Abstract
The epidemiology of Mycobacterium tuberculosis (Mtb) and M. africanum (Maf) suggest differences in their virulence, but the host immune profile to better understand the pathogenesis of tuberculosis (TB) have not been studied. We compared the transcriptomic and metabolic profiles between Mtb and Maf-infected TB cases to identify host biomarkers associated with lineages-specific pathogenesis and response to anti-TB chemotherapy. Venous blood samples from Mtb- and Maf-infected patients obtained before and after anti-TB treatment were analysed for cell composition, gene expression and metabolic profiles. Prior to treatment, similar transcriptomic profiles were seen in Maf- and Mtb-infected patients. In contrast, post-treatment, over 1600 genes related to immune responses and metabolic diseases were differentially expressed between the groups. Notably, the upstream regulator hepatocyte nuclear factor 4-alpha (HNF4α), which regulated 15% of these genes, was markedly enriched. Serum metabolic profiles were similar in both group pre-treatment, but the decline in pro-inflammatory metabolites post-treatment were most pronounced in Mtb-infected patients. Together, the differences in both peripheral blood transcriptomic and serum metabolic profiles between Maf- and Mtb-infected patients observed over the treatment period, might be indicative of intrinsic host factors related to susceptibility to TB and/or differential efficacy of the standard anti-TB treatment on the two lineages.
Introduction
Tuberculosis (TB) caused by lineages of the Mycobacterium tuberculosis (Mtb) complex (MTBC) remains a serious threat to public health globally. The MTBC comprises mycobacteria affecting humans and animals, amongst which Mtb and M. africanum (Maf) lineages are the most isolated in human. A plethora of studies have focused on the pathogen and shown distinct lineages within the MTBC; some lineages, such as Maf have evolved but remained region-specific, while others such as Mtb-Beijing and Mtb-Europe-America-Africa have successfully spread globally.1-3 In the Gambia, Maf West African type 2 causes up to 40% of all TB despite its relatively low virulence compared to Mtb.4,5 Analysis of the Maf genome has shown significant differences compared with Mtb that support its low virulence. These include the high number of pseudogenes6, impaired secretion of the virulence-associated protein early-secreted antigenic target 6 kDa (ESAT-6)7, and the high number of disrupted genes involved in bacterial carbohydrate, lipid and micronutrient metabolism.8
Few studies have investigated the human host response to different lineages of MTBC.9-11 In particular, the response of patients infected with different MTBC lineages to anti-TB drug treatment has been scarcely investigated12-14 and remains controversial. It has been suggested that different MTBC lineages have different rates of acquisition of drug resistance-associated mutations,15,16 and variable time to culture conversion.14 It has also been shown that host genetics, rather than the etiologic pathogen, also plays a major role in determining the response to anti-TB treatment12. Previous data from our laboratory have shown that the proportions of activated MTBC-specific T-cells are similar between Maf- and Mtb-infected patients before treatment, but significantly higher post-treatment in Maf- compared to Mtb-infected patients.17 This difference, after allegedly successful TB treatment, warrants further investigation into the possible underlying mechanism(s).
Transcriptomic and metabolic profiling are highly useful approaches towards a better understanding of host and pathogen interactions in disease and health.18 Studies describing peripheral blood cell transcriptional profiles have provided significant insights into TB pathogenesis19-27 and novel biomarkers for diagnosis and monitoring of the response to TB treatment.19-21,23 In this study, we perform an exploratory evaluation of transcriptomic and metabolic profiles of subjects infected with two different lineages of MTBC, Maf and Mtb, before and after anti-TB therapy in order to determine differences in host factors and/or biological processes associated with disease pathology and response to treatment.
