Abstract
Reduced nicotinamide adenine dinucleotide phosphate (NADPH) is an essential electron donor in all organisms. It provides the reducing power that drives numerous anabolic reactions, including those responsible for the biosynthesis of all major cell components and many products in biotechnology. The efficient synthesis of many of these products, however, is limited by the rate of NADPH regeneration. Hence, a thorough understanding of the reactions involved in the generation of NADPH is required to increase its turnover through rational strain improvement. Traditionally, the main engineering targets for increasing NADPH availability have included the dehydrogenase reactions of the oxidative pentose phosphate pathway and the isocitrate dehydrogenase step of the tricarboxylic acid (TCA) cycle. However, the importance of alternative NADPH-generating reactions has recently become evident. In the current review, the major canonical and non-canonical reactions involved in the production and regeneration of NADPH in prokaryotes are described, and their key enzymes are discussed. In addition, an overview of how different enzymes have been applied to increase NADPH availability and thereby enhance productivity is provided.
Keywords: NADPH regeneration, pentose phosphate pathway, isocitrate dehydrogenase, malic enzyme, transhydrogenase, GAPN, ferredoxin:NADP+ oxidoreductase, hydrogenase
Introduction
Reduced nicotinamide adenine dinucleotide phosphate (NADPH) is an essential electron donor in all eukaryotes, bacteria, and archaea. Not only is NADPH vital for the generation of reactive oxygen species (ROS) (Foreman et al., 2003; Brown and Griendling, 2009; Bylund et al., 2010; Nakamura et al., 2012) and the anti-oxidative defense mechanisms of most organisms (Nordberg and Arnér, 2001; Minard and Mcalister-Henn, 2005; Singh et al., 2008), most importantly, it is also the driving force of most biosynthetic enzymatic reactions, including those responsible for the biosynthesis of all major cell components, such as DNA and lipids (Arnér and Holmgren, 2000; Nordberg and Arnér, 2001; Hügler et al., 2002; Koh et al., 2004; Minard and Mcalister-Henn, 2005; Singh et al., 2008; Miller and Auchus, 2011). Owing to this essential role in biosynthesis, NADPH availability has been of major interest to industry (Papagianni, 2012; Lee et al., 2013b). Many natural products of industrial importance are complex secondary metabolites, the production of which often involves NADPH-dependent enzymes. To synthesize such products using purified enzyme systems in vitro would require the addition of huge amounts of NADPH in order to sustain production. From an industrial point of view, this would be too expensive. Hence, in situ NADPH regeneration from its oxidized counterpart (NADP+) is required (Chenault et al., 1988; Wandrey, 2004; Wichmann and Vasic-Racki, 2005; Zhang and Xu, 2010; Uppada et al., 2014).
Regeneration of NADPH in situ can be achieved by various strategies, including chemical, electrochemical, photochemical, and enzyme-based methods (Chenault and Whitesides, 1987; Zhao and Van Der Donk, 2003; Wichmann and Vasic-Racki, 2005; Liu and Wang, 2007; Zhang and Xu, 2010; Uppada et al., 2014). NADPH can be regenerated enzymatically by complementing the in vitro system with additional enzymatic reactions or by using substrate-coupled reaction systems. The latter system employs enzymes that use both NADP+ and NADPH and that are able to catalyze the synthesis of the desired product from one substrate and cofactor regeneration with another substrate (Chenault et al., 1988; Van Der Donk and Zhao, 2003; Liu and Wang, 2007). However, reduced productivity compared to systems without in situ regeneration and problems associated with enzyme stability make these options unattractive.
Microbial in vivo production systems also provide in situ NADPH regeneration and have several advantages when compared to in vitro systems. For example, microbes are able to grow on inexpensive renewable feedstocks that provide the organisms with reductant for the regeneration of NADPH. They also contain numerous pathways, involving stable and highly specific enzymes, thus obviating the need for expensive enzyme purification. In addition, our knowledge of natural metabolic pathways is rapidly advancing, allowing for rational design toward product formation (Chemler et al., 2010; Siedler et al., 2011; Papagianni, 2012; Lee et al., 2013b). Therefore, it is not surprising that microbial conversion is the preferred method for the synthesis of a range of products.
With the possibility of engineering microbial metabolism to facilitate product formation, it became clear that NADPH availability remains a major hurdle in the efficient generation of many products. These products range from medicinal compounds (Chemler et al., 2010; Siedler et al., 2011; Zhao et al., 2011) and (essential) amino acids (Becker et al., 2007; Papagianni, 2012) to molecules used as biofuels (Asadollahi et al., 2009; Kim et al., 2011; Peralta-Yahya et al., 2012) and building blocks for biodegradable plastic (Kabir and Shimizu, 2003). Given its involvement in a multitude of crucial biological functions and its importance in biosynthesis, NADPH is without question an essential molecule. Hence, a key question arises: what are the major NADPH-generating reactions and systems?
Traditionally, the dehydrogenase reactions of the oxidative pentose phosphate pathway (oxPPP), the Entner–Doudoroff (ED) pathway, and the isocitrate dehydrogenase step of the tricarboxylic acid (TCA) cycle have been considered the major sources of NADPH. However, the importance of other NADPH-generating enzymes, such as transhydrogenases, glucose dehydrogenases, and non-phosphorylating glyceraldehyde 3-phosphate dehydrogenase (GAPN), is becoming clear, indicating that the traditional view is over-simplistic (Sauer et al., 2004; Matsubara et al., 2011; Brasen et al., 2014).
In this review, we describe the major canonical and non-canonical biochemical mechanisms that are involved in the production and regeneration of NADPH in prokaryotes and discuss the key enzymes involved. We have divided the mechanisms into those that are directly coupled to central carbon metabolism and those that are not (Table 1). In addition, we briefly address how different enzymes have been applied to increase NADPH availability and thereby enhance NADPH-dependent biotransformation processes.
Table 1.
Overview of major canonical and non-canonical NADPH-generating enzymes.
| Enzyme | EC:number | Pathway | Bacteria* (%) | Archaea* (%) | Additional cofactors | Applied** | ΔrG'm*** (kJ/mol) | |
|---|---|---|---|---|---|---|---|---|
| NADPH-generating enzymes coupled to central carbon metabolism | G6PDH | EC:1.1.1.49 | oxPPP, ED | 66 | 0 | n.a. | yes | −2.3±2.6 |
| 6PGDH | EC:1.1.1.44 | oxPPP | 62 | 27 | n.a. | yes | −6.0±6.3 | |
| IDH | EC:1.1.1.42 | TCA cycle | 82 | 59 | n.a. | yes | −10.7±6.3 | |
| ME | EC:1.1.1.40 | Anaplerotic node | 47 | 25 | n.a. | no | −3.1±6.2 | |
| GAPN | EC:1.2.1.9 | EMP | 12 | 31 | n.a. | yes | −36.1±1.1 | |
| NADP+-GAPDH | EC:1.2.1.13 | EMP | n.d. | n.d. | n.a. | yes | 25.9±1.0 | |
| GDHs | EC:1.1.1.47,EC:1.1.1.119 | Modified EDs | 18 | 10 | n.a. | yes | −2.4±2.2 | |
| n.d. | n.d. | n.a. | ||||||
| NADPH-generating enzymes not coupled to central carbon metabolism | STH | EC:1.6.1.1 | n.a. | 19 | 0 | NADH, FAD | yes | 1.0±0.7 |
| H+-TH | EC:1.6.1.2 | n.a. | 50 | 5 | NADH | yes | 1.0±0.7 | |
| FNR | EC:1.18.1.2 | n.a. | 63 | 29 | FAD or FMN, Fdred | no | −15.6±11.7 | |
| SH | EC:1.12.1.3 | n.a. | 10 | 6 | FAD or FMN | no | −16.5±5.9 | |
| NADK | EC:2.7.1.23 | n.a. | 96 | 98 | NTP or poly(P), NAD+ or NADH | yes | n.a. |
Distribution of the enzyme in completely sequenced prokaryotic genomes. All available prokaryotic genomes were downloaded to Excel from the IMG database on 6/16/2015. The analysis included only finished genomes (198 archaeal and 3380 bacterial genomes) and only one representative genome from each genus (83 archaeal and 636 bacterial genomes), to correct for the many draft genomes and for the many sequences in closely related genomes, respectively. The obtained distribution is merely an indication of the true distribution of the respective enzymes among prokaryotes because the database is incomplete and gene functions are often automatically annotated.
Use of the enzyme in metabolic engineering to increase NADPH availability and enhance biotransformation processes.
Gibbs free energies (ΔrG′m) calculated using the biochemical thermodynamics calculator eQuilibrator 2.0 (Flamholz et al., 2012; Noor et al., 2012, 2013), which calculates ΔrG′m. This term describes the change in Gibbs free energy due to a chemical reaction at a particular pH and ionic strength (subscript r), using 1 mM concentrations for all reactants (superscript m). Default settings (pH 7 and ionic strength 0.1 M) were used for all reactions.
n.a., not applicable; n.d., no data available in the IMG database; G6PDH, glucose-6-phosphate dehydrogenase; 6PGDH, 6-phosphogluconate dehydrogenase; IDH, isocitrate dehydrogenase; ME, malic enzyme; GAPN, non-phosphorylating glyceraldehyde 3-phosphate dehydrogenase; NADP+-GAPDH, NADP+-dependent phosphorylating glyceraldehyde 3-phosphate dehydrogenase; GDH, glucose dehydrogenase; STH, energy-independent soluble transhydrogenase; H+-TH, energy-dependent or proton-translocating, membrane-bound transhydrogenase; FNR, ferredoxin:NADP+ oxidoreductase; SH, cytosolic NADP+-reducing hydrogenase; NADK, NAD+ kinase.
Biosynthesis of NADP+
To maintain a sufficient quantity of NADP+ for the generation of NADPH, NADP+ biosynthesis is essential. While the necessity for NADP+ synthesis during cell proliferation is clear, the requirement for NADP+ synthesis in non-dividing cells might not be obvious. NADP(H) is generally thought of solely as a redox carrier that facilitates the transfer of electrons between two redox couples, a role that does not account for the need for constant resynthesis. The constant need to resynthesize NADP+ arises from its participation in other crucial biological processes (Agledal et al., 2010). In eukaryotes, for example, NADP+ serves as the substrate in the synthesis of nicotinic acid-adenine dinucleotide phosphate (NAADP), an important intracellular Ca2+-mobilizing messenger (Lee, 1997; Chini and De Toledo, 2002; Churamani et al., 2004; Patel et al., 2011). Although NAADP has also been found in bacteria (Churamani et al., 2004), its physiological relevance in these organisms remains to be established.
The enzymes responsible for NADP+ resynthesis, best studied in eukaryotes, mainly include NAD(P)+ nucleosidase (Vu et al., 1996; Ying, 2008), NADP+ phosphatase, and NADPH phosphatase. The enzymes in prokaryotes are less well-studied, but similar activities and proteins have been found in this domain (Mather and Knight, 1972; Everse et al., 1975; Davis, 1980; Penyige et al., 1996; Pollak et al., 2007; Kawai and Murata, 2008; Ghosh et al., 2010). NADP+ phosphatase and NADPH phosphatase, together with NAD(H) kinase, regulate the intracellular balance of NAD(H) and NADP(H) (Kawai and Murata, 2008). Like NADPH, NADH mainly serves to transfer electrons from one molecule to another. However, unlike NADPH, NADH is primarily involved in catabolic reactions (Ying, 2008). In addition to its redox function, NAD+ serves as a substrate for mono- and poly-ADP ribosylation, participates in histone deacetylation, and contributes to the production of the signaling molecule cyclic ADP-ribose. Most of these reactions have been characterized in eukaryotes, but the ribosylation reactions also play a role in toxin production by pathogenic bacteria (Ziegler, 2000; Pollak et al., 2007). The consumption of NADP+ is thus connected to the consumption of NAD+ and to the regulation of various major biological activities such as DNA repair, gene expression, apoptosis, nitrogen fixation, and calcium homeostasis (Ziegler, 2000; Pollak et al., 2007; Kawai and Murata, 2008; Ying, 2008; Agledal et al., 2010).
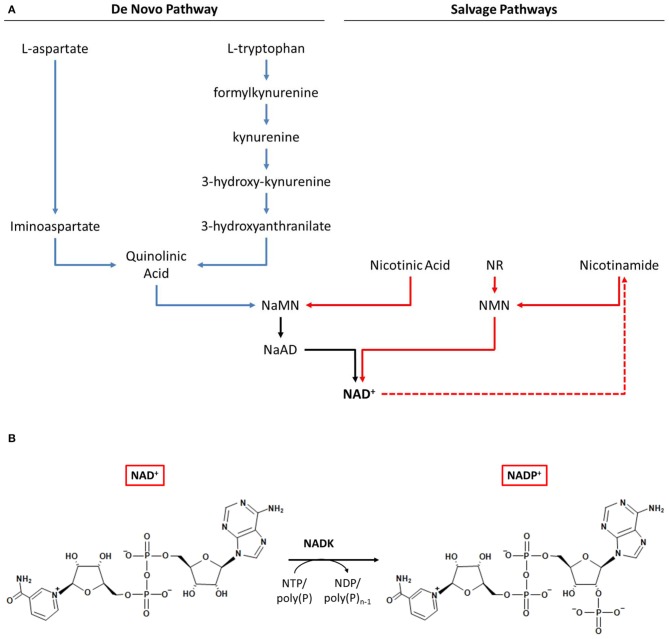
Given that the intracellular balance between NAD(H) and NADP(H) is regulated via the addition or removal of a phosphate group by NAD(H) kinase or NADP(H) phosphatase, respectively, it is clear that the biosynthesis of NAD+ plays a crucial role in the metabolism of NADP+. Two principal NAD+ biosynthesis pathways have been characterized: (1) the de novo pathway and (2) the salvage pathway (Magni et al., 1999, 2004; Begley et al., 2001). Both pathways have been reviewed recently (Pollak et al., 2007; Ying, 2008; Gazzaniga et al., 2009; Gossmann et al., 2012). In the de novo pathway, NAD+ is generated from quinolinic acid, which in prokaryotes is produced from either L-aspartate or L-tryptophan (Magni et al., 1999; Sakuraba et al., 2002; Kurnasov et al., 2003). In the salvage pathway, degradation products containing a pyridine ring, namely nicotinic acid and nicotinamide, are utilized to regenerate NAD+ (Figure 1A) (Magni et al., 1999; Gazzaniga et al., 2009).
Figure 1.
NAD+ and NADP+ biosynthetic pathways in prokaryotes. (A) De novo and salvage pathways for NAD+ biosynthesis. (B) NADP+ biosynthesis by NAD kinase. Abbreviations: NaMN, nicotinic acid mononucleotide; NaAD, nicotinic acid adenine dinucleotide; NR, nicotinamide riboside; NMN, nicotinamide mononucleotide; NAD, nicotinamide adenine dinucleotide; NADP, nicotinamide adenine dinucleotide phosphate; NADK, NAD kinase.
A portion of the synthesized NAD+ can be converted into NADP+ by NAD kinase (NADK) (Figure 1B). In the NAD+ synthesis pathways, several variations exist and multiple enzymes are involved (Bi et al., 2011; Gossmann et al., 2012). In contrast, NADK is the sole enzyme able to generate NADP+ de novo. NAD kinase is found in archaea (Sakuraba et al., 2005), bacteria (Kawai et al., 2000, 2001; Ochiai et al., 2004), and eukaryotes (Tseng et al., 1979; Butler and Mcguinness, 1982) and has been proven to be essential in prokaryotes (Kobayashi et al., 2003; Sassetti et al., 2003). According to the literature, only one species not able to synthesize NADP+ from NAD+ has been identified: Chlamydia trachomatis (Kawai and Murata, 2008; Szaszák et al., 2011). The intracellular parasite appears to lack NADK and hence relies completely on the metabolism of its host cell.
According to Kawai and Murata (2008), NADK orthologs can be classified into three types according to their substrate specificity: (I) NADKs that utilize both ATP and inorganic polyphosphate (poly(P)) as a phosphoryl donor and phosphorylate both NAD+ and NADH. This type of NADK has been identified in gram-positive bacteria and archaea. (II) NADKs that utilize ATP but not poly(P) and phosphorylate both NAD+ and NADH. This type has been identified in eukaryotes. (III) NADKs that utilize ATP but not poly(P) and phosphorylate NAD+ but not NADH. The latter type of NADK has been identified in gram-negative bacteria. Because of its vital role in NADP+ synthesis, NADK has received much attention since its discovery in 1950 (Kornberg, 1950). Hence, many papers about its structure, function, and application are available, including reviews by Kawai and Murata (2008), Shi et al. (2009), and Agledal et al. (2010).
Redox potential of NADP+/NADPH
The redox potential of NADP+/NADPH at standard physiological conditions is identical to that of NAD+/NADH (: −320 mV). However, the function of the cofactors is different. NADP+/NADPH is used for anabolic redox reactions, whereas NAD+/NADH is used for oxidation reactions. This is possible because NADP+/NADPH is generally maintained in a reduced state and NAD+/NADH in an oxidized state (Harold, 1986). The ratios for the electron carriers have been reported, and the values can differ by several orders of magnitude, depending on the organism and growth conditions. For example, reported NAD+/NADH values range from 3.74 to 31.3 for bacterial cells (Andersen and von Meyenburg, 1977; Thauer et al., 1977; Bennett et al., 2009; Amador-Noguez et al., 2011) and reach 1820 for mammalian cells (Veech et al., 1969), whereas NADP+/NADPH values range from 0.017 to 0.95 (Andersen and von Meyenburg, 1977; Thauer et al., 1977; Bennett et al., 2009; Amador-Noguez et al., 2011). This indicates that the actual redox potential of both redox couples can deviate significantly from the standard potential (i.e., generally more negative/positive than −320 mV for (NADP+/NADPH)/(NAD+/NADH), respectively). Such deviation affects the Gibbs energy of the NADPH-generating reactions and the feasibility of converting NADH into NADPH. To assess the feasibility of various NADPH-generating reactions, we calculated the Gibbs energies (ΔrG′m) using the biochemical thermodynamics calculator eQuilibrator 2.0 (Flamholz et al., 2012; Noor et al., 2012, 2013), setting all reactants at a physiological concentration of 1 mM (Table 1). However, even under these conditions, several reactions appeared to be at equilibrium or even slightly endergonic. At NADP+/NADPH ratios lower than 1, the reaction becomes even less feasible, indicating that the actual concentration of the reaction components must be substantially different for NADPH generation to proceed.
Systems for NADPH generation
NADP+ synthesized by NADK or generated by NADPH-oxidizing reactions is eventually reduced to NADPH. To ensure that the cellular redox balance is maintained in the absence of de novo NADP+ synthesis and NADP+ consumption, the catabolic fluxes through the NADPH-regenerating reactions must be matched to the anabolic demand. However, this is generally not the case (Fuhrer and Sauer, 2009). In addition, the precise NADPH formation rate depends on fluxes through the generating pathways, which in turn vary with different growth conditions (Dauner et al., 2001; Zhao et al., 2004a; Kanai et al., 2011). Therefore, prokaryotes must have other network-wide biochemical mechanisms that maintain the cellular redox balance (Fuhrer and Sauer, 2009). The exact mechanisms are not fully understood and are beyond the scope of this review, but papers about the topic are available (Singh et al., 2008; Fuhrer and Sauer, 2009).
Although numerous reactions reduce NADP+ to NADPH (at least 143 reactions according to the MetaCyc database, accessed on 03/30/15), only a few have been thought to contribute significantly. However, new studies have demonstrated the importance of other NADPH-generating reactions. The present review provides a general overview of the known major NADPH-generating reactions and discusses the key enzymes involved. We have divided the reactions into (I) those that are directly coupled to central carbon metabolism and (II) those that are not (Table 1). The enzymes that comprise the first group are the oxPPP enzymes glucose-6-phosphate dehydrogenase and 6-phosphogluconate dehydrogenase; isocitrate dehydrogenase of the TCA cycle; malic enzyme; and three enzymes involved in non-canonical NADPH-generating reactions: phosphorylating GAP dehydrogenase (NADP+-GAPDH), non-phosphorylating GAP dehydrogenase (GAPN), and glucose dehydrogenase. The enzymes in the second group are transhydrogenases (NADH:NADP+), ferredoxin:NADP+ oxidoreductase, hydrogenases (H2:NADP+), and NAD(H) kinase, although the latter is a de novo NADP+/NADPH-synthesizing enzyme rather than an NADPH-regenerating enzyme. NAD(H) kinase is included because it plays an essential role in NADP+ biosynthesis and the regulation of the NAD(H)/NADP(H) balance offers a potential strategy for improving the biosynthesis of industrially valuable metabolites.
NADPH-generating reactions coupled to central carbon metabolism
Glucose-6-phosphate dehydrogenase and 6-phosphogluconate dehydrogenase
The oxidative pentose phosphate pathway (oxPPP) enzymes glucose-6-phosphate dehydrogenase (G6PDH, EC:1.1.1.49) and 6-phosphogluconate dehydrogenase (6PGDH, EC:1.1.1.44) have a central role in metabolism in many microbes (Table 1). The enzymes are involved in the conversion of glucose-6-phosphate into ribulose-5-phosphate (Figure 2), a precursor of important molecules such as nucleic acids, and are generally considered a major source of NADPH. The importance of the oxPPP enzymes as a source of NADPH has been demonstrated in various organisms (Summers et al., 1995; Christiansen et al., 2002; Sauer et al., 2004). Flux through these enzymes reportedly increases when NADPH requirements are high (Obanye et al., 1996; Christiansen et al., 2002; Bartek et al., 2011; Celton et al., 2012) and decreases when NADPH requirements are low (Marx et al., 1999; Jonsbu et al., 2001). This is consistent with the anticipated anabolic role of the oxPPP as a supplier of pentoses and NADPH for biosynthesis. The key enzyme with regard to the control of PPP flux is thought to be G6PDH (Kletzien et al., 1994; Moritz et al., 2000; Saliola et al., 2012). However, in addition to its role in the oxPPP, G6PDH can also participate in the classical phosphorylated version of the Entner–Doudoroff (ED) pathway (Figure 2). The ED pathway can be considered an alternative to the Embden–Meyerhof–Parnas (EMP) glycolytic pathway, because both catabolize glucose to pyruvate (Conway, 1992; Sato and Atomi, 2011). The ED pathway is mainly present in prokaryotes, although some eukaryotes possess a functional ED pathway as well (Fabris et al., 2012). A generally held view is that the ED pathway is less important than the EMP pathway with respect to glucose catabolism. However, Fuhrer et al. have shown that this might be a misconception and that the ED pathway might be a major pathway for glucose catabolism even in species that possess both pathways (Fuhrer et al., 2005).
Figure 2.
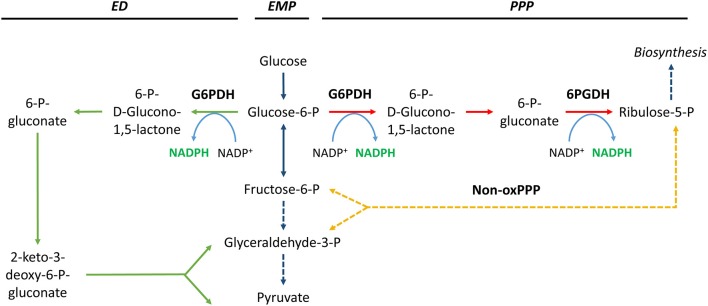
NADPH generation by glucose-6-phosphate dehydrogenase (G6PDH) and 6-phosphogluconate dehydrogenase (6PGDH). Solid and dashed lines indicate single and lumped reactions, respectively. Pathway abbreviations: ED, Entner–Doudoroff pathway (in green); EMP, Embden–Meyerhof–Parnas pathway (in blue); PPP, pentose phosphate pathway (oxidative phase in red, non-oxidative phase in yellow).
G6PDHs and 6PGDHs have been widely studied with respect to cofactor specificity. Although some bacterial G6PDHs and 6PGDHs show dual cofactor specificity (Stournaras et al., 1983; Sung and Lee, 2007; Fuhrer and Sauer, 2009) or have a strong preference for NAD+ (Stournaras et al., 1983; Sung and Lee, 2007) or other cofactors such as the flavin derivative F420 (Purwantini et al., 1997), most are NADP+-dependent (Levy, 1979; Fuhrer and Sauer, 2009; Olavarría et al., 2012). In addition, the dual cofactor specificity that some G6PDHs show is often observed in vitro under saturated conditions. However, physiological in vivo conditions often result in higher specificities for NADP+ (Fuhrer and Sauer, 2009). Therefore, G6PDHs and 6PGDHs are generally considered to catalyze the following reactions:
| (1) |
and
| (2) |
Like eukaryotes, some prokaryotes possess multiple G6PDH (Butler et al., 2002; Singh et al., 2005; Sung and Lee, 2007) or 6PGDH isozymes (Zamboni et al., 2004). In general, these isozymes are similar, with the same cofactor preferences, but exceptions exist. Mycobacterium smegmatis, for example, has two G6PDH isozymes, one of which is NADP+-dependent. The other is F420-dependent and does not utilize NAD+ or NADP+ (Purwantini and Daniels, 1996). In addition, cell-free extracts of several other Mycobacterium and Nocardia species contain both F420-dependent and NADP+-dependent G6PDH activity (Purwantini et al., 1997), indicating that the genomes of these species likely encode both variants. Similarly, exceptions among 6PGDH isozymes have been identified. The genome of Bacillus subtilis, for example, encodes three functional 6PGDH isozymes; two prefer NAD+, and one prefers NADP+ (Zamboni et al., 2004). As discussed by Zamboni et al., the presence of isozymes with different cofactor preferences would, in principle, enable NADP+, NAD+, and F420 reduction in the oxPPP to adjust to the overall metabolic requirements of the cell (Zamboni et al., 2004). However, the exact functional roles of these isozymes remains to be determined.
