Background: Yap1 regulates cadmium accumulation in the vacuole and mitigates cadmium-induced ROS.
Results: Yap1 induces the gene of the hypoxic repressor Rox1 that in turn represses FET4, avoiding cadmium uptake.
Conclusion: Repression of FET4, via Rox1, is a novel line of defense mediated by Yap1 against cadmium toxicity.
Significance: Evidence of cross-talk between oxidative and hypoxic regulators that results in increased tolerance to metal toxicity.
Keywords: gene regulation, iron, metal, stress response, yeast metabolism, Fet4, Rox1, Yap1, cadmium
Abstract
Cadmium is a well known mutagenic metal that can enter cells via nonspecific metal transporters, causing several cellular damages and eventually leading to death. In the yeast Saccharomyces cerevisiae, the transcription factor Yap1 plays a key role in the regulation of several genes involved in metal stress response. We have previously shown that Yap1 represses the expression of FET4, a gene encoding a low affinity iron transporter able to transport metals other than iron. Here, we have studied the relevance of this repression in cell tolerance to cadmium. Our results indicate that genomic deletion of Yap1 increases FET4 transcript and protein levels. In addition, the cadmium toxicity exhibited by this strain is completely reversed by co-deletion of FET4 gene. These data correlate well with the increased intracellular levels of cadmium observed in the mutant yap1. Rox1, a well known aerobic repressor of hypoxic genes, conveys the Yap1-mediated repression of FET4. We further show that, in a scenario where the activity of Yap1 or Rox1 is compromised, cells activate post-transcriptional mechanisms, involving the exoribonuclease Xrn1, to compensate the derepression of FET4. Our data thus reveal a novel protection mechanism against cadmium toxicity mediated by Yap1 that relies on the aerobic repression of FET4 and results in the impairment of cadmium uptake.
Introduction
Cadmium contamination poses a threat to the environment and human health, because cadmium is extremely toxic and carcinogenic even at low concentrations (1). Environmental contamination with this metal arises from both natural and anthropogenic sources. Human exposure to cadmium mainly occurs through contaminated dietary sources or by inhalation of tobacco smoke or polluted air, leading to its accumulation in the liver, kidney, and lungs (2).
The precise molecular mechanism of cadmium toxicity is not fully understood, but it is thought to cause injury primarily via oxidative-induced cellular damages. Cadmium is, however, unable to directly generate free radicals, but it is assumed to induce the formation of reactive oxygen and nitrogen species by two distinct mechanisms. One of those mechanisms involves the displacement of metals from proteins that, once unbound, may generate oxidative stress via Fenton reactions; the other relies on cadmium ability to inhibit the activity of antioxidant enzymes, such as superoxide dismutase and catalase (3, 4). Other forms of cadmium cellular toxicity include the inhibition of the DNA mismatch repair system (5), the induction of iron deficiency (6–8), and the disturbance of the homeostasis of other metals (9–11).
Cadmium enters cells through transporters evolved for the uptake of essential metals, such as iron, zinc, manganese, and calcium (7, 12–16). The budding yeast Saccharomyces cerevisiae has long been used as model organism to study the molecular mechanisms of cadmium toxicity and tolerance. The most relevant mechanism of cadmium detoxification in yeast relies on its accumulation in the vacuole, which is strongly dependent on Ycf1, a vacuolar membrane transporter of the ABC family. Ycf1 transports cadmium conjugated with glutathione into the vacuole (17). Two Ycf1 paralogues, Bpt1 and Vmr1, and the zinc transporter Zrc1 also play a minor role in vacuolar cadmium sequestration (18, 19). Another cadmium detoxification system in yeast relies on Pca1, a plasma membrane P-type ATPase involved in cadmium efflux (20). In addition, two other plasma membrane transporters, Alr1 and Yor1, have been implicated in cadmium detoxification; however, little is known regarding their mode of action (21, 22). Also the yeast internal Ca2+ transporters, Pmr1 and Pmc1 can act as ancillary pathways to cope with cadmium toxicity, particularly when Ycf1 activity is compromised (23, 24).
In yeast, the transcriptional regulator Yap1 plays a central role in cadmium stress response, by activating antioxidant genes and by inducing the expression of YCF1 (25). Yap1 senses cadmium by means of the direct interaction of its C-terminal cysteine rich domain with the drug (26). As a consequence, Yap1 nuclear export signal becomes masked, and the factor accumulates in the nucleus, activating its target genes (27).
We have recently shown that under normal growth conditions Yap1 is a negative regulator of FET4 (28). Fet4 is a cell surface low affinity iron transporter able to transport other divalent metals, including toxic cadmium ions (14). In this context, we put forward the hypothesis that Yap1-mediated negative regulation of FET4 may be a novel line of protection conferred by this regulator against cadmium insult. We show herewith that Yap1 represses FET4 gene expression via Rox1, which results in a reduction of cadmium uptake. We also show that, when challenged with cadmium, yap1 and rox1 mutant strains trigger post-transcriptional mechanisms, dependent on the 5′-3′ exoribonuclease Xrn1 that compensates the derepression of FET4 gene expression.
Experimental Procedures
Yeast Strains, Plasmids, and Growth Conditions
The yeast strains used in this study are listed in Table 1. All mutants constructed in this work were generated using the microhomology PCR method (30).
TABLE 1.
Saccharomyces cerevisiae strains used in this work
| Strain | Description | Source |
|---|---|---|
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 uraΔ0 | Euroscarf |
| BY4742 yap1 | MATα his3Δ1 leu2Δ0 lys2Δ0 uraΔ0 yap1Δ::kanMX4 | Euroscarf |
| BY4742 rox1 | MATα his3Δ1 leu2Δ0 lys2Δ0 uraΔ0 rox1Δ:: kanMX4 | Euroscarf |
| BY4742 fet4 | MATα his3Δ1 leu2Δ0 lys2Δ0 uraΔ0 fet4Δ:: kanMX4 | Euroscarf |
| BY4742 xrn1 | MATα his3Δ1 leu2Δ0 lys2Δ0 uraΔ0 xrn1Δ:: kanMX4 | This study |
| By4742 rnt1 | MATα his3Δ1 leu2Δ0 lys2Δ0 uraΔ0 rnt1Δ:: kanMX4 | This study |
| BY4742 yap1fet4 | MATα his3Δ1 leu2Δ0 lys2Δ0 uraΔ0 fet4Δ:: kanMX4 yap1Δ:: his3Δ1 | Ref. 28 |
| BY4742 yap1xrn1 | MATα his3Δ1 leu2Δ0 lys2Δ0 uraΔ0 xrn1Δ:: kanMX4 yap1Δ:: his3Δ1 | This study |
| BY4742 yap1rnt1 | MATα his3Δ1 leu2Δ0 lys2Δ0 uraΔ0 rnt1Δ:: kanMX4 yap1Δ:: his3Δ1 | This study |
| BY4742 yap1rox1 | MATα his3Δ1 leu2Δ0 lys2Δ0 uraΔ0 yap1Δ:: kanMX4 rox1Δ:: his3Δ1 | This study |
| BY4742 yap1xrn1fet4 | MATα his3Δ1 leu2Δ0 lys2Δ0 uraΔ0 xrn1Δ:: kanMX4 yap1Δ:: his3Δ1 fet4Δ:: ura | This study |
To construct the pROX1-lacZ plasmid, a fragment comprising a 1-kb sequence upstream of ROX1 ATG codon was amplified by PCR using the primers listed in Table 2. PCR product was first digested with BamHI, treated with Klenow, and next digested with SphI. The resulting fragment was cloned into the YEp356R vector previously digested with EcoRI, treated with Klenow, and cut with SphI.
TABLE 2.
