ABSTRACT
Clostridium thermocellum and Thermoanaerobacterium saccharolyticum are thermophilic bacteria that have been engineered to produce ethanol from the cellulose and hemicellulose fractions of biomass, respectively. Although engineered strains of T. saccharolyticum produce ethanol with a yield of 90% of the theoretical maximum, engineered strains of C. thermocellum produce ethanol at lower yields (∼50% of the theoretical maximum). In the course of engineering these strains, a number of mutations have been discovered in their adhE genes, which encode both alcohol dehydrogenase (ADH) and aldehyde dehydrogenase (ALDH) enzymes. To understand the effects of these mutations, the adhE genes from six strains of C. thermocellum and T. saccharolyticum were cloned and expressed in Escherichia coli, the enzymes produced were purified by affinity chromatography, and enzyme activity was measured. In wild-type strains of both organisms, NADH was the preferred cofactor for both ALDH and ADH activities. In high-ethanol-producing (ethanologen) strains of T. saccharolyticum, both ALDH and ADH activities showed increased NADPH-linked activity. Interestingly, the AdhE protein of the ethanologenic strain of C. thermocellum has acquired high NADPH-linked ADH activity while maintaining NADH-linked ALDH and ADH activities at wild-type levels. When single amino acid mutations in AdhE that caused increased NADPH-linked ADH activity were introduced into C. thermocellum and T. saccharolyticum, ethanol production increased in both organisms. Structural analysis of the wild-type and mutant AdhE proteins was performed to provide explanations for the cofactor specificity change on a molecular level.
IMPORTANCE This work describes the characterization of the AdhE enzyme from different strains of C. thermocellum and T. saccharolyticum. C. thermocellum and T. saccharolyticum are thermophilic anaerobes that have been engineered to make high yields of ethanol and can solubilize components of plant biomass and ferment the sugars to ethanol. In the course of engineering these strains, several mutations arose in the bifunctional ADH/ALDH protein AdhE, changing both enzyme activity and cofactor specificity. We show that changing AdhE cofactor specificity from mostly NADH linked to mostly NADPH linked resulted in higher ethanol production by C. thermocellum and T. saccharolyticum.
INTRODUCTION
Thermophilic organisms, Clostridium thermocellum in particular, hold great promise for the production of ethanol from lignocellulosic feedstocks (1, 2). C. thermocellum is a thermophilic, Gram-positive obligate anaerobe that rapidly consumes cellulose. Engineered strains of Thermoanaerobacterium saccharolyticum (3), a thermophilic anaerobe that ferments xylan and other sugars derived from biomass, have been shown to produce ethanol at >50 g/liter, a near-theoretical yield (4). While comparable concentrations of ethanol are tolerated by selected strains of C. thermocellum (5–7), the maximum reported concentration of ethanol produced by this organism is 23.6 g/liter (8) and the maximum ethanol yield achieved to date is 51% of the theoretical maximum (9) versus 92% in T. saccharolyticum (10). It is of interest to understand why ethanol production by T. saccharolyticum is thus far superior to that by C. thermocellum, in order to facilitate engineering of the C. thermocellum ethanol production pathway.
In microorganisms, fermentation of pyruvate to ethanol can proceed either with or without acetyl coenzyme A (acetyl-CoA) as an intermediate. In yeasts and Zymomonas mobilis, pyruvate is decarboxylated directly to acetaldehyde, which is then reduced to ethanol (11). In many other organisms, pyruvate is oxidatively decarboxylated to acetyl-CoA, which is reduced to acetaldehyde, which is further reduced to ethanol. This two-step conversion of acetyl-CoA to ethanol can be catalyzed by one protein, a bifunctional alcohol dehydrogenase (ADH), AdhE. AdhE consists of a C-terminal ADH domain and an N-terminal aldehyde dehydrogenase (ALDH) domain; the ADH domain is usually part of the iron-containing ADH superfamily (Fig. 1) (12). AdhE is present in a variety of mesophilic and thermophilic anaerobic bacteria capable of producing ethanol as a fermentation product (13–16). AdhE has also been found in parasitic eukaryotes (17), anaerobic fungi (18), and algae (19). In all of the organisms investigated thus far, the deletion of adhE is associated with loss of ethanol formation. When adhE was deleted from C. thermocellum and T. saccharolyticum, nearly 100% of their ethanol production was eliminated (20), demonstrating the importance of AdhE in ethanol formation by these two organisms.
FIG 1.
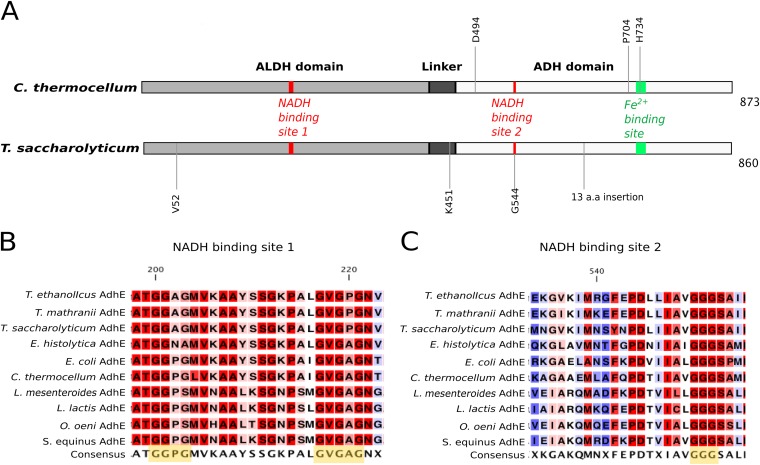
Primary structures of AdhE proteins of wild-type C. thermocellum and T. saccharolyticum. (A) The ALDH domain is at positions 1 to 423 for C. thermocellum and 1 to 420 for T. saccharolyticum, the ADH domain is at positions 463 to 873 for C. thermocellum and 460 to 860 for T. saccharolyticum, and the linker sequence is at positions 424 to 462 for C. thermocellum and 421 to 459 for T. saccharolyticum. NADH binding site 1 is at positions 200 to 221 for C. thermocellum and 199 to 220 for T. saccharolyticum; NADH binding site 2 is at positions 551 to 553 for C. thermocellum and 543 to 545 for T. saccharolyticum. Mutated residues discussed in this study are annotated at the appropriate positions as follows: D494G in LL350; P704L and H734R in LL346; V52A, K451N, and a 13-amino-acid (a.a.) insertion in LL1040; and G544D in LL1049. All elements are drawn to scale. Panels B and C show the sequence conservation of the NADH binding motifs (highlighted in yellow in the consensus sequence) of AdhE from Thermoanaerobacter ethanolicus, Thermoanaerobacter mathranii, T. saccharolyticum, Entamoeba histolytica, E. coli, C. thermocellum, Leuconostoc mesenteroides, Lactococcus lactis, Oenococcus oeni, and Streptococcus equinus. The residues highlighted in red are the most conserved, and those highlighted in blue are the least conserved. The numbering of amino acids is based on the AdhE sequence of C. thermocellum.
Numerous mutations in adhE have appeared during the course of engineering C. thermocellum and T. saccharolyticum for higher ethanol production and tolerance (Fig. 1; Table 1). In C. thermocellum strain LL346 [also known as adhE*(EA)], the mutations P704L and H734R in AdhE were associated with higher ethanol tolerance (21); in strain LL350, the D494G mutation in AdhE was associated with higher ethanol production (22). In T. saccharolyticum strains LL1040 (also known as ALK2) (10) and LL1049 (23), higher ethanol yield was achieved and mutations were also observed in AdhE. In wild-type C. thermocellum and T. saccharolyticum, the ADH activity in cell extracts was largely NADH linked (10, 21). In cell extracts from all of the engineered strains mentioned above, there was an increase in NADPH-linked ADH activity (10, 21, 22). However, it has not been unequivocally established whether this cofactor specificity change must be ascribed to mutations in AdhE, as cells contain multiple ADHs and measurements of cell extracts cannot distinguish between isoenzymes.
TABLE 1.
