ABSTRACT
The actinomycete Nonomuraea sp. strain ATCC 39727 produces the glycopeptide A40926, the precursor of dalbavancin. Biosynthesis of A40926 is encoded by the dbv gene cluster, which contains 37 protein-coding sequences that participate in antibiotic biosynthesis, regulation, immunity, and export. In addition to the positive regulatory protein Dbv4, the A40926-biosynthetic gene cluster encodes two additional putative regulators, Dbv3 and Dbv6. Independent mutations in these genes, combined with bioassays and liquid chromatography-mass spectrometry (LC-MS) analyses, demonstrated that Dbv3 and Dbv4 are both required for antibiotic production, while inactivation of dbv6 had no effect. In addition, overexpression of dbv3 led to higher levels of A40926 production. Transcriptional and quantitative reverse transcription (RT)-PCR analyses showed that Dbv4 is essential for the transcription of two operons, dbv14-dbv8 and dbv30-dbv35, while Dbv3 positively controls the expression of four monocistronic transcription units (dbv4, dbv29, dbv36, and dbv37) and of six operons (dbv2-dbv1, dbv14-dbv8, dbv17-dbv15, dbv21-dbv20, dbv24-dbv28, and dbv30-dbv35). We propose a complex and coordinated model of regulation in which Dbv3 directly or indirectly activates transcription of dbv4 and controls biosynthesis of 4-hydroxyphenylglycine and the heptapeptide backbone, A40926 export, and some tailoring reactions (mannosylation and hexose oxidation), while Dbv4 directly regulates biosynthesis of 3,5-dihydroxyphenylglycine and other tailoring reactions, including the four cross-links, halogenation, glycosylation, and acylation.
IMPORTANCE This report expands knowledge of the regulatory mechanisms used to control the biosynthesis of the glycopeptide antibiotic A40926 in the actinomycete Nonomuraea sp. strain ATCC 39727. A40926 is the precursor of dalbavancin, approved for treatment of skin infections by Gram-positive bacteria. Therefore, understanding the regulation of its biosynthesis is also of industrial importance. So far, the regulatory mechanisms used to control two other similar glycopeptides (balhimycin and teicoplanin) have been elucidated, and beyond a common step, different clusters seem to have devised different strategies to control glycopeptide production. Thus, our work provides one more example of the pitfalls of deducing regulatory roles from bioinformatic analyses only, even when analyzing gene clusters directing the synthesis of structurally related compounds.
INTRODUCTION
The glycopeptide antibiotic A40926 (Fig. 1A) is the precursor for dalbavancin, a semisynthetic lipoglycopeptide approved for clinical use in 2014 by the U.S. Food and Drug Administration to treat acute bacterial skin and skin structure infections caused by bacteria, such as Staphylococcus aureus (methicillin-susceptible and methicillin-resistant strains) and Streptococcus pyogenes. Compared with the structurally related glycopeptide teicoplanin, dalbavancin possesses better potency and pharmacokinetic properties.
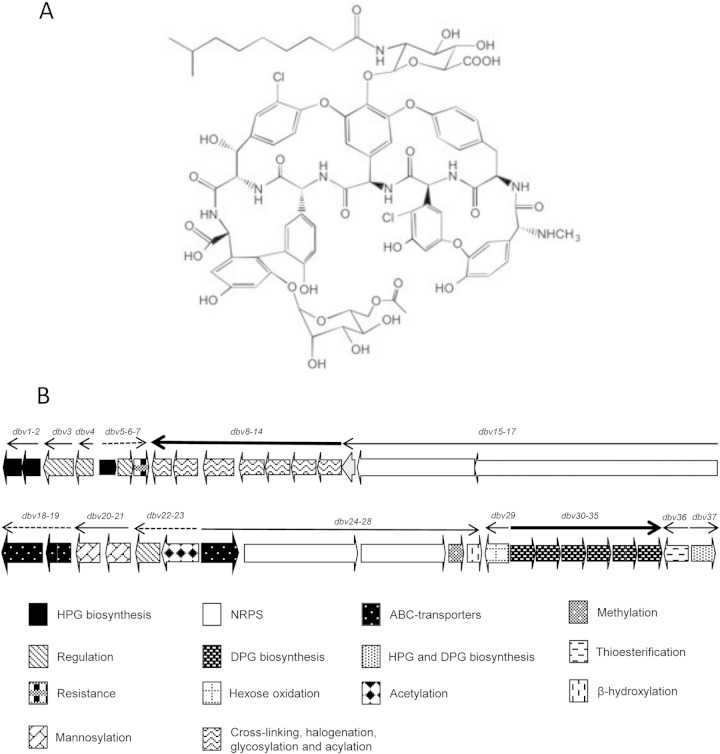
FIG 1.
Structures of A40926 (A) and of the dbv gene cluster (B). The arrows indicate operons, as previously determined experimentally (9). The thin arrows represent the transcriptional units controlled by Dbv3, the thick arrows indicate the Dbv4-controlled operons, and the dashed arrows indicate operons whose control is unknown. dbv genes are grouped by category as indicated.
A40926 is produced by the actinomycete Nonomuraea sp. strain ATCC 39727 as a complex of related compounds. It consists of a heptapeptide containing the proteinogenic amino acid tyrosine and the nonproteinogenic amino acids 3,5-dihydroxyphenylglycine (DPG) and 4-hydroxyphenylglycine (HPG) (1–4). The heptapeptide is assembled by a nonribosomal peptide synthetase (NRPS) and then modified by oxidative cross-linking of the aromatic side chains to yield a rigid peptide scaffold. Further tailoring steps include halogenation, glycosylation, oxidation, methylation, acetylation, and acylation (5). The A40926 components differ by the type of N-acyl chain attached to the glucuronic acid moiety and by the presence or absence of an O-linked acetyl residue at position 6 of the mannose (6–8). The form with the acetylated mannose was proposed to be the final biosynthetic product, which in turn exerts an inhibitory effect on A40926 production by a yet unknown mechanism (8). Detailed studies of A40926 biosynthesis and its regulation will be crucial for the development of rational approaches towards overproduction of the dalbavancin precursor.
The dbv gene cluster for A40926 biosynthesis contains 37 protein-coding sequences (Fig. 1B) that participate in antibiotic biosynthesis, regulation, immunity, and export (5). Previous reverse transcription (RT)-PCR analysis of the intergenic regions (9) revealed that the cluster contains five monocistronic transcription units (dbv3, dbv4, dbv29, dbv36, and dbv37) and nine operons (dbv2-dbv1, dbv5-dbv7, dbv14-dbv8, dbv17-dbv15, dbv18-dbv19, dbv20-dbv21, dbv22-dbv23, dbv24-dbv28, and dbv30-dbv35), indicated by arrows in Fig. 1B.
