Abstract
Phosphoinositide 3-kinases control many important aspects of immune cell development, differentiation and function. Mammals have eight PI3K catalytic subunits which are divided into 3 classes based on similarities in structure and function. Specific roles for the class I PI3Ks have been broadly investigated and are relatively well understood as is the function of their corresponding phosphatases. More recently, specific roles for the class II and class III PI3Ks have emerged. Through vertebrate evolution and in parallel with the evolution of adaptive immunity, there has been a dramatic increase not only in the genes for PI3K subunits but also in genes for phosphatases that act on 3-phopshoinositides and on 3-phosphoinositide binding proteins. Our understanding of the PI3Ks in immunity is guided by fundamental discoveries made in simpler model organisms as well as by appreciating new adaptions of this signalling module in mammals in general and immune cells in particular.
Keywords: PI3K, Akt, Foxo, mTOR, signalling, autophagy
Introduction
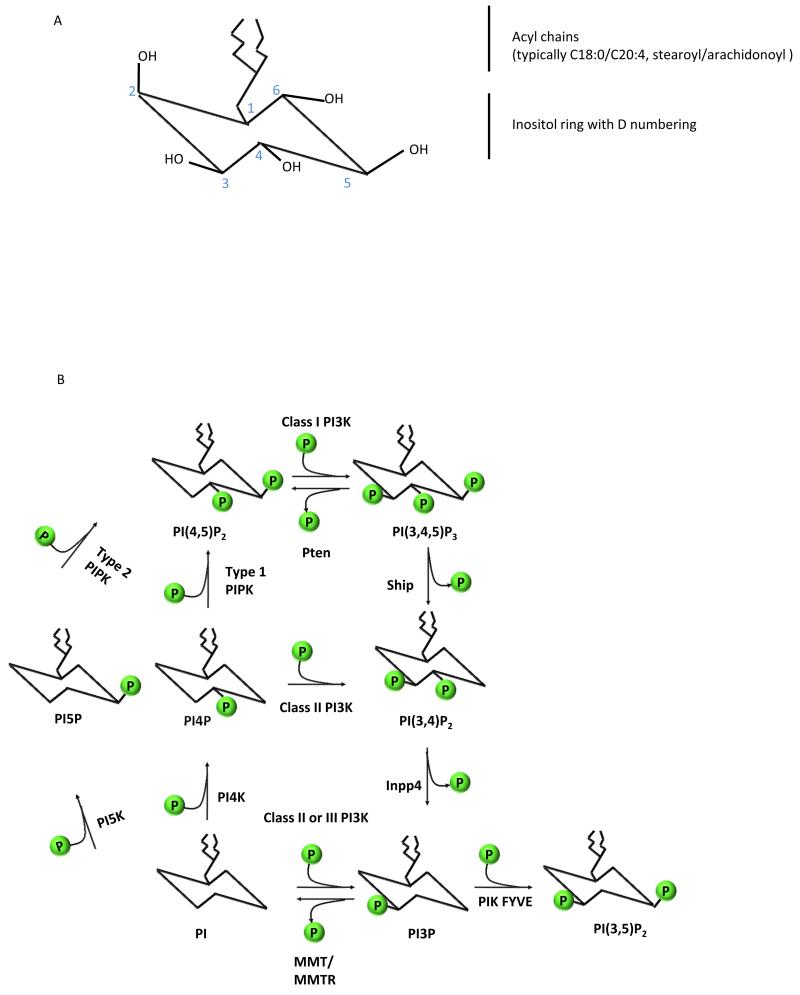
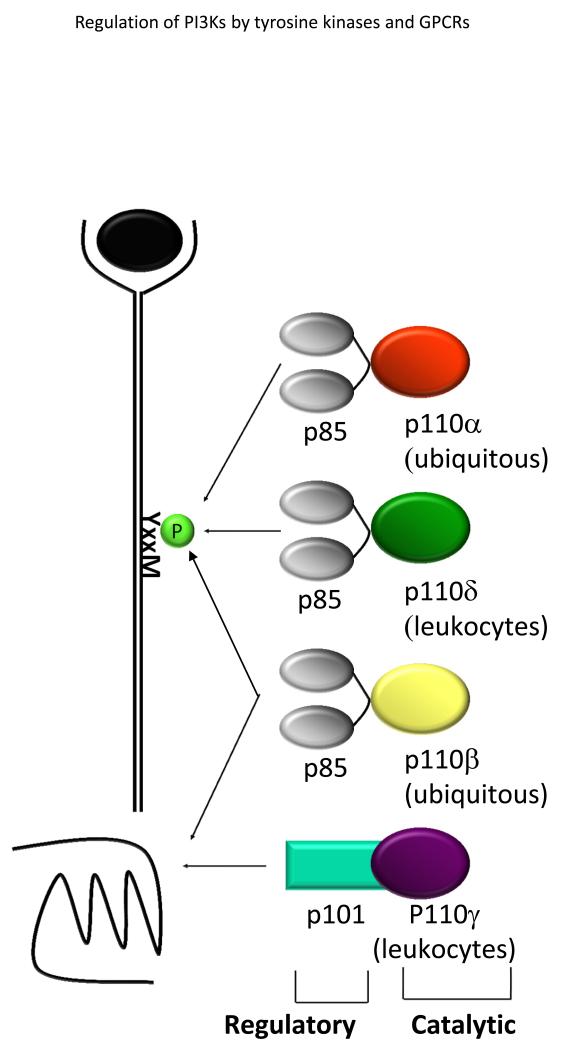
The phosphoinositides (PtdIns) represent a minor component of the plasma membrane, but play a major role in intracellular signalling. This review will focus on the role of PI3Ks which phosphorylate the inositol ring on the 3 position. The 3-phosphorylated inositol ring creates a docking site for proteins at the inner leaflet of the plasma membrane and on intracellular membranes. 3-phosphorylated phosphoinositides are subjected to the action of inositol phosphatases which either remove the 3-phosphate itself or additional phosphates on the 4 or 5 position. The asymmetry of the phosphorylated inositol ring and the highly regulated phosphorylation and de-phosphorylation of the 3, 4 and 5 positions has created a complex signalling code involving 7 differentially phosphorylated phosphoinositide species (1) (Fig 1).
Figure 1.
a) Numbering of the carbons of the myo-inositol ring.
b) Phosphatidyliositol and its 7 phosphorylated derivatives. The kinases and phosphatases shown are described in the text, except the PI4 and PI5 kinase that cooperate to generate PI(4,5)P2, the substrate of the class I PI3Ks.
The kinases and phosphatases, as well as the proteins that bind to phosphoinositides have been under strong selective pressure during vertebrate and mammalian evolution (2). Conserved functions of the PI3Ks include the control of intracellular vesicle trafficking, cellular motility and the regulation of nutrient metabolism. PI3Ks control these functions as well as more recently evolved and specialised aspects of cell biology necessary for the development and function of immune cells.
The PI3K family
There are 8 catalytic PI3K subunits encoded by vertebrate genomes and these are divided into three classes based on sequence alignment and domain structures (3, 4)(Fig 2). A number of tools have been developed over the last decade to probe the individual functions the PI3K catalytic and regulatory subunits. Gene targeted mice, selective small molecule inhibitors and crystal structures now exist for all the class I PI3Ks, for some of the class II and class III PI3Ks and also for some of the phosphatases that act on 3-phosphorylated phosphoinositides. These tools have contributed to a deep understanding of the isoform-selective functions among the class I PI3Ks and are beginning to unravel specific roles for class II and class III PI3Ks. This articles starts by reviewing what is known about the founding member of the family, Vps34, followed by a discussion on the class II PI3Ks. I then describe recent developments in our understanding class I PI3Ks in the latter half of the review, with particular focus on the role of the individual catalytic subunits.
Figure 2.
A. List of the genes and protein products of the mammalian PI3K family.
B. Phylogenetic tree of the 8 mammalian PI3K catalytic subunits. The tree was generated using ClustalW (http://www.ebi.ac.uk/Tools/phylogeny/clustalw2_phylogeny/)
C. The domain structures of the class III, II and I PI3Ks are represented by Vps34, CIIα and p110α, respectively. The domain structure of p85α is also shown. Not shown are class IB regulatory subunits and the Vps34 regulatory subunits. The Illustrations were generated using Prosite (http://prosite.expacy.org/scanprosite).
Class III PI3K: the founding member of the family
The class III PI3K Vps34 is the oldest PI3K and is the only PI3K found in yeast and plants as well as in metazoans (2). Vps34 phosphorylates PI to generate PI3P which is the most abundant of the phosphoinositides and which acts as a docking site for proteins that contain PX or FYVE domains (5)(Fig 1). Total PI3P levels do not change dramatically upon cell activation, however, local production can be significantly increased during the early stages of the formation of autophagic vesicles (omegasomes) or during endocytosis and phagocytosis (6-9). Vps34 was originally identified as gene required for protein sorting to the lysosome-like vacuole in yeast and subsequently shown to also control endocytosis, phagocytosis and autophagy in various cell types (10, 11). In yeast, Vps34 exists as part of two distinct complexes (12). A core complex consists of Vps15, Vps34 and Vps30 (also known as Atg6, the mammalian homologue of which is Beclin). Vps15 is a serine-threonine kinase which regulates the activity of Vps34 and which also brings Vps34 to lipid membranes (13). The role of Vps30 is less well understood but it is likely to act as an adapter protein. Vps34 complex I additionally contains Atg14 and is required for autophagy whereas Vps34 complex II contains Vps38 and is required for vacuolar sorting (12, 14). In mammalian cells, the picture is likely more complex, with the existence of three or more distinct Vps34-containing complexes (3, 5). Vps34 is also required for amino acid stimulation of mTOR activity in mammalian cells in mammalian cells (15, 16).
Currently, no selective inhibitors are available for Vps34, though 3-methyl adenine (3MA) is often used in conjunction with Wortmannin as an initial approach to implicate or exclude the role of Vps34 in a particular process (5). Such experiment need to be interpreted with caution since both Wortmannin and 3MA inhibits other PI3K isoforms as well (5). A crystal structure of Vps34 was recently published which not only revealed novel insight into regulation of Vps34 activity, but which should guide the development of more selective inhibitors (17). Vps34−/− mice die as embryos; however conditional knockouts are beginning to shed light on the role of Vps34 in selected mammalian cell types, including cells of the immune system (16, 18-20).
Role of PI3P in Phagocytosis and vesicle trafficking
In macrophages and neutrophils, PI3P accumulates on the phagocytic cup and is required for phagosome maturation (7, 8, 21). Vps34-produced PI3P is also required for the activation of the NOX2 NADPH oxidase which generates reactive oxygen species that contribute to the destruction of bacteria (22, 23). The p40 subunit of NOX2 contains a PI3P-binding PX domain and a mutation in this PX domain abrogates ROS production in responses to bacterial challenge of neutrophils (24). Recently, SLAM has been identified as a receptor on macrophages that can bind bacterial proteins and which is associated with a complex containing Vps15, Vps34 and beclin (25, 26). This is an interesting observation as the mechanism of recruitment of Vps34 to the membrane has been elusive (5). Phagosomes eventually fuse with lysosomes to which they deliver their cargo and therefore share some common features with autophagosomes, including their reliance on PI3P produced by Vps34.
Autophagosomes are PI3P-rich structures
Autophagy is generally thought of as a mechanism of organelle clearance during prolonged periods of starvation or when the organelles are no longer functional. Autophagosomes can be recognised by electron microscopy as double-membrane structures within the cells. Autophagosomes can also be detected by other methods using antibodies against LC3 (which become lipidated upon autophagy and forms puncta in the cytoplasm) or with probes that recognise PI3P (27, 28). Antigen presenting cells appear to have co-opted autophagy to recognise and destroy invading pathogens (29, 30). This insight was used to explain how viruses, which replicate in the cytoplasm, can be detected by TLRs in the lysosome of plasmacytoid dendritic cells (pDCs) where the autophagy pathway delivers virus particles from the cytoplasm to the lysosome (31). Both wortmannin and 3ME inhibited virus-induced autophagosome induction and TLR-dependent type 1 interferon response to the viruses (31). Moreover, these responses were absent in Atg5−/− pDCs suggesting that this process depends on autophagy. Autophagosomes can also target pathogens to MHC class II loading compartments providing a route for antigen cross-presentation (32, 33). A number of other studies have indicated that phagocytes more generally use autophagy to degrade intracellular pathogens and how pathogens have evolved to avoid autophagy (19, 29, 30). However, unequivocal evidence supporting a role for Vps34 in the recognition and destruction of pathogens awaits the development of selective inhibitors or gene-targeting of Vps34 in myeloid cells.
Vps34 is essential for peripheral T cell homeostasis
Autophagosomes can eliminate mitochondria by a process known as mitophagy. Normally, mitochondrial content is dramatically reduced as thymocytes differentiate to peripheral T cells, but T cells deficient in Atg proteins contain higher numbers of mitochondria and this is thought to negatively impact their survival in the periphery (34). Two recent publications have reached opposing conclusions regarding the role of Vps34 in T cell autophagy (19, 20). Both published reports showed relatively normal thymic development, but with fewer CD4 or CD8 single positive thymocytes and reduced numbers of T cells in the spleen and lymph nodes (19, 20). McLeod and colleagues showed stimulation with anti-CD3 and anti-CD28 induced the appearance of LC3 puncta in wild type and Vps34−/− T cells, but not in Atg3−/− T cells, suggesting that autophagosomes were formed independently of Vps34. Moreover, the appearance of LC3 puncta in Vps34−/− T cells could be blocked using the inhibitor 3MA (which also demonstrated that this reagent is not a specific inhibitor for Vps34). The authors argued that Vps34 was not required for autophagy in T cells, but rather for optimal IL-7 expression, as IL-7 was trapped in intracellular vesicles in Vps34−/− T cells (20). However, Willinger and Flavell argued against an intrinsic role for Vps34 in regulating IL-7R expression and instead showed that the reduced expression of IL7R on Vps34 T cells was secondary to the influence of a lymphopenic environment since normal IL7R expression was restored on Vps34−/− T cells in mixed bone marrow chimaeric mice (19). Although Willinger and Flavell detected lipidated LC3 protein in Vps34−/− T cells, they found that autophagic flux was impaired in Vps34−/− T cells. That is, basal autophagy was normal (as judged by appearance of lipidated LC3), but when autophagic vesicles fusion with the phagosomes was blocked by the toxin bafilomycin A, lipidated LC3 accumulated in WT cells to a greater extent than in Vps34−/− cells (19). How could the two reports reach such different conclusions? Part of the answer may involve differences in how the assays were carried out. Jaber and colleagues have shown that LC3 puncta can accumulate in Vps34−/− cells, but that these fail to localise to autophagosomes and instead exist as cytoplasmic protein aggregates (16). Thus Vps34 may be required for the targeting, rather than formation of LC3 puncta within cells. Consistent with this idea, Wipi2, which is a mammalian homologue of Atg18, binds PI3P and is required for the recruitment of LC3 to autophagosomes (35).
