Abstract
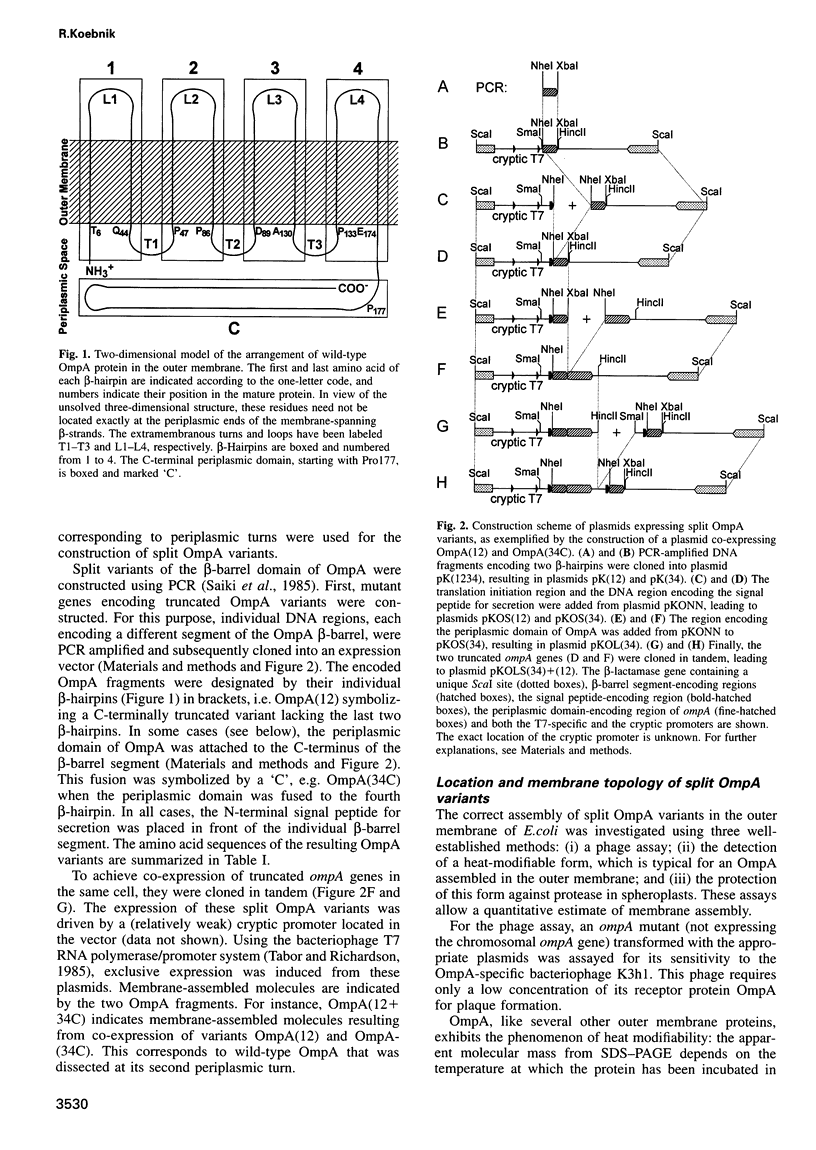
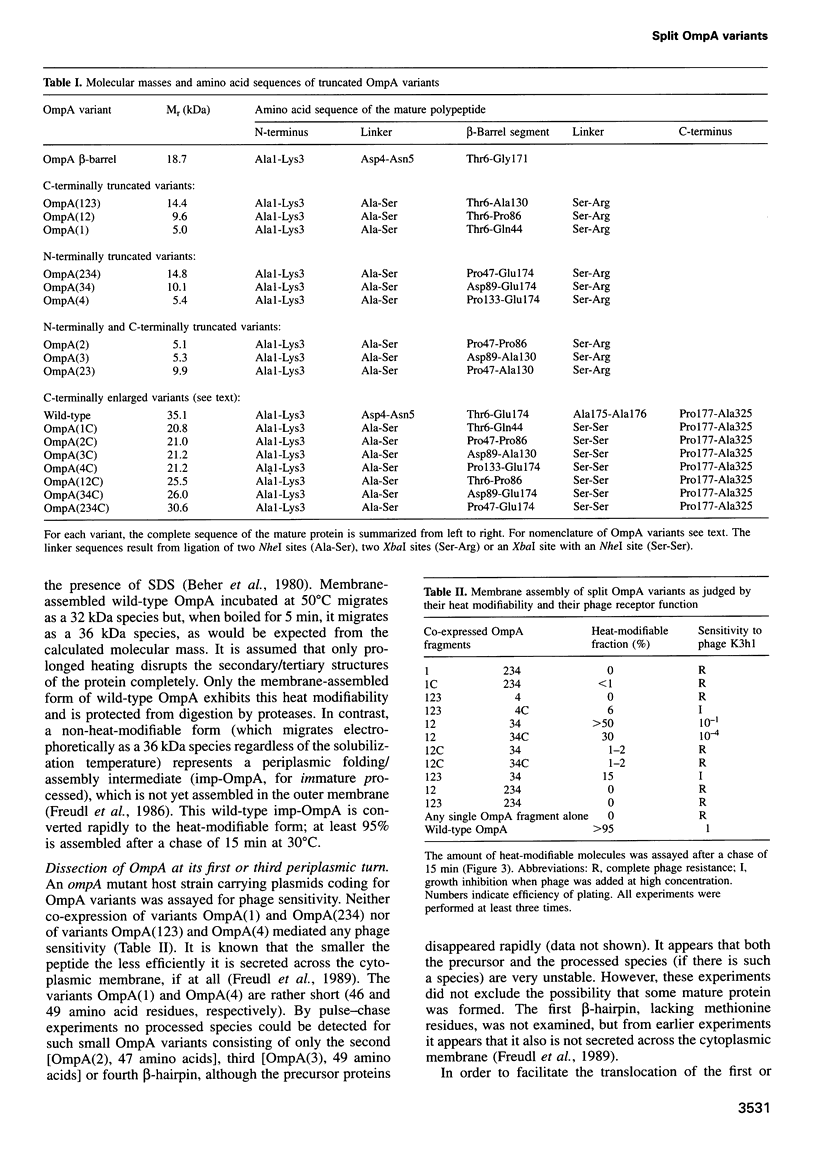
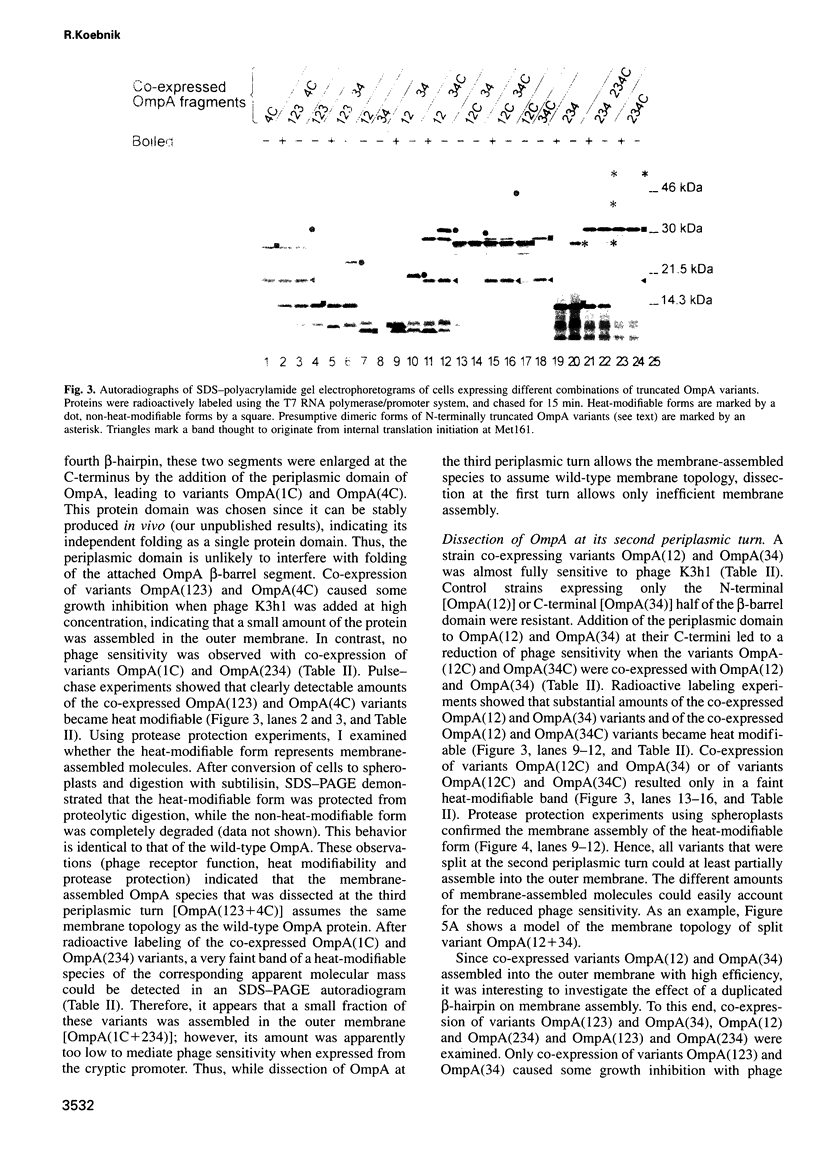
The two-domain, 325 residue outer membrane protein OmpA of Escherichia coli is a well-established model for the study of membrane assembly. The N-terminal domain, consisting of approximately 170 amino acid residues, is embedded in the membrane, presumably in the form of a beta-barrel consisting of eight antiparallel transmembrane beta-strands. A set of 16 gene variants carrying deletions in the membrane-embedded domain of OmpA was constructed. When pairs of these mutant genes were co-expressed in E.coli, it was found that a functional OmpA protein could be assembled efficiently from two complementary protein fragments. Assembly was found when the polypeptide chain was split at the second or third periplasmic turn. All four protein termini were located in the periplasmic space. Interestingly, duplication of transmembrane strands five and six led to a variant with an unusual topology: the N-terminus of one fragment and the C-terminus of the other fragment were exposed at the cell surface. This is the first demonstration of correct membrane assembly of split beta-structured membrane proteins. These findings are important for a better understanding of their folding/assembly pathway and may have implications for the development of artificial outer membrane proteins and for the cell surface display of heterologous peptides or proteins.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beher M. G., Schnaitman C. A., Pugsley A. P. Major heat-modifiable outer membrane protein in gram-negative bacteria: comparison with the ompA protein of Escherichia coli. J Bacteriol. 1980 Aug;143(2):906–913. doi: 10.1128/jb.143.2.906-913.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betton J. M., Hofnung M. In vivo assembly of active maltose binding protein from independently exported protein fragments. EMBO J. 1994 Mar 1;13(5):1226–1234. doi: 10.1002/j.1460-2075.1994.tb06372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch D., Scholten M., Verhagen C., Tommassen J. The role of the carboxy-terminal membrane-spanning fragment in the biogenesis of Escherichia coli K12 outer membrane protein PhoE. Mol Gen Genet. 1989 Mar;216(1):144–148. doi: 10.1007/BF00332243. [DOI] [PubMed] [Google Scholar]