The availability of NADPH is of principal importance for various industrially important classes of products, including amino acids, proteins, antibiotics, organic acids, and high-value metabolites. The apparent relationship between the rate of NADPH generation and actual carbon fluxes through the oxPPP suggests that the oxPPP can be a target for metabolic engineering for overproduction of NADPH. The oxPPP has been engineered to increase the NADPH/NADP+ ratio, through overexpression of oxPPP enzymes (Lim et al., 2002; Choi et al., 2003; Becker et al., 2007; Lee et al., 2007a) and through redirection of the carbon flux from glycolysis to the oxPPP, by disrupting phosphoglucose isomerase (Kabir and Shimizu, 2003; Marx et al., 2003; Chemler et al., 2010) or phosphofructokinase (Chin and Cirino, 2011; Wang et al., 2013d), overexpressing fructose 1,6-bisphosphatase (Becker et al., 2005), or introducing glucose dehydrogenase (Zhang et al., 2011). Both strategies have been applied successfully in various prokaryotes, but reduction in growth is a common side effect (Lim et al., 2002; Lee et al., 2003; Marx et al., 2003). However, the opposite effect was observed with an archaeal strain recently developed in our lab: introduction of the oxPPP increased the hydrogen yield and significantly improved the growth rate (unpublished data).
Although both strategies have been applied successfully in various prokaryotes, they are not effective in every organism (Poulsen et al., 2005; Smith et al., 2010). Moreover, as mentioned above, G6PDH is a key enzyme not only for control of the PPP flux but also for control of the ED pathway. Overexpressing G6PDH could therefore affect the flux of both pathways simultaneously. However, this is generally not observed. Cells apparently possess the ability to regulate fluxes to ensure a network-wide balancing of NADPH supply and demand (Nicolas et al., 2007; Fuhrer and Sauer, 2009).
The effects of G6PDH and 6PGDH disruption have also been investigated. The exact effects are strongly dependent on the organism, the genetic background of the parent strain, and the environmental growth conditions (Summers et al., 1995; Butler et al., 2002; Hua et al., 2003; Zamboni et al., 2004; Zhao et al., 2004b; Richhardt et al., 2013). However, in general, G6PDH-deficient and 6PGDH-deficient prokaryotes exhibit remarkably mild phenotypes. Their growth rates are similar to or somewhat lower than those of the parent strain, and they are more sensitive to oxidative stress. Flux through the oxPPP and ED pathways is lower in G6PDH-deficient strains. To compensate for this and to ensure a sufficient supply of anabolic precursors and NADPH, flux rerouting via the EMP pathway and the non-oxPPP is generally observed (Figure 2). In addition, glucose-grown mutants generally display enhanced TCA cycle activity. In contrast, acetate- or pyruvate-grown cells generally display decreased TCA cycle activity (Zhao et al., 2004a,b). Like G6PDH-deficient strains, 6PGDH-deficient strains exhibit a strong reduction in oxPPP flux, but not in ED pathway flux. In contrast, prokaryotes with a functional ED pathway generally respond to 6PGDH knockout by rerouting through the ED pathway and by reversing the direction of the non-oxPPP, slightly increasing the flux through the EMP pathway, and activating malic enzyme (Jiao et al., 2003; Zhao et al., 2004b). Prokaryotes that lack a functional ED pathway display a similar response, but instead of rerouting the flux through the ED pathway, they increase flux through the EMP pathway (Zamboni et al., 2004). Thus, in general, prokaryotes can compensate for disruption of G6PDH or 6PGDH.
Isocitrate dehydrogenase
Isocitrate dehydrogenase (IDH, EC:1.1.1.42) is a component of the TCA cycle that catalyzes the decarboxylation of isocitrate to 2-oxoglutarate (also known as α-ketoglutarate), with the release of CO2 and NADPH (Figure 3). The IDH reaction is important for the generation of reducing power. Through the generation of 2-oxoglutarate, the reaction also links nitrogen and carbon metabolism and plays an important role in the cellular defense against oxidative damage and detoxification of ROS (Muro-Pastor et al., 2005; Mailloux et al., 2007; Paul et al., 2007). Together with isocitrate lyase, IDH is a branching point between the TCA cycle and the glyoxylate shunt (Figure 3), a pathway needed for growth on non-fermentative carbon sources such as acetate and ethanol. IDH activity is important in controlling the metabolic flux between both pathways and is affected by various regulatory factors, such as metal ions (Murakami et al., 1997; Yoon et al., 2003), PEP concentration (Ogawa et al., 2007), and phosphorylation/dephosphorylation (Walsh and Koshland, 1985; Cozzone, 1998). In addition, IDH is a component of the reductive tricarboxylic acid (RTCA) cycle. The RTCA cycle is a CO2 fixation pathway, present in some bacteria and archaea, in which four molecules of CO2 are fixed to produce one molecule of oxaloacetate (Shiba et al., 1985; Beh et al., 1993; Kanao et al., 2002). Obviously, IDH cannot act as a source of NADPH under these conditions.
Figure 3.
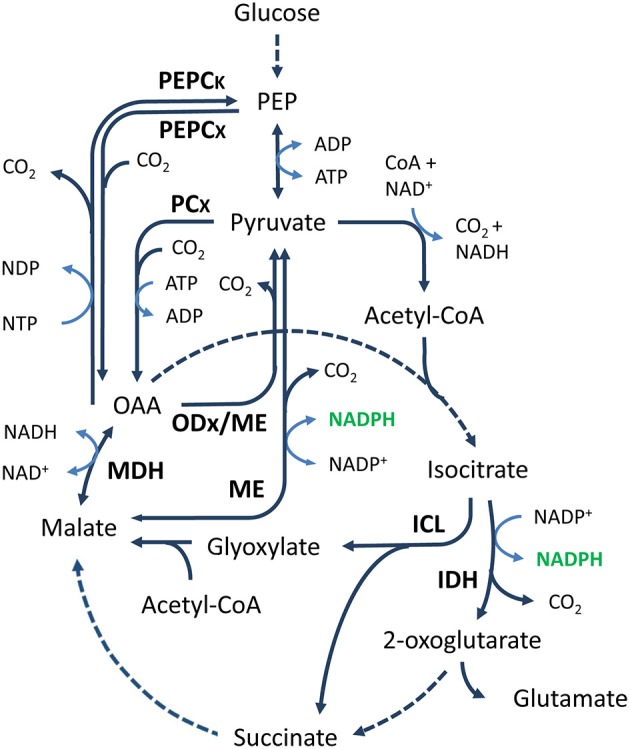
General overview of the TCA cycle and anaplerotic node. Solid and dashed arrows represent single and lumped enzymatic reactions, respectively. Malic enzyme is able to catalyze the decarboxylation of malate and OAA. Abbreviations for metabolites (normal text) and enzymes (bold text): PEP, phosphoenolpyruvate; OAA, oxaloacetate; PEPCK, phosphoenolpyruvate carboxykinase; PEPCx, phosphoenolpyruvate carboxylase; PCx, pyruvate carboxylase; ODx, oxaloacetate decarboxylase; MDH, malate dehydrogenase; ME, malic enzyme; ICL, isocitrate lyase; IDH, isocitrate dehydrogenase.
In contrast to eukaryotes, prokaryotes are generally thought to possess only NADP+-dependent IDHs. Although this is true for most prokaryotes, a small but increasing number of reports have described bacterial and archaeal NAD+-dependent IDHs, such as the one found in Hydrogenobacter thermophilus (Lloyd and Weitzman, 1988; Chen and Jeong, 2000; Steen et al., 2001; Inoue et al., 2002; Aoshima et al., 2004; Stokke et al., 2007; Wang et al., 2011b, 2012, 2013a). Moreover, a survey of sequenced prokaryotic genomes by Zhu et al. identified nine additional species that likely contain NAD+-dependent IDHs (Zhu et al., 2005). However, although the IDH of H. thermophilus was originally described as an NAD+-dependent IDH, Aoshima and Igarashi later showed it was not a conventional decarboxylating IDH but a novel non-decarboxylating enzyme that catalyzes the conversion between isocitrate and oxalosuccinate. They therefore proposed that the enzyme should not be categorized as an IDH (Aoshima and Igarashi, 2008).
With the exception of the few NAD+-preferring IDHs, prokaryotic IDHs generally catalyze the following decarboxylating reaction:
| (3) |
In addition to the cofactor NADP+, IDH also needs divalent cations, particularly Mg2+ or Mn2+. Although most IDHs have a strong requirement for either Mg2+ or Mn2+, substitution of other divalent cations, or even univalent ions such as Na+, is sometimes possible. However, in most cases, this leads to little or no activity (Murakami et al., 1997; Steen et al., 1997; Yoon et al., 2003; Banerjee et al., 2005).
Like G6PDH and 6PGDH, IDH is an important source of NADPH (Hua et al., 2003; Sauer et al., 2004). In wild-type Escherichia coli aerobically grown in chemostats under glucose- or ammonia-limited conditions, the IDH reaction was the major producer of NADPH, accounting for more than 60% of the total NADPH production (Hua et al., 2003). Accordingly, overexpression of IDH resulted in a 31% increase in the production of GDP-L-fucose, whose formation is NADPH-dependent. While this is a significant improvement, the biosynthesis of GDP-L-fucose in the IDH overexpression strain was not better than that in a G6PDH overexpression strain (Lee et al., 2011). This suggests that, under the applied conditions, IDH is not a better option than G6PDH for NADPH regeneration in biotransformation reactions (Lee et al., 2013b). The ability of IDH overexpression to enhance glutamate formation in Corynebacterium glutamicum has also been tested (Eikmanns et al., 1995). C. glutamicum secretes glutamate under certain conditions. Because IDH supplies 2-oxoglutarate, the direct precursor of glutamate, overexpression of IDH was an obvious choice. However, overexpression of IDH, even in combination with glutamate dehydrogenase, did not enhance glutamate production.
Disruption of IDH has also been investigated. The exact effects of IDH disruption were dependent on the specific conditions and species used, but some general trends were apparent (Mcdermott and Kahn, 1992; Muro-Pastor and Florencio, 1994; Eikmanns et al., 1995; Kabir and Shimizu, 2004; Becker et al., 2009). First, all IDH mutants investigated were glutamate auxotrophs. Second, although glutamate-containing cultivation media sustained the growth of IDH mutants, their growth was slower than that of the parental strains. The reduced specific growth rate was most likely due to a lower ATP/ADP ratio and a lower NADPH/NADP+ ratio, as shown in an E. coli IDH disruption strain grown on glucose under aerobic conditions (Kabir and Shimizu, 2004). Moreover, other significant effects observed in the mutant E. coli strain included upregulation of oxPPP enzymes and the anaplerotic glyoxylate pathway. Similarly, redirection from the TCA cycle to anaplerosis has also been reported for an IDH knockdown of C. glutamicum (Becker et al., 2009). However, with respect to citrate synthase, contradictory results have been obtained. Kabir and Shimizu (2004) found upregulation of citrate synthase, but an earlier study of E. coli IDH mutants (Lakshmi and Helling, 1976) and a study of Rhizobium meliloti (Mcdermott and Kahn, 1992) found that IDH mutants spontaneously gave rise to IDH-citrate synthase double mutants.
Malic enzyme
The TCA cycle-associated malic enzyme (ME, EC:1.1.1.40) catalyzes the NAD(P)+-dependent oxidative decarboxylation of malate to pyruvate (Figure 3). Confusingly, malic enzyme is also known as “malate dehydrogenase (oxaloacetate-decarboxylating),” while true malate dehydrogenases, enzymes that catalyze the non-decarboxylating reaction between malate and oxaloacetate (Figure 3), are sometimes referred to as malic enzyme. In addition, various classes of ME that differ in their preference for NAD+ or NADP+ and in their ability to decarboxylate oxaloacetate have also been characterized (Frenkel, 1975). Enzymes in the first ME class (EC:1.1.1.38) use NAD+ and can decarboxylate oxaloacetate as well as malate. Enzymes in the second class (EC:1.1.1.39) also prefer NAD+, but they are unable to decarboxylate oxaloacetate. Enzymes in the third class (EC:1.1.1.40) are NADP+-dependent; they can catalyze the decarboxylation of malate and oxaloacetate. Moreover, MEs that do not fit the general classification scheme exactly, such as those from the gram-negative bacterium Rhizobium meliloti and the hyperthermophilic archaeon Thermococcus kodakarensis, have been described as well (Voegele et al., 1999; Fukuda et al., 2005). The latter, for example, prefers NADP+ but, unlike normal NADP+-dependent MEs, is not able to decarboxylate oxaloacetate.
Although malic enzymes (MEs) are widely distributed amongst prokaryotes (Table 1), most ME-containing species possess an ME that belongs to the third class (Sauer and Eikmanns, 2005; Lerondel et al., 2006) and catalyzes the following reaction:
| (4) |
However, prokaryotes without an ME or with only an NAD+-dependent ME, such as lactic acid bacteria (London et al., 1971; Kawai et al., 1996; Landete et al., 2010; Espariz et al., 2011), or prokaryotes with both NAD+- and NADP+-dependent MEs have also been characterized (Iwakura et al., 1978; Voegele et al., 1999; Sauer and Eikmanns, 2005; Lerondel et al., 2006; Bologna et al., 2007). Moreover, Morimoto et al. recently showed that directed evolution could easily alter the cofactor preference of an ME from T. kodakarensis (Morimoto et al., 2014). Thus, NAD+/NADP+ specificity seems to depend only on a few specific, highly conserved residues (Kuo et al., 2000; Yang et al., 2002; Chang and Tong, 2003; Fukuda et al., 2005; Lerondel et al., 2006; Hsieh and Hung, 2009).
All MEs require a divalent cation as a cofactor. Maximum activity is generally observed in the presence of Mn2+, followed closely by Mg2+. Divalent cations such as Co2+ and Ni2+ are usually only partially able to replace Mn2+, while others such as Ca2+, Cu2+, or Sr2+ cannot replace Mn2+ at all (Gourdon et al., 2000; Fukuda et al., 2005; Wang et al., 2011a). In addition, the MEs of various prokaryotes also require a monovalent cation for activity, preferably and K+ (Lamed and Zeikus, 1981; Garrido-Pertierra et al., 1983; Kawai et al., 1996; Driscoll and Finan, 1997; Gourdon et al., 2000).
In many aerobic and facultative anaerobic organisms, the TCA cycle-associated ME is part of the PEP-pyruvate-oxaloacetate node, also referred to as the anaplerotic node (Figure 3). This node represents the metabolic link between glycolysis/gluconeogenesis and the TCA cycle (Owen et al., 2002; Sauer and Eikmanns, 2005). As such, ME is involved in the interconversion of C4 and C3 compounds, which is important for maintaining the levels of TCA cycle intermediates (anaplerotic reactions) and for growth on C4 and C3 compounds and substrates that enter central metabolism via acetyl-CoA, such as acetate, fatty acids, and ethanol. Although ME can catalyze C3-carboxylation and C4-decarboxylation reactions, it is generally involved in the latter (decarboxylation of malate to pyruvate with the concomitant formation of NADPH) (Voegele et al., 1999; Gourdon et al., 2000; Sauer and Eikmanns, 2005; Lerondel et al., 2006; Wang et al., 2011a). However, examples of prokaryotic MEs without a preference for either (Fukuda et al., 2005) and MEs with a preference for the C3-carboxylation reaction have been identified (Matula et al., 1969). To determine the exact contribution of ME to the cellular NADPH pool, the physiological direction and flux through the enzyme needs to be established. However, regulation of the carbon flux at the anaplerotic node involves a complex interplay between the enzymes involved and depends on the growth conditions and species (Petersen et al., 2000; Dauner et al., 2001; Fischer and Sauer, 2003; Netzer et al., 2004). A detailed discussion of this complex regulation is beyond the scope of this review, but comprehensive reviews, such as those by Owen et al. (2002) and Sauer and Eikmanns (2005), are available.
To understand the physiological role of ME in the metabolism of prokaryotes, the disruption of ME and alternative pathways has been studied. The significance of the latter is clearly demonstrated by the absence of a detectable phenotype in Campylobacter jejuni and E. coli ME disruption strains (Velayudhan and Kelly, 2002; Kao et al., 2005). The lack of a clear phenotype can be explained by the presence of alternative pathways and enzymes, such as the malate dehydrogenase-phosphoenolpyruvate carboxykinase pathway (mdh-PEPCK) and isocitrate dehydrogenase, that can fulfill the pyruvate/PEP and NADPH requirements, respectively (Figure 3) (Hansen and Juni, 1975; Oh et al., 2002; Velayudhan and Kelly, 2002; Wang et al., 2011a). Because of the wide distribution of such alternatives in prokaryotes, it is likely that ME gene disruption in other species will yield similar results. Nevertheless, species in which ME gene disruption yields a phenotype, such as Corynebacterium glutamicum, Bacillus subtilis, and various lactic acid bacteria, have been identified (Gourdon et al., 2000; Lerondel et al., 2006; Landete et al., 2010; Espariz et al., 2011).
Analysis of various disruption strains created to understand the physiological role of ME in prokaryotes has shown that it is crucial to take the genetic background of a strain into consideration because the exact physiological role of ME differs from species to species (Riedel et al., 2001; Velayudhan and Kelly, 2002; Kao et al., 2005; Landete et al., 2010; Meyer and Stulke, 2013). Analysis has also shown that the effect of ME disruption can be subtle and/or heavily dependent on the cultivation conditions (Gourdon et al., 2000; Lerondel et al., 2006). A C. glutamicum ME disruption strain, for example, showed no detectable phenotype during growth on either acetate or glucose, but showed a significantly modified growth behavior during lactate metabolism (Gourdon et al., 2000). Moreover, the physiological role of NAD+- and NADP+-dependent MEs appears to be different (Lerondel et al., 2006; Bologna et al., 2007; Landete et al., 2010; Espariz et al., 2011; Wang et al., 2011a; Meyer and Stulke, 2013). Whereas NAD+-dependent MEs generally play a role in malate catabolism, NADP+-dependent MEs function either as gluconeogenic enzymes, by supplying pyruvate from C4-dicarboxylic acids, or as NADPH-generating systems needed for various biosynthetic purposes. It is interesting in this respect that the concerted action of ME, MDH, and pyruvate carboxylase (PCx) can act as a kind of transhydrogenase. The combined action of these enzymes enables the ATP-dependent conversion of pyruvate to OAA and malate and back to pyruvate, while converting NADH into NADPH (Figure 3) (Sauer and Eikmanns, 2005). A similar reaction sequence can involve PEP carboxylase (PEPCx) instead of pyruvate carboxylase, which also enables conversion of NADH into NADPH. This so-called malate shunt was recently described in Clostridium thermocellum (Zhou et al., 2013).
NAD+- and NADP+-dependent MEs have also been used in metabolic engineering approaches to improve specific biotechnological applications. However, to our knowledge, no prokaryotic ME has been used to increase intracellular NADPH levels, as has been done in the yeast Saccharomyces cerevisiae (Moreira Dos Santos et al., 2004; Matsuda et al., 2013). Instead, prokaryotic MEs have been overexpressed for the formation of C4-dicarboxylic acids from glucose or pyruvate (Stols and Donnelly, 1997; Hong and Lee, 2001; Shin et al., 2007; Zheng et al., 2009). These acids are important intermediates in the production of tetrapyrroles (Shin et al., 2007) and amino acids (Sahm et al., 2000), for example, and they can be chemically converted into common intermediates in the petrochemical industry. Hence, C4-dicarboxylic acids are important intermediates in the production of commodity chemicals from renewable carbohydrate feedstocks (Jain et al., 1989; Szmant, 1989). However, to produce these acids from glucose or pyruvate, one has to reverse the physiological direction of ME. This has been accomplished in different ways (Stols and Donnelly, 1997; Hong and Lee, 2001; Zheng et al., 2009).
NADP+-dependent glyceraldehyde-3-phosphate dehydrogenases
NADP+-dependent glyceraldehyde-3-phosphate dehydrogenases belong to the aldehyde dehydrogenase (ALDH) superfamily, a divergently related group of enzymes widely distributed among all three domains of life (Sophos et al., 2001; Sophos and Vasiliou, 2003). Enzymes in this group metabolize a broad spectrum of endogenous and exogenous aldehydes, which are oxidized to the corresponding carboxylic acid, using NAD+ or NADP+ as a cofactor (Lindahl, 1992; Vasiliou et al., 2000):
As such, the enzymes play essential roles in processes such as intermediary metabolism and detoxification (Talfournier et al., 2011; Esser et al., 2013). Although many ALDHs are NADP+-dependent, only the NADP+-dependent glyceraldehyde-3-phosphate dehydrogenases will be addressed here, because these enzymes contribute significantly to NADPH levels in certain prokaryotes. Moreover, extensive reviews of the ALDH superfamily are available (Sophos et al., 2001; Sophos and Vasiliou, 2003; Marchitti et al., 2008; Jackson et al., 2011).
While most glyceraldehyde-3-phosphate dehydrogenases (GAPDH, EC:1.2.1.12) are NAD+-dependent enzymes responsible for the reversible oxidation of GAP into 1,3-bisphosphoglycerate (1,3-BPG) as part of the classical EMP pathway, NADP+-dependent variants exist as well. In fact, there are two different types of NADP+-dependent GAPDHs, a phosphorylating type (NADP+-GAPDH, EC:1.2.1.13), producing 1,3-BPG, and a non-phosphorylating type (GAPN, EC:1.2.1.9), unidirectionally producing 3-phosphoglycerate (3-PG) (Figure 4). However, the term NADP+-GAPDH is used inconsistently in different studies to denote either the phosphorylating or non-phosphorylating type or both types, thus generating confusion.
Figure 4.
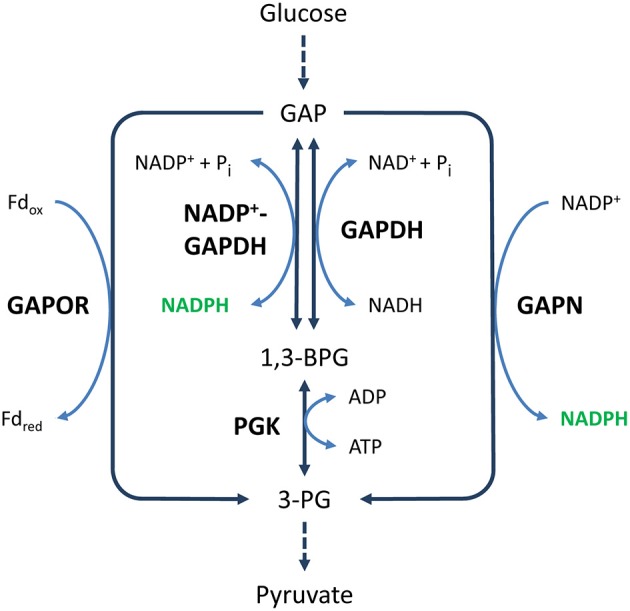
NADPH generation by GAP dehydrogenases. Solid and dashed arrows represent single and lumped enzymatic reactions, respectively. Abbreviations for metabolites (normal text) and enzymes (bold text): GAP, glyceraldehyde 3-phosphate; 1,3-BPG, 1,3-bisphosphoglycerate; 3-PG, 3-phosphoglycerate; GAPOR, GAP:ferredoxin oxidoreductase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GAPN, non-phosphorylating GAPDH; PGK, phosphoglycerate kinase.
GAPN was originally thought to be present only in photosynthetic eukaryotes (Kelly and Gibbs, 1973; Mateos and Serrano, 1992). However, early reports of GAPN activity in Streptococcus mutans were later confirmed by the characterization of the first prokaryotic GAPN (Crow and Wittenberger, 1979; Boyd et al., 1995). Since then, GAPN has also been found in various (hyper)thermophilic archaea, such as Thermococcus kodakarensis, Thermoproteus tenax, and Sulfolobus solfataricus (Brunner et al., 1998; Ettema et al., 2008; Matsubara et al., 2011), and several gram-positive bacteria, such as Clostridium acetobutylicum, Streptococcus pyogenes, and Bacillus halodurans (Table 1) (Iddar et al., 2002, 2003, 2005; Sophos and Vasiliou, 2003; Agledal et al., 2010). GAPN catalyzes the irreversible, non-phosphorylating oxidation of GAP to 3-PG with the concurrent reduction of NADP+ to NADPH:
| (5) |
In this reaction, 1,3-BPG is not formed, and the oxidation of GAP is not coupled to the generation of ATP (Figure 4). Such modifications of the classical EMP or ED pathway are regularly found in archaea (Ahmed et al., 2005; Siebers and Schonheit, 2005; Ettema et al., 2008; Matsubara et al., 2011; Brasen et al., 2014). Interestingly, GAPN has been reported to have a key function in the regulation of these modified pathways (Brunner et al., 1998; Brunner and Hensel, 2001; Lorentzen et al., 2004; Ettema et al., 2008).
With the exception of the GAPN in T. tenax, which has a preference for NAD+, prokaryotic GAPNs, in general, have a clear preference for NADP+. However, although the GAPN of T. tenax was shown to be NAD+-dependent, it was later shown to be allosterically regulated, shifting its cofactor preference from NAD+ to NADP+ in the presence of activators such as F6P and G1P (Lorentzen et al., 2004). Based on sequence similarity, similar allosteric regulation might also exist for GAPNs in a number of other hyperthermophiles (Lorentzen et al., 2004; Ito et al., 2012). Aside from NADP+, GAPNs do not require other cofactors or metals.