Oligonucleotides
| Primer | Sequence | Use |
|---|---|---|
| ACT1 | 5′-CTATTGGTAACGAAAGATTCAG-3′ | qRT-PCR |
| 5′-CCTTACGGACATCGACATCA-3′ | ||
| CTH2 | 5′-AGGTATGCTGCTGGAGCTGT-3′ | qRT-PCR |
| 5′-GAGGGCCATGAAGGTATCAA-3′ | ||
| FET4 | 5′-GGAGAACTGCCTGTGGAAAA-3′ | qRT-PCR |
| 5′-TTCTCCGGTGTAAGGTGGAG-3′ | ||
| FET3 | 5′-ACGGTGTGAATTACGCCTTC-3′ | qRT-PCR |
| 5′-TTGGAAAGCGTGACCATGTA-3′ | ||
| ROX1 | 5′-AGGGCTTACAACCGGAAGAT-3′ | qRT-PCR |
| 5′-GCTGTTGCTCGATTTCCTTC-3′ | ||
| CUP1 | 5′-TGAAGGTCATGAGTGCCAAT-3′ | qRT-PCR |
| 5′-GCATTTGTCGTCGCTGTTAC-3′ | ||
| ARN2 | 5′-AGGTATGCTGCTGGAGCTGT-3′ | qRT-PCR |
| 5′-GAGGGCCATGAAGGTATCAA-3′ | ||
| ACT1-ChIP | 5′-GATCCTTTCCTTCCCAATCTCTCTTG-3′ | ChIP |
| 5′-GCTCATGTAGTAGAAGATCCTATT-3′ | ||
| ARN2 | 5′-GGTATGCTGCTGGAGCTGT-3′ | ChIP |
| 5′-AGGGCCATGAAGGTATCAA-3′ | ||
| ROX1-414bp | 5′-GCAAAACAATTGGAAATCTGG-3′ | ChIP |
| 5′-GAACAACAAAAGAGGCAGCA-3′ | ||
| ROX1-897bp | 5′-TCTACATAATGCACGAAACTTGG-3′ | ChIP |
| 5′-CGCAGTGTGTGTTCCTGTCT-3′ | ||
| ROX1p'1000 | 5′-CTAGCATGCAGTTGACCTACATTCAAC-3′ | pROX1 |
| A4-ROX1 | 5′-GGATTTCGCATCCTAGACCA-3′ | |
| ROX1m Fw | 5′-TGGCGATTGAAGACAAAGAAGAAA-3′ | MUT-pROX1 |
| ROX1m Rv | 5′-TTTCTTCTTTGTCTTCAATCGCCA-3′ | |
| ROX1-ATG-codon | 5′-CTAGCATGCAGTTGACCTACATTCAAC-3′ | pROX1-lacZ |
| 5′-CTTGGATCCGGATTCATTGTTGATTGTC-3′ | ||
| FET4-ORF | 5′-GAATTCCTGCAGCCCCTGTGCTTGCTGTTC-3′ | pFET4-HA |
| 5′-ATGTACCCATACGATGTTCCAGATTACGCTTAGCTTCATTGAACA-3′ | ||
| 0.5Kb-FET4-terminator | 5′-AGCGTAATCTGGAACATCGTATGGGTACATTTTTTCCAACATCAT-3′ | pFET4-HA |
| 5′-ACTAGTGGATCCCCCGACATATAAGCGGAG-3′ | ||
| FET4m1 Fw | 5′-GCCTTCTTAATTGAGTTTAGCATC-3′ | MUT-pFET4-HA (Aft1 site 1 deletion) |
| FET4m1 Rv | 5′-GATGCTAAACTCAATTAAGAAGGC-3′ | |
| FET4m2 Fw | 5′-GTTCCGAAAACCCACTTTTTGTTC-3 | MUT-pFET4-HA (Aft1 site 2 deletion) |
| FET4m2 Rv | 5′-GAACAAAAAGTGGGTTTTCGGAAC-3′ |
To generate the C-terminal HA-tagged version of FET4 (FET4-HA), one fragment comprising 1 kb upstream from the ATG plus the FET4 coding region and another including 0.5 kb downstream from the stop codon were amplified by PCR using the primers listed in Table 2. The HA sequence was inserted in frame with FET4 coding region just before the TAG stop codon. Both fragments were inserted into the pRS416 vector, previously linearized with SmaI, by homologous recombination using the In-Fusion Advantage PCR cloning kit (Clontech).
To construct the p-ROX1 plasmid, ROX1 gene was amplified by PCR with specific primers (Table 2). The resulting fragment was inserted into the SmaI site of pRS416. The MUTp-ROX1 plasmid was generated using as template p-ROX1, and the primers depicted in Table 2 were used in a PCR-directed mutagenesis reaction to mutate the YRE2 site located at −414 bp, as detailed in Ref. 31. A similar and sequential strategy was used to construct MUTp-FET4-HA using the primers listed in Table 2.
Yeast strains were grown in synthetic medium (SC) or medium lacking specific requirements (SD), as previously described (31). Phenotypic growth assays were carried out by spotting 5 μl of cultures in early exponential phase (A600 = 0.4–0.5) sequentially diluted (∼5 × 103 to 50 cells) in medium containing the indicated concentrations of CdCl2 and supplemented or not with FeSO4. These assays were repeated at least twice. Cultures were grown for 2 days at 30 °C. The bacteria Escherichia coli strain XL1-Blue (Stratagene) was used as a host for routine cloning purposes. Standard methods were used for genetic analysis, cloning, and transformation.
Measurements of β-Galactosidase Activity
The BY4742 (wild-type) and the yap1 mutant strains were transformed with the pROX1-lacZ plasmid. Cells were grown in liquid SD medium until the early exponential phase in the presence or absence of 25 μm CdCl2 and harvested after 15 min. Relative β-galactosidase activity was monitored as described in Ref. 31. Enzymatic activity was assayed by following the degradation of the colorimetric substrate O-nitrophenyl-β-d-galactopyranoside at A420 and normalizing against A600. The results are the average of at least six biological replicates.
Immunoblot Assays
Wild-type, yap1 and rox1 mutant strains containing the FET4-HA or MUTp-FET4-HA plasmids were grown until the early exponential phase (A600 = 0.4–0.5) and exposed to 25 μm CdCl2. Cells were harvested at different time points after treatment with CdCl2. Total proteins were extracted from cell cultures as described in Ref. 31. Proteins were resolved in a 10% SDS-PAGE and immunoblotted with horseradish peroxidase-bound anti-HA IgG (Roche). In what concerns PMSF treatment, cells were grown until the early exponential phase, exposed to 1.2 mm of PMSF for 90 min, and supplemented (or not) with CdCl2 for 1 h. Pgk1 was used as loading control. Immunoblots were repeated at least twice with different protein extracts.
Quantitative Real Time RT-PCR Analyses
Cells were grown until the early exponential phase, cultures were left untreated or treated with 25 μm CdCl2. Cells were harvested at the indicated time points, and RNA was isolated. RNA samples were next treated with DNase (TURBOTM DNase-free; Ambion) according to the manufacturer's instructions and purified. Total RNA (1 μg) was reverse transcribed with transcriptor reverse transcriptase (Roche Diagnostics). Quantitative PCRs were performed in the Light Cycler 480 II real time PCR system (Roche), using Light Cycler 480 SYBR Green I Master (Roche). Relative standard curves were constructed for each gene, using triplicate serial dilutions of cDNA. The relative expression of the genes was calculated by the relative quantification method with efficiency correction, using the Light Cycler 480 software 1.5 (32). Actin gene was used as a reference gene. All assays were made using biological and technical triplicates. Primers used in this assay are listed in Table 2.
ChIP Analysis
ChIP assays were carried out as previously described (31). Cells transformed with a c-Myc-tagged version of Yap1 (27) and treated with cadmium were harvested at A600 0.6 and fixed with 1% formaldehyde. The cross-linking was stopped by addition of glycine, and cells were disrupted with a FastPrep®-24 instrument (MP Biomedical). Cell extracts were sonicated to yield DNA fragments with an average of 500 bp. c-Myc-tagged Yap1 was immunoprecipitated by incubating the cross-linked chromatin with a c-Myc antibody prebound to Dynabeads Pan mouse IgG (Invitrogen) for 16 h at 4 °C. Immunoprecipitated proteins were eluted from the beads by heating the samples in appropriate buffer, and fixation was reversed. Aliquots of total chromatin input (IN) and immunoprecipitated chromatin were simultaneously processed for subsequent normalization. After sample treatment with proteinase K and RNase A, DNA was purified. Quantification of specific DNA targets in the input and immunoprecipitated samples was performed by quantitative PCR. A standard curve, generated with a dilution series of the immunoprecipitated sample, was used to assess the PCR efficiency. The relative enrichment of a specific pRox-YRE in the immunoprecipitate was determined using the ΔΔCT method through the calculation of log2 (immunoprecipitated/input). The primer sequences used are listed in Table 2. The primers ROX1-414bp and ROX1-897bp were used to amplify the regions of the ROX1 promoter flanking both YRE sites located at −414 and −897 bp from the ATG, respectively. ARN2 was used as a negative control.
Measurement of Cadmium and Iron
Strains were grown to early exponential phase (A600 0.4–0.5) in SC medium and left untreated or treated with 25 μm CdCl2. Cells were harvested after 6 h of stress induction, collected by centrifugation, and washed with 10 mm EDTA and metal-free water. This time point was chosen because, in all strains, the maximum of growth inhibition was observed after 6 h of treatment with cadmium (data not shown). The total cadmium and iron content was measured by inductively coupled plasma atomic emission spectroscopy (ICP-AES). The data were normalized against A600. All assays were made using biological triplicates.
