Abstract
As the regional influenza reference centre operating within the Italian network InfluNet, here we report data on virological and epidemiological surveillance of influenza, as well as on the vaccination coverage rates achieved in Lombardy (Northern Italy) over 10 consecutive winter seasons (2004–2014).
Over the past 10 years, influenza vaccine coverage declined both in the general population (from 15.7% in 2004–2005 to 11.7% in 2013–2014) and in the vaccine-target population of individuals ≥65-y-of-age (from 65.3% in 2004–2005 to 48.6% in 2013–2014) and is far below the minimum planned threshold level (75%). The highest influenza-like illness (ILI) rates were recorded during the 2004–2005 and 2009–2010 epidemics (peak incidence: 12.04‰ and 13.28‰, respectively). Both seasons were characterised by the introduction of novel viral strains: A/Fujian/411/2002(H3N2) (a drifted hemagglutinin variant) and A/California/7/2009(H1N1) pandemic virus (a swine origin quadruple reassortant), respectively. Because the antigenic match between vaccine and circulating strains was good in both of these seasons, a relevant proportion of cases may have been prevented by vaccination. A different situation was observed during the 2011–2012 season, when ILI morbidity rates in individuals ≥65-y-of-age were 1.5–6-fold higher than those registered during the other epidemics under review. The higher morbidity resulted from the circulation during the 2011–2012 season of an A/Victoria/361/2011(H3N2)-like variant that presented a reduced genetic match with the A(H3N2) strain included in the 2011–2012 vaccine composition.
The continuous surveillance of the characteristics of circulating viruses is an essential tool for monitoring their matching with seasonal vaccine strains. Strategies to increase coverage rates are warranted.
Keywords: Influenza virus, influenza surveillance network, influenza vaccination, vaccine coverage, epidemiology, phylogeny
Introduction
Seasonal influenza epidemics caused by influenza A and B viruses occur annually during the winter in temperate regions, with an annual attack rate estimated at 5–10% in adults and 20–30% in children.1,2 Worldwide, these annual epidemics are estimated to result in 3–5 million cases of severe illness, and about 250 000 to 500 000 deaths.1,2 Influenza epidemics can impose a considerable economic burden in the form of hospital and other healthcare costs and lost productivity.1-3
This recurrent distinctive epidemiological feature rests on the high propensity of influenza viruses to change the genetic and antigenic characteristics of their surface antigens hemagglutinin (HA) and neuraminidase (NA)—the main targets for the human immune system.4
Control and prevention of influenza rely on the international cooperation of a wide range of public health partners brought together under the coordination of the World Health Organization (WHO) in the Global Influenza Surveillance and Response System (GISRS).5 The overarching goal of the WHO GISRS is to minimise the impact of the disease by providing information to public health authorities for planning appropriate control and intervention measures and health resource allocation. The specific goal is to provide timely, high-quality data to evaluate the impact and severity of each season, to determine the influenza burden, to monitor the evolution of influenza viruses, and to provide recommendations in areas including laboratory diagnostics, vaccines, antiviral susceptibility and risk assessment. The WHO GISRS also serves as a global alert mechanism for the emergence of influenza viruses with pandemic potential.5 Altogether, the surveillance activities performed by this network are crucial to make the annual update of vaccine composition a reliable option, rather than a risky bet, to use for the control and prevention of influenza at a global level.
As part of the Italian influenza surveillance network, the activity and circulation of influenza viruses in Northern Italy (specifically, Lombardy, which is the most populous Italian region and which accounts for nearly 10 million of the 60 million inhabitants of Italy) were investigated during 10 consecutive winter seasons (from 2004–2005 to 2013–2014). The aim of this paper is to describe data on epidemiological and virological surveillance of influenza, as well as the influenza vaccine coverage rates in Northern Italy over the last 10 influenza seasons.
Results
Epidemiological surveillance
In the surveyed area, the epidemic waves had different epidemiological profiles over the last 10 consecutive influenza seasons. Table 1 shows the incidence of ILI at its peak during the considered influenza epidemics in the general population and by age group. In the general population, the highest ILI morbidity rates were recorded during the 2004–2005 and 2009–2010 epidemics, with peak incidence of 12.04‰ and 13.28‰, respectively. Both seasons were characterised by the introduction of novel viral strains: a drift HA variant of previously circulating A(H3N2) virus—the A/Fujian/411/2002(H3N2)-like strain—and the 2009 A(H1N1) pandemic virus—A/California/7/2009(H1N1)-like virus. These two influenza epidemics exhibited different timing: the unusual event of the 2009 pandemic was characterised by a sharp wave and an early peak at week 44, while seasonal influenza activity usually peaked at week 5 or week 6 (i.e., early-to-mid-February).