While the physiological significance of GAPN might vary between organisms, the enzyme is an important and essential source of NADPH in the bacterium S. mutans and the archaeon T. kodakarensis, respectively (Boyd et al., 1995; Matsubara et al., 2011; Arutyunov et al., 2013). Moreover, the ability to use GAPN as a source of NADPH has been demonstrated in a mutant E. coli strain in which the endogenous NAD+-dependent glyceraldehyde-3-phosphate dehydrogenase (NAD+-GAPDH) was replaced by the GAPN of S. mutans. Transcriptional analysis of this strain revealed upregulation of the transhydrogenase and downregulation of the PPP and TCA cycle, which together indicate a response to avoid NADPH excess (Centeno-Leija et al., 2013). In addition, GAPN has been used successfully in metabolic engineering efforts intended to improve the production of metabolites whose biosynthesis depends on NADPH, such as L-lysine or poly-3-hydroxybutyrate, or to decrease the formation of undesired by-products such as glycerol in industrial ethanol production strains (Takeno et al., 2010; Guo et al., 2011; Centeno-Leija et al., 2014).
In contrast to GAPN, NADP+-GAPDH produces 1,3-BPG. It is therefore more similar to NAD+-GAPDH (Figure 4).
| (6) |
The generation of 1,3-BPG is an important difference between the reactions catalyzed by NADP+-GAPDH and GAPN, because further oxidation of 1,3-BPG to 3-PG generates ATP. Moreover, while the oxidation of GAP by NADP+-GAPDH is reversible, the oxidation by GAPN is not. Hence, NADP+-GAPDH, in contrast to GAPN, is active in both glycolysis and gluconeogenesis. However, in prokaryotes that also contain alternative GAP oxidation enzymes, such as NAD+-GAPDH, GAPN, or GAP:ferredoxin oxidoreductase (GAPOR) (Figure 4), NADP+-GAPDH is primarily involved in gluconeogenesis and does not appear to play a role in generating NADPH (Schäfer and Schönheit, 1993; Koksharova et al., 1998; Fillinger et al., 2000; Brunner et al., 2001; Matsubara et al., 2011; Ito et al., 2012).
As with GAPN, the significance of NADP+-GAPDH as a source of NADPH has been demonstrated. Overexpression of NADP+-GAPDH or replacement of the native NAD+-GAPDH with the NADP-variant improved the yield of NADPH-dependent products such as lycopene, ϵ-caprolactone, L-ornithine, or coenzyme Q10 (Martinez et al., 2008; Huang et al., 2011; Jiang et al., 2013; Wang et al., 2013c). Moreover, metabolic flux analysis of the exchange mutant revealed that the oxPPP branch and TCA fluxes were significantly reduced, presumably to avoid NADPH excess (Martinez et al., 2008). In addition, an attempt in C. glutamicum to alter the cofactor specificity of native NAD+-GAPDH to NADP+ through protein engineering resulted in a 60% increase in lysine production (Bommareddy et al., 2014).
NADP+-dependent glucose dehydrogenase
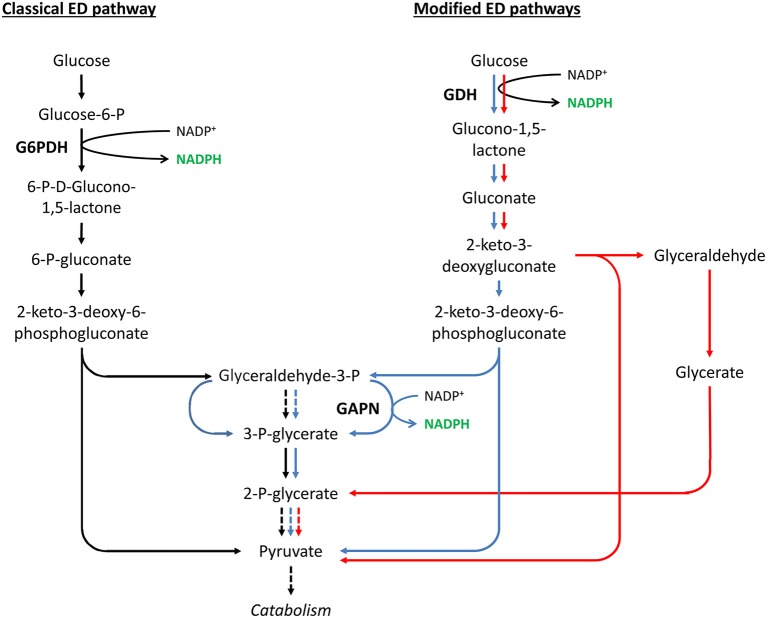
Similar to GAPN and GAPDH, other non-canonical NADP+-dependent enzymes, such as NADP+-dependent glucose dehydrogenase (NADP+-GDH), might make a larger contribution to NADPH generation than previously thought. Glucose dehydrogenases (GDHs, EC:1.1.1.47 and EC:1.1.1.119) are responsible for the first step of the modified ED pathways found in some archaea and bacteria (Table 1), i.e., the semiphosphorylated ED, the non-phosphorylated ED, and the branched ED pathways (Figure 5) (Brasen et al., 2014). GDHs are responsible for a main difference between modified ED pathways and the classical ED pathway. GDHs catalyze the direct oxidation of glucose into gluconolactone. In contrast, in the classical ED, glucose is phosphorylated before its oxidation into 6-P-gluconolactone (Brasen et al., 2014).
Figure 5.
Classical ED pathway compared to modified ED pathways. Solid and dashed arrows represent single and lumped enzymatic reactions, respectively. Shown are the classical ED pathway (in black), the semiphosphorylated ED pathway (in blue), the non-phosphorylated ED pathway (in red), and the branched ED pathway that includes the semi- and non-phosphorylated ED pathways. Only the glucose degradation directions are shown. Abbreviated enzymes (in bold text): G6PDH, glucose-6-phosphate dehydrogenase; GAPN, non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase; GDH, glucose dehydrogenase.
| (7) |
Two different classes of GDHs are capable of using NADP+ as cofactor: EC:1.1.1.47, which shows dual cosubstrate specificity with a preference for NADP+, and EC:1.1.1.119, which is strictly dependent on NADP+. GDHs belonging to these groups have been found in extremely halophilic and thermoacidophilic archaea, as well as in bacteria such as Bacillus subtilis, Bacillus megaterium, and Gluconobacter oxydans (Pauly and Pfleiderer, 1975; Adachi and Ameyama, 1981; Lampel et al., 1986; Brasen et al., 2014).
NADP+-GDHs have been used for NADPH regeneration in various production systems, such as the in vitro synthesis of poly(3-hydroxybutyrate) or L-leucovorin (Eguchi et al., 1992; Satoh et al., 2003). In addition, NADP+-GDHs are important components of many commercial glucose-sensing assays. As such, they have been utilized for blood glucose monitoring (Ferri et al., 2011). Moreover, NADP+-GDHs have also been used as NADPH regenerators in various whole-cell, or in vivo, production systems. The GDHs of B. megaterium and B. subtilis, for example, have been expressed in several E. coli strains for the synthesis of various products, including indigo, S-sulfoxide, and ethyl (R/S)-4-chloro-3-hydroxybutanoate (Kataoka et al., 1998, 1999; Kizaki et al., 2001; Yun et al., 2005; Lu and Mei, 2007; Xu et al., 2007; Park et al., 2010; Zhang et al., 2011).
NADPH-generating reactions not coupled to carbon metabolism
Transhydrogenases
Pyridine nucleotide transhydrogenase (TH) directly catalyzes the reversible hydride transfer between NAD(H) and NADP(H). There are two different isoforms: the energy-independent soluble transhydrogenase (STH, EC:1.6.1.1) and the energy-dependent, or proton-translocating, membrane-bound transhydrogenase (H+-TH, EC:1.6.1.2) (Figure 6). Soluble transhydrogenases from Gammaproteobacteria such as Escherichia coli, Azotobacter vinelandii, Pseudomonas fluorescens, and Pseudomonas aeruginosa have been studied in some detail (Cohen and Kaplan, 1970; Voordouw et al., 1979; French et al., 1997; Boonstra et al., 1999). In contrast, proton-translocating, membrane-bound transhydrogenases have been investigated extensively, reviewed in (Bizouarn et al., 2000, 2002; Jackson, 2003, 2012; Pedersen et al., 2008; Jackson and Obiozo, 2009; Lee et al., 2013b). They are widely distributed in the mitochondria of eukaryotes and in certain bacteria, but are very rare in archaea (Table 1) (Jackson and Obiozo, 2009; Jackson, 2012). Bacteria with multiple copies of the transhydrogenase genes are not uncommon, and some species, particularly among the Enterobacteriaceae, contain both isoforms (Sauer et al., 2004).
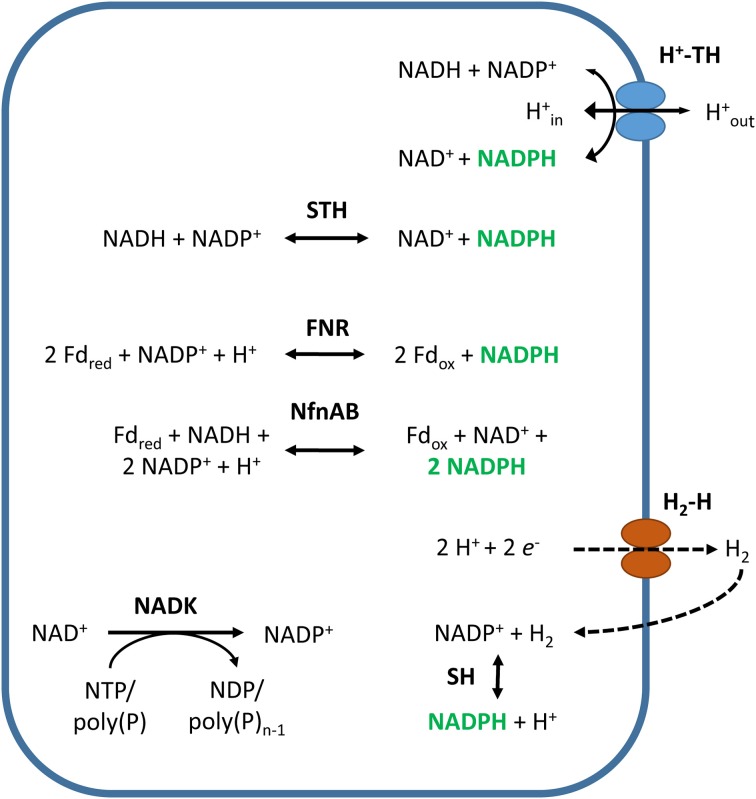
Figure 6.
NADPH-generating reactions not coupled to carbon metabolism. Abbreviated enzymes (in bold): H2-H, hydrogen-evolving hydrogenase; SH, cytosolic NADP+-reducing hydrogenase; NfnAB, electron-bifurcating NADH-dependent reduced ferredoxin:NADP+ oxidoreductase; FNR, ferredoxin:NADP+ oxidoreductase; STH, energy-independent soluble transhydrogenase; H+-TH, energy-dependent or proton-translocating, membrane-bound transhydrogenase.
The two transhydrogenase isoforms transfer electrons between NAD(H) and NADP(H) in the following reaction:
| (8) |
However, in contrast to STHs, H+-THs couple the electron transfer reaction to the translocation of a proton across the membrane, according to the reaction:
| (9) |
Where “out” and “in” denote the periplasmic space and cytosol, respectively. Although the reaction is in principle reversible, the electrochemical proton gradient (Δp) across the membrane under most physiological conditions strongly favors NADPH formation (Hoek and Rydstrom, 1988; Bizouarn et al., 2000; Jackson, 2003; Pedersen et al., 2008).
Since the discovery of transhydrogenases, there has been much debate about their physiological role (Bragg et al., 1972; Voordouw et al., 1983; Harold, 1986; Hoek and Rydstrom, 1988; Jackson et al., 1998; Pedersen et al., 2008). Given the type of reaction they catalyze, transhydrogenases might play an important role in maintaining the redox balance within microorganisms. Various studies have shown that several bacterial species require STH for growth under metabolic conditions with excess NADPH formation (Canonaco et al., 2001; Hua et al., 2003; Sauer et al., 2004; Zhao et al., 2008; Fuhrer and Sauer, 2009). Moreover, mutant bacteria suffering from a disturbed redox balance due to an increased NADPH concentration often benefit from the heterologous expression of STH (Boonstra et al., 2000; Angermayr et al., 2012; Qi et al., 2014; Reddy et al., 2015). These observations support the idea that the physiological role of STH is to convert NADPH into NADH to prevent an excess of NADPH (Voordouw et al., 1983), a notion that is generally accepted and is supported by actual NAD+/NADH and NADP+/NADPH ratios (see above), which indicate that the formation of NADPH from NADH will generally require energy input.
However, Sanchez et al. have reported that the NADPH-requiring production of poly(3-hydroxybutyrate) in E. coli benefitted from the overexpression of STH (Sanchez et al., 2006). This result is contrary to the generally accepted physiological role of STH and suggests that the enzyme catalyzes the opposite reaction. Moreover, recent reports have shown that STH is able to catalyze the opposite reaction and that overexpression of the enzyme can improve NADPH-dependent production yields (Lee et al., 2010; Decorosi et al., 2011; Jan et al., 2013; He et al., 2014). Additionally, although STH disruption and overexpression mutants often have disturbed or improved redox balances, respectively, similar mutants that do not show any effect have also been described (Chin et al., 2009; Yamauchi et al., 2014). Together, these examples show that we have yet to achieve a thorough understanding of the physiological role of STH and that its role is strongly dependent on species, culture conditions, and genotypes.
Similarly, the physiological role of H+-TH has been debated in the literature. It has often been reported that the H+-TH isoform is a major source of NADPH, needed for the biosynthesis of amino acids, for example, or the reduction of glutathione (required to minimize oxidative damage caused by free radicals generated in the respiratory chain) (Bragg et al., 1972; Voordouw et al., 1983; Hoek and Rydstrom, 1988; Hickman et al., 2002; Hua et al., 2003; Sauer et al., 2004; Fuhrer and Sauer, 2009; Imam et al., 2013; He et al., 2014). In E. coli, H+-TH has been shown to provide about 40% of the total NADPH during growth on glucose in batch cultures (Sauer et al., 2004). Moreover, its expression is induced when there is demand for NADPH, and it is required for optimal growth on carbon sources whose metabolism does not directly generate NADPH (Sauer et al., 2004; Imam et al., 2013). Additionally, Fuhrer and Sauer calculated that it is very unlikely, from a thermodynamic point of view, that H+-THs catalyze NADPH oxidation. They concluded that the NADP+ reduction catalyzed by H+-THs is irreversible under physiological conditions in various bacterial species (Fuhrer and Sauer, 2009). Reports such as these contribute to the general view that the physiological role of H+-TH is reduction of NADP+ at the expense of NADH. However, other reports suggest that H+-TH catalyzes the opposite reaction or show that disruption or overexpression has no measurable effect (Chin et al., 2009; Jan et al., 2013; Yamauchi et al., 2014).
THs require the cofactors NAD(H) and NADP(H) for activity. In addition, and in contrast to H+-THs, STHs also require a flavin (flavin adenine dinucleotide, FAD) cofactor (Middleditch et al., 1972; Boonstra et al., 1999; Cao et al., 2011). THs generally do no required other cofactors or metal ions. In fact, metal ions have even been reported to inhibit TH activity (Whitehead et al., 2005, 2009; Cao et al., 2011). Nevertheless, submillimolar concentrations of Ca2+ and Mg2+ have been reported to stimulate the H+-THs of Rhodobacter capsulatus and E. coli (Cotton et al., 1989; Lever et al., 1991; Chang et al., 1992). Similarly, saturating concentrations of Ca2+ have been reported to release the phosphate-induced inhibition of the NADP+ reducing reaction and to cause full activation of the enzyme in Azotobacter vinelandii and Pseudomonas aeruginosa, respectively (Hojeberg and Rydstrom, 1977; Voordouw et al., 1980).
Because of their ability to generate NADPH, STHs and H+-THs have been overexpressed to improve the production yields of NADPH-dependent metabolites. For example, overexpression of STH increased NADPH-dependent poly(3-hydroxybutyrate) production by 82% in an engineered E. coli strain (Sanchez et al., 2006). Moreover, the overexpression of STH in two other engineered E. coli strains increased (S)-2-chloropropionate and thymidine production 150 and 100%, respectively (Lee et al., 2010; Jan et al., 2013). Similarly, overexpression of H+-TH has significantly improved the yields of various NADPH-dependent products, such as chiral alcohols, isobutanol, and 3-hydroxypropionic acid in E. coli (Weckbecker and Hummel, 2004; Bastian et al., 2011; Rathnasingh et al., 2012; Shi et al., 2013) and L-valine and L-lysine in C. glutamicum (Kabus et al., 2007; Bartek et al., 2011).
Ferredoxin:NADP+ oxidoreductase
Ferredoxin:NADP+ oxidoreductases (FNRs, EC:1.18.1.2) are ubiquitous flavoenzymes (Table 1) that catalyze the reversible transfer of reducing equivalents between the one-electron carrier ferredoxin (Fd) and the two-electron carrying NADP(H), according to the following reaction (Figure 6):
| (10) |
FNRs are present in all three domains of life (Ma and Adams, 2001a; Ceccarelli et al., 2004; Santangelo et al., 2011; Yan et al., 2014). They are mainly known for their essential role in photosynthetic organisms (plants, algae, and cyanobacteria), in which they connect the Fdred-generating reactions of photosynthesis to NADPH-requiring carbon assimilation (Shin and Arnon, 1965; Goss and Hanke, 2014). In addition to enabling the flux of electrons from the photosystem into other metabolic pathways, FNRs might also avert this efflux of electrons and mediate the return of Fd-bound electrons to the plastoquinone pool of the photosynthetic system. By doing so, FNRs contribute to the so-called cyclic electron flow, which is essential for balancing the ATP/NADPH ratio generated by the photosystems (Munekage et al., 2004; Shikanai, 2007; Kramer and Evans, 2011; Mulo, 2011; Goss and Hanke, 2014).
In non-photosynthetic tissues and organisms, FNRs participate in the electron transfer chains of various other metabolic processes such as nitrogen fixation, sulfate assimilation, isoprenoid biosynthesis, osmotic and oxidative stress responses, and iron-sulfur cluster biogenesis (Arakaki et al., 1997; Krapp et al., 2002; Seeber et al., 2005; Giro et al., 2006; Lee et al., 2007b; Balconi et al., 2009; Tang et al., 2010; Lewis et al., 2013). However, in contrast to the FNRs involved in photosynthesis, the physiological direction of FNRs involved in these processes is toward the production of Fdred. As a result, FNRs are sometimes classified as autotrophic (all photosynthetic FNRs) and heterotrophic (all other FNRs) (Arakaki et al., 1997), which, from a protein-family point of view, is not entirely correct.
FNRs are generally classified into two phylogenetically and structurally unrelated protein families that are subdivided into four subclasses: the plant-type FNRs comprising the plastidic-type and bacterial-type subclasses and the glutathione (GR)-type FNRs comprising the mitochondrial adrenodoxin reductases (AdR-like) and oxygenase-coupled NADH:ferredoxin reductases (ONFR-like) (Carrillo and Ceccarelli, 2003; Ceccarelli et al., 2004; Karplus and Faber, 2004; Aliverti et al., 2008; Musumeci et al., 2012). In addition, a third type of FNR has been reported, the so-called thioredoxin reductase-like FNR (TRLF), which functions as an FNR but has structural homology to NADPH-dependent thioredoxin reductases (Seo and Sakurai, 2002; Seo et al., 2004; Komori et al., 2010; Muraki et al., 2010; Yan et al., 2014). Interestingly, a novel type of FNR has recently been reported, the electron-bifurcating NADH-dependent reduced ferredoxin:NADP+ oxidoreductase (NfnAB), which couples the exergonic reduction of NADP+ with Fdred and the endergonic reduction of NADP+ with NADH in a reversible reaction (reaction 11 and Figure 6) (Wang et al., 2010; Huang et al., 2012; Rydzak et al., 2014). Genome analyses have revealed the NfnAB genes are present in many anaerobic bacteria and archaea (Huang et al., 2012; Buckel and Thauer, 2013), a finding that might reflect the importance of a postulated NfnAB function: NfnAB and other electron-bifurcating reactions have been implicated as a third mode of energy conservation, in addition to substrate level phosphorylation and electron transport phosphorylation (Buckel and Thauer, 2013).
| (11) |
Despite what the names imply, both subdivisions of the plant-type FNRs (plastidic-type and bacterial-type) as well as FNRs of the GR-type, TRLF-type, and “NfnAB-type” are found in prokaryotes (Ceccarelli et al., 2004; Aliverti et al., 2008; Komori et al., 2010). Unfortunately, the literature does not always make clear the class to which an FNR belongs, generating confusion and making it difficult to judge the true NADPH-generating potential of FNRs found in different prokaryotes. Nevertheless, under normal physiological conditions, it appears that only the plastidic-type FNRs, found in cyanobacteria, for example, and the novel “NfnAB-type” have significant effects on NADPH availability in prokaryotes.
The reaction catalyzed by the different FNR types is in principle reversible, as demonstrated by various in vitro studies, as well as by in vivo studies in cyanobacteria and the thermophilic bacterium Hydrogenobacter thermophilus TK-6. Cyanobacteria contain a single FNR that is responsible for NADP+ reduction in vegetative cells as well as for Fd reduction in heterocysts (Razquin et al., 1996). A similar situation exists in H. thermophilus TK-6, where a single FNR catalyzes either the forward or reverse reaction depending on the specific type of Fd involved in the reaction. A [4Fe-4S]-containing Fd is involved in the forward (NADP+ reducing) reaction, whereas a [2Fe-2S]-containing Fd is involved in the reverse reaction (Ikeda et al., 2009). Such insights might facilitate the development of rational engineering approaches aimed at gaining directional control of the reaction. Elucidating the reaction mechanisms of various FNR-types is a topic of many studies and has been reviewed extensively (Arakaki et al., 1997; Onda et al., 2000; Aliverti et al., 2001, 2008; Carrillo and Ceccarelli, 2003; Medina and Gomez-Moreno, 2004; Martinez-Julvez et al., 2009; Yeom et al., 2009; Musumeci et al., 2012).
FNRs, in general, contain a non-covalently bound flavin (FAD) cofactor as a prosthetic group, but a flavin mononucleotide (FMN)-containing FNR has also been characterized (Ikeda et al., 2009). The FAD group can alternate between an oxidized, one-electron-reduced semiquinone and a fully reduced hydroquinone state, thereby mediating the reversible electron change between the one-electron carrier Fd and the two-electron carrying NADP(H) (Carrillo and Ceccarelli, 2003). The iron-sulfur-containing Fd can sometimes be substituted by the flavin mononucleotide (FMN)-containing flavodoxin (under iron-depleted conditions in some bacteria) or, in the case of the GR-type FNRs, by the mitochondrial iron-sulfur-containing adrenodoxin (Yamazaki and Ichikawa, 1990; Ceccarelli et al., 2004; Sancho, 2006; Ewen et al., 2011). Moreover, a novel rubrerythrin-like protein, named ferriperoxin (Fpx), has been described that exhibits NADPH- and FNR-dependent peroxidase activity in the thermophilic hydrogen-oxidizing bacterium H. thermophilus (Sato et al., 2012). Whereas most FNRs are NADP(H)-dependent, an NAD(H)-dependent variant also exists, the ONFR-like subtype of the GR-type FNRs (EC:1.18.1.3) (Aliverti et al., 2008). However, because this group of FNRs is not involved in NADPH availability, it will not be discussed further in this review.
NADP+-dependent hydrogenase
Hydrogenases (H2ases) are metalloenzymes that catalyze the reversible oxidation of hydrogen gas into two protons and two electrons:
H2ases enable organisms to utilize H2 as a source of reducing power or to use protons as terminal electron acceptors, generating H2. According to the metal atoms at their active site, H2ases can be classified into three types: [NiFe]-H2ases, [FeFe]-H2ases, and [Fe]-H2ases. The latter were formerly known as metal-free or iron-sulfur-free H2ases; they are involved in CO2 reduction with H2 to methane and are only found in methanogenic archaea (Lyon et al., 2004; Vignais and Colbeau, 2004; Shima and Thauer, 2007; Trchounian et al., 2012; Trchounian and Gary Sawers, 2014; Peters et al., 2015). [NiFe]-H2ases are generally responsible for H2 oxidation and are widely distributed among both aerobic and anaerobic bacteria and archaea; they have been identified in 28.9 and 57.8% of the available complete bacterial and archaeal genomes, respectively. [FeFe]-H2ases, in contrast, are generally responsible for H2 evolution and are only found in anaerobic bacteria; they have been identified in 9.8% of the available complete bacterial genomes (Peters et al., 2015). Alignment of the sequence motifs that coordinate the active site, separates [NiFe]-H2ases into at least four, and possibly five, functional groups that are, in general, consistent with the physiological roles of the enzymes (Vignais et al., 2001; Vignais and Colbeau, 2004; Vignais and Billoud, 2007; Peters et al., 2015). Thus, H2ases are a diverse group of enzymes. They are cytosolic or membrane bound, involved in H2 oxidation and/or evolution, and able to use a variety of electron acceptors and donors. It is beyond the scope of this review to address all of the different types of H2ases. We will focus on those involved in NADPH generation in prokaryotes. For more information about H2ases in general, the reader is referred to various excellent reviews, such as those by Vignais and Colbeau (2004), Shima and Thauer (2007), and Peters et al. (2015).
