Abstract
As a result of ancestral whole-genome and small-scale duplication events, the genomes of Saccharomyces cerevisiae and many eukaryotes still contain a substantial fraction of duplicated genes. In all investigated organisms, metabolic pathways, and more particularly glycolysis, are specifically enriched for functionally redundant paralogs. In ancestors of the Saccharomyces lineage, the duplication of glycolytic genes is purported to have played an important role leading to S. cerevisiae's current lifestyle favoring fermentative metabolism even in the presence of oxygen and characterized by a high glycolytic capacity. In modern S. cerevisiae strains, the 12 glycolytic reactions leading to the biochemical conversion from glucose to ethanol are encoded by 27 paralogs. In order to experimentally explore the physiological role of this genetic redundancy, a yeast strain with a minimal set of 14 paralogs was constructed (the “minimal glycolysis” [MG] strain). Remarkably, a combination of a quantitative systems approach and semiquantitative analysis in a wide array of growth environments revealed the absence of a phenotypic response to the cumulative deletion of 13 glycolytic paralogs. This observation indicates that duplication of glycolytic genes is not a prerequisite for achieving the high glycolytic fluxes and fermentative capacities that are characteristic of S. cerevisiae and essential for many of its industrial applications and argues against gene dosage effects as a means of fixing minor glycolytic paralogs in the yeast genome. The MG strain was carefully designed and constructed to provide a robust prototrophic platform for quantitative studies and has been made available to the scientific community.
INTRODUCTION
Gene duplication plays a key role in evolution by providing DNA templates for evolutionary innovation, while preventing interference with the cellular function of the original genes (1–3). Immediately after gene duplication, the resulting paralog pairs are usually identical and therefore functionally redundant. Unless duplication confers a selective advantage, either via gene dosage effects or via mutational acquisition of modified or new functions, duplicated genes will eventually be pseudogenized and/or lost from the genome (2, 3).
Gene duplication played a key role in the evolutionary history of the yeast Saccharomyces cerevisiae, an intensively investigated model in eukaryotic evolutionary biology. A whole-genome duplication event (WGD) in an ancestor of S. cerevisiae, ca. 100 million years ago, was followed by loss of ca. 90% of the resulting gene duplications (4, 5). Despite the long time interval that separates current S. cerevisiae strains from the WGD, many surviving paralog pairs still exhibit a substantial degree of functional redundancy, as indicated by neutral or weak phenotypes of single-paralog deletion mutants (6–9). However, the selective pressures that caused the long-term retention of these functionally redundant paralogs remain elusive (1, 10).
The Embden-Meyerhof-Parnas (EMP) pathway, the main route for oxidation of glucose to pyruvate in all eukaryotes and in many other organisms, is among the most slowly evolving metabolic pathways (11, 12). In all taxa, paralog families are found for the structural genes that encode the EMP enzymes (12–14). Under conditions of oxygen limitation and/or sugar excess, S. cerevisiae couples the EMP pathway to the fermentative production of ethanol via pyruvate decarboxylase and NAD+-dependent alcohol dehydrogenase (15, 16). In this article, we use the term “glycolysis” to indicate the set of 12 enzyme-catalyzed reactions in yeast that convert intracellular glucose to ethanol.
