Abstract
We isolated several related but distinct cDNA clones encoding novel structure proteins (NSP) when screening a cDNA library. Analysis revealed that these cDNAs and several similar ESTs in the public databases are derived from a single gene of 17 exons that span a minimum of 227-kb region. This gene is located at chromosome 17p11.2, a region frequently amplified in human gliomas and osteosarcomas, and involved in Birt–Hogg–Dube syndrome, a tumor-prone syndrome. The major coding sequences shared by all isolated transcripts are predicted to encode SMC (structural maintenance of chromosome)/SbcC ATPase motifs and coiled-coil domains commonly seen in motor or structure proteins. Two 5′-end and two 3′-end variants (type 5α/β and 3α/β, respectively) were identified, making a total of four possible transcripts. Both 5α and 5β variants were detected in human testis mRNA, but only type 5α was detectable in RNA samples extracted from HeLa cells. The unique carboxyl-terminus of 3β contains a Ca2+-dependent actin-binding domain. Immunohistochemistry studies revealed that NSPs were mostly localized to nuclei. Northern blot analysis demonstrated two major bands and the expression levels are tremendously high in testis while barely detectable in other normal tissues examined. Interestingly, NSP5α3α is highly expressed in some tumor cell lines. These results suggest that NSPs represent a new family of structure proteins with a possible role in nuclear dynamics during cell division, and that NSP5α3α may serve as a tumor marker.
Keywords: NSP, SMC, ATPase, cell division, actin-binding protein, coiled-coil domain
Introduction
Cell division is a complex process that involves multiple procedures including DNA replication, DNA repair, the condensation of DNA, and separation of sister chromatids. Faithful segregation of genetic materials into two daughter cells is the key of the cell division (Hirano, 1999, 2000). Failure to duplicate the chromosome, repair damaged DNA, or segregate chromosomes leads to genetic instability and neoplasm. The retinoblastoma protein family and p53, collaborating with other cell cycle regulatory proteins, regulate the cell cycle and guard the genetic stability by triggering growth arrest and DNA repair when needed (Sang et al., 1995).
SMC (structural maintenance of chromosomes) is a family of chromosome-associated proteins that play essential roles in chromosome dynamics (Cobbe and Heck, 2000). It has been proposed that SMC has DNA-dependent ATPase activity and is involved in DNA recombination repair, chromosome condensation, and sister-chromatid cohesion, during mitosis and meiosis (Hirano and Hirano, 1998; Hopfner et al., 2000, Hirano et al., 2001). Structurally, SMC contains several functional domains including ATP-binding domain, ATP hydrolysis domain, coiled-coil domains, and a flexible hinge (Cobbe and Heck, 2000). Eucaryotic cells have at least four different SMC family members. A member may heterodimerize with a specific partner member, and this dimer further recruits other non-SMC proteins to form a functional complex. Null mutation of SMC gene in Bacillus subtilis leads to accumulation of anucleate cells, disruption of nucleoid structure, and misassembly of a protein complex required for chromosome partitioning (Britton et al., 1998).
Targeting proteins recognized by a monoclonal antibody against pRb, the retinoblastoma tumor suppressor (Sang et al., 1995), we identified a gene encoding several novel structure proteins (NSPs) that share conserved motifs with the SMC family. Interestingly, this gene is expressed at a very low or null level in normal tissues except testis, but is highly expressed in some tumor cell lines examined.
Results
Cloning of cDNA isoforms from HeLa cell line
When HeLa cell lysates were used in Western blot and immunoprecipitation, a monoclonal antibody against pRb (XZ77) (Hu et al., 1992) recognized additional bands. To identify the molecular nature of these bands, we performed an expression screening (see Material and methods). After several rounds of purification, 21 putative clones were identified. Among them, 10 clones were readily recognized by both XZ37 and another monoclonal antibody against a different epitope of pRb, and thus excluded for further analysis. Sequence analysis of the remaining 11 clones revealed an additional five clones that were derived from the Rb gene. Interestingly, four none-Rb-related clones were found to contain overlapping, but not identical cDNA fragments (Figure 1a). One of the four clones had a 3′-untranslated region (3-UTR) shorter than that of the other three, representing polyadenylation at an alternative site. The others differ from each other only at the 5′-extension and the length of the poly-A tails. Clone #8 and #10 (Figure 1a) were used as templates for in vitro transcription and translation in the presence of [35S]methionine (Sang et al., 2001). Western blot and immunoprecipitation confirmed that the in vitro translated products are recognizable by the antibody XZ77 (not shown). However, predicted amino-acid sequences showed no significant homology with pRb, except for a limited region that shares high similarity with the pRb-epitope recognized by the monoclonal antibody used.
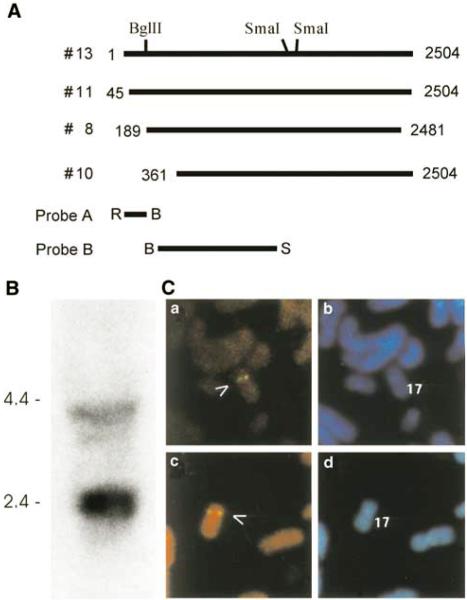
Figure 1.