The cluster contains the positive regulatory gene dbv4, the putative regulatory gene dbv3, and the two-component system-encoding genes dbv6-dbv22 (5). Most glycopeptide gene clusters encode a homolog of Dbv4, as well as conserved Dbv4-binding sites (10–12). Indeed, our previous analysis revealed that Dbv4 is a DNA-binding protein that acts as a positive regulator of A40926 biosynthesis by controlling expression of just two dbv operons: dbv14-dbv8, encoding the four cross-linking oxygenases, the halogenase, the N-acetylglucosamine transferase, and the N-acylase, and dbv30-dbv35, encoding the four enzymes involved in DPG biosynthesis, as well as the sodium-proton antiporter and a protein of unknown function (9). In previous work, we also showed that expression of dbv4 is increased under phosphate-limiting conditions (9).
The putative regulator Dbv3 belongs to the LAL (large ATP-binding regulators of LuxR) family. Recently, several regulators of the LAL family have been identified in antibiotic and other secondary-metabolite gene clusters from actinomycetes (13–16). The last two positive regulators, dbv6 and dbv22, encode a response regulator and a histidine sensory kinase, respectively, homologous to the Streptomyces lividans CutR/CutS pair that negatively regulates secondary metabolism (17).
In this study, we characterize the functions of dbv3, dbv4, and dbv6 in regulating A40926 biosynthesis in Nonomuraea sp. strain ATCC 39727 and demonstrate that most of the dbv genes are under the control of Dbv3.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli strains were grown in Luria broth liquid medium at 37°C and were supplemented with 50 μg ml−1 apramycin when necessary to maintain plasmids. Frozen cell stocks of Nonomuraea strains were prepared by storing individual aliquots of the desired strain from midexponential cultures in R3 medium [10 g/liter glucose, 5 g/liter yeast extract, 0.1 g/liter Casamino Acids, 3 g/liter l-proline, 10 g/liter MgCl2 · 6H2O, 4 g/liter CaCl2 · 2H2O, 0.2 g/liter K2SO4, 0.05 g/liter KH2PO4, 5.6 g/liter N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid (TES), 2 ml/liter trace elements, pH 7.2] at −80°C. Liquid cultures of Nonomuraea strains were prepared by inoculating 0.2 ml of the frozen cell stock into 20 ml of fresh R3 medium; after 120 h of growth, 1 ml of this culture was inoculated into 50 ml of fresh R3 medium in an orbital shaker (500 × g) in 250-ml baffled flasks at 30°C. The cultivation media were supplemented with 50 μg ml−1 apramycin to select for strains carrying integrated antibiotic resistance genes. For solid media, R3 was supplemented with 1.8% agar. To detect glycopeptide production, Nonomuraea strains were incubated in R3 liquid medium without apramycin, and aliquots of spent media or culture broths of the strains at different time points were withdrawn for analysis and stored at −80°C. To study the effect of dbv3 expression in the dbv3-overexpressing (Oe-dbv3) strain, Nonomuraea sp. strain ATCC 39727 and the Oe-dbv3 strain were grown in R3 liquid medium with 10−8 M thiostrepton. This concentration was found to have no effect on Nonomuraea sp. strain ATCC 39727 growth and dbv3 transcription, and it did not inhibit Micrococcus luteus ATCC 9341 (18) and thus did not interfere with the assay of antibiotic activity due to A40926.
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Nonomuraea strains | ||
| ATCC 39727 | Wild type | ATCC |
| Δdbv3 mutant | Strain in which dbv3 gene was replaced with an apramycin cassette | This work |
| Δdbv4 mutant | Strain in which dbv4 gene was replaced with an apramycin cassette | This work |
| Δdbv6 mutant | Strain in which dbv6 gene was replaced with an apramycin cassette | This work |
| Oe-dbv3 | Strain carrying pIJ8600-dbv3 integrated into the attB site in which an additional copy of the dbv3 gene is under tipA promoter control | This work |
| Micrococcus luteus ATCC 9341 | Used for bioassay of A40926 | ATCC |
| E. coli strains | ||
| DH10B | F− endA1 recA1 galE15 galK16 nupG rpsL ΔlacX74 ϕ80lacZΔM15 araD139 Δ(ara-leu)7697 mcrA Δ(mrr-hsdRMS-mcrBC) λ−; host for routine subcloning experiments | Invitrogen |
| ET12567(pUZ8002) | (dam-13::Tn9 dcm-6) pUZ8002+ (ΔoriT); used for conjugative transfer of DNA and for demethylating plasmid DNA | 39 |
| BW25113/pIJ790 | Δ(araD-araB)567 ΔlacZ4787(::rrnB-4) lacIp-4000(lacIq) λ− rpoS369(Am) rph-1 Δ(rhaD-rhaB)568 hsdR514: used to generate recombinant cosmid 11A5 | 40 |
| Plasmids | ||
| 11A5 | Cosmid containing part of the dbv cluster from dbv1 to dbv16 | 9 |
| 11A5 Δdbv3 | Derivative of 11A5 with the inactivated dbv3 gene | This work |
| 11A5 Δdbv4 | Derivative of 11A5 with the inactivated dbv4 gene | This work |
| 11A5 Δdbv6 | Derivative of 11A5 with the inactivated dbv6 gene | This work |
| pIJ773 | pUC19 containing the aac(3)IV-oriT cassette; source of the aac(3) gene | 22 |
| pIJ790 | λ Red plasmid; contains the resistance marker cat and a temperature-sensitive origin of replication | 40 |
| pIJ8600 | Actinomycete integrative vector with the tipA promoter | 21 |
| pIJ8600-dbv3 | pIJ8600 derivative containing dbv3 under tipA promoter control | This work |
Analysis of antibiotic production.
A paper disc diffusion method was used with M. luteus as the assay organism as described previously (19). Nonomuraea strains were grown in selected liquid media under the conditions specified above. After cultivation, 100 μl of spent medium was applied to Whatman 3 MM Chr paper discs (Whatman, Maidstone, United Kingdom). The wet discs were placed on the surface of LB soft agar inoculated with 100 μl of M. luteus (optical density at 600 nm [OD600] = 1.2), and inhibition zones were measured after overnight incubation at 37°C. In parallel bioassays, the specific competitor d-Ala–d-Ala (20 μg/μl) was added to the spent medium of broth cultures to confirm A40926 antibacterial activity. For high-performance liquid chromatography (HPLC) and liquid chromatography-mass spectrometry (LC-MS) analyses, 50 μl of broth cultures was collected at the time points indicated and mixed briefly with 2 volumes of methanol followed by 1 h of incubation at 40°C. After centrifugation to remove the cells, the supernatant was lyophilized and redissolved in 100 μl 10% dimethyl sulfoxide (DMSO). HPLC analyses were performed using an LC 2010A-HT (Shimadzu) equipped with a Merck LiChrosphere (5-μm; 4.6-mm by 100-mm) C18 column. Elution was performed at 1 ml min−1 at 50°C with a linear gradient from 10% to 90% phase B in 30 min. Phase A was 0.1% (vol/vol) trifluoroacetic acid (TFA) in H2O, and phase B was CH3CN. UV detection was set at 230 nm. LC-electrospray ionization (ESI)-MS data were recorded on an Ion Trap equipped with an LC Agilent 1100, using an Ascentis express Supelco RP18, 2.7-μm (50- by 4.6-mm) column kept at 40°C, eluting at 1 ml min−1 with a 7-min linear gradient from 95:5 phase A (0.05% [vol/vol] TFA in water)-phase B (0.05% [vol/vol] TFA in acetonitrile) to 100% phase B.