Results
Yap1 Regulation of FET4 Contributes to S. cerevisiae Cadmium Tolerance
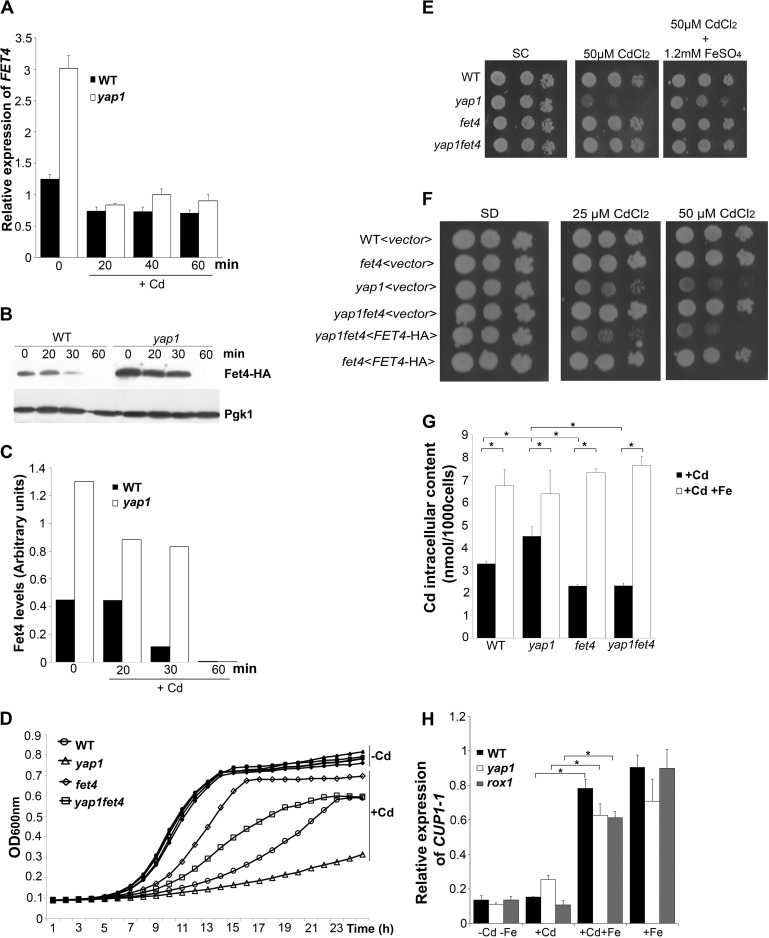
Our previous work indicated that Yap1 is a negative regulator of FET4 (28), a gene encoding a low affinity iron transporter also involved in the transport of zinc, cobalt, manganese, and cadmium (14, 33). To evaluate whether the uptake of cadmium via FET4 could contribute to the well known sensitivity exhibited by the yap1 mutant to this metal, we first tested FET4 dependence on Yap1 by qRT-PCR (Fig. 1A). As illustrated in Fig. 1A, FET4 expression is induced in yap1 unstressed cells. A sharp decrease of FET4 levels was, however, observed after cadmium challenge. Indeed, after 20 min of cadmium exposure, the levels of FET4 transcripts in the yap1 mutant and WT strains were comparable. We have also assessed the protein expression of a HA-tagged version of Fet4 driven by its native promoter. In agreement with the gene expression data, we observed that protein levels were consistently higher in the yap1 mutant compared with the WT strain, being rapidly reduced after cadmium exposure (Fig. 1, B and C).
FIGURE 1.
Yap1 mediates FET4 repression. A, WT and yap1 strains were challenged with 25 μm CdCl2 and harvested at the indicated time points. The expression of FET4 was monitored by qRT-PCR. B, the WT strain and the yap1 mutant were transformed with a plasmid containing FET4-HA, cultures were treated with 25 μm CdCl2, and Fet4 expression was analyzed by Western blot with an anti-HA antibody. C, Fet4 protein levels were normalized to Pgk1, from B. D, growth exhibited by the WT, yap1, fet4, and yap1fet4 strains in SC medium was recorded over a period of 24 h. The open and closed symbols represent strains grown in media containing or not 25 μm CdCl2, respectively. E, growth sensitivity exhibited by the WT, yap1, fet4, and yap1fet4 strains in SC plates containing 50 μm CdCl2 and supplemented with 1.2 mm FeSO4. F, exponentially growing WT, yap1, fet4, and yap1fet4 cells were transformed with the FET4-HA plasmid (<FET4-HA>) or with the empty vector (<vector>), harvested, serially diluted, and spotted onto SD plates or SD plates supplemented with 25 or 50 μm CdCl2. G, cadmium content of WT, yap1, fet4, and yap1fet4 strains were determined by ICP-AES, after treatment with 25 μm CdCl2 (+Cd) or after treatment with 25 μm CdCl2 and 1.2 mm of FeSO4 (+Cd +Fe) for 6 h. H, WT, yap1 and rox1 strains were grown in medium left untreated (−Cd−Fe) or treated with 25 μm CdCl2 (+Cd); with 25 μm CdCl2 and 1.2 mm of FeSO4 (+Cd +Fe) or 1.2 mm of FeSO4 (+Fe). CUP1 expression levels were assessed by qRT-PCR after 15 min of treatment. In this figure, all values are the means of at least biological triplicates (n = 3) ± S.D. Significance of differences was calculated with the t test. *, p < 0.05.
We next examined the growth phenotype of the double mutant yap1fet4 in the presence of cadmium (Fig. 1, D and E). If repression of FET4 by Yap1 is required to prevent cadmium toxicity, one would expect the double mutant to be more resistant than the single yap1 mutant to this metal. As anticipated, the double mutant yap1fet4 grew better than the yap1 strain in the presence of cadmium (Fig. 1, D and E). In addition, reintroduction of FET4 into the yap1fet4 mutant restores cadmium sensitivity (Fig. 1F). Accordingly, intracellular cadmium levels are increased ∼30% in the yap1 mutant when compared with the WT, whereas in the fet4 and yap1fet4 mutants, these values are 50–60% lower than in the yap1 mutant (Fig. 1G). Moreover, supplementation of the growth medium with iron attenuates the yap1 mutant sensitivity, suggesting the involvement of an iron transporter in this process (Fig. 1E). To test this, we also measured cadmium contents when cells were simultaneously treated with iron (Fig. 1G). Contrary to our expectations, we noticed that in the presence of iron, cadmium uptake increases. In an attempt to understand this apparently contradictory finding, we revisited our data on the genome-wide transcriptional analysis of S. cerevisiae exposed to high iron conditions (31) and searched for genes whose expression was induced by high iron and that may play a role in cadmium detoxification. We found that metallothionein genes CUP1-1 and CUP1-2 were up-regulated in response to high iron (Table S1 in Ref. 31). Cup1-1 overexpression confers resistance to cadmium ions (34), and by qRT-PCR we found that CUP1 genes are highly expressed when cells are treated simultaneously with cadmium and iron (Fig. 1H). This finding strongly suggests that iron attenuates cadmium toxicity by inducing CUP1 expression, which possibly binds and sequesters cadmium. Altogether, these data shed light on a novel Yap1-mediated mechanism of cadmium stress tolerance involving the Fet4 low affinity iron transporter.
Yap1 Is a Direct Regulator of the Repressor ROX1
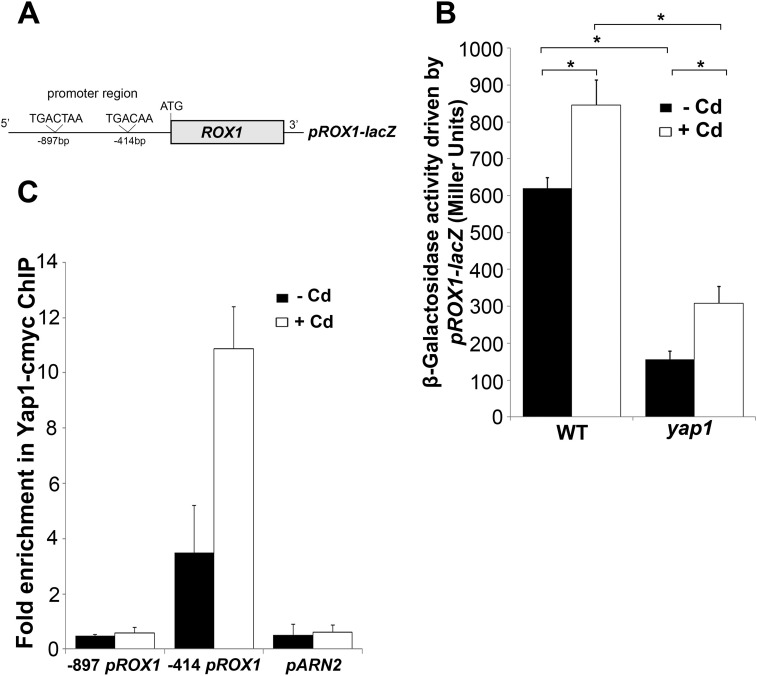
The promoter region of FET4 does not contain any canonical YREs (35), a fact that strongly suggests that the regulation of FET4 by Yap1 involves another regulator. Strikingly, our previous microarray data indicate that Yap1 is a positive regulator of ROX1 gene (28). Rox1 is a repressor of anaerobic genes and also represses FET4 expression under oxygenated conditions (29, 36). Remarkably, microarray data equally revealed that all the genes up-regulated in the yap1 mutant are also known Rox1 targets (see Ref. (28) and Table 3). Furthermore, using YEASTRACT (37), we verified that the promoter region of ROX1 possesses two YRE sites, located at −414 and −897 bp upstream the ATG codon (Fig. 2A).