Expression screening identifies cDNA fragments in HeLa cells. (a) Schematic illustration of several overlapping cDNA clones recognized by an anti-pRb antibody. (b) Northern blot shows the existence of more than one transcript in HeLa cells. Total RNA was extracted from HeLa cells and separated on denaturing agarose gel with molecular weight marker. Probe B indicated in (a) was used for the detection. (c) FISH analysis reveals that NSP is a single-copy gene localized at chromosome 17p12.2. Panels a and c show the FISH signals on chromosome 17; panels b and d indicate the same mitotic figure stained with DAPI to identify chromosome 17
Database analysis revealed that part of amino-acid sequence deduced from the cDNA share a high similarity (almost identical), to a region of a sperm antigen with unknown function (HCMOGT-1, GB Accession #AB041533), suggesting the existence of homologous genes or differentially spliced isoforms from a single gene. To confirm the existence of different transcripts and to estimate the approximate length of the transcripts, we extracted total RNA from HeLa cells and performed Northern blot analysis. Not surprisingly, the result in Figure 1b revealed two bands:one is around 4.0 kb and the other one is around 2.5 kb. To determine if these transcripts were derived from homologous genes or from a single gene and the chromosome location, we utilized fluorescence in situ hybridization (FISH) technology. The longest isolate (clone #13 in Figure 1a) was used to make a probe for FISH analysis. Lymphocytes were isolated from human blood and cultured. DAPI banding was used to identify the specific chromosome, and the assignment between signal from probe and the short arm of chromosome 17 was obtained. Examples of FISH mapping results are given in Figure 1c, which demonstrate a single signal localized at chromosome 17p11.2. There was no additional locus picked by FISH under the hybridization conditions used. These results suggest that these transcripts are most likely derived from a single gene located at human chromosome 17p11.2. This was consistent with subsequently released human genome data.
Expression in multiple normal tissues
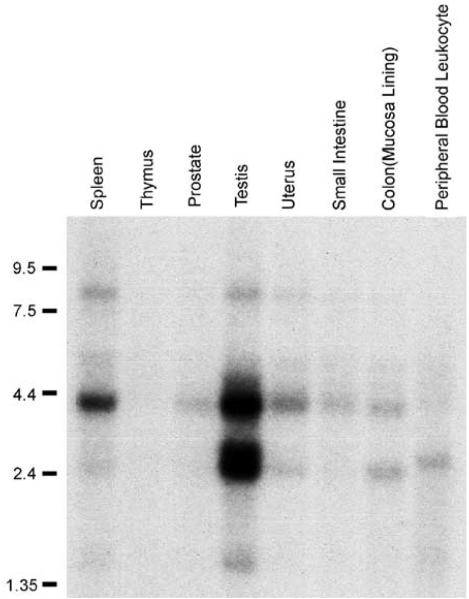
A database search revealed NSP gene represents HCMOGT-1 locus that has been previously reported to encode a human sperm antigen (Accession #NM152904 and #AB041533). Several transcript variants or ESTs have been identified from libraries derived from testis (GB Accession #AB041533) and brain tissues (hippocampus:GB Accession #BC050058; cerebellum: GB Accession #AK094274), suggesting that this gene is, at least, expressed in brain tissues as well. To further investigate the expression pattern and levels in various tissues, we obtained two multiple tissue Northern blot membranes from Clontech. An BglII–SmaI fragment (Figure 1a, probe B) was used to prepare the probe. Results of the first blot shown in Figure 2 indicate that two transcripts are highly expressed in testis. The larger transcript was also readily detected in the spleen. However, in other tissue or cells, including thymus, prostate, uterus, small intestine, colon, and peripheral blood leukocytes, both are barely detectable. The other blot probed at the same condition revealed lack or very low levels of expression in human heart, brain, placenta, lung, liver, kidney, skeleton muscle, and pancreas (not shown). Low-level or a lack of expression of this gene in normal tissues except for testis was further confirmed by a multitissue dot-blot (not shown). Taken together, these data suggest that high-level expression of this gene is specific for testis among normal tissues, consistent with the notion that it encodes a sperm antigen.
Figure 2.
Expression pattern of NSP in normal tissues. Multitissue Northern blots were purchased from Clontech (BD Science) and detected by Probe B indicated in Figure 1a
Overexpression in tumor cell lines
Several ESTs with certain variations were isolated from various tumors including melanotic melanoma (GB Accession #BC021123), embryonic carcinoma (GB Accession #BC33618), and HeLa cell line (this study), suggesting that corresponding transcript variants are present in these tumor cell lines. In addition, the chromosome 17p11.2 has been implicated in amplification in human osteosarcoma (van Dartel et al., 2002) and glioma (van Dartel et al., 2003). We investigated the expression status of this gene in a collection of tumor cell lines including Saos-2 (osteosarcoma), T98G (glioma), HT29 (colonic carcinoma), HeLa (cervical carcinoma), MCF7 (breast carcinoma), ML1 (myeloma), Manca (lymphoma), CEM (lymphoid cells), WERI (retinoblastoma), and 293 (adenovirus transformed kidney cells). We observed that the short form is highly expressed in HeLa, Saos-2, MCF7, T98G, and WERI cell lines (Figure 3a and b). The large transcript was also detected in these lines, but at relatively lower levels. Neither transcript was detected in other tumor cell lines examined.
Figure 3.
