Abstract
The pharmacological actions of morphine and morphine-like drugs such as heroin mediate primarily through the mu opioid receptor (MOR). It represents the target of the most valuable painkiller in contemporary medicine. Here we report that poly(ADP-ribose) polymerase 1 (PARP-1) binds to the double-stranded poly(C) element essential for the MOR promoter and represses promoter activity at the transcriptional level. We identified PARP-1 by affinity column chromatography using the double-stranded poly(C) element, followed by two-dimensional gel electrophoresis and MALDI-TOF mass spectrometry. PARP-1 binding to the poly(C) sequence of the MOR gene was sequence-specific as confirmed by the supershift assay. In cotransfection studies, PARP-1 repressed the MOR promoter only when the poly(C) sequence was intact. When PARP-1 was disrupted in NS20Y cells using siRNA, transcription of the endogenous target MOR gene increased significantly. Chromatin immunoprecipitation assays showed specific binding of PARP-1 to the double-stranded poly(C) element essential for the MOR promoter. Inhibition of PARP-1's catalytic domain with 3-aminobenzamide increased endogenous MOR mRNA levels in cultured NS20Y cells, suggesting that automodification of PARP-1 regulates MOR transcription. Our data suggest that PARP-1 can function as a repressor of MOR transcription dependent on the MOR poly(C) sequence. We demonstrate for the first time a role of PARP-1 as a transcriptional repressor in MOR gene regulation.
Keywords: Mu opioid receptor, transcriptional repressor, poly(C) element, poly(ADP-ribose) polymerase, 3-aminobenzamide
Introduction
Opioids are used as potent clinical analgesics, but have serious limitations such as tolerance and dependence. The pharmacological effects of morphineas a pain killer are mainly mediated by, and dependent on, the number of mu opioid receptors (MOR) in the cell surface membrane, suggesting the importance of studying MOR gene regulation [1, 2]. The opioid receptors are classified into three major types (δ, κ and μ), and have been studied in numerous pharmacological studies and by molecular cloning [3]. All three types of opioid receptors belong to the superfamily of G-protein-coupled receptors [1]. MOR is known to play roles in morphine-induced analgesia, tolerance and dependence, as reported in pharmacological studies and analyses of MOR knockout mice [4, 5]. Upon the binding of opioids, MOR is able to couple to G-proteins and to regulate adenylyl cyclase, intracellular calcium, inwardly-rectifying potassium channels, MAP kinase and other messengers, which trigger cascades of further intracellular events [6]. MOR is expressed mainly in the central nervous system, with receptors varying in density in different regions and perhaps playing different roles [7, 8]. To achieve its unique spatial expression pattern, expression of MOR must be tightly regulated. The mouse MOR gene spans about 250 kb and consists of multiple exons [9]. Several MOR isoforms have been reported [10, 11].
Two different promoters (distal and proximal) of the mouse MOR gene have been reported, located within 1 kb upstream of the ATG translational start site [12]. The distal promoter initiates MOR transcription from a single transcription initiation site located 794 bp upstream of the translation start site. The proximal promoter initiates MOR transcription from four major transcription initiation sites located in a region ranging from 291 to 268 bp upstream of the translation start site. The mouse MOR promoter contains a 5′-distal promoter regulatory sequence: a 34-bp cis-acting element that possesses a strong inhibitory effect against the transcrip-tional function of the distal promoter [13]. Both promoters exhibit characteristics of housekeeping genes lacking a TATA box. The distal promoter is known to be 20-fold less active than the proximal promoter, based on quantitative RT-PCR using adult and embryonic mouse brains [14]. The proximal promoter appears to be the predominant promoter for directing MOR gene transcription in the adult mouse brain, as well as during embryonic development [12, 14].
The mammalian poly(ADP-ribose) polymerase 1 (PARP-1)— the major isoform of the PARP-1 family—is comprised of 1014 amino acids (114 kD) and is expressed continuously in eukaryotes. It has a 46-kD DNA-binding domain at the N-terminal containing a DEVD sequence, which acts as a target for caspase-3 during apoptosis. When cleaved by caspase-3, PARP-1 is inactivated, resulting in the formation of two proteolytic fragments: a 29-kD amino-terminal fragment and an 85-kD carboxyl-terminal fragment. A 54-kD domain of PARP-1 located in the carboxyl terminus represents the β-nicotinamide adenine dinucleotide (NAD+)-binding domain [15, 16]. Between the DNA-binding domain and the catalytic domain is a 22-kD automodification domain that facilitates the homo/heterodimerization of PARP-1 with other proteins [15, 17].
PARP-1's basal enzymatic activity is very low, but is stimulated dramatically in the presence of a variety of allosteric activators, including damaged DNA [18]. The targets of PARP-1's enzymatic activity include PARP-1 itself (the primary target in vivo), core histones, the linker histone H1 and a variety of transcription-related factors (e.g. p53, fos, NF-κB, RNA polymerase I and II) [19–24]. Kraus et al.[25] described the nucleosome binding properties of PARP-1 that promote the formation of compact transcriptionally repressed chromatin structure. PARP-1 binds in a specific manner to nucleosomes and modulates chromatin structure through NAD+-dependent automodification. The automodification activity of PARP-1 is potently stimulated by nucleosome, causing the release of PARP-1 from chromatin. Thus, PARP-1 functions both as a structural component of chromatin and as a modulator of chromatin structure through its intrinsic enzymatic activity [25].
PARP-1 modulates gene expression in both a positive and a negative fashion, with the final effects depending on the cell type, the gene and the transcription factor involved. For example, when bound to DNA, PARP-1 impairs RNA polymerase II activity [26], However, conflicting results suggest that PARP-1 is localized preferentially in regions of actively transcribed chromatin and able to enhance transcription by promoting protein complex formation on the enhancer template. PARP-1 facilitates the detection of transcription initiation DNA sequences and promotes the activity of transcription factors such as activator protein 2 (AP-2), Oct-1. NF-κB and p53 [27–30].
In this study, we report the identification and characterization of a poly(C)-sequence-binding protein that regulates mouse MOR gene regulation. We employed affinity column chromatography containing a specific competitor, two-dimensional gel elec-trophoresis (2-DE) and mass spectrometry to purify and identify the specific factor interacting between the poly(C) sequence of the MOR proximal promoter region and its binding proteins in mouse NS20Y neuroblastoma cells. PARP-1 was identified as a poly(C)-binding protein. PARP-1 was able to bind specifically to the mouse MOR poly(C) sequence. This protein served as a transcription repressor in the proximal promoter of the mouse MOR gene. This study demonstrates that PARP-1 can function as a repressor of MOR transcription dependent on the MOR poly(C) sequence and presents for the first time a role for PARP-1 in MOR gene regulation as atranscriptional repressor.
Materials and Methods
Plasmid construction
A luciferase fusion plasmid (pGL450; −450 to +1 bp, relative to the translation start site of the mouse MOR [+1]) was generated by ligating the PCR product (-450 to +1) into the Sad and Hindi 11 sites of pGL3-basic (Promega, Madison, Wl). The PCR reaction was performed using genomic DNA from mouse NS20Y cells as a template and an upstream sense primer (5′-ATTGAGCTCCTGCAGCATCCCCGCTTCTGC-3′) containing a Sad site (underlined), and a downstream antisense primer (5′-ATAAAGCTTTGGTTCTGAATGCTTGCTGCG-3′) containing a Hindlll site (underlined). The pGL450mut construct was generated by ligating the mutated PCR product (-450 to +1) into the Sad and Hindlll sites of pGL3-basic (Promega, Madison, Wl, USA). The PCR reaction was performed using pGL450 plasmid as a template and an upstream sense primer (5′-ATTGAGCTC CTTCTGCTCCCTTCCGGCCTACCC-3′; mutated nucleotides are indicated in italics) containing a Sacl site (underlined), and a downstream antisense primer (5′-ATAAAGCTTTGGTTCTGAATGCTTGCTGCG-3′) containing a Hindlll site (underlined). pCMV-PARP-1 was obtained cordially from Dr. S.C. Lee [31].
Cell culture, transient transfection and reporter gene assay
Mouse neuroblastoma NS20Y cells were routinely grown in Dulbecco's Minimum Essential Medium supplemented with 10% heat-inactivated fetal bovine serum at 37°C in a humidified atmosphere of 5% CO2. For the inhibition of PARP-1, 3-aminobenzamide (3-AB) was added to the culture. The NS20Y cells were plated in 6-well dishes at a concentration of 0.5 ± 106 cells/well and cultured overnight before transfection. Various plasmids at equimolar concentrations were used with Effectene transfection reagent (Qiagen, Valencia, CA) as described previously [32]. Briefly, for luciferase analysis of MOR promoters, 0.5 pg of the reporter plasmids were mixed with the Effectene transfection reagent for 10 min before being added to the NS20Y cells. Forty-eight hours after transfection, cells grown to confluence were washed once with 1 × phosphate-buffered saline and lysed with lysis buffer (Promega, Madison, Wl, USA). To correct for differences in transfection efficiency, a one-fifth molar ratio of pCH110 (Amersham, Piscataway, NJ) containing the β-galactosidase gene under the SV40 promoter was included in each transfection for normalization. The luciferase and β-galactosidase activities of each lysate were determined according to the manufacturer's recommendations (Promega and Tropics, respectively).