Table 1. Incidence of influenza-like illness (ILI) (×1000 inhabitants) at peak in the general population and by age group in the last 10 influenza epidemics (from 2004–2005 to 2013–2014) in Lombardy.
| Season | Incidence of ILI (×1000 inhabitants) at epidemic peak | |||||
|---|---|---|---|---|---|---|
| General population | Age group | |||||
| 0–4 y | 5–14 y | 15–64 y | ≥65 y | |||
| 2004–2005 | 12.04 | 26.86 | 29.81 | 11.85 | 5.16 | |
| (2005–06) | (2005–07) | (2005–05) | (2005-–6) | (2005–07) | ||
| 2005–2006 | 2.46 | 10.49 | 9.22 | 1.77 | 0.54 | |
| (2006–12) | (2006–12) | (2006–12) | (2006–11) | (2006–11) | ||
| 2006–2007 | 5.14 | 22.80 | 12.56 | 4.40 | 1.78 | |
| (2007–06) | (2007–06) | (2007–05) | (2007–06) | (2007–07) | ||
| 2007–2008 | 5.16 | 14.54 | 10.84 | 5.88 | 1.69 | |
| (2008–02) | (2008–04) | (2008–04) | (2008–02) | (2008–02) | ||
| 2008–2009 | 5.55 | 16.60 | 12.03 | 5.21 | 2.18 | |
| (2009–05) | (2009–05) | (2009–04) | (2009–05) | (2009–06) | ||
| 2009–2010 | 13.28 | 20.41 | 43.68 | 8.85 | 1.63 | |
| (2009–44) | 2009–45 | 2009–44 | (2009–45) | (2009–45) | ||
| 2010–2011 | 8.13 | 17.90 | 20.22 | 5.94 | 1.68 | |
| (2011–05) | (2011–04) | (2011–05) | (2011–05) | (2011–05) | ||
| 2011–2012 | 8.47 | 24.85 | 16.93 | 6.60 | 3.22 | |
| (2012–05) | (2012–04) | (2012–04) | (2012–05) | (2012–06) | ||
| 2012–2013 | 8.09 | 17.38 | 19.57 | 6.2 | 2.19 | |
| (2013–05) | (2013–05) | (2013–05) | (2013–06) | (2013–06) | ||
| 2013–2014 | 4.94 | 12.93 | 7.96 | 5.1 | 1.79 | |
| (2014–06) | (2014–06) | (2014–06) | (2014–06) | (2014–02) | ||
The weeks during which peak incidence was reported are in brackets..
During the 2005–2006 season, ILI incidence exceeded slightly the threshold value of 2‰ late in March and only for few weeks (weeks 11 and 12 of 2006), thus indicating a very limited influenza activity during this season. The following three epidemics (from 2006–2007 to 2008–2009) were characterised by a comparable impact, with an ILI peak incidence of about 5‰. A similar picture was observed during the three post-pandemic seasons (from 2010–2011 to 2012–2013) when the epidemic waves almost overlapped in terms of impact (an ILI incidence at peak of nearly 8‰) and time frame (peak at week 5). These three seasons were characterised by the alternation of predominant viral type/subtype: A(H1N1)pdm09 virus in 2010–2011, A(H3N2) virus in 2011–2012, and influenza B virus in 2012–2013. The latter influenza season considered (2013–2014) was characterised by a lower influenza activity compared with the eight previous ones, with a peak ILI morbidity below 5‰.
