Abstract
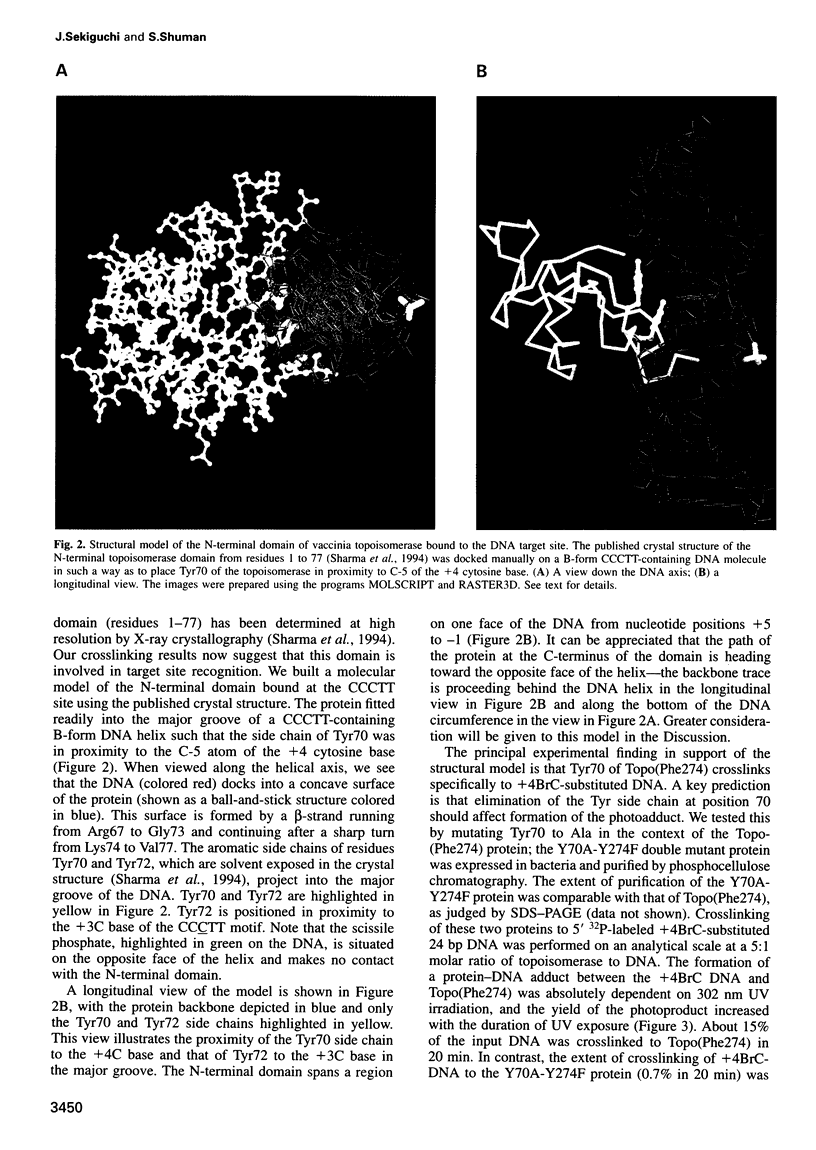
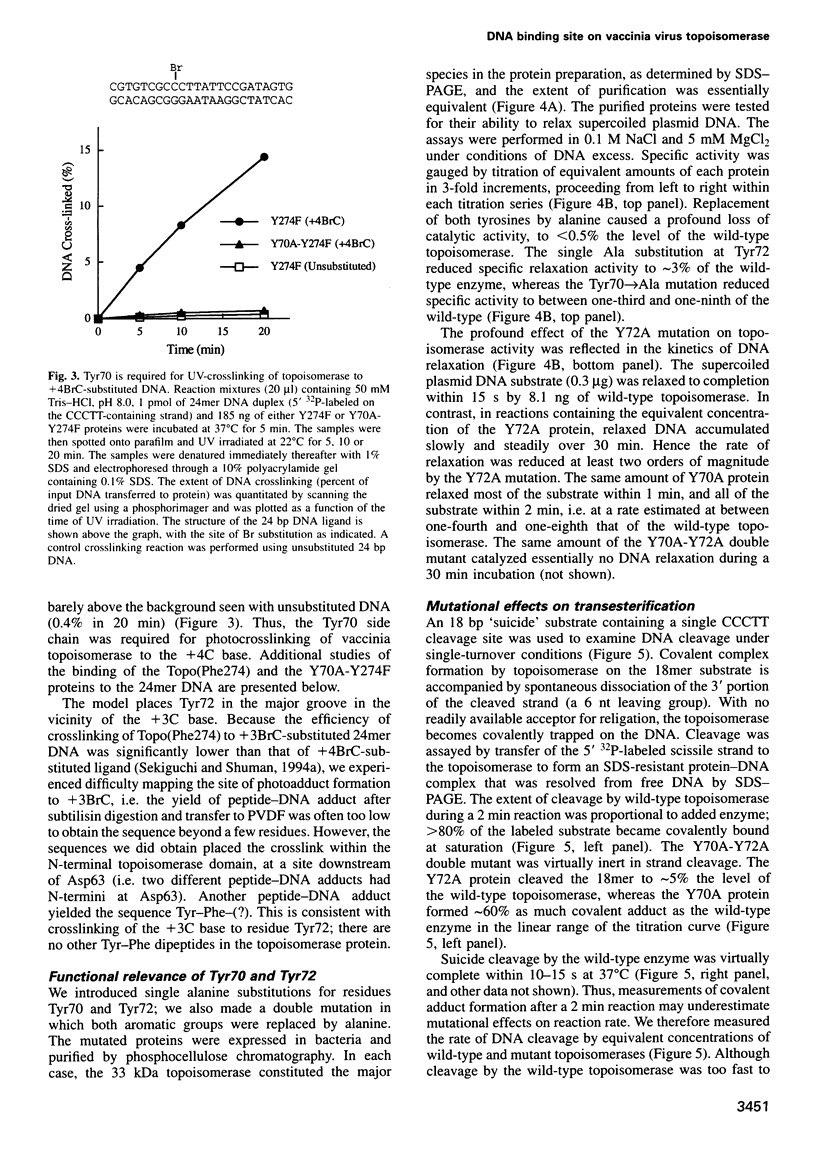
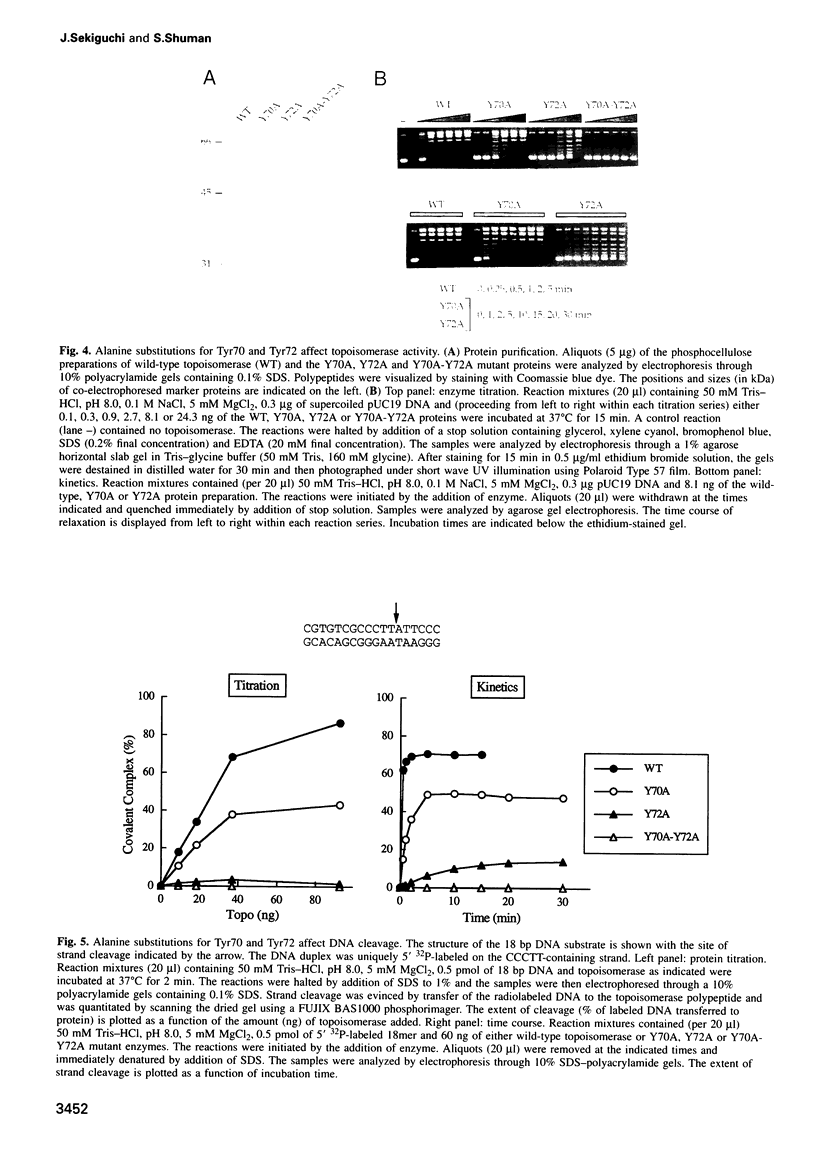
Vaccinia DNA topoisomerase, a eukaryotic type I enzyme, binds and cleaves duplex DNA at sites containing the sequence 5'-(T/C)CCTT. We report the identification of Tyr70 as the site of contact between the enzyme and the +4C base of its target site. This was accomplished by UV-crosslinking topoisomerase to bromocytosine-substituted DNA, followed by isolation and sequencing of peptide-DNA photoadducts. A model for the topoisomerase-DNA interface is proposed, based on the crystal structure of a 9 kDa N-terminal tryptic fragment. The protein domain fits into the DNA major groove such that Tyr70 is positioned close to the +4C base and Tyr72 is situated near the +3C base. Mutational analysis indicates that Tyr70 and Tyr72 contribute to site recognition during covalent catalysis. We propose, based on this and other studies of the vaccinia protein, that DNA backbone recognition and reaction chemistry are performed by a relatively well-conserved 20 kDa C-terminal portion of the vaccinia enzyme, whereas discrimination of the DNA sequence at the cleavage site is accomplished by a separate N-terminal domain, which is less conserved between viral and cellular proteins. Division of function among distinct structural modules may explain the different site specificities of the eukaryotic type I topoisomerases.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Been M. D., Burgess R. R., Champoux J. J. Nucleotide sequence preference at rat liver and wheat germ type 1 DNA topoisomerase breakage sites in duplex SV40 DNA. Nucleic Acids Res. 1984 Apr 11;12(7):3097–3114. doi: 10.1093/nar/12.7.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonven B. J., Gocke E., Westergaard O. A high affinity topoisomerase I binding sequence is clustered at DNAase I hypersensitive sites in Tetrahymena R-chromatin. Cell. 1985 Jun;41(2):541–551. doi: 10.1016/s0092-8674(85)80027-1. [DOI] [PubMed] [Google Scholar]