- Brok R. G., Brinkman E., van Boxtel R., Bekkers A. C., Verheij H. M., Tommassen J. Molecular characterization of enterobacterial pldA genes encoding outer membrane phospholipase A. J Bacteriol. 1994 Feb;176(3):861–870. doi: 10.1128/jb.176.3.861-870.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbit A., Ronco J., Michel V., Werts C., Hofnung M. Permissive sites and topology of an outer membrane protein with a reporter epitope. J Bacteriol. 1991 Jan;173(1):262–275. doi: 10.1128/jb.173.1.262-275.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan S. W., Schirmer T., Rummel G., Steiert M., Ghosh R., Pauptit R. A., Jansonius J. N., Rosenbusch J. P. Crystal structures explain functional properties of two E. coli porins. Nature. 1992 Aug 27;358(6389):727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- Freudl R., Schwarz H., Degen M., Henning U. A lower size limit exists for export of fragments of an outer membrane protein (OmpA) of Escherichia coli K-12. J Mol Biol. 1989 Feb 20;205(4):771–775. doi: 10.1016/0022-2836(89)90321-5. [DOI] [PubMed] [Google Scholar]
- Freudl R., Schwarz H., Stierhof Y. D., Gamon K., Hindennach I., Henning U. An outer membrane protein (OmpA) of Escherichia coli K-12 undergoes a conformational change during export. J Biol Chem. 1986 Aug 25;261(24):11355–11361. [PubMed] [Google Scholar]
- Fsihi H., Kottwitz B., Bremer E. Single amino acid substitutions affecting the substrate specificity of the Escherichia coli K-12 nucleoside-specific Tsx channel. J Biol Chem. 1993 Aug 15;268(23):17495–17503. [PubMed] [Google Scholar]
- Kippen A. D., Fersht A. R. Analysis of the mechanism of assembly of cleaved barnase from two peptide fragments and its relevance to the folding pathway of uncleaved barnase. Biochemistry. 1995 Jan 31;34(4):1464–1468. doi: 10.1021/bi00004a042. [DOI] [PubMed] [Google Scholar]
- Klose M., Schwarz H., MacIntyre S., Freudl R., Eschbach M. L., Henning U. Internal deletions in the gene for an Escherichia coli outer membrane protein define an area possibly important for recognition of the outer membrane by this polypeptide. J Biol Chem. 1988 Sep 15;263(26):13291–13296. [PubMed] [Google Scholar]
