Abstract
Axon degeneration is an intrinsic self-destruction program that underlies axon loss during injury and disease. Sterile alpha and TIR motif containing 1 (SARM1) protein is an essential mediator of axon degeneration. We report that SARM1 initiates a local destruction program involving rapid breakdown of NAD+ after injury. We used an engineered protease-sensitized SARM1 to demonstrate that SARM1 activity is required after axon injury to induce axon degeneration. Dimerization of the Toll-Interleukin Receptor (TIR) domain of SARM1 alone was sufficient to induce locally-mediated axon degeneration. Formation of the SARM1 TIR dimer triggered rapid breakdown of NAD+, whereas SARM1-induced axon destruction could be counteracted by increased NAD+ synthesis. SARM1-induced depletion of NAD+ may explain the potent axon protection in Wallerian Degeneration slow (Wlds) mutant mice.
Cells undergo regulated self-destruction during development and in response to stresses (1). Axons, the longest cellular structures in the body, have a locally-mediated self-destruction program that removes damaged axons but also promotes axon loss in the setting of neurological disorders (2). Axon degeneration is antagonized by the Wlds chimeric protein (3). The active moiety of Wlds is the enzyme nicotinamide mononucleotide adenyltransferase 1 (Nmnat1), which synthesizes the essential cofactor nicotinamide adenine dinucleotide (NAD+) (4), but the function of Nmnat1 and NAD+ in axon protection remains unclear (2). The protein SARM1 is an essential mediator of axon degeneration (5, 6). SARM1 is a negative regulator of Toll-Like Receptor-activated transcriptional programs (7), but its mechanism for axon degeneration is unknown.
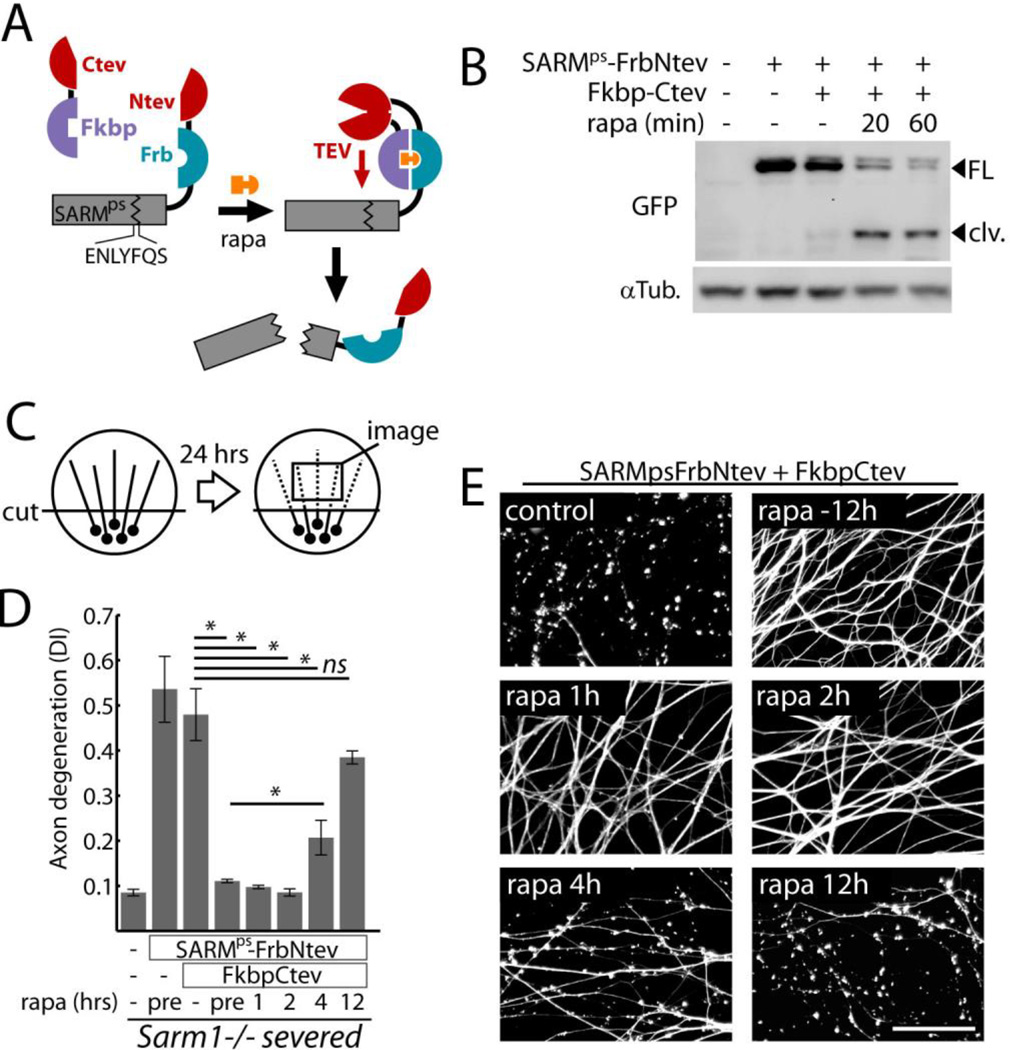
To investigate whether SARM1 functions before or after injury, we engineered a system to inactivate SARM1 with pharmacologic control. Protease sensitized SARM1 (SARMps) contains a tobacco etch virus (TEV) protease consensus sequence between the Sterile Alpha Motif (SAM) and TIR domains, which are both essential for SARM1 function (6). SARMps is thus cleaved and inactivated by TEV protease. SARMps was fused to the rapamycin binding domain Frb and the N-terminal portion of split TEV protease (Ntev) (8) and co-expressed with C-terminal split TEV fused to FK866 Binding Protein (Fkbp-Ctev), allowing rapamycin-induced cleavage (Fig 1A, Fig S1). In dorsal root ganglion (DRG) neurons, cleavage of SARMps was mostly complete within 60 minutes of rapamycin treatment (Fig 1B, Fig S2A). SARMps functionality was verified by expression of SARMps in isolated Sarm1−/− DRG neurons. When Sarm1−/− axons were severed (diagrammed in Fig 