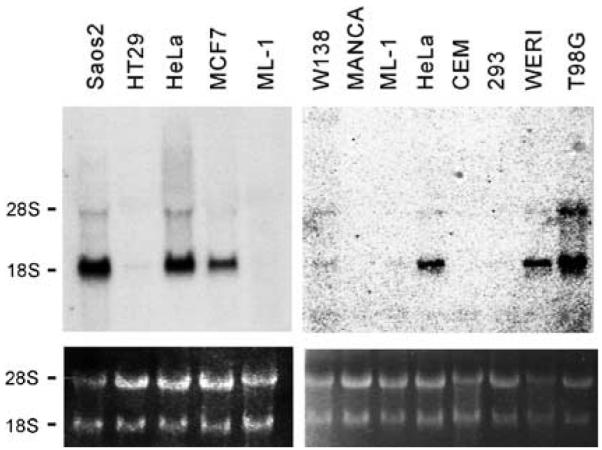
NSP5α3α is overexpressed in several tumor cell lines. Total RNA samples were extracted from the cell lines indicated. The samples were fractionated and transferred onto nylon membranes and hybridized with Probe B indicated in Figure 1a
Molecular cloning and characterization of full-length cDNA isoforms
To obtain the full-length cDNA of these transcripts, we screened a 5′-stretched HeLa library with a fragment derived from clone #13 (5′-end of BglII site, probe A in Figure 1a). This screening extended the cDNA sequence about 1200 bp toward the 5′-end. We next used RT–PCR, 5′ and 3′ RACE–PCR techniques to investigate the 5′ and 3′ variations. Both forms of 3′-ends were confirmed in mRNA samples derived from HeLa cells and testis (Figure 4a). However, from testis mRNA, an additional form of 5′-end was found (Figure 4b). For clarity in description, we named the 5′-end originally isolated for HeLa cell isoform α and the additional form from testis isoform β. Similarly, the two forms of 3′-ends were named isoforms 3α and 3β. Summarizing the data generated by various approaches, we compiled four putative full-length cDNAs with sizes consistent with the transcripts shown in Northern blots. The entire cDNA sequences and predicted amino-acid sequences are shown in Figure 4a and b. Based on the cDNA sequences, predicted characteristics of these transcripts and peptides were summarized in Table 1. Finally, using the DNA fragments from different clones, we constructed full-length cDNA of NSP5α3α and NSP5α3β by standard DNA recombination methods. In vitro transcription/translation resulted in two peptides with predicted sizes (Figure 4c).
Figure 4.
Nucleotide and amino-acid sequences of NSPs. Putative translation start codons are in bold face. Putative nuclear localization signal is underlined. An alternative poly-A-tail site is underlined. (a) NSP5α3α and 5α3β. (b) Nucleotide and amino-acid sequences of the 5′-end variants from testis (NSP5α). (c) In vitro transcription and translation of NSP5α3α and NSP5α3β. SDS–PAGE shows that the longest translational products have the expected sizes. Faster migration species represent products translated from internal initiating codons
Table 1.
Predicted characteristics of the NSP gene products
Conserved motifs and features of the predicted peptides
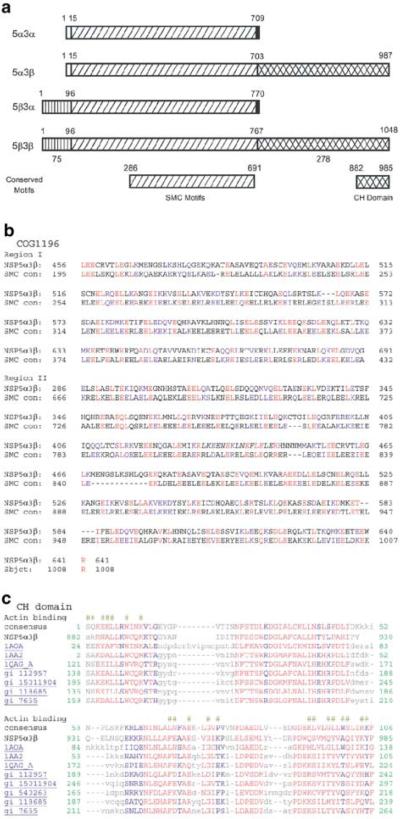
To seek clues of the function of NSP, we searched protein database (Swiss-prot) for sequence similarities with other proteins. Results indicate that major part of the middle region shared by both putative peptides has a primary structure similar to various motor proteins, including a variety of myosins, at the coiled-coil region, but lacks the myosin head domain. The representative myosin with the highest similarity is the skeletal muscle heavy polypeptide 2 (GB Accession #NP_659210) (Allen and Leinwand, 2002). Interestingly, it also shows significant similarity to NUF1 or SPC110 (yeast spindle poly-body spacer protein, Swiss-prot Accession #P32380) (Mirzayan et al., 1992; Kilmartin et al., 1993), centrosome protein 2 (CEP250/c-Nap1, Swiss-prot Accession #Q9BV73; GB Accession #NP_009117) (Fry et al., 1998; Mayor et al., 2000, 2002), centromeric protein E (Accession #Q02224) (Yen et al., 1992; Thrower et al., 1995; Chan et al., 1998), kinesin heavy chain (Synkin, Accession #O43093) (Grummt et al., 1998), and Spo15p (S. pombe sporulating protein, Accession #Q10411) (Ikemoto et al., 2000). A conserved domain search revealed that this region has a coiled-coil structure and forms a conserved chromosome segregation ATPase domain similar to that of SMC family members (Figure 5a and b). SMC is a family of chromosome-associated ATPases that play important roles in chromosome maintenance, dynamics, and partitioning (Hirano, 1999, 2000; Cobbe and Heck, 2000). It is also involved in DNA repair and recombination. Finally, this region also shares significant similarity with SbcC of Escherichia coli (Swiss-prot Accession #Q97FK1) (Connelly et al., 1997, 1998) and RAD50 of S. cerevisiae (Sharples and Leach, 1995; Hopfner et al., 2000). SbcC is an ATP-driven nuclease involved in DNA repair and belongs to the SMC superfamily (Connelly et al., 1998). The 153 kDa RAD50 protein is required for DNA repair during vegetative growth and is needed in prophase for chromosome synapsis and recombination during meiosis. NSP3β has a unique CH domain at the carboxyl-terminus (Carugo et al., 1997; Banuelos et al., 1998) (Figure 5a), which is similar to that found at the N-terminus of beta-spectrin and various alpha-actinins, and predicted to be a Ca2+-dependent actin-binding domain (Figure 5c). The actin-binding domain at the C-terminus suggests a potentially unique biological role.