A further possibility that could explain differences between the two studies concerns the Vps34 gene targeting strategy. MacLeod et al used mice with Vps34 exon 17 and 18 were conditionally deleted (floxed), whereas Willinger and Flavells had floxed exon 4. Deletion of Exon 4 led to the disappearance of the entire protein whereas deletion of exons 17 and 18 could theoretically lead to the production of a truncated protein that would lack kinase activity, but which could stabilise beclin, for instance. The levels of Beclin were dramatically reduced in the mice with floxed exon 4 which might have had a strong impact on autophagy independently of the Vps34 kinase activity (19). However, to accept this explanation, one would have to argue that Vps34 plays a more important role as a scaffolding protein than as a kinase that decorates early autophagosomes (omegasomes) with PI3P. This notion is inconsistent with our understanding of how Vps34 works as PI3P is considered to be a key membrane tether for proteins involved in autophagy (35).
The most parsimonious explanation therefore seems to be that autophagic components such as LC3 can be assembled in cells in absence of Vps34 activity, but that Vps34 kinase activity is required for the correct targeting of LC3-containing protein complexes to the autophagosome and for autophagosome maturation. However, this interpretation does not exclude a key role for Vps34 in controlling endocytosis or normal trafficking of other membrane compartments and their cargo in T cells.
Regulation of potassium channels by the Class II PI3K C2β in T cells
Mammalians express three class II PI3Ks: C2α, C2β and C2γ. Although the C2 PI3Ks can phosphorylate PI or PI4P in vitro, their favoured substrate is thought to be PI (Refs 3, 36). Different receptors, such as insulin receptor, EGF receptor, GPCRs and TNF family receptors have been reported to activate C2 PI3Ks. The precise mechanism coupling these receptors to the C2 PI3K enzyme is unclear, but is likely to involve induced translocation of the enzyme to the plasma membrane where they interact with phospholipids, clathrin and calcium (3, 36). C2α−/− mice suffer from kidney failure and increased IgA depositions in the kidneys were observed. However, bone marrow reconstitution experiments suggested the kidney failure was not caused by altered immune responses in the C2α−/− mice (37). C2β−/− have so far not revealed any phenotypic abnormalities (38, 39). Nevertheless, Skolnik and colleagues have shown that C2β, but not C2α, can move towards the immune synapse formed by a Jurkat T cell and anti-CD3 antibodies embedded in a lipid bilayer (40). C2β colocalised with CD3 in the central supramolecular activation cluster suggesting that its translocation was under control of TCR signalling. Moreover, they found that PI3P generated by C2β could activate the potassium cannel KCa3.1 in CD4 T cells (40-44). Potassium channels such as KCa3.1 maintain negative membrane potential by facilitating the efflux of K+. This is required for the cells to maximise their intake of Ca2+ once the plasma membrane calcium channels have been opened. C2β regulation of KCa3.1 channels appears to involve the activation of nucleoside diphosphate kinase b (NDPK-b) which is required for the phosphorylation of a histidine residue within KCa3.1 (44). However, NDPK-b does not contain any recognised PI3P binding motive, so exactly how it is activated by C2β is unclear.
PIKfyve and PI(3,5)P2
PI3P is not only a tether for PX and FYVE domains, but also a substrate for the kinase PIK-FYVE, known as Fab1 in yeast where it was first identified (45). Fab1 phosphorylates PI3P to generate PI(3,5)P2. Fab1 yeast mutants fail to recycle proteins and membranes from the vacuoles to other compartments and consequently have very large vacuoles (46). Little is known about the role of PI(3,5)P2 in immune cells; however a mouse knockout has been generated and these mice die early during embryogenesis (47). Moreover, inhibition of PIKfyve has also been shown to limit infection by Salmonella in epithelial cells (48). It would not be surprising if PIKfyve also turns out to play important roles in APCs and other immune cells.
Myotubularins are PI3P phosphatases
PI3P is dephosphorylated by the myotubularins (MTM and the related MTMRs) of which there are 14 members in mammals (49, 50). Intriguingly, 6 of these lack phosphatase activity, but pair up with phosphatase active members to increase their activity and/or target them to the correct compartment. Mutations in genes in the MTM family are the cause of various neuromuscular diseases (49). MTMR6 can inhibit the Kca3.1 channel in T cells (43), but little is known more generally about the role of the MTMRs in immune cells. However, in Drosophila, one of the 7 MTMRs, Mtm was found to antagonise the function of In haemocytes (insect immune cells) under circumstances where Vps34 appeared not to contribute significantly (51). Further work is required to determine whether some MTMRs preferentially antagonise Vps34 signalling and other C2 PI3K signalling, but given the abundance of the MTMR subunits and the number of possible heterodimeric complexes, this will not be a simple task.
The Class I PI3Ks generate PI(3,4,5)P3 in response to receptor activation
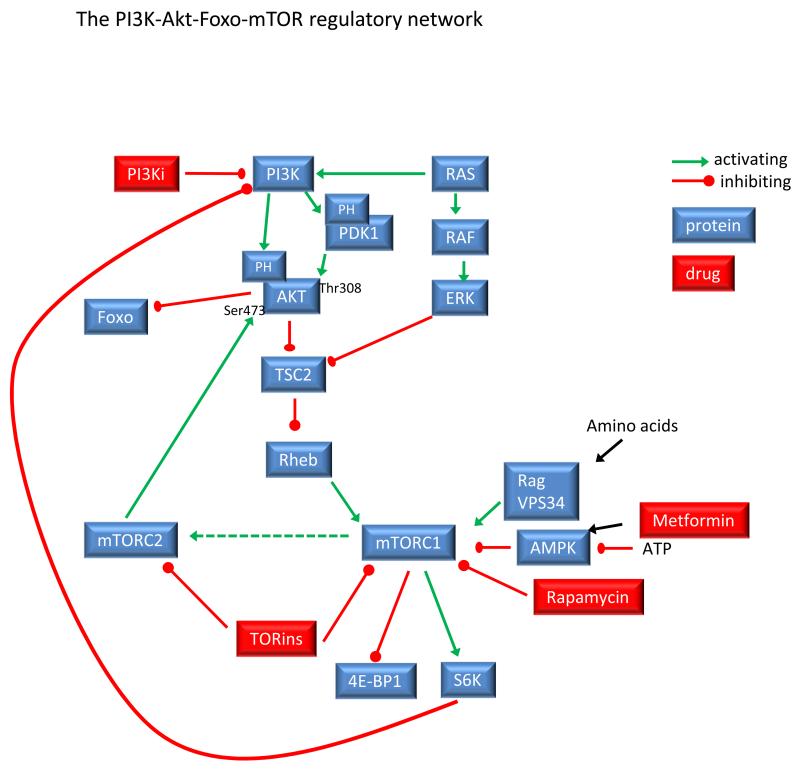
The class I PI3Ks are heterodimeric proteins containing and adapter subunit and a catalytic subunit. C. elegans and Drosophila have one class I isoform each, whereas in vertebrates, the class I PI3K subunits are further subdivided into class IA (p110α, p110β and p110δ) and class IB (p110γ) (2, 3). The class IA subunits are associated with SH2 containing regulatory subunit, of which there are several types: p85α, p55α, p50α, p85β and p55γ (collectively referred to as p85 in this review; a detailed review on the distinct role of the class IA PI3K-regulatory subunits was published recently (52)). The p85 SH2 domains recruit p110α, p110β or p110δ to proteins that have been phosphorylated on YXXM motifs by tyrosine kinases. By contrast p110γ is found as a heterodimer with p101 or p84, both of which bind Gβγ-subunits and therefore recruit p110γ to activated G protein coupled receptors (GPCRs). At the plasma membrane, PI3K activity is further enhanced by the interaction between GTP-Ras and the Ras binding domain in the p110 subunit (53). All the class I PI3Ks have in common that their preferred substrate is PI(4,5)P2 which is converted to PI(3,4,5)P3 (simply known as PIP3 since this is the only tris-phosphorylated phosphoinositide) (1). PIP3 acts as a membrane-tether for a subset of proteins containing one or more PH domains (described in greater detail below).
Why do immune cells express 4 class I PI3K catalytic subunits?
Individual roles for each of the class I PI3Ks have been described in immune cells using knockout mice, kinase-dead knockin mice and specific inhibitors (54-63). P110α and p110β are broadly expressed in most cell types, whereas p110γ and p110δ expression is more limited to cells of the immune system (3). It is easy to rationalise why p110γ plays a unique role in many settings by virtue of its distinct p101 and p84 regulatory subunits (Fig 3). Accordingly, p110γ is a key signalling protein engaged by chemokine receptors in the immune system (64). The surprising finding, especially for the class IA PI3Ks, is there is often lack of redundancy between these subunits in different cellular contexts. If the class IA PI3Ks bind the same adapter proteins and phosphorylate the same substrate, then what makes them different? Some clues are emerging. By a mechanism that has yet to be elucidated, p110β can also be activated by GPCRs in many cell types, especially those that do not express high levels of p110γ (65-67). Furthermore, p110β was shown to integrate signals from tyrosine-kinase linked FcγR receptor and the GPCR coupled leukotriene receptor BLT1 in neutrophils suggesting a specialised function for p110β as a coincident tyrosine kinase and GPCR signalling detector (56)(Fig 3). Surprisingly, In NK cells and B cells, p110δ is activated by chemokine receptors (68, 69); however, in general, p110δ is less effectively activated by GPCRs than are p110γ and p110β. If one considers that p110β compensates for the lack of p110γ in non-immune cells and in addition is especially adept at integrating tyrosine kinase and GPCR signals then this can be considered as an adaption that distinguishes p110β from p110α and p110δ. What then are the differential roles of p110α and p110δ? How are these activated differently by different receptors in different cell types? Before we consider this question, it is worth reviewing some recent developments that have shed new light on how class IA PI3Ks are activated.
Figure 3.
Schematic illustrating the recruitment of class I PI3Ks to tyrosine kinase-linked receptors and GPCRs. P110β is illustrated as a coincidence detector for tyrosine kinase and GPCR signalling.
The unique role of p110α in human cancers
PIK3CA, the gene for p110α is the one of the most frequently mutated oncogenes in human cancer (70). The mutations in PIK3CA centre around three hotspots where E542K, E545K and H1047R are the most common amino acid substitutions, respectively (71-73). In the p85-p110 heterodimer, the p85 SH2 and iSH2 domains make several contacts with the p110 kinase domain (72, 74-76). These are inhibitory contacts that are relieved upon engagement of the SH2 domain by a tyrosine phosphorylated peptide (such as would be found on an activated receptor or its associated adapter proteins)(77-79). The PIK3CA E542K and E545K mutants exhibits higher basal kinase activity which is not further increased by the binding of pY peptides to the p85 SH2 domain (80). Corresponding oncogenic mutations in p85 have also been identified which abrogate the inhibitory effect on p110 (81, 82).
The ability of oncogenic mutants of Ras to transform cells depends, at least in part, on their ability to activate p110α (53, 83). Moreover, the Ras binding domain of p110α is required for normal development of lymphatic vesicles (83). It is therefore of interest that the H1047R mutation increases the association of p110α with the plasma membrane and also renders p110α less dependent on Ras activity (80, 84). The high penetrance of the PIK3CA E545K, H1047R and similar mutations in human cancers, and how these mutations bypass the requirements for upstream activating events, has thus revealed how important dual input of p85 and Ras are for optimal activation of p110α under normal physiological conditions.
More stringent criteria for the activation of p110β and p110δ than for p110α
Structure function experiments have revealed interesting differences in how p110α, p110β and p110δ are regulated by p85 and Ras. Some of the greatest sequence divergence between the class I PI3K isoforms is found within their Ras binding domains (85). The Ras binding domain of p110α and p110γ can bind most members of the Ras family, including H-Ras, K-Ras and N-Ras which are key signal transducers by tyrosine-kinase linked receptors and frequently mutated oncogenes (86). P110β showed very limited activation by Ras in in vitro assays, whereas P110δ was activated by R-Ras and TC21 (also known as R-Ras2), but not by the other Ras proteins (86). Indeed, gene targeting studies have shown that TC21 contributes to p110δ activation in B cells and T cells (87). Whereas the optimal activation of p110α by p85 requires the engagement of the N terminal SH2 domain of p85 by a tyrosine phosphorylated peptide, both the N-terminal and C-terminal SH2 domains of p85 need to be thus engaged for optimal activation of p110β or p110δ (79, 88). Therefore, whether the activation signal comes from p85 or from Ras, the activation requirements for p110δ and p110β appear to be more stringent than for p110α. Indeed, in fibroblasts, insulin responsive tissues, endothelial cells and epithelial tumour cells, p110α is the main isoform that generates PIP3 in response to activation by tyrosine-kinase linked receptors (89-92). In immature pre-B cells, p110α and p110δ show complete redundancy (55). However, p110α deletion has minimal effect on the phosphorylation of Akt in response to activation of the BCR, whereas p110δ inhibition nearly completely ablates Akt phosphorylation in mature B cells in which both isoforms are expressed (54, 55, 61, 93, 94). Similarly, p110δ is the main isoform activated by various tyrosine kinase-linked receptors on T cells, mast cells, macrophages, and dendritic cells (95). How cells of the immune system evolved such exquisite dependence on p110δ remains an unresolved conundrum – made even more difficult to understand given that p110δ in principle should be more difficult to activate than p110α which is also expressed in cells of the immune system, albeit at slightly lower levels (79, 86, 96). One interesting observation is that in a cellular transformation assay where the oncogenic potential of the p110 units were tested, p110α was dependent on its RBD to cause transformation, whereas p110δ was not (80, 84). These observations raise the possibility that p110δ activity is less dependent on Ras. Consistent with this possibility, the Ras binding domain of p110δ is differently orientated relative to the kinase domain than in p110α (97). Additional questions remain about how the p85 SH2 domains are productively engaged in immune cells (95). Bidentate YXXM…YXXM motifs which could simultaneously engage both SH2 domains of p85 and which are deemed to be required for optimal activation of p110δ (79) are not commonly found in mature immune cells (a notable exception being CD19 in B cells (98)). Indeed, in many contexts, the proteins and the specific tyrosine residues involved in the recruitment of p85-p110 heterodimers in cells of the immune system remain incompletely characterised (95).
PIP3 and PI(3,4)P2 are lipid second messenger signalling molecules
PIP3 is a short lived lipid species that is rapidly degraded by one of two pathways (Fig 1). The lipid phosphatase Pten is the only enzyme known to be able to convert PIP3 back to PI(4,5)P2 (99). Ship1 and Ship2 are members of a larger family of inositol 5 phosphatases that can convert PIP3 to PI(3,4)P2 (99). Ship phosphatases are of particular interest to immunologists as they can be recruited by their SH2 domains to receptors that contain immune tyrosine inhibitory motifs (ITIMs) found in FcγRIIB (100). The Akt PH domain binds non-discriminately to PI(3,4)P2 and PIP3, whereas other PH domains bind selectively to either lipid (101-103). As a consequence, PI(3,4)P2 may have unique signalling properties by virtue of its selective recruitment or proteins such as Bam32 and TAP (104). The deletion of Pten in thymocytes leads to aggressive T cell lymphoma and autoimmunity (105, 106). However, Pten can be deleted in mature T cells without causing transformation and instead renders the T cells hypersensitive to stimulation (105, 107, 108). This hypersensitivity can be attenuated by p110δ inhibition (108). Deletion of Pten alone in B cells prevents the induction of anergy, leads to increased production of natural antibodies, but also prevents the B cells from undergoing class switching (109-111). The latter phenomenon will be described further in a subsequent section. The effect of Pten deletion in B cells is partially restored by simultaneous deletion or inhibition of p110δ (93, 109). Deletion of Ship leads to lethal autoimmunity caused by the aberrant activation of T cells, B cells and myeloid cells (112). Moreover, the deletion of both Pten and Ship in B cells leads to leukaemic transformation (113). The phosphatase INPP4B removes the 4 phosphate from PI(3,4)P2. Solid tumours lacking both PTEN and INPP4B have found, suggesting that these phosphatases work in concert to extinguish class I PI3K signals (i.e. by degrading PIP3 and PI(3,4)P2, respectively)(114, 115). These results also suggest the PIP3 and PI(3,4)P2 work together for optimal activation of downstream pathways (112). The role of INPP4B or the related INPP4A in immune cells is not yet known.
