Significance
Substituted d-galactopyranosides, particularly those in the α configuration and/or with hydrophobic constituents at the anomeric position, bind to LacY with higher affinity than the physiological substrate lactose that has a β configuration. The structure of a conformationally restricted LacY mutant with bound p-nitrophenyl-α-d-galactopyranoside (α-NPG), a high-affinity lactose analog, is described. Higher affinity, gained by nonspecific hydrophobic interaction of the nitrophenyl group, shows identical interaction at the key galactosyl moiety as in thio-digalactoside and so validates the highly specific, oriented set of hydrogen bonds with the key galactosyl moiety of substrates. Confirmation of galactose-specific binding interactions delineates a directional hydrogen-bonding network that couples the binding site to sites that are sensitive to protonation in the mechanism.
Keywords: X-ray structure, membrane protein, transport, major facilitator superfamily, conformational change
Abstract
The X-ray crystal structure of a conformationally constrained mutant of the Escherichia coli lactose permease (the LacY double-Trp mutant Gly-46→Trp/Gly-262→Trp) with bound p-nitrophenyl-α-d-galactopyranoside (α-NPG), a high-affinity lactose analog, is described. With the exception of Glu-126 (helix IV), side chains Trp-151 (helix V), Glu-269 (helix VIII), Arg-144 (helix V), His-322 (helix X), and Asn-272 (helix VIII) interact directly with the galactopyranosyl ring of α-NPG to provide specificity, as indicated by biochemical studies and shown directly by X-ray crystallography. In contrast, Phe-20, Met-23, and Phe-27 (helix I) are within van der Waals distance of the benzyl moiety of the analog and thereby increase binding affinity nonspecifically. Thus, the specificity of LacY for sugar is determined solely by side-chain interactions with the galactopyranosyl ring, whereas affinity is increased by nonspecific hydrophobic interactions with the anomeric substituent.
The lactose permease of Escherichia coli (LacY) binds and catalyzes the coupled stoichiometric transport of d-galactose or β-d-galactopyranosides and H+ (galactoside/H+ symport) but does not interact with glucopyranosides. Biochemical studies (1–7) indicate that affinity and specificity are distinct properties determined by different interactions with LacY. Specificity is determined entirely by interactions with the galactopyranosyl ring, whereas affinity is better with α- than β-galactopyranosides (anomeric at C1) and can be increased dramatically by hydrophobic anomeric substituents with no effect on specificity.
By using the free energy released from the energetically downhill movement of H+ in response to the electrochemical H+ gradient (∆µ̃H+), LacY catalyzes uphill (active) transport of galactosides against a concentration gradient. Because coupling between sugar and H+ translocation is obligatory, in the absence of ∆µ̃H+, LacY can also transduce the free energy released from the downhill transport of sugar to drive uphill H+ transport with the generation of ∆µ̃H+, the polarity of which depends upon the direction of the sugar gradient (reviewed in refs. 8–10).
Rates of equilibrium exchange and counterflow (exchange of one substrate molecule for another labeled molecule from the other side of the membrane) are unaffected by imposition of ∆µ̃H+. Therefore, it is apparent that alternating accessibility of sugar- and H+-binding sites to either side of the membrane is the result of galactoside binding and dissociation and not ∆µ̃H+ (reviewed in refs. 8–10). Moreover, downhill lactose/H+ symport from a high to a low lactose concentration in the absence of ∆µ̃H+ exhibits a primary deuterium isotope effect that is not observed for ∆µ̃H+-driven lactose/H+ symport, equilibrium exchange, or counterflow (11, 12). Thus, it is likely that the rate-limiting step for downhill symport is deprotonation (13, 14), whereas in the presence of ∆µ̃H+, opening of a cavity on the other side of the membrane after dissociation of sugar and H+ is limiting (15). Based on these and other findings, a detailed mechanism for symport by LacY has been proposed (10).
Initial X-ray structures of LacY without bound sugar exhibit two pseudosymmetrical bundles of mostly irregular transmembrane helices surrounding a large aqueous cavity in the middle of the molecule; these initial structures were open on the cytoplasmic side and sealed on the periplasmic side (an inward-open conformation) (16–19). However, our recent X-ray crystallography studies (20) of the conformationally trapped double-Trp mutant G46W/G262W cocrystallized with β-d-galactopyranosyl-1-thio-β-d-galactopyranoside (TDG) reveal an almost occluded conformation with a narrowly outward (periplasmic)-open conformation and a tightly sealed cytoplasmic side [Protein Data Bank (PDB) ID code 4OAA]. In addition, a molecule of TDG is bound in a central cavity. The evidence shows that specific galactoside binding is consistent with prior findings from mutagenesis (21–23) and uses induced fit to interact with the surrounding protein (20). The findings also provide a strong indication that the transport mechanism of LacY involves a substrate-bound, occluded, intermediate conformation.
Lactose has only one galactopyranosyl ring. Similarly, one galactopyranosyl ring of TDG lies against Trp-151 (helix V), confirming hydrophobic stacking between the bottom of the galactopyranosyl ring and the aromatic indole ring as suggested (24, 25). Glu-269 (helix VIII) is the acceptor of hydrogen bonds from the C4-OH group of the galactopyranosyl ring (21, 26). The η1 NH2 group of Arg-144 (helix V) donates a hydrogen bond to O5 in the ring and is within hydrogen-bond distance of the C6-OH, whereas the η2 NH2 group of Arg-144 donates hydrogen bonds to the C2′-OH (the other galactopyranosyl ring) of TDG and to Glu-126 Oε2 (27–29). Glu-126 (helix IV) acts as hydrogen-bond acceptor from the C2′-OH of TDG (27–29). His-322 (helix X) acts as a hydrogen-bond donor/acceptor between the εNH of the imidazole ring and the C3-OH of TDG (29–33) and is stabilized by a hydrogen-bond donor/acceptor interaction with the δNH of the imidazole and the OH of Tyr-236 (29, 34, 35). Finally, Asn-272 (helix VIII) donates a hydrogen bond to the C4-OH of TDG (23). These interactions define the specificity of LacY (summarized in ref. 20).