H2ases that use the cofactor NADP+ as an electron acceptor are not common, but have been identified in some bacteria and archaea (Table 1). Cytosolic NADP+-reducing hydrogenases (EC:1.12.1.3, SH) are present in the sulfate-reducing bacterium Desulfovibrio fructosovorans and the hyperthermophilic archaea Pyrococcus furiosus and Thermococcus kodakarensis (Malki et al., 1995; Kanai et al., 2003; Nouailler et al., 2006; Van Haaster et al., 2008). Moreover, cytosolic bidirectional NAD(P)+-H2ases and a membrane-bound NAD(P)+-H2ase, known to use both NAD+ and NADP+ as substrates, have been found in cyanobacteria and Klebsiella pneumonia, respectively (Steuber et al., 1999; Schmitz et al., 2002; Wells et al., 2011). Many of these H2ases recycle the H2 produced by H2-evolving H2ases by coupling the oxidation of H2 to the reduction of NADP+ (Figure 6) (Steuber et al., 1999; Silva et al., 2000; Van Haaster et al., 2008; Kanai et al., 2011; Santangelo et al., 2011; Schut et al., 2012), according to the following reaction:
| (12) |
H2 recycling might have a distinct energetic advantage because the NADP+-reducing H2ase could provide reductant in the form of NADPH without interfering with the energy balance through electron transport phosphorylation (Schut et al., 2012). However, various studies have shown that disruption of NADP+-reducing H2ases has minor effects, suggesting that the role of the enzymes is not that important for overall growth (Casalot et al., 2002; Kanai et al., 2011; Lipscomb et al., 2011; Santangelo et al., 2011; Schut et al., 2012; Lauterbach et al., 2013).
Like most H2ases, NADP+-reducing H2ases require metal ions and cofactors for their activity. They contain multiple iron–sulfur clusters ([Fe–S]), which are coordinated in the active site by carbon monoxide (CO) and cyanide (CN−) groups. Moreover, various H2ases capable of reducing NADP+ belong to the [NiFe]-H2ases, making the availability of Ni essential as well (Ma and Adams, 2001b; Kanai et al., 2003; Germer et al., 2009; Peters et al., 2015). In addition, the enzymes have been reported to contain flavin cofactors: FAD in the case of T. kodakarensis and P. furiosus (Ma et al., 2000; Ma and Adams, 2001b; Kanai et al., 2003) and FMN in the case of D. fructosovorans (Malki et al., 1995).
The requirement for these different ligands, metals, and cofactors makes the biosynthesis of multisubunit H2ases a complex and dynamic process, involving various maturation proteins (Bock et al., 2006; Forzi and Sawers, 2007; Watanabe et al., 2012; Peters et al., 2015). It is thus no surprise that reports about the practical applications of NADP+-reducing H2ases for NADPH generation are limited. Nevertheless, the potential of these enzymes to improve the production yields of NADPH-dependent metabolites has been demonstrated. For example, two prochiral model substrates, acetophenone and (2S)-hydroxy-1-phenyl-propanone, were quantitatively reduced to the corresponding (S)-alcohol and (1R,2S)-diol by an NADPH-dependent alcohol dehydrogenase in an in vitro system containing H2 and the partially purified NADP+-reducing H2ase of P. furiosus (Mertens et al., 2003). Although in vitro examples like this demonstrate the potential of NADP+-H2ases, expression of heterologous [NiFe]-H2ases outside of a closely related host is still challenging (English et al., 2009). It often results in an inactive enzyme or activity that is detectable only in vitro (Voordouw et al., 1987; Grzeszik et al., 1997; Sun et al., 2010). Nevertheless, successful expression of heterologous [NiFe]-H2ase has been reported (Wells et al., 2011).
NAD+ kinase and NADH kinase
NAD+ kinase (NADK, EC:2.7.1.23) catalyzes the conversion of NAD+ to NADP+. It is the sole enzyme leading to de novo NADP+ biosynthesis (Figures 1, 6). As such, NADK is essential for NADP+ and NADPH availability. Together with NADP+ phosphatase (NADPase), which is thought to catalyze the opposite reaction (the conversion of NADP+ to NAD+), NADK is crucial for the regulation of the intracellular balance of NAD(H) and NADP(H) (Kawai and Murata, 2008; Shi et al., 2009). Although NADPase activity has been detected in various organisms (Reidl et al., 2000; Kawai et al., 2004), to our knowledge, no enzyme that functions solely as an NADPase has been isolated to date. However, some enzymes that exhibit NADPase activity in addition to another activity have been isolated, such as the bifunctional NADP+ phosphatase/NAD+ kinase, discovered in the archaeon Methanococcus jannaschii (Kawai et al., 2005; Fukuda et al., 2007).
NADK activity was enriched in extracts from the yeast Saccharomyces cerevisiae for the first time in 1950 (Kornberg, 1950) and has since been purified from various organisms (Mcguinness and Butler, 1985; Magni et al., 1999). However, it was not until 2000 that the gene encoding NADK was identified by Kawai et al. in Micrococcus flavus and Mycobacterium tuberculosis H37Rv (Kawai et al., 2000). Since then, NADKs from various organisms have been studied in detail, and although the exact physiological functions, related pathways, and regulatory mechanisms have not always clearly been elucidated, some reviews of NADK structure, function, and potential applications are available (Magni et al., 2006; Pollak et al., 2007; Kawai and Murata, 2008; Shi et al., 2009; Agledal et al., 2010).
NADKs are ubiquitous enzymes (Table 1). Homologs, according to literature, can be found in almost all eukaryotes and prokaryotes, with the exception of the intracellular parasite Chlamydia trachomatis (Grose et al., 2006; Kawai and Murata, 2008; Szaszák et al., 2011). The discrepancy between the distribution shown in Table 1 and that described in the literature might reflect the rapidly increasing availability of prokaryotic genomes and errors inherent to automated genome annotation (Bakke et al., 2009). Functionally and structurally, NADKs are weakly related to other kinases, suggesting that they belong to a kinase superfamily (Labesse et al., 2002). With the exception of the NADKs of gram-negative bacteria, the enzymes accept NAD+ and its reduced form, NADH (Kawai and Murata, 2008). Nevertheless, they are generally referred to as NAD+ kinases (EC:2.7.1.23) because they usually have a strong preference for the oxidized form. NADKs with a preference for the reduced form exist as well (EC:2.7.1.86) (Outten and Culotta, 2003; Strand et al., 2003; Turner et al., 2005), but have not to our knowledge been found in prokaryotes.
Aside from NAD(H), NADKs also need a phosphoryl donor for catalysis. The preferred phosphoryl donor often is a nucleoside triphosphate, particularly ATP, but many bacterial and archaeal NADKs can also utilize the ancient inorganic energy carrier poly(P) (Kawai et al., 2000, 2005; Garavaglia et al., 2003; Sakuraba et al., 2005). In addition, some bacterial enzymes can utilize ADP or glucose-6-phosphate (Bark et al., 1993; Lindner et al., 2010). However, the use of glucose-6-phosphate as a phosphoryl donor has not been investigated further. Finally, NADKs often also require divalent cations such as Mg2+, Mn2+, Zn2+, or Ca2+ for their activity (Kawai et al., 2000, 2001; Garavaglia et al., 2003; Sakuraba et al., 2005). The NADK of Corynebacterium glutamicum, however, does not require divalent cations when using ATP as phosphoryl donor, but the enzyme is stimulated in their presence. When using poly(P) as phosphoryl donor, divalent cations are required for activity (Lindner et al., 2010).
According to the phosphoryl donor specificity, NADK orthologs are subdivided into several types. The classification typically applied divides the NADK orthologs into: (I) poly(P)/ATP NADKs, which can use poly(P) and ATP as phosphoryl donors and (II) ATP NADKs, which are specific for ATP (Mori et al., 2005; Nakamichi et al., 2013). Poly(P)/ATP NADKs have been identified in gram-positive bacteria such as Mycobacterium tuberculosis, Micrococcus flatus, and Bacillus subtilis (Kawai et al., 2000; Garavaglia et al., 2003) and in archaea such as Methanococcus jannaschii and Pyrococcus horikoshii (Kawai et al., 2005; Sakuraba et al., 2005). ATP NADKs have been found in gram-negative α- and γ-Proteobacteria such as Sphingomonas sp. A1, Salmonella enterica, and E. coli and in eukaryotes (Kawai et al., 2001; Ochiai et al., 2004; Grose et al., 2006; Nakamichi et al., 2013). A slightly different classification system is sometimes applied, dividing the NADK orthologs into: (I) ATP NADKs, which are strictly dependent on ATP, (II) NTP NADKs, which use ATP in addition to other nucleoside triphosphates, and (III) poly(P)/NTP NADKs, which use nucleoside triphosphates and poly(P) (Shi et al., 2009). The existence of these slightly different classification systems can make it difficult to ascertain the type of NADK being discussed. However, because no strict ATP NADKs (as defined in the latter system) have been identified in prokaryotes to date, it is not an issue in this review. The reaction catalyzed by NADKs can be described as either:
| (13) |
or
| (14) |
NADK is the sole enzyme leading to de novo NADP+ biosynthesis. It has a central role in the regulation of the NAD(H)/NADP(H) balance because it can directly phosphorylate NADH to form NADPH or cooperate with various NADP+ reducing enzymes to form NAD(H)/NADP(H) metabolic networks that ultimately convert NADH into NADPH (Singh et al., 2007, 2008; Shi et al., 2009). In view of the importance of NADK, it is not surprising that disruption of NADK is lethal in various prokaryotes (Gerdes et al., 2002; Thanassi et al., 2002; Kobayashi et al., 2003; Sassetti et al., 2003; Zalacain et al., 2003; Grose et al., 2006; Poncet-Montange et al., 2007).
Because of its essential role in NADP+ biosynthesis and the regulation of the NAD(H)/NADP(H) balance, NADKs have attracted attention as a potential strategy by which to improve the biosynthesis of industrially valuable metabolites. Several studies have shown that overexpression of NADK-encoding genes can increase the production yields of various NADPH-dependent biosynthetic pathways, such as poly(3-hydroxybutyrate) production and thymidine production in E. coli (Lee et al., 2009, 2010; Li et al., 2009), lysine and isoleucine production in C. glutamicum (Lindner et al., 2010; Shi et al., 2012), and arginine production in Corynebacterium crenatum (Rahman et al., 2012). These results suggest that increased NADP+ availability, resulting from NADK overexpression, disturbs the intracellular redox balance, subsequently affecting the expression of NADP+-reducing enzymes (Lee et al., 2009; Li et al., 2009; Lee et al., 2010). However, a comparable attempt to improve isobutanol production in E. coli showed that NADK overexpression alone is not always sufficient and that the co-(over)expression of an NADP+ reducing enzyme might be required to increase NADPH availability (Shi et al., 2013). Similarly, combined overexpression of the NADK gene from E. coli and the NADP+-GAPDH gene from Bacillus subtilis in E. coli increased 2-chloropropionic acid production yield (Wang et al., 2013c). In addition to increasing NADPH availability indirectly by overexpressing NADK, a more direct approach has also been applied. For example, the production yields of GDP-L-fucose and ε-caprolactone in E. coli were increased by introducing the NADH kinase (EC:2.7.1.86) gene from the yeast S. cerevisiae (Lee et al., 2013a).
Final remarks and conclusion
NADPH is an essential reducing agent in all organisms. It plays an important role in various biological processes and is the driving force behind numerous biosynthetic enzymatic reactions of importance to industry. However, efficient NADPH regeneration is often a bottleneck that limits the productivity of such biotransformation processes. A thorough understanding of the reactions involved in the generation of NADPH is thus essential to increase the availability of NADPH and enhance productivity in engineered production strains.
This review has provided an overview of the major canonical NADPH-generating reactions as well as recently described NADPH-generating reactions (Table 1), and it has summarized how these different reactions have been applied to improve NADPH availability. However, additional NADPH-generating enzymes, such as gluconate 5-dehydrogenase or the recently discovered NADP+-specific electron-bifurcating [FeFe]-hydrogenase might also contribute to the overall NADPH supply (Klasen et al., 1995; Wang et al., 2013b). Similarly, enzymes generally considered to consume NADPH or enzymes newly developed to regenerate NADPH, such as glutamate dehydrogenase and F420-dependent NADP+ reductases or phosphite dehydrogenase, respectively, can occasionally become important NADPH suppliers (Berk and Thauer, 1997; Lee et al., 2013b; Yokooji et al., 2013).
Although various NADPH-generating enzymes have been overexpressed successfully, no general strategy that guarantees improved NADPH supply exists. Cells usually contain various integrated, network-wide, redox balancing systems, such that engineering of NADPH-generating pathways can result in unexpected metabolic perturbations. Thus, strategies often need to be optimized individually, taking the culture conditions and genetic background of the strains into consideration. Future research involving a systems biology approach is thus required to improve our understanding of the individual NADPH-generating enzymes and their effect on various metabolic backgrounds, in order to predict which approaches will increase NADPH availability in various biotransformation processes.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work is part of the Open Programme (“Biological hydrogen production beyond current limits” file number 820.02.011), which is financed by the Netherlands Organisation for Scientific Research (NWO).
References
- Adachi O., Ameyama M. (1981). d-Glucose dehydrogenase from Gluconobacter suboxydans: solubilization, purification and characterization. Agric. Biol. Chem. 45, 159–163. [Google Scholar]
- Agledal L., Niere M., Ziegler M. (2010). The phosphate makes a difference: cellular functions of NADP. Redox Rep. 15, 2–10. 10.1179/174329210X12650506623122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed H., Ettema T. J. G., Tjaden B., Geerling A. C. M., van der Oost J., Siebers B. (2005). The semi-phosphorylative Entner-Doudoroff pathway in hyperthermophilic archaea: a re-evaluation. Biochem. J. 390, 529–540. 10.1042/BJ20041711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aliverti A., Faber R., Finnerty C. M., Ferioli C., Pandini V., Negri A., et al. (2001). Biochemical and crystallographic characterization of ferredoxin-NADP(+) reductase from nonphotosynthetic tissues. Biochemistry 40, 14501–14508. 10.1021/bi011224c [DOI] [PubMed] [Google Scholar]
- Aliverti A., Pandini V., Pennati A., de Rosa M., Zanetti G. (2008). Structural and functional diversity of ferredoxin-NADP+ reductases. Arch. Biochem. Biophys. 474, 283–291. 10.1016/j.abb.2008.02.014 [DOI] [PubMed] [Google Scholar]
- Amador-Noguez D., Brasg I. A., Feng X. J., Roquet N., Rabinowitz J. D. (2011). Metabolome remodeling during the acidogenic-solventogenic transition in Clostridium acetobutylicum. Appl. Environ. Microbiol. 77, 7984–7997. 10.1128/AEM.05374-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen K. B., von Meyenburg K. (1977). Charges of nicotinamide adenine nucleotides and adenylate energy charge as regulatory parameters of the metabolism in Escherichia coli. J. Biol. Chem. 252, 4151–4156. [PubMed] [Google Scholar]
- Angermayr S. A., Paszota M., Hellingwerf K. J. (2012). Engineering a cyanobacterial cell factory for production of lactic acid. Appl. Environ. Microbiol. 78, 7098–7106. 10.1128/AEM.01587-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoshima M., Igarashi Y. (2008). Nondecarboxylating and decarboxylating isocitrate dehydrogenases: oxalosuccinate reductase as an ancestral form of isocitrate dehydrogenase. J. Bacteriol. 190, 2050–2055. 10.1128/JB.01799-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoshima M., Ishii M., Igarashi Y. (2004). A novel biotin protein required for reductive carboxylation of 2-oxoglutarate by isocitrate dehydrogenase in Hydrogenobacter thermophilus TK-6. Mol. Microbiol. 51, 791–798. 10.1046/j.1365-2958.2003.03863.x [DOI] [PubMed] [Google Scholar]
- Arakaki A. K., Ceccarelli E. A., Carrillo N. (1997). Plant-type ferredoxin-NADP+ reductases: a basal structural framework and a multiplicity of functions. FASEB J. 11, 133–140. [DOI] [PubMed] [Google Scholar]
- Arnér E. S. J., Holmgren A. (2000). Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 267, 6102–6109. 10.1046/j.1432-1327.2000.01701.x [DOI] [PubMed] [Google Scholar]
- Arutyunov D., Schmalhausen E., Orlov V., Rahuel-Clermont S., Nagradova N., Branlant G., et al. (2013). An unusual effect of NADP+ on the thermostability of the nonphosphorylating glyceraldehyde-3-phosphate dehydrogenase from Streptococcus mutans. Biochem. Cell Biol. 91, 295–302. 10.1139/bcb-2012-0104 [DOI] [PubMed] [Google Scholar]
- Asadollahi M. A., Maury J., Patil K. R., Schalk M., Clark A., Nielsen J. (2009). Enhancing sesquiterpene production in Saccharomyces cerevisiae through in silico driven metabolic engineering. Metab. Eng. 11, 328–334. 10.1016/j.ymben.2009.07.001 [DOI] [PubMed] [Google Scholar]
- Bakke P., Carney N., Deloache W., Gearing M., Ingvorsen K., Lotz M., et al. (2009). Evaluation of three automated genome annotations for Halorhabdus utahensis. PLoS ONE 4:e6291. 10.1371/journal.pone.0006291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balconi E., Pennati A., Crobu D., Pandini V., Cerutti R., Zanetti G., et al. (2009). The ferredoxin-NADP+ reductase/ferredoxin electron transfer system of Plasmodium falciparum. FEBS J. 276, 3825–3836. 10.1111/j.1742-4658.2009.07100.x [DOI] [PubMed] [Google Scholar]
- Banerjee S., Nandyala A., Podili R., Katoch V., Hasnain S. (2005). Comparison of Mycobacterium tuberculosis isocitrate dehydrogenases (ICD-1 and ICD-2) reveals differences in coenzyme affinity, oligomeric state, pH tolerance and phylogenetic affiliation. BMC Biochem. 6, 1–14. 10.1186/1471-2091-6-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bark K., Kampfer P., Sponner A., Dott W. (1993). Polyphosphate-dependent enzymes in some coryneform bacteria isolated from sewage sludge. FEMS Microbiol. Lett. 107, 133–138. 10.1111/j.1574-6968.1993.tb06019.x [DOI] [PubMed] [Google Scholar]
- Bartek T., Blombach B., Lang S., Eikmanns B. J., Wiechert W., Oldiges M., et al. (2011). Comparative 13C metabolic flux analysis of pyruvate dehydrogenase complex-deficient, L-valine-producing Corynebacterium glutamicum. Appl. Environ. Microbiol. 77, 6644–6652. 10.1128/AEM.00575-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastian S., Liu X., Meyerowitz J. T., Snow C. D., Chen M. M. Y., Arnold F. H. (2011). Engineered ketol-acid reductoisomerase and alcohol dehydrogenase enable anaerobic 2-methylpropan-1-ol production at theoretical yield in Escherichia coli. Metab. Eng. 13, 345–352. 10.1016/j.ymben.2011.02.004 [DOI] [PubMed] [Google Scholar]
- Becker J., Klopprogge C., Herold A., Zelder O., Bolten C. J., Wittmann C. (2007). Metabolic flux engineering of l-lysine production in Corynebacterium glutamicum—over expression and modification of G6P dehydrogenase. J. Biotechnol. 132, 99–109. 10.1016/j.jbiotec.2007.05.026 [DOI] [PubMed] [Google Scholar]
- Becker J., Klopprogge C., Schroder H., Wittmann C. (2009). Metabolic engineering of the tricarboxylic acid cycle for improved lysine production by Corynebacterium glutamicum. Appl. Environ. Microbiol. 75, 7866–7869. 10.1128/AEM.01942-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J., Klopprogge C., Zelder O., Heinzle E., Wittmann C. (2005). Amplified expression of fructose 1,6-Bisphosphatase in Corynebacterium glutamicum increases in vivo flux through the pentose phosphate pathway and lysine production on different carbon sources. Appl. Environ. Microbiol. 71, 8587–8596. 10.1128/AEM.71.12.8587-8596.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley T. P., Kinsland C., Mehl R. A., Osterman A., Dorrestein P., Gerald Litwack T. B. (2001). The biosynthesis of nicotinamide adenine dinucleotides in bacteria. Vitam. Horm. 61, 103–119. 10.1016/s0083-6729(01)61003-3 [DOI] [PubMed] [Google Scholar]
- Beh M., Strauss G., Huber R., Stetter K.-O., Fuchs G. (1993). Enzymes of the reductive citric acid cycle in the autotrophic eubacterium Aquifex pyrophilus and in the archaebacterium Thermoproteus neutrophilus. Arch. Microbiol. 160, 306–311. 10.1007/BF00292082 [DOI] [Google Scholar]
- Bennett B. D., Kimball E. H., Gao M., Osterhout R., Van Dien S. J., Rabinowitz J. D. (2009). Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 5, 593–599. 10.1038/nchembio.186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk H., Thauer R. K. (1997). Function of coenzyme F420-dependent NADP reductase in methanogenic archaea containing an NADP-dependent alcohol dehydrogenase. Arch. Microbiol. 168, 396–402. 10.1007/s002030050514 [DOI] [PubMed] [Google Scholar]
- Bi J., Wang H., Xie J. (2011). Comparative genomics of NAD(P) biosynthesis and novel antibiotic drug targets. J. Cell. Physiol. 226, 331–340. 10.1002/jcp.22419 [DOI] [PubMed] [Google Scholar]
- Bizouarn T., Althage M., Pedersen A., Tigerström A., Karlsson J., Johansson C., et al. (2002). The organization of the membrane domain and its interaction with the NADP(H)-binding site in proton-translocating transhydrogenase from E. coli. Biochim. Biophys. Acta Bioenerg. 1555, 122–127. 10.1016/S0005-2728(02)00266-9 [DOI] [PubMed] [Google Scholar]
- Bizouarn T., Meuller J., Axelsson M., Rydström J. (2000). The transmembrane domain and the proton channel in proton-pumping transhydrogenases. Biochim. Biophys. Acta Bioenerg. 1459, 284–290. 10.1016/S0005-2728(00)00163-8 [DOI] [PubMed] [Google Scholar]
- Bock A., King P. W., Blokesch M., Posewitz M. C. (2006). Maturation of hydrogenases. Adv. Microb. Physiol. 51, 1–71. 10.1016/S0065-2911(06)51001-X [DOI] [PubMed] [Google Scholar]
- Bologna F. P., Andreo C. S., Drincovich M. F. (2007). Escherichia coli malic enzymes: two isoforms with substantial differences in kinetic properties, metabolic regulation, and structure. J. Bacteriol. 189, 5937–5946. 10.1128/JB.00428-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommareddy R. R., Chen Z., Rappert S., Zeng A.-P. (2014). A de novo NADPH generation pathway for improving lysine production of Corynebacterium glutamicum by rational design of the coenzyme specificity of glyceraldehyde 3-phosphate dehydrogenase. Metab. Eng. 25, 30–37. 10.1016/j.ymben.2014.06.005 [DOI] [PubMed] [Google Scholar]