Signalling by class I PI3Ks
PH domains must have sufficient to affinity for PIP3 relative to other phosphoinositides to be selectively regulated by class I PI3Ks. For instance, a PH domain that binds equivalently to PI(4,5)P2 and PIP3 is unlikely to be influenced by PI3K signalling since PI(4,5)P2 is much more abundant in the lipid membrane (116). Instead, such proteins may use phosphoinositides more generally to facilitate their interactions with plasma membranes (117, 118). In mammals, about 40 of the 200+ proteins containing PH domains can potentially be controlled by PIP3 (116, 119, 120). However, most work to date has focused on the regulation of the Akt pathway and its role in controlling the activation of mTOR and transcription by Foxo (Figure)(121). There is a strong rationale for focusing on this pathway given that it is conserved through evolution and because the Foxo transcription factors and mTOR play important roles in immune responses (121-124). Akt can also phosphorylate a plethora of other substrates that can influence the function of immune cells (125). Vertebrates express three Akt isoforms, Akt1, Akt2 and Akt3 (also known as PKBα, PKBβ and PKBγ). Akt activity is controlled by the binding of PIP3 or PI3,4P2 to its PH domain and by phosphorylation on Thr308 by Pdk1 and Thr473 by mTORC2 (125). The different Akt isoforms share some common substrates, but evidence is also emerging for isoform selective substrates (126). Each of the Akt isoforms have shown to be required for haematopoietic stem cell survival, T cell development and/or B cell development (127-131).
Akt-Foxo
Phosphorylation of Foxo1, Foxo3 and Foxo4 by Akt leads to their nuclear exclusion and degradation (121). Key Foxo target genes in lymphocytes include the Rag recombinases, Ikaros, the Il7r, Foxp3, Ccr7 and Cd62l (121, 132-143). By activating class I PI3Ks, certain Foxo target genes can be turned off. It follows that class I PI3K-Akt signalling needs to be inhibited to turn Foxo target genes on or off. Hence, the temporal activation and inhibition of class I PI3Ks and Akt during immune development is of key importance and this is a theme we will return to in context of B cell development and Treg differentiation. Akt also controls gene expression by regulating mRNA stability. In a human glioma cell line, 40% of genes whose expression was down-regulated upon PI3K inhibition showed reduced mRNA stability (144). mRNA stability is controlled by so-called ARE-binding proteins which bind to AU-rich sequences in the 3′ untranslated region of mRNAs (145). Phosphorylation of Foxo proteins and ARE-binding proteins may represent the major mechanisms through which class I PI3Ks control gene expression.
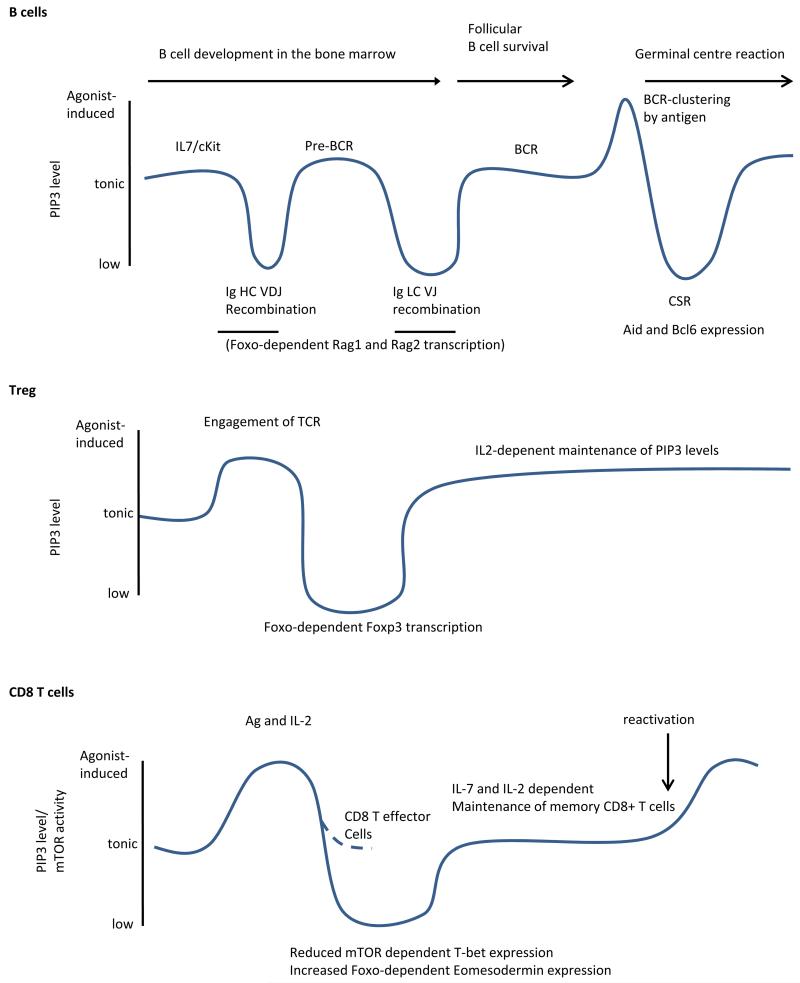
Akt-mTOR
mTOR is a serine threonine kinase with whose catalytic domain shows homology to the PI3Ks, and which along with DNAPK and ATM is sometime referred to as a class IV PI3K (even though neither protein can phosphorylate phosphoinositides) (2). mTOR exists as part of two protein complexes, the rapamycin sensitive complex1 (mTORC1 which contains Raptor) and the rapamycin insensitive complex 2 (mTORC2 which contains Rictor) (122, 146)(Fig 4). mTORC1 activity is controlled by the small GTPase Rheb. The GTPase activating domain of Tuberin (Tsc2) increase the rate of hydrolysis of Rheb-bound GTP, returning Rheb to the inactive GDP-bound form. Thus, by phosphorylating and inhibiting Tsc2, Akt releases Rheb from the inhibition by Tsc2 and allows GTP-Rheb to activate mTORC1. In addition to Akt, both Erk and Rsk can phosphorylate Tsc2, and hence activate the mTOR pathway (147). Thus while mTOR requires receptor initiated signals (e.g. from the TCR or BCR), there is redundancy in the pathway leading to its activation (132, 148). Moreover mTOR will only be activated if there is sufficient amino acid availability and ATP in the cells (otherwise initiating protein or lipid synthesis, both of which are key consequences of mTOR activation, would be futile). AMPK, which is active when AMP/ATP ratios are high, inhibits mTOR whereas amino acids activate mTOR through a mechanism that has yet to be fully elucidated, but which involves the assembly of proteins of the Rag family at the lysosome (149, 150). Exactly how mTORc2 gets activated is not known. However, its substrate Ser473 on Akt is of key interest for the current discussion on class I PI3K signalling.
Figure 4.
Diagram illustrating the circular network that connects PI3K signalling to MTOR activation. Regulatory subunits are not shown. How Vps34 contributes to amino acid –dependent mTOR activity is incompletely understood. PI3Ki indicates various PI3K inhibitors, some of which can also inhibit mTOR. TORins describes ATP-competitive mTOR inhibitors. Rapamycin blocks mTORC1 specifically via its association with FKBP12 (not shown).
The best characterised substrates for mTORC1 are S6K and 4EBP1 (146). These in turn contribute to protein synthesis by phosphorylating ribosomal S6 and by releasing the inhibition of elongation factors, respectively. A more comprehensive analysis of mTORc1 substrates has recently been published, but the function of most of these have yet to be established explicitly in immune cells (151, 152). Among the key genes whose expression is dependent on mTORc1 is HIF1a which is required to increase glycolysis in lymphocytes (153, 154). mTORC1 can also negatively regulate class I PI3K signalling via different mechanisms including phosphorylation of receptors. Thus acute inhibition mTORc1 can enhance class I PI3K signalling (146). However, persistent administration of Rapamycin has a knock-on effect on mTORc2 and hence leads also to the inhibition of Akt (155). This dual effect of mTORC1 inhibition was recently demonstrated in CD8 T cells where Rapamycin enhanced Akt Ser473 phosphorylation for the first 12 hours, but subsequently suppressed Akt Ser473 phosphorylation (thus enhancing Foxo activity) (156). ATP-analogues that inhibit mTOR regardless of which complex it finds itself in have been developed and these will also inhibit Akt Ser473 phosphorylation regardless of the duration of inhibition. The complexity of this circular pathway, where Akt is both upstream and downstream of mTOR, means that on has to take some care when interpreting experiments using Rapamycin or other mTOR inhibitors as these can also affect Akt substrates, including Foxo (Fig 4).
PIP3-dependent activation of Tec-family tyrosine kinases
Another important family of PIP3-bining proteins are the Tec kinases represented by Itk, Btk and Tec (157). A fourth member, Rlk lacks a PIP3-binding PH domain. Drosophila express a Btk orthologue which also has a PIP3-binding PH domain suggesting an evolutionary conserved role for PI3K regulation of Tec kinases (119). It is important to appreciate, however, that Tec kinases also contain SH2 and SH3 domains that can recruit these kinases to protein complexes at the plasma membrane. Thus PI3Ks may not be absolutely required for the recruitment and activation of Tec kinases, but rather make more subtle contributions to their function once recruited by other mechanisms. The Xid mutation in mice is caused by a mutation in the PH domain which renders Btk insensitive to PIP3 (158). This was seen as strong genetic evidence that Btk requires PIP3 for its full activation. However, PI3K and Btk-deficient phenotypes are non-overlapping and subsequent studies by Koyasu and colleagues showed that the Xid mutation causes Btk to be less stable and as such acts as a hypomorphic mutation (159, 160). Direct evidence that PI3K is required for the other Tec kinases (Tec and Itk) in primary immune cells is lacking and may require the generation of new knockin mutations in their PH domain that do not affect protein stability.
PIP3-dependent activation and inhibition of small GTPases
In many cell types, class PI3Ks control the activation the small GTPase Rac (161). P-Rex1 was purified as a protein that controlled the activation of Rac in neutrophils (162). Sequential activation of p110γ and p110δ contributes to many important facets of neutrophil activation, including migration and ROS production (64, 163). P-Rex contains a PH domain which binds PIP3 with high selectivity and a GEF domain that catalyses the exchange of Rac-bound GDP for GDP. P-Rex is in part redundant with Vav proteins in neutrophils where these proteins regulate chemotaxis, ROS production and adhesion (164). The Vav proteins also contain PH domains that can bind PIP3, but the Vav PH domain also binds PIP2 and it seems less clear that Vav activation is acutely regulated by PI3K signalling (117, 118). Rather, increasing evidence suggests that Vav is an important activator of PI3K as both the PH domain of Vav and its catalytic activity are required for optimal PI3K activity (117, 165).
Curiously PI3K is also required for the activation of the Rac-GAP ArhGAP15 which contains a PH domain and which is recruited to the plasma membrane in a PIP3-depedent manner (166, 167). ArhGAP15−/− neutrophils show enhanced chemotaxis, ROS production and phagocytosis (167). A major outstanding question is how the positive and negative effect of PIP3 via P-Rex1 and ArhGAP15, respectively, are balanced. Do they control distinct pools of Rac or are the respective PI3K-dependent GEF and GAP activities separated temporally?
Arap3 is a Rho-GAP which contains a remarkable 5 PH domains, two of which bind PIP3 synergistically (120, 168). In addition, Arap3 is activated by Rap1. ARAP3−/− neutrophils adhere more strongly to integrin ligands, thus interfering with normal chemotaxis (169). Thus emerging evidence suggests that in addition to be controlled by small GTPases of the Ras family, PI3Ks can both positively and negatively control the activation of diverse small GTPases by stimulating GEFs and GAPS.
Role of individual class I PI3K p110 catalytic subunits and opposing phosphatases in the immune system
Several reviews have considered the role of class I PI3K in the immune system more generally (170-175); here I will focus on more recent studies that have elucidated individual roles for each of the catalytic subunits as well as studies that have emphasised the requirement to increase or suppress PI3K activity at particular stages during development and differentiation.
Class I PI3K signalling promotes HSC differentiation
Haematopoiesis in p110δ and p110γ deficient mice appears normal, however, mice lacking all class I PI3K activity in HSCs have yet to be reported. The role of PI3K activity in HSCs has been investigated using complementary approaches, however. Deletion of Pten led to increased proliferation of HSCs at the expense of long-term repopulation capacity. There was reduced retention of white blood cells in the bone marrow and, instead, blood and spleens were enriched in myeloid cells, eventually leading to myeloma and T cell lymphomas (176, 177). Remarkably, treatment of the mice with the mTOR inhibitor rapamycin not only depleted the leukemic-initiating cells, but also restored the HSC compartment (177). These experiments therefore revealed a differential dependence on the PI3K pathway for normal HSC maintenance (requiring low flux through class I PI3K to mTOR) and leukaemia initiating cells (favoured by elevated class I PI3K/mTOR signalling). The deletion of Foxo-1, -3 and -4 also led to depletion of repopulating HSCs and an increase in myelopoiesis (178). The effects could be reversed by the anti-oxidant N-acetyl-L-cysteine (NAC), consistent with the observation that genes that regulate ROS are enriched among those controlled by Foxo transcription factors. By contrast, HSCs lacking Akt1 and Akt2 showed increased quiescence, reduced ROS, and again, reduced HSC repopulation capacity (128). Together, these reports emphasise that the class I PI3K/Akt/mTOR/Foxo pathways need to be carefully balanced for optimal HSC renewing capacity and hence homeostasis of the haematopoietic system.
