Abstract
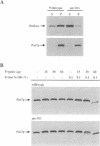
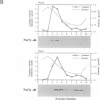
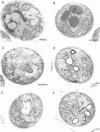
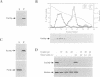
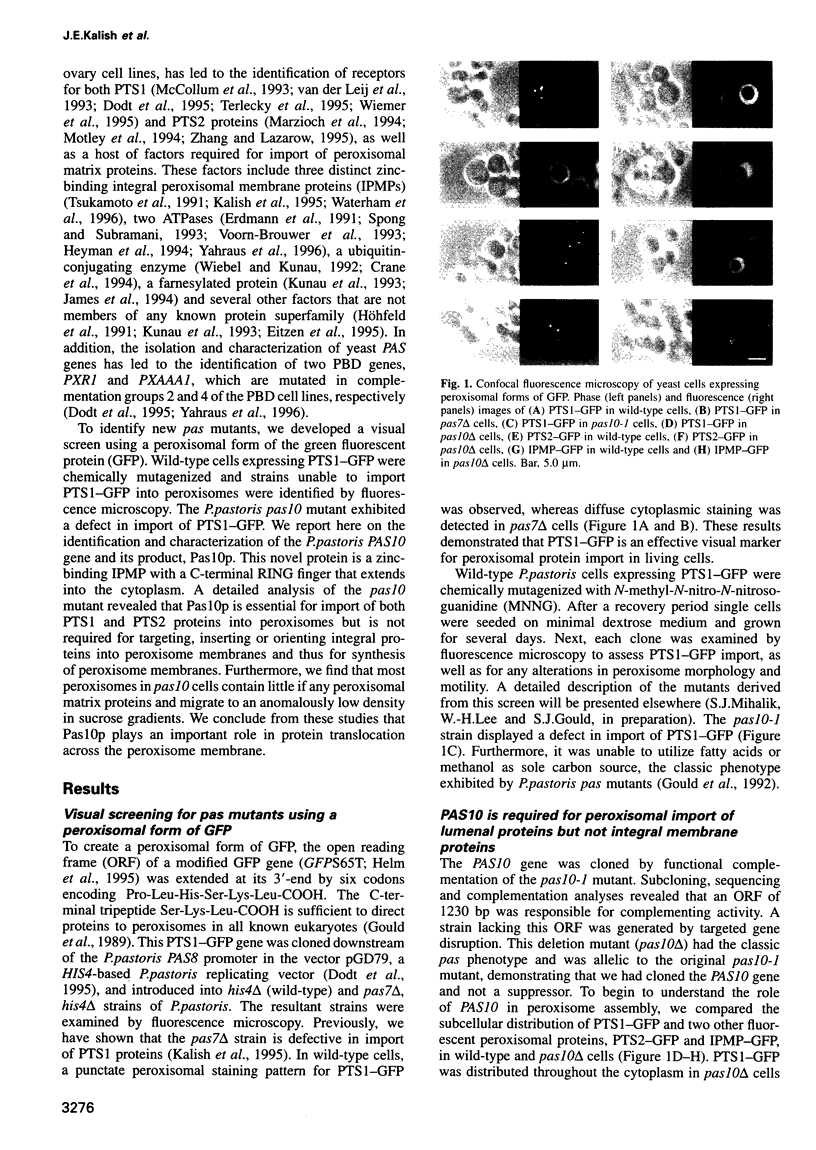
Fluorescent peroxisomal probes were developed by fusing green fluorescent protein (GFP) to the matrix peroxisomal targeting signals PTS1 and PTS2, as well as to an integral peroxisomal membrane protein (IPMP). These proteins were used to identify and characterize novel peroxisome assembly (pas) mutants in the yeast Pichia pastoris. Mutant cells lacking the PAS10 gene mislocalized both PTS1-GFP and PTS2-GFP to the cytoplasm but did incorporate IPMP-GFP into peroxisome membranes. Similar distributions were observed for endogenous peroxisomal matrix and membrane proteins. While peroxisomes from translocation-competent pas mutants sediment in sucrose gradients at the density of normal peroxisomes, >98% of peroxisomes from pas10 cells migrated to a much lower density and had an extremely low ratio of matrix:membrane protein. These data indicate that Pas10p plays an important role in protein translocation across the peroxisome membrane. Consistent with this hypothesis, we find that Pas10p is an integral protein of the peroxisome membrane. In addition, Pas10p contains a cytoplasmically-oriented C3HC4 zinc binding domain that is essential for its biological activity.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aitchison J. D., Murray W. W., Rachubinski R. A. The carboxyl-terminal tripeptide Ala-Lys-Ile is essential for targeting Candida tropicalis trifunctional enzyme to yeast peroxisomes. J Biol Chem. 1991 Dec 5;266(34):23197–23203. [PubMed] [Google Scholar]
- Aitchison J. D., Szilard R. K., Nuttley W. M., Rachubinski R. A. Antibodies directed against a yeast carboxyl-terminal peroxisomal targeting signal specifically recognize peroxisomal proteins from various yeasts. Yeast. 1992 Sep;8(9):721–734. doi: 10.1002/yea.320080905. [DOI] [PubMed] [Google Scholar]
- Barlow P. N., Luisi B., Milner A., Elliott M., Everett R. Structure of the C3HC4 domain by 1H-nuclear magnetic resonance spectroscopy. A new structural class of zinc-finger. J Mol Biol. 1994 Mar 25;237(2):201–211. doi: 10.1006/jmbi.1994.1222. [DOI] [PubMed] [Google Scholar]
