Abstract
Recombinant VLP-based vaccines have been successfully used against 3 diseases caused by viral infections: Hepatitis B, cervical cancer and hepatitis E. The VLP approach is attracting increasing attention in vaccine design and development for human and veterinary use. This review summarizes the clinically relevant epitopes on the VLP antigens in successful human vaccines. These virion-like epitopes, which can be delineated with molecular biology, cryo-electron microscopy and x-ray crystallographic methods, are the prerequisites for these efficacious vaccines to elicit functional antibodies. The critical epitopes and key factors influencing these epitopes are discussed for the HEV, HPV and HBV vaccines. A pentamer (for HPV) or a dimer (for HEV and HBV), rather than a monomer, is the basic building block harboring critical epitopes for the assembly of VLP antigen. The processing and formulation of VLP-based vaccines need to be developed to promote the formation and stabilization of these epitopes in the recombinant antigens. Delineating the critical epitopes is essential for antigen design in the early phase of vaccine development and for critical quality attribute analysis in the commercial phase of vaccine manufacturing.
Keywords: capsid protein, conformation-dependent epitope, human vaccine, neutralizing antibodies, immunodominant epitopes, virus-like particle
Introduction
Virus-like particles (VLPs) as a passport to immune system recognition have been widely used in vaccine development for prophylaxis against viral infection.1 VLPs are safe because of their non-replicating nature and are strong immunogens attributing to their particulate nature.2-4 In addition, VLPs are highly ordered macromolecular assemblies of the viral protein(s), presenting virion-like epitopes that can be uptaken by antigen presenting cells. The VLP antigen with regular and repetitive epitopes has been demonstrated to more effectively stimulate the immune response than subunit proteins or peptides with much smaller molecular weights.5-7
Over the past 3 decades, VLP-based vaccines have made a great impact in viral disease prevention.4 VLP-based vaccines against 3 viral infections have been approved for human use to date: Recombivax HB® and Engerix®-B against hepatitis B virus (HBV), Gardasil® and Cervarix® against human papillomavirus (HPV), and Hecolin® against hepatitis E virus (HEV) (Table 1). All of the 5 vaccines, listed in Table 1, have demonstrated a significant clinical effect with good safety and efficacy profiles. Currently, numerous VLP-based vaccine candidates have entered preclinical testing or clinical trials.
Table 1.
Representative recombinant VLP-based human vaccines on the market
| Trade name | Manufacture (year of licensure) | Expression host | Structure | Potency marker | Reference |
|---|---|---|---|---|---|
| Recombivax HB (HBV) | Merck (1986) | Saccharomyces cerevisiae | Octahedral | RF-1, A1.2, 5F11 | 104,105,111,114,115,152 |
| Engerix-B (HBV) | GlaxoSmithKline (1988) | Saccharomyces cerevisiae | N/A | RF-1 | 153 |
| Gardasil (HPV) | Merck (2006) | Saccharomyces cerevisiae | Icosahedral | H16.V5, H18.R5, H11.B2, H6.B10.5 | 20,68,154,155 |
| Cervarix (HPV) | GlaxoSmithKline (2009) | Trichoplusiani/baculovirus | Icosahedral | H16.V5 | 156 |
| Hecolin (HEV) | Xiamen Innovax Biotech (2011) | Escherichia coli | N/A | 8C11, 8H3 | 8,21,31 |
The evaluation of vaccine potency can be conducted by animal testing and clinical trials.8-13 Generation of neutralizing antibodies against specific epitopes is the underlying mechanism of efficacious prophylactic vaccines. In general, titer of neutralizing antibodies elicited by vaccination correlates with the efficacy of a given vaccine. Numerous studies have demonstrated that the generation of neutralizing antibodies is dependent on the correct antigen conformation and native-like epitope presentation on the VLP surface. Recombinant VLPs maintain the 3D structural features of key epitopes on antigen surface. Effective antigen presentation with regularly arrayed epitopes on the nanometer scale bionanoparticles and high local antigen concentration (depot effect for an adjuvanted vaccine) could account for this effective Th2 response. For Th1 response involving cell-mediated immune response, the antigens are processed into peptide fragments prior to binding to MHCs, making VLPs to be less advantageous than smaller subunits in stimulating immune system. Making the native-like epitopes on VLP surface is of utmost importance in vaccine design and vaccine bioprocessing.
In this study, the importance of neutralizing epitopes localized on the VLP surface for vaccine potency is reviewed for VLP-based human vaccines. Neutralizing epitopes are the structural basis of an antigen to elicit functional and neutralizing antibodies. These epitopes or surface features on VLPs can be identified by various methods including cryo-EM and crystal structural determination.4 For HEV, some representative neutralizing epitopes on the VLP surface have been identified with cryo-EM using 3D structural reconstruction and/or crystal structure analysis in combination with site-directed mutagenesis to confirm the epitope structure on the viral capsid.14-18 In contrast, due to lack of high-resolution crystal structures or 3D reconstruction structures of the VLPs, the type-specific neutralizing epitopes localized in the hypervariable loops of the HPV VLP surface which determine the type specificity for genotypes and serotypes have been identified mainly by molecular biology methods such as loop swapping, foreign epitopes grafting into the surface loops and site-directed mutagenesis.19,20
Full exposure of the key neutralizing epitopes on the particle surface is essential for VLP-based vaccines to induce effective humoral response and to confer immunity against viral infection.4 The accurate mapping of neutralizing epitopes on the whole viral capsid or on subunits would aid in better understanding of the vaccine potency and in facilitating the rational design and refinement of VLP-based vaccines. Increased knowledge on epitopes would lead to improved epitope-based assays, which are useful in process control. This is critically important to have well designed assays during vaccine manufacturing owing to the biochemical or biophysical complexity in vaccine products.
Mapping of immuno dominant and conformation-dependent neutralizing epitopes of VLPs using molecular biology methods
Prophylactic vaccines are efficacious against virus infection. VLP-based vaccines exert their function by inducing the humoral immune response to generate neutralizing antibodies, which are raised against specific epitopes presented by the spatial structure of the immunogen. The integrity of these clinically relevant epitopes localized on the surface of VLPs is critically important for inducing effective titers of functional antibodies, thus conferring immunity against a certain pathogen. The dominant neutralizing epitopes are identified using a method of antibody cross-blocking of a given mAb versus a clinical serum or serum from an immunized animal. The detection of virus neutralizing activity is usually achieved by PCR-based virus capturing neutralization assay for HEV-specific antibodies or a pseudovirus neutralization model for anti-HPV antibodies.21,22 Additionally, the neutralizing effect of serum antibodies against a human viral pathogen can be tested by passive immunity of transferring polyclonal antibodies or administration of one or more monoclonal antibodies followed by virus challenge in animal models.8
Mapping of the immuno dominant neutralizing epitopes of HEV VLPs
Two VLP-based vaccines against HEV have been developed, both containing aa 458–607 as a core antigenic region of pORF2 (the capsid protein encoded by ORF2). This region harbors the key neutralizing epitopes against the virus.23 The VLP antigen used in the first vaccine is assembled from a 56 kDa peptide that was expressed in insect cells and encoded by ORF2.24,25 The results of a phase II trial conducted in Nepal demonstrated that the vaccine was highly efficacious with efficacy against the HEV infection of 95.5% and well tolerated (Fig. 1).11 The other vaccine was a further truncated version of pORF2, designated as p239. p239, a shorter version of pORF2, self-assembles into VLPs with a total of 239 aa (MW ˜26 kDa). The p239 antigen was expressed in an E.coli system and self-assembled into particles with a diameter of 20–30 nm in purified antigen preparations.8 A total of 112,604 healthy adult participants were enrolled in a randomized, controlled phase III clinical trial. The vaccine was also well-tolerated, protected against hepatitis E disease with an efficacy of 100% and had 78% efficacy against HEV infection.9 This VLP-based vaccine was licensed for human use in China in 2011 (Fig. 1 and Table 1).26
Figure 1.
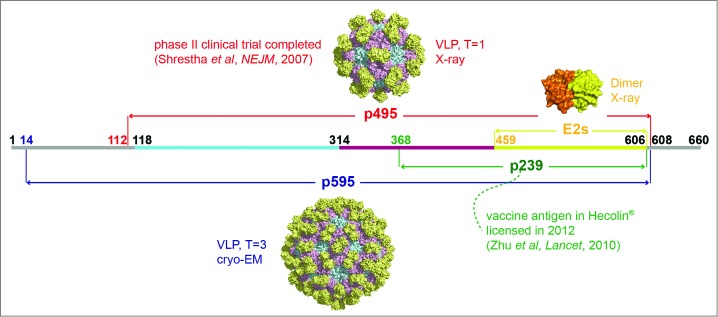
Presentation of different truncated versions of HEV pORF2. E2s (aa 459–606), the shortest version to form dimer, harbors the major neutralizing epitopes, and its crystal structure was determined with a high resolution of 2.0 Å. p239 (aa 368–606) is the vaccine antigen in Hecolin® in a particulate form.8,9 p495 (aa 112–608), which can form VLP with T = 1 icosahedral symmetry, is used in the other HEV vaccine which has been tested in a phase II clinical trial.17,91 p595 (aa 14–608) also can form a particle (T = 3) which is more similar to the native virions.15 The structure of T = 3 VLP was acquired by using the T = 1 VLP (PDB ID: 2ZTN) as a template to build the homology model with the spatial information of side chain because the reconstruction model from cryo-EM data of T = 3 VLP (PDB ID: 3IYO) only recorded the information of αC atoms. The peptide located at aa 368–459 is supposed to play a key role in inducing multimerization of p239.
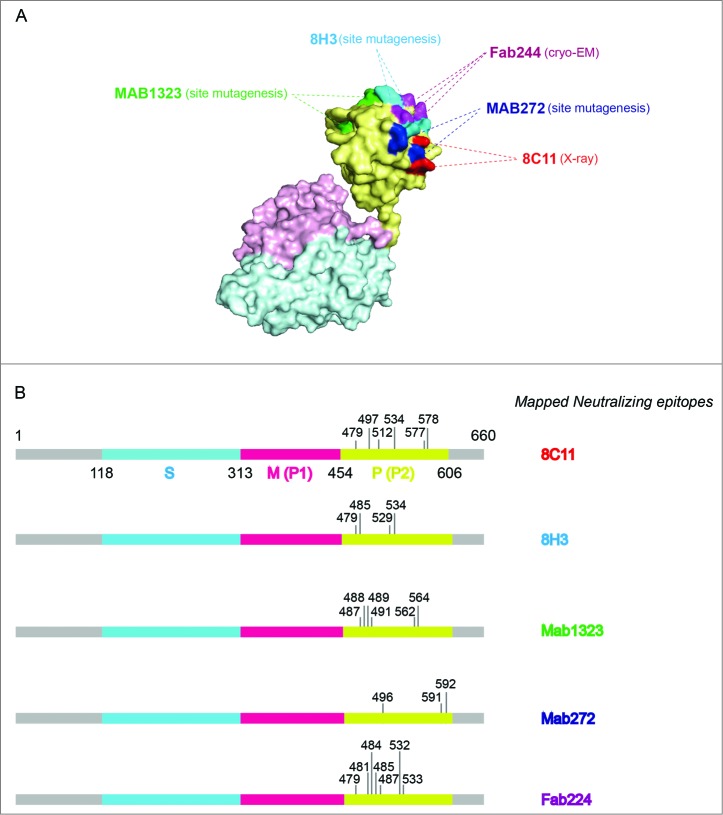
HEV vaccine could play an important role in preventing HEV infection which is responsible for acute viral hepatitis in the developing world, especially resulting in the high mortality in pregnant women about 20–30%.23 HEV infection in animal models can elicit a strong humoral immune response that results in effective immune protection.8 Furthermore, clinical cases have shown that the serum antibodies raised against specific epitopes can protect humans from severe HEV infection-related illness.10,27,28 The HEV genome is approximately 7.2 kb long, single stranded, positive sense and includes 3 ORFs, among which the ORF2 encodes the viral capsid protein (Fig. 1 and Table 1).29 A series of B cell epitopes have been identified, particularly for the neutralizing mAb 8C11, which improves the understanding of the neutralization mechanism (Figs. 2 and 3).16,30 More importantly, because of the importance of the 8C11 epitope, this neutralizing mAb was used to design assays to assess the p239 particle integrity and antigen stability.21,31 Monitoring product consistency and process reproducibility is essential in the lifetime management of a licensed vaccine.
Figure 2.
The binding sites of representative neutralizing antibodies on the HEV VLP surface. (A) The pORF2 monomer is divided into 3 sections named the S domain (aa 118–313), the P1 domain (aa 314–453) or P domain (aa 320–455) and the P2 domain (aa 454–606) or P domain (aa 456–606), which are shown in color blue, purple, and yellow, respectively. The P2 or P domain is dimeric and harbors all of the identified neutralizing epitopes. The neutralizing epitopes determined by different methods against several neutralizing antibodies are shown in different colors. Such as E479, D481, T484, Y485, S487, Y532 and S533 for FAB244;17 S487, S488, T489, P491, N562 and T564 for MAB1323;15 D496, G591 and P592 for MAB272;15 E479, Y485, I529 and K534 for 8H3; E479, S497, R512, K534, H577 and R578 for 8C11. These antibodies are useful serological markers for evaluating the clinical efficacy of the vaccine. (B) Key neutralizing epitopes on P (P2) domain of pORF2. The S domain, M (P1) domain and P (P2) domain are colored in blue, purple, and yellow, respectively.
Figure 3.
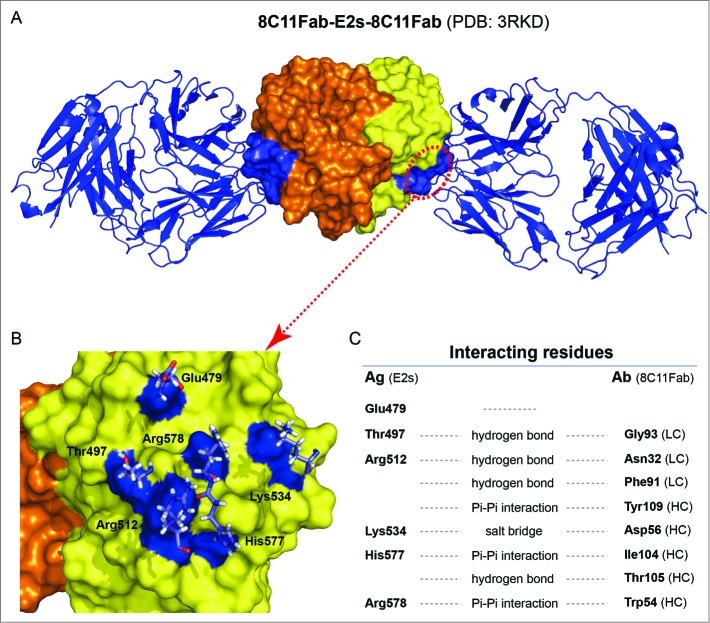
The crystal structure of 8C11Fab in complex with the E2s domain. (A) The structures of the E2s-8C11Fab complexes. (B) The epitope of mAb 8C11 on E2s, including E479, T497, R512, K534, H577 and R578 on the recombinant HEV capsid protein at the interface with an interacting Fab of the 8C11. (C) Interacting residues at the interface between E2s and 8C11Fab. The interactions include hydrogen bonds, Pi-Pi interaction and salt bridge.