- Boonstra B., French C. E., Wainwright I., Bruce N. C. (1999). The udhA gene of Escherichia coli encodes a soluble pyridine nucleotide transhydrogenase. J. Bacteriol. 181, 1030–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonstra B., Rathbone D. A., French C. E., Walker E. H., Bruce N. C. (2000). Cofactor regeneration by a soluble pyridine nucleotide transhydrogenase for biological production of hydromorphone. Appl. Environ. Microbiol. 66, 5161–5166. 10.1128/AEM.66.12.5161-5166.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd D. A., Cvitkovitch D. G., Hamilton I. R. (1995). Sequence, expression, and function of the gene for the nonphosphorylating, NADP-dependent glyceraldehyde-3-phosphate dehydrogenase of Streptococcus mutans. J. Bacteriol. 177, 2622–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragg P. D., Davies P. L., Hou C. (1972). Function of energy-dependent transhydrogenase in Escherichia coli. Biochem. Biophys. Res. Commun. 47, 1248–1255. 10.1016/0006-291X(72)90969-2 [DOI] [PubMed] [Google Scholar]
- Brasen C., Esser D., Rauch B., Siebers B. (2014). Carbohydrate metabolism in Archaea: current insights into unusual enzymes and pathways and their regulation. Microbiol. Mol. Biol. Rev. 78, 89–175. 10.1128/MMBR.00041-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D. I., Griendling K. K. (2009). Nox proteins in signal transduction. Free Radic. Biol. Med. 47, 1239–1253. 10.1016/j.freeradbiomed.2009.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner N. A., Brinkmann H., Siebers B., Hensel R. (1998). NAD+-dependent glyceraldehyde-3-phosphate dehydrogenase from Thermoproteus tenax. The first identified archaeal member of the aldehyde dehydrogenase superfamily is a glycolytic enzyme with unusual regulatory properties. J. Biol. Chem. 273, 6149–6156. 10.1074/jbc.273.11.6149 [DOI] [PubMed] [Google Scholar]
- Brunner N. A., Hensel R. (2001). Nonphosphorylating glyceraldehyde-3-phosphate dehydrogenase from Thermoproteus tenax. Methods Enzymol. 331, 117–131. 10.1016/S0076-6879(01)31051-0 [DOI] [PubMed] [Google Scholar]
- Brunner N. A., Siebers B., Hensel R. (2001). Role of two different glyceraldehyde-3-phosphate dehydrogenases in controlling the reversible Embden-Meyerhof-Parnas pathway in Thermoproteus tenax: regulation on protein and transcript level. Extremophiles 5, 101–109. 10.1007/s007920100181 [DOI] [PubMed] [Google Scholar]
- Buckel W., Thauer R. K. (2013). Energy conservation via electron bifurcating ferredoxin reduction and proton/Na(+) translocating ferredoxin oxidation. Biochim. Biophys. Acta 1827, 94–113. 10.1016/j.bbabio.2012.07.002 [DOI] [PubMed] [Google Scholar]
- Butler J. R., Mcguinness E. T. (1982). Candida utilis NAD+ kinase: purification, properties and affinity gel studies. Int. J. Biochem. 14, 839–844. 10.1016/0020-711X(82)90106-9 [DOI] [PubMed] [Google Scholar]
- Butler M. J., Bruheim P., Jovetic S., Marinelli F., Postma P. W., Bibb M. J. (2002). Engineering of primary carbon metabolism for improved antibiotic production in Streptomyces lividans. Appl. Environ. Microbiol. 68, 4731–4739. 10.1128/AEM.68.10.4731-4739.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bylund J., Brown K. L., Movitz C., Dahlgren C., Karlsson A. (2010). Intracellular generation of superoxide by the phagocyte NADPH oxidase: how, where, and what for? Free Radic. Biol. Med. 49, 1834–1845. 10.1016/j.freeradbiomed.2010.09.016 [DOI] [PubMed] [Google Scholar]
- Canonaco F., Hess T. A., Heri S., Wang T., Szyperski T., Sauer U. (2001). Metabolic flux response to phosphoglucose isomerase knock-out in Escherichia coli and impact of overexpression of the soluble transhydrogenase UdhA. FEMS Microbiol. Lett. 204, 247–252. 10.1111/j.1574-6968.2001.tb10892.x [DOI] [PubMed] [Google Scholar]
- Cao Z., Song P., Xu Q., Su R., Zhu G. (2011). Overexpression and biochemical characterization of soluble pyridine nucleotide transhydrogenase from Escherichia coli. FEMS Microbiol. Lett. 320, 9–14. 10.1111/j.1574-6968.2011.02287.x [DOI] [PubMed] [Google Scholar]
- Carrillo N., Ceccarelli E. A. (2003). Open questions in ferredoxin-NADP+ reductase catalytic mechanism. Eur. J. Biochem. 270, 1900–1915. 10.1046/j.1432-1033.2003.03566.x [DOI] [PubMed] [Google Scholar]
- Casalot L., De Luca G., Dermoun Z., Rousset M., de Philip P. (2002). Evidence for a fourth hydrogenase in Desulfovibrio fructosovorans. J. Bacteriol. 184, 853–856. 10.1128/JB.184.3.853-856.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccarelli E. A., Arakaki A. K., Cortez N., Carrillo N. (2004). Functional plasticity and catalytic efficiency in plant and bacterial ferredoxin-NADP(H) reductases. Biochim. Biophys. Acta 1698, 155–165. 10.1016/j.bbapap.2003.12.005 [DOI] [PubMed] [Google Scholar]
- Celton M., Goelzer A., Camarasa C., Fromion V., Dequin S. (2012). A constraint-based model analysis of the metabolic consequences of increased NADPH oxidation in Saccharomyces cerevisiae. Metab. Eng. 14, 366–379. 10.1016/j.ymben.2012.03.008 [DOI] [PubMed] [Google Scholar]
- Centeno-Leija S., Huerta-Beristain G., Giles-Gomez M., Bolivar F., Gosset G., Martinez A. (2014). Improving poly-3-hydroxybutyrate production in Escherichia coli by combining the increase in the NADPH pool and acetyl-CoA availability. Antonie Van Leeuwenhoek 105, 687–696. 10.1007/s10482-014-0124-5 [DOI] [PubMed] [Google Scholar]
- Centeno-Leija S., Utrilla J., Flores N., Rodriguez A., Gosset G., Martinez A. (2013). Metabolic and transcriptional response of Escherichia coli with a NADP(+)-dependent glyceraldehyde 3-phosphate dehydrogenase from Streptococcus mutans. Antonie Van Leeuwenhoek 104, 913–924. 10.1007/s10482-013-0010-6 [DOI] [PubMed] [Google Scholar]
- Chang D. Y., Hou C., Bragg P. D. (1992). Anomalous effect of uncouplers on respiratory chain-linked transhydrogenation in Escherichia coli membranes: evidence for a localized proton pathway? Arch. Biochem. Biophys. 293, 246–253. 10.1016/0003-9861(92)90392-A [DOI] [PubMed] [Google Scholar]
- Chang G.-G., Tong L. (2003). Structure and function of malic enzymes, a new class of oxidative decarboxylases. Biochemistry 42, 12721–12733. 10.1021/bi035251+ [DOI] [PubMed] [Google Scholar]
- Chemler J. A., Fowler Z. L., Mchugh K. P., Koffas M. A. G. (2010). Improving NADPH availability for natural product biosynthesis in Escherichia coli by metabolic engineering. Metab. Eng. 12, 96–104. 10.1016/j.ymben.2009.07.003 [DOI] [PubMed] [Google Scholar]
- Chen R., Jeong S. S. (2000). Functional prediction: identification of protein orthologs and paralogs. Protein Sci. 9, 2344–2353. 10.1110/ps.9.12.2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenault H. K., Simon E. S., Whitesides G. M. (1988). Cofactor regeneration for enzyme-catalysed synthesis. Biotechnol. Genet. Eng. Rev. 6, 221–270. 10.1080/02648725.1988.10647849 [DOI] [PubMed] [Google Scholar]
- Chenault H. K., Whitesides G. M. (1987). Regeneration of nicotinamide cofactors for use in organic synthesis. Appl. Biochem. Biotechnol. 14, 147–197. 10.1007/BF02798431 [DOI] [PubMed] [Google Scholar]
- Chin J. W., Cirino P. C. (2011). Improved NADPH supply for xylitol production by engineered Escherichia Coli with glycolytic mutations. Biotechnol. Prog. 27, 333–341. 10.1002/btpr.559 [DOI] [PubMed] [Google Scholar]
- Chin J. W., Khankal R., Monroe C. A., Maranas C. D., Cirino P. C. (2009). Analysis of NADPH supply during xylitol production by engineered Escherichia coli. Biotechnol. Bioeng. 102, 209–220. 10.1002/bit.22060 [DOI] [PubMed] [Google Scholar]
- Chini E. N., De Toledo F. G. S. (2002). Nicotinic acid adenine dinucleotide phosphate: a new intracellular second messenger? Am. J. Physiol. Cell Physiol. 282, C1191–C1198. 10.1152/ajpcell.00475.2001 [DOI] [PubMed] [Google Scholar]
- Choi J.-C., Shin H.-D., Lee Y.-H. (2003). Modulation of 3-hydroxyvalerate molar fraction in poly(3-hydroxybutyrate-3-hydroxyvalerate) using Ralstonia eutropha transformant co-amplifying phbC and NADPH generation-related zwf genes. Enzyme Microb. Technol. 32, 178–185. 10.1016/S0141-0229(02)00274-0 [DOI] [Google Scholar]
- Christiansen T., Christensen B., Nielsen J. (2002). Metabolic network analysis of Bacillus clausii on minimal and semirich medium using (13)C-labeled glucose. Metab. Eng. 4, 159–169. 10.1006/mben.2001.0219 [DOI] [PubMed] [Google Scholar]
- Churamani D., Carrey E. A., Dickinson G. D., Patel S. (2004). Determination of cellular nicotinic acid-adenine dinucleotide phosphate (NAADP) levels. Biochem. J. 380, 449–454. 10.1042/BJ20031754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P. T., Kaplan N. O. (1970). Purification and properties of the pyridine nucleotide transhydrogenase from Pseudomonas aeruginosa. J. Biol. Chem. 245, 2825–2836. [PubMed] [Google Scholar]
- Conway T. (1992). The entner-doudoroff pathway: history, physiology and molecular biology. FEMS Microbiol. Rev. 9, 1–27. 10.1111/j.1574-6968.1992.tb05822.x [DOI] [PubMed] [Google Scholar]
- Cotton N. P., Lever T. M., Nore B. F., Jones M. R., Jackson J. B. (1989). The coupling between protonmotive force and the NAD(P)+ transhydrogenase in chromatophores from photosynthetic bacteria. Eur. J. Biochem. 182, 593–603. 10.1111/j.1432-1033.1989.tb14868.x [DOI] [PubMed] [Google Scholar]
- Cozzone A. J. (1998). Regulation of acetate metabolism by protein phosphorylation in enteric bacteria. Annu. Rev. Microbiol. 52, 127–164. 10.1146/annurev.micro.52.1.127 [DOI] [PubMed] [Google Scholar]
- Crow V. L., Wittenberger C. L. (1979). Separation and properties of NAD+- and NADP+-dependent glyceraldehyde-3-phosphate dehydrogenases from Streptococcus mutans. J. Biol. Chem. 254, 1134–1142. [PubMed] [Google Scholar]
- Dauner M., Storni T., Sauer U. (2001). Bacillus subtilis metabolism and energetics in carbon-limited and excess-carbon chemostat culture. J. Bacteriol. 183, 7308–7317. 10.1128/JB.183.24.7308-7317.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis W. B. (1980). Identification of a nicotinamide adenine dinucleotide glycohydrolase and an associated inhibitor in isoniazid-susceptible and -resistant Mycobacterium phlei. Antimicrob. Agents Chemother. 17, 663–668. 10.1128/AAC.17.4.663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decorosi F., Lori L., Santopolo L., Tatti E., Giovannetti L., Viti C. (2011). Characterization of a Cr(VI)-sensitive Pseudomonas corrugata 28 mutant impaired in a pyridine nucleotide transhydrogenase gene. Res. Microbiol. 162, 747–755. 10.1016/j.resmic.2011.06.014 [DOI] [PubMed] [Google Scholar]
- Driscoll B. T., Finan T. M. (1997). Properties of NAD(+)- and NADP(+)-dependent malic enzymes of Rhizobium (Sinorhizobium) meliloti and differential expression of their genes in nitrogen-fixing bacteroids. Microbiology 143(Pt 2), 489–498. 10.1099/00221287-143-2-489 [DOI] [PubMed] [Google Scholar]
- Eguchi T., Kuge Y., Inoue K., Yoshikawa N., Mochida K., Uwajima T. (1992). NADPH regeneration by glucose dehydrogenase from Gluconobacter scleroides for l-leucovorin synthesis. Biosci. Biotechnol. Biochem. 56, 701–703. 10.1271/bbb.56.701 [DOI] [PubMed] [Google Scholar]
- Eikmanns B. J., Rittmann D., Sahm H. (1995). Cloning, sequence analysis, expression, and inactivation of the Corynebacterium glutamicum icd gene encoding isocitrate dehydrogenase and biochemical characterization of the enzyme. J. Bacteriol. 177, 774–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English C. M., Eckert C., Brown K., Seibert M., King P. W. (2009). Recombinant and in vitro expression systems for hydrogenases: new frontiers in basic and applied studies for biological and synthetic H2 production. Dalton Trans. 9970–9978. 10.1039/b913426n [DOI] [PubMed] [Google Scholar]
- Espariz M., Repizo G., Blancato V., Mortera P., Alarcon S., Magni C. (2011). Identification of malic and soluble oxaloacetate decarboxylase enzymes in Enterococcus faecalis. FEBS J. 278, 2140–2151. 10.1111/j.1742-4658.2011.08131.x [DOI] [PubMed] [Google Scholar]
- Esser D., Kouril T., Talfournier F., Polkowska J., Schrader T., Bräsen C., et al. (2013). Unraveling the function of paralogs of the aldehyde dehydrogenase super family from Sulfolobus solfataricus. Extremophiles 17, 205–216. 10.1007/s00792-012-0507-3 [DOI] [PubMed] [Google Scholar]
- Ettema T. J. G., Ahmed H., Geerling A. C. M., Van Der Oost J., Siebers B. (2008). The non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) of Sulfolobus solfataricus: a key-enzyme of the semi-phosphorylative branch of the Entner-Doudoroff pathway. Extremophiles 12, 75–88. 10.1007/s00792-007-0082-1 [DOI] [PubMed] [Google Scholar]
- Everse J., Griffin J. B., Kaplan N. O. (1975). The pyridine nucleosidase from Bacillus subtilis. Kinetic properties and enzyme-inhibitor interactions. Arch. Biochem. Biophys. 169, 714–723. 10.1016/0003-9861(75)90216-7 [DOI] [PubMed] [Google Scholar]
- Ewen K. M., Kleser M., Bernhardt R. (2011). Adrenodoxin: the archetype of vertebrate-type [2Fe–2S] cluster ferredoxins. Biochim. Biophys. Acta Proteins Proteomics 1814, 111–125. 10.1016/j.bbapap.2010.06.003 [DOI] [PubMed] [Google Scholar]
- Fabris M., Matthijs M., Rombauts S., Vyverman W., Goossens A., Baart G. J. E. (2012). The metabolic blueprint of Phaeodactylum tricornutum reveals a eukaryotic Entner-Doudoroff glycolytic pathway. Plant J. 70, 1004–1014. 10.1111/j.1365-313X.2012.04941.x [DOI] [PubMed] [Google Scholar]
- Ferri S., Kojima K., Sode K. (2011). Review of glucose oxidases and glucose dehydrogenases: a bird's eye view of glucose sensing enzymes. J. Diabetes Sci. Technol. 5, 1068–1076. 10.1177/193229681100500507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillinger S., Boschi-Muller S., Azza S. D., Dervyn E., Branlant G., Aymerich S. (2000). Two Glyceraldehyde-3-phosphate dehydrogenases with opposite physiological roles in a nonphotosynthetic bacterium. J. Biol. Chem. 275, 14031–14037. 10.1074/jbc.275.19.14031 [DOI] [PubMed] [Google Scholar]
- Fischer E., Sauer U. (2003). A novel metabolic cycle catalyzes glucose oxidation and anaplerosis in hungry Escherichia coli. J. Biol. Chem. 278, 46446–46451. 10.1074/jbc.M307968200 [DOI] [PubMed] [Google Scholar]
- Flamholz A., Noor E., Bar-Even A., Milo R. (2012). eQuilibrator—the biochemical thermodynamics calculator. Nucleic Acids Res. 40, D770–D775. 10.1093/nar/gkr874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foreman J., Demidchik V., Bothwell J. H. F., Mylona P., Miedema H., Torres M. A., et al. (2003). Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422, 442–446. 10.1038/nature01485 [DOI] [PubMed] [Google Scholar]
- Forzi L., Sawers R. G. (2007). Maturation of [NiFe]-hydrogenases in Escherichia coli. Biometals 20, 565–578. 10.1007/s10534-006-9048-5 [DOI] [PubMed] [Google Scholar]
- French C. E., Boonstra B., Bufton K. A., Bruce N. C. (1997). Cloning, sequence, and properties of the soluble pyridine nucleotide transhydrogenase of Pseudomonas fluorescens. J. Bacteriol. 179, 2761–2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel R. (1975). Regulation and physiological functions of malic enzymes. Curr. Top. Cell. Regul. 9, 157–181. 10.1016/B978-0-12-152809-6.50012-3 [DOI] [PubMed] [Google Scholar]
- Fuhrer T., Fischer E., Sauer U. (2005). Experimental identification and quantification of glucose metabolism in seven bacterial species. J. Bacteriol. 187, 1581–1590. 10.1128/JB.187.5.1581-1590.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrer T., Sauer U. (2009). Different biochemical mechanisms ensure network-wide balancing of reducing equivalents in microbial metabolism. J. Bacteriol. 191, 2112–2121. 10.1128/JB.01523-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda C., Kawai S., Murata K. (2007). NADP(H) Phosphatase activities of archaeal inositol monophosphatase and eubacterial 3′-Phosphoadenosine 5′-Phosphate phosphatase. Appl. Environ. Microbiol. 73, 5447–5452. 10.1128/AEM.02703-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda W., Ismail Y. S., Fukui T., Atomi H., Imanaka T. (2005). Characterization of an archaeal malic enzyme from the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. Archaea 1, 293–301. 10.1155/2005/250757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garavaglia S., Galizzi A., Rizzi M. (2003). Allosteric regulation of Bacillus subtilis NAD kinase by quinolinic acid. J. Bacteriol. 185, 4844–4850. 10.1128/JB.185.16.4844-4850.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido-Pertierra A., Martinez Marcos C., Martin Fernandez M., Ruiz-Amil M. (1983). Properties and function of malate enzyme from Pseudomonas putida. Biochimie 65, 629–635. 10.1016/S0300-9084(84)80026-7 [DOI] [PubMed] [Google Scholar]
- Gazzaniga F., Stebbins R., Chang S. Z., Mcpeek M. A., Brenner C. (2009). Microbial NAD metabolism: lessons from comparative genomics. Microbiol. Mol. Biol. Rev. 73, 529–541. 10.1128/MMBR.00042-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes S. Y., Scholle M. D., D'souza M., Bernal A., Baev M. V., Farrell M., et al. (2002). From genetic footprinting to antimicrobial drug targets: examples in cofactor biosynthetic pathways. J. Bacteriol. 184, 4555–4572. 10.1128/JB.184.16.4555-4572.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germer F., Zebger I., Saggu M., Lendzian F., Schulz R., Appel J. (2009). Overexpression, isolation, and spectroscopic characterization of the bidirectional [NiFe] hydrogenase from Synechocystis sp. PCC 6803. J. Biol. Chem. 284, 36462–36472. 10.1074/jbc.M109.028795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh J., Anderson P. J., Chandrasekaran S., Caparon M. G. (2010). Characterization of Streptococcus pyogenes beta-NAD+ glycohydrolase: re-evaluation of enzymatic properties associated with pathogenesis. J. Biol. Chem. 285, 5683–5694. 10.1074/jbc.M109.070300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giro M., Carrillo N., Krapp A. R. (2006). Glucose-6-phosphate dehydrogenase and ferredoxin-NADP(H) reductase contribute to damage repair during the soxRS response of Escherichia coli. Microbiology 152, 1119–1128. 10.1099/mic.0.28612-0 [DOI] [PubMed] [Google Scholar]
- Goss T., Hanke G. (2014). The end of the line: can ferredoxin and ferredoxin NADP(H) oxidoreductase determine the fate of photosynthetic electrons? Curr. Protein Pept. Sci. 15, 385–393. 10.2174/1389203715666140327113733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossmann T. I., Ziegler M., Puntervoll P., De Figueiredo L. F., Schuster S., Heiland I. (2012). NAD+ biosynthesis and salvage - a phylogenetic perspective. FEBS J. 279, 3355–3363. 10.1111/j.1742-4658.2012.08559.x [DOI] [PubMed] [Google Scholar]
- Gourdon P., Baucher M. F., Lindley N. D., Guyonvarch A. (2000). Cloning of the malic enzyme gene from Corynebacterium glutamicum and role of the enzyme in lactate metabolism. Appl. Environ. Microbiol. 66, 2981–2987. 10.1128/AEM.66.7.2981-2987.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grose J. H., Joss L., Velick S. F., Roth J. R. (2006). Evidence that feedback inhibition of NAD kinase controls responses to oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 103, 7601–7606. 10.1073/pnas.0602494103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzeszik C., Lubbers M., Reh M., Schlegel H. G. (1997). Genes encoding the NAD-reducing hydrogenase of Rhodococcus opacus MR11. Microbiology 143(Pt 4), 1271–1286. 10.1099/00221287-143-4-1271 [DOI] [PubMed] [Google Scholar]
- Guo Z.-P., Zhang L., Ding Z.-Y., Wang Z.-X., Shi G.-Y. (2011). Improving ethanol productivity by modification of glycolytic redox factor generation in glycerol-3-phosphate dehydrogenase mutants of an industrial ethanol yeast. J. Ind. Microbiol. Biotechnol. 38, 935–943. 10.1007/s10295-010-0864-9 [DOI] [PubMed] [Google Scholar]
- Hansen E. J., Juni E. (1975). Isolation of mutants of Escherichia coli lacking NAD- and NADP-linked malic. Biochem. Biophys. Res. Commun. 65, 559–566. 10.1016/S0006-291X(75)80183-5 [DOI] [PubMed] [Google Scholar]
- Harold F. M. (1986). The Vital Force: A Study of Bioenergetics. New York, NY: W. H. Freeman. [Google Scholar]
- He L., Xiao Y., Gebreselassie N., Zhang F., Antoniewicz M. R., Tang Y. J., et al. (2014). Central metabolic responses to the overproduction of fatty acids in Escherichia coli based on 13C-metabolic flux analysis. Biotechnol. Bioeng. 111, 575–585. 10.1002/bit.25124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman J. W., Barber R. D., Skaar E. P., Donohue T. J. (2002). Link between the membrane-bound pyridine nucleotide transhydrogenase and glutathione-dependent processes in Rhodobacter sphaeroides. J. Bacteriol. 184, 400–409. 10.1128/JB.184.2.400-409.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek J. B., Rydstrom J. (1988). Physiological roles of nicotinamide nucleotide transhydrogenase. Biochem. J. 254, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hojeberg B., Rydstrom J. (1977). Ca2+-dependent allosteric regulation of nicotinamide nucleotide transhydrogenase from Pseudomonas aeruginosa. Eur. J. Biochem. 77, 235–241. 10.1111/j.1432-1033.1977.tb11662.x [DOI] [PubMed] [Google Scholar]
- Hong S. H., Lee S. Y. (2001). Metabolic flux analysis for succinic acid production by recombinant Escherichia coli with amplified malic enzyme activity. Biotechnol. Bioeng. 74, 89–95. 10.1002/bit.1098 [DOI] [PubMed] [Google Scholar]
- Hsieh J.-Y., Hung H.-C. (2009). Engineering of the cofactor specificities and isoform-specific inhibition of malic enzyme. J. Biol. Chem. 284, 4536–4544. 10.1074/jbc.M807008200 [DOI] [PubMed] [Google Scholar]
- Hua Q., Yang C., Baba T., Mori H., Shimizu K. (2003). Responses of the central metabolism in Escherichia coli to phosphoglucose isomerase and glucose-6-phosphate dehydrogenase knockouts. J. Bacteriol. 185, 7053–7067. 10.1128/JB.185.24.7053-7067.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Wang S., Moll J., Thauer R. K. (2012). Electron bifurcation involved in the energy metabolism of the acetogenic bacterium Moorella thermoacetica growing on glucose or H2 plus CO2. J. Bacteriol. 194, 3689–3699. 10.1128/JB.00385-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M., Wang Y., Liu J., Mao Z. (2011). Multiple strategies for metabolic engineering of Escherichia coli for efficient production of coenzyme. Chin. J. Chem. Eng. 19, 316–326. 10.1016/S1004-9541(11)60171-7 [DOI] [Google Scholar]
- Hügler M., Menendez C., Schägger H., Fuchs G. (2002). Malonyl-coenzyme a reductase from Chloroflexus aurantiacus, a key enzyme of the 3-Hydroxypropionate cycle for autotrophic CO2 Fixation. J. Bacteriol. 184, 2404–2410. 10.1128/JB.184.9.2404-2410.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iddar A., Valverde F., Assobhei O., Serrano A., Soukri A. (2005). Widespread occurrence of non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase among gram-positive bacteria. Int. Microbiol. 8, 251–258. [PubMed] [Google Scholar]
