Abstract
Research into lipid droplets is rapidly expanding, and new cellular and organismal roles for these lipid storage organelles are continually being discovered. The early Drosophila embryo is particularly well suited for addressing certain questions in lipid-droplet biology and combines technical advantages with unique biological phenomena. This review summarizes key features of this experimental system and the techniques available to study it, in order to make it accessible to researchers outside this field. It then describes the two topics most heavily studied in this system, lipid-droplet motility and protein sequestration on droplets, discusses what is known about the molecular players involved, points to open questions, and compares the results from Drosophila embryo studies to what it is known about lipid droplets in other systems.
Keywords: Drosophila embryo, lipid droplet, microtubule motors, protein sequestration
Graphical Abstract
I. Introduction
Lipid droplets are the main cellular site to store neutral lipids, such as triglycerides and sterol esters. They are ubiquitous organelles in fungi, plants, and animals and play crucial roles in lipid metabolism [1, 2]. Droplet accumulation removes potentially toxic lipids and stores them safely away from other cellular compartments. Lipid droplets also provide a reservoir of lipids for generating energy, membrane components, and signaling molecules. Due to their role in lipid homeostasis, lipid droplets are implicated in human diseases from obesity, diabetes, and cardiovascular disease, to neutral lipid storage diseases, hepatic steatosis, neurodegeneration, and certain pathogen infections [3-10].
Lipid droplets also play critical roles in the processing and storage of specific proteins. Lipid droplets have been usurped by a number of viruses as an assembly platform for new viral particles [11-14]. In Drosophila embryos, droplets transiently store specific histones to support rapid embryonic development [15, 16]. In certain mammalian cell lines, lipid droplets temporarily sequester ApoB protein destined for degradation [17]. Lipid droplets can also accumulate antiviral and antibacterial proteins [18, 19]. In different species and cell types, numerous proteins from other cellular compartment can be rerouted to lipid droplets; these observations lead to the proposal that lipid droplets may generally serve as sites to sequester proteins [20, 21]. Finally, lipid droplets appear to be involved in how immune cells present antigens to T cells: they somehow redirect peptides derived from phagocytosed material into a pathway usually reserved for endogenous peptides [22].
The structure of lipid droplets is unique among organelles: a core of neutral lipids is surrounded by a monolayer of polar lipids and proteins [2] (Fig. 1A,B). Despite this simple structure, droplets are functionally complex, containing dozens, if not hundreds, of different lipids and proteins, that can vary dramatically between cell types. Lipid droplets have functional and physical interactions with many other cellular compartments, including the ER, mitochondria, and phagosomes, and their composition, size, and intracellular distribution are actively regulated [1, 23-26]. They are increasingly recognized as highly dynamic, multi-functional organelles, with the number of publications focused on lipid droplets growing ever faster [27]. Yet even basic aspects of droplet cell biology are only beginning to be understood, such as how droplets originate, grow, and shrink, how proteins are targeted to them, how they move and interact with other organelles, and how these processes are regulated [23, 25, 26, 28-34].
Figure 1. The life cycle of Drosophila embryonic lipid droplets.
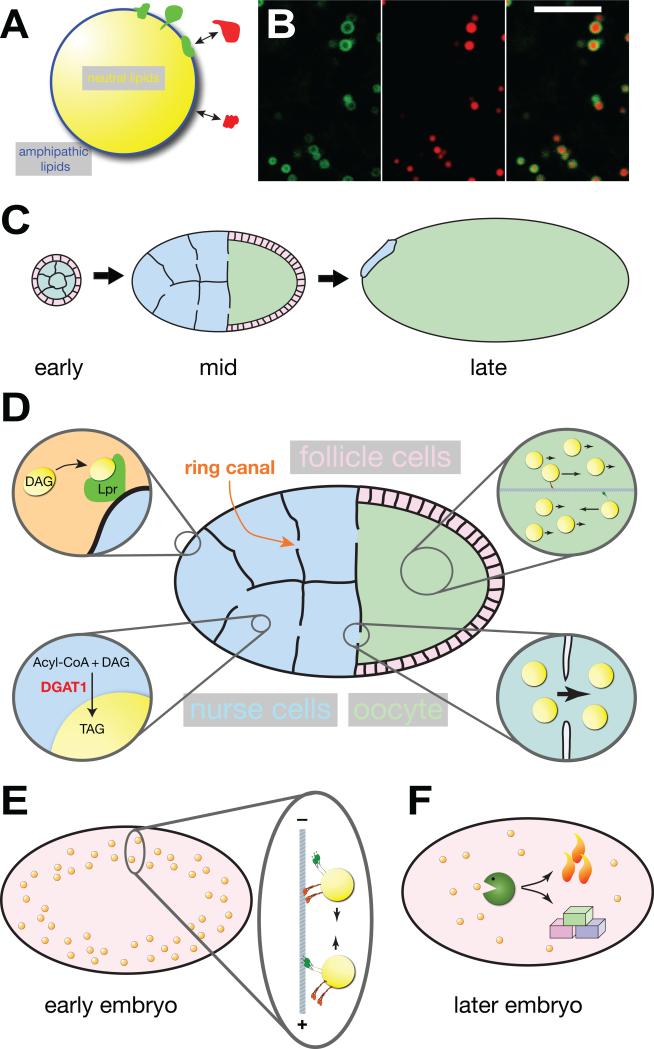
(A) Structure of lipid droplets: A core of neutral lipids (triglycerides (TAG), sterol esters, retinol esters) is surrounded by a monolayer of amphipathic lipids (such as phosphoglycerides and sterols). Proteins can be stably embedded (green) in this monolayer or reversibly bound (red) to other proteins or lipid head groups. (B) Colocalization of a GFP fusion (green) that targets to lipid droplets (GFP-LD [56]) and neutral lipids (red, detected with the dye Nile Red). The fusion protein surrounds a core of neutral lipids. Note that this particular fusion protein labels only a subset of lipid droplets. Scale bar = 5 μm. Image modified from [56]. (C) Overview of oogenesis (for color code of cell types, see D). In early egg chambers, the 16-daughter cells of the germ-line cytoblast (the future nurse cells and oocytes) are surrounded by a layer of somatic follicle cells. In mid-stage egg chambers, nurse cells supply the growing oocyte with nutrients, proteins and RNAs; follicle cells have migrated to cover the oocyte. In late stages, nurse cells have transferred most of their contents to the oocyte and have undergone apoptosis. The oocyte is surrounded by an eggshell (not shown) that was produced by the follicle cells. The various stages are not drawn to scale; by the end of oogenesis the oocyte volume has increased more than a hundred fold. (D) Lipid droplets in mid-stage egg chambers. One of the ring canals connecting nurse cells and oocyte is indicated. Top left: lipophorin particles (diacylglycerol (DAG) rich components of the hemolymph) are taken up by nurse cells via lipophorin receptors (Lpr). Bottom left: in the nurse cells, DGAT1 converts acyl-CoA and DAG into TAG, which contributes to the growth of the LD core. Bottom right: cytoplasmic streaming transports lipid droplets from nurse cells through ring canals into the oocyte. Top right: in the oocyte cytoplasm, most lipid droplets move passively by cytoplasmic streaming; a subset is actively transported by motors along microtubules. (E) In early embryos, lipid droplets move bidirectionally along microtubules. (F) Later in embryogenesis, lipid droplets are thought to be broken down to generate energy (via oxidative phosphorylation) and building blocks (for biomass production).
The initial cell biological and molecular characterization of lipid droplets focused on mammalian systems. Subsequent studies of fungi, worms, and insects have revealed that many aspects of lipid-droplet biology are highly conserved. Now a diverse range of model systems is employed in droplet research, including the fruit fly Drosophila melanogaster. For example, functional genomic screens in cultured Drosophila cells have identified many conserved genes involved in droplet biogenesis and turnover [35, 36]. Studies on fat storage in Drosophila larvae and adults have elucidated how lipid droplets contribute to organismal energy homeostasis and how droplet structure is controlled in distinct cell types [37]. And studies in Drosophila embryos have unraveled how droplet motility is regulated [24] and lead to the proposal of lipid droplets as general protein sequestration sites [15, 20].
There are a number of excellent recent reviews on lipid droplets in general and on the Drosophila studies in cultured cells, larvae and adults [2, 31, 32, 37-41]. However, there has been little systematic description of the lipid-droplet research in Drosophila embryos, even though this system is particularly well suited to unravel certain problems in droplet biology and combines technical advantages with unique biological phenomena. This review attempts to fill this gap. It summarizes our current knowledge about lipid droplets in Drosophila embryos, compares their properties to those of lipid droplets in other systems, and points out open questions and frontiers for future research. My goal is to make this intriguing experimental system accessible to researchers interested in lipid-droplet biology and to encourage them to take advantage of the unique approaches possible here.
II. Background
The lipid droplets of the early Drosophila embryo are generated during oogenesis and provided by the mother fly to the developing egg. Indirect evidence suggests that these lipid droplets provide the major energy source for the developing embryo and a large fraction of the stored lipid is used up during the course of embryogenesis. Thus, just like lipid droplets present in the adipose tissue of vertebrates and insects, the embryonic lipid droplets in Drosophila are a place to transiently store energy and to provide fuel when the organism lacks a source of food (adipose droplets) or when the organism itself is not yet capable of feeding (embryonic droplets).
Origin during oogenesis
In the Drosophila ovary, germline stem cells give rise to cystoblasts that undergo four incomplete mitotic divisions [42]; the resulting sixteen daughter cells remain connected via ring canals through which they can exchange molecules. One of the daughters becomes the oocyte, while its fifteen sisters, the nurse cells, are dedicated to providing the growing oocytes with nutrients, organelles, and signaling molecules (Fig. 1C,D). During mid-oogenesis (starting with stage 9), the nurse cells also generate massive amounts of lipid droplets [43]. Lipid accumulation is under hormonal control: the steroid hormone ecdysone activates, via the SREBP transcription factor, genes involved in lipid metabolism [44]. Like in other insects, the lipids needed for this massive droplet assembly are ultimately derived from lipoproteins (here called lipophorins) circulating in the hemolymph, the insect blood [45, 46]; lipophorins shuttle lipids between the gut, the fat body (the fly adipose tissue), and peripheral tissues, including the ovary [47]. Uptake of the neutral lipids into nurse cells is mediated by lipophorin receptors [48] (Fig. 1D), whose expression is also under ecdysone/SREBP control [44].
During the course of oogenesis, most of the contents of the nurse cells are transferred, via the ring canals, into the oocyte. For example, many ribonucleoprotein (RNP) particles critical for oocyte axis determination travel early on via active, microtubule based transport [49], while actin-based constriction of the nurse cells later in oogenesis promotes bulk transfer via cytoplasmic streaming [50]. Lipid droplets remain abundant in nurse cells until stage 11 [51], and they still accumulate in oocytes when microtubules are disrupted pharmacologically [52]. These observations suggest that transfer of lipid droplets is largely via actin-mediated cytoplasmic streaming. To my knowledge, there has not yet been an attempt to discern whether all lipid droplets found in oocytes have been transferred from nurse cells or if a subset is generated de novo in the oocyte. Interestingly, total triglyceride levels in mature oocytes varied little for females raised on diets of dramatically different nutrient content, suggesting the existence of homeostatic mechanisms (likely involving steroid signaling and SREBP) to keep oocyte lipid content constant even in environments in which the mothers encounter a fluctuating food supply [44].
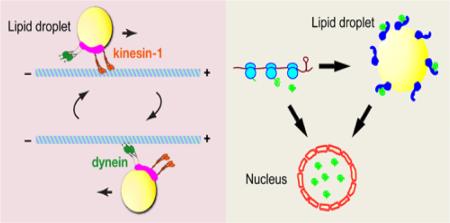
In the oocyte itself, lipid droplets are highly motile, in a microtubule-dependent manner [53, 54] (For movies of lipid-droplet motion in oocytes, see entries 4 and 5 in Table 1). During stages 10B-12 of oogenesis, the oocyte cytoplasm displays extensive streaming, dependent on microtubules and the microtubule motor kinesin-1 [52, 55]. Most lipid droplets seem to be carried along with the bulk flow of ooplasm, but some move actively along microtubule tracks, faster than the bulk flow or even against it (Fig. 1D). These droplets are thus likely moved by plus- and minus-end directed microtubule motors. These motors have not yet been directly identified, but based on analogy to motion in early embryos (below), kinesin-1 and cytoplasmic dynein are promising candidates.
Table 1.
Movies of Drosophila lipid droplets
| # | Label used to detect droplets | Detection method | Reference | Movie number |
|---|---|---|---|---|
| Bidirectional motion of embryonic droplets | ||||
| 1 | GFP-LD | Confocal microscopy | [56] | S1 and S2 |
| 2 | no label | fSRL microscopy | [104] | S2 |
| 3 | H2Av-GFP | Confocal microscopy | [15] | S9 |
| Droplet motion in oocytes | ||||
| 4 | No label | Confocal reflection microscopy | [53] | 1, 2, 3, 4 |
| 5 | H2Av-GFP | Confocal microscopy - regular and reflection | [53] | 10 |
| Net transport of lipid droplets in embryos | ||||
| 6 | No label | DIC microscopy | [76] | S1 |
| 7 | No label | fSRL microscopy | [104] | S1 |
Examples of movies from the literature that show lipid-droplet motion in ovaries and embryos. The label and detection method employed to visualize droplets are indicated. The movies listed can typically be found in the online supplemental/supporting material of the paper listed under “reference”.
The lipid droplets in oocytes and embryos are quite uniform in size, with diameters around 0.5 μm [56-59]. This is a typical size for small lipid droplets across eukaryotes. In contrast, lipid droplets in the larval fat body vary by over an order of magnitude [25, 60, 61]. Substantial size variations in lipid droplets are also characteristic of droplets in the adult fat body [25] and in cultured cells [62]. Why droplet size varies only within a narrow range in oocytes and embryos is not clear. One possibility is that the ring canals provide an upper limit for droplet transfer from nurse cells to oocytes. However, by stage 10, ring canals have an inner diameter of several microns and entire mitochondria can pass through them [63]; in addition, if ring canals were the size-determining bottleneck, droplets might still grow further once they have reached the oocyte, via local triglyceride synthesis or via droplet fusion. The limited droplet size in oocytes and embryos might therefore be functionally important, e.g., to minimize the drag experienced by motile droplets or to maximize droplet surface, for sequestering proteins or for interacting with other organelles.
The biochemical pathway responsible for droplet formation in nurse cells is not well characterized, though one key enzyme has been identified. Like mammals, flies express two enzymes catalyzing the final step of triglyceride biosynthesis, the conversion of diacylglycerol and fatty acyl CoA into triglycerides [64]. These enzymes, DGAT1 and DGAT2, have recently been shown to mediate lipid-droplet size control [62], with the DGAT1 pathway generating only small droplets, and the DGAT2 pathway larger droplets. Unlike DGAT1, DGAT2 can relocalize from the ER to the droplet surface and thus locally promote growth of droplets even after they detach from the ER. DGAT1 and DGAT2 are expressed throughout Drosophila development [65] and apparently function partially redundantly, as DGAT1 null mutants do contain triglycerides in larval and adult stages, yet at reduced levels [25]. However, nurse cells and oocytes of DGAT1 null mutants lack lipid droplets almost entirely [66], suggesting that triglyceride synthesis in ovaries is dominated by DGAT1 (Fig. 1D). Oocyte may thus achieve the narrow variation in droplet size by relying predominately on the DGAT1 pathway specific for small lipid droplets.
Lipid-droplet size in nurse cells is also under the control of insulin signaling [67]. Loss of PTEN, a negative regulator of insulin signaling, results in the formation of giant lipid droplets as well as in upregulation of LSD-2 [67]. LSD-2, also known as PLIN2, is a member of the Perilipin family of lipid-droplet proteins [68]. Whether LSD-2 overexpression accounts for the increased droplet size is unknown, but lack of LSD-2 has been linked to impaired droplet formation in nurse cells, with neutral lipid inappropriately retained in the ER [51].
Embryonic lipid droplets as nutrient stores
In many insects, lipids - in particular neutral lipids – are the main energy source for embryogenesis [69, 70]. For example, mosquito embryos derive about 90% of their energy from the breakdown of lipids [71]. In such embryos, triglycerides are also thought to provide precursors for phospholipids to generate new membranes [69]. Although these issues have not yet been directly addressed in Drosophila, indirect evidence suggests that breakdown of neutral lipids is also essential for Drosophila embryogenesis. Embryos from mothers lacking the lipophorin receptor Lpr2 in the germline have very few lipid droplets, and they die before they complete development [48]. In addition, adult females lacking LSD-2 lay embryos with reduced triglyceride content, and a fraction of these embryos dies in the middle of embryogenesis [51].
The mechanisms by which triglycerides and lipid droplets are turned over during embryogenesis remain to be identified (Fig. 1F). As these processes are much better understood in cultured cells and for the larval and adult fat body [2, 37, 39], many candidate pathways are already known and their contribution in embryos is in principle readily testable. For example, Brummer lipase (an ortholog of mammalian ATGL) makes major contributions to triglyceride breakdown in both cultured cells and in adult flies [35, 72]. It likely plays a similar role in embryos: Embryos lacking Brummer have increased triglyceride levels late in embryogenesis relative to wild type and fail to hatch [72].
Triglyceride breakdown during embryogenesis appears to be temporally regulated, though the control mechanisms remain to be elucidated. There is a dramatic upregulation of genes involved in fatty acid breakdown around mid-embryogenesis [73], implying that triglyceride hydrolysis provides ample substrates for these enzymes. In other insects, physiologically studies have found that the initial energy needs of embryos are met by the breakdown of carbohydrates, but the rest of embryogenesis is driven largely by catabolism of lipids [69]. A similar pattern probably also holds for Drosophila: in the first two hours of embryogenesis, total carbohydrate levels drop dramatically [74]; in parallel, the glycogen-storage depots of the early embryos, the so called β-spheres, undergo major structural rearrangements [74]. In contrast, the number and size of lipid droplets remains unchanged for the first three hours [57], consistent with initially limited consumption of lipids. During later stages of embryogenesis, both glycogen and triglyceride levels continue to decrease, but it has not yet been mapped out to what extent turnover of these storage molecules serves the energy needs of the embryo and to what extent they are channeled into the production of new biomass [73].
In addition to lipids, embryonic droplets also store specific proteins to support embryogenesis. Certain histones are abundant on lipid droplets, and they can be transferred to nuclei and contribute to proper chromatin assembly [15, 16, 75] (Fig. 4B,D,E). Histones are also released when bacteria are present in the cytoplasm; this contributes to bacterial killing and promotes embryo survival in the presence of pathogens [19] (Fig. 4G). As the lipid-droplet proteome is complex [15] and contains many additional proteins from other compartments, this storage role may be even more general.
Figure 4. Histone sequestration on droplets.
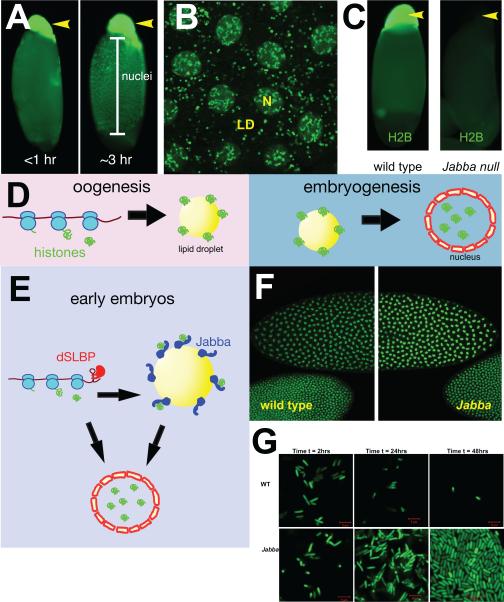
(A) Centrifuged embryos stained for histone H2Av. In Phase 0 embryos, H2Av signal is almost exclusively associated with the lipid-droplet layer (yellow arrow). By Phase II, H2Av signal is found both on the droplet layer and in nuclei. Image modified from [16]. (B) In Phase I embryos, H2Av-GFP (green) is present in nuclei (large blobs, N) and on lipid droplets (small rings in the cytoplasm, LD). Image courtesy of Zhihuan Li. (C) In-vivo centrifugation demonstrates that histone H2B is present on lipid droplets in a Jabba-dependent manner. Both panels show Phase 0 embryos after centrifugation; lipid-droplet layers are indicated by yellow arrows. Left: wild-type embryo stained for H2B (H2B is highly enriched on the droplet layer). Right: Jabba mutant embryo stained for H2B (H2B is absent from the droplet layer). Image modified from [16]. (D) Storage of histones on lipid droplets allows temporal uncoupling of histone production and usage: During oogenesis, newly synthesized histones are sequestered on lipid droplets. The sequestered histones are released during embryogenesis and are relocated to the nucleus to package chromatin. (E) Histone dynamics in early embryos. Early embryos contain both histone mRNAs and histone proteins provided from the mother. The translation of the messages for canonical histones is regulated by the Drosophila stem loop binding protein (dSLBP). Maternal histone proteins are stored on lipid droplets via binding to Jabba. Both newly translated histones and droplet-stored histones contribute to chromatin assembly in the nucleus. In addition, excess newly synthesized histone can be sequestered on droplets, thus buffering the histone supply. (F) Wild-type and Jabba mutant embryos stained for H2Av. At certain stages, Jabba mutant embryos display overaccumulation of H2Av in their nuclei. Image modified from [75]. (G) Wild-type and Jabba mutant embryos injected with E. coli expressing GFP. While over time the bacterial population declines in the wild-type embryos, it dramatically increases in Jabba mutants. Image from [19].
Droplet motion during early embryogenesis
In early Drosophila embryos, lipid droplets are highly motile: they move in linear paths, with average speeds on the order of 0.5 μm/s and uninterrupted motion in a given direction for up to several microns [76-78]. Both travel velocities and travel distances vary across broad ranges, and droplets that travel for shorter distances tend to move more slowly [77]. Motion has been first detected by ~1.5 hrs of embryogenesis, and it continues for many hours, only interrupted during mitoses. At any one moment, almost all of the droplets are in motion, and pauses in motility are short (on average ~ 0.6 s [77]). For movies of lipid-droplet motion in embryos, see entries 1, 2, and 3 in Table 1.
These lipid droplets move along microtubules, powered by the molecular motors kinesin-1 and cytoplasmic dynein [76, 77] (Figs. 1E, 2D). Microtubules are polar filaments, with distinct plus and minus ends. Microtubule motors recognize this polarity and typically transport cargo unidirectionally along the microtubule tracks, i.e., either towards the plus or towards the minus end. However, many cargoes, including the lipid droplets in Drosophila embryos, carry both plus- and minus-end directed motors and move in a characteristic bidirectional fashion: after travel in the plus-end direction, they move in the minus-end direction for a while, then switch back to plus-end motion, and so on [79, 80]. In Drosophila embryos, lipid droplets switch the direction of motion every few seconds [57].
Figure 2. Lipid droplet motility in the early embryos.
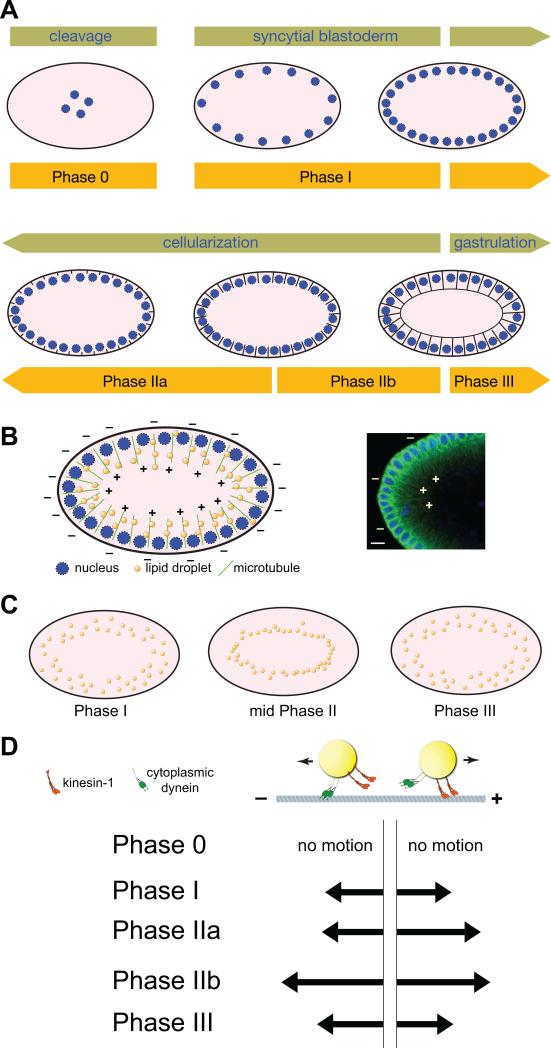
(A) Overview of early embryogenesis. During cleavage stages nuclei divide in the embryo interior. By syncytial blastoderm (nuclear cycle 10-13), a subset of nuclei has reached the surface and continues to undergo mitosis. During cellularization (nuclear cycle 14), plasma membranes grow in between the nuclei, converting the syncytial embryo with thousands of nuclei into a monolayer of cells with one nucleus per cell. Gastrulation movements start shortly thereafter. The orange bars represent the extent of the phases of lipid-droplet transport relative to other morphological events. Embryos are drawn schematically and not to scale; in reality, there are some ~6000 nuclei present at the periphery at cellularization. In addition, from syncytial blastoderm onward, the center of the embryo also contains polyploidy yolk nuclei; for clarity, they have been omitted from the cartoon. (B) Left: Schematic representation of the distribution of nuclei, lipid droplets, and microtubules in early embryos (not drawn to scale). Nuclei are present close to the embryo surface, all around the embryo. Microtubules are oriented radially, with their minus ends close to the surface and their plus ends pointing into the embryo interior. Right: Micrograph of parts of an early embryo, depicting DNA (blue, to highlight nuclei) and microtubules (green). Microtubule polarity (+ and – ends) is indicated in a few instances. Scale bar = 10 μm. Image modified from [76]. (C) Schematic representation of the global distribution of lipid droplets during early embryogenesis (not drawn to scale). In Phase I and III, droplets are found all over the peripheral cytoplasm. In Phase II, droplets relocate inward and accumulate around the central yolk (not shown, but see Fig. 3B). (D) Cartoon of lipid-droplet motion: Droplets move back and forth along microtubules, powered by the plus-end motor kinesin-1 and the minus-end motor cytoplasmic dynein. The arrows show the relative lengths of movements in the minus- and plus-end directions in various phases (based largely on the measurements in [83], but see also [86] and [57]; the exact run-length values vary with the tracking method employed; the figure summarizes the relative balance of motion in the two directions).
Over time, this bidirectional droplet motion results in dramatic shifts in the global distribution of lipid droplets. These shifts are due to the peculiar arrangement of microtubules in early embryos. Understanding this geometry requires a brief review of early embryogenesis [81] (Fig. 2A). Drosophila embryos are roughly football shaped, with a long axis of ~500 μm and a short axis of ~150 μm. Lipid droplets are found throughout the peripheral region of the embryo, from the surface to roughly 40 μm deep (Fig. 2B). During the first 2.5 hours after fertilization, nuclei divide every 8-20 min, but there is no cytokinesis (with one exception, the formation of primordial germ cells, a process not relevant for this discussion); this process yields a syncytial embryo with thousands of nuclei in a shared cytoplasm. The nuclei divide near synchronously, making the total number of mitoses that have occurred since fertilization a convenient measure for the developmental stage of the embryo: the period from mitosis 12 to mitosis 13 is called “nuclear cycle 13”, etc. The nuclei are initially present internally (“cleavage stages”), but then a fraction migrates out to the surface, arriving there in cycle 10; this forms the syncytial blastoderm (Fig. 2A). The nuclear density at the cortex doubles with each of the next four mitoses. During the ensuing cycle 14, a highly synchronized cytokinesis occurs (a process called cellularization), generating a single layer of cells (35-40 μm tall and with one diploid nucleus each) that surrounds a giant central yolk cell with many polyploidy nuclei. Right at the end of cellularization, gastrulation movements start, converting the single sheet of cells into a complex 3D structure.
Because the nuclei are associated with microtubule organizing centers (MTOCs), this distribution of nuclei also governs the arrangement of microtubules (Fig. 2B). Microtubule minus ends are associated with the MTOCs close to the plasma membrane, and plus ends point into the embryo interior, resulting in a radial array of microtubules with largely uniform polarity [57, 82]. By cycle 14, nuclei are densely packed at the surface, and microtubules are arranged grossly parallel around the whole embryo periphery. With the cell movements of gastrulation, the global parallelity of microtubules is lost.
Droplet motion is regulated temporally, and dramatic changes in transport parameters occur over very short time spans, and reproducibly at specific developmental transitions [57, 77, 83] (Fig. 2A,D). After an initial period of immobility before syncytial stages (Phase 0), lipid droplets start moving in both directions, and – on average – the distance moved in the plus-end direction is the same as in the minus-end direction (Phase I). In early embryonic cycle 14 (Phase IIa; ~2.5 hrs of embryogenesis), plus-end travel distances are upregulated while minus-end distances remain unchanged. Over the next hour, transport parameters switch twice more, late in cycle 14 (Phase IIb, travel in both directions upregulated), and at the beginning of gastrulation (Phase III, travel in both directions downregulated). Transitions between phases occur over just 10 min (or less). Originally, Phases IIa and IIb were not distinguished from each other, and were lumped together as Phase II. This review uses the nomenclature IIa and IIb when it is clear from the literature which subdivision is being analyzed. I will generically use “Phase II” if a statement applies to both Phases IIa and IIb or if it is not clear from the literature exactly which part of Phase II was analyzed.
These changes in motility result in global redistribution of the overall droplet population (Fig. 2C, see also movie 7 in Table 1). Recall that nuclei and microtubule minus ends are close to the embryo surface and microtubule plus ends point towards the interior. During Phase I, when plus-end and minus-end travel distances are balanced, there is no net change in droplet distribution, though individual droplets constantly trade places. Upregulation of plus-end transport in Phase IIa results in net inward transport: The droplet population shifts towards the plus ends, away from the embryo surface; the average distance of droplets from the surface increases by 10μm [57], which represents ~ one fourth of the height of the cells that form during cellularization. At any given point in time, most droplets are accumulated around the central yolk, with only a few remaining in the periphery. However, this is a dynamic distribution as the droplets continue to move bidirectionally: as some peripheral droplets join the pool around the yolk, some droplets from interior regions move back out into the periphery. In Phases IIb and III, minus-end transport predominates, and the droplet population shifts back towards the periphery, again in a dynamic fashion, as individual droplets continue to move bidirectionally. Although it is clear that droplet motion is elaborately regulated in the early embryo, what biological roles this motion serves remains a matter of speculation. Some possibilities are discussed in section VIII.
