaSHV naturally present in K. pneumoniae, antibiotic resistance genes were detected in only one hvKP isolate in our collection. This CC23 isolate carried genes mediating resistance against aminoglycosides, sulfonamide, and trimethoprim. Alarmingly, multidrug-resistant hypervirulent isolates have recently been reported with increasing frequency, including CC23 isolates carrying the extended-spectrum beta-lactamase CTX-15 and the KPC-2 carbapenemase (31, 66, 67). The development of multidrug resistance in hvKP strains is truly disturbing, leaving few treatment options for the severe infections caused by these strains.
In summary, using whole-genome sequencing (WGS), we demonstrated a close genetic relationship among hvKP CC23 isolates, which form a monophyletic clade clearly distinct from nonhypervirulent strains. Our data suggest that CC23 has recently evolved by clonal expansion, perhaps after acquisition of a virulence plasmid. Indeed, the conservation of the aerobactin- and RmpA-encoding pLVPK plasmid in the otherwise-unrelated hvKP K2 lineages indicates a key role of this plasmid in K. pneumoniae hypervirulence. The success of the CC23 lineage compared to other hypervirulent clonal lineages is likely multifactorial and could be related to specific genetic determinants associated with CC23. These include the distinct CC23 variant of the KPHPI208 genomic island encoding microcin E492 and colibactin, the allS region, and the novel genomic regions identified as unique for CC23 in this study. Future studies should elucidate the exact role of these factors in the virulence and emergence of this successful hypervirulent clone.
MATERIALS AND METHODS
Bacterial isolates.
A total of 67 K. pneumoniae clinical isolates sampled in Canada, Denmark, Norway, South Africa, Taiwan, and the United States in the period from 1996 to 2012 and two reference strains of serotype K1 strains were included (see Table S1 in the supplemental material). Thirty of the isolates were isolated from cases of community-acquired liver abscess or pneumonia and were therefore considered hypervirulent.
Capsule serotyping.
Capsule serotyping of the hvKP isolates was performed at the WHO Collaborating Centre for Reference and Research on Escherichia and Klebsiella by countercurrent immunoelectrophoresis (CCIE) using a modified version of the method described by Palfreyman (68). An extract was used as antigen instead of a whole-cell suspension. The extract was heated only once for 1 h at 100°C before centrifugation.
MLST.
Multilocus sequence typing (MLST) was performed according to the work of Diancourt et al. (69). Alleles and sequence types were assigned by using the MLST database (http://www.pasteur.fr/mlst/Kpneumoniae.html).
Genome sequencing.
The genomes of the 69 K. pneumoniae isolates were sequenced using Illumina GAIIX (47 samples) or MiSeq (22 samples) sequencing technologies as described previously (70, 71). DNA samples were extracted using the DNeasy Blood and Tissue kit as described by the manufacturer (Qiagen, Valencia, CA). For sequencing preparation on the Illumina GAIIX system, 47 samples had approximately 2 µg of DNA sheared to an average size of 500 bp using the Sonicman sonicator (Brooks Automation, Spokane, WA, USA) and prepared for sequencing using the Kapa Biosystems library preparation kit protocol (catalog no. KK8201; Woburn, MA) as described by the manufacturer. These libraries were quantified prior to sequencing using quantitative PCR (qPCR) with the Kapa library quantification kit (catalog no. KK4835; Woburn, MA) on the ABI 7900HT PCR system (Life Technologies Corporation, Carlsbad, CA). Libraries were sequenced to a read length of 100 bp on the Illumina GAIIX system. For sequencing preparation on the MiSeq system, fragment libraries were constructed using the Nextera kit (Illumina, San Diego, CA) followed by 251-bp paired-end sequencing on a MiSeq sequencer (Illumina) according to the manufacturer’s instructions.
The sequencing reads were assembled using CLC Genomics Workbench 7.5 (Qiagen, Aarhus, Denmark) with default parameters to include only contigs of ≥500 nucleotides and with over 10-fold average coverage. A summary of genome sequencing and assembly is found in Table S4 in the supplemental material.
Identifications of SNPs.