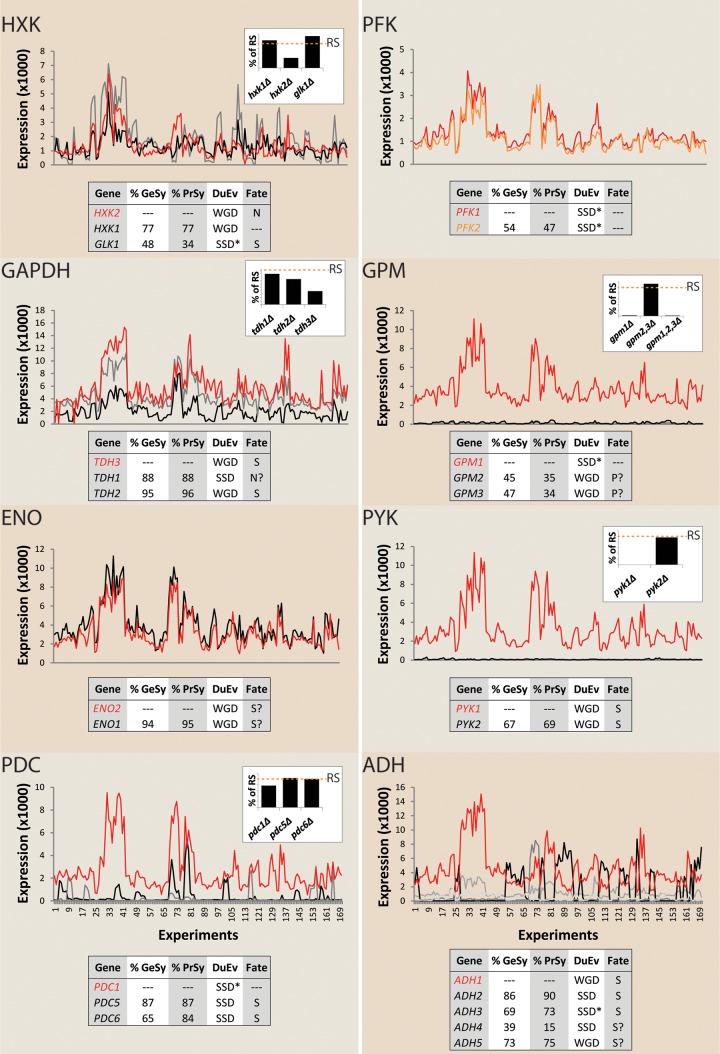
In S. cerevisiae, no fewer than 8 of the 12 enzyme reactions in glycolysis are represented by multiple paralogous genes (Fig. 1). This incidence represents a significant overrepresentation (P = 1.9 × 10−10, based on hypergeometric distribution analysis) relative to the ca. 26% of the yeast genome that consists of parologous combinations (17). The WGD event and resulting duplication of genes involved in central metabolism have been implicated in the appearance of several key physiological characteristics of S. cerevisiae. In particular, the duplication of glycolytic genes has been proposed to have contributed to the strong tendency of S. cerevisiae to produce ethanol under aerobic conditions (Crabtree effect) and its high glycolytic capacity (18–20). However, the impact of reducing the number of glycolytic paralogs on these and other physiological characteristics of S. cerevisiae has not been systematically explored. In all paralogous gene sets in yeast glycolysis, with the notable exception of the phosphofructokinase gene, gene expression and gene deletion studies support the definition of a single major paralog and one to four minor paralogs (Fig. 1). Except for the pseudogenes GPM2 and GPM3, all paralogs have retained their original catalytic function, although their context-dependent expression profiles differ (Fig. 1). Deletion of minor paralogs for individual glycolytic enzymes has minor effects on enzyme activities in cell extracts and on the specific growth rate under standard laboratory conditions (usually shake flask cultivation on yeast extract, peptone, and glucose) (21–25) (Fig. 1). Several hypotheses have been forwarded to explain the neutral effect of paralog deletion (6, 26–31). These include experimental limitations, such as the poor sensitivity of fitness screens (32) and the narrow range of cultivation condition tested (generally complex medium) (33). Additionally, analysis of deletion mutants in which paralogs that encode a single glycolytic enzyme are inactivated cannot reveal synergistic effects of the minor paralogs of different glycolytic enzymes. Such synergistic effects might, for example, arise from the well-documented phenomenon of distribution of metabolic control over multiple enzymes in metabolic pathways (34, 35) or from other regulatory or catalytic interactions.
FIG 1.
Separation of major and minor glycolytic paralogs in Saccharomyces cerevisiae. The following eight enzymes in yeast glycolysis are encoded by parologous genes: hexokinase/glucokinase (HXK), phosphofructokinase (PFK), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), phosphoglycerate mutase (GPM), enolase (ENO), pyruvate kinase (PYK), pyruvate decarboxylase (PDC), and alcohol dehydrogenase (ADH). Percentages of gene and protein similarity (GeSy and PrSy, respectively) between paralogs, the type of duplication event (DuEv), whole-genome duplication (WGD) or other small scale duplication (SSD), and the proposed fate (pseudogenization [P], subfunctionalization [S], or neofunctionalization [N]) of the different paralogs are indicated in the tables in each panel. Separation between WGD and small-scale duplications pre-WGD (SSD*) and post-WGD (SSD) was based on the information in the Yeast Gene Order Browser (YGOB; http://ygob.ucd.ie/) (119). Scatter plots show the comparisons between expression levels of the different glycolytic paralogs in S. cerevisiae measured under 170 different conditions (75) (see Table S6 in the supplemental material). Except for PFK, where both paralogs PFK1 and PFK2 were considered paralogs with equivalent contributions, the expression levels of the major glycolytic paralogs (labeled in red) are indicated by a red line. Bar graphs display in vitro enzyme activities of glycolytic enzymes in mutants carrying individual and multiple glycolytic gene knockouts. Values are presented as percentages of in vitro enzyme activities measured in reference strains (RS [denoted by an orange dotted line]) as reported in the literature (21–25).