DNA manipulation.
Standard genetic techniques with E. coli and in vitro DNA manipulations were performed as described previously (20). Isolation of Streptomyces total DNA was performed by the salting-out procedure (21). Southern hybridization was carried out with probes labeled with digoxigenin by using the DIG DNA labeling kit (Roche Biochemicals). PstI- or PvuII (Invitrogen)-digested pIJ8600 was used as a probe. Chromosomal DNAs were digested with BamHI and PstI (Invitrogen).
Construction of the Δdbv3, Δdbv4, Δdbv6, and Oe-dbv3 strains.
dbv3, dbv4, and dbv6 of Nonomuraea sp. strain ATCC 39727 were deleted by replacing the gene with a cassette containing an apramycin-selectable marker using PCR targeting (22). pIJ773 containing the apramycin resistance gene [aac(3)IV] and the oriT replication origin was used as a template. The PCR primers used to amplify the resistance cassette for deletion of the dbv genes are reported in Table S1 in the supplemental material. E. coli BW25113/pIJ790 bearing cosmid 11A5 (5) was electrotransformed with the deletion cassettes. The isolated mutant cosmids were first introduced into the nonmethylating E. coli strain ET12567 containing pUZ8002 and then transferred to Nonomuraea sp. strain ATCC 39727 by intergeneric conjugation. Briefly, Nonomuraea sp. strain ATCC 39727 was grown in 30 ml R3 medium in a 250-ml baffled flask and incubated at 30°C on a rotary shaker at 200 rpm for 40 h. Mycelium was collected, washed twice with ice-cold 10% (vol/vol) glycerol, and suspended in 10 ml of 10% (vol/vol) glycerol. Then, 0.5 ml of the mycelium was added to 0.5 ml E. coli donor cells prepared as described previously (21). The mixtures were plated out on R3 agar plates. The conjugation plates were incubated for 18 to 20 h at 30°C and then overlaid with 1 ml water containing 1 mg nalidixic acid and 1 mg apramycin. The plates were incubated at 30°C for 10 to 15 days. Exconjugants were picked off and analyzed by PCR (data not shown), using primers reported in Table S1 in the supplemental material. One clone (out of 3 analyzed) contained the interrupted dbv3 gene, 3 clones (out of 12 analyzed) contained the interrupted dbv4 gene, and 1 clone (out of 9) carried the interrupted dbv6 gene. The fidelity of the PCR products was confirmed by sequencing (BMR Genomics).
To construct the Oe-dbv3 strain, a PCR fragment prepared using the primers dbv3 over for and dbv3 over rev (see Table S1 in the supplemental material) was cloned into pGEM T-Easy (Promega), digested with BglII (Invitrogen) and NdeI (Invitrogen), and then inserted between the BamHI (Invitrogen) and NdeI (Invitrogen) sites of pIJ8600 (21) under the control of the thiostrepton-inducible tipA promoter. Cloning was confirmed by sequencing the insert. The plasmid was transferred into Nonomuraea sp. strain ATCC 39727 by conjugation from E. coli ET12567(pUZ8002) as described above, generating a strain called Oe-dbv3. As a control, pIJ8600 was inserted into the Nonomuraea sp. strain ATCC 39727 chromosome. Southern hybridization using PstI-digested pIJ8600 as a probe was carried out (see Fig. S1 in the supplemental material). As expected for integration at the chromosomal attB site, hybridizing bands of about 4.3 and 3.9 kb were observed with BamHI-digested chromosomal DNA (see Fig. S1, lane 1, in the supplemental material) and four bands of about 6, 4.3, 2, and 0.8 kb (see Fig. S2, lane 2, in the supplemental material) were observed with PstI-digested chromosomal DNA. No hybridization signal was detected for the parental strain (see Fig. S1, lanes 3 and 4, in the supplemental material).
Total RNA isolation and qRT-PCR analysis.
To perform transcriptional analysis of selected genes, mycelium was harvested from 6 ml of culture, resuspended in 1 ml P buffer (21) containing lysozyme (50 mg/ml), and then incubated for 20 min at 37°C. RNA was extracted by using the RNeasy midi kit (Qiagen) according to the manufacturer's instructions. DNase I (Roche) treatment was performed at 37°C for 1 h, and RNA was precipitated with 2 volumes of ethanol in the presence of 0.1 volume of 3 M sodium acetate. After a washing step with 70% (vol/vol) ethanol and drying in air, the RNA pellet was resuspended in water. As a control for RNA quality, an RT-PCR with 0.1 μg of total RNA and primer pairs internal to hrdB, encoding a vegetative sigma factor, was carried out using the Superscript One-Step RT-PCR kit (Invitrogen) under the conditions indicated by the supplier. PCRs using the same primers were performed on 0.5 μg of RNA samples using 40 cycles to exclude the presence of genomic DNA. For quantitative RT (qRT)-PCR, a two-step protocol was used. The High-Capacity cDNA Archive kit (Applied Biosystems) was used to retrotranscribe 2 μg of total extracted RNA in a total volume of 100 μl. Primer pairs for the genes analyzed by qRT-PCR were designed with Primer3web version 4.0.0 (http://bioinfo.ut.ee/primer3/) to fulfill the following criteria: length, 20 ± 2 bp; melting temperature (Tm), 60 ± 1°C; GC content, >50%; and amplicon lengths ranging from 200 to 500 bp (see Table S1 in the supplemental material). Primer specificity was controlled by BLAST analysis. The PCR efficiency was estimated by using a free Web-based service (http://srvgen.upct.es/efficiency.html) (23). Gene expression was analyzed quantitatively by using the Applied Biosystems 7300 real-time PCR system (Applied Biosystems) with SYBR green PCR master mix (Applied Biosystems) in 96-well plates. Two microliters of cDNA was added to 20 μl of PCR mixture. Amplification required activation of AmpErase UNG at 50°C for 2 min, followed by denaturation at 95°C for 10 min and then 40 cycles at 95°C for 15 s and 60°C for 1 min. Melting curves were performed from 60 to 98°C to validate the specificity of the PCR. Three independent measurements were performed for each gene. The threshold cycle (CT) values were determined with the baseline set automatically. The results were analyzed using the comparative critical threshold (ΔΔCT) method, in which the amount of target RNA is adjusted to an internal reference (24). hrdB, encoding a vegetative sigma factor, and a 16S rRNA gene did not show significant expression variation, as revealed by absolute quantitative RT-PCR using RNA extracted after 24, 48, and 72 h from Nonomuraea sp. strain ATCC 39727 (see Fig. S2 in the supplemental material). Standard deviations were calculated from three independent experiments. The values were calculated using a standard curve obtained by measuring the fluorescence of known amounts (10, 100, 1,000, and 10,000 μg) of cDNA and were used as internal references to normalize the results. Two different sets of experiments were carried out using one of the endogenous genes; the relative transcription values were compared and reported as the average of these two sets of experiments. Expression ratios were expressed as 2ΔΔCT. Each run included negative controls.
RESULTS
Transcript abundance of regulatory genes during growth.