In order to gain insight into the genetic relationships between isolates, whole-genome SNP typing was performed as previously described (70, 72, 73). Briefly, an SNP phylogeny was constructed based on an SNP matrix that tabulates all SNPs among the isolates by their location within the genome using NASP (http://github.com/TGenNorth/NASP/releases/tag/v0.9.6). The matrix was generated by aligning sequence data from the sequenced isolates with a reference genome, NTUH-K2044 (NCBI accession no. AP006725) using Novoalign 3.00.03 (Novocraft Technologies, Selangor, Malaysia). Reads that mapped to multiple locations within the reference genome were excluded from the alignments, as were reads containing insertions or deletions. SNPs were identified using the Genome Analysis Toolkit (GATK) Unified Genotyper v2.4 (74). Only SNP loci found throughout all samples were used for the phylogenetic analysis. Additionally, SNPs were excluded if they did not meet a minimum coverage of 10-fold coverage or if the variant was present in less than 90% of the base calls for that position. MUMmer version 3.22 (75) was used to identify duplicated regions within NTUH-K2044, and SNPs located within these regions were removed from the final analysis. Furthermore, regions of high SNP density indicative of recombination were identified and removed using Gubbins (34) with default parameters. Maximum-likelihood inferred trees based on the SNP matrix were constructed using MEGA6.06 with the GTR (generalized time-reversible) nucleotide substitution model (76). Figure 1 was rooted by including the near-neighbor species Klebsiella variicola in a separate analysis tree.
Comparative genomics.
To identify unique and shared genomic regions in CC23, four CC23 genomes (CAS695 and CAS701 and the closed genomes of strains NTUH-K2044 and 1084) and four non-CC23 genomes (the closed genomes of strains HS11286, CG43, KCTC 2242, and MGH78578) were analyzed using the progressiveMauve algorithm implemented in Mauve 2.3.1 using default settings (77). Genomic regions conserved in at least two of the CC23 genomes but absent in at least two of the non-CC23 genomes were submitted to local BLASTN searches among the assembled genomes of all 30 CC23 and 50 non-CC23 strains included in the study using CLC bio’s Genomic Workbench 7.5 (Qiagen, Aarhus, Denmark). The presence or absence of genomic regions was determined using thresholds of 90% nucleotide identity, 90% coverage of the query sequence length, and a sequence depth of >10×. Genomic regions conserved in 90% of CC23 strains but absent from 90% of non-CC23 strains were considered to be CC23 associated. BLASTN atlases of the pNTUH-K2044 virulence plasmid and the KPHPI208 genomic island were constructed using BLAST Ring Image Generator v0.95 with default parameters (78).
Antibiotic resistance genes.
The presence of antibiotic resistance coding genes in the hvKP isolates was investigated by use of the web-based tool ResFinder at http://cge.cbs.dtu.dk/services/ResFinder.
Nucleotide sequence accession numbers.
The accession numbers for the Illumina sequences generated from the 67 K. pneumoniae isolates sequenced in this study are available in the European Nucleotide Archive (ENA; http://www.ebi.ac.uk/ena) under the primary identification number PRJEB7967.
SUPPLEMENTAL MATERIAL
Rooted maximum-likelihood phylogeny of 80 K. pneumoniae isolates after recombination filtering by Gubbins (34). Hypervirulent clonal lineages (ST14, CC23, and ST86) are marked with shadings. Download
Scatterplot of SNP number in CAS686 related to position in the NTUH-K2044 chromosome. Of 11,519 SNPs identified, only 254 SNPs were located in the region corresponding to position 1.0 to 3.7 Mbp in the NTUH-K2044 chromosome, revealing a composition of CAS686 as a chimera between a CC23 strain and a strain of unknown clonal origin. Download
Phylogenetic analysis of the non-CC23 segments of the genome of the ST260 strain CAS686 (orange marking). Download
BLASTN atlas illustrating the presence of the pNTUH-K2044 virulence plasmid in hypervirulent (A) and nonhypervirulent (B) K. pneumoniae isolates. Annotated regions indicate genes encoding the virulence factors RmpA, RmpA2, aerobactin (iucABCD-iutA), and salmochelin (iroBCDN) and gene clusters encoding silver (sil) and copper (pco) resistance. Red rings in panel A are the 28 CC23-related isolates in the same order from inside to outside as outlined in Table S1 (CAS683 to CAS686). Blue rings in panel A are, from inside to outside, the hypervirulent K2 isolates CAS688 and CAS689 and the virulence plasmid pLVPK from the K2 strain CG43. Green rings in panel B are the 39 nonhypervirulent isolates in the same order from inside to outside as outlined in Table S1 (A5054 to 3091). The purple ring is pKCTC2242 from strain KCTC 2242 of unknown virulence potential. Download
BLASTN atlas illustrating the presence of the different genomic modules (GM1 to GM8) of KPHPI208 in hypervirulent and nonhypervirulent K. pneumoniae isolates. Red rings are, from inside to outside, the CC23-related isolates CAS683, CAS685, CAS686, CAS687, CAS690, CAS691, CAS694, CAS695, CAS698, CAS699, CAS701, CAS726, CAS727, CAS813, CAS951, CAS983, CAS984, CAS985, CAS986, CAS987, CAS988, CAS989, CAS990, CAS991, and CAS992. The orange ring is NTUH-K2044. Blue rings are, from inside to outside, the hypervirulent K2 isolates CAS688, hvKP1, and Kp52.145. Green rings are, from inside to outside, the nonhypervirulent isolates TG12354, TG21545, TG21982, TG28459, SB2390, HS11286, and Kp13. Download
Characteristics of the 69 isolates sequenced in this study.