In all considered seasons, the highest ILI incidence rates were recorded in children ≤4-y-of-age and in school-aged children (5–14 y). Children ≤4-y-of-age were usually affected more during the epidemics sustained by the A(H3N2) virus (i.e., seasons 2006–2007, 2008–2009, 2011–2012, and 2013–2014), whilst school-aged children were particularly affected in those seasons characterised by the prevalent circulation of A(H1N1)pdm09 virus (seasons 2009–2010 and 2010–2011) and in the 2012–2013 season, when influenza B virus was predominant. The lowest ILI morbidity rates were usually reported in individuals ≥65-y-of-age, with some exceptions. In fact, during the 2004–2005 and 2011–2012 seasons, the ILI morbidity rates in the population over 65-y-of-age were higher than those registered in the other epidemics (including the pandemic one). Both of these seasons were characterised by the predominant circulation of A(H3N2) viruses with specific antigenic and genetic features.6,7
Virological surveillance
Respiratory samples (nasal swabs) were collected from outpatients with a clinical presentation of ILI. Overall, 5952 nasal swabs were collected and analysed during the study period. The percentage of samples that resulted in positive influenza virus identification ranged between 31.8% and 46.8%. As shown in Table 2, each influenza season was characterised by a specific virological profile. In all considered epidemics except two (2007–2008 and 2012–2013 seasons), influenza virus type A detections predominated over type B detections in the surveyed area. Within type A, the H3 subtype predominated over H1 in the 2004–2005 (H3:H1 ratio = 6.4), 2006–2007 (H3:H1 ratio = 4.9), 2008–2009 and 2011–2012 (no H1 identified), and 2013–2014 (H3:H1 ratio = 2.3) seasons.
Table 2. Proportion (%) of influenza virus types and subtypes out of total influenza virus detections in Lombardy by season (from 2004–2005 to 2013–2014).
| Season | Influenza A(H1N1) virus* | Influenza A(H3N2) virus | Influenza B virus |
|---|---|---|---|
| 2004–2005 | 9% | 54% | 38% |
| 2005–2006 | 40% | 40% | 20% |
| 2006–2007 | 16% | 78% | 6% |
| 2007–2008 | 36% | n.d. | 64% |
| 2008–2009 | n.d. | 92% | 8% |
| 2009–2010 | 78% | n.d. | 22% |
| 2010–2011 | 60% | 5% | 35% |
| 2011–2012 | 4% | 87% | 9% |
| 2012–2013 | 19% | 3% | 78% |
| 2013–2014 | 30% | 70% | n.d. |
Referred to as A(H1N1)pdm09 from 2009 onwards; n.d., not detected.
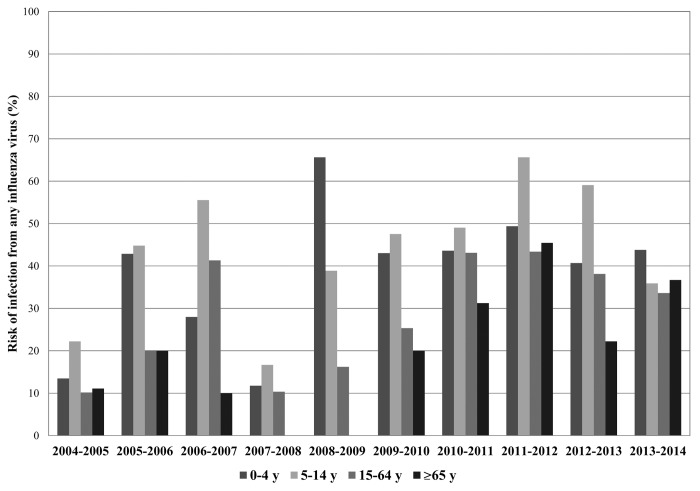
The risk of infection from any influenza virus by age group and by season was calculated as the number of laboratory-confirmed influenza infections out of the total number of subjects with ILI. As shown in Figure 1, the risk of infection was always relevant among the younger subjects (0–14 y), but increased in the last seasons even among the elderly, probably as a consequence of the decrease of influenza vaccine coverage rate in this age group.
Figure 1. Risk of infection from any influenza virus by age group and by season (from 2004–2005 to 2013–2014). Risk of infection is expressed as a percentage (number of individuals with a laboratory-confirmed influenza virus infection out of the total number of subjects with ILI).
Molecular characteristics of circulating influenza viruses and match with vaccine strains
A spatial-temporal representative number of influenza-positive specimens collected during the analysed seasons was genetically characterised, and the phylogenetic analysis of the HA gene was performed.