In addition to the 8C11 epitope determined by the crystal structure, additional important neutralizing epitopes localized in the pORF2 were studied using classical molecular biology techniques. Zhou et al. indicated that a truncated ORF2 protein (aa 112–607) contains the most antigenic epitopes region in the pORF2 (Figs. 1 and 2).30 Three distinct antigenic regions were identified, localized at aa 25–38, aa 341–354 and aa 517–530 of the pORF2. The same group also demonstrated that the C- (aa 12–147) and N- (aa 573–660) termini of pORF2 contain strong IgG and IgM epitopes by using synthetic peptides.32 Zhang et al. reported that the recombinant capsid protein p166Chn (amino acids 464–629) harbors the major antigenic epitopes of pORF2.33 Another study showed that in the main structural protein encoded by ORF2, 2 immuno dominant antigenic regions were identified at aa 394–470 and aa 546–580.34 Li et al. utilized some GST-ORF2 fusion proteins to identify that aa 394–660 of the pORF2 was strongly reactive with both acute and convalescent sera but aa 394–473 alone was poorly reactive.35 These findings suggest that the reactive epitopes are discontinuous and conformational (Fig. 2).
The immuno dominant and shorter epitope region was further identified. Riddell et al. suggested that the sequence spanning aa 394 to 457 of the capsid protein participates in the formation of strongly immuno dominant epitopes on the surface of HEV particles. The presence of those epitopes may be important in developing immunity to HEV infection.36 Meng et al. showed that the truncated version of pORF2 containing aa 452–617 elicited antibodies with specific HEV neutralizing activity.37 This was the first identification of the smallest fragment of pORF2 that can efficiently present virion-like neutralizing epitopes. Zhang et al. expressed a 23 kDa peptide, E2, mapping to aa 394–604 of pORF2 using an E.coli expression system (Figs. 1 and 2). This peptide was shown to form homodimers and the dimeric form strongly reacted with human sera from HEV infected individuals.38 Strikingly, the serum reactivity to E2 was abrogated when the dimer was dissociated into monomers. Furthermore, the dimeric form of the peptide elicited a strong immune response in animals. The antisera were capable of neutralizing HEV. Emerson et al. further narrowed down the dominant neutralizing epitopes localized at aa 458–607 of pORF2 (Fig. 2).39 More specifically, Zhang et al. reported that Leu 477 and Leu 613 in pORF2 are critical to the formation of neutralizing antigenic epitopes by constructing the truncated proteins and assessing their reactivity with the identified neutralizing mAbs.40
HPV vaccine and the key epitopes on VLPs
The antigens in HPV vaccines (Gardasil® and Cervarix®) are also in the VLP form. These VLPs are based on the HPV major capsid protein, L1 (Fig. 4). The type-specific immune response induced by HPV infection is mainly directed against the L1 protein. Although the capsomere harbors most of the neutralizing epitopes, the VLP form is still superior to the capsomere because the immunogenicity of VLPs is 20–40 times higher than that of the capsomere form.41,42 Thus, VLPs are promising vaccine immunogens for efficacious vaccines. The efficacy of the vaccines is critically dependent upon the integrity of type-specific conformational epitopes. Studies in animal models have shown systemic immunization with a VLP-based vaccine. In early studies, Harro et al. and Koutsky et al. reported the safety and immunogenicity of an HPV16 VLP-based vaccine in healthy adults.43,44 The vaccine is well tolerated and is highly immunogenic with the majority of the recipients achieving high functional antibody titers that were 40-fold higher than those in natural infection.44 As a result, the HPV16 vaccine reduced the incidence of both HPV16 infection and the related cervical intraepithelial neoplasia.43
Figure 4.
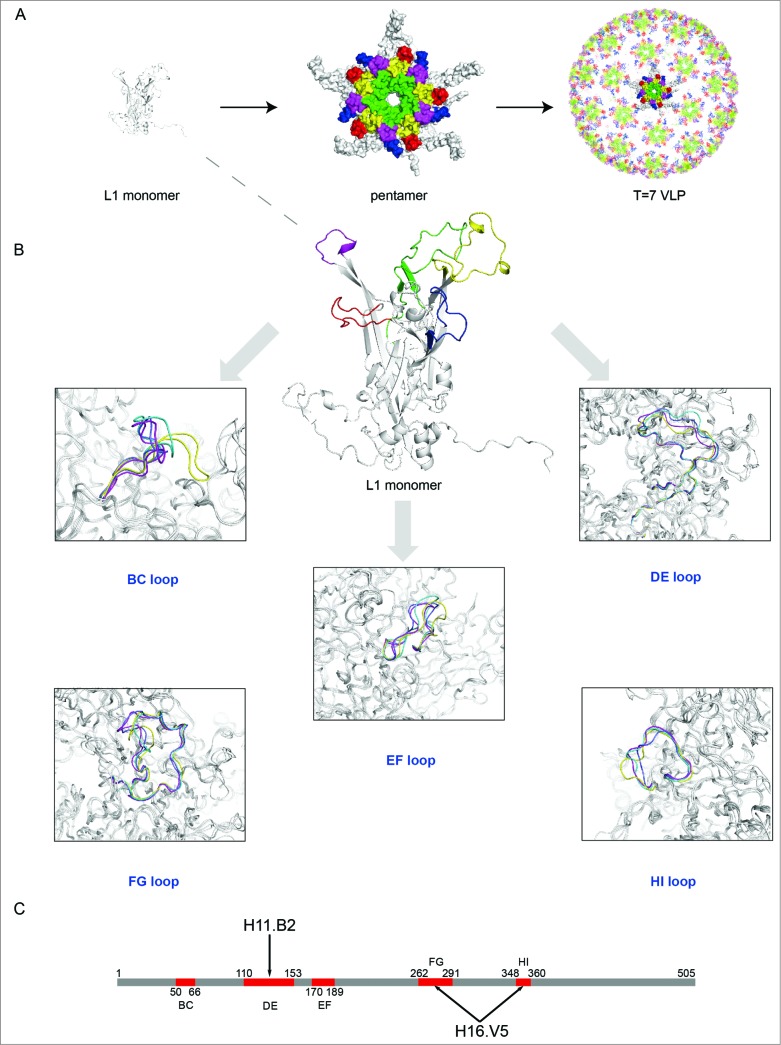
(See previous page) Comparison of different loops, harboring key neutralizing epitopes, of 4 genotypes HPV11, HPV16, HPV18 and HPV35 (for which X-ray structures are available).96 (A) The BC loop, DE loop, EF loop, FG loop and HI loop colored in red, green, blue, yellow and magenta, respectively, are shown in L1 monomer, pentamer and T = 7 VLP. (B) Comparison of the L1 surface loops, BC loop, DE loop, EF loop, FG loop and HI loop respectively. HPV11, HPV16, HPV18 and HPV35 are shown in different colors (blue, magenta, yellow and green, respectively). (C) Localization of the different loops in the full length of HPV16 L1 protein. The epitope regions of 2 type-specific neutralizing mAbs are labeled with an arrow (H16.V5 binding to the FG and HI loop, H11.B2 binding to the DE loop).
Two VLP-based HPV prophylactic vaccines are licensed globally: these were developed and manufactured by Merck & Co., Inc. and GlaxoSmithKline (GSK) (Table 1).4 One is a quadrivalent HPV vaccine produced in yeast (Saccharomyces cerevisiae), Gardasil®, for preventing HPV6/11/16/18 infection and was licensed for use in the USA in June 2006. The other bivalent (16/18) HPV vaccine, which is produced in a baculovirus-based insect cell expression system, is Cervarix®, which is also licensed in the USA. Both vaccines have satisfactory safety profiles and good efficacy at preventing infection and the diseases associated with the relevant types.13,45-51 The efficacy of the quadrivalent HPV vaccine was demonstrated by clinical studies that recruited women from over 30 countries.47 A 56% decrease was found in vaccine type (HPV6/11/16/18) prevalence among females aged 14–19 y from 11.5% in the prevaccine era (2003–2006) to 5.1% in the vaccine era (2007–2010).52 For the bivalent vaccine, at 4.5 y of follow-up, the vaccine is still highly immunogenic and safe and induces a high degree of protection against HPV16/18 infection and the related cervical lesions.48 The serological markers of clinical efficacy are neutralizing functional antibody titers in serum samples. The functional titers are evaluated by competition (with a neutralizing mAb such as V5 for HPV16) based immuno assays, or with pseudovirion-based neutralization assays.22,53
HPVs are small DNA viruses with a circular double stranded DNA genome. Owing to the difficulty of growing and propagating native HPV virions in culture, the pseudovirion system, which includes 2 genes encoding the structural proteins L1 and L2, was developed for neutralization assays.22 The major capsid protein of papillomavirus, L1, successfully expressed in insect cells, yeast, and E.coli, is a ∼55 kDa protein which can spontaneously self-assemble into VLPs with L1 pentamers as the basic building blocks. These recombinant L1-based VLPs have diameters of 50–60 nm and a strong resemblance to native virions (Fig. 4). The VLPs are potent immunogens because the well-defined epitopes are regularly displayed on the VLP surface where they can be easily recognized by B cell receptors and induce high-titer, conformation-dependent neutralizing antibodies. The characterization of the regions of HPVs that can elicit a neutralizing immune response provides insight for the development of prophylactic vaccines and for processing controls during bioprocessing for vaccine production.
In early studies, site mutation and loop swapping were the main methods of delineating neutralizing domains. The development of the athymic mouse xenograft system facilitated serologic studies on the response to HPV infection and verified the presence of specific neutralizing antibodies in the serum of patients with the related clinical diseases.54-56 HPV16 and HPV18 are high-risk types of HPV that associated with cervical and other cancers. Some of the neutralizing epitopes were determined by molecular biological methods, and some of the identified neutralizing mAbs were used to monitor the critical quality attributes of the vaccines.20,57-60
Three neutralizing mAbs (H16.E70, H16.U4 and H16.V5) that are uniquely reactive to surface conformational epitopes on intact HPV16 VLPs were identified by Christensen et al.61 The epitopes of H16.V5 and H16.E70 were primarily mapped to the FG loop and HI loop of HPV16 L1 with high neutralization efficiency (Fig. 4 and Table 3). Roden et al. and White et al. also indicated that in HPV16 the L1 residues 50, 266, 282 are vital for the binding of the neutralizing mAbs H16.V5 and H16.E70.62,63 In addition, Carpentier et al. suggested that amino acid 270 in the FG loop is part of the H16.E70 epitope.64 Carter et al. found that the sequence at both ends of the FG loop (aa 260–290) of L1 was required for the mAbs H16.V5 and H16.E70 (Fig. 4 and Table 3).65,66 Furthermore, a new antibody binding site was identified on the C-terminal arm of L1, between residues 427–445, that was recognized by the mAb H16.U4.65 Recently, Cardone et al. also indicated that the epitope recognized by H16.U4 was mapped to “the suspended bridge” in a recently determined higher-resolution structure of HPV16 mature capsid.67 HPV18-neutralizing mAbs R5 and J4 have been reported and used in the quality control of the 2 licensed HPV vaccines (Table 1 and Table 3).20,68
Table 3.
The key epitope information of neutralizing mAbs against HEV, HPV and HBV
| Virus | mAb | Epitope information | Notes | References |
|---|---|---|---|---|
| HEV | 8C11 | Glu479, Ser497, Arg512, His577, Arg578, Lys534 | Key epitopes located in the dimeric E2s domain which is formed by the P2 (aa 454–606) or P (aa 456–606) domain with hydrophobic interaction and hydrogen bond and presents a single β-barrel structure | 16 |
| 8H3 | Glu479, Tyr485, Ile529, Lys534 | 14 | ||
| MAB272 | Asp496, Gly591, Pro592 | 15 | ||
| MAB1323 | Ser487, Ser488, Thr489, Pro491, Asn562, Thr564 | 15 | ||
| Fab244 | Glu479, Asp481, Thr484, Tyr485, Ser487, Tyr532, Ser533 | 17 | ||
| HPV | V5 (HPV16) | FG loop (aa 266–297) and HI loop (aa 339–365) | Key epitopes located on the 5 flexible surface loops: BC, DE, EF, FG and HI | 20 |
| R5 (HPV18) | NA | 68 | ||
| B10.5 (HPV6) | BC loop (aa 49–54) and DE loop (aa 169–178) | 157 | ||
| B2 (HPV11) | DE loop (aa 123–141) | 73 | ||
| A6 (HPV31) | EF loop | 81 | ||
| E12 (HPV33) | DE (aa 132–140) and FG loop (aa 260–270 and aa 282–291) | 82 | ||
| O3 (HPV35) | NA | 20 | ||
| C1 (HPV45) | NA | 20 | ||
| 8D11, 9F7 (HPV52) | NA | 83 | ||
| 2C3, 6F11 (HPV58) | NA | 83 | ||
| B1.A1(BPV1) | NA | 83 | ||
| HBV | RF1 | aa 124–137 | Key epitopes are sensitive to redox treatment and are located near the dimeric interface | 113 |
| A1.2 | NA | 110 | ||
| 5F11 | NA | 111 |
The type-specific neutralizing epitopes of the low-risk HPV types HPV6 and HPV11 have also been identified. HPV6 and HPV11 are responsible for condylomata acuminata, and antigens from these viruses have also been included in vaccine development, with significant efficacy in reducing the risk of HPV6/11-related genital warts.69-72 Christensen et al. identified three conformation-dependent HPV6-specific antibodies H6.B10.5, H6.M48 and H6.N8 (Table 3).73 Following this study, McClements et al. mapped the binding sites of these 3 mAbs by making HPV11-based amino acid substitutions in the HPV6 major capsid protein L1. The region defined by residues 49–54 in the BC loop is critical for recognition by all 3 of the HPV6-specific, conformation-dependent antibodies. Additionally, the other domain defined by the L1 residues 169–178 in the EF loop also contributes to H6.B10.5 and H6.M48 binding (Table 3).74 It has been demonstrated that specific antibodies (H11.B2, H11.F1, H11.G5, H11.H3 and H11G131S.G3) against HPV11 can neutralize the virus by binding to 2 distinct domains of DE and HI loops on the particle surface.75–78 Specifically, the epitope for H11.B2 was mapped to residues 123–141 in the DE loop of L1, and another 3 mAbs, H11.F1, H11.B2, H11.G5, recognize a dominant epitope centered on residue 132 (Fig. 4 and Table 3). The mAb H11.H3 has been shown to bind to the region that includes residues 346–349 in the HI loop. Another highly neutralizing mAb, H11G131S.G3, has been shown to bind the region encompassing residues 263–290 in the FG loop, in addition to residues 346–349 (Table 3).