Characteristics of publicly available K. pneumoniae genomes included in this study.
Genomic regions unique to or highly associated with CC23.
Summary of genome sequencing and assembly.
ACKNOWLEDGMENTS
We are grateful to the following for providing K. pneumoniae isolates: F. C. Fang, S. Libby, S. Jenkins, S. K. Söbirk, A. Fostervold, and J. C. Holter.
This work was supported in part by the U.S. NIH R01 award AI090782.
Footnotes
Citation Struve C, Roe CC, Stegger M, Stahlhut SG, Hansen DS, Engelthaler DM, Andersen PS, Driebe EM, Keim P, Krogfelt KA. 2015. Mapping the evolution of hypervirulent Klebsiella pneumoniae. mBio 6(4):e00630-15. doi:10.1128/mBio.00630-15.
Contributor Information
Julie Segre, National Human Genome Research Institute.
David A. Relman, VA Palo Alto Health Care System.
REFERENCES
- 1.Podschun R, Ullmann U. 1998. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev 11:589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang FY, Chou MY. 1995. Comparison of pyogenic liver abscesses caused by Klebsiella pneumoniae and non-K. pneumoniae pathogens. J Formos Med Assoc 94:232–237. [PubMed] [Google Scholar]
- 3.Ko W-C, Paterson DL, Sagnimeni AJ, Hansen DS, Von Gottberg A, Mohapatra S, Casellas JM, Goossens H, Mulazimoglu L, Trenholme G, Klugman KP, McCormack JG, Yu VL. 2002. Community-acquired Klebsiella pneumoniae bacteremia: global differences in clinical patterns. Emerg Infect Dis 8:160–166. doi: 10.3201/eid0802.010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang JH, Liu YC, Lee SS, Yen MY, Chen YS, Wann SR, Lin HH. 1998. Primary liver abscess due to Klebsiella pneumoniae in Taiwan. Clin Infect Dis 26:1434–1438. doi: 10.1086/516369. [DOI] [PubMed] [Google Scholar]
- 5.Shon AS, Bajwa RP, Russo TA. 2013. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence 4:107–118. doi: 10.4161/viru.22718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsai F-C, Huang Y-T, Chang L-Y, Wang J-T. 2008. Pyogenic liver abscess as endemic disease, Taiwan. Emerg Infect Dis 14:1592–1600. doi: 10.3201/eid1410.071254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung DR, Lee SS, Lee HR, Kim HB, Choi HJ, Eom JS, Kim JS, Choi YH, Lee JS, Chung MH, Kim YS, Lee H, Lee MS, Park CK, Korean Study Group for Liver Abscess . 2007. Emerging invasive liver abscess caused by K1 serotype Klebsiella pneumoniae in Korea. J Infect 54:578–583. doi: 10.1016/j.jinf.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 8.Siu LK, Yeh K-M, Lin J-C, Fung C-P, Chang F-Y. 2012. Klebsiella pneumoniae liver abscess: a new invasive syndrome. Lancet Infect Dis 12:881–887. doi: 10.1016/S1473-3099(12)70205-0. [DOI] [PubMed] [Google Scholar]
- 9.Lederman ER, Crum NF. 2005. Pyogenic liver abscess with a focus on Klebsiella pneumoniae as a primary pathogen: an emerging disease with unique clinical characteristics. Am J Gastroenterol 100:322–331. doi: 10.1111/j.1572-0241.2005.40310.x. [DOI] [PubMed] [Google Scholar]
- 10.Rahimian J, Wilson T, Oram V, Holzman RS. 2004. Pyogenic liver abscess: recent trends in etiology and mortality. Clin Infect Dis 39:1654–1659. doi: 10.1086/425616. [DOI] [PubMed] [Google Scholar]
- 11.Fang C-T, Chuang Y-P, Shun C-T, Chang S-C, Wang J-T. 2004. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med 199:697–705. doi: 10.1084/jem.20030857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu VL, Hansen DS, Ko WC, Sagnimeni A, Klugman KP, von Gottberg A, Goossens H, Wagener MM, Benedi VJ, International Klebsiella Study Group . 2007. Virulence characteristics of Klebsiella and clinical manifestations of K. pneumoniae bloodstream infections. Emerg Infect Dis 13:986–993. doi: 10.3201/eid1307.070187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu W-L, Ko W-C, Cheng K-C, Lee C-C, Lai C-C, Chuang Y-C. 2008. Comparison of prevalence of virulence factors for Klebsiella pneumoniae liver abscesses between isolates with capsular K1/K2 and non-K1/K2 serotypes. Diagn Microbiol Infect Dis 62:1–6. doi: 10.1016/j.diagmicrobio.