Abstract
Methanococcus maripaludis is a strictly anaerobic, methane-producing archaeon. Aromatic amino acids (AroAAs) are biosynthesized in this autotroph either by the de novo pathway, with chorismate as an intermediate, or by the incorporation of exogenous aryl acids via indolepyruvate oxidoreductase (IOR). In order to evaluate the roles of these pathways, the gene that encodes the third step in the de novo pathway, 3-dehydroquinate dehydratase (DHQ), was deleted. This mutant required all three AroAAs for growth, and no DHQ activity was detectible in cell extracts, compared to 6.0 ± 0.2 mU mg−1 in the wild-type extract. The growth requirement for the AroAAs could be fulfilled by the corresponding aryl acids phenylacetate, indoleacetate, and p-hydroxyphenylacetate. The specific incorporation of phenylacetate into phenylalanine by the IOR pathway was demonstrated in vivo by labeling with [1-13C]phenylacetate. M. maripaludis has two IOR homologs. A deletion mutant for one of these homologs contained 76, 74, and 42% lower activity for phenylpyruvate, p-hydoxyphenylpyruvate, and indolepyruvate oxidation, respectively, than the wild type. Growth of this mutant in minimal medium was inhibited by the aryl acids, but the AroAAs partially restored growth. Genetic complementation of the IOR mutant also restored much of the wild-type phenotype. Thus, aryl acids appear to regulate the expression or activity of the de novo pathway. The aryl acids did not significantly inhibit the activity of the biosynthetic enzymes chorismate mutase, prephenate dehydratase, and prephenate dehydrogenase in cell extracts, so the inhibition of growth was probably not due to an effect on these enzymes.
Methanococcus maripaludis is a strictly anaerobic, methane-producing archaeon. It is a mesophile that utilizes CO2 as the sole carbon source during autotrophic growth in minimal medium but assimilates acetate and some amino acids as carbon sources when they are present (28, 46, 47, 66). Regardless of the carbon source, the reduction of carbon dioxide to methane by H2 or formate is a required energy source for the growth of these obligate lithotrophs.
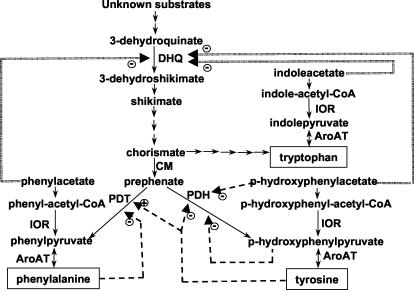
Several lines of evidence suggest that the aromatic amino acids (AroAAs) can be biosynthesized by the de novo pathway in methanogens and other archaea (for a review, see reference 67). The de novo pathway starts from the common pathway leading to chorismate and then splits to biosynthesize tryptophan, tyrosine, or phenylalanine (Fig. 1). This pathway is sufficient to explain the pattern of isotope incorporation in the AroAAs in many methanogens (11, 12, 53). Bioinformatic analyses of genomic sequences further demonstrate homologs for the genes for the de novo pathway in many archaea (44, 52, 67). However, homologs have not been found for the first two steps in the methanogens, which may utilize different precursors (60). A few enzymes in the pathway have also been biochemically characterized. In the common pathway, only shikimate kinase from Methanocaldococcus jannaschii has been characterized in archaea (9). From chorismate to tyrosine and phenylalanine, chorismate mutase (CM) from M. jannaschii, prephenate dehydratase (PDT) from Halobacterium vallismortis, and aromatic aminotransferases (AroAT) from Methanococcus aeolicus and Pyrococcus furiosus have been characterized (27, 34, 62, 69). Most of the enzymes for the biosynthesis of tryptophan from chorismate have been characterized in archaea, including some Methanococcus species (14, 24, 29, 38, 48, 55, 61).
FIG. 1.
Pathways for the biosynthesis of AroAAs in M. maripaludis. The precursors for the de novo pathway in methanogens are not known, but homologs for genes at all the steps after 3-dehydroquinate are present in the genomic sequence (see the text). After chorismate, the pathway splits into three branches, one of which leads to tryptophan. At prephenate, the branches leading to phenylalanine and tyrosine form. The aryl acid pathway starts from phenylacetate, p-hydroxyphenylacetate, and indoleacetate, precursors for phenylalanine, tyrosine, and tryptophan, respectively. The enzymes shown are DHQ, CM, PDT, PDH, AroAT, and IOR. Possible transcriptional regulation of DHQ is indicated by double dashed lines. Inhibition (−) or activation (+) of the PDT and PDH enzyme activities are indicated by single dashed lines.
In addition to the de novo pathway, Methanothermobacter marburgensis has been proposed to form the AroAAs from the corresponding aryl acids (56). In this pathway, the aryl acids are activated to the coenzyme A (CoA) thioesters indoleacetyl-CoA, phenylacetyl-CoA, and p-hydroxyphenylacetyl-CoA, which are then reductively carboxylated by indolepyruvate oxidoreductase (IOR) to indolepyruvate, phenylpyruvate or p-hydroxyphenylpyruvate, respectively (Fig. 1). These compounds are substrates for the AroAA aminotransferases (69). The presence of this pathway in M. marburgensis is supported by characterization of the key enzyme IOR and incorporation of radiolabeled phenylacetate into phenylalanine (56). In contrast, a similar pathway in the hyperthermophilic peptide-fermenting archaeon P. furiosus is involved in AroAA degradation to aryl acids (36). Thus, the presence of the enzymes of this pathway is not proof of its physiological function.
The present work provides genetic and biochemical evidence for both of these pathways for AroAA biosynthesis in M. maripaludis.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and culture conditions.
The bacterial strains and plasmids used in this work are listed in Table 1. Escherichia coli was grown in Luria-Bertani medium with ampicillin (100 μg/ml) when needed. M. maripaludis was grown with 276 kPa of H2-CO2 gas (80:20 [vol/vol]) at 37°C in the mineral medium McN, McNA (McN plus 10 mM sodium acetate), or McC (McNA plus 0.2% [wt/vol] Casamino Acids and 0.2% [wt/vol] yeast extract) as described previously (65). Puromycin (2.5 μg/ml) or neomycin (500 μg/ml) was added when needed. Under these growth conditions, logarithmic growth is observed only at low cell densities because the rate of transfer of H2 gas to the liquid medium limits the rate of growth at even moderate cell densities. For this reason, only linear growth curves are presented. For preparation of the cell extracts, M. maripaludis was grown in bottles with 100 ml of McNA medium and 138 kPa of H2-CO2 gas (80:20 [vol/vol]) at 37°C. The cells were harvested, and the cell extract was prepared as previously described (33, 47).
TABLE 1.
Bacterial strains and plasmids
| Bacterial strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| M. maripaludis | ||
| S2 | Wild type | 65 |
| S83 | ΔaroD::pac | This work |
| S87 | ΔaroD::pac | This work |
| S122 | ΔiorA2::pac | This work |
| S151 | ΔiorA2::pac/pMEV2-iorAB2 | This work |
| S153 | ΔiorA2::pac/pMEV2-iorAB2 | This work |
| S155 | ΔiorA2::pac/pMEV2-iorAB2 | This work |
| E. coli DH5α | F− (φ80dlacZΔM15) recA1 endA1 gyrA96 thi-1 hsdR17 (rK− mK+) supE44 Δ(lacZYA-argF) U169 | 19 |
| Plasmids | ||
| pIJA03 | Purr methanogen integration vector | 54 |
| pIJA03-aroD | pIJA03 with the upstream and downstream regions of the aroD gene | This work |
| pIJA03-iorA2 | pIJA03 with the upstream and downstream regions of the iorA2 gene | |
| pMEV2 | Neomycin shuttle vector | 32 |
| pMEV2-iorAB2 | pMEV2 with iorAB2 genes | This work |
Construction of mutants.
The ΔaroD::pac and ΔiorA2::pac mutants were made by transformation with suicide vectors based upon pIJA03. Upon transformation and selection, these vectors exchange the internal portion of the aroD or iorA2 gene with the pac cassette, which encodes puromycin resistance in methanococci (15, 54). In pIJA03, the pac cassette is flanked by two multicloning regions that allow directed cloning of genomic DNA. The upstream and downstream regions of the aroD gene were PCR amplified from genomic DNA using the primers U1-U2 and D1-D2, respectively. pIJA03-aroD was constructed in E. coli DH5α (19) by cloning the U1-U2 PCR products into the BglII-XbaI sites and the D1-D2 products into the KpnI and NheI sites of pIJA03. Similarly, the plasmid pIJA03-iorA2 was constructed using the primers U3-U4 and D3-D4. Upon insertion into the genome, this plasmid would be expected to delete the 3′ end of iorA2, as well as the first codon of iorB2. The pIJA03-aroD or pIJA03-iorA2 plasmid was transformed into M. maripaludis S2 by the polyethylene glycol method (59). After transformation, cultures were plated on McC medium plus puromycin, random puromycin-resistant colonies were restreaked, and representative isolates were picked into tubes containing broth of the same composition. After growth, 2.5 ml of the 5-ml culture was used for determination of the genotype and phenotype. The remaining culture was used for preparation of frozen stocks. First, the culture tube was transferred to an anaerobic glove box, and the remaining culture was centrifuged and resuspended in 0.4 ml of 30% glycerol plus McC medium (58). The suspension was distributed into 0.2-ml fractions, sealed in airtight cryogenic vials (Corning), and stored at −70°C.