Figure 5.
Structure analysis of NSP proteins. (a) Schematic illustration of conserved domains of NSP. Amino-acid numbering of conserved domains was based on NSP5α3β. (b) Alignment of the core homologous region of NSP with the conserved domain of SMC (KOG0161). (c) Alignment of the CH domains of NSP3β with other CH containing proteins. Surface required for actin binding is indicated by ‘#’
Structure of NSP gene
We analysed the genomic structure of the NSP gene by comparing the cDNA sequences with the updated genomic database. Blast search revealed that the entire cDNA sequences are derived from three genomic clones of chromosome 17 (clone #CTD-2010G8, Accession #AC007963; clone #HRPC836L9, Accession #AC004702; and clone #RP11-121A13, Accession #AC008088.8). Since these three clones overlap with one another, we were able to compile the genomic sequences of NSPs from the cDNAs and these three clones (GB Accession #BK005598). Based on these sequences, the NSP gene contains 17 exons, which are distributed over a minimum of 227-kb region. The distribution of exons and introns and selective usage of the exons by the transcripts were illustrated in Figure 6, with details listed in Table 2.
Figure 6.
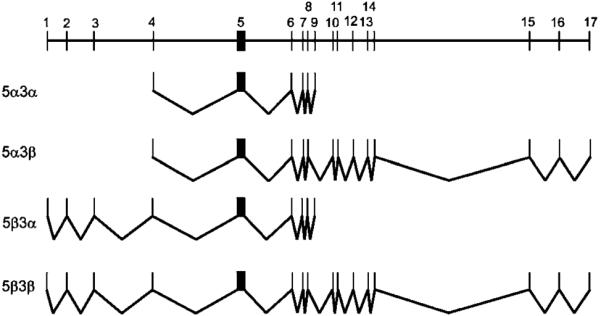
Genomic organization of NSP gene and the splicing schemes
Table 2.
Genomic arrangement of exons and the components of splicing variants
| Exons | Genomic | Length | 5 α 3 α | 5 α 3 β | 5 β 3 α | 5 β 3 β |
|---|---|---|---|---|---|---|
| 1 | 1–30 | 30 | – | – | 1–30 | 1–30 |
| 2 | 9610–9777 | 168 | — | — | 31–198 | 31–198 |
| 3 | 23 406–23 541 | 136 | — | — | 199–334 | 199–334 |
| 4 | 69 068–69 215 | 148 | 1–148 | 1–148 | — | — |
| 5 | 117 312–118 891 | 1580 | 149–1728 | 149–1728 | 335–1914 | 335–1914 |
| 6 | 140 392–140 599 | 208 | 1729–1936 | 1729–1936 | 1915–2122 | 1915–2122 |
| 7 | 144 737–144 810 | 74 | 1937–2010 | 1937–2010 | 2123–2196 | 2123–2196 |
| 8 | 145 179–145 384 | 206 | 2011–2216 | 2011–2216 | 2197–2402 | 2197–2402 |
| 9a.b | 149 839–150 158 | 320 | 2217–2537 | — | 2403–2723 | — |
| 10 | 158 905–159 050 | 146 | — | 2217–2362 | — | 2403–2548 |
| 11 | 160 198–160 298 | 101 | — | 2363–2463 | — | 2549–2649 |
| 12 | 166 484–166 565 | 82 | — | 2464–2545 | — | 2650–2731 |
| 13 | 170 430–170 586 | 157 | — | 2516–2702 | — | 2732–2888 |
| 14 | 173 171–173 273 | 103 | — | 2703–2805 | — | 2889–2991 |
| 15 | 209 940–210 056 | 117 | — | 2806–2922 | — | 2992–3108 |
| 16 | 219 002–219 061 | 60 | — | 2923–2982 | — | 3109–3168 |
| 17 | 226 955–227 734 | 780 | — | 2983–3762 | — | 3169–3948 |
One isolate (Accession # BC033618) from the embryonic carcinoma shows an alternative end (nt 152 406) of this exon
In this study, sequence analysis of cDNAs indicates an additional “c” between nt 149918–149919 in the genomic sequence
Subcellular localization
Each of the predicted peptides contains a putative nuclear localization signal (Figure 4a). To further characterize the NSP proteins, we detected the expression and subcellular distribution of NSP, which shares an antigenic epitope with pRb. Since Saos-2 expresses NSP mRNA (Figure 3) but lacks pRb (Sang et al., 1997), we grew Saos-2 cells in chamber slides and proceeded for immunohistochemical studies. When XZ77 antibody was used, a clear nucleus-staining pattern was observed (Figure 7a). Monoclonal antibody against adenovirus E1A oncoprotein was used as a negative control (Figure 7b). Another monoclonal antibody against pRb, XZ37, showed a negative staining in Saos-2 cells (Figure 7c) but clear positive nuclear staining in HeLa cells (Figure 7d), confirming that pRb is absent in the Saos-2 cells used. Taken together, these data suggest that NSPs may be nuclear proteins.
