Abstract
The activity of all mitogen-activated protein kinases (MAPKs) is stimulated via phosphorylation by upstream MAPK kinases (MAPKK), which are in their turn activated via phosphorylation by MAPKK kinases (MAPKKKs). The cells ensure the specificity of signaling in these cascades by employing a variety of scaffolding proteins that bind matching MAPKKKs, MAPKKs, and MAPKs. All four vertebrate arrestin subtypes bind JNK3, but only arrestin-3 serves as a scaffold, promoting JNK3 activation in intact cells. Arrestin-3-mediated JNK3 activation does not depend on arrestin-3 interaction with G protein-coupled receptors (GPCRs), as demonstrated by the ability of some arrestin mutants that cannot bind receptors to activate JNK3, whereas certain mutants with enhanced GPCR binding fail to promote JNK3 activation. Recent findings suggest that arrestin-3 directly binds both MAPKKs necessary for JNK activation and facilitates JNK3 phosphorylation at both Thr (by MKK4) and Tyr (by MKK7). JNK3 is expressed in a limited set of cell types, whereas JNK1 and JNK2 isoforms are as ubiquitous as arrestin-3. Recent study showed that arrestin-3 facilitates the activation of JNK1 and JNK2, scaffolding MKK4/7-JNK1/2/3 signaling complexes. In all cases, arrestin-3 acts by bringing the kinases together: JNK phosphorylation shows biphasic dependence on arrestin-3, being enhanced at lower and suppressed at supraoptimal concentrations. Thus, arrestin-3 regulates the activity of multiple JNK isoforms, suggesting that it might play a role in survival and apoptosis of all cell types.
Keywords: Arrestin, JNK, Scaffold, Cell signaling, Protein phosphorylation, Apoptosis
1 The Discovery of the Role of Arrestins in JNK Activation
Arrestins are best known for their ability to specifically bind active phosphorylated forms of their cognate G protein-coupled receptors (GPCRs) (Carman and Benovic 1998; Gurevich and Gurevich 2006b; Gurevich et al. 2011). Arrestin-3 binding to MAP kinase c-Jun N-terminal kinase 3 (JNK3) and its upstream MAP kinase kinase kinase (MAP3K) apoptosis signal-regulating kinase 1 (ASK1) was described in 2000 (McDonald et al. 2000), within a few years of the discovery of the first non-receptor-binding partner of arrestin, clathrin (Goodman et al. 1996).
Using co-immunoprecipitation (co-IP), the authors of the original study (McDonald et al. 2000) demonstrated that one of the nonvisual subtypes, β-arrestin2 (systematic name arrestin-31), is found in a complex with JNK3, mitogen-activated protein kinase kinase (MAP2K) 4 (MKK4), and ASK1, which together constitute one of the typical three-kinase MAPK activation modules, ASK1–MKK4–JNK3. The results of co-IP from differentially transfected cells suggested that arrestin-3 bound ASK1 and JNK3, whereas MKK4 was brought to the complex via interactions with the other two kinases. The authors also found that the stimulation of the angiotensin II type 1A receptor increased JNK3 phosphorylation in transfected cells and triggered the colocalization of arrestin-3 and active phospho-JNK3 to cytoplasmic vesicles (McDonald et al. 2000). This leads to the hypothesis that arrestins function as receptor-regulated MAPK scaffolds, promoting JNK3 phosphorylation and localizing active JNK3 to ligand-activated GPCRs. However, follow-up study from the same group showed that receptor is not obligatory for this function of arrestin-3: it effectively facilitated JNK3 phosphorylation in cells overexpressing ASK1 in the absence of receptor activation (Miller et al. 2001).
Both of these studies suggested that arrestin-3 is the only nonvisual subtype that binds JNK3 and its upstream kinases, so the fact that arrestin-2 does not promote JNK3 activation was explained by the lack of binding (McDonald et al. 2000; Miller et al. 2001). However, arrestin-2 and arrestin-3 are highly homologous (Gurevich and Gurevich 2006a), and both appear to bind numerous GPCRs (Gurevich et al. 1995; Barak et al. 1997), clathrin (Goodman et al. 1996), clathrin adaptor AP2 (Laporte et al. 1999), and protein kinase c-Src (Luttrell et al. 1999) comparably. Therefore, the issue of the binding of different arrestin subtypes to JNK3 and upstream kinases was further investigated (Song et al. 2006, 2007, 2009).
2 Which Kinases Bind Which Arrestin Subtypes?
Earlier observations showed that arrestin-3 redistributes JNK3 from the nucleus to the cytoplasm (Scott et al. 2002) and that this phenomenon requires functional nuclear export signal (NES) in the C terminus of arrestin-3 (Wang et al. 2003). Based on these findings, it was demonstrated that a single amino acid substitution in the C terminus of arrestin-2 that creates functional NES results in similar removal of JNK3 from the nucleus in the presence of NES-positive arrestin-2, demonstrating that arrestin-2 actually binds JNK3 (Song et al. 2006). Moreover, visual arrestin-1 (Song et al. 2006) and arrestin-4 (Song et al. 2007) were found to redistribute JNK3 in the cell as efficiently as nonvisual subtypes, contradicting the idea that JNK3 binding is a unique feature of arrestin-3 (McDonald et al. 2000) and demonstrating that all vertebrate arrestins bind JNK3. Obviously, JNK3 activation requires simultaneous recruitment of upstream kinases, so inability of other subtypes to bind ASK1 and MKK4/7 could explain why only arrestin-3 promotes JNK3 activation. However, this explanation also did not survive experimental testing: both arrestin-2 (Song et al. 2009; Seo et al. 2011) and arrestin-1 (Gurevich et al. 2011) were shown to bind ASK1 and MKK4, similar to arrestin-3. Reconstitution of MKK4-JNK3α2 signaling module with arrestin-2 and arrestin-3 from purified proteins suggested that the difference might be in affinity: arrestin-3 showed tighter binding of both JNK3α2 and MKK4 than arrestin-2 (Zhan et al. 2011b). However, a subsequent study described arrestin-3 mutant with higher affinity for JNK3α2 than WT, which also comparably bound MKK4 and ASK1, but failed to promote JNK3α2 phosphorylation in cells (Breitman et al. 2012). Thus, while difference in affinity might play a role in differential effects of scaffolding of MAP kinase cascades by the two nonvisual arrestins, it is certainly not the only factor. Extensive structure–function studies of the ability of arrestin-3 to facilitate JNK3α2 phosphorylation in intact cells (Seo et al. 2011; Breitman et al. 2012) suggest that an arrestin can bind all required kinases, but hold them in “wrong” orientation, which is not conducive to signaling.
3 Does Arrestin Conformation Affect Its Interactions with JNK3 and Upstream Kinases?
Nonvisual arrestins interact with a fairly diverse group of non-receptor-binding partners, including proteins involved in receptor trafficking, protein kinases, E3 ubiquitin ligases and deubiquitinating enzymes, small G proteins, etc. (see chapters “β-arrestins and G Protein-Coupled Receptor Trafficking,” “Arrestin Interaction with E3 Ubiquitin Ligases and Deubiquitinases: Functional and Therapeutic Implications,” “Arrestin-Dependent Activation of ERK and Src Family Kinases,” “Arrestin-Dependent Activation of JNK Family Kinases,” “Arrestin-Mediated Activation of p38 MAPK: Molecular Mechanisms and Behavioral Consequences,” “Arrestin-Dependent Localization of Phosphodiesterases,” “Arrestins in Apoptosis,” “Molecular Mechanisms Underlying Beta-Arrestin-Dependent Chemotaxis and Actin-Cytoskeletal Reorganization,” “Arrestins in Host-Pathogen Interactions,” and “Arrestin Regulation of Small GTPases”). Ever-expanding repertoire of arrestin-binding proteins has been identified at a fast pace since the discovery of the first non-receptor partner, clathrin, in 1996 (Goodman et al. 1996). In a recent proteomic analysis, 71 proteins were reported to bind arrestin-2, 162 proteins bound arrestin-3, and 102 proteins interacted with both nonvisual arrestins (Xiao et al. 2007). Interestingly, some proteins prefer receptor-bound arrestin conformation and others preferentially bind arrestins in their basal state, whereas some do not have an obvious preference (Luttrell et al. 2001; Song et al. 2006, 2009; Ahmed et al. 2011; Coffa et al. 2011b). Although arrestin often functions as scaffold or adaptor by tethering multiple components together, a single receptor-bound arrestin is not big enough to accommodate more than 4–6 partners simultaneously (Gurevich and Gurevich 2006b). Thus, many of the binding partners have to compete with each other for arrestin. How arrestin makes the “decision” to interact with the “right” set of partners remains a very interesting and challenging question. Arrestins undergo significant conformational changes upon binding different partners (Schleicher et al. 1989; Hanson et al. 2006a; Kim et al. 2012; Zhuang et al. 2013). Arrestins are known to exist in at least three distinct conformations: free, revealed by most crystal structures (Granzin et al. 1998; Hirsch et al. 1999; Han et al. 2001; Milano et al. 2002; Sutton et al. 2005; Zhan et al. 2011a), receptor-bound (Palczewski et al. 1991; Kim et al. 2012; Zhuang et al. 2013), and microtubule-bound (Hanson et al. 2006a, 2007). Upon binding to the receptor, arrestin undergoes a global conformational change (Schleicher et al. 1989; Gurevich and Benovic 1993; Hanson et al. 2006b; Kim et al. 2012; Zhuang et al. 2013). This rearrangement alters the set of exposed elements, thereby affecting the association of arrestins with their non-receptor-binding partners (Ahmed et al. 2011). Binding-induced conformational changes in arrestin could play decisive role in arrestin-mediated assembly of distinct signaling complexes in various physiological conditions (Ahmed et al. 2011). Two recent structures, one of truncated form of arrestin-2 [first described as “pre-activated” mutant in Celver et al. (2002)] associated with multi-phosphorylated C-terminus of vasopressin V2 receptor (Shukla et al. 2013) and the other of short arrestin-1 splice variant p44 also lacking the C-tail (Kim et al. 2013), are remarkably similar. Importantly, arrestin in both is not in complex with the receptor, for which it has sub-nanomolar to nanomolar affinity (Gurevich et al. 1993, 1995; Bayburt et al. 2011). Instead, it is either associated with phosphopeptide (Shukla et al. 2013), which has orders of magnitude lower affinity for arrestin, or not associated with any part of the receptor (Kim et al. 2013). It has been previously shown that the more “pre-activating” mutations are introduced into arrestin, the less stable the protein becomes (Schleicher et al. 1989; Hirsch et al. 1999; Sutton et al. 2005), suggesting that it is highly unlikely that fully activated arrestin conformation can be stable without bound receptor. Therefore, these structures likely do not represent the active receptor-bound conformation, but an intermediate step on the way to it. Nonetheless, these structures are very informative. Relative to the basal state, both feature a significant movement of the “139-loop,” previously shown to change position in response to receptor binding (Schleicher et al. 1989; Hirsch et al. 1999; Kim et al. 2012) and act as a “brake” reducing arrestin binding to non-preferred forms of the receptor (Vishnivetskiy et al. 2013), and the twisting of the two domains relative to each other by 20–21° (Kim et al. 2013; Shukla et al. 2013). Previous studies showed that an extended inter-domain hinge is necessary for receptor binding of arrestin-1 (Vishnivetskiy et al. 2002), as well as nonvisual arrestin-2 and arrestin-3 (Hanson et al. 2007). However, proposed on the basis of these data, clam-like domain movement (Gurevich and Gurevich 2004) was not detected by intramolecular distance measurements using pulse EPR technique DEER (Kim et al. 2012). The twisting of the two domains revealed by these structures (Kim et al. 2013; Shukla et al. 2013) represents an alternative movement that explains the requirement of a certain length of the hinge, which was suggested by reported deleterious effect of hinge deletions on receptor binding (Vishnivetskiy et al. 2002; Hanson et al. 2007).