Influenza A(H1N1) viruses
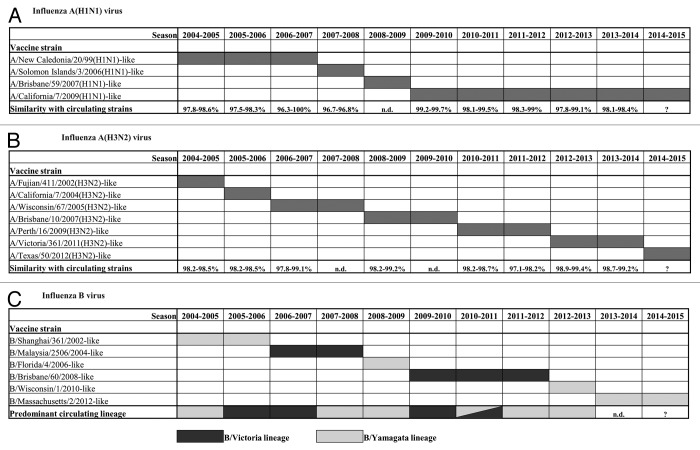
Influenza A(H1N1) viruses were scaled-down during the 2004–2005 and 2006–2007 seasons (accounting for 9% and 16% of all influenza virus detections, respectively), though these viruses circulated at a greater extent during the 2005–2006 and 2007–2008 epidemics, accounting for nearly 40% of all detections. The A(H1N1) viruses circulating during these seasons shared high similarity with viral strains included in vaccine formulations for the related epidemics (Fig. 2A).
Figure 2. Viral strains included in seasonal influenza vaccine formulations from 2004–2005 to 2013–2014 seasons and their genetic match (identity range) with influenza viruses circulating in Lombardy by season and by type/subtype. (A) A(H1N1) influenza virus (referred to as A[H1N1]pdm09 from 2009 onwards); (B) A(H3N2) influenza virus, and (C) influenza B virus. (n.d., not detected).
In 2009, a novel A(H1N1) virus was introduced into the human population. Since then, no former seasonal influenza A(H1N1) viruses were detected in the surveyed area. A(H1N1)pdm09 viruses circulated almost exclusively during the pandemic (2009–2010) and post-pandemic season (2010–2011). These viruses were identified only sporadically (4% of all detections) in the 2011–2012 season, though accounted for 19% and 30% of all influenza detections in the latter two seasons (2012–2013 and 2013–2014, respectively). The phylogenetic analysis of HA sequences revealed that A(H1N1)pdm09 viruses detected between 2009 and 2014 fell into different phylogenetic groups, according to WHO classification.8 The HA sequences of the A(H1N1)pdm09 viruses identified in the post-pandemic season (2010–2011) segregated in different genetic groups with respect to those identified during the 2009 pandemic and were mainly dispersed among group 5 (A/Astrakhan/1/2011-like, characterised by amino acid mutations D97N, R205K, I216V, and V249L) and group 6 (A/St Petersburg/27/2011-like, characterised by amino acid changes D97N and S185T). A(H1N1)pdm09 viruses identified during the 2012–2013 season fell into group 7 (A/St. Petersburg/100/2011-like, with amino acid mutations S124N, S143G, S185T, and A197T) or group 6 (A/St Petersburg/27/2011-like, characterised by amino acid changes D97N, S124N, and S185T). In particular, these latter were further dispersed in subgroup 6A (A/Tennessee/09/2012-like) or 6B (A/Hong Kong/5659/2012-like). All HA sequences of A(H1N1)pdm09 identified in the 2013–2014 season belonged to this latter subgroup—characterised by the amino acid changes K163Q, A256T, and K283E—and shared high similarity with the reference strain A/South Africa/3626/2013 (identity range: 98.9–99.5%). Thus, although several viral variants have been circulating since its emergence in the human population, the post-pandemic A(H1N1)pdm09 viruses remain closely related to the reference vaccine viral strain (A/California/7/2009) (Fig. 2A).
Influenza A(H3N2) viruses
Influenza A(H3N2) viruses were detected with significant proportions in most analysed seasons (2004–2005, 2005–2006, 2006–2007, 2008–2009, 2011–2012, and 2013–2014); were only sporadically detected in the 2010–2011 and 2012–2013 seasons; and undetected in the remaining seasons (2007–2008 and 2009–2010).