Strains used in this study
| Organism | Strain | Description | Accession no.a | Source or reference(s) |
|---|---|---|---|---|
| C. thermocellum | LL1004 | Wild-type C. thermocellum strain DSM 1313; low ethanol productionb | CP002416 | DSMZc |
| C. thermocellum | LL346 | Also known as adhE*(EA); evolved C. thermocellum strain, tolerates ethanol at 40 g/liter, has mutations P704L and H734R in AdhE; low ethanol production | SRX030164.1 | 21 |
| C. thermocellum | LL350 | Also known as ΔhydG mutant; C. thermocellum Δhpt ΔhydG strain with mutation D494G in AdhE; moderate ethanol productiond | NAf | 22 |
| C. thermocellum | LL1111 | C. thermocellum Δhpt ΔadhE; no ethanol production | SRX744221 | 20 |
| C. thermocellum | LL1160 | LL1111 C. thermocellum strain with adhE reintroduced into the original adhE locus; low ethanol production | NA | This study |
| C. thermocellum | LL1161 | LL1160 with mutation D494G in AdhE; moderate ethanol production | NA | This study |
| T. saccharolyticum | LL1025 | Wild-type T. saccharolyticum strain JW/SL-YS485; moderate ethanol production | CP003184 | 3 |
| T. saccharolyticum | LL1049 | Also known as M1442 or MO1442; evolved T. saccharolyticum Δ(pta-ack) Δldh Δor796 ure metE Δeps strain with mutation G544D in AdhE; high ethanol productione | SRA233073 | 23, 51 |
| T. saccharolyticum | LL1040 | Also known as strain ALK2; evolved T. saccharolyticum Δldh::erm Δ(pta-ack)::kan strain with mutations V52A and K451N and 13-amino-acid insertion in AdhE; high ethanol production | SRA233066 | 10 |
| T. saccharolyticum | LL1076 | ΔadhE::(pta-ack kan); no ethanol production | SRX744220 | Mascoma Corp. |
| T. saccharolyticum | LL1193 | adhE::kan; differs from wild type only by kan marker downstream of AdhE; moderate ethanol production; also known as M2203 | NA | 23 |
| T. saccharolyticum | LL1194 | LL1193 with G544D mutation in AdhE; also known as M2202 | NA | 23 |
Accession numbers starting with CP refer to finished genome sequences in GenBank; accession numbers starting with SR refer to raw sequencing data from the Joint Genome Institute.
Produces ethanol at a 0 to 40% theoretical yield.
DSMZ, German Collection of Microorganisms and Cell Cultures, Leibniz Institute, Braunschweig, Germany.
Produces ethanol at a 40 to 80% theoretical yield.
Produces ethanol at an 80 to 90% theoretical yield.
NA, not applicable.
It was therefore of interest to investigate purified preparations of AdhE and mutant forms thereof to determine whether ALDH and/or ADH activity has changed in cofactor specificity and whether it is a general trend that the change in AdhE cofactor specificity from NADH to NADPH is associated with more favorable features for ethanol production.
MATERIALS AND METHODS
Plasmid and strain construction.
The adhE genes from strains LL1004 (wild-type C. thermocellum), LL346, LL350, LL1025 (wild-type T. saccharolyticum), LL1040, and LL1049 (Table 1 contains descriptions of these strains) were cloned into plasmid pEXP5-NT/TOPO (Invitrogen) by standard molecular biology techniques, generating the respective Escherichia coli expression plasmids (see Table S1 in the supplemental material). Cloning of the adhE genes into plasmid pEXP5-CT/TOPO instead of pEXP5-NT/TOPO generated native AdhE proteins without His tags. The plasmids were Sanger sequenced (Genewiz) to confirm correct insertion of the target gene and then transformed into chemically competent lysY/Iq E. coli cells (New England BioLabs). Control plasmid pNT-CALML3 (Invitrogen) was also transformed into E. coli. The resulting E. coli strains were used for protein expression. C. thermocellum strains LL1160 and LL1161 were constructed by transforming respective integration plasmids pSH016 and pSH019 into strain LL1111 (Table 1; see Table S1); transformation and colony selection were carried out as previously described (24). T. saccharolyticum strains LL1193 and LL1194 were constructed by transforming the respective vectors pCP14 and pCP14* into wild-type T. saccharolyticum by using a natural-competence-based system (25) (Table 1; see Table S1), and transformants were selected by resistance to the antibiotic kanamycin.
Media and growth conditions.
For biochemical characterization and transformation, C. thermocellum and T. saccharolyticum strains were grown anaerobically to exponential phase (optical density at 600 nm [OD600] of ∼0.5) in the appropriate medium. For C. thermocellum, CTFUD rich medium at pH 7.0 was used as previously described (24); for T. saccharolyticum, CTFUD rich medium at pH 6.0 was used. E. coli strains were grown in LB broth Miller (Acros) with the appropriate antibiotic. Fermentation end products were measured by high-pressure liquid chromatography as previously described (26). For end product analysis, C. thermocellum and T. saccharolyticum strains were grown in the appropriate medium. For C. thermocellum, chemically defined MTC medium was used as previously described (27); for T. saccharolyticum, the MTC medium used was modified by adding thiamine to a final concentration of 4 mg/liter and replacing urea with ammonium chloride at a final concentration of 1.5 g/liter. In preparation for fermentation end product analysis, cultures were grown at 55°C in 150-ml serum bottles with a 50-ml working volume and a 100-ml headspace for 72 h. Ethanol concentrations were calculated from biological duplicates and are reported in Table 2; other end products are reported in Table S2 in the supplemental material.
TABLE 2.
ALDH and ADH activities in C. thermocellum and T. saccharolyticum cell extracts
| Strain | Description | Ethanol yielda | ALDH activityb |
ADH activity |
||
|---|---|---|---|---|---|---|
| NADH | NADPH | NADH | NADPH | |||
| C. thermocellum LL1004 | Wild type | 0.16 | 2.20 ± 0.05c | 0.21 ± 0.03 | 6.73 ± 0.72 | 0.04 ± 0.00 |
| C. thermocellum LL346 | Ethanol tolerant | 0.11 | 0.27 ± 0.13 | 0.05 ± 0.03 | 0.66 ± 0.20 | 0.38 ± 0.02 |
| C. thermocellum LL350 | Moderate ethanol production | 0.22 | 2.00 ± 0.49 | 0.05 ± 0.01 | 6.71 ± 0.93 | 5.90 ± 0.27 |
| C. thermocellum LL1111 | adhE deletion | 0.01 | 0.05 ± 0.00 | 0.13 ± 0.02 | 0.10 ± 0.09 | 0.22 ± 0.18 |
| T. saccharolyticum LL1025 | Wild type | 0.26 | 0.41 ± 0.13 | 0.05 ± 0.04 | 7.06 ± 0.50 | 0.95 ± 0.58 |
| T. saccharolyticum LL1049 | High ethanol production | 0.43 | 0.09 ± 0.02 | 0.50 ± 0.05 | 0.18 ± 0.14 | 1.10 ± 0.42 |
| T. saccharolyticum LL1040 | High ethanol production | 0.45 | 0.08 ± 0.02 | 0.30 ± 0.02 | 0.08 ± 0.05 | 1.55 ± 0.72 |
| T. saccharolyticum LL1076 | adhE deletion | 0.01 | 0.00 ± 0.18d | 0.018 ± 0.09 | 0.04 ± 0.18 | 1.78 ± 0.56 |
Ethanol yield is in grams per gram of cellobiose produced from cellobiose at 5 g/liter.
Activity is expressed in units per milligram of protein (see Materials and Methods).
Error represents 1 standard deviation (n = 4 to 8).
When activity is very low and the background activity is higher than the measured activity, the value is negative (see Materials and Methods) and is shown here as zero activity.
Expression of various adhE genes.
A 500-μl volume of an E. coli culture containing a plasmid with the adhE gene of interest was inoculated into 100 ml of sterile LB broth Miller (Acros) with the appropriate antibiotic and grown aerobically to an OD600 of 0.5 with shaking at 200 rpm at 37°C (Eppendorf Innova 42 shaker). The E. coli strain harboring the pNT-CALML3 control plasmid (Invitrogen) was used as a negative control to measure native E. coli ADH or ALDH activity. Because E. coli AdhE was shown to be sensitive to oxygen (13) and C. thermocellum cell extracts lost ADH activity after exposure to air for 30 min (data not shown), AdhE protein expression and all subsequent experiments were carried out anaerobically. The E. coli cultures were then transferred to sterile serum bottles, and 40 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was used to induce protein expression. The serum bottles were purged with nitrogen to generate an anaerobic protein expression environment, and the cells were cultured for an additional 2 h at 37°C before being harvested.