Cys-148 (helix V), well known with respect to substrate protection against alkylation (reviewed in ref. 22), is also close to bound TDG but not sufficiently close to interact directly. Similarly, replacement of Ala-122 (helix IV) with a bulky side chain or alkylation of A122C with bulky thiol reagents causes LacY to become specific for the monosaccharide galactose. Disaccharide binding and transport are blocked sterically (36). Although Ala-122 does not make direct contact with TDG, bulky substituents at position 122 would clearly impact disaccharide binding.
p-Nitrophenyl-α-d-galactopyranoside (α-NPG) is a FRET acceptor from Trp-151 (37) and binds to LacY with ∼eight times higher affinity than the twofold symmetric TDG, ∼two orders of magnitude better than β-NPG, and ∼three orders of magnitude better than the physiological substrate lactose or the monosaccharide galactose (6, 7, 38, 39). As we show here, the side-chain interactions with the galactopyranosyl moiety of α-NPG that provide specificity are almost identical to those described for TDG. In contrast, the increased affinity of α-NPG versus TDG is probably attributable primarily to hydrophobic interactions between the nitrophenyl group of NPG and Phe-20, Met-23, and Phe-27 from helix I. In addition, the nitro group is in close contact with polar groups that can sustain polar interactions of the type that also pertain to the glucose moiety of lactose.
Results
Global Fold.
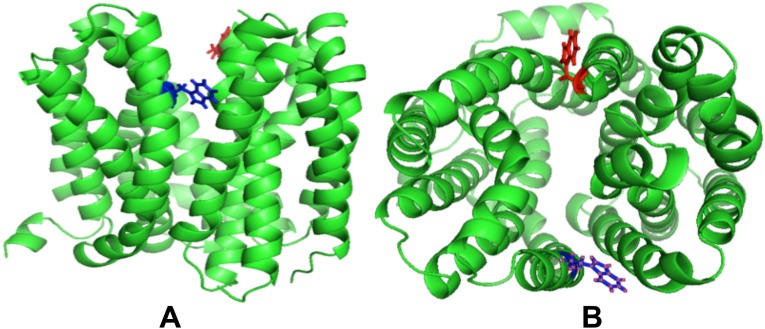
The X-ray crystal structure of LacY G46W/G262W cocrystallized in the presence of 1 mM α-NPG was determined to 3.3-Å resolution (Fig. 1). Data collection and refinement statistics are summarized in Table 1. Crystals of the G46W/G262W mutant with α-NPG are in space group C2221 with the cell dimensions a = 103.17 Å, b = 122.01 Å, c = 266.13 Å. The structure was determined by molecular replacement using the almost occluded outward-open structure of G46W/G262W LacY alone after removing the ligand TDG from PDB ID code 4OAA (20) as the search molecule. The structure of the protein with bound α-NPG is essentially the same as the TDG-bound structure (PDB ID code 4OAA) (rmsd, 0.52 Å), in contrast to the inward-facing WT LacY structure (19) (PDB ID code 2V8N).
Fig. 1.
Electron density map of the α-NPG ligand site in molecule A, LacY G46W/G262W. Density contoured at 1.3σ (golden yellow mesh) is superimposed on the cartoon representation of the structure. Residues surrounding the ligand are labeled. Carbon atoms in α-NPG are shown in orange, nitrogen in blue, and oxygen in red. Side-chain atoms are shown as carbon in yellow, nitrogen in blue, oxygen in red, and sulfur in light yellow. Broken blue lines show hydrogen bonds with lengths labeled in Å. The cytoplasmic side is upward, and the normal to the membrane plane is approximately vertical.
Table 1.
Data collection and refinement statistics
| Wavelength, Å | 1.116 |
| Resolution range, Å | 37–3.31 (3.42–3.31) |
| Space group | C 2 2 21 |
| Unit cell | 103.2 122.0 266.1 |
| Total reflections | 114045 (10755) |
| Unique reflections | 25009 (2429) |
| Multiplicity | 4.6 (4.4) |
| Completeness, % | 97.6 (97.2) |
| Mean I/σ(I) | 7.8 (0.4) |
| Wilson B-factor | 126 |
| R-merge | 0.16 (4.06) |
| R-measured | 0.18 |
| CC1/2 | 0.998 (0.082) |
| CC* | 0.999 (0.390) |
| R-work | 0.25 (0.40) |
| R-free | 0.28 (0.41) |
| Nonhydrogen atoms | 6157 |
| Ligand atoms | 84 |
| No. waters | 0 |
| Protein residues | 772 |
| rms (bonds), Å | ±0.004 |
| rms (angles), ° | ±0.85 |
| Ramachandran favored, % | 95 |
| Ramachandran outliers, % | 0.39 |
| Clash score | 14.20 |
| Average B-factor | 142 |
| Macromolecules | 142 |
| Ligands | 141 |
There are two independent molecules of LacY in the asymmetric unit that are adjacent to one another but of opposite-facing orientation relative to the putative plane of the membrane (Fig. S1). The protein occupies 28% of the unit cell volume. The Trp replacements G46W/G262W on the periplasmic side induce the transition to an almost occluded, outward-facing conformation and allow substrate binding in an almost identical manner to TDG with respect to the galactopyranosyl moiety (Fig. S2).
Fig. S1.
The 2Fo-Fc electron density for one asymmetric unit, each molecule with one NPG substrate bound. (A) The crystallographic asymmetric unit showing two LacY G46WG262W molecules antiparallel to each other. Two molecules of β-nonyl glucoside can be seen in the interface between LacY molecules; NPG is shown in the centers. Chain A (blue ribbons) has the cytoplasmic surface upward. Chain B (yellow) has the periplasmic face upward. (B) Shown is the density around bound NPG in chain A, with carbon in yellow and oxygen in red.
Fig. S2.
LacY G46WG262W. The partially occluded state is facing outward. The figure shows Trp replacement at position 46 in red and Trp replacement at positions 262 in blue. (A) Side view with the periplasmic side upward. (B) View from the periplasmic side.
The two independent α-NPG–bound LacY molecules in the unit cell (termed A and B) are both in an almost-occluded, outward-open conformation with a single molecule of α-NPG in the central sugar-binding site, suggesting that they are in their most stable solution structure (Fig. 2). The two structures differ slightly from each other only in the orientation of the substrate and the slight rearrangement of the interacting side chains (Table S1). There are also two molecules of nonyl-β-d-glucopyranoside bound in the interface between the LacY molecules. (Fig. S1).
Fig. 2.
The galactoside-binding site in LacY G46W/G262W, molecule A. The cytoplasmic side is upward, and the normal to the membrane plane is approximately vertical. Atoms in α-NPG are shown with carbon in orange, nitrogen in blue, and oxygen in red. Side-chain atoms are shown with carbon in yellow, nitrogen in blue, oxygen in red, and sulfur in light yellow. Dotted blue lines identify hydrogen bonds.
Table S1.
