Abstract
Studies of gene–environment (G × E) interactions require effective characterization of all environmental exposures from conception to death, termed the exposome. The exposome includes environmental exposures that impact health. Improved metabolic profiling methods are needed to characterize these exposures for use in personalized medicine. In the present study, we compared the analytic capability of dual chromatography-Fourier-transform mass spectrometry (DC-FTMS) to previously used liquid chromatography-FTMS (LC-FTMS) analysis for high-throughput, top-down metabolic profiling. For DC-FTMS, we combined data from sequential LC-FTMS analyses using reverse phase (C18) chromatography and anion exchange (AE) chromatography. Each analysis was performed with electrospray ionization in the positive ion mode and detection from m/z 85 to 850. Run time for each column was 10 min with gradient elution; 10 µl extracts of plasma from humans and common marmosets were used for analysis. In comparison to analysis with the AE column alone, addition of the second LC-FTMS analysis with the C18 column increased m/z feature detection by 23–36%, yielding a total number of features up to 7,000 for individual samples. Approximately 50% of the m/z matched to known chemicals in metabolomic databases, and 23% of the m/z were common to analyses on both columns. Database matches included insecticides, herbicides, flame retardants, and plasticizers. Modularity clustering algorithms applied to MS-data showed the ability to detection clusters and ion interactions. DC-FTMS thus provides improved capability for high-performance metabolic profiling of the exposome and development of personalized medicine.
Keywords: Metabolomics, LC/MS, Anion exchange, Reverse phase, Exposome, Personalized medicine, Predictive health, FT-ICR, Plasma
1 Introduction
The metabolome includes the complete set of low-molecular-weight chemicals in biological samples (Ellis et al. 2007) such as metabolic intermediates, hormones and other signaling molecules, and secondary metabolites. Study of the metabolome is facilitated by cumulative databases containing assemblies of knowledge of thousands of chemicals of human metabolism (Kind et al. 2009). Unfortunately, a large extent of the metabolome is still unidentified, especially many of the chemicals in the diet, as well as persistent organic pollutants and chemicals derived from commercial products, household products and health behaviors. Links between such exposures and human diseases have already been identified (Howell and Mangum 2010; Peng et al. 2010; Lewis et al. 2007; Ziech et al. 2010; Gilmour et al. 2006), and systematic collection of information on these exposures by improved metabolic profiling could be instrumental to characterization of the summation of these exposures, termed the “exposome” (Wild 2005) and use in personalized medicine and predictive health (Loscalzo et al. 2007; Brigham 2010; Voit 2009). Therefore, the aim of this study was to increase overall metabolic detection in order to capture a larger extent of the exposome. Associations of exposures to disease and interactions with genetic, epigenetic and other risk factors will facilitate personalized medicine.
Metabolite detection varies greatly depending on the biological sample, extraction method, separation technique and mass spectrometer used for analysis. Plasma is frequently used for biological sampling because it is relatively accessible and includes metabolites indicative of both short-term (Haouala et al. 2009; Hodel et al. 2009) and long-term environmental exposures (Aronson et al. 2010; Wang et al. 2007; Sandanger et al. 2006). However, only about one-half of the metabolites detected in plasma match chemicals included in available databases (Evans et al. 2009; Takahashi et al. 2008); whereas, a large proportion of these unknown metabolites may be related to the exposome. Much of the best human exposure information is derived from measures of hydrophilic products in urine; however, many environmental chemicals are hydrophobic. Hydrophobic chemicals are not readily excreted into urine, so there is a possibility that the body burden for hydrophobic chemicals is better measured in plasma.
To enhance metabolite detection, many laboratories have begun using combination of separation techniques. For example, it is common to analyze samples through both liquid chromatography (LC) and gas chromatography (GC) technologies, which results in a chemically diverse set of metabolites totaling between 300 and 1,500 (Lawton et al. 2008; Evans et al. 2009; Sreekumar et al. 2009). Overall, LC/MS has become the standard approach for metabolomics analyses due its ability to separate, ionize and detect the widest range of chemicals (Buscher et al. 2009; Nordstrom et al. 2008). Species detection and diversity is also influenced by column selection. Untargeted metabolomics studies have used reverse phase C18 columns (Evans et al. 2009; Crews et al. 2009), RP with polar end-caps (Lawton et al. 2008; Evans et al. 2009), and hydrophilic interaction chromatography (HILIC) (Buscher et al. 2009) to separate a range of chemicals and increase chemical detection. Further improvement of LC with ultra performance LC (UPLC) can provide high-resolution chromatograms while consuming fewer samples in a faster analysis time (Evans et al. 2009). Column switching valves allow convenient sequential analysis of replicate aliquots of samples following separation on different columns. We describe the latter as “dual chromatography” to allow distinction from the common tandem or 2-dimensional chromatography (2D-LC, or LC2), that is used in peptide analyses of proteomics.
High-resolution mass spectrometers, such as FT-ICR, time-of-flight (TOF), and Orbitrap instruments (Marshall and Hendrickson 2008); can decrease the demand for extensive chromatographic separation. Crews et al. report detection of over 3,600 and 4,800 metabolic features in biological replicates of human plasma and CSF, respectively, using LC-TOF/MS (Crews et al. 2009). The TOF instruments can scan rapidly but have less resolving power than the FT-ICR and Orbitrap instruments (Marshall and Hendrickson 2008) and retain greater demand for chromatographic separation. Resolving power of the FT-ICR and Orbitrap is increased with a longer scan time, but this decreases the number of spectra over a chromatographic peak. Both of these instruments detect thousands of chemicals. The currently available Orbitrap requires twice the time to achieve the resolution of the FT-ICR. A new Orbitrap has profiling capabilities comparable to the FT-ICR (Olsen et al. 2009).
Metabolomics is often divided between targeted and non-targeted analysis, although a hybrid analysis is also possible. In targeted analyses, known chemicals are quantified relative to internal or external standards. For non-targeted analysis, unidentified ions are identified as m/z features in which the retention time (RT) and m/z are used as identifiers in the absence of knowledge of chemical identity. Even without chemical identity, m/z features can be associated with disease or other characteristics of biological interest. An advantage of high-resolution and high mass accuracy instruments is that elemental composition often can be predicted from the accurate m/z values. This information can be used along with MS/MS and other approaches, such as relative abundance of isotopic peaks, to aid in identification of the m/z features (Miura et al. 2010). Prediction of elemental composition from high-resolution mass spectrometry can facilitate identification of exposures including halogenated hydrocarbons and sulfonated chemicals.
We recently described a high-performance metabolic profiling method using LC-FTMS with anion exchange (AE) chromatography and electrospray ionization in the positive mode (Johnson et al. 2010). The method was designed to provide a practical approach for metabolic profiling in personalized medicine. The report established detection of 2,124 m/z in a 10-min analysis of a 10-µl extract of plasma. The method used a one-step sample preparation for deproteination, a C18-precolumn to minimize ion suppression and automated analysis for high throughput. In the present study, we have used an improved version of apLCMS (http://www.sph.emory.edu/apLCMS), incorporated internal standards into the extraction, and evaluated the extent of improvement provided by combining the AE analysis with a comparable analysis using a C18 column. For this, we used a switching valve to alternately collect data from AE and C18; this allowed the entire process to be performed in an automated DC-FTMS format. We hypothesized that addition of the C18 column would improve chemical coverage to detect more metabolic features (from the 2,124 reported previously) thereby enhancing the ability to study the exposome. We applied the DC-FTMS to a population of free-living humans and captive marmosets. The results show that with an improved version of apLCMS software (Yu et al. 2009), measurement provides up to 7,000 metabolites in human plasma in 20 min. Further, a number of environmental chemicals were detected in human and marmoset plasma, including pesticides, flame retardants, and plasticizers, indicating that this approach is suitable for study of the exposome.