Preparation of cell extracts.
C. thermocellum, T. saccharolyticum, and E. coli cultures were grown as described above. Cells were harvested by centrifugation at 3,000 × g for 30 min at 4°C, the supernatant was decanted, and the pellet was stored anaerobically at −80°C. All cell extracts were generated anaerobically. Prior to cell extract generation, the frozen pellets were thawed on ice and resuspended in 0.5 ml of lysis buffer consisting of 1× BugBuster reagent (EMD Millipore) at pH 7.0 in phosphate buffer (100 mM) with 5 μM FeSO4. Dithiothreitol (DTT) was added to a final concentration of 0.1 mM. For the T. saccharolyticum cell extracts used in ALDH activity measurements, ubiquinone-0 (Sigma catalog number D9150) was added to a final concentration of 2 mM to relieve the possible inhibition of ALDH activity as previously reported (20). The cells were lysed with Ready-Lyse Lysozyme (Epicentre), and DNase I (New England BioLabs) was added to reduce viscosity. The resulting solution was centrifuged at 10,000 × g for 5 min at room temperature, and the supernatant was used as cell-free extract for enzyme assays.
Protein purification.
The E. coli crude extracts described above were incubated at 50°C anaerobically for 20 min to denature E. coli proteins, and the denatured proteins were separated by centrifugation. The ADH activity in cell extracts of E. coli strains LL346 and LL1040 expressing AdhE was sensitive to heat and lost activity after incubation at 50°C, so these cell extracts were applied directly to the purification column without heating. E. coli cells expressing control plasmid pNT-CALML3 were subjected to the same treatment as described above, and their ALDH and ADH activities before and after heat treatment were measured (see Table S3 in the supplemental material). The cell extracts containing His-tagged AdhE were then subjected to anaerobic affinity column purification (Ni-nitrilotriacetic acid [NTA] spin columns; Qiagen). The purification was carried out according to the Qiagen protocol for Ni-NTA–agarose purification of 6×His-tagged proteins from E. coli under native conditions, with some modifications, as described below. The column was first equilibrated with equilibrium buffer (50 mM NaH2PO4, 300 mM NaCl, 5 mM imidazole, 5 μM FeSO4, pH 7), cell extracts were applied to the column, and then the column was washed twice with wash buffer (50 mM NaH2PO4, 300 mM NaCl, 50 mM imidazole, 20% ethanol, 5 μM FeSO4, pH 7). The His-tagged AdhE protein was eluted by the addition of 200 μl of elution buffer (50 mM NaH2PO4, 300 mM NaCl, 500 mM imidazole, 5 μM FeSO4, pH 7); this is eluent 1 in Tables S3 and S4 in the supplemental material. Repetition of this elution step sequentially generated more-purified eluents 2 and 3. C. thermocellum and T. saccharolyticum AdhE activities were measured at various stages during purification (see Table S4). Electrophoresis results showed that eluent 3 had the least amount of contaminating bands; thus, eluent 3 was used for enzyme assays. The degree of protein purity was estimated by gel densitometry with the image analysis software ImageJ, where the density of each visible gel band from eluent 3 was plotted against its size in the gel. The area of each resulting peak was then integrated and compared to the total of all of the peaks to estimate the percentage of protein that can be attributed to each band (see Table S5 in the supplemental material). E. coli cell extracts with native AdhE expressed (i.e., without the His tag) were used directly without purification (see Table S6 in the supplemental material).
ALDH and ADH activity assays.
All of the ALDH activity measurements mentioned in this report refer to the reaction in the acetaldehyde-producing direction. All of the ADH activity measurements mentioned in this report refer to the reaction in the ethanol-producing direction. For ADH (acetaldehyde reduction) reactions, the anaerobic reaction mixture contained 0.24 mM NADH or NADPH, 17.6 mM acetaldehyde, 1 mM DTT, 100 mM Tris-HCl, 5 μM FeSO4, and cell extract or purified protein solution (the protein amount is indicated separately for each assay). The final volume was 850 μl, the assay temperature was 55°C, and the assay was started by the addition of acetaldehyde. For ALDH (acetyl-CoA reduction) reactions, the acetaldehyde in the above-described anaerobic reaction mixture was replaced with 0.35 mM acetyl-CoA. Background activity was recorded before the start of the reaction (addition of acetaldehyde or acetyl-CoA) and was subtracted from the reaction activity recorded. In the inhibition assays (see Table S8 in the supplemental material), the inhibitor was added before the start of the reaction at the following concentration: 1 M ethanol or 2.35 mM NAD(P)+. The decrease in absorbance at 340 nm caused by NAD(P)H oxidation was monitored with an Agilent 8453 UV-Vis spectrophotometer with Peltier temperature control (28). Protein concentration was determined by the Bradford method with bovine serum albumin (Thermo Scientific) as the standard. Specific activities are expressed in units per milligram of protein. One unit of activity equals the formation of 1 μmol of product per minute. Specific activities in Tables 2 and 3 are reported for at least two biological replicates. t tests were performed to analyze the cofactor specificity in Tables 2 and 3: NADH-linked and NADPH-linked activities were analyzed with the unpaired two-tailed t test, and differences between the two cofactors were considered significant if the P value was <0.05. Standard deviations and raw data for all enzyme assays are presented in Table S7 in the supplemental material. The software Visual Enzymics (SoftZymics) was used for nonlinear regression to calculate the apparent Km and kcat values in Table 4. kcat was calculated on the basis of a molecular mass of 96 kDa for LL1004 AdhE and LL350 AdhE.
TABLE 3.
Cofactor specificities of purified AdhE proteins
| Source of AdhE | Description | ALDH activitya |
ADH activity |
||
|---|---|---|---|---|---|
| NADH | NADPH | NADH | NADPH | ||
| C. thermocellum LL1004 | Wild type | 18.02 ± 2.45b | 2.03 ± 0.46 | 42.23 ± 3.13 | 1.96 ± 1.25 |
| C. thermocellum LL346 | Ethanol tolerant | 13.50 ± 3.54 | 0.43 ± 0.42 | 4.02 ± 1.48 | 0.03 ± 0.34 |
| C. thermocellum LL350 | Moderate ethanol production | 31.50 ± 5.80 | 5.76 ± 0.16 | 42.67 ±7.46 | 42.30 ± 4.28 |
| T. saccharolyticum LL1025 | Wild type | 10.63 ± 1.15 | 4.46 ± 1.63 | 17.43 ± 2.45 | 2.43 ± 2.43 |
| T. saccharolyticum LL1049 | High ethanol production | 1.63 ± 0.30 | 11.13 ± 1.35 | 0.66 ± 1.97 | 12.70 ± 2.18 |
| T. saccharolyticum LL1040 | High ethanol production | 0.07 ± 0.55 | 5.03 ± 2.05 | 0.01 ± 1.34 | 12.96 ± 4.54 |
Activity is expressed in units per milligram of protein (see Materials and Methods).
Error represents 1 standard deviation (n = 4 to 8).
TABLE 4.
Apparent Km and kcat values of purified C. thermocellum wild-type and LL350 AdhE proteins
| C. thermocellum straina (characteristic) and enzyme | Substrate | Km (mM) | kcat (S−1) | kcat/Km (S−1 M−1) |
|---|---|---|---|---|
| LL1004 (wild type) | ||||
| ALDH | Acetyl-CoA | 0.0084 ± 0.0002e | 280 ± 2 | 3.3E + 07 |
| NADH | 0.052 ± 0.003 | 150 ± 3 | 3.0E + 06 | |
| ADH | Acetaldehyde | 1.6 ± 0.05 | 240 ± 4 | 1.5E + 05 |
| NADH | 0.088 ± 0.006 | 240 ± 6 | 2.7E + 06 | |
| LL350 (moderate ethanol production) | ||||
| ALDH | Acetyl-CoA | 0.0042 ± 0.0004d | 230 ± 2f | 5.4E + 07 |
| NADH | 0.026 ± 0.0002d | 62 ± 0.1f | 2.4E + 06d | |
| ADH | Acetaldehydeb | 1.4 ± 0.05 | 220 ± 2d | 1.5E + 05 |
| Acetaldehydec | 1.4 ± 0.04 | 210 ± 3d | 1.5E + 05 | |
| NADH | 0.17 ± 0.01d | 220 ± 5 | 1.3E + 06d | |
| NADPH | 0.075 ± 0.01 | 280 ± 7 | 3.7E + 06 |
Source of AdhE, which was expressed in E. coli and affinity purified.