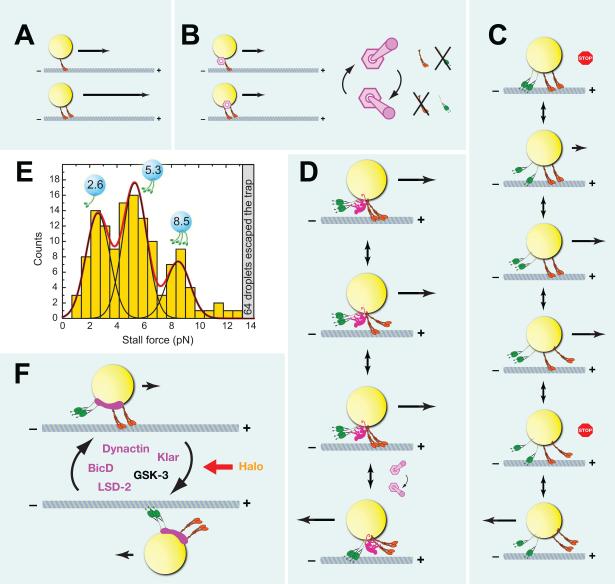
Genetic and biochemical approaches have shed light on the molecular mechanisms driving droplet motion and mediating its temporal regulation. Transport is powered by the plus-end motor kinesin-1 and the minus-end motor cytoplasmic dynein [76, 77] (Fig. 2D). The dynein cofactor Dynactin, the kinase GSK-3, and the nesprin ortholog Klarsicht are important for proper force production during transport and may activate motors and/or prevent futile competition between opposing motors [57, 84, 85] (Figs. 5F, 6A). Temporal regulation is largely driven by expression of the novel protein Halo and has been linked to changes in the droplet levels of the dynein cofactor BicD and the phosphorylation state of the Perilipin LSD-2 [83, 86, 87] (Fig. 6D). More details on these molecules and how they act in droplet transport are provided in sections IX through XI.
Figure 5. The motors driving lipid droplet motion.
(A) Motor behavior in vitro: Cargo moved by two motors can travel for considerable longer distances than cargo moved by a single motor. Arrows indicate distance traveled. (B) Proposed “switch” model for lipid-droplet transport in vivo. Left: a switch mechanism terminates motion independent of motor number; both cargoes move the same distance. Arrows indicate distance traveled. Right: For bidirectional transport, the switch toggles between two states: “kinesin-1 ON, cytoplasmic dynein OFF” and “kinesin-1 OFF, cytoplasmic dynein ON” (right). (C) Bidirectional transport as a result of a tug-of-war between opposing motors. Arrows indicate travel velocity. If the numbers and forces of opposing motors are well balanced, cargoes will frequently be stalled, in severe motor competition (panel 1). As motors attach and detach stochastically, motor imbalance will arise that allows slow motion in a particular direction (panel 2). If motors under load release more readily, this imbalance will quickly resolve itself into only motors for one direction being actively engaged on the microtubule (panel 3). Stochastic binding/release of motors will re-establish the paused state and can even result in reversal of direction (panels 4, 5, 6). (D) Bidirectional transport as the result of the still hypothetical coordination machinery (pink): The coordination machinery keeps cytoplasmic dynein off (possibly by sterically preventing binding to the track) while the opposing kinesin-1 motors are on. Once the switch is triggered, the coordination machinery turns kinesin-1 off and simultaneously makes cytoplasmic dynein active. (E) Stall force measurements for plus-end directed lipid droplets in Drosophila embryos show peaks at multiples of ~2.6 pN. This pattern indicates the action of 1, 2, or 3 kinesins per droplet. Image from [59]. (F) Factors known to regulate lipid-droplet motion. Lipid droplets constantly switch between motion dominated by the plus-end motor (top) and motion dominated by the minus-end motor (bottom). The pink blob represents a hypothesized switching complex. Dynactin and Klar have been proposed to act as integral parts of the switch mechanism involved. BicD, GSK-3, and LSD-2 also affect the distance traveled in one or both directions, and thus may be involved in flipping the switch. Halo acts as transacting signal that mediates the temporal pattern of switching frequency. Klar, LSD-2, Dynactin, and BicD are localized to lipid droplets and may be part of the switching complex. Whether GSK-3 or Halo are physically present on the droplets is unknown.
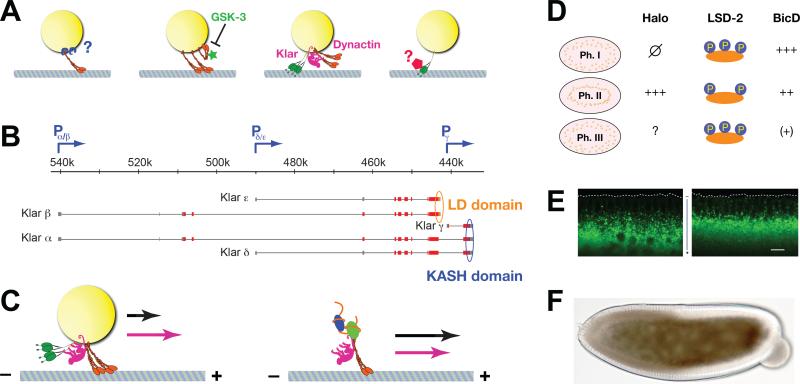
Figure 6. Regulators of lipid-droplet transport.
(A) Proposed models of force regulation during lipid-droplet transport (from left to right): 1) Motor number per droplet might be controlled by the availability of docking sites or the number of motors available for docking. The cargo adaptor for motors on droplets is not yet known [76]. 2) The activity of motors might be controlled after docking; GSK-3 has been proposed to restrict the activity of docked kinesin-1 [85]. 3) Motor coordinators may allow full force production by keeping opposite-polarity motors inactive, as proposed for dynactin and Klar [57, 84]. 4) Cytoplasmic dynein can exist in low- and high-force states [312], and these states can be controlled by transacting factors [214]. So force production by cytoplasmic dynein on lipid droplets might be regulated in vivo, an idea that has not yet been tested. (B) The complex klar locus encodes five different protein isoforms, α, β, γ, δ and ε. Promoters are indicated by blue arrows, non-coding exons by gray bars, and coding exons by red/orange/blue bars. LD domain is shown in orange, KASH domain in blue. Map modified after [266]. (C) Comparison of the effect of Klar on lipid droplet (left) and mRNA (right) transport. Arrows symbolize run lengths in the presence (pink) or absence (black) of Klar. Presence of Klar increases plus-end travel lengths for lipid droplets, but reduces them for RNP particles [57, 235]. (D) Differences in Halo, LSD-2, and BicD proteins between phases of droplet transport. Halo is absent in Phase I and expressed in Phase II; its status in Phase III is unknown, but circumstantial evidence and halo's mRNA expression pattern has led to the proposal that Halo is degraded by this time [79, 86]. LSD-2 is highly phosphorylated in Phase I and III, but less so in Phase II [87]. Droplet levels of BicD protein drop progressively from Phase I to II to III [83]. (E) Halo acts as a directionality determinant for transport. GFP-labeled lipid droplets in late Phase IIa embryos in which Halo is either expressed (left) or missing (right). The dotted line outlines the embryo surface. In the presence of Halo, net transport is plus-end directed (inward); in the absence of Halo, net transport is minus-end direction (outward). Scale bar = 10 μm. Image modified from [56]. (F) Acute effect of Halo on lipid-droplet distribution and embryo transparency. Bright-field image of a Phase IIa embryo mutant for Halo in which in-vitro generated halo mRNA was injected on the right. In the left half of the embryo, Halo activity was absent and lipid droplets are spread throughout the periphery, resulting in a broad brown “halo” around the central yolk. In the right half of the embryo, Halo activity was present, and lipid droplets accumulated around the central yolk (as a narrow dark band), leaving the periphery depleted of droplets. As a result, the peripheral cytoplasm is transparent. Image from [86].
The molecular machinery driving droplet motion in Drosophila embryos is shared with many other transport processes. Kinesin-1 and cytoplasmic dynein are two widely employed motors for cargo transport, and they frequently work together in bidirectional motion [79]. BicD, Dynactin, Klar, and GSK-3 regulate the transport of a diverse set of cargoes; these factors (alone or in combination) have been shown to modulate the motion of RNP particles, different vesicles, mitochondria, and even whole nuclei. Finally, motion of lipid droplets is characterized by frequent, sharp reversals, few pauses, and an exponential distribution of run lengths [77]; similar patterns of motions have been described for pigment granules, viruses, and RNP particles [88-90]. Thus, a mechanistic understanding of droplet motion is likely relevant for the study of other intracellular transport systems, and vice versa.
Embryonic droplets in other species
Lipid droplets have been observed in the eggs and oocytes of many animals, including insects, spiders, annelids, frogs, fish, marsupials, and placental mammals [91-100]. Presumably, they provide a critical, maternally generated energy source for the developing embryo not yet able to feed. Similar strategies are used in other organisms to transmit energy to the next generation: the mammalian milk that feeds the newborns is rich in fat droplets that ultimately derive from lipid droplets produced in mammary glands [101], and many plant seeds contain abundant lipid droplets, here called oil bodies [102], that accumulate during seed development and that support development of the embryonic plant before the onset of photosynthesis.
Like in Drosophila, embryonic droplets in many species are highly motile: for example, lipid droplets in both mouse and fish embryos display active motion dependent on the cytoskeleton [92, 94], and for fish, annelids, and moths, massive redistribution of the droplet population accompanies early development [95, 97, 98]. Very little is known about mechanism and function of droplet motility in these species.
III. Studying embryonic lipid droplets
Imaging droplets
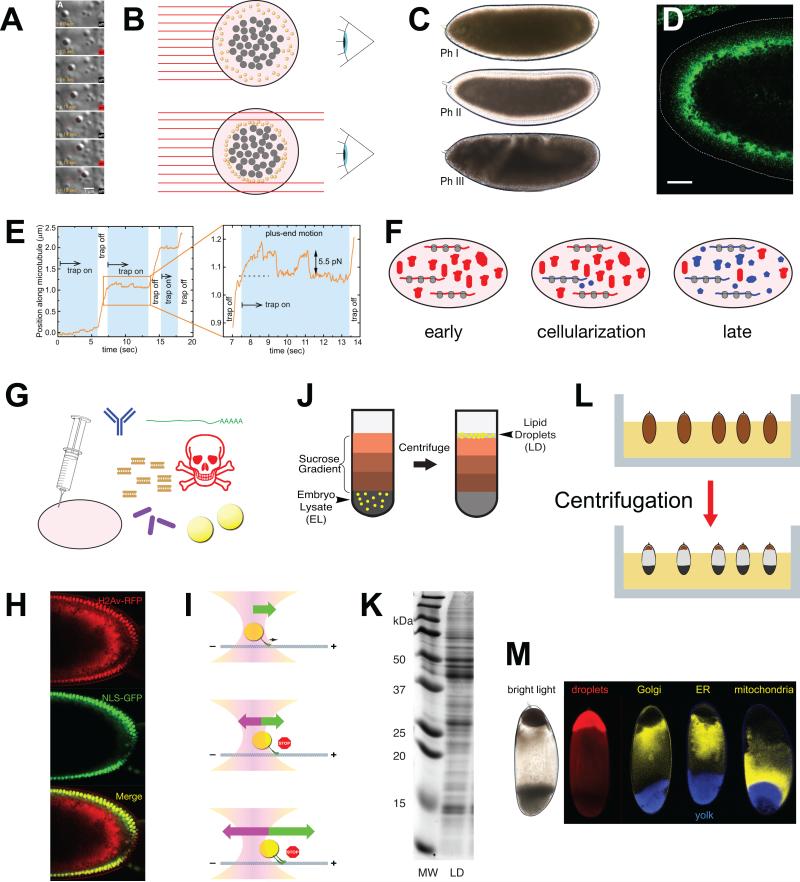
In contrast to many other intracellular structures, lipid droplets can often be unambiguously detected in living cells, even without the application of stains or exogenously introduced expression constructs. Of the many label-free methods that have been developed to detect lipid droplets, two have been reported for the analysis of Drosophila embryos: lipid droplets provide the major contrast in embryos for both third-harmonic generation microscopy [103] and femtosecond Stimulated Raman Loss (fSRL) microscopy [104]. In oocytes, lipid droplets have been specifically detected using confocal reflection microscopy [53]. Such label-free methods make it possible to analyze droplets in vivo in many genetic backgrounds without further manipulations, and they avoid possible artifacts due to the expression of exogenous proteins. For examples of visualization of lipid-droplet motion by fSRL microscopy, see entries 2 and 7 in Table 1.
Lipid droplets are also easily visualized by differential interference contrast microscopy, and their size and round shape makes it possible to distinguish them from other cellular structures [57, 77] (Fig. 3A). To increase image clarity and resolution, it is possible to gently flatten the embryos until they burst open; in undisturbed regions, these ex-vivo preparations preserve many, if not all, characteristics of droplet motion for tens of minutes [57]. Improvements in mounting techniques achieve similarly well-contrasted images in intact embryos that continue to develop normally for hours [59]. Using differential interference microscopy, it is possible to quantify motion at high temporal and spatial resolution: the position of the droplet center can be determined at nanometer precision, and acquisition rates of 30 images per second are routinely achieved [57] (Fig. 3A, E). Because contrast is best near the edge of the embryo, most tracking is done with droplets moving within a few micrometers of the plasma membrane. fSRL microscopy is currently not quite as fast or precise, but – unlike the differential interference approach – it can detect droplet motion deep in the embryo and thus can examine whether parameters of droplet motion depend on the distance from the plasma membrane [104].
Figure 3. Methods to study Drosophila embryonic droplets.
(A) Lipid-droplet motion visualized by differential interference contrast (DIC) microscopy and manipulated by an optical tweezer/trap. Time in seconds is indicated as well as whether the laser (centered on the droplet) is turned on or off. Red line shows the position of the center of the lipid droplet tracked over time. The droplet proceeded steadily along a linear path (inferred to be a microtubule) and its progress was impeded by the optical trap. For details of the tracking and laser trap analysis, see E. (Image from [59]). (B) How lipid droplet distribution affects embryo transparency. Shown is an embryo cross-section perpendicular to the long axis of the embryo. Light (red lines) from a source on the left passes through the embryo and is collected by an observer on the right. Yolk granules (gray) and lipid droplets (yellow) scatter light and thus prevent light from passing through the cytoplasm. In the top embryo, lipid droplets are spread out all over the periphery and block light evenly. In the bottom embryo, lipid droplets have moved away from the periphery and are accumulated around the central yolk; hence, light can pass through the periphery. (C) Changes in lipid-droplet distribution cause altered embryo transparency. In Phase I and III, the embryo periphery is opaque because the abundant lipid droplets scatter light. In Phase II, the periphery is transparent because it is depleted of lipid droplets. Image from [76]. (D) Jabba immunostaining (green) to highlight lipid droplets. The dotted line outlines the embryo surface. In this Phase II embryo, the droplets have accumulated basally, clustering around the central yolk. Scale bar = 25 μm. Image modified from [16]. (E) Movement of a lipid droplet along a microtubule as a function of time, in the presence or absence of an opposing force from an optical trap. This image is from [59] and represents the quantitation of the experiments shown in (A). The enlarged portion shows that the droplet stalls when the trap is switched on and then drops to the trap center. Another movement attempt again results in a stall. When the trap is switched off, the motors are able to continue to move the droplet. The distance at which the stall occurred is a measure for the force generated by the motor(s) moving the droplet, in this case ~5.5 pN. (F) Early embryos contain proteins directly inherited from the mother (red blobs) as well as proteins generated in the embryo from translation of maternal messaged (red). By cellularization, the zygote has started transcribing its own genes, generating its own messages and proteins (shown in blue). Later in embryogenesis, translation is driven by zygotic messages and most, but not all, proteins are the product of zygotic transcription. (G) By microinjection, various substances can be introduced into embryos, including antibodies, mRNAs, dsRNAs for RNA interference, inhibitors, bacteria, and lipid droplets. (H) Transplantation of H2Av-RFP covered lipid droplets into recipient embryos in which nuclei are marked by NLS-GFP. Merged image reveals the transplanted lipid droplets in red; some of the droplet-bound H2Av-RFP was released from droplets and was transferred into nuclei. Image from [15]. (I) Principles of optical trapping. Top: Lipid droplet in the center of an optical trap (optical tweezer). It is propelled along the microtubule by the force generated by the microtubule motor. The trap does not yet exert any force on the droplet. Middle: Once the droplet is displaced from the laser center, the trap exerts a force pulling the droplet back towards the center. At some point, the force from laser and motor are balanced, resulting in stalled motion. Bottom: If the droplet is pulled by multiple motors, this force balance occurs at a distance further from the center of the laser. (J) Schematic representation of lipid-droplet purification by floatation. Embryo lysate in high-density buffer is overlaid by sucrose solutions of increasingly lower density. After centrifugation, lipid droplets can be recovered at the very top of the gradient. Image from [16]. (K) Protein content of lipid-droplet fraction after sucrose gradient. Proteins from droplet fraction (LD) were analyzed by SDS PAGE. MW = molecular weight markers. Image from [16]. (L) Schematic depiction of in-vivo centrifugation of embryos. Embryos before cellularization are embedded in agar to keep them in a fixed orientation (top). After centrifugation, the contents of each embryo are separated by density (bottom). Image from [127]. (M) Separation of organelles by in-vivo centrifugation. Living embryos were centrifuged as in (L), which results in distinct stratification visible by bright-field microscopy (left). Distribution of various organelles was detected by fluorescence microscopy. Image originally from [15], as modified in [127].
The global distribution of lipid droplets can be easily determined in living embryos by standard transmitted light microscopy (Fig. 3C, see also 6F). The embryo contents that contribute most to light scattering are yolk vesicles and lipid droplets. Since by the end of Phase I yolk vesicles have redistributed to the center of the embryo, the opacity of the embryo periphery in Phases IIa, IIb, and III is dominated by lipid droplets [57]. As a result, the net transport of droplets causes reproducible changes in overall embryo transparency [57] (Fig. 3 B,C; see also movie 6 in Table 1). In the wild type, the embryo periphery is full of lipid droplets in Phase I and therefore appears opaque under transmitted light. The periphery becomes transparent during Phase IIa as lipid droplets deplete from it, and turns opaque again starting in Phases IIb and fully in Phase III. Net inward (plus-end) transport is therefore also referred to as “clearing” and net outward transport as “clouding”. That these changes in transparency are indeed due to lipid-droplet redistribution was confirmed with mutants that prevent the net inward transport in Phase IIa [86] or the net outward transport in Phase III [57]. Equivalent transparency changes reveal droplet accumulation in oogenesis: the nurse cell cytoplasm turns opaque as it fills with lipid droplets beginning with stage 9.
In fixed material, embryonic lipid droplets can be detected with neutral lipid-specific dyes commonly used in other systems, such as Nile Red [57] and BODIPY493/503 [105] (Fig. 1B). However, care must be taken to not inadvertently delipidate the embryo; many standard fixation protocols for Drosophila embryos include a heptane/methanol step to disrupt the vitelline membrane; this step also removes neutral lipids. Such delipidation also largely abolishes light scattering by lipid droplets and thus interferes with using embryo transparency as a read-out for global droplet distribution. As an alternative, lipid droplets can be detected in fixed material with antibodies against proteins highly enriched or exclusively present on lipid droplets: antibodies against Jabba [16] (Fig. 3D), Klar [105], and LSD-2 [87] are suitable for such immunodetection.
To fluorescently label lipid droplets in vivo, several transgenic constructs are available that express fluorescent proteins targeted to lipid droplets. GFP-LD (Fig. 1B) incorporates the droplet-targeting domain of Klar and is almost exclusively present on lipid droplets [56], while H2Av-GFP or H2Av-RFP marks both lipid droplets and nuclei [15, 16] (see movies 1, 3, and 5 in Table 1). A GFP trap line in LSD-2 [106] can also be used to detect lipid droplets (M. A. W., unpublished observations).
Genetics
Drosophila research can take advantage of rich genetic and genomic resources, including many classical mutants, chromosomal deletions and rearrangements, and transgenes for overexpression studies and RNAi [107, 108]. Using both forward and reverse genetic strategies, a number of proteins have been confirmed to play roles in lipid-droplet biology in embryos, in particular for droplet motion [56, 57, 76, 77, 83-87, 105, 109, 110] and for protein sequestration [16, 75].
The early embryo contains many gene products derived from the mother, very few from the father, and some that result from new transcription in the embryo itself (Fig 3F). Early embryogenesis is almost exclusively driven by maternal gene products, and zygotic transcription is massively upregulated only during the mid-blastula transition (around cellularization). The same pattern holds true for droplet transport: When zygotic transcription is inhibited with the RNA polymerase inhibitor alpha amanitin, the first evidence for abnormal distribution of lipid droplets occurs in Phase IIa [111]. This effect was subsequently mapped to zygotic expression of the gene encoding Halo [86], a temporal regulator of droplet transport (see section X). So far, no other genes have been identified whose zygotic expression is important for droplet motion or any other droplet characteristics. In addition, there is no evidence for gene products from the father affecting droplet phenotypes.
Thus, almost all proteins important for droplet biology in the early embryo are maternally expressed, e.g. Klar and LSD-2 (affect droplet transport) and Jabba (for histone sequestration) [16, 57, 87]. Maternal contribution poses a particular challenge for the analysis of essential proteins, like the motors kinesin-1 and cytoplasmic dynein. Animals lacking these motors completely do not reach adult stages and thus cannot produce eggs. For kinesin-1, this problem has been circumvented using germline clone technology [76], which makes it possible to generate nurse cells and oocytes homozygous for the mutation of interest in an otherwise heterozygous animal. Tissue-specific RNA interference is an alternative promising strategy, and a new generation of vectors allows efficient knockdown in the female germ line [112]. Yet these approaches are challenging for the analysis of cytoplasmic dynein since this motor is required for multiple steps during oogenesis, and in its absence no mature oocytes are produced [113]. In some cases, the use of hypomorphic or dominant-negative mutations is a work-around, allowing the production of viable embryos in which protein function is sufficiently impaired to give phenotypes [77 , 84]. Alternatively, some phenomena, like droplet motion and protein sequestration, already occur in oocytes [53, 54], and thus could be studied even in mutants that only support oogenesis, but not embryogenesis.
Inhibitor studies
Studies of lipid droplets in cultured Drosophila cells benefit greatly from the ability to directly manipulate the cells with pharmacological agents and double-stranded RNAs [35, 36]; these reagents can simply be added to the culture medium. For embryos, the multilayered eggshell provides a formidable barrier to exogenously applied reagents: although the outer chorion can be physically or chemically removed, the waxy layer surrounding the inner vitelline membrane is essentially impermeable to most molecules. Some techniques for permeabilization exist [114, 115], and recently have been greatly improved to allow application of small molecules to embryos [116, 117]; however, they are not yet routinely used.
An alternative means of introducing molecules is by microinjection, in which a needle pierces both eggshell and plasma membrane and delivers substances directly into the cytoplasm (Fig. 3G, see also 6F). When performed with syncytial embryos, i.e., before widespread cytokinesis, the introduced substance can in principle diffuse throughout the whole embryo. For lipid-droplet studies, microinjection has been used to introduce double-stranded RNAs for RNA interference [86, 109], function-inhibiting antibodies [76], transcriptional inhibitors [86], bacterial pathogens [19] as well as mRNAs to rescue mutant phenotypes [86].
Physical manipulation of droplets
Microinjections have also been used to physically move lipid droplets from one embryo to another: a donor embryo, centrifuged to concentrate droplets, is pricked with a needle; then cytoplasm with lipid droplets is taken up into the needle and transplanted into a host (Fig. 3H). Such transplanted droplets retain the ability to exchange proteins with nuclei [15] and to move bidirectionally (MAW, unpublished observations). This strategy makes it possible to study the behavior of one type of droplet in a new cellular context, such as a different genetic background or phase of transport.
Lipid droplets can also be manipulated within the embryo cytoplasm using optical traps (also called optical tweezers) (Fig. 3I). When small dielectric objects are placed into a highly focused laser beam, they experience a force that pulls them towards the center of the beam [118]. In vitro, such optical traps have been used extensively to move small particles, to measure the forces produced by molecular motors (when attached to glass or plastic beads), and to apply forces to deform individual molecules and polymers like microtubules. The use of optical traps in vivo has been much more limited, in part because applying them in a quantitative manner in vivo is technically challenging [118, 119]. Lipid droplets, however, are ideal candidates for quantitative optical trapping in vivo: they are perfectly round, have uniform refractive indices that differ significantly from those of the surrounding cytoplasm, and they are in the right size range to apply biologically meaningful forces. In the Drosophila embryo, moving droplets can indeed be trapped repeatedly, without appreciable photodamage [57] (Fig. 3A,E). Initial attempts to measure the forces generated by moving embryonic droplets used a few constant laser forces and determined which fraction of the droplets can escape from the trap. This method provides an estimate for the forces generated across a population of droplets, e.g., the force sufficient to stall all/most of the moving droplets [57, 77, 84, 86]. Advances in detecting the exact position of the stalled droplet made it possible to greatly refine this approach, as the displacement of the droplet relative to the center of the laser beam is a measure for the force experienced by the droplet (Fig. 3E, I). This refinement now makes it possible to perform force measurements on single droplets and to follow changes in force production by the same droplet over time [59, 76, 85]. Such quantitative analysis has revealed that plus-end moving lipid droplets are typically propelled by the simultaneous activity of 2-3 copies of kinesin-1 [59, 76] (Fig. 5E); the situation for minus-end directed motion is likely similar [76].
Until recently, Drosophila embryonic droplets were essentially the only in vivo system that allowed quantifiable force measurements of moving cargoes. More recent work has shown that similar force measurements are possible with lipid droplets in cultured mammalian cells [120, 121] or droplets isolated from liver whose motion has been reconstituted in vitro [122]. By allowing cultured cells to phagocytose latex beads, it is now also possible to trap phagosomes and related vesicles and measure the forces generated during their motion [123, 124]. Like for embryonic lipid droplets, these vesicles appear to move by the combined activity of a small number of motors.
Biochemical purification
One of the distinguishing characteristics of lipid droplets is their high neutral lipid content; as a consequence they have a much lower buoyant density than any other cellular structures, including vesicles, mitochondria, or membrane fragments. This property has been used extensively to purify lipid droplets by floatation, from a variety of sources: bacteria, fungi, plants, cultured animal cells, and various animal body parts and tissues. This technique has been adapted to Drosophila embryos (Fig. 3J), and droplets can be isolated to high purity, as determined by visual inspection and markers for other organelles [15, 87]. Such preparations of purified droplets have subsequently been analyzed by SDS PAGE (Fig. 3K), western blotting, and proteomics approaches, to reveal their protein content [15, 16, 19, 56, 76, 83, 85, 87], and by immunoprecipitation, to uncover physical interactions between droplet-localized proteins [16]. To isolate lipid droplets corresponding to specific developmental stages, adult females are allowed to lay eggs only for a short period of time, and then these collections are aged to the appropriate stage. Using ~1hr long laying times, it was uncovered that droplet protein content changes between phases of transport [15, 83, 87].
For other motile organelles, studies into the mechanism of motion have greatly benefitted from the ability to reconstitute motility in vitro [125]. For lipid droplets, in particular, such a system would allow powerful comparisons with isolated motors on the one hand (e.g., it is possible to purify kinesin-1 from Drosophila embryos [126]) and in-vivo motion on the other hand. Droplets isolated from rat liver are quite motile in vitro [122], although motion is dominated by kinesin-1 and not obviously bidirectional. Droplets purified from Drosophila embryos have kinesin-1, cytoplasmic dynein, and dynactin attached to them and can exhibit long-range unidirectional motility [58]. They also show frequent short-range (a few hundred nanometers) back-and-forth motion, but not yet the sustained bidirectional runs of 1 μm or more observed in vivo [58]. Thus, robust bidirectional in-vitro motility has not yet been achieved, but even with the partial reconstitution possible so far, one can ask how impairing various known transport regulators (e.g., by isolating droplets from mutant embryos) affects various parameters, like run length and travel velocity, and whether this reproduces the effect of the mutations in vivo.
In-vivo centrifugation
The low buoyant density of lipid droplets can also be used to separate droplets from other cellular components in vivo [127] (Fig. 3L,M). Embryos, if still surrounded by their eggshell or even just the vitelline membrane, can withstand substantial centrifugal forces without deformation. During cleavage and syncytial blastoderm stages, the embryo is essentially a single cell, and upon centrifugation, intracellular constituents should be free to move and arrange according to their density. Major organelles indeed separate reproducibly along the heavy-light axis [15]. This assay has been used to enrich droplets for transplantation [15] and to determine localization of proteins to lipid droplets [15, 16, 56, 87, 105].
Separating cellular constituents via centrifugation also works well in other large cells, like nurse cells, oocytes, and the eggs of other species [15, 16, 127, 128]. It even has been used successfully for the unicellular alga Euglena [129] and for hyphae of the bread mold Neurospora [130]. Finally, for fission yeast, centrifugation is employed to displace the nucleus within the cell to study the mechanism by which nuclei find their way back to the center [131]. Thus, in-vivo centrifugation might work in many cells or tissues to spatially enrich lipid droplets.
IV. The droplet lipidome
The lipid component of lipid droplets can be quite complex, with hundreds of lipid species [132]. Lipid composition can also vary dramatically between cells: mammalian adipocytes are triglyceride rich, while testis droplets and macrophage droplets prominently contain sterol esters. In addition, the exact mix of phospholipids present at the droplet surface controls the droplet's surface properties and plays a role in size control [133].