Insight into the importance, under laboratory conditions, of the minor glycolytic paralogs is essential for understanding an apparent genetic redundancy in a key ubiquitous metabolic pathway in an important model organism and industrial platform. In addition, analysis of the extent of genetic redundancy in central metabolism is highly relevant for the complete redesign and construction of entirely synthetic yeast genomes, as pursued in the Synthetic Yeast 2.0 initiative (36). Moreover, if complexity in yeast glycolysis can be significantly reduced by elimination of “redundant” isoenzymes, this could eliminate uncertainties and thereby facilitate the formulation and validation of mathematical models that describe the kinetics of this key metabolic pathway (37, 38).
The goals of the present study are to experimentally explore genetic redundancy in yeast glycolysis by cumulative deletion of minor paralogs and to provide a new experimental platform for fundamental yeast research by constructing a yeast strain with a functional “minimal glycolysis” (MG). To this end, we deleted 13 minor paralogs, leaving only the 14 major paralogs for the S. cerevisiae glycolytic pathway. The cumulative impact of deletion of all minor paralogs was investigated by two complementary approaches. A first, quantitative analysis focused on the impact on glycolytic flux under a number of controlled cultivation conditions that in wild-type strains result in different glycolytic fluxes. These quantitative growth studies were combined with transcriptome, enzyme activity, and intracellular metabolite assays to capture potential small phenotypic effects. A second, semiquantitative characterization explored the phenotype of the “minimal glycolysis” (MG) strain under a wide array of experimental conditions to identify potential context-dependent phenotypes.
MATERIALS AND METHODS
Strains and strain construction.
Plasmid propagation and isolation were performed with chemically competent Escherichia coli DH5α (Z-competent transformation kit; Zymo Research, Orange, CA) cultivated in lysogeny broth (LB) medium (39, 40) supplemented with 100 mg liter−1 ampicillin (LBAmp) when required. All yeast strains are derived from the CEN.PK family (41–43) and are listed in Table S3 in the supplemental material. All strains were stored at −80°C in 1-ml aliquots of 30% glycerol and the appropriate medium. CEN.PK102-12A was selected as the parental strain for the minimal glycolysis (MG) strain. The order of gene deletions that led to the final MG strain IMX372 was GLK1, HXK1, TDH1, TDH2, GPM2, GPM3, ENO1, PYK2, PDC5, PDC6, ADH2, ADH5, and ADH4. The genes GLK1, HXK1, TDH1, and TDH2 were deleted using the auxotrophic and dominant markers Sphis5 (44, 45), KlLEU2 (45, 46), KanMX (47, 48), and hphNT1 (49, 50), respectively. These markers remained in the genome during the whole strain construction process. GPM2 to ADH4 deletions were performed using a strategy of selection and counterselection with the KlURA3/5-fluoroorotic acid (5-FOA) system (51) for the recovery of the marker module. KlURA3 was removed seamlessly as previously described (52, 53). The dominant marker modules KanMX and hphNT1 were removed using deletion cassettes containing KlURA3 and amdSYM, respectively. These markers were sequentially removed as reported previously (54). To obtain a prototrophic strain, a cassette containing the marker module KlURA3 was integrated in the TDH1 locus; this generated the MG strain. All genetic modifications were confirmed by PCR and later by whole-genome sequencing.
Molecular biology techniques.