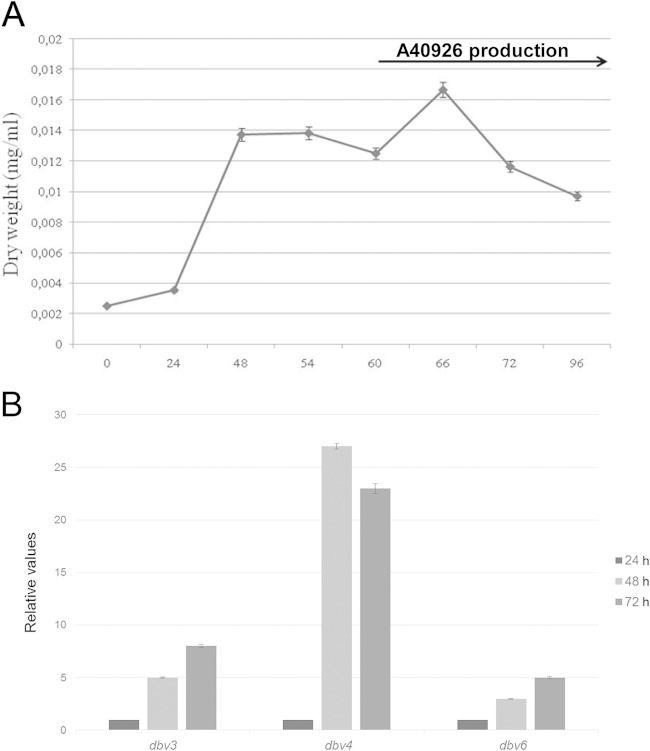
We evaluated the transcript levels of three regulatory genes present in the dbv cluster, dbv3, dbv4, and dbv6, during growth and in respect to A40926 production. Nonomuraea sp. strain ATCC 39727 growth and A40926 production were monitored in R3 liquid medium for 96 h. Microbiological bioassays indicated that A40926 biosynthesis was detectable after 60 h of growth (Fig. 2A). Total RNA was extracted before (at 24 and 48 h) and during (at 72 h) production and was analyzed for gene expression by qRT-PCR (Fig. 2B) using hrdB, encoding a vegetative sigma factor, as a control. The transcript levels for dbv3, dbv4, and dbv6 were ∼5-, ∼20, and ∼3-fold higher, respectively, at 48 and 72 h than at 24 h, indicating that transcription of all the regulatory genes is upregulated and largely precedes the detection of A40926 in the medium (Fig. 2). The largest increase was observed for dbv4.
FIG 2.
(A) Growth curve of Nonomuraea sp. strain ATCC 39727 grown in R3 medium. The arrow indicates A40926 detection in the supernatant. (B) qRT-PCR analysis of dbv3, dbv4, and dbv6 after 24, 48, and 72 h of growth. mRNA levels are expressed relative to hrdB transcripts, with the ratio values for the 24-h sample arbitrarily set to 1. The standard deviations (indicated by error bars) were calculated from three independent determinations of mRNA abundance in each sample.
Effects of inactivation of the dbv regulatory genes.
Disruption of dbv3, dbv4, or dbv6 did not significantly alter the growth kinetics or the morphology of the mutants on the different media used (data not shown), indicating that these genes are not critical for bacterial growth and differentiation under the conditions used. In contrast, bioassays showed that inactivation of dbv3 and dbv4 abolished A40926 biosynthesis, while the absence of dbv6 did not have a measurable effect on production (Fig. 3A). HPLC analysis confirmed that the Δdbv4 and Δdbv3 mutants produced undetectable levels of precursors and/or final product. In contrast, the metabolite profile of the Δdbv6 mutant was indistinguishable from that of Nonomuraea sp. strain ATCC 39727 (data not shown). These results indicate that both Dbv3 and Dbv4 are necessary for antibiotic production, whereas Dbv6 is not. Since Dbv6 did not show a role in antibiotic biosynthesis, we further characterized only the Δdbv3 and Δdbv4 strains.
FIG 3.
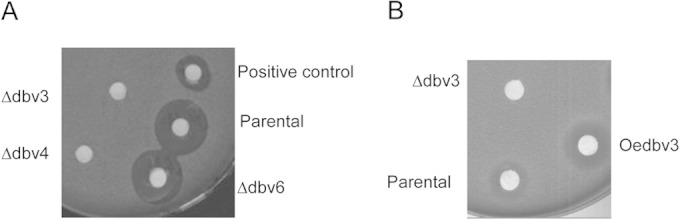
Bioassays of spent media from parental, Δdbv3, Δdbv4, and Δdbv6 strains (A) and from the Δdbv3 and Oe-dbv3 strains (B) after 72 h of growth. The positive control was spent medium collected from the parental strain after 120 h.
Interplay between the regulators.
In order to establish any hierarchical control, qRT-PCR of the dbv regulatory genes was performed in the Δdbv3 and Δdbv4 mutant strains. In the Δdbv4 strain, dbv3 and dbv6 were transcribed as in the parental strain (Fig. 4), indicating that their expression is not strictly dependent on Dbv4. In contrast, qRT-PCR analysis of the Δdbv3 strain revealed that dbv4 transcription was 10-fold reduced with respect to the parental strain, while dbv6 transcription was not changed significantly (Fig. 4). These results suggest that Dbv3 controls dbv4 transcription but does not have a major role, if any, in regulating dbv6 transcription.
FIG 4.
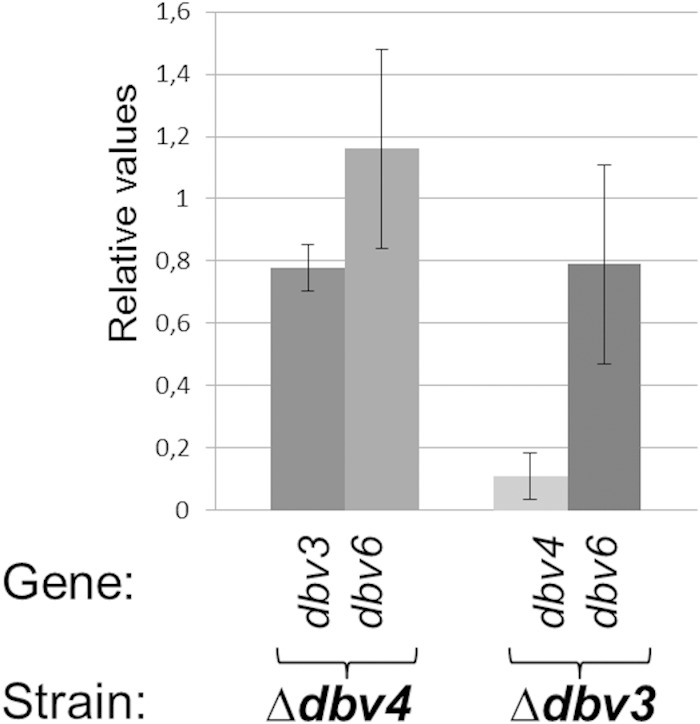
qRT-PCR analysis of dbv3 and dbv6 in the Δdbv4 mutant and of dbv4 and dbv6 in the Δdbv3 mutant after 60 h of growth. The transcription levels in the mutants were compared to those of the parental strain, taken as 1.0. The standard deviations (indicated by error bars) were calculated from three independent determinations of mRNA abundance in each sample. hrdB was used as an endogenous control.