The quadrivalent HPV vaccine (types 6, 11, 16, and 18), Gardasil®, can prevent 70% of cervical cancer and high-grade cervical intraepithelial neoplasia and 90% of anogenital warts. Inclusion of more types of HPV in the vaccine would provide even wider protection. Merck has developed a 9-valent HPV vaccine, which has been approved for the US market by the FDA (V503) in which 5 additional HPV types (31/33/45/52/58) are included (licensed in the US in late 2014). The vaccines also use recombinant VLPs as antigens.79 Therefore, information about the neutralizing epitopes of more HPV types is desired. A variety of neutralizing mAbs recognizing immuno dominant epitopes are needed for vaccine quality control and serological antibody analysis. Three type-specific neutralizing mAbs, H31.F16, H31.H12 and H31.B1, which recognize conformational epitopes, were developed and characterized by Fleury et al.80 The type-specific epitopes for these antibodies are located at aa positions 264–272 in the FG loop of L1 for H31.B1, aa 264–269 for H31.H12 and aa 265–273 for H31.F16. In addition, another 2 conformation-dependent neutralizing mAbs (H31.F7 and H31.C19) react with conformational epitopes.81 The epitopes of these 2 mAbs have been shown to be located in the FG loop of H31 L1. The epitope of another antibody, H31.A6, was also investigated and this antibody was shown recognize a conformation-dependent epitope on the EF loop (Table 3).81
To characterize the epitopes of HPV33-specific antibodies, hybrid VLPs were designed in which the type-specific sequence of the surface loops of the HPV33 VLP was replaced by the corresponding amino acids of HPV16. The major residues contributing to the binding sites of 3 mAbs (H33.B6, H33.E12 and H33.J3) were identified by Roth et al.82 The sequences at 2 hyper loops, DE (aa 132–140) and FGb (aa 282–291) were determined to be essential for the binding of H33.B6. The epitope of H33.E12 was even more complex with an overlap with that of H33.B6 in the DE and FGb loops (Table 3). In addition, the binding of this antibody also required aa 260–270, localized in the FGa loop. The epitope of H33.J3 was determined to reside in the BC loop (aa 51–58). For quality control purposes in the Merck's project of V503, Brown et al. developed neutralizing mAbs for additional oncogenic HPV types: 31, 33, 45, 52 and 58 (H31.5F12 and H31.5D10, H33.5D4 and H33.6G9, H45.6G6 and H45.10B4, H52.8D11 and H52.9F7, H58.2C3 and H58.6E11).83 These elite mAbs displayed high affinity, type-specificity, high neutralization efficiency and recognition of conformational epitopes. These characteristics make these mAbs suitable as analytical tools to monitor the production process, to analyze potency in vitro and to assess the induced in vivo immune response of a vaccine (Table 3).
For the HPV-neutralizing mAbs for which epitopes have been identified, the immuno dominant epitopes have been shown to localize in the different flexible loops on the VLP surface (Table 3 and Fig. 4).84-86 Therefore, the correct spatial conformation of these loops and the structural integrity of the VLPs are essential for inducing effective neutralizing antibodies for vaccine efficacy. From a practical aspect, these representative neutralizing antibodies can be used to develop assays for process monitoring and production consistency during vaccine manufacturing.
Particle assembly and structural determination
In general, VLPs are assembled from the related viral capsid protein(s). Different truncated versions of the capsid protein can form particles with different sizes. Most VLPs retain native-like epitopes. Characterization of the assembled VLPs is essential for vaccine development, especially in regard to the size and morphology. Various biophysical and biochemical methods are used to characterize these properties, including high performance liquid chromatography (HPLC), analytical ultracentrifugation (AUC), dynamic light scattering (DLS), transmission electron microscopy (TEM), Cryo-EM and atomic force microscopy (AFM). The structural determination of the VLPs and/or their subunits can provide more details to aid in the in-depth understanding of the assembly mechanism and the preservation of epitopes on the VLPs surface.
The sole structural protein of HEV (pORF2) is encoded by ORF2. Different truncated versions of the capsid protein can assemble into distinctly sized spherical VLPs.8,24,87-89 The N- and C-terminal truncated version of aa 112–608 can form T = 1 VLPs (Fig. 1) and another version that contains aa 14–608 form T = 3 VLPs, which are analogous to the native virus (Fig. 1).15,90–92 The p239, with aa 368–606 of pORF2, can also self-assemble into VLPs with a diameter of 20–30 nm, with the key epitopes properly presented (Fig. 1).8,21,31 The HEV T = 1 VLPs have been visualized by electron microscopy .93,94 The overall spatial structures of the T = 1 and T = 3 VLPs have been elucidated using cryo-EM and crystallography with the development of structural analysis techniques (Table 2).15,90–92 The full length of ORF2 protein in T = 1 VLPs can be divided into 3 sections: the S domain (aa 118–313), the P1 domain (aa 314–453) or M domain (aa 320-455) and the P2 domain (aa 454–606) or P domain (aa 456–606) (Fig. 3).15,90 The P2 or P domain is localized on the outer surface, named E2s, and harbors the major neutralizing epitopes. A high-resolution crystal structure of the E2s domain was determined and its structural analysis verified the existence of this functional domain in a homodimeric form. The E2s protrusion projecting from the surface of the viral shell plays a key role in the interaction of HEV with host cells. Maintenance of the correct conformation and the dimeric form are essential for eliciting an effective immune response.14
Table 2.
Structure determination of recombinant virus-like particles or the subunit antigens of prophylactic vaccines against 3 viruses: HEV, HPV and HBV
| Methods of structural determination | ||||
|---|---|---|---|---|
| Virus | VLP or capsid protein | X-ray | Cryo-EM | Reference |
| HEV | E2s (I) | √ | × | 14 |
| E2s (IV) | √ | × | 16 | |
| p239 | × | × | 8 | |
| T = 1 VLP (I) | √ | √ | 91,92 | |
| T = 1 VLP (III) | √ | × | 15 | |
| T = 1 VLP (IV) | √ | × | 90 | |
| T = 3 VLP (III) | × | √ | 92 | |
| HPV | Pentamer (HPV11,16,18,35) | √ | × | 96 |
| T = 1 VLP (HPV16) | √ | × | 95 | |
| T = 7 VLP (HPV11,16) | × | √ | 19,67 | |
| HBV | HBsAg (Octahedral) | × | √ | 108 |
HPV vaccine are another type of vaccine for human use and have played a major role in human health since their development in the past decade. HPV vaccines utilize VLPs, which are assembled from the major capsid protein L1, as the immunogen. Electron microscopy has shown that the HPV11 VLPs have a similar surface topography to the infectious viruses75. The assembly of VLPs from HPV16 L1 pentamers indicated that full length L1 can assemble into T = 7 particles resembling the 72-pentamer native virions, whereas a truncated L1 lacking 10 N-terminal residues can only assemble into T = 1 particles including 12 pentamers.95 Several reports have indicated that the hypervariable loops on the surface of VLP, which determine the type specificity for genotypes and serotypes, are essential for binding by most of the identified neutralizing mAbs and for the integrity of the key neutralizing epitopes. Vaccination generating high levels of type-specific neutralizing antibodies is the primary mechanism of protection against infection. The structural determination of L1 in its pentameric form or in the T = 1 small VLP form provided insight into the type specificity of neutralizing antibodies.95,96
Chen and colleagues prepared L1 pentamers of 4 different HPV types, HPV11, HPV16, HPV18, HPV35 and determined their crystal structures with high resolution (Fig. 4 and Table 2).96 Comparison of the crystal structures of the 4 types revealed that the L1 structure among different types of HPV shared a conserved core domain and highly flexible loops on the capsid surface which harbor the major neutralizing epitopes and determine the type specificity for genotype and serotype (Fig. 4). The distinct loop structures provide an explanation for the mechanism of binding and elicitation of type-specific neutralizing antibodies. The version of the capsid protein L1 that is truncated by 10 N-terminal residues can assemble into small T = 1 icosahedral particles.95 Chen et al. determined the crystal structure of T = 1 HPV16 VLPs (Table 2) and this structure provided a starting point for understanding the particle (˜20 nm) assembly mechanism and the importance of the loops in determining the type specificity. These particles are smaller than those used in the vaccines (˜40–60 nm) and have different intercapsomeric contacts from the native virions, whereas the epitopes on the capsomeres are largely identical.
The spatial structure of HPV virions or VLPs can also be determined by cryo-EM and 3D reconstruction. Bovine papillomavirus and human papillomavirus are in the same genus and are highly similar to each other, especially in regard to their structure. BPV and HPV both consist of 72 capsomeres arranged on a T = 7 icosahedral lattice.97 Baker et al. determined the structure of BPV1 and HPV1 at 25 Å resolution using Cryo-EM and a 3 dimensional image reconstruction technique.97 The surface representation of the BPV1 and HPV1 reconstructions shows that the capsids of the 2 viruses consist of a layer of nearly continuous density from which the capsomeres project radially. It can be seen that the capsomeres have clear 5-fold symmetry in the reconstructed structures.
A higher resolution BPV structure (9 Å) was characterized by Trus et al. using cryo-EM.98 With the improvement in resolution, finer structural features were resolved and differences between the hexavalent and pentavalent capsomeres were observed and analyzed. Holes were clearly seen in the center of the hexavalent capsomeres but not in the pentavalent capsomeres. The intercapsomere connections were clearly resolved, which indicates that the protrusions emanate from the facets of the pentavalent capsomeres and extend toward a vertex of each of the 5 neighboring hexavalent capsomeres. An atomic model was generated for HPV from a combination of cryo-EM and crystallographic data, and this model was named the ‘invading arm’ model.99 A reconstruction of BPV at approximately 3.6 Å resolution has permitted reasonable fitting and refinement of the atomic model for the L1 shell utilizing the HPV16 L1 pentamer as a homology template.100,101 The new model corrects one feature of the earlier model in which the critical contacts are in the C-terminal arm. The loops emanate from the core of the subunit, contact 2 subunits in a neighboring pentamer and reinsert into the pentamer from which they emanate.101 A recent pseudoatomic model based on a 9 Å resolution reconstruction of fully mature capsids of HPV16 using cryo-EM and 3D image analysis showed strikingly high degree of similarity to the high-resolution cryo-EM-based model of BPV1.67 The whole atomic model provides the precise location and composition of the dominant epitopes. Therefore, cryo-EM and 3D reconstruction technique could be a useful tool to determine the precise spatial structure of biomolecules (VLP or VLP-Ab complexes) and monitor dynamic processes of VLP assembly and maturation.67 Ideally, high-resolution structures of VLP-antibody complexes will be determined so that the epitopes can be observed in the complexed form with near atomic level resolution.
Structure analysis of antigen-antibody complexes
Efforts have been made to characterize the neutralizing epitopes using mAbs by cryo-EM (3D-structure reconstruction), X-ray crystal structure determination and site-directed mutagenesis.
For HEV, to date, all of the identified neutralizing epitopes are conformational and all have been mapped to the E2s domain, which consists of a discontinuous amino acid sequence (Fig. 2 and Table 3).23 Xing et al. prepared Fab244 and the HEV VLP-Fab244 for Cryo-EM determination and showed the general structure of the Ag-Ab complex.17 The results indicated that the Fab244 recognizes a conformational epitopes that includes the residues E479, D481, T484, Y485, S487, Y532 and S533. This is consistent with the results of Western blot assays indicating that residues 597–601, localized at C- terminal region, are critical for Fab244 binding to the ORF2 protein. Separately, Yamashita et al. identified two neutralizing antibodies and determined their binding sites at the surface of the P (P2) domain.15 The MAB1323 recognizes a discontinuous epitope consisting of residues, S487, S488, T489, P491, N562 and T564. The other mAb, MAB272, binds to the residues D496, G591 and P592. The neutralization activity of the 2 antibodies was determined using mutation analysis. These results further support the notion that the P (P2) domain is the major dominant neutralizing epitope region.
Additional epitopes of highly neutralizing mAbs located at the P or P2 domain have also been determined. Li et al. suggested that the groove region on the surface of the E2s (P or P2) domain is the most likely antibody-binding region.14 A series of mutants targeting the groove region were constructed and the binding activity with the neutralizing antibody 8H3 was studied. The data showed that the mutants E479A, Y485A, I529A and K534A independently abrogated the reactivity of mAb 8H3. A study by Tang et al. was the first to report a crystal structure of a truncated HEV capsid protein and a dominant type specific (referred to genotype 1) neutralizing antibody (8C11).16 The sites of the interaction were determined and include the residues E479, S497, R512, K534, H577 and R578. Of the residues identified in the complex structure as contact points, R512 is the most crucial site for the interaction of 8C11 with the E2s domain and for the neutralization function, as determined by mutations analysis and cell model assays (Fig. 2, Fig. 3A and B).16 Identification of the neutralizing epitopes on the surface of the E2s protrusion facilitates the understanding of the effective immune response against HEV and provides important information about the epitopes for the design and improvement of the vaccine.