Given the importance of various lipid species for both structure and function of the droplets, it is surprising that no characterization of the lipid content of Drosophila embryonic lipid droplets has yet been published. Triglycerides presumably are a major component since early embryos are rich in these neutral lipids [16, 51, 73] and need them for energy production and membrane synthesis. Sterol esters likely also make a contribution: the embryo needs sterols as membrane precursors as well as for steroid hormone signaling (e.g., [134]), yet insects cannot synthesize sterols and have to take them up with the food. Thus, the embryonic sterols must be maternally provided, and are presumably stored as sterol esters in lipid droplets; indeed, early embryos are rich in sterol esters [135]. Lipidomics has revealed a rich diversity of lipids across various life stages of flies [135-137], and similar studies on the lipid droplets of embryos should provide an important source of new information on the biological roles of the droplets, and may shed light on the mechanisms of droplet motility and of protein sequestration.
In mammalian cells, sterol esters and triglycerides have been reported to be stored in distinct droplets, even within the same cell, droplets that also differ in their protein complement [138]. This issue has not yet been examined in Drosophila embryos, and it remains unknown whether embryos contain droplets of distinct identity or composition: the droplet population is not differentiated into obvious size classes, and the few droplet proteins that have been examined by imaging have not shown dramatic variation between droplets [15, 16, 87]. The only exception is a GFP fusion of the droplet-targeting domain of Klar (GFP-LD; [56]), whose levels vary dramatically even on neighboring droplets. But the nature of this variation is obscure, and may not reflect different composition of the droplets but rather their origin [56].
V. The droplet proteome
Proteomic studies suggest that lipid droplets can be associated with hundreds of proteins [21, 139]. The lipid droplets of Drosophila are no exception. The one published analysis of early embryonic droplets isolated droplets by sucrose-gradient centrifugation and uncovered over 500 candidate droplet proteins [15]. A priori, it is not clear, which of these proteins are bona fide lipid-droplet proteins and which might simply be due to unavoidable contamination during the biochemical isolation. One study estimated that the fraction of contaminants in droplet preparations using conventional proteomics approaches may be as high as 90% [140]. Thus, as for other lipid-droplet proteomes, most of the candidate proteins of embryonic droplets await validation by independent tests. It is important, however, not to prematurely dismiss candidates as false positives: Histones, for example, are abundant cellular proteins and frequent contaminants in various proteomic analyses; yet for Drosophila embryos follow-up studies demonstrated that histones are true droplet components [15] with important biological functions [16, 19] (see section VI).
Validation is possible by a comprehensive set of techniques. They include standard approaches employed in many other systems, such as western analysis of isolated droplets [76, 87] and colocalization with droplet markers (Fig. 1B), (either in intact embryos [16, 56, 83] or squash preparations [77, 87]). In addition, a number of mutants are available that predictably alter the global distribution of lipid droplets; a genuine droplet protein has to show the same changes in distribution [16, 56] (Fig. 6E). In addition, co-localization is easily demonstrated by in-vivo centrifugation [15, 127] as droplet proteins are highly enriched in the lipid-droplet layer [15, 56, 87, 105] (Fig. 4A,C). Finally, if mutants in candidate proteins are available, they can be used to probe for phenotypic effects on droplet properties, such as changes in motility parameters [76, 77, 85]. New validation strategies that employ global proteomic comparisons across many different biochemical fractions [140] have yet to be applied to embryonic droplets.
Validation has so far been achieved for just a small number of proteins (see Table 2). This is a highly selected group of proteins involved in lipid-droplet motion and protein sequestration since these are the processes most often studied in early Drosophila embryos. A number of other proteins found by mass spectrometry are also highly likely to be true droplet proteins since they have been verified to be droplet-associated in cultured fly cells or at other developmental stages or their orthologs are known to localize to lipid droplets (for example, CG9186 [141] or the Drosophila ortholog of CGI-58 [142, 143]).
Table 2.
Confirmed lipid-droplet proteins of early Drosophila embryos
| Protein | Evidence for droplet localization | Amount |
|---|---|---|
| Kinesin | Western analysis [76] Colocalization on purified droplets [58] Mutants and antibody injection alter droplet motility [76] |
Minor |
| Dynein | Western analysis [76, 87] Colocalization in disrupted embryos [77] Colocalization on purified droplets [58] Mutants alter droplet motility [77, 84] Mutants for known dynein cofactors alter motility [83, 84] |
Minor |
| Dynactin | Colocalization on purified droplets [58] Mutant alters droplet motility [84] |
Minor |
| LSD-2 | Western analysis [87] Colocalization in disrupted embryos [87] Centrifuged embryos [87] Mutants alter droplet motility [87] |
Likely exclusively on droplets |
| BicD | Western analysis [83] Mutants alter droplet motility [83] |
Minor |
| Klar | Colocalization in intact embryos [105] Centrifuged embryos [56, 76, 105] Mutants alter droplet motility [56, 57, 105] Mutants that alter droplet distribution alter Klar distribution accordingly [76, 105] |
Large fraction |
| H2A | Western analysis [15, 16] Centrifuged embryos [15, 16] |
Large fraction in early embryos |
| H2B | Western analysis [15, 16] Centrifuged embryos [15, 16] |
Large fraction in early embryos |
| H2Av | Western analysis [16] Centrifuged embryos [15, 16, 75] Colocalization in intact embryos [15] Mutants that alter droplet distribution alter H2Av distribution accordingly [15] |
Large fraction in early embryos |
| Jabba | Western analysis [16] Centrifuged embryos [16] Colocalization in intact embryos [16] Mutants abolish histone sequestration on droplets [16] Mutants that alter droplet distribution alter Jabba distribution accordingly [16] |
Large fraction, possibly exclusively on droplets |
Amount refers to the fraction of the protein detected on lipid droplets compared to the total levels of the protein in the embryo.
There are two other published droplet proteomes from Drosophila sources, one using droplets from larval fat bodies [61], the other droplets from cells grown in culture [140] (the widely used fly S2 cell line). The overlap between these sets is surprisingly small (as compiled in Fig. 5A of [140]): Only four proteins were present in all three sets. The differences observed might simply be due to distinct levels of contamination in the three sets or more or less stringent criteria for including a candidate. For example, for the droplets from cultured cells, protein correlation profiling was used to exclude proteins predominantly localizing in non-droplet fractions [140]; of the 20 candidates analyzed by microscopy, 18 displayed (exclusively or in part) localization in the ring-like pattern characteristic for lipid-droplet proteins (Fig. 1B), suggesting a very low false positive rate [140]. Alternatively, the difference between the three sets might reflect differences in expression levels of those candidates in different types of cells or might be due to tissue-specific targeting to droplets. For example, three types of histones are among the most abundant lipid-droplet proteins in embryos, and their droplet localization has been abundantly confirmed by many independent approaches [15, 16]. Yet, these histones are not detectable on lipid droplets in S2 cells [15, 140]. For detailed lists of the proteins shared between the embryo and cultured-cell sets and embryo and fat body sets, respectively, see [140] and [37].
Some proteins, like certain Perilipins, localize constitutively to lipid droplets [68, 144]; for others, droplet localization is conditional; e.g., the lipase HSL translocates to lipid droplets in response to lipolytic stimuli [145] while the enzyme CCT1 relocates to droplets when they expand [36]. For the proteins in Table 2, changes in protein levels during the first few hours of embryogenesis occur for the histones H2A, H2B, and H2Av [15, 16] and the dynein co-factor BicD [83] (Fig. 6D), whose droplet levels go down as development proceeds. In addition, the Perilipin LSD-2 transiently changes its phosphorylation state [87] (Fig. 6D). Over longer time frames, the droplet protein content likely changes more dramatically: proteomic analyses of larval fat-body and of embryonic droplets show massive differences [15, 37, 61], and direct comparisons of the droplet proteome of different life stages of the moth Manduca sexta [146] reveal major remodeling across development.
For a number of lipid-droplet proteins, there have been tremendous advances in our understanding of how they are targeted to lipid droplets [2, 26, 147]. For the proteins in Table 2, however, very little is known in this regard. Histones are anchored to droplets via Jabba [16], presumably by electrostatic interactions, since salt washes can remove histones from droplets [15]. Although histones and Jabba have been found to co-immunoprecipitate and thus apparently are present in common protein complexes [16], it remains unknown whether they directly interact and which regions of Jabba, a novel protein, mediate the interaction. For Klar, targeting to lipid droplets is mediated by its C-terminal LD-domain, possibly via an amphipathic helix [56, 105]. LSD-2 binding to droplets is dominated by hydrophobic interactions [15], and a model for droplet binding has been proposed by modeling and analysis of GST fusions [148].
VI. Histone sequestration
When lipid droplets are biochemically purified from early Drosophila embryos, three types of histones are among the most prominent proteins present, the core histones H2A and H2B and the variant histone H2Av [15, 16]. In newly laid embryos, the histone content of the droplets is equivalent to that in thousands of nuclei [15]. This does not represent contamination with chromatin since other core histones (H3 and H4) were not found. Indeed, a combination of biochemical, genetic, and in-vivo imaging studies established conclusively that histones are prominent lipid-droplet proteins in these embryos [15], using the validation criteria discussed in the previous section (Fig. 4A, B).
Histone association with lipid droplets is developmentally controlled [15]. It can already be detected in nurse cells and oocytes, is massive during early stages, and declines during the first few hours of embryogenesis, so that by 12 hrs histones were no longer detected on droplets [15]. By mass spectrometry, certain histones (H2Av, H2B, and H4) were also found in purified droplet samples from larval fat body [61]. In addition, mutations in Jabba, the histone anchor on lipid droplets (see below), partially reduce a cytoplasmic histone H2B pool in extracts from adult flies [19]; the nature of this histone pool is unknown, but may represent lipid droplets since Jabba is a droplet protein. In contrast, in S2 cells, a common Drosophila cultured cell line, histones were not detected on lipid droplets [15, 140]. Finally, proteomic analysis of various life stages of the moth Manduca sexta [146] revealed the presence of H2A, H2B, and H4, but relative amounts varied with developmental stage.
In the early embryo, droplet association of histones is transient. Bulk measurements of histone levels on droplets reveal that histones leave lipid droplets during the first few hours of embryogenesis [15]. When droplets with H2Av-RFP were transplanted into recipient embryos corresponding to Phases I or II, H2Av-RFP appeared in nuclei within tens of minutes [15], leading to the proposal that droplet-bound histones are a storage site for histone used later on for chromatin assembly. Whether histones leave lipid droplets with similarly rapid kinetics at other developmental stages remains to be examined.
Jabba anchors histones to lipid droplets
Histones are bound to lipid droplets via electrostatic interactions [15], and the novel protein Jabba has been identified as necessary for recruiting histones to droplets, likely acting as histone anchor [16]. Jabba localizes to lipid droplets in early embryos and in ovaries [16] and is one of the most abundant proteins in purified droplet samples from such embryos [15, 16], rivaling or exceeding histones in abundance. GFP fusions of Jabba also localize to lipid droplets in cultured Drosophila cells [16].
Jabba mutants have abundant lipid droplets, but compared to the wild type these droplets lack a number of proteins, including histones [16]. Absence of histones was confirmed by western analysis of purified droplets, immunostaining of centrifuged embryos (Fig. 4C), and observation of live embryos expressing H2Av-GFP. Thus, Jabba is necessary for the presence of histones on lipid droplets.
Two lines of evidence suggest that Jabba anchors histones to droplets by physical interactions [16]. First, when H2Av-GFP is immunoprecipitated from droplet preparations, Jabba – but not other lipid-droplet proteins – are found in the pellet. Second, reduction in Jabba levels results in a parallel decrease of droplet-bound histones. Whether Jabba directly contacts histones or via intermediary proteins is currently under investigation.
The Jabba locus has the potential to encode seven protein isoforms that share a common 320 aa N-terminus and vary C-terminally [16]. Western analysis reveals multiple Jabba-specific bands [16], but which bands correspond to which isoform(s) remains to be established. Not all of these versions of Jabba behave identically: some are highly enriched on purified lipid droplets, while others are undetectable. It is unknown if those latter forms of Jabba localize elsewhere in the embryo or are lost during purification; however, immunolocalization in undisturbed and in centrifuged embryos has detected Jabba unequivocally on lipid droplets.
Jabba is required for high histone levels in the embryo
Across eukaryotes, histone expression levels are typically carefully balanced. If too few histones are available, replication is slowed down, transcription patterns are altered, and cells are more sensitive to DNA damage [149-152]. Histones present in excess also disrupt gene expression patterns, increase DNA damage sensitivity, and result in chromosome loss during mitosis and lethality [153-156]. As a result, transcriptional and post-transcriptional regulation typically limits histone biosynthesis to times of need [157, 158], and excess histones are proteolytically degraded [159, 160].
Drosophila embryos are an exception to this rule, as the newly laid embryo contains a thousand fold excess of histone proteins [15]. In other animal eggs, e.g. the frog Xenopus [161], a similar maternally provided histone pool is present, and it had been proposed that this pool provides the building blocks for rapid chromatin assembly in the earliest embryonic stages. In Drosophila, Jabba plays a crucial role in maintaining the maternal histone protein pool since it is severely compromised in newly laid Jabba mutant embryos: levels for histones H2A, H2B, and H2Av are dramatically reduced compared to wild type; in contrast, levels of H3, not found on lipid droplets, are normal [16].
Indirect evidence suggests that this deficit in the maternal histone pool is not due to problems with histone biosynthesis, but with maintaining this pool. In Jabba mutant embryos, histone mRNA levels are unaltered, and these histone messages can be translated. In yeast and mammalian cells, excess histones are eliminated by proteasome-mediated degradation [149, 153, 162]. It was therefore proposed that wild-type Drosophila embryos accumulate extra-nuclear histones because droplet binding, via Jabba, protects the histones from degradation. The mechanisms for histone turnover in Drosophila remain unexplored, but recent insights in yeast into the molecular machinery mediating histone turnover [159, 163] should lead to direct tests whether histone degradation is indeed increased in the absence of Jabba.
Droplet-bound histones provide a largely redundant source of histones to support early embryogenesis
Early embryos contain not only the histone protein deposit associated with lipid droplets, but also histones mRNAs, provided maternally and later replenished by zygotic transcription [158, 164] (Fig. 4E). Although new translation of histones was initially thought to make little contribution before cycle 14 [158, 165], histone levels do indeed rise after fertilization [75] and lack of zygotic transcription of histones has been linked to increased risk of DNA damage [166]. In Jabba mutant embryos, new synthesis of H2A/H2B/H2Av proteins is able to compensate for the lack of the maternal histone deposit so that by 3.5 hrs of development, they have caught up to wild-type levels [16].
This rescue of normal histone protein levels is likely the reason why Jabba mutant embryos are viable and grossly normal, even though they lack the maternally provided histone supply. Indeed, Jabba mutants were initially thought to develop entirely normally [16]. However, when raised at elevated temperatures to speed up development, they display mildly reduced survival and excessive “nuclear falling” [75], an embryo-specific DNA damage response that eliminates defective nuclei during blastoderm stages [167]. These findings suggest that Jabba and histone-sequestration on droplets contribute to the thermal robustness of embryogenesis, which in the wild type is remarkably consistent across a broad temperature range [168].
Massive nuclear falling is also observed in embryos with a severely compromised histone supply [164], suggesting that it is the lack of maternal histones that causes this defect in Jabba mutants. However, these mutants also lack other droplet proteins, and thus the connection to histones remained tentative. This issue was addressed when it was discovered that specifically compromising new histones synthesis in a Jabba mutant background dramatically enhances nuclear falling [16]. The mRNAs of core histones do not carry PolyA tails, but instead end in characteristic stem loops. This stem loop is bound by the stem loop binding protein (the Drosophila version is called dSLBP) (Fig. 4E), an interaction that promotes correct processing and stability of these mRNAs [164]. Partial reduction of dSLBP levels has no detectable effects on the development of wild-type embryos, but in Jabba mutants it results in loss-of-histone phenotypes, including abnormal mitoses, DNA damage responses, and almost complete embryo lethality [16]. These observations suggest that the droplet-bound histones become essential for life when new histone biosynthesis is even mildly impaired. Consistent with the idea that droplets serve as a redundant source of histones for early development, the loss of histones from droplets is accelerated when new histone biosynthesis is impaired [16]. These findings suggest that the embryonic lipid droplets provide a redundant pool of histones to support early embryogenesis and thus are indeed histone storage sites.
Droplets as histone buffers
Immunostaining of early Jabba mutant embryos revealed a striking imbalance of histone accumulation in the nuclei: While the nuclear signal for the canonical histones H2A, H2B, and H3 was indistinguishable from the wild type, nuclear signal of the variant H2Av was two to three fold increased, but only in specific nuclear cycles [75] (Fig. 4F). Because developmental westerns suggest that in the early embryo H2Av and canonical histones are synthesized in an imbalanced manner, it was proposed that H2Av produced in relative excess could be captured by lipid droplets and prevented from being imported into the nuclei. This mechanism would ensure proper nuclear balance between variant and canonical histones in the wild type, even if levels in the cell overall are imbalanced; in Jabba mutants, lack of this sequestration then results in improper histone ratios in the nucleus. Indeed, when lipid droplets were transplanted between embryos, histones from the recipient embryo could be recruited to the droplets from the donor, indicating that histones can indeed be loaded onto droplets in the embryo [75]. These observations indicate that lipid droplets do not only serve as sites for long-term storage of histones produced during oogenesis, but that they also provide short-term buffering ability in the embryo. This buffering function is likely biologically important as Jabba mutant embryos are hypersensitive to H2Av overexpression [75].
Droplet-bound histones as antibacterial defense
In vitro, histones have potent bactericidal activity [169]. Lipid droplets purified from Drosophila embryos are also highly toxic to both gram-positive and gram-negative bacteria [19]. This killing activity is – largely or completely - due to the histones bound to the droplet surface: for example, it is abolished when histones are stripped from droplets by salt washes, when anti-histone antibodies are included with the purified droplets, or when droplets from Jabba mutants are used [19]. To test whether this killing activity was relevant in vivo, GFP-labeled E. coli were microinjected into both wild-type and Jabba mutant early Drosophila embryos. The presence of histones on lipid droplets had a profound effect on bacterial proliferation: at the employed dosage, bacterial load decreased in the wild type, but increased dramatically in the mutants [19] (Fig. 4G). When challenged with pathogenic bacteria via microinjection, wild-type embryos also survived several-fold better than Jabba mutants. These observations suggest that droplet-bound histones can serve as a potent antibacterial defense and may constitute a novel form of innate immunity against intracellular pathogenic bacteria [19].
Is has not yet been established whether this defense strategy contributes to embryo survival in the wild. Embryos are protected by a multi-layer eggshell that is impermeable to even medium-sized molecules and thus they would not seem to be at risk from bacterial pathogens. However, later embryonic stages can induce a strong classical immune response after experimental bacterial exposure [170, 171], implying a real risk of infection in nature. Possibly, pathogens could enter the egg if the eggshell is mechanically damaged or chemically compromised, e.g., by nematode or insect predators. Alternatively, the droplet-bound histones – already present in oocytes [15] – might prevent transmission of bacterial pathogens from the mother to the embryo.
Jabba-mediated anti-bacterial protection also appears to be active in adult flies [19]. Jabba mutants exposed to the intracellular pathogen Listeria monocytogenes build up larger bacterial loads than wild-type controls and die much more readily. The mechanism of this protection remains to be explored in detail, but a non-nuclear histone pool – presumably associated with lipid droplets – is reduced in Jabba mutant adults [19]. Intriguingly, Jabba is also among a group of 229 Drosophila immune-regulated genes that are significantly upregulated in adults following a bacterial challenge [172, 173].
A moonlighting role for histones on droplets?
There is now strong evidence that droplet-bound histones can leave the droplets and make profound contributions to chromatin assembly and antibacterial defense. It is an open question whether the histones are stored on the droplets solely for such use elsewhere in the cell or whether they also function on the droplets. In Jabba mutant embryos, the embryonic triglyceride content is normal, yet droplets display abnormal clustering [16]. Although this anti-clustering activity of Jabba may be unrelated to histones, histones on the droplet surface might possibly provide a positively charged shell that prevents the droplets from approaching too closely and thus does not allow them to stick to each other. In cultured cells, droplets have a tendency to cluster [36], a tendency that may need to be suppressed in the early embryo to allow the droplets to be highly motile.
VII. Lipid droplets as general protein sequestration sites
Histones on lipid droplets are widespread
Available evidence suggests that association between lipid droplets and histones is not restricted to Drosophila embryos but occurs from fungi to mammals: By immunolocalization, histones have been detected on lipid droplets in Drosophila ovaries [15], in housefly embryos [15] and in mouse oocytes [93], as well as on the lipid-droplet–related microvesicles of mammalian sebocytes [174]. Western analysis has detected specific histones on lipid droplets isolated from mouse liver [19]. Finally, specific histones have been reported as part of the lipid-droplet proteome isolated from unicellular eukaryotes [175, 176], nematodes [104], various insect tissues [61, 146], and several mammalian cell types [177-179].
The biological role of these droplet-bound histones remains to be elucidated. On the one hand, they may act in innate immunity like in Drosophila. Intriguingly, lipid droplets from mouse liver carry histones, and histone levels go up after a simulated infection [19], possibly indicating an adaptive antibacterial response. The liver is indeed involved in protection against bacterial pathogens [180]. On the other hand, rerouting histones to lipid droplets prior to chromatin assembly might control histone levels post-translationally, balance the supply of different histones, or store histones for contingencies. Such roles could have broad implications since abnormal histone levels are linked to cancer progression and aging and can cause altered transcription, mitotic defects, and increased DNA damage sensitivity [149-151, 153-155, 160, 181].
For Drosophila larvae, there is an intriguing, though at the moment only highly speculative link for a connection between lipid droplets and chromatin assembly: In mammalian cells, the metalloprotease invadolysin localizes to lipid droplets [182]. In Drosophila, animals mutant for invadolysin die as third-instar larvae, with defects in mitotic chromosome packaging [183]. Genetic interaction studies suggest that these phenotypes result from a pathway controlling histone ubiquitination, and Invadolysin mutants indeed display overaccumulation of mono-ubiquitinated histone H2B [184]. However, whether invadolysin is present on droplets in Drosophila larvae and whether it is there it interacts with H2B is not yet known.
Refugee proteins
Besides histones, there are many other proteins with known functions elsewhere in the cell that associate with lipid droplets in certain cell types or under certain conditions, e.g., the cytosolic enzyme Inosine-5’monophosphate dehydrogenase (IMPDH) [185], the spliceosome activator Prp19p [186], the membrane protein caveolin [187, 188], ApoB, a component of secreted lipoprotein particles [17], and possibly HMG CoA reductase [189] (but see [190]). These proteins might play novel roles on lipid droplets; alternatively, they might be transiently sequestered on lipid droplets before or after acting elsewhere in the cell [15, 20], a notion captured by the term “refugee protein”. It has been speculated that droplet sequestration might inactivate toxic proteins, prevent aggregation of proteins lacking binding partners, promote assembly of macromolecular complexes, or deliver proteins intracellularly, via motile droplets [20, 21, 191].
This notion of lipid droplets as protein sequestration sites has been critically examined only in a few cases: several viral proteins transiently accumulate on lipid droplets, and this association promotes assembly of the final viral particle [11, 12]; and maternal histones are stored on lipid droplets of Drosophila embryos [16]. But many of the reported lipid-droplet proteomes contain numerous proteins from other cellular compartments. If even a small fraction of these proteins are not contaminants or moonlighting proteins, then lipid droplets might have important cellular roles far beyond lipid metabolism. In turn, altered lipid metabolism could affect many other cellular processes via overstorage (such as in obesity) or understorage (as in lipodystrophies) of lipids, as the extent of cellular lipid storage would presumably influence the cell's protein sequestration potential.
The droplets of early Drosophila embryo might carry refugee proteins other than histones, as proteomic analysis suggests the presence of many proteins from other compartments [15]. One prominent example includes selected subunits of ATP synthase, also found in lipid-droplet preparations from CHO cells [192], adipocytes [143], Drosophila larval fat body [61], several life stages of the moth Manduca sexta [146], yeast [193], and a dinoflagellate [176]. That an enzyme ordinarily targeted to the mitochondrial matrix may be present on lipid droplets is certainly surprising, but there is a precedent for a non-canonical intracellular distribution of ATP synthase subunits: in some cells, they have been shown to localize to and function at the plasma membrane [194, 195].
A role of droplets in protein sequestration may also provide an explanation for a puzzling link between lipid metabolism and early development: Females mutant for the long-chain acyl-CoA synthetase (Acsl/CG8732) produce embryos with characteristic segmentation defects, i.e., embryos lack adjacent morphological features along the head-to-tail axis [196]. Acyl-CoA synthetases activate free fatty acids, and different family members direct fatty acids into different metabolic pathways [197]. How lack of activation of fatty acids might bring about such specific developmental defects is obscure, but one of the hypotheses proposed [196] was that aberrant formation of lipid droplets in the mutants might influence the delivery or generation of signaling molecules or the storage of maternal determinants. Acsl is indeed implicated in droplet function: In cultured fly cells, Acsl localizes to LDs [140], and knockdown results in smaller droplets [36]. Among the mammalian family members, Acsl is most similar to ACLS3 and 4: in the fly nervous system, ACLS4 is functionally analogous to Acsl [198]; ACSL3 is essential for the correct initiation of droplet formation from the ER [199].
VIII. Droplet motion – general considerations
As discussed in section II, lipid droplets are highly motile in Drosophila oocytes and embryos. In the embryos, almost all droplets are in constant motion, moving bidirectionally along microtubules [57]. In stage 10B oocytes, lipid droplets display both passive flow due to cytoplasmic streaming and active motion along microtubules, in both directions [53]. Unlike in the embryo, individual droplets have not been observed to display bidirectional motion. Here, reversals of travel direction are rare; rather droplets move unidirectionally and then diffuse away, presumably after detaching from the microtubules. It is unknown how the switch from predominantly unidirectional motion in oocytes to an entirely bidirectional mode of transport in embryos is accomplished.
There has not yet been a systematic attempt to determine to what extent droplet motion occurs in other tissues and life stages. The massive lipid droplets predominant in the fat body, the adipose tissue of the fly, are probably too bulky for extensive motion, if any. But many other tissues (e.g., wing imaginal discs, larval salivary glands, and follicle cells [48, 56, 200]) contain numerous tiny lipid droplets, and even in the fat body, there are subpopulations of small droplets [201]. To my knowledge, no attempts have been made to determine whether any of these droplets move and in what manner. Time-lapse movies of Drosophila S2 cells during fatty acid feeding display abundant and seemingly chaotic small-scale excursions, and over long time periods droplets accumulate in clusters [36], though it is unclear whether these movements are microtubule-based or represent Brownian motion.
The biological reasons for the massive and highly regulated droplet movements in early embryos remain largely a mystery. Under laboratory conditions, the exact global distribution of lipid droplets is not critical for survival since mutations that disrupt net inward transport in Phase IIa [86, 87] or net outward transport in Phase III [57, 105] allow embryos to develop into viable and fertile adults. Because the major net inward transport of droplets occurs just before cellularization, it is tempting to speculate that the droplets need to be moved out of the way for this massive and highly synchronous assembly of new cell membranes to occur correctly. However, in halo mutants, lipid droplets fail to display net inward transport in Phase IIa (Fig. 6E), yet no defects in cellularization have been observed [86]. The overall pattern of net inward and outward transport (Fig. 2C) is similar among many different Drosophila species and even in house flies (cited in [15]), suggesting that this process has been conserved for at least one hundred million years of evolution. This conservation strongly suggests that regulated redistribution of lipid droplets is somehow advantageous for long-term survival in nature. Yet, the only organismal consequences of aberrant droplet distribution that has been noted so far is a subtle change in histone gene expression, when lipid droplets fail to move back into the periphery in Phase III [15]. For studying the mechanism of droplet motion, the lack of strong organismal phenotypes is beneficial, because this feature makes it possible to disrupt the transport system genetically, even severely, without having to be concerned about secondary effects of lethality.
There is an additional puzzle: Why do the droplets move bidirectionally rather than unidirectionally? If the main purpose of droplet transport is to change the intracellular distribution of droplets, achieving this net transport via slightly imbalanced back-and-forth motions appears inefficient and energetically incredibly costly. This puzzle also applies to other example of bidirectional transport along microtubules, a major class of intracellular motion [79, 80]. One possibility is that bidirectional motion enables quick changes of transport direction and makes it possible to set up graded intracellular distributions rather than to simply deliver the cargo to a fixed destination. Bidirectional transport may also increase the rate of physical encounters with other organelles and thus might facilitate the exchange of proteins and lipids between compartments. For lipid droplets in embryos, the extensive bidirectional motion may allow droplets to deliver refugee proteins throughout the embryonic periphery or soak up toxic lipids and proteins, sweeping through the cytoplasm like a vacuum cleaner. Although intriguing, these ideas are currently difficult to test, in part because it is not yet possible to abolish droplet motion selectively: The only described example that completely abrogates bidirectional droplet transport in the embryo is genetic ablation of kinesin-1; but kinesin-1 has many other functions, and such embryos fail to cellularize and die in mid-Phase II [76], which presumably masks more subtle effects of the absence of droplet motion.
Droplet motion is not a peculiarity of Drosophila. Lipid droplets move in many cells, from fungi to humans [24]. Droplet motion has been linked to nutrient transport, biogenesis and breakdown of lipid droplets, protein and lipid exchange between cellular compartments, and even the maturation of viruses. In most of the cases that have been analyzed in detail, droplet motion depends on microtubules, like in Drosophila embryos [24], but in some cases, droplet motion is partially or entirely driven by actin-based mechanisms [50, 92, 202], and droplet movements in fission yeast have been proposed to be entirely due to Brownian motion [203].
IX. The droplet motors
The droplet plus-end motor: Kinesin-1
Kinesin-1, also called conventional kinesin, is a well-characterized molecular motor that uses the hydrolysis of ATP to “walk” along microtubules. Kinesin-1 is a hetero-tetrameric molecule, made of two heavy chains (Khc) and two light chains (Klc). The motor domain, encoded by the heavy chain, binds to microtubules and has ATPase activity, while the tail of the motor, formed jointly by heavy and light chains, attaches to the cargo to be transported. Kinesin-1 is responsible for a wide range of intracellular transport processes [204]; it powers the transport of RNAs, mitochondria, and vesicles, in animals from squid to flies to mammals. It has particularly prominent roles in axonal transport, and its malfunction has been linked to neurodegenerative diseases such as Alzheimer's Disease [205].