Nuclear extract preparation
Nuclear extracts were prepared from NS20Y cells as described previously [33]. Briefly cells were grown to confluence, harvested and washed with phosphate-buffered saline. All of the following steps were performed at 4°C. The cells were resuspended in sucrose buffer (0.32 M sucrose, 3 mM CaCl2, 2 mM magnesium acetate, 0.1 mM Ethylene Diamine Tetra-acetic Acid (EDTA), 10 mM Tris-HCI [pH 8.0], 1 mM Dithiothreitol (DTT), 0.5 mM Phenyl methylsulfonyl fluoride (PMSF), 0.5% NP-40). The lysate was microcentrifuged at 500 g for 5 min to pellet the nuclei, which were washed with sucrose buffer. The nuclei were resuspended in low-salt buffer (20 mM HEPES [pH 7.9], 25% glycerol, 20 mM KCI, 1.5 mM MgCI2, 0.2 mM EDTA, 0.5 mM DTT, 0.5 mM PMSF), followed by addition of high-salt buffer and incubation for 20 min on a rotary platform to extract the nuclei. Two-and-a-half volumes of a diluent (25 mM HEPES [pH 7.6], 25% glycerol, 0.1 mM EDTA, 0.5 mM DTT, 0.5 mM PMSF) were added and the sample was microcentrifuged at 13,000 g. Aliquots of the supernatant containing the nuclear extracts were stored at −80°C.
DNA-affinity purification of poly (C)-binding protein using a competitor
The following procedure is based on the interaction between biotin and strep-tavidin. Oligonucleotides were synthesized and purified using HPLC. In a sterile tube, 500 pmoles of biotinylated sense oligonucleotide (5′-CTTCTGCTCCCCCCCCCCCTACCC-3′) and 500 pmoles of non-biotinylated antisense oligonucleotide (5′-GGGTAGGGGGGGGGGGAGCAGAAG-3′) were combined. The two oligonucleotides were annealed in a total volume of 20 μl by incubating in a heating block at 95°C for 10 min, then allowed to cool completely to room temperature. A total of 500 μl of 0.5× SSC were added to 500 pmoles of the 5′-terminal-biotinylated double-stranded DNA. Meanwhile, 500 pmoles of streptavidin-paramagnetic particles (Promega, Madison, Wl, USA) were resuspended by gently flicking the bottom of the tube until they were completely dispersed, then captured by placing the tube in a magnetic stand. The supernatant was carefully removed. The magnetic particles were washed three times with 0.5× SSC and resuspended in 100 μl of 0.5× SSC.
Five hundred pmoles of biotinylated double-stranded DNA and 500 pmoles of the streptavidin-paramagnetic particles were combined and incubated for 15 min at room temperature. Samples were mixed by gentle inversion every 2 min. The magnetic beads were captured using a magnetic stand. The particles were washed three times with 300 μl of buffer A (5 mM HEPES, 26% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 0.5 mM PMSF, 300 mM NaCl), pH 7.9. One milligram of nuclear proteins was added to the affinity particles and incubated for 1 hr at 4°C. The particles were washed three times with buffer A, buffer B (5 mM HEPES, 26% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 0.5 mM PMSF, 100 mM NaCI), pH 7.9 and buffer C (5 mM HEPES, 26% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 0.5 mM PMSF), pH 7.9. Fifty micro-liters of 1× SDS sample buffer were added and proteins bound to the particles were released by incubation in a heating block for 10 min at 95°C.
In order to eliminate nuclear proteins that might bind non-specifically. control experiments were performed as follows: 2500 pmoles of non-biotinylated double-stranded DNA (5× competitor) were mixed with 1 mg of nuclear proteins for 15 min on ice. The nuclear extracts containing the 5× competitor were added to the affinity particles and incubated for 1 hr at 4°C. The remainder of the procedure was performed as above. The resultant protein solutions with (Control) and without (Sample) competitor were elec-trophoresed on two-dimensional gels and stained with Coomassie blue.
Two-dimensional gel electrophoresis (2-DE), in-gel tryptic digestion and MALDI-TOF mass spectrometric analysis of poly(C)-binding protein
Purified proteins were resolved by 2-DE as described by Görg et al., with minor modifications [34]. Control and sample 2-DE gels were run under identical conditions. Immobilized pH gradient (IPG) strips were used according to the manufacturer's instruction. Isoelectric focusing (IEF) as the first dimension was carried out on a Protean IEF cell (Bio-Rad: Hercules, CA). Briefly, purified samples were mixed with an aliquot (185 μl) of rehydration solution (7 M urea, 2 M thiourea, 4% CHAPS [w/v], 60 mM DTT, a trace of bromophenol blue, 0.5% IPG buffer [v/v]; Amersham Pharmacia Biotech, Piscataway, NJ) then applied to the IPG strips. After rehydration for 12 hrs, IEF was carried out for 500 V for 1 hr, 1000 V for 1 hr, and a gradient to 8000 V for a total of 50,000 volt-hours. The IPG strips were then incubated for 15 min with an equilibration solution (50 mM Tris-HCl [pH 8.8], 6 M urea, 30% glycerol [v/v], 2% SDS [w/v], 2% DTT [w/v]), followed by equilibration for another 15 min in the same buffer containing 2.5% iodoacetamide (w/v) instead of DTT. SDS-PAGE as the second dimension was carried out at 90 V constant for 3 hrs. Molecular masses were determined by running standard protein markers (DualColor PrecisionPlus Protein(tm) standard; Bio-Rad). Gels were stained with colloidal Coomassie (GelCode® Blue stain reagent; Pierce Biotechnology Rockford, IL) to visualize protein spots. Gel slices of interest (differential bands) were subjected to in-gel tryptic digestion as described previously [35]. Tryptic peptides were extracted with 5% acetic acid, followed by 5% acetic acid and 50% acetonitrile. Samples were dissolved in 5% acetic acid and desalted using ZipTip(tm) C18 reverse-phase desalting Eppendorf tips (Millipore, Billerica, MA, USA). The peptides were eluted with 2% acetonitrile containing 0.1% trifluoroacetates (TFA) in a volume of 20 μl. Samples were analysed using a MALDI-TOF mass spectrometer (Applied Biosystems, Foster City, CA, USA). The masses of monoisotopic peaks were used for comparison to a theoretical digestion of the protein by trypsin. The Mascot database-searching software (Matrix Science, http://www.matrixscience.com) was used to identify binding proteins.
Western blot analysis
Proteins purified from NS20Y cells were incubated with treatment buffer (62.5 mM Tris-HCI [pH 6.8], 2% SDS, 10% glycerol, 5% 2-mercaptoethanol) and boiled for 5 min. Treated extracts were resolved by SDS-PAGE using a 12% polyacrylamide gel. Gels were electroblotted onto polyvinylidene diflu-oride membranes (Amersham Biosciences) in transfer buffer (48 mM Tris-HCl, 39 mM glycine, 20% methanol). Membranes were blocked in blocking solution (10% dry milk, 0.1% Tween-20 in Tris-buffered saline) overnight at 4°C. Western blotting with anti-PARP-1 (Santa Cruz, Santa Cruz, CA) and anti-β-actin antibodies (Cell Signaling, Beverly, MA) was performed according to the manufacturer's instructions (Amersham Biosciences, Piscataway, NJ, USA). Signals were detected using a Storm 840 Phosphorlmager system (Amersham Biosciences, Piscataway, NJ, USA).
Autopoly(ADP-ribosyl)ation of PARP-1
Two micrograms of recombinant PARP-1 (R&D Systems, Inc., Minneapolis, MN) were incubated with 0.2 μg sonicated DNA (as an activator) in reaction buffer (10 mM Tris-HCI, [pH 8.0], 1 mM MgCI2, 1 mM DTT) at 37°C for 10 min. The reactions were started by adding NAD+ to a final concentration of 1 mM and incubated at 37°C for 10 min. SDS sample buffer was added to stop the reaction. Samples were boiled and analysed by SDS-PAGE. Poly(ADP-ribose) was detected by immunoblot analysis using anti-PAR (Alexis Biochemicals, Billerica, MA, USA).