The HA sequences of the A(H3N2) viruses isolated in the surveyed area during the considered epidemics fell into different phylogenetic groups. Those identified in the 2004–2005 season shared a 98.2–98.6% identity with vaccine strain A/Fujian/411/2002(H3N2), which was replaced by the A/California/7/2004(H3N2) strain in the following season. The HA sequence of A(H3N2) viruses identified during 2006–2007 epidemic, when this subtype was predominant, were similar to vaccine strain A/Wisconsin/67/2005 (identity range: 97.8–99.1%). The 2008–2009 season was characterised by the extensive circulation of A(H3N2) viruses that shared a high similarity (range: 98.2–99.2%) with the vaccine strain A/Brisbane/10/2007(H3N2). Most of the HA sequences from A(H3N2) viruses detected during the 2010–2011 season, which was characterised by a low circulation of this viral subtype, fell into the A/Victoria/208/2009 genetic clade, and segregated into group 7 (A/Alabama/04/2011-like). The identity of these HA sequences with those of the vaccine strain A/Perth/16/2009(H3N2) ranged between 98.2% and 98.7%. All HA sequences of the A(H3N2) viruses isolated in the 2011–2012 season fell predominantly into group 6 (A/Iowa/19/2010-like) or group 3 of the A/Victoria/208/2009 genetic clade, with a similarity of 97.1–98.2% to the vaccine virus A/Perth/16/2009(H3N2). All HA sequences of A(H3N2) detected in the 2012–2013 season belonged to the genetic group 3C (A/Victoria/361/2011-like, which is characterised by the amino acid mutations S45N, T48I, S145N, A198S, V223I, and N312S). The HA sequences of influenza A(H3N2) identified in the latter epidemic fell into the genetic subgroup 3C. Overall, the viruses molecularly characterised during this season were similar (range: 98.5–99%) to the A/Texas/50/2012(H3N2) strain (Fig. 2B).
Influenza B viruses
In the surveyed area, influenza virus type B detections predominated over type A only during two epidemics (i.e., 2007–2008 and 2012–2013). In the other seasons, the proportion of influenza B virus identified ranged between 6% to 38% of all influenza virus detections. During seasons in which co-circulation of influenza A and B viruses was observed, B viruses usually circulated later in the season (end of March) than influenza A viruses (which generally circulate in January–February).
The phylogenetic analysis of the HA sequences of influenza B viruses clearly showed that B viruses presented with a mixed circulation of viral variants, since viruses belonging to both B/Victoria and B/Yamagata lineages co-circulated in different proportions (Fig. 2C). This peculiar epidemiological feature was observed in the 2007–2008 season, when viruses of the B/Yamagata lineage predominated by a 3-fold margin over those of the B/Victoria lineage, while the vaccine for the 2007–2008 season contained the B/Malaysia/2508/2004 virus of the B/Victoria lineage. On the other hand, the last influenza B season (2012–2013) was characterised by the almost exclusive circulation of B viruses belonging to the B/Yamagata lineage (included in the 2012–2013 vaccine formulation).
Overall, a complete (or nearly complete) match between circulating B viruses and vaccine strains was observed in 5 out of 10 analysed seasons, a partial match (50%) in one season (2010–2011), and a mismatch in three seasons (2005–2006, 2007–2008, and 2011–2012).
Influenza vaccine coverage rate
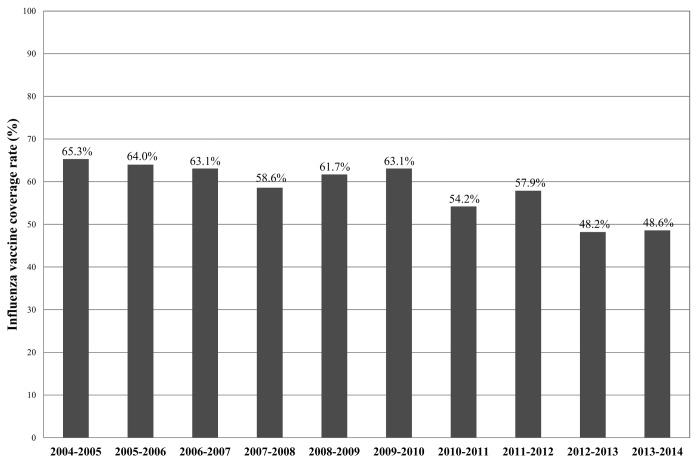
In Lombardy, influenza vaccine coverage rate in the general population was 16% in the 2005–2006 season and decreased to 11.7% in 2013–2014. Among individuals over 65 y, who comprise the main vaccine-target population, the coverage rate was 65.3% in 2004–2005 and, starting from 2010, decreased over the years, and was 48.6% in the 2013–2014 season (Fig. 3). This decrease in coverage is likely due to a loss of confidence in the influenza vaccine after the 2009 pandemic. These coverage rates are far below the minimum planned threshold level of 75%.9
Figure 3. Influenza vaccine coverage rates in individuals ≥65 y (vaccine target population) in Lombardy by season (from 2004–2005 to 2013–2014).