2 Materials and methods
2.1 Materials
Acetonitrile (HPLC grade), formic acid (puriss. p.a. 98%), water (HPLC grade) and caffeine were obtained from Sigma–Aldrich (St. Louis) and the reverse phase test mix (Cat#: 47641-U) was from Supelco Analytical (Bellefonte, PA). Trimethyl-[13C3]-caffeine (Cat#: CLM-514-0) and [15N]-l-tyrosine (Cat#: NLM-590-0) were obtained from Cambridge Isotope Laboratories, Inc (Andover, PA).
2.2 Samples
For the purpose of method development and characterization in the present study, plasma samples were derived from four sources (Supplementary S1). Samples from common marmosets (Callithrix jacchus) and humans were used to obtain information on the coverage of the exposome in captive animals and free-living humans. The marmosets were housed at the New England Primate Research Center and maintained in accordance with the Guide for the Care and Use of Laboratory Animals (Institute of Laboratory Animal Resources 1996). The facility is AAALAC-accredited, and all work was approved by Harvard Medical School’s Standing Committee on Animals. Animals received commercial marmoset chow (New World Primate Chow 8791, Harlan Teklad, Indianapolis, IN) supplemented daily with a combination of fresh fruits, vegetables, seeds, eggs, and/or mealworms. Water was provided ad libitum in polycarbonate water bottles, with fresh water provided daily. Cage pans were cleaned three times each week, and cages were sanitized every other week. Each animal received environmental enrichment consisting of food enrichment, toys, nest boxes, and music. Room temperature was maintained at 25.6 ± 2.2°C, with a relative humidity of 30–70%. After sedation with 0.2 ml of ketamine IM (trade name: Ketaset®, Fort Dodge Animal Health; Fort Dodge, IA) during the marmosets’ quarterly physical examinations, body composition was evaluated using EchoMRI and blood was sampled. Plasma was obtained in EDTA and stored at −80°C (Johnson et al. 2008). Due to the small size of the common marmoset (350–400 g) and limited blood volume that may be obtained at each phlebotomy (3 ml), EDTA plasma is collected to allow greater flexibility in the types of assays that can be run concurrently. For example, FACS analysis and intracellular cytokine staining are important for study of immunosenescence and these assays have been optimized for EDTA samples.
Human plasma consisted of pooled human reference plasma from two sources and samples from a previously published study (Jones et al. 2011), reviewed and approved by the Emory Institutional Review Board. In the latter study, samples, collected with informed consent for metabolic profiling, were in lithium heparin and stored at −80°C. Four samples were randomly selected for analysis; characteristics of individuals are provided in Supplementary S1. We find that inclusion of pooled human reference samples with internal standards provide means to evaluate variation in the chromatography and instrument response characteristics. One reference sample consisted of two separate aliquots of pooled plasma donated from the National Institute of Standards and Technology (NIST). This was pooled plasma from an equal number of healthy men and women aged 40–50 years. The second reference sample consisted of two separate aliquots of pooled plasma from Equitech-Bio, Inc (Kerrville, TX). This consisted of plasma pooled from an unknown number of males and females without demographic information. For the present study, these pooled samples were useful to obtain information on the number of chemicals detected in plasma by the DC-FTMS approach.
An internal standard mix consisting of 14 stable isotopic chemicals that cover a broad range of chemical properties represented in small molecules was added to samples prior to analysis by DC-FTMS. These chemicals included [13C6]-d-glucose, [15N]-indole, [2-15N]-l-lysine dihydrochloride, [13C5]-l-glutamic acid, [13C7]-benzoic acid, [3,4-13C2]-cholesterol, [15N]-l-tyrosine, [trimethyl-13C3]-caffeine, [15N2]-uracil, [3,3-13C2]-cystine, [1,2-13C2]-palmitic acid, [15N,13C5]-l-methionine, [15N]-choline chloride, and 2’-deoxyguanosine-15N2,13C10-5’-monophosphate. Only two of these chemicals, [15N]-l-tyrosine and [trimethyl-13C3]-caffeine, were detected on AE and C18 columns in all human and marmoset analyses.
2.3 Chromatography
Aliquots (50 µl) were treated with acetonitrile (2:1, v/v), spiked with 2.5 µl internal standard mix and centrifuged at 14,000 × g for 5 min at 4°C remove protein (Want et al. 2006); supernatants were then loaded onto a Shimadzu® (Sil-20AC Prominence) autosampler and maintained at 4°C until injection. Previous validation experiments showed minimal changes in metabolic patterns over a 24-h period due to maintenance in the autosampler (Johnson et al. 2010). Injection volume was 10 µl for each run of the DC-FTMS analysis. Analyses were performed alternately between AE and C18 by use of a Switchos (LC Packings) control valve. This approach allowed one column to undergo a wash cycle while separation was performed on the other column. For the off-line wash periods, 2% (v/v) formic acid in acetonitrile was used at 0.5 ml/min. Columns were equilibrated to initial conditions for 2 min prior to the next sample injection.
The AE analysis was essentially that of Johnson, et al. (Johnson et al. 2010). Briefly, analyte separation was performed by AE (Hamilton PRP-X110S, 2.1 × 10 cm) with a formic acid gradient run at 0.35 ml/min. A short, end-capped C18 pre-column (Higgins Analytical, Targa guard) was included for desalting and improved separation. Solution A was 2% (v/v) formic acid in water. Equilibration and the first 2-min period consisted of 5% A, 45% water and 50% acetonitrile. This was followed by a 5-min linear gradient to 50%A, 50% acetonitrile. The final 3-min period was maintained at 50% A, 50% acetonitrile prior to the wash cycle.
Reverse phase analysis was performed with a C18 column (Higgins Analytical, Targa, 2.1 × 10 cm) and an acetonitrile gradient. The C18 chromatography used a flow rate of 0.35 ml/min for the first 6 min and then changed to 0.5 ml/min for the remaining 4 min. The first 2-min period consisted of 5% A, 60% water, 35% acetonitrile, followed by a 4-min linear gradient to 5% A, 0% water, 95% acetonitrile. The final 4-min period was maintained at 5% A, 95% acetonitrile.
2.4 Mass spectrometry
A Thermo LTQ-FT mass spectrometer (Thermo Fisher, San Diego, CA) was set to collect data from m/z 85 to 850. Optimizations of the operating conditions were detailed previously (Johnson et al. 2010). In brief, a spray voltage of 6 kV, sheath gas of 60 (arbitrary units), capillary temperature of 275°C, capillary voltage of 44 V and tube lens of 120 V were used. Ion transfer optics were optimized automatically. Maximum injection time was 500 ms, and the maximum number of ions collected for each scan was 3 × 106. The number of ions per scan exceeds that recommended by the manufacturer and represents an operational condition that improved detection of m/z features with minimal deterioration of mass resolution and accuracy. A wide range scan was used for the FT-ICR with mass resolution of 50,000.
2.5 Data collection and processing
Data were collected continuously over the 10-min chromatographic separation and stored as raw files. These files were converted using Xcalibur file converter software (Thermo Fisher, San Diego, CA) to cdf files for further data processing. An adaptive processing software package (apLCMS, http://www.sph.emory.edu/apLCMS) designed for use with LC-FTMS data was used for peak extraction and quantification of ion intensities (Yu et al. 2009). This software provided m/z feature tables containing m/z values, retention time, and integrated ion intensity for each m/z feature, obtained through five major processing steps: (1) noise filter, (2) peak identification, (3) retention time correction, (4) m/z peak alignment across multiple spectra, and (5) re-analysis to capture peaks originally missed because of weak signal relative to the signal to noise filter. Features were matched between the AE and C18 columns using m/z within a 10 ppm window.