- Berteaux-Lecellier V., Picard M., Thompson-Coffe C., Zickler D., Panvier-Adoutte A., Simonet J. M. A nonmammalian homolog of the PAF1 gene (Zellweger syndrome) discovered as a gene involved in caryogamy in the fungus Podospora anserina. Cell. 1995 Jun 30;81(7):1043–1051. doi: 10.1016/s0092-8674(05)80009-1. [DOI] [PubMed] [Google Scholar]
- Blattner J., Dörsam H., Clatyon C. E. Function of N-terminal import signals in trypanosome microbodies. FEBS Lett. 1995 Mar 6;360(3):310–314. doi: 10.1016/0014-5793(95)00128-v. [DOI] [PubMed] [Google Scholar]
- Blattner J., Swinkels B., Dörsam H., Prospero T., Subramani S., Clayton C. Glycosome assembly in trypanosomes: variations in the acceptable degeneracy of a COOH-terminal microbody targeting signal. J Cell Biol. 1992 Dec;119(5):1129–1136. doi: 10.1083/jcb.119.5.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden K. L., Boddy M. N., Lally J., O'Reilly N. J., Martin S., Howe K., Solomon E., Freemont P. S. The solution structure of the RING finger domain from the acute promyelocytic leukaemia proto-oncoprotein PML. EMBO J. 1995 Apr 3;14(7):1532–1541. doi: 10.1002/j.1460-2075.1995.tb07139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane D. I., Gould S. J. The Pichia pastoris HIS4 gene: nucleotide sequence, creation of a non-reverting his4 deletion mutant, and development of HIS4-based replicating and integrating plasmids. Curr Genet. 1994 Nov-Dec;26(5-6):443–450. doi: 10.1007/BF00309932. [DOI] [PubMed] [Google Scholar]
- Cregg J. M., Barringer K. J., Hessler A. Y., Madden K. R. Pichia pastoris as a host system for transformations. Mol Cell Biol. 1985 Dec;5(12):3376–3385. doi: 10.1128/mcb.5.12.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didion T., Roggenkamp R. Targeting signal of the peroxisomal catalase in the methylotrophic yeast Hansenula polymorpha. FEBS Lett. 1992 Jun 1;303(2-3):113–116. doi: 10.1016/0014-5793(92)80500-g. [DOI] [PubMed] [Google Scholar]
- Distel B., Gould S. J., Voorn-Brouwer T., van der Berg M., Tabak H. F., Subramani S. The carboxyl-terminal tripeptide serine-lysine-leucine of firefly luciferase is necessary but not sufficient for peroxisomal import in yeast. New Biol. 1992 Feb;4(2):157–165. [PubMed] [Google Scholar]
- Dodt G., Braverman N., Wong C., Moser A., Moser H. W., Watkins P., Valle D., Gould S. J. Mutations in the PTS1 receptor gene, PXR1, define complementation group 2 of the peroxisome biogenesis disorders. Nat Genet. 1995 Feb;9(2):115–125. doi: 10.1038/ng0295-115. [DOI] [PubMed] [Google Scholar]