Results
Study participants
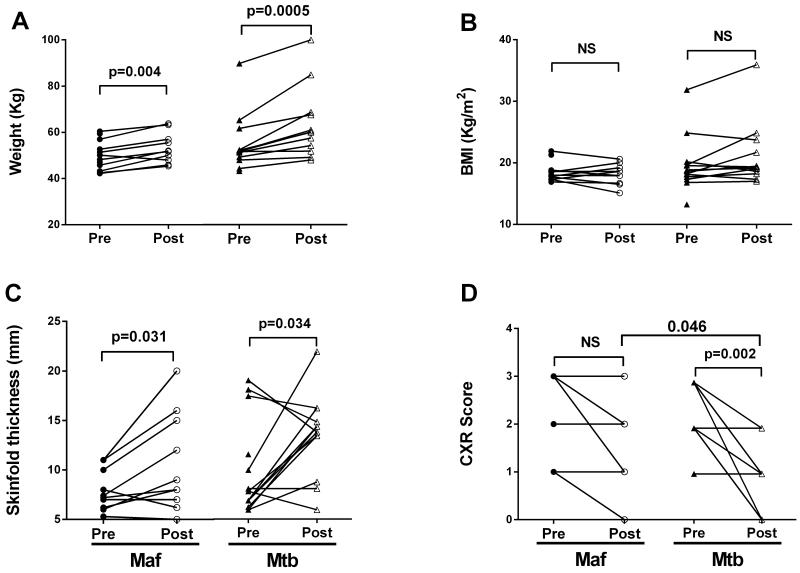
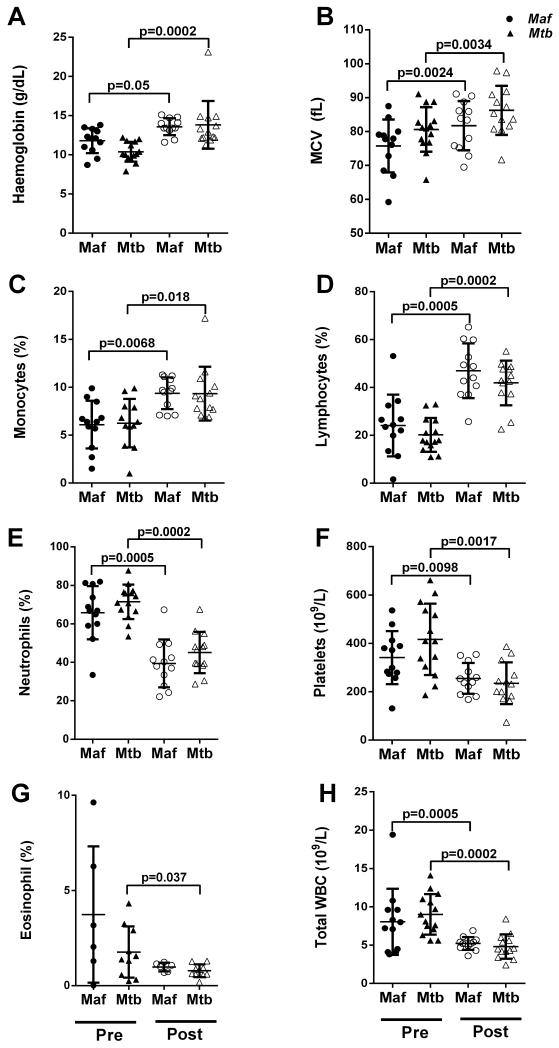
Twenty-six HIV-negative TB patients with drug-sensitive bacterial isolates were included in this study. Following genotyping of sputum isolates, 12 patients were classified as Maf West African type 2 (lineage 6) infected and 14 as infected with Mtb Europe-America-Africa (lineage 4), as previously described.17 Both groups had similar age, sex and ethnic distributions (Table 1). Although the smear microscopy grade was significantly higher in Mtb- than Maf-infected patients (Table 1; p=0.012), there was no significant difference in the disease severity as shown by TB-related clinical parameters such as duration of cough, body mass index (BMI), skin-fold thickness and chest X-ray (CXR) score prior to treatment (Table 1; Figure 1). However, Mtb-infected patients showed greater improvement than Maf-infected patients in all clinical parameters following a similar length of anti-TB therapy and this was significant for the CXR score (Table 1, Figure 1 D; p=0.046). Following treatment, the haemoglobin concentration, red blood cells (MCV), lymphocyte and monocyte counts were significantly increased in the two groups while neutrophil, platelet, eosinophil and total white blood cell (WBC) counts were significantly decreased, there was no significant differences between the two groups (Figure 2).
Table 1. Demographic and clinical characteristics of M. africanum and M. tuberculosis patients.
| Maf | Mtb | p-value | |||
|---|---|---|---|---|---|
| n | Positive (%) | n | Positive (%) | ||
| Number of cases a | 12 | 1 (46) | 14 | 14 (54) | |
| Demographics | |||||
| Age in years, median (range) | 12 | 27.5 (19–58) | 14 | 26 (15–79) | 0.683 |
| Sex (Female) | 12 | 3 (25) | 14 | 5 (36) | 1.0 |
| Ethnicity | 12 | 14 | 0.452 | ||
| Mandinka | 4 (33) | 8 (57) | |||
| Jola | 2 (17) | 2 (14) | |||
| Other | 6 (50) | 4 (29) | |||
| Clinical characteristics | |||||
| Duration of cough (>3 weeks) | 12 | 11 (92) | 14 | 11 (79) | 1 |
| Days of treatment prior recruitment | 12 | 6 (0–21) | 14 | 15 (0–21) | 0.155 |
| BMI, median (range) | |||||
| Enrolment | 12 | 18.2 (17–22) | 14 | 18.6 (13–32) | 0.643 |
| 6 months | 10 | 18.2 (15–21) | 12 | 19.1 (17–36) | 0.092 |
| Skinfold thickness mm median (range) | |||||
| Enrolment | 12 | 7.3 (5–11) | 14 | 8 (6–20) | 0.289 |
| 6 months | 10 | 8.5 (5–20) | 13 | 14 (6-23) | 0.113 |
| CXRc) Extent of disease | 12 [10]b | 14 [13] | 0.673 [0.046]b | ||
| Minimal | 3(25) [2(20)]b | 2(14) [8(61)]b | |||
| Moderate | 3(25) [5(50)]b | 6(43) [2(15)]b | |||
| Severe | 6(50) [2(20)]b | 6(43) [0(0)]b | |||
| Maximum smear grade | 12 | 14 | 0.012 | ||
| 1 | 3 (25) | 0 (0) | |||
| 2 | 6 (50) | 3 (21) | |||
| 3 | 3 (25) | 11 (79) |
Total number of patients recruited = 26,
[ ], 6-month CXR scores. Abbreviations: BMI, body mass index; CXR, chest X-ray; Maf, M. africanum; Mtb, M. tuberculosis
Figure 1. Changes in clinical parameters in Maf- and Mtb-infected patients pre- and post-treatment.
Line graphs representing variation of weight (A), body mass index (BMI; B), skinfold thickness (C) and chest X-ray score (CXR score; D) in Maf- (circle) and Mtb- (triangle) infected patients pre- and post-treatment. P-values shown derived using a Wilcoxon matched-pairs signed rank test.
Figure 2. Comparison of peripheral blood cell populations between Maf- and Mtb-infected patients pre- and post-treatment.
Each dot represents an individual patient in the Maf- (circle) or Mtb- (triangle)-infected groups pre- and post-treatment. %: percentage of peripheral blood cells, MCV: mean corpuscular volume (red blood cells), WBC: white blood cell. The horizontal bars indicate median and interquartile responses. A two-tailed Mann Whitney U test was used for comparison between groups at each time point, and a two-tailed Wilcoxon matched-pairs signed-rank test for comparison between pre- and post-treatment results within each group. P-values for significant differences are shown.