Role of p110α and p110δ in B cell development and function
B cell development occurs through several stages characterised by checkpoints for quality control (179). The first such checkpoint occurs after the rearrangement of the immunoglobulin heavy chain. Only pre-B cells which functionally rearranged the Igh locus and express the heavy chain in conjunction with surrogate light chains develop further. Mice lacking p110α and p110δ activity in lymphocytes show a complete block at this stage of development, whereas p110β and p110γ do not contribute significantly (55). The precise reason for this block in development is not clear, but may reflect requirement PI3K to transmit signals from the pre-BCR promoting further differentiation. Defective B cell development in p110α-p110δ deficient mice could also be caused by impaired IL7R signalling. Phosphorylation of Tyr449 in the IL7Ra chain can lead to recruitment and activation of PI3K via p85; however a Tyr449Phe mutation does not block early B cell development (180, 181). Nevertheless, signalling via the pre-BCR can sensitise the developing B cells to limiting amounts of IL-7. Thus, even if recruitment of p85/p110 to the IL7R itself is not essential, IL7 responsiveness could be affected by defective pre-BCR signalling in absence of p110α and p110δ. (182). Indeed, p110α-p110-deficient pre-B cells showed impaired proliferative responses to IL-7 in culture and addition of antibodies against the pre-BCR failed to rescue development of p110α-p110δ deficient pre-B cells in vivo suggesting that both pre-BCR signalling and IL7 responsiveness are compromised in p110α-p110δ double deficient pre-B cells (55). Blocked class I PI3K signalling can also lead to the failure to turn the expression of Rag off (133, 134). Attenuation of Rag expression is necessary to allow for clonal expansion before Rag is re-expressed to rearrange the κ or λ light chains (179). Indeed, pre B cells lacking p110α and p110δ activity show high expression of Rag, had rearranged Igh genes and expressed the heavy chain (55). By contrast, pre B cells that lack Foxo protein fail to express Rag proteins and the IL7-R and are therefore blocked in their development (133, 134). The latter results indicate the constitutively high PI3K activity would also be detrimental for normal B cell development as it would prevent the expression of important Foxo target genes. Jumaa et al have proposed a model whereby the pre-BCR-associated adapter protein SLP65 eventually leads to the suppression of class I PI3K activity and that this is required for B cells to progress towards a stage of development when the light chains are rearranged to form the mature BCR with Igh (179, 183, 184). After the light chain rearrangement is completed, class I PI3K activity may yet again be required for further development. Basal levels of class I PI3K activity in mature B cells is also required to suppress RAG expression and prevent spurious Ig gene rearrangements in immature and mature B cells (185-187). Because neither p110α or p110δ deficiency alone significantly affect B cell development in the bone marrow, yet double deficient cells are blocked at pre-B cell stage, p110α and p110δ can be considered redundant with regards to early B cell development (55). In fact, the expression of p110α from a single allele was sufficient to promote normal B cell development in the bone marrow in absence of p110δ, though the spleen was nearly devoid of B cells in these mice (55).
The different roles of p110α and p110δ in tonic and agonist induced BCR signalling
Tonic class I PI3K signalling by the BCR is required to keep mature B cells alive (188-190). The requirement for tonic signalling generally was initially demonstrated by the loss of B cells after induced deletion of the BCR or the associated Igα signalling adapter (188, 189). Strikingly, expression of a membrane targeted p110α transgene, the deletion of Foxo or the deletion of Pten were each able to rescue the survival of B cells that had lost BCR expression (190). Moreover, gradual loss p110α and p110δ activities led to a dose-dependent loss of follicular B cells although the loss of p110δ activity had a more profound affect than the loss of p110α activity (55). Therefore, in terms of B cell survival, some redundancy between p110α and p110δ was evident.
However, BCR-dependent Akt phosphorylation, proliferation, development of marginal zone B cells and the production of natural antibodies were blocked by p110δ deficiency, but were unaffected by p110α deficiency (55). Why can p110α no longer compensate for the loss of p110δ under these circumstances? Pre B cell selection and follicular B cell survival depend on the pre BCR and mature BCR, respectively, but there is no requirement for crosslinking of the BCR by antigen. The development of MZ B cells, by contrast, is facilitated by self-antigen recognition by the BCR, as do strong phosphorylation of Akt (some Akt phosphorylation can be detected in resting B cells). Indeed, transgenic expression of p110α only moderately increased phosphorylation of Akt (190). It appears therefore that tonic BCR signalling leading to low levels of Akt phosphorylation can be mediated by either p110α or p110δ, whereas agonist induced signalling resulting in high levels of Akt phosphorylation is effectively mediated by p110δ, but not p110α. The reason why agonist induced BCR signalling does effectively engage p110α remains unknown.
GC reaction, class Switching, affinity maturation
During the germinal centre reaction B cells with intermediate affinities for antigen are selected to undergo class switch recombination and somatic hyper-mutation of their Ig genes to generate high affinity IgG antibodies. P110δ deficient mice show impaired T cell dependent and T cell independent immune responses (54, 60, 61, 191). These results combined with a requirement for p110δ to promote B cell proliferation in response to BCR crosslinking, could be interpreted as requirement for p110δ activity within B cells to promote the differentiation of B cells to antibody producing plasma cells. This is indeed the case for the production of natural antibodies during T cell-independent immune responses. These are mostly of the IgM class, have low to intermediated affinity for self-antigens and common bacterial epitopes. Natural antibodies are much reduced in p110δ-deficient mice due in large part to the lack of marginal zone B cells (54, 61, 192). By contrast, deletion Pten in B cell increases marginal zone B cells and natural antibody production, both of which can be attenuated by concomitant deletion of p110δ (93, 111). Surprisingly, however, deletion of Pten in B cells interferes with T cell-dependent Ig class switching and affinity maturation, whereas the deletion of p110δ in B cells has little impact on these events (109, 111, 193). It appears that PI3K signalling antagonises the expression of key proteins involved in class switching and plasma cell differentiation, including Aid and Bcl6 (109, 111, 194, 195). Therefore, as was the case after heavy chain Ig rearrangements, PI3K signalling needs to be actively suppressed in the germinal centre reaction to produce class-switched IgG producing plasma cells. How p110δ activity is suppressed at this stage is not clear, but the recent observation that cycling B cells in the GC express high levels of Shp1 protein (196). It is interesting to speculate that Shp1 dephosphorylates proteins that activate p110δ and hence reduce the production of PIP3. Why do p110δ-deficient mice produce reduced IgG levels after immunisation? The answer to this question is that follicular helper T cells (TFH) require p110δ for their development and/or maintenance (193). Therefore, deletion of p85 or p110δ in T cells reduces the production of high affinity class switched antibodies, whereas deletion of Pten in activated T cells as the opposite effect (193, 197).
Role of T cell development and function
T cell development proceeds through various checkpoints, the first of which is to ensure that the TCRβ chain is productively rearranged (the β-checkpoint) during which T cells progress from CD4−CD8− double negative to CD4+CD8+ double positive thymocytes (198). In addition to signals generated by the pre-TCR composed of TCRβ and the surrogate preTα chain, the Notch and IL7 receptors are also required for the development of DP thymocytes. However, the requirement for the pre-TCR or IL-7 can be bypassed by deleting Pten which highlights the requirement for PI3K signalling for the development of DP cells (199). By contrast to B cells, mature T cell do develop in mice lacking p110α and p110δ in thymocytes (55). Instead, the combined loss of p110δ and p110γ leads to a near complete block in T cell development at the β-checkpoint (200-203). This is somewhat of a surprise, because p110δ and p110γ are activated by distinct receptors in T cells and hence redundancy would not necessarily be expected. This conundrum was in part resolved through the realisation that signalling via CXCR4 is required at the β-checkpoint (200). Thus while the pre-TCR engages p110δ, the CXCR4 receptor preferentially activates p110γ, with some contribution also from p110δ (200). The observation that some PI3K activity provided either by the pre-TCR or CXCR4 via p110δ or p110γ is sufficient for the development of DP T cells raises some interesting questions. If some class I PI3K signalling is sufficient, as suggested by the ability of Pten deletion to bypass β-selection, then what prevents the CXCR4 receptor from promoting differentiation of DP T cells from progenitors that have not productively rearranged their TCRβ chain? One possibility is that pre-TCR signalling is required to render DN thymocytes cells responsive to CXCL12. Notch also requires class I PI3K activity to promote thymocyte development (204, 205). Notch signals by releasing its intracellular domain which acts as a transcription factor in the nucleus. Recent evidence suggest that Notch increases class I PI3K signalling through its target gene Hes1 which suppresses the expression of Pten (206). Notch may also regulate class I PI3K signalling via the interaction between the notch intracellular domain and the mTORc2 associated protein Rictor (207).
Mice lacking two or more Akt isoforms or Pdk1 in T cells show near complete block in T cell development (129, 130, 208, 209). However, caution is needed before one concludes that these are the only PI3K dependent signals required since DP thymocytes do develop when both Pten and Pdk1 are deleted in thymocytes, suggesting that other PIP3 effectors also play a critical role during development (210). The precise nature of these PIP3 effectors is not known, but Pten deletion could not rescue the development of thymocytes in which Rho has been inactivated suggesting that a PIP3-dependent RhoGEF may be important for T cell development (210).
The mature TCR activates p110δ, and this can be enhanced by CD28 through a mechanism that does not involve direct recruitment of p85 to the CD28 YXXM motif (191). Rather, CD28 may act to boost and sustain signalling by the TCR more generally (211, 212). The related costimulatory receptor ICOS, by contrast, appears to depend entirely on its YXXM motif to signal via PI3K to promote T cell help to B cells (213). ICOS plays a key role in the development and survival of TFH cells and which are virtually absent in mice where the ICOS YXXM motif has been mutated to FXXM or where p110δ was conditionally deleted (193, 213). P110γ also plays an important role in mature T cells by promoting the survival of memory T cells (214). While p110γ is largely dispensable for the chemotaxis of naïve T cells, activated p110γ−/− T cells show reduced chemotaxis towards inflammatory chemokines such as CCL5, CCL22 and LTB4 (69, 215-217). An intriguing possibility is that these chemokines also provide important survival signals to tissues resident memory T cells.
PI3K contributes to the differentiation of T cell subsets
PI3K signalling is required for the differentiation of native CD4 T cells towards either of the main T helper (Th) subsets, Th1, Th2, Th17 and TFH (108, 193, 218-221). Recent studies have indicated that Foxo proteins can positively regulate the Ifnγ gene and that PI3K can control IL17 transcription via mTOR-dependent reciprocal regulation of Gfi1 and Rorγ (132, 222). Another possibility is that Foxo prevents T cells from differentiation such that class I PI3K-dependent removal of Foxo from the nucleus is a pre-requisite for further differentiation. The ability of p110δ inhibitors to attenuate Th differentiation and production offers therapeutic opportunities to alleviate autoimmune and inflammatory diseases using small molecule inhibitors (223, 224). However, this needs to be balanced against the effect of p110δ inhibition on Treg and the effect of inhibiting p110δ in dendritic cells (171, 225, 226). Class I PI3Ks can both promote and inhibit the development of Treg (227). Accordingly, there are increased numbers of Treg in the thymus of p110δ-deficient mice, but reduced numbers in the spleen and lymph nodes (226). The dual effect of p110δ inhibition on Treg numbers may be explained in part by the observation that delayed PI3K and mTOR inhibition after T cell activation supports enhanced Treg differentiation (228). Rapamycin and derivatives are some of the most widely used inhibitors in kidney transplantation. Yet, Rapamycin has recently been shown to enhance the generation of memory CD8 T cells (229). This is thought to reflect a change in the balance between the transcription factors T-bet and Eomesodermin (156, 230, 231). mTOR activity is required for T-bet induction, but not for emomesodermin and whereas effector CD8 T cells express high levels of T-bet, memory T cells express higher levels of emomesodermin. By shifting the balance between the two, Rapamycin promotes memory T cell development. This implies that at or just before the peak of the immune response there must be a mechanism for shutting of class I PI3K and/or mTOR signalling in a subset of cells to promote the formation of memory T cells precursor. This is reminiscent of the attenuation of PI3K signalling required during the germinal centre reaction in B cells.
PI3K inhibition in dendritic cells enhances type I immune responses
The deletion of p85α resulted in increased IL-12 production by dendritic cells, and consequently enhanced type 1 immune responses against Leishmania major characterised by increased T cell-derived IFNγ production (225). P110δ-deficient mice, by contrast, show impaired T cell IFNγ responses in response to Leishmania infection despite elevated IL-12 production by DCs and macrophages (232). These differences may be explained by the different effect of targeting the p110δ subunit and targeting the p85α subunit. Deletion of p85α alone has little effect on T cell activation, whereas deletion of both p85α and p85b in T cell ablates TCR-induced Akt phosphorylation, similar to that seen in p110δ-deficient T cells (197). Therefore, in the p85α−/− mice, DCs provided an excess of IL-12 to phenotypically normal T cells, whereas in p110δD910A mice, the T cell were unable to produce high levels of IFNγ, despite the excess IL-12 produced by the myeloid cells. Interestingly, p110δD910A mice controlled Leismania infection more effectively than did wild type mice (232). This is likely to be due, at least in part, to defective Treg function in p110δ mice (218, 227).
Therapeutic use of PI3K inhibitors
At the time of writing, the p110δ inhibitors GS-1101 is entering phase III clinical trials in to test their efficacy in CLL and iNHL after showing promising results in phase I and II trials, where a large proportion of patients responded by showing a shrinkage of the lymph nodes (233-235). The effectiveness of p110δ inhibitors alone in B cell leukaemias may be explained in part by the lack of involvement of p110γ in GPCR signalling which instead is dependent on p110δ in B cells. By contrast, in mouse a mouse model of T cell leukaemia and in human T-ALL cells, the p110δ-p110γ dual inhibitor CAL-263 was much more potent than inhibition of p110γ or p110δ alone, perhaps reflecting the redundancy between these subunits in T cell precursors which give rise to T cell leukaemias (236).
Perspectives
The roles played by the class II and III PI3Ks in immune cells are only beginning to be uncovered. Further development of gene targeted mice and small molecule inhibitors will no doubts accelerate these efforts, as they did for the class I PI3Ks. Such studies are likely to provide further insights into how PI3P-dependent pathways control the uptake and killing of pathogens by antigen presenting cells and how PI3P promotes immune cell homeostasis more generally.
The role of the individual PI3K isoforms in lymphocyte biology is still incompletely understood, yet rapid developments are supported by gene targeted mouse lines and the development of isoform selective inhibitors. However, one concept observed in B cell development, germinal centre reactions and Treg differentiation, is that transient suppression of class I PI3K can be essential to facilitate expression of Foxo and possibly other genes. Once these genes have been induced, PI3K signalling may again be required to promote growth and proliferation. The requirement for temporal suppression of a signalling pathway is not always readily appreciated by knockout studies and can also be difficult to probe using small molecule inhibitors, especially in vivo. Methods to accurately monitor signalling output over time are also lacking. Nevertheless, this dual nature of PI3K signalling may have led authors to reach opposite conclusions about the role of the pathway, depending on the experimental model system they used and manner they used to inhibit or boost PI3K signalling. Key challenges in this area include tools to switch pathways on and off at will, a deeper understanding of how this happens within cells (e.g. removal of ligands, dephosphorylation of proteins at the membrane or activation of phosphatases).
Finally, elucidating the role of non-canonical PIP3-binding proteins which have shown great increase in numbers and diversity during vertebrate evolution, will no-doubt continue to reveal new and important insights into the role of class I PI3Ks in immunity.
Figure 5.