1C), they remained intact after 24 hrs, whereas axons of neurons expressing SARMps showed degeneration measured by axon morphometry (Fig. 1D), similar to wild-type axons. SARMps function was lost upon cleavage triggered by rapamycin in the presence of Fkbp-Ctev (Fig. 1D–E) or by expression of full-length TEV (Fig. S2B). Cleavage of SARMps initiated 12 hours before- or up to 2 hours after axon transection fully suppressed axon degeneration measured 24 hours after axotomy. Because cleavage of SARMps after axons were disconnected from cell bodies resulted in protection, SARM1 must function after injury to promote degeneration.
Fig. 1.
SARM1 functions following axon injury to promote destruction. A) Schematic showing how expression of SARMps-Frb-Ntev with Fkbp-Ctev allows rapamycin-induced complementation of split TEV and concomitant SARMps cleavage. B) Gel electrophoresis with anti-GFP immunoblot showing SARMps cleavage in DRG neurons induced by 100 nM rapamycin (rapa); FL=full length SARMps-Frb-Ntev-Cerulean; clv = cleaved form. α-Tubulin (αTub) was a loading control. C) Diagram of in vitro injury model: isolated DRG neurons were severed and axon degeneration was quantified after 24 hours from axon images. D) Requirement for SARM1 activity after axotomy to induce axon degeneration.. Axon degeneration is reported as the degeneration index (DI), a morphometric ratio of fragmented axon area to total axon area (13). Sarm1−/− DRG neurons treated with expression lentiviruses (control, SARMps-FrbNtev, and Fkbp-Ctev) were severed and treated with 100 nM rapamycin at various times (pre = 12 hours pre-injury). E) Micrographs show representative α-Tubulin stained axons corresponding to select treatment groups in (D). Scale bar = 50 micrometers. Error bars = SEM; * p < 0.01; one-way analysis of variance (ANOVA) with Tukey’s post-hoc test.
SARM1 has no predicted enzymatic function but contains a TIR domain, which is the effector domain of Toll-Like Receptors (TLRs). Activation of TLRs results in dimerization of TIR domains that transmit a signal to cytosolic effector proteins (9). We tested whether multimerization of the TIR domain of SARM1 (sTIR) might induce axon degeneration. A minimal region of human SARM1 comprising sTIR and the adjacent multimerization (SAM) domains but lacking the auto-inhibitory N-terminus (SAM-TIR) is constitutively active and promotes cell and axon destruction in cultured DRG neurons (6). Expression of this activated form of SARM1 in vivo in Drosophila motor (Fig 2A) or sensory neurons (Fig S3) also caused cell and axon destruction. This degeneration was not observed in Drosophila expressing SAM-TIR harboring a disruptive sTIR mutation.