Electrophoretic mobility shift assay (EMSA)
The EMSA was performed as described previously [36]. The upper and lower strands of each probe (5′-CTTCTGCTCCCCCCCCCCCTACCC-3′ and 5′-GGGTAGGGGGGGGGGGAGCAGAAG-3′) were annealed and the double-strand oligonucleotides were then end-labelled with [(-32p]dATP. Free nucleotides were separated by centrifugation through a G-25 column (Roche, Indianapolis, IN). The end-labelled DNA probes were incubated with recombinant PARP-1 (0.5 μg) and purified proteins in a final volume of 20 μl EMSA buffer (10 mM Tris [pH 7.5], 5% glycerol, 1 mM EDTA [pH 7.1], 50 mM NaCl, 1 mM DTT, 0.1 mg/ml poly [dl-dC]) at room temperature for 20 min. For oligonucleotide competition analyses, a 100-fold molar excess of cold competitor oligonucleotide was added to the mixture prior to adding the probe. The reactions were then incubated at 4°C for 30 min. The reaction mixtures were electrophoresed on a nondenaturing 4% polyacrylamide gel in 0.5× TBE (45 mM Tris-borate, 1 mM EDTA) at 4°C and visualized by autoradiography.
Chromatin immunoprecipitation (ChIP) assay and real-time quantitative PCR (qPCR)
Chromatin immunoprecipitation (ChIP) assays were performed by using a modified protocol from Upstate Biotechnology as previously reported [2]. Cells were treated for 10 min with 1% formaldehyde at room temperature. The cells were lysed in SDS lysis buffer (50 mM Tris-HCI [pH 8.1], 10 mM EDTA, 1% SDS). The lysates were sonicated under conditions yielding fragments ranging from 200 to 500 bp. Two percent of each lysate was used for input, and samples of the residual lysates (about 25 μg chromatin after determination of the amount of protein) were subsequently precleared at 4°C with recombinant protein G-agarose beads (Upstate Biotechnology Richmond, CA, USA) coated with salmon sperm DNA. Precleared lysates (100 μl) diluted in immunoprecipitation buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl [pH 8.1], 167 mM NaCl) were immunoprecipitated overnight at 4°C with 2 μg of antibodies against anti-PARP (from R&D). All ChIP assays were controlled by performing parallel experiments with no antibody or with gal4 antibody (sc-577; Santa Cruz). The complexes were collected for 1 hr by using recombinant protein G-agarose beads coated in salmon sperm DNA. After thorough washing of the beads and elution, formaldehyde cross-linking was reversed by 6 hrs of incubation at 65°C. DNA was extracted with phenol-chloroform, precipitated in ethanol, and dissolved in 50 μl of TE buffer (10 mM Tris-HCI and 1 mM EDTA, pH 8.0). A total of 2 μl of each immunoprecipitated chromatin was used for real-time qPCR analysis using SYBR Green I (QuantitectSYBR Green PCR kit; Qiagen) performed in an iCycler (Bio-Rad). To calculate the number of gene copies, amplification curves of standard samples that contained 101 to 108 molecules of the gene of interest (e.g. the MOR plasmid pmMuEG constructed in our lab) were monitored and the number of target molecules in the test sample was analysed using qCalculator ver. 1.0 software (http://www.gene-quantification.de/download.html#qcalculator) based on the mathematical model of Pfaffl [37]. Primers for mouse MOR were: 5′-CCAATTTACACTCCTTTACACG-3′ (sense) and 5′-GGGCTGTGAGGGATCCAGAGGCTAG-3′ (antisense). Primers for mouse β-actin were: 5′-TGGCCTTAGGGTGCAGGGGG-3′ (sense) and 5′-GTGGGCCGCTCTAGGCACCA-3′ (antisense). The specificity of real qPCR primers was determined using a melt curve after the amplification to show that only a single species of qPCR product resulted from the reaction. Single PCR products were also verified on a 2% agarose gel.
Total RNA Preparation and RT-PCR Analysis
Total RNA was isolated using TRI Reagent (Molecular Research Center Inc., Cincinnati, OH) according to the supplier's protocol. Two micrograms of total RNA were used for the RT-PCR reaction using the OneStep RT-PCR reagent (Qiagen). The PCR cycle conditions consisted of 95°C for 1 min, 60°C for 1 min and 72°C for 1 min, followed by a 10-min extension at 72°C, for 38 cycles. Primers specific to total mouse MOR mRNA were: 5′-CATCAAAGCACTGATCACGATTCC-3′ (sense, located at exon 3) and 5′-TAGGGCAATGGAGCAGTTTCTGC-3′ (antisense, located at exon 4). Intron 3 (between exons 3 and 4 of the mouse MOR genome) is about 20 kb, indicating that the RT-PCR for the MOR gene using the above primers produced only MOR mRNA. Similar reactions were carried out using 5′-TGGCCTTAGGGTGCAGGGGG-3′ (sense) and 5′-GTGGGCCGCTCTAGGCACCA-3′ (antisense) primers for mouse β-actin as an internal control, except that the number of cycles was reduced to 25. The resulting PCR products were analysed on a 2% agarose gel. The DNA sequences of PCR products were confirmed by automated DNA sequencing. Quantitative analyses were done using ImageQuant 5.2 (Amersham Biosciences) software.
Up-regulation of the MOR gene by silencing PARP-1 with siRNA
To confirm the role of PARP-1 in MOR regulation, we used siRNAs to inhibit the expression of PARP-1 and examined the subsequent effects on MOR expression levels. One hundred picomoles of siRNA duplexes for PARP-1 (r[GGCUAUAGUUCUCAAUUAA]dTdT [sense] and (r[UUAAUUGAGAACUAUAGCC]dCdA [antisense]; Qiagen, HP Genomewide siRNA) were transfected into NS20Y cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Control and scrambled (i.e. non-targeting oligos designed by Qiagen) transfections were included as negative controls. RNA was isolated 48 hrs after the transfections and the expression levels of MOR were determined by RT-PCR using the same PCR primers and conditions as described above. The protein levels of PARP-1 and β-actin were determined by western blotting using anti-PARP-1 (Santa Cruz Biotechnology, Santa Cruz, CA, USA) and anti-β-actin (Cell Signaling) antibodies.
Results
Isolation and identification of a new transcription factor that interacts with poly(C) DNA sequences in the mouse MOR proximal promoter
Studies from our laboratory have shown that expression of mouse MOR is driven by two promoters, distal and proximal [12]. Previously, we reported that MOR transcription is regulated by a cis-acting poly(C) sequence in the mouse MOR promoter through the binding of Sp1 and Sp3 [38]. The poly(C) sequence (Fig.1A) is essential for promoter activity of the mouse MOR. We report here the discovery of another regulator for this MOR promoter through binding to the poly(C) sequence, identified using the following new procedure (Fig.1B).
Figure 1.
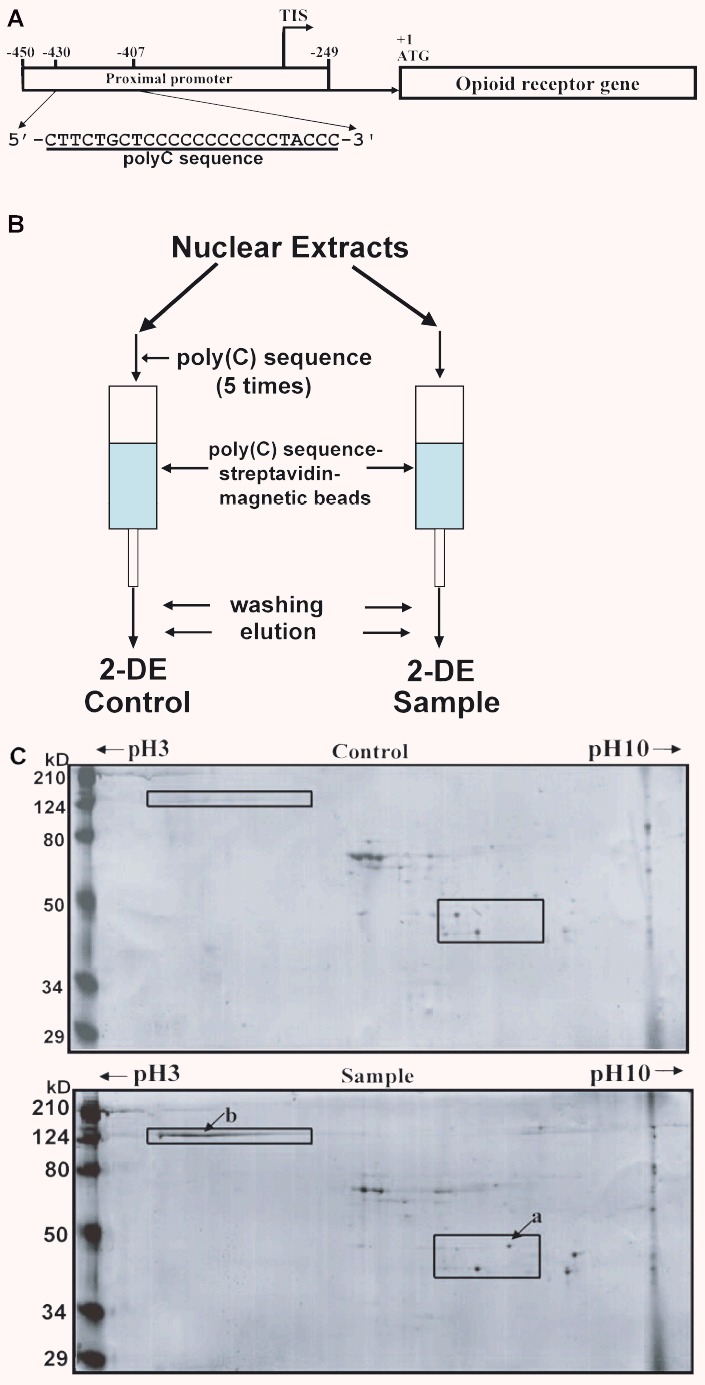
Schematic representation of the mouse MOR gene and one-step purification of poly(C)-binding proteins using an affinity column and two-dimensional gel electrophoresis. (A) The poly(C) sequence of the mouse MOR proximal promoter. TIS, transcriptional initiation site. (B) Double-stranded oligonucleotides biotinylated on the 5′-terminal were used as affinity particles. For control experiments to eliminate non-specific binding of nuclear proteins, nuclear extracts were preincubated with 2500 pmoles of non-biotinylated double-stranded DNA (5× competitor) prior to affinity purification. (C) Coomassie-stained images of poly(C)-binding proteins purified using an affinity column (Control and Sample) and separated by two-dimensional gel electrophoresis. Molecular weight markers are indicated on the left and pl values across the top. The arrows (a, b) indicate the 36-kD protein (α-CP3) and the 124-kD band used for mass spectrometry identification, respectively.