All integrative cassettes were constructed using Phusion Hot Start II high-fidelity polymerase (Thermo Scientific, Landsmeer, The Netherlands) following the manufacturer's recommendations and with the primer pairs and plasmids listed in Tables S4 and S5 in the supplemental material, respectively, as the templates. Correct integrations, deletions, and marker excision were confirmed by PCR using Dreamtaq polymerase (Thermo Scientific) and following the manufacturer's recommendations. The primers used for confirmation are listed in Table S4. Genomic DNA that served as the template for PCR was obtained by extraction with 0.05 N NaOH directly from single colonies or by purification using the YeaStar genomic DNA kit (Zymo Research, Orange, CA) following the manufacturer's recommendations. All PCR products were loaded on gels containing 1% (wt/vol) agarose (Thermo Scientific) and 1× Tris-acetate-EDTA (TAE) buffer (Thermo Scientific). Integrative cassettes were gel purified using the Zymoclean gel DNA recovery kit (Zymo Research). Yeast strain transformations were performed with the lithium acetate protocol as previously described (55). Plasmids were isolated from E. coli with the GenElute plasmid miniprep kit (Sigma-Aldrich, St. Louis, MO).
Media.
Complex and nonselective media for growth rate determination and propagation contained 10 g liter−1 yeast extract, 20 g liter−1 peptone, and 20 g liter−1 glucose (YPD). When selection was required, YPD was supplemented with 200 mg liter−1 G418 (YPD+G418) or 200 mg liter−1 hygromycin (YPD+Hyg). Synthetic medium (SM) containing 3 g liter−1 KH2PO4, 0.5 g liter−1 MgSO4·7H2O, 5 g liter−1 (NH4)2SO4, 1 ml liter−1 of a trace element solution, and 1 ml liter−1 of a vitamin solution as previously described (56). SM was supplemented with 20 g liter−1 glucose (SMG) for propagation, growth rate determination, and batch cultures. When auxotrophic strains were cultivated, SMG was supplemented with 150 mg liter−1 uracil, 125 mg liter−1 histidine, and/or 500 mg liter−1 leucine, according to strain auxotrophy (57). Selection of strains lacking the KlURA3 marker was done in SMG supplemented with 150 mg liter−1 uracil and 1 g liter−1 5-fluoroorotic acid (SMG–5-FOA). Solid media were obtained by addition of 20 g liter−1 agar or agarose. Unless otherwise stated, liquid cultures were performed in 500-ml shake flasks with a working volume of 100 ml and incubated at 30°C and 250 rpm, the targeted starting optical density at 660 nm (OD660), measured using a Libra S11 spectrophotometer (Biocrom, Cambridge, United Kingdom), being 0.5. Unless otherwise stated, cultures on solid media were incubated for 3 days at 30°C.
Semiquantitative phenotypic characterization on plates.
Growth of the MG strain and of the prototrophic control strain CEN.PK113-7D was performed on agar plates containing YPD or SM plus 2% glucose (SMG), SM plus 2% galactose (SMGal), SM plus 2% maltose (SMMal), SM plus 2% sucrose (SMSuc), SM plus 3% (vol/vol) ethanol (SMEtOH), SM plus 6% glycerol (SMGlyc), SM plus 5% glucose (SMHG), SM plus 0.5% glucose (SMLG), SMG plus 2.5, 5, or 10 mM LiCl (SMG-Lit), SMG plus 200 or 500 mM NaCl (SMGSa), SMG plus 50, 100, or 200 μM CdCl2 (SMGCad), SMG plus 1, 1.5, or 2 M sorbitol (SMGSor), SMG at pH 4, 6, or 7.5 (SMGpH), or SMG plus 1 mM H2O2 (SMGOx). Spot plates were inoculated with 101 to 105 cells obtained by serial dilutions of liquid cultures grown to the exponential phase on SMG. All plates were incubated at 30°C for 3 days, except for plates containing SMGlyc, which were incubated for 6 days.
Quantitative characterization of the MG strain's physiology in shake flasks.
Growth rate determination was performed by OD660 measurement of cultures grown in 500-ml shake flasks containing 100 ml of YPD, SMG, or SMEtOH. Cultures for growth rate measurements were inoculated with precultures grown until the late exponential phase under identical conditions (i.e., the same medium and temperature).
Glucose/ethanol/glucose switches were started by inoculation of SMG shake flasks with a preculture grown overnight on the same medium. At the mid-exponential phase (OD660 of ca. 4) samples were taken, washed twice with sterile demineralized water, and used to inoculate shake flasks containing 100 ml of SMEtOH. When reaching an OD660 of 3, a sample from the culture was washed twice with sterile demineralized water and used to inoculate a 500-ml shake flask containing 100 ml SMG. Growth was then monitored until the late exponential phase.