Conclusion
As part of the Italian influenza surveillance network, the activity and circulation of influenza viruses in Lombardy were investigated during the past 10 consecutive seasons. In this surveyed area, the epidemic waves had different profiles with the highest ILI morbidity rates recorded during the 2004–2005 and 2009–2010 seasons. Both of these epidemics were characterised by the introduction of novel viral strains: a drift HA variant of previously circulating A(H3N2) virus and the 2009 A(H1N1) pandemic virus.
Each influenza season was characterised by a specific virological profile; influenza virus type A detections predominated over type B in all considered epidemics except those that occurred in 2007–2008 and in 2012–2013. Within type A, the H3 subtype predominated over H1 in five out of the eight influenza A seasons. However, although a predominant influenza virus was identified in each season, every epidemic was characterised by composite epidemiological patterns where several viral types/subtypes circulated to a different extent.
From an epidemiological point of view, all considered influenza seasons were characterised by higher morbidity rates in children (≤14-y-of-age), whereas the lowest ILI morbidity rates were usually reported in individuals ≥65 y. This latter figure was observed in all seasons except the 2004–2005 and 2011–2012 seasons, when higher morbidity rates were reported among the elderly. It is interesting to note that both of these latter seasons were characterised by the predominant circulation of A(H3N2) viruses with specific antigenic and genetic features.6,7
The vaccine coverage rate among the elderly reached its peak (65.3%) in 2004–2005 and decreased consistently from 2010. This decrease is likely due to a loss of confidence in the influenza vaccine after the 2009 pandemic.10
The molecular characterisation of circulating influenza viruses showed a general good match between seasonal vaccine strains and circulating influenza A viruses over the past 10 y. A different epidemiological profile was observed for influenza B viruses. Such viruses presented with a mixed circulation of viral variants, since viruses belonging to both B/Victoria and B/Yamagata lineages co-circulated during the considered seasons in different proportions. As previously reported by other authors,11-13 this epidemiological feature suggests that the inclusion of strains belonging to both B lineages in influenza vaccines may be advisable. Licensed trivalent influenza vaccines (TIV) contain antigens from only a single influenza B virus, thus providing limited immunity against circulating influenza B strains of the lineage not present in the vaccine. In recent years, predictions about which B lineage will predominate in an upcoming influenza season have been no better than casual—correct in only 5 of the 10 seasons from 2001 to 2011.14 Seasonal influenza vaccines may be improved by inclusion of influenza B strains of both lineages. The resulting quadrivalent influenza vaccines (QIV) would allow influenza vaccination campaigns to respond more effectively to current global influenza epidemiology.15,16 The WHO GISRS has recommended and encouraged further research and clinical evaluation of QIV.17
In conclusion, epidemiological and virological surveillance of influenza is an essential informative tool for tuning vaccination strategies. The continuous surveillance of the characteristics of circulating viruses is crucial to monitor their matching with seasonal vaccine strains.
Strategies to increase vaccination coverage are warranted to control and prevent influenza in individuals at risk of complications, such as the elderly, as well as in the population at large.
Materials and Methods
The Influenza Surveillance Network methodology
The influenza epidemiological and virological surveillance network (InfluNet)18 consists of a number of paediatricians and general practitioners, who survey approximately 2% of the general population, reporting weekly on the number of new cases of influenza-like illness (ILI), and collecting respiratory samples for virological evaluation.9 The standard case definition of ILI is abrupt onset of fever (>38 °C), one or more respiratory symptoms (non-productive cough, sore throat, or rhinitis), and one or more systemic symptoms (myalgia, headache, or severe malaise).9
Epidemiological data are collected from week 42 to week 17 of the following year, while virological surveillance begins on week 46 and ends on week 17 of the following year. Epidemiological and virological data are collected at a regional level and are aggregated subsequently at a national level on a weekly basis.9,18
Influenza viruses identification and molecular characterisation
RNA was extracted from respiratory samples using the Invisorb® Spin Virus RNA Mini kit (Stratec Molecular GmbH, catalogue number: 1040300300). A one-step real-time retro-transcription (RT) multiplex polymerase chain reaction (PCR) assay was performed to simultaneously detect influenza A and B viruses using primer/probe sets for two different genome regions: the matrix region of influenza type A virus and the nucleoprotein region of influenza type B virus.19 The subtyping of influenza A positive samples was performed by a one-step real-time RT-PCR assay using specific primer/probe sets for the HA gene.19
The molecular characterisation of influenza viruses was performed by sequence analysis of the globular head region of the HA protein (HA1 subunit) specific for A(H1N1) influenza virus (nucleotide, nt., 64–1058), A(H3N2) influenza virus (nt. 174–1056), and B (nt. 12–1169) influenza virus.11,20,21 The HA nucleotide sequences were obtained by automated DNA sequencing (ABI PRISM® 3100 Genetic Analyzer, Life Technologies). Multiple sequence alignment was conducted using ClustalX, version 2.0.10.