The plasmid pMEV2-iorAB2 for complementation of the IOR mutant was constructed by cloning the iorAB2 genes into the NsiI and XbaI sites of the methanococcal expression vector pMEV2 (32). The iorAB2 genes were PCR amplified from genomic DNA using the primers E1 and E2. The plasmid pMEV2-iorAB2 was constructed in E. coli DH5α (19) and transformed into M. maripaludis S122 (59). Transformants were screened on McC plates containing neomycin. Isolated colonies were restreaked and stored as described above. All subcultures of this strain also contained neomycin.
The sequences of the primers used in this work will be provided upon request.
Southern hybridization.
Southern hybridizations were performed using the DIG High Prime DNA Labeling and Detection Starter Kit I (Roche, Mannheim, Germany). The probe for aroD was made by PCR amplification with primers U1 and U2. The probe for the iorA2 gene was made by PCR amplification using primers U3 and U4.
Enzymatic assays.
The activity of 3-dehydroquinate dehydratase (DHQ) was assayed by monitoring the formation of 3-dehydroshikimate at 234 nm (ɛ = 12 × 103 M−1 cm−1) at 37°C. The standard assay mixture (1 ml) contained potassium-PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] buffer (50 mM; pH 7.0) and 10 mM 3-dehydroquinate (6). Dehydroquinate was prepared by the oxidation of quinic acid (Sigma, St. Louis, Mo.) with nitric acid and separation of the products by ion-exchange chromatography on Dowex 1 (Sigma) resin (17). The fractions that contained dehydroquinate were collected, and the dehydroquinate was precipitated as previously described (22).
Pyruvate oxidoreductase (POR) activity was assayed anaerobically as pyruvate- and CoA-dependent methyl viologen reduction, as previously reported (33). IOR activity was assayed similarly, except that pyruvate was replaced with phenylpyruvate (1 mM), indolepyruvate (0.5 mM), or p-hydroxyphenylpyruvate (1 mM) as the substrate of the reaction. Stock solutions of the last two substrates were prepared in ethanol. An ethanol-only control had no activity.
CM activity was assayed by measuring the transformation of chorismate to prephenate after the chemical conversion of prephenate to phenylpyruvate in acid (1). Unless otherwise stated, the concentration of chorismate was 1 mM. PDT was assayed by measuring the conversion of prephenate to phenylpyruvate as previously described (16), except that the enzymatic reaction was performed for 10 min with 0.5 mM prephenate. For the kinetics experiments for PDT, the concentrations of KCl, phenylalanine, and tyrosine were 725, 0.1, and 0.1 mM, respectively. Prephenate dehydrogenase (PDH) activity was assayed by following the formation of NADH upon the oxidative decarboxylation of prephenate (10). The concentration of prephenate was 1 mM.
All specific activities are given in milliunits per milligram (or nanomoles per minute per milligram of protein). Protein concentrations were determined using the bicinchoninic acid protein assay kit (Pierce, Rockford, Ill.) after incubation at 90°C in 0.1 M NaOH for 30 min.
13C labeling and isolation of amino acids from M. maripaludis proteins.
The AroAA auxotroph S87 was grown in 6.5 liters of modified McNA medium in H2-CO2 gas as previously described (46). In addition to the usual components, the McNA medium contained 0.1 mM [1-13C]phenylacetate and p-hydroxyphenylacetate and 0.02 mM indoleacetate. The aryl acids were flushed with N2 gas and sterilized by filtration. The fermentor was inoculated with 400 ml of culture grown in the same medium with unlabeled phenylacetate. The cells were harvested by centrifugation as previously described (46). The proteins from the cell pellets were extracted as described previously (64), except that the cell paste (8.2 g [wet weight]) was suspended in 326 ml of ice-cold 5% (wt/vol) trichloroacetic acid and the protein pellet was washed twice with acetone before being dried in a vacuum desiccator. For acid hydrolysis, the dried proteins were suspended in 6 N HCl (40 μl/mg of dry protein) in an acid-cleaned, anaerobic glass culture tube (51). The tube was sealed with a butyl rubber stopper and aluminum seal, and the solution was frozen in an ethanol-dry ice bath. The tube was flushed with N2 gas for 15 min and incubated in a sand bath at 110°C for 20 h. Then, the acid was diluted with an equal volume of water. Rotary evaporation was used for removing the acid, and the amino acid mixture was concentrated to 0.5 ml by lyophilization. The concentrated mixture of amino acids was then diluted to 1.5 ml in water, and multivalent cations were removed during passage through a 3-ml column of iminodiacetic acid chelating resin (Sigma). The column was treated with 500 ml of 1 N HCl and 500 ml of water before the sample was loaded. After the sample was loaded, the column was eluted with distilled water. The amino acids were detected by spotting drops from the column on filter paper. After the drops dried, a drop of ninhydrin reagent (100 ml of butanol, 3 ml of acetic acid, and 0.3 g of ninhydrin) was added and dried. The formation of an orange color indicated the presence of amino acids. The amino acids eluted within 13 ml and were taken to dryness by lyophilization.
NMR experiment.
1H-decoupled 13C nuclear magnetic resonance (NMR) data were acquired at 20°C on a 400-MHz spectrometer (399.8 MHz; 1H), while 1H detected spectra were recorded on a 500-MHz spectrometer (499.8 MHz; 1H). 1H chemical shifts at 20°C were referenced to 2,2-dimethyl-2-silapentane-5-sulfonic acid via the HDO resonance frequency at 4.76 ppm, and 13C chemical shifts were referenced to tetramethylsilane at 0.0 ppm. All 2D phase-sensitive gradient-enhanced and 1D version 1H-13C HMQC-clean-TOCSY (HMQC, heteronuclear multiquantum coherence; TOCSY, total correlation spectroscopy) (31) experiments were acquired with GARP decoupling (45) during acquisition. For all 2D experiments, quadrature detection in the indirectly observed dimensions was obtained using the time-proportional phase increment method (37). The 2D data were acquired with an acquisition time of 100 ms, with four scans for each of 256 free induction decays. The 1D HMQC-clean-TOCSY data were recorded with 64 scans.
Phylogenetic analysis.
The genes encoding the α subunits of the two IORs from M. maripaludis were used for a BLASTP search of the National Center for Biotechnology Information (National Institutes of Health) database. The amino acid sequences were aligned using Clustal X (57). This alignment was manually edited prior to construction of the phylogenetic trees with PHYLIP (13). Evolutionary distances were determined with PROTDIST, and the neighbor-joining and Fitch-Margoliash dendrograms were generated with NEIGHBOR and FITCH, respectively. Parsimony analysis was performed with PROTPARS. The SEQBOOT program was used to calculate bootstrap values based upon 100 replicate trees.
RESULTS
Biosynthesis of AroAAs by the de novo pathway.
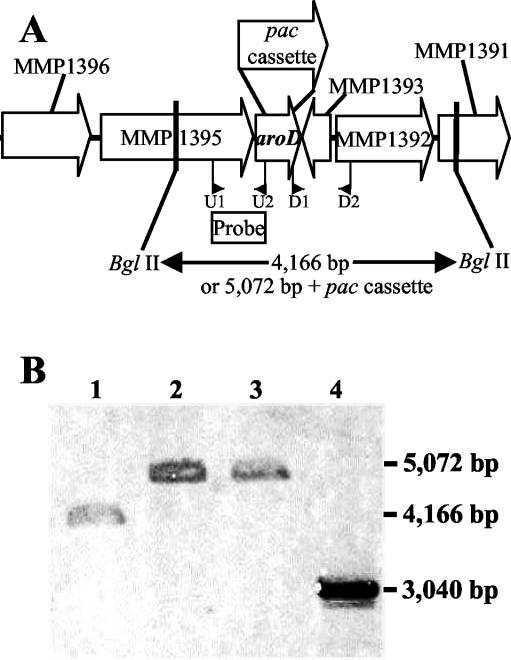
The genome of M. maripaludis possesses homologs for five of the seven genes of the de novo pathway of AroAA biosynthesis up to chorismate (J. Leigh, personal communication). The ORF MMP1394 is the homolog to aroD, which encodes the third enzyme in the pathway, DHQ. This is the first step for which a homolog has been identified. MMP1394 shows ∼30% amino acid identity to the bacterial type I DHQs. Because there is no direct biochemical evidence for the identity of aroD in the archaea, a deletion strain of MMP1394 was constructed to confirm its role in AroAA biosynthesis (Fig. 2A). The genotypes of the resulting mutants, S83 and S87, were confirmed by Southern blotting. The replacement of an internal portion of MMP1394 with the pac cassette resulted in an increase in size of the BglII fragment from 4.2 kb in the wild type to 5.1 kb in the mutants (Fig. 2B). The mutants were auxotrophic for all three AroAAs (Fig. 3A and data not shown). In cell extracts of the mutant S87, the DHQ activity was below the limit of detection, or <0.5 mU mg−1. In contrast, the specific activity in the wild-type strain was 6.0 ± 0.2 mU mg−1. These results confirmed the identity of MMP1394 as aroD and its role in the pathway of AroAA biosynthesis.