TABLE 3.
Rox1 target genes that appear upregulated in yap1 mutant cells
This information is adapted from Ref. 28.
| Systematic name | Gene name | Fold change | Description |
|---|---|---|---|
| YLR034C | SMF3 | 1.3 | Member of the Nramp family of the metal transport proteins |
| YMR319c | FET4 | 4.5 | Low affinity Fe(II) transporter of the plasma membrane |
| YER014w | HEM14 | 1.5 | Protoporphyrinogen oxidase |
| YKL008c | LAC1 | 1.6 | Ceramide synthase component |
| YHR007c | ERG11 | 1.7 | Lanosterol 14-α-demethylase |
| YDR297w | SUR2 | 1.7 | Sphinganine C4-hydroxylase |
| YDR004w | HEM13 | 2.2 | Coproporphyrinogen III oxidase |
| YIL11w | COX5B | 1.7 | Subunit Vb of cytochrome c oxidase |
| YBR085w | AAC3 | 2.2 | Mitochondrial inner membrane ADP/ATP translocator |
| YEL047c | NA | 1.6 | Soluble fumarate reductase |
| YDR518w | EUG1 | 1.4 | Disulfide isomerase of the endoplasmatic reticulum lumen |
| YHR179w | OYE2 | 1.4 | Conserved NADPH oxidoreductase |
| YAL028w | FRT2 | 1.5 | Tail-anchored endoplasmic reticulum membrane protein |
| YNR014w | NA | 1.4 | Putative protein of unknown function |
FIGURE 2.
Yap1 is a direct regulator of ROX1. A, schematic representation of the ROX1 promoter region containing the two YREs sites located at −897 and −414 bp upstream the ATG. B, WT and yap1 mutant strains were transformed with the pROX1-lacZ plasmid. Cells were grown exponentially in SD medium and challenged with 25 μm CdCl2 for 15 min. β-Galactosidase activity was assayed as detailed under “Experimental Procedures.” The values are the means of biological decaplicates ± S.D. Significance of differences was calculated with the t test. *, p < 0.05. C, yap1 mutant cells transformed with a plasmid containing a c-Myc-tagged version of Yap1 were grown exponentially in SD medium and treated with 25 μm CdCl2 for 10 min. ChIP analyses combined with quantitative PCR were used to determine the fold enrichment of each YRE. The promoter region of ARN2 was used as a negative control, because it does not possess YREs. The values are the means of at least biological triplicates ± S.D.
In line with these data, we bring forward the hypothesis that Yap1 might repress FET4 via ROX1 induction. As a first approach to test this, a plasmid including the promoter region of ROX1 fused to the lacZ reporter gene (pROX1-lacZ) was generated and used to transform the WT and yap1 mutant strains. We observed that β-galactosidase activity was higher in the WT compared with the mutant strain, even in the absence of cadmium (Fig. 2B). A slight but significant increase of the β-galactosidase activity was noticed in both strains upon cadmium stress. These results were further confirmed by the analysis of the levels of ROX1 transcripts in both strains, in either the absence or the presence of cadmium, by qRT-PCR (Fig. 3B).
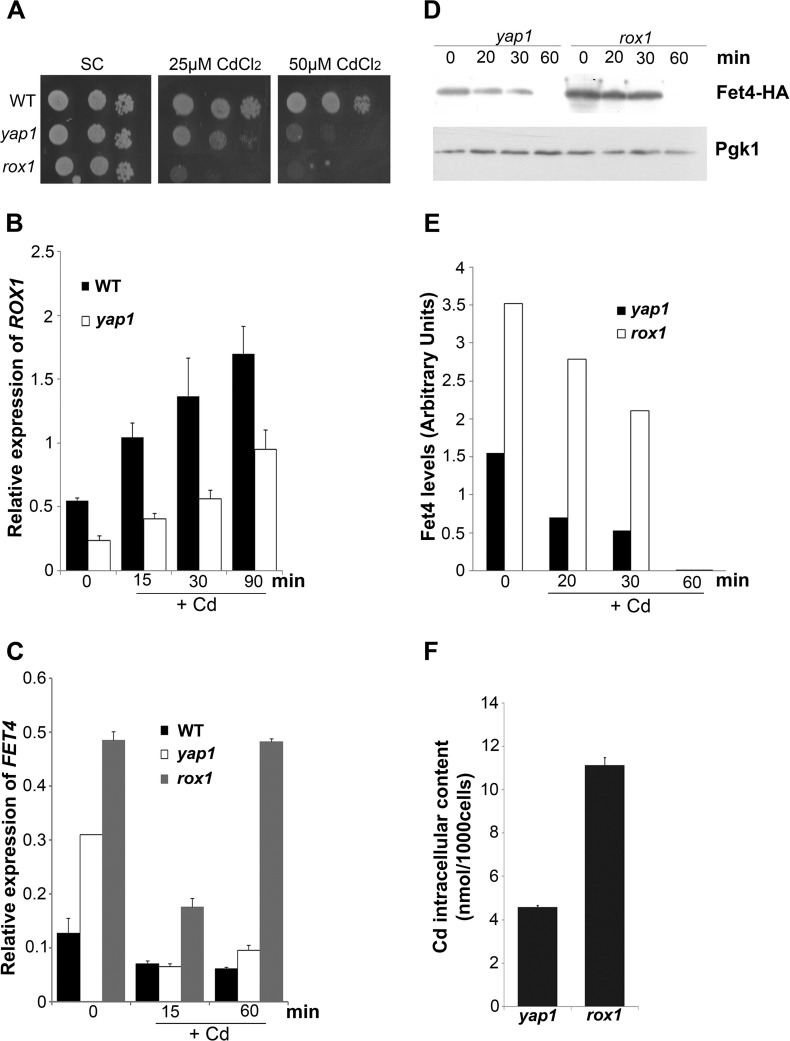
FIGURE 3.
Yap1 partially regulates ROX1. A, growth sensitivity exhibited by WT, yap1, and rox1 strains in SC plates containing 25 or 50 μm CdCl2. B, ROX1 expression in the WT and yap1 mutant was analyzed by qRT-PCR at the indicated time points, after treatment with 25 μm CdCl2. C, FET4 expression in the WT and yap1 and rox1 mutants was analyzed by qRT-PCR, at the indicated time points after treatment with 25 μm CdCl2. D, yap1 and rox1 mutants were transformed with FET4-HA plasmid and treated with 25 μm CdCl2. Fet4 protein levels were analyzed by Western blot, at the indicated time points. E, Fet4 protein levels were normalized to Pgk1, from D. F, cadmium intracellular content in yap1 and rox1 mutant strains was determined by ICP-AES, after treatment with 25 μm CdCl2 for 6 h. The values are the means of at least biological triplicates ± S.D.
We next examined whether Yap1 is a direct regulator of ROX1 by carrying out ChIP analyses. A Yap1 c-Myc-tagged version was used to test whether each of the YRE sites found in the ROX1 promoter region (Fig. 2A) was recognized by this factor. As depicted in Fig. 2C, after immunoprecipitation of the chromatin bound to Yap1 c-Myc, an enrichment of ROX1 promoter harboring the YRE located at −414 bp was observed. No enrichment was noticed in the ROX1 region comprising the YRE located at −897 bp or in the promoter region of the ARN2 gene (used as a negative control). The increased enrichment in the −414-bp YRE-containing sequence after cadmium treatment correlates well with Yap1 nuclear accumulation kinetics (26). Although the above data clearly indicate that Yap1 is a direct regulator of ROX1, we observed that rox1 strain is more sensitive to cadmium than yap1 (Fig. 3A). This finding suggests that Yap1 cannot be the only regulator of ROX1. In agreement, ROX1 transcripts are not fully dependent on Yap1 (Fig. 3B), and FET4 mRNA (Fig. 3C), Fet4 protein levels (Fig. 3, D and E), and cadmium intracellular contents (Fig. 3F) are higher in the rox1 mutant than in the yap1 strain. The results here described clearly show that, although partially, Yap1 directly regulates ROX1 expression.