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 14.Fung C-P, Chang F-Y, Lee S-C, Hu B-S, Kuo BI, Liu C-Y, Ho M, Siu LK. 2002. A global emerging disease of Klebsiella pneumoniae liver abscess: is serotype K1 an important factor for complicated endophthalmitis? Gut 50:420–424. doi: 10.1136/gut.50.3.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fang C-T, Lai S-Y, Yi W-C, Hsueh P-R, Liu K-L, Chang S-C. 2007. Klebsiella pneumoniae genotype K1: an emerging pathogen that causes septic ocular or central nervous system complications from pyogenic liver abscess. Clin Infect Dis 45:284–293. doi: 10.1086/519262. [DOI] [PubMed] [Google Scholar]
- 16.Pomakova DK, Hsiao C-B, Beanan JM, Olson R, MacDonald U, Keynan Y, Russo TA. 2012. Clinical and phenotypic differences between classic and hypervirulent Klebsiella pneumoniae: an emerging and under-recognized pathogenic variant. Eur J Clin Microbiol Infect Dis 31:981–989. doi: 10.1007/s10096-011-1396-6. [DOI] [PubMed] [Google Scholar]
- 17.Wu K-M, Li L-H, Yan J-J, Tsao N, Liao T-L, Tsai H-C, Fung C-P, Chen H-J, Liu Y-M, Wang J-T, Fang C-T, Chang S-C, Shu H-Y, Liu T-T, Chen Y-T, Shiau Y-R, Lauderdale T-L, Su I-J, Kirby R, Tsai S-F. 2009. Genome sequencing and comparative analysis of Klebsiella pneumoniae NTUH-K2044, a strain causing liver abscess and meningitis. J Bacteriol 191:4492–4501. doi: 10.1128/JB.00315-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y-T, Chang H-Y, Lai Y-C, Pan C-C, Tsai S-F, Peng H-L. 2004. Sequencing and analysis of the large virulence plasmid pLVPK of Klebsiella pneumoniae CG43. Gene 337:189–198. doi: 10.1016/j.gene.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 19.Lin T-L, Lee C-Z, Hsieh P-F, Tsai S-F, Wang J-T. 2008. Characterization of integrative and conjugative element ICEKp1-associated genomic heterogeneity in a Klebsiella pneumoniae strain isolated from a primary liver abscess. J Bacteriol 190:515–526. doi: 10.1128/JB.01219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu C-C, Huang Y-J, Fung C-P, Peng H-L. 2010. Regulation of the Klebsiella pneumoniae Kpc fimbriae by the site-specific recombinase KpcI. Microbiology 156:1983–1992. doi: 10.1099/mic.0.038158-0. [DOI] [PubMed] [Google Scholar]
- 21.Chou H-C, Lee C-Z, Ma L-C, Fang C-T, Chang S-C, Wang J-T. 2004. Isolation of a chromosomal region of Klebsiella pneumoniae associated with allantoin metabolism and liver infection. Infect Immun 72:3783–3792. doi: 10.1128/IAI.72.7.3783-3792.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma L-C, Fang C-T, Lee C-Z, Shun C-T, Wang J-T. 2005. Genomic heterogeneity in Klebsiella pneumoniae strains is associated with primary pyogenic liver abscess and metastatic infection. J Infect Dis 192:117–128. doi: 10.1086/430619. [DOI] [PubMed] [Google Scholar]
- 23.Brisse S, Fevre C, Passet V, Issenhuth-Jeanjean S, Tournebize R, Diancourt L, Grimont P. 2009. Virulent clones of Klebsiella pneumoniae: identification and evolutionary scenario based on genomic and phenotypic characterization. PLoS One 4:e4982. doi: 10.1371/journal.pone.0004982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo Y, Wang Y, Ye L, Yang J. 2014. Molecular epidemiology and virulence factors of pyogenic liver abscess causing Klebsiella pneumoniae in China. Clin Microbiol Infect 20:O818–O824. doi: 10.1111/1469-0691.12664. [DOI] [PubMed] [Google Scholar]