FIG. 2.
Construction of the ΔaroD::pac mutation. (A) M. maripaludis aroD (MMP1394) gene region. The ORFs MMP1396, MMP1395 and MMP1391 were annotated as aminotransferase, DEAD/DEAH box helicase and aspartate-semialdehyde dehydrogenase, respectively. The ORFs MMP1393 and MMP1392 were annotated as hypothetical proteins (J. Leigh, personal communication). The locations of the primers U1, U2, D1, and D2 used to clone the upstream and downstream regions flanking MMP1394 are shown. (B) Confirmation of the genotypes of the wild-type S2 and mutants S83 and S87 by Southern hybridization. The genomic DNA (3.2 μg) was digested with BglII prior to hybridization with the probe indicated in panel A. Lanes 1, 2, and 3, genomic DNAs of S2, S83, and S87, respectively, digested with BglII. Lane 4, pJA03-aroD digested with PvuII and NheI as a positive control.
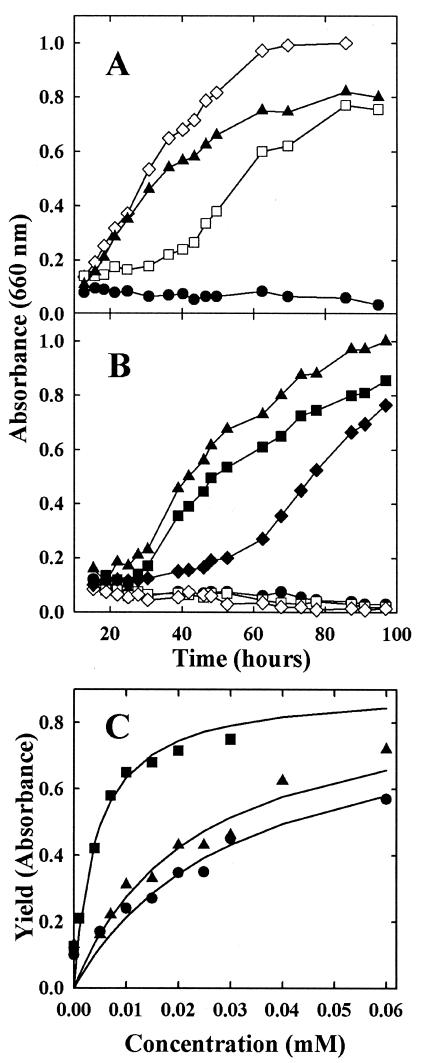
FIG. 3.
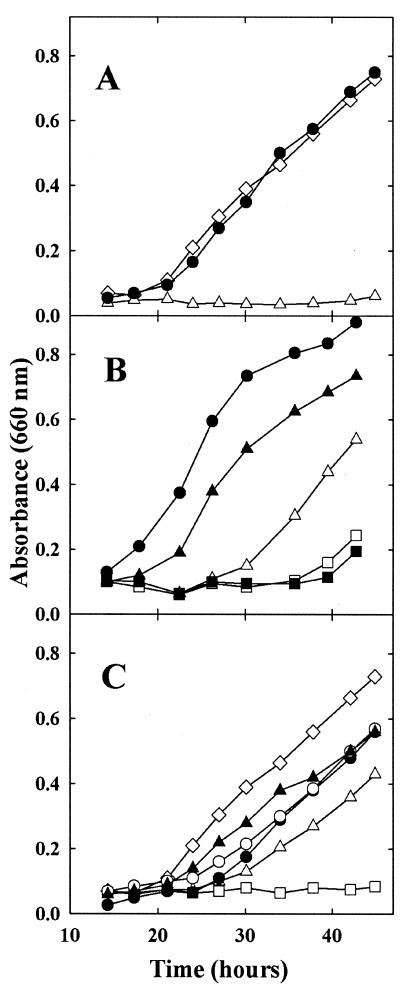
Growth requirement of the auxotroph S87 for AroAAs. The McNA medium contained 1 mM AroAAs or aryl acids, except when otherwise indicated. (A) Growth in the presence of AroAAs or aryl acids. The inoculum was 105 cells. Shown are growth of the wild-type S2 with or without acids (◊) and growth of S87 with all three aryl acids (▴), with all three AroAAs (□), and without any addition (•). (B) Assimilation of phenylalanine and tyrosine limit the growth rate of the mutant S87. The inoculum was 107 cells washed in McNA medium. Shown are growth of S87 without any addition (•); with all three aryl acids (▴); with phenylalanine, indoleacetate, and p-hydroxyphenylacetate (▪); with tyrosine, indoleacetate, and phenylacetate (⧫); with indoleacetate and p-hydroxyphenylacetate alone (□); and with indoleacetate and phenylacetate alone (◊). (C) Growth yield of the mutant S87 with limiting concentrations of indoleacetate (▪), p-hydroxyphenyacetate (▴), and phenylacetate (•). The inoculum was 107 cells washed in McNA medium.
Biosynthesis of AroAAs from the aryl acids.
The three aryl acids, phenylacetate, p-hydroxyphenylacetate, and indoleacetate, fulfilled the AroAA requirement for growth of the aroD mutant S87 (Fig. 3A). These results suggested that M. maripaludis was also able to biosynthesize the AroAAs from the aryl acids using the IOR pathway.
Aryl acids are readily available in typical methanogenic environments, where they are formed from the AroAAs by anaerobic heterotrophs (3, 4). In contrast, strains of M. maripaludis poorly assimilate low concentrations of the AroAAs themselves, as well many other amino acids (66). The rapid growth of the auxotroph with aryl acids suggested that these acids might be assimilated at environmentally relevant concentrations (Fig. 3A). In fact, replacement of phenylacetate and p-hydroxyphenylacetate with phenylalanine and tyrosine, respectively, actually inhibited growth, confirming that the aryl acids were assimilated more readily than the amino acids (Fig. 3B). The aryl acids were assimilated quantitatively. For instance, the concentrations of indoleacetate, phenylacetate, and p-hydroxyphenylacetate required for growth of S87 were close to the values expected from the abundance of the AroAAs in proteins (Fig. 3C). The cellular yields for phenylacetate, p-hydroxyphenylacetate, and indoleacetate were 3.7, 4.2, and 23 g (dry weight) mmol−1, respectively. From the compositions of AroAAs in E. coli, the expected yields were very similar: 5.7, 7.6, and 19 g (dry weight) mmol−1 (39). These results demonstrated that low concentrations of aryl acids could fulfill the growth requirement for AroAAs.
In bacteria, chorismate is an intermediate in the biosynthesis of p-aminobenzoate, quinones, and the catechol siderophores (5). Although methanogens are not known to possess the last two compounds, p-aminobenzoate is required for methanopterin biosynthesis in Methanobrevibacter sp. (63). Although labeling of p-aminobenzoate was consistent with its formation from chorismate, this biosynthetic route was not demonstrated conclusively. Therefore, it was interesting that aryl acids alone were sufficient to support growth of the ΔaroD::pac mutant. To confirm that no other compounds were required for growth, this mutant was serially transferred five times in minimal medium with acetate and aryl acids, for a total dilution of ∼108-fold. After five transfers, the growth of the mutant closely resembled that of the wild type in this medium (data not shown). From this result, methanococci must have an alternative pathway of p-aminobenzoate biosynthesis during growth on aryl acids. It is also possible that chorismate is an intermediate during growth without these AroAA precursors.
In vivo incorporation of phenylacetate into phenylalanine.