Multiple lines of evidence demonstrate that kinesin-1 also drives the plus-end motion of embryonic lipid droplets (Fig. 2D). Embryos lacking the kinesin heavy chain (Khc) entirely display dramatic disruption in droplet motion: there is no net inward transport in Phase IIa, and at the single particle level all directed motion is abolished [76]. This complete lack of motion, in the plus- as well as the minus-end direction, likely reflects motor matchmaking [206], the little understood interdependence of the opposing motors on bidirectionally moving cargoes (see below), but might potentially indicate that kinesin-1's role is indirect, e.g., being responsible for earlier assembly of the motor machinery during oogenesis, and that another motor powers droplet motion. However, both Khc and Klc can be detected by western analysis in purified droplet preparations [76, 85], and association of Khc and droplets was observed after immunostaining of purified droplets [58]. In addition, acute kinesin-1 inhibition via antibody injection into the embryo disrupts droplet motion within minutes [76], and Khc alleles that impair, but do not abolish kinesin-1 function support droplet motion, but with altered characteristics [76]. Finally, reduction in Khc levels results in concomitant reduction of droplet stall forces [76]. Taken together, these observations argue convincingly that kinesin-1's effect is direct and that kinesin-1 is the motor responsible for plus-end motion of lipid droplets. And although for some cargoes the kinesin heavy chain can function without the light chain, e.g., in mitochondrial transport [207], droplet transport requires the canonical kinesin-1 since Klc is present on droplets and klc mutants abolish droplet motion [76].
How kinesin-1 is linked to its cargoes is well established in some cases [208, 209]. In contrast, the molecular machinery that links kinesin-1 to lipid droplets remains unknown. However, how much kinesin-1 is attached apparently depends on the total amount of kinesin-1 present in the embryo: Reduced Khc dosage results in lower Khc protein levels both in the embryo as a whole and on lipid droplets [76].
The available evidence suggests that kinesin-1 also powers plus-end motion of lipid droplets in other systems. Lipid droplets isolated from rat liver display active motion along microtubules in vitro, mostly in the plus-end direction [122]. Since western analysis detects kinesin-1 on these purified droplets and a peptide inhibitor of kinesin-1 reduces droplet motility in a dosage-dependent manner, droplet motion is apparently largely driven by kinesin-1 [122]. Using mass spectrometry, kinesin light chain had already been identified as a candidate droplet protein of rat hepatocyte lipid droplets [210]. HEK293 cells, derived from human embryonic kidney cells, display clustered lipid droplets when PLIN1 is ectopically expressed, and in response to increased cAMP levels these droplets disaggregate and disperse, in a process accompanied by bidirectional transport along microtubules [211]. These droplets show significant colocalization with kinesin-1 family members. Finally, lipid droplets in COS-1 cells, fibroblast-like cells derived from African green monkey kidneys, also show active motion, and knockdown of casein kinase 2 (CK-2) severely reduces droplet stall forces, as measured by in-vivo optical trapping [121]. As CK2 can bind to and activate kinesin-1 in vitro [121] (by inducing a conformational change [212]), these observations are consistent with the notion that kinesin-1 also powers droplet transport in COS-1 cells. Proteomic analysis identified CK2 in preparation of purified Drosophila embryonic lipid droplets [15], though its functional significance has not yet been tested.
The droplet minus-end motor: cytoplasmic dynein
Cytoplasmic dynein is structurally more complex than kinesin family members: it contains two heavy chains (responsible for force production), intermediate chains and numerous light chains [213]. Like kinesin-1, it uses the hydrolysis of ATP to walk processively along microtubules. In vivo, cytoplasmic dynein acts in a dizzying range of processes, from chromosome segregation to nuclear positioning to the transport of myriads of cargoes, including lysosomes, endosomes, mRNA particles, mitochondria, and viruses. In fact, cytoplasmic dynein is responsible for the vast majority of all minus-end cargo transport in cells. Although in vitro dynein displays robust motor activity on its own, in vivo it requires a large array of cofactors, including Dynactin, Bicaudal D, and Lis-1; individual cofactors contribute to different subset of dynein's cellular roles. Some cofactors target dynein to specific cargoes [213], while others modulate dynein's force production [214].
There is good evidence that cytoplasmic dynein also powers the minus-end transport of lipid droplets in Drosophila embryos (Fig. 2D). On the one hand, dynein intermediate chain is present on droplets, as judged by immunostaining of squash preparations of embryos [77] or purified droplets [58] as well as by western analysis of purified droplets [87]. On the other hand, mutations in dynein heavy chain as well as in the known dynein regulators BicD and Dynactin alter specific parameters of droplet motion [77, 83, 84]; in addition, severe disruptions of dynein heavy chain and of BicD also prevent net minus end transport in Phase III, resulting in abnormally transparent embryos [77, 83]. Since dynein plays essential roles in oogenesis [113], it has not been possible to examine how full disruption of dynein function might impact droplet motion. However, acute disruption of dynein activity via injection of function-blocking antibodies is in principle possible and has been used to study the role of cytoplasmic dynein in RNA localization in early embryos [215, 216].
Given the many distinct roles of dynein, it is not unexpected that there are many different ways to link dynein to cargo [213]. In some cases, dynein subunits directly contact proteins on the cargo, though more frequently dynein partners act as the linking proteins, such as Dynactin or others. For lipid droplets, very little is known about the linking mechanism. Dramatic reduction of BicD levels results in much fewer droplets whose motion can detected, suggesting that BicD contributes to the recruitment of motors to droplets [83]. This is consistent with the observations that mislocalization of BicD to other organelles can ectopically recruit dynein there [217]. However, it is unlikely that BicD is the sole factor linking dynein to droplets, since there is no correlation between droplet levels of dynein and BicD. In particular, in Phase III wild-type embryos BicD is undetectable on droplets, even though motion in both directions is robust and dynein is readily detected [83]. It has been speculated that BicD may participate in the initial recruitment of dynein, but – once linkage has been established – BicD is not longer needed for dynein binding, but instead serves to regulate dynein's activity [83].
Cytoplasmic dynein is also implicated as a lipid-droplet motor in other systems. Dynein intermediate chain has been identified by proteomic analysis in lipid-droplet preparations purified from mouse mammary glands [218] and was found, using western analysis, to be associated with droplets immunopurified from NIH 3T3 cells, a mouse embryo fibroblast cell line [219]. Cytoplasmic dynein also colocalizes with droplets in HEK293 cells expressing PLIN1 [211]. In hepatocytes, lipid droplets move bidirectionally [220], and expression of the Hepatitis C core protein induces droplet relocalization towards the microtubule organizing center (MTOC), i.e., the minus ends of microtubules [221]. Dynein appears to be one of the motors involved as microinjection of antibodies against the dynein intermediate chain prevents relocalization to the MTOC [221]. Microinjection of similar antibodies into NIH 3T3 cells compromises lipid-droplet fusion [222]; analysis of a cell-free system derived from 3T3 cells suggests that under these conditions droplet fusion requires microtubules and dynein activity [219]. In these cells, recruitment of dynein is under the control of ERK2 (extracellular-signal-regulated kinase 2) [222]. Finally, overexpression of the Dynactin subunit dynamitin to disrupt Dynactin function abolishes the bidirectional motion of lipid droplets in fibroblasts [223].
How many motors?
In vitro, optical trapping has been used extensively to study the force production by microtubule motors and to determine the number of motors active per cargo [224] (Fig. 3I). Lipid droplets were the first endogenous microtubule-motor cargo for which precise measurements of in-vivo stall forces became possible [57]. These measurements revealed a force balance between plus- and minus-end motion, and that the maximal force produced was developmentally controlled: relatively low in Phase I, much higher in Phase IIa, and intermediate in Phase III [57]. Refinement in technology made it possible to measure the stall force on individual droplets, even repeatedly on the same droplet [76] (Fig. 3A,E). These measurements result in distinct, equally spaced force peaks, suggesting that they read out multiple motors, similar to in vitro measurements (Fig. 5E). They also suggest that most droplets are actively moved by one or two motors, though peaks corresponding to three motors have been observed and some droplets escape the trap, consistent with even higher motor numbers [76, 85]. Once stalled, droplets typically fall back to the center of the trap, but then frequently start moving again (Fig. 3E). By focusing on droplets that switched direction after such a stall, it was possible to compare the forces generated by the same droplet for both directions of motion: in the vast majority of cases, forces were similar, suggesting that forces are not only balanced for the entire droplet population [57, 76], but even at the single droplet level [59].
For motors moving cargo without much external resistance, motor number has only slight effects on the speed of transport. In fact, more motors can lead to slower transport [225]. However, if there is significant drag or an opposing force, then motor number has considerable effect on transport, with more motors going increasingly faster. Thus, under certain conditions, velocities might serve as a proxy for motor numbers, circumventing the need for direct force measurements. In many systems, transport shows distinct velocity classes, for example for peroxisomes and axonal vesicles, and these have been interpreted as representing different numbers of active motors. For axonal cargoes, for example, reduction in kinesin levels indeed causes a shift to slower peaks, consistent with this model [226]. However, for embryonic lipid droplets, this does not seem to apply [76, 227]: in particular, when the kinesin dosage was reduced, it resulted in lower forces produced during plus-end travel, indicating that indeed fewer kinesins were active per droplet. However, this reduction in the number of active motors did not result in slower motion of droplets [76]; instead, droplet velocity was slightly increased [76], consistent with a theoretical model that more motors can slow down transport [225].
In a number of mutant backgrounds, the measured droplet stall forces are altered relative to the wild type. In embryos lacking Klar, forces are dramatically reduced, to a similar degree for both directions of motion [57]. In contrast, in mutants lacking Halo or with defective Dynein/Dynactin or with reduced levels of the kinase GSK-3, the forces in the two directions become imbalanced [85, 86, 89]. Such changes in force might arise because the number of active motors has changed, because the force produced per individual motor is altered, or because opposing motors compete with each other (Fig. 6A). Since the studies for dynein, Dynactin, Halo, and Klar employed older technology and could only report maximal stall forces of the droplet population as a whole, it is difficult to distinguish between these models given the available data. However, for the GSK-3 mutants, it was clearly shown that the location of force peaks was not changed, but that droplets moved by 3 or more motors are more frequently observed, implying an increase in the number of motors active per droplet [85].
Can droplet motion be explained by a tug of war?
For many cases of bidirectional transport, there is good evidence that – even as the cargo undergoes excursions in both directions – plus-end as well as minus-end directed motors are always present on the cargo. If all of these motors are actively engaged with the microtubule track, their simultaneous presence suggests that they mechanically compete with each other and that at any one moment the direction of transport might depend on which set of motors is stronger or more active. Indeed, many features of bidirectional transport can be explained by such a tug-of-war between opposing motors [228]. Extended periods of uninterrupted motion in one direction can still arise from such a tug-ofwar (Fig. 5C): Even if motors are initially competing against each other, this competition will quickly be resolved if a force imbalance is present: motors have an increased tendency to fall of their tracks if they have to work against an opposing force (but see [59, 119, 229]), and thus the non-favored direction will more quickly lose active motors, leading to a (brief) interval of unrestricted motion in the opposite, favored direction (see also [230]).
These ideas have been developed into detailed quantitative models that predict the behavior of bidirectionally moving cargoes from the known – or in principle measurable – properties of the individual motors [228, 231]. These models accurately describe important properties of a number of bidirectional transport processes, qualitatively and even quantitatively [125, 232, 233]. In other cases, however, these models break down, failing to quantitatively account for critical motion properties [229], a failure that suggests that beyond motor competition there exists some kind of active motor regulation. Currently, there is a lively debate in the field to understand to what extent tug-of-war by itself explains bidirectional motion and for which cargoes. For an excellent recent review, see [231].
For embryonic lipid droplets, genetic impairment of cytoplasmic dynein results in mild inhibition of minus-end motion, but much stronger inhibition of plus-end motion [84]. At first glance, a straightforward prediction of a tug-of-war model is that impairment of the minus-end motor should relieve competition and enhance plus-end motion. Since just the opposite was observed, these results were initially interpreted as excluding a tug of war [84]. However, predicting how altered motor parameters affect the outcome of a tug-of-war is not simple, and detailed quantitative modeling revealed that even in a tug-of-war situation impairment of dynein could, in principle, cause the observed changes in plus- and minus-end motility [228]. This match, however, has to be taken with a grain of salt since it requires that dynein's single motor properties were affected by the mutants in a very specific manner, and those parameters have yet to be measured for these particular mutants.
Although tug-of-war models can describe many of the features of droplet motion, sometimes with impressive quantitative agreement [228], there is doubt as to what extent the originally published models realistically capture the properties of the motor machinery. In particular, explicit and implicit assumptions of those models have been challenged [231]: about how motor detachment from the microtubule tracks depends on the opposing force, about properties of the motor-cargo tether, and about synchronized stepping of multiple motors. Second-generation models addressing these criticisms were developed, incorporating experimentally measured detachment rates, and one class of models was found to describe unidirectional multi-motor transport highly accurately [225, 229]. This stochastic model was then extended to develop a refined tug-of-war model of bidirectional transport. Despite extensive fitting of parameters and allowing for broad deviations from the parameters measured in vitro, this model failed to reproduce critical aspects of droplet motion as observed in vivo, and in particular failed to predict correctly the previously experimentally determined response of the system to reduction in kinesin-1 levels [76]. These discrepancies were not just quantitative, but failed to capture important qualitative features; it was therefore concluded that unregulated tug-of-war cannot explain lipid-droplet motion and that a regulated higher-order switching mechanism must control transport in vivo [229].
Evidence against a pure tug-of-war mechanism for droplet transport also emerges from a detailed analysis of stall force measurements in vivo [59]. After being stalled by an optical trap, droplets typically fall back to the center and start moving again, presumably after reattaching to the track randomly. If so, a tug-of-war model predicts that – for equal numbers of motors – the direction of motion afterwards should be equal and independent of the direction of motion before the stall. However, in vivo, there was a very strong bias for droplets to keep moving in the direction they pursued before the stall [59]. These observations argue for a “memory” of directionality and are consistent with a mechanism that allows only one set of motors to be active at any one time [59].
Coordinators and switches
In summary, there is accumulating evidence that a tug-of-war mechanism is inadequate to fully explain droplet motion, and higher levels of regulation have been invoked [76, 77, 84]. The mechanistic basis for this regulation remains highly speculative, but there are several hints about what sort of control has to be involved and which molecules might contribute to it.
For lipid-droplet motion, two distinct, but related concepts have been proposed to capture critical features of this inferred regulation. First, the activity of opposing motors is controlled such that while one set of motors is active, the other set of motors is off [57]. Such “coordination” prevents competition between motors and thus avoids a tug of war (Fig. 5D). Second, since the inherent processivity of motors in vitro is higher than the observed travel lengths in vivo, a “switch” mechanism actively ends travel in one direction and simultaneously turns on travel in the opposite direction [77] (Fig. 5B).
Coordination implies that there is a machinery that can detect the activity of one set of motors and, in response, controls the activity of the opposing motor (Fig. 5D). Coordination, thus, prevents a tug of war, and if the motors on lipid droplets are indeed coordinated, it would provide an explanation for why tug-of-war models have proven inadequate to describe droplet motion. In particular, coordination reduces – or eliminates – periods in which droplets are stalled due to motor competition; such periods are frequent for tug-of-war models (Fig. 5C, panel 1), substantially more frequent than observed in vivo [229]. While direct evidence for a coordination mechanism is as-of-yet lacking, two molecules, Dynactin and Klar, have been proposed to act as coordinators, based on mutant phenotypes that suggest enhanced competition between motors (e.g., reduced stall forces and travel velocities) [57, 77] (Fig. 6A). Consistent with the idea that a coordinator should somehow bridge between the opposing motors, Dynactin has been shown, in other systems, to bind to cytoplasmic dynein as well as to plus-end motors [234]. Similarly, Klar shows, direct or indirect, physical interactions with kinesin-1 in oocytes [235], and its likely ortholog in C. elegans, Unc-83, [236] can bind to both cytoplasmic dynein and kinesin-1 [237].
A switch mechanism was initially proposed to explain the relationship between the distance traveled by a droplet and the number of motors powering motion [77] (Fig. 5A,B). In vitro, microtubule motors like kinesin-1 and cytoplasmic dynein are highly processive (i.e., they can take many steps before falling off the track), and multiple copies of these motors greatly increase how far the cargo travels before it (and the motors) fall off the track [119, 238]. In vivo, droplet stall forces change between Phases II and III, but minus-end travel distances do not, which is unexpected if travel distances were simply controlled by motor number [77]. In addition, reduction of kinesin-1 levels using genetics reduces stall forces and the number of active motors per droplets, but – if anything – travel distances go up [76]. Finally, travel distances in vivo are much shorter than expected from the in vitro behavior of multiple motors [77]. Taken together, these observations argue that in vivo, travel distances are not determined by the inherent processivity of the motors, but that periods of uninterrupted travel in a given direction (“runs”) are cut short by a different mechanism, a mechanism that is apparently insensitive to motor number (Fig. 5B). Because typically runs in one direction are followed immediately by runs in the opposite direction (i.e., pauses are relatively rare [77, 78]), this mechanism was termed a switch: it turns one set of motors off and, simultaneously, turns on the opposite set of motors.
The molecular basis for the switch is unknown. In principle, runs could be ended by competition from the opposing motors. In tug-of-war models, runs are indeed much shorter than for the same number of motors engaged in unidirectional transport, on the same order as observed in vivo [228, 229]. However, since tug-of-war is not compatible with other aspects of motion [229], additional mechanisms must (also) be at work. The mechanisms may include the activities of the motor regulators Dynactin, Klar, BicD, and LSD-2 (Fig. 5F): these molecules have been shown to be present on lipid droplets and when they are absent or impaired run lengths are changed.
Motor co-dependence
One of the deficiencies of pure tug-of-war models is the observation that in many instances, the opposing motors appear to depend on each other for activity [231]. In a simple tug-of-war situation, lack of one set of motors should enhance motion in the opposite direction, as competition is relieved. This enhanced motion has indeed been observed in many instances; however, in numerous other cases, the opposite was observed: inhibiting the motor for one direction simultaneously impairs motion in the opposite direction (for example, [239-242]). This phenomenon has been referred to in the literature by multiple names, including motor coupling, motor matching, and the paradox of co-dependence [76, 206, 231]. The underlying mechanism remains unclear, though multiple models have been proposed [179]. For peroxisomes in Drosophila cultured cells, it was shown that the ability of the motor to generate motion was critical for the activation of the opposing partner and that molecularly quite different motors could substitute for each other [243]. These results imply that the ability of one motor to produce force is somehow transduced into activation of the opposite-polarity motor attached to the same cargo.
The motors on embryonic lipid droplets show a similar co-dependence. If kinesin-1 is genetically depleted, the droplets fail to show any directed motion (neither towards the plus nor the minus ends) and are not even tethered to their microtubules tracks; instead they display diffusive behavior [76]. Simple reduction in kinesin-1 activity (by reducing the dosage of the gene encoding Kinesin heavy chain) resulted in a parallel reduction in the forces for minus-end travel [76], suggesting that this interdependence leads to a delicate matching of motor activity in the two directions. Whether this interdependence is reciprocal (i.e., whether loss of dynein activity results in a corresponding reduction of plus-end motion) has not yet been investigated.
There are intriguing hints that the mechanism that matches motor activity acts in a time-delayed manner. While chronic ablation of kinesin-1 during oogenesis results in lack of lipid-droplet motion in both directions, acute inhibition of kinesin-1 by antibody injections leads to a net minus-end shift of the droplet population [244]; these results imply that cytoplasmic dynein is still active even as kinesin-1 has ceased to function. In a similar experiment, function-inhibiting antibodies were injected into mammalian cultured cells to acutely interfere with cytoplasmic dynein; lysosomes and late endosomes displayed immediate dramatic reduction in minus-end motility, while plus-end motility also decreased, but with a much longer time course [245].
X. Components of the droplet transport machinery
The dynein cofactor Dynactin may coordinate the opposing motors
Dynactin is a large protein complex that acts as a critical cofactor in essentially all processes powered by cytoplasmic dynein. In particular, dynactin helps target dynein to specific intracellular locations, can recruit dynein to cargoes, and promotes increased run lengths for travel along microtubules [246, 247]. Dynactin has an elaborate rod-and-sidearm structure and is made up of 11 different polypeptides, some present in multiple copies. It engages in a dizzying array of physical and functional interactions: it can directly bind to microtubules, to various motors, and to other dynein cofactors. Binding to microtubules and to dynein (in particular, the dynein intermediate chain) is mediated by dynactin's p150Glued subunit.
Given its role as essential dynein cofactor, it is not surprising that dynactin also participates in lipid-droplet transport in embryos, though insights into its role here are limited. Dynactin has been detected on purified droplets by immunostaining [58]. In embryos expressing a dominant-negative form of p150Glued, minus-end motion is somewhat compromised, but plus-end motion is much more severely affected [84]. In particular, stall forces in the minus-end direction are normal, suggesting that dynein can pull the cargo unimpeded; however, stall forces in the plus-end direction are reduced. These observations lead to the proposal that wild-type dynactin keeps dynein off while the plus-end motor is active, i.e., that dynactin acts as a coordinator (Figs. 6A, 5D). Later work on dynactin's role in axonal transport supports this model: lack of the critical dynactin subunit Arp1 (actin-related protein 1) did not reduce dynein attachment to membranous cargo, but severely compromised motion in both minus- and plus-end directions [248].
How dynactin might promote motor coordination is still unclear. However, recent in-vitro work identified two domains in p150Glued with distinct effects on cytoplasmic dynein: one domain stimulates dynein processivity; the other activates a diffusive state in dynein characterized by back-and-forth drift along the track and minimal force production [249]. It was proposed that this latter ability of dynactin to turn off dynein may minimize the occurrence of tug-of-war interactions during bidirectional transport. The proposed concept of a coordinator argues that motors are turned off when the opposite-polarity motors are active (Fig. 5D), implying that the coordinator can somehow monitor the activity of the opposing motors. Consistent with this idea, dynactin can not only bind to cytoplasmic dynein, but also to plus-end motors [234], though to my knowledge direct interactions of dynactin with kinesin-1 in particular have not yet been reported.
Glycogen synthase kinase controls plus-end force production
Glycogen synthase kinase 3 (GSK-3) is a serine/threonine kinase with diverse cellular roles, and proposed substrates range from transcription factors and signaling proteins to metabolic enzymes and microtubule-associated proteins [250]. Studies using mammalian cells, squid axoplasm, and Drosophila larvae have uncovered a critical role of GSK-3 in axonal transport, in particular as regulator of kinesin-1 [85, 251, 252]. In Drosophila segmental nerves, GSK-3 acts as negative regulator of the transport of amyloid precursor protein (APP) vesicles, as reduction of GSK-3 levels lead to an increase in plus-end (kinesin-1 mediated) directed motion [85]; in contrast, plus-end motion of synaptic vesicle precursors (SVP vesicles), powered by a different member of the kinesin superfamily, is normal. Intriguingly, minus-end (cytoplasmic dynein-mediated) directed motion of APP vesicles [85] was also increased. Because dynein-mediated transport of SVP vesicles was unaffected, it was proposed that the minus-end effects for APP vesicles arise indirectly, via misregulation of kinesin-1.
GSK-3 is maternally contributed to the early Drosophila embryo [253], and in embryos from mothers with reduced GSK-3 dosage, lipid-droplet motion is significantly stimulated [85]. In those mutants, run lengths and velocities increase for both the plus-end and minus-end travel direction; in addition, stall forces in the plus-end – but not in the minus-end – direction are higher than in the wild type. Thus, like in the axons, GSK-3 acts as a negative regulator of transport. It remains to be determined whether, as proposed for axonal vesicles, the primary target of regulation is kinesin-1 and dynein properties are altered as a consequence. It has not been reported whether these changes in droplet motility affect the global distribution of droplets or the normal developmental sequence of net in- and outward transport.
Although plus-end stall forces are only modestly increased upon GSK-3 reduction, they are significantly different from the wild type and higher than those reported in any other mutant [85]. The increase in stall forces was not due to altered force production at the single-motor level; rather, in the mutants, the droplets are – on average – pulled by a higher number of active kinesins. The authors did not detect changes in the levels of kinesin on biochemically purified droplets, suggesting that GSK-3 does not alter docking of kinesin to droplets, but rather the activity state of the motors once docked [85] (Fig. 6A). Such activation after docking represents a new mechanism for regulating bidirectional transport.
The molecular mechanism underlying this negative regulation by GSK-3 remains unknown. It was proposed that GSK-3 might increase the rate at which motors detach from the tracks by weakening microtubule-motor interactions [85], either by modification of the motor or a motor co-factor or by changing the amount of microtubule-binding proteins attached to the tracks. It will be critical to identify the biologically relevant targets of GSK-3 among the droplet-transport machinery.
Klarsicht, a proposed motor coordinator and switch component
Loss-of-function mutations in klarsicht prevent net minus-end transport in Phase III, resulting in embryos with few lipid droplets in the periphery [57, 105]. These embryos therefore have an unusually transparent periphery; this phenotype lead to the name for the gene: the German word “Klarsicht” means “clear view”. Biophysical analysis revealed severe defects in droplet motion from Phase I through III, suggesting that Klar is a crucial component of the motor machinery [57]. It was proposed that Klar protein physically interacts with both the plus- and minus-end motors on droplets and coordinates their activity [57]. The Klar protein indeed localizes to lipid droplets [105] and, at least in oocytes, is complexed with kinesin-1 [235].
Klar `s role is not restricted to lipid-droplet motion in embryos. Klar is also involved in several nuclear positioning events: Klar is required for nuclei in larval photoreceptors as well as developing cone cells to migrate from basal to apical positions [254, 255], for the multiple nuclei in embryonic muscle cells to arrange in two parallel rows [256], and for nuclei to spread equally throughout larval muscle cells [256]. Klar is also important for the proper growth of the apical membrane in embryonic salivary glands and has been proposed to modulate dynein-mediated delivery of secretory vesicles [257]. In mid-stage oocytes, Klar modulates the kinesin-1–driven transport of RNP particles, restraining their motility; in the absence of Klar, oskar RNPs are delivered prematurely to the posterior pole and cannot be anchored correctly to the oocyte cortex [235]. In testes, Klar is necessary for the asymmetric sister-chromatid segregation of sex chromosomes [258, 259]; it was proposed that sister-chromatids in the nucleus are linked to centrosomes via cytoplasmic microtubules and Klar-containing LINC complexes in the nuclear envelope. And in a fly model of the neurodegenerative disease Amyotrophic Lateral Sclerosis (ALS), reduction in Klar expression potently suppressed the disease phenotypes [260]. Although the nature of this suppression has yet to be characterized at the molecular level, it is provocative than many other genetic modifiers recovered affect proteins physically or functionally connected to lipid droplets [260]. Furthermore, indirect evidence suggests that Klar may also function in branch migration in trachea [261], in wing development [262], in the remodeling of neuroendocrine cell [263], and in starvation stress resistance [264]. Finally, in cultured cells, Klar suppresses microtubule shrinkage events and promotes microtubule stability [265]. Although the molecular mechanism by which Klar functions remains obscure, in many cases it has been proposed to somehow link cargoes (vesicles, nuclei, lipid droplets, RNP particles, chromosomes) to microtubules and/or microtubule motors.
Klar is encoded by a complex locus (Fig. 6B). The klar locus spans ~100 kb in the genome [255], and encodes messages for five different isoforms: α, β, γ, δ and ε [266]. These messages are generated by the use of three distinct promoters and regulated 3’ end formation [105]. The most 3’ exon of klar is present in only a subset of isoforms (α, γ, and δ) and encodes a C-terminal KASH domain, a signature of the mammalian nesprins and their invertebrate orthologs. KASH domain proteins are present in the outer nuclear envelope and are crucial components of LINC complexes that connect the cytoskeleton to the nucleoskeleton [236, 267, 268]. In animals, these proteins are typically encoded by complex loci that also give rise to KASH-less isoforms localized to the cytoplasm where they modulate interactions between the cytoskeleton and various organelles, including mitochondria [256, 269], RNP particles [235, 270], and lipid droplets [105]. Although for many Klar functions it remains to be teased out which isoform(s) play a role, it is clear that nuclear migration in larval photoreceptors depends on the α isoform [255, 271], where it works together with the SUN protein Klaroid [254], its partner in the inner nuclear envelope, and with nuclear lamins [271]. In contrast, motion of RNP particles in oocytes and of lipid droplets in embryos is regulated by the β isoform [56, 105, 235, 266].
In embryos, Klar β localizes to lipid droplets, as shown by immunolocalization in intact and centrifuged embryos [105]. Many other droplet proteins are present all over the droplet surface, including LSD-2, Jabba and histones [15, 16, 87]. In contrast, Klar is concentrated in one or a few dots per droplet [105], a distribution resembling that of dynein [77]. Klar's droplet localization does not require Halo [105], Kinesin-1 [76] or Jabba [16]; thus those proteins cannot be the essential link between Klar and droplets. Droplet localization is mediated by Klar β's C-terminal LD domain, a domain shared with isoform ε, but absent from the other isoforms (Fig. 6B). This domain is both necessary and sufficient to localize Klar to lipid droplets, in embryos as well as in cultured cells [56, 105]. The 114 aa LD domain is conserved across Diptera and contains a putative amphipathic helix [56]. Amphipathic helices are known to mediate attachment to lipid droplets in other droplet proteins [2, 26], likely by direct interactions with the hydrophobic core. Whether it is this helix that mediates Klar's droplet localization remains to be tested. In mid-stage oocytes, however, Klar β has not been detected on lipid droplets, even though lipid droplets are abundantly present; Klar β is instead associated with RNP particles, and its intracellular distribution is independent of the LD domain [235]. Whether this switch in targeting is due to functional inactivation of the LD domain or absence of a partner on lipid droplets is not yet known.
Apart from the KASH domain, the primary sequence of Klar is not obviously conserved beyond arthropods. However, functional comparisons suggest that Klar α is the Drosophila analog of mammalian Nesprin-4 and C. elegans Unc-83 [236], which also regulate specific nuclear migration events. This parallel might provide clues into Klar's molecular function during lipid-droplet transport as both Nesprin-4 and Unc-83 physically interact with kinesin-1 and/or cytoplasmic dynein [237, 272]. Klar β and kinesin-1 can indeed be detected in common complexes in ovaries [235]. Nesprin-4 and Unc-83 have been proposed to anchor the motors to the nuclear envelope or possibly coordinate the activity of the opposing motors [237, 272]. Similarly, Klar α has been suggested to anchor dynein to the nuclear envelope and thus promote correct nuclear migration in photoreceptors [271, 273]. In photoreceptors, disruption of dynein function indeed results in a similar disruption of nuclear positioning as lack of Klar [274]. However, such an anchoring function of Klar α has not yet been directly demonstrated. And for lipid droplets, motor anchoring cannot be exclusively mediated by Klar since in embryos lacking Klar β (and Klar α) entirely, lipid droplets still move bidirectionally [57, 105], and thus must have both plus- and minus-end motors attached.