Measurements were done with NADH as a cofactor.
Measurements were done with NADPH as a cofactor.
Compared to value obtained with wild-type AdhE, two-tailed P value of <0.05.
Error represents 1 standard error, at a confidence level of 0.95 (n = 10 to 16, depending on the sample).
Compared to value obtained with wild-type AdhE, two-tailed P value of <0.01.
Homology modeling and molecular dynamics.
The homology models corresponding to the ADH domains of the AdhE proteins from LL1004, LL346, LL350, LL1025, LL1040, and LL1049 were constructed with the bioinformatics toolkit SWISS-MODEL (Swiss Institute of Bioinformatics). The recent 2.5 Å resolution X-ray structure of the ADH of Geobacillus thermoglucosidasius (3ZDR) (12) and 1.3 Å resolution X-ray structure of the ADH from Thermotoga maritima (1O2D) (29) were used as templates for their high level of homology and the presence of NADP cofactor and iron ion, respectively. The resulting structures were inspected for proper phi/psi angles. All resulting structures were submitted to molecular dynamics simulations with the program CHARMM with the CHARMM 36 force field and the TIP3P water model (30). The systems were generated via the CHARMM-GUI web server (31), and the parameters for NADP were generated by ParamChem. The structures were initially minimized in vacuo with the steepest descent for 1,000 steps and then solvated in a cubic water box with a minimum of 10 Å from the edge of the box; sodium cations were added to neutralize the system. The resulting systems were minimized by using the steepest descent for 1,000 steps, followed by Newton-Raphson minimization for 100 steps. They were then submitted to 1 ns of equilibration in the NPT ensemble at 298 K and 105 Pa, followed by 10 ns of simulation in the NVT ensemble with an integration time step of 2 fs. All simulations were conducted in duplicate with different starting seeds and analyzed with carma (32).
Nucleotide sequence accession numbers.
The accession numbers of the adhE genes of strains LL1004, LL346, LL350, LL1025, LL1040, and LL1049 are KR632757, KR632758, KR632759, KR632761, KR632760, and KR632762, respectively.
RESULTS
Cofactor specificity of wild-type and mutant AdhE.
ALDH and ADH activities were determined in cell extracts of C. thermocellum and T. saccharolyticum strains, as well as in the affinity-purified AdhE expressed in E. coli. In purified preparations of AdhE, gel densitometry results showed that the proteins were all >70% pure (see Table S5 in the supplemental material). Furthermore, negative E. coli controls all showed <0.4 U/mg specific activity for ALDH and ADH (see Table S3 in the supplemental material), indicating that the small amount of contaminating protein observed on the gel did not substantially interfere with ADH or ALDH activity measurements. With respect to cofactor preference, the cell extracts of the T. saccharolyticum high ethanol producers (LL1040, LL1049) has changed their ADH and ALDH cofactor specificity to mostly NADPH linked, compared to mostly NADH linked in the wild type (LL1025) (Table 2). The difference between NADH- and NADPH-linked activities was significant (P < 0.05) for all three strains, except for ADH activity in LL1049. Cell extracts from wild-type (LL1004) and ethanol-tolerant (LL346) C. thermocellum strains both showed a greater preference for NADH as a cofactor, but the preference was statistically significant for the wild-type strain and nonsignificant for the ethanol-tolerant strain. The C. thermocellum moderate ethanol producer (LL350) had significantly increased NADPH-linked ADH activity while maintaining significant NADH linkage in ALDH activity. The results for affinity-purified AdhE enzymes (Table 3) also showed higher NADPH-linked AdhE activity in evolved strains. The purified AdhE enzymes were all significantly linked to either NADH or NADPH (P < 0.05), with the exception of LL350 AdhE, where, as in cell extracts, NADPH-linked ADH activity increased to an amount comparable to that of NADH-linked ADH activity. In all cases, the change in cofactor specificity for NADPH was much more complete in T. saccharolyticum AdhE than in C. thermocellum AdhE (Table 3). Additionally, strains exhibiting this increase in NADPH-linked cofactor specificity in AdhE also generally showed greater ethanol production than their parent strains (Table 2 and 3).
Because the Asp-494-Gly (D494G) mutation (22) in AdhE of the C. thermocellum moderate ethanol producer (LL350) enabled the enzyme to use both NADH and NADPH as cofactors, the apparent kcat and Km values were measured with purified protein from LL350 and wild-type C. thermocellum expressed in E. coli (Table 4). Unpaired t tests were conducted to compare the kinetic parameters of wild-type AdhE and the D494G mutant form. In terms of Km, kcat, and catalytic efficiency (kcat/Km), the newly acquired NADPH-linked activity in the D494G mutant form was not significantly different from the NADH-linked ADH activity in the wild-type protein (P > 0.05). For NADH-linked activity, however, a significantly lower (P < 0.05) catalytic efficiency was observed for NADH in the D494G mutant protein than in the wild type.
Product inhibition [ethanol or NAD(P)+] of purified AdhE proteins from C. thermocellum and T. saccharolyticum was measured (see Table S8 in the supplemental material). AdhE of the C. thermocellum ethanol-tolerant strain (LL346) was significantly different from other AdhE proteins in both ethanol and NAD(P)+ inhibition. It retained 98% of its ADH activity and 92% of its ALDH activity in the presence of 2.35 mM NAD+, while the other five AdhE proteins, on average, retained only 30% of their ADH activity and 33% of their ALDH activity. Interestingly, in the presence of 1 M ethanol, LL346 AdhE showed a 2-fold increase in ADH activity, while the other five AdhE proteins, on average, retained only 40% of their ADH activity.
Effects of AdhE mutations on ethanol production.
The physiological effects of two selected point mutations were investigated by reintroducing those mutations into either C. thermocellum or T. saccharolyticum. For C. thermocellum, the D494G mutation (22) was chosen. This mutation could not be introduced directly into the wild-type strain (because of limits of existing genetic tools), so instead, adhE was deleted (strain LL1111) and replaced with D494G mutant adhE (strain LL1161). A control strain (LL1160) was made by the reintroducing wild-type adhE into strain LL1111. Fermentation of cellobiose at 5 g/liter resulted in ethanol yields of 0.14 g/g of cellobiose for strain LL1160 and 0.24 g/g of cellobiose for strain LL1161, a 1.7-fold increase. This increase is larger than the 1.4-fold increase in ethanol yield over that of wild-type C. thermocellum in moderate ethanol producer LL350, which has the D494G AdhE mutation but also other genetic modifications.
For T. saccharolyticum, the AdhE G544D mutation (23) was chosen. In this organism, the mutation could be introduced directly into the wild-type strain, although a kanamycin antibiotic resistance marker (kan) had to be added downstream of adhE. The resulting strain was LL1194. A control strain (LL1193) was made by inserting only the kan marker downstream of adhE. Fermentation of cellobiose at 5 g/liter resulted in ethanol production of 0.21 g/liter for strain LL1193 and 0.32 g/liter for strain LL1194, a 1.5-fold increase. This increase is comparable to the 1.7-fold increase in ethanol production from wild-type T. saccharolyticum to high ethanol producer LL1049, which has the G544D AdhE mutation but also other genetic modifications.
AdhE protein structure prediction.