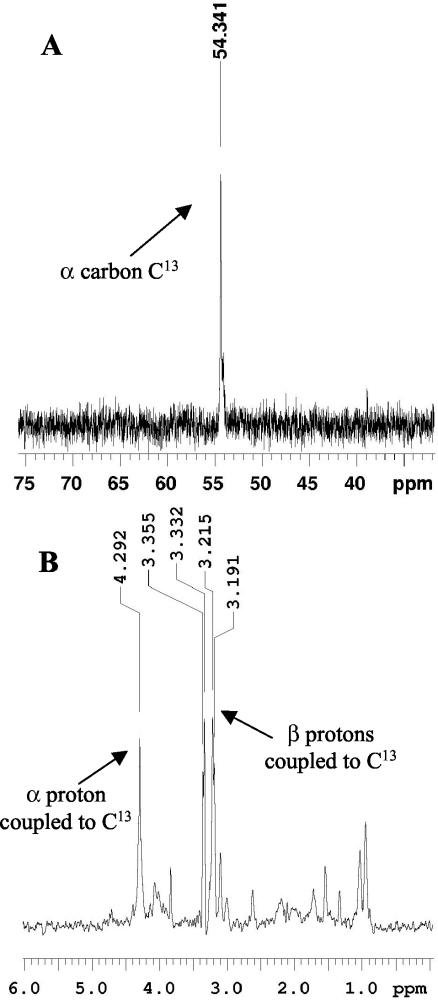
To confirm the incorporation of aryl acids into the AroAAs, the mutant S87 was cultured with [1-13C]phenylacetate, unlabeled p-hydroxyphenylacetate, and indoleacetate. After extraction of the cellular proteins and acid hydrolysis to produce a mixture of amino acids, only one amino acid carbon was specifically enriched by the 13C label (Fig. 4A). This carbon was identified as the Cα of phenylalanine based in part upon its 13C chemical shift in a 1H-decoupled 13C experiment. However, this criterion alone was not sufficient, and the identification was confirmed by the 1Hα and 1Hβ resonances obtained in 1H-13C HMQC-TOCSY experiments of the mixture and those of amino acid standards prepared at the same pH. The observed 1H chemical shifts in the mixture were 4.29 ppm for the 1Hα proton and 3.34 and 3.20 ppm for the 1Hβ protons (Fig. 4B). These matched the resonances of the phenylalanine standard observed at 4.25 ppm for the 1Hα proton and 3.33 and 3.18 ppm for the 1Hβ protons. In contrast, the 1H chemical shifts of other candidate standards had significant deviations from that observed for the mixture. For example, tryptophan had the closest chemical shifts and resonances for the 1Hα proton at 4.24 ppm and the 1Hβ protons at 3.49 and 3.37 ppm.
FIG. 4.
NMR spectra of the mixture of amino acids produced by acid hydrolysis of labeled proteins. The amino acids (10 mg) were isolated by Robert's method from S87 cells grown in the presence of 0.1 mM [1-13C]phenylacetate. (A) 13C spectrum of the mixture of amino acids produced by acid hydrolysis of the labeled proteins. (B) Proton spectrum of the 13C-labeled amino acids demonstrating coupling to 13C.
IOR from M. maripaludis.
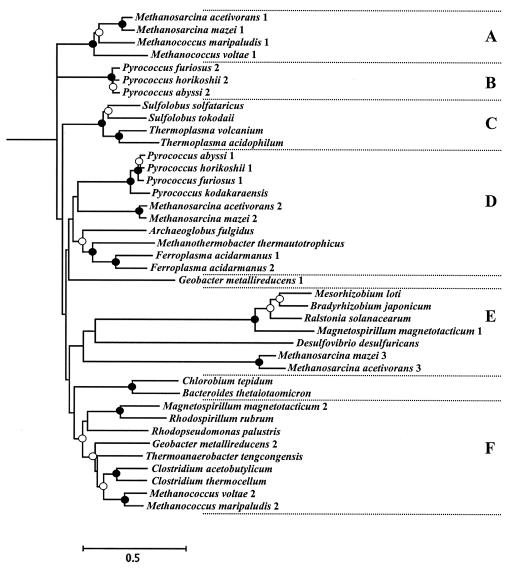
The IORs from M. marburgensis and Pyrococcus spp. have been purified and found to contain two subunits (35, 49, 50, 56). Two homologs of the IORs are present in the genomes of M. maripaludis (J. Leigh, personal communication), each one encoded by adjacent ORFs: MMP0316 and MMP0315, and MMP0713 and MMP0714 for the α and β subunits, respectively. A phylogenetic tree of the α subunits from the prokaryotes grouped the IOR homologs into six clades (Fig. 5). While all six clades were observed by the Fitch-Margoliash, neighbor-joining, and parsimony algorithms, only some were strongly supported by bootstrap analyses. Moreover, it was not possible to confidently assign the genes from Chlorobium tepidum, Bacteroides thetaiotaomicron, and homolog 1 of Geobacter metallireducens to a clade or to determine the deep branching order among the clades (Fig. 5). Four clades (A to D) contained only archaeal genes, and two clades (E and F) contained bacterial and archaeal sequences. The biochemically characterized IORs from Pyrococcus spp. and M. marburgensis, which is closely related to Methanothermobacter thermoautotrophicus (56), were all in clade D, which included only archaeal genes but not the methanococcal homologs. Instead the M. maripaludis homologs were found in clades A and F, along with the homologs from the closely related mesophile Methanococcus voltae (W. B. Whitman, R. A. Feldman, and R. Overbeek, unpublished observation). Clade A was composed entirely of archaeal genes. In addition to the methanococcal homologs, it also included homolog 1 of Methanosarcina spp. In contrast, the methanococcal homologs were the only archaeal genes in clade F, which also contained genes from a variety of facultative and strictly anaerobic bacteria. Because of the low similarity of the methanococcal homologs to the biochemically characterized enzymes and the broad distribution among different types of prokaryotes, it was not possible to infer the physiological properties of the IOR homologs from this phylogeny.
FIG. 5.
Phylogeny of the iorA genes that encode the α subunit of IOR. The phylogenetic tree was constructed by using the PHYLIP package based upon an alignment of conserved positions using the Fitch-Margoliash algorithm. Similar phylogenetic trees were generated by neighbor-joining and parsimony algorithms (data not shown). The bootstrap values for all three algorithms were very close and are labeled in the tree by the symbols at the branch points: •, values of >90%; ○, values of >60%; unlabeled, values of ≤60%. The scale bar is 0.5 expected amino acid substitutions per site. The six clades found are labeled A to F. The accession numbers for protein sequences from the National Center for Biotechnology Information database (from top to bottom in the tree) are: AAM05134, AAM32330, CAF29872.1, AAR21228, AAL80969, NP_143041, NP_126757, C90374, BAB65740, BAB59718, CAC12141, Q9UZ57, O58495, AAL80657, BAA20528 [formerly named Pyrococcus sp KOD1(43)], NP_615972, NP_634117, O28783, AAB86318, ZP_00000830, ZP_00001160, ZP_00080126, NP_106112, BAC48676, CAD15531, ZP_00056522, ZP_00129724, AAM32484, AAM05385, NP_661020, AAO75537, ZP_00054515, ZP_00015617, ZP_00010808, ZP_00079304, NP_623747, NP_348620, ZP_00059944, AAR21230, and CAF30269.1.
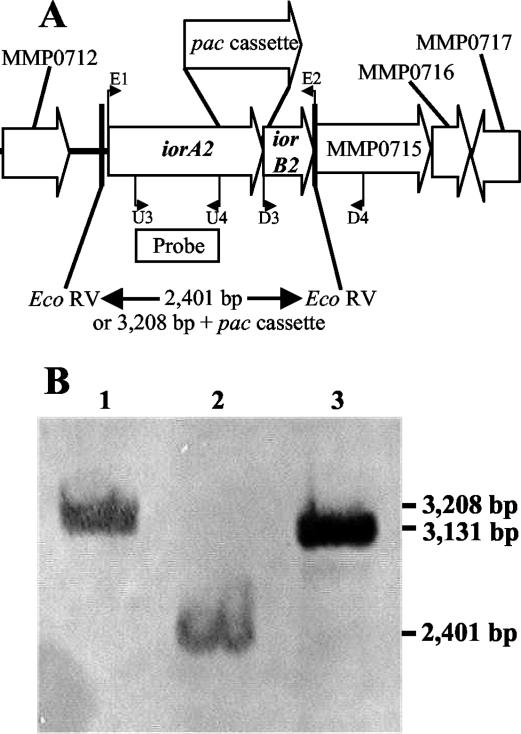
In order to further understand the role of the IORs in M. maripaludis, the iorA2 (MMP0713) gene was deleted by replacement with the pac cassette (Fig. 6A). The genotype of the resulting mutant, S122, was confirmed by Southern blotting. The replacement of the internal portion of MMP0713 with the pac cassette resulted in an increase in size of the EcoRV fragment from 2.4 kb in the wild type to 3.2 kb in the mutant S122 (Fig. 6B). The IOR specific activity in the mutant was greatly reduced, from 162 mU mg−1 in the wild type to 39 mU mg−1 with phenylpyruvate as the substrate (Table 2). The genes iorAB2 were then cloned next to a strong promoter in the methanococcal expression vector pMEV2 (32), and the vector was transformed into the mutant S122 to produce strains S151, S153, and S155. The complemented strains all possessed elevated levels of IOR activity, confirming that these genes encoded IOR (Table 2 and data not shown). Interestingly, the specific activity for POR in these complemented strains was reduced, from 390 mU mg−1 in the wild type and strain S122 to 180 to 220 mU mg−1 in the complementation mutants. Presumably, biosynthesis of elevated levels of IOR depleted the coenzymes needed for biosynthesis of normal levels of POR. Although the methanococcal IORs were not purified, the properties of the mutants suggested that the enzymes had different specificities for the aryl acids. Compared to the wild type, extracts of the ΔiorA2::pac mutant S122 possessed greatly reduced activity with phenylpyruvate and p-hydroxyphenylpyruvate (Table 2), suggesting that IOR2 preferentially utilized these substrates. Similarly, complemented strains possessed very high activities for phenylpyruvate and p-hydroxyphenylpyruvate compared to the activity with indolepyruvate (Table 2). Presumably, the residual activity in the mutant S122, which was nearly the same for all three substrates, represented IOR1. Thus, the substrate specificities of IOR1 and IOR2 appeared to differ.