- Eitzen G. A., Aitchison J. D., Szilard R. K., Veenhuis M., Nuttley W. M., Rachubinski R. A. The Yarrowia lipolytica gene PAY2 encodes a 42-kDa peroxisomal integral membrane protein essential for matrix protein import and peroxisome enlargement but not for peroxisome membrane proliferation. J Biol Chem. 1995 Jan 20;270(3):1429–1436. doi: 10.1074/jbc.270.3.1429. [DOI] [PubMed] [Google Scholar]
- Elgersma Y., van den Berg M., Tabak H. F., Distel B. An efficient positive selection procedure for the isolation of peroxisomal import and peroxisome assembly mutants of Saccharomyces cerevisiae. Genetics. 1993 Nov;135(3):731–740. doi: 10.1093/genetics/135.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann R., Veenhuis M., Mertens D., Kunau W. H. Isolation of peroxisome-deficient mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1989 Jul;86(14):5419–5423. doi: 10.1073/pnas.86.14.5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann R., Wiebel F. F., Flessau A., Rytka J., Beyer A., Fröhlich K. U., Kunau W. H. PAS1, a yeast gene required for peroxisome biogenesis, encodes a member of a novel family of putative ATPases. Cell. 1991 Feb 8;64(3):499–510. doi: 10.1016/0092-8674(91)90234-p. [DOI] [PubMed] [Google Scholar]
- Faber K. N., Keizer-Gunnink I., Pluim D., Harder W., Ab G., Veenhuis M. The N-terminus of amine oxidase of Hansenula polymorpha contains a peroxisomal targeting signal. FEBS Lett. 1995 Jan 3;357(2):115–120. doi: 10.1016/0014-5793(94)01317-t. [DOI] [PubMed] [Google Scholar]
- Freemont P. S., Hanson I. M., Trowsdale J. A novel cysteine-rich sequence motif. Cell. 1991 Feb 8;64(3):483–484. doi: 10.1016/0092-8674(91)90229-r. [DOI] [PubMed] [Google Scholar]
- Fujiki Y., Fowler S., Shio H., Hubbard A. L., Lazarow P. B. Polypeptide and phospholipid composition of the membrane of rat liver peroxisomes: comparison with endoplasmic reticulum and mitochondrial membranes. J Cell Biol. 1982 Apr;93(1):103–110. doi: 10.1083/jcb.93.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y., Hubbard A. L., Fowler S., Lazarow P. B. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J Cell Biol. 1982 Apr;93(1):97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietl C., Faber K. N., van der Klei I. J., Veenhuis M. Mutational analysis of the N-terminal topogenic signal of watermelon glyoxysomal malate dehydrogenase using the heterologous host Hansenula polymorpha. Proc Natl Acad Sci U S A. 1994 Apr 12;91(8):3151–3155. doi: 10.1073/pnas.91.8.