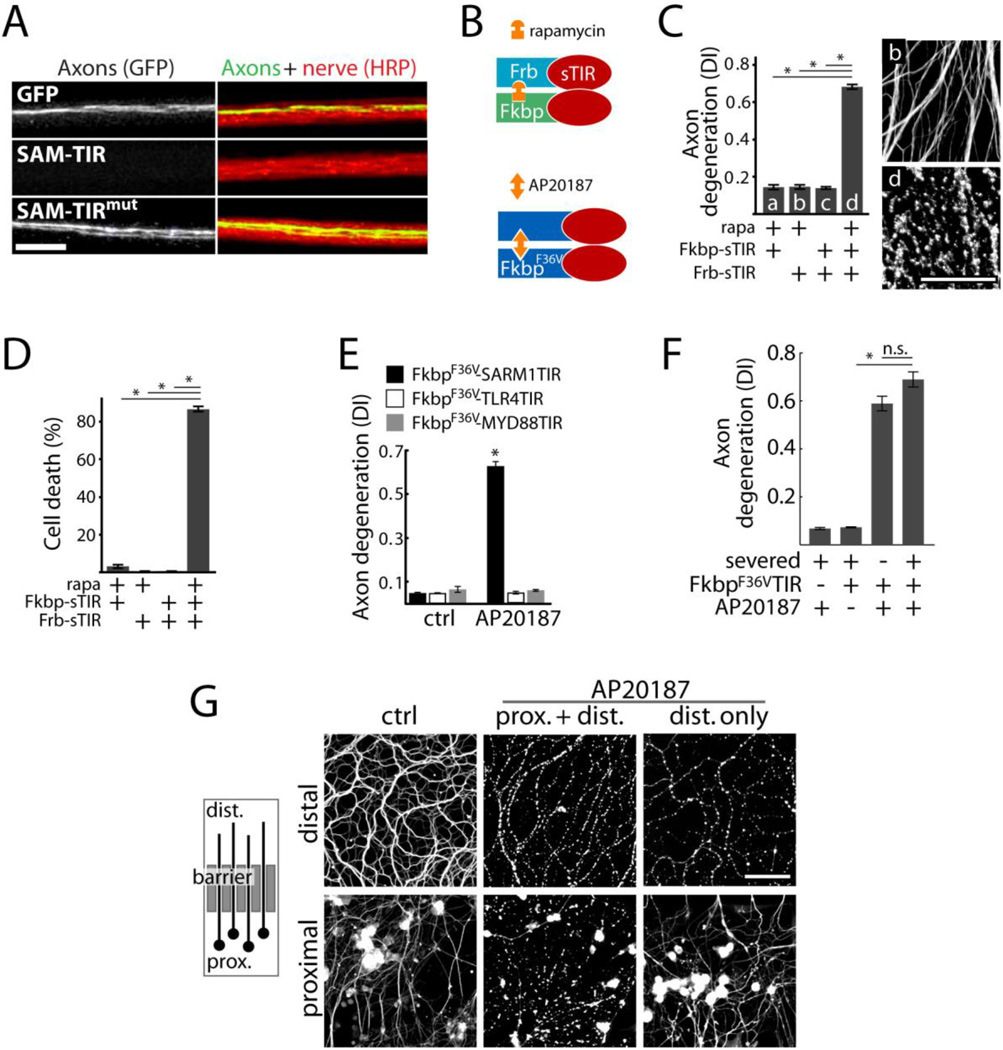
Fig. 2.
Axon degeneration and neuronal death induced by sTIR dimerization. A) Micrograph showing motor nerves of third instar Drosophila larvae. M12-Gal4 drives expression from mCD8-GFP (green) alone or with either UAS-SAM-TIR or UAS-SAM-TIRmut in single motor axons in each nerve (red=HRP). UAS-SAM-TIR expression caused axon loss in 49/49 nerves as shown; whereas SAM-TIR with a disruptive TIR mutation led to degeneration in 0/70 nerves (χ2=119; p<0.001); scale bar=10 micrometers. B) Schematic showing sTIR dimerization by rapamycin or AP20187. C) Effect of sTIR, dimerized sTIR, and rapamycin on axon degeneration. α-Tubulin stained axons correspond to bars b and d. D) Effect of sTIR dimerization on neuronal viability quantified by ethidium homodimer exclusion after 24 hours. E) Effects of dimerization of sTIR or TIR domains of MYD88 or TLR4 on axon degeneration. F) Effects of sTIR dimerization on degeneration of Sarm1−/− axons physically disconnected from cell bodies. G) Left: diagram of axons growing through a diffusion barrier into an isolated fluid compartment. Right: micrographs of isolated distal axon segments after application of AP20187 globally or selectively to distal axons. Scale bar = 50 micrometers. Error bars = SEM; * p < 0.01; one-way ANOVA with Tukey’s post-hoc test.
To evaluate the sufficiency of sTIR dimerization in axon destruction, we engineered a pharmacologically-controlled dimerizable sTIR by fusing it to the rapamycin-binding domains Frb and Fkbp (Fig 2B) (10). We expressed Frb-sTIR and Fkbp-sTIR in DRG neurons and found that sTIR dimerization by rapamycin induced axon fragmentation within 12 hrs (Fig 2C) and neuronal cell death within 24 hrs (Fig 2D). sTIR-induced toxicity did not require inhibition of mammalian target of rapamycin (mTOR) because the rapamycin analogue AP20187, which does not target mTOR, also stimulated axon degeneration in cells expressing the homodimerizable FkbpF36V-sTIR (10). SARM1 activation is thus sufficient to elicit axonal and neuronal destruction. Cell and axon degeneration is not induced upon dimerization of the TIR domains of Toll-Like Receptor 4 (TLR4) or the adaptor MYD88 (Fig 2E).