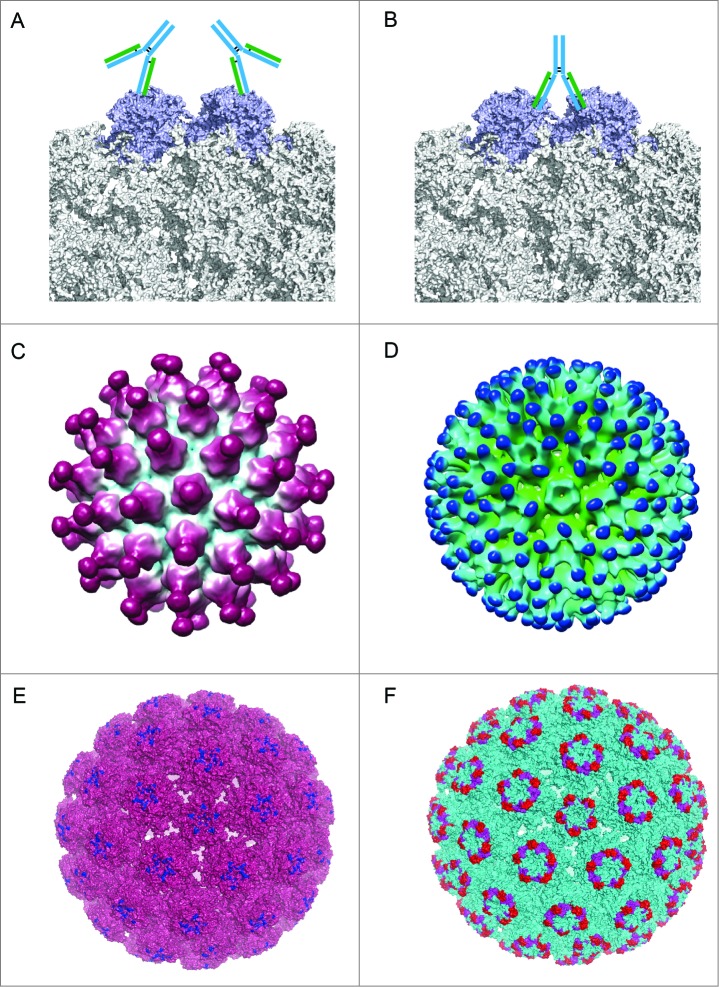
Owing to the lack of a high-resolution crystal structure of an Ag-Ab complex, precise epitope mapping data are still missing for HPV. Complexes of BPV and 2 neutralizing mAbs have been determined by cryo-EM and 3D image reconstruction to 13 Å resolution (Fig. 5).102 The two mAbs had 2 distinct binding patterns. The reconstructions revealed that mAb #9 binds to the L1 protein of both pentavalent and hexavalent capsomeres. However, the mAb 5B6 only binds to each of the L1 molecules in the hexavalent capsomeres. Epitope localization shows that mAb #9 binds monovalently to the tips of the capsomeres (Fig. 5A), whereas 5B6 binds both monovalently and bivalently to the sides of hexavalent capsomeres about 2-thirds of the way down from the outer tips yet to none of the L1 molecules in the pentavalent capsomeres because of steric hindrance (Fig. 5B). The binding patterns observed in the complexes of BPV and antibodies might shed light on the patterns in HPV-antibody complexes. For recombinant vaccine VLPs, Zhao et al. also determined 2 structures of HPV VLPs and Fabs, with H11.B2 binding to HPV11 VLPs and H16.V5 binding to HPV16 VLPs, using cryo-EM and 3D reconstruction techniques.19 For HPV11, the mAb H11.B2 binds to the center of the capsomeres (DE loop), with 72 potential binding sites on the VLP surface (Fig. 5C and E). The mAb H16.V5 has a different binding site (the FG and HI loops) that allows it to bind to the capsomeres at the 3-fold axes of symmetry, but not at the 5-fold axes of symmetry. Thus, HPV16 VLPs provide 300 potential binding sites per VLP for the H16.V5 antibody (Fig. 5D and F).
Figure 5.
Different binding patterns of antibodies to BPV and HPV.19,102 (A) mAb #9 binding to the outer surface of the hexavalent capsomere. (B) mAb 5B6 binding to the 2 L1 molecules of the adjacent hexavalent capsomeres. (C) A 3D reconstruction structure of mAb H11.B2 binding to HPV11 VLP (the DE loops on L1, Fig. 4) indicates that the binding sites are located at the center of the capsomere. (D) A 3D reconstruction structure based on cryo-EM data of mAb H16.V5 binding to HPV16 VLP (FG and HI loops on L1, Fig. 4) shows that H16.V5 only binds to the hexavalent capsomeres but not the pentavalent capsomeres.19 Recently, atomic model of the V5 epitope were built with higher resolution 3D reconstruction of cryo-EM data, demonstrating certain conformational changes induced by V5 binding to its epitope and confirming the preferential binding to the hexavalent capsomeres.151 The different binding models demonstrate different neutralization mechanisms for neutralizing antibodies exerting their effects against the viral infection. (E) The 3D structural model of mAb H11.B2 binding to HPV VLP in which the binding site of H11.B2 was indicated in blue and warmpink for VLP. (F) The 3D structural model of mAb H16.V5 binding to HPV16 VLP, with magenta for H16.V5 binding site on FG loop and red on HI loop, and cyan for VLP.
Discussion
Identification of the viral neutralization sites on the major capsid protein may be useful for serologically characterizing the immune response serologically of both vaccine recipients and naturally exposed individuals. This knowledge would provide insights into the structure-function relationships of vaccines and virus-host interactions. The presence of conformational and virion-like epitopes in a vaccine antigen is essential for eliciting neutralizing antibodies to confer immunity. Although most neutralizing epitopes are conformational, some linear neutralizing epitopes exposed on the surface of HPV VLPs have also been identified.73 Combita et al. noted that the HPV capsid protein L1 contains common linear neutralizing epitopes and suggested that some degree of cross-protection could occur.103 However, the level of cross-protection is very low after immunization, representing less than 1% of the homologous neutralizing activity.103 Furthermore, there is neither virological nor epidemiological evidence of natural cross-protection between related HPV types.103 Therefore, cross-type epitopes may exist, but they are unlikely to be any of the immuno dominant antigenic determinants on the viral capsid.
The determination of the antigenic and immunogenic structure of a viral protein is very important for the design and development of prophylactic vaccines. The related immunochemical tools, such as mAbs, are important for quality control of different batches of VLP antigens during vaccine manufacturing. Determining the structure of VLP-based vaccine antigens, whose molecular weight values are of the order of millions of daltons, is not trivial. The first licensed VLP-based human vaccine, the HBV vaccine, contains the HBsAg protein, which has 226 amino acids per monomer and with 96 monomers per particle.104,105 Owing to their intimate association with lipids and the presence of multiple disulfide bonds, no crystal structure for HBsAg particles has been determined to date.106 In addition, because of the high cysteine content in the major hydrophilic region (aa 101–170), which is the main antigenic region (with 8 cysteines of approximately 70 amino acids), the subunit structure of HBsAg particles has not been solved.107 The only cryo-EM structure of HBsAg particles shows that the hydrophilic regions of 4 subunits form protrusions from the particle surface, so this region contains 32 cysteine residues which result in the complex disulfide cross-linking.105,108 This region is postulated to be immuno dominant and to generate most of the neutralizing antibodies (Table 2). However, a high-resolution structure of the major neutralizing epitopes region has not been determined, making an analysis of antigen binding to neutralizing mAbs even more critical. Three neutralizing antibodies, RF-1, A1.2 and 5F11, have been identified. They recognize the immuno dominant region on the surface loops of HBsAg near the dimer interface with a high sensitivity to the epitope integrity on the VLP surface (Table 3).109–111 RF-1 is the only mAb specific for HBsAg that has been demonstrated to have the protective activity in a viral challenge experiment in chimpanzees.112 The epitope of RF-1 is composed of a stretch of 14 amino acids that includes 3 cysteines (aa 124–137), and the mAb binds to an intramolecularly disulfide-bonded cyclic peptide, as shown by ELISA and Western blot.113 RF-1 has been used as a competitor of clinical sera to validate the immune effects of a prophylactic vaccine.105,114,115 The mAb 5F11 also has a high sensitivity to the disulfide integrity in HBsAg in addition to a high in vitro neutralizing efficiency for HBV. All of the 3 mAbs are used as molecular probes to monitor the particle integrity and to assess the correct conformation of HBsAg, a prerequisite for an efficacious vaccine.111
Similar to the critical nature of the disulfides in HBsAg, disulfide bond formation plays an important role in facilitating the assembly and in maintaining the conformational stability of HPV VLPs.116 HPV16 L1 consists of 505 amino acids, including 12 cysteines. When the intermolecular disulfide bonds are disrupted in the capsid, the VLPs assembled from the L1 protein dissociate into individual capsomeres. Thus, the presence of the disulfide bonds is critical to the assembly and structural integrity of VLPs. A study by Ishii et al. indicated that Cysteines 175, 185, and 428 are involved in intercapsomeric disulfide bonding and in the normal assembly of the VLPs.117,118 In addition, the Cysteines 229 and 379 are most likely involved in intramolecular disulfide bonding, which may also contribute to the integrity and stability of the VLPs as well. A change in the redox environment can influence the assembly and disassembly of VLPs.119,120 Capsid maturation (from procapsids to capsids) was observed and characterized related largely to disulfide formation. The conformation of DE loop (near the center of the capsomere) was stabilized while the axial region of the capsomere formed, whereas the spatial structure of another 4 exterior loops (BC, EF, FG and HI loop, Fig. 4) were gradually shaped after a dynamic process of the capsid maturation with the consolidation induced by the disulfide cross-linking of the adjacent capsomeres [67]. The stability and homogeneity of HPV VLPs can be improved by disassembly and reassembly [119,120]. With these structural improvements, immuno reactivity with conformation-dependent neutralizing antibodies was markedly enhanced. However, the capacity of the well-formed and fully closed VLPs to bind to antibodies recognizing linear epitopes was greatly reduced.
Unlike HBV and HPV VLP assemblies, redox conditions are not critical for the bioprocessing of HEV VLPs. There is no cysteine in p239, which is the vaccine antigen and the basic structural unit in dimeric form maintained by hydrophobic interaction and which harbors the major neutralizing epitopes.14,23,121 As long as the E2s fragment (or the P/P2 domain) is expressed and purified, its intrinsic properties drive the formation of the dimer form through strong hydrophobic interactions and hydrogen bonds, and this form properly presents neutralizing epitopes such as those recognized by the mAbs 8C11 and 8H3.14,16
More recently, using a similar recombinant VLP approach, a vaccine against influenza virus produced by Protein Sciences, Inc, (Meriden, CT), FluBlok®, was recently licensed in 2013 in the USA.122,123 FluBlok® is a recombinant trivalent hemagglutinin-based vaccine produced in the baculovirus expression system. These multimeric, rosette-like particles with a diameter of 20–40 nm properly present the virion-like epitopes.124–127 FluBlok® has been tested in multiple clinical trials and shown to be safe and well tolerated. It displayed strong immunogenicity and elicited a long lasting immune response in these studies.128-133 This vaccine was shown to provide cross-protection against genetic drift in influenza virus strains.134 The effective protection was mainly dependent on the correct subunit folding for effective epitope formation, the correct spatial conformation and the presence of biologically active rHA antigens in the assembled form. FluBlok® contains 3 times more HA protein than the egg-based trivalent inactivated influenza vaccine. The HA antigens in FluBlok® are full-length proteins, including HA1, HA2 and transmembrane domain. rHA antigens with the correct conformation and native-like epitopes can induce protective immune responses, as evidenced by the presence of both hemagglutination-inhibiting and virus-neutralizing antibodies.
Another influenza VLP-based vaccine is being developed by Novavax using insect cell cultures. This VLP consists of HA and the matrix protein (M1) with or without neuroamidase.135 Thus, this vaccine can mimic the native virus in its stimulation of the immune response. The particle form can be more readily up taken by dendritic cells and in turn activates these dendritic cells.136 The results of clinical trials indicate that the vaccine can elicit cross-protection in animals and humans vaccinated with H5 HA-containing VLPs.137,138 The use of a liposomal adjuvant can further enhance the immunogenicity.139
In addition, another VLP-based vaccine against mosquito borne chikungunya virus (CHIKV) is being developed.140-142 CHIKV, a re-emerged, arthritogenic and mosquito-borne virus, causes severe diseases in human including fever, rash, myalgia, fatigue and the joint symtoms, often debilitating.143-145 The native virus contains 240 heterodimers of E1/E2 arranged as trimeric spikes on the virus surface. The study by Akataha et al. showed a self-assembled VLP of viral structural protein expressed in 239F cells. The preclinical data indicated that the VLP-based vaccine induced high neutralization titers in Balb/C or in nonhuman primates.140 Furthermore, the vaccine was evaluated in a Phase I dose-escalation clinical trial. After a follow-up study for 44 weeks, the presence of neutralization antibodies were detected, and the cross-type protection was achieved.146 To study the early neutralizing response to CHIKV in infected patients, immunoglobulin G3 targeting a dominant linear epitope on the E2 glycoprotein was shown to be associated with the virus clearance and long-term clinical protection.147,148 Two human mAbs, 5F10 and 8B10, with neutralization activity was identified and characterized. The structural analysis indicated that the 5F10 epitope was located at the tip of the E2 domain B (containing the residue of V216) whereas the epitope recognized by 8B10 was proposed to be a transient epitope which was only exposed under acidic conditions.149,150 Insightful findings on epitopes would aid in the vaccine design and development for CHIKV.
Vaccines can confer protection against viruses, and this protection depends mainly on inducing the immune system to generate neutralizing antibodies. Therefore, monitoring the major neutralizing epitopes on the VLP surface is an essential aspect of the development and quality control of vaccines. During bioprocessing, conditions should be provided to promote the formation of the virion-like epitopes. Accordingly, formulations should also be developed to stabilize the antigens by maintaining these epitopes during the shelf-life of the vaccines. A panel of monoclonal antibodies with the identified desired characteristics makes up a powerful tool box for monitoring the production process and characterizing the vaccine-induced clinical response. A panel of representative monoclonal antibodies is need for vaccine quality analysis and in the detection assay of vaccine release. The precise epitope mapping of a neutralizing antibody will aid in understanding the relationship between structure and function. In turn, this knowledge will also aid in the optimization and development of novel vaccines and in the quality assessment of licensed vaccines during life-cycle management to deliver safe and efficacious vaccine products to consumers.