FIG. 6.
Construction of ΔiorA2::pac mutation. (A) M. maripaludis iorA2 (MMP0713) gene region. The ORFs MMP0712, MMP715, MMP0716, and MMP0717 were annotated as solute-binding protein/glutamate receptor, coenzyme F390 synthetase II, acetohydroxyacid synthase small-subunit related, and hypothetical protein, respectively (J. Leigh, personal communication). The primers U3, U4, D3 and D4 were used to clone the flanking regions for construction of pIJA03-iorA. The primers E1 and E2 were used for cloning iorA2 and iorB2 during the construction of pMEV2-iorAB2. (B) Confirmation of the genotype by Southern hybridization with the wild-type S2 and the iorA2 mutant S122. The genomic DNA (3.3 μg) was digested with EcoRV prior to hybridization to the probe indicated in panel A. Lanes 1 and 2, S122 and S2 genomic DNAs, respectively; lane 3, pJA03-iorA2 DNA digested with PvuII and NheI as a positive control.
TABLE 2.
Indolepyruvate oxidoreductase specific activities of the wild-type S2 and mutant S122 and S155 strainsa
| Strain | Genotype | Sp act (mU mg−1) with:
|
||
|---|---|---|---|---|
| Phenylpyruvate | p-Hydroxyphenylpyruvate | Indole-3-pyruvate | ||
| S2 | Wild type | 162 ± 33 | 101 ± 31 | 43 ± 12 |
| S122 | ΔiorA2::pac | 39 ± 5 | 26 ± 7 | 25 ± 10 |
| S155b | ΔiorA2::pac/pMEV2-iorAB2 | 519 ± 44 | 381 ± 10 | 88 ± 2 |
Except as noted, specific activities were determined in triplicate from four independent cultures grown in McNA medium. The concentrations of the substrates phenylpyruvate and p-hydroxyphenylpyruvate were 1 mM. The concentration of the substrate indolepyruvate was 0.5 mM. Means and one standard deviation are reported.
Specific activities were determined in duplicate. The two additional complementation strains S151 and S153 gave results similar to these with S155.
Regulation of de novo AroAA biosynthesis by aryl acids.
The growth of the ΔiorA2::pac mutant S122 was indistinguishable from that of the wild type in medium with (McC) or without (McNA or McN) amino acids. However, the aryl acids phenylacetate and p-hydroxyphenylacetate, either alone or in combination, severely inhibited growth of the mutant in minimal medium (Fig. 7A and data not shown). In some experiments, growth of the ΔiorA2::pac mutant resumed after a long lag in the presence of phenylacetate or p-hydroxyphenylacetate (Fig. 7B). Growth in these cases was best explained by selection for revertants. Growth was seldom seen when very small inocula were used, suggesting that growth occurred due to selection for mutants arising spontaneously. Moreover, the aryl acids no longer inhibited the growth of these cultures upon subsequent transfers in the same medium (data not shown). The inhibition by the individual aryl acids suggested that phenylacetate and p-hydroxyphenylacetate inhibited the de novo pathway of AroAA biosynthesis. The inhibition by all three aryl acids together further indicated that the level of IOR activity remaining in the mutant was insufficient to provide AroAAs from the aryl acids.
FIG. 7.
Effects of aryl acids on growth of the ΔiorA2::pac mutant S122. The McNA medium contained 1 mM aryl acids or AroAAs as indicated. The inoculum was ∼2 × 105 cells. (A) Inhibition of growth of strain S122 by phenylacetate or p-hydroxyphenylacetate. ◊, wild-type S2 without any addition; •, S122 without any addition; ▵, S122 with phenylacetate alone, p-hydroxyphenylacetate alone, both phenylacetate and p-hydroxyphenylacetate, or all three aryl acids. (B) Restoration of growth by AroAAs. Shown are S122 without any addition (•), with phenylacetate (▵), with phenylacetate and phenylalanine (▴), with p-hydroxyphenylacetate (□), and with p-hydroxyphenylacetate and tyrosine (▪). (C) Complementation of the ΔiorA2::pac mutant with pMEV2-iorAB2. Shown are growth of wild-type S2 without any addition (◊), the complemented strain S155 without any addition (•), S155 with phenylacetate (▵), S155 with p-hydroxyphenylacetate (□), S155 with phenylacetate and p-hydroxyphenylacetate (○), and S155 with the three aryl acids (▴).
In support of these hypotheses, phenylalanine provided partial protection against inhibition by phenylacetate, indicating that limitation for AroAAs caused at least some of the growth inhibition (Fig. 7B). However, tyrosine failed to protect against inhibition by p-hydroxyphenylacetate. Tyrosine was assimilated poorly by M. maripaludis. For instance, tyrosine supported only poor growth of the ΔaroD::pac mutant S87 after a lag of >2 days (Fig. 3B). Therefore, the failure of tyrosine to protect against aryl acid inhibition probably reflected its poor uptake. In addition, p-hydroxyphenylacetate appeared to be a stronger inhibitor than phenylacetate (see below), so its effects would be more difficult to reverse upon the addition of the AroAAs. Because phenylacetate was less inhibitory, phenylalanine could provide partial protection, presumably by sparing the chorismate requirement for biosynthesis of the other AroAAs. In contrast, indoleacetate did not inhibit the growth of the ΔiorA2::pac mutant (data not shown). Presumably, it was either not a strong inhibitor of AroAA biosynthesis or the levels of IOR activity were sufficient for tryptophan biosynthesis.
In strain S155, where the chromosomal ΔiorA2::pac mutation was complemented by iorAB2 expression from a plasmid, the relative growth inhibition by the aryl acids was greatly reduced. First, p-hydroxyphenylacetate or phenylacetate alone was still inhibitory, as expected if they inhibited the de novo pathway. However, phenylacetate and p-hydroxyphenylacetate together or all three aryl acids together were no longer inhibitory, as expected if the IOR activity was now sufficient to provide AroAAs (Fig. 7C). This result confirmed the hypothesis that the failure of S122 to grow in the presence of the aryl acids was due to low levels of IOR activity and not due to a polar effect on other genes at this locus. Compared to the wild type, strain S155 also grew slowly in McNA medium without the addition of the aryl acids (Fig. 7C). Presumably, this result reflected the reduced levels of POR activity.
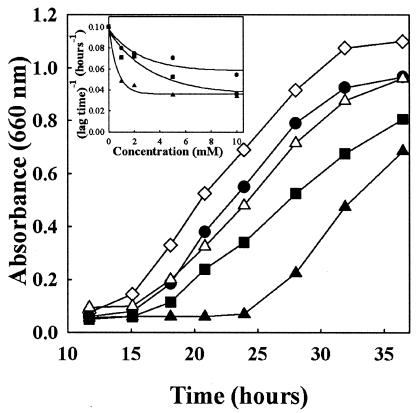
If the aryl acids inhibited de novo AroAA biosynthesis, they might also inhibit the growth of the wild type. While even low concentrations of p-hydroxyphenylacetate were inhibitory, higher concentrations of phenylacetate or indoleacetate were required (Fig. 8). In contrast to the mutant S122, the aryl acids were inhibitory only when present individually, and combinations did not inhibit. Presumably, when the three aryl acids were present together, the AroAAs were formed from the aryl pathway and the requirement for the de novo pathway was spared. The absence of inhibition by all three aryl acids strongly suggested that inhibition by aryl acids was due to an effect on AroAA biosynthesis. However, because tyrosine was assimilated poorly, it was not possible to demonstrate protection by the AroAAs themselves.
FIG. 8.
Effects of aryl acids on growth of wild type S2. The McNA medium contained 1 mM aryl acids where indicated. The inoculum was ∼2 × 105 cells. Shown is the S2 strain growing without any addition (◊), with phenylacetate alone (•), with indoleacetate alone (▪), with p-hydroxyphenylacetate alone (▴), and with the three aryl acids (▵). The inset shows the growth lag of strain S2 during growth with increasing concentrations of phenylacetate (•), indoleacetate (▪), and p-hydroxyphenylacetate (▴).
Effect of aryl acids on enzymes of the de novo pathway.