- 25.Chung DR, Lee HR, Lee SS, Kim SW, Chang H-H, Jung S-I, Oh MD, Ko KS, Kang C-I, Peck KR, Song J-H. 2008. Evidence for clonal dissemination of the serotype K1 Klebsiella pneumoniae strain causing invasive liver abscesses in Korea. J Clin Microbiol 46:4061–4063. doi: 10.1128/JCM.01577-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siu LK, Fung C-P, Chang F-Y, Lee N, Yeh K-M, Koh TH, Ip M. 2011. Molecular typing and virulence analysis of serotype K1 Klebsiella pneumoniae strains isolated from liver abscess patients and stool samples from noninfectious subjects in Hong Kong, Singapore, and Taiwan. J Clin Microbiol 49:3761–3765. doi: 10.1128/JCM.00977-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liao CH, Huang YT, Chang CY, Hsu HS, Hsueh PR. 2014. Capsular serotypes and multilocus sequence types of bacteremic Klebsiella pneumoniae isolates associated with different types of infections. Eur J Clin Microbiol Infect Dis 33:365–369. doi: 10.1007/s10096-013-1964-z. [DOI] [PubMed] [Google Scholar]
- 28.Turton JF, Englender H, Gabriel SN, Turton SE, Kaufmann ME, Pitt TL. 2007. Genetically similar isolates of Klebsiella pneumoniae serotype K1 causing liver abscesses in three continents. J Med Microbiol 56:593–597. doi: 10.1099/jmm.0.46964-0. [DOI] [PubMed] [Google Scholar]
- 29.Decré D, Verdet C, Emirian A, Le Gourrierec T, Petit J-C, Offenstadt G, Maury E, Brisse S, Arlet G. 2011. Emerging severe and fatal infections due to Klebsiella pneumoniae in two university hospitals in France. J Clin Microbiol 49:3012–3014. doi: 10.1128/JCM.00676-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin J-C, Koh TH, Lee N, Fung C-P, Chang F-Y, Tsai Y-K, Ip M, Siu LK. 2014. Genotypes and virulence in serotype K2 Klebsiella pneumoniae from liver abscess and non-infectious carriers in Hong Kong, Singapore and Taiwan. Gut Pathog 6:21. doi: 10.1186/1757-4749-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bialek-Davenet S, Criscuolo A, Ailloud F, Passet V, Jones L, Delannoy-Vieillard A-S, Garin B, Le Hello S, Arlet G, Nicolas-Chanoine M-H, Decré D, Brisse S. 2014. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg Infect Dis 20:1812–1820. doi: 10.3201/eid2011.140206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo C, Yang X, Wu Y, Yang H, Han Y, Yang R, Hu L, Cui Y, Zhou D. 2015. MLST-based inference of genetic diversity and population structure of clinical Klebsiella pneumoniae, China. Sci Rep 5:7612. doi: 10.1038/srep07612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen L, Mathema B, Pitout JD, DeLeo FR, Kreiswirth BN. 2014. Epidemic Klebsiella pneumoniae ST258 is a hybrid strain. mBio 5(3):e01355-14. doi: 10.1128/mBio.01355-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, Parkhill J, Harris SR. 2015. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res 43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lai Y-C, Lin A-C, Chiang M-K, Dai Y-H, Hsu C-C, Lu M-C, Liau C-Y, Chen Y-T. 2014. Genotoxic Klebsiella pneumoniae in Taiwan. PLoS One 9:e96292. doi: 10.1371/journal.pone.0096292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fung C-P, Lin Y-T, Lin J-C, Chen T-L, Yeh K-M, Chang F-Y, Chuang H-C, Wu H-S, Tseng C-P, Siu LK. 2012. Klebsiella pneumoniae in gastrointestinal tract and pyogenic liver abscess. Emerg Infect Dis 18:1322–1325. doi: 10.3201/eid1808.111053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chung DR, Lee H, Park MH, Jung S-I, Chang H-H, Kim Y-S, Son JS, Moon C, Kwon KT, Ryu SY, Shin SY, Ko KS, Kang C-I, Peck KR, Song J-H. 2012. Fecal carriage of serotype K1 Klebsiella pneumoniae ST23 strains closely related to liver abscess isolates in Koreans living in Korea. Eur J Clin Microbiol Infect Dis 31:481–486. doi: 10.1007/s10096-011-1334-7. [DOI] [PubMed] [Google Scholar]