Rox1 Mediates Yap1 Repression of FET4
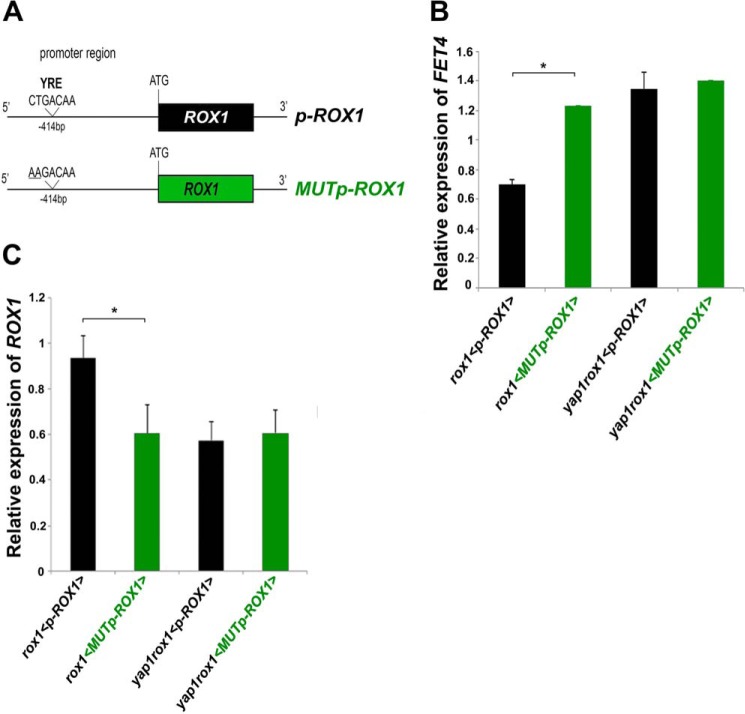
To confirm whether Yap1 regulates FET4 expression via Rox1, we cloned ROX1 gene and mutated the functional YRE located at −414 bp (Figs. 2C and 4A). The resulting constructs (p-ROX1 and MUTp-ROX1) were next used to transform the double mutant strain yap1rox1 and the single mutant strain rox1. FET4 expression was thereafter assayed by qRT-PCR (Fig. 4B).
FIGURE 4.
Rox1 mediates Yap1 repression of FET4. A, schematic representation of the constructs p-ROX1 and MUTp-ROX1. MUTp-ROX1 possesses two mutations in the functional YRE (CTGACAA to AAGACAA). B and C, FET4 (B) and ROX1 (C) expression in rox1 and yap1rox1 mutant strains transformed with p-ROX1 (black; <p-ROX1>) or MUTp-ROX1 (green; < MUTp-ROX1>) was analyzed by qRT-PCR. The values are the means of at least biological triplicates ± S.D. Significance of differences was calculated with the t test. *, p < 0.05.
Supporting our hypothesis, we noticed that the expression of FET4 was higher in rox1 cells transformed with MUTp-ROX1 than with p-ROX1 (Fig. 4B). The levels of FET4 transcripts in the former were close to those exhibited by the double mutant yap1rox1 transformed with p-ROX1. As expected, ROX1 levels were decreased in rox1 strain transformed with MUTp-ROX1 and in the double mutant transformed with p-ROX1 (Fig. 4C). These data support that Yap1 mediates the repression of FET4 via Rox1.
The Exoribonuclease 5′-3′ Xrn1 Alleviates the FET4 Derepression Observed in the yap1 Mutant
Xrn1 is an exoribonuclease responsible for the degradation of mRNAs from the 5′ to the 3′ end. This protein is conserved in all eukaryotes and is involved in the normal mRNA decay (38). We have recently shown that arsenate stress triggers Xrn1-mediated degradation of FET3 transcripts (39).
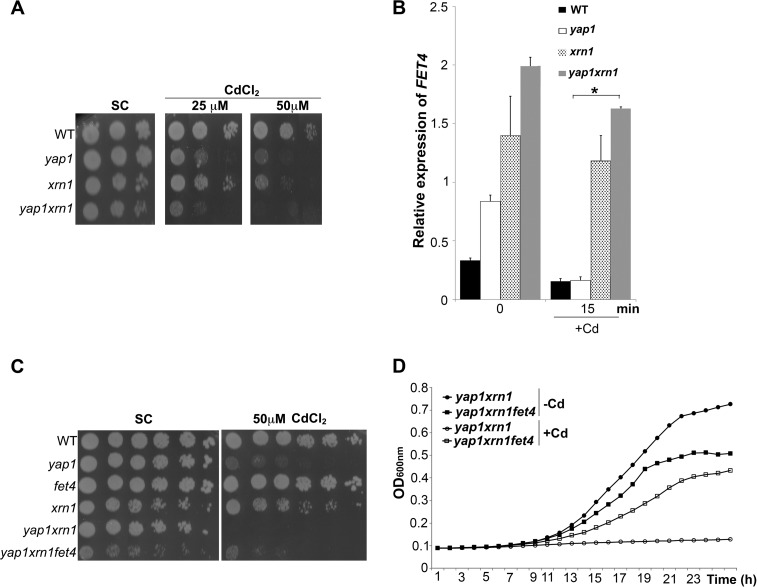
In an attempt to determine whether Xrn1 is also involved in the decrease of FET4 mRNAs levels observed in the yap1 mutant after cadmium treatment (see Figs. 3C and 1A), we first constructed the double mutant yap1xrn1 and assayed its cadmium sensitivity. We observed that the double mutant is more sensitive to cadmium than the yap1 or xrn1 single mutants (Fig. 5A). We next monitored FET4 mRNA levels in these strains and observed that in both xrn1 and yap1xrn1 mutants the accentuated drop of FET4 levels after stress was no longer evident (Fig. 5B). To assess whether the higher sensitivity of the yap1xrn1 mutant to cadmium was due to higher levels of FET4 expression, we constructed the triple mutant yap1xrn1fet4 and observed that it is more tolerant to cadmium compared with the double mutant (Fig. 5, C and D). Overall, these data strongly suggests that a post-transcriptional mechanism involving Xrn1 may counteract the derepression of FET4 observed in the yap1 mutant after cadmium insult.
FIGURE 5.
The exoribonuclease Xrn1 controls FET4 transcript levels and has a role in cadmium tolerance. A, growth sensitivity exhibited by WT, yap1, xrn1, and yap1xrn1 strains in SC plates containing 25 and 50 μm CdCl2. B, FET4 expression in WT, yap1, xrn1, and yap1xrn1 strains was assessed by qRT-PCR at the indicated time points, after treatment with 25 μm CdCl2. The values are the means of at least biological triplicates ± S.D. *, p < 0.05. C, deletion of FET4 from the yap1xrn1 background partially renders cells more tolerant to cadmium. The indicated strains were spotted onto SC plates supplemented or not with 50 μm CdCl2. D, this effect is also observed when cultures are grown in liquid SC medium with a lower cadmium concentration (25 μm CdCl2).
Degradation of FET4 by Xrn1 Appears to Be Stress-specific
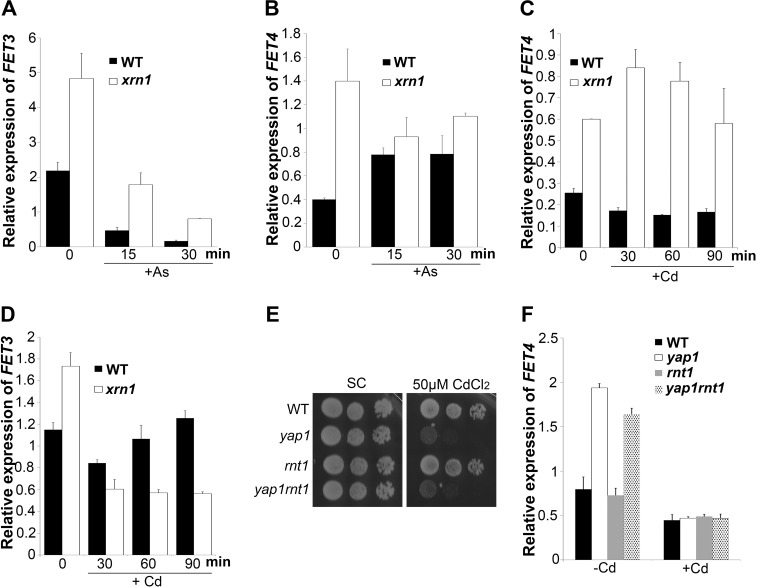
To understand whether FET4 degradation mediated by Xrn1 was stress-specific, we examined and compared the levels of FET4 and FET3 in the mutant xrn1 after treatment with cadmium or arsenate. In the absence of stress, FET3 and FET4 mRNA levels are dependent on Xrn1 (Fig. 6, A and B, time point 0). As we have previously described (39) and herein confirmed by qRT-PCR, the drop of FET3 mRNA levels in response to arsenate is dependent on Xrn1 (Fig. 6A). Contrary to FET3, FET4 mRNA levels are no longer dependent on Xrn1 after arsenate treatment (Fig. 6B). In response to cadmium, however, FET4 mRNA levels continue to be strongly dependent on Xrn1 (Fig. 6C), whereas FET3 transcripts become insensitive to this exoribonuclease (Fig. 6D).
FIGURE 6.