References
- 1.Hawkins PT, Anderson KE, Davidson K, Stephens LR. Signalling through Class I PI3Ks in mammalian cells. Biochem Soc Trans. 2006;34:647–62. doi: 10.1042/BST0340647. [DOI] [PubMed] [Google Scholar]
- 2.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–19. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 3.Vanhaesebroeck B, Guillermet-Guibert J, Graupera M, Bilanges B. The emerging mechanisms of isoform-specific PI3K signalling. Nat Rev Mol Cell Biol. 2010;11:329–41. doi: 10.1038/nrm2882. [DOI] [PubMed] [Google Scholar]
- 4.Vanhaesebroeck B, Leevers SJ, Panayotou G, Waterfield MD. Phosphoinositide 3-kinases: a conserved family of signal transducers. Trends Biochem Sci. 1997;22:267–72. doi: 10.1016/s0968-0004(97)01061-x. [DOI] [PubMed] [Google Scholar]
- 5.Backer JM. The regulation and function of Class III PI3Ks: novel roles for Vps34. Biochem J. 2008;410:1–17. doi: 10.1042/BJ20071427. [DOI] [PubMed] [Google Scholar]
- 6.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellson CD, Anderson KE, Morgan G, Chilvers ER, Lipp P, et al. Phosphatidylinositol 3-phosphate is generated in phagosomal membranes. Curr Biol. 2001;11:1631–5. doi: 10.1016/s0960-9822(01)00447-x. [DOI] [PubMed] [Google Scholar]
- 8.Vieira O, Botelho R, Rameh L, Brachmann S, Matsuo T, et al. Distinct roles of class I and class III phosphatidylinositol 3-kinases in phagosome formation and maturation. J Cell Biol. 2001 Oct 1;155:19–25. doi: 10.1083/jcb.200107069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simonsen A, Lippe R, Christoforidis S, Gaullier JM, Brech A, et al. EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature. 1998;394:494–8. doi: 10.1038/28879. [DOI] [PubMed] [Google Scholar]
- 10.Schu PV, Takegawa K, Fry MJ, Stack JH, Waterfield MD, et al. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science. 1993;260:88–91. doi: 10.1126/science.8385367. [DOI] [PubMed] [Google Scholar]
- 11.Simonsen A, Wurmser AE, Emr SD, Stenmark H. The role of phosphoinositides in membrane transport. Curr Opin Cell Biol. 2001;13:485–92. doi: 10.1016/s0955-0674(00)00240-4. [DOI] [PubMed] [Google Scholar]
- 12.Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001;152:519–30. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stack JH, Herman PK, Schu PV, Emr SD. A membrane-associated complex containing the Vps15 protein kinase and the Vps34 PI 3-kinase is essential for protein sorting to the yeast lysosome-like vacuole. EMBO J. 1993;12:2195–204. doi: 10.1002/j.1460-2075.1993.tb05867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Obara K, Ohsumi Y. PtdIns 3-Kinase Orchestrates Autophagosome Formation in Yeast. J Lipids. 2011;2011:498768. doi: 10.1155/2011/498768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nobukuni T, Joaquin M, Roccio M, Dann SG, Kim SY, et al. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc Natl Acad Sci U S A. 2005;102:14238–43. doi: 10.1073/pnas.0506925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaber N, Dou Z, Chen JS, Catanzaro J, Jiang YP, et al. Class III PI3K Vps34 plays an essential role in autophagy and in heart and liver function. Proc Natl Acad Sci U S A. 2012;109:2003–8. doi: 10.1073/pnas.1112848109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller S, Tavshanjian B, Oleksy A, Perisic O, Houseman BT, et al. Shaping development of autophagy inhibitors with the structure of the lipid kinase Vps34. Science. 2010;327:1638–42. doi: 10.1126/science.1184429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou X, Takatoh J, Wang F. The mammalian class 3 PI3K (PIK3C3) is required for early embryogenesis and cell proliferation. PLoS One. 2011;6:e16358. doi: 10.1371/journal.pone.0016358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Willinger T, Flavell RA. Canonical autophagy dependent on the class III phosphoinositide-3 kinase Vps34 is required for naive T-cell homeostasis. Proc Natl Acad Sci U S A. 2012;109:8670–5. doi: 10.1073/pnas.1205305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McLeod IX, Zhou X, Li QJ, Wang F, He YW. The class III kinase Vps34 promotes T lymphocyte survival through regulating IL-7Ralpha surface expression. J Immunol. 2011;187:5051–61. doi: 10.4049/jimmunol.1100710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson KE, Chessa TA, Davidson K, Henderson RB, Walker S, et al. PtdIns3P and Rac direct the assembly of the NADPH oxidase on a novel, pre-phagosomal compartment during FcR-mediated phagocytosis in primary mouse neutrophils. Blood. 2010 doi: 10.1182/blood-2010-03-275602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanai F, Liu H, Field SJ, Akbary H, Matsuo T, et al. The PX domains of p47phox and p40phox bind to lipid products of PI(3)K. Nat Cell Biol. 2001;3:675–8. doi: 10.1038/35083070. [DOI] [PubMed] [Google Scholar]
- 23.Ellson CD, Gobert-Gosse S, Anderson KE, Davidson K, Erdjument-Bromage H, et al. PtdIns(3)P regulates the neutrophil oxidase complex by binding to the PX domain of p40(phox) Nat Cell Biol. 2001;3:679–82. doi: 10.1038/35083076. [DOI] [PubMed] [Google Scholar]
- 24.Ellson C, Davidson K, Anderson K, Stephens LR, Hawkins PT. PtdIns3P binding to the PX domain of p40phox is a physiological signal in NADPH oxidase activation. EMBO J. 2006;25:4468–78. doi: 10.1038/sj.emboj.7601346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berger SB, Romero X, Ma C, Wang G, Faubion WA, et al. SLAM is a microbial sensor that regulates bacterial phagosome functions in macrophages. Nat Immunol. 2010;11:920–7. doi: 10.1038/ni.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma C, Wang N, Detre C, Wang G, O’Keeffe M, et al. Receptor signaling lymphocyte-activation molecule family 1 (Slamf1) regulates membrane fusion and NADPH oxidase 2 (NOX2) activity by recruiting a Beclin-1/Vps34/ultraviolet radiation resistance-associated gene (UVRAG) complex. J Biol Chem. 2012;287:18359–65. doi: 10.1074/jbc.M112.367060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ktistakis NT, Manifava M, Schoenfelder P, Rotondo S. How phosphoinositide 3-phosphate controls growth downstream of amino acids and autophagy downstream of amino acid withdrawal. Biochem Soc Trans. 2012;40:37–43. doi: 10.1042/BST20110684. [DOI] [PubMed] [Google Scholar]
- 28.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–75. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–35. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yordy B, Iwasaki A. Autophagy in the control and pathogenesis of viral infection. Curr Opin Virol. 2011;1:196–203. doi: 10.1016/j.coviro.2011.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- 32.Paludan C, Schmid D, Landthaler M, Vockerodt M, Kube D, et al. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science. 2005;307:593–6. doi: 10.1126/science.1104904. [DOI] [PubMed] [Google Scholar]
- 33.Lee HK, Mattei LM, Steinberg BE, Alberts P, Lee YH, et al. In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity. 2010;32:227–39. doi: 10.1016/j.immuni.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He MX, McLeod IX, Jia W, He YW. Macroautophagy in T lymphocyte development and function. Front Immunol. 2012;3:22. doi: 10.3389/fimmu.2012.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Polson HE, de Lartigue J, Rigden DJ, Reedijk M, Urbe S, et al. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy. 2010:6. doi: 10.4161/auto.6.4.11863. [DOI] [PubMed] [Google Scholar]
- 36.Falasca M, Maffucci T. Regulation and cellular functions of class II phosphoinositide 3-kinases. Biochem J. 2012;443:587–601. doi: 10.1042/BJ20120008. [DOI] [PubMed] [Google Scholar]
- 37.Harris DP, Vogel P, Wims M, Moberg K, Humphries J, et al. Requirement for class II phosphoinositide 3-kinase C2alpha in maintenance of glomerular structure and function. Mol Cell Biol. 2011;31:63–80. doi: 10.1128/MCB.00468-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harada K, Truong AB, Cai T, Khavari PA. The class II phosphoinositide 3-kinase C2beta is not essential for epidermal differentiation. Mol Cell Biol. 2005;25:11122–30. doi: 10.1128/MCB.25.24.11122-11130.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anderson KE, Boyle KB, Davidson K, Chessa TA, Kulkarni S, et al. CD18-dependent activation of the neutrophil NADPH oxidase during phagocytosis of Escherichia coli or Staphylococcus aureus is regulated by class III but not class I or II PI3Ks. Blood. 2008;112:5202–11. doi: 10.1182/blood-2008-04-149450. [DOI] [PubMed] [Google Scholar]
- 40.Srivastava S, Di L, Zhdanova O, Li Z, Vardhana S, et al. The class II phosphatidylinositol 3 kinase C2beta is required for the activation of the K+ channel KCa3.1 and CD4 T-cells. Mol Biol Cell. 2009;20:3783–91. doi: 10.1091/mbc.E09-05-0390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Srivastava S, Choudhury P, Li Z, Liu G, Nadkarni V, et al. Phosphatidylinositol 3-phosphate indirectly activates KCa3.1 via 14 amino acids in the carboxy terminus of KCa3.1. Mol Biol Cell. 2006;17:146–54. doi: 10.1091/mbc.E05-08-0763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Srivastava S, Ko K, Choudhury P, Li Z, Johnson AK, et al. Phosphatidylinositol-3 phosphatase myotubularin-related protein 6 negatively regulates CD4 T cells. Mol Cell Biol. 2006;26:5595–602. doi: 10.1128/MCB.00352-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Srivastava S, Li Z, Lin L, Liu G, Ko K, et al. The phosphatidylinositol 3-phosphate phosphatase myotubularin- related protein 6 (MTMR6) is a negative regulator of the Ca2+-activated K+ channel KCa3.1. Mol Cell Biol. 2005;25:3630–8. doi: 10.1128/MCB.25.9.3630-3638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feske S, Skolnik EY, Prakriya M. Ion channels and transporters in lymphocyte function and immunity. Nat Rev Immunol. 2012;12:532–47. doi: 10.1038/nri3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dove SK, Cooke FT, Douglas MR, Sayers LG, Parker PJ, et al. Osmotic stress activates phosphatidylinositol-3,5-bisphosphate synthesis. Nature. 1997;390:187–92. doi: 10.1038/36613. [DOI] [PubMed] [Google Scholar]
- 46.Dove SK, Dong K, Kobayashi T, Williams FK, Michell RH. Phosphatidylinositol 3,5-bisphosphate and Fab1p/PIKfyve underPPIn endo-lysosome function. Biochem J. 2009;419:1–13. doi: 10.1042/BJ20081950. [DOI] [PubMed] [Google Scholar]
- 47.Ikonomov OC, Sbrissa D, Delvecchio K, Xie Y, Jin JP, et al. The phosphoinositide kinase PIKfyve is vital in early embryonic development: preimplantation lethality of PIKfyve−/− embryos but normality of PIKfyve+/− mice. J Biol Chem. 2011;286:13404–13. doi: 10.1074/jbc.M111.222364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kerr MC, Wang JT, Castro NA, Hamilton NA, Town L, et al. Inhibition of the PtdIns(5) kinase PIKfyve disrupts intracellular replication of Salmonella. EMBO J. 2010;29:1331–47. doi: 10.1038/emboj.2010.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hnia K, Vaccari I, Bolino A, Laporte J. Myotubularin phosphoinositide phosphatases: cellular functions and disease pathophysiology. Trends Mol Med. 2012;18:317–27. doi: 10.1016/j.molmed.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 50.Robinson FL, Dixon JE. Myotubularin phosphatases: policing 3-phosphoinositides. Trends Cell Biol. 2006;16:403–12. doi: 10.1016/j.tcb.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 51.Velichkova M, Juan J, Kadandale P, Jean S, Ribeiro I, et al. Drosophila Mtm and class II PI3K coregulate a PI(3)P pool with cortical and endolysosomal functions. J Cell Biol. 2010;190:407–25. doi: 10.1083/jcb.200911020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fruman DA. Regulatory subunits of class IA PI3K. Curr Top Microbiol Immunol. 2010;346:225–44. doi: 10.1007/82_2010_39. [DOI] [PubMed] [Google Scholar]
- 53.Castellano E, Downward J. RAS Interaction with PI3K: More Than Just Another Effector Pathway. Genes Cancer. 2011;2:261–74. doi: 10.1177/1947601911408079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Okkenhaug K, Bilancio A, Farjot G, Priddle H, Sancho S, et al. Impaired B and T cell antigen receptor signaling in p110delta PI 3-kinase mutant mice. Science. 2002;297:1031–4. doi: 10.1126/science.1073560. [DOI] [PubMed] [Google Scholar]
- 55.Ramadani F, Bolland DJ, Garcon F, Emery JL, Vanhaesebroeck B, et al. The PI3K isoforms p110alpha and p110delta are essential for pre-B cell receptor signaling and B cell development. Sci Signal. 2010;3:ra60. doi: 10.1126/scisignal.2001104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kulkarni S, Sitaru C, Jakus Z, Anderson KE, Damoulakis G, et al. PI3Kbeta Plays a Critical Role in Neutrophil Activation by Immune Complexes. Sci Signal. 2011;4:ra23. doi: 10.1126/scisignal.2001617. [DOI] [PubMed] [Google Scholar]
- 57.Hirsch E, Katanaev VL, Garlanda C, Azzolino O, Pirola L, et al. Central role for G protein-coupled phosphoinositide 3-kinase gamma in inflammation. Science. 2000;287:1049–53. doi: 10.1126/science.287.5455.1049. [DOI] [PubMed] [Google Scholar]
- 58.Sasaki T, Irie-Sasaki J, Jones RG, Oliveira-dos-Santos AJ, Stanford WL, et al. Function of PI3Kγ in thymocyte development, T cell activation, and neutrophil migration. Science. 2000;287:1040–6. doi: 10.1126/science.287.5455.1040. [DOI] [PubMed] [Google Scholar]
- 59.Li Z, Jiang H, Xie W, Zhang Z, Smrcka AV, et al. Roles of PLC-beta2 and -beta3 and PI3Kgamma in chemoattractant-mediated signal transduction. Science. 2000;287:1046–9. doi: 10.1126/science.287.5455.1046. [DOI] [PubMed] [Google Scholar]
- 60.Jou ST, Carpino N, Takahashi Y, Piekorz R, Chao JR, et al. Essential, nonredundant role for the phosphoinositide 3-kinase p110δ in signaling by the B-cell receptor complex. Mol Cell Biol. 2002;22:8580–91. doi: 10.1128/MCB.22.24.8580-8591.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Clayton E, Bardi G, Bell SE, Chantry D, Downes CP, et al. A Crucial Role for the p110δ Subunit of Phosphatidylinositol 3-Kinase in B Cell Development and Activation. Journal of Experimental Medicine. 2002;196:753–63. doi: 10.1084/jem.20020805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sadhu C, Masinovsky B, Dick K, Sowell CG, Staunton DE. Essential Role of Phosphoinositide 3-Kinase δ in Neutrophil Directional Movement. Journal of Immunology. 2003;170:2647–54. doi: 10.4049/jimmunol.170.5.2647. [DOI] [PubMed] [Google Scholar]