- 39.Bilal S, Volz MS, Fiedler T, Podschun R, Schneider T. 2014. Klebsiella pneumoniae-induced liver abscesses, Germany. Emerg Infect Dis 20:1939–1940. doi: 10.3201/eid2011.140149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gundestrup S, Struve C, Stahlhut SG, Hansen DS. 2014. First case of liver abscess in Scandinavia due to the international hypervirulent Klebsiella pneumoniae clone ST23. Open Microbiol J 8:22–24. doi: 10.2174/1874285801408010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holmås K, Fostervold A, Stahlhut SG, Struve C, Holter JC. 2014. Emerging K1 serotype Klebsiella pneumoniae primary liver abscess: three cases presenting to a single university hospital in Norway. Clin Case Rep 2:122–127. doi: 10.1002/ccr3.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCabe R, Lambert L, Frazee B. 2010. Invasive Klebsiella pneumoniae infections, California, USA. Emerg Infect Dis 16:1490–1491. doi: 10.3201/eid1609.100386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pope JV, Teich DL, Clardy P, McGillicuddy DC. 2011. Klebsiella pneumoniae liver abscess: an emerging problem in North America. J Emerg Med 41:e103–e105. doi: 10.1016/j.jemermed.2008.04.041. [DOI] [PubMed] [Google Scholar]
- 44.Vila A, Cassata A, Pagella H, Amadio C, Yeh K-M, Chang F-Y, Siu LK. 2011. Appearance of Klebsiella pneumoniae liver abscess syndrome in Argentina: case report and review of molecular mechanisms of pathogenesis. Open Microbiol J 5:107–113. doi: 10.2174/1874285801105010107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang HY, Lee JH, Deng WL, Fu TF, Peng HL. 1996. Virulence and outer membrane properties of a galU mutant of Klebsiella pneumoniae CG43. Microb Pathog 20:255–261. doi: 10.1006/mpat.1996.0024. [DOI] [PubMed] [Google Scholar]
- 46.Russo TA, Gill SR. 2013. Draft genome sequence of the hypervirulent Klebsiella pneumoniae strain hvKP1, isolated in Buffalo, New York. Genome Announc 1(2):e0006513. doi: 10.1128/genomeA.00065-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nassif X, Fournier JM, Arondel J, Sansonetti PJ. 1989. Mucoid phenotype of Klebsiella pneumoniae is a plasmid-encoded virulence factor. Infect Immun 57:546–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nassif X, Sansonetti PJ. 1986. Correlation of the virulence of Klebsiella pneumoniae K1 and K2 with the presence of a plasmid encoding aerobactin. Infect Immun 54:603–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lery LM, Frangeul L, Tomas A, Passet V, Almeida AS, Bialek-Davenet S, Barbe V, Bengoechea JA, Sansonetti P, Brisse S, Tournebize R. 2014. Comparative analysis of Klebsiella pneumoniae genomes identifies a phospholipase D family protein as a novel virulence factor. BMC Biol 12:41. doi: 10.1186/1741-7007-12-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang H-L, Chiang M-K, Liou W-J, Chen Y-T, Peng H-L, Chiou C-S, Liu K-S, Lu M-C, Tung K-C, Lai Y-C. 2010. Correlation between Klebsiella pneumoniae carrying pLVPK-derived loci and abscess formation. Eur J Clin Microbiol Infect Dis 29:689–698. doi: 10.1007/s10096-010-0915-1. [DOI] [PubMed] [Google Scholar]
- 51.Lin Y-C, Lu M-C, Tang H-L, Liu H-C, Chen C-H, Liu K-S, Lin C, Chiou C-S, Chiang M-K, Chen C-M, Lai Y-C. 2011. Assessment of hypermucoviscosity as a virulence factor for experimental Klebsiella pneumoniae infections: comparative virulence analysis with hypermucoviscosity-negative strain. BMC Microbiol 11:50. doi: 10.1186/1471-2180-11-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin A-C, Liao T-L, Lin Y-C, Lai Y-C, Lu M-C, Chen Y-T. 2012. Complete genome sequence of Klebsiella pneumoniae 1084, a hypermucoviscosity-negative K1 clinical strain. J Bacteriol 194:6316. doi: 10.1128/JB.01548-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheng HY, Chen YS, Wu CY, Chang HY, Lai YC, Peng HL. 2010. RmpA regulation of capsular polysaccharide biosynthesis in Klebsiella pneumoniae CG43. J Bacteriol 192:3144–3158. doi: 10.1128/JB.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lai YC, Peng HL, Chang HY. 2003. RmpA2, an activator of capsule biosynthesis in Klebsiella pneumoniae CG43, regulates K2 cps gene expression at the transcriptional level. J Bacteriol 185:788–800. doi: 10.1128/JB.185.3.788-800.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hsu C-R, Lin T-L, Chen Y-C, Chou H-C, Wang J-T. 2011. The role of Klebsiella pneumoniae rmpA in capsular polysaccharide synthesis and virulence revisited. Microbiology 157:3446–3457. doi: 10.1099/mic.0.050336-0. [DOI] [PubMed] [Google Scholar]