FET4 transcripts dependence on Xrn1 is stress-specific. A–D, FET3 (A) and FET4 (B) expression in WT and xrn1 strains were assessed by qRT-PCR at the indicated time points, after treatment with 1 mm AsV (+As) or and after treatment (C and D) with 25 μm CdCl2 (+Cd). The values are the means of at least biological triplicates ± S.D. E, growth sensitivity exhibited by WT, yap1, rnt1, and yap1rnt1 strains spotted on SC plates supplemented or not with 50 μm CdCl2. F, FET4 expression levels in WT, yap1, rnt1, and yap1rnt1 strains were examined by qRT-PCR after treatment with 25 μm CdCl2 (+Cd) for 15 min. The values are the means of at least biological triplicates ± S.D.
Some of the mRNAs degraded by Xrn1 are first cleaved by Rnt1, a double-stranded RNA endonuclease (40). Rnt1 specifically recognizes particular RNA hairpins and therefore recognizes its targets (40). Although FET4 transcripts do not contain such hairpin structures and are not affected by Rnt1 under normal growth conditions (41), we could not certainly rule out the possibility that a different scenario occurs in response to cadmium. Indeed, if Rnt1 specifically recognizes FET4 transcripts in the presence of cadmium, one would expect FET4 mRNA levels to be highly dependent on Xrn1. To test this, we constructed the single and the double mutants rnt1 and yap1rnt1 and examined their growth phenotype in the presence of cadmium, as well as FET4 gene expression by qRT-PCR. We observed that cells lacking Rnt1 are not sensitive to cadmium toxicity and that deletion of RNT1 gene from the yap1 background does not aggravate the sensitive growth phenotype exhibited by the yap1 mutant to cadmium (Fig. 6E). In addition, our results indicate that the absence of Rnt1 does not affect FET4 mRNA levels either under normal (as previously shown in Ref. 41) or under cadmium stress conditions (Fig. 6F). Overall, our results show that the mechanism by which Xrn1 controls FET4 transcripts is stress-dependent and does not rely on the endoribonuclease Rnt1.
Iron Homeostasis Is Perturbed by Cadmium Treatment in the Mutant Strains yap1 and rox1
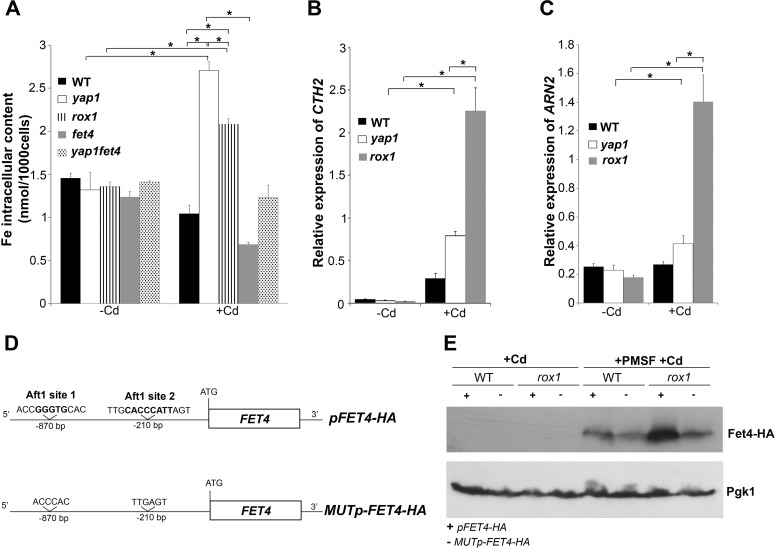
Remarkably, we found that FET4 transcript levels in the rox1 mutant tend to increase after prolonged cadmium treatment (Fig. 3C). The fact that cadmium induces iron deficiency (36), together with the knowledge that the iron sensing transcription factor, Aft1, controls FET4 expression in response to depletion of cellular iron levels (42), led us to examine whether iron homeostasis could be over affected in rox1 strain after extended cadmium treatment. To this end, we have first evaluated iron contents in WT, yap1, rox1, fet4, and yap1fet4 strains, after and before cadmium stress (Fig. 7A). In the absence of treatment, iron levels did not differ among the strains. In the presence of cadmium, however, iron levels were significantly increased in the yap1 and rox1 mutants, as compared with the WT and control conditions (no cadmium). Among the tested strains, yap1 was the one that exhibited the highest iron content after cadmium addition (Fig. 7A). Because deletion of FET4 from the yap1 background restored iron contents to control levels, we conclude that the derepression of FET4 observed in this strain (Fig. 1, A–C) was mediating iron increase.
FIGURE 7.
Iron homeostasis is perturbed in the yap1 and rox1 mutants. A, iron intracellular contents in WT, yap1, rox1, fet4, and yap1fet4 strains were determined by ICP-AES in unstressed cultures (−Cd) and after culture treatment with 25 μm CdCl2 (+Cd) for 6 h. B and C, CTH2 (B) and ARN2 (C) expression in WT, yap1, and rox1 strains was analyzed by qRT-PCR after treatment with 25 μm CdCl2 for 60 min (+Cd). The values are the means of at least biological triplicates ± S.D. Significance of differences was calculated with the t test. *, p < 0.05. D, schematic representation of the constructs pFET4-HA and MUTp-FET4-HA. In the MUTp-FET4-HA construct, both Aft1 sites (1 and 2) were deleted. E, WT and rox1 mutants transformed with pFET4-HA or MUTp-FET4-HA constructs were treated with Cd for 1 h (+Cd) or treated with 1.2 mm of PMSF for 90 min and then induced with 25 μm CdCl2 for 1 h (+PMSF+Cd). Fet4 protein levels were analyzed by Western blot. Pgk1 was used as a loading control.
We next monitored Aft1 activity in WT, yap1 and rox1 strains by evaluating the expression of CTH2 and ARN2, two target genes of Aft1 (43, 44). After prolonged cadmium exposure (60 min), the expression of these genes was increased in all the strains, but in a more pronounced way in the rox1 mutant (Fig. 7, B and C), suggesting that Aft1 was more active in this strain. If this was the case, then the increase of FET4 levels observed in rox1 cells after prolonged cadmium treatment (Fig. 3C) could be ascribed to Aft1 activity. To test this hypothesis, we have deleted the two Aft1 consensus sites (36) from the promoter region of the construct pFET4-HA. The resulting plasmid (MUTp-FET4-HA; Fig. 7D) was used to transform WT and rox1 cells, and Fet4 protein levels were assessed after extended cadmium treatment. Because Fet4-HA protein levels were rapidly reduced after cadmium exposure (Fig. 3, D and E), we performed these experiments in the presence of PMSF to block the activity of vacuolar proteases (and as such, the vacuolar degradation pathway) and in the presence of MG132, a drug that blocks the proteolytic activity of the proteasome complex. Our results indicate that Fet4 degradation induced by cadmium is not dependent on the proteasome (data not shown) but rather on the vacuolar degradation pathway (Fig. 7E). Moreover our data clearly show that, in the presence of both PMSF and cadmium, deletion of Aft1 consensus sites from FET4 gene compromises Fet4 protein levels in the rox1 but not in the WT strain, indicating that in the rox1 strain FET4 gene is up-regulated by Aft1 when cells are exposed to prolonged cadmium treatment.
Discussion
In S. cerevisiae, eight stress-responsive transcription factors, Yap1 to Yap8, orchestrate the regulation of gene expression in response to a plethora of environmental cues (reviewed in Ref. 45). Yap1, the master regulator of oxidative stress, plays a pivotal role in cell tolerance against metal toxicity, mainly by inducing genes coding for proteins involved in (i) vacuolar metal sequestration, (ii) metal reduction and extrusion, or (iii) detoxification of reactive oxygen species generated by metal-catalyzed Fenton chemistry.
In this work, we have identified a new line of action of Yap1 toward cadmium toxicity. We showed that the negative regulation of the low affinity iron transporter gene, FET4, mediated by Yap1 (Fig. 1), is important for yeast resistance to cadmium. Indeed, the yap1 mutant accumulates higher cadmium levels compared with the WT strain, whereas the deletion of FET4 gene from the yap1 background restores cadmium tolerance (Fig. 1). Other authors have also reported an increase of cadmium levels in yap1 cells, but didn't clarify the underlying mechanism (15).
Another set of data clearly indicates that repression of FET4 by Yap1 is exerted via Rox1, an aerobic repressor of hypoxic genes, previously implicated in cadmium toxicity through a mechanism involving the repression of FET4 (36). We have in fact shown that Yap1 directly regulates ROX1 expression through the recognition of an YRE located 414 bp upstream of its ATG codon (Fig. 2) and that this consensus is relevant for Rox1-mediated FET4 repression (Fig. 4). The mutant yap1, however, is more tolerant to cadmium than the rox1 strain (Fig. 3). Accordingly, ROX1 expression is not fully dependent on Yap1 and FET4 mRNA, and protein levels are therefore consistently higher in the rox1 mutant than in the yap1 strain (Fig. 3). This observation is in agreement with the fact that Rox1 is also regulated by Hap1 (46), a heme-dependent transcription factor (47). Moreover, ROX1 transcript levels and the expression of Rox1 target genes were reported to be only moderately decreased in the hap1 mutant, suggesting the presence of another regulator (46, 48).