Figure 7.
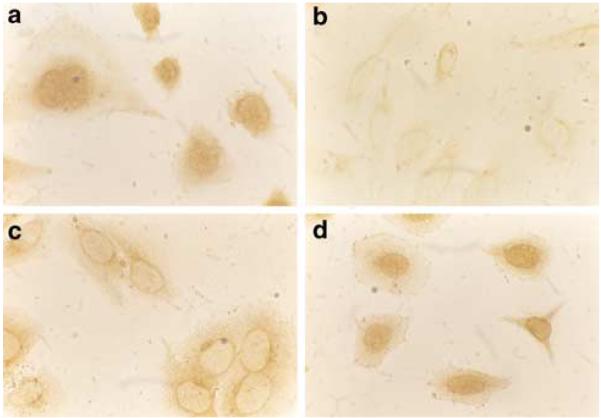
Subcellular localization of NSP gene products. Saos-2 and HeLa cells were cultured in chamber slides and proceeded for immunohistochemistry studies as described in Material and methods. The crossreactive antibody recognized nuclear signals in the absence of pRb (a). (b) Normal mouse serum gave no signal. (c) XZ37 failed to stain the nuclei of Saos-2 cells (c), but detected pRb in HeLa cells (d)
Discussion
Nuclear structure proteins play essential roles in the regulation of chromosomal structure, DNA replication, gene expression, cell kinetics, and cell division. In this study, we identified several differentially spliced transcripts encoding novel family of nuclear structure proteins. Search of databases revealed that the NSP3α is very similar (almost identical) to the human sperm antigen (HCMGOT-1) in the database with unknown function. A small discrepancy between the amino-acid sequence of NSP3α isoforms and the sperm antigen in the database may represent either natural mutations or human errors. Analysis of the genomic structure of the gene revealed that the cDNA ecoding sperm antigen in the Genebank might represent isoform 5β3α. The expression of this gene in other tissues, demonstrated by this study and by the fact that ESTs were isolated from other tissue sources, suggests that its expression is not strictly limited to sperm. It is at least expressed at a low level in brain tissues. However, the special expression pattern in different tissues suggests that this gene is not a housekeeping gene encoding cellular structure proteins common to all cell types. Instead, it may have a specific function required only for certain types of cells and/or during a specific development stages. Nevertheless, homologous sequences are present in Mus musculus (Accession #XM_205737), Rattus norvegicus (Accession #XM_220544), Danio rerio (AL928675), Caenorhabditis elegans (Accession #U58747), and Drosophila melanogaster (Accession #T13410), suggesting that this gene is highly conserved and thus may have a fundamental function for a broad range of organisms.
Analysis of the amino-acid sequences revealed that the major part shared by all isoforms forms a coiled-coil structure. Similarity between NSP and some nuclear motor proteins that play essential roles in chromosome or organelle partitioning suggests that NSP may function as nuclear force-producing proteins. The unique carboxyl-terminus of NSP3β (103 amino acids) forms a CH domain (Carugo et al., 1997; Banuelos et al., 1998) that is predicted to serve as a Ca2+-dependent actin-binding domain similar to that located at the N-termini of alpha-actinins and spectrin beta-chain (Rozycki et al., 1994; Banuelos et al., 1998; Viel, 1999). This domain is predicted to interact with actin or other proteins to generate motion (Rozycki et al., 1994; Ample and Vandekerckhove, 1994; Volkmann et al., 2001). In addition, predicted polypeptides contain a nuclear localization signal (Figure 4a). In agreement with this prediction, immunohistochemistry studies showed that NSP is generally localized to the nuclei (Figure 7). Taken together, these data suggest that NSP3β are nuclear motor proteins that may be functionally involved in chromosome or organelle partitioning during cell division, especially in meiosis. NSP3α, which lacks the unique actin-binding domain of NSP3β, may functionally interplay with NSP3β.
Similarity was also observed between the conserved SMC region of NSP and use of E. coli SbcC (Connelly et al., 1997, 1998) and S. cerevisiae Rad50 (Hopfner et al., 2000), both are involved in ATP-driven DNA recombination and double-strand break repair. If NSP is involved in a similar function, abnormal expression of NSP in some tumor cell lines may contribute to the genetic instability that is commonly observed in tumor cells. While the precise biological function of this gene remains to be investigated, this gene and its products could serve as new molecular targets for cancer therapy, especially if expression of this gene is confirmed to be a promoting factor for certain types of tumors.
Northern blot analysis reveals that the 3β transcript is expressed at barely detectable levels in normal human tissues except for testis, where both 3α and 3β transcripts are highly expressed. However, the 3α form is expressed at a significantly high level in tumor cell lines including HeLa, Saos-2, T98G, and WERI (retinoblastoma). Interestingly, NSP/CMGOT gene is within the chromosome region that was amplified in osteosarcomas and gliomas (van Dartel et al., 2002, 2003). If a relationship between the expression of NSP5α3α and tumor phenotypes can be established, NSP5α3α may serve as a novel tumor marker, thus providing another molecular tool for the diagnosis and prognosis of certain tumors, and a molecular target for cancer therapy.