Influenza vaccine coverage
Influenza vaccine coverage rates are presented as the percentage of individuals immunised with influenza vaccine per year. The record of the number of individuals vaccinated in Lombardy was collected from local health authorities and then put in the data warehouse managed and coordinated by the Directorate General for Health, Regione Lombardia. To estimate vaccine coverage rates, data on population were obtained from the Italian National Institute of Statistics (Istat) database.22
Statistical analysis
The risk of infection is expressed as number of subjects with a laboratory-confirmed infection out of the total number of subjects with ILI. Data were analysed by the Open Source Epidemiologic Statistics for Public Health (OpenEpi), version 3.01.23
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by grants from Directorate General for Health, Regione Lombardia (grant no. 5988 and no. 17616).
Glossary
Abbreviations:
- GISRS
Global Influenza Surveillance and Response System
- HA
Hemagglutinin
- ILI
Influenza-like Illness
- PCR
Polymerase Chain Reaction
- QIV
Quadrivalent Influenza Vaccine
- RT
Retro Transcription
- TIV
Trivalent Influenza Vaccine
- WHO
World Health Organization
References
- 1.Nicholson KG, Wood JM, Zambon M.. Influenza. Lancet 2003; 362:1733 - 45; http://dx.doi.org/ 10.1016/S0140-6736(03)14854-4; PMID: 14643124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization (WHO). Influenza (Seasonal) - Fact sheet 211. Available from: http://www.who.int/mediacentre/factsheets/fs211/en/. (Last access: 06/25/2014).
- 3.European Centre for Disease Prevention and Control (ECDC). Seasonal human influenza and vaccination - the facts. Available from: http://ecdc.europa.eu/en/healthtopics/documents/0712_seasonal_human_influenza_vaccination.pdf. (Last access: 06/25/2014).
- 4.Hay AJ, Gregory V, Douglas AR, Lin YP.. The evolution of human influenza viruses. Philos Trans R Soc Lond B Biol Sci 2001; 356:1861 - 70; http://dx.doi.org/ 10.1098/rstb.2001.0999; PMID: 11779385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization (WHO) Global Influenza Surveillance and Response System. (GISRS). Available from: http://www.who.int/influenza/gisrs_laboratory/en/ (Last access: 06/25/2014).
- 6.Jin H, Zhou H, Liu H, Chan W, Adhikary L, Mahmood K, Lee MS, Kemble G.. Two residues in the hemagglutinin of A/Fujian/411/02-like influenza viruses are responsible for antigenic drift from A/Panama/2007/99. Virology 2005; 336:113 - 9; http://dx.doi.org/ 10.1016/j.virol.2005.03.010; PMID: 15866076 [DOI] [PubMed] [Google Scholar]
- 7.Pariani E, Amendola A, Ebranati E, Ranghiero A, Lai A, Anselmi G, Zehender G, Zanetti A.. Genetic drift influenza A(H3N2) virus hemagglutinin (HA) variants originated during the last pandemic turn out to be predominant in the 2011-2012 season in Northern Italy. Infect Genet Evol 2013; 13:252 - 60; http://dx.doi.org/ 10.1016/j.meegid.2012.10.019; PMID: 23174527 [DOI] [PubMed] [Google Scholar]
- 8.World Health Organization (WHO) Influenza Centre London. February 2014 interim report. Report prepared for the WHO annual consultation on the composition of influenza vaccine for the Northern Hemisphere 2014/2015. 17th–19th February 2014. Available from: http://www.nimr.mrc.ac.uk/documents/about/NIMR-report-Feb2014-web.pdf (Last access: 06/25/2014).