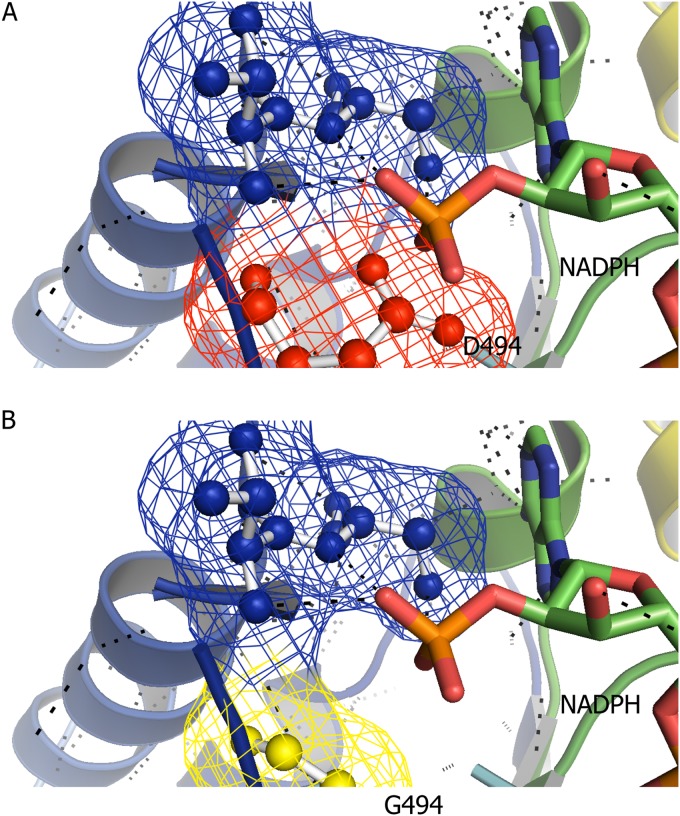
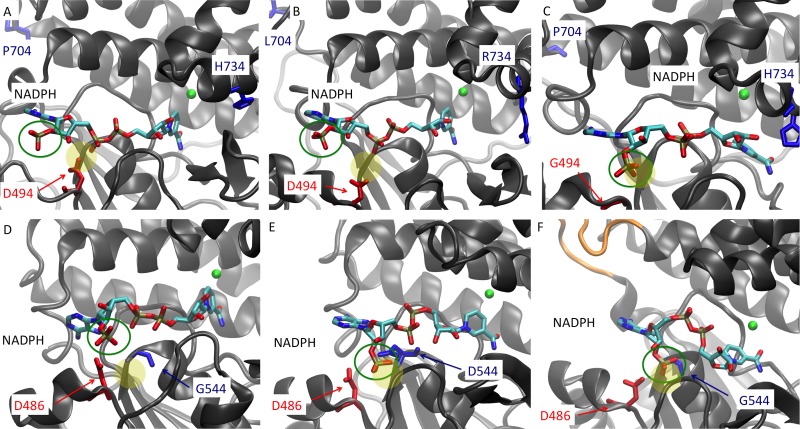
To understand the impact of mutations on cofactor specificity, we performed homology modeling and docking. The average structure of the ADH domains from the wild-type and D494G mutant forms of C. thermocellum AdhE were compared to identify potential explanations for the switch in cofactor specificity (Fig. 2). In wild-type C. thermocellum AdhE, Asp-494 interferes with the 2′-phosphate group of NADPH because of electrostatic repulsion (both are negatively charged) and steric hindrance. Molecular dynamics simulation was conducted to compare the average structures of the six different ADH domains (Fig. 3), including the previously mentioned D494G mutant form, to evaluate if the observations from homology modeling and docking were correct. The conformation of NADPH in the binding pocket of the ADH domain varied in the AdhE mutants. In the case of C. thermocellum, NADPH behaves similarly in wild-type C. thermocellum (LL1004) AdhE and ethanol-tolerant C. thermocellum (LL346) AdhE, where the 2′-phosphate group of NADPH does not have access to the binding pocket (Fig. 3). In the C. thermocellum moderate ethanol producer (LL350) AdhE protein, the D494G mutation changed NADPH binding significantly, as mentioned above, allowing the 2′-phosphate group of NADPH access to the binding pocket. A similar trend was observed in the case of T. saccharolyticum AdhE, where the mutations in the high ethanol producers LL1049 and LL1040 seem to change the conformation of NADPH in the binding pocket, allowing NADPH to bind.
FIG 2.
Homology modeling and docking analysis of the phosphate in NADPH interacting with C. thermocellum wild-type AdhE (A) and the D494G mutant form (B). The dotted lines represent hydrogen bonds. The red residue is D494, the yellow residue is G494, and the blue residues are N495 and F496.
FIG 3.
Average structure of the ADH domains of AdhE from C. thermocellum wild-type LL1004 (A), the ethanol-tolerant LL346 strain (B), moderate ethanol producer LL350 (C), T. saccharolyticum wild-type LL1025 (D), high ethanol producer LL1049 (E), and high ethanol producer LL1040 (F). The amino acids of interest are shown in blue and red; the NADPH cofactor is shown color coded by elements, with its 2′-phosphate group highlighted (green open circle); and the iron ions (green) are also shown. Additionally, the 39-bp insertion in the high ethanol producer LL1040 is shown in orange (F). The yellow-filled circle represents the binding pocket where the additional 2′-phosphate of NADPH is commonly found in NADPH-dependent AdhE proteins. The locations of D494 and D486 are usually the recognition sites for NADH and do not allow the 2′-phosphate group of NADPH (green circle) to access the preferred binding pocket (yellow circle). When the green open circle and the yellow filled circle overlap, that indicates that the NADPH molecule is able to access its preferred binding pocket. This is present in panels C, E, and F but not in the other panels. Panels C, E, and F correspond to enzymes that can use NADPH as a cofactor. Note that NADPH was used for modeling purposes and does not reflect the actual cofactor specificity of the enzymes but rather was used to explain the observed levels of affinity of the enzymes for NADPH.
DISCUSSION
Primary structure of AdhE.
The ALDH and ADH domains of AdhE are highly conserved and connected by a linker sequence. There is a disagreement in the literature on the number of NADH binding sites in AdhE. Some studies predict a single NADH binding site located within or near the linker region of AdhE (13, 33–37), suggesting that the ALDH and ADH domains share one nicotinamide binding site. Other studies have predicted an additional NADH binding site in the ALDH domain (19, 28, 38, 39). Fungal AdhE enzymes have been shown to have three putative NADH binding sites (18). In our analysis, we focused on glycine-rich regions and relied on structural information from our homology models or closely related structural homologs. In both the C. thermocellum and T. saccharolyticum AdhE proteins, we found two strong NADH binding sites (Fig. 1A) and an additional putative nucleotide binding region with a GXGXXG motif in the linker region between the ALDH and ADH domains, which has been reported in previous studies as a potential recognition locus (13, 18, 19, 28, 33–39). This putative binding region within the linker is almost identical to one identified in another iron-dependent ADH from E. coli (40). In this study, the glycine at the center of the locus was mutated but resulted in only a marginal loss of NAD+ binding. However, mutations in the other glycine-rich locus with the motif GGG located in the ALDH domain resulted in complete loss of NAD+ binding (40), indicating that the GGG motif is important in NAD+ binding. The GXGXXG motif located in the linker is conserved in several AdhE enzymes but does not seem to contribute to nucleotide binding directly; however, it might be important in nucleotide channeling or recognition before entering the binding pocket. The NADH binding site in the ALDH domain appears to be a combination of a conventionally accepted binding motif (GXGXG) and another glycine-rich helix turn (Fig. 1B). The NADH binding site in the ALDH domain has also been suggested to have acetyl-CoA binding abilities (28). We see a high level of conservation of both strong binding sites across different organisms (Fig. 1B and C). The prediction of two binding sites (one in each domain) agrees with our observation that D494G mutant AdhE gained NADPH specificity for ADH activity but not for ALDH activity. If D494G mutant AdhE shared a single NADH binding site in the linker region, then one would expect to find NADPH cofactor specificity in ALDH activity as well.
Effect of AdhE cofactor specificity on ethanol production.
Thus far, most of the bifunctional AdhE enzymes investigated are NADH linked; however, there are some exceptions. Thermoanaerobacter mathranii AdhE showed a small amount of NADPH-linked activity in addition to NADH-linked activity for both ALDH and ADH (37), and Thermoanaerobacter ethanolicus JW200 AdhE showed NADH-linked ALDH activity and small amounts of NADPH-linked ADH activity (36). In all of the T. saccharolyticum strains investigated in this study, higher ethanol production was associated with an increase in NADPH specificity and a decrease in NADH specificity for both ADH and ALDH activities (Tables 2 and 3). In C. thermocellum, this association is not as consistent; although NADPH-linked ADH activity significantly increased, NADH-linked ADH and ALDH activities were maintained at wild-type levels in the moderate ethanol producer LL350 strain (Tables 2 and 3). When mutations that were previously shown to increase NADPH-linked ADH activity were reintroduced into wild-type C. thermocellum and T. saccharolyticum AdhE, ethanol production increased in the resulting strains (LL1161 and LL1194), showing that these mutations (both of which caused changes in cofactor specificity) are responsible for at least some of the increased ethanol production in the high-ethanol-producing strains. The effect of reintroducing the adhE mutations depends somewhat on medium composition. Another study comparing the same mutants found almost no difference in ethanol production (M2202 versus M2203 in Table 3 in reference 23). We suspect that the difference between our results and those of Shaw et al. is due to the presence of large quantities of yeast extract in the medium they used.