We tested whether SARM1 promotes axon degeneration through a local mechanism. sTIR-induced degeneration does not require a physical connection between the axon and soma: Sarm1−/− axons persisted after severing; however, sTIR dimerization by AP20187 caused fragmentation of these severed segments within 12 hours (Fig 2F). Dimerization of sTIR locally within axons also led to selective axon destruction. We grew DRG neurons in adjacent fluid compartments: one containing the cell bodies and proximal axons and the other containing only distal axons (Fig 2G). Application of AP20187 to both compartments led to destruction of proximal and distal axons, whereas selective application to the distal chamber elicited selective distal axon degeneration after 24 hours (Fig 2G).
SARM1 TIR dimerization elicited rapid pathophysiologic changes: axon degeneration and neuronal death were evident within 1.5 and 6 hours respectively (Fig S4A and B), and neuronal mitochondrial membrane potential dissipated and calcium accumulated with similar kinetics (Fig S4C–E). These measurements indicate early energetic failure. We thus focused on biochemical events leading from SARM1 activation to axonal demise. Axon degeneration is antagonized by the NAD+ synthetic enzyme Nmnat1, which, like SARM1, functions locally within axons (11). Injured axons exhibit declining levels of NAD+ before morphologic changes (12), but it is unknown whether this is a cause or consequence of axon destruction. Although Wlds/Nmnat1 does not increase steady-state abundance of NAD+ (13), in the setting of acute NAD+ depletion it might maintain sufficient levels of NAD+ for viability (diagrammed in Fig 3A). We thus tested whether SARM1 activation leads to depletion of NAD+.
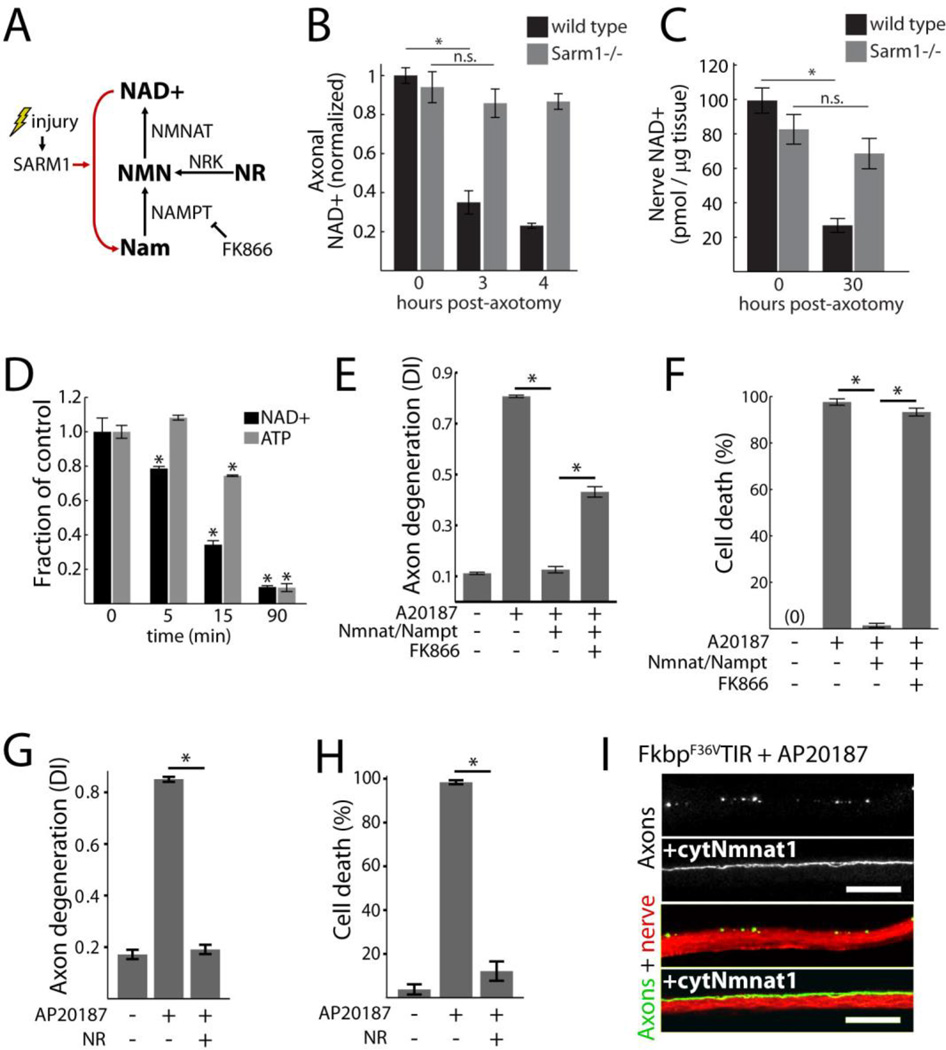
Fig. 3.