Bond distances between side chain atoms and sugar atoms (NPG and TDG) in two different binding sites in the asymmetric unit (calculated in COOT)
| Protein side-chain atom | NPG atom | Distance in chain A (Å) | Distance in chain B (Å) | TDG atom | Distance in chain A (Å) | Distance in chain B (Å) |
| E269 OE1 | O4 | 2.9 | 3.2 | O4 | 2.7 | (3.6) |
| E269 OE2 | O4 | 3.2 | (3.8) | O4 | 3.4 | (4.3) |
| E269 OE2 | O3 | 2.8 | 2.8 | O3 | 3.1 | (4.3) |
| R144 NH1 | O6 | 3.0 | 2.8 | O6 | 2.5 | 2.6 |
| R144 NH2 | O6 | (3.7) | 3.0 | O6 | (4.1) | 3.5 |
| R144 NH2 | E126 OE2 | 3.2 | 3.5 | E126 OE2 | 2.8 | 2.8 |
| R144 NH2 | O5 | (3.6) | 3.0 | O5 | (4.1) | 3.2 |
| R144 NE | E126 OE2 | (3.6) | 3.0 | E126 OE2 | (3.9) | 2.6 |
| H322 NE2 | O2 | 3.4 | (3.7) | O2 | (4.2) | (4.1) |
| H322 NE2 | O3 | 3.1 | (3.9) | O3 | (3.5) | 3.2 |
| H322 ND1 | E325 OE2 | 3.2 | (3.7) | E325 OE2 | 3.3 | 3.5 |
| H322 ND1 | Y236O | 3.3 | 3.0 | Y236O | 3.1 | 2.8 |
| N272 ND2 | O4 | 3.3 | 2.9 | O4 | 3.1 | 3.2 |
| F27 CE2 | C11 | (4.0) | (3.6) | C6′ | (3.8) | (3.7) |
| M23 SD | C10 | 3.5 | (4.3) | C5′ | (4.5) | (5.4) |
| C148 SG | O4 | (3.9) | 3.1 | O4 | (4.6) | (5.2) |
| C148 SG | O6 | 3.1 | 3.2 | O6 | (4.1) | (4.3) |
Bold numbers indicate distances that are longer than hydrogen bonding.
Compared with WT LacY [without substrate bound (PDB ID code 2V8N)], the overall rmsd between 390 Cαs of the G46W/G262W is 4.5 Å, reflecting the major conformational change to the almost occluded, outward-facing conformation (leaving out the ligand and immediate surroundings).
The Substrate-Binding Site.
The conformation of a higher-affinity α-NPG molecule (Kd, ∼2 µM) is apparent in the almost occluded central cavity (Figs. 1 and 2 and Fig. S1) and allows assignment of probable hydrogen-bond interactions with the protein (Table S1). Although at 3.3-Å resolution interatomic distances are an approximation, their mutual orientation is key to defining hydrogen-bonding interactions. Electron density for the α-NPG is also detected in an unbiased omit map after refinement without ligand clearly showing its orientation and interactions.
Cys-148, which is protected from alkylation by substrate binding, is somewhat closer to the galactopyranosyl ring here (3.1 Å versus 4.6 Å) than observed previously with the TDG-bound structure (PDB ID code 4OAA). Therefore, Cys-148 may be hydrogen bonded to C6-OH of the galactoside in the primary specificity site. (Table S1).
His-322 in both the TDG and α-NPG bound structures provides a hydrogen-bonded bridge between the C2-OH and C3-OH of the galactoside on one side of the imidazole ring and Glu-325 and Tyr-236 on the other side (Fig. S3). In turn, Tyr-236 accepts a hydrogen bond from donor Arg-302. Without substrate bound, this complex internal site could rearrange to redistribute charge, decrease the effective pKa of the site around Glu-325, and account for coupled H+ transport.
Fig. S3.
Shown is the density around His-322, which is in close proximity to Arg-302, Tyr-236, and Glu-325. Carbon is depicted in yellow, nitrogen in blue, and oxygen in red.
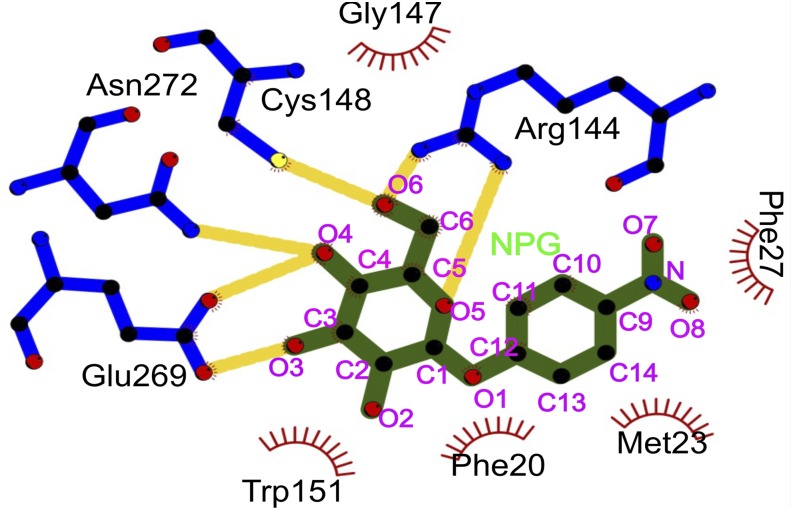
α-NPG is tilted toward Met-23, and the p-nitrophenyl group is in van der Waals contact with it (Fig. 2); the sulfur atom in Met-23 is ∼3.5 Å from the C10 (Fig. S4 and Table S1) of α-NPG. Together, Phe-20, Phe-27, and Met-23 form a hydrophobic cavity. A polar contact from the carbonyl oxygen of Phe-118 (helix IV) is rotated toward one nitrophenyl oxygen (O7) of α-NPG at a distance of ∼3 Å. The side chain of Asn-119 (helix IV) also contributes to the polar environment in the vicinity of the nitro moiety of the nitrophenyl ring at a distance of ∼3.7–4.0 Å.
Fig. S4.
Shown is numbering of atoms in NPG in the galactoside binding site. Solid yellow lines represent hydrogen bonds, and semicircle with red rays depict hydrophobic interaction. The figure was made in Ligplot. The black solid circle represents carbon; red, oxygen; blue, nitrogen; and yellow, sulfur.
Discussion
In order for LacY to transport galactopyranosides sufficiently rapidly for the cell to grow at optimal rates, binding affinity cannot be too high. Otherwise, the dissociation rate of the sugar would be limiting for growth. Thus, lactose, the physiological substrate for LacY, binds with millimolar affinity but is transported at an optimal rate. However, the low affinity for lactose precludes direct determination of binding, and in this regard, protection of Cys-148 against alkylation by substrate is particularly useful for measuring relative affinities of low-affinity substrates (5–7, 38, 40, 41). By using this technique, it was shown that despite having the lowest affinity, the monosaccharide galactose binds as specifically as substituted galactopyranosides that bind orders of magnitude better, but glucose or glucopyranosides have no effect. It has also been shown (5, 6) that although the specificity of LacY is strongly directed toward the C4-OH of the galactopyranosyl ring, the OH groups are important in the following order: C4-OH >> C6-OH > C3-OH > C2-OH. These interactions seen in the structure broadly explain the relative preferences in terms of the number of hydrogen bonding groups that coordinate the galactopyranosyl hydroxyls.