Gene expression profiles in Maf- and Mtb-infected patients pre- and post-treatment
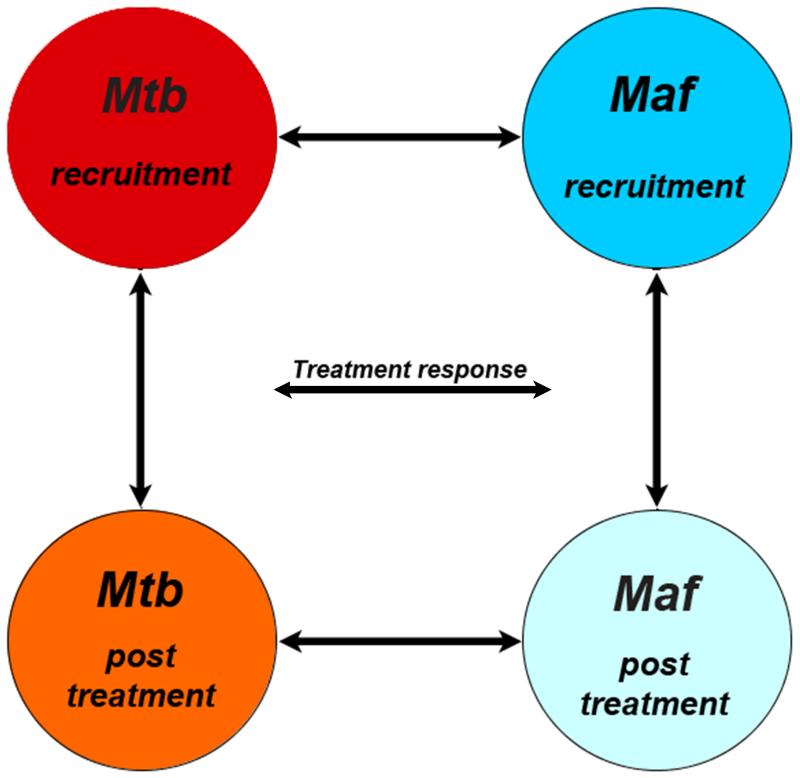
The analysis strategy to ascertain differences in gene expression between Maf- and Mtb-infected patients pre- and post-treatment is summarized in Figure 3. Firstly, we combined the 2 groups and performed a comparison between pre- and post-treatment time points. We found that over 6000 genes were differentially expressed between the pre- and post-treatment, with enrichment in Gene Ontology (GO) terms related to changes in metabolism and immune processes. Secondly, gene expression profiles in peripheral blood from all patients were compared pre- and post-treatment in direct group wise comparisons (Figure 3). Prior to treatment, gene expression profiles in Maf- and Mtb-infected patients were highly similar, with only 51 genes showing significant differential expression. Strikingly, after completion of the standard 6-month anti-TB chemotherapy, over 1600 gene probes, representing ~1400 annotated transcripts, were differentially expressed between Maf- and Mtb-infected patients (see Supplementary Table 1A for complete list); two thirds of these genes were significantly higher, while one-third was lower expressed in Maf- compared with Mtb-infected patients (Supplementary Figure 1). Very few genes (n=13) showed significant differences in the interaction between treatment time points and mycobacterial infecting lineage.
Figure 3. Analysis strategy of Maf- and Mtb-infected patients’ transcriptomic data.
Circles represent the different groups at each treatment time point. Arrows indicate the different analytical comparisons described herein where a) is the comparison between the two lineages at each time point and b) the comparison within each lineage between recruitment and post-treatment time point.
Differential expression post-treatment is associated with nuclear transcription factor HNF4A
A possible explanation for this differential gene expression between the groups after treatment could be incomplete resolution of pathology in one of the groups. We checked for an overlap with genes in the disease signature (i.e. the > 6.000 genes differentially expressed between pre- and post-treatment). Only 175 annotated transcripts were present in both comparisons and these did not relate to any GO enrichment, indicating that the differential gene expression post-treatment might not be a residual disease signature.
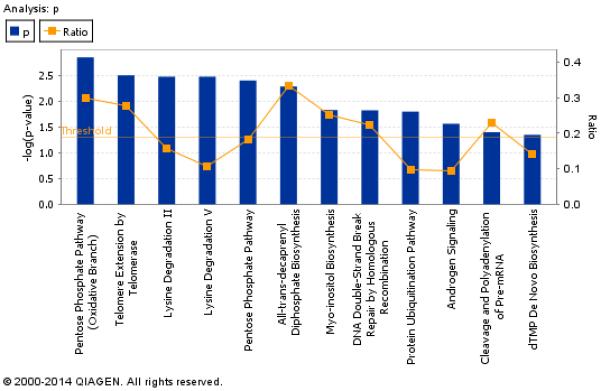
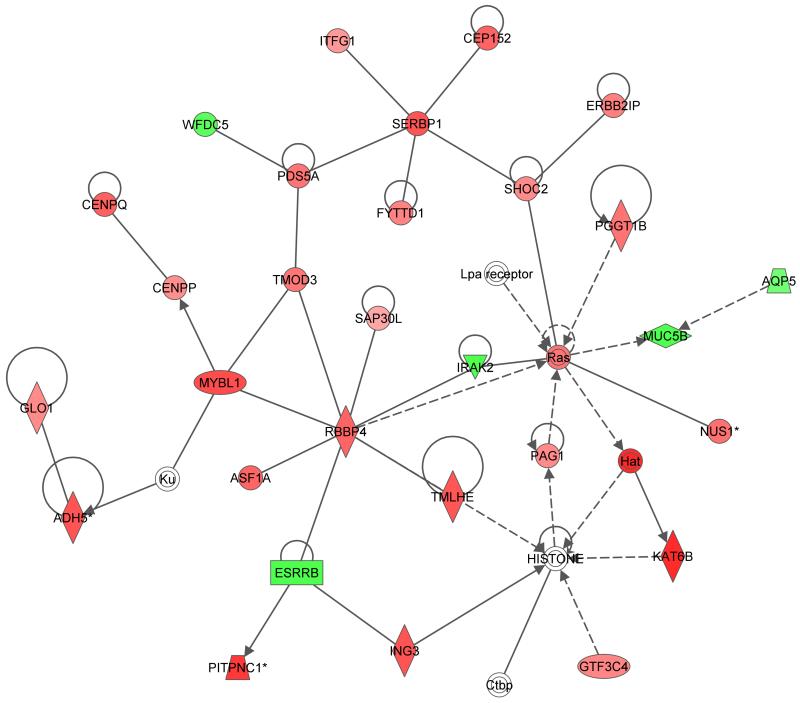
To further understand the biological processes associated with the differentially expressed genes between the two groups post-treatment, a pathway analysis was performed using Ingenuity Pathway Analysis (IPA) software. Ingenuity canonical pathway analysis revealed high enrichment in general metabolic processes such as the pentose phosphate pathway (Figure 4A). In addition, a network of immune response and metabolic disease-related genes, including Ras, MYBL1, SERBP1, SHOC2, PDS5A, RBBP4, Hat and TMOD3 showed an overall higher expression in Maf- than in Mtb-infected patients (Figure 4B). Further analysis of the gene set revealed a highly significant enrichment (p= 1.0−7) for the upstream regulator hepatocyte nuclear factor 4 alpha (HNF4α), which regulates almost 200 genes (~ 15% of genes) in the list of differentially expressed genes between the two groups post-treatment (Supplementary Table 1B).