We had developed a new one-step method for purification of transcription factors that bind to specific DNA sequences; contamination by non-specifically binding nuclear proteins is eliminated using a competitor. Using this protocol, we were able to purify and identify a protein from NS20Y nuclear extracts that binds specifically to the poly(C) sequence. We recently reported a repressor protein (αCP3) in the sequence [39]. However, no other proteins binding to the poly(C) sequence were identified using SDS-PAGE alone. We therefore used 2-DE to find a new major transcription factor binding to the poly(C) sequence. A unique band migrating at 120 kD was visualized by Coomassie staining (‘b’ in Fig.1C), cut out of the gel, and identified by MALDI-TOF mass spectrometric analysis. Based on its high score (111) on the Mascot search results (Fig.2), the protein was identified as PARP-1. In general, PARP-1 exists in cells in both unribosylated and ribosylated forms [40, 41]. The pi of unribosylated PARP-1 is basic (10–11) and the pl of ribosylated PARP-1 is acidic. The low pl (i.e. 4) of the purified PARP-1 indicates that the poly(C)-binding form of PARP-1 is ribosylated.
Figure 2.

Identification of PARP-1 as a poly(C)-binding protein. Mascot results of the mass spectrometry identification of the 124-kD protein band. The value with the highest score (111) identifies the protein as PARP-1.
PARP-1 binds specifically to poly(C) DNA sequences of the mouse MOR proximal promoter
Before performing EMSA, we prepared both poly(ADP-ribosyl)ated and unpoly(ADP-ribosyl)ated forms of the PARP-1 protein. Under control conditions (i.e. a nonenzymatic reaction without NAD+), electrophoresed PARP-1 protein appears as a band of approximately 125 kD and is unpoly(ADP-ribosyl)ated (Fig.3B, lane 1). After enzymatic activation in the presence of NAD+, electrophoresed PARP-1 proteins appear as both an unpoly(ADP-ribosyl)ated form (∼125 kD) and a hyperpoly(ADP-ribosyl)ated form (>210kD) (Fig.3B, lane 2). When the enzymatic reaction was performed in the presence of 3-AB, both the 125 kD and 210 kD forms were present, although autopoly(ADP-ribosyl)ation of PARP-1 was inhibited (Fig.3B, lane 3).
Figure 3.

Auto-poly(ADP-ribosyl)ation of PARP-1 in vitvo w’froand EMSA of poly(C)-binding sequence with recombinant PARP-1 and purified proteins. (A) The MOR poly(C) sequence (NS). (B) Auto-poly(ADP-ribosyl)ation of PARP-1 in vitro. Recombinant PARP-1 was incubated in the absence or presence of 10 mM 3-AB for 20 min. PARP-1 and poly(ADP-ribosyl)ated PARP-1 were detected using anti-PARP-1 and anti-poly(ADP-ribose) (anti-PAR). Lane 1: control (nonenzymatic reaction without NAD+); lane 2: enzymatic reaction with NAD+; lane 3: inhibited enzymatic reaction with NAD+ and 3-AB. (C) EMSAs were performed using the labelled poly(C) sequence (NS) and unpoly(ADP-ribosyl)ated or poly(ADP-ribosyl)ated PARP-1. Lane 1: probe alone; lane 2: unpoly(ADP-ribosyl)ated PARP-1; lane 3: poly(ADP-ribosyl)ated PARP-1; lane 4: poly(ADP-ribosyl)ation of PARP-1 inhibited by 3-AB; lane 5: unpoly(ADP-ribosyl)ated PARP-1 in the presence of competitor; lane 6: poly(ADP-ribosyl)ation of PARP-1 in the presence of competitor; lane 7: poly(ADP-ribosyl)ation of PARP-1 in the presence of competitor and 3-AB inhibitor. The PARP-1-poly(C) sequence complex is indicated by an arrow. (D) Coomassie-stained gel of poly(C)-binding proteins purified from NS20Y nuclear extracts and western blot analysis of purified poly(C)-binding proteins probed with anti-PARP-1 and anti-PAR antibodies. Arrows indicate PARP-1, poly(ADP-ribosyl)ated PARP-1 and poly(ADP-ribosyl)ated proteins. (E) EMSA of purified poly(C)-binding proteins using anti-PARP and anti-PAR antibody. EMSAs were performed using the 32p-labelled MOR poly(C) sequence (NS) as a probe with purified poly(C)-binding proteins. Lane 1: Self-competitor without antibody; lane 2: EMSA reaction without antibody; lane 3: EMSA with anti-PARP antibody (1 μg); lane 4: EMSA with anti-PARP antibody (2 μg); lane 5: EMSA with anti-PAR antibody. Supershifted bands of PARP antibody and PAR antibody are indicated by arrows.
To determine the physical interaction of PARP-1 with the mouse MOR promoter and verify its contribution to promoter activity, EMSAs were performed using recombinant PARP-1 and a regulatory sequence (NS; Fig.3A) from the MOR poly(C) sequence as a probe. Only one major band (Fig.3C, arrow), indicating the PARP-1/NS complex, was detected (Fig.3C, lane 2). In contrast, hyper-poly(ADP-ribosyl)ated PARP-1 did not bind the regulatory sequence (Fig.3C, lane 3). When the enzymatic reaction was inhibited by 3-AB, binding of PARP-1 to NS was slightly increased, likely due to the decrease in the hyper-poly(ADP-ribosyl)ated form of PARP-1 (Fig.3C, lane 4). A 100-fold molar excess of unlabelled NS oligonucleotide (Fig.3C, lanes 5–7) completely inhibited complex formation. These results demonstrate that PARP-1 binds specifically to the MOR poly(C) sequence, while hyper-poly(ADP-ribosyl)ated PARP-1 does not.
SDS-PAGE and western blots with anti-PARP-1 and anti-PAR detected the presence of both poly(ADP-ribosyl)ated PARP-1 and poly(ADP-ribosyl)ated proteins in affinity-purified samples (Fig.3D). This suggested that PARP-1 might form part of a repressor complex with poly(ADP-ribosyl)ated proteins. To determine if poly(ADP-ribosyl)ated PARP-1 interacted physically with the poly(C) sequence of the mouse MOR proximal promoter, EMSAs were performed using purified poly(ADP-ribosyl)ated PARP-1 and the NS regulatory sequence from the MOR poly(C) sequence as a probe. The complex was present in the absence of antibody (Fig.3E, asterisk). One microgram of anti-PARP-1 produced a minor supershifted band (arrow), while 2 μg of anti-PARP-1 produced a supershifted band (arrow) with concomitant reduction in the intensity of the complex band (asterisk). To determine if poly(ADP-ribosyl)ated PARP and poly(ADP-ribosyl)ated proteins bound DNA directly, we carried out gel shift assays using the anti-PAR (poly(ADP)-ribose) antibody. That antibody produce supershifted bands (arrow) containing poly(ADP-ribosyl)ated PARP-1 and poly(ADP-ribosyl)ated proteins (Fig.3E).