- Klose M., Störiko A., Stierhof Y. D., Hindennach I., Mutschler B., Henning U. Membrane assembly of the outer membrane protein OmpA of Escherichia coli. J Biol Chem. 1993 Dec 5;268(34):25664–25670. [PubMed] [Google Scholar]
- Koebnik R., Braun V. Insertion derivatives containing segments of up to 16 amino acids identify surface- and periplasm-exposed regions of the FhuA outer membrane receptor of Escherichia coli K-12. J Bacteriol. 1993 Feb;175(3):826–839. doi: 10.1128/jb.175.3.826-839.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koebnik R., Krämer L. Membrane assembly of circularly permuted variants of the E. coli outer membrane protein OmpA. J Mol Biol. 1995 Jul 28;250(5):617–626. doi: 10.1006/jmbi.1995.0403. [DOI] [PubMed] [Google Scholar]
- Kreusch A., Schulz G. E. Refined structure of the porin from Rhodopseudomonas blastica. Comparison with the porin from Rhodobacter capsulatus. J Mol Biol. 1994 Nov 11;243(5):891–905. doi: 10.1006/jmbi.1994.1690. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Liao M. J., Huang K. S., Khorana H. G. Regeneration of native bacteriorhodopsin structure from fragments. J Biol Chem. 1984 Apr 10;259(7):4200–4204. [PubMed] [Google Scholar]
- Morona R., Klose M., Henning U. Escherichia coli K-12 outer membrane protein (OmpA) as a bacteriophage receptor: analysis of mutant genes expressing altered proteins. J Bacteriol. 1984 Aug;159(2):570–578. doi: 10.1128/jb.159.2.570-578.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popot J. L., Gerchman S. E., Engelman D. M. Refolding of bacteriorhodopsin in lipid bilayers. A thermodynamically controlled two-stage process. J Mol Biol. 1987 Dec 20;198(4):655–676. doi: 10.1016/0022-2836(87)90208-7. [DOI] [PubMed] [Google Scholar]
- Ried G., Koebnik R., Hindennach I., Mutschler B., Henning U. Membrane topology and assembly of the outer membrane protein OmpA of Escherichia coli K12. Mol Gen Genet. 1994 Apr;243(2):127–135. doi: 10.1007/BF00280309. [DOI] [PubMed] [Google Scholar]