- 63.Camps M, Ruckle T, Ji H, Ardissone V, Rintelen F, et al. Blockade of PI3Kγ suppresses joint inflammation and damage in mouse models of rheumatoid arthritis. Nature Medicine. 2005;11:936–43. doi: 10.1038/nm1284. [DOI] [PubMed] [Google Scholar]
- 64.Andrews S, Stephens LR, Hawkins PT. PI3K class IB pathway. Sci STKE. 2007;2007:cm2. doi: 10.1126/stke.4072007cm2. [DOI] [PubMed] [Google Scholar]
- 65.Guillermet-Guibert J, Bjorklof K, Salpekar A, Gonella C, Ramadani F, et al. The p110beta isoform of phosphoinositide 3-kinase signals downstream of G protein-coupled receptors and is functionally redundant with p110gamma. Proc Natl Acad Sci U S A. 2008;105:8292–7. doi: 10.1073/pnas.0707761105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ciraolo E, Iezzi M, Marone R, Marengo S, Curcio C, et al. Phosphoinositide 3-kinase p110beta activity: key role in metabolism and mammary gland cancer but not development. Sci Signal. 2008;1:ra3. doi: 10.1126/scisignal.1161577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jia S, Liu Z, Zhang S, Liu P, Zhang L, et al. Essential roles of PI(3)K-p110beta in cell growth, metabolism and tumorigenesis. Nature. 2008 doi: 10.1038/nature07091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Saudemont A, Garcon F, Yadi H, Roche-Molina M, Kim N, et al. p110gamma and p110delta isoforms of phosphoinositide 3-kinase differentially regulate natural killer cell migration in health and disease. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0808594106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reif K, Okkenhaug K, Sasaki T, Penninger JM, Vanhaesebroeck B, et al. Cutting Edge: Differential Roles for Phosphoinositide 3-Kinases, p110γ and p110δ, in Lymphocyte Chemotaxis and Homing. J Immunol. 2004;173:2236–40. doi: 10.4049/jimmunol.173.4.2236. [DOI] [PubMed] [Google Scholar]
- 70.Samuels Y, Waldman T. Oncogenic mutations of PIK3CA in human cancers. Curr Top Microbiol Immunol. 2010;347:21–41. doi: 10.1007/82_2010_68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 72.Vadas O, Burke JE, Zhang X, Berndt A, Williams RL. Structural basis for activation and inhibition of class I phosphoinositide 3-kinases. Sci Signal. 2011;4:re2. doi: 10.1126/scisignal.2002165. [DOI] [PubMed] [Google Scholar]
- 73.Project TCG. Cosmic. 2012. [Google Scholar]
- 74.Gabelli SB, Huang CH, Mandelker D, Schmidt-Kittler O, Vogelstein B, et al. Structural effects of oncogenic PI3Kalpha mutations. Curr Top Microbiol Immunol. 2010;347:43–53. doi: 10.1007/82_2010_53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang CH, Mandelker D, Schmidt-Kittler O, Samuels Y, Velculescu VE, et al. The structure of a human p110alpha/p85alpha complex elucidates the effects of oncogenic PI3Kalpha mutations. Science. 2007;318:1744–8. doi: 10.1126/science.1150799. [DOI] [PubMed] [Google Scholar]
- 76.Miled N, Yan Y, Hon WC, Perisic O, Zvelebil M, et al. Mechanism of two classes of cancer mutations in the phosphoinositide 3-kinase catalytic subunit. Science. 2007;317:239–42. doi: 10.1126/science.1135394. [DOI] [PubMed] [Google Scholar]
- 77.Yu J, Wjasow C, Backer JM. Regulation of the p85/p110alpha phosphatidylinositol 3′-kinase. Distinct roles for the n-terminal and c-terminal SH2 domains. J Biol Chem. 1998;273:30199–203. doi: 10.1074/jbc.273.46.30199. [DOI] [PubMed] [Google Scholar]
- 78.Yu J, Zhang Y, McIlroy J, Rordorf-Nikolic T, Orr GA, et al. Regulation of the p85/p110 phosphatidylinositol 3′-kinase: stabilization and inhibition of the p110alpha catalytic subunit by the p85 regulatory subunit. Mol Cell Biol. 1998;18:1379–87. doi: 10.1128/mcb.18.3.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Burke JE, Vadas O, Berndt A, Finegan T, Perisic O, et al. Dynamics of the phosphoinositide 3-kinase p110delta interaction with p85alpha and membranes reveals aspects of regulation distinct from p110alpha. Structure. 2011;19:1127–37. doi: 10.1016/j.str.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhao L, Vogt PK. Helical domain and kinase domain mutations in p110alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc Natl Acad Sci U S A. 2008;105:2652–7. doi: 10.1073/pnas.0712169105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sun M, Hillmann P, Hofmann BT, Hart JR, Vogt PK. Cancer-derived mutations in the regulatory subunit p85alpha of phosphoinositide 3-kinase function through the catalytic subunit p110alpha. Proc Natl Acad Sci U S A. 2010;107:15547–52. doi: 10.1073/pnas.1009652107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu H, Shekar SC, Flinn RJ, El-Sibai M, Jaiswal BS, et al. Regulation of Class IA PI 3-kinases: C2 domain-iSH2 domain contacts inhibit p85/p110alpha and are disrupted in oncogenic p85 mutants. Proc Natl Acad Sci U S A. 2009;106:20258–63. doi: 10.1073/pnas.0902369106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gupta S, Ramjaun AR, Haiko P, Wang Y, Warne PH, et al. Binding of ras to phosphoinositide 3-kinase p110alpha is required for ras-driven tumorigenesis in mice. Cell. 2007;129:957–68. doi: 10.1016/j.cell.2007.03.051. [DOI] [PubMed] [Google Scholar]
- 84.Kang S, Denley A, Vanhaesebroeck B, Vogt PK. Oncogenic transformation induced by the p110beta, -gamma, and -delta isoforms of class I phosphoinositide 3-kinase. Proc Natl Acad Sci U S A. 2006;103:1289–94. doi: 10.1073/pnas.0510772103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vanhaesebroeck B, Welham MJ, Kotani K, Stein R, Warne PH, et al. P110delta, a novel phosphoinositide 3-kinase in leukocytes. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:4330–5. doi: 10.1073/pnas.94.9.4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rodriguez-Viciana P, Sabatier C, McCormick F. Signaling specificity by Ras family GTPases is determined by the full spectrum of effectors they regulate. Mol Cell Biol. 2004;24:4943–54. doi: 10.1128/MCB.24.11.4943-4954.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Delgado P, Cubelos B, Calleja E, Martinez-Martin N, Cipres A, et al. Essential function for the GTPase TC21 in homeostatic antigen receptor signaling. Nat Immunol. 2009;10:880–8. doi: 10.1038/ni.1749. [DOI] [PubMed] [Google Scholar]
- 88.Zhang X, Vadas O, Perisic O, Anderson KE, Clark J, et al. Structure of lipid kinase p110beta/p85beta elucidates an unusual SH2-domain-mediated inhibitory mechanism. Mol Cell. 2011;41:567–78. doi: 10.1016/j.molcel.2011.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Foukas LC, Claret M, Pearce W, Okkenhaug K, Meek S, et al. Critical role for the p110α phosphoinositide-3-OH kinase in growth and metabolic regulation. Nature. 2006;441:366–70. doi: 10.1038/nature04694. [DOI] [PubMed] [Google Scholar]
- 90.Knight ZA, Gonzalez B, Feldman ME, Zunder ER, Goldenberg DD, et al. A Pharmacological Map of the PI3-K Family Defines a Role for p110alpha in Insulin Signaling. Cell. 2006;125:733–47. doi: 10.1016/j.cell.2006.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Graupera M, Guillermet-Guibert J, Foukas LC, Phng LK, Cain RJ, et al. Angiogenesis selectively requires the p110alpha isoform of PI3K to control endothelial cell migration. Nature. 2008;453:662–6. doi: 10.1038/nature06892. [DOI] [PubMed] [Google Scholar]
- 92.Zhao JJ, Cheng H, Jia S, Wang L, Gjoerup OV, et al. The p110alpha isoform of PI3K is essential for proper growth factor signaling and oncogenic transformation. Proc Natl Acad Sci U S A. 2006;103:16296–300. doi: 10.1073/pnas.0607899103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Janas ML, Hodson D, Stamataki Z, Hill S, Welch K, et al. The effect of deleting p110delta on the phenotype and function of PTEN-deficient B cells. J Immunol. 2008;180:739–46. doi: 10.4049/jimmunol.180.2.739. [DOI] [PubMed] [Google Scholar]
- 94.Bilancio A, Okkenhaug K, Camps M, Emery JL, Ruckle T, et al. Key role of the p110δ isoform of PI3K in B-cell antigen and IL-4 receptor signaling: comparative analysis of genetic and pharmacologic interference with p110δ function in B cells. Blood. 2006;107:642–50. doi: 10.1182/blood-2005-07-3041. [DOI] [PubMed] [Google Scholar]
- 95.Okkenhaug K, Ali K, Vanhaesebroeck B. Antigen receptor signalling: a distinctive role for the p110δ isoform of PI3K. Trends Immunol. 2007;28:80–7. doi: 10.1016/j.it.2006.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Geering B, Cutillas PR, Nock G, Gharbi SI, Vanhaesebroeck B. Class IA phosphoinositide 3-kinases are obligate p85-p110 heterodimers. Proc Natl Acad Sci U S A. 2007;104:7809–14. doi: 10.1073/pnas.0700373104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Berndt A, Miller S, Williams O, Le DD, Houseman BT, et al. The p110 delta structure: mechanisms for selectivity and potency of new PI(3)K inhibitors. Nat Chem Biol. 2010;6:117–24. doi: 10.1038/nchembio.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tuveson DA, Carter RH, Soltoff SP, Fearon DT. CD19 of B cells as a surrogate kinase insert region to bind phosphatidylinositol 3-kinase. Science. 1993;260:986–9. doi: 10.1126/science.7684160. [DOI] [PubMed] [Google Scholar]
- 99.Leslie NR, Dixon MJ, Schenning M, Gray A, Batty IH. Distinct inactivation of PI3K signalling by PTEN and 5-phosphatases. Adv Enzyme Regul. 2011 doi: 10.1016/j.advenzreg.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 100.Ward SG, Blunt MD. Pharmacological Targeting of Phosphoinositide Lipid Kinases and Phosphatases in the Immune System: Success, Disappointment and New Opportunities. Front Immunol. 2012:3. doi: 10.3389/fimmu.2012.00226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rameh LE, Arvidsson A, Carraway KL, 3rd, Couvillon AD, Rathbun G, et al. A comparative analysis of the phosphoinositide binding specificity of pleckstrin homology domains. J Biol Chem. 1997;272:22059–66. doi: 10.1074/jbc.272.35.22059. [DOI] [PubMed] [Google Scholar]
- 102.Franke TF, Kaplan DR, Cantley LC, Toker A. Direct regulation of the Akt proto-oncogene product by phosphatidylinositol-3,4-bisphosphate. Science. 1997;275:665–8. doi: 10.1126/science.275.5300.665. [DOI] [PubMed] [Google Scholar]
- 103.Lemmon MA, Ferguson KM. Signal-dependent membrane targeting by pleckstrin homology (PH) domains. Biochem J. 2000;350(Pt 1):1–18. [PMC free article] [PubMed] [Google Scholar]
- 104.Pauls SD, Lafarge ST, Landego I, Zhang T-t, Marshall AJ. The phosphoinositide 3-kinase signalling pathway in normal and malignant B cells: activation mechanisms, regulation and impact on cellular functions. Front Immunol. 2012:3. doi: 10.3389/fimmu.2012.00224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Newton RH, Turka LA. Regulation of T Cell Homeostasis and Responses by Pten. Front Immunol. 2012:3. doi: 10.3389/fimmu.2012.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Suzuki A, Yamaguchi MT, Ohteki T, Sasaki T, Kaisho T, et al. T cell-specific loss of Pten leads to defects in central and peripheral tolerance. Immunity. 2001;14:523–34. doi: 10.1016/s1074-7613(01)00134-0. [DOI] [PubMed] [Google Scholar]
- 107.Buckler JL, Walsh PT, Porrett PM, Choi Y, Turka LA. Cutting edge: T cell requirement for CD28 costimulation is due to negative regulation of TCR signals by PTEN. J Immunol. 2006;177:4262–6. doi: 10.4049/jimmunol.177.7.4262. [DOI] [PubMed] [Google Scholar]
- 108.Soond DR, Garcon F, Patton DT, Rolf J, Turner M, et al. Pten loss in CD4 T cells enhances their helper function but does not lead to autoimmunity or lymphoma. J Immunol. 2012;188:5935–43. doi: 10.4049/jimmunol.1102116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Omori SA, Cato MH, Anzelon-Mills A, Puri KD, Shapiro-Shelef M, et al. Regulation of Class-Switch Recombination and Plasma Cell Differentiation by Phosphatidylinositol 3-Kinase Signaling. Immunity. 2006 doi: 10.1016/j.immuni.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 110.Browne CD, Del Nagro CJ, Cato MH, Dengler HS, Rickert RC. Suppression of phosphatidylinositol 3,4,5-trisphosphate production is a key determinant of B cell anergy. Immunity. 2009;31:749–60. doi: 10.1016/j.immuni.2009.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Suzuki A, Kaisho T, Ohishi M, Tsukio-Yamaguchi M, Tsubata T, et al. Critical roles of Pten in B cell homeostasis and immunoglobulin class switch recombination. J Exp Med. 2003;197:657–67. doi: 10.1084/jem.20021101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kerr WG. Inhibitor and activator: dual functions for SHIP in immunity and cancer. Ann N Y Acad Sci. 2011;1217:1–17. doi: 10.1111/j.1749-6632.2010.05869.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Miletic AV, Anzelon-Mills AN, Mills DM, Omori SA, Pedersen IM, et al. Coordinate suppression of B cell lymphoma by PTEN and SHIP phosphatases. J Exp Med. 2010;207:2407–20. doi: 10.1084/jem.20091962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fedele CG, Ooms LM, Ho M, Vieusseux J, O’Toole SA, et al. Inositol polyphosphate 4-phosphatase II regulates PI3K/Akt signaling and is lost in human basal-like breast cancers. Proc Natl Acad Sci U S A. 2010;107:22231–6. doi: 10.1073/pnas.1015245107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gewinner C, Wang ZC, Richardson A, Teruya-Feldstein J, Etemadmoghadam D, et al. Evidence that inositol polyphosphate 4-phosphatase type II is a tumor suppressor that inhibits PI3K signaling. Cancer Cell. 2009;16:115–25. doi: 10.1016/j.ccr.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- 117.Prisco A, Vanes L, Ruf S, Trigueros C, Tybulewicz VL. Lineage-specific requirement for the PH domain of Vav1 in the activation of CD4+ but not CD8+ T cells. Immunity. 2005;23:263–74. doi: 10.1016/j.immuni.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 118.Rapley J, Tybulewicz VL, Rittinger K. Crucial structural role for the PH and C1 domains of the Vav1 exchange factor. EMBO Rep. 2008;9:655–61. doi: 10.1038/embor.2008.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Park WS, Heo WD, Whalen JH, O’Rourke NA, Bryan HM, et al. Comprehensive identification of PIP3-regulated PH domains from C. elegans to H. sapiens by model prediction and live imaging. Mol Cell. 2008;30:381–92. doi: 10.1016/j.molcel.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Krugmann S, Anderson KE, Ridley SH, Risso N, McGregor A, et al. Identification of ARAP3, a novel PI3K effector regulating both Arf and Rho GTPases, by selective capture on phosphoinositide affinity matrices. Mol Cell. 2002;9:95–108. doi: 10.1016/s1097-2765(02)00434-3. [DOI] [PubMed] [Google Scholar]