Defining the core PARP-1-binding motif of the poly(C) sequence
To determine the PARP-1 binding motif within the poly(C) sequence of the proximal promoter, EMSAs were carried out using recombinant PARP-1 and labelled NS sequence with sequences mutated as indicated (Fig.4A; M1–M8) as competitors. Major complexes formed with labelled NS in the presence of M3, M5 and M7 (Fig.4B, lanes 5, 7 and 9, respectively), although at reduced levels relative to samples without competitor (Fig.4B, lane 1). No PARP-1 complexes were observed when unlabelled NS was used as a competitor (Fig.4B, lanes 2). Reduced PARP-1 binding to labelled NS was observed using M1, M2, M4, M6 and M8 (Fig.4B, lanes 3,4, 6,8 and 10, respectively). Based on these observations, we determined that the poly(C) sequences 5′-CCCC-3′ (underlined in Fig.4C) serve as PARP-1 binding motifs within the mouse MOR proximal promoter.
Figure 4.
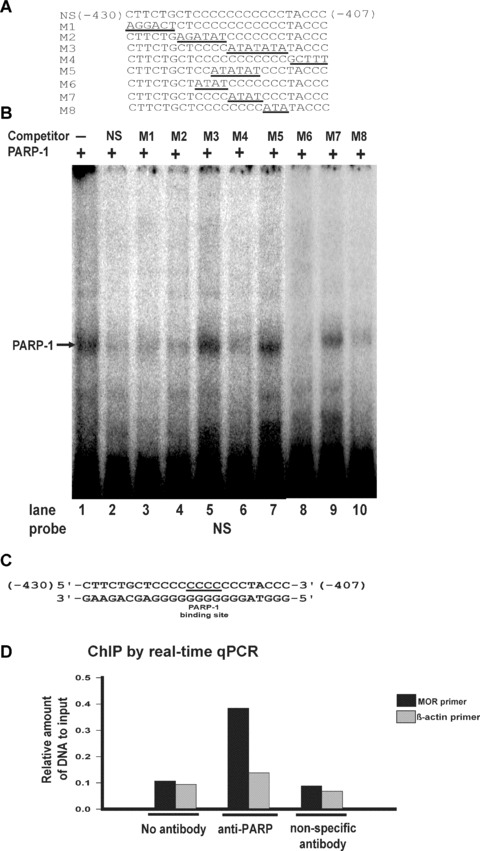
EMSA analysis of the PARP-1-binding motif using mutant oligonucleotide sequences and ChIP assay. (A) Representation of the double-stranded oligonucleotide sequence (NS) and mutant oligonucleotide sequences (M1–M8). (B) EMSAs were performed using unlabelled poly(C) sequence (NS; lane 2) or unlabelled poly(C) mutated sequences (M1–M8; lanes 3–10) as competitors for recombinant PARP-1 protein binding to a labelled poly(C) sequence. Lane 1: Negative control (no unlabelled poly(C) sequence). The PARP-1-poly(C) sequence complex is indicated by an arrow. (C) The PARP-1-binding motif of the poly(C) sequence (NS). (D) ChIP analysis by real-time qPCR for PARP-1 binding interaction with the MOR promoter poly(C) sequence. Interactions were examined by ChIP assay with anti-PARP antibody and nonspecific antibody (anti-gal4). Precipitated DNAs were amplified using mouse MOR and β-actin (negative control) primers.
ChIP allowed further confirmation in vivo of the specific binding of PARP-1 to the poly(C) sequence of the mouse MOR promoter (Fig.4D). ChIP experiments using anti-PARP-1 in NS20Y cells showed that PARP-1 was densely localized to the mouse MOR proximal promoter. In contrast, chromatin from NS20Y cells immunoprecipitated with no antibody or anti-gal4 displayed little enrichment in the mouse MOR proximal promoter (Fig.4D). These results indicate that PARP-1 binds to the poly(C) sequence of the mouse MOR promoter in vivo and that such binding is specific.
PARP-1 represses mouse MOR proximal promoter activity through the PARP-1 binding motif
To examine the functional role of the PARP-1 protein in mouse MOR gene regulation, we used the MOR proximal promoter fused to a PARP-1 expression plasmid with a luciferase reporter (Fig.5A). When this plasmid was cotransfected into the NS20Y cells with the mouse proximal promoter construct pGL450, PARP-1 repressed about 50% of MOR proximal promoter activity, compared to cells transfected with the pcDNA4 vector alone (Fig.5B). However, PARP-1 could not repress the promoter activity (Fig.5C) of the pGL450mut construct (containing a mutated PARP-1 binding motif; Fig.5A). The results suggest that PARP-1 regulates MOR transcription through the PARP-1 binding motif of the MOR proximal promoter.
Figure 5.
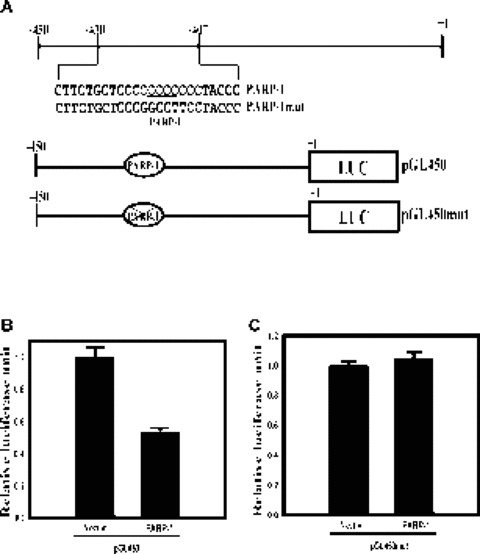
PARP-1 represses the proximal promoter of the mouse MOR gene. (A) Schematic representations of the mouse MOR proximal promoter region (the PARP-1 -binding motif is underlined), the pGL450 (wild-type) promoter construct and the pGL450mut construct (containing a mutated PARP-1 binding site). The ‘X’ in the filled ovals indicates the mutation, which includes the PARP-1 binding site and its flanking sequence. Nucleotide +1 corresponds to the translation start site (ATG). (B, C) Neuronal NS20Y cells endogenously expressing the MOR gene were co-transfected with 2 pg of the PARP-1 constructs and 1 pg of the MOR-promoter luciferase-reporter constructs, pGL450 and pGL450mut. The activities of the luciferase reporter were expressed as n-fold relative to the activity of each corresponding luciferase reporter transfected with vector alone, which was assigned an activity value of 1.0. Transfection efficiencies were normalized by β-galactosidase activity. The data shown are the mean and standard errors of three independent experiments with at least two different plasmid preparations.
The role of PARP-1 in the regulation of the endogenous MOR gene was tested using a siRNA, RT-PCR and western blot analysis. Mouse NS20Y cells expressing PARP-1 endogenously were transfected with 100 pmoles of mouse PARP-1 siRNA or scrambled siRNA. After total RNA was isolated from the transfected cell lines, RT-PCR was performed. Each mRNA signal was quantified using ImageQuant 5.2 software and normalized against β-actin. PARP-1 siRNA effectively repressed endogenous PARP-1 expression to 30% of the levels seen in untreated controls or scramble-transfected samples (Fig.6A). However, RT-PCR revealed that in the presence of PARP-1 siRNA, MOR mRNA levels increased 80% relative to the controls (Fig.6C), while β-actin mRNA levels were unchanged (Fig.6B). Silencing of PARP-1 in PARP-1-expressing NS20Y cells with siRNA results in an increase of endogenous MOR mRNA transcription levels. These results indicate that the PARP-1 protein regulates endogenous MOR gene expression at the tran-scriptional level.
Figure 6.
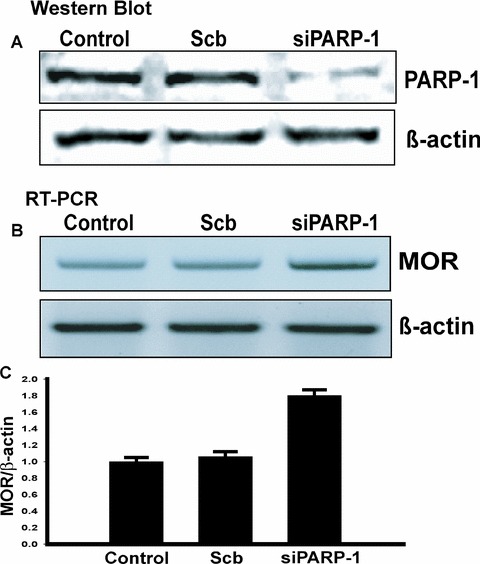
Analysis of mouse MOR gene regulation by PARP-1 in vivo using siRNA. PARP-1 siRNA increases MOR transcription in NS20Y cells. (A) NS20Y cells were trans-fected with either scrambled siRNA (Scb) or PARP-1 siRNA (siPARP-1). Whole-cell extracts were made after incubation with the siRNAs for 48 hrs. Immunoblot analyses for PARP-1 and β-actin were performed. This figure is a representative of three separate experiments. (B) Quantification of MOR transcripts was performed by RT-PCR. Total RNA from NS20Y cells was prepared and treated with DNase I, and primer pairs specific for the coding sequence of each gene were used for RT-PCR. (C) Quantitative analysis using ImageQuant 5.2 software. The MOR mRNA levels from Control, scrambled (Scb) or siRNA-treated (siPARP-1) cells were normalized against β-actin levels. The values were obtained from triplicate data points and changes in transcript levels for Scb or siPARP-1-treated samples were compared to Control, which was assigned a value of 1.0. Bars indicate the range of standard error.