Loss of NAD+ underlies SARM1-induced destruction. A) Diagram of NAD+ synthesis and inhibition by FK866; Nam=nicotinamide, NR = nicotinamide riboside, Nrk=nicotinamide riboside kinase, NMN=nicotinamide mononucleotide. B) Axonal NAD+ concentration in cultured wild-type and Sarm1−/− DRG neurons following axotomy; normalized to wild-type control. C) NAD+ concentration in distal sciatic nerve segments from wild-type or Sarm1−/− animals following transection; wild-type n=5; Sarm1−/− n=9. D) Neuronal NAD+ and ATP concentrations following sTIR dimerization by AP20187; comparisons made to zero minutes control. E–F) Axon degeneration (E) and neuronal cell death (F) induced by sTIR homodimerization (AP20187) and inhibition by NAD+ synthetic enzymes with or without the Nampt inhibitor FK866 (10 nM); measured 24 hours after sTIR dimerization and FK866 application. G–H) Effect of NR (1 mM) on axon degeneration (G) and neuronal cell death (H) induced by sTIR homodimerization (AP20187) for 24 hours with or without NR. I) Micrographs showing sTIR-induced motor axon fragmentation in third instar Drosophila larvae blocked by cytosolic Nmnat1 (cytNmnat1) expression. M12-Gal4 drives expression from UAS-mCD8-GFP (green) and UAS-FkF36VTIR with or without UAS-cytNmnat1 in single motor axons in each nerve (red=HRP). Degeneration score = 76+/−4% (control) vs 11+/−2% (cytNmnat1); p<0.001 (t-test); scale bar=20 micrometers. Error bars = SEM; * p < 0.01; one-way ANOVA with Tukey’s post-hoc test.
To test whether endogenous SARM1 is necessary for axonal loss of NAD+ after axotomy, we isolated axons from cultured wild-type and Sarm1−/− DRG neurons 3 and 4 hours after injury, a time when they remain morphologically intact, and measured abundance of NAD+ using high performance liquid chromatography (HPLC). Abundance of NAD+ decreased after injury in wild-type axons but remained stable in Sarm1−/− axons (Fig 3B). Loss of ATP, an expected consequence of NAD+ depletion, was also SARM1-dependent (Fig S5A). To determine whether SARM1 is also necessary for axotomy-induced loss of NAD+ in vivo, we compared concentrations of NAD+ in distal sciatic nerve segments from adult wild-type and Sarm1−/− mice. At 30 hours after injury, amounts of NAD+ were decreased in wild-type nerves but remained stable in Sarm1−/− nerves (Fig 3C). At this time, injured nerves remained morphologically intact (Fig S5C) and amounts of ATP were stable (Fig S5B).
We tested whether SARM1 activation was sufficient to elicit loss of NAD+ by measuring neuronal NAD+ after sTIR dimerization. sTIR dimerization by addition of AP20187 caused rapid loss of NAD+; within 15 minutes abundance of NAD+ was reduced by 66% and by 90 minutes 90% of the NAD+ was lost (Fig 3D). Abundance of ATP also declined after sTIR dimerization, but its depletion was slower than that of NAD+.
Together these data implicate NAD+ loss as a critical step in SARM1-mediated axon destruction. We therefore examined whether increased NAD+ synthesis could counteract the destruction program activated by sTIR dimerization. In DRG neurons, both axon degeneration and cell death initiated by sTIR dimerization was completely blocked by expression of Nmnat1 and nicotinamide phosphoribosyltransferase (Nampt), which together synthesize NAD+ (Fig 3A). Protection appeared to require NAD+ synthesis because concurrent treatment with the Nampt inhibitor FK866 blocked the protection afforded by these enzymes (Fig 3E–F). Similarly, sTIR-induced axon degeneration and cell death were blocked by supplementation with the cell-permeable NAD+ precursor Nicotinamide Riboside (NR) (Fig 3G–H) (14). Drosophila larvae expressing the dimerizable FkbpF36V-sTIR in motor neurons that were fed AP20187 showed extensive axon degeneration that was blocked by co-expression of cytosolic Nmnat1 (Fig 3I).
To extend our analysis of biochemical events following SARM1 activation, we created a heterologous human embryonic kidney (HEK293T) cell line (HTir) that stably expresses Frb-sTIR and Fkbp-sTIR. Following 12 hours of sTIR dimerization in HTir cells, toxicity was evident as loss of ATP (Fig S6A) and altered morphology (Fig S6B). Both effects were blocked by NR supplementation. Inhibition of NAD+ synthesis with FK866 increased loss of ATP, whereas FK866 was not toxic in the absence of sTIR dimerization (Fig S6A, B).
To evaluate whether NAD+ depletion alone is sufficient to induce axon destruction, we stimulated direct intracellular breakdown of NAD+ by dimerization of the Poly ADP-Ribose Polymerase (PARP) domain of Tankyrase 1 (Tnkp; diagrammed in Fig 4A). We generated dimerizable FkbpF36V-Tnkp and showed that AP20187 treatment of cells expressing this construct led to loss of NAD+ and formation of Poly ADP-Ribose (PAR) (Fig S7A, B). In the presence of FK866, which inhibits de novo NAD+ synthesis, Tnkp dimerization in dividing cells led to rapid energetic failure (ATP loss) that was blocked by the Tankyrase inhibitor XAV939 (Fig S7C). NR supplementation blocked toxicity but not PAR formation, indicating NAD+ loss and not PAR formation caused cell death (Fig S7A, C). In neurons, Tnkp-induced depletion of NAD+ caused degeneration of uninjured wild-type and Sarm1−/− neurons (Fig 4B, C). Moreover, NAD+ depletion from isolated (pre-severed) Sarm1−/− axons led to degeneration (Fig 4C). Thus, rapid NAD+ depletion is sufficient to cause rapid axon loss.