2.6 Bioinformatics
Principal component analysis was performed using Pirouette version 4.0 (InfoMetrix), and modulated modularity clustering (MMC) was performed using the publicly available version via the web (http://mmc.gnets.ncsu.edu/) and described in detail in (Stone and Ayroles 2009). The output from MMC lists the module numbers with degree of clustering for each m/z feature and average correlation for each module in descending order. The heat maps generated by MMC show modules of clustered m/z features along the diagonal (upper left to lower right), where strongly associated and positively correlated m/z features are sorted together and shown in dark red and strongly-associated but negatively-correlated m/z features are visualized in blue. Although MMC was originally developed to detect associations between genes and then mapped to genomic pathways, this technique is also useful in metabolic pathway analysis based on the different strengths of associations.
3 Results and discussion
3.1 Characterization of metabolic profiles of marmoset plasma by DC-FTMS
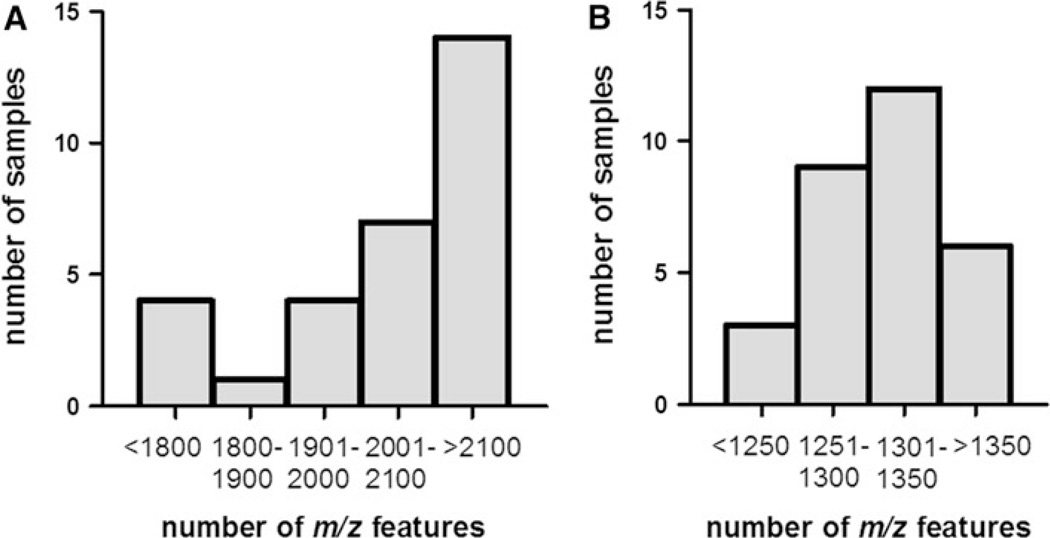
The number of m/z features detected by LC-FTMS was remarkably different when using the AE chromatography compared to the C18 chromatography. For analysis of marmoset samples, the AE detected a total of 2,594 m/z features, similar to that previously reported for this method (Johnson et al. 2010). In contrast, the C18 method detected a total of 1,558 m/z features. In both cases, there was variability in the number of m/z detected among samples (Fig. 1). In the AE analysis, the median metabolic profile consisted of between 2,100 and 2,200 m/z features (83% of AE total); whereas, in the C18 analysis, the median profile consisted of between 1,300 and 1,350 m/z features (85% of C18 total). Notably, the samples with the fewest number of m/z features in both columns were from those oldest marmosets (>3.6 years), which coincides with our previous finding that the abundance of many m/z features declines with age (Soltow et al. 2010).
Fig. 1.
Histogram showing distribution of features detected in 30 young, healthy marmoset samples. a Anion exchange column, median 2100–2200 features; b reverse phase column, median 1300–1350 features
3.2 Unique and common features of chromatographic separations
The internal standards had mean retention times of 2.63 ± 0.39 min (C18) and 1.18 ± 0.04 min (AE) for [trimethyl-13C3]-caffeine and 1.13 ± 0.04 min (C18) and 0.97 ± 0.06 min (AE) for [15N]-l-tyrosine. The between-sample coefficient of variation (CV) of the measured ion intensities for [trimethyl-13C3]-caffeine was 22% and for [15N]-l-tyrosine was 29%. These CV are greater than the within-sample CV (Johnson et al. 2010), indicating variation in addition of internal standards. Thus, inclusion of internal standards in sample preparation provides ability to evaluate reproducibility of analysis.
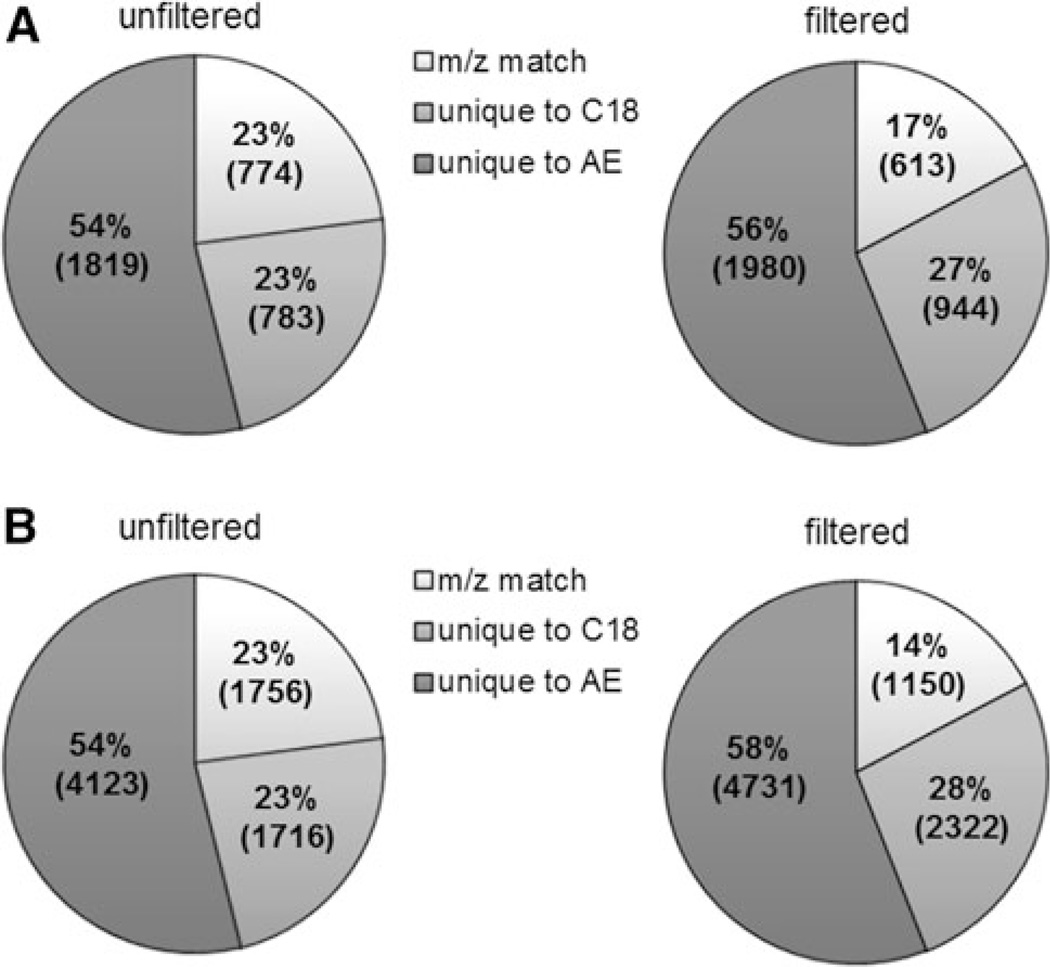
To determine the contribution of the C18 analysis to the overall profile of metabolites detected, m/z were matched between the AE and C18 feature tables (Fig. 2a, unfiltered). A total of 3376 m/z were found in the overall marmoset profile, 23% of which were detected in both the AE and C18. The majority of the m/z features were unique to the AE analysis (54%). The C18 analysis added 783 m/z features that were undetected by the AE separation, i.e., approximately 23%.