Effect of PARP-1 inhibition on mouse MOR gene expression
PARP-1 catalyzes the transfer of multiple ADP-ribose units to target nuclear proteins. 3-AB, a specific PARP-1 inhibitor, blocks poly(ADP-ribosyl)ation of PARP-1 and nuclear proteins. Mouse neuronal cells were treated with 2 mM 3-AB for 48 hrs in order to monitor the effects of PARP-1 inhibition on mouse MOR gene transcription. Treatment with 3-AB up-regulated mouse MOR mRNA levels nearly 2.5-fold over untreated cells (Fig.7B). These data suggest that PARP-1-mediated poly(ADP-ribosyl)ation is essential for mouse MOR gene expression, and that PARP-1 plays an important role in the regulation of mouse MOR gene expression.
Figure 7.
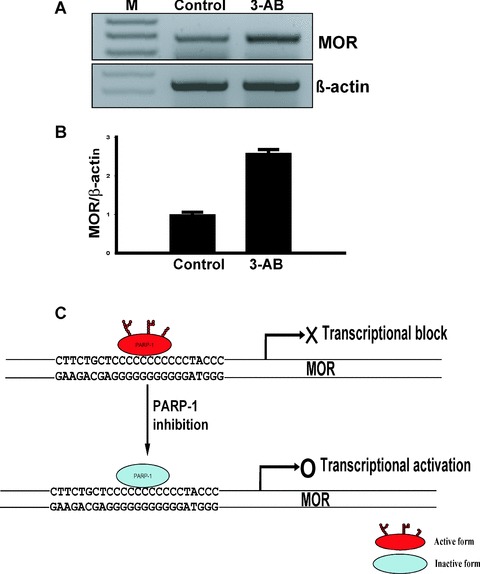
3-AB inhibits mouse MOR mRNA expression in NS20Y cells and schematic model for PARP-1 in modulation of mouse MOR transcription. (A) Quantification of transcripts was performed by RT-PCR. Total RNA from NS20Y cells treated with 2 mM 3-AB was prepared and treated with DNase I. Primer pairs specific for the coding sequence of each gene were used for RT-PCR. PCR products were visualized in a 2% agarose gel. Lane 1: Molecular weight markers (M); lane 2: Control; lane 3: 3-AB-treated cells. (B) Quantitative analysis using ImageQuant 5.2 software. The MOR mRNA levels from Control and 3-AB-treated cells were normalized against β-actin levels. The values were obtained from triplicate data points. Changes in transcript levels for 3-AB-treated samples were compared to Control, which was assigned a value of 1.0. Bars indicate the range of standard error. (C) Schematic model for the role of PARP-1 in modulation of mouse MOR gene transcription. In neuronal cells, enzymatically active PARP-1 interacts strongly with the poly(C) sequence of the mouse MOR promoter and aids in the formation of tran-scriptionally inactive chromatin. Enzymatic inhibition of PARP-1 by 3-AB results in non-poly(ADP-ribosyl)ated PARP-1 and subsequently, an increase in the levels of MOR mRNA in mouse NS20Y cells.
Discussion
Precise transcriptional regulation of opioid receptor genes in the brain is crucial for normal neuropharmacological function. Several classes of nuclear proteins are intricately involved in controlling expression of these genes [1]. The 5′-flanking region of the mouse MOR has two promoters, distal and proximal, and the activities of each differ in the brain [14]. The proximal promoter is the major promoter of mouse MOR gene activity. It is regulated by various cis-elements and trans-factors, all of which are important for its activity [12]. Our earlier studies showed that mouse MOR transcription was regulated by a cis-acting element, a poly(C) sequence that was essential for the activity of the mouse MOR promoter through the binding of Sp1 and Sp3 [38].
We have developed an efficient method to purify transcription factors [39]. This simple method has many advantages, including a smaller population of cells required for analysis, rapidity (<5 hrs), and a one-step process that eliminates the need for additional column chromatography. Additionally, this method is very effective at removing nuclear proteins that bind non-specifi-cally. We have used this new procedure to purify new transcription factors that bind to single-stranded DNA [42] of the MOR proximal promoter (unpublished data). We also previously used this method to purify αCP3, a poly(C) sequence-binding protein [39]. In this study, using a combination of the one-step purification method, 2-DE and MALDI-TOF mass spectrometry, we identified PARP-1 as another protein that binds to the poly(C) sequence of the MOR promoter.
PARP-1 is a 114-kD nuclear protein widely known for its DNA binding properties and for its unique enzyme activity: It catalyzes the covalent attachment of ADP-ribose units from its NAD+ to several nuclear-acceptor proteins, including itself. Initial studies implicated this enzyme in many crucial biological functions, including DNA repair, recombination, apoptosis and cancer [43, 44]. Transcriptional regulation of PARP-1 occurs by at least two mechanisms that are not mutually exclusive: first, modifying his-tones to alter chromatin structure and second, functioning as part of enhancer/promoter binding complexes in conjunction with other DNA binding factors and coactivators [18, 25]. There is evidence that PARP-1 can act as a transcription activator, but other data show that PARP-1 may repress transcription. Recently, PARP-1 was shown to have a distinctly dualistic role, with opposing effects in AP-2α-mediated transcriptional regulation. It was demonstrated that distinct regions of PARP-1 interact with AP-2α differently and independently control its transcriptional activation; while PARP-1's middle region enhances transcription, its catalytic domain functions to repress transcription [45, 46].
Here we report that PARP-1 binds to the double-stranded poly(C) element essential for activity of the MOR promoter and represses promoter activity at the transcriptional level. Specific interaction between PARP-1 and the poly(C) sequence of the mouse MOR promoter was first observed during one-step purification using an affinity column. EMSA further revealed the characteristics of the sequence-specific interaction between PARP-1 and the poly(C) sequence of the mouse MOR promoter. In particular, a four-base sequence (5′-CCCC-3′; −418 to −415) is critical for PARP-1-poly(C) complex formation. Our gel shift assay showed that poly(ADP-ribosyl)ated PARP and poly(ADP-ribosyl)ated proteins bound to the poly(C) sequence of the mouse MOR promoter.
This implies that poly(ADP-ribosyl)ated PARP-1 might be part of a protein complex that interacts with the poly(C) sequence. PARP-1 could be part of repressor complex. While the poly(C) sequence of the mouse MOR promoter binds both poly(ADP-ribosyl)ated PARP and poly(ADP-ribosyl)ated proteins, interactions between poly(ADP-ribosyl)ated PARP and poly(ADP-ribosyl)ated proteins were not detected (data not shown). Intriguingly, in EMSAs, unribosylated PARP-1 and ribosylated PARP-1 did bind to poly(C) sequence, while hyper-ribosylated PARP-1 apparently did not bind. It is possible that in circumstances where high levels of PARP-1 activation occur (i.e. hyper-poly(ADP-ribosyl)ation of PARP-1), electrostatic repulsion by the negatively charged polymers separates PARP-1 from DNA, resulting in reduced binding efficiency.
Recent findings demonstrate a key role for PARP-1 binding to cis-acting elements in transcriptional activation and repression. PARP-1 promotes the expression of CXCL1 chemokine by recognizing an element upstream of the Cxcl1 promoter (5′-TCGAT-3′), and regulates Reg expression by recognizing the transcriptional start region of the Reg gene (5′-CCCCTCCC-3′), cardiac troponin T gene (cTnT) expression by recognizing the MCAT1 element (5’-TGTTG-3’), and insulin-producing β-cell regeneration [47–49]. PARP-1 represses the expression of the α-synuclein gene SNCA by binding to the polymorphic site upstream of the gene. In this study, the mouse MOR gene also was repressed by PARP-1 binding to the poly(C) sequences in the proximal promoter of the MOR gene.