To evaluate tolerance of MG to oxidative stress, cells grown to the exponential phase in shake flasks containing SMG were transferred to a shake flask containing SMG supplemented with 4.5 mM H2O2. Samples were then taken every 20 min for 1 h, diluted, and plated on SMG plates to reach approximately 100 cells per plate. CFU were used to determine the percentage of surviving cells.
Quantitative characterization of the MG strain's physiology in the bioreactor.
Aerobic and anaerobic batch cultures were performed in 2-liter laboratory fermenters (Applikon, Schiedam, The Netherlands) with a 1-liter working volume. SM was used and set at pH 5 before autoclaving at 121°C. Antifoam emulsion C (Sigma) was added to a concentration of 0.2 g liter−1 from a 20% (vol/vol) solution autoclaved separately (121°C). Prior to inoculation, glucose was added to a final concentration of 20 g liter−1 from a sterile (110°C) 50% glucose solution and 1 ml of a vitamin solution (56). Anaerobic cultures were supplemented with 0.01 g liter−1 ergosterol and 0.42 g liter−1 Tween 80 dissolved in ethanol as previously described (56). The pH was maintained at 5 by the automatic addition of 2 M KOH. Compressed air or gaseous nitrogen (quality, 4.5; >99.995% vol N2, <10 volumes per million [vpm] O2 pollution) (Linde Gas, Schiedam, The Netherlands) for aerobic and anaerobic cultures, respectively, was sparged to the bioreactor at a rate of 500 ml min−1 via an Ion Science Saga digital flow meter (Cambridge, United Kingdom). The temperature in the fermenters was kept at 30°C. Complete mixing of the medium was ensured by stirring at 800 rpm. Four independent batch cultures were performed for each condition and each strain. Precultures were started by inoculation of SMG shake flasks with 1 stock vial of the appropriate strain. After ca. 16 h of incubation, these cultures were used to inoculate new SMG shake flasks. The starting OD660 was tightly controlled to reach an OD660 of 1 after 8 h of incubation. These exponentially growing cultures were washed twice with demineralized water (spinning at 5,000 rpm for 4 min) and resuspended in 100 ml demineralized water. This was the cell suspension that was used to inoculate fermenters with a starting OD660 of 0.1.
The OD660 of diluted cell suspensions was measured as described above. Biomass dry weight was determined by filtration as previously described (56). The concentration of extracellular metabolite in culture supernatants was measured by high-performance liquid chromatography (HPLC) analysis using a Aminex HPX-87H ion-exchange column operated at 60°C with 5 mM H2SO4 as the mobile phase at a flow rate of 0.6 ml min−1.
Chromosome separation using CHEF electrophoresis.
Agarose plugs for the different strains were prepared using the contour-clamped homogenous electric field (CHEF) yeast genomic DNA plugs kit (Bio-Rad, Richmond, CA), following the manufacturer's recommendations, and used for CHEF electrophoresis. CHEF electrophoresis was performed as previously described (54).
Vacuole staining.
MG and CEN.PK113-7D vacuoles were stained with the red fluorescent dye FM4-64 (excitation/emission, ∼515/640 nm) (Thermo Fisher Scientific) following the manufacturer's recommendations. Yeast cells and vacuoles were visualized with an Imager-Z1 microscope equipped with an AxioCam MR camera, an EC Plan-Neofluar 100×/1.3 oil Ph3 M27 objective, and the filter set BP 535/25, FT 580, and LP 590 (Carl-Zeiss, Oberkochen, Germany).
Enzyme activities.
Samples equivalent to 62.5 mg of dry weight biomass taken at the mid-exponential phase (ca. 10 h after inoculation) of aerobic batch cultures were used to obtain cell extracts as previously described (58). Measurement of the activity of glycolytic enzymes was carried out as previously described (59), except for phosphofructokinase, the activity of which was determined as described in reference 60. Enzyme activities are expressed as micromoles of substrate per minute per milligram of protein or units per milligram of protein. Protein concentrations in cell extracts were determined according to reference 61 with bovine serum albumin as a standard.
Intracellular metabolite determination.