The physiological reason for the increase in NADPH-linked activity in strains LL350, LL1040, and LL1049 remains to be elucidated. NADPH is generally thought to be the reducing equivalent for anabolism, whereas NADH is generally thought to be the reducing equivalent in anaerobic catabolism. Apparently, these strains use NADPH for both anabolic and catabolic processes. This may be related to changes in the fluxes of NADH and NADPH generation elsewhere in metabolism. There are several possible sources in C. thermocellum and T. saccharolyticum that can provide NADPH for ethanol production. The nfnAB genes, which are present in both C. thermocellum and T. saccharolyticum, encode the NfnAB complex that catalyzes the reaction 2NADP+ + NADH + ferredoxinred2− + H+ → 2NADPH + NAD+ + ferredoxinox (41). Other T. saccharolyticum enzymes that generate NADPH for catabolic purposes include the glucose-6-phosphate dehydrogenase and the phosphogluconate dehydrogenase. These T. saccharolyticum genes are both highly expressed (42). Although C. thermocellum does not have the above two enzymes present in the pentose phosphate cycle, the malic enzyme in C. thermocellum, which catalyzes the formation of pyruvate from l-malate and generates NADPH, is very active (26).
Enzymes in the T. saccharolyticum ethanol production pathway.
Although the T. saccharolyticum LL1040 and LL1049 strains are able to produce ethanol at high yield, purified AdhE proteins from these mutant strains showed comparable or lower ADH activity (Table 3). Furthermore, one-way analysis of variance showed that the ADH activity in cell extracts of LL1040 and LL1049 (Table 2) is not significantly different from that in T. saccharolyticum adhE deletion strain LL1076 (P value 0.56). This suggests that the T. saccharolyticum high ethanol producers do not largely rely on the ADH activity from AdhE for ethanol production and that another ADH may be the main ADH in these strains. The cell extract activity measurements in Table 2 suggest that this other ADH is NADPH linked and may have higher ADH activity than AdhE. It has been reported that an NADPH-linked primary ADH, AdhA, is present in the Thermoanaerobacter species and may be part of the ethanol production pathway (43, 44). Sequence analysis shows that T. saccharolyticum JW/SL-YS485 has a gene (Tsac_2087) encoding an ADH that is 86% identical (at the protein level) to T. mathranii and T. ethanolicus AdhA. Other reported NADPH-linked ADHs involved in ethanol production include the AdhB enzyme, such as the secondary ADH reported in T. ethanolicus 39E (45). However, sequence analysis showed that T. saccharolyticum does not possess an adhB gene. Therefore, AdhA may be responsible for the observed NADPH-linked ADH activity in T. saccharolyticum cell extracts and also may be important in ethanol production by T. saccharolyticum high ethanol production strains LL1040 and LL1049.
Unique characteristics of ethanol-tolerant C. thermocellum strain LL346.
The mutations introduced into the AdhE protein from the ethanol-tolerant strain of C. thermocellum, LL346 [also known as adhE*(EA)], are notably different from the other mutations that we have described thus far with respect to inhibition. It has been reported that small amounts of NAD+ and ethanol inhibit ADH activity in cell extracts of C. thermocellum (46). High inhibition by NAD(P)+ of purified AdhE proteins of C. thermocellum was observed (at least 70% of the activity was inhibited), with the exception of LL346, in which inhibition by NAD+ was less than 10% (see Table S8 in the supplemental material). Another unexpected property of AdhE from strain LL346 was increased activity in the presence of ethanol. A similar phenomenon has been observed with the Z. mobilis ZADH-2 enzyme, which was also stimulated by ethanol (47). The authors proposed ethanol-induced acceleration of NAD+ dissociation as a mechanism for the observed activation by ethanol, because nicotinamide dissociation is presumed to be the rate-limiting step in most dehydrogenases.
It has been reported by Brown et al. (21) that mutations in the adhE gene of C. thermocellum LL346 (P704L and H734R) are the sole basis for the alcohol tolerance of this mutant. As the mutations coincided with a change from NADH-linked to NADPH-linked ADH activity in cell extracts, they concluded that these mutations are responsible for a change in cofactor specificity from NADH to NADPH in the ADH part of AdhE. An increase in NADPH-linked ADH activity was observed in the cell extracts of LL346 compared to those of the wild type (Table 2), but in assays done with purified AdhE from LL346, nearly 100% of the activity for both ALDH and ADH was NADH linked (Table 3). This changes the interpretation of the effect of the mutation, and suggests that it reduced enzyme activity instead of changing cofactor specificity. Thus, the small increase in NADPH-linked ADH activity observed in the cell extracts of LL346 may be the result of another enzyme.
AdhE cofactor specificity at the molecular level.
Several factors can explain the changes in cofactor specificity described in AdhE of the moderate ethanol producer C. thermocellum strain LL350. It is clearly not energetically favorable to accommodate the extra 2′-phosphate group in wild-type C. thermocellum AdhE because of the negative charge of Asp-494. This 2′-phosphate group is absent from NADH, which may, in fact, be stabilized by hydrogen bonding interactions with this residue. This evidence suggests that Asp-494 is important in distinguishing nicotinamide cofactors as previously described. As shown in Fig. 2, substitution of glycine for Asp-494 removes the interference between Asp-494 and NADPH, thus enabling the ADH to use both NADH and NADPH as cofactors. The low Km value for NADPH in D494G mutant AdhE (Table 4) agrees with the structural prediction, as it suggests that this mutation resulted in an increase in the affinity of the enzyme for NADPH. Aspartic acid residues have been shown to play an important role in regulating the binding of NADH over NADPH and are potential targets for mutations to change cofactor specificity. For example, the D38N mutation in the NADH recognition motif of an NADH-dependent Drosophila ADH allowed the enzyme to use both NADH and NADPH (48). A similar study was conducted with an ADH yielding the same results (40). The positions of these aspartic acids are almost identical to that of D494 in wild-type C. thermocellum (LL1004) AdhE.
Regarding LL346, the mutations would likely lead to a loss of enzymatic activity in AdhE. Even though the LL346 mutations H734R and P704L both occurred in the ADH domain, the ALDH activity may also be affected. The H734R mutation has been studied in E. histolytica AdhE (also known as EhADH2), where it resulted in reduced ALDH and ADH activities (28). Those results suggested that alterations in the ADH domain, especially within the putative iron binding domain where H734R resides, could affect ALDH domain activity.
Helical assemblies of AdhE proteins named “spirosomes” have been observed in many other organisms (12, 28, 34, 49), as well as in recombinant AdhE following His purification (50). The formation of such structures has been suggested to influence enzyme activity (28). The formation of this quaternary structure offers a potential explanation for how mutations in one domain of AdhE could impact the activity of the other domain.
In wild-type T. saccharolyticum AdhE (from strain LL1025), Asp-486 is the equivalent of Asp-494 in C. thermocellum AdhE and, as mentioned above, may selectively mediate the binding of NADH over NADPH. The G544D mutation in LL1049 replaces a glycine residue with a charged aspartic acid across from Asp-486, and the 2′-phosphate group of NADPH appears sandwiched between these two amino acid residues (Fig. 3E). There are several hydrogen bonds shared between this phosphate group and the two aspartic acids that could help relieve their overall repulsion based on their respective charges. In the case of the LL1040 variant, there is a large loop of 13 amino acids introduced in the ADH domain, and given its flexibility and close proximity to the NADH binding site in the linker sequence, it could induce subtle changes in the binding site that would result in the observed cofactor specificity change (Fig. 3F).
Regarding cofactor change in the ALDH domain of the LL1040 and LL1049 mutants, this domain either possesses a mutation far from the NADH binding site (LL1040) or lacks such a mutation (LL1049). It is possible that spirosome formation (12, 28, 34, 49) not only influences enzyme activity but also affects cofactor specificity; thus, cofactor changes in the ADH domain may cause cofactor changes in the ALDH domain through the formation of such superstructures.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Mascoma Corporation for their gift of T. saccharolyticum strains LL1076 (also known as M3223), LL1040 (also known as ALK2 or M0001), LL1049 (also known as M1442 or MO1442), LL1193 (also known as M2203), and LL1194 (also known as M2202). We thank Sean Jean-Loup Murphy for his contribution in end product analysis.