- Caron P. R., Wang J. C. Appendix. II: Alignment of primary sequences of DNA topoisomerases. Adv Pharmacol. 1994;29B:271–297. doi: 10.1016/s1054-3589(08)61143-6. [DOI] [PubMed] [Google Scholar]
- Champoux J. J. DNA is linked to the rat liver DNA nicking-closing enzyme by a phosphodiester bond to tyrosine. J Biol Chem. 1981 May 25;256(10):4805–4809. [PubMed] [Google Scholar]
- Christiansen K., Svejstrup A. B., Andersen A. H., Westergaard O. Eukaryotic topoisomerase I-mediated cleavage requires bipartite DNA interaction. Cleavage of DNA substrates containing strand interruptions implicates a role for topoisomerase I in illegitimate recombination. J Biol Chem. 1993 May 5;268(13):9690–9701. [PubMed] [Google Scholar]
- Edwards K. A., Halligan B. D., Davis J. L., Nivera N. L., Liu L. F. Recognition sites of eukaryotic DNA topoisomerase I: DNA nucleotide sequencing analysis of topo I cleavage sites on SV40 DNA. Nucleic Acids Res. 1982 Apr 24;10(8):2565–2576. doi: 10.1093/nar/10.8.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng W. K., Pandit S. D., Sternglanz R. Mapping of the active site tyrosine of eukaryotic DNA topoisomerase I. J Biol Chem. 1989 Aug 15;264(23):13373–13376. [PubMed] [Google Scholar]
- Hanai R., Wang J. C. Protein footprinting by the combined use of reversible and irreversible lysine modifications. Proc Natl Acad Sci U S A. 1994 Dec 6;91(25):11904–11908. doi: 10.1073/pnas.91.25.11904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzberg R. P., Caranfa M. J., Hecht S. M. On the mechanism of topoisomerase I inhibition by camptothecin: evidence for binding to an enzyme-DNA complex. Biochemistry. 1989 May 30;28(11):4629–4638. doi: 10.1021/bi00437a018. [DOI] [PubMed] [Google Scholar]
- Jaxel C., Capranico G., Kerrigan D., Kohn K. W., Pommier Y. Effect of local DNA sequence on topoisomerase I cleavage in the presence or absence of camptothecin. J Biol Chem. 1991 Oct 25;266(30):20418–20423. [PubMed] [Google Scholar]