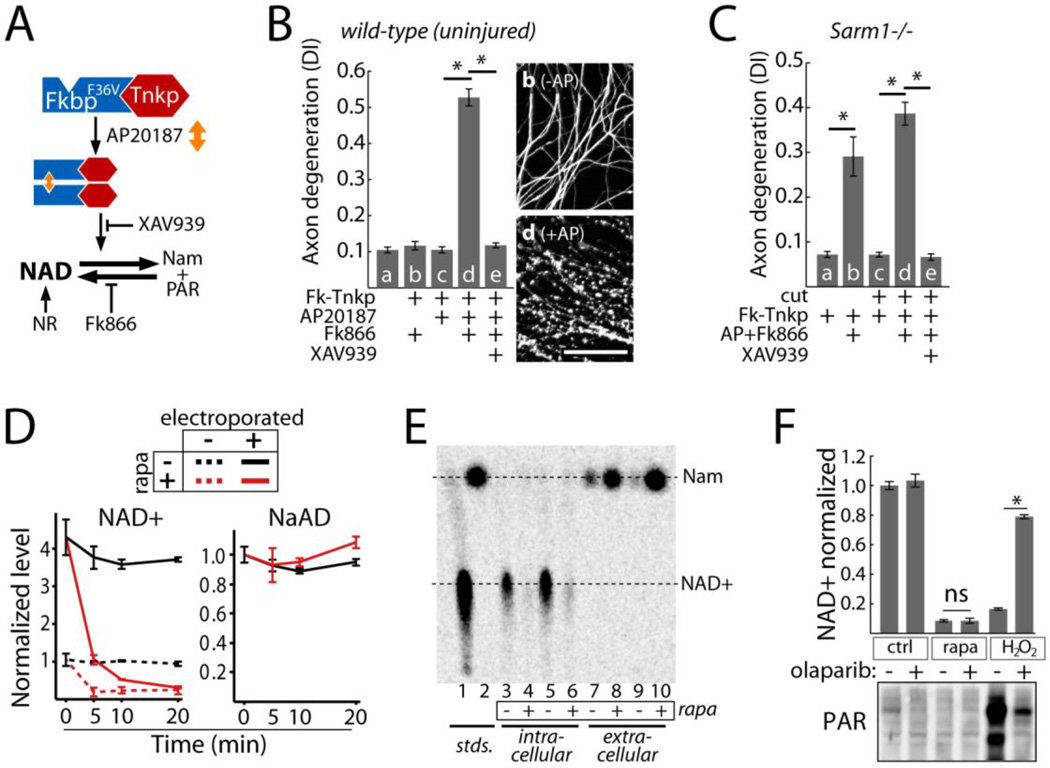
Fig. 4.
Effects of NAD+ breakdown on axon degeneration. A) Diagram of NAD+ manipulation using Tnkp dimerization. NAD+ loss induced by FkbpF36V-Tnkp dimerization is blocked by Tankyrase inhibitor XAV939 or NR and is exacerbated by FK866. B) Axon degeneration in response to NAD+ depletion by dimerized Tnkp and FK866 after 24 hours (bar d) and inhibition by Tankyrase inhibitor XAV939 (100 nM; bar e). Representative α-Tubulin-stained axons corresponding to bars b and d are shown; scale bar = 50 micrometers. C) Effect of NAD+ depletion by dimerized Tnkp + FK866 on axon degeneration in Sarm1−/− uninjured axons (bar b) or isolated (cut) Sarm1−/− axons (bar d). D) Effect of sTIR dimerization on endogenous (dotted lines) and exogenously introduced (solid lines) NAD+ or NaAD (control) in HTir cells. NaAD is undetectable in non-electroporated cells. E) Conversion of 14C-NAD+ in HTir cells to Nam 15 minutes after SARM1 TIR dimerization. NAD+ and Nam from cell extracts and extracellular media were resolved by thin layer chromatography. F) Top: effect of the PARP inhibitor olaparib (100 nM) on NAD+ loss induced by 1 mM H2O2 (10 min) or sTIR dimerization (10 min). Bottom: PAR formation after H2O2 treatment or sTIR dimerization in HTir cells expressing PARG shRNA and inhibition by olaparib. Error bars = SEM; * p < 0.01; one way ANOVA with Tukey’s post-hoc test.