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover J. R., Andrews D. W., Subramani S., Rachubinski R. A. Mutagenesis of the amino targeting signal of Saccharomyces cerevisiae 3-ketoacyl-CoA thiolase reveals conserved amino acids required for import into peroxisomes in vivo. J Biol Chem. 1994 Mar 11;269(10):7558–7563. [PubMed] [Google Scholar]
- Gould S. G., Keller G. A., Subramani S. Identification of a peroxisomal targeting signal at the carboxy terminus of firefly luciferase. J Cell Biol. 1987 Dec;105(6 Pt 2):2923–2931. doi: 10.1083/jcb.105.6.2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould S. J., Keller G. A., Hosken N., Wilkinson J., Subramani S. A conserved tripeptide sorts proteins to peroxisomes. J Cell Biol. 1989 May;108(5):1657–1664. doi: 10.1083/jcb.108.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould S. J., Keller G. A., Schneider M., Howell S. H., Garrard L. J., Goodman J. M., Distel B., Tabak H., Subramani S. Peroxisomal protein import is conserved between yeast, plants, insects and mammals. EMBO J. 1990 Jan;9(1):85–90. doi: 10.1002/j.1460-2075.1990.tb08083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould S. J., Keller G. A., Subramani S. Identification of peroxisomal targeting signals located at the carboxy terminus of four peroxisomal proteins. J Cell Biol. 1988 Sep;107(3):897–905. doi: 10.1083/jcb.107.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould S. J., Krisans S., Keller G. A., Subramani S. Antibodies directed against the peroxisomal targeting signal of firefly luciferase recognize multiple mammalian peroxisomal proteins. J Cell Biol. 1990 Jan;110(1):27–34. doi: 10.1083/jcb.110.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould S. J., McCollum D., Spong A. P., Heyman J. A., Subramani S. Development of the yeast Pichia pastoris as a model organism for a genetic and molecular analysis of peroxisome assembly. Yeast. 1992 Aug;8(8):613–628. doi: 10.1002/yea.320080805. [DOI] [PubMed] [Google Scholar]