- Lynn R. M., Bjornsti M. A., Caron P. R., Wang J. C. Peptide sequencing and site-directed mutagenesis identify tyrosine-727 as the active site tyrosine of Saccharomyces cerevisiae DNA topoisomerase I. Proc Natl Acad Sci U S A. 1989 May;86(10):3559–3563. doi: 10.1073/pnas.86.10.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morham S. G., Shuman S. Covalent and noncovalent DNA binding by mutants of vaccinia DNA topoisomerase I. J Biol Chem. 1992 Aug 5;267(22):15984–15992. [PubMed] [Google Scholar]
- Sekiguchi J., Shuman S. Proteolytic footprinting of vaccinia topoisomerase bound to DNA. J Biol Chem. 1995 May 12;270(19):11636–11645. doi: 10.1074/jbc.270.19.11636. [DOI] [PubMed] [Google Scholar]
- Sekiguchi J., Shuman S. Requirements for noncovalent binding of vaccinia topoisomerase I to duplex DNA. Nucleic Acids Res. 1994 Dec 11;22(24):5360–5365. doi: 10.1093/nar/22.24.5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi J., Shuman S. Vaccinia topoisomerase binds circumferentially to DNA. J Biol Chem. 1994 Dec 16;269(50):31731–31734. [PubMed] [Google Scholar]
- Sharma A., Hanai R., Mondragón A. Crystal structure of the amino-terminal fragment of vaccinia virus DNA topoisomerase I at 1.6 A resolution. Structure. 1994 Aug 15;2(8):767–777. doi: 10.1016/s0969-2126(94)00077-8. [DOI] [PubMed] [Google Scholar]
- Shuman S., Golder M., Moss B. Characterization of vaccinia virus DNA topoisomerase I expressed in Escherichia coli. J Biol Chem. 1988 Nov 5;263(31):16401–16407. [PubMed] [Google Scholar]
- Shuman S., Kane E. M., Morham S. G. Mapping the active-site tyrosine of vaccinia virus DNA topoisomerase I. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9793–9797. doi: 10.1073/pnas.86.24.9793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman S., Moss B. Identification of a vaccinia virus gene encoding a type I DNA topoisomerase. Proc Natl Acad Sci U S A. 1987 Nov;84(21):7478–7482. doi: 10.1073/pnas.84.21.7478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman S., Prescott J. Specific DNA cleavage and binding by vaccinia virus DNA topoisomerase I. J Biol Chem. 1990 Oct 15;265(29):17826–17836. [PubMed] [Google Scholar]