TDG appears to be unique as a relatively high-affinity ligand, because the anomeric functional group is hydrophilic rather than hydrophobic. However, the hydrophilic moiety is another galactopyranosyl molecule, and because the two moieties are linked one-to-one, TDG is perfectly symmetrical. Therefore, the molecule can associate with the binding site in LacY from either end. This being the case, the symmetrical digalactopyranoside TDG may not be able to dissociate readily from the binding site, which causes TDG to bind almost three orders of magnitude better than lactose but also results in much lower transport activity relative to lactose (26, 42).
Furthermore, galactopyranosides in the α-anomeric (C1) configuration bind with better affinity than β-anomeric (C1) galactopyranosides, and affinity can be increased dramatically by hydrophobic additions to the anomeric position C1 of galactose with no effect on specificity (5). For example, LacY exhibits no demonstrable affinity for p-nitrophenyl-α-d-glucopyranoside in which the C4-OH has the alternate chirality (43, 44). These results also emphasize that the interactions seen in the structure determine specificity.
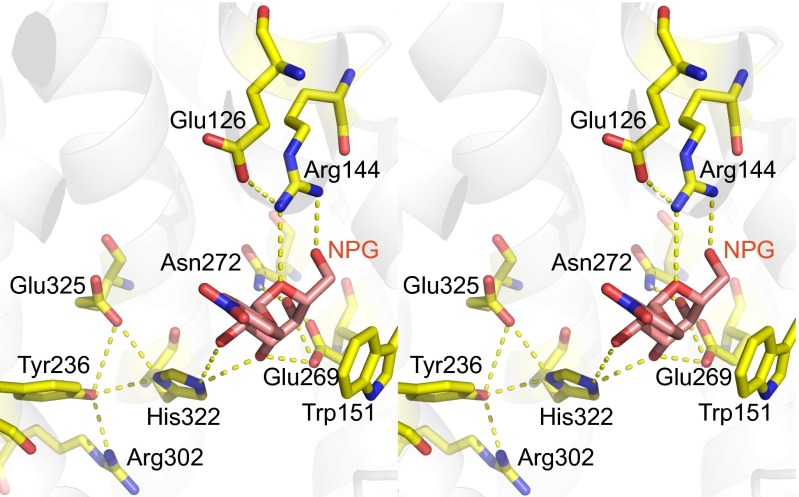
The galactopyranosyl rings of α-NPG and TDG in the binding site are in an essentially identical configuration, although they differ in chirality at C1, α in NPG (an α-galactoside, downward in Fig. 3) versus β in TDG (a β-thio di-galactoside, upward in Fig. 3). This difference in chirality is offset by the nitrophenyl in α-NPG lying in a more hydrophobic cavity; with more polar surroundings of the second galactose ring in TDG, where Glu-126 (helix IV) and the η2 NH1 of Arg-144 are hydrogen-bonded to the C2′-OH. The Arg-144 η2 NH1 can no longer hydrogen bond to the nitrophenyl group, and instead hydrogen bonds to O5 of the galactopyranosyl ring (Fig. 3). However, Glu-126 Oε2 remains as a hydrogen bond acceptor from the η2 NH2 group of Arg-144 (Figs. 1–4). The hydrogen bond of the η1 NH2 group of Arg-144 (helix V) to C6-OH is maintained with both ligands. Thus, Arg-144 makes two hydrogen bonds with the galactopyranosyl ring in α-NPG (Fig. 4).
Fig. 3.
Comparison of the sugar-binding site in α-NPG-bound with TDG-bound LacY G46WG262W. The view is crossed-eyed stereo. The TDG structure (PDB ID code 4OAA chain A) [light blue ribbons and side chains as sticks (carbon in blue, oxygen in red, nitrogen in dark blue, and sulfur in yellow) with TDG (shown as carbon in blue, oxygen in red, nitrogen in dark blue, and sulfur in yellow) hydrogen bonds in dashed red] is superimposed on the α-NPG structure [green ribbons with side chains as sticks (carbon in green, oxygen in red, nitrogen in dark blue, and sulfur in yellow) with NPG (shown as carbon in green, oxygen in red, and nitrogen in dark blue) hydrogen bonds in dashed black].
Fig. 4.
Connection between the ligand-binding site and residues known to be involved in protonation/deprotonation during the transport cycle are indicated. The figure is in crossed-eyed stereo. The cytoplasmic surface is upward, and the normal to the membrane plane is approximately vertical. The figure shows the hydrophobic contact of the galactoside ring with the indole ring of Trp-151. O2 and O3 of the galactose are hydrogen-bonded to the imidazole of His-322 that in turn forms hydrogen bonds with Glu-325 and Tyr-236. The hydroxyl of Tyr-236 is also acceptor to a hydrogen bond from Arg-302.
In both α-NPG and TDG, the primary galactopyranosyl ring lies against the aromatic face of Trp-151 (helix V), providing hydrophobic stacking, as found in other sugar-binding sites. Glu-269 (helix VIII) is acceptor of two hydrogen bonds, each with almost ideal geometry, from the C3-OH and C4-OH groups of the galactopyranosyl ring and so provides selectivity-determining interactions. The εN2 of His-322 (helix X) forms a bifurcated hydrogen bond to both C2-OH and C3-OH. If Glu-269 is unprotonated and negatively charged it would imply that C3-OH provides the donor hydrogen to Glu-269. Therefore, the His εN2 must be protonated and a binary donor to two oxygen substituents of the pyranosyl ring. On the other side of the imidazole ring, the δNH of the imidazole of His-322 forms a critical linkage with groups that are essential for protonation/deprotonation particularly the carboxyl of Glu-325 and also the hydroxyl of Tyr-236 (Fig. 4). If Glu-269 is charged, it will act as acceptor from C4-OH of the pyranose, therefore Asn-272 (helix VIII) most probably donates a hydrogen bond to the oxygen of C4-OH in α-NPG.