- Ruiz-Sanz J., de Prat Gay G., Otzen D. E., Fersht A. R. Protein fragments as models for events in protein folding pathways: protein engineering analysis of the association of two complementary fragments of the barley chymotrypsin inhibitor 2 (CI-2). Biochemistry. 1995 Feb 7;34(5):1695–1701. doi: 10.1021/bi00005a026. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Scharf S., Faloona F., Mullis K. B., Horn G. T., Erlich H. A., Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985 Dec 20;230(4732):1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- Steidler L., Remaut E., Fiers W. LamB as a carrier molecule for the functional exposition of IgG-binding domains of the Staphylococcus aureus protein A at the surface of Escherichia coli K12. Mol Gen Genet. 1993 Jan;236(2-3):187–192. doi: 10.1007/BF00277111. [DOI] [PubMed] [Google Scholar]
- Struyvé M., Moons M., Tommassen J. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J Mol Biol. 1991 Mar 5;218(1):141–148. doi: 10.1016/0022-2836(91)90880-f. [DOI] [PubMed] [Google Scholar]
- Surrey T., Jähnig F. Refolding and oriented insertion of a membrane protein into a lipid bilayer. Proc Natl Acad Sci U S A. 1992 Aug 15;89(16):7457–7461. doi: 10.1073/pnas.89.16.7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniuchi H., Parr G. R., Juillerat M. A. Complementation in folding and fragment exchange. Methods Enzymol. 1986;131:185–217. doi: 10.1016/0076-6879(86)31042-5. [DOI] [PubMed] [Google Scholar]
- Vogel H., Jähnig F. Models for the structure of outer-membrane proteins of Escherichia coli derived from raman spectroscopy and prediction methods. J Mol Biol. 1986 Jul 20;190(2):191–199. doi: 10.1016/0022-2836(86)90292-5. [DOI] [PubMed] [Google Scholar]
- Weiss M. S., Abele U., Weckesser J., Welte W., Schiltz E., Schulz G. E. Molecular architecture and electrostatic properties of a bacterial porin. Science. 1991 Dec 13;254(5038):1627–1630. doi: 10.1126/science.1721242. [DOI] [PubMed] [Google Scholar]
- Wetlaufer D. B. Folding of protein fragments. Adv Protein Chem. 1981;34:61–92. doi: 10.1016/s0065-3233(08)60518-5. [DOI] [PubMed] [Google Scholar]
- Zen K. H., McKenna E., Bibi E., Hardy D., Kaback H. R. Expression of lactose permease in contiguous fragments as a probe for membrane-spanning domains. Biochemistry. 1994 Jul 12;33(27):8198–8206. doi: 10.1021/bi00193a005. [DOI] [PubMed] [Google Scholar]