To identify the site of action of the aryl acids, the activities for some of the key steps were examined in cell extracts. In many prokaryotes and yeasts, the initial reaction, DAHP synthase, is a key regulatory step (18, 30, 40, 41). However, in methanococci, the initial step of the de novo pathway is not known (67), and it was not possible to examine that reaction. For the enzymes of the de novo pathway tested, growth in the presence of the aryl acids had only small effects on the specific activities in cell extracts. DHQ is the first enzyme in the pathway that is known. Its specific activity in wild-type cells grown with the aryl acids was one-third (2.3 ± 0.1 mU mg−1) of the level in cells grown without additions (6.0 ± 0.2 mU mg−1). For PDH, the specific activity of cells grown with the aryl acids (23 ± 1.2 mU mg−1) was nearly the same as the level in cells grown without additions (18 ± 2.1 mU mg−1). For CM and PDT, the specific activities were the same (see below). Therefore, the presence of aryl acids did not have a large affect on the expression of these enzymes.
The effect of phenylacetate and p-hydroxyphenylacetate on the activities of the phenylalanine or tyrosine biosynthetic enzymes from chorismate were also examined. CM is the first common enzyme for the biosynthesis of both phenylalanine and tyrosine (Fig. 1). The CM from M. jannaschii has been categorized as a monofunctional AroQ (7, 34). The aroQ gene of many organisms is often fused with either other AroAA biosynthetic genes or a regulatory domain (7). M. maripaludis possesses a homolog of the M. jannaschii CM (encoded by aroQ), MMP0578. In cell extracts, the CM specific activity was affected very little by the AroAAs, phenylacetate, or p-hydroxyphenylacetate, which was expected because a regulatory domain was not present in the open reading frame (ORF) (Table 3). Thus, CM was unlikely to be a major site of inhibition by the aryl acids.
TABLE 3.
CM, PDT, and PDH specific activities of the wild-type strain S2
| Additiona | Sp actb (mU mg−1) of:
|
||
|---|---|---|---|
| CM | PDT | PDH | |
| None | 1.2 | 1.7 | 17.8 |
| Phenylalanine | 1.9 | 0.5 | 15.1 |
| Tyrosine | 1.5 | 3.8 | 4.4 |
| Phenylacetate | 1.1 | 1.6 | 18.2 |
| p-Hydroxyphenylacetate | 1.5 | 1.7 | 7.6 |
| p-Hydroxyphenylpyruvate | NTc | NTc | 5.3 |
The concentrations of the potential effectors were 5 mM for CM and 0.5 mM for PDT and PDH.
Specific activities are the averages of duplicate assays. All duplicates were within 10% of the mean value.
NT, not tested.
PDT is the first enzyme in the phenylalanine branch (Fig. 1). Like the PDT from H. vallismortis (27), the activity of PDT from wild-type M. maripaludis was inhibited ∼70% by phenylalanine and activated ∼2-fold by tyrosine (Table 3). In one set of experiments, the Vmax and Km for prephenate were 5.3 mU min−1 and 1.2 mM, respectively. The apparent Vmax and Km for prephenate in the presence of the inhibitor phenylalanine at 0.1 mM were 2.8 mU min−1 and 0.96 mM, respectively, and the Vmax and Km for prephenate in the presence of the activator tyrosine at 0.1 mM were 6.0 mU min−1 and 0.63 mM, respectively. Thus, phenylalanine affected mostly the Vmax, while tyrosine affected mostly the Km for prephenate. Methanococci contain high intracellular concentrations of potassium, which is an activator for some biosynthetic enzymes (70). However, PDT activity from M. maripaludis was inhibited by KCl, as was previously found for PDT activity from E. coli (16). In this series of experiments, the Vmax and Km for prephenate with physiological concentrations of KCl of 0.725 M (25) were 7.8 mU min−1 and 3.5 mM, respectively. In the absence of KCl, the Vmax and Km for prephenate were 6.8 mU min−1 and 1.1 mM, respectively, in this experiment. Thus, KCl largely affected the Km for prephenate. Phenylacetate and p-hydroxyphenylacetate had no effect on the activity of PDT from M. maripaludis, even when the assay contained well below the Km concentration of prephenate (Table 3). Therefore, it is unlikely that this enzyme was the site of inhibition by the aryl acids.
Finally, PDH, the first committed enzyme of tyrosine biosynthesis, was tested. PDH activity from wild-type M. maripaludis was inhibited by ∼70% by either tyrosine or p-hydroxyphenylpyruvate and 55% by p-hydroxyphenylacetate (Table 3). Phenylalanine had a small effect, and phenylacetate had no effect on the PDH activity. This inhibition pattern was consistent with its role in tyrosine biosynthesis, and it would not explain the growth inhibition by aryl acids.
DISCUSSION
Evidence has been presented for two pathways of AroAA biosynthesis in M. maripaludis. The presence of the de novo pathway from dehydroquinate is supported by construction of an AroAA auxotroph via deletion of the aroD homolog in the genome. Moreover, activities of four enzymes in this pathway were demonstrated directly in cell extracts. Evidence for the IOR pathway from aryl acids includes growth experiments where the aryl acids fulfilled the requirement for AroAAs by the auxotroph and the specific incorporation of the isotopically labeled aryl acid phenylacetate into phenylalanine. The role of IOR2 in this pathway is further supported by the apparent inability of the deletion mutant S122 to incorporate aryl acids.
Anaerobic heterotrophs ferment AroAAs to the aryl acids to provide energy under starvation conditions (4, 20, 21). Thus, the aryl acids may be more abundant in anaerobic environments than the amino acids themselves. Consistent with this idea is the observation that methanococci utilized the aryl acids more readily than the amino acids themselves. Moreover, the aryl acids down regulated the de novo biosynthetic pathway sufficiently to inhibit growth in the ΔiorA2::pac mutant. Because the AroAAs were taken up poorly, it was not possible to reverse this inhibition completely by the addition of the amino acids. Thus, it might be that chorismate was a precursor for additional cellular components in addition to the AroAAs. The inability to demonstrate an additional nutritional requirement following extensive subculture of the aroD::pac mutant with only the aryl acids does not support this hypothesis. In addition, the individual aryl acids phenylacetate and p-hydroxyphenylacetate inhibited the growth of the wild-type S2, the ΔiorA2::pac mutant S122, and the iorAB2 complementation strain S155. However, when the aryl acids were provided together, only the ΔiorA2::pac mutant was inhibited. These results suggested that AroAA biosynthesis, which is the product of the IOR pathway, was the major site of aryl acid inhibition. These results further imply that the aryl acids are physiologically relevant precursors for the AroAAs.
Although it was not possible to identify the major site of regulation by aryl acids, some regulatory features of the de novo pathway were elucidated. These features are summarized in the working model proposed in Fig. 1. Growth in the presence of the aryl acids lowered the expression of DHQ but not the levels of CM, PDH, and PDT activity. Thus, regulation of levels of expression appeared to be a relatively minor factor in the regulation of this pathway in methanococci. Similarly, the presence of branched-chain amino acids had only small effects on the specific activities of their biosynthetic enzymes (W. L. Gardner and W. B. Whitman, unpublished data).
In contrast, feedback inhibition in the methanococcal enzymes closely resembles that found in other prokaryotes. Like the enzyme from the halophilic archaeon H. vallismortis, the M. maripaludis PDT activity is inhibited by phenylalanine and activated by tyrosine (27). Additionally the H. vallismortis PDT is inhibited by tryptophan and activated by methionine, leucine, and isoleucine. This type of interpathway regulation was discovered in Bacillus subtilis and named metabolic interlock (26). Feedback inhibition of PDT by phenylalanine is highly conserved in gram-negative and gram-positive bacteria and yeast (16, 26, 30). In E. coli, the bifunctional P protein has been mapped, and the PDT, CM, and regulatory domains have been clearly identified (72). Two highly conserved motifs in the regulatory domain that are involved in the binding of phenylalanine are also present in the M. maripaludis PDT sequence (reference 42 and data not shown). These observations agree with the proposal for an ancient origin of PDT regulation (27).
The feedback inhibition of PDH by tyrosine is also highly conserved among gram-positive and gram-negative bacteria (2, 8, 23). The B. subtilis PDH is also inhibited by phenylalanine, tryptophan, and p-hydroxyphenylpyruvate (8). The M. maripaludis PDH is inhibited by tyrosine, p-hydroxyphenylpyruvate, and p-hydroxyphenylacetate, connecting it to the aryl acid pathway. However, the magnitude of the inhibition by p-hydroxyphenylacetate is not sufficient to explain the growth inhibition by this aryl acid. Presumably, the aryl acids have other sites of inhibition in the de novo pathway. In any case, feedback inhibition was also observed in the methanococcal acetohydroxyacid synthase, the first enzyme in branched-chain amino acid biosynthesis (68, 71), suggesting that this regulatory mechanism is widely conserved among the prokaryotes.
Acknowledgments
This work was supported by grant DE-FG02-01ER15262 from the Department of Energy.
We thank Andrew Leech for his recommendations on the preparation of dehydroquinate and Robert S. Phillips and John A. Leigh for helpful discussions.