Glu-325 (helix X) and Arg-302 (helix IX) are directly involved in coupled H+ translocation; neutral replacement of either residue yields mutants that are defective in all transport reactions that involve lactose/H+ symport but bind substrate well and catalyze equilibrium exchange and/or counterflow as well or better than WT (45–47). The mutants therefore undergo the conformational change allowing alternate access to either side of the membrane and hence translocation of galactopyranoside across the membrane, a fundamental property of the symporter. However, the mutants are no longer coupled to the pKa changes in Glu-325 that must occur during coupled H+ transport (10).
The phenyl ring of α-NPG derives its affinity primarily from hydrophobic interactions. The side chains of Met-23, Phe-20, and Phe-27 in helix I (∼3.6 Å) provide a hydrophobic cavity accounting for higher affinity. α-NPG is tilted toward Met-23, and the p-nitrophenyl group is in van der Waals contact; the sulfur atom in Met-23 is ∼3.5 Å from the C10 of α-NPG (Fig. 2, Fig. S4, and Table S1). A polar contact from the carbonyl oxygen of Phe-118 (helix IV) is rotated toward one nitrophenyl oxygen (O7) of α-NPG at a distance of ∼3 Å. The side chain of Asn-119 (helix IV) also contributes to the polar environment in the vicinity of the nitro moiety of the nitrophenyl ring at a distance of ∼3.7–4.0 Å. The large cavity around the nitrophenyl group provides both hydrophobic and polar counterparts in what would be the vicinity of the glucose moiety of lactose. Notably, glucose is often bound using hydrophobic and polar groups in proteins.
α-NPG binds 10 times better than phenyl-α-d-galactopyranoside (lacking the nitro group), and the nitro group provides ∼three times higher affinity in the para position, as opposed to the ortho or meta position (7). This effect could thus be entropic because of the higher entropic cost of ordering the ortho and meta substituents or to the plausible solvent linkage to Asn-119. The methyl group of Ala-122 (helix IV) is ∼4.5 Å from O7 of the nitrophenyl ring, and mutational analysis is also consistent with α-NPG being relatively distant from Ala-122.
In addition to the nitrophenyl group, methyl, allyl, cyclohexyl, phenyl, methylphenyl, napththyl (7), or dansyl (48) increase affinity to varying degrees. However, these hydrophobic interactions not only make no contribution to specificity, but individual mutation of the surrounding side chains in LacY has little or no effect on the ability of LacY to catalyze lactose/H+ symport (21, 49). Thus, they are not involved in the symport mechanism. Perhaps surprising is the large volume around the substituents of the primary galactosyl moiety, suggesting a role for water in the coordination of substrate and, particularly, a role for water after the galactoside dissociates, when LacY must return empty in a state that deprotonates before transitioning into the empty occluded state.
In the mechanism proposed recently (10), Glu-325 plays the key role in coupled H+ translocation. To bind galactoside, Glu-325 must be protonated, and because this side chain appears to have a pKa of ∼10.5 for binding substrates (29, 50, 51), LacY is protonated over the physiological pH range. Furthermore, replacement of any single side chain that interacts directly with the galactosyl moiety of substrate drastically decreases affinity (29). As shown in the current structure (Fig. 4), Glu-325 is linked to the galactoside-binding site via a hydrogen bond with His-322Nδ. The His-322NεH is hydrogen donor to 3OH of the galactopyranose, because if Glu-269 is negatively charged, the 3OH in turn acts as hydrogen donor to Glu-269. If the site returns to the occluded state without substrate bound, it must lose an H+, implying a drop in the effective pKa. It does so presumably through interaction of protonated Glu-325 with Arg-302 and water molecules and loses the H+ to the side that is closing. This proposal can then account for the vectorial nature of the deprotonation, determined primarily by the water access of the closing side, whichever that may be. Although Glu-325 and Arg-302 are essential for protonation/deprotonation and His-322 is essential for binding, all three may be required for the pKa change in Glu-325 upon closing to the occluded, substrate-free conformation.
Materials and Methods
Materials.
NPG and buffers were from Sigma-Aldrich. Talon superflow resin was from BD Clontech. Dodecyl-β-d-maltopyranoside (DDM) and n-nonyl-β-d-glucoside (NG) were from Affymetrix. All other materials were of reagent grade and obtained from commercial sources.
Growth, Expression, and Purification.
Plasmid pT7-5 encoding LacY G46W/G262W/His6 tag was expressed in E. coli C41. A detailed procedure is described in SI Materials and Methods. Membranes were prepared and solubilized in 2% (wt/vol) DDM, and LacY was purified on a Co (II) column and eluted in 20 mM Hepes/0.2% NG/200 mM imidazole (pH 6.5). Details are given in SI Materials and Methods.
Crystallization, Data Collection, and Structure Determination.
NPG was added (1 mM, final concentration) to a protein solution [10 mg/mL in 20 mM Hepes (pH 6.5)/0.2% NG] before crystallization trials. Crystallization screens were performed using the hanging-drop vapor-diffusion method on a Mosquito Crystal Robot (TTP Labtech) in a 96-well plate with a drop ratio of 100 nL of well solution:300 nL of protein. Crystals appeared in 24 h and grew to 600 × 200 × 150 µm in a week. Crystals were harvested after 7 d and screened for diffraction. Crystals were reproducible and appeared in wide range of PEG concentrations. They also appeared at different pH values and with different buffers. The best diffracting crystals grew in 1.0 M NaCl/0.05 M Tris (pH 8.0)/26% PEG 600. No cryoprotectant was added before data collection.
Diffraction data were collected at the Lawrence Berkeley National Laboratory Advanced Light Source Beamline 8.3.1, at −170 °C at a wavelength of 1.115 Å. The highest-resolution crystals diffracted to 3.3 Å. Data were processed with XDS (52) and HKL2000 (53).
The structure was determined by molecular replacement using LacYG46WG262W (PDB ID code 4OAA) as the search model, in the program suite PHENIX (54). The program COOT was used for density fitting (55), and refinement was carried out using PHENIX and Refmac from the CCP4 suite (56). All covalent hydrogen atoms were added in expected positions. Table 1 shows data and refinement statistics. The MolProbity server (57) was used for structural validation leading to improvement. Restrained refinement was performed with noncrystallographic symmetry (NCS) and stereochemistry restraint for all of the refinement, at later stage crystallographic refinement server developed by Haddadian et al. (58) (godzilla.uchicago.edu) was used for further refinement.
SI Materials and Methods
α-NPG and buffers were purchased from Sigma-Aldrich. Talon superflow resin was purchased from BD Clontech. DDM and NG were obtained from Affymetrix. All other materials were of reagent grade and obtained from commercial sources.
Growth and Expression.