Figure 4. Ingenuity pathways analysis of differentially expressed genes between Maf- and Mtb-infected patients post-treatment.
(A) Bar chart shows gene enrichment in general metabolic processes and orange points represent ratio of significantly expressed genes calculated by dividing the number of genes in a given pathway that meet the cut-off criteria by the total number of genes that make up the pathway and that are in the reference gene set. The threshold is at p=0.05. (B) Shows the Ingenuity network containing immune response and metabolic disease-related genes that were differentially expressed between Maf- and Mtb- infected patients post-treatment. Genes that were higher expressed in Maf than in Mtb are depicted in red, those lower expressed in Maf in green.
Serum metabolic profiles of Maf- and Mtb-infected patients
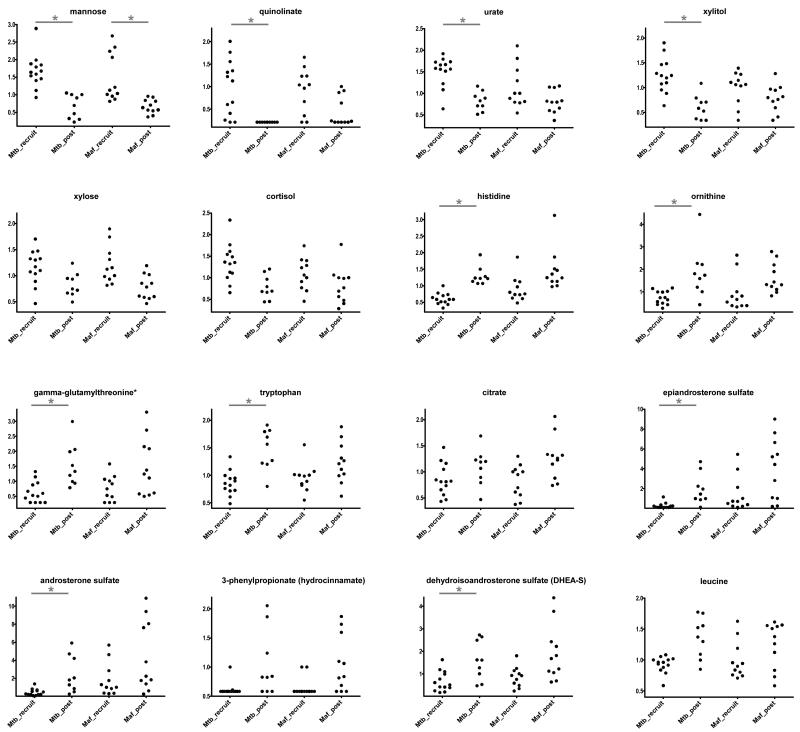
Given that the differentially expressed genes were predominantly those involving metabolic pathways, we investigated the serum metabolic profiles of the same patients. In general, there was no statistically significant difference in the serum metabolites between Mtb and Maf-infected patients at either pre- or post-treatment time points. In accordance, analysis of all patients combined showed that the abundances of 51 compounds were significantly different between the pre- and post-treatment time points (Supplementary Table 2). However, Maf-infected patients showed less pronounced differences than Mtb-infected patients between pre- and post-treatment (Figure 5). Mannose was the only metabolite that showed significant differences between pre- and post-treatment in Maf-infected patients. The hypergeometric test for pathway enrichment amongst the 51 compounds showed a statistically significant enrichment of the “carbohydrate” category (p < 0.03, 3.3-fold enrichment).
Figure 5. Some metabolites with statistically significant different serum concentrations between pre- and post-treatment time points.
Changes in relative abundance in selected serum metabolites pre- (recruit) and post-treatment in Maf- and Mtb-infected patients are shown. Each dot represents a patient. Stars indicate significant differences between profiles for each lineage separately (results from Wilcoxon rank sum test corrected for multiple testing (Benjamini-Hochberg); * Corr p<0.05), y-axis indicates normalized values (ion counts rescaled to median = 1).
Correlation between peripheral blood metabolites and gene expression profiles
We analyzed for correlations between the serum metabolites and gene expression to identify possible biological relationships. To obtain a conservative and robust list of potential correlations between metabolic and transcriptomic profiles, we included only 25 % of genes with the highest interquartile range (IQR) and metabolites with an IQR higher than 0, resulting in 505 metabolites. Only intra-group correlations between peripheral blood metabolites and gene expression were considered so as to remove the effect of the inter-group correlations. We fitted a linear regression model for each of the data with the groups (pre- and post-treatment) as the predictor, and used the residuals from these models to calculate Spearman rank correlation coefficients. We found that among the 21 strongest correlations (with correlation coefficient rho > 0.65), 8 corresponded to correlations between particular genes and the metabolite arabinose, which was present at significantly higher levels in patients from the pre-treatment group. Also, 4 of the strongest correlations were between X- or Y-chromosome-linked genes and steroid hormones. To obtain a better understanding of the biological link between arabinose and gene expression profiles, we performed GO enrichment analysis (using GOrilla) of genes sorted according to their absolute correlation with arabinose. Interestingly, we found a significant enrichment in genes involved in bacterial defense and acute inflammatory responses (Supplementary Figure 2).