Dbv4 was previously shown to bind to the upstream regions of dbv14 and dbv30 (9). Consistently, transcription of these two genes was strongly reduced in the Δdbv4 mutant (data not shown), indicating that Dbv4 is essential for transcription of dbv14 and dbv30 and, presumably, of the dbv14-dbv8 and dbv30-dbv35 operons (Fig. 1B).
Identification of Dbv3 target genes.
To identify Dbv3 target genes, total RNA was extracted from Nonomuraea sp. strain ATCC 39727 and the Δdbv3 strain, grown in R3 medium for 60 h, when antibiotic becomes detectable in the parental strain, and expression of dbv genes was analyzed by qRT-PCR. We concentrated on a subset of dbv genes, selecting them on the basis of transcriptional organization (Fig. 1B): the two monocistronic genes dbv4 and dbv29 and dbv1, dbv6, dbv14, dbv15, dbv20, and dbv32 as representatives of the dbv1-dbv2, dbv5-dbv7, dbv14-dbv8, dbv17-dbv15, dbv20-dbv21, and dbv30-dbv35 operons, respectively (Fig. 1B). The results clearly demonstrated that dbv1, dbv4, dbv14, dbv15, dbv20, dbv29, and dbv32 either are not transcribed or are 10-fold less transcribed in the Δdbv3 strain than in the parental strain (Fig. 5). In contrast, dbv6 transcript levels in the Δdbv3 strain were similar to those in the parental strain, indicating again that Dbv3 does not control dbv6 transcription.
FIG 5.
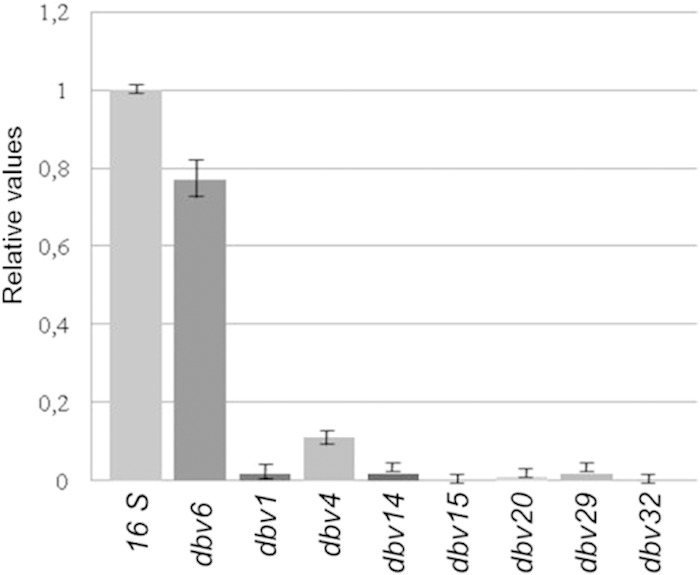
qRT-PCR of selected dbv genes in the Δdbv3 mutant after 60 h of growth. The transcription levels in the mutant were compared to those of the parental strain, taken as 1.0. The standard deviations (indicated by error bars) were calculated from three independent qRT-PCR experiments. The values were normalized using an endogenous 16S rRNA gene.
Increasing A40926 production by overexpression of dbv3.
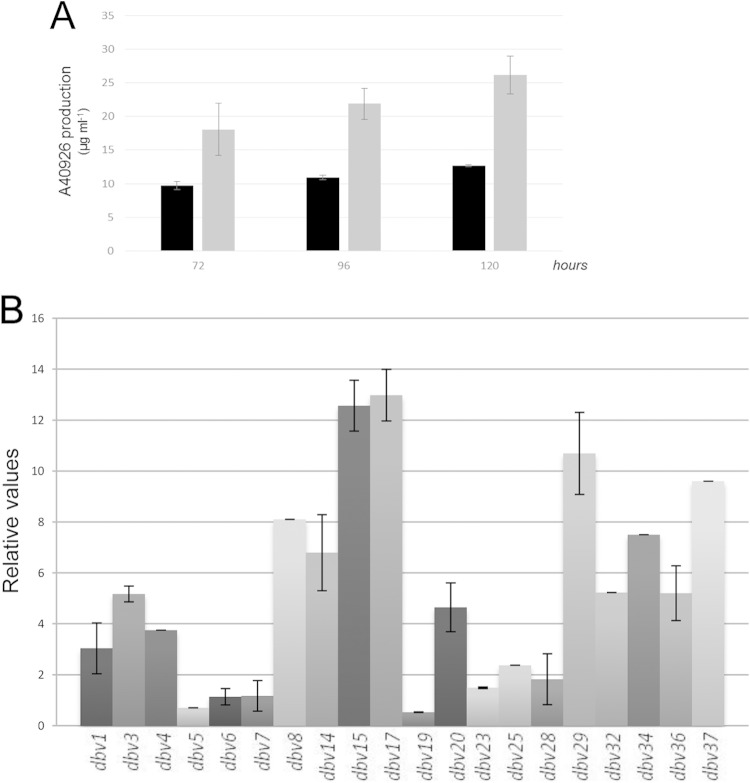
A dbv3-overexpressing strain (Oe-dbv3) was constructed by placing dbv3 under the control of the thiostrepton-inducible tipA promoter and integrating the construct at the chromosomal attB site (see Fig. S1 in the supplemental material). In agreement with the results obtained with the Δdbv3 mutant, no differences were observed between growth of the parental and Oe-dbv3 strains in liquid R3 medium (data not shown). To evaluate the effect of dbv3 overexpression on A40926 production, bioassays were performed using spent culture medium collected at different time points. The Oe-dbv3 strain produced more A40926 than the parental strain (Fig. 3B). HPLC and LC-MS analyses confirmed that the Oe-dbv3 strain produced 1.7- to 2.2-fold more A40926 than the parental strain (Fig. 6A). (The control strain, containing a copy of the empty vector, showed production profiles similar to those of the parental strain, thus excluding an effect of pIJ8600 integration on antibiotic production [data not shown].)
FIG 6.
A40926 production and qRT-PCR of selected dbv genes in the Oe-dbv3 strain. (A) A40926 production (micrograms per milliliter) by the parental (black bars) and Oe-dbv3 (gray bars) strains after 72, 96, and 120 h in R3 liquid medium. (B) qRT-PCR of selected dbv genes in the Oe-dbv3 strain after 60 h in R3 liquid medium. The transcript levels in the Oe-dbv3 strain were compared to those of the parental strain, taken as 1.0. The relative values were determined by comparison to the transcript levels of endogenous hrdB and 16S rRNA genes. Error bars indicate standard deviations.
Transcriptional control of A40926 production by Dbv3.