Materials and methods
cDNA cloning
E. coli BB4 and XL-1 strains, Lambda gt11 HeLa library, and helper phages were purchased from Stratagene. Monoclonal antibodies against pRb (XZ37, XZ77, and XZ131) were described previously (Hu et al., 1992). The library titer was first determined by sequential dilution of the original stock and plated out each dilution with BB4 cells on NZY agar plates (Sambrook and Russell, 2001). To screen the library, 1.2 × 106 plaque-forming units (PFU) were plated out with BB4 cells on NZY agarose (bottom and top). The plates were first incubated in 42°C for 4 h and then transferred to 37°C, and covered with filters that had been saturated with 10 mm IPTG and dried. After 6 h, the filters were carefully removed, washed three times with 1 × TBS (Sang et al., 2002) with 0.05% Tween 20 for 15 min each. The filters were then blocked in 1 × TBS with 5% milk, 0.1% Tween-20 for 2 h. The supernatant of XZ131 (1 : 10) and XZ77 (1 : 10) were mixed with 1 × TBS containing 1 × 3% fat-free milk and used as in immunodetection. After incubation with the antibodies over night at 4°C, the filters were washed three times with 1 × TBST, 15 min each, and incubated with HRP-conjugated rabbit anti-mouse IgG antibody, 1 : 10000 dilution in 3% fat-free milk TBS at room temperature for 2 h. Finally, after three washes with 1 × TBST, the filters were developed with ECL kit. The positive plaques were cored and put in 300 μl of SM buffer (Sambrook and Russell, 2001), followed by adding 20 μl of chloroform, incubated at 37°C for 30 min, and centrifuged. Samples of the supernatant were diluted and plated out for further purification using similar procedures.
For in vivo pBluescript excision, the purified phages and 10 μl of helper phages were mixed with XL-1 cells and incubated for 37°C for 15 min. After the addition of 5 ml of LB medium, incubation was continued for 5 h. After being heated at 68°C for 15 min, the mixture was centrifuged at 4°C and the supernatant was diluted with SM to 10−2. A measure of 20 μl of 10−2 dilution of rescued phagmids were mixed with 200 μl of XL-1 and incubated at 37°C for 20 min. Finally, the samples were diluted to 10−2 and plated with top agarose on LB plates containing ampicillin. After overnight incubation, colonies formed were picked up for plasmid preparation and further characterization.
Subsequently, a cDNA fragment (BamHI–BglII) derived from the 5′-end of one of the initial positive clone was labeled with α-dCTP by a Random Primer kit (Roche) following the manufacturer’s instruction. This probe was used to screen a 5′-extended HeLa library in Lambda gt10 (Stratagene). Briefly, the library was diluted, plated out with NZY medium (both top and bottom agarose), and tittered initially. Then 1.2 × 106 PFU were plated out and screened at low stringency (38% formamide, 5 × Denhart’s solution, 5 × SSPE, 0.1%SDS, and 150 μg/ml of herring sperm DNA). Hybridization was performed at 42°C overnight and the membranes were washed three times in 2 × SSC, 0.1% SDS at 42°C. The subsequent excision procedure was identical to above.
RACE–PCR and RT-PCR
The 3 × 106 PFU of the 5-stretched HeLa library was plated out on 50 plates (150 mm). After overnight incubation, the recombinant phage DNA was purified from plaques and used for RACE–PCR amplification. The primers used were as follows: PR1, 5′-CTG TGG CTT TCT GCT TCT CAC CCT G-3′ (rev); PR2, 5′-CCT TCC AAG GTA ACT CTA CAC TCT TC-3′ (rev); and 5FD, CTG GTT CGG CCC ACC TCT GAA GGT TCC AGA ATC GAT AG-3′ (fwd). For RT–PCR: total RNA from HeLa cells was reverse transcribed first with a reverse transcription kit (Roche), followed by PCR amplification. For 5′-end amplification, PCR was performed with PR3: 5′-CCC CAA GCT TAG AAG CAG AAA AGG TTG GAT A-3′; and PR4: 5′-TGG TTC CGG ATC CTC GGG C-3′, as primers. For 3′-end amplification of NSP2, PCR was performed with PR5: 5′-CGG TTG CAG AAG GAG CTG G-3′; and PR6: 5′-CAG AAC TTC ATT TTA TCC TC-3′. The PCR product was cloned into pGEM-easy (Promega) vector and sequenced subsequently. The above primers were synthesized either by Nuclear Acid Facility at Kimmel Cancer Institute, Thomas Jefferson University, or by Integrated DNA Inc.
RNA extraction and Northern blot analysis
Adult human multitissue Northern blots and dot-blot membranes were purchased from Clontech. Each lane contained 2 μg of poly-(A)+RNA. For Northern blot analysis of tumor cell lines, total RNA was extracted with the RNAzol B method (CINNA/BIOTECX, Friendswood, TX, USA). The total RNAs were quantitated spectrophotometrically and the integrity was confirmed by fractionation of 2 μg of each RNA sample on a 1% agarose gel, followed by ethidium bromide staining and observation under UV light.
Total RNA (25 μg) of each sample was subjected to electrophoresis in through a 1% denaturing agarose gel containing formaldehyde. RNAs were transferred overnight to an Immobilon N-nylon membrane (Millipore) with 20 × SSC and RNAs were crosslinked to the membrane by UV irradiation. The membranes were prehybridized at 42°C first in a solution containing: 5 × SSPE, 10 × Denhardt’s solution, 100 μg/ml of fresh denatured, sheared salmon sperm DNA, 50% formamide, 2% SDS for 12 h, and hybridized with α-dCTP-labeled cDNA probe (1.5 × 106 c.p.m./ml). After overnight hybridization, membranes were washed twice with 2 × SSC, 0.2% SDS at room temperature for 20 min, followed by one wash with 0.2 × SSC, 0.1% SDS at 42°C for 10 min, and exposed to a Kodak X-ray film at −80°C with intensifying screens for 3 days.