The genome sequencing work conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy under contract DE-AC02-05CH11231. The BioEnergy Science Center is a U.S. Department of Energy Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science. The manuscript was authored by Dartmouth College under subcontract 4000115284 and contract DE-AC05-00OR22725 with the U.S. Department of Energy.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00232-15.
REFERENCES
- 1.Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS. 2002. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol Mol Biol Rev 66:506–577. doi: 10.1128/MMBR.66.3.506-577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blumer-Schuette SE, Brown SD, Sander KB, Bayer EA, Kataeva I, Zurawski JV, Conway JM, Adams MWW, Kelly RM. 2014. Thermophilic lignocellulose deconstruction. FEMS Microbiol Rev 38:393–448. doi: 10.1111/1574-6976.12044. [DOI] [PubMed] [Google Scholar]
- 3.Mai V, Lorenz WW, Wiegel J. 1997. Transformation of Thermoanaerobacterium sp. strain JW/SL-YS485 with plasmid pIKM1 conferring kanamycin resistance. FEMS Microbiol Lett 148:163–167. doi: 10.1111/j.1574-6968.1997.tb10283.x. [DOI] [Google Scholar]
- 4.Shaw AJ, Covalla SF, Miller BB, Firliet BT, Hogsett DA, Herring CD. 2012. Urease expression in a Thermoanaerobacterium saccharolyticum ethanologen allows high titer ethanol production. Metab Eng 14:528–532. doi: 10.1016/j.ymben.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 5.Shao XJ, Raman B, Zhu MJ, Mielenz JR, Brown SD, Guss AM, Lynd LR. 2011. Mutant selection and phenotypic and genetic characterization of ethanol-tolerant strains of Clostridium thermocellum. Appl Microbiol Biotechnol 92:641–652. doi: 10.1007/s00253-011-3492-z. [DOI] [PubMed] [Google Scholar]
- 6.Williams TI, Combs JC, Lynn BC, Strobel HJ. 2007. Proteomic profile changes in membranes of ethanol-tolerant Clostridium thermocellum. Appl Microbiol Biotechnol 74:422–432. doi: 10.1007/s00253-006-0689-7. [DOI] [PubMed] [Google Scholar]
- 7.Rani KS, Swamy M, Sunitha D, Haritha D, Seenayya G. 1996. Improved ethanol tolerance and production in strains of Clostridium thermocellum. World J Microbiol Biotechnol 12:57–60. [DOI] [PubMed] [Google Scholar]
- 8.Sato K, Tomita M, Yonermura S, Goto S, Sekine K, Okuma E, Takagi Y, Hon-nami K, Saiki T. 1993. Characterization of and ethanol hyper production by Clostridium thermocellum I-1-B. Biosci Biotechnol Biochem 57:2116–2121. [Google Scholar]
- 9.Argyros DA, Tripathi SA, Barrett TF, Rogers SR, Feinberg LF, Olson DG, Foden JM, Miller BB, Lynd LR, Hogsett DA, Caiazza NC. 2011. High ethanol titers from cellulose by using metabolically engineered thermophilic, anaerobic microbes. Appl Environ Microbiol 77:8288–8294. doi: 10.1128/AEM.00646-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shaw AJ, Podkaminer KK, Desai SG, Bardsley JS, Rogers SR, Thorne PG, Hogsett DA, Lynd LR. 2008. Metabolic engineering of a thermophilic bacterium to produce ethanol at high yield. Proc Natl Acad Sci U S A 105:13769–13774. doi: 10.1073/pnas.0801266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.König S. 1998. Subunit structure, function and organisation of pyruvate decarboxylases from various organisms. Biochim Biophys Acta 1385:271–286. doi: 10.1016/S0167-4838(98)00074-0. [DOI] [PubMed] [Google Scholar]
- 12.Extance J, Crennell SJ, Eley K, Cripps R, Hough DW, Danson MJ. 2013. Structure of a bifunctional alcohol dehydrogenase involved in bioethanol generation in Geobacillus thermoglucosidasius. Acta Crystallogr Sect D Biol Crystallogr 69(Pt 10):2104–2115. doi: 10.1107/S0907444913020349. [DOI] [PubMed] [Google Scholar]
- 13.Membrillo-Hernandez J, Echave P, Cabiscol E, Tamarit J, Ros J, Lin ECC. 2000. Evolution of the adhE gene product of Escherichia coli from a functional reductase to a dehydrogenase. J Biol Chem 275:33869–33875. doi: 10.1074/jbc.M005464200. [DOI] [PubMed] [Google Scholar]
- 14.Yao S. 2008. PhD thesis Technical University of Denmark, Kongens Lyngby, Denmark. [Google Scholar]
- 15.Peng H, Wu G, Shao W. 2008. The aldehyde/alcohol dehydrogenase (AdhE) in relation to the ethanol formation in Thermoanaerobacter ethanolicus JW200. Anaerobe 14:125–127. doi: 10.1016/j.anaerobe.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 16.Bhandiwad A, Shaw AJ, Guss A, Guseva A, Bahl H, Lynd LR. 2014. Metabolic engineering of Thermoanaerobacterium saccharolyticum for n-butanol production. Metab Eng 21:17–25. doi: 10.1016/j.ymben.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 17.Pineda E, Encalada R, Olivos-García A, Néquiz M, Moreno-Sánchez R, Saavedra E. 2013. The bifunctional aldehyde-alcohol dehydrogenase controls ethanol and acetate production in Entamoeba histolytica under aerobic conditions. FEBS Lett 587:178–184. doi: 10.1016/j.febslet.2012.11.020. [DOI] [PubMed] [Google Scholar]
- 18.Boxma B, Voncken F, Jannink S, van Alen T, Akhmanova A, van Weelden SWH, van Hellemond JJ, Ricard G, Huynen M, Tielens AGM, Hackstein JHP. 2004. The anaerobic chytridiomycete fungus Piromyces sp. E2 produces ethanol via pyruvate:formate lyase and an alcohol dehydrogenase E. Mol Microbiol 51:1389–1399. doi: 10.1046/j.1365-2958.2003.03912.x. [DOI] [PubMed] [Google Scholar]
- 19.Atteia A, van Lis R, Mendoza-Hernández G, Henze K, Martin W, Riveros-Rosas H, González-Halphen D. 2003. Bifunctional aldehyde/alcohol dehydrogenase (ADHE) in chlorophyte algal mitochondria. Plant Mol Biol 53:175–188. doi: 10.1023/B:PLAN.0000009274.19340.36. [DOI] [PubMed] [Google Scholar]
- 20.Lo J, Zheng T, Hon S, Olson DG, Lynd LR. 2015. The bifunctional alcohol and aldehyde dehydrogenase gene, adhE, is necessary for ethanol production in Clostridium thermocellum and Thermoanaerobacterium saccharolyticum. J Bacteriol 197:1386–1393. doi: 10.1128/JB.02450-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown SD, Guss AM, Karpinets TV, Parks JM, Smolin N, Yang S, Land ML, Klingeman DM, Bhandiwad A, Rodriguez M Jr, Raman B, Shao X, Mielenz JR, Smith JC, Keller M, Lynd LR. 2011. Mutant alcohol dehydrogenase leads to improved ethanol tolerance in Clostridium thermocellum. Proc Natl Acad Sci U S A 108:13752–13757. doi: 10.1073/pnas.1102444108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biswas R, Zheng T, Olson DG, Lynd LR, Guss AM. 2015. Elimination of hydrogenase active site assembly blocks H2 production and increases ethanol yield in Clostridium thermocellum. Biotechnol Biofuels 8:20. doi: 10.1186/s13068-015-0204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaw AJ, Miller BB, Rogers SR, Kenealy WR, Meola A, Bhandiwad A, Sillers WR, Shikhare I, Hogsett DA, Herring CD. 2015. Anaerobic detoxification of acetic acid in a thermophilic ethanologen. Biotechnol Biofuels 8:75. doi: 10.1186/s13068-015-0257-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olson DG, Lynd LR. 2012. Transformation of Clostridium thermocellum by electroporation. Methods Enzymol 510:317–330. doi: 10.1016/B978-0-12-415931-0.00017-3. [DOI] [PubMed] [Google Scholar]