- Grant S. G., Jessee J., Bloom F. R., Hanahan D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen H., Didion T., Thiemann A., Veenhuis M., Roggenkamp R. Targeting sequences of the two major peroxisomal proteins in the methylotrophic yeast Hansenula polymorpha. Mol Gen Genet. 1992 Nov;235(2-3):269–278. doi: 10.1007/BF00279370. [DOI] [PubMed] [Google Scholar]
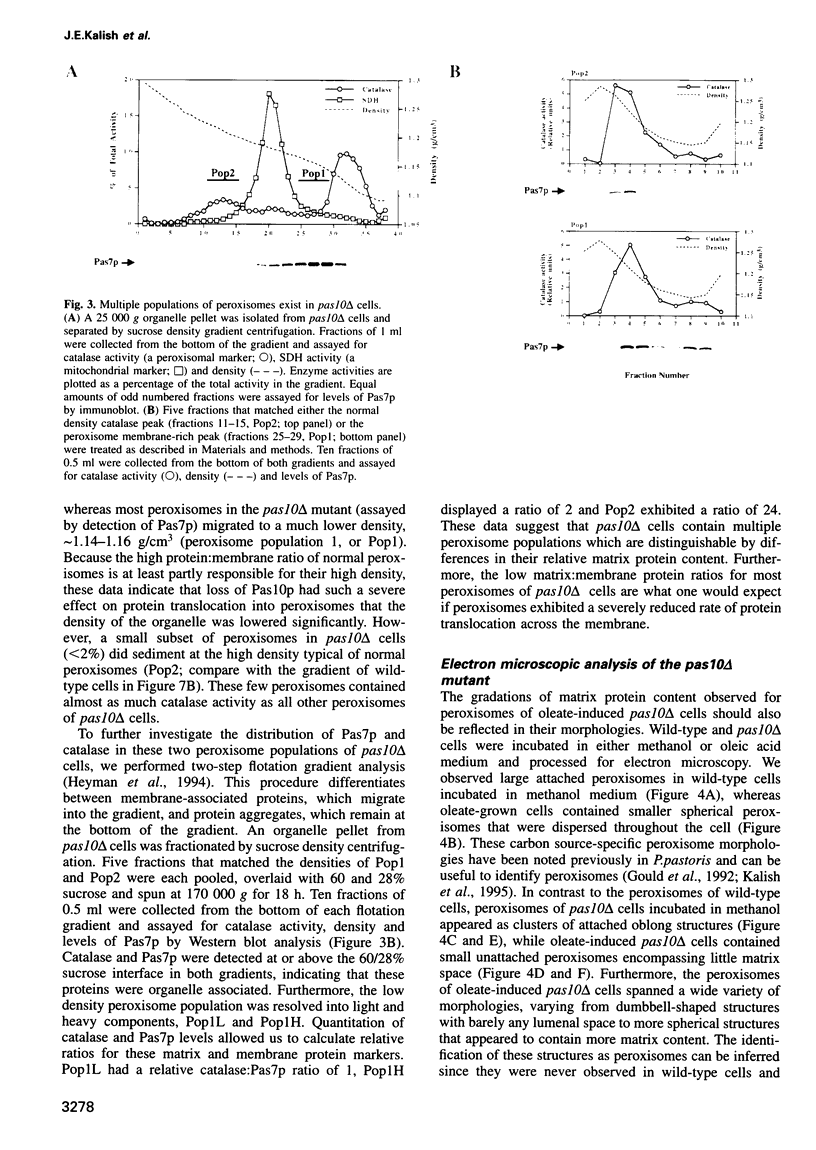
- Heim R., Cubitt A. B., Tsien R. Y. Improved green fluorescence. Nature. 1995 Feb 23;373(6516):663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- Heyman J. A., Monosov E., Subramani S. Role of the PAS1 gene of Pichia pastoris in peroxisome biogenesis. J Cell Biol. 1994 Dec;127(5):1259–1273. doi: 10.1083/jcb.127.5.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman C. S., Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57(2-3):267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Höhfeld J., Veenhuis M., Kunau W. H. PAS3, a Saccharomyces cerevisiae gene encoding a peroxisomal integral membrane protein essential for peroxisome biogenesis. J Cell Biol. 1991 Sep;114(6):1167–1178. doi: 10.1083/jcb.114.6.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igual J. C., Matallaná E., Gonzalez-Bosch C., Franco L., Pérez-Ortin J. E. A new glucose-repressible gene identified from the analysis of chromatin structure in deletion mutants of yeast SUC2 locus. Yeast. 1991 May-Jun;7(4):379–389. doi: 10.1002/yea.320070408. [DOI] [PubMed] [Google Scholar]
- James G. L., Goldstein J. L., Pathak R. K., Anderson R. G., Brown M. S. PxF, a prenylated protein of peroxisomes. J Biol Chem. 1994 May 13;269(19):14182–14190. [PubMed] [Google Scholar]
- Kalish J. E., Theda C., Morrell J. C., Berg J. M., Gould S. J. Formation of the peroxisome lumen is abolished by loss of Pichia pastoris Pas7p, a zinc-binding integral membrane protein of the peroxisome. Mol Cell Biol. 1995 Nov;15(11):6406–6419. doi: 10.1128/mcb.15.11.6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller G. A., Gould S., Deluca M., Subramani S. Firefly luciferase is targeted to peroxisomes in mammalian cells. Proc Natl Acad Sci U S A. 1987 May;84(10):3264–3268. doi: 10.1073/pnas.84.10.3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller G. A., Krisans S., Gould S. J., Sommer J. M., Wang C. C., Schliebs W., Kunau W., Brody S., Subramani S. Evolutionary conservation of a microbody targeting signal that targets proteins to peroxisomes, glyoxysomes, and glycosomes. J Cell Biol. 1991 Sep;114(5):893–904. doi: 10.1083/jcb.114.5.