Fig. 2.
a Percentage of chemicals detected by each column and by both columns in the marmoset samples; m/z match: the number of m/z that were detected by both chromatographic separations; unique to AE: only detected by the AE column; unique to C18: only detected by the C18 column. A total of 3,376 m/z were detected in the marmoset samples. The ion intensity ratios of the m/z detected by the AE to that of the C18 were calculated, and the ion intensities of stable isotopic chemical standards (13C3-caffeine and 15N-tyrosine) were found to vary up to 11-fold between the two columns. Of the 774 matched m/z in the marmoset samples, 613 m/z were within this 11-fold range. After filtering the 161 matched m/z varying in ion intensity >11-fold, the “filtered” pie chart demonstrates the new distribution of chemicals. b Percentage of chemicals detected by each column and by both columns in the human samples. A total of 7597 m/z were detected. The ion intensity ratios [AE/C18] of stable isotopic chemical standards were found to vary up to 5.2-fold between the two columns. Of the 1756 matched m/z in the human samples, 1150 m/z were within this 5.2-fold range. After filtering the 606 matched m/z varying in ion intensity over 5.2-fold, the “filtered” pie chart demonstrates the new distribution of chemicals
Due to the presence of a large number of isomers and chemicals with the same elemental composition, an m/z detected by AE analysis may not be the same chemical as the same m/z detected by C18 analysis, and this would underestimate the additional metabolites detected. Consequently, we used the ion intensity for each m/z on the AE compared to the respective ion intensity on the C18, as a basis to estimate how many additional chemicals could be present but not discriminated solely based upon m/z. To provide cutoff values, we used the ratio of ion intensities for internal standards detected by both AE and C18 analyses. Of the 14 internal standards added to the samples, only [15N]-l-tyrosine and [trimethyl-13C3]-caffeine were detected on both columns in all marmoset samples; thus, these two chemicals were used to generate cutoff values. While the average [AE]/[C18] ion intensity ratio for 15N-tyrosine was 1.07 ± 0.09 (1.07-to-1 ratio), the ratio for 13C3-caffeine was 0.09 ± 0.02 (1-to-11 ratio). Consequently, we used the more conservative value for 13C3-caffeine, i.e., an 11-fold ratio, as the cutoff for whether identical m/z values for the columns represented the same chemical. With this criterion, 161 of the m/z matches with greater than 11-fold difference in intensities were considered to be additional chemicals. A graphical representation of the reallocated m/z features is demonstrated in Fig. 2a-filtered, and the new totals show that the C18 analysis adds about 36% more metabolic coverage to the marmoset profiles than with the AE analysis alone.
3.3 Characteristics of human plasma analysis by DC-FTMS
Similar data were obtained for analysis of human samples. A total of 7,597 m/z were found in the overall profile, 23% of which were found in both AE and C18 (Fig. 2b, unfiltered). The majority of the m/z features were unique to the AE analysis (54%). The C18 analysis added 1,716 m/z features that were undetected by the AE separation (23%). Using the same stable isotopic standards to determine whether identical m/z for the columns represented the same chemical, a 5.2-fold ion intensity ratio was found for 15N-tyrosine and was used as the cutoff ratio. With this criterion, 606 of the m/z matches with greater than 5.2-fold difference in intensities were considered to be additional chemicals detected (Fig. 2b, filtered). New totals show that the C18 analysis adds 39% more metabolic coverage to the human profiles than with the AE analysis alone.
A number of factors could have affected the variation in the total number of detected features between the marmoset and human sample sets. Other uses of the instrument in the shared use facility could affect response characteristics and at least partially account for differences. Additionally, differences in the total number of features between marmoset and human samples may be due to (1) the use of EDTA plasma in marmosets versus heparin plasma in humans; (2) the use of DMSO as a solvent for the internal standards in the marmosets versus the use of water in the humans; and/or (3) the environmental exposures and dietary variations of captive marmosets being less than in free-living humans. A comparison of four human samples collected in heparin and EDTA from the same individuals showed a similar number of features (5,618 features using EDTA vs. 5,394 features with heparin), indicating that the anticoagulant can only account for a minor difference in the total features. A previous report showed that DMSO results in a significant reduction in gas phase ion formation during the electrospray ionization process due to its low vapor pressure and can decrease signal intensity by over 70% (Tjernberg et al. 2006). Our direct analysis of the effect of DMSO showed only a minor effect in feature detection compared to water (3,376 vs. 3,386 features, respectively). On the other hand, environmental exposures from the diet, cookware, plastic ware, clothing, pharmaceuticals, cosmetics, and other sources may contribute to a greater number of metabolites in human samples. Specific environmental chemicals detected in both species are discussed further in Sect. 3.4, and additional studies may be required to define this.
Comparison of the number of m/z detected for each column used a cutoff based on the ion intensity range of the internal standards. The ion intensity ratios of individual internal standards ranged up to 11-fold in the marmoset samples and 5.2-fold in the human samples. The reason for this difference is not clear but may be a consequence of differences in instrument response characteristics on the days of analysis or dependent upon the different solvents for internal standards, as mentioned above. Another alternative is that different column types may provide superior quantification for some chemicals. If the latter is true, then algorithms could be written to use data from analyses on different columns to improve reliability of identification and quantification of m/z features.
3.4 Matches to metabolomics databases
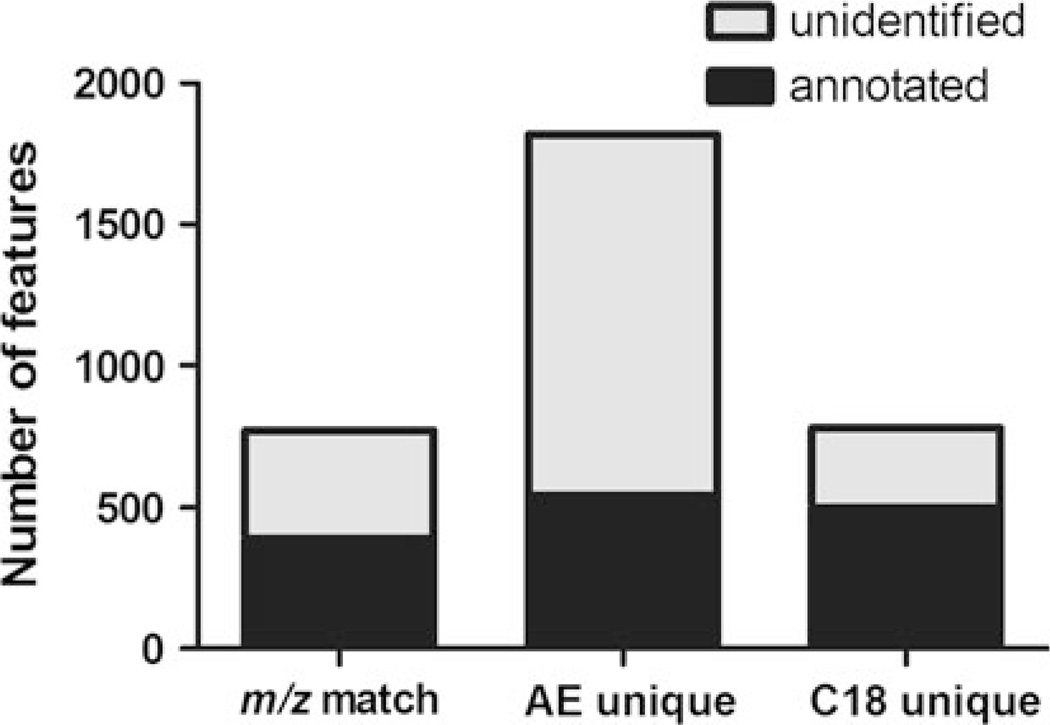
Many metabolomics-based studies rely on mapping of m/z to known chemicals and/or the identification of m/z features, so we sought to determine the number of m/z detected in the marmoset samples that correspond to known chemicals using two online metabolomics databases: Madison Metabolomics Consortium Database (MMCD) and the Metlin database (Fig. 3). Each database was searched for the [M + H]+ and [M + Na]+ species within ±5 ppm of the detected m/z. Only MMCD was used to search [13C] species because the Metlin database does not provide this capability. In Fig. 3, the totals for all annotated m/z are displayed, which includes the [M + H]+, [M + Na]+, and 13C[M + H]+ species. Only 3–5% of m/z matched to more than one chemical species. About half of the m/z detected by both chromatographic separations matched known chemicals; whereas, only 30% of features detected by the AE analysis were matched. Notably, over 60% of m/z detected by the C18 analysis matched known chemicals, but because fewer m/z were detected, the absolute numbers were similar; 522 versus 570 features, respectively. Reverse phase chromatography is widely used among laboratories involved in metabolomics database construction (Yanes et al. 2011; Brown et al. 2009); thus, more extensive characterization of metabolites separated by C18 has been reported. Because more than twice as many features were detected by AE, this may be a worthwhile addition to MS-based metabolomics platforms. More extensive use of AE analysis would further enhance the knowledge base for metabolomics research.