- 56.Nougayrède J-P, Homburg S, Taieb F, Boury M, Brzuszkiewicz E, Gottschalk G, Buchrieser C, Hacker J, Dobrindt U, Oswald E. 2006. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science 313:848–851. doi: 10.1126/science.1127059. [DOI] [PubMed] [Google Scholar]
- 57.Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, Nougayrède J-P. 2010. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc Natl Acad Sci U S A 107:11537–11542. doi: 10.1073/pnas.1001261107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guerra L, Guidi R, Frisan T. 2011. Do bacterial genotoxins contribute to chronic inflammation, genomic instability and tumor progression? FEBS J 278:4577–4588. doi: 10.1111/j.1742-4658.2011.08125.x. [DOI] [PubMed] [Google Scholar]
- 59.Nowrouzian FL, Oswald E. 2012. Escherichia coli strains with the capacity for long-term persistence in the bowel microbiota carry the potentially genotoxic pks island. Microb Pathog 53:180–182. doi: 10.1016/j.micpath.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 60.Johnson JR, Johnston B, Kuskowski MA, Nougayrede J-P, Oswald E. 2008. Molecular epidemiology and phylogenetic distribution of the Escherichia coli pks genomic island. J Clin Microbiol 46:3906–3911. doi: 10.1128/JCM.00949-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.De Lorenzo V. 1984. Isolation and characterization of microcin E492 from Klebsiella pneumoniae. Arch Microbiol 139:72–75. doi: 10.1007/BF00692715. [DOI] [PubMed] [Google Scholar]
- 62.Hetz C, Bono MR, Barros LF, Lagos R. 2002. Microcin E492, a channel-forming bacteriocin from Klebsiella pneumoniae, induces apoptosis in some human cell lines. Proc Natl Acad Sci U S A 99:2696–2701. doi: 10.1073/pnas.052709699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang W-K, Chang JW, See L-C, Tu H-T, Chen J-S, Liaw C-C, Lin Y-C, Yang T-S. 2012. Higher rate of colorectal cancer among patients with pyogenic liver abscess with Klebsiella pneumoniae than those without: an 11-year follow-up study. Colorectal Dis 14:e794–e801. doi: 10.1111/j.1463-1318.2012.03174.x. [DOI] [PubMed] [Google Scholar]
- 64.Russo TA, Shon AS, Beanan JM, Olson R, MacDonald U, Pomakov AO, Visitacion MP. 2011. Hypervirulent K. pneumoniae secretes more and more active iron-acquisition molecules than “classical” K. pneumoniae thereby enhancing its virulence. PLoS One 6:e26734. doi: 10.1371/journal.pone.0026734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Russo TA, Olson R, Macdonald U, Metzger D, Maltese LM, Drake EJ, Gulick AM. 2014. Aerobactin mediates virulence and accounts for increased siderophore production under iron-limiting conditions by hypervirulent (hypermucoviscous) Klebsiella pneumoniae. Infect Immun 82:2356–2367. doi: 10.1128/IAI.01667-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cejas D, Fernández Canigia L, Rincón Cruz G, Elena AX, Maldonado I, Gutkind GO, Radice MA. 2014. First isolate of KPC-2-producing Klebsiella pneumoniae sequence type 23 from the Americas. J Clin Microbiol 52:3483–3485. doi: 10.1128/JCM.00726-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Su S-C, Siu LK, Ma L, Yeh K-M, Fung C-P, Lin J-C, Chang F-Y. 2008. Community-acquired liver abscess caused by serotype K1 Klebsiella pneumoniae with CTX-M-15-type extended-spectrum beta-lactamase. Antimicrob Agents Chemother 52:804–805. doi: 10.1128/AAC.01269-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Palfreyman JM. 1978. Klebsiella serotyping by counter-current immunoelectrophoresis. J Hyg (Lond) 81:219–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol 43:4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gillece JD, Schupp JM, Balajee SA, Harris J, Pearson T, Yan Y, Keim P, DeBess E, Marsden-Haug N, Wohrle R, Engelthaler DM, Lockhart SR. 2011. Whole genome sequence analysis of