Biophysical characterization of droplet motion reveals severe disruptions in the absence of Klar: droplets move for shorter distances, with reduced speeds, and with greatly reduced forces. The defects are observed in all phases examined and for both directions of motion, indicating that the function of both kinesin-1 and cytoplasmic dynein are greatly impaired [57]. How Klar acts as a crucial co-factor for both motors has not yet been determined. But the severe disruption in both directions can be explained if absence of Klar induces a tug-of-war that is normally avoided in the wild type: motor competition, for example, would reduce the net force for transport in a given direction and thus might explain the extremely low stall forces measured in klar mutants [57] (compare Figs. 5C and 5D). As described above, there is currently an ongoing debate to what extent motors in bidirectional transport engage in a tug-of-war. This debate remains to be resolved, but if the presence of Klar indeed allows opposing motors to avoid a futile tug-of-war then Klar is a good candidate for the proposed, but so far elusive coordinators, i.e., molecules that keep one set of motors “off” while the opposing motors are actively transporting the cargo (Figs. 5D, 6A).
The effect of lack of Klar on motility has also been determined for RNP particles in oocytes [235]. Oskar RNPs are transported to the posterior pole of mid-stage oocytes by kinesin-1 [275, 276]. For these RNPs, lack of Klar is just the opposite of that observed for lipid droplets: run lengths in the klar loss-of-function mutants are longer, relatively more particles move, and net transport to the posterior is enhanced compared to the wild type. These observations suggest that Klar is a versatile regulator of motors and can, depending on context, act either as positive or negative regulator of run lengths (Fig. 6C). For example, Klar may have an inherent ability to restrain kinesin-1 activity, and thus for the mostly unidirectionally moving RNP particles absence of Klar results in longer runs. For lipid droplets, this ability to restrain kinesin-1 may only be activated when the opposing motor is engaged with the microtubules; as a result of lack of Klar, kinesin-1 would remain inappropriately active during dynein-based movements, impairing minus-end motion. Whatever the detailed molecular mechanisms, the analysis of the droplet and RNP particle motion in klar mutants, when taken together, suggest that Klar is an integral component of the postulated switching mechanism that turns motor activity on or off.
The analysis of RNP transport uncovered another surprising feature of Klar β-based regulation: Lack of Klar causes much more severe defects in the posterior localization of RNPs at 18°C than at 25°C [235]. It was proposed that Klar adjusts the rate of RNP delivery to compensate for temperature-induced changes in other linked cellular processes, such as translation and anchoring, so that they remain balanced. If this model is correct, Klar is one of the very few molecularly identified robustness factors that allow organisms to develop consistently in variable temperature environments. Whether Klar itself is directly responsive to temperature and if other Klar-dependent processes, including embryonic droplet motion, show similar temperature compensation remains an exciting problem for the future.
For embryonic lipid droplets, lack of Klar does not only lead to severe reduction of many motion parameters, it also alters the temporal regulation of transport [57]. Unlike in the wild type, plus-end directed motion continues to predominate in Phase III in klar mutant embryos, explaining the failure of lipid droplets to move back towards the periphery. This lack of correct temporal regulation of transport may simply be a consequence of the severe disruption of motility, e.g., if the motor machinery is locked in an unnatural tug-of-war, it may not be able to respond correctly to the signals that usually modulate the temporal progression of transport. Alternatively, the failure to switch to net minus-end transport may be due to a separate function of Klar that mediates temporal regulation. These issues will likely only be resolved once the functional domains of Klar have been identified and impaired individually.
X. Temporal regulation
From Phase 0 to Phase III, lipid-droplet motility undergoes stereotypic changes that result in dramatic alterations in net transport (Fig. 2C,D). Throughout these phases, the plus-end motor kinesin-1 and the minus-end motor cytoplasmic dynein are present on droplets; for kinesin-1, it has been directly demonstrated that its levels on droplets change very little during this period [76]. Thus, there must be some non-motor molecules that cause the observed changes in motility. Of all the transitions, that from Phase I to Phase IIa is best understood; it involves new production of the directionality determinant Halo [86] and dephosphorylation of the Perilipin LSD-2 [87]. In addition, the droplet levels of the dynein co-factor BicD change dramatically between Phases [83], and these changes might be responsible for the changes in motion parameters associated with these developmental transitions.
The directionality determinant Halo promotes the transition from Phase I to Phase IIa
When transcription in the early embryo is inhibited pharmacologically, embryos develop morphologically almost entirely normally until the beginning of cellularization [277]. However, in Phase IIa, inward transport of lipid droplets fails [86], suggesting that zygotic expression of one or more genes is required to promote net plus-end transport. Using chromosomal deletions and microinjection of double stranded RNAs, it was shown that a single gene is responsible for these droplet transport defects [86, 109, 111]. This gene was named halo: in its absence, the peripheral cytoplasm remains opaque due to the high concentration of lipid droplets (Fig. 6F, left); the resulting embryos thus display a peripheral brown ring (or halo) around the dark central yolk [86]. During cycle 14, wild-type and halo mutant embryos can easily be distinguished by visual inspection. This property has been used to recognize embryos of particular genotypes at very early stages [278-281], when the use of other markers (such as GFP-based transgenes) is not practical.
Lack of Halo profoundly affects the intracellular distribution of lipid droplets, but not of any other organelle [86]. In Phase IIa wild-type embryos, plus-end run lengths are longer than minus-end run lengths, resulting in net plus-end (inward) transport (Fig. 2D). In the absence of Halo, this balance of run lengths is reversed, and lipid droplets display net minus-end (outward transport); they accumulate between and just under the nuclei at the cell surface, and are depleted from around the central yolk [86, 111] (Fig. 6E). Thus, Halo acts as a directionality determinant: without Halo, net transport in Phase IIa is minus-end directed; in the presence of Halo, plus-end directed.
Halo mRNA expression is highly dynamic [86]. By in-situ hybridization, halo mRNA is barely above background in Phases 0 and I, and is highly upregulated in Phase IIa. At the transition to Phase IIb, halo signal drops dramatically, implying rapid turnover of the message. Halo mRNA is also among a small set of mRNAs that is retained inside nuclei when damaged blastoderm nuclei are eliminated by nuclear falling [166]; the physiological significance of halo mRNA retention is unknown.
Three factors have been implicated in the regulation of halo transcription. First, the zinc-finger protein Zelda is a key activator for many genes expressed in the early zygote [282]. There are two Zelda binding sites within 200 bp of the halo transcription start site, and, in embryos lacking Zelda, halo mRNA levels are dramatically depressed (~15fold) [282]. Second, Lilliputian is an AF4/FMR2-related transcription factor. Embryos lacking Lilliputian show reduced extent of net inward droplet transport in Phase IIa [283], similar to embryos carrying only a single functional copy of halo [86]. Whether, as proposed [283], lack of Lilliputian reduces halo expression remains to be tested. Third, halo was found to be induced in early embryos by ionizing radiation in a p53–dependent manner [284], one of only 29 genes thus identified. The consequences of this upregulation for droplet transport remain to be examined.
The pattern of Halo protein expression has not yet been published, but has been inferred from functional studies (Fig. 6D). Lack of halo in the mother has no detectable consequences for transport, and thus the protein is presumably not maternally provided and not present in Phases 0 and I. As halo promotes net plus-end transport in Phase IIa, Halo protein can be inferred to accumulate then, consistent with the spike in halo mRNA levels. To explain the return to net minus-end transport in Phase III, it has been proposed that, like the mRNA, Halo protein is also rapidly degraded once Phases IIb or III start [86].
Halo is a small protein (109 aa) and belongs to a novel protein family [86, 285] of unknown function (DUF733). All sequenced Drosophila genomes contain multiple members of this family, but no representatives are apparent in other animals, including other insects or lower Diptera. Three divergent clues exist to explain how Halo might act mechanistically. 1) One family member, SNCF, has been shown to interact with the transcription factor SoxNeuro by yeast-two-hybrid assays and to enhance SoxNeuro-mediated transcription in transfected cells [285]. Thus, Halo might similarly act as transcription factor and regulate the expression of molecules that control motor activity. 2) Data from high-throughput screens available through the Drosophila Interactions Database [286, 287] suggest that other family members interact with microtubule motors. HL6 expressed in cultured cells co-precipitates with dynein components, and HL2 has a yeast-two-hybrid interaction with the dynein light chain Tctex-1. Halo might similarly physically contact the motors on lipid droplets and modulate their behavior. 3) Halo is required for the developmentally controlled dephosphorylation of the Perilipin LSD-2 in Phase II [87] (Fig. 6D). There is currently no information about whether this is a direct effect of Halo on LSD-2, but it raises the possibility that Halo acts by activating a phosphatase or inhibiting a kinase. Since droplet transport in LSD-2 mutants is less severely affected than in halo mutants [86, 87], LSD-2 is probably only one of multiple targets modified by Halo.
Overall lipid-droplet distribution is similar, if not identical, in embryos lacking Halo specifically and in embryos in which zygotic transcription has been entirely abolished [86]. This comparison suggests that Halo is the major zygotic contribution to the Phase I to Phase IIa transition. However, this transition also requires input not dependent on transcription; halo mutant embryos still display changes between Phase I and Phase IIa, both in the motility of individual droplets and in overall droplet distribution [86].
The phospho-state of the Perilipin family member LSD-2 changes in a phase dependent manner
One of the targets of Halo is the Perilipin family member LSD-2, also known as PLIN2 [87]. Perilipins play central roles in the regulation of lipid homeostasis in animals and fungi [68, 144]. Perilipins localize constitutively or facultatively to the lipid-droplet surface and control lipid metabolism and interactions with other cellular compartments. In Drosophila, there are two family members, LSD-1 (PLIN1) and LSD-2 (PLIN2), that cooperate to control overall cellular lipid content, in a partially overlapping, partially complementary manner [25, 288].
The role of LSD-2 in lipid-droplet transport was discovered by an unbiased proteomics approach [87]. Lipid-droplet proteins from Phase I, II, and III embryos were compared by 2D gel electrophoresis. One prominent spot dramatically changed in intensity and was identified as a particular phospho-isoform of LSD-2; it is the most highly phosphorylated form of LSD-2 detectable and is specifically absent in Phase II. Although the kinases and phosphates acting on LSD-2 remain to be identified, in embryos lacking Halo the Phase II-specific dephosphorylation of LSD-2 fails to occur. As the only known role of Halo is in the regulation of droplet transport [86] and in embryos LSD-2 localizes exclusively to lipid droplets [87], these findings suggest that the LSD-2 phosphorylation state plays an important role in controlling motor activity on droplets.
To critically test the role of LSD-2 phosphorylation in droplet transport, it will be necessary to identify the phosphorylation sites and probe their contribution via mutational analysis. However, analysis of embryos lacking LSD-2 entirely has already revealed that LSD-2 plays an important role in transport regulation. In such embryos, there is little, if any, net plus-end transport in Phase IIa and overall lipid-droplet distribution resembles that of Phase I. This is corroborated by tracking of individual droplets: in the mutant embryos, motion parameters change little from Phase I to Phase IIa, and run lengths remain balanced. However, in contrast to the phenotype observed in embryos lacking Klar [57] or kinesin-1 [76], travel velocities are normal and run lengths are overall of wild-type magnitude [87]. Thus, LSD-2 is not required for motor activity per se, but “simply” for correct temporal progression [87]. It was therefore proposed that LSD-2 transmits the temporal information provided by Halo to the core transport machinery.
How could LSD-2 impact motor activity? So far, we have just two intriguing hints. First, LSD-2 has a yeast-two-hybrid interaction with the LD domain of Klar β [87], a core component of the droplet transport machinery, which can, in turn, exist in common complexes with kinesin-1 [235]. Thus, conformational changes in LSD-2 might be transmitted via Klar to kinesin-1. Second, a mammalian member of the Perilipin family, PLIN2 (formerly known as ADRP; note that mammalian PLIN2 is not the direct ortholog of fly PLIN2 [25]) can co-precipitate dynein from cell lysates, and this interaction is phosphorylation-dependent [222]. If there is a similar interaction between LSD-2 and dynein, it apparently does not mediate recruitment of dynein to lipid droplets, as amounts of dynein on purified droplets are similar for wild-type and LSD-2 null embryos [87].
Perilipins are also implicated in droplet motion in other systems. In hepatocytes, the dynein-dependent relocalization of lipid droplets upon expression of Hepatitis C virus core protein is accompanied by loss of PLIN2 from droplets, and knockdown of PLIN2 is sufficient to induce similar droplet relocalization [221]. Follow-up studies showed that expression of the core protein does not alter net directionality of droplet motion, but reduces travel speeds [220]; how these motility parameters are altered by PLIN2 knockdown has not yet been reported. In fibroblasts and HEK293 cells, ectopic expression of PLIN1 causes clustering of lipid droplets near the MTOC and dispersion along microtubules in response to increased cAMP levels. Dispersion is apparently mediated by PLIN1 phosphorylation as it is abolished when PLIN1 phosphorylation sites are mutated [211, 289].
Levels of Bicaudal D change in a phase-dependent manner
Cytoplasmic dynein has a myriad of cellular roles, powering the vast majority of minus-end directed transport in the cytoplasm as well as participating in multiple steps of mitosis. To be able to recruit dynein to particular cargoes and specifically regulate its activity, the cell has a number of dynein cofactors that adapt the motor to particular cellular functions [247] and typically are involved in a subset of dynein-based processes. One such factor is Bicaudal D (BicD) [290], a protein first characterized for its role in Drosophila oogenesis and early embryogenesis (special BicD alleles result in bicaudal (two-tailed) embryos). Drosophila BicD and its mammalian orthologs, BicD1 and BicD2, have since been shown to be involved in localization of specific RNAs, transport of Golgi vesicles, certain nuclear migration events, and microtubule organization [247, 290]. As BicD can physically interact with dynein and Dynactin as well as with molecules involved in cargo binding (such as Egalitarian and Rab6), BicD may recruit dynein to the cargo or may provide one of the links by which dynein activity is controlled in a cargo-specific manner, for example by tuning the velocity of dynein-based movements [291].
Like the motors, LSD-2, and Klar, BicD is provided to the early embryo from the mother. Using a combination of BicD null alleles and transient expression from a BicD transgene, it was possible to generate embryos whose BicD levels are massively reduced (to less than 1% of the wild-type amount) [83]. In such embryos, lipid-droplet transport is severely affected: In Phase III, droplets fail to undergo net outward (plus-end) transport, resulting in embryos with a transparent periphery. Overall, much fewer droplets displayed motion, consistent with the notion that one of the functions of BicD is to recruit motors to the droplets. But even the droplets able to move and thus apparently carrying motors showed defects: Tracking of individual moving droplets revealed that minus-end run lengths were reduced in the mutant embryos in Phases IIa, IIb and III, indicating severe impairment of dynein-based motion. In addition, plus-end run lengths were reduced in Phases IIa and IIb, but not Phase III [83]. At the moment, it is unclear whether the effect on plus-end transport is an indirect consequence of altered dynein activity or represents an independent role of BicD in controlling kinesin-1. The latter possibility is supported by analysis of mammalian BicD2; it can physically interact with kinesin-1 [292] and can promote kinesin-1 activity to help properly position the nucleus relative to the centrosome prior to mitosis [293].
These studies also uncovered that high levels of BicD are critical for proper developmental regulation of lipid-droplet transport. In embryos with wild-type levels of BicD, run lengths for both directions are dramatically upregulated in Phase IIb and downregulated again in Phase III (Fig. 2D). This temporal regulation is lost in the BicD mutant embryos, and run lengths remain similar throughout [83].
Four additional pieces of evidence implicate BicD in the temporal regulation of droplet transport: First, BicD levels on purified droplets fall precipitously from Phase I to Phase II to Phase III [83] (Fig. 6D). This decrease is apparently a droplet-specific event since overall BicD levels in the embryo are fairly stable during the same time period. There is no parallel decrease in dynein levels on droplets, suggesting that most of the BicD does not simply act to tether dynein to the droplets. Second, overexpression of BicD prevents net plus-end droplet transport in Phase IIa [294], presumably because BicD droplet levels remain too high. Third, overexpression of the mRNA binding protein Egalitarian (Egl) results in excessive inward droplet transport in Phase II. Since Egl is a known binding partner of BicD and mild co-overexpression of BicD suppresses the excessive inward transport, it was proposed that excess Egl sequesters BicD, thus causing premature BicD depletion from droplets [294]. Finally, a BicD allele with a point mutation in the putative dynein-binding region displays an abnormal pattern of run-length changes during development: run lengths are reduced in Phase IIa, close to normal in Phase IIb, and increased relative to wild-type in Phase III [83].
XII. Other molecules with roles in droplet transport
Mutations in a number of molecules result in a failure of the switch in net droplet transport from Phase II to Phase III; the resulting embryos retain an inward accumulation of lipid droplets and are thus abnormally transparent during gastrulation and germ-band extension. Such disruption of “clouding” can be observed when Dynein heavy chain, BicD, Klar, Wech, or Dop are impaired [57, 77, 83, 105, 109, 295]. A clouding defect might point to problems with the temporal regulation of the switch [83], might reflect compromised dynein function [77, 84], or possibly both (as may be the case for Klar). For mutations in Wech and Dop, not enough is yet known to propose into which of these categories they fall.
The RBCC/TRIM protein Wech
Wech was identified as a candidate regulator of lipid-droplet motion in a genome-wide microarray screen for RNAs specifically upregulated around cellularization, i.e. embryonic stages around Phase IIa and IIb of droplet transport [109] (Fig. 2A). Disrupting Wech by RNA interferenceimpaired net outward droplet transport in Phase III. The transcription unit disrupted in this RNAi experiment was initially annotated as dappled, a gene involved in tumor suppression [296]. Subsequent work established that dappled mutations are due to disruption of cytochrome b5 [297]. The transcription unit linked to droplet transport is now called CG42396 or Wech [298].
Wech is a member of the RBCC/TRIM protein family. Its orthologs mediate important developmental decisions in C. elegans and mice [299, 300] and have been shown to exhibit multiple molecular activities, from RNA binding [301] and translational repression [302] to E3 ligase activity and regulation of protein stability [303]. Which, if any, of these activities are relevant for understanding the function of Drosophila Wech remains to be determined.
Although Wech is broadly expressed during development, its functional characterization has been limited. Ectopic Wech expression interferes with the development of the peripheral nervous system [304]. Knockdown of Wech by injection of dsRNAs into embryos prevents net outward transport of lipid droplets [109]. Finally, embryos homozygous for wech loss-of-function alleles die near the end of embryogenesis, with muscles detached from the body wall, apparently due to a failure of integrin-mediated cell adhesion [298]. It has been proposed that in this context Wech acts as scaffolding protein that links integrins and the actin cytoskeleton [298].
There are only limited clues regarding how Wech acts in droplet transport. In the early embryo, Wech was detected as puncta in the cytoplasm and at the membrane [298]; it will be interesting to determine if some of these puncta colocalize with lipid droplets. Wech RNA levels rise late in Phase IIa, due to zygotic transcription [296], consistent with a model that rising Wech protein levels drive the transition to Phase IIb or Phase III of droplet transport. Thus, while the phenotypic data suggest that Wech is in some way crucial for proper regulation of droplet transport, a molecular understanding is missing.
The MAST kinase Drop-out
Dop (drop-out) mutations were first identified because they induce nuclear falling in early embryos [305]. They were subsequently found to affect a plethora of processes in the early embryo, including membrane growth during cellularization, distribution of endosomes, RNA motility, and transport of lipid droplets [110, 306]. Dop also functions during eye and wing development [306]. The initial proposal that dop phenotypes are due to misregulation of the RNAi pathway component Ago2 [110] was later shown to be incorrect [295]. Rather, Dop encodes the sole Drosophila member of the Microtubule-Associated Serine/Threonine (MAST) kinase family [306], a kinase family implicated in a number of human diseases [307].
The available evidence suggests that Dop acts by controlling the activity of cytoplasmic dynein [306]. In dop mutant embryos, the phosphorylation of the dynein subunit Dynein Intermediate Chain is reduced, and single particle tracking revealed that minus-end motion of RNA particles is impaired while plus-end motion is normal. In addition, dop mutants synergize with mutants in dynein pathway components. Indeed, many of the dop mutant phenotypes can at least in principle be explained by reduced dynein function. It remains to be investigated whether Dop directly phosphorylates dynein or acts in an indirect manner.
A disruption of dynein function is also consistent with the known effects of Dop on lipid-droplet motion. In embryos from dop mutant mothers, lipid droplets fail to undergo net minus-end transport in Phase III, resulting in abnormally clear embryos [110]. The available evidence suggests that net plus-end transport in Phase IIa is normal, and that the first differences to the wild-type arise in Phase IIb [110], potentially implicating Dop in this developmental transition. An attractive possibility is that Dop mediates the transient upregulation of dynein run lengths during Phase IIb (Fig. 2D); lack of Dop would leave plus-end motion dominant and thus result in continued net plus-end motion. To test this – and other – models, it will be necessary to quantify both plus- and minus-end droplet motility at the single-particle level.
XIII. Conclusions and outlook
Lipid droplets have been noted by microscopists since the late nineteenth century (see [23]), but for many years received only sporadic attention. That changed when in the 1990s pioneering work by Constantine Londos and coworkers identified the first protein specifically localized to lipid droplets, perilipin, and provided evidence for a crucial role in the breakdown of triglycerides [308]. Now, lipid droplets are recognized as central players in lipid metabolism and energy homeostasis, with profound relevance for human health. It has also become clear that droplets are extremely dynamic organelles that play even broader roles in biology [309], including viral assembly and protein trafficking, and I suspect that new cellular and organismal roles of lipid droplets remain to be discovered.
Research in Drosophila embryos has made critical contributions to lipid-droplet research. So far, the focus has been on droplet motility and protein sequestration, two generally important phenomena that are particularly prominent in early Drosophila embryos. Although for both processes a number of molecular players have been identified (Figs. 4, 5, 6), many issues remain unresolved, promising exciting new research avenues for the future. In addition, many other aspects of lipid-droplet biology can, in principle, be addressed using Drosophila embryos, and I hope many researchers will be inspired to exploit the unique biology of this system as well as the broad range of available tools (section III; Fig. 3). Such investigations should not only yield insights into fundamental properties of lipid droplets, but they also have the potential to reveal their significance for whole organism physiology and for development.
Regulation of droplet motion
The machinery powering and controlling droplet motion is known in broad strokes (sections IX through XII): the droplets are transported by kinesin-1 and cytoplasmic dynein, and motion is regulated by dynactin, GSK-3, Klar, Halo, LSD-2, BicD, Wech, and Dop (Figs. 5F, 6A, 6D). Likely many more regulators remain to be discovered, but at the moment we do not even know how far we are from a comprehensive inventory. Thus, one of the challenges for the future will be to identify additional regulators, such as molecules that physically or genetically interact with known transport components or the orthologs of regulators characterized in other systems of bidirectional transport [79, 80, 231, 310].
However, a complete inventory will just be the beginning. It is as important to understand which of these proteins are present on lipid droplets or elsewhere, if they are localized to droplets statically or dynamically, and which other components they interact with, both physically and functionally. For example, in ovaries, kinesin-1 is present in complexes with Klar [235], and in yeast, LSD-2 and Klar interact in two-hybrid assays [87]. Do these interactions also occur on lipid droplets in embryos, do they vary between phases, and are all three molecules present in one single complex or distinct binary complexes? Similarly, do the two kinases identified, GSK-3 and Dop, act during droplet transport in embryos to modulate motor properties on lipid droplets, or do they control transport indirectly, e.g., by regulating assembly of the transport machinery during oogenesis? And do the temporal regulators identified, Halo, BicD, and LSD-2 (Fig. 6D), all act in a single pathway or do they provide (partially) independent inputs that control different motor properties or the same ones, but at different phase transitions? Answering these questions will require combining physical interaction assays with live tracking, super-resolution imaging and simultaneous genetic manipulation of multiple components.
In other systems, the identification of motor cargo adaptors has provided a framework for how regulation might occur [311]. However, apart from some evidence that BicD promotes motor loading [83], essentially nothing is known about the mechanisms that targets kinesin-1 and cytoplasmic dynein to lipid droplets. Identifying the unknown cargo adaptors has the potential to greatly clarify our understanding of droplet motion: In principle, such adaptors might control the number of motors per cargo (Fig. 6A, left), might mediate coordination (e.g., via physically contacting both types of motors), and might be the target for the action of regulators (e.g., if they contribute to the proposed switch; Fig. 5B,D).
The molecular nature of the adaptors may also illuminate how motor forces are controlled in vivo. Such forces are a read-out for the activity of the motors driving transport. For embryonic lipid droplets, these forces are intricately regulated: they first increase and then decrease as development proceeds [57]; these changes occur in parallel for the plus- and minus-end directions, so that forces remain balanced [57]. In principle, forces could be controlled by multiple pathways (Fig. 6A): First, the number of motors on the cargo might be altered by control of motor-cargo docking; indeed reduced amounts of kinesin-1 on droplets (achieved by lowered Khc dosage) results in lowered forces [76]. Biochemical analysis has so far not found any dramatic changes in the amount of kinesin-1 on droplets between phases [76], though the temporal resolution of those experiments would not have detected a transient spike in the very short Phase IIb. And the time course for dynein levels on droplets remains to be investigated. Second, the activity of some of the docked motors may be switched on and off, so that only a subset of all motors physically present are active. Some such mechanism is likely at play since forces change dramatically across phases [57], while kinesin-1 levels do not [76]. GSK-3 is involved in restricting kinesin-1 activity [85], but whether it acts directly and how kinesin-1 is turned off is unclear. As casein-kinase 2 can activate kinesin-1 in vitro [121, 212] and has been detected in droplet preparations [15], it might mediate this regulation. Third, motor coordinators may allow full force production by keeping opposite-polarity motors inactive, as has been proposed for dynactin and Klar [57, 84]. Finally, cytoplasmic dynein can operate in both a low- and a high-force state [312]. These states can be controlled by transacting factors [214], possibly acting through a C-terminal inhibitory domain in the dynein heavy chain [313]. Whether such modulation of the inherent dynein force contributes to the force regulation observed for droplets in vivo is not yet known. Because lipid droplets are one of the few in-vivo systems in which the forces driving transport can be measured, this experimental system provides a unique opportunity for dissecting the underlying mechanisms. Such an analysis will also establish whether there is a connection to the switch mechanism; available evidence has so far not revealed any link between run-length control and number of active motors [76, 77].
One of the central issues for any bidirectional transport system is understanding what controls run lengths, since the balance of runs in the plus- and minus-end directions controls the net direction of transport and global distribution of cargo. This is a difficult question, and tug-of-war models, although quite promising [228], seem not to adequately describe a number of bidirectional transport systems [231], including lipid droplets [229]. The alternative concepts of coordinators and switches, while conceptually attractive, remain so far largely speculative and vague. However, we have a genetic inroad into this problem because a number of regulators have been identified where mutations alter run lengths (Fig. 5F): Halo, Klar, Dynactin, BicD, LSD-2, GSK-3 [57, 83-87]. In addition, mutations in Dop and Wech alter net transport of droplets, and quantitative tracking of individual droplets should reveal if this is the result of changed run length. Finally, the dynein cofactor Lis1 was recently shown to play an important role in run-length control for dynein in RNA transport during embryonic stages corresponding to IIa and IIb [82]; it is not yet clear if Lis1 might also influence run length for droplet motion: overall distribution of droplets was normal, suggestive of little effect, but quantitation of the motion at the single droplet level has yet to be reported. Thus, even without a comprehensive list of proteins involved in droplet motion, there are already many potential leads available to probe the molecular nature of the switch.
Molecularly dissecting the function of the known transport regulators should provide critical clues into the mechanism of run-length control and thus to a better understanding of the proposed switch. For example, Klar and kinesin-1 exist in common protein complexes [235]. Once the molecular basis for this interaction is known (e.g., via a particular domain of Klar or a specific bridging protein), one can abolish it genetically and then ask which properties of transport are affected. If Klar is indeed part of a coordinator complex that pushes kinesin away from the track when dynein is active (Fig. 5D), then lack of the kinesin/Klar interaction should induce motor competition specifically during minus-end motion. For dynactin, a candidate domain for switching dynein “off” has already been identified: at least in vitro, a particular domain of the dynactin subunit p150Glued can promote a diffusive state with minimal force production [249]. It will be interesting to test if mutating this domain - or overexpressing it in embryos – will alter lipid-droplet motion.
One issue that has not yet received attention is to what extent lipid-droplet dynamics is controlled by properties of the microtubule network. First, microtubules can be profoundly affected by a large number of posttranslational modifications [314]. In particular, acetylation of tubulin is known to promote kinesin-1–driven transport [315]. As tubulin acetylation is first detected during cellularization [316], it might contribute to the Phase I to IIa transition. Second, the presence of microtubule-binding proteins can also change run lengths, with differential effects on kinesin-1 and cytoplasmic dynein [317, 318]. Finally, runs might also be impeded by various obstacles or if motors jump from one track to another; these issues might be particularly important in the region apical to the nuclei with its dense and potentially crisscrossing cytoskeletal network (Fig. 2B). It will therefore be important to establish to what extent the architecture and properties of the tracks modulate droplet motion across the phases; i.e., the switch mechanism may not only act at the level of the lipid droplet (as implied by the cartoons in Figs. 5D,F), but also via altering the tracks.
Protein sequestration
Histones are sequestered on lipid droplets and are released to assemble chromatin in the nucleus as embryogenesis proceeds (Fig. 4D). Sequestration is mediated by Jabba (Fig. 4C,E), but whether – and how - recruitment and release of histones is regulated remains unknown. Histones might constantly equilibrate between cytoplasm and lipid droplets, and the cytoplasmic pool might provide the histones for the assembly of new chromatin after replication. Alternatively, histones may be tightly bound to the droplets, and release in times of need may require developmental signals and/or feedback from the replication machinery. To resolve this issue, it will be important to follow the trafficking of histones from and to lipid droplets in real time and identify histones’ partners in various locations. E.g., are classical histone chaperones needed to unload histones from the lipid droplets or can some Jabba isoforms leave droplets and travel with the histones into the nucleus? And are histones stored as monomers, as H2A-H2B dimers, or complexed with other proteins, in addition to Jabba?