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunau W. H., Beyer A., Franken T., Götte K., Marzioch M., Saidowsky J., Skaletz-Rorowski A., Wiebel F. F. Two complementary approaches to study peroxisome biogenesis in Saccharomyces cerevisiae: forward and reversed genetics. Biochimie. 1993;75(3-4):209–224. doi: 10.1016/0300-9084(93)90079-8. [DOI] [PubMed] [Google Scholar]
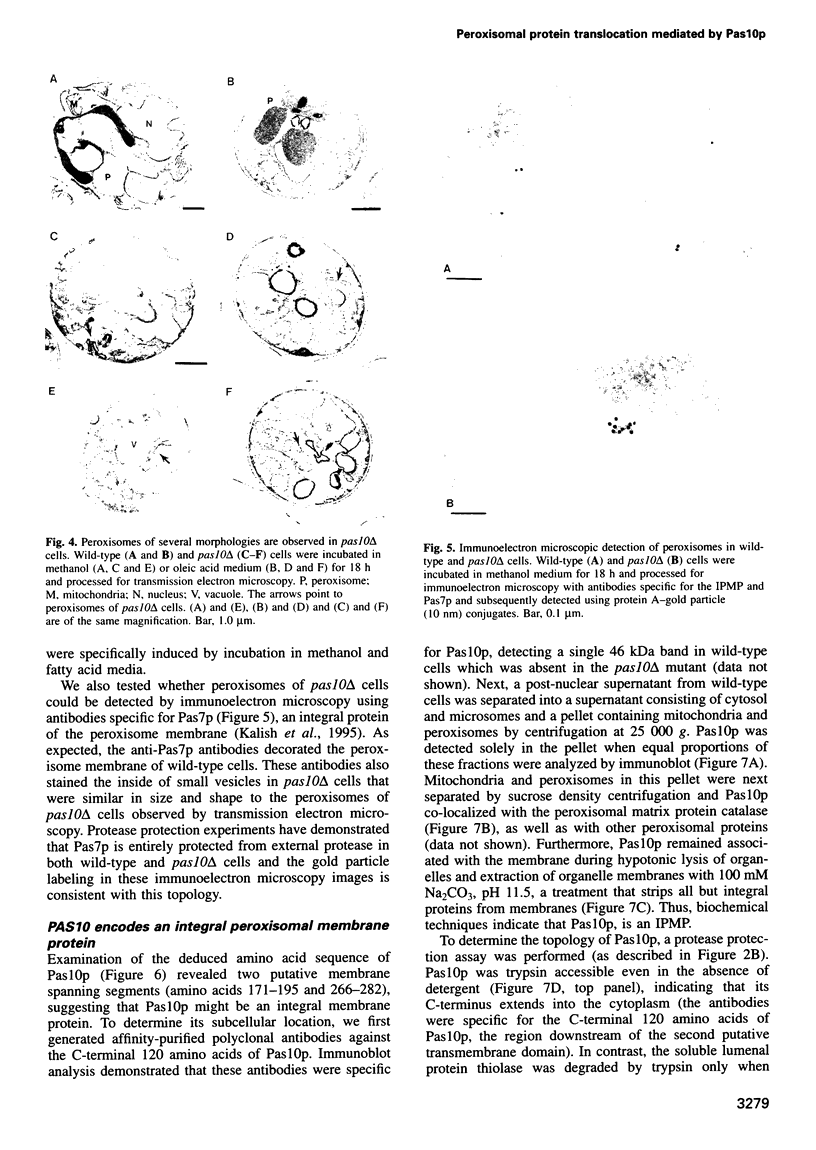
- Lazarow P. B., Fujiki Y. Biogenesis of peroxisomes. Annu Rev Cell Biol. 1985;1:489–530. doi: 10.1146/annurev.cb.01.110185.002421. [DOI] [PubMed] [Google Scholar]
- Liu H., Tan X., Veenhuis M., McCollum D., Cregg J. M. An efficient screen for peroxisome-deficient mutants of Pichia pastoris. J Bacteriol. 1992 Aug;174(15):4943–4951. doi: 10.1128/jb.174.15.4943-4951.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzioch M., Erdmann R., Veenhuis M., Kunau W. H. PAS7 encodes a novel yeast member of the WD-40 protein family essential for import of 3-oxoacyl-CoA thiolase, a PTS2-containing protein, into peroxisomes. EMBO J. 1994 Oct 17;13(20):4908–4918. doi: 10.1002/j.1460-2075.1994.tb06818.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCammon M. T., McNew J. A., Willy P. J., Goodman J. M. An internal region of the peroxisomal membrane protein PMP47 is essential for sorting to peroxisomes. J Cell Biol. 1994 Mar;124(6):915–925. doi: 10.1083/jcb.124.6.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura S., Kasuya-Arai I., Mori H., Miyazawa S., Osumi T., Hashimoto T., Fujiki Y. Carboxyl-terminal consensus Ser-Lys-Leu-related tripeptide of peroxisomal proteins functions in vitro as a minimal peroxisome-targeting signal. J Biol Chem. 1992 Jul 15;267(20):14405–14411. [PubMed] [Google Scholar]
- Motley A., Hettema E., Distel B., Tabak H. Differential protein import deficiencies in human peroxisome assembly disorders. J Cell Biol. 1994 May;125(4):755–767. doi: 10.1083/jcb.125.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osumi T., Tsukamoto T., Hata S., Yokota S., Miura S., Fujiki Y., Hijikata M., Miyazawa S., Hashimoto T. Amino-terminal presequence of the precursor of peroxisomal 3-ketoacyl-CoA thiolase is a cleavable signal peptide for peroxisomal targeting. Biochem Biophys Res Commun. 1991 Dec 31;181(3):947–954. doi: 10.1016/0006-291x(91)92028-i. [DOI] [PubMed] [Google Scholar]
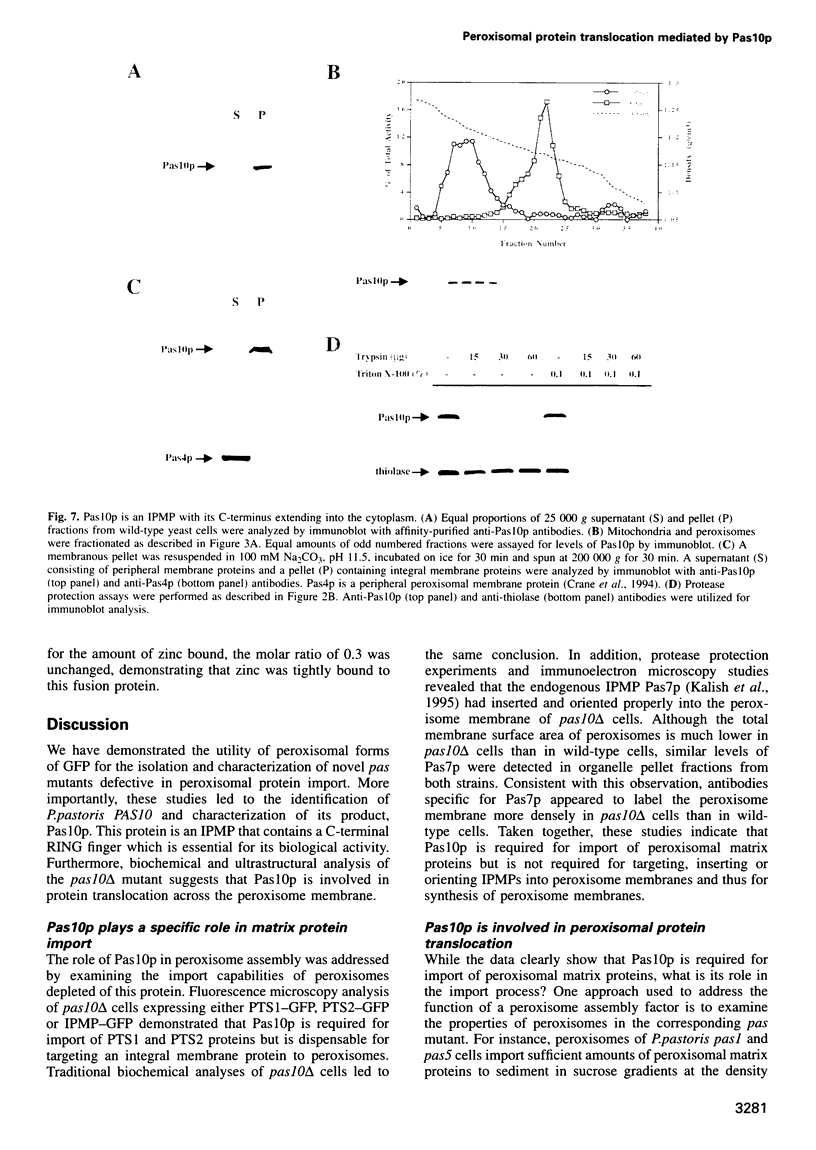