- 121.Hedrick SM. The cunning little vixen: Foxo and the cycle of life and death. Nat Immunol. 2009;10:1057–63. doi: 10.1038/ni.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Powell JD, Pollizzi KN, Heikamp EB, Horton MR. Regulation of immune responses by mTOR. Annu Rev Immunol. 2012;30:39–68. doi: 10.1146/annurev-immunol-020711-075024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Finlay D, Cantrell DA. Metabolism, migration and memory in cytotoxic T cells. Nat Rev Immunol. 2011;11:109–17. doi: 10.1038/nri2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fruman DA, Limon JJ. Akt and mTOR in B cell activation and differentiation. Front Immunol. 2012:3. doi: 10.3389/fimmu.2012.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–74. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chin YR, Toker A. Akt isoform-specific signaling in breast cancer: uncovering an anti-migratory role for palladin. Cell Adh Migr. 2011;5:211–4. doi: 10.4161/cam.5.3.15790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Calamito M, Juntilla MM, Thomas M, Northrup DL, Rathmell J, et al. Akt1 and Akt2 promote peripheral B-cell maturation and survival. Blood. 2010;115:4043–50. doi: 10.1182/blood-2009-09-241638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Juntilla MM, Patil VD, Calamito M, Joshi RP, Birnbaum MJ, et al. AKT1 and AKT2 maintain hematopoietic stem cell function by regulating reactive oxygen species. Blood. 2010;115:4030–8. doi: 10.1182/blood-2009-09-241000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Juntilla MM, Wofford JA, Birnbaum MJ, Rathmell JC, Koretzky GA. Akt1 and Akt2 are required for alphabeta thymocyte survival and differentiation. Proc Natl Acad Sci U S A. 2007;104:12105–10. doi: 10.1073/pnas.0705285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Fayard E, Gill J, Paolino M, Hynx D, Hollander GA, et al. Deletion of PKBalpha/Akt1 affects thymic development. PLoS ONE. 2007;2:e992. doi: 10.1371/journal.pone.0000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mao C, Tili EG, Dose M, Haks MC, Bear SE, et al. Unequal contribution of Akt isoforms in the double-negative to double-positive thymocyte transition. J Immunol. 2007;178:5443–53. doi: 10.4049/jimmunol.178.9.5443. [DOI] [PubMed] [Google Scholar]
- 132.Macintyre AN, Finlay D, Preston G, Sinclair LV, Waugh CM, et al. Protein kinase B controls transcriptional programs that direct cytotoxic T cell fate but is dispensable for T cell metabolism. Immunity. 2011;34:224–36. doi: 10.1016/j.immuni.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Amin RH, Schlissel MS. Foxo1 directly regulates the transcription of recombination-activating genes during B cell development. Nat Immunol. 2008;9:613–22. doi: 10.1038/ni.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Dengler HS, Baracho GV, Omori SA, Bruckner S, Arden KC, et al. Distinct functions for the transcription factor Foxo1 at various stages of B cell differentiation. Nat Immunol. 2008;9:1388–98. doi: 10.1038/ni.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fabre S, Carrette F, Chen J, Lang V, Semichon M, et al. FOXO1 regulates L-Selectin and a network of human T cell homing molecules downstream of phosphatidylinositol 3-kinase. J Immunol. 2008;181:2980–9. doi: 10.4049/jimmunol.181.5.2980. [DOI] [PubMed] [Google Scholar]
- 136.Yusuf I, Kharas MG, Chen J, Peralta RQ, Maruniak A, et al. KLF4 is a FOXO target gene that suppresses B cell proliferation. Int Immunol. 2008;20:671–81. doi: 10.1093/intimm/dxn024. [DOI] [PubMed] [Google Scholar]
- 137.Dejean AS, Beisner DR, Ch’en IL, Kerdiles YM, Babour A, et al. Transcription factor Foxo3 controls the magnitude of T cell immune responses by modulating the function of dendritic cells. Nat Immunol. 2009;10:504–13. doi: 10.1038/ni.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kerdiles YM, Beisner DR, Tinoco R, Dejean AS, Castrillon DH, et al. Foxo1 links homing and survival of naive T cells by regulating L-selectin, CCR7 and interleukin 7 receptor. Nat Immunol. 2009;10:176–84. doi: 10.1038/ni.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ouyang W, Beckett O, Flavell RA, Li MO. An essential role of the Forkhead-box transcription factor Foxo1 in control of T cell homeostasis and tolerance. Immunity. 2009;30:358–71. doi: 10.1016/j.immuni.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Merkenschlager M, von Boehmer H. PI3 kinase signalling blocks Foxp3 expression by sequestering Foxo factors. J Exp Med. 2010;207:1347–50. doi: 10.1084/jem.20101156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ouyang W, Beckett O, Ma Q, Paik JH, DePinho RA, et al. Foxo proteins cooperatively control the differentiation of Foxp3+ regulatory T cells. Nat Immunol. 2010;11:618–27. doi: 10.1038/ni.1884. [DOI] [PubMed] [Google Scholar]
- 142.Yang JY, Chang CJ, Xia W, Wang Y, Wong KK, et al. Activation of FOXO3a is sufficient to reverse mitogen-activated protein/extracellular signal-regulated kinase kinase inhibitor chemoresistance in human cancer. Cancer Res. 2010;70:4709–18. doi: 10.1158/0008-5472.CAN-09-4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Alkhatib A, Werner M, Hug E, Herzog S, Eschbach C, et al. FoxO1 induces Ikaros splicing to promote immunoglobulin gene recombination. J Exp Med. 2012;209:395–406. doi: 10.1084/jem.20110216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Graham JR, Hendershott MC, Terragni J, Cooper GM. mRNA degradation plays a significant role in the program of gene expression regulated by phosphatidylinositol 3-kinase signaling. Mol Cell Biol. 2010;30:5295–305. doi: 10.1128/MCB.00303-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Turner M, Hodson D. Regulation of lymphocyte development and function by RNA-binding proteins. Curr Opin Immunol. 2012;24:160–5. doi: 10.1016/j.coi.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 146.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–93. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Shaw RJ, Cantley LC. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature. 2006;441:424–30. doi: 10.1038/nature04869. [DOI] [PubMed] [Google Scholar]
- 148.Salmond RJ, Emery J, Okkenhaug K, Zamoyska R. MAPK, phosphatidylinositol 3-kinase, and mammalian target of rapamycin pathways converge at the level of ribosomal protein S6 phosphorylation to control metabolic signaling in CD8 T cells. J Immunol. 2009;183:7388–97. doi: 10.4049/jimmunol.0902294. [DOI] [PubMed] [Google Scholar]
- 149.Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332:1317–22. doi: 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Yu Y, Yoon SO, Poulogiannis G, Yang Q, Ma XM, et al. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science. 2011;332:1322–6. doi: 10.1126/science.1199484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Shi LZ, Wang R, Huang G, Vogel P, Neale G, et al. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J Exp Med. 2011;208:1367–76. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Dang EV, Barbi J, Yang HY, Jinasena D, Yu H, et al. Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell. 2011;146:772–84. doi: 10.1016/j.cell.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Molecular Cell. 2006;22:159–68. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 156.Rao RR, Li Q, Gubbels Bupp MR, Shrikant PA. Transcription factor Foxo1 represses T-bet-mediated effector functions and promotes memory CD8(+) T cell differentiation. Immunity. 2012;36:374–87. doi: 10.1016/j.immuni.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Berg LJ, Finkelstein LD, Lucas JA, Schwartzberg PL. TEC FAMILY KINASES IN T LYMPHOCYTE DEVELOPMENT AND FUNCTION*. Annual Review of Immunology. 2005;23:549–600. doi: 10.1146/annurev.immunol.22.012703.104743. [DOI] [PubMed] [Google Scholar]
- 158.Li Z, Wahl MI, Eguinoa A, Stephens LR, Hawkins PT, et al. Phosphatidylinositol 3-kinase-gamma activates Bruton’s tyrosine kinase in concert with Src family kinases. Proc Natl Acad Sci U S A. 1997;94:13820–5. doi: 10.1073/pnas.94.25.13820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Suzuki H, Matsuda S, Terauchi Y, Fujiwara M, Ohteki T, et al. PI3K and Btk differentially regulate B cell antigen receptor-mediated signal transduction. Nat Immunol. 2003 doi: 10.1038/ni890. [DOI] [PubMed] [Google Scholar]
- 160.Matsuda S, Mikami Y, Ohtani M, Fujiwara M, Hirata Y, et al. Critical role of class IA PI3K for c-Rel expression in B lymphocytes. Blood. 2009;113:1037–44. doi: 10.1182/blood-2008-06-163725. [DOI] [PubMed] [Google Scholar]
- 161.Welch HC, Coadwell WJ, Stephens LR, Hawkins PT. Phosphoinositide 3-kinase-dependent activation of Rac. FEBS Lett. 2003;546:93–7. doi: 10.1016/s0014-5793(03)00454-x. [DOI] [PubMed] [Google Scholar]
- 162.Welch HC, Coadwell WJ, Ellson CD, Ferguson GJ, Andrews SR, et al. P-Rex1, a PtdIns(3,4,5)P3- and Gbetagamma-regulated guanine-nucleotide exchange factor for Rac. Cell. 2002;108:809–21. doi: 10.1016/s0092-8674(02)00663-3. [DOI] [PubMed] [Google Scholar]
- 163.Condliffe AM, Davidson K, Anderson KE, Ellson CD, Crabbe T, et al. Sequential activation of class IB and class IA PI3K is important for the primed respiratory burst of human but not murine neutrophils. Blood. 2005;106:1432–40. doi: 10.1182/blood-2005-03-0944. [DOI] [PubMed] [Google Scholar]
- 164.Lawson CD, Donald S, Anderson KE, Patton DT, Welch HC. P-Rex1 and Vav1 cooperate in the regulation of formyl-methionyl-leucyl-phenylalanine-dependent neutrophil responses. J Immunol. 2011;186:1467–76. doi: 10.4049/jimmunol.1002738. [DOI] [PubMed] [Google Scholar]
- 165.Saveliev A, Vanes L, Ksionda O, Rapley J, Smerdon SJ, et al. Function of the nucleotide exchange activity of vav1 in T cell development and activation. Sci Signal. 2009;2:ra83. doi: 10.1126/scisignal.2000420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Costa C, Barberis L, Ambrogio C, Manazza AD, Patrucco E, et al. Negative feedback regulation of Rac in leukocytes from mice expressing a constitutively active phosphatidylinositol 3-kinase gamma. Proc Natl Acad Sci U S A. 2007;104:14354–9. doi: 10.1073/pnas.0703175104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Costa C, Germena G, Martin-Conte EL, Molineris I, Bosco E, et al. The RacGAP ArhGAP15 is a master negative regulator of neutrophil functions. Blood. 2011;118:1099–108. doi: 10.1182/blood-2010-12-324756. [DOI] [PubMed] [Google Scholar]
- 168.Craig HE, Coadwell J, Guillou H, Vermeren S. ARAP3 binding to phosphatidylinositol-(3,4,5)-trisphosphate depends on N-terminal tandem PH domains and adjacent sequences. Cell Signal. 2010;22:257–64. doi: 10.1016/j.cellsig.2009.09.025. [DOI] [PubMed] [Google Scholar]
- 169.Gambardella L, Anderson KE, Nussbaum C, Segonds-Pichon A, Margarido T, et al. The GTPase-activating protein ARAP3 regulates chemotaxis and adhesion-dependent processes in neutrophils. Blood. 2011;118:1087–98. doi: 10.1182/blood-2010-10-312959. [DOI] [PubMed] [Google Scholar]
- 170.So L, Fruman DA. PI3K signalling in B- and T-lymphocytes: new developments and therapeutic advances. Biochem J. 2012;442:465–81. doi: 10.1042/BJ20112092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Okkenhaug K, Fruman DA. PI3Ks in lymphocyte signaling and development. Curr Top Microbiol Immunol. 2010;346:57–85. doi: 10.1007/82_2010_45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Fruman DA, Bismuth G. Fine tuning the immune response with PI3K. Immunol Rev. 2009;228:253–72. doi: 10.1111/j.1600-065X.2008.00750.x. [DOI] [PubMed] [Google Scholar]
- 173.Okkenhaug K, Vanhaesebroeck B. PI3K in lymphocyte development, differentiation and activation. Nat Rev Immunol. 2003;3:317–30. doi: 10.1038/nri1056. [DOI] [PubMed] [Google Scholar]
- 174.Deane JA, Fruman DA. Phosphoinositide 3-kinase: diverse roles in immune cell activation. Annu Rev Immunol. 2004;22:563–98. doi: 10.1146/annurev.immunol.22.012703.104721. [DOI] [PubMed] [Google Scholar]
- 175.Koyasu S. The role of PI3K in immune cells. Nat Immunol. 2003;4:313–9. doi: 10.1038/ni0403-313. [DOI] [PubMed] [Google Scholar]
- 176.Yilmaz OH, Valdez R, Theisen BK, Guo W, Ferguson DO, et al. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 2006;441:475–82. doi: 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- 177.Zhang J, Grindley JC, Yin T, Jayasinghe S, He XC, et al. PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature. 2006;441:518–22. doi: 10.1038/nature04747. [DOI] [PubMed] [Google Scholar]
- 178.Tothova Z, Kollipara R, Huntly BJ, Lee BH, Castrillon DH, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128:325–39. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 179.Herzog S, Reth M, Jumaa H. Regulation of B-cell proliferation and differentiation by pre-B-cell receptor signalling. Nat Rev Immunol. 2009;9:195–205. doi: 10.1038/nri2491. [DOI] [PubMed] [Google Scholar]
- 180.Corcoran AE, Smart FM, Cowling RJ, Crompton T, Owen MJ, et al. The interleukin-7 receptor alpha chain transmits distinct signals for proliferation and differentiation during B lymphopoiesis. Embo J. 1996;15:1924–32. [PMC free article] [PubMed] [Google Scholar]
- 181.Osborne LC, Duthie KA, Seo JH, Gascoyne RD, Abraham N. Selective ablation of the YxxM motif of IL-7Ralpha suppresses lymphomagenesis but maintains lymphocyte development. Oncogene. 2010;29:3854–64. doi: 10.1038/onc.2010.133. [DOI] [PubMed] [Google Scholar]
- 182.Marshall AJ, Fleming HE, Wu GE, Paige CJ. Modulation of the IL-7 dose-response threshold during pro-B cell differentiation is dependent on pre-B cell receptor expression. J Immunol. 1998;161:6038–45. [PubMed] [Google Scholar]