Although arrestin was originally shown to act as a receptor-regulated scaffold for JNK3 activation (McDonald et al. 2000), the follow-up studies indicated that receptor stimulation is not obligatory for this arrestin function (Miller et al. 2001). JNK3 interactions with different arrestin conformations: basal, constitutively “pre-activated” form [3A mutant with forcibly detached C terminus (Gurevich 1998; Carter et al. 2005)]; and the mutant with the deletion of seven residues in the inter-domain hinge that is impaired in receptor binding (Δ7) (Vishnivetskiy et al. 2002; Hanson et al. 2007) were evaluated in a nuclear exclusion assay, based on the ability of arrestins with functional NES to remove their binding partners from the nucleus to the cytoplasm (Song et al. 2006, 2007). WT arrestins, as well as “pre-activated” 3A and non-receptor-binding Δ7 mutants, effectively relocated JNK3 (Song et al. 2006), indicating that JNK3 binding does not depend on arrestin conformation. Furthermore, the Δ7 mutant robustly promoted JNK3 activation, at least as well as WT (Song et al. 2009), whereas another mutant termed KNC, in which receptor interaction was precluded by the elimination of key GPCR-binding residues, demonstrated stronger binding to JNK3 than WT arrestin-3 (Breitman et al. 2012). Recently, we demonstrated direct JNK3 interaction with arrestins in the basal conformation using purified proteins in vitro (Zhan et al. 2011b, 2013). Importantly, in this strictly controlled system reconstructed from pure proteins, free arrestin-3 in the absence of any receptor functioned as a scaffold facilitating JNK3 phosphorylation by both MKK4 (Zhan et al. 2011b) and MKK7 (Zhan et al. 2013). However, these studies did not test the effect of arrestin binding to the receptor on its interactions with JNK3 or upstream kinases. To evaluate the effect of receptor on arrestin-JNK3 interaction, careful biochemical experiments with purified proteins need to be performed to measure the binding affinities of JNK3, MKK4, MKK7, and ASK1 for arrestin-3 in its basal conformation and bound to the receptor, as well as the efficiency of arrestin-3-mediated scaffolding this signaling cascade in the presence and absence of GPCRs.
Arrestins were reported to facilitate the activation of three main subfamilies of MAP kinases: JNK (McDonald et al. 2000), ERK (Luttrell et al. 2001), and p38 (Bruchas et al. 2006). In each cascade, in order to promote signaling, arrestins need to assemble appropriate combinations of MAPK, MAPKK, and MAPKKK. It is not clear how simultaneous association of mismatched kinases that would create unproductive complexes is prevented. One possibility, based on previous observations, is that the binding of one kinase to arrestin can significantly alter the recruitment of another. For example, MKK4, MAPKK in JNK3 activation cascade, demonstrates weaker binding to arrestin than ASK1 and JNK3 (McDonald et al. 2000; Song et al. 2009; Breitman et al. 2012). The binding of ASK1, JNK3, or both dramatically enhances MKK4 association with arrestin (McDonald et al. 2000; Song et al. 2009). Our recent studies showed that JNK3 binding differentially modulates the recruitments of the two upstream MAPKKs, MKK4 and MKK7, to arrestin-3, enhancing the binding of MKK4 while reducing that of MKK7 (Zhan et al. 2013). Although these results were obtained with kinases from the same module, the data suggest that interdependence of the binding of MAP kinases likely contributes to the assembly of signaling complexes containing matching kinases.
4 The Arrangement of ASK1, MKK4, and JNK3 on Arrestin
Arrestin-mediated JNK3 activation requires simultaneous recruitment of JNK3 and its upstream kinases. The original work (McDonald et al. 2000) proposed a model where both ASK1, the MAPKKK in JNK3 cascade, and JNK3 directly associate with arrestin-3, whereas MKK4 is recruited via ASK1 and/or JNK3 without directly interacting with arrestin-3. This model was mainly based on the observation that the interaction between MKK4 and arrestin-3 could not be detected by co-IP unless MKK4 was co-expressed with ASK1, JNK3, or both. However, direct interaction between MKK4 and arrestin-3 was demonstrated in follow-up studies by several different assays including co-IP (Song et al. 2009), direct pull-down using purified proteins (Zhan et al. 2011b, 2013; Kook et al. 2013), and MKK activity assay in reconstituted arrestin-3–MKK4–JNK3 (Zhan et al. 2011b), arrestin-3–MKK7–JNK3 (Zhan et al. 2013), as well as arrestin-3–MKK4/7–JNK1/2 (Kook et al. 2013) systems. Moreover, separated arrestin-3N- and C-domains were shown to bind each kinase in the ASK1–MKK4–JNK3 module (Song et al. 2009). Interestingly, the kinases of the ERK1/2 module (c-Raf1, MEK1, and ERK2) also bind equally well to both domains of arrestin-2 and arrestin-3 (Song et al. 2009). This appears to be a universal mode of assembly of the three kinases in MAPK cascades on arrestin scaffolds. Therefore, a different model of the MAPK cascade organized by arrestin has been proposed, which is based on the identification of multiple arrestin-binding elements in MAP kinases and localization of binding elements for each kinase on both N- and C-domains of nonvisual arrestins (Fig. 1). In this model, arrestin binds all three kinases directly, with each kinase interacting with both domains of arrestin. In fact, since the two arrestin domains apparently move relative to each other upon its binding to GPCRs (Schleicher et al. 1989; Gurevich and Gurevich 2004) and microtubules (Hanson et al. 2006a), the most straightforward mechanism that would make the binding of any protein sensitive to arrestin conformation is the engagement of elements in both arrestin domains. Although JNK3 is a rare example of a binding partner that does not show a clear preference for a particular arrestin conformation (Song et al. 2006, 2007), it appears to follow this general rule.
Fig. 1.