Samples (1.2 ml each) were taken from aerobic batch cultures using a rapid sampling setup (62) and placed directly into vials containing 5 ml 80% methanol precooled at −40°C. The samples were washed with precooled 80% methanol, and the extraction was performed with boiling ethanol as previously described (63). 13C-labeled cell extract was used as an internal standard for metabolite quantification (64). Intracellular glucose (Gluc), glucose-6-phosphate (Gluc-6-P), fructose-6-phosphate (Fruc-6-P), dihydroxyacetone phosphate (DHAP), glyceraldehyde-3-phosphate (GAP), 3-phosphoglycerate (3PG), 2-phosphoglycerate (2PG), and pyruvate (Pyr) were measured by gas chromatography-mass spectrometry (GC-MS) according to reference 65. Intracellular fructose-1,6-bisphosphate (Fruc-1,6-bP) and phosphoenolpyruvate (PEP) were measured by liquid chromatography-mass spectrometry (LC-MS) according to reference 66. Acetaldehyde determination was performed as previously described (67).
RNA-seq.
Samples for transcriptome sequencing (RNA-seq) were obtained from aerobic batch cultures at the mid-exponential phase of growth on glucose (ca. 10 h after inoculation). Sampling, rapid quenching in liquid nitrogen, and RNA extraction were performed as previously described (68). Sequencing was performed with the Illumina HiSeq 2500 and carried out by BaseClear (Leiden, The Netherlands). A data set of 51-bp single reads of at least 1 Gb was generated. The genome sequence of CEN.PK113-7D (42) was used for all analyses. The data were aligned to the reference using the Burrow-Wheeler alignment tool BWA (69, 70). Gene expression levels were estimated using fragments per kilobase per million (FPKM) values by the Cufflinks software (71). To identify differential gene expression between strains CEN.PK113-7D and IMX372, RNA-seq data comparison was performed and statistically assessed using Cuffdiff (71).
Whole-genome sequencing.
High-quality genomic DNA of the strains CEN.PK102-12A and IMX372 (MG) was obtained using the Qiagen 100/G kit (Qiagen, Hilden, Germany) following the manufacturer's recommendations. Libraries of 300-bp inserts were constructed and paired end sequenced (100-bp reads) using an illlumina HiSeq 2500 sequencer (Baseclear BV, Leiden, The Netherlands). A minimum data quantity of 950 Mb was generated, representing a minimum 80-fold coverage. The sequence reads were mapped onto CEN.PK113-7D genome (42) using the Burrows-Wheeler Alignment tool BWA and further processed using SAMtools (69, 70). Single-nucleotide variations were extracted from the mapping using SAMtools' varFilter. Default settings were used, except that the maximum read depth was set to 400× (−D400). To minimize false-positive mutation calls, custom Perl scripts were used for further mutation filtering as follows: (i) mutation calls containing ambiguous bases in mapping consensus were filtered out, (ii) only the single-nucleotide variations with a quality of at least 20 were kept (with variant quality defined as the Phred-scaled probability that the mutation call is incorrect [72, 73]), (iii) mutations with a depth of coverage below 10× were discarded, and (iv) the mutations found in CEN.PK102-12A were subtracted from the list sequence. Eventually, the single-nucleotide variations were physically positioned and functionally annotated according to the CEN.PK113-7D sequence annotation.
Microarray data accession number.
The RNA-seq data generated in this study have been submitted to the Genome Expression Omnibus database and assigned accession no. GSE63884. The sequence data generated in this study are searchable at NCBI–Entrez (http://www.ncbi.nlm.nih.gov/) under Bioproject PRJNA269221.
RESULTS
From 27 to 14 glycolytic genes: minimal yeast glycolysis.
To enable construction of a “minimal glycolysis” (MG) yeast strain, the first goal was to identify the major paralogs that should be retained in the final strain design. This assessment was based on information from the literature on phenotypes of relevant deletion mutants and on nonglycolytic roles (“moonlighting functions”) (74) of glycolytic isoenzymes. Furthermore, a compendium of S. cerevisiae transcriptome data obtained under a wide range of controlled cultivation conditions (75) was used to compare expression profiles of glycolytic paralogs. Major paralogs were elected based on the following, nonexclusive criteria (Fig. 1; see Fig. S1 in the supplemental material): (i) the highest transcript level over a range of growth conditions, (ii) the most extensive loss of enzyme activity in cell extracts upon deletion, (iii) moonlighting functions with a strong impact on specific growth rate or robustness (HXK2, ENO1, and ENO2) (76, 77), and (iv) the strongest decrease in specific growth rate upon deletion.