The observed drastic decrease of FET4 transcripts after cadmium addition to the medium, suggests that yap1 and rox1 mutant cells tend to counteract FET4 derepression (Fig. 3C). We further showed that the 5′-3′ exoribonuclease Xrn1 is mediating this reduction (Fig. 5). Although in vitro Xrn1 shows little specificity to particular mRNAs, in vivo this is not the case. Jones et al. (49) have proposed that binding of a specific RNA sequence by ncRNAs and/or RNA-binding proteins may recruit the 5′-3′ degradation complex. As such, it is tempting to speculate that cadmium can promote the binding of such an element to FET4 transcripts leading to their specific degradation. Moreover, it now seems likely that a translational/post-translational regulation of Fet4 activity occurs in response to cadmium, because in the WT strain, the protein levels decrease after cadmium insult (Fig. 1B), whereas the mRNA levels do not vary (Fig. 1A), and the blocking of the vacuolar degradation pathway restores protein levels (Fig. 7E).
The intriguing finding that FET4 mRNA levels in the rox1 mutant tend to increase over time after the initial cadmium-induced depression (Fig. 3C) may result from the combinatorial control of this gene by several transcription factors (36, 50). Aft1, the major regulator of the iron depletion response, also controls FET4 expression (42). Cadmium stress induces iron starvation (36), implying that differences in the cadmium status of both mutants (rox1 and yap1) may differently activate Aft1. After cadmium treatment, the yap1 strain accumulates lower cadmium levels compared with the rox1 strain (Fig. 3F), and accordingly, rox1 cells appear to be more iron-starved because their iron content is lower (Fig. 7A), and Aft1 is more active (Fig. 7, B, C, and E).
The fact that ROX1 regulation mediated by Yap1 occurs under normal growth conditions (Fig. 2) raises the question whether this regulation could serve a broader purpose, in addition to hindering cellular cadmium uptake via the repression of FET4. In line with this possibility, Liu and Barrientos (51) have recently reported that reactive oxygen species induce the expression of hypoxic genes in a Rox1-independent manner, although the levels of ROX1 transcripts are strongly increased upon oxidative stress. Induction of ROX1 expression was, however, shown to be Yap1-independent. The discrepancy between the data of Liu and Barrientos (51), and our own data are likely due to differences in the quantitative analysis of ROX1 transcripts (relative expression versus fold change). Here, we clearly show that Yap1 up-regulates ROX1 gene (Fig. 2), and this may act as a compensatory mechanism of Rox1-defective repression of hypoxic genes, under oxidative environments. Interestingly, controlled hypoxia is often used as a treatment to overcome the catastrophic effects observed after intoxication by ingestion with paraquat, a potent superoxide generator that accumulates in lungs (52, 53). In this case, hypoxia can be used to mitigate reactive oxygen species. As such, it is reasonable to hypothesize that Yap1 regulation of hypoxic genes through Rox1 may be relevant to overcome oxidative stress in a scenario where Yap1 activity is impaired and intracellular reactive oxygen species accumulate.
Author Contributions
C. P. and C. R.-P. conceived and designed the experiments; S. M. C., R. M., C. A., and C. P. performed the experiments; C. P. and S. M. C. analyzed the data; and C. P. and C. R.-P. wrote the paper.
Acknowledgment
We thank Prof. Jorge Pimentel (ICU, Hospitais da Universidade de Coimbra) for helpful discussions.
This work was supported by the Fundação para Ciência e Tecnologia through Grants EXPL/BIA-MIC/2525/2013 (to C. P.) and Pest-OE/EQB/LA0004/2011 (to Instituto de Tecnologia Química e Biológica) and Fellowships SFRH/BD/91077/2012 (to S. M. C.), SFRH/BPD/74294/2010 (to C. A.), and SFRH/BPD/90823/2012 (to C. P.). The authors declare that they have no conflicts of interest with the contents of this article.
- YRE
- Yap-responsive element
- ICP-AES
- inductively coupled plasma atomic emission spectroscopy
- qRT-PCR
- quantitative RT-PCR.
References
- 1. Huff J., Lunn R. M., Waalkes M. P., Tomatis L., Infante P. F. (2007) Cadmium-induced cancers in animals and in humans. Int. J. Occup. Environ. Health 13, 202–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Johri N., Jacquillet G., Unwin R. (2010) Heavy metal poisoning: the effects of cadmium on the kidney. Biometals 23, 783–792 [DOI] [PubMed] [Google Scholar]
- 3. Valko M., Rhodes C. J., Moncol J., Izakovic M., Mazur M. (2006) Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem. Biol. Interact. 160, 1–40 [DOI] [PubMed] [Google Scholar]
- 4. Huang Y. H., Shih C. M., Huang C. J., Lin C. M., Chou C. M., Tsai M. L., Liu T. P., Chiu J. F., Chen C. T. (2006) Effects of cadmium on structure and enzymatic activity of Cu, Zn-SOD and oxidative status in neural cells. J. Cell. Biochem. 98, 577–589 [DOI] [PubMed] [Google Scholar]
- 5. Jin Y. H., Clark A. B., Slebos R. J., Al-Refai H., Taylor J. A., Kunkel T. A., Resnick M. A., Gordenin D. A. (2003) Cadmium is a mutagen that acts by inhibiting mismatch repair. Nat. Genet. 34, 326–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Thorsen M., Perrone G. G., Kristiansson E., Traini M., Ye T., Dawes I. W., Nerman O., Tamás M. J. (2009) Genetic basis of arsenite and cadmium tolerance in Saccharomyces cerevisiae. BMC Genomics 10, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ruotolo R., Marchini G., Ottonello S. (2008) Membrane transporters and protein traffic networks differentially affecting metal tolerance: a genomic phenotyping study in yeast. Genome Biol. 9, R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Heo D. H., Baek I. J., Kang H. J., Kim J. H., Chang M., Jeong M. Y., Kim T. H., Choi I. D., Yun C. W. (2010) Cadmium regulates copper homoeostasis by inhibiting the activity of Mac1, a transcriptional activator of the copper regulon, in Saccharomyces cerevisiae. Biochem. J. 431, 257–265 [DOI] [PubMed] [Google Scholar]
- 9. Himeno S., Yanagiya T., Enomoto S., Kondo Y., Imura N. (2002) Cellular cadmium uptake mediated by the transport system for manganese. Tohoku J. Exp. Med. 196, 43–50 [DOI] [PubMed] [Google Scholar]
- 10. Gardarin A., Chédin S., Lagniel G., Aude J. C., Godat E., Catty P., Labarre J. (2010) Endoplasmic reticulum is a major target of cadmium toxicity in yeast. Mol. Microbiol. 76, 1034–1048 [DOI] [PubMed] [Google Scholar]
- 11. Clemens S. (2006) Toxic metal accumulation, responses to exposure and mechanisms of tolerance in plants. Biochimie 88, 1707–1719 [DOI] [PubMed] [Google Scholar]
- 12. Liu X. F., Supek F., Nelson N., Culotta V. C. (1997) Negative control of heavy metal uptake by the Saccharomyces cerevisiae BSD2 gene. J. Biol. Chem. 272, 11763–11769 [DOI] [PubMed] [Google Scholar]
- 13. Clemens S., Antosiewicz D. M., Ward J. M., Schachtman D. P., Schroeder J. I. (1998) The plant cDNA LCT1 mediates the uptake of calcium and cadmium in yeast. Proc. Natl. Acad. Sci. U.S.A. 95, 12043–12048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dix D. R., Bridgham J. T., Broderius M. A., Byersdorfer C. A., Eide D. J. (1994) The FET4 gene encodes the low affinity Fe(II) transport protein of Saccharomyces cerevisiae. J. Biol. Chem. 269, 26092–26099 [PubMed] [Google Scholar]
- 15. Gomes D. S., Fragoso L. C., Riger C. J., Panek A. D., Eleutherio E. C. (2002) Regulation of cadmium uptake by Saccharomyces cerevisiae. Biochim. Biophys. Acta 1573, 21–25 [DOI] [PubMed] [Google Scholar]
- 16. Gitan R. S., Shababi M., Kramer M., Eide D. J. (2003) A cytosolic domain of the yeast Zrt1 zinc transporter is required for its post-translational inactivation in response to zinc and cadmium. J. Biol. Chem. 278, 39558–39564 [DOI] [PubMed] [Google Scholar]