Why the supernumerary histones in Drosophila embryos are tethered to lipid droplets rather than to other organelles or why they are not simply present as protein complexes in the cytoplasm remains an intriguing puzzle. For example, if Jabba binding physically protects histones from attack by the degradation machinery, protection should be similarly effective wherever Jabba is located. One possibility is that tethering of histones to a large cytoplasmic organelle prevents their uncontrolled import into the nucleus. And the tens of thousands of embryonic lipid droplets may provide an extensive surface for storage, a surface that remains fairly constant during early developmental stages. If Jabba has distinct domains for binding to lipid droplets and to histones (as imagined in Fig. 4E), it should be possible to redirect the histone-binding domain of Jabba to distinct intracellular locations (say, the outer mitochondrial membrane or the cytoplasm) and test whether histone storage and buffering are compromised.
In Jabba mutant embryos, the maternal deposit of H2A, H2B, and H2Av is essentially gone [16]. The available evidence suggests that histones are synthesized normally, but are degraded. However, enhanced histone turnover has not yet been demonstrated directly. It remains conceivable that, in addition to its role as histone anchor, Jabba also activates histone translation during oogenesis. It will therefore be important to determine if in the absence of Jabba histone degradation is indeed enhanced and what mechanisms mediate turnover. Such studies will test one of the main proposed models of why refugee proteins are targeted to droplets, namely to shield them from degradation [20]. In addition, they will provide insights into the surveillance mechanisms that protect cells from toxic histone overaccumulation, mechanisms that remain poorly characterized in animals [162].
Lack of Jabba causes aberrant nuclear accumulation of histones, nuclear falling (presumably as a result of DNA damage), and embryonic death [16, 75]. How these phenotypes are connected is not yet clear, e.g., incorrect histone supply might impair transcription, replication, or chromosome condensation during mitosis. In yeast, oversupply of histones has been shown to have multiple unrelated consequences [181]. Understanding which nuclear function(s) are altered when droplets cannot sequester histones is important not only for determining the dynamics of histone metabolism and chromatin assembly in early embryos, but also to provide molecular markers that can serve as read-outs of failed sequestration and/or buffering. Such markers will be particularly useful for investigating if droplet-mediated histone buffering is an embryo-specific phenomenon or occurs also at other stages.
The droplet-sequestered histones support not only chromatin assembly, but can also act as antibacterial defense (Fig. 4G). Many details of this defense mechanisms are not yet known: how are histones released in response to bacteria (in vitro, bacterial cell wall/membrane components trigger release [19]), how do histones kill the bacterial invaders, and against which types of organisms is this defense effective? Most important will be to establish whether Jabba-mediated protection against infection in adults also depends on stored histones or other properties of Jabba and if this type of innate immunity exists in other species, as suggested [19].
Finally, droplet-bound refugee proteins are likely common [20, 21], but because the mechanism of droplet targeting is unknown, this phenomenon has been hard to study. In Drosophila embryos, not only histones are missing from lipid-droplet preparations of Jabba mutants, but also a number of other abundant proteins [16]. If it can be confirmed that these proteins are also anchored to droplets via Jabba, dissecting their mode of sequestration becomes feasible. It will be interesting to determine the fate of these proteins in Jabba mutants (e.g., are they degraded or do they now localize elsewhere in the cell?) and whether they are recruited via interactions distinct from the ones mediating histone binding.
Beyond motility and sequestration
It would be a missed opportunity if droplet research in Drosophila embryos were restricted to motility and sequestration, ignoring other aspects of droplet biology. For example, we know that the biogenesis of lipid droplets during oogenesis is highly developmentally regulated and occurs on a massive scale, but so far only the broad outlines and a few molecular components are understood (Fig. 1D); and our knowledge of the turnover of lipid droplets during embryogenesis is extremely fragmentary (Fig. 1F). As much is already known about droplet biogenesis and turnover from studies in cultured cells and fungi [23, 25, 26, 28-34], it will be relatively straightforward to develop hypotheses about which pathways may drive these processes in ovaries and embryos, hypotheses that should be readily testable with the well-developed genetic tools available in Drosophila. If known pathways are insufficient to explain what is going on, a broad range of molecular genetic approaches are available to uncover novel components [107, 108]. Once the relevant pathways, expected or novel, are identified, they can then be analyzed using the unique toolkit for Drosophila embryos (Fig. 3).
More broadly, studying the lipid droplets of Drosophila embryos has the potential to link fundamental droplet processes to the biology of the whole organism. Studies aimed at understanding the cell biology of lipid droplets will often allow the generation of embryos in which droplet abundance, composition, distribution, or function is altered, providing insight into the mechanisms underlying these droplet properties. But these embryos can then also be used to determine if the embryos with these altered droplets can develop and function normally. For example, mutations in the histone anchor Jabba revealed the mechanism by which histones are recruited to lipid droplets and that droplet binding is apparently necessary to stably maintain the maternal histone deposit [16]. These same mutations could then also be used to uncover that the stored histones are important for normal development and embryo survival [16].
It is becoming increasingly clear that metabolism makes important contributions to both normal and pathological development (e.g., [73, 319, 320]). Given the central role of lipid droplets in lipid and energy homeostasis as well as their myriad additional functions, I anticipate that they can modulate development in many different ways. Drosophila embryogenesis is well characterized molecularly and cell-biologically, and many processes, from cell and tissue structure to gene expression, are understood in exquisite mechanistic detail (e.g., [281, 282, 321]). It is therefore in principle possible to determine even subtle consequences of altered droplet composition and function on development. Given the many cellular roles of lipid droplets and the deep analysis of development possible in Drosophila, I am confident that many exciting discoveries about Drosophila embryonic lipid droplets and their organismal roles will be made in the future. And because Drosophila has emerged as an excellent model to analyze broadly conserved aspects of energy metabolism [322], such studies may also illuminate the link between droplet biology and development in other organisms.
Highlights.
Drosophila embryos are a powerful model system for studying lipid droplets
Key biological and technical features of this experimental system are discussed
Focus is on lipid-droplet motility and droplet-mediated protein sequestration
Includes a discussion of open questions and cross-organism comparisons
Acknowledgements
Research in my laboratory on lipid droplets has been funded by NIH grants GM102155 and GM64687 and by a grant from the Schmitt Program on Integrative Brain Research. I would like to thank three anonymous reviewers for carefully reading the manuscript and providing excellent suggestions. I am grateful to Lili Chen, Matthew Johnson, Gurpreet Arora and Sean Lindley for comments on figures and to Zhihuan Li for generating Fig. 4B.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Martin S, Parton RG. Lipid droplets: a unified view of a dynamic organelle. Nat Rev Mol Cell Biol. 2006;7:373–378. doi: 10.1038/nrm1912. [DOI] [PubMed] [Google Scholar]
- 2.Walther TC, Farese RV., Jr. Lipid droplets and cellular lipid metabolism. Annu Rev Biochem. 2012;81:687–714. doi: 10.1146/annurev-biochem-061009-102430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Igal RA, Coleman RA. Neutral lipid storage disease: a genetic disorder with abnormalities in the regulation of phospholipid metabolism. J Lipid Res. 1998;39:31–43. [PubMed] [Google Scholar]
- 4.Greenberg AS, Coleman RA, Kraemer FB, McManaman JL, Obin MS, Puri V, Yan QW, Miyoshi H, Mashek DG. The role of lipid droplets in metabolic disease in rodents and humans. J Clin Invest. 2011;121:2102–2110. doi: 10.1172/JCI46069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paul A, Chan L, Bickel PE. The PAT family of lipid droplet proteins in heart and vascular cells. Curr Hypertens Rep. 2008;10:461–466. doi: 10.1007/s11906-008-0086-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patterson MC. A riddle wrapped in a mystery: understanding Niemann-Pick disease, type C. Neurologist. 2003;9:301–310. doi: 10.1097/01.nrl.0000094627.78754.5b. [DOI] [PubMed] [Google Scholar]
- 7.Cohen JC, Horton JD, Hobbs HH. Human fatty liver disease: old questions and new insights. Science. 2011;332:1519–1523. doi: 10.1126/science.1204265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saka HA, Valdivia R. Emerging roles for lipid droplets in immunity and host-pathogen interactions. Annu Rev Cell Dev Biol. 2012;28:411–437. doi: 10.1146/annurev-cellbio-092910-153958. [DOI] [PubMed] [Google Scholar]
- 9.Liu L, Zhang K, Sandoval H, Yamamoto S, Jaiswal M, Sanz E, Li Z, Hui J, Graham BH, Quintana A, Bellen HJ. Glial Lipid Droplets and ROS Induced by Mitochondrial Defects Promote Neurodegeneration. Cell. 2015;160:177–190. doi: 10.1016/j.cell.2014.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inloes JM, Hsu KL, Dix MM, Viader A, Masuda K, Takei T, Wood MR, Cravatt BF. The hereditary spastic paraplegia-related enzyme DDHD2 is a principal brain triglyceride lipase. Proc Natl Acad Sci U S A. 2014;111:14924–14929. doi: 10.1073/pnas.1413706111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, Bartenschlager R, Wakita T, Hijikata M, Shimotohno K. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol. 2007;9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- 12.Herker E, Harris C, Hernandez C, Carpentier A, Kaehlcke K, Rosenberg AR, Farese RV, Jr., Ott M. Efficient hepatitis C virus particle formation requires diacylglycerol acyltransferase-1. Nat Med. 2010;16:1295–1298. doi: 10.1038/nm.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samsa MM, Mondotte JA, Iglesias NG, Assuncao-Miranda I, Barbosa-Lima G, Da Poian AT, Bozza PT, Gamarnik AV. Dengue virus capsid protein usurps lipid droplets for viral particle formation. PLoS Pathog. 2009;5:e1000632. doi: 10.1371/journal.ppat.1000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheung W, Gill M, Esposito A, Kaminski CF, Courousse N, Chwetzoff S, Trugnan G, Keshavan N, Lever A, Desselberger U. Rotaviruses associate with cellular lipid droplet components to replicate in viroplasms, and compounds disrupting or blocking lipid droplets inhibit viroplasm formation and viral replication. J Virol. 2010;84:6782–6798. doi: 10.1128/JVI.01757-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cermelli S, Guo Y, Gross SP, Welte MA. The lipid droplet proteome reveals that droplets are a protein storage depot. Curr Biol. 2006;16 doi: 10.1016/j.cub.2006.07.062. [DOI] [PubMed] [Google Scholar]
- 16.Li Z, Thiel K, Thul PJ, Beller M, Kühnlein RP, Welte MA. Lipid droplets control the maternal histone supply of Drosophila embryos. Curr Biol. 2012;22:2104–2113. doi: 10.1016/j.cub.2012.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohsaki Y, Cheng J, Fujita A, Tokumoto T, Fujimoto T. Cytoplasmic lipid droplets are sites of convergence of proteasomal and autophagic degradation of apolipoprotein B. Mol Biol Cell. 2006;17:2674–2683. doi: 10.1091/mbc.E05-07-0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hinson ER, Cresswell P. The antiviral protein, viperin, localizes to lipid droplets via its N-terminal amphipathic alpha-helix. Proc Natl Acad Sci U S A. 2009;106:20452–20457. doi: 10.1073/pnas.0911679106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anand P, Cermelli S, Li Z, Kassan A, Bosch M, Sigua R, Huang L, Ouellette AJ, Pol A, Welte MA, Gross SP. A novel role for lipid droplets in the organismal antibacterial response. Elife. 2012;1:e00003. doi: 10.7554/eLife.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Welte MA. Proteins under new management: lipid droplets deliver. Trends Cell Biol. 2007;17:363–369. doi: 10.1016/j.tcb.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 21.Hodges BD, Wu CC. Proteomic insights into an expanded cellular role for cytoplasmic lipid droplets. J Lipid Res. 2011;51:262–273. doi: 10.1194/jlr.R003582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bougneres L, Helft J, Tiwari S, Vargas P, Chang BH, Chan L, Campisi L, Lauvau G, Hugues S, Kumar P, Kamphorst AO, Dumenil AM, Nussenzweig M, MacMicking JD, Amigorena S, Guermonprez P. A role for lipid bodies in the cross-presentation of phagocytosed antigens by MHC class I in dendritic cells. Immunity. 2009;31:232–244. doi: 10.1016/j.immuni.2009.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farese RV, Jr., Walther TC. Lipid droplets finally get a little R-E-S-P-E-C-T. Cell. 2009;139:855–860. doi: 10.1016/j.cell.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Welte MA. Fat on the move: intracellular motion of lipid droplets. Biochem Soc Trans. 2009;37:991–996. doi: 10.1042/BST0370991. [DOI] [PubMed] [Google Scholar]
- 25.Beller M, Bulankina AV, Hsiao HH, Urlaub H, Jäckle H, Kühnlein RP. PERILIPIN-dependent control of lipid droplet structure and fat storage in Drosophila. Cell Metab. 2010;12:521–532. doi: 10.1016/j.cmet.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 26.Thiele C, Spandl J. Cell biology of lipid droplets. Curr Opin Cell Biol. 2008;20:378–385. doi: 10.1016/j.ceb.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 27.Reue K. A thematic review series: lipid droplet storage and metabolism: from yeast to man. J Lipid Res. 2011;52:1865–1868. doi: 10.1194/jlr.E020602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo Y, Cordes KR, Farese RV, Jr., Walther TC. Lipid droplets at a glance. J Cell Sci. 2009;122:749–752. doi: 10.1242/jcs.037630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Digel M, Ehehalt R, Fullekrug J. Lipid droplets lighting up: insights from live microscopy. FEBS Lett. 2010;584:2168–2175. doi: 10.1016/j.febslet.2010.03.035. [DOI] [PubMed] [Google Scholar]
- 30.Zehmer JK, Huang Y, Peng G, Pu J, Anderson RG, Liu P. A role for lipid droplets in inter-membrane lipid traffic. Proteomics. 2009;9:914–921. doi: 10.1002/pmic.200800584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pol A, Gross SP, Parton RG. Review: biogenesis of the multifunctional lipid droplet: lipids, proteins, and sites. J Cell Biol. 2014;204:635–646. doi: 10.1083/jcb.201311051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thiam AR, Farese RV, Jr., Walther TC. The biophysics and cell biology of lipid droplets. Nat Rev Mol Cell Biol. 2013;14:775–786. doi: 10.1038/nrm3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hashemi HF, Goodman JM. The life cycle of lipid droplets. Curr Opin Cell Biol. 2015;33C:119–124. doi: 10.1016/j.ceb.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rutkowski JM, Stern JH, Scherer PE. The cell biology of fat expansion. J Cell Biol. 2015;208:501–512. doi: 10.1083/jcb.201409063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beller M, Sztalryd C, Southall N, Bell M, Jackle H, Auld DS, Oliver B. COPI complex is a regulator of lipid homeostasis. PLoS Biol. 2008;6:e292. doi: 10.1371/journal.pbio.0060292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo Y, Walther TC, Rao M, Stuurman N, Goshima G, Terayama K, Wong JS, Vale RD, Walter P, Farese RV. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature. 2008;453:657–661. doi: 10.1038/nature06928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kühnlein RP. The contribution of the Drosophila model to lipid droplet research. Prog Lipid Res. 2011;50:348–356. doi: 10.1016/j.plipres.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 38.Kühnlein RP. Drosophila as a lipotoxicity model organism--more than a promise? Biochim Biophys Acta. 2010;1801:215–221. doi: 10.1016/j.bbalip.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 39.Kühnlein RP. Thematic review series: Lipid droplet synthesis and metabolism: from yeast to man. Lipid droplet-based storage fat metabolism in Drosophila. J Lipid Res. 2012;53:1430–1436. doi: 10.1194/jlr.R024299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee H, Peng Y, Guo Y. Analysis of lipid droplet dynamics and functions in Drosophila melanogaster. Methods Cell Biol. 2013;116:53–69. doi: 10.1016/B978-0-12-408051-5.00004-8. [DOI] [PubMed] [Google Scholar]
- 41.Yang HR, Li P. Lipid Droplets: Methods in Cell Biology. Vol. 116. Academic Press; San Diego, CA, USA: 2013. p. 287. [Google Scholar]
- 42.Fuller MT, Spradling AC. Male and female Drosophila germline stem cells: two versions of immortality. Science. 2007;316:402–404. doi: 10.1126/science.1140861. [DOI] [PubMed] [Google Scholar]
- 43.King RC. Ovarian development in Drosophila melanogaster. Academic Press, Inc.; New York, NY: 1970. [Google Scholar]
- 44.Sieber MH, Spradling AC. Steroid Signaling Establishes a Female Metabolic State and Regulates SREBP to Control Oocyte Lipid Accumulation. Curr Biol. 2015 doi: 10.1016/j.cub.2015.02.019. in press http://dx.doi.org/10.1016/j.cub.2015.1002.1019. [DOI] [PMC free article] [PubMed]
- 45.Arrese EL, Soulages JL. Insect fat body: energy, metabolism, and regulation. Annu Rev Entomol. 2010;55:207–225. doi: 10.1146/annurev-ento-112408-085356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ziegler R, Van Antwerpen R. Lipid uptake by insect oocytes. Insect Biochem Mol Biol. 2006;36:264–272. doi: 10.1016/j.ibmb.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 47.Palm W, Sampaio JL, Brankatschk M, Carvalho M, Mahmoud A, Shevchenko A, Eaton S. Lipoproteins in Drosophila melanogaster--assembly, function, and influence on tissue lipid composition. PLoS Genet. 2012;8:e1002828. doi: 10.1371/journal.pgen.1002828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parra-Peralbo E, Culi J. Drosophila lipophorin receptors mediate the uptake of neutral lipids in oocytes and imaginal disc cells by an endocytosis-independent mechanism. PLoS Genet. 2011;7:e1001297. doi: 10.1371/journal.pgen.1001297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roth S, Lynch JA. Symmetry breaking during Drosophila oogenesis. Cold Spring Harb Perspect Biol. 2009;1:a001891. doi: 10.1101/cshperspect.a001891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gutzeit HO. The role of microfilaments in cytoplasmic streaming in Drosophila follicles. J Cell Sci. 1986;80:159–169. doi: 10.1242/jcs.80.1.159. [DOI] [PubMed] [Google Scholar]
- 51.Teixeira L, Rabouille C, Rorth P, Ephrussi A, Vanzo NF. Drosophila Perilipin/ADRP homologue Lsd2 regulates lipid metabolism. Mech Dev. 2003;120:1071–1081. doi: 10.1016/s0925-4773(03)00158-8. [DOI] [PubMed] [Google Scholar]
- 52.Gutzeit H. The role of microtubules in the differentiation of ovarian follicles during vitellogenesis in Drosophila. Roux's Arch Dev Biol. 1986;195:173–181. doi: 10.1007/BF02439435. [DOI] [PubMed] [Google Scholar]
- 53.Gaspar I, Szabad J. In vivo analysis of MT-based vesicle transport by confocal reflection microscopy. Cell Motil Cytoskeleton. 2009;66:68–79. doi: 10.1002/cm.20334. [DOI] [PubMed] [Google Scholar]
- 54.Gaspar I, Szabad J. Glu415 in the alpha-tubulins plays a key role in stabilizing the microtubule-ADP-kinesin complexes. J Cell Sci. 2009;122:2857–2865. doi: 10.1242/jcs.050252. [DOI] [PubMed] [Google Scholar]
- 55.Serbus LR, Cha BJ, Theurkauf WE, Saxton WM. Dynein and the actin cytoskeleton control kinesin-driven cytoplasmic streaming in Drosophila oocytes. Development. 2005;132:3743–3752. doi: 10.1242/dev.01956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu YV, Li Z, Rizzo NP, Einstein J, Welte MA. Targeting the motor regulator Klar to lipid droplets. BMC Cell Biol. 2011;12:9. doi: 10.1186/1471-2121-12-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Welte MA, Gross SP, Postner M, Block SM, Wieschaus EF. Developmental regulation of vesicle transport in Drosophila embryos: forces and kinetics. Cell. 1998;92:547–557. doi: 10.1016/s0092-8674(00)80947-2. [DOI] [PubMed] [Google Scholar]
- 58.Bartsch TF, Longoria RA, Florin EL, Shubeita GT. Lipid droplets purified from Drosophila embryos as an endogenous handle for precise motor transport measurements. Biophys J. 2013;105:1182–1191. doi: 10.1016/j.bpj.2013.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leidel C, Longoria RA, Gutierrez FM, Shubeita GT. Measuring molecular motor forces in vivo: implications for tug-of-war models of bidirectional transport. Biophys J. 2012;103:492–500. doi: 10.1016/j.bpj.2012.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miura S, Gan JW, Brzostowski J, Parisi MJ, Schultz CJ, Londos C, Oliver B, Kimmel AR. Functional conservation for lipid storage droplet association among Perilipin, ADRP, and TIP47 (PAT)-related proteins in mammals, Drosophila, and Dictyostelium. J Biol Chem. 2002;277:32253–32257. doi: 10.1074/jbc.M204410200. [DOI] [PubMed] [Google Scholar]
- 61.Beller M, Riedel D, Jansch L, Dieterich G, Wehland J, Jackle H, Kuhnlein RP. Characterization of the Drosophila lipid droplet subproteome. Mol Cell Proteomics. 2006;5:1082–1094. doi: 10.1074/mcp.M600011-MCP200. [DOI] [PubMed] [Google Scholar]
- 62.Wilfling F, Wang H, Haas JT, Krahmer N, Gould TJ, Uchida A, Cheng JX, Graham M, Christiano R, Frohlich F, Liu X, Buhman KK, Coleman RA, Bewersdorf J, Farese RV, Jr., Walther TC. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev Cell. 2013;24:384–399. doi: 10.1016/j.devcel.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Robinson DN, Cant K, Cooley L. Morphogenesis of Drosophila ovarian ring canals. Development. 1994;120:2015–2025. doi: 10.1242/dev.120.7.2015. [DOI] [PubMed] [Google Scholar]
- 64.Yen CL, Stone SJ, Koliwad S, Harris C, Farese RV., Jr. Thematic review series: glycerolipids. DGAT enzymes and triacylglycerol biosynthesis. J Lipid Res. 2008;49:2283–2301. doi: 10.1194/jlr.R800018-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chintapalli VR, Wang J, Dow JA. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- 66.Buszczak M, Lu X, Segraves WA, Chang TY, Cooley L. Mutations in the midway gene disrupt a Drosophila acyl coenzyme A: diacylglycerol acyltransferase. Genetics. 2002;160:1511–1518. doi: 10.1093/genetics/160.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vereshchagina N, Wilson C. Cytoplasmic activated protein kinase Akt regulates lipid-droplet accumulation in Drosophila nurse cells. Development. 2006;133:4731–4735. doi: 10.1242/dev.02659. [DOI] [PubMed] [Google Scholar]
- 68.Bickel PE, Tansey JT, Welte MA. PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim Biophys Acta. 2009;1791:419–440. doi: 10.1016/j.bbalip.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kinsella JE. General metabolism of the hexapod embryo with particular reference to lipids. Comp Biochem Physiol. 1966;19:291–304. doi: 10.1016/0010-406x(66)90568-8. [DOI] [PubMed] [Google Scholar]
- 70.Beenakkers AMT, Van der Horst DJ, van Marrewijk WJA. Role of lipids in energy metabolism. In: Downer RGH, editor. Energy Metabolism in Insects. Plenum; New York: 1981. pp. 53–100. [Google Scholar]
- 71.Van Handel E. Fuel metabolism of the mosquito (Culex quinquefasciatus) embryo. J. Insect Physiol. 1993;39:831–833. [Google Scholar]
- 72.Grönke S, Mildner A, Fellert S, Tennagels N, Petry S, Müller G, Jäckle H, Kühnlein RP. Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metab. 2005;1:323–330. doi: 10.1016/j.cmet.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 73.Tennessen JM, Bertagnolli NM, Evans J, Sieber MH, Cox J, Thummel CS. Coordinated metabolic transitions during Drosophila embryogenesis and the onset of aerobic glycolysis. G3 (Bethesda) 2014;4:839–850. doi: 10.1534/g3.114.010652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gutzeit HO, Zissler D, Grau V, Liphardt M, Heinrich UR. Glycogen stores in mature ovarian follicles and young embryos of Drosophila: ultrastructural changes and some biochemical correlates. Eur J Cell Biol. 1994;63:52–60. [PubMed] [Google Scholar]
- 75.Li Z, Johnson MR, Ke Z, Chen L, Welte MA. Drosophila lipid droplets buffer the H2Av supply to protect early embryonic development. Curr Biol. 2014;24:1485–1491. doi: 10.1016/j.cub.2014.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shubeita GT, Tran SL, Xu J, Vershinin M, Cermelli S, Cotton SL, Welte MA, Gross SP. Consequences of motor copy number on the intracellular transport of kinesin-1-driven lipid droplets. Cell. 2008;135:1098–1107. doi: 10.1016/j.cell.2008.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gross S, Welte M, Block S, Wieschaus E. Dynein-mediated cargo transport in vivo: a switch controls travel distance. J Cell Biol. 2000;148:945–956. doi: 10.1083/jcb.148.5.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Petrov DY, Mallik R, Shubeita GT, Vershinin M, Gross SP, Yu CC. Studying molecular motor-based cargo transport: what is real and what is noise? Biophys J. 2007;92:2953–2963. doi: 10.1529/biophysj.106.097253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Welte MA. Bidirectional Transport along Microtubules. Curr Biol. 2004;14:R525–537. doi: 10.1016/j.cub.2004.06.045. [DOI] [PubMed] [Google Scholar]
- 80.Gross SP. Hither and yon: a review of bi-directional microtubule-based transport. Physical Biology. 2004;1:R1–11. doi: 10.1088/1478-3967/1/2/R01. [DOI] [PubMed] [Google Scholar]
- 81.Foe VE, Odell GM, Edgar BA. Mitosis and morphogenesis in the Drosophila embryo: point and counterpoint. In: Bate M, Martinez-Arias A, editors. The Development of Drosophuila melanogaster. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1993. pp. 149–300. [Google Scholar]
- 82.Dix CI, Soundararajan HC, Dzhindzhev NS, Begum F, Suter B, Ohkura H, Stephens E, Bullock SL. Lissencephaly-1 promotes the recruitment of dynein and dynactin to transported mRNAs. J Cell Biol. 2013;202:479–494. doi: 10.1083/jcb.201211052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Larsen KS, Xu J, Cermelli S, Shu Z, Gross SP. BicaudalD actively regulates microtubule motor activity in lipid droplet transport. PLoS ONE. 2008;3:e3763. doi: 10.1371/journal.pone.0003763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gross SP, Welte MA, Block SM, Wieschaus EF. Coordination of opposite-polarity microtubule motors. J Cell Biol. 2002;156:715–724. doi: 10.1083/jcb.200109047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Weaver C, Leidel C, Szpankowski L, Farley NM, Shubeita GT, Goldstein LS. Endogenous GSK-3/shaggy regulates bidirectional axonal transport of the amyloid precursor protein. Traffic. 2013;14:295–308. doi: 10.1111/tra.12037. [DOI] [PubMed] [Google Scholar]
- 86.Gross SP, Guo Y, Martinez JE, Welte MA. A determinant for directionality of organelle transport in Drosophila embryos. Curr Biol. 2003;13:1660–1668. doi: 10.1016/j.cub.2003.08.032. [DOI] [PubMed] [Google Scholar]
- 87.Welte MA, Cermelli S, Griner J, Viera A, Guo Y, Kim DH, Gindhart JG, Gross SP. Regulation of lipid-droplet transport by the Perilipin homologue LSD2. Curr Biol. 2005;15:1266–1275. doi: 10.1016/j.cub.2005.06.062. [DOI] [PubMed] [Google Scholar]
- 88.Bullock SL, Nicol A, Gross SP, Zicha D. Guidance of bidirectional motor complexes by mRNA cargoes through control of dynein number and activity. Curr Biol. 2006;16:1447–1452. doi: 10.1016/j.cub.2006.05.055. [DOI] [PubMed] [Google Scholar]
- 89.Gross SP, Tuma MC, Deacon SW, Serpinskaya AS, Reilein AR, Gelfand VI. Interactions and regulation of molecular motors in Xenopus melanophores. J Cell Biol. 2002;156:855–865. doi: 10.1083/jcb.200105055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Smith GA, Gross SP, Enquist LW. Herpesviruses use bidirectional fast-axonal transport to spread in sensory neurons. Proc Natl Acad Sci U S A. 2001;98:3466–3470. doi: 10.1073/pnas.061029798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Callaini G. Cleavage and membrane formation in the blastoderm of the dipteran Ceratitis capitata wied. J. Morphology. 1987;193:305–315. doi: 10.1002/jmor.1051930308. [DOI] [PubMed] [Google Scholar]