Funding
We acknowledge the support of the China Ministry of Science and Technology via the Major Project (2012AA02A408), National Science Foundation of China (81373061 and 81471934), and Institute Reconstruction Fund (2011FU125Z04).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Grgacic EV, Anderson DA. Virus-like particles: passport to immune recognition. Methods 2006; 40:60-5; PMID:16997714; http://dx.doi.org/ 10.1016/j.ymeth.2006.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chuan YP, Rivera-Hernandez T, Wibowo N, Connors NK, Wu Y, Hughes FK, Lua LH, Middelberg AP. Effects of pre-existing anti-carrier immunity and antigenic element multiplicity on efficacy of a modular virus-like particle vaccine. Biotechnol Bioeng 2013; 110:2343-51; PMID:23532896; http://dx.doi.org/ 10.1002/bit.24907 [DOI] [PubMed] [Google Scholar]
- 3.Steinmetz NF. Viral nanoparticles as platforms for next-generation therapeutics and imaging devices. Nanomedicine 2010; 6:634-41; PMID:20433947; http://dx.doi.org/ 10.1016/j.nano.2010.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao Q, Li S, Yu H, Xia N, Modis Y. Virus-like particle-based human vaccines: quality assessment based on structural and functional properties. Trend Biotechnol 2013; 31:654-63; PMID:24125746; http://dx.doi.org/ 10.1016/j.tibtech.2013.09.002 [DOI] [PubMed] [Google Scholar]
- 5.Ma Y, Nolte RJ, Cornelissen JJ. Virus-based nanocarriers for drug delivery. Adv Drug Deliv Rev 2012; 64:811-25; PMID:22285585; http://dx.doi.org/ 10.1016/j.addr.2012.01.005 [DOI] [PubMed] [Google Scholar]
- 6.Tan M, Jiang X. Subviral particle as vaccine and vaccine platform. Curr Opin Virol 2014; 6C:24-33; PMID:24662314; http://dx.doi.org/ 10.1016/j.coviro.2014.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeltins A. Construction and characterization of virus-like particles: a review. Mol Biotechnol 2013; 53:92-107; PMID:23001867; http://dx.doi.org/ 10.1007/s12033-012-9598-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li SW, Zhang J, Li YM, Ou SH, Huang GY, He ZQ, Ge SX, Xian YL, Pang SQ, Ng MH, et al.. A bacterially expressed particulate hepatitis E vaccine: antigenicity, immunogenicity and protectivity on primates. Vaccine 2005; 23:2893-901; PMID:15780738; http://dx.doi.org/ 10.1016/j.vaccine.2004.11.064 [DOI] [PubMed] [Google Scholar]
- 9.Zhu FC, Zhang J, Zhang XF, Zhou C, Wang ZZ, Huang SJ, Wang H, Yang CL, Jiang HM, Cai JP, et al.. Efficacy and safety of a recombinant hepatitis E vaccine in healthy adults: a large-scale, randomised, double-blind placebo-controlled, phase 3 trial. Lancet 2010; 376:895-902; PMID:20728932; http://dx.doi.org/ 10.1016/S0140-6736(10)61030-6 [DOI] [PubMed] [Google Scholar]
- 10.Zhang J, Liu CB, Li RC, Li YM, Zheng YJ, Li YP, Luo D, Pan BB, Nong Y, Ge SX, et al.. Randomized-controlled phase II clinical trial of a bacterially expressed recombinant hepatitis E vaccine. Vaccine 2009; 27:1869-74; PMID:19168109; http://dx.doi.org/ 10.1016/j.vaccine.2008.12.061 [DOI] [PubMed] [Google Scholar]
- 11.Shrestha MP, Scott RM, Joshi DM, Mammen MP Jr., Thapa GB, Thapa N, Myint KS, Fourneau M, Kuschner RA, Shrestha SK, et al.. Safety and efficacy of a recombinant hepatitis E vaccine. N Engl J Med 2007; 356:895-903; PMID:17329696; http://dx.doi.org/ 10.1056/NEJMoa061847 [DOI] [PubMed] [Google Scholar]
- 12.Joura EA, Leodolter S, Hernandez-Avila M, Wheeler CM, Perez G, Koutsky LA, Garland SM, Harper DM, Tang GW, Ferris DG, et al.. Efficacy of a quadrivalent prophylactic human papillomavirus (types 6, 11, 16, and 18) L1 virus-like-particle vaccine against high-grade vulval and vaginal lesions: a combined analysis of three randomised clinical trials. Lancet 2007; 369:1693-702; PMID:17512854; http://dx.doi.org/ 10.1016/S0140-6736(07)60777-6 [DOI] [PubMed] [Google Scholar]
- 13.Paavonen J, Naud P, Salmeron J, Wheeler CM, Chow SN, Apter D, Kitchener H, Castellsague X, Teixeira JC, Skinner SR, et al.. Efficacy of human papillomavirus (HPV)-16/18 AS04-adjuvanted vaccine against cervical infection and precancer caused by oncogenic HPV types (PATRICIA): final analysis of a double-blind, randomised study in young women. Lancet 2009; 374:301-14; PMID:19586656; http://dx.doi.org/ 10.1016/S0140-6736(09)61248-4 [DOI] [PubMed] [Google Scholar]
- 14.Li S, Tang X, Seetharaman J, Yang C, Gu Y, Zhang J, Du H, Shih JW, Hew CL, Sivaraman J, et al.. Dimerization of hepatitis E virus capsid protein E2s domain is essential for virus-host interaction. PLoS Pathog 2009; 5:e1000537; PMID:19662165; http://dx.doi.org/ 10.1371/journal.ppat.1000537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamashita T, Mori Y, Miyazaki N, Cheng RH, Yoshimura M, Unno H, Shima R, Moriishi K, Tsukihara T, Li TC, et al.. Biological and immunological characteristics of hepatitis E virus-like particles based on the crystal structure. Proc Natl Acad Sci U S A 2009; 106:12986-91; PMID:19620712; http://dx.doi.org/ 10.1073/pnas.0903699106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang X, Yang C, Gu Y, Song C, Zhang X, Wang Y, Zhang J, Hew CL, Li S, Xia N, et al.. Structural basis for the neutralization and genotype specificity of hepatitis E virus. Proc Natl Acad Sci U S A 2011; 108:10266-71; PMID:21642534; http://dx.doi.org/ 10.1073/pnas.1101309108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xing L, Wang JC, Li TC, Yasutomi Y, Lara J, Khudyakov Y, Schofield D, Emerson SU, Purcell RH, Takeda N, et al.. Spatial configuration of hepatitis E virus antigenic domain. J Virol 2011; 85:1117-24; PMID:21068233; http://dx.doi.org/ 10.1128/JVI.00657-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu Y, Tang X, Zhang X, Li S, Xia N Structural Basis for the Neutralization of Hepatitis E Virus by a Cross-genotype Antibody. Cell Research 2015; 34:xxx-xxx. doi: 10.1038/cr.2015.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao Q, Potter CS, Carragher B, Lander G, Sworen J, Towne V, Abraham D, Duncan P, Washabaugh MW, Sitrin RD. Characterization of virus-like particles in GARDASIL(R) by cryo transmission electron microscopy. Hum Vaccin Immunother 2013; 10:734-9; PMID:24299977; http://dx.doi.org/ 10.4161/hv.27316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christensen ND, Dillner J, Eklund C, Carter JJ, Wipf GC, Reed CA, Cladel NM, Galloway DA. Surface conformational and linear epitopes on HPV-16 and HPV-18 L1 virus-like particles as defined by monoclonal antibodies. Virology 1996; 223:174-84; PMID:8806551; http://dx.doi.org/ 10.1006/viro.1996.0466 [DOI] [PubMed] [Google Scholar]
- 21.Wei M, Zhang X, Yu H, Tang ZM, Wang K, Li Z, Zheng Z, Li S, Zhang J, Xia N, et al.. Bacteria expressed hepatitis E virus capsid proteins maintain virion-like epitopes. Vaccine 2014; 32:2859-65; PMID:24662711; http://dx.doi.org/ 10.1016/j.vaccine.2014.02.025 [DOI] [PubMed] [Google Scholar]
- 22.Pastrana DV, Buck CB, Pang YY, Thompson CD, Castle PE, FitzGerald PC, Kruger Kjaer S, Lowy DR, Schiller JT. Reactivity of human sera in a sensitive, high-throughput pseudovirus-based papillomavirus neutralization assay for HPV16 and HPV18. Virology 2004; 321:205-16; PMID:15051381; http://dx.doi.org/ 10.1016/j.virol.2003.12.027 [DOI] [PubMed] [Google Scholar]
- 23.Zhang J, Li SW, Wu T, Zhao Q, Ng MH, Xia NS. Hepatitis E virus: neutralizing sites, diagnosis, and protective immunity. Rev Med Virol 2012; 22:339-49; PMID:22645002; http://dx.doi.org/ 10.1002/rmv.1719 [DOI] [PubMed] [Google Scholar]
- 24.Robinson RA, Burgess WH, Emerson SU, Leibowitz RS, Sosnovtseva SA, Tsarev S, Purcell RH. Structural characterization of recombinant hepatitis E virus ORF2 proteins in baculovirus-infected insect cells. Protein Expr Purif 1998; 12:75-84; PMID:9473460; http://dx.doi.org/ 10.1006/prep.1997.0817 [DOI] [PubMed] [Google Scholar]
- 25.Zhang M, Emerson SU, Nguyen H, Engle RE, Govindarajan S, Gerin JL, Purcell RH. Immunogenicity and protective efficacy of a vaccine prepared from 53 kDa truncated hepatitis E virus capsid protein expressed in insect cells. Vaccine 2001; 20:853-7; PMID:11738749; http://dx.doi.org/ 10.1016/S0264-410X(01)00399-1 [DOI] [PubMed] [Google Scholar]
- 26.Wu T, Li SW, Zhang J, Ng MH, Xia NS, Zhao Q. Hepatitis E vaccine development: a 14 year odyssey. Hum Vacci Immunother 2012; 8:823-7; PMID:22699438; http://dx.doi.org/ 10.4161/hv.20042 [DOI] [PubMed] [Google Scholar]
- 27.Wu T, Zhu FC, Huang SJ, Zhang XF, Wang ZZ, Zhang J, Xia NS. Safety of the hepatitis E vaccine for pregnant women: a preliminary analysis. Hepatology 2012; 55:2038; PMID:22161542; http://dx.doi.org/ 10.1002/hep.25522 [DOI] [PubMed] [Google Scholar]
- 28.Wu T, Huang SJ, Zhu FC, Zhang XF, Ai X, Yan Q, Wang ZZ, Yang CL, Jiang HM, Liu XH, et al.. Immunogenicity and safety of hepatitis E vaccine in healthy hepatitis B surface antigen positive adults. Hum Vacci Immunother 2013; 9:2474-9; PMID:23887167; http://dx.doi.org/ 10.4161/hv.25814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krain LJ, Nelson KE, Labrique AB. Host immune status and response to hepatitis E virus infection. Clin Microbiol Rre 2014; 27:139-65; PMID:24396140; http://dx.doi.org/ 10.1128/CMR.00062-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou YH, Purcell RH, Emerson SU. A truncated ORF2 protein contains the most immunogenic site on ORF2: antibody responses to non-vaccine sequences following challenge of vaccinated and non-vaccinated macaques with hepatitis E virus. Vaccine 2005; 23:3157-65; PMID:15837215; http://dx.doi.org/ 10.1016/j.vaccine.2004.12.020 [DOI] [PubMed] [Google Scholar]
- 31.Zhang X, Wei M, Pan H, Lin Z, Wang K, Weng Z, Zhu Y, Xin L, Zhang J, Li S, et al.. Robust manufacturing and comprehensive characterization of recombinant hepatitis E virus-like particles in Hecolin((R)). Vaccine 2014; 32:4039-50; PMID:24892250; http://dx.doi.org/ 10.1016/j.vaccine.2014.05.064 [DOI] [PubMed] [Google Scholar]
- 32.Khudyakov YE, Lopareva EN, Jue DL, Crews TK, Thyagarajan SP, Fields HA. Antigenic domains of the open reading frame 2-encoded protein of hepatitis E virus. J Clin Microbiol 1999; 37:2863-71; PMID:10449466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang H, Dai X, Shan X, Meng J. Characterization of antigenic epitopes of the ORF2 protein from hepatitis E virus genotype 4. Virus Res 2009; 142:140-3; PMID:19428747; http://dx.doi.org/ 10.1016/j.virusres.2009.02.002 [DOI] [PubMed] [Google Scholar]
- 34.Khudyakov Yu E, Favorov MO, Jue DL, Hine TK, Fields HA. Immunodominant antigenic regions in a structural protein of the hepatitis E virus. Virology 1994; 198:390-3; PMID:8259678; http://dx.doi.org/ 10.1006/viro.1994.1048 [DOI] [PubMed] [Google Scholar]
- 35.Li F, Torresi J, Locarnini SA, Zhuang H, Zhu W, Guo X, Anderson DA. Amino-terminal epitopes are exposed when full-length open reading frame 2 of hepatitis E virus is expressed in Escherichia coli, but carboxy-terminal epitopes are masked. J Med Virol 1997; 52:289-300; PMID:9210039; http://dx.doi.org/ 10.1002/(SICI)1096-9071(199707)52:3%3c289::AID-JMV10%3e3.0.CO;2-E [DOI] [PubMed] [Google Scholar]
- 36.Riddell MA, Li F, Anderson DA. Identification of immunodominant and conformational epitopes in the capsid protein of hepatitis E virus by using monoclonal antibodies. J Virol 2000; 74:8011-7; PMID:10933710; http://dx.doi.org/ 10.1128/JVI.74.17.8011-8017.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meng J, Dai X, Chang JC, Lopareva E, Pillot J, Fields HA, Khudyakov YE. Identification and characterization of the neutralization epitope(s) of the hepatitis E virus. Virology 2001; 288:203-11; PMID:11601892; http://dx.doi.org/ 10.1006/viro.2001.1093 [DOI] [PubMed] [Google Scholar]
- 38.Zhang JZ, Ng MH, Xia NS, Lau SH, Che XY, Chau TN, Lai ST, Im SW. Conformational antigenic determinants generated by interactions between a bacterially expressed recombinant peptide of the hepatitis E virus structural protein. J Med Virol 2001; 64:125-32; PMID:11360244; http://dx.doi.org/ 10.1002/jmv.1027 [DOI] [PubMed] [Google Scholar]
- 39.Emerson SU, Clemente-Casares P, Moiduddin N, Arankalle VA, Torian U, Purcell RH. Putative neutralization epitopes and broad cross-genotype neutralization of Hepatitis E virus confirmed by a quantitative cell-culture assay. J Gen Virol 2006; 87:697-704; PMID:16476993; http://dx.doi.org/ 10.1099/vir.0.81545-0 [DOI] [PubMed] [Google Scholar]
- 40.Zhang H, Dai X, Shan X, Meng J. The Leu477 and Leu613 of ORF2-encoded protein are critical in forming neutralization antigenic epitope of hepatitis E virus genotype 4. Cell Mol Immunol 2008; 5:447-56; PMID:19118511; http://dx.doi.org/ 10.1038/cmi.2008.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rose RC, Reichman RC, Bonnez W. Human papillomavirus (HPV) type 11 recombinant virus-like particles induce the formation of neutralizing antibodies and detect HPV-specific antibodies in human sera. J Gen Virol 1994; 75 ( Pt 8):2075-9; PMID:8046412; http://dx.doi.org/ 10.1099/0022-1317-75-8-2075 [DOI] [PubMed] [Google Scholar]