REFERENCES
- 1.Ahmad, S., and R. A. Jensen. 1988. Phylogenetic distribution of components of the overflow pathway to l-phenylalanine within the enteric lineage of bacteria. Curr. Microbiol. 16:295-302. [Google Scholar]
- 2.Ahmad, S., and R. A. Jensen. 1988. The phylogenetic origin of the bifunctional tyrosine-pathway protein in the enteric lineage of bacteria. Mol. Biol. Evol. 5:282-297. [DOI] [PubMed] [Google Scholar]
- 3.Bader, J., P. Rauschenbach, and H. Simon. 1982. On a hitherto unknown fermentation path of several amino acids by proteolytic clostridia. FEBS Lett. 140:67-72. [DOI] [PubMed] [Google Scholar]
- 4.Barker, H. A. 1981. Amino acid degradation by anaerobic bacteria. Annu. Rev. Biochem. 50:23-40. [DOI] [PubMed] [Google Scholar]
- 5.Bentley, R. 1990. The shikimate pathway—a metabolic tree with many branches. Crit. Rev. Biochem. Mol. Biol. 25:307-384. [DOI] [PubMed] [Google Scholar]
- 6.Bottomley, J. R., C. L. Clayton, P. A. Chalk, and C. Kleanthous. 1996. Cloning, sequencing, expression, purification and preliminary characterization of a type II dehydroquinase from Helicobacter pylori. Biochem. J. 319:559-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calhoun, D. H., C. A. Bonner, W. Gu, G. Xie, and R. A. Jensen. 27. July 2001, posting date. The emerging periplasm-localized subclass of AroQ chorismate mutases, exemplified by those from Salmonella typhimurium and Pseudomonas aeruginosa. Genome Biol. 2:RESEARCH0030. [Online.] http://genomebiology.com/2001/2/8/RESEARCH/0030. [DOI] [PMC free article] [PubMed]
- 8.Champney, W. S., and R. A. Jensen. 1970. The enzymology of prephenate dehydrogenase in Bacillus subtilis. J. Biol. Chem. 245:3763-3770. [PubMed] [Google Scholar]
- 9.Daugherty, M., V. Vonstein, R. Overbeek, and A. Osterman. 2001. Archaeal shikimate kinase, a new member of the GHMP-kinase family. J. Bacteriol. 183:292-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davidson, B. E., and G. S. Hudson. 1987. Chorismate mutase-prephenate dehydrogenase from Escherichia coli. Methods Enzymol. 142:440-450. [DOI] [PubMed] [Google Scholar]
- 11.Ekiel, I., K. F. Jarrell, and G. D. Sprott. 1985. Amino acid biosynthesis and sodium-dependent transport in Methanococcus voltae, as revealed by 13C NMR. Eur. J. Biochem. 149:437-444. [DOI] [PubMed] [Google Scholar]
- 12.Ekiel, I., I. C. Smith, and G. D. Sprott. 1983. Biosynthetic pathways in Methanospirillum hungatei as determined by 13C nuclear magnetic resonance. J. Bacteriol. 156:316-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Felsenstein, J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 14.Gast, D. A., U. Jenal, A. Wasserfallen, and T. Leisinger. 1994. Regulation of tryptophan biosynthesis in Methanobacterium thermoautotrophicum Marburg. J. Bacteriol. 176:4590-4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gernhardt, P., O. Possot, M. Foglino, L. Sibold, and A. Klein. 1990. Construction of an integration vector for use in the archaebacterium Methanococcus voltae and expression of a eubacterial resistance gene. Mol. Gen. Genet. 221:273-279. [DOI] [PubMed] [Google Scholar]
- 16.Gething, M. J., B. E. Davidson, and T. A. Dopheide. 1976. Chorismate mutase/prephenate dehydratase from Escherichia coli K12. 1. The effect of NaCl and its use in a new purification involving affinity chromatography on sepharosyl-phenylalanine. Eur. J. Biochem. 71:317-325. [DOI] [PubMed] [Google Scholar]
- 17.Grewe, R., and H. Haendler. 1966. 5-Dehydroquinic acid. Biochem. Prep. 11:21-26. [Google Scholar]
- 18.Hall, G. C., M. B. Flick, and R. A. Jensen. 1983. Regulation of the aromatic pathway in the cyanobacterium Synechococcus sp. strain Pcc6301 (Anacystis nidulans). J. Bacteriol. 153:423-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 20.Harwood, C. S., and E. Canale-Parola. 1981. Branched-chain amino acid fermentation by a marine spirochete: strategy for starvation survival. J. Bacteriol. 148:109-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harwood, C. S., and E. Canale-Parola. 1984. Ecology of spirochetes. Annu. Rev. Microbiol. 38:161-192. [DOI] [PubMed] [Google Scholar]
- 22.Haslam, E., R. D. Haworth, and P. F. Knowles. 1963. The preparation and identification of 5-dehydroquinic and 5-dehydroshikimic acids. Methods Enzymol. 6:498-501. [Google Scholar]
- 23.Hudson, G. S., V. Wong, and B. E. Davidson. 1984. Chorismate mutase/prephenate dehydrogenase from Escherichia coli K12: purification, characterization, and identification of a reactive cysteine. Biochemistry 23:6240-6249. [DOI] [PubMed] [Google Scholar]
- 24.Ivens, A., O. Mayans, H. Szadkowski, M. Wilmanns, and K. Kirschner. 2001. Purification, characterization and crystallization of thermostable anthranilate phosphoribosyltransferase from Sulfolobus solfataricus. Eur. J. Biochem. 268:2246-2252. [DOI] [PubMed] [Google Scholar]
- 25.Jarrell, K. F., and G. D. Sprott. 1984. Intracellular potassium concentration and relative acidity of the ribosomal proteins of methanogenic bacteria. Can. J. Microbiol. 30:663-668. [Google Scholar]
- 26.Jensen, R. A. 1969. Metabolic interlock. Regulatory interactions exerted between biochemical pathways. J. Biol. Chem. 244:2816-2823. [PubMed] [Google Scholar]
- 27.Jensen, R. A., T. A. d'Amato, and L. I. Hochstein. 1988. An extreme-halophile archaebacterium possesses the interlock type of prephenate dehydratase characteristic of the Gram-positive eubacteria. Arch. Microbiol. 148:365-371. [DOI] [PubMed] [Google Scholar]
- 28.Jones, W. J., M. J. B. Paynter, and R. Gupta. 1983. Characterization of Methanococcus maripaludis sp. nov., a new methanogen isolated from salt marsh sediment. Arch. Microbiol. 135:91-97. [Google Scholar]
- 29.Knochel, T., A. Ivens, G. Hester, A. Gonzalez, R. Bauerle, M. Wilmanns, K. Kirschner, and J. N. Jansonius. 1999. The crystal structure of anthranilate synthase from Sulfolobus solfataricus: functional implications. Proc. Natl. Acad. Sci. USA 96:9479-9484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koll, P., R. Bode, and D. Birnbaum. 1988. Regulation of metabolic branch points of aromatic amino acid biosynthesis in Pichia guilliermondii. J. Basic Microbiol. 28:619-627. [DOI] [PubMed] [Google Scholar]
- 31.Lerner, L., and A. Bax. 1986. Sensitivity-enhanced two-dimensional heteronuclear relayed coherence transfer NMR spectroscopy. J. Magn. Reson. 69:375-380. [Google Scholar]
- 32.Lin, W., and W. B. Whitman. 2004. The importance of porE and porF in the anabolic pyruvate oxidoreductase of Methanococcus maripaludis. Arch. Microbiol. 181:68-73. [DOI] [PubMed] [Google Scholar]
- 33.Lin, W. C., Y. L. Yang, and W. B. Whitman. 2003. The anabolic pyruvate oxidoreductase from Methanococcus maripaludis. Arch. Microbiol. 179:444-456. [DOI] [PubMed] [Google Scholar]
- 34.MacBeath, G., P. Kast, and D. Hilvert. 1998. A small, thermostable, and monofunctional chorismate mutase from the archaeon Methanococcus jannaschii. Biochemistry 37:10062-10073. [DOI] [PubMed] [Google Scholar]
- 35.Mai, X., and M. W. Adams. 1994. Indolepyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon Pyrococcus furiosus. A new enzyme involved in peptide fermentation. J. Biol. Chem. 269:16726-16732. [PubMed] [Google Scholar]
- 36.Mai, X., and M. W. Adams. 1996. Purification and characterization of two reversible and ADP-dependent acetyl coenzyme A synthetases from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 178:5897-5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marion, D., and K. Wuthrich. 1983. Application of phase sensitive two-dimensional correlated spectroscopy (COSY) for measurements of 1H-1H spin-spin coupling constants in proteins. Biochem. Biophys. Res. Commun. 113:967-974. [DOI] [PubMed] [Google Scholar]