- Iddar A., Valverde F., Serrano A., Soukri A. (2002). Expression, purification, and characterization of recombinant nonphosphorylating NADP-dependent glyceraldehyde-3-phosphate dehydrogenase from Clostridium acetobutylicum. Protein Expr. Purif. 25, 519–526. 10.1016/S1046-5928(02)00032-3 [DOI] [PubMed] [Google Scholar]
- Iddar A., Valverde F., Serrano A., Soukri A. (2003). Purification of recombinant non-phosphorylating NADP-dependent glyceraldehyde-3-phosphate dehydrogenase from Streptococcus pyogenes expressed in E. coli. Mol. Cell. Biochem. 247, 195–203. 10.1023/A:1024112027440 [DOI] [PubMed] [Google Scholar]
- Ikeda T., Nakamura M., Arai H., Ishii M., Igarashi Y. (2009). Ferredoxin-NADP reductase from the thermophilic hydrogen-oxidizing bacterium, Hydrogenobacter thermophilus TK-6. FEMS Microbiol. Lett. 297, 124–130. 10.1111/j.1574-6968.2009.01667.x [DOI] [PubMed] [Google Scholar]
- Imam S., Noguera D. R., Donohue T. J. (2013). Global insights into energetic and metabolic networks in Rhodobacter sphaeroides. BMC Syst. Biol. 7:89. 10.1186/1752-0509-7-89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H., Tamura T., Ehara N., Nishito A., Nakayama Y., Maekawa M., et al. (2002). Biochemical and molecular characterization of the NAD(+)-dependent isocitrate dehydrogenase from the chemolithotroph Acidithiobacillus thiooxidans. FEMS Microbiol. Lett. 214, 127–132. 10.1111/j.1574-6968.2002.tb11335.x [DOI] [PubMed] [Google Scholar]
- Ito F., Chishiki H., Fushinobu S., Wakagi T. (2012). Comparative analysis of two glyceraldehyde-3-phosphate dehydrogenases from a thermoacidophilic archaeon, Sulfolobus tokodaii. FEBS Lett. 586, 3097–3103. 10.1016/j.febslet.2012.07.059 [DOI] [PubMed] [Google Scholar]
- Iwakura M., Tokushige M., Katsuki H., Muramatsu S. (1978). Studies on regulatory functions of malic enzymes. V. Comparative studies of malic enzymes in bacteria. J. Biochem. 83, 1387–1394. [DOI] [PubMed] [Google Scholar]
- Jackson B., Brocker C., Thompson D. C., Black W., Vasiliou K., Nebert D. W., et al. (2011). Update on the aldehyde dehydrogenase gene (ALDH) superfamily. Hum. Genomics 5, 283–303. 10.1186/1479-7364-5-4-283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson J. B. (2003). Proton translocation by transhydrogenase. FEBS Lett. 545, 18–24. 10.1016/S0014-5793(03)00388-0 [DOI] [PubMed] [Google Scholar]
- Jackson J. B. (2012). A review of the binding-change mechanism for proton-translocating transhydrogenase. Biochim. Biophys. Acta 1817, 1839–1846. 10.1016/j.bbabio.2012.04.006 [DOI] [PubMed] [Google Scholar]
- Jackson J. B., Obiozo U. M. (2009). Proton-translocating transhydrogenase in photosynthetic bacteria, in The Purple Phototrophic Bacteria, eds Hunter C. N., Daldal F., Thurnauer M., Beatty J. T. (Dordrecht: Springer; ), 495–508. [Google Scholar]
- Jackson J. B., Quirk P. G., Cotton N. P., Venning J. D., Gupta S., Bizouarn T., et al. (1998). Interdomain hydride transfer in proton-translocating transhydrogenase. Biochim. Biophys. Acta 1365, 79–86. 10.1016/S0005-2728(98)00046-2 [DOI] [PubMed] [Google Scholar]
- Jain M. K., Datta R., Zeikus J. G. (1989). High-value organic acids fermentation - emerging processes and products, in Bioprocess Engineering: The First Generation, ed Ghose T. K. (Chichester: Ellis Horwood Ltd; ), 366–389. [Google Scholar]
- Jan J., Martinez I., Wang Y., Bennett G. N., San K.-Y. (2013). Metabolic engineering and transhydrogenase effects on NADPH availability in Escherichia coli. Biotechnol. Prog. 29, 1124–1130. 10.1002/btpr.1765 [DOI] [PubMed] [Google Scholar]
- Jiang L.-Y., Zhang Y.-Y., Li Z., Liu J.-Z. (2013). Metabolic engineering of Corynebacterium glutamicum for increasing the production of L-ornithine by increasing NADPH availability. J. Ind. Microbiol. Biotechnol. 40, 1143–1151. 10.1007/s10295-013-1306-2 [DOI] [PubMed] [Google Scholar]
- Jiao Z., Baba T., Mori H., Shimizu K. (2003). Analysis of metabolic and physiological responses to gnd knockout in Escherichia coli by using C-13 tracer experiment and enzyme activity measurement. FEMS Microbiol. Lett. 220, 295–301. 10.1016/S0378-1097(03)00133-2 [DOI] [PubMed] [Google Scholar]
- Jonsbu E., Christensen B., Nielsen J. (2001). Changes of in vivo fluxes through central metabolic pathways during the production of nystatin by Streptomyces noursei in batch culture. Appl. Microbiol. Biotechnol. 56, 93–100. 10.1007/s002530100613 [DOI] [PubMed] [Google Scholar]
- Kabir M. M., Shimizu K. (2003). Fermentation characteristics and protein expression patterns in a recombinant Escherichia coli mutant lacking phosphoglucose isomerase for poly(3-hydroxybutyrate) production. Appl. Microbiol. Biotechnol. 62, 244–255. 10.1007/s00253-003-1257-z [DOI] [PubMed] [Google Scholar]
- Kabir M. M., Shimizu K. (2004). Metabolic regulation analysis of icd-gene knockout Escherichia coli based on 2D electrophoresis with MALDI-TOF mass spectrometry and enzyme activity measurements. Appl. Microbiol. Biotechnol. 65, 84–96. 10.1007/s00253-004-1627-1 [DOI] [PubMed] [Google Scholar]
- Kabus A., Georgi T., Wendisch V. F., Bott M. (2007). Expression of the Escherichia coli pntAB genes encoding a membrane-bound transhydrogenase in Corynebacterium glutamicum improves L-lysine formation. Appl. Microbiol. Biotechnol. 75, 47–53. 10.1007/s00253-006-0804-9 [DOI] [PubMed] [Google Scholar]
- Kanai T., Ito S., Imanaka T. (2003). Characterization of a cytosolic NiFe-hydrogenase from the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Bacteriol. 185, 1705–1711. 10.1128/JB.185.5.1705-1711.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai T., Matsuoka R., Beppu H., Nakajima A., Okada Y., Atomi H., et al. (2011). Distinct physiological roles of the three [NiFe]-hydrogenase orthologs in the hyperthermophilic archaeon Thermococcus kodakarensis. J. Bacteriol. 193, 3109–3116. 10.1128/JB.01072-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanao T., Kawamura M., Fukui T., Atomi H., Imanaka T. (2002). Characterization of isocitrate dehydrogenase from the green sulfur bacterium Chlorobium limicola. A carbon dioxide-fixing enzyme in the reductive tricarboxylic acid cycle. Eur. J. Biochem./FEBS 269, 1926–1931. 10.1046/j.1432-1033.2002.02849.x [DOI] [PubMed] [Google Scholar]
- Kao K. C., Tran L. M., Liao J. C. (2005). A global regulatory role of gluconeogenic genes in Escherichia coli revealed by transcriptome network analysis. J. Biol. Chem. 280, 36079–36087. 10.1074/jbc.M508202200 [DOI] [PubMed] [Google Scholar]
- Karplus P. A., Faber H. R. (2004). Structural aspects of plant ferredoxin: NADP(+) oxidoreductases. Photosyn. Res. 81, 303–315. 10.1023/B:PRES.0000036884.57303.2e [DOI] [PubMed] [Google Scholar]
- Kataoka M., Sri Rohani L. P., Wada M., Kita K., Yanase H., Urabe I., et al. (1998). Escherichia coli transformant expressing the glucose dehydrogenase gene from Bacillus megaterium as a cofactor regenerator in a chiral alcohol production system. Biosci. Biotechnol. Biochem. 62, 167–169. 10.1271/bbb.62.167 [DOI] [PubMed] [Google Scholar]
- Kataoka M., Yamamoto K., Kawabata H., Wada M., Kita K., Yanase H., et al. (1999). Stereoselective reduction of ethyl 4-chloro-3-oxobutanoate by Escherichia coli transformant cells coexpressing the aldehyde reductase and glucose dehydrogenase genes. Appl. Microbiol. Biotechnol. 51, 486–490. 10.1007/s002530051421 [DOI] [PubMed] [Google Scholar]
- Kawai S., Fukuda C., Mukai T., Murata K. (2005). MJ0917 in archaeon Methanococcus jannaschii is a novel NADP phosphatase/NAD kinase. J. Biol. Chem. 280, 39200–39207. 10.1074/jbc.M506426200 [DOI] [PubMed] [Google Scholar]
- Kawai S., Mori S., Mukai T., Hashimoto W., Murata K. (2001). Molecular characterization of Escherichia coli NAD kinase. Eur. J. Biochem. 268, 4359–4365. 10.1046/j.1432-1327.2001.02358.x [DOI] [PubMed] [Google Scholar]
- Kawai S., Mori S., Mukai T., Murata K. (2004). Cytosolic NADP phosphatases I and II from Arthrobacter sp. strain KM: implication in regulation of NAD+/NADP+ balance. J. Basic Microbiol. 44, 185–196. 10.1002/jobm.200310362 [DOI] [PubMed] [Google Scholar]
- Kawai S., Mori S., Mukai T., Suzuki S., Yamada T., Hashimoto W., et al. (2000). Inorganic Polyphosphate/ATP-NAD Kinase of Micrococcus flavus and Mycobacterium tuberculosis H37Rv. Biochem. Biophys. Res. Commun. 276, 57–63. 10.1006/bbrc.2000.3433 [DOI] [PubMed] [Google Scholar]
- Kawai S., Murata K. (2008). Structure and function of NAD kinase and NADP phosphatase: key enzymes that regulate the intracellular balance of NAD(H) and NADP(H). Biosci. Biotechnol. Biochem. 72, 919–930. 10.1271/bbb.70738 [DOI] [PubMed] [Google Scholar]
- Kawai S., Suzuki H., Yamamoto K., Inui M., Yukawa H., Kumagai H. (1996). Purification and characterization of a malic enzyme from the ruminal bacterium Streptococcus bovis ATCC 15352 and cloning and sequencing of its gene. Appl. Environ. Microbiol. 62, 2692–2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly G. J., Gibbs M. (1973). Nonreversible d-glyceraldehyde 3-phosphate dehydrogenase of plant tissues. Plant Physiol. 52, 111–118. 10.1104/pp.52.2.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. M., Cho H.-S., Jung G. Y., Park J. M. (2011). Engineering the pentose phosphate pathway to improve hydrogen yield in recombinant Escherichia coli. Biotechnol. Bioeng. 108, 2941–2946. 10.1002/bit.23259 [DOI] [PubMed] [Google Scholar]
- Kizaki N., Yasohara Y., Hasegawa J., Wada M., Kataoka M., Shimizu S. (2001). Synthesis of optically pure ethyl (S)-4-chloro-3-hydroxybutanoate by Escherichia coli transformant cells coexpressing the carbonyl reductase and glucose dehydrogenase genes. Appl. Microbiol. Biotechnol. 55, 590–595. 10.1007/s002530100599 [DOI] [PubMed] [Google Scholar]
- Klasen R., Bringer-Meyer S., Sahm H. (1995). Biochemical characterization and sequence analysis of the gluconate:NADP 5-oxidoreductase gene from Gluconobacter oxydans. J. Bacteriol. 177, 2637–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kletzien R. F., Harris P. K., Foellmi L. A. (1994). Glucose-6-phosphate dehydrogenase: a “housekeeping” enzyme subject to tissue-specific regulation by hormones, nutrients, and oxidant stress. FASEB J. 8, 174–181. [DOI] [PubMed] [Google Scholar]
- Kobayashi K., Ehrlich S. D., Albertini A., Amati G., Andersen K. K., Arnaud M., et al. (2003). Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. U.S.A. 100, 4678–4683. 10.1073/pnas.0730515100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh H.-J., Lee S.-M., Son B.-G., Lee S.-H., Ryoo Z. Y., Chang K.-T., et al. (2004). Cytosolic NADP+-dependent isocitrate dehydrogenase plays a key role in lipid metabolism. J. Biol. Chem. 279, 39968–39974. 10.1074/jbc.M402260200 [DOI] [PubMed] [Google Scholar]
- Koksharova O., Schubert M., Shestakov S., Cerff R. (1998). Genetic and biochemical evidence for distinct key functions of two highly divergent GAPDH genes in catabolic and anabolic carbon flow of the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol. Biol. 36, 183–194. 10.1023/A:1005925732743 [DOI] [PubMed] [Google Scholar]
- Komori H., Seo D., Sakurai T., Higuchi Y. (2010). Crystal structure analysis of Bacillus subtilis ferredoxin-NADP(+) oxidoreductase and the structural basis for its substrate selectivity. Protein Sci. 19, 2279–2290. 10.1002/pro.508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg A. (1950). Enzymatic synthesis of triphosphopyridine nucleotide. J. Biol. Chem. 182, 805–813. [Google Scholar]
- Kramer D. M., Evans J. R. (2011). The importance of energy balance in improving photosynthetic productivity. Plant Physiol. 155, 70–78. 10.1104/pp.110.166652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp A. R., Rodriguez R. E., Poli H. O., Paladini D. H., Palatnik J. F., Carrillo N. (2002). The flavoenzyme ferredoxin (flavodoxin)-NADP(H) reductase modulates NADP(H) homeostasis during the soxRS response of Escherichia coli. J. Bacteriol. 184, 1474–1480. 10.1128/JB.184.5.1474-1480.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C. C., Tsai L. C., Chin T. Y., Chang G. G., Chou W. Y. (2000). Lysine residues 162 and 340 are involved in the catalysis and coenzyme binding of NADP(+)-dependent malic enzyme from pigeon. Biochem. Biophys. Res. Commun. 270, 821–825. 10.1006/bbrc.2000.2502 [DOI] [PubMed] [Google Scholar]
- Kurnasov O., Goral V., Colabroy K., Gerdes S., Anantha S., Osterman A., et al. (2003). NAD biosynthesis: identification of the tryptophan to quinolinate pathway in bacteria. Chem. Biol. 10, 1195–1204. 10.1016/j.chembiol.2003.11.011 [DOI] [PubMed] [Google Scholar]
- Labesse G., Douguet D., Assairi L., Gilles A. M. (2002). Diacylglyceride kinases, sphingosine kinases and NAD kinases: distant relatives of 6-phosphofructokinases. Trends Biochem. Sci. 27, 273–275. 10.1016/S0968-0004(02)02093-5 [DOI] [PubMed] [Google Scholar]
- Lakshmi T. M., Helling R. B. (1976). Selection for citrate synthase deficiency in icd mutants of Escherichia coli. J. Bacteriol. 127, 76–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamed R., Zeikus J. G. (1981). Thermostable, ammonium-activated malic enzyme of Clostridium thermocellum. Biochim. Biophys. Acta 660, 251–255. 10.1016/0005-2744(81)90167-4 [DOI] [PubMed] [Google Scholar]
- Lampel K. A., Uratani B., Chaudhry G. R., Ramaley R. F., Rudikoff S. (1986). Characterization of the developmentally regulated Bacillus subtilis glucose dehydrogenase gene. J. Bacteriol. 166, 238–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landete J. M., Garcia-Haro L., Blasco A., Manzanares P., Berbegal C., Monedero V., et al. (2010). Requirement of the Lactobacillus casei MaeKR two-component system for L-malic acid utilization via a malic enzyme pathway. Appl. Environ. Microbiol. 76, 84–95. 10.1128/AEM.02145-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauterbach L., Lenz O., Vincent K. A. (2013). H(2)-driven cofactor regeneration with NAD(P)(+)-reducing hydrogenases. FEBS J. 280, 3058–3068. 10.1111/febs.12245 [DOI] [PubMed] [Google Scholar]
- Lee H. C. (1997). Mechanisms of calcium signaling by cyclic ADP-ribose and NAADP. Physiol. Rev. 77, 1133–1164. [DOI] [PubMed] [Google Scholar]
- Lee H. C., Kim J. S., Jang W., Kim S. Y. (2009). Thymidine production by overexpressing NAD+ kinase in an Escherichia coli recombinant strain. Biotechnol. Lett. 31, 1929–1936. 10.1007/s10529-009-0097-z [DOI] [PubMed] [Google Scholar]
- Lee H. C., Kim J. S., Jang W., Kim S. Y. (2010). High NADPH/NADP+ ratio improves thymidine production by a metabolically engineered Escherichia coli strain. J. Biotechnol. 149, 24–32. 10.1016/j.jbiotec.2010.06.011 [DOI] [PubMed] [Google Scholar]
- Lee J.-N., Shin H.-D., Lee Y.-H. (2003). Metabolic engineering of pentose phosphate pathway in Ralstonia eutropha for enhanced biosynthesis of poly-β-hydroxybutyrate. Biotechnol. Prog. 19, 1444–1449. 10.1021/bp034060v [DOI] [PubMed] [Google Scholar]
- Lee W.-H., Chin Y.-W., Han N. S., Kim M.-D., Seo J.-H. (2011). Enhanced production of GDP-L-fucose by overexpression of NADPH regenerator in recombinant Escherichia coli. Appl. Microbiol. Biotechnol. 91, 967–976. 10.1007/s00253-011-3271-x [DOI] [PubMed] [Google Scholar]
- Lee W.-H., Kim J.-W., Park E.-H., Han N. S., Kim M.-D., Seo J.-H. (2013a). Effects of NADH kinase on NADPH-dependent biotransformation processes in Escherichia coli. Appl. Microbiol. Biotechnol. 97, 1561–1569. 10.1007/s00253-012-4431-3 [DOI] [PubMed] [Google Scholar]
- Lee W. H., Kim M. D., Jin Y. S., Seo J. H. (2013b). Engineering of NADPH regenerators in Escherichia coli for enhanced biotransformation. Appl. Microbiol. Biotechnol. 97, 2761–2772. 10.1007/s00253-013-4750-z [DOI] [PubMed] [Google Scholar]
- Lee W.-H., Park J.-B., Park K., Kim M.-D., Seo J.-H. (2007a). Enhanced production of ε-caprolactone by overexpression of NADPH-regenerating glucose 6-phosphate dehydrogenase in recombinant Escherichia coli harboring cyclohexanone monooxygenase gene. Appl. Microbiol. Biotechnol. 76, 329–338. 10.1007/s00253-007-1016-7 [DOI] [PubMed] [Google Scholar]
- Lee Y., Yeom J., Kang Y.-S., Kim J., Sung J.-S., Jeon C. O., et al. (2007b). Molecular characterization of fprB (ferredoxin-NADP+ reductase) in Pseudomonas putida KT2440. J. Microbiol. Biotechnol. 17, 1504–1512. [PubMed] [Google Scholar]
- Lerondel G., Doan T., Zamboni N., Sauer U., Aymerich S. (2006). YtsJ has the major physiological role of the four paralogous malic enzyme isoforms in Bacillus subtilis. J. Bacteriol. 188, 4727–4736. 10.1128/JB.00167-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lever T. M., Palmer T., Cunningham I. J., Cotton N. P., Jackson J. B. (1991). Purification and properties of the H(+)-nicotinamide nucleotide transhydrogenase from Rhodobacter capsulatus. Eur. J. Biochem. 197, 247–255. 10.1111/j.1432-1033.1991.tb15905.x [DOI] [PubMed] [Google Scholar]
- Levy H. R. (1979). Glucose-6-phosphate dehydrogenases. Adv. Enzymol. Relat. Areas Mol. Biol. 48, 97–192. 10.1002/9780470122938.ch3 [DOI] [PubMed] [Google Scholar]
- Lewis T. A., Glassing A., Harper J., Franklin M. J. (2013). Role for ferredoxin:NAD(P)H oxidoreductase (FprA) in sulfate assimilation and siderophore biosynthesis in Pseudomonads. J. Bacteriol. 195, 3876–3887. 10.1128/JB.00528-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.-J., Cai L., Wu Q., Chen G.-Q. (2009). Overexpression of NAD kinase in recombinant Escherichia coli harboring the phbCAB operon improves poly(3-hydroxybutyrate) production. Appl. Microbiol. Biotechnol. 83, 939–947. 10.1007/s00253-009-1943-6 [DOI] [PubMed] [Google Scholar]
- Lim S.-J., Jung Y.-M., Shin H.-D., Lee Y.-H. (2002). Amplification of the NADPH-related genes zwf and gnd for the oddball biosynthesis of PHB in an E. coli transformant harboring a cloned phbCAB operon. J. Biosci. Bioeng. 93, 543–549. 10.1016/S1389-1723(02)80235-3 [DOI] [PubMed] [Google Scholar]
- Lindahl R. (1992). Aldehyde dehydrogenases and their role in carcinogenesis. Crit. Rev. Biochem. Mol. Biol. 27, 283–335. 10.3109/10409239209082565 [DOI] [PubMed] [Google Scholar]
- Lindner S. N., Niederholtmeyer H., Schmitz K., Schoberth S. M., Wendisch V. F. (2010). Polyphosphate/ATP-dependent NAD kinase of Corynebacterium glutamicum: biochemical properties and impact of ppnK overexpression on lysine production. Appl. Microbiol. Biotechnol. 87, 583–593. 10.1007/s00253-010-2481-y [DOI] [PubMed] [Google Scholar]
- Lipscomb G. L., Stirrett K., Schut G. J., Yang F., Jenney F. E., Jr., Scott R. A., et al. (2011). Natural competence in the hyperthermophilic archaeon Pyrococcus furiosus facilitates genetic manipulation: construction of markerless deletions of genes encoding the two cytoplasmic hydrogenases. Appl. Environ. Microbiol. 77, 2232–2238. 10.1128/AEM.02624-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Wang P. (2007). Cofactor regeneration for sustainable enzymatic biosynthesis. Biotechnol. Adv. 25, 369–384. 10.1016/j.biotechadv.2007.03.002 [DOI] [PubMed] [Google Scholar]
- Lloyd A. J., Weitzman D. (1988). Purification and characterization of NAD-linked isocitrate dehydrogenase from Methylophilus methylotrophus. Biochem. Soc. Trans. 16, 871–872. 10.1042/bst0160871 [DOI] [Google Scholar]
- London J., Meyer E. Y., Kulczyk S. (1971). Comparative biochemical and immunological study of malic enzyme from two species of lactic acid bacteria: evolutionary implications. J. Bacteriol. 106, 126–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorentzen E., Hensel R., Knura T., Ahmed H., Pohl E. (2004). Structural Basis of allosteric regulation and substrate specificity of the non-phosphorylating glyceraldehyde 3-phosphate dehydrogenase from Thermoproteus tenax. J. Mol. Biol. 341, 815–828. 10.1016/j.jmb.2004.05.032 [DOI] [PubMed] [Google Scholar]
- Lu Y., Mei L. (2007). Co-expression of P450 BM3 and glucose dehydrogenase by recombinant Escherichia coli and its application in an NADPH-dependent indigo production system. J. Ind. Microbiol. Biotechnol. 34, 247–253. 10.1007/s10295-006-0193-1 [DOI] [PubMed] [Google Scholar]
- Lyon E. J., Shima S., Buurman G., Chowdhuri S., Batschauer A., Steinbach K., et al. (2004). UV-A/blue-light inactivation of the 'metal-free' hydrogenase (Hmd) from methanogenic archaea. Eur. J. Biochem. 271, 195–204. 10.1046/j.1432-1033.2003.03920.x [DOI] [PubMed] [Google Scholar]
- Ma K., Adams M. W. (2001a). Ferredoxin:NADP oxidoreductase from Pyrococcus furiosus. Meth. Enzymol. 334, 40–45. 10.1016/S0076-6879(01)34456-7 [DOI] [PubMed] [Google Scholar]
- Ma K., Adams M. W. W. (2001b). Hydrogenases I and II from Pyrococcus furiosus, in Methods in Enzymology, eds Michael R. M. K., Adams W. W. (Academic Press; ), 208–216. [DOI] [PubMed] [Google Scholar]
- Ma K., Weiss R., Adams M. W. W. (2000). Characterization of hydrogenase II from the hyperthermophilic archaeon Pyrococcus furiosus and assessment of its role in sulfur reduction. J. Bacteriol. 182, 1864–1871. 10.1128/JB.182.7.1864-1871.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magni G., Amici A., Emanuelli M., Orsomando G., Raffaelli N., Ruggieri S. (2004). Enzymology of NAD+ homeostasis in man. Cell. Mol. Life Sci. 61, 19–34. 10.1007/s00018-003-3161-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magni G., Amici A., Emanuelli M., Raffaelli N., Ruggieri S. (1999). Enzymology of NAD+ synthesis. Adv. Enzymol. Relat. Areas Mol. Biol. 73, 135–182. 10.1002/9780470123195.ch5 [DOI] [PubMed] [Google Scholar]
- Magni G., Orsomando G., Raffaelli N. (2006). Structural and functional properties of NAD kinase, a key enzyme in NADP biosynthesis. Mini Rev. Med. Chem. 6, 739–746. 10.2174/138955706777698688 [DOI] [PubMed] [Google Scholar]
- Mailloux R. J., Bériault R., Lemire J., Singh R., Chénier D. R., Hamel R. D., et al. (2007). The tricarboxylic acid cycle, an ancient metabolic network with a novel twist. PLoS ONE 2:e690. 10.1371/journal.pone.0000690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malki S., Saimmaime I., De Luca G., Rousset M., Dermoun Z., Belaich J. P. (1995). Characterization of an operon encoding an NADP-reducing hydrogenase in Desulfovibrio fructosovorans. J. Bacteriol. 177, 2628–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchitti S. A., Brocker C., Stagos D., Vasiliou V. (2008). Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Exp. Opin. Drug Metab. Toxicol. 4, 697–720. 10.1517/17425255.4.6.697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez I., Zhu J., Lin H., Bennett G. N., San K.-Y. (2008). Replacing Escherichia coli NAD-dependent glyceraldehyde 3-phosphate dehydrogenase (GAPDH) with a NADP-dependent enzyme from Clostridium acetobutylicum facilitates NADPH dependent pathways. Metab. Eng. 10, 352–359. 10.1016/j.ymben.2008.09.001 [DOI] [PubMed] [Google Scholar]