Discussion
In this study, we investigated the differences in TB pathogenesis and response to drug treatment between two MTBC lineages, Maf and Mtb, infecting a sympatric host population by comparing peripheral blood gene expression and serum metabolic profiles before and after anti-TB chemotherapy. Discovery and exploitation of MTBC lineages-specific host biomarkers could further our understanding of TB pathogenesis as well as improve diagnosis, treatment or vaccine development for TB. Peripheral blood gene expression profiles were not different between Maf- and Mtb-infected patients pre-treatment but differed significantly post-treatment, and these were predominantly associated with immune responses and metabolic diseases. Intriguingly, the hepatocyte nuclear factor 4 alpha (HNF4α) regulated about 15% of the genes differentially expressed between the groups post-treatment. The serum metabolic profiles were similar between Maf- and Mtb-infected patients both pre- and post-treatment, but significantly different between pre- and post-treatment, particularly in Mtb- than in Maf-infected groups.
Although both groups had very similar clinical presentations at baseline and no significant interaction between treatment and lineages was found, the machine learning models performed better for Mtb samples than Maf samples, suggesting that a better clinical resolution of the disease was achieved for Mtb-infected groups. The similarity in peripheral blood transcriptomic and metabolic profiles before treatment suggests uniformity in host responses after succumbing to infection, irrespective of the infecting lineage of MTBC. This is also supported by the similar clinical scores and activated T cells proportions between the groups previously described before start of treatment.17 Differences observed after completing 6-month treatment could reflect differences in the mycobacterial lineage response to anti-TB drugs and/or host immune response for recovery. 19,23,26 Although we did not identify a strong association or enrichment of inflammatory response-related genes, clinical data suggest slower recovery in Maf-infected patients. The poor resolution of CXR, and higher proportion of activated T cells17 following completion of anti-TB treatment in Maf-infected patients likely indicate persistence of the bacilli, possibly due to lower in-vivo drug sensitivity or inability of the host anti-TB immune response to have resolved within this period post-treatment or persistence of the bacilli in Maf-patients. The similarity of the peripheral blood cell counts between the two groups both pre- and post-treatment, suggesting that differences in transcriptomic and metabolic profiles might be functional rather than structural support that it takes longer time for the inflammatory responses in Maf to resolve.
HNF4α is a highly conserved member of the nuclear receptor superfamily expressed in liver, kidney, intestine and pancreas of mammals,28 and was found to regulate about 15% of the genes differentially expressed between the groups post-treatment in this study. It is considered a global regulator of hundreds of genes involved in intermediary metabolism, liver functions, inflammatory responses,28,29 and glucose, fat and drug metabolism.30 Further, HNF4α is directly linked with different human diseases including diabetes, a major risk factor for TB,31,32 hepatitis B virus infections, atherosclerosis and hemophilia.33,34 Thus, our results suggest that Maf-infected patients may have a subclinical metabolic or liver dysfunction that is associated with susceptibility to this relatively low virulent lineage of MTBC. Moreover the role of HNF4α in xenobiotic and drug metabolism28 may support defective anti-TB drug metabolism and in-vivo efficacy leading to poor clearance of the bacilli. HNF4α also interacts closely with liver X receptor (LXR), another master regulator of lipid metabolism,35 which contributes to the protective immune response against Mtb.36 A recent study showed that polymorphisms in the lxr gene are associated with increased susceptibility to TB in a Chinese population.37 Taken together, our results indicate that HNF4A is potentially associated with susceptibility to TB. Further research is needed to contest this hypothesis.
Interestingly, the metabolites that changed significantly following treatment are consistent with those previously identified to differ between active TB and healthy individuals.38 The difference in the pattern of changes of these metabolites between the time points in each group suggests subtle differences in the response of the MTBC lineages to anti-TB treatment. For example, the serum tryptophan concentration significantly increased in Mtb-infected patients after treatment, while this effect was not detected in Maf-infected patients. Serum tryptophan concentration has been shown to be lower in active TB patients compared to treated patients and healthy controls, and has been suggested as a prognostic marker for pulmonary TB.39 Tryptophan is actively degraded into kynurenine by indoleamine 2, 3-dioxygenase, which is induced by the bacteria during active TB,40 thereby driving an increase in kynurenine abundance with a reverse scenario in healthy individuals.38,39 However, no differences were detected in kynurenine concentration post-treatment in either group probably because kynurenine metabolism constitutes only a small part of tryptophan catabolism.41 In contrast to the transcriptomic data, the metabolic profiles did not show any significant differences between the two groups post-treatment or any significant interactions between treatment (pre and post) and bacterial lineage.
The concentration of serum arabinose, a pro-inflammatory molecule, which significantly decreased in Mtb compared to Maf patients following treatment, showed higher correlation with genes associated with host defense. In addition, xylitol shown to have anti-microbial activity against S. pneumonia and Influenza virus in animal models,42,43 was more significantly decreased in Maf than Mtb-infected patients post-treatment. The differences in these metabolites are indicative of variance in the resolution of pro-inflammatory responses between Maf and Mtb after treatment.44
The weak or absence of statistical significance in some comparisons in our study may be in part due to the small group sizes. However, a number of important findings that could be further explored to get more understanding of the pathogenesis of TB have been identified in this exploratory finding.
Very few studies have investigated the impact of MTBC lineage differences on human host peripheral blood transcriptomic and metabolic profiles.12 Our study is among the first to address this question using clinical data and high-throughput techniques to compare host responses between Maf- and Mtb-infected patients on the global level. Changes in both peripheral blood cell transcriptomic and serum metabolomic profiles during treatment and differences in gene expression between the two groups after treatment, suggest that host-intrinsic differences contribute to TB susceptibility and/or differences in response to standard anti-TB therapy.
Methods
Ethics statement
Ethical approval for this study was obtained from The Gambian Government/Medical Research Council (MRC) Joint Ethics Committee in The Gambia, and the London School of Hygiene & Tropical Medicine Ethics Committee in the UK. Written informed consent was obtained from all study participants.