Cell lines and culture
In this study, following cell lines were used: Saos-2 (osteosarcoma), HT-29 (colon adenocarcinoma), Hela (cervical carcinoma), TG98 (glioma), MCF-7 (breast adenocarcinoma) WERI (retinoblastoma), ML1 (myeloid leukemia), Manca (lymphoma), CEM (lymphoid cells), and 293 (adenovirus transformed kidney cells). All these cell lines were obtained from ATCC and cultured at conditions recommended by ATCC.
Immunohistochemistry
Saos-2 cell were cultured in chamber slides (Fisher Scientific). The slides were washed with 1 × PBS for 20 min, embedded in 0.3% of H2O2 for 20 min, and washed with 1 × PBS again for 20 min. The slides then were incubated with 150 μl of normal goat serum diluted in 10 ml of 1 × PBS for 1 h. After washing with PBS for 10 min, the slides were incubated with primary antibody for 2 h and washed again with 1 × PBS for 10 min. Then the slides were incubated with 150 μl of normal horse serum diluted in 10 ml of 1 × PBS for 1 h. After a 10-min wash with 1 × PBS, the slides were incubated with biotinylated secondary antibody (goat against mouse IgG) for 1 h. Then, the slides were washed with 1 × PBS for 10 min, followed by incubation with ABC for 1 h (100 μl of A 100 μl of B diluted in 5 ml 1 × PBS). After 10 min incubation with 1 × PBS, the slides were exposed to DAB for 2–5 min (100 μl DAB, 100 μl H2O2, and 100 μl of buffer in 5 ml 1 × PBS). Finally, the slides were washed with water and examined under a microscope.
In situ hybridization and FISH detection
To prepare the slides for FISH analysis, lymphocytes isolated from human blood were cultured in a minimal essential medium (MEM) supplemented with 10% fetal calf serum and phytohemagglutinin at 37°C for 68–72 h. The lymphocyte cultures were treated with BrdU (0.18 mg/ml Sigma) to synchronize the cell population. The synchronized cells were washed three times with serum-free medium to release the block and recultured at 37°C for 6 h in alpha-MEM with thymidine (2.5 μg/ml; Sigma). Cells were harvested and slides were made by using standard procedures including hypotonic treatment, fixation, etc. (Heng et al., 1992).
cDNA probes were biotinylated with dATP using the BRL BioNick labeling kit (15°C, l h) (Heng et al., 1992, 1994). The procedure for FISH detection was performed according to Heng et al. (1992) and Heng and Tsui (1993). Briefly, slides were baked at 55°C for 1 h. After RNase treatment, the slides were denatured in 70% formamide in 2 × SSC for 2 min at 70°C followed by dehydrated with ethanol. Probes were denatured at 75°C for 5 min in a hybridization mix consisting of 50% formamide, 10% dextran sulfate, and human Cot1 DNA. Probes were incubated with the denatured slides. After overnight hybridization, slides were washed and detected as well as amplified using published method (Heng et al., 1994). FISH signals and the DAPI banding patterns were recorded separately by taking photographs, and the assignment of the FISH mapping data with chromosomal bands was achieved by superimposing FISH signals with DAPI-banded chromosomes (Heng and Tsui, 1993).
Acknowledgements
This work was initiated in Dr Giordano’s laboratory in Temple University as part of the thesis research of NS and supported by various grants (NIH and Searro Health Research Organization www.shro.org) to AG Late work of this project was partly supported by fund from Cardeza Foundation and Howard Temin award from NIH/NCI to NS. NS acknowledges the support from Dr J Caro and Dr S Mckenzie. We thank Dr HHQ Heng for his expertise in FISH analysis and Mr Likens for his professional assistance in preparation of illustrations.