- Martinez-Julvez M., Medina M., Velazquez-Campoy A. (2009). Binding thermodynamics of ferredoxin:NADP+ reductase: two different protein substrates and one energetics. Biophys. J. 96, 4966–4975. 10.1016/j.bpj.2009.02.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx A., Eikmanns B. J., Sahm H., De Graaf A. A., Eggeling L. (1999). Response of the central metabolism in Corynebacterium glutamicum to the use of an NADH-dependent glutamate dehydrogenase. Metab. Eng. 1, 35–48. 10.1006/mben.1998.0106 [DOI] [PubMed] [Google Scholar]
- Marx A., Hans S., Mockel B., Bathe B., De Graaf A. A., Mccormack A. C., et al. (2003). Metabolic phenotype of phosphoglucose isomerase mutants of Corynebacterium glutamicum. J. Biotechnol. 104, 185–197. 10.1016/S0168-1656(03)00153-6 [DOI] [PubMed] [Google Scholar]
- Mateos M., Serrano A. (1992). Occurrence of phosphorylating and non-phosphorylating NADP+-dependent glyceraldehyde-3-phosphate dehydrogenases in photosynthetic organisms. Plant Sci. 84, 163–170. 10.1016/0168-9452(92)90130-E1764059 [DOI] [Google Scholar]
- Mather L. H., Knight M. (1972). A heat-stable nicotinamide-adenine dinucleotide glycohydrolase from Pseudomonas putida KB1. Partial purification and some properties of the enzyme and an inhibitory protein. Biochem. J. 129, 141–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara K., Yokooji Y., Atomi H., Imanaka T. (2011). Biochemical and genetic characterization of the three metabolic routes in Thermococcus kodakarensis linking glyceraldehyde 3-phosphate and 3-phosphoglycerate. Mol. Microbiol. 81, 1300–1312. 10.1111/j.1365-2958.2011.07762.x [DOI] [PubMed] [Google Scholar]
- Matsuda F., Ishii J., Kondo T., Ida K., Tezuka H., Kondo A. (2013). Increased isobutanol production in Saccharomyces cerevisiae by eliminating competing pathways and resolving cofactor imbalance. Microb. Cell Fact. 12:119. 10.1186/1475-2859-12-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matula T. I., Mcdonald I. J., Martin S. M. (1969). CO2 fixation by malic enzyme in a species of Micrococcus. Biochem. Biophys. Res. Commun. 34, 795–802. 10.1016/0006-291X(69)90250-2 [DOI] [PubMed] [Google Scholar]
- Mcdermott T. R., Kahn M. L. (1992). Cloning and mutagenesis of the Rhizobium meliloti isocitrate dehydrogenase gene. J. Bacteriol. 174, 4790–4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcguinness E. T., Butler J. R. (1985). NAD+ kinase–a review. Int. J. Biochem. 17, 1–11. 10.1016/0020-711X(85)90079-5 [DOI] [PubMed] [Google Scholar]
- Medina M., Gomez-Moreno C. (2004). Interaction of Ferredoxin-NADP(+) Reductase with its substrates: optimal interaction for efficient electron transfer. Photosyn. Res. 79, 113–131. 10.1023/B:PRES.0000015386.67746.2c [DOI] [PubMed] [Google Scholar]
- Mertens R., Greiner L., Van Den Ban E. C. D., Haaker H. B. C. M., Liese A. (2003). Practical applications of hydrogenase I from Pyrococcus furiosus for NADPH generation and regeneration. J. Mol. Catal. B 24–25, 39–52. 10.1016/S1381-1177(03)00071-7 [DOI] [Google Scholar]
- Meyer F. M., Stulke J. (2013). Malate metabolism in Bacillus subtilis: distinct roles for three classes of malate-oxidizing enzymes. FEMS Microbiol. Lett. 339, 17–22. 10.1111/1574-6968.12041 [DOI] [PubMed] [Google Scholar]
- Middleditch L. E., Atchison R. W., Chung A. E. (1972). Pyridine nucleotide transhydrogenase from Azotobacter vinelandii. Some aspects of its structure. J. Biol. Chem. 247, 6802–6809. [PubMed] [Google Scholar]
- Miller W. L., Auchus R. J. (2011). The molecular biology, biochemistry, and physiology of human steroidogenesis and its disorders. Endocr. Rev. 32, 81–151. 10.1210/er.2010-0013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minard K. I., Mcalister-Henn L. (2005). Sources of NADPH in yeast vary with carbon source. J. Biol. Chem. 280, 39890–39896. 10.1074/jbc.M509461200 [DOI] [PubMed] [Google Scholar]
- Moreira Dos Santos M., Raghevendran V., Kotter P., Olsson L., Nielsen J. (2004). Manipulation of malic enzyme in Saccharomyces cerevisiae for increasing NADPH production capacity aerobically in different cellular compartments. Metab. Eng. 6, 352–363. 10.1016/j.ymben.2004.06.002 [DOI] [PubMed] [Google Scholar]
- Mori S., Kawai S., Shi F., Mikami B., Murata K. (2005). Molecular conversion of NAD kinase to NADH kinase through single amino acid residue substitution. J. Biol. Chem. 280, 24104–24112. 10.1074/jbc.M502518200 [DOI] [PubMed] [Google Scholar]
- Morimoto Y., Honda K., Ye X., Okano K., Ohtake H. (2014). Directed evolution of thermotolerant malic enzyme for improved malate production. J. Biosci. Bioeng. 117, 147–152. 10.1016/j.jbiosc.2013.07.005 [DOI] [PubMed] [Google Scholar]
- Moritz B., Striegel K., De Graaf A. A., Sahm H. (2000). Kinetic properties of the glucose-6-phosphate and 6-phosphogluconate dehydrogenases from Corynebacterium glutamicum and their application for predicting pentose phosphate pathway flux in vivo. Eur. J. Biochem. 267, 3442–3452. 10.1046/j.1432-1327.2000.01354.x [DOI] [PubMed] [Google Scholar]
- Mulo P. (2011). Chloroplast-targeted ferredoxin-NADP(+) oxidoreductase (FNR): structure, function and location. Biochim. Biophys. Acta 1807, 927–934. 10.1016/j.bbabio.2010.10.001 [DOI] [PubMed] [Google Scholar]
- Munekage Y., Hashimoto M., Miyake C., Tomizawa K.-I., Endo T., Tasaka M., et al. (2004). Cyclic electron flow around photosystem I is essential for photosynthesis. Nature 429, 579–582. 10.1038/nature02598 [DOI] [PubMed] [Google Scholar]
- Murakami K., Iwata S., Haneda M., Yoshino M. (1997). Role of metal cations in the regulation of NADP-linked isocitrate dehydrogenase from porcine heart. Biometals 10, 169–174. 10.1023/A:1018395510334 [DOI] [PubMed] [Google Scholar]
- Muraki N., Seo D., Shiba T., Sakurai T., Kurisu G. (2010). Asymmetric dimeric structure of ferredoxin-NAD(P)+ oxidoreductase from the green sulfur bacterium Chlorobaculum tepidum: implications for binding ferredoxin and NADP+. J. Mol. Biol. 401, 403–414. 10.1016/j.jmb.2010.06.024 [DOI] [PubMed] [Google Scholar]
- Muro-Pastor M. I., Florencio F. J. (1994). NADP (+)-isocitrate dehydrogenase from the Cyanobacterium Anabaena sp. strain PCC 7120: purification and characterization of the enzyme and cloning, sequencing, and disruption of the icd gene. J. Bacteriol. 176, 2718–2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro-Pastor M. I., Reyes J. C., Florencio F. J. (2005). Ammonium assimilation in cyanobacteria. Photosyn. Res. 83, 135–150. 10.1007/s11120-004-2082-7 [DOI] [PubMed] [Google Scholar]
- Musumeci M. A., Ceccarelli E. A., Catalano-Dupuy D. L. (2012). The plant–type ferredoxin-NADP+ reductases, in Advances in Photosynthesis - Fundamental Aspects, ed Najafpour D. M. (Rijeka: InTech; ), 539–562. 10.5772/28665 [DOI] [Google Scholar]
- Nakamichi Y., Yoshioka A., Kawai S., Murata K. (2013). Conferring the ability to utilize inorganic polyphosphate on ATP-specific NAD kinase. Sci. Rep. 3, 2632. 10.1038/srep02632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M., Bhatnagar A., Sadoshima J. (2012). Overview of pyridine nucleotides review series. Circ. Res. 111, 604–610. 10.1161/CIRCRESAHA.111.247924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netzer R., Krause M., Rittmann D., Peters-Wendisch P. G., Eggeling L., Wendisch V. F., et al. (2004). Roles of pyruvate kinase and malic enzyme in Corynebacterium glutamicum for growth on carbon sources requiring gluconeogenesis. Arch. Microbiol. 182, 354–363. 10.1007/s00203-004-0710-4 [DOI] [PubMed] [Google Scholar]
- Nicolas C., Kiefer P., Letisse F., Kromer J., Massou S., Soucaille P., et al. (2007). Response of the central metabolism of Escherichia coli to modified expression of the gene encoding the glucose-6-phosphate dehydrogenase. FEBS Lett. 581, 3771–3776. 10.1016/j.febslet.2007.06.066 [DOI] [PubMed] [Google Scholar]
- Noor E., Bar-Even A., Flamholz A., Lubling Y., Davidi D., Milo R. (2012). An integrated open framework for thermodynamics of reactions that combines accuracy and coverage. Bioinformatics 28, 2037–2044. 10.1093/bioinformatics/bts317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor E., Haraldsdóttir H. S., Milo R., Fleming R. M. T. (2013). Consistent estimation of gibbs energy using component contributions. PLoS Comput. Biol. 9:e1003098. 10.1371/journal.pcbi.1003098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordberg J., Arnér E. S. J. (2001). Reactive oxygen species, antioxidants, and the mammalian thioredoxin system. Free Radic. Biol. Med. 31, 1287–1312. 10.1016/S0891-5849(01)00724-9 [DOI] [PubMed] [Google Scholar]
- Nouailler M., Morelli X., Bornet O., Chetrit B., Dermoun Z., Guerlesquin F. (2006). Solution structure of HndAc: a thioredoxin-like domain involved in the NADP-reducing hydrogenase complex. Protein Sci. 15, 1369–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obanye A. I. C., Hobbs G., Gardner D. C. J., Oliver S. G. (1996). Correlation between carbon flux through the pentose phosphate pathway and production of the antibiotic methylenomycin in Streptomyces coelicolor A3(2). Microbiology (Reading Engl). 142, 133–137. 10.1099/13500872-142-1-133 [DOI] [PubMed] [Google Scholar]
- Ochiai A., Mori S., Kawai S., Murata K. (2004). Overexpression, purification, and characterization of ATP-NAD kinase of Sphingomonas sp. A1. Protein Expr. Purif. 36, 124–130. 10.1016/j.pep.2004.03.012 [DOI] [PubMed] [Google Scholar]
- Ogawa T., Murakami K., Mori H., Ishii N., Tomita M., Yoshin M. (2007). Role of phosphoenolpyruvate in the NADP-isocitrate dehydrogenase and isocitrate lyase reaction in Escherichia coli. J. Bacteriol. 189, 1176–1178. 10.1128/JB.01628-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh M.-K., Rohlin L., Kao K. C., Liao J. C. (2002). Global expression profiling of acetate-grown Escherichia coli. J. Biol. Chem. 277, 13175–13183. 10.1074/jbc.M110809200 [DOI] [PubMed] [Google Scholar]
- Olavarría K., Valdés D., Cabrera R. (2012). The cofactor preference of glucose-6-phosphate dehydrogenase from Escherichia coli – modeling the physiological production of reduced cofactors. FEBS J. 279, 2296–2309. 10.1111/j.1742-4658.2012.08610.x [DOI] [PubMed] [Google Scholar]
- Onda Y., Matsumura T., Kimata-Ariga Y., Sakakibara H., Sugiyama T., Hase T. (2000). Differential interaction of maize root ferredoxin:NADP(+) oxidoreductase with photosynthetic and non-photosynthetic ferredoxin isoproteins. Plant Physiol. 123, 1037–1045. 10.1104/pp.123.3.1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Outten C. E., Culotta V. C. (2003). A novel NADH kinase is the mitochondrial source of NADPH in Saccharomyces cerevisiae. EMBO J. 22, 2015–2024. 10.1093/emboj/cdg211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen O. E., Kalhan S. C., Hanson R. W. (2002). The key role of anaplerosis and cataplerosis for citric acid cycle function. J. Biol. Chem. 277, 30409–30412. 10.1074/jbc.R200006200 [DOI] [PubMed] [Google Scholar]
- Papagianni M. (2012). Recent advances in engineering the central carbon metabolism of industrially important bacteria. Microb. Cell Fact. 11:50. 10.1186/1475-2859-11-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H. J., Jung J., Choi H., Uhm K.-N., Kim H. K. (2010). Enantioselective bioconversion using Escherichia coli cells expressing Saccharomyces cerevisiae reductase and Bacillus subtilis glucose dehydrogenase. J. Microbiol. Biotechnol. 20, 1300–1306. 10.4014/jmb.1003.03025 [DOI] [PubMed] [Google Scholar]
- Patel S., Ramakrishnan L., Rahman T., Hamdoun A., Marchant J. S., Taylor C. W., et al. (2011). The endo-lysosomal system as an NAADP-sensitive acidic Ca(2+) store: role for the two-pore channels. Cell Calcium 50, 157–167. 10.1016/j.ceca.2011.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul L., Mishra P., Blumenthal R., Matthews R. (2007). Integration of regulatory signals through involvement of multiple global regulators: control of the Escherichia coli gltBDF operon by Lrp, IHF, Crp, and ArgR. BMC Microbiol. 7:2. 10.1186/1471-2180-7-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauly H. E., Pfleiderer G. (1975). D-glucose dehydrogenase from Bacillus megaterium M 1286: purification, properties and structure. Hoppe-Seyler's Z. Physiol. Chem. 356, 1613–1623. 10.1515/bchm2.1975.356.2.1613 [DOI] [PubMed] [Google Scholar]
- Pedersen A., Karlsson G. B., Rydstrom J. (2008). Proton-translocating transhydrogenase: an update of unsolved and controversial issues. J. Bioenerg. Biomembr. 40, 463–473. 10.1007/s10863-008-9170-x [DOI] [PubMed] [Google Scholar]
- Penyige A., Deak E., Kalmanczhelyi A., Barabas G. (1996). Evidence of a role for NAD+-glycohydrolase and ADP-ribosyltransferase in growth and differentiation of Streptomyces griseus NRRL B-2682: inhibition by m-aminophenylboronic acid. Microbiology 142(Pt 8), 1937–1944. 10.1099/13500872-142-8-1937 [DOI] [PubMed] [Google Scholar]
- Peralta-Yahya P. P., Zhang F., Del Cardayre S. B., Keasling J. D. (2012). Microbial engineering for the production of advanced Biofuels 488, 320–328. 10.1038/nature11478 [DOI] [PubMed] [Google Scholar]
- Peters J. W., Schut G. J., Boyd E. S., Mulder D. W., Shepard E. M., Broderick J. B., et al. (2015). [FeFe]- and [NiFe]-hydrogenase diversity, mechanism, and maturation. Biochim. Biophys. Acta Mol. Cell Res. 1853, 1350–1369. 10.1016/j.bbamcr.2014.11.021 [DOI] [PubMed] [Google Scholar]
- Petersen S., De Graaf A. A., Eggeling L., Möllney M., Wiechert W., Sahm H. (2000). In vivo quantification of parallel and bidirectional fluxes in the anaplerosis of Corynebacterium glutamicum. J. Biol. Chem. 275, 35932–35941. 10.1074/jbc.M908728199 [DOI] [PubMed] [Google Scholar]
- Pollak N., Dölle C., Ziegler M. (2007). The power to reduce: pyridine nucleotides – small molecules with a multitude of functions. Biochem. J. 402, 205–218. 10.1042/BJ20061638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poncet-Montange G., Assairi L., Arold S., Pochet S., Labesse G. (2007). NAD kinases use substrate-assisted catalysis for specific recognition of NAD. J. Biol. Chem. 282, 33925–33934. 10.1074/jbc.M701394200 [DOI] [PubMed] [Google Scholar]
- Purwantini E., Daniels L. (1996). Purification of a novel coenzyme F420-dependent glucose-6-phosphate dehydrogenase from Mycobacterium smegmatis. J. Bacteriol. 178, 2861–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purwantini E., Gillis T. P., Daniels L. (1997). Presence of F420-dependent glucose-6-phosphate dehydrogenase in Mycobacterium and Nocardia species, but absence from Streptomyces and Corynebacterium species and methanogenic Archaea. FEMS Microbiol. Lett. 146, 129–134. 10.1111/j.1574-6968.1997.tb10182.x [DOI] [PubMed] [Google Scholar]
- Qi H., Li S., Zhao S., Huang D., Xia M., Wen J. (2014). Model-driven redox pathway manipulation for improved isobutanol production in Bacillus subtilis complemented with experimental validation and metabolic profiling analysis. PLoS ONE 9:e93815. 10.1371/journal.pone.0093815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M. M. Q., Zhao Q., Dou W., Zhiming R., Xu Z. (2012). Over-expression of NAD kinase in Corynebacterium crenatum and its impact on L-Arginine biosynthesis. Tropic. J. Pharm. Res. 11, 909–916. 10.4314/tjpr.v11i6.6 [DOI] [Google Scholar]
- Rathnasingh C., Raj S. M., Lee Y., Catherine C., Ashok S., Park S. (2012). Production of 3-hydroxypropionic acid via malonyl-CoA pathway using recombinant Escherichia coli strains. J. Biotechnol. 157, 633–640. 10.1016/j.jbiotec.2011.06.008 [DOI] [PubMed] [Google Scholar]
- Razquin P., Fillat M. F., Schmitz S., Stricker O., Bohme H., Gomez-Moreno C., et al. (1996). Expression of ferredoxin-NADP+ reductase in heterocysts from Anabaena sp. Biochem. J. 316(Pt 1), 157–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy G. K., Lindner S. N., Wendisch V. F. (2015). Metabolic Engineering an ATP-neutral EMP pathway in Corynebacterium glutamicum: adaptive point mutation in NADH dehydrogenase restores growth. Appl. Environ. Microbiol. 81, 1996–2005. 10.1128/AEM.03116-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidl J., Schlor S., Kraiss A., Schmidt-Brauns J., Kemmer G., Soleva E. (2000). NADP and NAD utilization in Haemophilus influenzae. Mol. Microbiol. 35, 1573–1581. 10.1046/j.1365-2958.2000.01829.x [DOI] [PubMed] [Google Scholar]
- Richhardt J., Bringer S., Bott M. (2013). Role of the pentose phosphate pathway and the Entner-Doudoroff pathway in glucose metabolism of Gluconobacter oxydans 621H. Appl. Microbiol. Biotechnol. 97, 4315–4323. 10.1007/s00253-013-4707-2 [DOI] [PubMed] [Google Scholar]
- Riedel C., Rittmann D., Dangel P., Mockel B., Petersen S., Sahm H., et al. (2001). Characterization of the phosphoenolpyruvate carboxykinase gene from Corynebacterium glutamicum and significance of the enzyme for growth and amino acid production. J. Mol. Microbiol. Biotechnol. 3, 573–583. [PubMed] [Google Scholar]
- Poulsen B. R., Nohr J., Douthwaite S., Hansen L. V., Iversen J. J. L., Visser J., et al. (2005). Increased NADPH concentration obtained by metabolic engineering of the pentose phosphate pathway in Aspergillus niger. FEBS J. 272, 1313–1325. 10.1111/j.1742-4658.2005.04554.x [DOI] [PubMed] [Google Scholar]
- Rydzak T., Grigoryan M., Cunningham Z. J., Krokhin O. V., Ezzati P., Cicek N., et al. (2014). Insights into electron flux through manipulation of fermentation conditions and assessment of protein expression profiles in Clostridium thermocellum. Appl. Microbiol. Biotechnol. 98, 6497–6510. 10.1007/s00253-014-5798-0 [DOI] [PubMed] [Google Scholar]
- Sahm H., Eggeling L., De Graaf A. A. (2000). Pathway analysis and metabolic engineering in Corynebacterium glutamicum. Biol. Chem. 381, 899–910. 10.1515/BC.2000.111 [DOI] [PubMed] [Google Scholar]
- Sakuraba H., Kawakami R., Ohshima T. (2005). First Archaeal Inorganic Polyphosphate/ATP-Dependent NAD Kinase, from Hyperthermophilic Archaeon Pyrococcus horikoshii: cloning, expression, and characterization. Appl. Environ. Microbiol. 71, 4352–4358. 10.1128/AEM.71.8.4352-4358.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuraba H., Satomura T., Kawakami R., Yamamoto S., Kawarabayasi Y., Kikuchi H., et al. (2002). L-Aspartate oxidase is present in the anaerobic hyperthermophilic archaeon Pyrococcus horikoshii OT-3: characteristics and role in the de novo biosynthesis of nicotinamide adenine dinucleotide proposed by genome sequencing. Extremophiles 6, 275–281. 10.1007/s00792-001-0254-3 [DOI] [PubMed] [Google Scholar]
- Saliola M., Tramonti A., Lanini C., Cialfi S., De Biase D., Falcone C. (2012). Intracellular NADPH levels affect the oligomeric state of the glucose 6-phosphate dehydrogenase. Eukaryot. Cell 11, 1503–1511. 10.1128/EC.00211-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez A. M., Andrews J., Hussein I., Bennett G. N., San K.-Y. (2006). Effect of overexpression of a soluble pyridine nucleotide transhydrogenase (UdhA) on the production of poly(3-hydroxybutyrate) in Escherichia coli. Biotechnol. Prog. 22, 420–425. 10.1021/bp050375u [DOI] [PubMed] [Google Scholar]
- Sancho J. (2006). Flavodoxins: sequence, folding, binding, function and beyond. Cell. Mol. Life Sci. 63, 855–864. 10.1007/s00018-005-5514-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo T. J., Cubonova L., Reeve J. N. (2011). Deletion of alternative pathways for reductant recycling in Thermococcus kodakarensis increases hydrogen production. Mol. Microbiol. 81, 897–911. 10.1111/j.1365-2958.2011.07734.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassetti C. M., Boyd D. H., Rubin E. J. (2003). Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48, 77–84. 10.1046/j.1365-2958.2003.03425.x [DOI] [PubMed] [Google Scholar]
- Sato T., Atomi H. (2011). Novel metabolic pathways in Archaea. Curr. Opin. Microbiol. 14, 307–314. 10.1016/j.mib.2011.04.014 [DOI] [PubMed] [Google Scholar]
- Sato Y., Kameya M., Fushinobu S., Wakagi T., Arai H., Ishii M., et al. (2012). A novel enzymatic system against oxidative stress in the thermophilic hydrogen-oxidizing bacterium Hydrogenobacter thermophilus. PLoS ONE 7:e34825. 10.1371/journal.pone.0034825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh Y., Tajima K., Tannai H., Munekata M. (2003). Enzyme-catalyzed poly(3-hydroxybutyrate) synthesis from acetate with CoA recycling and NADPH regeneration in vitro. J. Biosci. Bioeng. 95, 335–341. 10.1016/S1389-1723(03)80064-6 [DOI] [PubMed] [Google Scholar]
- Sauer U., Canonaco F., Heri S., Perrenoud A., Fischer E. (2004). The soluble and membrane-bound transhydrogenases UdhA and PntAB have divergent functions in NADPH metabolism of Escherichia coli. J. Biol. Chem. 279, 6613–6619. 10.1074/jbc.M311657200 [DOI] [PubMed] [Google Scholar]
- Sauer U., Eikmanns B. J. (2005). The PEP–pyruvate–oxaloacetate node as the switch point for carbon flux distribution in bacteria. FEMS Microbiol. Rev. 29, 765–794. 10.1016/j.femsre.2004.11.002 [DOI] [PubMed] [Google Scholar]
- Schäfer T., Schönheit P. (1993). Gluconeogenesis from pyruvate in the hyperthermophilic archaeon Pyrococcus furiosus: involvement of reactions of the Embden-Meyerhof pathway. Arch. Microbiol. 159, 354–363. 10.1007/BF00290918 [DOI] [Google Scholar]
- Schmitz O., Boison G., Salzmann H., Bothe H., Schutz K., Wang S.-H., et al. (2002). HoxE–a subunit specific for the pentameric bidirectional hydrogenase complex (HoxEFUYH) of cyanobacteria. Biochim. Biophys. Acta 1554, 66–74. 10.1016/S0005-2728(02)00214-1 [DOI] [PubMed] [Google Scholar]
- Schut G. J., Nixon W. J., Lipscomb G. L., Scott R. A., Adams M. W. W. (2012). Mutational analyses of the enzymes involved in the metabolism of hydrogen by the hyperthermophilic archaeon Pyrococcus furiosus. Front. Microbiol. 3:163. 10.3389/fmicb.2012.00163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeber F., Aliverti A., Zanetti G. (2005). The plant-type ferredoxin-NADP+ reductase/ferredoxin redox system as a possible drug target against apicomplexan human parasites. Curr. Pharm. Des. 11, 3159–3172. 10.2174/1381612054864957 [DOI] [PubMed] [Google Scholar]
- Seo D., Kamino K., Inoue K., Sakurai H. (2004). Purification and characterization of ferredoxin-NADP+ reductase encoded by Bacillus subtilis yumC. Arch. Microbiol. 182, 80–89. 10.1007/s00203-004-0701-5 [DOI] [PubMed] [Google Scholar]
- Seo D., Sakurai H. (2002). Purification and characterization of ferredoxin-NAD(P)(+) reductase from the green sulfur bacterium Chlorobium tepidum. Biochim. Biophys. Acta 1597, 123–132. 10.1016/S0167-4838(02)00269-8 [DOI] [PubMed] [Google Scholar]
- Shi A., Zhu X., Lu J., Zhang X., Ma Y. (2013). Activating transhydrogenase and NAD kinase in combination for improving isobutanol production. Metab. Eng. 16, 1–10. 10.1016/j.ymben.2012.11.008 [DOI] [PubMed] [Google Scholar]