Study participants
The 26 sputum smear- and culture-positive TB patients included in this study were recruited at the TB clinic of the Medical Research Council (MRC) Unit, in The Gambia as previously described.17 All patients were HIV-negative with no history of previous TB disease and were consecutively enrolled before starting anti-TB treatment and followed-up at 6 months of treatment. All patients underwent a routine CXR that was analyzed as previously described,45 were treated according to the Gambian National Leprosy and Tuberculosis Programme treatment guidelines (2HRZE/4HR), and were sputum smear-negative post-treatment. The infecting lineage genotype was determined using standard spoligotyping analysis, and by assessing the presence or absence of lineage defining large sequence polymorphisms (LSP) RD702 and TbD1, as previously described.17,46 Drug susceptibility tests for first-line anti-TB drugs were performed using the BACTEC MGIT 960 SIRE kit (Becton-Dickinson). All patients had fully drug-susceptible TB isolates at recruitment. At each time point, peripheral whole blood (2.5 mL) was collected from every patient in PAXgene tubes (PreAnalaytiX) and stored at −80 °C before processing. Serum samples were stored at −20 °C.
RNA extraction and microarray procedure
RNA was isolated from PAXgene tubes using the PAXgene blood RNA kit (PreAnalytix). RNA was labeled with the Fluorescent Linear Amplification Kit (Agilent Technologies) according to manufacturer’s instructions. Quantity and labeling efficiency were verified before hybridization of the samples to whole-genome 4×44k human expression arrays (Agilent) and scanned at 5 μm using an Agilent scanner. Microarray data comply with MIAME (minimal information about microarray experiment) guidelines and have been deposited in the Gene Expression Omnibus (GEO) database under accession # GSE62147.
Data and Statistical Analysis strategy
Analysis of the scanned images was performed with Feature Extraction software (version 10.5.1, Agilent Technologies). Gene expression data were analyzed using Agilent’s GeneSpring software (version 12.1). Raw microarray data were quantiles normalized without baseline transformation. Metabolomic profiles and correlation analyses were performed using the R package Limma. We followed 2 main analysis strategies. First, 2-way ANOVA was used to analyze differences in expression between the groups of patients infected with Maf and Mtb at recruitment and post-treatment (Figure 3). This strategy includes the analysis of differences in anti-TB treatment effect between Maf and Mtb-infected patients. Second, moderated T-tests were applied for direct group wise comparisons, which may give a clearer picture of differences between patient groups (Maf and Mtb) at the time of recruitment or post-treatment. Benjamini-Hochberg correction for multiple testing was applied in all tests and corrected p-values < 0.01 were considered significant. Since no gene expression or metabolomics data on similar patients groups are available, no power estimates could be performed to test the used parameters on our group sizes. Enrichment analysis of differentially expressed genes based on Gene Ontology (GO) terms was performed using the web-based tool GOrilla (http://cbl-gorilla.cs.technion.ac.il). Ingenuity Pathway Analysis (IPA) was used to study the gene interaction network and enrichment of upstream regulators of expressed genes. Continuous clinical data were compared between the groups using a two-tailed Mann Whitney U test and Wilcoxon matched-pairs signed-rank test, while categorical data was compared using a Fisher’s exact test.
Serum metabolic profiling procedure
Metabolites in serum from the study group were analyzed using non-targeted mass spectrometry (MS) analysis, which was performed at Metabolon, Inc (North Carolina, USA) as described previously.47 Metabolites were identified by automated comparison of ion features in the experimental samples to a reference library of chemical standard entries, including retention time, molecular weight (m/z), preferred adducts, and in-source fragments as well as associated MS spectra. Identification of known chemical entities is based on comparison to metabolomic library entries of purified standards. Statistical analysis of metabolomic data was done using the R package limma.48 Machine learning was performed using the Random Forest package from R.49
Supplementary Material
Figure 6.
Acknowledgments
We thank the Gambian National Leprosy and Tuberculosis Programme for their continuing collaboration. We are also grateful to study participants, field workers, especially K. Kanyi and O. Ceesay for performing sample collection, MRC TB clinical staff and P. Camara for obtaining consent and enrolling participants, TB immunology and TB diagnostic laboratory staff, and M. Antonio, P. Owiafe, A. Bojang, J. Mendy, M. Daramy, J. Otu and F. Mendy for laboratory assistance. We also thank T. Togun for clinical examination of study patients and M.L. Grossman for editorial support. Finally, we thank Metabolon Inc. for generating the metabolic profiles.
Funding
The study was funded by the MRC Unit, The Gambia as a PhD fellowship awarded to L.D.T, the European Commission Advanced Immunization Technologies (ADITEC) Grant FP7-HEALTH-2011-280873, and the Max Planck Institute for Infection Biology in Berlin, Germany.
Footnotes
Conflict of interest: The authors declare no financial or commercial conflict of interest.