To study the influence of Dbv3 on transcription of a subset of 20 dbv genes, RNA was extracted from Nonomuraea sp. strain ATCC 39727 and from the Oe-dbv3 mutant grown in the presence of 10−8 M thiostrepton for 60 h and analyzed by qRT-PCR. With the exception of dbv19; the three cotranscribed genes dbv5, dbv6, and dbv7; and perhaps dbv23, the expression of all of the analyzed genes was significantly enhanced in the Oe-dbv3 strain compared with the parental strain (Fig. 6B), with enhancement levels ranging from 2-fold (e.g., dbv1, dbv25, and dbv28) to 12-fold (dbv15 and dbv17). Thus, from the established operon structure of the dbv gene cluster (Fig. 1B), we infer that Dbv3 may act as a positive regulator of 29 out of 37 dbv genes: those belonging to the six operons (dbv2-dbv1, dbv14-dbv8, dbv17-dbv15, dbv21-dbv20, dbv24-dbv28, and dbv30-dbv35) and the four monocistronic transcription units dbv4, dbv29, dbv36, and dbv37.
DISCUSSION
The present study was designed to elucidate the regulatory network for A40926 biosynthesis in the actinomycete Nonomuraea sp. strain ATCC 39727. Our experiments demonstrated that the transcriptional activators Dbv3 and Dbv4 are both required for A40926 biosynthesis, while Dbv6 does not influence A40926 biosynthesis (Fig. 3).
Disruption of any of these regulators did not significantly alter growth in Nonomuraea sp. strain ATCC 39727. This is in accordance with recent work showing that inactivation of tei15* (dbv4-like) or tei16* (dbv3-like) in Actinoplanes teichomyceticus does not influence growth but leads to a complete block of teicoplanin production (12).
qRT-PCR analysis showed that, in the absence of dbv4, transcription of dbv3 and dbv6 is not affected (Fig. 4), whereas transcription of the dbv14-dbv8 and dbv30-dbv35 operons is (data not shown), confirming that Dbv4 acts as a positive pathway-specific regulator of A40926 biosynthesis. Moreover, dbv4, dbv14-dbv8, and dbv30-dbv35 transcription was previously demonstrated to be activated by phosphate limitation, and mobility shift assays demonstrated that a recombinant His-Dbv4 binds to a consensus sequence present upstream of dbv14 and dbv30 (9). Similarly to Tei15* (12) from the teicoplanin gene cluster and differently from Bbr (11) and StrR (25) from the balhimycin- and streptomycin-biosynthetic gene clusters, respectively, Dbv4 was reported previously not to control its own expression (9).
Expanding on previous results, we demonstrate that dbv4 transcription is controlled by Dbv3, a regulator belonging to the LAL protein family. We found that Dbv3 plays a positive role in A40926 production, since the Δdbv3 strain completely lost the ability to produce the antibiotic (Fig. 3) and transcription of dbv4 and of many other dbv genes was drastically inhibited (Fig. 5). Several members of the LAL family are pathway-specific transcriptional activators of secondary-metabolite gene clusters in a range of actinomycetes (13, 14, 26–32). Recently, two LAL proteins were identified as pleiotropic regulators affecting various cellular processes in Streptomyces coelicolor (15).
Improvement of A40926 production by Nonomuraea sp. strain ATCC 39727 has been successfully obtained using different growth conditions (33–36). One example of successful genetic manipulation has also been reported, since deletion of dbv23, responsible for adding the acetyl group to the mannose moiety, led to a 2-fold increase in A40926 yield (8). In our studies, overexpression of dbv3 also resulted in higher antibiotic production, providing a potentially useful example of knowledge-based strain improvement.
Taken together, our results demonstrate that both Dbv3 and Dbv4 act as pathway-specific activators of A40926 biosynthesis, with Dbv3 hierarchically controlling dbv4 transcription. Our data suggest a cascade-like regulatory mechanism (Fig. 7) in which Dbv3 triggers transcription of dbv4 and, as a consequence, positively controls DPG biosynthesis and some key reactions that are the hallmark of glycopeptides (the four cross-links, halogenation, glycosylation, and acylation); in addition, Dbv3 controls HPG biosynthesis and heptapeptide backbone biosynthesis, as well as other tailoring reactions (mannosylation and hexose oxidation) and A40926 export. To our knowledge, the transcriptional activation of the NRPS genes by Dbv3 (Fig. 6B) represents the first example of a LAL regulator positively controlling the expression of NRPS genes, while a few examples of LAL regulators acting on polyketide synthase (PKS) gene expression have been reported (14).
FIG 7.
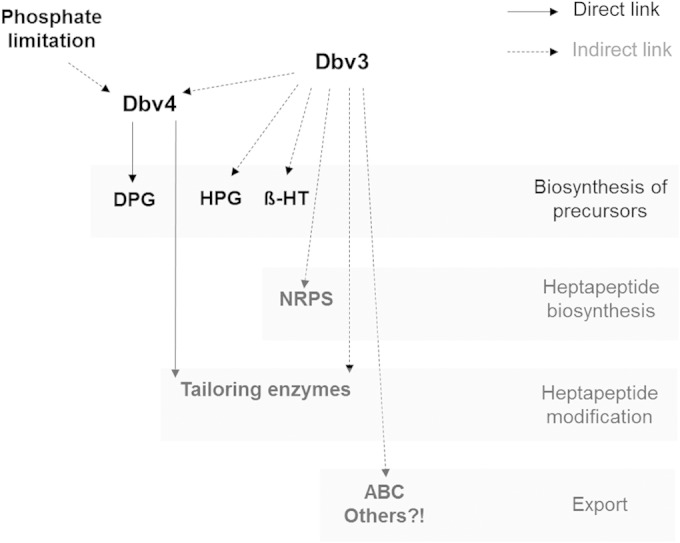
Model proposed for regulation of A40926 biosynthesis by Dbv3 and Dbv4. The solid and dashed arrows indicate direct and likely indirect links, respectively. β-HT, β-hydroxytyrosine.
Dbv4 and Dbv3 appear to be controlled by different stimuli, i.e., dbv4 transcription and the two Dbv4-controlled biosynthetic steps were demonstrated to be negatively regulated by phosphate, while the Dbv3-controlled genes were not (9). Surprisingly, while the intergenic regions preceding Dbv4-controlled genes are highly conserved (9, 10), we could not identify any conserved sequences that could be the target of Dbv3, nor has a binding site for Tei16* been reported (12). Thus, it remains to be determined whether Dbv3 exerts its influence by direct binding to regulatory regions or if other mechanisms are involved. The absence of conserved DNA sequences in all of the dbv genes whose transcript levels are higher after dbv3 overexpression suggests that Dbv3 control of A40926 biosynthesis might be indirect.
The dbv cluster was reported to have a mosaic structure (10) in which the various subclusters, responsible for different functional portions of the pathway, originated from different sources. Remarkably, the highly conserved Dbv4 regulator controls the expression of two operons devoted to important checkpoints along the A40926-biosynthetic pathway: one for synthesis of the specialized precursor DPG and the other for cross-linking, halogenation, and N-acetylglucosamine addition and acylation. The latter set of reactions contribute to the characteristic features of glycopeptides (multiple cross-links of the aromatic residues) and of the teicoplanin family in particular (acylated glucosamine attached to amino acid 4). It should also be noted that the P450 mono-oxygenases act during heptapeptide synthesis (37), and thus, lack of expression of the dbv14-dbv8 operon, in which the mono-oxygenase genes reside, is likely to result in truncated peptides devoid of antibacterial activity, as seen with balhimycin (38).