Plasmid pT7-5 encoding LacY G46W/G262W/His6 tag was expressed in E. coli C41. Transformed cells (colonies from LB/Amp plates) were inoculated into 200 mL of LB/Amp (0.1 mg/mL ampicillin, final concentration) and grown overnight at 37 °C on a shaker; 200-mL cultures were inoculated into 10 l fermenters, and when the OD600 reached 1 (∼6 h after inoculation), 2 mM IPTG was added to induce. Cells were grown for another 4–5 h before harvesting. Cells were harvested, and 200 g of cells were resuspended to 800 mL in 50 mM NaPi (pH 7.5)/2 mM DTT/25 mg 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF)/two tablets Complete Protease Inhibitor (Sigma-Aldrich). A Bead Beater was the used to lyse the cells at 0 °C using pulses of 1 min on and 3 min off for 10 cycles. Preparations were centrifuged at ∼12,000 × g for 15 min, and the supernatant was centrifuged in a Ti45 rotor at 42,000 rpm for 3 h. The membrane pellets were resuspended in 100 mL of 50 mM cold NaPi (pH 7.5) using a Teflon and glass homogenizer. The membrane suspension (25 mL aliquots in 50-mL tubes) were flash-frozen in liquid nitrogen and stored at −80 °C until use.
Purification.
Thawed membrane suspensions (50 mL) were placed in a glass beaker in an ice bath with stirring. DDM powder (1 g) was added to 50 mL of membrane suspension (2% DDM, final concentration), and stirring was continued on ice for 30 min. The sample was then filtered thru 5- and 2-µm filters to remove insoluble material. To the combined supernatants (50 mL), 2.7 mL of 4 M NaCl (200 mM, final concentration) and 0.9 mL of 300 mM imidazole (5 mM, final concentration) were added, and the pH was adjusted to pH 7.6 by the addition of 0.5 M Na2HPO4 (pH 10). The extract was then loaded onto a Talon column (1.5 × 5 cm) that had been equilibrated with 50 mM NaPi/0.01% DDM/200 mM NaCl/5 mM imidazole (pH 7.6) (column buffer). Nonabsorbed material was washed from the column with column buffer, followed by washing with 50 mM NaPi/0.01% DDM/200 mM NaCl/10 mM imidazole (pH 7.6) (wash buffer 2) and 50 mM NaPi/0.2% NG (pH 7.6) (wash buffer 3) to background UV absorption. Purified protein was eluted from the column with 20 mM Hepes (pH 6.5)/0.2% NG/200 mM imidazole. The protein was finally concentrated with Amicon Ultra centrifugal filters (molecular mass cutoff, 50 KDa) at 3,500 rpm in an Eppendorf centrifuge 5810R swinging-bucket rotor to 0.2–0.5 mL and diluted in 15 mL of 20 mM Hepes /0.2% NG (pH 6.5) and concentrated again to ∼10 mg/mL.
Crystallization and Data Collection.
NPG (1 mM final) was added to the protein [10 mg/mL in 20 mM Hepes (pH 6.5)/0.2% NG)] before setting up trays. Crystallization trials were carried out using the Mosquito robot in a 96-well plate with a drop ratio of 100 nL of well solution to 100 nL of protein or 100 nL of well solution to 300 nL of protein. Crystals appeared in 24 h and grew to 600 × 200 × 150 µm in a week. Crystals were harvested after 7 d and screened for diffraction. Crystals were reproducible and appear in wide range of PEG concentrations. They also appeared at different pH values and in different buffers. The best diffracting crystals were found in 1 M NaCl/0.05 M Tris (pH 8.0)/26% PEG 600. Cryoprotectant was not used to harvest the crystals.
Diffraction data were collected at the Lawrence Berkeley National Laboratory Advanced Light Source Beamline 8.3.1 in a −170 °C nitrogen cryostream at a wavelength of 1.115 Å.
Molecular Replacement and Refinement.
The structure was solved by molecular replacement using LacYG46WG262W PDB ID code 4OAA (20) as a search model. The highest resolution crystals diffracted to 3.3 Å. Data processing was performed either using XDS (52) or HKL2000 (53). Phasing by molecular replacement was performed using Phaser from the PHENIX suite of programs (54). COOT was used for model building (55), and refinement was accomplished by using COOT, PHENIX, and CCP4 (56). The MolProbity (57) server and PHENIX was used for structural validation and improvement.
Restrained refinement was performed with NCS, and stereochemistry restraint for all of the refinement, at a later-stage crystallographic refinement server developed by Haddadian et al. (58) (godzilla.uchicago.edu), was used for further refinement.
Refinement proceeded with manual rebuilding in loop region 98–111 and 33–46 in COOT with the assistance of σA-weighted 2Fo-Fc and Fo-Fc maps by maximum-likelihood–based energy minimization and isotropic B-factor refinement in PHENIX. This model and a 2Fo-Fc map were then submitted to a crystallographic refinement server at godzilla.uchicago.edu. The resulting model had improved Rwork and Rfree to 0.2538 and 0.2775, respectively. Final refinement was done in PHENIX, refining isotropic B-factor, coordinates, and occupancies with NCS and stereochemistry restraint. The final round improved stereochemistry and geometry of the structure, with 95% of residues in the favored region.
Acknowledgments
We thank Meseret Tessema (University of California, San Francisco) for cell growth; Joseph D. O’Connell III (University of California, San Francisco) for help in initial crystal trials; Jun-yong Choe (Rosalind Franklin University of Medicine and Science) for screening crystals for diffraction at the Advanced Photon Source, Chicago; Dr. James Holton and George Meigs for their help in X-ray data collection at Beamline 8.3.1 of the Lawrence Berkeley National Laboratory Advanced Light Source. We are grateful to University of California Office of the President, Multicampus Research Programs and Initiatives Grant MR‐15‐328599 and the Program for Breakthrough Biomedical Research, which is partially funded by the Sandler Foundation, for support for Beamline 8.3.1. This work was supported by National Institutes of Health Grant DK069463 and National Science Foundation Grants MCB-1129551 (to H.R.K.) and R37GM024485 (to R.M.S.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4ZYR).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1509854112/-/DCSupplemental.