References
- 1.Gagneux S. Host-pathogen coevolution in human tuberculosis. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2012;367:850–859. doi: 10.1098/rstb.2011.0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Comas I, Chakravartti J, Small PM, Galagan J, Niemann S, Kremer K, et al. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat Genet. 2010;42:498–503. doi: 10.1038/ng.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Niemann S, Kubica T, Bange FC, Adjei O, Browne EN, Chinbuah MA, et al. The species Mycobacterium africanum in the light of new molecular markers. J Clin Microbiol. 2004;42:3958–3962. doi: 10.1128/JCM.42.9.3958-3962.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Jong BC, Adetifa I, Walther B, Hill PC, Antonio M, Ota M, et al. Differences between tuberculosis cases infected with Mycobacterium africanum, West African type 2, relative to Euro-American Mycobacterium tuberculosis: an update. FEMS Immunol Med Microbiol. 2010;58:102–105. doi: 10.1111/j.1574-695X.2009.00628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Via LE, Weiner DM, Schimel D, Lin PL, Dayao E, Tankersley SL, et al. Differential virulence and disease progression following Mycobacterium tuberculosis complex infection of the common marmoset (Callithrix jacchus) Infect Immun. 2013;81:2909–2919. doi: 10.1128/IAI.00632-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bentley SD, Comas I, Bryant JM, Walker D, Smith NH, Harris SR, et al. The genome of Mycobacterium africanum West African 2 reveals a lineage-specific locus and genome erosion common to the M. tuberculosis complex. PLoS Negl Trop Dis. 2012;6:e1552. doi: 10.1371/journal.pntd.0001552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bold TD, Davis DC, Penberthy KK, Cox LM, Ernst JD, de Jong BC. Impaired fitness of Mycobacterium africanum despite secretion of ESAT-6. J Infect Dis. 2012;205:984–990. doi: 10.1093/infdis/jir883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gehre F, Otu J, DeRiemer K, de Sessions PF, Hibberd ML, Mulders W, et al. Deciphering the growth behaviour of Mycobacterium africanum. PLoS Negl Trop Dis. 2013;7:e2220. doi: 10.1371/journal.pntd.0002220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Varahram M, Farnia P, Nasiri MJ, Karahrudi MA, Dizagie MK, Velayati AA. Association of Mycobacterium Tuberculosis Lineages with IFN-gamma and TNF-alpha Gene Polymorphisms among Pulmonary Tuberculosis Patient. Mediterranean journal of hematology and infectious diseases. 2014;6:e2014015. doi: 10.4084/MJHID.2014.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Portevin D, Gagneux S, Comas I, Young D. Human macrophage responses to clinical isolates from the Mycobacterium tuberculosis complex discriminate between ancient and modern lineages. PLoS Pathog. 2011;7:e1001307. doi: 10.1371/journal.ppat.1001307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang C, Peyron P, Mestre O, Kaplan G, van Soolingen D, Gao Q, et al. Innate immune response to Mycobacterium tuberculosis Beijing and other genotypes. PLoS One. 2010;5:e13594. doi: 10.1371/journal.pone.0013594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coussens AK, Wilkinson RJ, Nikolayevskyy V, Elkington PT, Hanifa Y, Islam K, et al. Ethnic variation in inflammatory profile in tuberculosis. PLoS Pathog. 2013;9:e1003468. doi: 10.1371/journal.ppat.1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pareek M, Evans J, Innes J, Smith G, Hingley-Wilson S, Lougheed KE, et al. Ethnicity and mycobacterial lineage as determinants of tuberculosis disease phenotype. Thorax. 2013;68:221–229. doi: 10.1136/thoraxjnl-2012-201824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Click ES, Winston CA, Oeltmann JE, Moonan PK, Mac Kenzie WR. Association between Mycobacterium tuberculosis lineage and time to sputum culture conversion. Int J Tuberc Lung Dis. 2013;17:878–884. doi: 10.5588/ijtld.12.0732. [DOI] [PubMed] [Google Scholar]
- 15.Ford CB, Shah RR, Maeda MK, Gagneux S, Murray MB, Cohen T, et al. Mycobacterium tuberculosis mutation rate estimates from different lineages predict substantial differences in the emergence of drug-resistant tuberculosis. Nat Genet. 2013;45:784–790. doi: 10.1038/ng.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gumbo T. Biological variability and the emergence of multidrug-resistant tuberculosis. Nat Genet. 2013;45:720–721. doi: 10.1038/ng.2675. [DOI] [PubMed] [Google Scholar]
- 17.Tientcheu LD, Sutherland JS, de Jong BC, Kampmann B, Jafali J, Adetifa IM, et al. Differences in T-cell responses between Mycobacterium tuberculosis and Mycobacterium africanum-infected patients. Eur J Immunol. 2014;44:1387–1398. doi: 10.1002/eji.201343956. [DOI] [PubMed] [Google Scholar]
- 18.Weiner J, Maertzdorf J, Kaufmann SH. The dual role of biomarkers for understanding basic principles and devising novel intervention strategies in tuberculosis. Ann N Y Acad Sci. 2013;1283:22–29. doi: 10.1111/j.1749-6632.2012.06802.x. [DOI] [PubMed] [Google Scholar]
- 19.Bloom CI, Graham CM, Berry MP, Wilkinson KA, Oni T, Rozakeas F, et al. Detectable changes in the blood transcriptome are present after two weeks of antituberculosis therapy. PLoS One. 2012;7:e46191. doi: 10.1371/journal.pone.0046191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson ST, Kaforou M, Brent AJ, Wright VJ, Banwell CM, Chagaluka G, et al. Diagnosis of childhood tuberculosis and host RNA expression in Africa. N Engl J Med. 2014;370:1712–1723. doi: 10.1056/NEJMoa1303657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaforou M, Wright VJ, Oni T, French N, Bangani N, Banwell CM, et al. Detection of tuberculosis in HIV-infected and -uninfected African adults using whole blood RNA expression signatures: a case-control study. PLoS Med. 2013;10:e1001538. doi: 10.1371/journal.pmed.1001538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maertzdorf J, Weiner J, 3rd, Mollenkopf HJ, Network TB, Bauer T, Prasse A, et al. Common patterns and disease-related signatures in tuberculosis and sarcoidosis. Proc Natl Acad Sci U S A. 2012;109:7853–7858. doi: 10.1073/pnas.1121072109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berry MP, Graham CM, McNab FW, Xu Z, Bloch SA, Oni T, et al. An interferon inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466:973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maertzdorf J, Ota M, Repsilber D, Mollenkopf HJ, Weiner J, Hill PC, et al. Functional correlations of pathogenesis-driven gene expression signatures in tuberculosis. PLoS One. 2011;6:e26938. doi: 10.1371/journal.pone.0026938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maertzdorf J, Repsilber D, Parida SK, Stanley K, Roberts T, Black G, et al. Human gene expression profiles of susceptibility and resistance in tuberculosis. Genes Immun. 2011;12:15–22. doi: 10.1038/gene.2010.51. [DOI] [PubMed] [Google Scholar]