- 42.Thones N, Herreiner A, Schadlich L, Piuko K, Muller M. A direct comparison of human papillomavirus type 16 L1 particles reveals a lower immunogenicity of capsomeres than viruslike particles with respect to the induced antibody response. J Virol 2008; 82:5472-85; PMID:18385253; http://dx.doi.org/ 10.1128/JVI.02482-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koutsky LA, Ault KA, Wheeler CM, Brown DR, Barr E, Alvarez FB, Chiacchierini LM, Jansen KU, Proof of Principle Study I. A controlled trial of a human papillomavirus type 16 vaccine. N Engl J Med 2002; 347:1645-51; PMID:12444178; http://dx.doi.org/ 10.1056/NEJMoa020586 [DOI] [PubMed] [Google Scholar]
- 44.Harro CD, Pang YY, Roden RB, Hildesheim A, Wang Z, Reynolds MJ, Mast TC, Robinson R, Murphy BR, Karron RA, et al.. Safety and immunogenicity trial in adult volunteers of a human papillomavirus 16 L1 virus-like particle vaccine. J Natl Cancer Inst 2001; 93:284-92; PMID:11181775; http://dx.doi.org/ 10.1093/jnci/93.4.284 [DOI] [PubMed] [Google Scholar]
- 45.Villa LL, Costa RL, Petta CA, Andrade RP, Paavonen J, Iversen OE, Olsson SE, Hoye J, Steinwall M, Riis-Johannessen G, et al.. High sustained efficacy of a prophylactic quadrivalent human papillomavirus types 6/11/16/18 L1 virus-like particle vaccine through 5 years of follow-up. Br J Cancer 2006; 95:1459-66; PMID:17117182; http://dx.doi.org/ 10.1038/sj.bjc.6603469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Safaeian M, Porras C, Pan Y, Kreimer A, Schiller JT, Gonzalez P, Lowy DR, Wacholder S, Schiffman M, Rodriguez AC, et al.. Durable antibody responses following one dose of the bivalent human papillomavirus L1 virus-like particle vaccine in the Costa Rica Vaccine Trial. Cancer Prevent Res 2013; 6:1242-50; PMID:24189371; http://dx.doi.org/ 10.1158/1940-6207.CAPR-13-0203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perez G, Lazcano-Ponce E, Hernandez-Avila M, Garcia PJ, Munoz N, Villa LL, Bryan J, Taddeo FJ, Lu S, Esser MT, et al.. Safety, immunogenicity, and efficacy of quadrivalent human papillomavirus (types 6, 11, 16, 18) L1 virus-like-particle vaccine in Latin American women. Int J Cancer 2008; 122:1311-8; PMID:18000825; http://dx.doi.org/ 10.1002/ijc.23260 [DOI] [PubMed] [Google Scholar]
- 48.Harper DM, Franco EL, Wheeler CM, Moscicki AB, Romanowski B, Roteli-Martins CM, Jenkins D, Schuind A, Costa Clemens SA, Dubin G, et al.. Sustained efficacy up to 4.5 years of a bivalent L1 virus-like particle vaccine against human papillomavirus types 16 and 18: follow-up from a randomised control trial. Lancet 2006; 367:1247-55; PMID:16631880; http://dx.doi.org/ 10.1016/S0140-6736(06)68439-0 [DOI] [PubMed] [Google Scholar]
- 49.Munoz N, Manalastas R Jr., Pitisuttithum P, Tresukosol D, Monsonego J, Ault K, Clavel C, Luna J, Myers E, Hood S, et al.. Safety, immunogenicity, and efficacy of quadrivalent human papillomavirus (types 6, 11, 16, 18) recombinant vaccine in women aged 24-45 years: a randomised, double-blind trial. Lancet 2009; 373:1949-57; PMID:19493565; http://dx.doi.org/ 10.1016/S0140-6736(09)60691-7 [DOI] [PubMed] [Google Scholar]
- 50.Paavonen J, Jenkins D, Bosch FX, Naud P, Salmeron J, Wheeler CM, Chow SN, Apter DL, Kitchener HC, Castellsague X, et al.. Efficacy of a prophylactic adjuvanted bivalent L1 virus-like-particle vaccine against infection with human papillomavirus types 16 and 18 in young women: an interim analysis of a phase III double-blind, randomised controlled trial. Lancet 2007; 369:2161-70; PMID:17602732; http://dx.doi.org/ 10.1016/S0140-6736(07)60946-5 [DOI] [PubMed] [Google Scholar]
- 51.Villa LL, Costa RL, Petta CA, Andrade RP, Ault KA, Giuliano AR, Wheeler CM, Koutsky LA, Malm C, Lehtinen M, et al.. Prophylactic quadrivalent human papillomavirus (types 6, 11, 16, and 18) L1 virus-like particle vaccine in young women: a randomised double-blind placebo-controlled multicentre phase II efficacy trial. Lancet Oncol 2005; 6:271-8; PMID:15863374; http://dx.doi.org/ 10.1016/S1470-2045(05)70101-7 [DOI] [PubMed] [Google Scholar]
- 52.Markowitz LE, Hariri S, Lin C, Dunne EF, Steinau M, McQuillan G, Unger ER. Reduction in human papillomavirus (HPV) prevalence among young women following HPV vaccine introduction in the United States, National Health and Nutrition Examination Surveys, 2003-2010. J Infect Dis 2013; 208:385-93; PMID:23785124; http://dx.doi.org/ 10.1093/infdis/jit192 [DOI] [PubMed] [Google Scholar]
- 53.Robbins HA, Kemp TJ, Porras C, Rodriguez AC, Schiffman M, Wacholder S, Gonzalez P, Schiller J, Lowy D, Poncelet S, et al.. Comparison of antibody responses to human papillomavirus vaccination as measured by three assays. Front Oncol 2014; 3:328; PMID:24455487; http://dx.doi.org/ 10.3389/fonc.2013.00328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bryan JT, Jansen KU, Lowe RS, Fife KH, McClowry T, Glass D, Brown DR. Human papillomavirus type 11 neutralization in the athymic mouse xenograft system: correlation with virus-like particle IgG concentration. J Med Virol 1997; 53:185-8; PMID:9365880; http://dx.doi.org/ 10.1002/(SICI)1096-9071(199711)53:3%3c185::AID-JMV1%3e3.0.CO;2-4 [DOI] [PubMed] [Google Scholar]
- 55.Christensen ND, Kreider JW, Cladel NM, Patrick SD, Welsh PA. Monoclonal antibody-mediated neutralization of infectious human papillomavirus type 11. J Virol 1990; 64:5678-81; PMID:2170694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kreider JW, Howett MK, Wolfe SA, Bartlett GL, Zaino RJ, Sedlacek T, Mortel R. Morphological transformation in vivo of human uterine cervix with papillomavirus from condylomata acuminata. Nature 1985; 317:639-41; PMID:2997616; http://dx.doi.org/ 10.1038/317639a0 [DOI] [PubMed] [Google Scholar]
- 57.Olcese VA, Chen Y, Schlegel R, Yuan H. Characterization of HPV16 L1 loop domains in the formation of a type-specific, conformational epitope. BMC Microbiol 2004; 4:29; PMID:15260888; http://dx.doi.org/ 10.1186/1471-2180-4-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Slupetzky K, Shafti-Keramat S, Lenz P, Brandt S, Grassauer A, Sara M, Kirnbauer R. Chimeric papillomavirus-like particles expressing a foreign epitope on capsid surface loops. J Gen Virol 2001; 82:2799-804; PMID:11602792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen HS, Bromberg-White J, Conway MJ, Alam S, Meyers C. Study of infectious virus production from HPV18/16 capsid chimeras. Virology 2010; 405:289-99; PMID:20598725; http://dx.doi.org/ 10.1016/j.virol.2010.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sadeyen JR, Tourne S, Shkreli M, Sizaret PY, Coursaget P. Insertion of a foreign sequence on capsid surface loops of human papillomavirus type 16 virus-like particles reduces their capacity to induce neutralizing antibodies and delineates a conformational neutralizing epitope. Virology 2003; 309:32-40; PMID:12726724; http://dx.doi.org/ 10.1016/S0042-6822(02)00134-4 [DOI] [PubMed] [Google Scholar]
- 61.Christensen ND, Cladel NM, Reed CA, Budgeon LR, Embers ME, Skulsky DM, McClements WL, Ludmerer SW, Jansen KU. Hybrid papillomavirus L1 molecules assemble into virus-like particles that reconstitute conformational epitopes and induce neutralizing antibodies to distinct HPV types. Virology 2001; 291:324-34; PMID:11878901; http://dx.doi.org/ 10.1006/viro.2001.1220 [DOI] [PubMed] [Google Scholar]
- 62.Roden RB, Armstrong A, Haderer P, Christensen ND, Hubbert NL, Lowy DR, Schiller JT, Kirnbauer R. Characterization of a human papillomavirus type 16 variant-dependent neutralizing epitope. J Virol 1997; 71:6247-52; PMID:9223527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.White WI, Wilson SD, Palmer-Hill FJ, Woods RM, Ghim SJ, Hewitt LA, Goldman DM, Burke SJ, Jenson AB, Koenig S, et al.. Characterization of a major neutralizing epitope on human papillomavirus type 16 L1. J Virol 1999; 73:4882-9; PMID:10233949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carpentier GS, Fleury MJ, Touze A, Sadeyen JR, Tourne S, Sizaret PY, Coursaget P. Mutations on the FG surface loop of human papillomavirus type 16 major capsid protein affect recognition by both type-specific neutralizing antibodies and cross-reactive antibodies. J Med Virol 2005; 77:558-65; PMID:16254978; http://dx.doi.org/ 10.1002/jmv.20492 [DOI] [PubMed] [Google Scholar]
- 65.Carter JJ, Wipf GC, Benki SF, Christensen ND, Galloway DA. Identification of a human papillomavirus type 16-specific epitope on the C-terminal arm of the major capsid protein L1. J Virol 2003; 77:11625-32; PMID:14557648; http://dx.doi.org/ 10.1128/JVI.77.21.11625-11632.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carter JJ, Wipf GC, Madeleine MM, Schwartz SM, Koutsky LA, Galloway DA. Identification of human papillomavirus type 16 L1 surface loops required for neutralization by human sera. J Virol 2006; 80:4664-72; PMID:16641259; http://dx.doi.org/ 10.1128/JVI.80.10.4664-4672.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cardone G, Moyer AL, Cheng N, Thompson CD, Dvoretzky I, Lowy DR, Schiller JT, Steven AC, Buck CB, Trus BL. Maturation of the human papillomavirus 16 capsid. mBio 2014; 5:e01104-14; PMID:25096873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marais DJ, Sampson C, Jeftha A, Dhaya D, Passmore JA, Denny L, Rybicki EP, Van Der Walt E, Stephen LX, Williamson AL. More men than women make mucosal IgA antibodies to Human papillomavirus type 16 (HPV-16) and HPV-18: a study of oral HPV and oral HPV antibodies in a normal healthy population. BMC Infect Dis 2006; 6:95; PMID:16762074; http://dx.doi.org/ 10.1186/1471-2334-6-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Group FIIS, Dillner J, Kjaer SK, Wheeler CM, Sigurdsson K, Iversen OE, Hernandez-Avila M, Perez G, Brown DR, Koutsky LA, et al.. Four year efficacy of prophylactic human papillomavirus quadrivalent vaccine against low grade cervical, vulvar, and vaginal intraepithelial neoplasia and anogenital warts: randomised controlled trial. BMJ 2010; 341:c3493; PMID:20647284; http://dx.doi.org/ 10.1136/bmj.c3493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ali H, Donovan B, Wand H, Read TR, Regan DG, Grulich AE, Fairley CK, Guy RJ. Genital warts in young Australians five years into national human papillomavirus vaccination programme: national surveillance data. Bmj 2013; 346:f2032; PMID:23599298; http://dx.doi.org/ 10.1136/bmj.f2032 [DOI] [PubMed] [Google Scholar]
- 71.Munoz N, Kjaer SK, Sigurdsson K, Iversen OE, Hernandez-Avila M, Wheeler CM, Perez G, Brown DR, Koutsky LA, Tay EH, et al.. Impact of human papillomavirus (HPV)-6/11/16/18 vaccine on all HPV-associated genital diseases in young women. J Natl Cancer Inst 2010; 102:325-39; PMID:20139221; http://dx.doi.org/ 10.1093/jnci/djp534 [DOI] [PubMed] [Google Scholar]
- 72.Garland SM, Hernandez-Avila M, Wheeler CM, Perez G, Harper DM, Leodolter S, Tang GW, Ferris DG, Steben M, Bryan J, et al.. Quadrivalent vaccine against human papillomavirus to prevent anogenital diseases. N Engl J Med 2007; 356:1928-43; PMID:17494926; http://dx.doi.org/ 10.1056/NEJMoa061760 [DOI] [PubMed] [Google Scholar]
- 73.Christensen ND, Reed CA, Cladel NM, Hall K, Leiserowitz GS. Monoclonal antibodies to HPV-6 L1 virus-like particles identify conformational and linear neutralizing epitopes on HPV-11 in addition to type-specific epitopes on HPV-6. Virology 1996; 224:477-86; PMID:8874508; http://dx.doi.org/ 10.1006/viro.1996.0554 [DOI] [PubMed] [Google Scholar]
- 74.McClements WL, Wang XM, Ling JC, Skulsky DM, Christensen ND, Jansen KU, Ludmerer SW. A novel human papillomavirus type 6 neutralizing domain comprising two discrete regions of the major capsid protein L1. Virology 2001; 289:262-8; PMID:11689049; http://dx.doi.org/ 10.1006/viro.2001.1146 [DOI] [PubMed] [Google Scholar]
- 75.Christensen ND, Hopfl R, DiAngelo SL, Cladel NM, Patrick SD, Welsh PA, Budgeon LR, Reed CA, Kreider JW. Assembled baculovirus-expressed human papillomavirus type 11 L1 capsid protein virus-like particles are recognized by neutralizing monoclonal antibodies and induce high titres of neutralizing antibodies. J Gen Virol 1994; 75 ( Pt 9):2271-6; PMID:7521393; http://dx.doi.org/ 10.1099/0022-1317-75-9-2271 [DOI] [PubMed] [Google Scholar]