- PENNINGTON R. J. Biochemistry of dystrophic muscle. Mitochondrial succinate-tetrazolium reductase and adenosine triphosphatase. Biochem J. 1961 Sep;80:649–654. doi: 10.1042/bj0800649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters T. J., Müller M., De Duve C. Lysosomes of the arterial wall. I. Isolation and subcellular fractionation of cells from normal rabbit aorta. J Exp Med. 1972 Nov 1;136(5):1117–1139. doi: 10.1084/jem.136.5.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slawecki M. L., Dodt G., Steinberg S., Moser A. B., Moser H. W., Gould S. J. Identification of three distinct peroxisomal protein import defects in patients with peroxisome biogenesis disorders. J Cell Sci. 1995 May;108(Pt 5):1817–1829. doi: 10.1242/jcs.108.5.1817. [DOI] [PubMed] [Google Scholar]
- Spong A. P., Subramani S. Cloning and characterization of PAS5: a gene required for peroxisome biogenesis in the methylotrophic yeast Pichia pastoris. J Cell Biol. 1993 Nov;123(3):535–548. doi: 10.1083/jcb.123.3.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramani S. Protein import into peroxisomes and biogenesis of the organelle. Annu Rev Cell Biol. 1993;9:445–478. doi: 10.1146/annurev.cb.09.110193.002305. [DOI] [PubMed] [Google Scholar]
- Swinkels B. W., Gould S. J., Bodnar A. G., Rachubinski R. A., Subramani S. A novel, cleavable peroxisomal targeting signal at the amino-terminus of the rat 3-ketoacyl-CoA thiolase. EMBO J. 1991 Nov;10(11):3255–3262. doi: 10.1002/j.1460-2075.1991.tb04889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinkels B. W., Gould S. J., Subramani S. Targeting efficiencies of various permutations of the consensus C-terminal tripeptide peroxisomal targeting signal. FEBS Lett. 1992 Jun 29;305(2):133–136. doi: 10.1016/0014-5793(92)80880-p. [DOI] [PubMed] [Google Scholar]
- Tan X., Waterham H. R., Veenhuis M., Cregg J. M. The Hansenula polymorpha PER8 gene encodes a novel peroxisomal integral membrane protein involved in proliferation. J Cell Biol. 1995 Feb;128(3):307–319. doi: 10.1083/jcb.128.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terlecky S. R., Nuttley W. M., McCollum D., Sock E., Subramani S. The Pichia pastoris peroxisomal protein PAS8p is the receptor for the C-terminal tripeptide peroxisomal targeting signal. EMBO J. 1995 Aug 1;14(15):3627–3634. doi: 10.1002/j.1460-2075.1995.tb00032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolbert N. E. Metabolic pathways in peroxisomes and glyoxysomes. Annu Rev Biochem. 1981;50:133–157. doi: 10.1146/annurev.bi.50.070181.001025. [DOI] [PubMed] [Google Scholar]