Functional analyses suggest that PARP-1 binds specifically to, and is able to repress, the MOR proximal promoter containing the wild-type poly(C) sequence, but not mutated poly(C) sequences. In addition, transfection with PARP-1 siRNA led to a marked increase in endogenous MOR transcription in NS20Y cells. These data suggest that PARP-1 acts as a transcriptional repressor. Inhibition of PARP-1's enzymatic activity with 3-AB abrogated this repressive effect and led to elevated levels of endogenous mouse MOR mRNA in NS20Y cells. A number of models could be proposed for how inhibition of PARP-1's enzymatic activity might directly or indirectly up-regulate mouse MOR gene expression. First, enzymatically active PARP-1 might alter chromatin structure in the promoter region containing the poly(C) sequence to a transcriptionally inaccessible form of DNA, while inhibition of the PARP-1 catalytic domain results in an open DNA conformation and transcriptional up-regulation. Alternatively, enzymatically active PARP-1 might recruit transcriptional repressors and prevent the binding and interaction of transcriptional activators; inhibition of PARP-1 would have the opposite effect, leading to up-regulation of MOR gene expression (Fig.7C).
It has been reported that the poly(C) sequence of the mouse MOR promoter regulates the MOR gene through the binding of Sp1 and Sp3 [38]. However, in this study, the quantity of Sp1 or Sp3 binding to the poly(C) region was very small [39]. Indeed, the major protein binding to the poly(C) region was PARP-1. It is possible that binding of Sp proteins to the poly(C) sequence is regulated by competition with PARP-1 for binding sites, and that such competition is important for negative regulation of the mouse MOR gene. We conclude that PARP-1 acts as a repressor of MOR transcription in neuronal cells, via a mechanism dependent on the poly(C) site of the MOR proximal promoter.
Several G-protein-coupled receptor genes, including the MOR gene, are also controlled by a promoter with constitutive activity. Thus, gene activity must be modulated via sequence-specific enhancer- and/or silencer-binding proteins in order to produce restricted patterns of expression in the nervous system. Tissue- or cell-specific regulatory factors [33, 50–52] presumably modulate the ability of ubiquitous factors (such as PARP-1 or other, as yet unidentified, factors) to regulate the MOR gene promoter activity. In summary, our findings may promote a better understanding of the molecular mechanisms underlying MOR gene expression.
Acknowledgments
This work was supported by National Institutes of Health research grants DA00564, DA01583, DA11806, DA11190, K05-DA00153 and K02-DA13926, and by the F&A Stark Fund of the Minnesota Medical Foundation. We thank Dr. S.C. Lee for providing PARP-1 plasmid and the staffs of the Mass Spectrometry Consortium for Life Science, University of Minnesota, Department of Biochemistry, Molecular Biology and Biophysics at St. Paul, MN for recording the mass spectra for the protein samples.
References
- 1.Wei LN, Loh HH. Regulation of opioid receptor expression. Curr Opin Pharmacol. 2002;2:69–75. doi: 10.1016/s1471-4892(01)00123-0. [DOI] [PubMed] [Google Scholar]
- 2.Hwang CK, Song KY, Kim CS, Choi HS, Guo XH, Law PY, Wei LN, Loh HH. Evidence of endogenous mu opioid receptor regulation by epigenetic control of the promoters. Mol Cell Biol. 2007;27:4720–36. doi: 10.1128/MCB.00073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Min BH, Augustin LB, Felsheim RF, Fuchs JA, Loh HH. Genomic structure analysis of promoter sequence of a mouse mu opioid receptor gene. Proc Natl Acad Sci USA. 1994;91:9081–5. doi: 10.1073/pnas.91.19.9081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kieffer BL. Recent advances in molecular recognition and signal transduction of active peptides: receptors for opioid peptides. Cell Mol Neurobiol. 1995;15:615–35. doi: 10.1007/BF02071128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kieffer BL. Opioids: first lessons from knockout mice. Trends Pharmacol Sci. 1991;20:19–26. doi: 10.1016/s0165-6147(98)01279-6. [DOI] [PubMed] [Google Scholar]
- 6.Law PY, Wong YH, Loh HH. Molecular mechanisms and regulation of opioid receptor signaling. Annu Rev Pharmacol Toxicol. 2000;40:389–430. doi: 10.1146/annurev.pharmtox.40.1.389. [DOI] [PubMed] [Google Scholar]
- 7.Maldonado R, Saiardi A, Valverde O, Samad TA, Roques BP, Borrelli E. Absence of opiate rewarding effects in mice lacking dopamine D2 receptors. Nature. 1997;388:586–9. doi: 10.1038/41567. [DOI] [PubMed] [Google Scholar]
- 8.Mansour A, Fox CA, Akil H, Watson SJ. Opioid-receptor mRNA expression in the rat CNS: anatomical and functional implications. Trends Neurosci. 1995;18:22–9. doi: 10.1016/0166-2236(95)93946-u. [DOI] [PubMed] [Google Scholar]
- 9.Pan YX, Xu J, Mahurter L, Bolan E, Xu M, Pasternak GW. Generation of the mu opioid receptor (MOR-1) protein by three new splice variants of the Oprm gene. Proc Natl Acad Sci USA. 2001;98:14084–9. doi: 10.1073/pnas.241296098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan YX. Identification and characterization of a novel promoter of the mouse mu opioid receptor gene (Oprm) that generates eight splice variants. Gene. 2002;295:97–108. doi: 10.1016/s0378-1119(02)00825-9. [DOI] [PubMed] [Google Scholar]
- 11.Choi HS, Kim CS, Hwang CK, Song KY, Wang W, Qiu Y, Law PY, Wei LN, Loh HH. The opioid ligand binding of human mu-opioid receptor is modulated by novel splice variants of the receptor. Biochem Biophys Res Commun. 2006;343:1132–40. doi: 10.1016/j.bbrc.2006.03.084. [DOI] [PubMed] [Google Scholar]
- 12.Ko JL, Minnerath SR, Loh HH. Dual promoters of mouse mu-opioid receptor gene. Biochem Biophys Res Commun. 1997;234:351–7. doi: 10.1006/bbrc.1997.6640. [DOI] [PubMed] [Google Scholar]
- 13.Hwang CK, Kim CS, Choi HS, McKercher SR, Loh HH. Transcriptional regulation of mouse mu opioid receptor gene by PU.1. J Biol Chem. 2004;279:19764–74. doi: 10.1074/jbc.M400755200. [DOI] [PubMed] [Google Scholar]
- 14.Ko JL, Chen HC, Loh HH. Differential promoter usage of mouse mu-opioid receptor gene during development. Brain Res Mol Brain Res. 2002;104:184–93. doi: 10.1016/s0169-328x(02)00357-1. [DOI] [PubMed] [Google Scholar]
- 15.Alvarez-Gonzalez R, Spring H, Muller M, Burkle A. Selective loss of poly(ADP-ribose) and the 85-kDa fragment of poly(ADP-ribose) polymerase in nucleoli during alkylation-induced apoptosis of HeLa cells. J Biol Chem. 1999;274:32122–6. doi: 10.1074/jbc.274.45.32122. [DOI] [PubMed] [Google Scholar]
- 16.Smulson ME, Simbulan-Rosenthal CM, Boulares AH, Yakovlev A, Stoica B, Iyer S, Luo R, Haddad B, Wang ZQ, Pang T. Roles of poly(ADP-ribosyl)ation and PARP in apoptosis, DNA repair, genomic stability and functions of p53 and E2F-1. Adv Enzyme Regul. 2000;40:183–215. doi: 10.1016/s0065-2571(99)00024-2. [DOI] [PubMed] [Google Scholar]
- 17.Amiri Kl, Ha HC, Smulson ME, Richmond A. Differential regulation of CXC ligand 1 transcription in melanoma cell lines by poly(ADP-ribose) polymerase-1. Oncogene. 2006;25:7714–22. doi: 10.1038/sj.onc.1209751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim MY, Zhang T, Kraus WL. Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying;NAD+ into a nuclear signal. Genes Dev. 2005;19:1951–67. doi: 10.1101/gad.1331805. [DOI] [PubMed] [Google Scholar]
- 19.Wesierska-Gadek J, Schmid G. Poly(ADP-ribose) polymerase-1 regulates the stability of the wild-type p53 protein. Cell Mol Biol Lett. 2001;6:117–40. [PubMed] [Google Scholar]
- 20.Amstad PA, Krupitza G, Cerutti PA. Mechanism of c-fos induction by active oxygen. Cancer Res. 1992;52:3952–60. [PubMed] [Google Scholar]