- 26.Cliff JM, Lee JS, Constantinou N, Cho JE, Clark TG, Ronacher K, et al. Distinct phases of blood gene expression pattern through tuberculosis treatment reflect modulation of the humoral immune response. J Infect Dis. 2013;207:18–29. doi: 10.1093/infdis/jis499. [DOI] [PubMed] [Google Scholar]
- 27.Joosten SA, Goeman JJ, Sutherland JS, Opmeer L, de Boer KG, Jacobsen M, et al. Identification of biomarkers for tuberculosis disease using a novel dual-color RT-MLPA assay. Genes Immun. 2012;13:71–82. doi: 10.1038/gene.2011.64. [DOI] [PubMed] [Google Scholar]
- 28.Hwang-Verslues WW, Sladek FM. HNF4alpha--role in drug metabolism and potential drug target? Current opinion in pharmacology. 2010;10:698–705. doi: 10.1016/j.coph.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Babeu JP, Boudreau F. Hepatocyte nuclear factor 4-alpha involvement in liver and intestinal inflammatory networks. World journal of gastroenterology : WJG. 2014;20:22–30. doi: 10.3748/wjg.v20.i1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol. 2001;21:1393–1403. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jimenez-Corona ME, Cruz-Hervert LP, Garcia-Garcia L, Ferreyra-Reyes L, Delgado-Sanchez G, Bobadilla-Del-Valle M, et al. Association of diabetes and tuberculosis: impact on treatment and post-treatment outcomes. Thorax. 2013;68:214–220. doi: 10.1136/thoraxjnl-2012-201756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Restrepo BI, Schlesinger LS. Impact of diabetes on the natural history of tuberculosis. Diabetes research and clinical practice. 2014 doi: 10.1016/j.diabres.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mohlke KL, Boehnke M. The role of HNF4A variants in the risk of type 2 diabetes. Current diabetes reports. 2005;5:149–156. doi: 10.1007/s11892-005-0043-y. [DOI] [PubMed] [Google Scholar]
- 34.Jafar-Mohammadi B, Groves CJ, Gjesing AP, Herrera BM, Winckler W, Stringham HM, et al. A role for coding functional variants in HNF4A in type 2 diabetes susceptibility. Diabetologia. 2011;54:111–119. doi: 10.1007/s00125-010-1916-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crestani M, De Fabiani E, Caruso D, Mitro N, Gilardi F, Vigil Chacon AB, et al. LXR (liver X receptor) and HNF-4 (hepatocyte nuclear factor-4): key regulators in reverse cholesterol transport. Biochem Soc Trans. 2004;32:92–96. doi: 10.1042/bst0320092. [DOI] [PubMed] [Google Scholar]
- 36.Korf H, Vander Beken S, Romano M, Steffensen KR, Stijlemans B, Gustafsson JA, et al. Liver X receptors contribute to the protective immune response against Mycobacterium tuberculosis in mice. J Clin Invest. 2009;119:1626–1637. doi: 10.1172/JCI35288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han M, Liang L, Liu LR, Yue J, Zhao YL, Xiao HP. Liver X receptor gene polymorphisms in tuberculosis: effect on susceptibility. PLoS One. 2014;9:e95954. doi: 10.1371/journal.pone.0095954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weiner J, 3rd, Parida SK, Maertzdorf J, Black GF, Repsilber D, Telaar A, et al. Biomarkers of inflammation, immunosuppression and stress with active disease are revealed by metabolomic profiling of tuberculosis patients. PLoS One. 2012;7:e40221. doi: 10.1371/journal.pone.0040221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki Y, Suda T, Asada K, Miwa S, Suzuki M, Fujie M, et al. Serum indoleamine 2,3-dioxygenase activity predicts prognosis of pulmonary tuberculosis. Clin Vaccine Immunol. 2012;19:436–442. doi: 10.1128/CVI.05402-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blumenthal A, Nagalingam G, Huch JH, Walker L, Guillemin GJ, Smythe GA, et al. M. tuberculosis induces potent activation of IDO-1, but this is not essential for the immunological control of infection. PLoS One. 2012;7:e37314. doi: 10.1371/journal.pone.0037314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sedlmayr P, Blaschitz A, Stocker R. The role of placental tryptophan catabolism. Frontiers in immunology. 2014;5:230. doi: 10.3389/fimmu.2014.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Renko M, Valkonen P, Tapiainen T, Kontiokari T, Mattila P, Knuuttila M, et al. Xylitol-supplemented nutrition enhances bacterial killing and prolongs survival of rats in experimental pneumococcal sepsis. BMC Microbiol. 2008;8:45. doi: 10.1186/1471-2180-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yin SY, Kim HJ, Kim HJ. Protective effect of dietary xylitol on influenza A virus infection. PLoS One. 2014;9:e84633. doi: 10.1371/journal.pone.0084633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ammons MC, Copie V. Mini-review: Lactoferrin: a bioinspired, anti-biofilm therapeutic. Biofouling. 2013;29:443–455. doi: 10.1080/08927014.2013.773317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Jong BC, Hill PC, Aiken A, Jeffries DJ, Onipede A, Small PM, et al. Clinical presentation and outcome of tuberculosis patients infected by M. africanum versus M. tuberculosis. Int J Tuberc Lung Dis. 2007;11:450–456. [PubMed] [Google Scholar]
- 46.de Jong BC, Antonio M, Awine T, Ogungbemi K, de Jong YP, Gagneux S, et al. Use of spoligotyping and large sequence polymorphisms to study the population structure of the Mycobacterium tuberculosis complex in a cohort study of consecutive smear-positive tuberculosis cases in The Gambia. J Clin Microbiol. 2009;47:994–1001. doi: 10.1128/JCM.01216-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem. 2009;81:6656–6667. doi: 10.1021/ac901536h. [DOI] [PubMed] [Google Scholar]
- 48.Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. Springer; New York: 2005. pp. 397–420. [Google Scholar]
- 49.Liaw AWM. Classification and regression by random-forest. R News. 2002;2:18–22. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.