- 76.Ludmerer SW, Benincasa D, Mark GE 3rd. Two amino acid residues confer type specificity to a neutralizing, conformationally dependent epitope on human papillomavirus type 11. J Virol 1996; 70:4791-4; PMID:8676509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ludmerer SW, Benincasa D, Mark GE 3rd, Christensen ND. A neutralizing epitope of human papillomavirus type 11 is principally described by a continuous set of residues which overlap a distinct linear, surface-exposed epitope. J Virol 1997; 71:3834-9; PMID:9094659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ludmerer SW, McClements WL, Wang XM, Ling JC, Jansen KU, Christensen ND. HPV11 mutant virus-like particles elicit immune responses that neutralize virus and delineate a novel neutralizing domain. Virology 2000; 266:237-45; PMID:10639310; http://dx.doi.org/ 10.1006/viro.1999.0083 [DOI] [PubMed] [Google Scholar]
- 79.Serrano B, Alemany L, Tous S, Bruni L, Clifford GM, Weiss T, Bosch FX, de Sanjose S. Potential impact of a nine-valent vaccine in human papillomavirus related cervical disease. Infect Agent Cancer 2012; 7:38; PMID:23273245; http://dx.doi.org/ 10.1186/1750-9378-7-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fleury MJ, Touze A, Maurel MC, Moreau T, Coursaget P. Identification of neutralizing conformational epitopes on the human papillomavirus type 31 major capsid protein and functional implications. Protein Sci 2009; 18:1425-38; PMID:19533761; http://dx.doi.org/ 10.1002/pro.156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fleury MJ, Touze A, Alvarez E, Carpentier G, Clavel C, Vautherot JF, Coursaget P. Identification of type-specific and cross-reactive neutralizing conformational epitopes on the major capsid protein of human papillomavirus type 31. Arch Virol 2006; 151:1511-23; PMID:16508703; http://dx.doi.org/ 10.1007/s00705-006-0734-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Roth SD, Sapp M, Streeck RE, Selinka HC. Characterization of neutralizing epitopes within the major capsid protein of human papillomavirus type 33. Virol J 2006; 3:83; PMID:17014700; http://dx.doi.org/ 10.1186/1743-422X-3-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Brown MJ, Seitz H, Towne V, Muller M, Finnefrock AC. Development of neutralizing monoclonal antibodies for oncogenic human papillomavirus types 31, 33, 45, 52, and 58. Clin Vaccine Immunol 2014; 21:587-93; PMID:24574536; http://dx.doi.org/ 10.1128/CVI.00773-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Giroglou T, Sapp M, Lane C, Fligge C, Christensen ND, Streeck RE, Rose RC. Immunological analyses of human papillomavirus capsids. Vaccine 2001; 19:1783-93; PMID:11166904; http://dx.doi.org/ 10.1016/S0264-410X(00)00370-4 [DOI] [PubMed] [Google Scholar]
- 85.Joshi H, Cheluvaraja S, Somogyi E, Brown DR, Ortoleva P. A molecular dynamics study of loop fluctuation in human papillomavirus type 16 virus-like particles: a possible indicator of immunogenicity. Vaccine 2011; 29:9423-30; PMID:22027487; http://dx.doi.org/ 10.1016/j.vaccine.2011.10.039 [DOI] [PubMed] [Google Scholar]
- 86.Orozco JJ, Carter JJ, Koutsky LA, Galloway DA. Humoral immune response recognizes a complex set of epitopes on human papillomavirus type 6 l1 capsomers. J VirolJ Virol 2005; 79:9503-14; PMID:16014913; http://dx.doi.org/ 10.1128/JVI.79.15.9503-9514.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li TC, Takeda N, Miyamura T, Matsuura Y, Wang JC, Engvall H, Hammar L, Xing L, Cheng RH. Essential elements of the capsid protein for self-assembly into empty virus-like particles of hepatitis E virus. J Virol 2005; 79:12999-3006; PMID:16189002; http://dx.doi.org/ 10.1128/JVI.79.20.12999-13006.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li TC, Yamakawa Y, Suzuki K, Tatsumi M, Razak MA, Uchida T, Takeda N, Miyamura T. Expression and self-assembly of empty virus-like particles of hepatitis E virus. J Virol 1997; 71:7207-13; PMID:9311793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li TC, Yoshimatsu K, Yasuda SP, Arikawa J, Koma T, Kataoka M, Ami Y, Suzaki Y, Mai TQ, Hoa NT, et al.. Characterization of self-assembled virus-like particles of rat hepatitis E virus generated by recombinant baculoviruses. J Gen Virol 2011; 92:2830-7; PMID:21865442; http://dx.doi.org/ 10.1099/vir.0.034835-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Guu TS, Liu Z, Ye Q, Mata DA, Li K, Yin C, Zhang J, Tao YJ. Structure of the hepatitis E virus-like particle suggests mechanisms for virus assembly and receptor binding. Proc Natl Acad Sci U S A 2009; 106:12992-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xing L, Kato K, Li T, Takeda N, Miyamura T, Hammar L, Cheng RH. Recombinant hepatitis E capsid protein self-assembles into a dual-domain T = 1 particle presenting native virus epitopes. Virology 1999; 265:35-45; PMID:10603315; http://dx.doi.org/ 10.1006/viro.1999.0005 [DOI] [PubMed] [Google Scholar]
- 92.Xing L, Li TC, Mayazaki N, Simon MN, Wall JS, Moore M, Wang CY, Takeda N, Wakita T, Miyamura T, et al.. Structure of hepatitis E virion-sized particle reveals an RNA-dependent viral assembly pathway. J Biol Chem 2010; 285:33175-83; PMID:20720013; http://dx.doi.org/ 10.1074/jbc.M110.106336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Huang CC, Nguyen D, Fernandez J, Yun KY, Fry KE, Bradley DW, Tam AW, Reyes GR. Molecular cloning and sequencing of the Mexico isolate of hepatitis E virus (HEV). Virology 1992; 191:550-8; PMID:1448913; http://dx.doi.org/ 10.1016/0042-6822(92)90230-M [DOI] [PubMed] [Google Scholar]
- 94.Sreenivasan MA, Arankalle VA, Sehgal A, Pavri KM. Non-A, non-B epidemic hepatitis: visualization of virus-like particles in the stool by immune electron microscopy. J Gen Virol 1984; 65 ( Pt 5):1005-7; PMID:6427404; http://dx.doi.org/ 10.1099/0022-1317-65-5-1005 [DOI] [PubMed] [Google Scholar]
- 95.Chen XS, Garcea RL, Goldberg I, Casini G, Harrison SC. Structure of small virus-like particles assembled from the L1 protein of human papillomavirus 16. Mol Cell 2000; 5:557-67; PMID:10882140; http://dx.doi.org/ 10.1016/S1097-2765(00)80449-9 [DOI] [PubMed] [Google Scholar]
- 96.Bishop B, Dasgupta J, Klein M, Garcea RL, Christensen ND, Zhao R, Chen XS. Crystal structures of four types of human papillomavirus L1 capsid proteins: understanding the specificity of neutralizing monoclonal antibodies. J Biol Chem 2007; 282:31803-11; PMID:17804402; http://dx.doi.org/ 10.1074/jbc.M706380200 [DOI] [PubMed] [Google Scholar]
- 97.Baker TS, Newcomb WW, Olson NH, Cowsert LM, Olson C, Brown JC. Structures of bovine and human papillomaviruses. Analysis by cryoelectron microscopy and three-dimensional image reconstruction. Biophys J 1991; 60:1445-56; PMID:1663794; http://dx.doi.org/ 10.1016/S0006-3495(91)82181-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Trus BL, Roden RB, Greenstone HL, Vrhel M, Schiller JT, Booy FP. Novel structural features of bovine papillomavirus capsid revealed by a three-dimensional reconstruction to 9 A resolution. Nat Struct Biol 1997; 4:413-20; PMID:9145113; http://dx.doi.org/ 10.1038/nsb0597-413 [DOI] [PubMed] [Google Scholar]
- 99.Modis Y, Trus BL, Harrison SC. Atomic model of the papillomavirus capsid. EMBO J 2002; 21:4754-62; PMID:12234916; http://dx.doi.org/ 10.1093/emboj/cdf494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wolf M, Garcea RL, Grigorieff N, Harrison SC. Subunit interactions in bovine papillomavirus. Proc Natl Acad Sci U S A 2010; 107:6298-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Grigorieff N, Harrison SC. Near-atomic resolution reconstructions of icosahedral viruses from electron cryo-microscopy. Curr Opin Struct Biol 2011; 21:265-73; PMID:21333526; http://dx.doi.org/ 10.1016/j.sbi.2011.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Booy FP, Roden RB, Greenstone HL, Schiller JT, Trus BL. Two antibodies that neutralize papillomavirus by different mechanisms show distinct binding patterns at 13 A resolution. J Mol Biol 1998; 281:95-106; PMID:9680478; http://dx.doi.org/ 10.1006/jmbi.1998.1920 [DOI] [PubMed] [Google Scholar]
- 103.Combita AL, Touze A, Bousarghin L, Christensen ND, Coursaget P. Identification of two cross-neutralizing linear epitopes within the L1 major capsid protein of human papillomaviruses. J Virol 2002; 76:6480-6; PMID:12050360; http://dx.doi.org/ 10.1128/JVI.76.13.6480-6486.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Greiner VJ, Manin C, Larquet E, Ikhelef N, Greco F, Naville S, Milhiet PE, Ronzon F, Klymchenko A, Mely Y. Characterization of the structural modifications accompanying the loss of HBsAg particle immunogenicity. Vaccine 2014; 32:1049-54; PMID:24440114; http://dx.doi.org/ 10.1016/j.vaccine.2014.01.012 [DOI] [PubMed] [Google Scholar]
- 105.Mulder AM, Carragher B, Towne V, Meng Y, Wang Y, Dieter L, Potter CS, Washabaugh MW, Sitrin RD, Zhao Q. Toolbox for non-intrusive structural and functional analysis of recombinant VLP based vaccines: a case study with hepatitis B vaccine. PloS one 2012; 7:e33235; PMID:22493667; http://dx.doi.org/ 10.1371/journal.pone.0033235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Greiner VJ, Egele C, Oncul S, Ronzon F, Manin C, Klymchenko A, Mely Y. Characterization of the lipid and protein organization in HBsAg viral particles by steady-state and time-resolved fluorescence spectroscopy. Biochimie 2010; 92:994-1002; PMID:20420879; http://dx.doi.org/ 10.1016/j.biochi.2010.04.014 [DOI] [PubMed] [Google Scholar]
- 107.Mangold CM, Streeck RE. Mutational analysis of the cysteine residues in the hepatitis B virus small envelope protein. J Virol 1993; 67:4588-97; PMID:8392600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gilbert RJ, Beales L, Blond D, Simon MN, Lin BY, Chisari FV, Stuart DI, Rowlands DJ. Hepatitis B small surface antigen particles are octahedral. Proc Natl Acad Sci U S A 2005; 102:14783-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Iwarson S, Tabor E, Thomas HC, Goodall A, Waters J, Snoy P, Shih JW, Gerety RJ. Neutralization of hepatitis B virus infectivity by a murine monoclonal antibody: an experimental study in the chimpanzee. J Med Virol 1985; 16:89-96; PMID:2413167; http://dx.doi.org/ 10.1002/jmv.1890160112 [DOI] [PubMed] [Google Scholar]
- 110.Shearer MH, Sureau C, Dunbar B, Kennedy RC. Structural characterization of viral neutralizing monoclonal antibodies to hepatitis B surface antigen. Mol Immunol 1998; 35:1149-60; PMID:10199389; http://dx.doi.org/ 10.1016/S0161-5890(98)00110-2 [DOI] [PubMed] [Google Scholar]
- 111.Zhu Y, Zhang T, Zhao J, Weng Z, Yuan Q, Li S, Zhang J, Xia N, Zhao Q. Toward the development of monoclonal antibody-based assays to probe virion-like epitopes in hepatitis B vaccine antigen. Hum Vacci Immunother 2014; 10:1013-23; PMID:24499806; http://dx.doi.org/ 10.4161/hv.27753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Waters J, Pignatelli M, Galpin S, Ishihara K, Thomas HC. Virus-neutralizing antibodies to hepatitis B virus: the nature of an immunogenic epitope on the S gene peptide. J Gen Virol 1986; 67 ( Pt 11):2467-73; PMID:2431101; http://dx.doi.org/ 10.1099/0022-1317-67-11-2467 [DOI] [PubMed] [Google Scholar]
- 113.Waters JA, Brown SE, Steward MW, Howard CR, Thomas HC. Analysis of the antigenic epitopes of hepatitis B surface antigen involved in the induction of a protective antibody response. Virus Res 1992; 22:1-12; PMID:1371369; http://dx.doi.org/ 10.1016/0168-1702(92)90085-N [DOI] [PubMed] [Google Scholar]
- 114.Zhao Q, Towne V, Brown M, Wang Y, Abraham D, Oswald CB, Gimenez JA, Washabaugh MW, Kennedy R, Sitrin RD. In-depth process understanding of RECOMBIVAX HB(R) maturation and potential epitope improvements with redox treatment: multifaceted biochemical and immunochemical characterization. Vaccine 2011; 29:7936-41; PMID:21871939; http://dx.doi.org/ 10.1016/j.vaccine.2011.08.070 [DOI] [PubMed] [Google Scholar]
- 115.Zhao Q, Wang Y, Abraham D, Towne V, Kennedy R, Sitrin RD. Real time monitoring of antigenicity development of HBsAg virus-like particles (VLPs) during heat- and redox-treatment. Biochem Biophys Res Commun 2011; 408:447-53; PMID:21527246; http://dx.doi.org/ 10.1016/j.bbrc.2011.04.048 [DOI] [PubMed] [Google Scholar]
- 116.Li M, Beard P, Estes PA, Lyon MK, Garcea RL. Intercapsomeric disulfide bonds in papillomavirus assembly and disassembly. J Virol 1998; 72:2160-7; PMID:9499072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ishii Y, Kondo K, Matsumoto T, Tanaka K, Shinkai-Ouchi F, Hagiwara K, Kanda T. Thiol-reactive reagents inhibits intracellular trafficking of human papillomavirus type 16 pseudovirions by binding to cysteine residues of major capsid protein L1. Virol J 2007; 4:110; PMID:17961263; http://dx.doi.org/ 10.1186/1743-422X-4-110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ishii Y, Tanaka K, Kanda T. Mutational analysis of human papillomavirus type 16 major capsid protein L1: the cysteines affecting the intermolecular bonding and structure of L1-capsids. Virology 2003; 308:128-36; PMID:12706096; http://dx.doi.org/ 10.1016/S0042-6822(02)00099-5 [DOI] [PubMed] [Google Scholar]