When comparing transcriptional regulation in different glycopeptide gene clusters, it is striking that the StrR-like regulators Dbv4, Bbr, and Tei15* all control one common biosynthetic step, DPG biosynthesis. However, beyond this common step, different clusters seem to have devised different regulatory strategies to control glycopeptide production: a single StrR-like regulator for balhimycin production, a StrR-like regulator controlling a LAL regulator in teicoplanin biosynthesis, and a LAL regulator controlling a StrR-like regulator for A40926 production. Our results not only enlarge our knowledge of the regulatory mechanisms used to control glycopeptide biosynthesis but also provide a basis for rational approaches for the generation of overproducers of the dalbavancin precursor.
Supplementary Material
ACKNOWLEDGMENTS
L.L.G. was partially supported by contract number PON01_02093. This work was financially supported by the University of Palermo, Fondo Finalizzato alla Ricerca (FFR/2012/2013), and the Biotechnological and Biological Sciences Research Council (BBSRC) Institute Strategic Programme Grant Understanding and Exploiting Plant and Microbial Secondary Metabolism (BB/J004561/1).
We thank Matteo Massimi for HPLC analyses.
We declare that we have no competing interests.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00262-15.
REFERENCES
- 1.Chen H, Tseng CC, Hubbard BK, Walsh CT. 2001. Glycopeptide antibiotic biosynthesis: enzymatic assembly of the dedicated amino acid monomer (S)-3,5-dihydroxyphenyl glycine. Proc Natl Acad Sci U S A 98:14901–14906. doi: 10.1073/pnas.221582098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pfeifer V, Nicholson GJ, Ries J, Recktenwald J, Schefer Shawky R, Schroder J, Wohlleben W, Pelzer S. 2001. A polyketide synthase in glycopeptide biosynthesis: the biosynthesis of the non-proteinogenic amino acid (S)-3,5-dihydroxyphenylglycine. J Biol Chem 276:38370–38377. doi: 10.1074/jbc.M106580200. [DOI] [PubMed] [Google Scholar]
- 3.Hubbard BK, Thomas MG, Walsh CT. 2000. Biosynthesis of L-p-hydroxyphenylglycine, a non-proteinogenic amino acid constituent of peptide antibiotics. Chem Biol 7:931–942. doi: 10.1016/S1074-5521(00)00043-0. [DOI] [PubMed] [Google Scholar]
- 4.Li TL, Choroba OW, Charles EH, Sandercock AM, Williams DH, Spencer JB. 2001. Characterisation of a hydroxymandelate oxidase involved in the biosynthesis of two unusual amino acids occurring in the vancomycin group of antibiotics. Chem Commun 18:1752–1753. [DOI] [PubMed] [Google Scholar]
- 5.Sosio M, Stinchi S, Beltrametti F, Lazzarini A, Donadio S. 2003. The gene cluster for the biosynthesis of the glycopeptide antibiotic A40926 by Nonomuraea species. Chem Biol 10:541–549. doi: 10.1016/S1074-5521(03)00120-0. [DOI] [PubMed] [Google Scholar]
- 6.Goldstein BP, Selva E, Gastaldo L, Berti M, Pallanza R, Ripamonti F, Ferrari P, Denaro M, Arioli V, Cassani G. 1987. A40926, a new glycopeptide antibiotic with anti-Neisseria activity. Antimicrob Agents Chemother 31:1961–1966. doi: 10.1128/AAC.31.12.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waltho JP, Williams DH, Selva E, Ferrari P. 1987. Structure elucidation of the glycopeptide antibiotic complex A40926. J Chem Soc Perkin Trans 1:2103–2107. [Google Scholar]
- 8.Sosio M, Canavesi A, Stinchi S, Donadio S. 2010. Improved production of A40926 by Nonomuraea sp. through deletion of a pathway-specific acetyltransferase. Appl Microbiol Biotechnol 87:1633–1638. doi: 10.1007/s00253-010-2579-2. [DOI] [PubMed] [Google Scholar]
- 9.Alduina R, Lo Piccolo L, D'Alia D, Ferraro C, Gunnarsson N, Donadio S, Puglia AM. 2007. Phosphate-controlled regulator for the biosynthesis of the dalbavancin precursor A40926. J Bacteriol 189:8120–8129. doi: 10.1128/JB.01247-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donadio S, Sosio M, Stegmann E, Weber T, Wohlleben W. 2005. Comparative analysis and insights into the evolution of gene clusters for glycopeptide antibiotic biosynthesis. Mol Genet Genomics 274:40–50. doi: 10.1007/s00438-005-1156-3. [DOI] [PubMed] [Google Scholar]
- 11.Shawky RM, Puk O, Wietzorrek A, Pelzer S, Takano E, Wohlleben W, Stegmann E. 2007. The border sequence of the balhimycin biosynthesis gene cluster from Amycolatopsis balhimycina contains bbr, encoding a StrR-like pathway-specific regulator. J Mol Microbiol Biotechnol 13:76–88. doi: 10.1159/000103599. [DOI] [PubMed] [Google Scholar]
- 12.Horbal L, Kobylyanskyy A, Truman AW, Zaburranyi N, Ostash B, Luzhetskyy A, Marinelli F, Fedorenko V. 2014. The pathway-specific regulatory genes, tei15* and tei16*, are the master switches of teicoplanin production in Actinoplanes teichomyceticus. Appl Microbiol Biotechnol 98:9295–9309. doi: 10.1007/s00253-014-5969-z. [DOI] [PubMed] [Google Scholar]
- 13.Kuscer E, Coates N, Challis I, Gregory M, Wilkinson B, Sheridan R, Petković H. 2007. Roles of rapH and rapG in positive regulation of rapamycin biosynthesis in Streptomyces hygroscopicus. J Bacteriol 189:4756–4763. doi: 10.1128/JB.00129-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He W, Lei J, Liu Y, Wang Y. 2008. The LuxR family members GdmRI and GdmRII are positive regulators of geldanamycin biosynthesis in Streptomyces hygroscopicus 17997. Arch Microbiol 189:501–510. doi: 10.1007/s00203-007-0346-2. [DOI] [PubMed] [Google Scholar]
- 15.Guerra SM, Rodríguez-García A, Santos-Aberturas J, Vicente CM, Payero TD, Martín JF, Aparicio JF. 2012. LAL regulators SCO0877 and SCO7173 as pleiotropic modulators of phosphate starvation response and actinorhodin biosynthesis in Streptomyces coelicolor. PLoS One 7:e31475. doi: 10.1371/journal.pone.0031475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pan W, Kang Q, Wang L, Bai L, Deng Z. 2013. Asm8, a specific LAL-type activator of 3-amino-5-hydroxybenzoate biosynthesis in ansamitocin production. Sci China Life Sci 56:601–608. doi: 10.1007/s11427-013-4502-4. [DOI] [PubMed] [Google Scholar]
- 17.Chang HM, Chen MY, Shieh YT, Bibb MJ, Chen CW. 1996. The cutRS signal transduction system of Streptomyces lividans represses the biosynthesis of the polyketide antibiotic actinorhodin. Mol Microbiol 21:1075–1085. [PubMed] [Google Scholar]