- 38.Meile, L., R. Stettler, R. Banholzer, M. Kotik, and T. Leisinger. 1991. Tryptophan gene cluster of Methanobacterium thermoautotrophicum Marburg: molecular cloning and nucleotide sequence of a putative trpEGCFBAD operon. J. Bacteriol. 173:5017-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neidhardt, F. C., and H. E. Umbarger. 1996. Chemical composition of Escherichia coli, p. 13-16. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 40.Panina, E. M., A. G. Vitreschak, A. A. Mironov, and M. S. Gelfand. 2001. Regulation of aromatic amino acid biosynthesis in gamma-proteobacteria. J. Mol. Microbiol. Biotechnol. 3:529-543. [PubMed] [Google Scholar]
- 41.Panina, E. M., A. G. Vitreschak, A. A. Mironov, and M. S. Gelfand. 2003. Regulation of biosynthesis and transport of aromatic amino acids in low-GC Gram-positive bacteria. FEMS Microbiol. Lett. 222:211-220. [DOI] [PubMed] [Google Scholar]
- 42.Pohnert, G., S. Zhang, A. Husain, D. B. Wilson, and B. Ganem. 1999. Regulation of phenylalanine biosynthesis. Studies on the mechanism of phenylalanine binding and feedback inhibition in the Escherichia coli P-protein. Biochemistry 38:12212-12217. [DOI] [PubMed] [Google Scholar]
- 43.Rahman, R. N., S. Fujiwara, H. Nakamura, M. Takagi, and T. Imanaka. 1998. Ion pairs involved in maintaining a thermostable structure of glutamate dehydrogenase from a hyperthermophilic archaeon. Biochem. Biophys. Res. Commun. 248:920-926. [DOI] [PubMed] [Google Scholar]
- 44.Selkov, E., N. Maltsev, G. J. Olsen, R. Overbeek, and W. B. Whitman. 1997. A reconstruction of the metabolism of Methanococcus jannaschii from sequence data. Gene 197:GC11-GC26. [DOI] [PubMed] [Google Scholar]
- 45.Shaka, A. J., P. B. Barker, and R. Freeman. 1985. Computer-optimized decoupling scheme for wideband applications and low-level operation. J. Magn. Reson. 64:547-552. [Google Scholar]
- 46.Shieh, J., and W. B. Whitman. 1988. Autotrophic acetyl coenzyme A biosynthesis in Methanococcus maripaludis. J. Bacteriol. 170:3072-3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shieh, J. S., and W. B. Whitman. 1987. Pathway of acetate assimilation in autotrophic and heterotrophic methanococci. J. Bacteriol. 169:5327-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sibold, L., and M. Henriquet. 1988. Cloning of the trp genes from the archaebacterium Methanococcus voltae: nucleotide sequence of the trpBA genes. Mol. Gen. Genet. 214:439-450. [DOI] [PubMed] [Google Scholar]
- 49.Siddiqui, M. A., S. Fujiwara, and T. Imanaka. 1997. Indolepyruvate ferredoxin oxidoreductase from Pyrococcus sp. KOD1 possesses a mosaic structure showing features of various oxidoreductases. Mol. Gen. Genet. 254:433-439. [DOI] [PubMed] [Google Scholar]
- 50.Siddiqui, M. A., S. Fujiwara, M. Takagi, and T. Imanaka. 1998. In vitro heat effect on heterooligomeric subunit assembly of thermostable indolepyruvate ferredoxin oxidoreductase. FEBS Lett. 434:372-376. [DOI] [PubMed] [Google Scholar]
- 51.Simpson, P. G. 1993. Investigation of amino acid metabolism in Methanococcus voltae. Ph.D. thesis. University of Georgia, Athens.
- 52.Smith, D. R., L. A. Doucette-Stamm, C. Deloughery, H. Lee, J. Dubois, T. Aldredge, R. Bashirzadeh, D. Blakely, R. Cook, K. Gilbert, D. Harrison, L. Hoang, P. Keagle, W. Lumm, B. Pothier, D. Qiu, R. Spadafora, R. Vicaire, Y. Wang, J. Wierzbowski, R. Gibson, N. Jiwani, A. Caruso, D. Bush, J. N. Reeve, et al. 1997. Complete genome sequence of Methanobacterium thermoautotrophicum δH: functional analysis and comparative genomics. J. Bacteriol. 179:7135-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sprott, G. D., I. Ekiel, and G. B. Patell. 1993. Metabolic pathways in Methanococcus jannaschii and other methanogenic bacteria. Appl. Environ. Microbiol. 59:1092-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stathopoulos, C., W. Kim, T. Li, I. Anderson, B. Deutsch, S. Palioura, W. Whitman, and D. Soll. 2001. Cysteinyl-tRNA synthetase is not essential for viability of the archaeon Methanococcus maripaludis. Proc. Natl. Acad. Sci. USA 98:14292-14297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tang, X. F., S. Ezaki, H. Atomi, and T. Imanaka. 2000. Biochemical analysis of a thermostable tryptophan synthase from a hyperthermophilic archaeon. Eur. J. Biochem. 267:6369-6377. [DOI] [PubMed] [Google Scholar]
- 56.Tersteegen, A., D. Linder, R. K. Thauer, and R. Hedderich. 1997. Structures and functions of four anabolic 2-oxoacid oxidoreductases in Methanobacterium thermoautotrophicum. Eur. J. Biochem. 244:862-868. [DOI] [PubMed] [Google Scholar]
- 57.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tumbula, D. L., J. Keswani, J. S. Shieh, and W. B. Whitman. 1995. Maintenance of methanogen stock cultures in glycerol at −70°C, p. 85-87. In F. T. Robb, K. R. Sowers, S. DasSarma, A. R. Place, H. J. Schreier, and E. M. Fleischmann (ed.), Archaea—a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 59.Tumbula, D. L., R. A. Makula, and W. B. Whitman. 1994. Transformation of Methanococcus maripaludis and identification of a PstI-like restriction system. FEMS Microbiol. Lett. 121:309-314. [Google Scholar]
- 60.Tumbula, D. L., Q. Teng, M. G. Bartlett, and W. B. Whitman. 1997. Ribose biosynthesis and evidence for an alternative first step in the common aromatic amino acid pathway in Methanococcus maripaludis. J. Bacteriol. 179:6010-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tutino, M. L., A. Tosco, G. Marino, and G. Sannia. 1997. Expression of Sulfolobus solfataricus trpE and trpG genes in E. coli. Biochem. Biophys. Res. Commun. 230:306-310. [DOI] [PubMed] [Google Scholar]
- 62.Ward, D. E., W. M. de Vos, and J. van der Oost. 2002. Molecular analysis of the role of two aromatic aminotransferases and a broad-specificity aspartate aminotransferase in the aromatic amino acid metabolism of Pyrococcus furiosus. Archaea 1:133-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.White, R. H. 1985. Biosynthesis of 5-(p-aminophenyl)-1,2,3,4-tetrahydroxypentane by methanogenic bacteria. Arch. Microbiol. 143:1-5. [Google Scholar]
- 64.Whitman, W. B., E. Ankwanda, and R. S. Wolfe. 1982. Nutrition and carbon metabolism of Methanococcus voltae. J. Bacteriol. 149:852-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Whitman, W. B., J. S. Shieh, S.-H. Sohn, D. S. Caras, and U. Premachandran. 1986. Isolation and characterization of 22 mesophilic methanococci. Syst. Appl. Microbiol. 7:235-240. [Google Scholar]
- 66.Whitman, W. B., S. Sohn, and R. Y. Xing. 1987. Role of amino acids and vitamins in nutrition of mesophilic Methanococcus spp. Appl. Environ. Microbiol. 53:2373-2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xie, G., N. O. Keyhani, C. A. Bonner, and R. A. Jensen. 2003. Ancient origin of the tryptophan operon and the dynamics of evolutionary change. Microbiol. Mol. Biol. Rev. 67:303-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xing, R., and W. B. Whitman. 1994. Purification and characterization of the oxygen-sensitive acetohydroxy acid synthase from the archaebacterium Methanococcus aeolicus. J. Bacteriol. 176:1207-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xing, R. Y., and W. B. Whitman. 1992. Characterization of amino acid aminotransferases of Methanococcus aeolicus. J. Bacteriol. 174:541-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xing, R. Y., and W. B. Whitman. 1991. Characterization of enzymes of the branched-chain amino acid biosynthetic pathway in Methanococcus spp. J. Bacteriol. 173:2086-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xing, R. Y., and W. B. Whitman. 1987. Sulfometuron methyl-sensitive and -resistant acetolactate synthases of the archaebacteria Methanococcus spp. J. Bacteriol. 169:4486-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang, S., G. Pohnert, P. Kongsaeree, D. B. Wilson, J. Clardy, and B. Ganem. 1998. Chorismate mutase-prephenate dehydratase from Escherichia coli. Study of catalytic and regulatory domains using genetically engineered proteins. J. Biol. Chem. 273:6248-6253. [DOI] [PubMed] [Google Scholar]