References
- 1.Sandermann H., Jr β-D-Galactoside transport in Escherichia coli: Substrate recognition. Eur J Biochem. 1977;80(2):507–515. doi: 10.1111/j.1432-1033.1977.tb11906.x. [DOI] [PubMed] [Google Scholar]
- 2.Rudnick G, Schildiner S, Kaback HR. Equilibrium between two forms of the lac carrier protein in energized and nonenergized membrane vesicles from Escherichia coli. Biochemistry. 1976;15(23):5126–5131. doi: 10.1021/bi00668a028. [DOI] [PubMed] [Google Scholar]
- 3.Schuldiner S, Kerwar GK, Kaback HR, Weil R. Energy-dependent binding of dansylgalactosides to the beta-galactoside carrier protein. J Biol Chem. 1975;250(4):1361–1370. [PubMed] [Google Scholar]
- 4.Schuldiner S, Weil R, Kaback HR. Energy-dependent binding of dansylgalactoside to the lac carrier protein: Direct binding measurements. Proc Natl Acad Sci USA. 1976;73(1):109–112. doi: 10.1073/pnas.73.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sahin-Tóth M, Akhoon KM, Runner J, Kaback HR. Ligand recognition by the lactose permease of Escherichia coli: Specificity and affinity are defined by distinct structural elements of galactopyranosides. Biochemistry. 2000;39(17):5097–5103. doi: 10.1021/bi0000263. [DOI] [PubMed] [Google Scholar]
- 6.Sahin-Tóth M, Lawrence MC, Nishio T, Kaback HR. The C-4 hydroxyl group of galactopyranosides is the major determinant for ligand recognition by the lactose permease of Escherichia coli. Biochemistry. 2001;40(43):13015–13019. doi: 10.1021/bi011233l. [DOI] [PubMed] [Google Scholar]
- 7.Sahin-Tóth M, Gunawan P, Lawrence MC, Toyokuni T, Kaback HR. Binding of hydrophobic D-galactopyranosides to the lactose permease of Escherichia coli. Biochemistry. 2002;41(43):13039–13045. doi: 10.1021/bi0203076. [DOI] [PubMed] [Google Scholar]
- 8.Guan L, Kaback HR. Lessons from lactose permease. Annu Rev Biophys Biomol Struct. 2006;35:67–91. doi: 10.1146/annurev.biophys.35.040405.102005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Madej MG, Kaback HR. 2014. The life and times of Lac permease: Crystals ain’t everything, but they certainly do help. Membrane Transport Mechanism: 3D Structure and Beyond, Springer Series in Biophysics: Transporters, eds Ziegler C, Kraemer R (Springer, Heidelberg), Vol 17, pp 121–158.
- 10.Kaback HR. A chemiosmotic mechanism of symport. Proc Natl Acad Sci USA. 2015;112(5):1259–1264. doi: 10.1073/pnas.1419325112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaiko O, Bazzone A, Fendler K, Kaback HR. Electrophysiological characterization of uncoupled mutants of LacY. Biochemistry. 2013;52(46):8261–8266. doi: 10.1021/bi4013269. [DOI] [PubMed] [Google Scholar]
- 12.Viitanen P, Garcia ML, Foster DL, Kaczorowski GJ, Kaback HR. Mechanism of lactose translocation in proteoliposomes reconstituted with lac carrier protein purified from Escherichia coli. 2. Deuterium solvent isotope effects. Biochemistry. 1983;22(10):2531–2536. doi: 10.1021/bi00279a034. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Celma JJ, Smirnova IN, Kaback HR, Fendler K. Electrophysiological characterization of LacY. Proc Natl Acad Sci USA. 2009;106(18):7373–7378. doi: 10.1073/pnas.0902471106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia-Celma JJ, Ploch J, Smirnova I, Kaback HR, Fendler K. Delineating electrogenic reactions during lactose/H+ symport. Biochemistry. 2010;49(29):6115–6121. doi: 10.1021/bi100492p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smirnova I, Kasho V, Sugihara J, Kaback HR. Opening the periplasmic cavity in lactose permease is the limiting step for sugar binding. Proc Natl Acad Sci USA. 2011;108(37):15147–15151. doi: 10.1073/pnas.1112157108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abramson J, et al. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301(5633):610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 17.Mirza O, Guan L, Verner G, Iwata S, Kaback HR. Structural evidence for induced fit and a mechanism for sugar/H+ symport in LacY. EMBO J. 2006;25(6):1177–1183. doi: 10.1038/sj.emboj.7601028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaptal V, et al. Crystal structure of lactose permease in complex with an affinity inactivator yields unique insight into sugar recognition. Proc Natl Acad Sci USA. 2011;108(23):9361–9366. doi: 10.1073/pnas.1105687108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guan L, Mirza O, Verner G, Iwata S, Kaback HR. Structural determination of wild-type lactose permease. Proc Natl Acad Sci USA. 2007;104(39):15294–15298. doi: 10.1073/pnas.0707688104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar H, et al. Structure of sugar-bound LacY. Proc Natl Acad Sci USA. 2014;111(5):1784–1788. doi: 10.1073/pnas.1324141111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frillingos S, Sahin-Tóth M, Wu J, Kaback HR. Cys-scanning mutagenesis: A novel approach to structure function relationships in polytopic membrane proteins. FASEB J. 1998;12(13):1281–1299. doi: 10.1096/fasebj.12.13.1281. [DOI] [PubMed] [Google Scholar]
- 22.Kaback HR, Sahin-Tóth M, Weinglass AB. The kamikaze approach to membrane transport. Nat Rev Mol Cell Biol. 2001;2(8):610–620. doi: 10.1038/35085077. [DOI] [PubMed] [Google Scholar]
- 23.Jiang X, Villafuerte MK, Andersson M, White SH, Kaback HR. Galactoside-binding site in LacY. Biochemistry. 2014;53(9):1536–1543. doi: 10.1021/bi401716z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guan L, Hu Y, Kaback HR. Aromatic stacking in the sugar binding site of the lactose permease. Biochemistry. 2003;42(6):1377–1382. doi: 10.1021/bi027152m. [DOI] [PubMed] [Google Scholar]
- 25.Vázquez-Ibar JL, Guan L, Svrakic M, Kaback HR. Exploiting luminescence spectroscopy to elucidate the interaction between sugar and a tryptophan residue in the lactose permease of Escherichia coli. Proc Natl Acad Sci USA. 2003;100(22):12706–12711. doi: 10.1073/pnas.1835645100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ujwal ML, Sahin-Tóth M, Persson B, Kaback HR. Role of glutamate-269 in the lactose permease of Escherichia coli. Mol Membr Biol. 1994;11(1):9–16. doi: 10.3109/09687689409161024. [DOI] [PubMed] [Google Scholar]
- 27.Frillingos S, Gonzalez A, Kaback HR. Cysteine-scanning mutagenesis of helix IV and the adjoining loops in the lactose permease of Escherichia coli: Glu126 and Arg144 are essential. off. Biochemistry. 1997;36(47):14284–14290. doi: 10.1021/bi972314d. [DOI] [PubMed] [Google Scholar]