- 18.Kovács G, Burghardt J, Pradella S, Schumann P, Stackebrandt E, Màrialigeti K. 1999. Kocuria palustris sp. nov. and Kocuria rhizophila sp. nov., isolated from the rhizoplane of the narrow-leaved cattail (Typha angustifolia). Int J Syst Bacteriol 49:167–173. doi: 10.1099/00207713-49-1-167. [DOI] [PubMed] [Google Scholar]
- 19.Giardina A, Alduina R, Gallo G, Monciardini P, Sosio M, Puglia A. 2014. Inorganic phosphate is a trigger factor for Microbispora sp. ATCC-PTA-5024 growth and NAI-107 production. Microb Cell Fact 13:133. doi: 10.1186/s12934-014-0133-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 21.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. 2000. Practical Streptomyces genetics. John Innes Foundation, Norwich, England. [Google Scholar]
- 22.Gust B, Kieser T, Chater KF. 2002. REDIRECT technology: PCR targeting system in Streptomyces coelicolor. The John Innes Centre, Norwich, United Kingdom. [Google Scholar]
- 23.Mallona I, Weiss J, Egea-Cortines M. 2011. PCR Efficiency: a Web tool for PCR amplification efficiency prediction. BMC Bioinformatics 12:404. doi: 10.1186/1471-2105-12-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmittgen TD, Zakrajsek BA, Mills AG, Gorn V, Singer MJ, Reed MW. 2000. Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: comparison of endpoint and real-time methods. Anal Biochem 285:194–204. doi: 10.1006/abio.2000.4753. [DOI] [PubMed] [Google Scholar]
- 25.Retzlaff L, Distler J. 1995. The regulator of streptomycin gene expression, StrR, of Streptomyces griseus is a DNA binding activator protein with multiple recognition sites. Mol Microbiol 18:151–162. doi: 10.1111/j.1365-2958.1995.mmi_18010151.x. [DOI] [PubMed] [Google Scholar]
- 26.Wilson DJ, Xue Y, Reynolds KA, Sherman DH. 2001. Characterization and analysis of the PikD regulatory factor in the pikromycin biosynthetic pathway of Streptomyces venezuelae. J Bacteriol 183:3468–3475. doi: 10.1128/JB.183.11.3468-3475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aparicio JF, Molnár I, Schwecke T, König A, Haydock SF, Khaw LE, Staunton J, Leadlay PF. 1996. Organization of the biosynthetic gene cluster for rapamycin in Streptomyces hygroscopicus: analysis of the enzymatic domains in the modular polyketide synthase. Gene 169:9–16. doi: 10.1016/0378-1119(95)00800-4. [DOI] [PubMed] [Google Scholar]
- 28.Sekurova ON, Brautaset T, Sletta H, Borgos SE, Jakobsen M ØM, Ellingsen TE, Strøm AR, Valla S, Zotchev SB. 2004. In vivo analysis of the regulatory genes in the nystatin biosynthetic gene cluster of Streptomyces noursei ATCC 11455 reveals their differential control over antibiotic biosynthesis. J Bacteriol 186:1345–1354. doi: 10.1128/JB.186.5.1345-1354.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carmody M, Byrne B, Murphy B, Breen C, Lynch S, Flood E, Finnan S, Caffrey P. 2004. Analysis and manipulation of amphotericin biosynthetic genes by means of modified phage KC515 transduction techniques. Gene 343:107–115. doi: 10.1016/j.gene.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 30.Caffrey P, Aparicio JF, Malpartida F, Zotchev SB. 2008. Biosynthetic engineering of polyene macrolides towards generation of improved antifungal and antiparasitic agents. Curr Top Med Chem 8:639–653. doi: 10.2174/156802608784221479. [DOI] [PubMed] [Google Scholar]
- 31.Li Q, Wang L, Xie Y, Wang S, Chen R, Hong B. 2013. SsaA, a member of a novel class of transcriptional regulators, controls sansanmycin production in Streptomyces sp. strain SS through a feedback mechanism. J Bacteriol 195:2232–2243. doi: 10.1128/JB.00054-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goranovič D, Blažič M, Magdevska V, Horvat J, Kuščer E, Polak T, Santos-Aberturas J, Martínez-Castro M, Barreiro C, Mrak P, Kopitar G, Kosec G, Fujs S, Martín JF, Petković H. 2012. FK506 biosynthesis is regulated by two positive regulatory elements in Streptomyces tsukubaensis. BMC Microbiol 12:238. doi: 10.1186/1471-2180-12-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gunnarsson N, Bruheim P, Nielsen J. 2003. Production of the glycopeptide antibiotic A40926 by Nonomuraea sp. ATCC 39727: influence of medium composition in batch fermentation. J Ind Microbiol Biotechnol 30:150–156. [DOI] [PubMed] [Google Scholar]
- 34.Beltrametti F, Jovetic S, Feroggio M, Gastaldo L, Selva E, Marinelli F. 2004. Valine influences production and complex composition of glycopeptide antibiotic A40926 in fermentations of Nonomuraea sp. ATCC 39727. J Antibiot (Tokyo) 57:37–44. doi: 10.7164/antibiotics.57.37. [DOI] [PubMed] [Google Scholar]
- 35.Technikova-Dobrova Z, Damiano F, Tredici SM, Vigliotta G, di Summa R, Palese L, Abbrescia A, Labonia N, Gnoni GV, Alifano P. 2004. Design of mineral medium for growth of Actinomadura sp. ATCC 39727, producer of the glycopeptide A40926: effects of calcium ions and nitrogen sources. Appl Microbiol Biotechnol 65:671–677. [DOI] [PubMed] [Google Scholar]
- 36.Jovetic S, Feroggio M, Marinelli F, Lancini G. 2008. Factors influencing cell fatty acid composition and A40926 antibiotic complex production in Nonomuraea sp. ATCC 39727. J Ind Microbiol Biotechnol 35:1131–1138. doi: 10.1007/s10295-008-0392-z. [DOI] [PubMed] [Google Scholar]
- 37.Woithe K, Geib N, Zerbe K, Li DB, Heck M, Fournier-Rousset S, Meyer O, Vitali F, Matoba N, Abou-Hadeed K, Robinson JA. 2007. Oxidative phenol coupling reactions catalyzed by OxyB: a cytochrome P450 from the vancomycin producing organism. Implications for vancomycin biosynthesis. J Am Chem Soc 129:6887–6895. [DOI] [PubMed] [Google Scholar]
- 38.Stegmann E, Frasch HJ, Wohlleben W. 2010. Glycopeptide biosynthesis in the context of basic cellular functions. Curr Opin Microbiol 13:595–602. doi: 10.1016/j.mib.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 39.MacNeil DJ, Gewain KM, Ruby CL, Dezeny G, Gibbons PH, MacNeil T. 1992. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 111:61–68. doi: 10.1016/0378-1119(92)90603-M. [DOI] [PubMed] [Google Scholar]
- 40.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.