- 17. Li Z. S., Lu Y. P., Zhen R. G., Szczypka M., Thiele D. J., Rea P. A. (1997) A new pathway for vacuolar cadmium sequestration in Saccharomyces cerevisiae: YCF1-catalyzed transport of bis(glutathionato)cadmium. Proc. Natl. Acad. Sci. U.S.A. 94, 42–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wawrzycka D., Sobczak I., Bartosz G., Bocer T., Ulaszewski S., Goffeau A. (2010) Vmr 1p is a novel vacuolar multidrug resistance ABC transporter in Saccharomyces cerevisiae. FEMS Yeast Res. 10, 828–838 [DOI] [PubMed] [Google Scholar]
- 19. Sharma K. G., Mason D. L., Liu G., Rea P. A., Bachhawat A. K., Michaelis S. (2002) Localization, regulation, and substrate transport properties of Bpt1p, a Saccharomyces cerevisiae MRP-type ABC transporter. Eukaryotic Cell 1, 391–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Adle D. J., Sinani D., Kim H., Lee J. (2007) A cadmium-transporting P1B-type ATPase in yeast Saccharomyces cerevisiae. J. Biol. Chem. 282, 947–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kern A. L., Bonatto D., Dias J. F., Yoneama M. L., Brendel M., Pêgas Henriques J. A. (2005) The function of Alr1p of Saccharomyces cerevisiae in cadmium detoxification: insights from phylogenetic studies and particle-induced X-ray emission. Biometals 18, 31–41 [DOI] [PubMed] [Google Scholar]
- 22. Nagy Z., Montigny C., Leverrier P., Yeh S., Goffeau A., Garrigos M., Falson P. (2006) Role of the yeast ABC transporter Yor1p in cadmium detoxification. Biochimie 88, 1665–1671 [DOI] [PubMed] [Google Scholar]
- 23. Mielniczki-Pereira A. A., Hahn A. B., Bonatto D., Riger C. J., Eleutherio E. C., Henriques J. A. (2011) New insights into the Ca2+-ATPases that contribute to cadmium tolerance in yeast. Toxicol. Lett. 207, 104–111 [DOI] [PubMed] [Google Scholar]
- 24. Lauer Júnior C. M., Bonatto D., Mielniczki-Pereira A. A., Schuch A. Z., Dias J. F., Yoneama M. L., Pêgas Henriques J. A. (2008) The Pmr1 protein, the major yeast Ca2+-ATPase in the Golgi, regulates intracellular levels of the cadmium ion. FEMS Microbiol. Lett. 285, 79–88 [DOI] [PubMed] [Google Scholar]
- 25. Wemmie J. A., Szczypka M. S., Thiele D. J., Moye-Rowley W. S. (1994) Cadmium tolerance mediated by the yeast AP-1 protein requires the presence of an ATP-binding cassette transporter-encoding gene, YCF1. J. Biol. Chem. 269, 32592–32597 [PubMed] [Google Scholar]
- 26. Azevedo D., Tacnet F., Delaunay A., Rodrigues-Pousada C., Toledano M. B. (2003) Two redox centers within Yap1 for H2O2 and thiol-reactive chemicals signaling. Free Radic. Biol. Med. 35, 889–900 [DOI] [PubMed] [Google Scholar]
- 27. Delaunay A., Isnard A. D., Toledano M. B. (2000) H2O2 sensing through oxidation of the Yap1 transcription factor. EMBO J. 19, 5157–5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pimentel C., Caetano S. M., Menezes R., Figueira I., Santos C. N., Ferreira R. B., Santos M. A., Rodrigues-Pousada C. (2014) Yap1 mediates tolerance to cobalt toxicity in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1840, 1977–1986 [DOI] [PubMed] [Google Scholar]
- 29. Lowry C. V., Zitomer R. S. (1988) ROX1 encodes a heme-induced repression factor regulating ANB1 and CYC7 of Saccharomyces cerevisiae. Mol. Cell. Biol. 8, 4651–4658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gueldener U., Heinisch J., Koehler G. J., Voss D., Hegemann J. H. (2002) A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 30, e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pimentel C., Vicente C., Menezes R. A., Caetano S., Carreto L., Rodrigues-Pousada C. (2012) The role of the Yap5 transcription factor in remodeling gene expression in response to Fe bioavailability. PLoS One 7, e37434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tellmann G. (2006) The E-Method: a highly accurate technique for gene-expression analysis. Nat. Methods 3, 3 [Google Scholar]
- 33. Li L., Kaplan J. (1998) Defects in the yeast high affinity iron transport system result in increased metal sensitivity because of the increased expression of transporters with a broad transition metal specificity. J. Biol. Chem. 273, 22181–22187 [DOI] [PubMed] [Google Scholar]
- 34. Jeyaprakash A., Welch J. W., Fogel S. (1991) Multicopy CUP1 plasmids enhance cadmium and copper resistance levels in yeast. Mol. Gen. Genet. 225, 363–368 [DOI] [PubMed] [Google Scholar]
- 35. Fernandes L., Rodrigues-Pousada C., Struhl K. (1997) Yap, a novel family of eight bZIP proteins in Saccharomyces cerevisiae with distinct biological functions. Mol. Cell. Biol. 17, 6982–6993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jensen L. T., Culotta V. C. (2002) Regulation of Saccharomyces cerevisiae FET4 by oxygen and iron. J. Mol. Biol. 318, 251–260 [DOI] [PubMed] [Google Scholar]
- 37. Teixeira M. C., Monteiro P., Jain P., Tenreiro S., Fernandes A. R., Mira N. P., Alenquer M., Freitas A. T., Oliveira A. L., Sá-Correia I. (2006) The YEASTRACT database: a tool for the analysis of transcription regulatory associations in Saccharomyces cerevisiae. Nucleic Acids Res. 34, D446–D451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Newbury S. F. (2006) Control of mRNA stability in eukaryotes. Biochem. Soc. Trans. 34, 30–34 [DOI] [PubMed] [Google Scholar]
- 39. Batista-Nascimento L., Toledano M. B., Thiele D. J., Rodrigues-Pousada C. (2013) Yeast protective response to arsenate involves the repression of the high affinity iron uptake system. Biochim. Biophys. Acta 1833, 997–1005 [DOI] [PubMed] [Google Scholar]
- 40. Chanfreau G., Buckle M., Jacquier A. (2000) Recognition of a conserved class of RNA tetraloops by Saccharomyces cerevisiae RNase III. Proc. Natl. Acad. Sci. U.S.A. 97, 3142–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee A., Henras A. K., Chanfreau G. (2005) Multiple RNA surveillance pathways limit aberrant expression of iron uptake mRNAs and prevent iron toxicity in S. cerevisiae. Mol. Cell 19, 39–51 [DOI] [PubMed] [Google Scholar]
- 42. Kaplan C. D., Kaplan J. (2009) Iron acquisition and transcriptional regulation. Chem. Rev. 109, 4536–4552 [DOI] [PubMed] [Google Scholar]
- 43. Puig S., Askeland E., Thiele D. J. (2005) Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell 120, 99–110 [DOI] [PubMed] [Google Scholar]
- 44. Puig S., Vergara S. V., Thiele D. J. (2008) Cooperation of two mRNA-binding proteins drives metabolic adaptation to iron deficiency. Cell Metab. 7, 555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rodrigues-Pousada C., Menezes R. A., Pimentel C. (2010) The Yap family and its role in stress response. Yeast 27, 245–258 [DOI] [PubMed] [Google Scholar]
- 46. Keng T. (1992) HAP1 and ROX1 form a regulatory pathway in the repression of HEM13 transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 12, 2616–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zitomer R. S., Lowry C. V. (1992) Regulation of gene expression by oxygen in Saccharomyces cerevisiae. Microbiol. Rev. 56, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Deckert J., Perini R., Balasubramanian B., Zitomer R. S. (1995) Multiple elements and auto-repression regulate Rox1, a repressor of hypoxic genes in Saccharomyces cerevisiae. Genetics 139, 1149–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jones C. I., Zabolotskaya M. V., Newbury S. F. (2012) The 5′ → 3′ exoribonuclease XRN1/Pacman and its functions in cellular processes and development. Wiley Interdiscip. Rev. RNA 3, 455–468 [DOI] [PubMed] [Google Scholar]
- 50. Waters B. M., Eide D. J. (2002) Combinatorial control of yeast FET4 gene expression by iron, zinc, and oxygen. J. Biol. Chem. 277, 33749–33757 [DOI] [PubMed] [Google Scholar]
- 51. Liu J., Barrientos A. (2013) Transcriptional regulation of yeast oxidative phosphorylation hypoxic genes by oxidative stress. Antioxid. Redox Signal. 19, 1916–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chollet A., Muszynsky J., Bismuth C., Pham J., El Khouly M., Surugue R. (1983) [Hypo-oxygenation in paraquat poisoning. Apropos of 6 cases]. Toxicol. Eur. Res. 5, 71–75 [PubMed] [Google Scholar]
- 53. Rhodes M. L., Zavala D. C., Brown D. (1976) Hypoxic protection in paraquat poisoning. Lab. Invest. 35, 496–500 [PubMed] [Google Scholar]