- 92.Watanabe T, Thayil A, Jesacher A, Grieve K, Debarre D, Wilson T, Booth M, Srinivas S. Characterisation of the dynamic behaviour of lipid droplets in the early mouse embryo using adaptive harmonic generation microscopy. BMC Cell Biol. 2010;11:38. doi: 10.1186/1471-2121-11-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kan R, Jin M, Subramanian V, Causey CP, Thompson PR, Coonrod SA. Potential role for PADI-mediated histone citrullination in preimplantation development. BMC Dev Biol. 2012;12:19. doi: 10.1186/1471-213X-12-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Webb TA, Kowalski WJ, Fluck RA. Microtubule-based movements during ooplasmic segregation in the Medaka fish egg (Oryzias latipes) Biol. Bull. 1995;188:146–156. doi: 10.2307/1542080. [DOI] [PubMed] [Google Scholar]
- 95.Abraham VC, Gupta S, Fluck RA. Ooplasmic segration in the Medaka (Oryzias latipes) egg. Biol. Bull. 1993;184:115–124. doi: 10.2307/1542222. [DOI] [PubMed] [Google Scholar]
- 96.Jedrzejowska I, Kubrakiewicz J. Yolk nucleus--the complex assemblage of cytoskeleton and ER is a site of lipid droplet formation in spider oocytes. Arthropod Struct Dev. 2010;39:350–359. doi: 10.1016/j.asd.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 97.Yamahama Y, Seno K, Hariyama T. Changes in lipid droplet localization during embryogenesis of the silkworm, Bombyx mori. Zoolog Sci. 2008;25:580–586. doi: 10.2108/zsj.25.580. [DOI] [PubMed] [Google Scholar]
- 98.Shimizu T. Cytoskeletal mechanisms of ooplasmic segregation in annelid eggs. Int J Dev Biol. 1999;43:11–18. [PubMed] [Google Scholar]
- 99.Au PC, Selwood L, Familari M. Cloning and characterization of a new gene from the PAT protein family, in a marsupial, the stripe-faced dunnart (Sminthopsis macroura) Mol Reprod Dev. 2010;77:373–383. doi: 10.1002/mrd.21158. [DOI] [PubMed] [Google Scholar]
- 100.Hausen P, Riebesell M. The early development of Xenopus laevis: An atlas of the histology. Springer-Verlag; Berlin: 1991. [Google Scholar]
- 101.Heid HW, Keenan TW. Intracellular origin and secretion of milk fat globules. Eur J Cell Biol. 2005;84:245–258. doi: 10.1016/j.ejcb.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 102.Huang AH. Oleosins and oil bodies in seeds and other organs. Plant Physiol. 1996;110:1055–1061. doi: 10.1104/pp.110.4.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Debarre D, Supatto W, Pena AM, Fabre A, Tordjmann T, Combettes L, Schanne-Klein MC, Beaurepaire E. Imaging lipid bodies in cells and tissues using third-harmonic generation microscopy. Nat Methods. 2006;3:47–53. doi: 10.1038/nmeth813. [DOI] [PubMed] [Google Scholar]
- 104.Dou W, Zhang D, Jung Y, Cheng JX, Umulis DM. Label-free imaging of lipid-droplet intracellular motion in early Drosophila embryos using femtosecond-stimulated Raman loss microscopy. Biophys J. 2012;102:1666–1675. doi: 10.1016/j.bpj.2012.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Guo Y, Jangi S, Welte MA. Organelle-specific Control of Intracellular Transport: Distinctly Targeted Isoforms of the Regulator Klar. Mol Biol Cell. 2005;16:1406–1416. doi: 10.1091/mbc.E04-10-0920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Buszczak M, Paterno S, Lighthouse D, Bachman J, Planck J, Owen S, Skora AD, Nystul TG, Ohlstein B, Allen A, Wilhelm JE, Murphy TD, Levis RW, Matunis E, Srivali N, Hoskins RA, Spradling AC. The carnegie protein trap library: a versatile tool for Drosophila developmental studies. Genetics. 2007;175:1505–1531. doi: 10.1534/genetics.106.065961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Southall TD, Elliott DA, Brand AH. The GAL4 System: A Versatile Toolkit for Gene Expression in Drosophila. CSH Protoc. 2008 doi: 10.1101/pdb.top49. doi:10.1101/pdb.top1149. [DOI] [PubMed] [Google Scholar]
- 108.Mohr SE, Hu Y, Kim K, Housden BE, Perrimon N. Resources for functional genomics studies in Drosophila melanogaster. Genetics. 2014;197:1–18. doi: 10.1534/genetics.113.154344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pilot F, Philippe JM, Lemmers C, Chauvin JP, Lecuit T. Developmental control of nuclear morphogenesis and anchoring by charleston, identified in a functional genomic screen of Drosophila cellularisation. Development. 2006;133:711–723. doi: 10.1242/dev.02251. [DOI] [PubMed] [Google Scholar]
- 110.Meyer WJ, Schreiber S, Guo Y, Volkmann T, Welte MA, Muller HA. Overlapping Functions of Argonaute Proteins in Patterning and Morphogenesis of Drosophila Embryos. PLoS Genet. 2006;2:1224–1239. doi: 10.1371/journal.pgen.0020134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Merrill PT, Sweeton D, Wieschaus E. Requirements for autosomal gene activity during precellular stages of Drosophila melanogaster. Development. 1988;104:495–509. doi: 10.1242/dev.104.3.495. [DOI] [PubMed] [Google Scholar]
- 112.Ni JQ, Zhou R, Czech B, Liu LP, Holderbaum L, Yang-Zhou D, Shim HS, Tao R, Handler D, Karpowicz P, Binari R, Booker M, Brennecke J, Perkins LA, Hannon GJ, Perrimon N. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat Methods. 2011;8:405–407. doi: 10.1038/nmeth.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gepner J, Li M, Ludmann S, Kortas C, Boylan K, Iyadurai SJ, McGrail M, Hays TS. Cytoplasmic dynein function is essential in Drosophila melanogaster. Genetics. 1996;142:865–878. doi: 10.1093/genetics/142.3.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Limbourg B, Zalokar M. Permeabilization of Drosophila eggs. Developmental biology. 1973;35:382–387. doi: 10.1016/0012-1606(73)90034-1. [DOI] [PubMed] [Google Scholar]
- 115.Mazur P, Cole KW, Mahowald AP. Critical factors affecting the permeabilization of Drosophila embryos by alkanes. Cryobiology. 1992;29:210–239. doi: 10.1016/0011-2240(92)90021-s. [DOI] [PubMed] [Google Scholar]
- 116.Rand MD, Kearney AL, Dao J, Clason T. Permeabilization of Drosophila embryos for introduction of small molecules. Insect Biochem Mol Biol. 2010;40:792–804. doi: 10.1016/j.ibmb.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rand MD. A method of permeabilization of Drosophila embryos for assays of small molecule activity. J Vis Exp. 2014:e51634. doi: 10.3791/51634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gross SP. Application of optical traps in vivo. Methods Enzymol. 2003;361:162–174. doi: 10.1016/s0076-6879(03)61010-4. [DOI] [PubMed] [Google Scholar]
- 119.Mallik R, Rai AK, Barak P, Rai A, Kunwar A. Teamwork in microtubule motors. Trends Cell Biol. 2013;23:575–582. doi: 10.1016/j.tcb.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 120.Sims PA, Xie XS. Probing dynein and kinesin stepping with mechanical manipulation in a living cell. Chemphyschem. 2009;10:1511–1516. doi: 10.1002/cphc.200900113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Xu J, Reddy BJ, Anand P, Shu Z, Cermelli S, Mattson MK, Tripathy SK, Hoss MT, James NS, King SJ, Huang L, Bardwell L, Gross SP. Casein kinase 2 reverses tail-independent inactivation of kinesin-1. Nat Commun. 2012;3:754. doi: 10.1038/ncomms1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Barak P, Rai A, Rai P, Mallik R. Quantitative optical trapping on single organelles in cell extract. Nat Methods. 2013;10:68–70. doi: 10.1038/nmeth.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hendricks AG, Holzbaur EL, Goldman YE. Force measurements on cargoes in living cells reveal collective dynamics of microtubule motors. Proc Natl Acad Sci U S A. 2012;109:18447–18452. doi: 10.1073/pnas.1215462109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rai AK, Rai A, Ramaiya AJ, Jha R, Mallik R. Molecular adaptations allow dynein to generate large collective forces inside cells. Cell. 2013;152:172–182. doi: 10.1016/j.cell.2012.11.044. [DOI] [PubMed] [Google Scholar]
- 125.Soppina V, Rai AK, Ramaiya AJ, Barak P, Mallik R. Tug-of-war between dissimilar teams of microtubule motors regulates transport and fission of endosomes. Proc Natl Acad Sci U S A. 2009;106:19381–19386. doi: 10.1073/pnas.0906524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sigua R, Tripathy S, Anand P, Gross SP. Isolation and purification of kinesin from Drosophila embryos. J Vis Exp. 2012:e3501. doi: 10.3791/3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tran SL, Welte MA. In-vivo centrifugation of Drosophila embryos. Journal of visualized experiments : JoVE. 2010:e2005. doi: 10.3791/2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Morgan TH. Experimental Embryology (Chapter XXII: The redistribution of the visible materials of the egg by centrifuging) Columbia University Press; New York: 1927. [Google Scholar]
- 129.Kempner ES, Miller JH. The molecular biology of Euglena gracilis. IV. Cellular stratification by centrifuging. Exp Cell Res. 1968;51:141–149. doi: 10.1016/0014-4827(68)90164-x. [DOI] [PubMed] [Google Scholar]
- 130.Zalokar M. Cytochemistry of centrifuged hyphae of Neurospora. Exp Cell Res. 1960;19:114–132. doi: 10.1016/0014-4827(60)90042-2. [DOI] [PubMed] [Google Scholar]
- 131.Daga RR, Chang F. Dynamic positioning of the fission yeast cell division plane. Proc Natl Acad Sci U S A. 2005;102:8228–8232. doi: 10.1073/pnas.0409021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bartz R, Li WH, Venables B, Zehmer JK, Roth MR, Welti R, Anderson RG, Liu P, Chapman KD. Lipidomics reveals that adiposomes store ether lipids and mediate phospholipid traffic. J Lipid Res. 2007;48:837–847. doi: 10.1194/jlr.M600413-JLR200. [DOI] [PubMed] [Google Scholar]
- 133.Krahmer N, Guo Y, Wilfling F, Hilger M, Lingrell S, Heger K, Newman HW, Schmidt-Supprian M, Vance DE, Mann M, Farese RV, Jr., Walther TC. Phosphatidylcholine synthesis for lipid droplet expansion is mediated by localized activation of CTP:phosphocholine cytidylyltransferase. Cell Metab. 2011;14:504–515. doi: 10.1016/j.cmet.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kozlova T, Thummel CS. Essential roles for ecdysone signaling during Drosophila mid-embryonic development. Science. 2003;301:1911–1914. doi: 10.1126/science.1087419. [DOI] [PubMed] [Google Scholar]
- 135.Guan XL, Cestra G, Shui G, Kuhrs A, Schittenhelm RB, Hafen E, van der Goot FG, Robinett CC, Gatti M, Gonzalez-Gaitan M, Wenk MR. Biochemical membrane lipidomics during Drosophila development. Dev Cell. 2013;24:98–111. doi: 10.1016/j.devcel.2012.11.012. [DOI] [PubMed] [Google Scholar]
- 136.Parisi M, Li R, Oliver B. Lipid profiles of female and male Drosophila. BMC Res Notes. 2011;4:198. doi: 10.1186/1756-0500-4-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Tortoriello G, Rhodes BP, Takacs SM, Stuart JM, Basnet A, Raboune S, Widlanski TS, Doherty P, Harkany T, Bradshaw HB. Targeted lipidomics in Drosophila melanogaster identifies novel 2-monoacylglycerols and N-acyl amides. PLoS One. 2013;8:e67865. doi: 10.1371/journal.pone.0067865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hsieh K, Lee YK, Londos C, Raaka BM, Dalen KT, Kimmel AR. Perilipin family members preferentially sequester to either triacylglycerol-specific or cholesteryl-ester-specific intracellular lipid storage droplets. J Cell Sci. 2012;125:4067–4076. doi: 10.1242/jcs.104943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yang L, Ding Y, Chen Y, Zhang S, Huo C, Wang Y, Yu J, Zhang P, Na H, Zhang H, Ma Y, Liu P. The proteomics of lipid droplets: structure, dynamics, and functions of the organelle conserved from bacteria to humans. J Lipid Res. 2012;53:1245–1253. doi: 10.1194/jlr.R024117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Krahmer N, Hilger M, Kory N, Wilfling F, Stoehr G, Mann M, Farese RV, Jr., Walther TC. Protein correlation profiles identify lipid droplet proteins with high confidence. Mol Cell Proteomics. 2013;12:1115–1126. doi: 10.1074/mcp.M112.020230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Thiel K, Heier C, Haberl V, Thul PJ, Oberer M, Lass A, Jackle H, Beller M. The evolutionarily conserved protein CG9186 is associated with lipid droplets, required for their positioning and for fat storage. J Cell Sci. 2013;126:2198–2212. doi: 10.1242/jcs.120493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Subramanian V, Rothenberg A, Gomez C, Cohen AW, Garcia A, Bhattacharyya S, Shapiro L, Dolios G, Wang R, Lisanti MP, Brasaemle DL. Perilipin A mediates the reversible binding of CGI-58 to lipid droplets in 3T3-L1 adipocytes. J Biol Chem. 2004;279:42062–42071. doi: 10.1074/jbc.M407462200. [DOI] [PubMed] [Google Scholar]
- 143.Brasaemle DL, Dolios G, Shapiro L, Wang R. Proteomic Analysis of Proteins Associated with Lipid Droplets of Basal and Lipolytically Stimulated 3T3-L1 Adipocytes. J Biol Chem. 2004;279:46835–46842. doi: 10.1074/jbc.M409340200. [DOI] [PubMed] [Google Scholar]
- 144.Brasaemle DL. Thematic review series: adipocyte biology. The perilipin family of structural lipid droplet proteins: stabilization of lipid droplets and control of lipolysis. J Lipid Res. 2007;48:2547–2559. doi: 10.1194/jlr.R700014-JLR200. [DOI] [PubMed] [Google Scholar]
- 145.Sztalryd C, Xu G, Dorward H, Tansey JT, Contreras JA, Kimmel AR, Londos C. Perilipin A is essential for the translocation of hormone-sensitive lipase during lipolytic activation. J Cell Biol. 2003;161:1093–1103. doi: 10.1083/jcb.200210169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Soulages JL, Firdaus SJ, Hartson S, Chen X, Howard AD, Arrese EL. Developmental changes in the protein composition of Manduca sexta lipid droplets. Insect Biochem Mol Biol. 2012;42:305–320. doi: 10.1016/j.ibmb.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wilfling F, Haas JT, Walther TC, Farese RV., Jr. Lipid droplet biogenesis. Curr Opin Cell Biol. 2014;29:39–45. doi: 10.1016/j.ceb.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Arrese EL, Rivera L, Hamada M, Soulages JL. Purification and characterization of recombinant lipid storage protein-2 from Drosophila melanogaster. Protein Pept Lett. 2008;15:1027–1032. doi: 10.2174/092986608785849191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Celona B, Weiner A, Di Felice F, Mancuso FM, Cesarini E, Rossi RL, Gregory L, Baban D, Rossetti G, Grianti P, Pagani M, Bonaldi T, Ragoussis J, Friedman N, Camilloni G, Bianchi ME, Agresti A. Substantial histone reduction modulates genomewide nucleosomal occupancy and global transcriptional output. PLoS Biol. 2011;9:e1001086. doi: 10.1371/journal.pbio.1001086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.O'Sullivan RJ, Kubicek S, Schreiber SL, Karlseder J. Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nat Struct Mol Biol. 2010;17:1218–1225. doi: 10.1038/nsmb.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Feser J, Truong D, Das C, Carson JJ, Kieft J, Harkness T, Tyler JK. Elevated histone expression promotes life span extension. Mol Cell. 2010;39:724–735. doi: 10.1016/j.molcel.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Mejlvang J, Feng Y, Alabert C, Neelsen KJ, Jasencakova Z, Zhao X, Lees M, Sandelin A, Pasero P, Lopes M, Groth A. New histone supply regulates replication fork speed and PCNA unloading. J Cell Biol. 2014;204:29–43. doi: 10.1083/jcb.201305017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Gunjan A, Verreault A. A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell. 2003;115:537–549. doi: 10.1016/s0092-8674(03)00896-1. [DOI] [PubMed] [Google Scholar]
- 154.Meeks-Wagner D, Hartwell LH. Normal stoichiometry of histone dimer sets is necessary for high fidelity of mitotic chromosome transmission. Cell. 1986;44:43–52. doi: 10.1016/0092-8674(86)90483-6. [DOI] [PubMed] [Google Scholar]
- 155.Herrero AB, Moreno S. Lsm1 promotes genomic stability by controlling histone mRNA decay. The EMBO journal. 2011;30:2008–2018. doi: 10.1038/emboj.2011.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Berloco M, Fanti L, Breiling A, Orlando V, Pimpinelli S. The maternal effect gene, abnormal oocyte (abo), of Drosophila melanogaster encodes a specific negative regulator of histones. Proc Natl Acad Sci U S A. 2001;98:12126–12131. doi: 10.1073/pnas.211428798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Marzluff WF, Duronio RJ. Histone mRNA expression: multiple levels of cell cycle regulation and important developmental consequences. Curr Opin Cell Biol. 2002;14:692–699. doi: 10.1016/s0955-0674(02)00387-3. [DOI] [PubMed] [Google Scholar]
- 158.Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nature reviews. Genetics. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Singh RK, Kabbaj MH, Paik J, Gunjan A. Histone levels are regulated by phosphorylation and ubiquitylation-dependent proteolysis. Nature cell biology. 2009;11:925–933. doi: 10.1038/ncb1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gunjan A, Paik J, Verreault A. Regulation of histone synthesis and nucleosome assembly. Biochimie. 2005;87:625–635. doi: 10.1016/j.biochi.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 161.Dilworth SM, Black SJ, Laskey RA. Two complexes that contain histones are required for nucleosome assembly in vitro: role of nucleoplasmin and N1 in Xenopus egg extracts. Cell. 1987;51:1009–1018. doi: 10.1016/0092-8674(87)90587-3. [DOI] [PubMed] [Google Scholar]
- 162.Singh RK, Paik J, Gunjan A. Generation and management of excess histones during the cell cycle. Front Biosci. 2009;14:3145–3158. doi: 10.2741/3441. [DOI] [PubMed] [Google Scholar]
- 163.Singh RK, Gonzalez M, Kabbaj MH, Gunjan A. Novel E3 ubiquitin ligases that regulate histone protein levels in the budding yeast Saccharomyces cerevisiae. PLoS One. 2012;7:e36295. doi: 10.1371/journal.pone.0036295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sullivan E, Santiago C, Parker ED, Dominski Z, Yang X, Lanzotti DJ, Ingledue TC, Marzluff WF, Duronio RJ. Drosophila stem loop binding protein coordinates accumulation of mature histone mRNA with cell cycle progression. Genes Dev. 2001;15:173–187. doi: 10.1101/gad.862801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Marzluff WF, Tatomer DC. Histones: sequestered by Jabba in fatty storehouse. Curr Biol. 2012;22:R951–953. doi: 10.1016/j.cub.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Iampietro C, Bergalet J, Wang X, Cody NA, Chin A, Lefebvre FA, Douziech M, Krause HM, Lecuyer E. Developmentally regulated elimination of damaged nuclei involves a Chk2-dependent mechanism of mRNA nuclear retention. Dev Cell. 2014;29:468–481. doi: 10.1016/j.devcel.2014.03.025. [DOI] [PubMed] [Google Scholar]
- 167.Sullivan W, Minden JS, Alberts BM. daughterless-abo-like, a Drosophila maternal-effect mutation that exhibits abnormal centrosome separation during the late blastoderm divisions. Development. 1990;110:311–323. doi: 10.1242/dev.110.2.311. [DOI] [PubMed] [Google Scholar]
- 168.Kuntz SG, Eisen MB. Drosophila embryogenesis scales uniformly across temperature in developmentally diverse species. PLoS Genet. 2014;10:e1004293. doi: 10.1371/journal.pgen.1004293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hirsch JG. Bactericidal action of histone. J Exp Med. 1958;108:925–944. doi: 10.1084/jem.108.6.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Tan KL, Vlisidou I, Wood W. Ecdysone mediates the development of immunity in the Drosophila embryo. Curr Biol. 2014;24:1145–1152. doi: 10.1016/j.cub.2014.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Vlisidou I, Dowling AJ, Evans IR, Waterfield N, ffrench-Constant RH, Wood W. Drosophila embryos as model systems for monitoring bacterial infection in real time. PLoS Pathog. 2009;5:e1000518. doi: 10.1371/journal.ppat.1000518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.De Gregorio E, Spellman PT, Tzou P, Rubin GM, Lemaitre B. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 2002;21:2568–2579. doi: 10.1093/emboj/21.11.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.De Gregorio E, Spellman PT, Rubin GM, Lemaitre B. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc Natl Acad Sci U S A. 2001;98:12590–12595. doi: 10.1073/pnas.221458698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Nagai A, Sato T, Akimoto N, Ito A, Sumida M. Isolation and identification of histone H3 protein enriched in microvesicles secreted from cultured sebocytes. Endocrinology. 2005;146:2593–2601. doi: 10.1210/en.2004-1478. [DOI] [PubMed] [Google Scholar]
- 175.Binns D, Januszewski T, Chen Y, Hill J, Markin VS, Zhao Y, Gilpin C, Chapman KD, Anderson RG, Goodman JM. An intimate collaboration between peroxisomes and lipid bodies. J Cell Biol. 2006;173:719–731. doi: 10.1083/jcb.200511125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Peng SE, Chen WN, Chen HK, Lu CY, Mayfield AB, Fang LS, Chen CS. Lipid bodies in coral-dinoflagellate endosymbiosis: proteomic and ultrastructural studies. Proteomics. 2011;11:3540–3555. doi: 10.1002/pmic.201000552. [DOI] [PubMed] [Google Scholar]
- 177.Wan HC, Melo RC, Jin Z, Dvorak AM, Weller PF. Roles and origins of leukocyte lipid bodies: proteomic and ultrastructural studies. FASEB J. 2007;21:167–178. doi: 10.1096/fj.06-6711com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Larsson S, Resjo S, Gomez MF, James P, Holm C. Characterization of the Lipid Droplet Proteome of a Clonal Insulin-producing beta-Cell Line (INS-1 832/13) J Proteome Res. 2012;11:1264–1273. doi: 10.1021/pr200957p. [DOI] [PubMed] [Google Scholar]
- 179.Zhang H, Wang Y, Li J, Yu J, Pu J, Li L, Zhang H, Zhang S, Peng G, Yang F, Liu P. Proteome of skeletal muscle lipid droplet reveals association with mitochondria and apolipoprotein a-I. J Proteome Res. 2011;10:4757–4768. doi: 10.1021/pr200553c. [DOI] [PubMed] [Google Scholar]
- 180.Mackay IR. Hepatoimmunology: a perspective. Immunol Cell Biol. 2002;80:36–44. doi: 10.1046/j.1440-1711.2002.01063.x. [DOI] [PubMed] [Google Scholar]
- 181.Singh RK, Liang D, Gajjalaiahvari UR, Kabbaj MH, Paik J, Gunjan A. Excess histone levels mediate cytotoxicity via multiple mechanisms. Cell Cycle. 2010;9:4236–4244. doi: 10.4161/cc.9.20.13636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Cobbe N, Marshall KM, Gururaja Rao S, Chang CW, Di Cara F, Duca E, Vass S, Kassan A, Heck MM. The conserved metalloprotease invadolysin localizes to the surface of lipid droplets. J Cell Sci. 2009;122:3414–3423. doi: 10.1242/jcs.044610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.McHugh B, Krause SA, Yu B, Deans AM, Heasman S, McLaughlin P, Heck MM. Invadolysin: a novel, conserved metalloprotease links mitotic structural rearrangements with cell migration. J Cell Biol. 2004;167:673–686. doi: 10.1083/jcb.200405155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Rao SG, Janiszewski MM, Duca E, Nelson B, Abhinav K, Panagakou I, Vass S, Heck MM. Invadolysin acts genetically via the SAGA complex to modulate chromosome structure. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv211. in press doi: 10.1093/nar/gkv1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Whitehead JP, Simpson F, Hill MM, Thomas EC, Connolly LM, Collart F, Simpson RJ, James DE. Insulin and oleate promote translocation of inosine-5' monophosphate dehydrogenase to lipid bodies. Traffic. 2004;5:739–749. doi: 10.1111/j.1600-0854.2004.00217.x. [DOI] [PubMed] [Google Scholar]
- 186.Cho SY, Shin ES, Park PJ, Shin DW, Chang HK, Kim D, Lee HH, Lee JH, Kim SH, Song MJ, Chang IS, Lee OS, Lee TR. Identification of mouse Prp19p as a lipid droplet-associated protein and its possible involvement in the biogenesis of lipid droplets. J Biol Chem. 2007;282:2456–2465. doi: 10.1074/jbc.M608042200. [DOI] [PubMed] [Google Scholar]
- 187.Martin S, Parton RG. Caveolin, cholesterol, and lipid bodies. Semin Cell Dev Biol. 2005;16:163–174. doi: 10.1016/j.semcdb.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 188.Pol A, Martin S, Fernandez MA, Ingelmo-Torres M, Ferguson C, Enrich C, Parton RG. Cholesterol and fatty acids regulate dynamic caveolin trafficking through the Golgi complex and between the cell surface and lipid bodies. Mol Biol Cell. 2005;16:2091–2105. doi: 10.1091/mbc.E04-08-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Hartman IZ, Liu P, Zehmer JK, Luby-Phelps K, Jo Y, Anderson RG, DeBose-Boyd RA. Sterol-induced dislocation of 3-hydroxy-3-methylglutaryl coenzyme A reductase from endoplasmic reticulum membranes into the cytosol through a subcellular compartment resembling lipid droplets. J Biol Chem. 2010;285:19288–19298. doi: 10.1074/jbc.M110.134213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Jo Y, Hartman IZ, DeBose-Boyd RA. Ancient ubiquitous protein-1 mediates sterol-induced ubiquitination of 3-hydroxy-3-methylglutaryl CoA reductase in lipid droplet-associated endoplasmic reticulum membranes. Mol Biol Cell. 2013;24:169–183. doi: 10.1091/mbc.E12-07-0564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Fujimoto T, Ohsaki Y, Cheng J, Suzuki M, Shinohara Y. Lipid droplets: a classic organelle with new outfits. Histochem Cell Biol. 2008 doi: 10.1007/s00418-008-0449-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Bartz R, Zehmer JK, Zhu M, Chen Y, Serrero G, Zhao Y, Liu P. Dynamic activity of lipid droplets: protein phosphorylation and GTP-mediated protein translocation. J Proteome Res. 2007;6:3256–3265. doi: 10.1021/pr070158j. [DOI] [PubMed] [Google Scholar]
- 193.Grillitsch K, Connerth M, Kofeler H, Arrey TN, Rietschel B, Wagner B, Karas M, Daum G. Lipid particles/droplets of the yeast Saccharomyces cerevisiae revisited: Lipidome meets Proteome. Biochim Biophys Acta. 2011 doi: 10.1016/j.bbalip.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Martinez LO, Jacquet S, Esteve JP, Rolland C, Cabezon E, Champagne E, Pineau T, Georgeaud V, Walker JE, Terce F, Collet X, Perret B, Barbaras R. Ectopic beta-chain of ATP synthase is an apolipoprotein A-I receptor in hepatic HDL endocytosis. Nature. 2003;421:75–79. doi: 10.1038/nature01250. [DOI] [PubMed] [Google Scholar]
- 195.Chi SL, Pizzo SV. Cell surface F1Fo ATP synthase: a new paradigm? Ann Med. 2006;38:429–438. doi: 10.1080/07853890600928698. [DOI] [PubMed] [Google Scholar]
- 196.Zhang Y, Zhang Y, Gao Y, Zhao X, Wang Z. Drosophila long-chain acyl-CoA synthetase acts like a gap gene in embryonic segmentation. Dev Biol. 2011;353:259–265. doi: 10.1016/j.ydbio.2011.02.030. [DOI] [PubMed] [Google Scholar]
- 197.Ellis JM, Frahm JL, Li LO, Coleman RA. Acyl-coenzyme A synthetases in metabolic control. Curr Opin Lipidol. 2010;21:212–217. doi: 10.1097/mol.0b013e32833884bb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Zhang Y, Chen D, Wang Z. Analyses of mental dysfunction-related ACSl4 in Drosophila reveal its requirement for Dpp/BMP production and visual wiring in the brain. Hum Mol Genet. 2009;18:3894–3905. doi: 10.1093/hmg/ddp332. [DOI] [PubMed] [Google Scholar]
- 199.Kassan A, Herms A, Fernandez-Vidal A, Bosch M, Schieber NL, Reddy BJ, Fajardo A, Gelabert-Baldrich M, Tebar F, Enrich C, Gross SP, Parton RG, Pol A. Acyl-CoA synthetase 3 promotes lipid droplet biogenesis in ER microdomains. J Cell Biol. 2013;203:985–1001. doi: 10.1083/jcb.201305142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Fauny JD, Silber J, Zider A. Drosophila Lipid Storage Droplet 2 gene (Lsd-2) is expressed and controls lipid storage in wing imaginal discs. Dev Dyn. 2005;232:725–732. doi: 10.1002/dvdy.20277. [DOI] [PubMed] [Google Scholar]
- 201.Diaconeasa B, Mazock GH, Mahowald AP, Dubreuil RR. Genetic studies of spectrin in the larval fat body of Drosophila melanogaster: evidence for a novel lipid uptake apparatus. Genetics. 2013;195:871–881. doi: 10.1534/genetics.113.155192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Wolinski H, Kolb D, Hermann S, Koning RI, Kohlwein SD. A role for seipin in lipid droplet dynamics and inheritance in yeast. J Cell Sci. 2011;124:3894–3904. doi: 10.1242/jcs.091454. [DOI] [PubMed] [Google Scholar]
- 203.Long AP, Manneschmidt AK, VerBrugge B, Dortch MR, Minkin SC, Prater KE, Biggerstaff JP, Dunlap JR, Dalhaimer P. Lipid droplet de novo formation and fission are linked to the cell cycle in fission yeast. Traffic. 2012;13:705–714. doi: 10.1111/j.1600-0854.2012.01339.x. [DOI] [PubMed] [Google Scholar]
- 204.Vale RD. The molecular motor toolbox for intracellular transport. Cell. 2003;112:467–480. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- 205.Stokin GB, Goldstein LS. Axonal transport and Alzheimer's disease. Annu Rev Biochem. 2006;75:607–627. doi: 10.1146/annurev.biochem.75.103004.142637. [DOI] [PubMed] [Google Scholar]
- 206.Welte MA. Bidirectional transport: matchmaking for motors. Curr Biol. 2010;20:R410–413. doi: 10.1016/j.cub.2010.03.018. [DOI] [PubMed] [Google Scholar]