- 28.Sahin-Tóth M, et al. Characterization of Glu126 and Arg144, two residues that are indispensable for substrate binding in the lactose permease of Escherichia coli. Biochemistry. 1999;38(2):813–819. doi: 10.1021/bi982200h. [DOI] [PubMed] [Google Scholar]
- 29.Smirnova I, Kasho V, Sugihara J, Choe JY, Kaback HR. Residues in the H+ translocation site define the pKa for sugar binding to LacY. Biochemistry. 2009;48(37):8852–8860. doi: 10.1021/bi9011918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Padan E, Sarkar HK, Viitanen PV, Poonian MS, Kaback HR. Site-specific mutagenesis of histidine residues in the lac permease of Escherichia coli. Proc Natl Acad Sci USA. 1985;82(20):6765–6768. doi: 10.1073/pnas.82.20.6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Püttner IB, Sarkar HK, Poonian MS, Kaback HR. lac permease of Escherichia coli: Histidine-205 and histidine-322 play different roles in lactose/H+ symport. Biochemistry. 1986;25(16):4483–4485. doi: 10.1021/bi00364a003. [DOI] [PubMed] [Google Scholar]
- 32.Püttner IB, Kaback HR. lac permease of Escherichia coli containing a single histidine residue is fully functional. Proc Natl Acad Sci USA. 1988;85(5):1467–1471. doi: 10.1073/pnas.85.5.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Püttner IB, Sarkar HK, Padan E, Lolkema JS, Kaback HR. Characterization of site-directed mutants in the lac permease of Escherichia coli. 1. Replacement of histidine residues. Biochemistry. 1989;28(6):2525–2533. doi: 10.1021/bi00432a027. [DOI] [PubMed] [Google Scholar]
- 34.Roepe PD, Kaback HR. Site-directed mutagenesis of tyrosine residues in the lac permease of Escherichia coli. Biochemistry. 1989;28(14):6127–6132. doi: 10.1021/bi00440a060. [DOI] [PubMed] [Google Scholar]
- 35.Vadyvaloo V, Smirnova IN, Kasho VN, Kaback HR. Conservation of residues involved in sugar/H(+) symport by the sucrose permease of Escherichia coli relative to lactose permease. J Mol Biol. 2006;358(4):1051–1059. doi: 10.1016/j.jmb.2006.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guan L, Sahin-Toth M, Kaback HR. Changing the lactose permease of Escherichia coli into a galactose-specific symporter. Proc Natl Acad Sci USA. 2002;99(10):6613–6618. doi: 10.1073/pnas.102178299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smirnova IN, Kasho VN, Kaback HR. Direct sugar binding to LacY measured by resonance energy transfer. Biochemistry. 2006;45(51):15279–15287. doi: 10.1021/bi061632m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sahin-Tóth M, Kaback HR. Functional conservation in the putative substrate binding site of the sucrose permease from Escherichia coli. Biochemistry. 2000;39(20):6170–6175. doi: 10.1021/bi000125g. [DOI] [PubMed] [Google Scholar]
- 39.Nie Y, Smirnova I, Kasho V, Kaback HR. Energetics of ligand-induced conformational flexibility in the lactose permease of Escherichia coli. J Biol Chem. 2006;281(47):35779–35784. doi: 10.1074/jbc.M607232200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frillingos S, Kaback HR. Probing the conformation of the lactose permease of Escherichia coli by in situ site-directed sulfhydryl modification. Biochemistry. 1996;35(13):3950–3956. doi: 10.1021/bi952601m. [DOI] [PubMed] [Google Scholar]
- 41.Venkatesan P, Kaback HR. The substrate-binding site in the lactose permease of Escherichia coli. Proc Natl Acad Sci USA. 1998;95(17):9802–9807. doi: 10.1073/pnas.95.17.9802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robertson DE, Kaczorowski GJ, Garcia ML, Kaback HR. Active transport in membrane vesicles from Escherichia coli: The electrochemical proton gradient alters the distribution of the lac carrier between two different kinetic states. Biochemistry. 1980;19(25):5692–5702. doi: 10.1021/bi00566a005. [DOI] [PubMed] [Google Scholar]
- 43.Smirnova I, et al. Sugar binding induces an outward facing conformation of LacY. Proc Natl Acad Sci USA. 2007;104(42):16504–16509. doi: 10.1073/pnas.0708258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Majumdar DS, et al. Single-molecule FRET reveals sugar-induced conformational dynamics in LacY. Proc Natl Acad Sci USA. 2007;104(31):12640–12645. doi: 10.1073/pnas.0700969104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carrasco N, Antes LM, Poonian MS, Kaback HR. lac permease of Escherichia coli: Histidine-322 and glutamic acid-325 may be components of a charge-relay system. Biochemistry. 1986;25(16):4486–4488. doi: 10.1021/bi00364a004. [DOI] [PubMed] [Google Scholar]
- 46.Carrasco N, et al. Characterization of site-directed mutants in the lac permease of Escherichia coli. 2. Glutamate-325 replacements. Biochemistry. 1989;28(6):2533–2539. doi: 10.1021/bi00432a028. [DOI] [PubMed] [Google Scholar]
- 47.Sahin-Toth M, Kaback HR. Arg-302 facilitates deprotonation of Glu-325 in the transport mechanism of the lactose permease from Escherichiacoli. Proc Natl Acad Sci USA. 2001;98(11):6068–6073. doi: 10.1073/pnas.111139698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schuldiner S, Kaback HR. Fluorescent galactosides as probes for the lac carrier protein. Biochim Biophys Acta. 1977;472(3-4):399–418. doi: 10.1016/0304-4157(77)90004-1. [DOI] [PubMed] [Google Scholar]
- 49.Sahin-Tóth M, Persson B, Schwieger J, Cohan P, Kaback HR. Cysteine scanning mutagenesis of the N-terminal 32 amino acid residues in the lactose permease of Escherichia coli. Protein Sci. 1994;3(2):240–247. doi: 10.1002/pro.5560030208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smirnova IN, Kasho V, Kaback HR. Protonation and sugar binding to LacY. Proc Natl Acad Sci USA. 2008;105(26):8896–8901. doi: 10.1073/pnas.0803577105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smirnova I, Kasho V, Sugihara J, Vázquez-Ibar JL, Kaback HR. Role of protons in sugar binding to LacY. Proc Natl Acad Sci USA. 2012;109(42):16835–16840. doi: 10.1073/pnas.1214890109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography, Part A, Methods in Enzymology, eds Abelson JN, Simon MI, Carter CW, Jr, Sweet RM (Academic, New York), Vol 276, pp 307–326.
- 54.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haddadian EJ, et al. Automated real-space refinement of protein structures using a realistic backbone move set. Biophys J. 2011;101(4):899–909. doi: 10.1016/j.bpj.2011.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336(6084):1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]