The arrangement of the three kinases on arrestin scaffold. MAPK activation modules consist of MAPKKK, MAPKK, and MAPK. The model shown is based on several lines of evidence. First, each of the three kinases in ASK1–MKK4–JNK3 and c-Raf1–MEK1–ERK2 signaling modules was shown to bind both arrestin domains (Song et al. 2009). Second, many purified kinases, such as JNK3 and MKK4 (Zhan et al. 2011b), MKK7 (Zhan et al. 2013), JNK1 and JNK2 (Kook et al. 2013), as well as MEK1 and ERK2 (Coffa et al. 2011a), were shown to bind arrestins directly. Receptor binding-deficient arrestin-3 mutant with 7-residue deletion in the inter-domain hinge facilitates ASK1-dependent JNK3 activation at least as efficiently as WT arrestin-3 (Song et al. 2009; Breitman et al. 2012), whereas arrestin-3-3A mutant with enhanced receptor binding (Celver et al. 2002) does not promote JNK3 activation (Breitman et al. 2012). Therefore, the model shows that not only receptor-bound but free arrestins can also scaffold MAP kinase cascades. Arr, arrestin scaffold; ASK, ASK1 (or any MAPKKK); MKK, MKK4/7 (or any MAPKK); J, JNK1/2/3 (or any MAPK)
5 Binding and Activation Are Two Distinct Phenomena
Earlier observations showed that arrestin-2 binds all three components in the ASK1–MKK4–JNK3 cascade (Song et al. 2009; Seo et al. 2011). However, in contrast to highly homologous arrestin-3 (Hanson et al. 2007; Ahmed et al. 2011), arrestin-2 and the two visual arrestins fail to facilitate the activation of JNK3 (Song et al. 2009). Thus, the ability to bind these kinase components simultaneously does not ensure the facilitation of JNK3 activation. The two nonvisual arrestins are highly homologous: bovine arrestin-2 and arrestin-3 have 78 % sequence identity and 88 % similarity (Sterne-Marr et al. 1993) and their crystal structures are also remarkably similar, particularly in the core arrestin fold (Han et al. 2001; Zhan et al. 2011a). Due to high structural similarity, arrestin chimeras usually remain fully functional (Gurevich et al. 1993, 1995; Vishnivetskiy et al. 2004; Hanson et al. 2006a; Ahmed et al. 2011). Several arrestin-2/3 chimeras and mutants constructed by swapping the elements between the two nonvisual arrestins were used to explore the structural basis of the ability of arrestin-3, but not arrestin-2, to facilitate JNK3 activation (Seo et al. 2011). Both domains of arrestin were found to contribute to JNK3 activation, with the C-domain being more important than the N-domain (Seo et al. 2011). In addition, it was shown that several residues on the non-receptor-binding side of arrestin-3 are critical for JNK3 activation: Val-343 is the key contributor to this function, whereas Leu-278, Ser-280, His-350, Asp-351, His-352, and Ile-353 play supporting roles (Seo et al. 2011). However, in contrast to many of arrestin-3 mutants containing arrestin-2 residues that have lost the ability to facilitate JNK3 phosphorylation, the efforts to build this function into arrestin-2 by replacing these critical residues with the corresponding ones from arrestin-3 were not particularly successful (Seo et al. 2011). Interestingly, virtually all arrestin-3 mutants deficient in their ability to promote JNK3 phosphorylation were shown to bind ASK1, MKK4, and JNK3 normally (Seo et al. 2011).
The observations that arrestin binding of kinases of JNK3 signaling cascade does not necessarily translate into the ability to facilitate JNK3 activation suggest that to promote signaling arrestin has to assemble the kinases in optimal relative orientation. Several arrestin-3 mutants have been generated which bind ASK1, MKK4, and JNK3 at least as well as WT arrestin-3 but fail to facilitate JNK3 activation (Seo et al. 2011; Breitman et al. 2012). Arguably, the most interesting and potentially useful of these is arrestin-3-KNC mutant, which binds JNK3 even better than WT and interacts with the two upstream kinases normally, but does not facilitate JNK3 activation in cells (Breitman et al. 2012). Because KNC mutant binds ASK1, MKK4, and JNK3 very effectively, it competes with WT arrestin-3 and other JNK scaffold proteins, thereby acting as a “silent scaffold,” suppressing JNK3 activation in the cell (Breitman et al. 2012).
To improve our understanding of the relationship between binding and scaffolding functions of arrestins, the interactions of arrestin-3 with each kinase in the cascade need to be characterized quantitatively. The relative orientation of the three kinases within the complex with arrestin-3 might be critical for JNK3 activation, but the differences in binding affinities can play an important role, as well. Most of the studies reporting the binding of arrestin to the kinases in JNK3 cascade used co-immunoprecipitation or cell-based BRET assays (McDonald et al. 2000; Song et al. 2009; Seo et al. 2011; Breitman et al. 2012). Considering that in intact cells arrestins can interact with wide variety of proteins (Xiao et al. 2007), it is hard to derive actual binding affinities from these largely qualitative observations or even ascertain that the interactions between arrestins and their partners are direct. To compare the interactions of WT arrestins and their mutants with each kinase, binding affinities should be measured directly using purified proteins. For example, relatively weak interactions of arrestins with MKK4 are hard to measure, and this binding is regulated significantly by the presence or absence of other kinases (Zhan et al. 2013). The MKK4 binding to the two nonvisual arrestins was believed to be comparable until the direct pull-down assay with purified proteins suggested otherwise: arrestin-3 binds MKK4 with higher affinity than arrestin-2 (Zhan et al. 2011b). These differences in binding affinity are consistent with the functional assay with reconstructed arrestin–MKK4–JNK3 modules, in which optimal concentration of arrestin-2 for maximal JNK3 activation was much higher than that of arrestin-3 (Zhan et al. 2011b). The optimal concentration of arrestins, and other scaffolds, is highly dependent on the binding affinities of the components (Burack and Shaw 2000).
Bringing all components into close proximity and holding them in optimal orientation are the two basic functions of scaffolding proteins that facilitate signal transduction (Dhanasekaran et al. 2007; Good et al. 2011). Tethering increases the effective local concentration of enzymes and their substrates, prevents the competition of other molecules, and keeps the specificity in signal transduction. Scaffold proteins can also direct the signal transduction through properly orienting target proteins with upstream enzymes, accelerating reactions manifold (Schlosshauer and Baker 2004). The evidence suggests that binding affinities along with the ability to ensure proper orientation of the kinases involved contribute to the subtype-specific facilitation of JNK3 activation by arrestin-3.
6 Reconstruction of Arrestin–MAP2K–MAPK Modules from Pure Proteins: Arrestins Are True Scaffolds
Although cell-based systems have been valuable tools to probe the roles of scaffold proteins in MAP kinase signaling, they have their limits. First, the number of exogenous components introduced into the cell is limited. As more cDNAs are introduced to study the signaling complex, co-transfection by all becomes less certain. Second, it is hard to control the expression level of each component, which changes upon co-expression of others, so that the amount of cDNA used needs to be adjusted for each combination. Besides, some proteins are less stable than others, which limits their expression levels, sometimes to the point of making their effects undetectable (Breitman et al. 2012). Third, it is virtually impossible to avoid the interference from unknown factors in the cellular milieu. Strictly controlled systems reconstructed from purified proteins avoid many inherent problems of cell-based assays, and often provide more direct and quantitative answers, helping to elucidate the underlying molecular mechanisms.
The reconstruction of arrestin-MKK4-JNK3 module from purified proteins has been reported recently, and the effects of arrestins on JNK3 phosphorylation by MKK4 have been carefully examined (Zhan et al. 2011b). JNK3α2 phosphorylation was shown to be a biphasic function of the concentration of both nonvisual arrestins. At lower arrestin concentrations, the JNK3α2 phosphorylation was enhanced, but it was inhibited by higher concentrations (Zhan et al. 2011b). Previous mathematical modeling suggested that the biphasic dependence of signaling efficiency on the concentration of a scaffold is observed when scaffold protein binds each component directly (Levchenko et al. 2000, 2004). Experiments with pure proteins proved that arrestin binds both MKK4 and JNK3 simultaneously, forming a ternary complex (Zhan et al. 2011b). In the reconstructed system (Fig. 2), JNK3 and MKK4 can exist in three states: associated in solution without scaffolds (JM), bound to scaffold protein not associated with the other kinase to form an incomplete complex (AM and AJ), and bound to scaffold simultaneously with the other kinase in an active signaling complex (JAM). Comparing to JM formed when JNK3 and MKK4 encounter each other by diffusion, the complete signaling complex JAM is a high output pathway, whereas the formation of incomplete complexes AM and AJ reduces the probability of the interaction of freely diffusing MKK4 and JNK3. An increase in scaffold concentration in the lower range enhances the formation of JAM complex, thereby facilitating the activation of the downstream kinase. In contrast, a further increase in scaffold concentration increases the probability of downstream and upstream kinase associating with the scaffold protein alone, forming incomplete inactive complexes (AM and AJ in Fig. 2). Therefore, the activation of MAPK by its upstream kinase can be inhibited by high concentrations of scaffolds. This has been demonstrated experimentally for kinase suppressor of Ras 1 (KSR1)-dependent ERK activation (Kortum and Lewis 2004), as well as JNK1/2/3 activation assisted by arrestin-3 (Zhan et al. 2011b, 2013; Kook et al. 2013). The optimal scaffold concentration for maximal JNK3 activation is highly dependent on several parameters. One is the binding affinity of the two kinases for arrestin and possible cooperativity between the binding of JNK3 and MKK4 to the scaffold. Stronger binding (lower KD) results in lower optimal scaffold concentration (Zhan et al. 2013). Therefore, higher optimal concentration of arrestin-2 than arrestin-3 observed in vitro was consistent with the direct binding assay, where arrestin-3 demonstrated stronger binding for MKK4 and JNK3 than arrestin-2 (Zhan et al. 2011b).
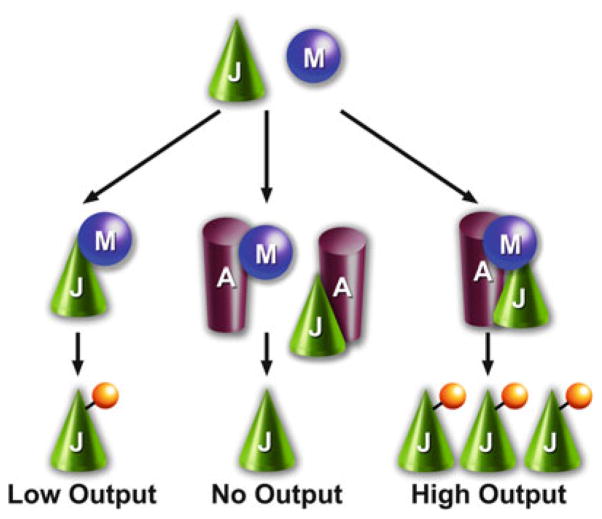
Fig. 2.