Fig. 3.
The number of m/z detected in the marmoset samples that were matched to a known metabolite in either Madison Metabolomics Consortium Database or Metlin database. Metabolites were considered a match if they were of a [M+H]+ species (i.e. no adducts); sodium adduct [M+Na]+; or 13carbon[M+H]+ species (i.e. no adducts) and within 5 ppm of the measured mass. Combined annotations from all species (i.e. [M+H], [M+Na], 13C[M+H]) are shown here for features detected by both columns (m/z match) and by each column individually (AE unique; C18 unique)
Identities of some m/z matches to metabolomics databases were confirmed using the internal standards and ion dissociation studies, including amino acids, and a relatively small number of phospholipids, nutrients, dietary compounds, intermediary metabolites, and environmental chemicals. However, the identification of the thousands of m/z detected was beyond the capability of the laboratory. Confirming the identity of the matches in the databases will require detailed verification. For some chemicals, e.g. amino acids, this fragmentation is straight forward; however, many chemicals have the same elemental composition and similar structures which makes detailed verification difficult. More critically, perhaps, half of the m/z do not match known chemicals in the metabolomics databases, and these could be very important as components of the exposome.
Database matches to environmental chemicals included pesticides, flame retardants, and plasticizers (Table 1). As expected, more exposure-related chemicals were detected in free-living humans than captive marmosets. Specifically, a higher number of plasticizers and pesticides were matched to m/z detected in human plasma. These include pesticides used on food crops and plasticizers that are used in consumer products, such as furnishings, cookware, food wrappers, nail polish, and polyvinyl chloride (PVC). Although we hypothesized that addition of the C18 column would enhance detection of hydrophobic chemicals related to exposures, the AE column actually detected a higher number of exposures in both human and marmoset plasma (Table 1).
Table 1.
Matches to environmental chemicals detected in marmoset (A) and human (B) plasma samples with DC-FTMS
| Environmental chemical | Detected m/z | Delta |
|---|---|---|
| A. Marmoset | ||
| Diethyl phthalate (plasticizer)ab | 223.0949 | 0.0015 |
| Diisopropyl phthalate (plasticizer)a | 251.1256 | 0.0021 |
| Dibutyl phthalate (plasticizer)ab | 279.1568 | 0.0023 |
| Butylbenzyl phthalate (plasticizer)ab | 313.1409 | 0.0026 |
| Triphenyl phosphate (flame retardant)a | 327.0762 | 0.0019 |
| Imazalil nitrateab | 359.0416 | 0.0017 |
| Di-n-heptyl phthalate (plasticizer)ab | 363.2497 | 0.0032 |
| Di(2-ethylhexyl) adipate (plasticizer)ab | 371.3126 | 0.0030 |
| Diisooctyl phthalate (plasticizer)ab | 391.2804 | 0.0039 |
| Macluraxanthone (insecticide)a | 395.1530 | −0.0041 |
| Diisononyl phthalate (plasticizer)ab | 419.3114 | 0.0042 |
| Diisodecyl phthalate (plasticizer)ab | 447.3431 | 0.0038 |
| Tetraethylene glycol (plasticizer)b | 195.1211 | 0.0016 |
| B. Human | ||
| Meta-tyrosine (herbicide)a | 182.0805 | 0.0007 |
| Carbendazim; mercarzole (carcinogen)a | 192.0741 | 0.0026 |
| N-butyl-benzenesulfonamide (plasticizer)ab | 214.0892 | 0.0004 |
| Diethyl phthalate (plasticizer)ab | 223.0937 | 0.0028 |
| Pirimicarb (insecticide)a | 239.1482 | 0.0021 |
| Diisopropyl phthalate (plasticizer)ab | 251.1270 | 0.0008 |
| Dibutyl phthalate (plasticizer)ab | 279.1574 | 0.0017 |
| Butylbenzyl phthalate (plasticizer)ab | 313.1417 | 0.0018 |
| Triphenyl phosphate (flame retardant)ab | 327.0766 | 0.0015 |
| Di-n-hexyl phthalate (plasticizer)ab | 335.2176 | 0.0041 |
| Chlorsulfuron (herbicide)a | 358.0372 | 0.0000 |
| Imazalil nitrateab | 359.0407 | 0.0027 |
| Di-n-heptyl phthalate (plasticizer)ab | 363.2503 | 0.0026 |
| Di(2-ethylhexyl) adipate (plasticizer)ab | 371.3134 | 0.0022 |
| Surfentrazone (herbicide)ab | 386.9885 | 0.0007 |
| Diisooctyl phthalate (plasticizer)ab | 391.2818 | 0.0025 |
| Macluraxanthone (insecticide)ab | 395.1526 | −0.0037 |
| Endosulfan (insecticide)a | 404.8213 | 0.0028 |
| Diisononyi phthalate (plasticizer)ab | 419.3134 | 0.0022 |
| Diisodecyl phthalate (plasticizer)ab | 447.3435 | 0.0034 |
Delta = theoretical m/z – detected m/z
indicates chemicals detected in the AE column,
indicates chemicals detected in the C18 column
The present data only compared two chromatography methods and gives no indication of other combinations that might provide better overall detection. In principle, a combination of multiple platforms is needed to effectively survey the chemical space occupied by environmental chemicals. Positive and negative electrospray ionization and atmospheric pressure chemical ionization (APCI) have different efficiencies for ionization of chemicals so that ionization methods can impact detection. In a platform using UPLC with a C18 column coupled to a LTQ-Orbitrap, negative ion mode analyses doubled the detection of features compared to positive ion mode for a range of biological specimens (Brown et al. 2009). Negative ion mode also increased the number of molecular formulas and annotations by up to 8.6%. This suggests that adding a negative ion mode analysis to the AE and C18 methods outlined in DC-FTMS would likely enhance chemical detection including those matching to known chemicals. Detection also can be enhanced by use of combination of cation exchange or HILIC (hydrophilic interaction chromatography) columns to reverse phase and anion exchange. With the combination illustrated in this report, adding C18 to the AE method in positive ion mode increased metabolic coverage by 36% and the number of annotated metabolites by 29%. If similar increases were observed with the other methods, greater than a 100% increase in feature detection may be possible by triplex or quadruplex chromatography with different separation and ionization combinations.
3.5 Modulated modularity clustering analysis
As indicated above, many chemicals exist in mass spectroscopy analysis in multiple forms that are detected as distinct m/z features. These include isotopomers containing 13C, which are useful to support definition of elemental formulas, but also ion pairs and ion clusters that can be difficult to identify. In the following, we consider bioinformatic data clustering methods as a means to identify the entities detected as ion clusters in mass spectrometry. To minimize confusion created by this unfortunate use of “clustering” with two different meanings, we use the term “data clustering” to refer to the bioinformatics method and “ion clusters” to refer to entities detected by mass spectrometry.