Based on these criteria, 11 of the 27 glycolytic genes in S. cerevisiae were assessed to be major paralogs: HXK2, PGI, FBA1, TPI1, TDH3, GPM1, ENO2, PGI1, PYK1 (CDC19), PDC1, and ADH1, while 13 minor paralogs (i.e., HXK1, GLK1, TDH1, TDH2, GPM2, GPM3, ENO1, PYK2, PDC5, PDC6, ADH2, ADH4, and ADH5) were selected for gene deletion. In S. cerevisiae, the PKF1 and PFK2 paralogs have evolved into subunits of a hetero-octameric phosphofructokinase (78). Since deletion of either gene substantially decreases fitness (79–81) (see Fig. S1 in the supplemental material), both were retained in the minimal glycolysis design. ADH3 encodes a mitochondrial alcohol dehydrogenase involved in an anaerobic redox shuttle across the mitochondrial inner membrane (82). To prevent reduced growth rates under anaerobic conditions, ADH3 was therefore also retained in the minimal glycolysis design.
Sequential deletion of the 13 minor paralogs in a haploid S. cerevisiae strain belonging to the CEN.PK family (41–43) yielded the prototrophic strain IMX372, which we will refer to as the “MG” (minimal glycolysis) strain. Resequencing of the genome of the MG strain confirmed the correct deletion of all 13 minor glycolytic genes. Although transformation of S. cerevisiae can be mutagenic (83, 84), only 20 single-nucleotide differences were detected in the MG strain relative to its ancestor strain, CEN.PK102-12A. Eleven of these differences occurred within open reading frames (ORFs), of which 10 resulted in amino acid changes (see Table S1 in the supplemental material). None of these mutations affected glycolytic genes or genes related to central carbon metabolism. Whole-genome sequencing and karyotyping indicated the duplication of two short sections of chromosome III (16.5 kbp, from YCR019W to YCR027C) and chromosome V (19.1 kbp, from YER093C-A to YER104W) (see Fig. S2 and Table S1 in the supplemental material). These regions do not carry genes involved in central carbon metabolism, and no interchromosomal rearrangements were observed.
Elimination of all minor glycolytic paralogs has minimal impacts on growth kinetics, intracellular metabolite concentrations, and gene expression.
To explore the physiological impact of the simultaneous deletion of all 13 minor glycolytic paralogs, we quantitatively compared specific growth rates, product formation, and gene expression in the MG strain and in a congenic, prototrophic reference strain with a full complement of glycolytic genes. In these studies, a synthetic, chemically defined medium was used, since complex media do not enable cells to express their full genetic potential (33) and complicate quantitative physiological analysis.
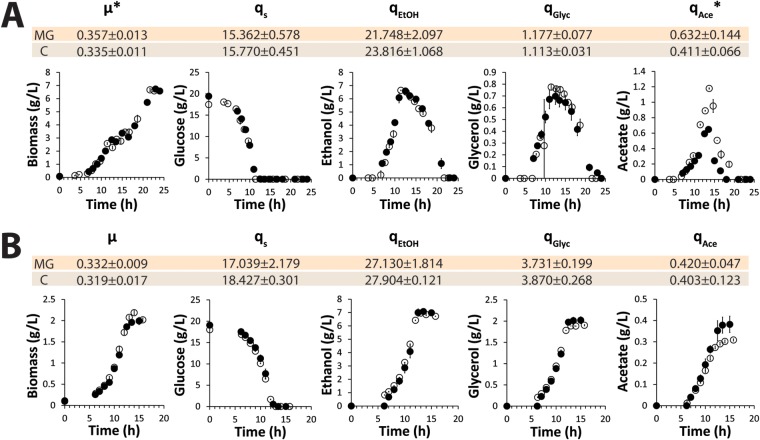
In aerobic, glucose-grown bioreactor batch cultures, specific rates of growth, substrate consumption, and product formation were not significantly affected by deletion of all 13 minor glycolytic paralogs (Fig. 2A). The only exception to this observation concerned acetate production, which was slightly higher in the MG strain. Also after the diauxic shift, where the glycolytic flux changes direction as the aerobic yeast cultures consumed the ethanol and acetate produced during the initial growth phase on glucose, the biomass formation and substrate consumption rates of the two strains were virtually identical. In anaerobic yeast cultures, the absence of oxidative phosphorylation makes glycolysis the only pathway for energy conservation. Also under these more demanding conditions, the specific growth rate and metabolic fluxes of the MG strain did not differ significantly from those of the congenic reference strain (Fig. 2B).