- 183.Herzog S, Hug E, Meixlsperger S, Paik JH, DePinho RA, et al. SLP-65 regulates immunoglobulin light chain gene recombination through the PI(3)K-PKB-Foxo pathway. Nat Immunol. 2008;9:623–31. doi: 10.1038/ni.1616. [DOI] [PubMed] [Google Scholar]
- 184.Werner M, Hobeika E, Jumaa H. Role of PI3K in the generation and survival of B cells. Immunol Rev. 2010;237:55–71. doi: 10.1111/j.1600-065X.2010.00934.x. [DOI] [PubMed] [Google Scholar]
- 185.Llorian M, Stamataki Z, Hill S, Turner M, Martensson IL. The PI3K p110delta is required for down-regulation of RAG expression in immature B cells. J Immunol. 2007;178:1981–5. doi: 10.4049/jimmunol.178.4.1981. [DOI] [PubMed] [Google Scholar]
- 186.Verkoczy L, Duong B, Skog P, Ait-Azzouzene D, Puri K, et al. Basal B cell receptor-directed phosphatidylinositol 3-kinase signaling turns off RAGs and promotes B cell-positive selection. J Immunol. 2007;178:6332–41. doi: 10.4049/jimmunol.178.10.6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Tze LE, Schram BR, Lam KP, Hogquist KA, Hippen KL, et al. Basal immunoglobulin signaling actively maintains developmental stage in immature B cells. PLoS Biol. 2005;3:e82. doi: 10.1371/journal.pbio.0030082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lam KP, Kuhn R, Rajewsky K. In vivo ablation of surface immunoglobulin on mature B cells by inducible gene targeting results in rapid cell death. Cell. 1997;90:1073–83. doi: 10.1016/s0092-8674(00)80373-6. [DOI] [PubMed] [Google Scholar]
- 189.Kraus M, Alimzhanov MB, Rajewsky N, Rajewsky K. Survival of resting mature B lymphocytes depends on BCR signaling via the Igalpha/beta heterodimer. Cell. 2004;117:787–800. doi: 10.1016/j.cell.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 190.Srinivasan L, Sasaki Y, Calado DP, Zhang B, Paik JH, et al. PI3 kinase signals BCR-dependent mature B cell survival. Cell. 2009;139:573–86. doi: 10.1016/j.cell.2009.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Garcon F, Patton DT, Emery JL, Hirsch E, Rottapel R, et al. CD28 provides T-cell costimulation and enhances PI3K activity at the immune synapse independently of its capacity to interact with the p85/p110 heterodimer. Blood. 2008;111:1464–71. doi: 10.1182/blood-2007-08-108050. [DOI] [PubMed] [Google Scholar]
- 192.Durand CA, Hartvigsen K, Fogelstrand L, Kim S, Iritani S, et al. Phosphoinositide 3-kinase p110 delta regulates natural antibody production, marginal zone and B-1 B cell function, and autoantibody responses. J Immunol. 2009;183:5673–84. doi: 10.4049/jimmunol.0900432. [DOI] [PubMed] [Google Scholar]
- 193.Rolf J, Bell SE, Kovesdi D, Janas ML, Soond DR, et al. Phosphoinositide 3-kinase activity in T cells regulates the magnitude of the germinal center reaction. J Immunol. 2010;185:4042–52. doi: 10.4049/jimmunol.1001730. [DOI] [PubMed] [Google Scholar]
- 194.Zhang TT, Makondo KJ, Marshall AJ. p110delta phosphoinositide 3-kinase represses IgE switch by potentiating BCL6 expression. J Immunol. 2012;188:3700–8. doi: 10.4049/jimmunol.1103302. [DOI] [PubMed] [Google Scholar]
- 195.Zhang TT, Okkenhaug K, Nashed BF, Puri KD, Knight ZA, et al. Genetic or pharmaceutical blockade of p110delta phosphoinositide 3-kinase enhances IgE production. J Allergy Clin Immunol. 2008;122:811–9. e2. doi: 10.1016/j.jaci.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 196.Khalil AM, Cambier JC, Shlomchik MJ. B cell receptor signal transduction in the GC is short-circuited by high phosphatase activity. Science. 2012;336:1178–81. doi: 10.1126/science.1213368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Deane JA, Kharas MG, Oak JS, Stiles LN, Luo J, et al. T-cell function is partially maintained in the absence of class IA phosphoinositide 3-kinase signaling. Blood. 2007;109:2894–902. doi: 10.1182/blood-2006-07-038620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Janas ML, Turner M. Stromal cell-derived factor 1alpha and CXCR4: newly defined requirements for efficient thymic beta-selection. Trends Immunol. 2010;31:370–6. doi: 10.1016/j.it.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 199.Hagenbeek TJ, Naspetti M, Malergue F, Garcon F, Nunes JA, et al. The loss of PTEN allows TCR alphabeta lineage thymocytes to bypass IL-7 and Pre-TCR-mediated signaling. J Exp Med. 2004;200:883–94. doi: 10.1084/jem.20040495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Janas ML, Varano G, Gudmundsson K, Noda M, Nagasawa T, et al. Thymic development beyond beta-selection requires phosphatidylinositol 3-kinase activation by CXCR4. J Exp Med. 2010;207:247–61. doi: 10.1084/jem.20091430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Webb LM, Vigorito E, Wymann MP, Hirsch E, Turner M. Cutting Edge: T Cell Development Requires the Combined Activities of the p110γ and p110δ Catalytic Isoforms of Phosphatidylinositol 3-Kinase. Journal of Immunology. 2005;175:2783–7. doi: 10.4049/jimmunol.175.5.2783. [DOI] [PubMed] [Google Scholar]
- 202.Ji H, Rintelen F, Waltzinger C, Bertschy Meier D, Bilancio A, et al. Inactivation of PI3Kγ and PI3Kδ distorts T-cell development and causes multiple organ inflammation. Blood. 2007;110:2940–7. doi: 10.1182/blood-2007-04-086751. [DOI] [PubMed] [Google Scholar]
- 203.Swat W, Montgrain V, Doggett TA, Douangpanya J, Puri K, et al. Essential role of PI3Kδ and PI3Kγ in thymocyte survival. Blood. 2006;107:2415–22. doi: 10.1182/blood-2005-08-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Kelly AP, Finlay DK, Hinton HJ, Clarke RG, Fiorini E, et al. Notch-induced T cell development requires phosphoinositide-dependent kinase 1. Embo J. 2007;26:3441–50. doi: 10.1038/sj.emboj.7601761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Ciofani M, Zuniga-Pflucker JC. Notch promotes survival of pre-T cells at the beta-selection checkpoint by regulating cellular metabolism. Nat Immunol. 2005;6:881–8. doi: 10.1038/ni1234. [DOI] [PubMed] [Google Scholar]
- 206.Wong GW, Knowles GC, Mak TW, Ferrando AA, Zuniga-Pflucker JC. HES1 opposes a PTEN-dependent check on survival, differentiation and proliferation of TCRbeta-selected mouse thymocytes. Blood. 2012 doi: 10.1182/blood-2011-12-395319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Perumalsamy LR, Marcel N, Kulkarni S, Radtke F, Sarin A. Distinct Spatial and Molecular Features of Notch Pathway Assembly in Regulatory T Cells. Sci. Signal. 2012;5:ra53. doi: 10.1126/scisignal.2002859. [DOI] [PubMed] [Google Scholar]
- 208.Juntilla MM, Koretzky GA. Critical roles of the PI3K/Akt signaling pathway in T cell development. Immunol Lett. 2008;116:104–10. doi: 10.1016/j.imlet.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Hinton HJ, Alessi DR, Cantrell DA. The serine kinase phosphoinositide-dependent kinase 1 (PDK1) regulates T cell development. Nat Immunol. 2004;5:539–45. doi: 10.1038/ni1062. [DOI] [PubMed] [Google Scholar]
- 210.Finlay DK, Sinclair LV, Feijoo C, Waugh CM, Hagenbeek TJ, et al. Phosphoinositide-dependent kinase 1 controls migration and malignant transformation but not cell growth and proliferation in PTEN-null lymphocytes. J Exp Med. 2009;206:2441–54. doi: 10.1084/jem.20090219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Acuto O, Michel F. CD28-mediated co-stimulation: a quantitative support for TCR signalling. Nat Rev Immunol. 2003;3:939–51. doi: 10.1038/nri1248. [DOI] [PubMed] [Google Scholar]
- 212.Holdorf AD, Green JM, Levin SD, Denny MF, Straus DB, et al. Proline residues in CD28 and the Src homology (SH)3 domain of Lck are required for T cell costimulation. J Exp Med. 1999;190:375–84. doi: 10.1084/jem.190.3.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Gigoux M, Shang J, Pak Y, Xu M, Choe J, et al. Inducible costimulator promotes helper T-cell differentiation through phosphoinositide 3-kinase. Proc Natl Acad Sci U S A. 2009;106:20371–6. doi: 10.1073/pnas.0911573106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Barber DF, Bartolome A, Hernandez C, Flores JM, Fernandez-Arias C, et al. Class IB-phosphatidylinositol 3-kinase (PI3K) deficiency ameliorates IA-PI3K-induced systemic lupus but not T cell invasion. Journal of Immunology. 2006;176:589–93. doi: 10.4049/jimmunol.176.1.589. [DOI] [PubMed] [Google Scholar]
- 215.Martin AL, Schwartz MD, Jameson SC, Shimizu Y. Selective regulation of CD8 effector T cell migration by the p110 gamma isoform of phosphatidylinositol 3-kinase. J Immunol. 2008;180:2081–8. doi: 10.4049/jimmunol.180.4.2081. [DOI] [PubMed] [Google Scholar]
- 216.Thomas MS, Mitchell JS, DeNucci CC, Martin AL, Shimizu Y. The p110gamma isoform of phosphatidylinositol 3-kinase regulates migration of effector CD4 T lymphocytes into peripheral inflammatory sites. J Leukoc Biol. 2008;84:814–23. doi: 10.1189/jlb.0807561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Nombela-Arrieta C, Lacalle RA, Montoya MC, Kunisaki Y, Megias D, et al. Differential requirements for DOCK2 and phosphoinositide-3-kinase γ during T and B lymphocyte homing. Immunity. 2004;21:429–41. doi: 10.1016/j.immuni.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 218.Okkenhaug K, Patton DT, Bilancio A, Garcon F, Rowan WC, et al. The p110delta isoform of phosphoinositide 3-kinase controls clonal expansion and differentiation of Th cells. J Immunol. 2006;177:5122–8. doi: 10.4049/jimmunol.177.8.5122. [DOI] [PubMed] [Google Scholar]
- 219.Soond DR, Bjorgo E, Moltu K, Dale VQ, Patton DT, et al. PI3K p110delta regulates T-cell cytokine production during primary and secondary immune responses in mice and humans. Blood. 2010;115:2203–13. doi: 10.1182/blood-2009-07-232330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Haylock-Jacobs S, Comerford I, Bunting M, Kara E, Townley S, et al. PI3Kdelta drives the pathogenesis of experimental autoimmune encephalomyelitis by inhibiting effector T cell apoptosis and promoting Th17 differentiation. J Autoimmun. 2011;36:278–87. doi: 10.1016/j.jaut.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 221.Nashed BF, Zhang T, Al-Alwan M, Srinivasan G, Halayko AJ, et al. Role of the phosphoinositide 3-kinase p110δ in generation of type 2 cytokine responses and allergic airway inflammation. Eur J Immunol. 2007;37:416–24. doi: 10.1002/eji.200636401. [DOI] [PubMed] [Google Scholar]
- 222.Kurebayashi Y, Nagai S, Ikejiri A, Ohtani M, Ichiyama K, et al. PI3K-Akt-mTORC1-S6K1/2 Axis Controls Th17 Differentiation by Regulating Gfi1 Expression and Nuclear Translocation of RORgamma. Cell Rep. 2012;1:360–73. doi: 10.1016/j.celrep.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 223.Rommel C, Camps M, Ji H. PI3K delta and PI3K gamma: partners in crime in inflammation in rheumatoid arthritis and beyond? Nat Rev Immunol. 2007;7:191–201. doi: 10.1038/nri2036. [DOI] [PubMed] [Google Scholar]
- 224.Banham-Hall E, Clatworthy MR, Okkenhaug K. The Therapeutic Potential for PI3K Inhibitors in Autoimmune Rheumatic Diseases. The Open Rheumatology Journal. 2012:6. doi: 10.2174/1874312901206010245. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Fukao T, Tanabe M, Terauchi Y, Ota T, Matsuda S, et al. PI3K-mediated negative feedback regulation of IL-12 production in DCs. Nat Immunol. 2002;3:875–81. doi: 10.1038/ni825. [DOI] [PubMed] [Google Scholar]
- 226.Patton DT, Garden OA, Pearce WP, Clough LE, Monk CR, et al. Cutting edge: the phosphoinositide 3-kinase p110 delta is critical for the function of CD4+CD25+Foxp3+ regulatory T cells. J Immunol. 2006;177:6598–602. doi: 10.4049/jimmunol.177.10.6598. [DOI] [PubMed] [Google Scholar]
- 227.Soond DR, Slack EC, Garden OA, Patton DT, Okkenhaug K. Does the PI3K pathway promote or antagonise regulatory T cell development and function? Front Immunol. 2012:3. doi: 10.3389/fimmu.2012.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Sauer S, Bruno L, Hertweck A, Finlay D, Leleu M, et al. T cell receptor signaling controls Foxp3 expression via PI3K, Akt, and mTOR. Proc Natl Acad Sci U S A. 2008;105:7797–802. doi: 10.1073/pnas.0800928105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Araki K, Turner AP, Shaffer VO, Gangappa S, Keller SA, et al. mTOR regulates memory CD8 T-cell differentiation. Nature. 2009;460:108–12. doi: 10.1038/nature08155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Li Q, Rao RR, Araki K, Pollizzi K, Odunsi K, et al. A central role for mTOR kinase in homeostatic proliferation induced CD8+ T cell memory and tumor immunity. Immunity. 2011;34:541–53. doi: 10.1016/j.immuni.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Rao RR, Li Q, Odunsi K, Shrikant PA. The mTOR kinase determines effector versus memory CD8+ T cell fate by regulating the expression of transcription factors T-bet and Eomesodermin. Immunity. 2010;32:67–78. doi: 10.1016/j.immuni.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Liu D, Zhang T, Marshall AJ, Okkenhaug K, Vanhaesebroeck B, et al. The p110delta isoform of phosphatidylinositol 3-kinase controls susceptibility to Leishmania major by regulating expansion and tissue homing of regulatory T cells. J Immunol. 2009;183:1921–33. doi: 10.4049/jimmunol.0901099. [DOI] [PubMed] [Google Scholar]
- 233.Gold MR, Puri KD. Selective inhibitors of phosphoinositide 3-kinase delta: Modulators of B-cell function with potential for treating autoimmune inflammatory diseases and B-cell malignancies. Front Immunol. 2012:3. doi: 10.3389/fimmu.2012.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Fruman DA, Rommel C. PI3Kdelta inhibitors in cancer: rationale and serendipity merge in the clinic. Cancer Discov. 2011;1:562–72. doi: 10.1158/2159-8290.CD-11-0249. [DOI] [PubMed] [Google Scholar]
- 235.Lannutti BJ, Meadows SA, Herman SE, Kashishian A, Steiner B, et al. CAL-101, a p110delta selective phosphatidylinositol-3-kinase inhibitor for the treatment of B-cell malignancies, inhibits PI3K signaling and cellular viability. Blood. 2011;117:591–4. doi: 10.1182/blood-2010-03-275305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Subramaniam PS, Whye DW, Efimenko E, Chen J, Tosello V, et al. Targeting nonclassical oncogenes for therapy in T-ALL. Cancer Cell. 2012;21:459–72. doi: 10.1016/j.ccr.2012.02.029. [DOI] [PubMed] [Google Scholar]