Data clustering analysis methods are commonly used in bioinformatic analyses to detect associations and co-regulated entities in complex biological systems (Mar et al. 2011; McLachlan et al. 2002; Hartigan and Wong 1979; Kohonen 1990; Kaufman and Rousseeuw 2005). In principle, such methods could be used to aid in detection of multiple forms of one chemical obtained by DC-FTMS, as well as to detect associations due to metabolic pathways in vivo.
For data clustering analysis, we used modulated modularity clustering (MMC) (Stone and Ayroles 2009) to generate Spearman rank correlations between m/z ion intensities and visualize these associations in terms of heat maps (Figs. 4, 5). In these heat maps, strongly associated m/z features are sorted together and shown in red. Strongly associated but negatively correlated m/z features are visualized in blue. The MMC method is capable of revealing m/z feature cluster structure with as little of four observations per m/z feature; however, the accuracy of data clustering increases with each additional observation. In the present analysis, m/z features with less than 70% non-zero ion intensities were excluded to minimize inaccurate data clustering (Stone and Ayroles 2009).
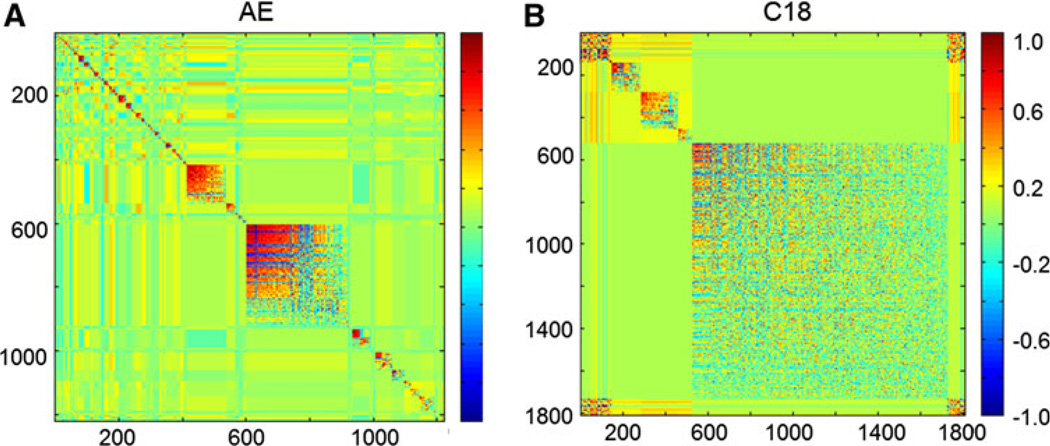
Fig. 4.
Modulated modularity clustering (MMC) heatmaps derived from marmoset samples with all m/z features with greater than 70% non-zero ion intensities in either the anion exchange column (1,809 m/z features) (a) or the reverse phase column (1222 m/z features) (b). Clusters of similar correlations are grouped as indicated in the legend (dark blue perfect anticorrelation, dark red perfect correlation)
Fig. 5.
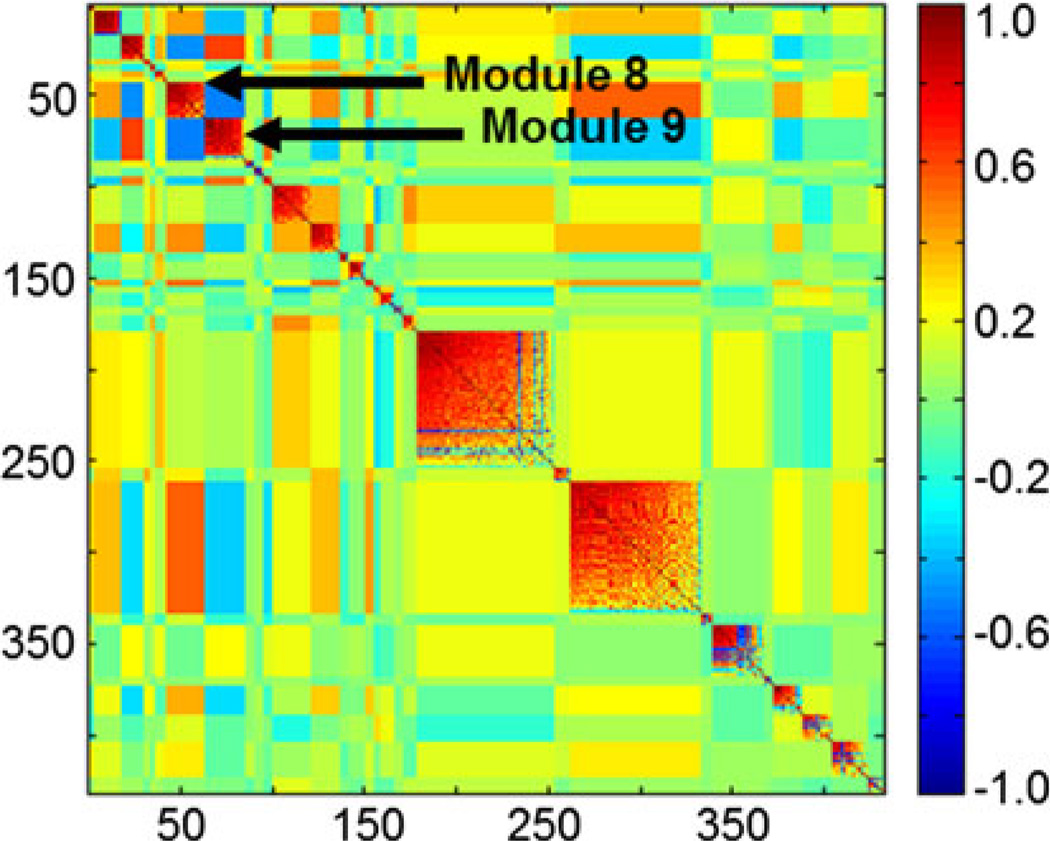
Off-diagonal negative correlations between two similarly correlated clusters (modules 8 and 9; shown by arrows) identify ion suppression by the cluster with higher ion intensity. The m/z features contained in modules 8 and 9 eluted at similar retention times between 50 and 60 s but with a twofold difference in ion intensity. The entire MMC heatmap was derived from the marmoset samples with all m/z features with a retention time between 0 and 150 s and greater than 70% non-zero ion intensities in the reverse phase column (432 m/z features). Clusters of similar correlations are grouped as indicated in the legend (dark blue perfect anticorrelation, dark red perfect correlation)
Because m/z features derived from a single chemical are likely to correlate, this analysis is expected to show strongly positive correlations for m/z that are forms of the same chemical, i.e., including ion clusters. For example, module 23 from an MMC analysis of C18 column data shows a cluster of ions related to ketamine (Table 2). Ketamine was used to anesthetize the marmosets prior to the blood draw, but the basis for associations of ketamine with the tripeptide of Cys-His-Gly and hydroxylauric acid is unknown. Thus, in development of high resolution metabolic profiling, data clustering algorithms can be used to simplify analysis of the extensive m/z data by identifying possible ion clusters of the same chemical species.
Table 2.