References
- Allen DL, Leinwand LA. J. Biol. Chem. 2002;277:45323–45330. doi: 10.1074/jbc.M208302200. [DOI] [PubMed] [Google Scholar]
- Ample C, Vandekerckhove J. Semin. Cell Biol. 1994;5:175–182. doi: 10.1006/scel.1994.1022. [DOI] [PubMed] [Google Scholar]
- Banuelos S, Saraste M, Garugo KD. Structure. 1998;6:1419–1431. doi: 10.1016/s0969-2126(98)00141-5. [DOI] [PubMed] [Google Scholar]
- Britton RA, Lin DC, Grossman AD. Genes Dev. 1998;12:1254–1259. doi: 10.1101/gad.12.9.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carugo KD, Banuelos S, Saraste M. Nat. Struct. Biol. 1997;4:175–179. doi: 10.1038/nsb0397-175. [DOI] [PubMed] [Google Scholar]
- Chan GK, Schaar BT, Yen TJ. J. Cell. Biol. 1998;143:49–63. doi: 10.1083/jcb.143.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobbe N, Heck MM. J. Struct. Biol. 2000;129:123–143. doi: 10.1006/jsbi.2000.4255. [DOI] [PubMed] [Google Scholar]
- Connelly JC, de Leau ES, Okely EA, Leach DR. J. Biol. Chem. 1997;272:19819–19826. doi: 10.1074/jbc.272.32.19819. [DOI] [PubMed] [Google Scholar]
- Connelly JC, Kirkham LA, Leach DR. Proc. Natl. Acad. Sci. USA. 1998;95:7969–7974. doi: 10.1073/pnas.95.14.7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry AM, Mayor T, Meraldi P, Stierhof YD, Tanaka K, Nigg EA. J. Cell Biol. 1998;141:1563–1574. doi: 10.1083/jcb.141.7.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt M, Pistor S, Lottspeich F, Schliwa M. FEBS Lett. 1998;427:79–84. doi: 10.1016/s0014-5793(98)00399-8. [DOI] [PubMed] [Google Scholar]
- Heng HHQ, Squire J, Tsui LC. Proc. Natl. Acad. Sci. USA. 1992;89:9509–9513. doi: 10.1073/pnas.89.20.9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng HHQ, Tsui LC. Chromosoma. 1993;102:325–332. doi: 10.1007/BF00661275. [DOI] [PubMed] [Google Scholar]
- Heng HHQ, Xiao H, Shi XM, Greenblatt J, Tsui LC. Hum. Mol. Genet. 1994;3:61–64. doi: 10.1093/hmg/3.1.61. [DOI] [PubMed] [Google Scholar]
- Hirano M, Anderson DE, Erickson HP, Hirano T. EMBO J. 2001;20:3238–3250. doi: 10.1093/emboj/20.12.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano M, Hirano T. EMBO J. 1998;17:7139–7148. doi: 10.1093/emboj/17.23.7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. Genes Dev. 1999;13:11–19. doi: 10.1101/gad.13.1.11. [DOI] [PubMed] [Google Scholar]
- Hirano T. Annu. Rev. Biochem. 2000;69:115–144. doi: 10.1146/annurev.biochem.69.1.115. [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Karcher A, Shin DS, Craig L, Arthur LM, Carney JP, Tainer JA. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- Hu Q, Lees JA, Buchkovich KJ, Harlow E. Mol. Cell. Biol. 1992;12:971–980. doi: 10.1128/mcb.12.3.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemoto S, Nakamura T, Kubo M, Shimoda C. J. Cell Sci. 2000;113:545–554. doi: 10.1242/jcs.113.3.545. [DOI] [PubMed] [Google Scholar]
- Kilmartin JV, Dyos SL, Kershaw D, Finch JT. J. Cell Biol. 1993;123:1175–1184. doi: 10.1083/jcb.123.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor T, Hacker U, Stierhof YD, Nigg EA. J. Cell Sci. 2002;115:3275–3284. doi: 10.1242/jcs.115.16.3275. [DOI] [PubMed] [Google Scholar]
- Mayor T, Stierhof YD, Tanaka K, Fry AM, Nigg EA. J. Cell Biol. 2000;151:837–846. doi: 10.1083/jcb.151.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzayan C, Copeland CS, Snyder M. J. Cell Biol. 1992;116:1319–1332. doi: 10.1083/jcb.116.6.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozycki MD, Myslik JC, Schutt CE, Lindberg U. Curr. Opin. Cell Biol. 1994;6:87–95. doi: 10.1016/0955-0674(94)90121-x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd CSHL press; New York: 2001. [Google Scholar]
- Sang N, Avantaggiatti ML, Giordano A. J. Cell. Biochem. 1997;66:277–285. doi: 10.1002/(sici)1097-4644(19970901)66:3<277::aid-jcb1>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Sang N, Baldi A, Giordano A. Mol. Cell. Differ. 1995;3:1–29. [Google Scholar]
- Sang N, Fang J, Srinivas V, Leshchinsky I, Caro J. Mol. Cell. Biol. 2002;22:2984–2992. doi: 10.1128/MCB.22.9.2984-2992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang N, Severino A, Russo P, Baldi A, Giordano A, Mileo AM, Paggi MG, De Luca A. J. Biol. Chem. 2001;276:27026–27033. doi: 10.1074/jbc.M010346200. [DOI] [PubMed] [Google Scholar]
- Sharples GJ, Leach DR. Mol. Microbiol. 1995;17:1215–1217. doi: 10.1111/j.1365-2958.1995.mmi_17061215_1.x. [DOI] [PubMed] [Google Scholar]
- Thrower DA, Jordan MA, Schaar BT, Yen TJ, Wilson L. EMBO J. 1995;14:918–926. doi: 10.1002/j.1460-2075.1995.tb07073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dartel M, Cornelissen PW, Redeker S, Tarkkanen M, Knuutila S, Hogendoorn PC, Westerveld A, Gomes I, Bras J, Hulsebos TJ. Cancer Genet. Cytogenet. 2002;139:91–96. doi: 10.1016/s0165-4608(02)00627-1. [DOI] [PubMed] [Google Scholar]
- Van Dartel M, Leenstra S, Troost D, Hulsebos TJ. Cancer Genet. Cytogenet. 2003;140:162–166. doi: 10.1016/s0165-4608(02)00683-0. [DOI] [PubMed] [Google Scholar]
- Viel A. FEBS Lett. 1999;460:391–394. doi: 10.1016/s0014-5793(99)01372-1. [DOI] [PubMed] [Google Scholar]
- Volkmann N, DeRosier D, Matsudaira P, Hanein D. J. Cell Biol. 2001;153:947–956. doi: 10.1083/jcb.153.5.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen TJ, Li G, Schaar BT, Szilak I, Cleveland DW. Nature. 1992;359:536–539. doi: 10.1038/359536a0. [DOI] [PubMed] [Google Scholar]