To define whether SARM1-mediated depletion of NAD+ results from increased consumption or decreased synthesis of NAD+, we introduced exogenous NAD+ and, as a control, nicotinic acid adenine dinucleotide (NaAD) into HTir cells by electroporation (15) followed by sTIR dimerization. Control cells show rapid loss of endogenous NAD+ within 5 minutes in response to sTIR dimerization. Electroporation in the presence of NAD+ increased the concentration of NAD+ by a factor of 4.3, but NAD+ was rapidly consumed upon sTIR dimerization. The specificity of this reaction is highlighted by the stability of the closely related analog NaAD (Fig 4D). sTIR-induced loss of NAD+ thus involves active consumption of NAD+. We next demonstrated that the consumed NAD+ is converted to nicotinamide (Nam). When radiolabeled 14C-NAD+ was introduced into cells, 15 minutes of sTIR dimerization elicited loss of 14C-NAD+ and concomitant increases in 14C Nam as detected by thin layer chromatography (Fig 4E). Similarly, sTIR dimerization in non-electroporated cells also elicited Nam release as detected by HPLC (Fig S8).
Rapid breakdown of NAD+ induced by SARM1 TIR is similar to that observed when Poly ADP-Ribose Polymerase (PARP) is activated in response to DNA damage (16). However, NAD+ breakdown induced by sTIR is PARP-independent. The PARP inhibitor olaparib reduced NAD+ loss induced by H2O2but had no effect on SARM1-induced loss of NAD+ (Fig 4F). Furthermore, H2O2 led to PARP-dependent accumulation of poly ADP-Ribose (PAR), whereas no PAR was detected after sTIR dimerization (Fig 4F). Finally, sTIR dimerization in Parp1−/− cells induced loss of NAD+, axon degeneration, and cell death (Fig S9). These cell-destruction phenotypes were also unaffected by genetic ablation of the NAD+ glycohydrolase CD38, another major consumer of NAD+ (17) (Fig S9). SARM1 therefore initiates an NAD+ breakdown program that drives axon destruction and cell death independently of Parp1 and CD38.
SARM1 and its orthologs promote axonal degeneration (5, 6) as well as neuronal (18–20) and non-neuronal (21, 22) cell death. SARM1-induced breakdown of NAD+ links axon degeneration to the axon-protective Wlds protein. The presence of Wlds or other sources of axonal Nmnat may allow for rapid re-synthesis of NAD+ and maintenance of metabolic function, thereby counteracting the destructive effects of NAD+ degradation by SARM1. Identification of a class of neuroprotective drugs that increase NAD+ biosynthesis through effects on Nampt has highlighted the therapeutic potential of augmented NAD+ synthesis in neurological disorders (23). Our study provides further biological rationale for NAD+ augmentation as a therapeutic approach. Inhibition of SARM1-mediated NAD+ loss may be an alternative or synergistic therapeutic strategy for the treatment of neurologic disorders.
Supplementary Material
Acknowledgements
Supported by the National Institutes of Health grants (RO1DA020812 (AD), RO1AG013730 (JM), RO1NS065053, RO1NS087632, and RO1NS078007 (JM and AD), F31NS074517 (JG) and a grant from Vertex Pharmaceuticals. We thank ChromaDex (Irvine, California) for providing nicotinamide riboside, A Strickland, T Farhner, and N Panchenko for technical assistance and members of the Milbrandt and DiAntonio labs for fruitful discussions.
References and Notes
- 1.Fuchs Y, Steller H. Programmed cell death in animal development and disease. Cell. 2011;147:742–758. doi: 10.1016/j.cell.2011.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Conforti L, Gilley J, Coleman MP. Wallerian degeneration: an emerging axon death pathway linking injury and disease. Nat. Rev. Neurosci. 2014;15:394–409. doi: 10.1038/nrn3680. [DOI] [PubMed] [Google Scholar]
- 3.Coleman MP, et al. An 85-kb tandem triplication in the slow Wallerian degeneration (Wlds) mouse. Proc. Natl. Acad. Sci. U. S. A. 1998;95:9985–9990. doi: 10.1073/pnas.95.17.9985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Araki T, Sasaki Y, Milbrandt J. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science (80-.) 2004;305:1010–1013. doi: 10.1126/science.1098014. [DOI] [PubMed] [Google Scholar]
- 5.Osterloh JM, et al. dSarm/Sarm1 is required for activation of an injury-induced axon death pathway. Science. 2012;337:481–484. doi: 10.1126/science.1223899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerdts J, Summers DW, Sasaki Y, DiAntonio A, Milbrandt J. Sarm1-mediated axon degeneration requires both SAM and TIR interactions. J. Neurosci. 2013;33:13569–13580. doi: 10.1523/JNEUROSCI.1197-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Neill LaJ, Fitzgerald Ka, Bowie AG. The Toll-IL-1 receptor adaptor family grows to five members. Trends Immunol. 2003;24:286–290. doi: 10.1016/s1471-4906(03)00115-7. [DOI] [PubMed] [Google Scholar]