- 25.Shaw AJ, Hogsett DA, Lynd LR. 2010. Natural competence in Thermoanaerobacter and Thermoanaerobacterium species. Appl Environ Microbiol 76:4713–4719. doi: 10.1128/AEM.00402-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou J, Olson DG, Argyros DA, Deng Y, van Gulik WM, van Dijken JP, Lynd LR. 2013. Atypical glycolysis in Clostridium thermocellum. Appl Environ Microbiol 79:3000–3008. doi: 10.1128/AEM.04037-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Lynd LR. 2003. Quantification of cell and cellulase mass concentrations during anaerobic cellulose fermentation: development of an enzyme-linked immunosorbent assay-based method with application to Clostridium thermocellum batch cultures. Anal Chem 75:3131–3139. doi: 10.1021/ac020271n. [DOI] [PubMed] [Google Scholar]
- 28.Espinosa A, Yan L, Zhang Z, Foster L, Clark D, Li E, Stanley SL Jr. 2001. The bifunctional Entamoeba histolytica alcohol dehydrogenase 2 (EhADH2) protein is necessary for amebic growth and survival and requires an intact C-terminal domain for both alcohol dehydrogenase and acetaldehyde dehydrogenase activity. J Biol Chem 276:20136–20143. doi: 10.1074/jbc.M101349200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwarzenbacher R, von Delft F, Canaves JM, Brinen LS, Dai X, Deacon AM, Elsliger MA, Eshaghi S, Floyd R, Godzik A, Grittini C, Grzechnik SK, Guda C, Jaroszewski L, Karlak C, Klock HE, Koesema E, Kovarik JS, Kreusch A, Kuhn P, Lesley SA, Mcmullan D, Mcphillips TM, Miller MA, Miller MD, Morse A, Moy K, Ouyang J, Page R, Robb A, Rodrigues K, Selby TL, Spraggon G, Stevens RC, van den Bedem H, Velasquez J, Vincent J, Wang X, West B, Wolf G, Hodgson KO, Wooley J, Wilson IA. 2004. Crystal structure of an iron-containing 1,3-propanediol dehydrogenase (TM0920) from Thermotoga maritima at 1.3 Å resolution. Proteins 54:174–177. doi: 10.1002/prot.10594. [DOI] [PubMed] [Google Scholar]
- 30.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. 1983. Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 31.Jo S, Kim T, Iyer VG, Im W. 2008. CHARMM-GUI: a web-based graphical user interface for CHARMM. J Comput Chem 29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 32.Koukos PI, Glykos NM. 2013. Grcarma: a fully automated task-oriented interface for the analysis of molecular dynamics trajectories. J Comput Chem 34:2310–2312. doi: 10.1002/jcc.23381. [DOI] [PubMed] [Google Scholar]
- 33.Arnau J, Jørgensen F, Madsen SM, Vrang A, Israelsen H. 1998. Cloning of the Lactococcus lactis adhE gene, encoding a multifunctional alcohol dehydrogenase, by complementation of a fermentative mutant of Escherichia coli. J Bacteriol 180:3049–3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bruchhaus I, Tannich E. 1994. Purification and molecular characterization of the NAD(+)-dependent acetaldehyde/alcohol dehydrogenase from Entamoeba histolytica. Biochem J 303:743–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dailly Y, Bunch P, Clark D. 2000. Comparison of the fermentative alcohol dehydrogenases of Salmonella typhimurium and Escherichia coli. Microbios 103:179–196. [PubMed] [Google Scholar]
- 36.Pei J, Zhou Q, Jiang Y, Le Y, Li H, Shao W, Wiegel J. 2010. Thermoanaerobacter spp. control ethanol pathway via transcriptional regulation and versatility of key enzymes. Metab Eng 12:420–428. doi: 10.1016/j.ymben.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 37.Yao S, Mikkelsen MJ. 2010. Identification and overexpression of a bifunctional aldehyde/alcohol dehydrogenase responsible for ethanol production in Thermoanaerobacter mathranii. J Mol Microbiol Biotechnol 19:123–133. doi: 10.1159/000321498. [DOI] [PubMed] [Google Scholar]
- 38.Koo OK, Jeong DW, Lee JM, Kim MJ, Lee JH, Chang HC, Kim JH, Lee HJ. 2005. Cloning and characterization of the bifunctional alcohol/acetaldehyde dehydrogenase gene (adhE) in Leuconostoc mesenteroides isolated from kimchi. Biotechnol Lett 27:505–510. doi: 10.1007/s10529-005-2541-z. [DOI] [PubMed] [Google Scholar]
- 39.Chen M, Li E, Stanley SL. 2004. Structural analysis of the acetaldehyde dehydrogenase activity of Entamoeba histolytica alcohol dehydrogenase 2 (EhADH2), a member of the ADHE enzyme family. Mol Biochem Parasitol 137:201–205. doi: 10.1016/j.molbiopara.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 40.Montella C, Bellsolell L, Perez-Luque R, Badia J, Baldoma L, Coll M, Aguilar J. 2005. Crystal structure of an iron-dependent group III dehydrogenase that interconverts l-lactaldehyde and l-1,2-propanediol in Escherichia coli. J Bacteriol 187:4957–4966. doi: 10.1128/JB.187.14.4957-4966.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang H, Wang S, Moll J, Thauer RK. 2012. Electron bifurcation involved in the energy metabolism of the acetogenic bacterium Moorella thermoacetica growing on glucose or H2 plus CO2. J Bacteriol 194:3689–3699. doi: 10.1128/JB.00385-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Currie DH, Guss AM, Herring CD, Giannone RJ, Johnson CM, Lankford PK, Brown SD, Hettich RL, Lynd LR. 2014. Profile of secreted hydrolases, associated proteins, and SlpA in Thermoanaerobacterium saccharolyticum during the degradation of hemicellulose. Appl Environ Microbiol 80:5001–5011. doi: 10.1128/AEM.00998-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verbeke TJ, Zhang X, Henrissat B, Spicer V, Rydzak T, Krokhin OV, Fristensky B, Levin DB, Sparling R. 2013. Genomic evaluation of Thermoanaerobacter spp. for the construction of designer co-cultures to improve lignocellulosic biofuel production. PLoS One 8:e59362. doi: 10.1371/journal.pone.0059362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Radianingtyas H, Wright PC. 2003. Alcohol dehydrogenases from thermophilic and hyperthermophilic archaea and bacteria. FEMS Microbiol Rev 27:593–616. doi: 10.1016/S0168-6445(03)00068-8. [DOI] [PubMed] [Google Scholar]
- 45.Burdette D, Zeikus JG. 1994. Purification of acetaldehyde dehydrogenase and alcohol dehydrogenases from Thermoanaerobacter ethanolicus 39E and characterization of the secondary-alcohol dehydrogenase (2 degrees Adh) as a bifunctional alcohol dehydrogenase-acetyl-CoA reductive thioesterase. Biochem J 302:163–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lamed R, Zeikus JG. 1980. Ethanol production by thermophilic bacteria: relationship between fermentation product yields of and catabolic enzyme activities in Clostridium thermocellum and Thermoanaerobium brockii. J Bacteriol 144:569–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neale AD, Scopes RK, Kelly JM, Wettenhall REH. 1986. The two alcohol dehydrogenases of Zymomonas mobilis purification by differential dye ligand chromatography, molecular characterisation and physiological roles. Eur J Biochem 154:119–124. doi: 10.1111/j.1432-1033.1986.tb09366.x. [DOI] [PubMed] [Google Scholar]
- 48.Chen Z, Lee WR, Chang SH. 1991. Role of aspartic acid 38 in the cofactor specificity of Drosophila alcohol dehydrogenase. Eur J Biochem 202:263–267. doi: 10.1111/j.1432-1033.1991.tb16371.x. [DOI] [PubMed] [Google Scholar]
- 49.Kessler D, Herth W, Knappe J. 1992. Ultrastructure and pyruvate formate-lyase radical quenching property of the multienzymic AdhE protein of Escherichia coli. J Biol Chem 267:18073–18073. [PubMed] [Google Scholar]
- 50.Extance JP. 2012. Ph.D. thesis University of Bath, Bath, United Kingdom. [Google Scholar]
- 51.Lee JM, Venditti RA, Jameel H, Kenealy WR. 2011. Detoxification of woody hydrolyzates with activated carbon for bioconversion to ethanol by the thermophilic anaerobic bacterium Thermoanaerobacterium saccharolyticum. Biomass Bioenergy 35:626–636. doi: 10.1016/j.biombioe.2010.10.021. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.