- 9.Ministero della Salute. Available from: http://www.salute.gov.it/ (Last access: 06/25/2014).
- 10.Poland GA.. The 2009-2010 influenza pandemic: effects on pandemic and seasonal vaccine uptake and lessons learned for seasonal vaccination campaigns. Vaccine 2010; 28:Suppl 4 D3 - 13; http://dx.doi.org/ 10.1016/j.vaccine.2010.08.024; PMID: 20713258 [DOI] [PubMed] [Google Scholar]
- 11.Ansaldi F, D’Agaro P, De Florentiis D, Puzelli S, Lin YP, Gregory V, Bennett M, Donatelli I, Gasparini R, Crovari P, et al.. Molecular characterization of influenza B viruses circulating in northern Italy during the 2001-2002 epidemic season. J Med Virol 2003; 70:463 - 9; http://dx.doi.org/ 10.1002/jmv.10418; PMID: 12767012 [DOI] [PubMed] [Google Scholar]
- 12.Puzelli S, Frezza F, Fabiani C, Ansaldi F, Campitelli L, Lin YP, Gregory V, Bennett M, D’Agaro P, Campello C, et al.. Changes in the hemagglutinins and neuraminidases of human influenza B viruses isolated in Italy during the 2001-02, 2002-03, and 2003-04 seasons. J Med Virol 2004; 74:629 - 40; http://dx.doi.org/ 10.1002/jmv.20225; PMID: 15484280 [DOI] [PubMed] [Google Scholar]
- 13.Chen GW, Shih SR, Hsiao MR, Chang SC, Lin SH, Sun CF, Tsao KC.. Multiple genotypes of influenza B viruses cocirculated in Taiwan in 2004 and 2005. J Clin Microbiol 2007; 45:1515 - 22; http://dx.doi.org/ 10.1128/JCM.02189-06; PMID: 17329451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ambrose CS, Levin MJ.. The rationale for quadrivalent influenza vaccines. Hum Vaccin Immunother 2012; 8:81 - 8; http://dx.doi.org/ 10.4161/hv.8.1.17623; PMID: 22252006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belshe RB.. The need for quadrivalent vaccine against seasonal influenza. Vaccine 2010; 28:Suppl 4 D45 - 53; http://dx.doi.org/ 10.1016/j.vaccine.2010.08.028; PMID: 20713260 [DOI] [PubMed] [Google Scholar]
- 16.Reed C, Meltzer MI, Finelli L, Fiore A.. Public health impact of including two lineages of influenza B in a quadrivalent seasonal influenza vaccine. Vaccine 2012; 30:1993 - 8; http://dx.doi.org/ 10.1016/j.vaccine.2011.12.098; PMID: 22226861 [DOI] [PubMed] [Google Scholar]
- 17.SAGE Working Group on Influenza Vaccines and Immunization. Background paper on influenza vaccines and immunization SAGE Working Group. 2012. http://www.who.int/immunization/sage/meetings/2012/april/1_Background_Paper_Mar26_v13_cleaned.pdf (Last access: 06/25/2014).
- 18.InfluNet website. Available from: http://www.iss.it/iflu/ (Last access: 06/25/2014).
- 19.World Health Organization (WHO) Global Influenza Surveillance Network. Manual for the laboratory diagnosis and virological surveillance of influenza. 2011. Available from: http://whqlibdoc.who.int/publications/2011/9789241548090_eng.pdf (Last access: 06/25/2014).
- 20.Ellis JS, Alvarez-Aguero A, Gregory V, Lin YP, Hay A, Zambon MC.. Influenza AH1N2 viruses, United Kingdom, 2001-02 influenza season. Emerg Infect Dis 2003; 9:304 - 10; http://dx.doi.org/ 10.3201/eid0903.020404; PMID: 12643824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ellis JS, Chakraverty P, Clewley JP.. Genetic and antigenic variation in the haemagglutinin of recently circulating human influenza A (H3N2) viruses in the United Kingdom. Arch Virol 1995; 140:1889 - 904; http://dx.doi.org/ 10.1007/BF01322680; PMID: 7503689 [DOI] [PubMed] [Google Scholar]
- 22.Italian National Institute of Statistics (Istat). Available from: http://www.istat.it/en/products/databases (Last access: 06/25/2014).
- 23.Open Source Epidemiologic Statistics for Public Health (OpenEpi). Available from: http://www.openepi.com (Last access: 06/25/2014).