- Shi F., Huan X., Wang X., Ning J. (2012). Overexpression of NAD kinases improves the L-isoleucine biosynthesis in Corynebacterium glutamicum ssp. lactofermentum. Enzyme Microb. Technol. 51, 73–80. 10.1016/j.enzmictec.2012.04.003 [DOI] [PubMed] [Google Scholar]
- Shi F., Li Y., Wang X. (2009). Molecular properties, functions, and potential applications of NAD kinases. Acta Biochim. Biophys. Sin. (Shanghai). 41, 352–361. 10.1093/abbs/gmp029 [DOI] [PubMed] [Google Scholar]
- Shiba H., Kawasumi T., Igarashi Y., Kodama T., Minoda Y. (1985). The CO2 assimilation via the reductive tricarboxylic acid cycle in an obligately autotrophic, aerobic hydrogen-oxidizing bacterium, Hydrogenobacter thermophilus. Arch. Microbiol. 141, 198–203. 10.1007/BF00408058 [DOI] [Google Scholar]
- Shikanai T. (2007). Cyclic electron transport around photosystem I: genetic approaches. Annu. Rev. Plant Biol. 58, 199–217. 10.1146/annurev.arplant.58.091406.110525 [DOI] [PubMed] [Google Scholar]
- Shima S., Thauer R. K. (2007). A third type of hydrogenase catalyzing H2 activation. Chem. Rec. 7, 37–46. 10.1002/tcr.20111 [DOI] [PubMed] [Google Scholar]
- Shin J.-A., Kwon Y. D., Kwon O.-H., Lee H. S., Kim P. (2007). 5-Aminolevulinic acid biosynthesis in Escherichia coli coexpressing NADP-dependent malic enzyme and 5-aminolevulinate synthase. J. Microbiol. Biotechnol. 17, 1579–1584. [PubMed] [Google Scholar]
- Shin M., Arnon D. I. (1965). Enzymic mechanisms of pyridine nucleotide reduction in chloroplasts. J. Biol. Chem. 240, 1405–1411. [PubMed] [Google Scholar]
- Siebers B., Schonheit P. (2005). Unusual pathways and enzymes of central carbohydrate metabolism in Archaea. Curr. Opin. Microbiol. 8, 695–705. 10.1016/j.mib.2005.10.014 [DOI] [PubMed] [Google Scholar]
- Siedler S., Bringer S., Bott M. (2011). Increased NADPH availability in Escherichia coli: improvement of the product per glucose ratio in reductive whole-cell biotransformation. Appl. Microbiol. Biotechnol. 92, 929–937. 10.1007/s00253-011-3374-4 [DOI] [PubMed] [Google Scholar]
- Silva P. J., Van Den Ban E. C., Wassink H., Haaker H., De Castro B., Robb F. T., et al. (2000). Enzymes of hydrogen metabolism in Pyrococcus furiosus. Eur. J. Biochem. 267, 6541–6551. 10.1046/j.1432-1327.2000.01745.x [DOI] [PubMed] [Google Scholar]
- Singh R., Beriault R., Middaugh J., Hamel R., Chenier D., Appanna V. D., et al. (2005). Aluminum-tolerant Pseudomonas fluorescens: ROS toxicity and enhanced NADPH production. Extremophiles 9, 367–373. 10.1007/s00792-005-0450-7 [DOI] [PubMed] [Google Scholar]
- Singh R., Lemire J., Mailloux R. J., Appanna V. D. (2008). A novel strategy involved anti-oxidative defense: the conversion of NADH into NADPH by a metabolic network. PLoS ONE 3:e2682. 10.1371/annotation/5fac086b-3806-4aa9-a5c5-2611b3355f8f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Mailloux R. J., Puiseux-Dao S., Appanna V. D. (2007). Oxidative stress evokes a metabolic adaptation that favors increased NADPH synthesis and decreased NADH production in Pseudomonas fluorescens. J. Bacteriol. 189, 6665–6675. 10.1128/JB.00555-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K. M., Cho K.-M., Liao J. C. (2010). Engineering Corynebacterium glutamicum for isobutanol production. Appl. Microbiol. Biotechnol. 87, 1045–1055. 10.1007/s00253-010-2522-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sophos N. A., Pappa A., Ziegler T. L., Vasiliou V. (2001). Aldehyde dehydrogenase gene superfamily: the 2000 update. Chem. Biol. Interact. 130–132, 323–337. 10.1016/S0009-2797(00)00275-1 [DOI] [PubMed] [Google Scholar]
- Sophos N. A., Vasiliou V. (2003). Aldehyde dehydrogenase gene superfamily: the 2002 update. Chem. Biol. Interact. 143–144, 5–22. 10.1016/S0009-2797(02)00163-1 [DOI] [PubMed] [Google Scholar]
- Steen I. H., Lien T., Birkeland N. K. (1997). Biochemical and phylogenetic characterization of isocitrate dehydrogenase from a hyperthermophilic archaeon, Archaeoglobus fulgidus. Arch. Microbiol. 168, 412–420. 10.1007/s002030050516 [DOI] [PubMed] [Google Scholar]
- Steen I. H., Madern D., Karlstrom M., Lien T., Ladenstein R., Birkeland N. K. (2001). Comparison of isocitrate dehydrogenase from three hyperthermophiles reveals differences in thermostability, cofactor specificity, oligomeric state, and phylogenetic affiliation. J. Biol. Chem. 276, 43924–43931. 10.1074/jbc.M105999200 [DOI] [PubMed] [Google Scholar]
- Steuber J., Krebs W., Bott M., Dimroth P. (1999). A Membrane-Bound NAD(P)(+)-Reducing hydrogenase provides reduced pyridine nucleotides during citrate fermentation by Klebsiella pneumoniae. J. Bacteriol. 181, 241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokke R., Madern D., Fedoy A.-E., Karlsen S., Birkeland N.-K., Steen I. H. (2007). Biochemical characterization of isocitrate dehydrogenase from Methylococcus capsulatus reveals a unique NAD+-dependent homotetrameric enzyme. Arch. Microbiol. 187, 361–370. 10.1007/s00203-006-0200-y [DOI] [PubMed] [Google Scholar]
- Stols L., Donnelly M. I. (1997). Production of succinic acid through overexpression of NAD(+)-dependent malic enzyme in an Escherichia coli mutant. Appl. Environ. Microbiol. 63, 2695–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stournaras C., Maurer P., Kurz G. (1983). 6-phospho-D-gluconate dehydrogenase from Pseudomonas fluorescens. Properties and subunit structure. Eur. J. Biochem. 130, 391–396. 10.1111/j.1432-1033.1983.tb07165.x [DOI] [PubMed] [Google Scholar]
- Strand M. K., Stuart G. R., Longley M. J., Graziewicz M. A., Dominick O. C., Copeland W. C. (2003). POS5 gene of Saccharomyces cerevisiae encodes a mitochondrial NADH kinase required for stability of mitochondrial DNA. Eukaryot. Cell 2, 809–820. 10.1128/EC.2.4.809-820.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers M. L., Wallis J. G., Campbell E. L., Meeks J. C. (1995). Genetic evidence of a major role for glucose-6-phosphate dehydrogenase in nitrogen fixation and dark growth of the cyanobacterium Nostoc sp. strain ATCC 29133. J. Bacteriol. 177, 6184–6194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Hopkins R. C., Jenney F. E., Jr., Mcternan P. M., Adams M. W. W. (2010). Heterologous expression and maturation of an NADP-Dependent [NiFe]-hydrogenase: a key enzyme in biofuel production. PLoS ONE 5:e10526. 10.1371/journal.pone.0010526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung J. Y., Lee Y. N. (2007). Isoforms of glucose 6-phosphate dehydrogenase in Deinococcus radiophilus. J. Microbiol. 45, 318–325. [PubMed] [Google Scholar]
- Szaszák M., Steven P., Shima K., Orzekowsky-Schröder R., Hüttmann G., König I. R., et al. (2011). Fluorescence lifetime imaging unravels C. trachomatis metabolism and its crosstalk with the host cell. PLoS Pathog 7:e1002108. 10.1371/journal.ppat.1002108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szmant H. H. (1989). Organic Building Blocks of the Chemical Industry. New York, NY: John Wiley & Sons. [Google Scholar]
- Takeno S., Murata R., Kobayashi R., Mitsuhashi S., Ikeda M. (2010). Engineering of Corynebacterium glutamicum with an NADPH-generating glycolytic pathway for L-lysine production. Appl. Environ. Microbiol. 76, 7154–7160. 10.1128/AEM.01464-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talfournier F., Stines-Chaumeil C., Branlant G. (2011). Methylmalonate-semialdehyde dehydrogenase from Bacillus subtilis: substrate specificity and coenzyme A binding. J. Biol. Chem. 286, 21971–21981. 10.1074/jbc.M110.213280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang K.-H., Yue H., Blankenship R. E. (2010). Energy metabolism of Heliobacterium modesticaldum during phototrophic and chemotrophic growth. BMC Microbiol. 10:150. 10.1186/1471-2180-10-150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanassi J. A., Hartman-Neumann S. L., Dougherty T. J., Dougherty B. A., Pucci M. J. (2002). Identification of 113 conserved essential genes using a high-throughput gene disruption system in Streptococcus pneumoniae. Nucleic Acids Res. 30, 3152–3162. 10.1093/nar/gkf418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thauer R. K., Jungermann K., Decker K. (1977). Energy conservation in chemotrophic anaerobic bacteria. Bacteriol. Rev. 41, 100–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trchounian A., Gary Sawers R. (2014). Novel insights into the bioenergetics of mixed-acid fermentation: can hydrogen and proton cycles combine to help maintain a proton motive force? IUBMB Life 66, 1–7. 10.1002/iub.1236 [DOI] [PubMed] [Google Scholar]
- Trchounian K., Poladyan A., Vassilian A., Trchounian A. (2012). Multiple and reversible hydrogenases for hydrogen production by Escherichia coli: dependence on fermentation substrate, pH and the F(0)F(1)-ATPase. Crit. Rev. Biochem. Mol. Biol. 47, 236–249. 10.3109/10409238.2012.655375 [DOI] [PubMed] [Google Scholar]
- Tseng Y.-M., Harris B. G., Jacobson M. K. (1979). Isolation and characterization of yeast nicotinamide adenine dinucleotide kinase. Biochim. Biophys. Acta 568, 205–214. 10.1016/0005-2744(79)90287-0 [DOI] [PubMed] [Google Scholar]
- Turner W. L., Waller J. C., Snedden W. A. (2005). Identification, molecular cloning and functional characterization of a novel NADH kinase from Arabidopsis thaliana (thale cress). Biochem. J. 385, 217–223. 10.1042/BJ20040292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppada V., Bhaduri S., Noronha S. B. (2014). Cofactor regeneration–an important aspect of biocatalysis. Curr. Sci. 106, 946. [Google Scholar]
- Van Der Donk W. A., Zhao H. (2003). Recent developments in pyridine nucleotide regeneration. Curr. Opin. Biotechnol. 14, 421–426. 10.1016/S0958-1669(03)00094-6 [DOI] [PubMed] [Google Scholar]
- Van Haaster D. J., Silva P. J., Hagedoorn P.-L., Jongejan J. A., Hagen W. R. (2008). Reinvestigation of the steady-state kinetics and physiological function of the soluble NiFe-hydrogenase I of Pyrococcus furiosus. J. Bacteriol. 190, 1584–1587. 10.1128/JB.01562-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiliou V., Pappa A., Petersen D. R. (2000). Role of aldehyde dehydrogenases in endogenous and xenobiotic metabolism. Chem. Biol. Interact. 129, 1–19. 10.1016/S0009-2797(00)00211-8 [DOI] [PubMed] [Google Scholar]
- Veech R. L., Eggleston L. V., Krebs H. A. (1969). The redox state of free nicotinamide–adenine dinucleotide phosphate in the cytoplasm of rat liver. Biochem. J. 115, 609–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velayudhan J., Kelly D. J. (2002). Analysis of gluconeogenic and anaplerotic enzymes in Campylobacter jejuni: an essential role for phosphoenolpyruvate carboxykinase. Microbiology 148, 685–694. [DOI] [PubMed] [Google Scholar]
- Vignais P. M., Billoud B. (2007). Occurrence, classification, and biological function of hydrogenases: an overview. Chem. Rev. 107, 4206–4272. 10.1021/cr050196r [DOI] [PubMed] [Google Scholar]
- Vignais P. M., Billoud B., Meyer J. (2001). Classification and phylogeny of hydrogenases. FEMS Microbiol. Rev. 25, 455–501. 10.1111/j.1574-6976.2001.tb00587.x [DOI] [PubMed] [Google Scholar]
- Vignais P. M., Colbeau A. (2004). Molecular biology of microbial hydrogenases. Curr. Issues Mol. Biol. 6, 159–188. [PubMed] [Google Scholar]
- Voegele R. T., Mitsch M. J., Finan T. M. (1999). Characterization of two members of a novel malic enzyme class. Biochim. Biophys. Acta 1432, 275–285. 10.1016/S0167-4838(99)00112-0 [DOI] [PubMed] [Google Scholar]
- Voordouw G., Hagen W. R., Kruse-Wolters K. M., Van Berkel-Arts A., Veeger C. (1987). Purification and characterization of Desulfovibrio vulgaris (Hildenborough) hydrogenase expressed in Escherichia coli. Eur. J. Biochem. 162, 31–36. 10.1111/j.1432-1033.1987.tb10537.x [DOI] [PubMed] [Google Scholar]
- Voordouw G., van der Vies S. M., Themmen A. P. (1983). Why are two different types of pyridine nucleotide transhydrogenase found in living organisms? Eur. J. Biochem. 131, 527–533. [DOI] [PubMed] [Google Scholar]
- Voordouw G., van der Vies S., Scholten J. W., Veeger C. (1980). Pyridine nucleotide transhydrogenase from Azotobacter vinelandii. Eur. J. Biochem. 107, 337–344. 10.1111/j.1432-1033.1980.tb06034.x [DOI] [PubMed] [Google Scholar]
- Voordouw G., Veeger C., Van Breemen J. F. L., Van Bruggen E. F. J. (1979). Structure of pyridine nucleotide transhydrogenase from Azotobacter vinelandii. Eur. J. Biochem. 98, 447–454. 10.1111/j.1432-1033.1979.tb13205.x [DOI] [PubMed] [Google Scholar]
- Vu C. Q., Lu P. J., Ghen C. S., Jacobson M. K. (1996). 2′-phospho-cyclic ADP-ribose, a calcium-mobilizing agent derived from NADP. J. Biol. Chem. 271, 4747–4754. 10.1074/jbc.271.9.4747 [DOI] [PubMed] [Google Scholar]
- Walsh K., Koshland D. E. (1985). Branch point control by the phosphorylation state of isocitrate dehydrogenase. a quantitative examination of fluxes during a regulatory transition. J. Biol. Chem. 260, 8430–8437. [PubMed] [Google Scholar]
- Wandrey C. (2004). Biochemical reaction engineering for redox reactions. Chem. Rec. 4, 254–265. 10.1002/tcr.20016 [DOI] [PubMed] [Google Scholar]
- Wang B., Wang P., Zheng E., Chen X., Zhao H., Song P., et al. (2011a). Biochemical properties and physiological roles of NADP-dependent malic enzyme in Escherichia coli. J. Microbiol. 49, 797–802. 10.1007/s12275-011-0487-5 [DOI] [PubMed] [Google Scholar]
- Wang P., Jin M., Su R., Song P., Wang M., Zhu G. (2011b). Enzymatic characterization of isocitrate dehydrogenase from an emerging zoonotic pathogen Streptococcus suis. Biochimie 93, 1470–1475. 10.1016/j.biochi.2011.04.021 [DOI] [PubMed] [Google Scholar]
- Wang P., Jin M., Zhu G. (2012). Biochemical and molecular characterization of NAD(+)-dependent isocitrate dehydrogenase from the ethanologenic bacterium Zymomonas mobilis. FEMS Microbiol. Lett. 327, 134–141. 10.1111/j.1574-6968.2011.02467.x [DOI] [PubMed] [Google Scholar]
- Wang P., Song P., Jin M., Zhu G. (2013a). Isocitrate dehydrogenase from Streptococcus mutans: biochemical properties and evaluation of a putative phosphorylation Site at Ser102. PLoS ONE 8:e58918. 10.1371/journal.pone.0058918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Huang H., Kahnt J., Mueller A. P., Kopke M., Thauer R. K. (2013b). NADP-specific electron-bifurcating [FeFe]-hydrogenase in a functional complex with formate dehydrogenase in Clostridium autoethanogenum grown on CO. J. Bacteriol. 195, 4373–4386. 10.1128/JB.00678-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Huang H., Moll J., Thauer R. K. (2010). NADP+ reduction with reduced ferredoxin and NADP+ Reduction with NADH are coupled via an electron-bifurcating enzyme complex in Clostridium kluyveri. J. Bacteriol. 192, 5115–5123. 10.1128/JB.00612-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., San K.-Y., Bennett G. N. (2013c). Improvement of NADPH bioavailability in Escherichia coli by replacing NAD(+)-dependent glyceraldehyde-3-phosphate dehydrogenase GapA with NADP (+)-dependent GapB from Bacillus subtilis and addition of NAD kinase. J. Ind. Microbiol. Biotechnol. 40, 1449–1460. 10.1007/s10295-013-1335-x [DOI] [PubMed] [Google Scholar]
- Wang Y., San K.-Y., Bennett G. N. (2013d). Improvement of NADPH bioavailability in Escherichia coli through the use of phosphofructokinase deficient strains. Appl. Microbiol. Biotechnol. 97, 6883–6893. 10.1007/s00253-013-4859-0 [DOI] [PubMed] [Google Scholar]
- Watanabe S., Sasaki D., Tominaga T., Miki K. (2012). Structural basis of [NiFe] hydrogenase maturation by Hyp proteins. Biol. Chem. 393, 1089–1100. 10.1515/hsz-2012-0197 [DOI] [PubMed] [Google Scholar]
- Weckbecker A., Hummel W. (2004). Improved synthesis of chiral alcohols with Escherichia coli cells co-expressing pyridine nucleotide transhydrogenase, NADP+-dependent alcohol dehydrogenase and NAD+-dependent formate dehydrogenase. Biotechnol. Lett. 26, 1739–1744. 10.1007/s10529-004-3746-2 [DOI] [PubMed] [Google Scholar]
- Wells M. A., Mercer J., Mott R. A., Pereira-Medrano A. G., Burja A. M., Radianingtyas H., et al. (2011). Engineering a non-native hydrogen production pathway into Escherichia coli via a cyanobacterial [NiFe] hydrogenase. Metab. Eng. 13, 445–453. 10.1016/j.ymben.2011.01.004 [DOI] [PubMed] [Google Scholar]
- Whitehead S. J., Iwaki M., Cotton N. P. J., Rich P. R., Jackson J. B. (2009). Inhibition of proton-transfer steps in transhydrogenase by transition metal ions. Biochim. Biophys. Acta 1787, 1276–1288. 10.1016/j.bbabio.2009.06.001 [DOI] [PubMed] [Google Scholar]
- Whitehead S. J., Rossington K. E., Hafiz A., Cotton N. P. J., Jackson J. B. (2005). Zinc ions selectively inhibit steps associated with binding and release of NADP(H) during turnover of proton-translocating transhydrogenase. FEBS Lett. 579, 2863–2867. 10.1016/j.febslet.2005.04.026 [DOI] [PubMed] [Google Scholar]
- Wichmann R., Vasic-Racki D. (2005). Cofactor regeneration at the lab scale. Adv. Biochem. Eng. Biotechnol. 92, 225–260. 10.1007/b98911 [DOI] [PubMed] [Google Scholar]
- Xu Z., Jing K., Liu Y., Cen P. (2007). High-level expression of recombinant glucose dehydrogenase and its application in NADPH regeneration. J. Ind. Microbiol. Biotechnol. 34, 83–90. 10.1007/s10295-006-0168-2 [DOI] [PubMed] [Google Scholar]
- Yamauchi Y., Hirasawa T., Nishii M., Furusawa C., Shimizu H. (2014). Enhanced acetic acid and succinic acid production under microaerobic conditions by Corynebacterium glutamicum harboring Escherichia coli transhydrogenase gene pntAB. J. Gen. Appl. Microbiol. 60, 112–118. 10.2323/jgam.60.112 [DOI] [PubMed] [Google Scholar]
- Yamazaki M., Ichikawa Y. (1990). Some properties of the apoenzyme of NADPH-adreno-ferredoxin reductase from bovine adrenocortical mitochondria. Int. J. Biochem. 22, 1147–1152. 10.1016/0020-711X(90)90113-H [DOI] [PubMed] [Google Scholar]
- Yan Z., Nam Y.-W., Fushinobu S., Wakagi T. (2014). Sulfolobus tokodaii ST2133 is characterized as a thioredoxin reductase-like ferredoxin:NADP+ oxidoreductase. Extremophiles 18, 99–110. 10.1007/s00792-013-0601-1 [DOI] [PubMed] [Google Scholar]
- Yang Z., Zhang H., Hung H.-C., Kuo C.-C., Tsai L.-C., Yuan H. S., et al. (2002). Structural studies of the pigeon cytosolic NADP(+)-dependent malic enzyme. Protein Sci. 11, 332–341. 10.1110/ps.38002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeom J., Jeon C. O., Madsen E. L., Park W. (2009). In vitro and in vivo interactions of ferredoxin-NADP+ reductases in Pseudomonas putida. J. Biochem. 145, 481–491. 10.1093/jb/mvn185 [DOI] [PubMed] [Google Scholar]
- Ying W. (2008). NAD+/NADH and NADP+/NADPH in cellular functions and cell death: regulation and biological consequences. Antioxid. Redox Signal. 10, 179–206. 10.1089/ars.2007.1672 [DOI] [PubMed] [Google Scholar]
- Yokooji Y., Sato T., Fujiwara S., Imanaka T., Atomi H. (2013). Genetic examination of initial amino acid oxidation and glutamate catabolism in the hyperthermophilic archaeon Thermococcus kodakarensis. J. Bacteriol. 195, 1940–1948. 10.1128/JB.01979-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J.-J., Hattori T., Shimada M. (2003). Purification and characterization of NADP-linked isocitrate dehydrogenase from the copper-tolerant wood-rotting basidiomycete Fomitopsis palustris. Biosci. Biotechnol. Biochem. 67, 114–120. 10.1271/bbb.67.114 [DOI] [PubMed] [Google Scholar]
- Yun H., Choi H.-L., Fadnavis N. W., Kim B.-G. (2005). Stereospecific synthesis of (R)-2-hydroxy carboxylic acids using recombinant E. coli BL21 overexpressing YiaE from Escherichia coli K12 and glucose dehydrogenase from Bacillus subtilis. Biotechnol. Prog. 21, 366–371. 10.1021/bp049694w [DOI] [PubMed] [Google Scholar]
- Zalacain M., Biswas S., Ingraham K. A., Ambrad J., Bryant A., Chalker A. F., et al. (2003). A global approach to identify novel broad-spectrum antibacterial targets among proteins of unknown function. J. Mol. Microbiol. Biotechnol. 6, 109–126. 10.1159/000076741 [DOI] [PubMed] [Google Scholar]
- Zamboni N., Fischer E., Laudert D., Aymerich S., Hohmann H.-P., Sauer U. (2004). The Bacillus subtilis yqjI gene encodes the NADP+-dependent 6-P-gluconate dehydrogenase in the pentose phosphate pathway. J. Bacteriol. 186, 4528–4534. 10.1128/JB.186.14.4528-4534.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.-D., Li A.-T., Yu H.-L., Imanaka T., Xu J.-H. (2011). Synthesis of optically pure S-sulfoxide by Escherichia coli transformant cells coexpressing the P450 monooxygenase and glucose dehydrogenase genes. J. Ind. Microbiol. Biotechnol. 38, 633–641. 10.1007/s10295-010-0809-3 [DOI] [PubMed] [Google Scholar]
- Zhang J., Xu J. H. (2010). Biocatalysis, cofactor regeneration. Encyclopedia Ind. Biotechnol. 1–9. 10.1002/9780470054581.eib073 [DOI] [Google Scholar]
- Zhao H., Van Der Donk W. A. (2003). Regeneration of cofactors for use in biocatalysis. Curr. Opin. Biotechnol. 14, 583–589. 10.1016/j.copbio.2003.09.007 [DOI] [PubMed] [Google Scholar]
- Zhao H., Wang P., Huang E., Ge Y., Zhu G. (2008). Physiologic roles of soluble pyridine nucleotide transhydrogenase in Escherichia coli as determined by homologous recombination. Ann. Microbiol. 58, 275–280. 10.1007/BF03175329 [DOI] [Google Scholar]
- Zhao J., Baba T., Mori H., Shimizu K. (2004a). Effect of zwf gene knockout on the metabolism of Escherichia coli grown on glucose or acetate. Metab. Eng. 6, 164–174. 10.1016/j.ymben.2004.02.004 [DOI] [PubMed] [Google Scholar]
- Zhao J., Baba T., Mori H., Shimizu K. (2004b). Global metabolic response of Escherichia coli to gnd or zwf gene-knockout, based on 13C-labeling experiments and the measurement of enzyme activities. Appl. Microbiol. Biotechnol. 64, 91–98. 10.1007/s00253-003-1458-5 [DOI] [PubMed] [Google Scholar]
- Zhao Z., Kuijvenhoven K., Van Gulik W. M., Heijnen J. J., Van Winden W. A., Verheijen P. J. T. (2011). Cytosolic NADPH balancing in Penicillium chrysogenum cultivated on mixtures of glucose and ethanol. Appl. Microbiol. Biotechnol. 89, 63–72. 10.1007/s00253-010-2851-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H., Ohno Y., Nakamori T., Suye S.-I. (2009). Production of L-malic acid with fixation of HCO3(-) by malic enzyme-catalyzed reaction based on regeneration of coenzyme on electrode modified by layer-by-layer self-assembly method. J. Biosci. Bioeng. 107, 16–20. 10.1016/j.jbiosc.2008.09.009 [DOI] [PubMed] [Google Scholar]
- Zhou J., Olson D. G., Argyros D. A., Deng Y., Van Gulik W. M., Van Dijken J. P., et al. (2013). Atypical glycolysis in Clostridium thermocellum. Appl. Environ. Microbiol. 79, 3000–3008. 10.1128/AEM.04037-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G., Golding G. B., Dean A. M. (2005). The selective cause of an ancient adaptation. Science 307, 1279–1282. 10.1126/science.1106974 [DOI] [PubMed] [Google Scholar]
- Ziegler M. (2000). New functions of a long-known molecule. Eur. J. Biochem. 267, 1550–1564. 10.1046/j.1432-1327.2000.01187.x [DOI] [PubMed] [Google Scholar]