Scaffold concentration matters. Signaling can occur in the absence of scaffolds (the formation of JM complex; low output) and optimum scaffold concentrations promote the formation of complete complexes (JAM), thereby facilitating signaling (high output), whereas supraoptimal levels of scaffold make the formation of incomplete complexes (JA and AM) more likely, suppressing signaling in the cascade (no output). Biphasic effect of scaffold concentration on signaling was predicted by mathematical modeling (Levchenko et al. 2000, 2004) and experimentally demonstrated in case of arrestin-3 scaffolding of signaling modules that activate JNK family kinases (Zhan et al. 2011b, 2013; Kook et al. 2013). A, arrestin-3; M, MKK4/7; J, JNK1/2/3
The reconstruction of signaling system from pure proteins also proved the existence of another relevant module scaffolded by arrestin-3: arrestin-3–MKK7–JNK3. JNK family kinases are activated by concomitant phosphorylation of a threonine and a tyrosine residue within a conserved Thr–Pro–Tyr (TPY) motif in the activation loop of the kinase domain. Two upstream MAP kinase kinases, MKK4 and MKK7, preferentially phosphorylate distinct JNK activation sites: MKK4 phosphorylates tyrosine, whereas MKK7 phosphorylates threonine (Lawler et al. 1998). Arrestin-3-dependent increase in the phosphorylation of Thr on JNK3α2 was demonstrated both in vitro and in intact cells (Zhan et al. 2013), which suggested that arrestin-3 can recruit MKK7 to activate JNK3 as well. Direct interaction between arresitn-3 and MKK7 in the absence of JNK3 has been confirmed by pull-down assay (Zhan et al. 2013). The binding to arrestin-3 of the active (phosphorylated) MKK4 and MKK7 was also evaluated in the reconstructed system. Both active MKKs bind arrestin-3 at the level comparable to that of inactive forms of these two kinases (Zhan et al. 2013). This is consistent with the model that these MKKs bind arrsetin-3 along with their upstream activator ASK1, which then phosphorylates them, whereupon they can phosphorylate their downstream substrates, JNKs. Unexpectedly, we found that the binding of downstream kinase JNK3 differentially affects the recruitment of these two MKKs, enhancing the binding of MKK4, while decreasing the binding of MKK7 (Zhan et al. 2013). As could be expected (Fig. 2), the activation of JNK3 by MKK7 demonstrated biphasic dependence on arrestin-3 concentration (Zhan et al. 2013), similar to that observed with MKK4 (Zhan et al. 2011b). However, arrestin-3 concentration optimal for JNK3 phosphorylation by MKK7 was eight- to tenfold higher than in case of MKK4 (Zhan et al. 2013). This finding is in excellent agreement with the negative cooperativity revealed by direct binding assay (Zhan et al. 2013). Mathematical models predict that positive binding cooperativity between the two components lowers the optimal scaffold concentration, whereas negative cooperativity increases the optimal scaffold concentration (Bray and Lay 1997; Ferrell 2000; Levchenko et al. 2000, 2004).
Thus, experiments with pure proteins yielded information that remained unattainable otherwise. Reconstruction of complete MAPKKK–MAPKK–MAPK modules with pure proteins in the presence or absence of arrestins must be performed next. The situation involving three kinases that sequentially phosphorylate and activate each other is more complex. Biochemical assays under strictly controlled conditions are powerful tools for elucidating the mechanisms of arrestin-mediated scaffolding of MAP kinase cascades. So far, these studies were limited to MAPKK–MAPK modules (Zhan et al. 2011b, 2013; Kook et al. 2013). The addition of MAPKKK and other regulators, such as GPCRs, will provide novel valuable information.
7 Suppression of JNK3 Activation by Arrestin-3-Based Silent Scaffold
JNK signaling is involved in the normal physiological processes of cell proliferation, apoptosis, differentiation, and cell migration (Davis 2000; Karin and Gallagher 2005). JNK3, a brain-specific isoform of JNKs, has been implicated in neurodegenerative diseases, such as Parkinson’s (Hunot et al. 2004), Alzheimer’s (Yoon et al. 2013), and Huntington’s (Morfini et al. 2009). These studies suggest that inhibition of JNK3 activity could be a promising therapeutic intervention for neurodegenerative diseases.
JNK3, like other MAP kinases, is activated by MAPKKs (concerted action of MKK4 or MKK7 is required in this case) (Lawler et al. 1998; Tournier et al. 2001), which in turn are activated by MAPKKKs (Davis 2000). To confer spatial and temporal regulation of the JNK signaling by multiple stimuli, cells have developed a class of proteins that function as specialized scaffolds. Protein scaffolds assemble signaling modules by binding multiple components of the MAPK cascade, bringing them into close proximity, thereby facilitating efficient propagation of the signal (Brown and Sacks 2009). Arrestin-3 was reported to function as scaffold protein for JNK3 signaling, promoting JNK3 phosphorylation by binding ASK1, MKK4, MKK7, and JNK3 (McDonald et al. 2000; Miller et al. 2001; Song et al. 2009; Seo et al. 2011; Zhan et al. 2011b, 2013). Several arrestin-3 mutants bind ASK1, MKK4, and JNK3 at least as well as WT arrestin-3, but do not facilitate JNK3 phosphorylation in cells (Seo et al. 2011). These data suggest that one can engineer an arrestin-3 mutant that suppresses the signaling in this cascade simply by recruiting the kinases into unproductive complexes, thereby keeping them away from productive scaffolds (Fig. 3).
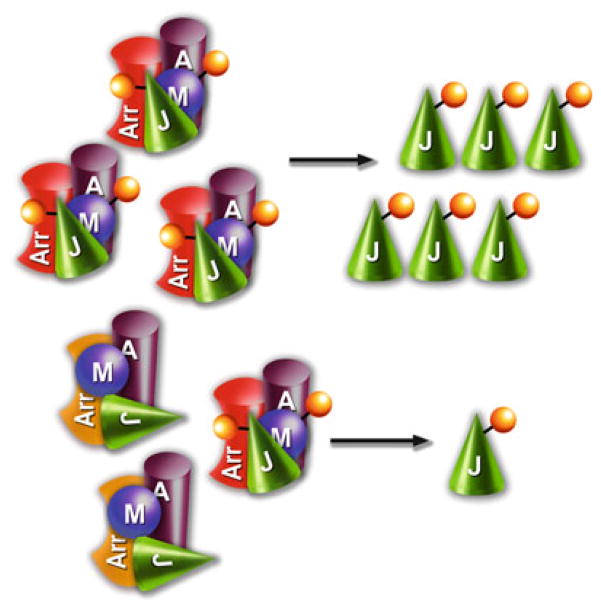
Fig. 3.
Silent scaffolds can suppress MAPK signaling. Upper panel. By assembling multi-kinase complexes, scaffolds can greatly enhance signaling in MAPK pathways (Brown and Sacks 2009). Lower panel. Scaffolding proteins with targeted mutations that bind all kinases but do not facilitate their activation by sequential phosphorylation recruit the kinases away from endogenous productive scaffolds, thereby acting as dominant-negative “silent scaffolds” that suppress the signaling in the cascade (Breitman et al. 2012). The simplest conceivable mechanism of this effect is that silent scaffolds bind kinases but hold them in “wrong” configuration that is not conducive to phosphorylation, which is reflected by different arrangement of the kinases on the silent scaffold shown. Arr red, WT arrestin (or any productive scaffold); Arr yellow, mutant arrestins (or any reengineered scaffold that binds kinases but does not promote the signaling); A, ASK1 (or any MAPKKK); M, MKK4/7 (or any MAPKK); J, JNK1/2/3 (or any MAPK)
This idea was recently tested and found to be correct (Breitman et al. 2012). Arrestin-3-KNC mutant, where 12 known receptor-binding residues were replaced with alanines (Vishnivetskiy et al. 2011; Gimenez et al. 2012), binds upstream kinases ASK1 and MKK4 normally and demonstrates stronger binding to JNK3 than WT arrestin-3 in cells, as shown by both arrestin-3-JNK3 BRET and co-immunoprecipitation, but does not have the ability to promote JNK3 activation (Breitman et al. 2012). As expected, arrestin-3-KNC mutant acts as a dominant-negative silent scaffold (Fig. 3): its increasing expression progressively decreases JNK3 phosphorylation in the cell induced by WT arrestin-3, apparently via sequestration of all three kinases into unproductive complexes (Breitman et al. 2012). Many experimental and modeling studies suggested that selective modulation of the interaction between scaffolds and individual targets might enable specific regulation of MAPK activity, directing the cellular response towards (or away from) a particular function, without attenuating global MAPK activity (Brown and Sacks 2009). However, arrestin-3-KNC is the first molecular tool that was developed on the basis of this concept, specifically for the suppression of JNK3 signaling in living cells.
Spatial and temporal changes in MAPK signaling affect cellular response to a specific stimulus and are very important for biological specificity of MAPKs. Many scaffold proteins appear to have a fundamental role in the spatial regulation of MAPK signaling (Morishima-Kawashima and Kosik 1996; Li et al. 2005; Brown and Sacks 2009). Most of activated JNK family kinases tend to translocate into the nucleus and phosphorylate nuclear substrates, such as c-Jun, anti-activating transcription factor (ATF-1), JunB, and JunD, thereby inducing JNK-dependent gene expression (Bogoyevitch and Kobe 2006). Interestingly, JNK3 activated by arrestin-3 remains in the cytosol (McDonald et al. 2000; Breitman et al. 2012), where it likely phosphorylates other cellular proteins, possibly those implicated in apoptotic cell death, such as Bcl2, p53, etc. (Fuchs et al. 1998; Yamamoto et al. 1999; Buschmann et al. 2001; Deng et al. 2001; Bogoyevitch and Kobe 2006). In cells expressing arrestin-3-KNC or arrestin3-3A mutants which do not facilitate JNK3 activation and likely inhibit its activity in the cytosol, most of phospho-JNK (likely activated via arrestin-independent mechanisms) was detected in nucleus (Breitman et al. 2012). Thus, arrestin-3-KNC and other cytoplasmic silent scaffolds appear to be suitable tools to regulate cellular JNK3 activity in compartment-specific manner, whereas similar dominant-negative scaffolds with nuclear localization can regulate MAPK signaling specifically in that compartment. Arrestin-3-KNC is not a perfect tool for targeted manipulation of JNK3-specific signaling, because it simultaneously decreases the activation of both JNK3 and ERK1/2 (Breitman et al. 2012). Nonetheless, the development and functional validation of the arrestin-3-based silent scaffold is an important proof-of-concept experiment. The construction of molecular tools of this type creates new methods for precisely targeted spatial and temporal regulation of MAPK signaling. Further experiments are necessary to test whether arrestin-3-KNC similarly reduces the activation of JNK1/2 in the cell and whether the suppression of JNK signaling by this mutant translates into increased cell survival in culture and in vivo.