Module 23 from the MMC analysis of features detected in the C18 column
| Module 23 containing eight m/z values (Average degree = 0.75) | ||||
|---|---|---|---|---|
| m/z | Formula | Name | Δppm | Species |
| 217.786 | C12H24O3 | Hydroxy lauric acid | 5 | [M + H]+ |
| 236.0819 | C13H14ClNO | Ketamine metabolite | 7 | [M + H]+ |
| 238.0975 | C13H16ClNO | Ketamine | 7 | [M + H]+ |
| 239.1006 | 13CC12H16ClNO | 13C of Ketamine | 9 | [M + H]+ |
| 240.0946 | C13H1637ClNO | 37Cl of Ketamine | 7 | [M + H]+ |
| 241.0977 | 13CC12H1637ClNO | 13C37Cl of Ketamine | 8 | [M + H]+ |
| 242.1011 | 13C2C11H1637ClNO | 13C237Cl of Ketamine | 6 | [M + H]+ |
| 316.1103 | C11H17N5O4S | Cys His Gly | 9 | [M + H]+ |
This module contains a cluster of ions that are forms of the same chemical, ketamine. Ketamine was used to anesthetize the marmosets prior to the blood draw. Annotations for the m/z were derived from matches to known metabolites in either Madison Metabolomics Consortium Database or Metlin database
In contrast to positive correlations of ions generated from a single chemical, ions that suppress detection of others are expected to cluster together but show strong negative correlations. Identification of these associations occurring as a consequence of the mass spectrometry analysis is important because they are superimposed upon the biological associations in metabolic pathways in vivo. Modules 8 and 9 in the C18 analysis demonstrate this phenomenon (Fig. 5). The entire heat map was generated with all m/z features with a retention time between 0 and 150 s and greater than 70% non-zero ion intensities in the reverse phase column (432 m/z features). Within this heat map, modules 8 and 9 were negatively associated with each other but show strong correlations within each respective cluster (r ≥ 0.7). The m/z features within these two modules eluted at similar retention times (50–60 s) but with a 2-fold difference in ion intensity. This suggests that a large number of ions eluted within this 10-s time frame causing ion suppression, such that ions in module 8 (mean ion intensity 4.61 × 105 ± 1.05 × 105) suppressed the ions in module 9 (mean ion intensity 2.43 × 105 ± 4.73 × 104). MMC could, therefore, prove useful to detect ion suppression from co-eluting chemicals and be used as a basis to adjust separation or ionization conditions to reduce ion suppression. Alternatively, it may be possible to develop algorithms to improve quantification by correcting for ion suppression.
MMC does not appear to have been applied to metabolic profiling data, but it offers a straightforward way to detect associations among m/z features. Multiple ions arising from the same chemical have posed a challenge for MS-based metabolomics, and application of MMC to MS data facilitate detection of such ions. MMC is based upon clustering of correlated elements so that ions that form proportionally during electrospray will be clustered within a module.
With DC-FTMS, over 3,300 features are unidentified so that MMC can be used to determine if unidentified ions are derived from known chemicals by looking at these associations and then performing MS2 to test for common fragmentation patterns. Further, MMC modules consisting of all unidentified ions will be worth future investigations to determine if they are related to environmental exposures, exposure metabolites, and/or exposure-related modifications to endogenous metabolites.
Comparisons of heat maps for MMC analysis of total features for AE showed very different characteristics than those obtained for C18 analyses (Fig. 4). AE show that about 600 of the features correlated with six separate and well-defined modules, while most of the other 1,200 features are present in a large module with weak correlations. C18 show a very different pattern, with two large, moderately correlated modules (mean r = 0.40) containing about 450 m/z features and 24 smaller, more highly correlated modules (mean r = 0.49). The basis for these differences is not clear but could potentially be used to separate analytic variation from biologic variation in complex data obtained by top-down metabolic profiling using mass spectrometry. Means to adjust for mass spectrometry-dependent analytic variation are critical to develop cumulative databases with data compiled from different mass spectrometers and obtained at different times.
The DC-FTMS method described here provides an improved high-throughput biomonitoring of chemicals in human plasma to support exposome research. Although many chemicals are biologically eliminated by metabolism and excretion, there are many unidentified chemicals in human plasma that have not been characterized. Hence, there is a need to improve metabolic coverage. The results show that improvement can be obtained by combining successive analysis on two chromatographic columns, i.e., a dual-chromatography method with AE and C18. The overlap in detection further suggests that the analytical rigor for detection of a subset of chemicals could be improved by writing software to extract data for these chemicals from two successive chromatographic separations. Additionally, some chemicals with the same elemental composition could separate on one column but not on the other so that with validation of chemical structures, the combination of data from two analyses could yield more reliable quantitative data.
4 Conclusions
A dual chromatography-high resolution mass spectrometry analysis protocol improves metabolic coverage by 36% and increases detection of m/z matching to known metabolites in available databases. Application of a bioinformatic data clustering method showed the ability to detect ion clusters derived from the same chemical, thereby simplifying chemical identification and providing a basis for improved feature detection and quantification. The results suggest a need for systematic analysis of combinations of chromatographic separations and ionization modes to maximize detection of chemicals for study of metabolic characteristics of diseases and environmental exposures in personalized medicine and predictive health.
Supplementary Material
Acknowledgments
The authors thank Jennifer M. Johnson, M.S., for her technical help with the internal standard and acquisition of human reference samples. This work was supported by research NIH grants P01ES016731 (DPJ), R01AG038746 (DPJ), R01ES011195 (DPJ) and P51RR000168 (KGM).
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s11306-011-0332-1) contains supplementary material, which is available to authorized users.
Contributor Information
Quinlyn A. Soltow, Department of Medicine, Division of Pulmonary, Allergy and Critical Care, Emory University, Atlanta, GA 30322, USA
Frederick H. Strobel, Mass Spectrometry Center, Emory University, Atlanta, GA 30322, USA
Keith G. Mansfield, Harvard Medical School, New England Primate Research Center, Southborough, MA 01772, USA
Lynn Wachtman, Harvard Medical School, New England Primate Research Center, Southborough, MA 01772, USA.
Youngja Park, Department of Medicine, Division of Pulmonary, Allergy and Critical Care, Emory University, Atlanta, GA 30322, USA; Clinical Biomarkers Laboratory, Emory University, Atlanta, GA 30322, USA.
Dean P. Jones, Email: dpjones@emory.edu, Department of Medicine, Division of Pulmonary, Allergy and Critical Care, Emory University, Atlanta, GA 30322, USA; Clinical Biomarkers Laboratory, Emory University, Atlanta, GA 30322, USA.