Cryptococcus gattii from the Pacific Northwest reveals unexpected diversity. PLoS One 6:e28550. doi: 10.1371/journal.pone.0028550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Etienne KA, Gillece J, Hilsabeck R, Schupp JM, Colman R, Lockhart SR, Gade L, Thompson EH, Sutton DA, Neblett-Fanfair R, Park BJ, Turabelidze G, Keim P, Brandt ME, Deak E, Engelthaler DM. 2012. Whole genome sequence typing to investigate the Apophysomyces outbreak following a tornado in Joplin, Missouri, 2011. PLoS One 7:e49989. doi: 10.1371/journal.pone.0049989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lazéra MS, Cavalcanti MA, Trilles L, Nishikawa MM, Wanke B. 1998. Cryptococcus neoformans var. gattii—evidence for a natural habitat related to decaying wood in a pottery tree hollow. Med Mycol 36:119–122. doi: 10.1080/02681219880000191. [DOI] [PubMed] [Google Scholar]
- 73.Engelthaler DM, Kelley E, Driebe EM, Bowers J, Eberhard CF, Trujillo J, Decruyenaere F, Schupp JM, Mossong J, Keim P, Even J. 2013. Rapid and robust phylotyping of spa t003, a dominant MRSA clone in Luxembourg and other European countries. BMC Infect Dis 13:339. doi: 10.1186/1471-2334-13-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol 5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Alikhan N-F, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST Ring image generator (Brig): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Rooted maximum-likelihood phylogeny of 80 K. pneumoniae isolates after recombination filtering by Gubbins (34). Hypervirulent clonal lineages (ST14, CC23, and ST86) are marked with shadings. Download
Scatterplot of SNP number in CAS686 related to position in the NTUH-K2044 chromosome. Of 11,519 SNPs identified, only 254 SNPs were located in the region corresponding to position 1.0 to 3.7 Mbp in the NTUH-K2044 chromosome, revealing a composition of CAS686 as a chimera between a CC23 strain and a strain of unknown clonal origin. Download
Phylogenetic analysis of the non-CC23 segments of the genome of the ST260 strain CAS686 (orange marking). Download
BLASTN atlas illustrating the presence of the pNTUH-K2044 virulence plasmid in hypervirulent (A) and nonhypervirulent (B) K. pneumoniae isolates. Annotated regions indicate genes encoding the virulence factors RmpA, RmpA2, aerobactin (iucABCD-iutA), and salmochelin (iroBCDN) and gene clusters encoding silver (sil) and copper (pco) resistance. Red rings in panel A are the 28 CC23-related isolates in the same order from inside to outside as outlined in Table S1 (CAS683 to CAS686). Blue rings in panel A are, from inside to outside, the hypervirulent K2 isolates CAS688 and CAS689 and the virulence plasmid pLVPK from the K2 strain CG43. Green rings in panel B are the 39 nonhypervirulent isolates in the same order from inside to outside as outlined in Table S1 (A5054 to 3091). The purple ring is pKCTC2242 from strain KCTC 2242 of unknown virulence potential. Download
BLASTN atlas illustrating the presence of the different genomic modules (GM1 to GM8) of KPHPI208 in hypervirulent and nonhypervirulent K. pneumoniae isolates. Red rings are, from inside to outside, the CC23-related isolates CAS683, CAS685, CAS686, CAS687, CAS690, CAS691, CAS694, CAS695, CAS698, CAS699, CAS701, CAS726, CAS727, CAS813, CAS951, CAS983, CAS984, CAS985, CAS986, CAS987, CAS988, CAS989, CAS990, CAS991, and CAS992. The orange ring is NTUH-K2044. Blue rings are, from inside to outside, the hypervirulent K2 isolates CAS688, hvKP1, and Kp52.145. Green rings are, from inside to outside, the nonhypervirulent isolates TG12354, TG21545, TG21982, TG28459, SB2390, HS11286, and Kp13. Download
Characteristics of the 69 isolates sequenced in this study.
Characteristics of publicly available K. pneumoniae genomes included in this study.
Genomic regions unique to or highly associated with CC23.
Summary of genome sequencing and assembly.