8 Arrestin-Dependent Activation of JNK1/2 Isoforms
The interaction of arrestin-3 and JNK3 was originally detected in yeast two-hybrid screen and confirmed using co-immunoprecipitation from cultured cells (McDonald et al. 2000). Subsequently, the ability of arrestin-3 to promote JNK3 phosphorylation was demonstrated by several groups using cell-based assays (Miller et al. 2001; Song et al. 2009; Seo et al. 2011) and in vitro reconstitution with purified proteins (Zhan et al. 2011b, 2013). Arrestin-3 is ubiquitously expressed (Attramadal et al. 1992; Sterne-Marr et al. 1993; Gurevich and Gurevich 2006a), whereas JNK3, which was first reported to be activated in arrestin-3-dependent manner, has more limited distribution, being expressed predominantly in neurons, heart, and testes (Gupta et al. 1996; Davis 2000). In contrast, different isoforms of JNK1 and JNK2 are expressed as ubiquitously as arrestin-3 (Gupta et al. 1996; Davis 2000). Even though JNK3 has unique extended N terminus of 38 amino acids that was reported to be the main arrestin-3-binding site (Guo and Whitmarsh 2008; Song et al. 2009), high level of sequence conservation among JNK isoforms raises the question whether arrestin-3 regulates the activity of other JNK family kinases. Twelve main isoforms of JNK are generated by alternative splicing of three genes (Gupta et al. 1996; Davis 2000). The analysis of the knockout of individual JNK genes in mice revealed that different isoforms have distinct, although partially overlapping functions (Yang et al. 1997; Tournier et al. 2000, 2001; Kuan et al. 2003; Hunot et al. 2004). To test whether arrestin-3 is involved in the regulation of JNK1/2 isoforms, direct interaction between JNK1/2 and arrestin-3 was probed using purified protein in vitro (Kook et al. 2013). These experiments showed that the amount of arrestin-3 retained by His-tagged JNK2α2 is similar to that retained by His-tagged JNK3α2, whereas JNK1α1 shows a weaker binding to arrestin-3 in this paradigm (Kook et al. 2013). These in vitro binding data were supported by co-immunoprecipitation of arrestin-3 with these JNKs from cells, suggesting that arrestin-3 might be involved in the regulation of ubiquitous JNK isoforms. Next, MKK4/7–JNK1α1/JNK2α2 signaling modules were reconstructed with pure proteins in vitro with and without arrestin-3. These experiments revealed that the phosphorylation of JNK1/2 by both MKK4 and MKK7 is enhanced in the presence of an optimal arrestin-3 concentration (Kook et al. 2013), similar to the effect of purified arrestin-3 on JNK3α2 phosphorylation by MKK4/7 (Zhan et al. 2011b, 2013). Importantly, the biphasic dependence of JNK1α1/ JNK2α2 phosphorylation by either MKK4 or MKK7 on arrestin-3 concentration in vitro suggests that arrestin-3 acts as a scaffold for MKK4/7-JNK1/2 signaling modules acting by bringing the two kinases to each other (Levchenko et al. 2000, 2004). Usually, it is hard to demonstrate the biphasic effect of scaffolding protein in intact cells, but in this case, it was shown that the activation of several isoforms of endogenous JNK1/2 by ASK1, MKK4, and MKK7 increases with the arrestin-3 expression up to a point, whereupon it is reduced by higher expression levels (Kook et al. 2013). These findings demonstrate that arrestin-3 promotes the activation of several isoforms of all three types of JNK kinases and can positively or negatively regulate JNK signaling in the majority of cell types.
9 Prospects of Manipulation of Cell Death and Survival by Signaling-Biased Arrestins
Despite remarkable recent progress in our understanding of arrestin-3 role in JNK activation (Zhan et al. 2011b, 2013; Kook et al. 2013), it remains to be elucidated how arrestin-3-dependent regulation affects cellular responses mediated by JNK signaling. JNK pathways regulate many vital cellular processes, including cell death and survival (Davis 2000; Tournier et al. 2000; Weston and Davis 2007). In particular, JNK3 activation is believed to play a key role in triggering cell death, thereby participating in the pathogenesis of several human neurodegenerative disorders (Hunot et al. 2004; Morfini et al. 2009; Yoon et al. 2013). Neurons derived from JNK3-deficient mice are more resistant to Aβ-induced apoptosis than neurons from wild-type mice (Morishima et al. 2001), and JNK phosphorylation (which reflects activity) in human postmortem brains from Alzheimer’s disease patients is markedly increased (Zhu et al. 2001). These reports suggest that modulation of JNK3 activation in Alzheimer’s disease is a possible therapeutic target. Parkinson’s disease (PD) is a neurodegenerative disorder characterized by the progressive loss of dopaminergic neurons in the substantia nigra. Increased activation of the downstream target of JNK3, transcription factor c-Jun, is detected in mice treated with the neurotoxin 1-methyl-4-phenyl-1,2,4,6-tetrahydropyridine (MPTP) that specifically destroys dopaminergic neurons (Hunot et al. 2004) and in human postmortem brains from PD patients (Hunot et al. 2004). Mice deficient in JNK2 and JNK3 are more resistant to MPTP-induced injury than WT littermates, whereas JNK1 knockout mice show the same susceptibility to MPTP as controls (Hunot et al. 2004). Single JNK3 knockout mice also show neuroprotection against brain injury after cerebral ischemia-hypoxia (Kuan et al. 2003) and against excitotoxicity of the glutamate receptor agonist kainic acid (Yang et al. 1997). This evidence suggests that the inhibition of JNK activity, particularly the activity of JNK3, is a promising therapeutic approach to the treatment of neurodegenerative diseases. Several research laboratories have pursued small molecule inhibitors of JNK for therapeutic purposes, but most of the inhibitors found so far are not sufficiently specific, as they inhibit other kinases besides JNKs (Bennett et al. 2001; Scapin et al. 2003; Carboni et al. 2004; Resnick and Fennell 2004; Wang et al. 2004; Sabapathy 2012) A cell-permeable peptide inhibitor containing a 21-amino acid element of JIP-1 that interacts with JNKs (Barr et al. 2002) protects against cerebral ischemic injury in rodent model (Borsello et al. 2003). This example strongly supports the idea that molecular tools based on modified proteins, such as arrestin-3 mutant that acts as a dominant-negative silent scaffold (Breitman et al. 2012), have therapeutic potential as negative regulators of proapoptotic JNK signaling.
Considering how many diverse cellular functions are controlled by JNKs, one of the major issues that need to be resolved to enable therapeutic manipulation of JNK signaling is how to ensure high specificity of JNK modulation and to avoid affecting multiple processes. Catalytic domains of all JNK isoforms of each subfamily are identical, and even in the three JNK subfamilies, they are highly homologous (Bogoyevitch and Kobe 2006), which suggests that it is virtually impossible to achieve specificity by targeting ATP-binding site or other elements of the catalytic domain with small molecules. In contrast, different JNKs have distinct regulatory elements on their N- and C-termini, which mediate their interactions with upstream kinases, substrates, and scaffolding proteins. Scaffolds are important spatial and temporal regulators of JNK signaling. Scaffolds with distinct subcellular distribution and the ability to bind selected JNK isoforms, which can be either productive or silent, have potential to enhance or inhibit JNK signaling in a specific cellular compartment to regulate distinct cellular responses. Productive scaffolds facilitate signaling by assembling individual components of MAPK cascade in correct orientation. However, supraoptimal levels of even productive scaffolds facilitate the formation of incomplete complexes, thereby blocking the signaling by sequestering these components away from potentially productive alternative signaling complexes, as was first suggested by mathematical modeling (Levchenko et al. 2000, 2004) and then recently shown experimentally (Zhan et al. 2011b, 2013; Kook et al. 2013). The identification of distinct binding sites for different JNK isoforms on arrestin-3 paves the way to designing small peptide inhibitors of the activation of individual JNKs. Signaling-biased arrestin-3 mutants, such as arrestin-3-KNC (Breitman et al. 2012) or other forms deficient in JNK activation (Seo et al. 2011), can be used to inhibit JNK signaling in cells. Moreover, arrestin-3- and other protein-based tools can be equipped with sequences that target them to specific subcellular compartments, such as cytosol or nucleus (Scott et al. 2002; Wang et al. 2003; Song et al. 2006). The construction of mutant scaffolds with signaling capability biased towards or against specific signaling pathways will allow more subtle and targeted modulation of MAPK pathways. This strategy can be used to correct signaling dysregulated in different pathological states, as well as to channel the signaling to pathways that have therapeutic potential even in cases where other molecular errors underlie the pathology.
Footnotes
Different systems of arrestin names are used in the field and in this book. We use systematic names of arrestin proteins: arrestin-1 (historic names S-antigen, 48 kDa protein, visual or rod arrestin), arrestin-2 (β-arrestin or β-arrestin1), arrestin-3 (β-arrestin2 or hTHY-ARRX), and arrestin-4 (cone or X-arrestin; for unclear reasons, its gene is called “arrestin-3” in the HUGO database).