- 119.Zhao Q, Allen MJ, Wang Y, Wang B, Wang N, Shi L, Sitrin RD. Disassembly and reassembly improves morphology and thermal stability of human papillomavirus type 16 virus-like particles. Nanomedicine 2012; 8:1182-9; PMID:22306156; http://dx.doi.org/ 10.1016/j.nano.2012.01.007 [DOI] [PubMed] [Google Scholar]
- 120.Zhao Q, Modis Y, High K, Towne V, Meng Y, Wang Y, Alexandroff J, Brown M, Carragher B, Potter CS, et al.. Disassembly and reassembly of human papillomavirus virus-like particles produces more virion-like antibody reactivity. VirolJ 2012; 9:52; PMID:22356831; http://dx.doi.org/ 10.1186/1743-422X-9-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yang C, Pan H, Wei M, Zhang X, Wang N, Gu Y, Du H, Zhang J, Li S, Xia N. Hepatitis E virus capsid protein assembles in 4M urea in the presence of salts. Protein Science 2013; 22:314-26; PMID:23281113; http://dx.doi.org/ 10.1002/pro.2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.McPherson CE. Development of a novel recombinant influenza vaccine in insect cells. Biologicals 2008; 36:350-3; PMID:18804387; http://dx.doi.org/ 10.1016/j.biologicals.2008.08.001 [DOI] [PubMed] [Google Scholar]
- 123.Cox MM. Recombinant protein vaccines produced in insect cells. Vaccine 2012; 30:1759-66; PMID:22265860; http://dx.doi.org/ 10.1016/j.vaccine.2012.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wang K, Holtz KM, Anderson K, Chubet R, Mahmoud W, Cox MM. Expression and purification of an influenza hemagglutinin-one step closer to a recombinant protein-based influenza vaccine. Vaccine 2006; 24:2176-85; PMID:16310896; http://dx.doi.org/ 10.1016/j.vaccine.2005.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cox MM, Hashimoto Y. A fast track influenza virus vaccine produced in insect cells. J Invertebr Pathol 2011; 107 Suppl:S31-41; PMID:21784229; http://dx.doi.org/ 10.1016/j.jip.2011.05.003 [DOI] [PubMed] [Google Scholar]
- 126.Cox MM, Karl Anderson D. Production of a novel influenza vaccine using insect cells: protection against drifted strains. Influenza Other Respir Viruses 2007; 1:35-40; PMID:19453478; http://dx.doi.org/ 10.1111/j.1750-2659.2006.00007.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Feshchenko E, Rhodes DG, Felberbaum R, McPherson C, Rininger JA, Post P, Cox MM. Pandemic influenza vaccine: characterization of A/California/07/2009 (H1N1) recombinant hemagglutinin protein and insights into H1N1 antigen stability. BMC Biotechnol 2012; 12:77; PMID:23110350; http://dx.doi.org/ 10.1186/1472-6750-12-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Keitel WA, Treanor JJ, El Sahly HM, Gilbert A, Meyer AL, Patriarca PA, Cox MM. Comparative immunogenicity of recombinant influenza hemagglutinin (rHA) and trivalent inactivated vaccine (TIV) among persons > or =65 years old. Vaccine 2009; 28:379-85; PMID:19879222; http://dx.doi.org/ 10.1016/j.vaccine.2009.10.037 [DOI] [PubMed] [Google Scholar]
- 129.King JC Jr., Cox MM, Reisinger K, Hedrick J, Graham I, Patriarca P. Evaluation of the safety, reactogenicity and immunogenicity of FluBlok trivalent recombinant baculovirus-expressed hemagglutinin influenza vaccine administered intramuscularly to healthy children aged 6-59 months. Vaccine 2009; 27:6589-94; PMID:19716456; http://dx.doi.org/ 10.1016/j.vaccine.2009.08.032 [DOI] [PubMed] [Google Scholar]
- 130.Baxter R, Patriarca PA, Ensor K, Izikson R, Goldenthal KL, Cox MM. Evaluation of the safety, reactogenicity and immunogenicity of FluBlok(R) trivalent recombinant baculovirus-expressed hemagglutinin influenza vaccine administered intramuscularly to healthy adults 50-64 years of age. Vaccine 2011; 29:2272-8; PMID:21277410; http://dx.doi.org/ 10.1016/j.vaccine.2011.01.039 [DOI] [PubMed] [Google Scholar]
- 131.Cox MM, Hollister JR. FluBlok, a next generation influenza vaccine manufactured in insect cells. Biologicals 2009; 37:182-9; PMID:19297194; http://dx.doi.org/ 10.1016/j.biologicals.2009.02.014 [DOI] [PubMed] [Google Scholar]
- 132.Cox MM, Patriarca PA, Treanor J. FluBlok, a recombinant hemagglutinin influenza vaccine. Influenza Other Respirat Viruses 2008; 2:211-9; PMID:19453397; http://dx.doi.org/ 10.1111/j.1750-2659.2008.00053.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Treanor JJ, El Sahly H, King J, Graham I, Izikson R, Kohberger R, Patriarca P, Cox M. Protective efficacy of a trivalent recombinant hemagglutinin protein vaccine (FluBlok(R)) against influenza in healthy adults: a randomized, placebo-controlled trial. Vaccine 2011; 29:7733-9; PMID:21835220; http://dx.doi.org/ 10.1016/j.vaccine.2011.07.128 [DOI] [PubMed] [Google Scholar]
- 134.Treanor JJ, Schiff GM, Hayden FG, Brady RC, Hay CM, Meyer AL, Holden-Wiltse J, Liang H, Gilbert A, Cox M. Safety and immunogenicity of a baculovirus-expressed hemagglutinin influenza vaccine: a randomized controlled trial. JAMA 2007; 297:1577-82; PMID:17426277; http://dx.doi.org/ 10.1001/jama.297.14.1577 [DOI] [PubMed] [Google Scholar]
- 135.Kang SM, Song JM, Quan FS, Compans RW. Influenza vaccines based on virus-like particles. Virus Res 2009; 143:140-6; PMID:19374929; http://dx.doi.org/ 10.1016/j.virusres.2009.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Song H, Wittman V, Byers A, Tapia T, Zhou B, Warren W, Heaton P, Connolly K. In vitro stimulation of human influenza-specific CD8+ T cells by dendritic cells pulsed with an influenza virus-like particle (VLP) vaccine. Vaccine 2010; 28:5524-32; PMID:20600506; http://dx.doi.org/ 10.1016/j.vaccine.2010.06.044 [DOI] [PubMed] [Google Scholar]
- 137.Khurana S, Wu J, Verma N, Verma S, Raghunandan R, Manischewitz J, King LR, Kpamegan E, Pincus S, Smith G, et al.. H5N1 virus-like particle vaccine elicits cross-reactive neutralizing antibodies that preferentially bind to the oligomeric form of influenza virus hemagglutinin in humans. J Virol 2011; 85:10945-54; PMID:21865396; http://dx.doi.org/ 10.1128/JVI.05406-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bright RA, Carter DM, Crevar CJ, Toapanta FR, Steckbeck JD, Cole KS, Kumar NM, Pushko P, Smith G, Tumpey TM, et al.. Cross-clade protective immune responses to influenza viruses with H5N1 HA and NA elicited by an influenza virus-like particle. PloS one 2008; 3:e1501; PMID:18231588; http://dx.doi.org/ 10.1371/journal.pone.0001501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Pushko P, Tumpey TM, Van Hoeven N, Belser JA, Robinson R, Nathan M, Smith G, Wright DC, Bright RA. Evaluation of influenza virus-like particles and Novasome adjuvant as candidate vaccine for avian influenza. Vaccine 2007; 25:4283-90; PMID:17403562; http://dx.doi.org/ 10.1016/j.vaccine.2007.02.059 [DOI] [PubMed] [Google Scholar]
- 140.Akahata W, Yang ZY, Andersen H, Sun S, Holdaway HA, Kong WP, Lewis MG, Higgs S, Rossmann MG, Rao S, et al.. A virus-like particle vaccine for epidemic Chikungunya virus protects nonhuman primates against infection. Nat Med 2010; 16:334-8; PMID:20111039; http://dx.doi.org/ 10.1038/nm.2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Metz SW, Gardner J, Geertsema C, Le TT, Goh L, Vlak JM, Suhrbier A, Pijlman GP. Effective chikungunya virus-like particle vaccine produced in insect cells. PLoS Negl Trop Dis 2013; 7:e2124; PMID:23516657; http://dx.doi.org/ 10.1371/journal.pntd.0002124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Metz SW, Martina BE, van den Doel P, Geertsema C, Osterhaus AD, Vlak JM, Pijlman GP. Chikungunya virus-like particles are more immunogenic in a lethal AG129 mouse model compared to glycoprotein E1 or E2 subunits. Vaccine 2013; 31:6092-6; PMID:24099875; http://dx.doi.org/ 10.1016/j.vaccine.2013.09.045 [DOI] [PubMed] [Google Scholar]
- 143.Pialoux G, Gauzere BA, Jaureguiberry S, Strobel M. Chikungunya, an epidemic arbovirosis. Lancet Infect Dis 2007; 7:319-27; PMID:17448935; http://dx.doi.org/ 10.1016/S1473-3099(07)70107-X [DOI] [PubMed] [Google Scholar]
- 144.Burt FJ, Rolph MS, Rulli NE, Mahalingam S, Heise MT. Chikungunya: a re-emerging virus. Lancet 2012; 379:662-71; PMID:22100854; http://dx.doi.org/ 10.1016/S0140-6736(11)60281-X [DOI] [PubMed] [Google Scholar]
- 145.Thiberville SD, Moyen N, Dupuis-Maguiraga L, Nougairede A, Gould EA, Roques P, de Lamballerie X. Chikungunya fever: epidemiology, clinical syndrome, pathogenesis and therapy. Antiviral Res 2013; 99:345-70; PMID:23811281; http://dx.doi.org/ 10.1016/j.antiviral.2013.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chang LJ, Dowd KA, Mendoza FH, Saunders JG, Sitar S, Plummer SH, Yamshchikov G, Sarwar UN, Hu Z, Enama ME, et al.. Safety and tolerability of chikungunya virus-like particle vaccine in healthy adults: a phase 1 dose-escalation trial. Lancet 2014; 384:2046-52; PMID:25132507; http://dx.doi.org/ 10.1016/S0140-6736(14)61185-5 [DOI] [PubMed] [Google Scholar]
- 147.Kam YW, Lum FM, Teo TH, Lee WW, Simarmata D, Harjanto S, Chua CL, Chan YF, Wee JK, Chow A, et al.. Early neutralizing IgG response to Chikungunya virus in infected patients targets a dominant linear epitope on the E2 glycoprotein. EMBO Mol Med 2012; 4:330-43; PMID:22389221; http://dx.doi.org/ 10.1002/emmm.201200213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kam YW, Simarmata D, Chow A, Her Z, Teng TS, Ong EK, Renia L, Leo YS, Ng LF. Early appearance of neutralizing immunoglobulin G3 antibodies is associated with chikungunya virus clearance and long-term clinical protection. J Infect Diss 2012; 205:1147-54; PMID:22389226; http://dx.doi.org/ 10.1093/infdis/jis033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lee CY, Kam YW, Fric J, Malleret B, Koh EG, Prakash C, Huang W, Lee WW, Lin C, Lin RT, et al.. Chikungunya virus neutralization antigens and direct cell-to-cell transmission are revealed by human antibody-escape mutants. PLoS Pathog 2011; 7:e1002390; PMID:22144891; http://dx.doi.org/ 10.1371/journal.ppat.1002390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Warter L, Lee CY, Thiagarajan R, Grandadam M, Lebecque S, Lin RT, Bertin-Maghit S, Ng LF, Abastado JP, Despres P, et al.. Chikungunya virus envelope-specific human monoclonal antibodies with broad neutralization potency. J Immunol 2011; 186:3258-64; PMID:21278338; http://dx.doi.org/ 10.4049/jimmunol.1003139 [DOI] [PubMed] [Google Scholar]
- 151.Lee H, Brendle SA, Bywaters SM, Guan J, Ashley RE, Yoder JD, Makhov AM, Conway JF, Christensen ND, Hafenstein S A cryo-electron microscopy study identifies the complete H16.V5 epitope and reveals global conformational changes initiated by binding of the neutralizing antibody fragment. Journal of virology 2015; 89:1428-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Schofield T. In vitro versus in vivo concordance: a case study of the replacement of an animal potency test with an immunochemical assay. Dev Biol 2002; 111:299-304; PMID:12678253 [PubMed] [Google Scholar]
- 153.Waters JA, Pignatelli M, Brown D, O'Rourke S, Lever A, Thomas HC. The immune response to hepatitis B virus. Postgrad Med J 1987; 63 Suppl 2:51-6; PMID:2446303 [PubMed] [Google Scholar]
- 154.Varsani A, Williamson AL, de Villiers D, Becker I, Christensen ND, Rybicki EP. Chimeric human papillomavirus type 16 (HPV-16) L1 particles presenting the common neutralizing epitope for the L2 minor capsid protein of HPV-6 and HPV-16. J Virol 2003; 77:8386-93; PMID:12857908; http://dx.doi.org/ 10.1128/JVI.77.15.8386-8393.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Shank-Retzlaff M, Wang F, Morley T, Anderson C, Hamm M, Brown M, Rowland K, Pancari G, Zorman J, Lowe R, et al.. Correlation between mouse potency and in vitro relative potency for human papillomavirus Type 16 virus-like particles and Gardasil vaccine samples. Hum Vaccin 2005; 1:191-7; PMID:17012876; http://dx.doi.org/ 10.4161/hv.1.5.2126 [DOI] [PubMed] [Google Scholar]
- 156.Deschuyteneer M, Elouahabi A, Plainchamp D, Plisnier M, Soete D, Corazza Y, Lockman L, Giannini S, Deschamps M. Molecular and structural characterization of the L1 virus-like particles that are used as vaccine antigens in Cervarix, the AS04-adjuvanted HPV-16 and -18 cervical cancer vaccine. Hum Vaccin 2010; 6:407-19; PMID:20953154; http://dx.doi.org/ 10.4161/hv.6.5.11023 [DOI] [PubMed] [Google Scholar]
- 157.Shank-Retzlaff ML, Zhao Q, Anderson C, Hamm M, High K, Nguyen M, Wang F, Wang N, Wang B, Wang Y, et al.. Evaluation of the thermal stability of Gardasil. Hum Vaccin 2006; 2:147-54; PMID:17012891; http://dx.doi.org/ 10.4161/hv.2.4.2989 [DOI] [PubMed] [Google Scholar]