- 21.Kameoka M, Ota K, Tetsuka T, Tanaka Y, Itaya A, Okamoto T, Yoshihara K. Evidence for regulation of NF-kappaB by poly(ADP-ribose) polymerase. Biochem J. 2000;346(Pt 3):641–9. [PMC free article] [PubMed] [Google Scholar]
- 22.Chang WJ, Alvarez-Gonzalez R. The sequence-specific DNA binding of NF-kappa B is reversibly regulated by the auto-modification reaction of poly (ADP-ribose) polymerase 1. J Biol Chem. 2001;276:47664–70. doi: 10.1074/jbc.M104666200. [DOI] [PubMed] [Google Scholar]
- 23.Muller WE, Zahn RK. Poly ADP-ribosyla-tion of DNA-dependent RNA polymerase I from quail oviduct. Dependence on progesterone stimulation. Mol Cell Blochem. 1976;12:147–59. doi: 10.1007/BF01741713. [DOI] [PubMed] [Google Scholar]
- 24.Taniguchi T, Suzuki S, Shizuta Y. Poly (ADP-ribosyl)ation of RNA polymerase II from wheat germ. Blochem Blophys Res Coininun. 1985;127:526–32. doi: 10.1016/s0006-291x(85)80191-1. [DOI] [PubMed] [Google Scholar]
- 25.Kim MY, Mauro S, Gevry N, Lis JT, Kraus WL. l\IAD+-dependent modulation of chromatin structure and transcription by nucle-osome binding properties of PARP-1. Cell. 2004;119:803–14. doi: 10.1016/j.cell.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 26.Vispe S, Yung TM, Ritchot J, Serizawa H, Satoh MS. A cellular defense pathway regulating transcription through poly(ADP-ribo-syl)ation in response to DNA damage. Proc Natl Acad Sci USA. 2000;97:9886–9891. doi: 10.1073/pnas.170280397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kannan P, Yu Y, Wankhade S, Tainsky MA. PolyADP-ribose polymerase is a coactivator for AP-2-mediated transcrip-tional activation. Nucleic Acids Res. 1999;27:866–74. doi: 10.1093/nar/27.3.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nie J, Sakamoto S, Song D, Qu Z, Ota K, Taniguchi T. Interaction of Oct-1 and auto-modification domain of poly(ADP-ribose) synthetase. FEBS Lett. 1998;424:27–32. doi: 10.1016/s0014-5793(98)00131-8. [DOI] [PubMed] [Google Scholar]
- 29.Hassa PO, Hottiger MO. A role of poly (ADP-ribose) polymerase in NF-kappaB transcriptional activation. J Biol Chem. 1999;380:953–9. doi: 10.1515/BC.1999.118. [DOI] [PubMed] [Google Scholar]
- 30.Wan X, Ohnishi K, Takahashi A, Ohnishi T. Poly(ADP-ribosyl)ation is required for p53-dependent signal transduction induced by radiation. Oncogene. 1998;17:2819–25. doi: 10.1038/sj.onc.1202216. [DOI] [PubMed] [Google Scholar]
- 31.Huang JY, Chen WH, Chang YL, Wang HT, Chuang WT, Lee SC. Modulation of nucleosome-binding activity of FACT by poly(ADP-ribosyl)ation. Nucleic Acids Res. 2006;34:2398–407. doi: 10.1093/nar/gkl241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi HS, Hwang CK, Kim CS, Song KY, Law PY, Wei LN, Loh HH. Transcriptional regulation of mouse mu opioid receptor gene: Sp3 isoforms (M1, M2) function as repressors in neuronal cells to regulate the mu opioid receptor gene. Mol Pharmacol. 2005;67:1674–83. doi: 10.1124/mol.104.008284. [DOI] [PubMed] [Google Scholar]
- 33.Kim CS, Hwang CK, Choi HS, Song KY, Law PY, Wei LN, Loh HH. Neuron-restrictive silencer factor (NRSF) functions as a repressor in neuronal cells to regulate the mu opioid receptor gene. J Biol Chem. 2004;279:46464–73. doi: 10.1074/jbc.M403633200. [DOI] [PubMed] [Google Scholar]
- 34.Görg A, Obermaier C, Boguth G, Harder A, Scheibe B, Wildgruber R, Weiss W. The current state of two-dimensional elec-trophoresis with immobilized pH gradients. Electrophoresis. 2000;21:1037–53. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1037::AID-ELPS1037>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 35.Patterson SD, Aebersold R. Mass spectrometric approaches for the identification of gel-separated proteins. Electrophoresis. 1995;16:1791–814. doi: 10.1002/elps.11501601299. [DOI] [PubMed] [Google Scholar]
- 36.Hwang CK, Wu X, Wang G, Kim CS, Loh HH. Mouse mu opioid receptor distal promoter transcriptional regulation by SOX proteins. J Biol Chem. 2003;278:3742–50. doi: 10.1074/jbc.M208780200. [DOI] [PubMed] [Google Scholar]
- 37.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ko JL, Liu HC, Loh HH. Role of an AP-2-like element in transcriptional regulation of mouse micro-opioid receptor gene. Brain Res Mol Brain Res. 2003;112:153–62. doi: 10.1016/s0169-328x(03)00086-x. [DOI] [PubMed] [Google Scholar]
- 39.Choi HS, Kim CS, Hwang CK, Song KY, Law PY, Wei LN, Loh HH. Novel function of the poly(C)-binding protein {alpha}CP3 as a transcriptional repressor of the mu opioid receptor gene. FASEB J. 2007;21:3963–73. doi: 10.1096/fj.07-8561com. [DOI] [PubMed] [Google Scholar]
- 40.Cohen-Armon M, Visochek L, Katzoff A, Levitan D, Susswein AJ, Klein R, Valbrun M, Schwartz JH. Long-term memory requires polyADP-ribosylation. Science. 2004;304:1820–2. doi: 10.1126/science.1096775. [DOI] [PubMed] [Google Scholar]
- 41.Visochek L, Steingart RA, Vulih-Shultzman I, Klein R, Priel E, Gozes I, Cohen-Armon M. PolyADP-ribosylation is involved in neurotrophic activity. J Neurosci. 2005;25:7420–8. doi: 10.1523/JNEUROSCI.0333-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ko JL, Loh HH. Single-stranded DNA-binding complex involved in transcriptional regulation of mouse mu-opioid receptor gene. J Biol Chem. 2001;276:788–95. doi: 10.1074/jbc.M004279200. [DOI] [PubMed] [Google Scholar]
- 43.D'Amours D, Desnoyers S, D'Silva I. Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Blochem J. 1999;342:249–68. [PMC free article] [PubMed] [Google Scholar]
- 44.Virag L, Szabo C. The therapeutic potential of poly(ADP-ribose) polymerase inhibitors. Pharmacol Rev. 2002;54:375–429. doi: 10.1124/pr.54.3.375. [DOI] [PubMed] [Google Scholar]
- 45.Li M, Naidu P, Yu Y, Berger NA, Kannan P. Dual regulation of AP-2alpha transcriptional activation by poly(ADP-ribose) poly-merase-1. Blochem J. 2004;382:323–9. doi: 10.1042/BJ20040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chiba-Falek O, Kowalak JA, Smulson ME, Nussbaum RL. Regulation of alphasynuclein expression by poly (ADP ribose) polymerase-1 (PARP-1) binding to the NACP-Rep1 polymorphic site upstream of the SNCA gene. Am J Hum Genet. 2005;76:478–92. doi: 10.1086/428655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nirodi C, NagDas S, Gygi SP, Olson G, Aebersold R, Richmond A. A role for poly(ADP-ribose) polymerase in the transcriptional regulation of the melanoma growth stimulatory activity (CXCL1) gene expression. J Biol Chem. 2001;276:9366–74. doi: 10.1074/jbc.M009897200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Akiyama T, Takasawa S, Nata K, Kobayashi S, Abe M, Shervani NJ, Ikeda T, Nakagawa K, Unno M, Matsuno S. Activation of Reg gene, a gene for insulin-producing beta-cell regeneration: poly(ADP-ribose) polymerase binds Reg promoter and regulates the transcription by autopoly(ADP-ribosyl)ation. Proc Natl Acad Sci USA. 2001;98:48–53. doi: 10.1073/pnas.240458597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Butler AJ, Ordahl CP. Poly(ADP-ribose) polymerase binds with transcription enhancer factor 1 to MCAT1 elements to regulate muscle-specific transcription. Mol Cell Biol. 1999;19:296–306. doi: 10.1128/mcb.19.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Korner M, Rattner A, Mauxion F, Sen R, Citri Y. A brain-specific transcription activator. Neuron. 1989;3:563–72. doi: 10.1016/0896-6273(89)90266-3. [DOI] [PubMed] [Google Scholar]
- 51.Kovarik A, Peat N, Wilson D, Gendler SJ, Taylor-Papadimitriou J. Analysis of the tissue-specific promoter of the MUC1 gene. J Biol Chem. 1993;268:9917–26. [PubMed] [Google Scholar]
- 52.Ogata H, Inoue N, Podolsky DK. Identification of a goblet cell-specific enhancer element in the rat intestinal trefoil factor gene promoter bound by a goblet cell nuclear protein. J Biol Chem. 1998;273:3060–7. doi: 10.1074/jbc.273.5.3060. [DOI] [PubMed] [Google Scholar]