Contributor Information
Xuanzhi Zhan, Email: xuanzhi.zhan@vanderbilt.edu.
Seunghyi Kook, Email: seunghyi.kook@vanderbilt.edu.
Eugenia V. Gurevich, Email: eugenia.gurevich@vanderbilt.edu.
Vsevolod V. Gurevich, Email: vsevolod.guervich@vanderbilt.edu.
References
- Ahmed MR, Zhan X, Song X, Kook S, Gurevich VV, Gurevich EV. Ubiquitin ligase parkin promotes Mdm2-arrestin interaction but inhibits arrestin ubiquitination. Biochemistry. 2011;50:3749–63. doi: 10.1021/bi200175q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attramadal H, Arriza JL, Aoki C, Dawson TM, Codina J, Kwatra MM, Snyder SH, Caron MG, Lefkowitz RJ. Beta-arrestin2, a novel member of the arrestin/beta-arrestin gene family. J Biol Chem. 1992;267:17882–90. [PubMed] [Google Scholar]
- Barak LS, Ferguson SS, Zhang J, Caron MG. A beta-arrestin/green fluorescent protein biosensor for detecting G protein-coupled receptor activation. J Biol Chem. 1997;272:27497–500. doi: 10.1074/jbc.272.44.27497. [DOI] [PubMed] [Google Scholar]
- Barr RK, Kendrick TS, Bogoyevich MA. Identification of the critical features of a small peptide inhibitor of JNK activity. J Biol Chem. 2002;277:10987–97. doi: 10.1074/jbc.M107565200. [DOI] [PubMed] [Google Scholar]
- Bayburt TH, Vishnivetskiy SA, McLean M, Morizumi T, C-c H, Tesmer JJ, Ernst OP, Sligar SG, Gurevich VV. Rhodopsin monomer is sufficient for normal rhodopsin kinase (GRK1) phosphorylation and arrestin-1 binding. J Biol Chem. 2011;286:1420–8. doi: 10.1074/jbc.M110.151043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett BL, Sasaki DT, Murray BW, O’Leary EC, Sakata ST, Xu W, Leisten JC, Motiwala A, Pierce S, Satoh Y, Bhagwat SS, Manning AM, Anderson DW. SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc Natl Acad Sci USA. 2001;98:13681–6. doi: 10.1073/pnas.251194298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogoyevitch MA, Kobe B. Uses for JNK: the many and varied substrates of the c-Jun N-terminal kinases. Microbiol Mol Biol Rev. 2006;70:1061–95. doi: 10.1128/MMBR.00025-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsello T, Clarke PG, Hirt L, Vercelli A, Repici M, Schorderet DF, Bogousslavsky J, Bonny C. A peptide inhibitor of c-Jun N-terminal kinase protects against excitotoxicity and cerebral ischemia. Nat Med. 2003;9:1180–6. doi: 10.1038/nm911. [DOI] [PubMed] [Google Scholar]
- Bray D, Lay S. Computer-based analysis of the binding steps in protein complex formation. Proc Natl Acad Sci USA. 1997;94:13493–8. doi: 10.1073/pnas.94.25.13493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitman M, Kook S, Gimenez LE, Lizama BN, Palazzo MC, Gurevich EV, Gurevich VV. Silent scaffolds: inhibition of c-Jun N-terminal kinase 3 activity in the cell by a dominant-negative arrestin-3 mutant. J Biol Chem. 2012;287:19653–64. doi: 10.1074/jbc.M112.358192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MD, Sacks DB. Protein scaffolds in MAP kinase signalling. Cell Signal. 2009;21:462–9. doi: 10.1016/j.cellsig.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruchas MR, Macey TA, Lowe JD, Chavkin C. Kappa opioid receptor activation of p38 MAPK is GRK3- and arrestin-dependent in neurons and astrocytes. J Biol Chem. 2006;281:18081–9. doi: 10.1074/jbc.M513640200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burack WR, Shaw AS. Signal transduction: hanging on a scaffold. Curr Opin Cell Biol. 2000;12:211–6. doi: 10.1016/s0955-0674(99)00078-2. [DOI] [PubMed] [Google Scholar]
- Buschmann T, Potapova O, Bar-Shira A, Ivanov VN, Fuchs SY, Henderson S, Fried VA, Minamoto T, Alarcon-Vargas D, Pincus MR, Gaarde WA, Holbrook NJ, Shiloh Y, Ronai Z. Jun NH2-terminal kinase phosphorylation of p53 on Thr-81 is important for p53 stabilization and transcriptional activities in response to stress. Mol Cell Biol. 2001;21:2743–54. doi: 10.1128/MCB.21.8.2743-2754.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carboni S, Hiver A, Szyndralewiez C, Gaillard P, Gotteland JP, Vitte PA. AS601245 (1,3-benzothiazol-2-yl (2-[[2-(3-pyridinyl) ethyl] amino]-4 pyrimidinyl) acetonitrile): a c-Jun NH2-terminal protein kinase inhibitor with neuroprotective properties. J Pharmacol Exp Ther. 2004;310:25–32. doi: 10.1124/jpet.103.064246. [DOI] [PubMed] [Google Scholar]
- Carman CV, Benovic JL. G-protein-coupled receptors: turn-ons and turn-offs. Curr Opin Neurobiol. 1998;8:335–44. doi: 10.1016/s0959-4388(98)80058-5. [DOI] [PubMed] [Google Scholar]
- Carter JM, Gurevich VV, Prossnitz ER, Engen JR. Conformational differences between arrestin2 and pre-activated mutants as revealed by hydrogen exchange mass spectrometry. J Mol Biol. 2005;351:865–78. doi: 10.1016/j.jmb.2005.06.048. [DOI] [PubMed] [Google Scholar]
- Celver J, Vishnivetskiy SA, Chavkin C, Gurevich VV. Conservation of the phosphate-sensitive elements in the arrestin family of proteins. J Biol Chem. 2002;277:9043–8. doi: 10.1074/jbc.M107400200. [DOI] [PubMed] [Google Scholar]
- Coffa S, Breitman M, Spiller BW, Gurevich VV. A single mutation in arrestin-2 prevents ERK1/2 activation by reducing c-Raf1 binding. Biochemistry. 2011a;50:6951–8. doi: 10.1021/bi200745k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffa S, Breitman M, Hanson SM, Callaway K, Kook S, Dalby KN, Gurevich VV. The Effect of Arrestin Conformation on the Recruitment of c-Raf1, MEK1, and ERK1/2 Activation. PLoS One. 2011b;6:e28723. doi: 10.1371/journal.pone.0028723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RJ. Signal transduction by the JNK group of MAP kinases. Cell. 2000;103:239–52. doi: 10.1016/s0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- Deng X, Xiao L, Lang W, Gao F, Ruvolo P, May WS., Jr Novel role for JNK as a stress-activated Bcl2 kinase. J Biol Chem. 2001;276:23681–8. doi: 10.1074/jbc.M100279200. [DOI] [PubMed] [Google Scholar]
- Dhanasekaran DN, Kashef K, Lee CM, Xu H, Reddy EP. Scaffold proteins of MAP-kinase modules. Oncogene. 2007;26:3185–202. doi: 10.1038/sj.onc.1210411. [DOI] [PubMed] [Google Scholar]
- Ferrell JEJ. What do scaffold proteins really do? Sci STKE 2000. 2000:pe1. doi: 10.1126/stke.2000.52.pe1. [DOI] [PubMed] [Google Scholar]
- Fuchs SY, Adler V, Pincus MR, Ronai Z. MEKK1/JNK signaling stabilizes and activates p53. Proc Natl Acad Sci USA. 1998;95:10541–6. doi: 10.1073/pnas.95.18.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez LE, Vishnivetskiy SA, Baameur F, Gurevich VV. Manipulation of very few receptor discriminator residues greatly enhances receptor specificity of non-visual arrestins. J Biol Chem. 2012;287:29495–505. doi: 10.1074/jbc.M112.366674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good MC, Zalatan JG, Lim WA. Scaffold proteins: hubs for controlling the flow of cellular information. Science. 2011;332:680–6. doi: 10.1126/science.1198701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman OB, Jr, Krupnick JG, Santini F, Gurevich VV, Penn RB, Gagnon AW, Keen JH, Benovic JL. Beta-arrestin acts as a clathrin adaptor in endocytosis of the beta2-adrenergic receptor. Nature. 1996;383:447–50. doi: 10.1038/383447a0. [DOI] [PubMed] [Google Scholar]
- Granzin J, Wilden U, Choe HW, Labahn J, Krafft B, Buldt G. X-ray crystal structure of arrestin from bovine rod outer segments. Nature. 1998;391:918–21. doi: 10.1038/36147. [DOI] [PubMed] [Google Scholar]
- Guo C, Whitmarsh AJ. The {beta}-arrestin-2 scaffold protein promotes c-jun N-terminal kinase-3 activation by binding to its nonconserved N terminus. J Biol Chem. 2008;283:15903–11. doi: 10.1074/jbc.M710006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Barrett T, Whitmarsh AJ, Cavanagh J, Sluss HK, Dérijard B, Davis RJ. Selective interaction of JNK protein kinase isoforms with transcription factors. EMBO J. 1996;15:2760–70. [PMC free article] [PubMed] [Google Scholar]
- Gurevich VV. The selectivity of visual arrestin for light-activated phosphorhodopsin is controlled by multiple nonredundant mechanisms. J Biol Chem. 1998;273:15501–6. doi: 10.1074/jbc.273.25.15501. [DOI] [PubMed] [Google Scholar]
- Gurevich VV, Benovic JL. Visual arrestin interaction with rhodopsin: sequential multisite binding ensures strict selectivity towards light-activated phosphorylated rhodopsin. J Biol Chem. 1993;268:11628–38. [PubMed] [Google Scholar]