- 207.Glater EE, Megeath LJ, Stowers RS, Schwarz TL. Axonal transport of mitochondria requires milton to recruit kinesin heavy chain and is light chain independent. J Cell Biol. 2006;173:545–557. doi: 10.1083/jcb.200601067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Kamal A, Stokin GB, Yang Z, Xia CH, Goldstein LS. Axonal transport of amyloid precursor protein is mediated by direct binding to the kinesin light chain subunit of kinesin-I. Neuron. 2000;28:449–459. doi: 10.1016/s0896-6273(00)00124-0. [DOI] [PubMed] [Google Scholar]
- 209.Akhmanova A, Hammer JA., 3rd Linking molecular motors to membrane cargo. Curr Opin Cell Biol. 2010;22:479–487. doi: 10.1016/j.ceb.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Turro S, Ingelmo-Torres M, Estanyol JM, Tebar F, Fernandez MA, Albor CV, Gaus K, Grewal T, Enrich C, Pol A. Identification and characterization of associated with lipid droplet protein 1: A novel membrane-associated protein that resides on hepatic lipid droplets. Traffic. 2006;7:1254–1269. doi: 10.1111/j.1600-0854.2006.00465.x. [DOI] [PubMed] [Google Scholar]
- 211.Orlicky DJ, Monks J, Stefanski AL, McManaman JL. Dynamics and molecular determinants of cytoplasmic lipid droplet clustering and dispersion. PLoS One. 2013;8:e66837. doi: 10.1371/journal.pone.0066837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Mattson-Hoss MK, Niitani Y, Gordon EA, Jun Y, Bardwell L, Tomishige M, Gross SP. CK2 activates kinesin via induction of a conformational change. Proc Natl Acad Sci U S A. 2014;111:7000–7005. doi: 10.1073/pnas.1321419111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Kardon JR, Reck-Peterson SL, Vale RD. Regulation of the processivity and intracellular localization of Saccharomyces cerevisiae dynein by dynactin. Proc Natl Acad Sci U S A. 2009;106:5669–5674. doi: 10.1073/pnas.0900976106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.McKenney RJ, Vershinin M, Kunwar A, Vallee RB, Gross SP. LIS1 and NudE induce a persistent dynein force-producing state. Cell. 2010;141:304–314. doi: 10.1016/j.cell.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Wilkie GS, Davis I. Drosophila wingless and pair-rule transcripts localize apically by dynein-mediated transport of RNA particles. Cell. 2001;105:209–219. doi: 10.1016/s0092-8674(01)00312-9. [DOI] [PubMed] [Google Scholar]
- 216.Delanoue R, Davis I. Dynein anchors its mRNA cargo after apical transport in the Drosophila blastoderm embryo. Cell. 2005;122:97–106. doi: 10.1016/j.cell.2005.04.033. [DOI] [PubMed] [Google Scholar]
- 217.Hoogenraad CC, Wulf P, Schiefermeier N, Stepanova T, Galjart N, Small JV, Grosveld F, de Zeeuw CI, Akhmanova A. Bicaudal D induces selective dynein-mediated microtubule minus end-directed transport. EMBO J. 2003;22:6004–6015. doi: 10.1093/emboj/cdg592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Wu CC, Howell KE, Neville MC, Yates JR, 3rd, McManaman JL. Proteomics reveal a link between the endoplasmic reticulum and lipid secretory mechanisms in mammary epithelial cells. Electrophoresis. 2000;21:3470–3482. doi: 10.1002/1522-2683(20001001)21:16<3470::AID-ELPS3470>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 219.Boström P, Rutberg M, Ericsson J, Holmdahl P, Andersson L, Frohman MA, Boren J, Olofsson SO. Cytosolic lipid droplets increase in size by microtubule-dependent complex formation. Arterioscler Thromb Vasc Biol. 2005;25:1945–1951. doi: 10.1161/01.ATV.0000179676.41064.d4. [DOI] [PubMed] [Google Scholar]
- 220.Lyn RK, Hope G, Sherratt AR, McLauchlan J, Pezacki JP. Bidirectional lipid droplet velocities are controlled by differential binding strengths of HCV core DII protein. PLoS One. 2013;8:e78065. doi: 10.1371/journal.pone.0078065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Boulant S, Douglas MW, Moody L, Budkowska A, Targett-Adams P, McLauchlan J. Hepatitis C Virus Core Protein Induces LIpid Droplet Redistribution in a Microtubule- and Dynein-Dependent Manner. Traffic. 2008;9:1268–1282. doi: 10.1111/j.1600-0854.2008.00767.x. [DOI] [PubMed] [Google Scholar]
- 222.Andersson L, Bostrom P, Ericson J, Rutberg M, Magnusson B, Marchesan D, Ruiz M, Asp L, Huang P, Frohman MA, Boren J, Olofsson SO. PLD1 and ERK2 regulate cytosolic lipid droplet formation. J Cell Sci. 2006;119:2246–2257. doi: 10.1242/jcs.02941. [DOI] [PubMed] [Google Scholar]
- 223.Valetti C, Wetzel DM, Schrader M, Hasbani MJ, Gill SR, Kreis TE, Schroer TA. Role of dynactin in endocytic traffic: effects of dynamitin overexpression and colocalization with CLIP-170. Mol Biol Cell. 1999;10:4107–4120. doi: 10.1091/mbc.10.12.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Svoboda K, Block SM. Biological applications of optical forces. Annu Rev Biophys Biomol Struct. 1994;23:247–285. doi: 10.1146/annurev.bb.23.060194.001335. [DOI] [PubMed] [Google Scholar]
- 225.Kunwar A, Vershinin M, Xu J, Gross SP. Stepping, strain gating, and an unexpected force-velocity curve for multiple-motor-based transport. Curr Biol. 2008;18:1173–1183. doi: 10.1016/j.cub.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Reis GF, Yang G, Szpankowski L, Weaver C, Shah SB, Robinson JT, Hays TS, Danuser G, Goldstein LS. Molecular motor function in axonal transport in vivo probed by genetic and computational analysis in Drosophila. Mol Biol Cell. 2012;23:1700–1714. doi: 10.1091/mbc.E11-11-0938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Martinez JE, Vershinin MD, Shubeita GT, Gross SP. On the use of in vivo cargo velocity as a biophysical marker. Biochem Biophys Res Commun. 2007;353:835–840. doi: 10.1016/j.bbrc.2006.12.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Müller MJ, Klumpp S, Lipowsky R. Tug-of-war as a cooperative mechanism for bidirectional cargo transport by molecular motors. Proc Natl Acad Sci U S A. 2008;105:4609–4614. doi: 10.1073/pnas.0706825105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Kunwar A, Tripathy SK, Xu J, Mattson MK, Anand P, Sigua R, Vershinin M, McKenney RJ, Yu CC, Mogilner A, Gross SP. Mechanical stochastic tug-of-war models cannot explain bidirectional lipid-droplet transport. Proc Natl Acad Sci U S A. 2011;108:18960–18965. doi: 10.1073/pnas.1107841108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Welte MA, Gross SP. Molecular motors: a traffic cop within? HFSP J. 2008;2:178–182. doi: 10.2976/1.2956447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Hancock WO. Bidirectional cargo transport: moving beyond tug of war. Nat Rev Mol Cell Biol. 2014;15:615–628. doi: 10.1038/nrm3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Schuster M, Lipowsky R, Assmann MA, Lenz P, Steinberg G. Transient binding of dynein controls bidirectional long-range motility of early endosomes. Proc Natl Acad Sci U S A. 2011;108:3618–3623. doi: 10.1073/pnas.1015839108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Hendricks AG, Perlson E, Ross JL, Schroeder HW, 3rd, Tokito M, Holzbaur EL. Motor coordination via a tug-of-war mechanism drives bidirectional vesicle transport. Curr Biol. 2010;20:697–702. doi: 10.1016/j.cub.2010.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Deacon SW, Serpinskaya AS, Vaughan PS, Fanarraga ML, Vernos I, Vaughan KT, Gelfand VI. Dynactin is required for bidirectional organelle transport. J Cell Biol. 2003 doi: 10.1083/jcb.200210066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Gaspar I, Yu YV, Cotton SL, Kim D-H, Ephrussi A, Welte MA. Klar ensures thermal robustness of oskar localization by restraining RNP motility. J Cell Biol. 2014;206:199–215. doi: 10.1083/jcb.201310010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Starr DA, Fridolfsson HN. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu Rev Cell Dev Biol. 2010;26:421–444. doi: 10.1146/annurev-cellbio-100109-104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Fridolfsson HN, Ly N, Meyerzon M, Starr DA. UNC-83 coordinates kinesin-1 and dynein activities at the nuclear envelope during nuclear migration. Dev Biol. 2010;338:237–250. doi: 10.1016/j.ydbio.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Gross SP, Vershinin M, Shubeita GT. Cargo transport: two motors are sometimes better than one. Curr Biol. 2007;17:R478–486. doi: 10.1016/j.cub.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 239.Barkus RV, Klyachko O, Horiuchi D, Dickson BJ, Saxton WM. Identification of an axonal kinesin-3 motor for fast anterograde vesicle transport that facilitates retrograde transport of neuropeptides. Mol Biol Cell. 2008;19:274–283. doi: 10.1091/mbc.E07-03-0261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Uchida A, Alami NH, Brown A. Tight functional coupling of kinesin-1A and dynein motors in the bidirectional transport of neurofilaments. Mol Biol Cell. 2009;20:4997–5006. doi: 10.1091/mbc.E09-04-0304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Pilling AD, Horiuchi D, Lively CM, Saxton WM. Kinesin-1 and Dynein are the primary motors for fast transport of mitochondria in Drosophila motor axons. Mol Biol Cell. 2006;17:2057–2068. doi: 10.1091/mbc.E05-06-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Ling SC, Fahrner PS, Greenough WT, Gelfand VI. Transport of Drosophila fragile X mental retardation protein-containing ribonucleoprotein granules by kinesin-1 and cytoplasmic dynein. Proc Natl Acad Sci U S A. 2004 doi: 10.1073/pnas.0408114101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Ally S, Larson AG, Barlan K, Rice SE, Gelfand VI. Opposite-polarity motors activate one another to trigger cargo transport in live cells. J Cell Biol. 2009;187:1071–1082. doi: 10.1083/jcb.200908075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Nan X, Potma EO, Xie XS. Nonperturbative chemical imaging of organelle transport in living cells with coherent anti-stokes Raman scattering microscopy. Biophys J. 2006;91:728–735. doi: 10.1529/biophysj.105.074534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Yi JY, Ori-McKenney KM, McKenney RJ, Vershinin M, Gross SP, Vallee RB. High-resolution imaging reveals indirect coordination of opposite motors and a role for LIS1 in high-load axonal transport. J Cell Biol. 2011;195:193–201. doi: 10.1083/jcb.201104076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Schroer TA. Dynactin. Annu Rev Cell Dev Biol. 2004;20:759–779. doi: 10.1146/annurev.cellbio.20.012103.094623. [DOI] [PubMed] [Google Scholar]
- 247.Kardon JR, Vale RD. Regulators of the cytoplasmic dynein motor. Nat Rev Mol Cell Biol. 2009;10:854–865. doi: 10.1038/nrm2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Haghnia M, Cavalli V, Shah SB, Schimmelpfeng K, Brusch R, Yang G, Herrera C, Pilling A, Goldstein LS. Dynactin is required for coordinated bidirectional motility, but not for dynein membrane attachment. Mol Biol Cell. 2007;18:2081–2089. doi: 10.1091/mbc.E06-08-0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Tripathy SK, Weil SJ, Chen C, Anand P, Vallee RB, Gross SP. Autoregulatory mechanism for dynactin control of processive and diffusive dynein transport. Nat Cell Biol. 2014;16:1192–1201. doi: 10.1038/ncb3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Jope RS, Johnson GV. The glamour and gloom of glycogen synthase kinase-3. Trends Biochem Sci. 2004;29:95–102. doi: 10.1016/j.tibs.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 251.Morfini G, Szebenyi G, Elluru R, Ratner N, Brady ST. Glycogen synthase kinase 3 phosphorylates kinesin light chains and negatively regulates kinesin-based motility. EMBO J. 2002;21:281–293. doi: 10.1093/emboj/21.3.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Morel M, Authelet M, Dedecker R, Brion JP. Glycogen synthase kinase-3beta and the p25 activator of cyclin dependent kinase 5 increase pausing of mitochondria in neurons. Neuroscience. 2010;167:1044–1056. doi: 10.1016/j.neuroscience.2010.02.077. [DOI] [PubMed] [Google Scholar]
- 253.Siegfried E, Chou TB, Perrimon N. wingless signaling acts through zeste-white 3, the Drosophila homolog of glycogen synthase kinase-3, to regulate engrailed and establish cell fate. Cell. 1992;71:1167–1179. doi: 10.1016/s0092-8674(05)80065-0. [DOI] [PubMed] [Google Scholar]
- 254.Kracklauer M, Banks S, Xie X, Wu Y, Fischer JA. Drosophila klaroid encodes a SUN domain protein required for Klarsicht localization to the nuclear envelope and nuclear migration in the eye. FLY. 2007;1:75–85. doi: 10.4161/fly.4254. [DOI] [PubMed] [Google Scholar]
- 255.Mosley-Bishop KL, Li Q, Patterson K, Fischer JA. Molecular analysis of the klarsicht gene and its role in nuclear migration within the differentiating cells of the Drosophila eye. Curr. Biol. 1999;9:1211–1220. doi: 10.1016/s0960-9822(99)80501-6. [DOI] [PubMed] [Google Scholar]
- 256.Elhanany-Tamir H, Yu YV, Shnayder M, Jain A, Welte M, Volk T. Organelle positioning in muscles requires cooperation between two KASH proteins and microtubules. J Cell Biol. 2012;198:833–846. doi: 10.1083/jcb.201204102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Myat MM, Andrew DJ. Epithelial tube morphology is determined by the polarized growth and delivery of apical membrane. Cell. 2002;111:879–891. doi: 10.1016/s0092-8674(02)01140-6. [DOI] [PubMed] [Google Scholar]
- 258.Yadlapalli S, Yamashita YM. Chromosome-specific nonrandom sister chromatid segregation during stem-cell division. Nature. 2013;498:251–254. doi: 10.1038/nature12106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Yamashita YM. Nonrandom sister chromatid segregation of sex chromosomes in Drosophila male germline stem cells. Chromosome Res. 2013;21:243–254. doi: 10.1007/s10577-013-9353-0. [DOI] [PubMed] [Google Scholar]
- 260.Sanhueza M, Andrea Chai A, Smith C, McCray BA, Simpson TI, Taylor JP, Pennetta G. Network Analyses Reveal Novel Aspects of ALS Pathogenesis. PLoS Genet. 2015 doi: 10.1371/journal.pgen.1005107. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Myat MM, Lightfoot H, Wang P, Andrew DJ. A molecular link between FGF and Dpp signaling in branch-specific migration of the Drosophila trachea. Dev Biol. 2005;281:38–52. doi: 10.1016/j.ydbio.2005.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Dworkin I, Kennerly E, Tack D, Hutchinson J, Brown J, Mahaffey J, Gibson G. Genomic consequences of background effects on scalloped mutant expressivity in the wing of Drosophila melanogaster. Genetics. 2009;181:1065–1076. doi: 10.1534/genetics.108.096453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Zhao T, Gu T, Rice HC, McAdams KL, Roark KM, Lawson K, Gauthier SA, Reagan KL, Hewes RS. A Drosophila gain-of-function screen for candidate genes involved in steroid-dependent neuroendocrine cell remodeling. Genetics. 2008;178:883–901. doi: 10.1534/genetics.107.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Harbison ST, Chang S, Kamdar KP, Mackay TF. Quantitative genomics of starvation stress resistance in Drosophila. Genome Biol. 2005;6:R36. doi: 10.1186/gb-2005-6-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Long JB, Bagonis M, Lowery LA, Lee H, Danuser G, Van Vactor D. Multiparametric analysis of CLASP-interacting protein functions during interphase microtubule dynamics. Mol Cell Biol. 2013;33:1528–1545. doi: 10.1128/MCB.01442-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Kim DH, Cotton SL, Manna D, Welte M. Novel isoforms of the transport regulator Klar. PLoS One. 2013;8:e55070. doi: 10.1371/journal.pone.0055070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Rothballer A, Kutay U. The diverse functional LINCs of the nuclear envelope to the cytoskeleton and chromatin. Chromosoma. 2013;122:415–429. doi: 10.1007/s00412-013-0417-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Razafsky D, Hodzic D. Bringing KASH under the SUN: the many faces of nucleo-cytoskeletal connections. J Cell Biol. 2009;186:461–472. doi: 10.1083/jcb.200906068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Starr DA, Han M. Role of ANC-1 in tethering nuclei to the actin cytoskeleton. Science. 2002;298:406–409. doi: 10.1126/science.1075119. [DOI] [PubMed] [Google Scholar]
- 270.Rajgor D, Mellad JA, Soong D, Rattner JB, Fritzler MJ, Shanahan CM. Mammalian microtubule P-body dynamics are mediated by nesprin-1. J Cell Biol. 2014;205:457–475. doi: 10.1083/jcb.201306076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Patterson K, Molofsky AB, Robinson C, Acosta S, Cater C, Fischer JA. The functions of Klarsicht and nuclear lamin in developmentally regulated nuclear migrations of photoreceptor cells in the Drosophila eye. Mol Biol Cell. 2004;15:600–610. doi: 10.1091/mbc.E03-06-0374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Roux KJ, Crisp ML, Liu Q, Kim D, Kozlov S, Stewart CL, Burke B. Nesprin 4 is an outer nuclear membrane protein that can induce kinesin-mediated cell polarization. Proc Natl Acad Sci U S A. 2009;106:2194–2199. doi: 10.1073/pnas.0808602106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Fischer JA, Acosta S, Kenny A, Cater C, Robinson C, Hook J. Drosophila klarsicht has distinct subcellular localization domains for nuclear envelope and microtubule localization in the eye. Genetics. 2004;168:1385–1393. doi: 10.1534/genetics.104.028662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Fan SS, Ready DF. Glued participates in distinct microtubule-based activities in Drosophila eye development. Development. 1997;124:1497–1507. doi: 10.1242/dev.124.8.1497. [DOI] [PubMed] [Google Scholar]
- 275.Brendza RP, Serbus LR, Duffy JB, Saxton WM. A function for kinesin I in the posterior transport of oskar mRNA and Staufen protein. Science. 2000;289:2120–2122. doi: 10.1126/science.289.5487.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Zimyanin VL, Belaya K, Pecreaux J, Gilchrist MJ, Clark A, Davis I, St Johnston D. In vivo imaging of oskar mRNA transport reveals the mechanism of posterior localization. Cell. 2008;134:843–853. doi: 10.1016/j.cell.2008.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Edgar BA, Kiehle CP, Schubiger G. Cell cycle control by the nucleo-cytoplasmic ratio in early Drosophila development. Cell. 1986;44:365–372. doi: 10.1016/0092-8674(86)90771-3. [DOI] [PubMed] [Google Scholar]
- 278.Mason FM, Tworoger M, Martin AC. Apical domain polarization localizes actin-myosin activity to drive ratchet-like apical constriction. Nat Cell Biol. 2013;15:926–936. doi: 10.1038/ncb2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.De Renzis S, Elemento O, Tavazoie S, Wieschaus EF. Unmasking activation of the zygotic genome using chromosomal deletions in the Drosophila embryo. PLoS Biol. 2007;5:e117. doi: 10.1371/journal.pbio.0050117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.De Renzis S, Yu J, Zinzen R, Wieschaus E. Dorsal-ventral pattern of Delta trafficking is established by a Snail-Tom-Neuralized pathway. Dev Cell. 2006;10:257–264. doi: 10.1016/j.devcel.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 281.He B, Doubrovinski K, Polyakov O, Wieschaus E. Apical constriction drives tissue-scale hydrodynamic flow to mediate cell elongation. Nature. 2014;508:392–396. doi: 10.1038/nature13070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Liang HL, Nien CY, Liu HY, Metzstein MM, Kirov N, Rushlow C. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature. 2008;456:400–403. doi: 10.1038/nature07388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Tang AH, Neufeld TP, Rubin GM, Muller HA. Transcriptional regulation of cytoskeletal functions and segmentation by a novel maternal pair-rule gene, lilliputian. Development. 2001;128:801–813. doi: 10.1242/dev.128.5.801. [DOI] [PubMed] [Google Scholar]
- 284.Akdemir F, Christich A, Sogame N, Chapo J, Abrams JM. p53 directs focused genomic responses in Drosophila. Oncogene. 2007;26:5184–5193. doi: 10.1038/sj.onc.1210328. [DOI] [PubMed] [Google Scholar]
- 285.Bonneaud N, Savare J, Berta P, Girard F. SNCF, a SoxNeuro interacting protein, defines a novel protein family in Drosophila melanogaster. Gene. 2003;319:33–41. doi: 10.1016/s0378-1119(03)00795-9. [DOI] [PubMed] [Google Scholar]
- 286.Murali T, Pacifico S, Yu J, Guest S, Roberts GG, 3rd, Finley RL., Jr. DroID 2011: a comprehensive, integrated resource for protein, transcription factor, RNA and gene interactions for Drosophila. Nucleic Acids Res. 2011;39:D736–743. doi: 10.1093/nar/gkq1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Yu J, Pacifico S, Liu G, Finley RL., Jr. DroID: the Drosophila Interactions Database, a comprehensive resource for annotated gene and protein interactions. BMC Genomics. 2008;9:461. doi: 10.1186/1471-2164-9-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Bi J, Xiang Y, Chen H, Liu Z, Gronke S, Kuhnlein RP, Huang X. Opposite and redundant roles of the two Drosophila perilipins in lipid mobilization. J Cell Sci. 2012;125:3568–3577. doi: 10.1242/jcs.101329. [DOI] [PubMed] [Google Scholar]
- 289.Marcinkiewicz A, Gauthier D, Garcia A, Brasaemle DL. The phosphorylation of serine 492 of perilipin a directs lipid droplet fragmentation and dispersion. J Biol Chem. 2006;281:11901–11909. doi: 10.1074/jbc.M600171200. [DOI] [PubMed] [Google Scholar]
- 290.Claussen M, Suter B. BicD-dependent localization processes: from Drosophilia development to human cell biology. Ann Anat. 2005;187:539–553. doi: 10.1016/j.aanat.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 291.Schlager MA, Serra-Marques A, Grigoriev I, Gumy LF, Esteves da Silva M, Wulf PS, Akhmanova A, Hoogenraad CC. Bicaudal d family adaptor proteins control the velocity of Dynein-based movements. Cell Rep. 2014;8:1248–1256. doi: 10.1016/j.celrep.2014.07.052. [DOI] [PubMed] [Google Scholar]
- 292.Grigoriev I, Splinter D, Keijzer N, Wulf PS, Demmers J, Ohtsuka T, Modesti M, Maly IV, Grosveld F, Hoogenraad CC, Akhmanova A. Rab6 regulates transport and targeting of exocytotic carriers. Dev Cell. 2007;13:305–314. doi: 10.1016/j.devcel.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 293.Splinter D, Tanenbaum ME, Lindqvist A, Jaarsma D, Flotho A, Yu KL, Grigoriev I, Engelsma D, Haasdijk ED, Keijzer N, Demmers J, Fornerod M, Melchior F, Hoogenraad CC, Medema RH, Akhmanova A. Bicaudal D2, dynein, and kinesin-1 associate with nuclear pore complexes and regulate centrosome and nuclear positioning during mitotic entry. PLoS Biol. 2010;8:e1000350. doi: 10.1371/journal.pbio.1000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Dienstbier M, Boehl F, Li X, Bullock SL. Egalitarian is a selective RNA-binding protein linking mRNA localization signals to the dynein motor. Genes Dev. 2009;23:1546–1558. doi: 10.1101/gad.531009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Hain D, Bettencourt B, Okamura K, Csorba T, Meyer W, Jin Z, Biggerstaff J, Siomi H, Hutvagner G, Lai E, Welte MA, Müller H-AJ. Natural variation of the amino-terminal glutamine-rich domain of Drosophila Argonaute2 is not associated with developmental defects. PLoS ONE. 2010;5:e15264. doi: 10.1371/journal.pone.0015264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Rodriguez A, Zhou Z, Tang ML, Meller S, Chen J, Bellen H, Kimbrell DA. Identification of immune system and response genes, and novel mutations causing melanotic tumor formation in Drosophila melanogaster. Genetics. 1996;143:929–940. doi: 10.1093/genetics/143.2.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Kleinhesselink K, Conway C, Sholer D, Huang I, Kimbrell DA. Regulation of Hemocytes in Drosophila Requires dappled Cytochrome b5. Biochem Genet. 2011;49:329–351. doi: 10.1007/s10528-010-9411-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Loer B, Bauer R, Bornheim R, Grell J, Kremmer E, Kolanus W, Hoch M. The NHL-domain protein Wech is crucial for the integrin-cytoskeleton link. Nat Cell Biol. 2008;10:422–428. doi: 10.1038/ncb1704. [DOI] [PubMed] [Google Scholar]
- 299.Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell. 2000;5:659–669. doi: 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- 300.Maller Schulman BR, Liang X, Stahlhut C, DelConte C, Stefani G, Slack FJ. The let-7 microRNA target gene, Mlin41/Trim71 is required for mouse embryonic survival and neural tube closure. Cell Cycle. 2008;7:3935–3942. doi: 10.4161/cc.7.24.7397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Loedige I, Stotz M, Qamar S, Kramer K, Hennig J, Schubert T, Loffler P, Langst G, Merkl R, Urlaub H, Meister G. The NHL domain of BRAT is an RNA-binding domain that directly contacts the hunchback mRNA for regulation. Genes Dev. 2014;28:749–764. doi: 10.1101/gad.236513.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Loedige I, Gaidatzis D, Sack R, Meister G, Filipowicz W. The mammalian TRIM-NHL protein TRIM71/LIN-41 is a repressor of mRNA function. Nucleic Acids Res. 2013;41:518–532. doi: 10.1093/nar/gks1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Rybak A, Fuchs H, Hadian K, Smirnova L, Wulczyn EA, Michel G, Nitsch R, Krappmann D, Wulczyn FG. The let-7 target gene mouse lin-41 is a stem cell specific E3 ubiquitin ligase for the miRNA pathway protein Ago2. Nat Cell Biol. 2009;11:1411–1420. doi: 10.1038/ncb1987. [DOI] [PubMed] [Google Scholar]
- 304.O'Farrell F, Kylsten P. A mis-expression study of factors affecting Drosophila PNS cell identity. Biochem Biophys Res Commun. 2008;370:657–662. doi: 10.1016/j.bbrc.2008.04.030. [DOI] [PubMed] [Google Scholar]
- 305.Galewsky S, Schulz RA. Drop out: a third chromosome maternal-effect locus required for formation of the Drosophila cellular blastoderm. Mol Reprod Dev. 1992;32:331–338. doi: 10.1002/mrd.1080320405. [DOI] [PubMed] [Google Scholar]
- 306.Hain D, Langlands A, Sonnenberg HC, Bailey C, Bullock SL, Muller HA. The Drosophila MAST kinase Drop out is required to initiate membrane compartmentalisation during cellularisation and regulates dynein-based transport. Development. 2014;141:2119–2130. doi: 10.1242/dev.104711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Pearce LR, Komander D, Alessi DR. The nuts and bolts of AGC protein kinases. Nat Rev Mol Cell Biol. 2010;11:9–22. doi: 10.1038/nrm2822. [DOI] [PubMed] [Google Scholar]
- 308.Greenberg AS, Egan JJ, Wek SA, Garty NB, Blanchette-Mackie EJ, Londos C. Perilipin, a major hormonally regulated adipocyte-specific phosphoprotein associated with the periphery of lipid storage droplets. J Biol Chem. 1991;266:11341–11346. [PubMed] [Google Scholar]
- 309.Welte MA. Expanding roles for lipid droplets. Curr Biol. 2015 doi: 10.1016/j.cub.2015.04.004. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Encalada SE, Goldstein LS. Biophysical challenges to axonal transport: motor-cargo deficiencies and neurodegeneration. Annu Rev Biophys. 2014;43:141–169. doi: 10.1146/annurev-biophys-051013-022746. [DOI] [PubMed] [Google Scholar]
- 311.Franker MA, Hoogenraad CC. Microtubule-based transport - basic mechanisms, traffic rules and role in neurological pathogenesis. J Cell Sci. 2013;126:2319–2329. doi: 10.1242/jcs.115030. [DOI] [PubMed] [Google Scholar]
- 312.Mallik R, Carter BC, Lex SA, King SJ, Gross SP. Cytoplasmic dynein functions as a gear in response to load. Nature. 2004;427:649–652. doi: 10.1038/nature02293. [DOI] [PubMed] [Google Scholar]
- 313.Nicholas MP, Hook P, Brenner S, Wynne CL, Vallee RB, Gennerich A. Control of cytoplasmic dynein force production and processivity by its C-terminal domain. Nat Commun. 2015;6:6206. doi: 10.1038/ncomms7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.MacRae TH. Tubulin post-translational modifications--enzymes and their mechanisms of action. Eur J Biochem. 1997;244:265–278. doi: 10.1111/j.1432-1033.1997.00265.x. [DOI] [PubMed] [Google Scholar]
- 315.Reed NA, Cai D, Blasius TL, Jih GT, Meyhofer E, Gaertig J, Verhey KJ. Microtubule acetylation promotes kinesin-1 binding and transport. Curr Biol. 2006;16:2166–2172. doi: 10.1016/j.cub.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 316.Wolf N, Regan CL, Fuller MT. Temporal and spatial pattern of differences in microtubule behaviour during Drosophila embryogenesis revealed by distribution of a tubulin isoform. Development. 1988;102:311–324. doi: 10.1242/dev.102.2.311. [DOI] [PubMed] [Google Scholar]
- 317.Vershinin M, Carter BC, Razafsky DS, King SJ, Gross SP. Multiple-motor based transport and its regulation by Tau. Proc Natl Acad Sci U S A. 2007;104:87–92. doi: 10.1073/pnas.0607919104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Dixit R, Ross JL, Goldman YE, Holzbaur EL. Differential regulation of dynein and kinesin motor proteins by tau. Science. 2008;319:1086–1089. doi: 10.1126/science.1152993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Homem CC, Steinmann V, Burkard TR, Jais A, Esterbauer H, Knoblich JA. Ecdysone and mediator change energy metabolism to terminate proliferation in Drosophila neural stem cells. Cell. 2014;158:874–888. doi: 10.1016/j.cell.2014.06.024. [DOI] [PubMed] [Google Scholar]
- 321.Blythe SA, Wieschaus EF. Zygotic genome activation triggers the DNA replication checkpoint at the midblastula transition. Cell. 2015;160:1169–1181. doi: 10.1016/j.cell.2015.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Padmanabha D, Baker KD. Drosophila gains traction as a repurposed tool to investigate metabolism. Trends Endocrinol Metab. 2014;25:518–527. doi: 10.1016/j.tem.2014.03.011. [DOI] [PubMed] [Google Scholar]