- 8.Wehr MC, et al. Monitoring regulated protein-protein interactions using split TEV. Nat. Methods. 2006;3:985–993. doi: 10.1038/nmeth967. [DOI] [PubMed] [Google Scholar]
- 9.Kang JY, Lee J-O. Structural biology of the Toll-like receptor family. Annu. Rev. Biochem. 2011;80:917–941. doi: 10.1146/annurev-biochem-052909-141507. [DOI] [PubMed] [Google Scholar]
- 10.Fegan A, White B, Carlson JCT, Wagner CR. Chemically controlled protein assembly: techniques and applications. Chem. Rev. 2010;110:3315–3336. doi: 10.1021/cr8002888. [DOI] [PubMed] [Google Scholar]
- 11.Sasaki Y, Milbrandt J. Axonal degeneration is blocked by nicotinamide mononucleotide adenylyltransferase (Nmnat) protein transduction into transected axons. J. Biol. Chem. 2010;285:41211–41215. doi: 10.1074/jbc.C110.193904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J, et al. A local mechanism mediates NAD-dependent protection of axon degeneration. J. Cell Biol. 2005;170:349–355. doi: 10.1083/jcb.200504028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sasaki Y, Vohra BPS, Lund FE, Milbrandt J. Nicotinamide mononucleotide adenylyl transferase-mediated axonal protection requires enzymatic activity but not increased levels of neuronal nicotinamide adenine dinucleotide. J. Neurosci. 2009;29:5525–5535. doi: 10.1523/JNEUROSCI.5469-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nikiforov A, Dölle C, Niere M, Ziegler M. Pathways and subcellular compartmentation of NAD biosynthesis in human cells: from entry of extracellular precursors to mitochondrial NAD generation. J. Biol. Chem. 2011;286:21767–21778. doi: 10.1074/jbc.M110.213298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elia MC, Motyka LE, Stamato TD. Electrotransfer of [32P]NAD allows labeling of ADP-ribosylated proteins in intact Chinese hamster ovary cells. Anal. Biochem. 1991;192:329–333. doi: 10.1016/0003-2697(91)90544-4. [DOI] [PubMed] [Google Scholar]
- 16.Kim MY, Zhang T, Kraus WL. Poly(ADP-ribosyl)ation by PARP-1: “PAR-laying” NAD+ into a nuclear signal. Genes Dev. 2005;19:1951–1967. doi: 10.1101/gad.1331805. [DOI] [PubMed] [Google Scholar]
- 17.Aksoy P, White TA, Thompson M, Chini EN. Regulation of intracellular levels of NAD: a novel role for CD38. Biochem. Biophys. Res. Commun. 2006;345:1386–1392. doi: 10.1016/j.bbrc.2006.05.042. [DOI] [PubMed] [Google Scholar]
- 18.Kim Y, et al. MyD88-5 links mitochondria, microtubules, and JNK3 in neurons and regulates neuronal survival. J. Exp. Med. 2007;204:2063–2074. doi: 10.1084/jem.20070868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mukherjee P, Woods TA, Moore RA, Peterson KE. Activation of the innate signaling molecule MAVS by bunyavirus infection upregulates the adaptor protein SARM1, leading to neuronal death. Immunity. 2013;38:705–716. doi: 10.1016/j.immuni.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Summers DW, DiAntonio A, Milbrandt J. Mitochondrial dysfunction induces sarm1-dependent cell death in sensory neurons. J. Neurosci. 2014;34:9338–9350. doi: 10.1523/JNEUROSCI.0877-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blum ES, Abraham MC, Yoshimura S, Lu Y, Shaham S. Control of nonapoptotic developmental cell death in Caenorhabditis elegans by a polyglutamine-repeat protein. Science. 2012;335:970–973. doi: 10.1126/science.1215156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panneerselvam P, et al. T-cell death following immune activation is mediated by mitochondria-localized SARM. Cell Death Differ. 2013;20:478–489. doi: 10.1038/cdd.2012.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang G, et al. P7C3 Neuroprotective Chemicals Function by Activating the Rate-Limiting Enzyme in NAD Salvage. Cell. 2014;158:1324–1334. doi: 10.1016/j.cell.2014.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerdts J, Sasaki Y, Vohra B, Marasa J, Milbrandt J. Image-based screening identifies novel roles for IkB kinase and Glycogen Synthase Kinase 3 in axonal degeneration. J. Biol. Chem. 2011;286:28011–28018. doi: 10.1074/jbc.M111.250472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiong X, et al. Protein turnover of the Wallenda/DLK kinase regulates a retrograde response to axonal injury. J. Cell Biol. 2010;191:211–223. doi: 10.1083/jcb.201006039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sasaki Y, Araki T, Milbrandt J. Stimulation of nicotinamide adenine dinucleotide biosynthetic pathways delays axonal degeneration after axotomy. J. Neurosci. 2006;26:8484–8491. doi: 10.1523/JNEUROSCI.2320-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshino J, Imai S-I. Accurate measurement of nicotinamide adenine dinucleotide (NAD+) with high-performance liquid chromatography. Methods Mol. Biol. 2013;1077:203–215. doi: 10.1007/978-1-62703-637-5_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.