- Gurevich VV, Gurevich EV. The molecular acrobatics of arrestin activation. Trends Pharmacol Sci. 2004;25:59–112. doi: 10.1016/j.tips.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Gurevich EV, Gurevich VV. Arrestins are ubiquitous regulators of cellular signaling pathways. Genome Biol. 2006a;7:236. doi: 10.1186/gb-2006-7-9-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich VV, Gurevich EV. The structural basis of arrestin-mediated regulation of G protein-coupled receptors. Pharmacol Ther. 2006b;110:465–502. doi: 10.1016/j.pharmthera.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich VV, Richardson RM, Kim CM, Hosey MM, Benovic JL. Binding of wild type and chimeric arrestins to the m2 muscarinic cholinergic receptor. J Biol Chem. 1993;268:16879–82. [PubMed] [Google Scholar]
- Gurevich VV, Dion SB, Onorato JJ, Ptasienski J, Kim CM, Sterne-Marr R, Hosey MM, Benovic JL. Arrestin interaction with G protein-coupled receptors. Direct binding studies of wild type and mutant arrestins with rhodopsin, b2-adrenergic, and m2 muscarinic cholinergic receptors. J Biol Chem. 1995;270:720–31. doi: 10.1074/jbc.270.2.720. [DOI] [PubMed] [Google Scholar]
- Gurevich VV, Hanson SM, Song X, Vishnivetskiy SA, Gurevich EV. The functional cycle of visual arrestins in photoreceptor cells. Prog Retin Eye Res. 2011;30:405–30. doi: 10.1016/j.preteyeres.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M, Gurevich VV, Vishnivetskiy SA, Sigler PB, Schubert C. Crystal structure of beta-arrestin at 1.9 A: possible mechanism of receptor binding and membrane translocation. Structure. 2001;9:869–80. doi: 10.1016/s0969-2126(01)00644-x. [DOI] [PubMed] [Google Scholar]
- Hanson SM, Francis DJ, Vishnivetskiy SA, Klug CS, Gurevich VV. Visual arrestin binding to microtubules involves a distinct conformational change. J Biol Chem. 2006a;281:9765–72. doi: 10.1074/jbc.M510738200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson SM, Francis DJ, Vishnivetskiy SA, Kolobova EA, Hubbell WL, Klug CS, Gurevich VV. Differential interaction of spin-labeled arrestin with inactive and active phosphor-hodopsin. Proc Natl Acad Sci USA. 2006b;103:4900–5. doi: 10.1073/pnas.0600733103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson SM, Cleghorn WM, Francis DJ, Vishnivetskiy SA, Raman D, Song S, Nair KS, Slepak VZ, Klug CS, Gurevich VV. Arrestin mobilizes signaling proteins to the cytoskeleton and redirects their activity. J Mol Biol. 2007;368:375–87. doi: 10.1016/j.jmb.2007.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch JA, Schubert C, Gurevich VV, Sigler PB. The 2.8 A crystal structure of visual arrestin: a model for arrestin’s regulation. Cell. 1999;97:257–69. doi: 10.1016/s0092-8674(00)80735-7. [DOI] [PubMed] [Google Scholar]
- Hunot S, Vila M, Teismann P, Davis RJ, Hirsch EC, Przedborski S, Rakic P, Flavell RA. JNK-mediated induction of cyclooxygenase 2 is required for neurodegeneration in a mouse model of Parkinson’s disease. Proc Natl Acad Sci USA. 2004;101:665–70. doi: 10.1073/pnas.0307453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M, Gallagher E. From JNK to pay dirt: jun kinases, their biochemistry, physiology and clinical importance. IUBMB Life. 2005;57:283–95. doi: 10.1080/15216540500097111. [DOI] [PubMed] [Google Scholar]
- Kim M, Vishnivetskiy SA, Van Eps N, Alexander NS, Cleghorn WM, Zhan X, Hanson SM, Morizumi T, Ernst OP, Meiler J, Gurevich VV, Hubbell WL. Conformation of receptor-bound visual arrestin. Proc Natl Acad Sci USA. 2012;109:18407–12. doi: 10.1073/pnas.1216304109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Hofmann KP, Ernst OP, Scheerer P, Choe HW, Sommer ME. Crystal structure of pre-activated arrestin p44. Nature. 2013;497:142–6. doi: 10.1038/nature12133. [DOI] [PubMed] [Google Scholar]
- Kook S, Zhan X, Kaoud TS, Dalby KN, Gurevich VV, Gurevich EV. Arrestin-3 binds JNK1α1 and JNK2α2 and facilitates the activation of these ubiquitous JNK isoforms in cells via scaffolding. J Biol Chem. 2013:288. doi: 10.1074/jbc.M113.510412. in revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortum RL, Lewis RE. The molecular scaffold KSR1 regulates the proliferative and oncogenic potential of cells. Mol Cell Biol. 2004;24:4407–16. doi: 10.1128/MCB.24.10.4407-4416.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuan CY, Whitmarsh AJ, Yang DD, Liao G, Schloemer AJ, Dong C, Bao J, Banasiak KJ, Haddad GG, Flavell RA, Davis RJ, Rakic P. A critical role of neural-specific JNK3 for ischemic apoptosis. Proc Natl Acad Sci USA. 2003;100:15184–9. doi: 10.1073/pnas.2336254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte SA, Oakley RH, Zhang J, Holt JA, Ferguson SG, Caron MG, Barak LS. The 2-adrenergic receptor/arrestin complex recruits the clathrin adaptor AP-2 during endocytosis. Proc Natl Acad Sci USA. 1999;96:3712–7. doi: 10.1073/pnas.96.7.3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawler S, Fleming Y, Goedert M, Cohen P. Synergistic activation of SAPK1/JNK1 by two MAP kinase kinases in vitro. Curr Biol. 1998;8:1387–90. doi: 10.1016/s0960-9822(98)00019-0. [DOI] [PubMed] [Google Scholar]
- Levchenko A, Bruck J, Sternberg PW. Scaffold proteins may biphasically affect the levels of mitogen-activated protein kinase signaling and reduce its threshold properties. Proc Natl Acad Sci USA. 2000;97:5818–23. doi: 10.1073/pnas.97.11.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levchenko A, Bruck J, Sternberg PW. Regulatory modules that generate biphasic signal response in biological systems. Syst Biol (Stevenage) 2004;1:139–48. doi: 10.1049/sb:20045014. [DOI] [PubMed] [Google Scholar]
- Li Z, McNulty DE, Marler KJ, Lim L, Hall C, Annan RS, Sacks DB. QGAP1 promotes neurite outgrowth in a phosphorylation-dependent manner. J Biol Chem. 2005;280:13871–8. doi: 10.1074/jbc.M413482200. [DOI] [PubMed] [Google Scholar]
- Luttrell LM, Ferguson SS, Daaka Y, Miller WE, Maudsley S, Della Rocca GJ, Lin F, Kawakatsu H, Owada K, Luttrell DK, Caron MG, Lefkowitz RJ. Beta-arrestin-dependent formation of beta2 adrenergic receptor-Src protein kinase complexes. Science. 1999;283:655–61. doi: 10.1126/science.283.5402.655. [DOI] [PubMed] [Google Scholar]
- Luttrell LM, Roudabush FL, Choy EW, Miller WE, Field ME, Pierce KL, Lefkowitz RJ. Activation and targeting of extracellular signal-regulated kinases by beta-arrestin scaffolds. Proc Natl Acad Sci USA. 2001;98:2449–54. doi: 10.1073/pnas.041604898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald PH, Chow CW, Miller WE, Laporte SA, Field ME, Lin FT, Davis RJ, Lefkowitz RJ. Beta-arrestin 2: a receptor-regulated MAPK scaffold for the activation of JNK3. Science. 2000;290:1574–7. doi: 10.1126/science.290.5496.1574. [DOI] [PubMed] [Google Scholar]
- Milano SK, Pace HC, Kim YM, Brenner C, Benovic JL. Scaffolding functions of arrestin-2 revealed by crystal structure and mutagenesis. Biochemistry. 2002;41:3321–8. doi: 10.1021/bi015905j. [DOI] [PubMed] [Google Scholar]
- Miller WE, McDonald PH, Cai SF, Field ME, Davis RJ, Lefkowitz RJ. Identification of a motif in the carboxyl terminus of beta -arrestin2 responsible for activation of JNK3. J Biol Chem. 2001;276:27770–7. doi: 10.1074/jbc.M102264200. [DOI] [PubMed] [Google Scholar]
- Morfini GA, You YM, Pollema SL, Kaminska A, Liu K, Yoshioka K, Björkblom B, Coffey ET, Bagnato C, Han D, Huang CF, Banker G, Pigino G, Brady ST. Pathogenic huntingtin inhibits fast axonal transport by activating JNK3 and phosphorylating kinesin. Nat Neurosci. 2009;12:864–71. doi: 10.1038/nn.2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishima Y, Gotoh Y, Zieg J, Barrett T, Takano H, Flavell R, Davis RJ, Shirasaki Y, Greenberg ME. Beta-amyloid induces neuronal apoptosis via a mechanism that involves the c-Jun N-terminal kinase pathway and the induction of Fas ligand. J Neurosci. 2001;21:7551–60. doi: 10.1523/JNEUROSCI.21-19-07551.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishima-Kawashima M, Kosik KS. The pool of map kinase associated with microtubules is small but constitutively active. Mol Biol Cell. 1996;7:893–905. doi: 10.1091/mbc.7.6.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K, Pulvermuller A, Buczylko J, Hofmann KP. Phosphorylated rhodopsin and heparin induce similar conformational changes in arrestin. J Biol Chem. 1991;266:18649–54. [PubMed] [Google Scholar]