- Shuman S. Site-specific DNA cleavage by vaccinia virus DNA topoisomerase I. Role of nucleotide sequence and DNA secondary structure. J Biol Chem. 1991 Jan 25;266(3):1796–1803. [PubMed] [Google Scholar]
- Shuman S. Site-specific interaction of vaccinia virus topoisomerase I with duplex DNA. Minimal DNA substrate for strand cleavage in vitro. J Biol Chem. 1991 Jun 15;266(17):11372–11379. [PubMed] [Google Scholar]
- Shuman S., Turner J. Site-specific interaction of vaccinia virus topoisomerase I with base and sugar moieties in duplex DNA. J Biol Chem. 1993 Sep 5;268(25):18943–18950. [PubMed] [Google Scholar]
- Stevnsner T., Mortensen U. H., Westergaard O., Bonven B. J. Interactions between eukaryotic DNA topoisomerase I and a specific binding sequence. J Biol Chem. 1989 Jun 15;264(17):10110–10113. [PubMed] [Google Scholar]
- Stivers J. T., Shuman S., Mildvan A. S. Vaccinia DNA topoisomerase I: kinetic evidence for general acid-base catalysis and a conformational step. Biochemistry. 1994 Dec 27;33(51):15449–15458. doi: 10.1021/bi00255a027. [DOI] [PubMed] [Google Scholar]
- Stivers J. T., Shuman S., Mildvan A. S. Vaccinia DNA topoisomerase I: single-turnover and steady-state kinetic analysis of the DNA strand cleavage and ligation reactions. Biochemistry. 1994 Jan 11;33(1):327–339. doi: 10.1021/bi00167a043. [DOI] [PubMed] [Google Scholar]
- Willis M. C., LeCuyer K. A., Meisenheimer K. M., Uhlenbeck O. C., Koch T. H. An RNA-protein contact determined by 5-bromouridine substitution, photocrosslinking and sequencing. Nucleic Acids Res. 1994 Nov 25;22(23):4947–4952. doi: 10.1093/nar/22.23.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittschieben J., Shuman S. Mutational analysis of vaccinia DNA topoisomerase defines amino acid residues essential for covalent catalysis. J Biol Chem. 1994 Nov 25;269(47):29978–29983. [PubMed] [Google Scholar]