References
- Aronson KJ, Wilson JW, Hamel M, et al. Plasma organochlorine levels and prostate cancer risk. Journal of Exposure Science and Environmental Epidemiology. 2010;20:434–445. doi: 10.1038/jes.2009.33. [DOI] [PubMed] [Google Scholar]
- Brigham KL. Predictive health: the imminent revolution in health care. Journal of the American Geriatrics Society. 2010;58(Suppl 2):S298–S302. doi: 10.1111/j.1532-5415.2010.03107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M, Dunn WB, Dobson P, et al. Mass spectrometry tools and metabolite-specific databases for molecular identification in metabolomics. The Analyst. 2009;134:1322–1332. doi: 10.1039/b901179j. [DOI] [PubMed] [Google Scholar]
- Buscher JM, Czernik D, Ewald JC, Sauer U, Zamboni N. Cross-platform comparison of methods for quantitative metabolomics of primary metabolism. Analytical Chemistry. 2009;81:2135–2143. doi: 10.1021/ac8022857. [DOI] [PubMed] [Google Scholar]
- Crews B, Wikoff WR, Patti GJ, et al. Variability analysis of human plasma and cerebral spinal fluid reveals statistical significance of changes in mass spectrometry-based metabolomics data. Analytical Chemistry. 2009;81:8538–8544. doi: 10.1021/ac9014947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis DI, Dunn WB, Griffin JL, Allwood JW, Goodacre R. Metabolic fingerprinting as a diagnostic tool. Pharmacogenomics. 2007;8:1243–1266. doi: 10.2217/14622416.8.9.1243. [DOI] [PubMed] [Google Scholar]
- Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Analytical Chemistry. 2009;81:6656–6667. doi: 10.1021/ac901536h. [DOI] [PubMed] [Google Scholar]
- Gilmour MI, Jaakkola MS, London SJ, Nel AE, Rogers CA. How exposure to environmental tobacco smoke, outdoor air pollutants, and increased pollen burdens influences the incidence of asthma. Environmental Health Perspectives. 2006;114:627–633. doi: 10.1289/ehp.8380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haouala A, Zanolari B, Rochat B, et al. Therapeutic Drug Monitoring of the new targeted anticancer agents imatinib, nilotinib, dasatinib, sunitinib, sorafenib and lapatinib by LC tandem mass spectrometry. Journal of Chromatography B. Analytical Technologies in the Biomedical and Life Sciences. 2009;877:1982–1996. doi: 10.1016/j.jchromb.2009.04.045. [DOI] [PubMed] [Google Scholar]
- Hartigan JA, Wong MA. A k-means clustering algorithm. Applied Statistics. 1979;28:100–108. [Google Scholar]
- Hodel EM, Zanolari B, Mercier T, et al. A single LC-tandem mass spectrometry method for the simultaneous determination of 14 antimalarial drugs and their metabolites in human plasma. Journal of Chromatography B. Analytical Technologies in the Biomedical and Life Sciences. 2009;877:867–886. doi: 10.1016/j.jchromb.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Howell G, III, Mangum L. Exposure to bioaccumulative organochlorine compounds alters adipogenesis, fatty acid uptake, and adipokine production in NIH3T3-L1 cells. Toxicology in Vitro. 2010;25(1):394–402. doi: 10.1016/j.tiv.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Institute of Laboratory Animal Resources. Guide for the care and use of laboratory animals. Washington, DC: National Academy Press; 1996. [Google Scholar]
- Johnson JM, Strobel FH, Reed M, Pohl J, Jones DP. A rapid LC-FTMS method for the analysis of cysteine, cystine and cysteine/cystine steady-state redox potential in human plasma. Clinica Chimica Acta. 2008;396:43–48. doi: 10.1016/j.cca.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JM, Yu T, Strobel FH, Jones DP. A practical approach to detect unique metabolic patterns for personalized medicine. Analyst. 2010;135:2864–2870. doi: 10.1039/c0an00333f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DP, Park Y, Gletsu-Miller N, et al. Dietary sulfur amino acid effects on fasting plasma cysteine/cystine redox potential in humans. Nutrition. 2011;27:199–205. doi: 10.1016/j.nut.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman L, Rousseeuw PJ. Finding groups in data: an introduction to cluster analysis. New York: Wiley; 2005. [Google Scholar]
- Kind T, Scholz M, Fiehn O. How large is the metabolome? a critical analysis of data exchange practices in chemistry. PLoS One. 2009;4:e5440. doi: 10.1371/journal.pone.0005440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohonen T. The self-organizing map. Proceedings of the IEEE. 1990;78:1464–1480. [Google Scholar]
- Lawton KA, Berger A, Mitchell M, et al. Analysis of the adult human plasma metabolome. Pharmacogenomics. 2008;9:383–397. doi: 10.2217/14622416.9.4.383. [DOI] [PubMed] [Google Scholar]
- Lewis CM, Whitwell SC, Forbes A, et al. Estimating risks of common complex diseases across genetic and environmental factors: the example of Crohn disease. Journal of Medical Genetics. 2007;44:689–694. doi: 10.1136/jmg.2007.051672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loscalzo J, Kohane I, Barabasi AL. Human disease classification in the postgenomic era: a complex systems approach to human pathobiology. Molecular systems biology. 2007;3:124. doi: 10.1038/msb4100163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mar JC, Wells CA, Quackenbush J. Defining an informativeness metric for clustering gene expression data. Bioinformatics. 2011;27(8):1094–1100. doi: 10.1093/bioinformatics/btr074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall AG, Hendrickson CL. High-resolution mass spectrometers. Annual review of Analytical Chemistry (Palo Alto Calif) 2008;1:579–599. doi: 10.1146/annurev.anchem.1.031207.112945. [DOI] [PubMed] [Google Scholar]
- McLachlan GJ, Bean RW, Peel D. A mixture model-based approach to the clustering of microarray expression data. Bioinformatics. 2002;18:413–422. doi: 10.1093/bioinformatics/18.3.413. [DOI] [PubMed] [Google Scholar]
- Miura D, Tsuji Y, Takahashi K, Wariishi H, Saito K. A strategy for the determination of the elemental composition by fourier transform ion cyclotron resonance mass spectrometry based on isotopic peak ratios. Analytical Chemistry. 2010;82:5887–5891. doi: 10.1021/ac902931x. [DOI] [PubMed] [Google Scholar]
- Nordstrom A, Want E, Northen T, Lehtio J, Siuzdak G. Multiple ionization mass spectrometry strategy used to reveal the complexity of metabolomics. Analytical Chemistry. 2008;80:421–429. doi: 10.1021/ac701982e. [DOI] [PubMed] [Google Scholar]
- Olsen JV, Schwartz JC, Griep-Raming J, et al. A dual pressure linear ion trap Orbitrap instrument with very high sequencing speed. Molecular and Cellular Proteomics. 2009;8:2759–2769. doi: 10.1074/mcp.M900375-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Oo ML, Andersen JK. Synergistic effects of environmental risk factors and gene mutations in Parkinson’s disease accelerate age-related neurodegeneration. Journal of Neurochemistry. 2010;115(6):1363–1373. doi: 10.1111/j.1471-4159.2010.07036.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandanger TM, Brustad M, Sandau CD, Lund E. Levels of persistent organic pollutants (POPs) in a coastal northern Norwegian population with high fish-liver intake. Journal of Environmental Monitoring. 2006;8:552–557. doi: 10.1039/b600046k. [DOI] [PubMed] [Google Scholar]
- Soltow QA, Jones DP, Promislow DE. A network perspective on metabolism and aging. Integrative and comparative biology. 2010;50:844–854. doi: 10.1093/icb/icq094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreekumar A, Poisson LM, Rajendiran TM, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–914. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Stone EA, Ayroles JF. Modulated modularity clustering as an exploratory tool for functional genomic inference. PLoS Genetics. 2009;5:e1000479. doi: 10.1371/journal.pgen.1000479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Kai K, Shinbo Y, et al. Metabolomics approach for determining growth-specific metabolites based on Fourier transform ion cyclotron resonance mass spectrometry. Analytical and Bioanalytical Chemistry. 2008;391:2769–2782. doi: 10.1007/s00216-008-2195-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjernberg A, Markova N, Griffiths WJ, Hallen D. DMSO-related effects in protein characterization. Journal of Biomolecular Screening: the Official Journal of the Society for Biomolecular Screening. 2006;11:131–137. doi: 10.1177/1087057105284218. [DOI] [PubMed] [Google Scholar]
- Voit EO. A systems-theoretical framework for health and disease: inflammation and preconditioning from an abstract modeling point of view. Mathematical Biosciences. 2009;217:11–18. doi: 10.1016/j.mbs.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang XB, Liu AL, et al. Significant positive correlation of plasma BPDE-albumin adducts to urinary 1-hydroxypyrene in coke oven workers. Biomedical and Environmental Sciences. 2007;20:179–183. [PubMed] [Google Scholar]
- Want EJ, O’Maille G, Smith CA, et al. Solvent-dependent metabolite distribution, clustering, and protein extraction for serum profiling with mass spectrometry. Analytical Chemistry. 2006;78:743–752. doi: 10.1021/ac051312t. [DOI] [PubMed] [Google Scholar]
- Wild CP. Complementing the genome with an “exposome”: the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer epidemiology, Biomarkers and Prevention. 2005;14:1847–1850. doi: 10.1158/1055-9965.EPI-05-0456. [DOI] [PubMed] [Google Scholar]
- Yanes O, Tautenhahn R, Patti GJ, Siuzdak G. Expanding coverage of the metabolome for global metabolite profiling. Analytical Chemistry. 2011;83:2152–2161. doi: 10.1021/ac102981k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Park Y, Johnson JM, Jones DP. apLCMS–adaptive processing of high-resolution LC/MS data. Bioinformatics. 2009;25:1930–1936. doi: 10.1093/bioinformatics/btp291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziech D, Franco R, Pappa A, et al. The role of epigenetics in environmental and occupational carcinogenesis. Chemico Biological Interactions. 2010;188:340–349. doi: 10.1016/j.cbi.2010.06.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.