- Resnick L, Fennell M. Targeting JNK3 for the treatment of neurodegenerative disorders. Drug Discov Today. 2004;9:932–9. doi: 10.1016/S1359-6446(04)03251-9. [DOI] [PubMed] [Google Scholar]
- Sabapathy K. Role of the JNK pathway in human diseases. Prog Mol Biol Transl Sci. 2012;106:145–69. doi: 10.1016/B978-0-12-396456-4.00013-4. [DOI] [PubMed] [Google Scholar]
- Scapin G, Patel SB, Lisnock J, Becker JW, LoGrasso PV. The structure of JNK3 in complex with small molecule inhibitors: structural basis for potency and selectivity. Chem Biol. 2003;10:705–12. doi: 10.1016/s1074-5521(03)00159-5. [DOI] [PubMed] [Google Scholar]
- Schleicher A, Kuhn H, Hofmann KP. Kinetics, binding constant, and activation energy of the 48-kDa protein-rhodopsin complex by extra-metarhodopsin II. Biochemistry. 1989;28:1770–5. doi: 10.1021/bi00430a052. [DOI] [PubMed] [Google Scholar]
- Schlosshauer M, Baker D. Realistic protein–protein association rates from a simple diffusional model neglecting long-range interactions, free energy barriers, and landscape ruggedness. Protein Sci. 2004;13:1660–9. doi: 10.1110/ps.03517304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott MG, Le Rouzic E, Perianin A, Pierotti V, Enslen H, Benichou S, Marullo S, Benmerah A. Differential nucleocytoplasmic shuttling of beta-arrestins. Characterization of a leucine-rich nuclear export signal in beta-arrestin2. J Biol Chem. 2002;277:37693–701. doi: 10.1074/jbc.M207552200. [DOI] [PubMed] [Google Scholar]
- Seo J, Tsakem EL, Breitman M, Gurevich VV. Identification of arrestin-3-specific residues necessary for JNK3 activation. J Biol Chem. 2011;286:27894–901. doi: 10.1074/jbc.M111.260448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla AK, Manglik A, Kruse AC, Xiao K, Reis RI, Tseng WC, Staus DP, Hilger D, Uysal S, Huang LY, Paduch M, Tripathi-Shukla P, Koide A, Koide S, Weis WI, Kossiakoff AA, Kobilka BK, Lefkowitz RJ. Structure of active β-arrestin-1 bound to a G-protein-coupled receptor phosphopeptide. Nature. 2013;497:137–41. doi: 10.1038/nature12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Raman D, Gurevich EV, Vishnivetskiy SA, Gurevich VV. Visual and both non-visual arrestins in their “inactive” conformation bind JNK3 and Mdm2 and relocalize them from the nucleus to the cytoplasm. J Biol Chem. 2006;281:21491–9. doi: 10.1074/jbc.M603659200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Gurevich EV, Gurevich VV. Cone arrestin binding to JNK3 and Mdm2: conformational preference and localization of interaction sites. J Neurochem. 2007;103:1053–62. doi: 10.1111/j.1471-4159.2007.04842.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Coffa S, Fu H, Gurevich VV. How does arrestin assemble MAPKs into a signaling complex? J Biol Chem. 2009;284:685–95. doi: 10.1074/jbc.M806124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterne-Marr R, Gurevich VV, Goldsmith P, Bodine RC, Sanders C, Donoso LA, Benovic JL. Polypeptide variants of beta-arrestin and arrestin3. J Biol Chem. 1993;268:15640–8. [PubMed] [Google Scholar]
- Sutton RB, Vishnivetskiy SA, Robert J, Hanson SM, Raman D, Knox BE, Kono M, Navarro J, Gurevich VV. Crystal structure of cone arrestin at 2.3Å: evolution of receptor specificity. J Mol Biol. 2005;354:1069–80. doi: 10.1016/j.jmb.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Tournier C, Hess P, Yang DD, Xu J, Turner TK, Nimnual A, Bar-Sagi D, Jones SN, Flavell RA, Davis RJ. Requirement of JNK for stress-induced activation of the cytochrome c-mediated death pathway. Science. 2000;288:870–4. doi: 10.1126/science.288.5467.870. [DOI] [PubMed] [Google Scholar]
- Tournier C, Dong C, Turner TK, Jones SN, Flavell RA, Davis RJ. MKK7 is an essential component of the JNK signal transduction pathway activated by proinflammatory cytokines. Genes Dev. 2001;15:1419–26. doi: 10.1101/gad.888501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishnivetskiy SA, Hirsch JA, Velez MG, Gurevich YV, Gurevich VV. Transition of arrestin into the active receptor-binding state requires an extended interdomain hinge. J Biol Chem. 2002;277:43961–7. doi: 10.1074/jbc.M206951200. [DOI] [PubMed] [Google Scholar]
- Vishnivetskiy SA, Hosey MM, Benovic JL, Gurevich VV. Mapping the arrestin-receptor interface: structural elements responsible for receptor specificity of arrestin proteins. J Biol Chem. 2004;279:1262–8. doi: 10.1074/jbc.M308834200. [DOI] [PubMed] [Google Scholar]
- Vishnivetskiy SA, Gimenez LE, Francis DJ, Hanson SM, Hubbell WL, Klug CS, Gurevich VV. Few residues within an extensive binding interface drive receptor interaction and determine the specificity of arrestin proteins. J Biol Chem. 2011;286:24288–99. doi: 10.1074/jbc.M110.213835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishnivetskiy SA, Baameur F, Findley KR, Gurevich VV. Critical role of the central 139-loop in stability and binding selectivity of arrestin-1. J Biol Chem. 2013;288:11741–50. doi: 10.1074/jbc.M113.450031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Wu Y, Ge X, Ma L, Pei G. Subcellular localization of beta-arrestins is determined by their intact N domain and the nuclear export signal at the C terminus. J Biol Chem. 2003;278:11648–53. doi: 10.1074/jbc.M208109200. [DOI] [PubMed] [Google Scholar]
- Wang W, Shi L, Xie Y, Ma C, Li W, Su X, Huang S, Chen R, Zhu Z, Mao Z, Han Y, Li M. SP600125, a new JNK inhibitor, protects dopaminergic neurons in the MPTP model of Parkinson’s disease. Neurosci Res. 2004;48:195–202. doi: 10.1016/j.neures.2003.10.012. [DOI] [PubMed] [Google Scholar]
- Weston CR, Davis RJ. The JNK signal transduction pathway. Curr Opin Cell Biol. 2007;19:142–9. doi: 10.1016/j.ceb.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Xiao K, McClatchy DB, Shukla AK, Zhao Y, Chen M, Shenoy SK, Yates JR, Lefkowitz RJ. Functional specialization of beta-arrestin interactions revealed by proteomic analysis. Proc Natl Acad Sci USA. 2007;104:12011–6. doi: 10.1073/pnas.0704849104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Ichijo H, Korsmeyer SJ. BCL-2 is phosphorylated and inactivated by an ASK1/Jun N-terminal protein kinase pathway normally activated at G(2)/M. Mol Cell Biol. 1999;19:8469–78. doi: 10.1128/mcb.19.12.8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DD, Kuan CY, Whitmarsh AJ, Rincón M, Zheng TS, Davis RJ, Rakic P, Flavell RA. Absence of excitotoxicity-induced apoptosis in the hippocampus of mice lacking the Jnk3 gene. Nature. 1997;389:865–70. doi: 10.1038/39899. [DOI] [PubMed] [Google Scholar]
- Yoon SO, Park DJ, Ryu JC, Ozer HG, Tep C, Shin YJ, Lim TH, Pastorino L, Kunwar AJ, Walton JC, Nagahara AH, Lu KP, Nelson RJ, Tuszynski MH, Huang K. JNK3 perpetuates metabolic stress induced by Aβ peptides. Neuron. 2013;75:824–37. doi: 10.1016/j.neuron.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X, Kaoud TS, Kook S, Dalby KN, Gurevich VV. JNK3 binding to arrestin-3 differentially affects the recruitment of upstream MAP kinase kinases. J Biol Chem. 2013;288:28535–28547. doi: 10.1074/jbc.M113.508085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X, Gimenez LE, Gurevich VV, Spiller BW. Crystal structure of arrestin-3 reveals the basis of the difference in receptor binding between two non-visual arrestins. J Mol Biol. 2011a;406:467–78. doi: 10.1016/j.jmb.2010.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X, Kaoud TS, Dalby KN, Gurevich VV. Non-visual arrestins function as simple scaffolds assembling MKK4–JNK3α2 signaling complex. Biochemistry. 2011b;50:10520–9. doi: 10.1021/bi201506g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Raina AK, Rottkamp CA, Aliev G, Perry G, Boux H, Smith MA. Activation and redistribution of c-jun N-terminal kinase/stress activated protein kinase in degenerating neurons in Alzheimer’s disease. J Neurochem. 2001;76:435–41. doi: 10.1046/j.1471-4159.2001.00046.x. [DOI] [PubMed] [Google Scholar]
- Zhuang T, Chen Q, Cho M-K, Vishnivetskiy SA, Iverson TI, Gurevich VV, Hubbell WL. Involvement of distinct arrestin-1 elements in binding to different functional forms of rhodopsin. Proc Natl Acad Sci USA. 2013;110:942–7. doi: 10.1073/pnas.1215176110. [DOI] [PMC free article] [PubMed] [Google Scholar]