- Tsukamoto T., Miura S., Fujiki Y. Restoration by a 35K membrane protein of peroxisome assembly in a peroxisome-deficient mammalian cell mutant. Nature. 1991 Mar 7;350(6313):77–81. doi: 10.1038/350077a0. [DOI] [PubMed] [Google Scholar]
- Van der Leij I., Franse M. M., Elgersma Y., Distel B., Tabak H. F. PAS10 is a tetratricopeptide-repeat protein that is essential for the import of most matrix proteins into peroxisomes of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1993 Dec 15;90(24):11782–11786. doi: 10.1073/pnas.90.24.11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorn-Brouwer T., van der Leij I., Hemrika W., Distel B., Tabak H. F. Sequence of the PAS8 gene, the product of which is essential for biogenesis of peroxisomes in Saccharomyces cerevisiae. Biochim Biophys Acta. 1993 Nov 16;1216(2):325–328. doi: 10.1016/0167-4781(93)90166-b. [DOI] [PubMed] [Google Scholar]
- Waterham H. R., Titorenko V. I., Swaving G. J., Harder W., Veenhuis M. Peroxisomes in the methylotrophic yeast Hansenula polymorpha do not necessarily derive from pre-existing organelles. EMBO J. 1993 Dec;12(12):4785–4794. doi: 10.1002/j.1460-2075.1993.tb06167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterham H. R., de Vries Y., Russel K. A., Xie W., Veenhuis M., Cregg J. M. The Pichia pastoris PER6 gene product is a peroxisomal integral membrane protein essential for peroxisome biogenesis and has sequence similarity to the Zellweger syndrome protein PAF-1. Mol Cell Biol. 1996 May;16(5):2527–2536. doi: 10.1128/mcb.16.5.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiebel F. F., Kunau W. H. The Pas2 protein essential for peroxisome biogenesis is related to ubiquitin-conjugating enzymes. Nature. 1992 Sep 3;359(6390):73–76. doi: 10.1038/359073a0. [DOI] [PubMed] [Google Scholar]
- Zhang J. W., Lazarow P. B. PEB1 (PAS7) in Saccharomyces cerevisiae encodes a hydrophilic, intra-peroxisomal protein that is a member of the WD repeat family and is essential for the import of thiolase into peroxisomes. J Cell Biol. 1995 Apr;129(1):65–80. doi: 10.1083/jcb.129.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]