FIG 2.
Biomass production and extracellular metabolite profiles from aerobic and anaerobic batch cultures in bioreactors of the minimal glycolysis (MG) strain and a congenic reference strain (indicated by “C”). Shown are biomass and extracellular metabolite profiles from aerobic (A) and anaerobic (B) controlled batch cultures of the minimal glycolysis strain (MG) (open circles) and the prototrophic reference strain CEN.PK113-7D (closed circles). The data shown in the graphs are the average and average deviation of the mean from two independent cultures for each strain. The specific growth rate (μ [per hour]) and biomass-specific rates of glucose consumption (qs), ethanol production (qEtOH), glycerol production (qGlyc), and acetate production (qAce) (all expressed in millimoles per gram dry weight per hour) represent the average and standard deviation of data from four independent cultures for each strain. *, statistically significant (by two-tailed t test assuming the same variance in the populations) differences between the two tested strains (P = 0.03 and P = 0.02 for μ and qAce, respectively, in aerobic cultures).
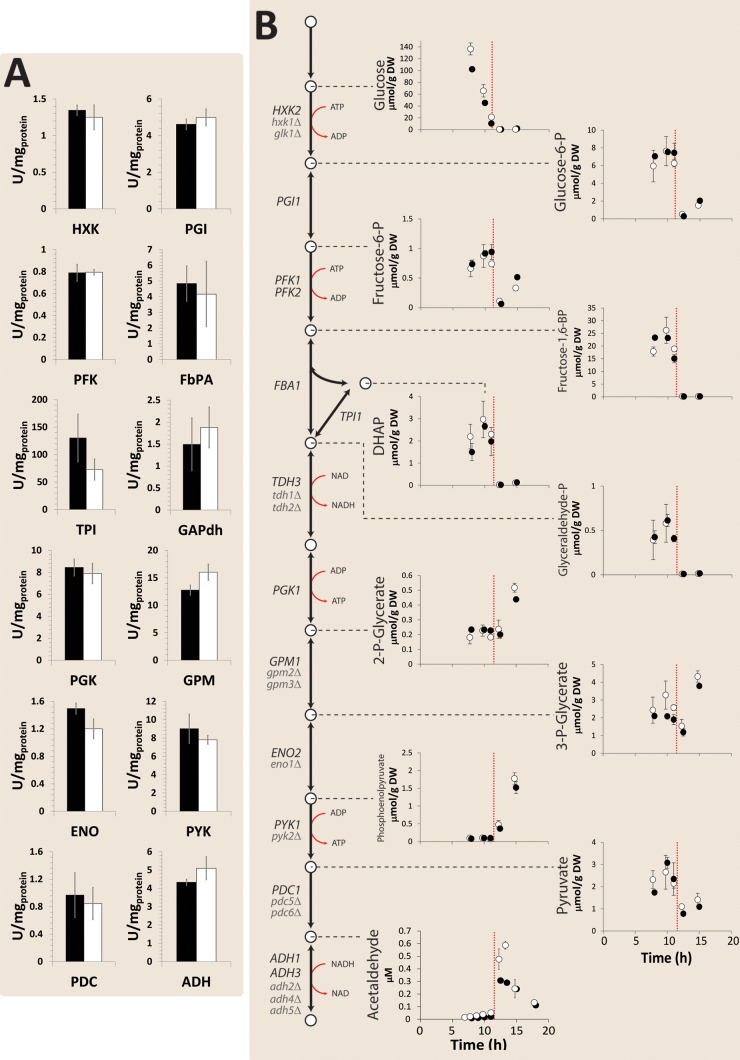
To further compare growth of the MG strain in aerobic bioreactor batch cultures, its glycolytic enzyme activities in cell extracts, intracellular metabolite concentrations, and transcriptome were compared with those of a congenic reference strain. Remarkably, no significant differences were observed in the transcript levels of any of the major glycolytic paralogs or in vitro glycolytic enzyme activities (Fig. 3). With the exception of slightly higher intracellular concentrations of acetaldehyde, concentrations of glycolytic intermediates in the MG strain did not significantly differ from those in the reference strain. To explore potential impacts of the deletion of 13 minor glycolytic paralogs outside glycolysis, genome-wide transcript levels of the MG strain and the reference strain were compared. As few as 17 genes showed a significantly different transcript level (see Table S2 in the supplemental material), 12 of which were located on the duplicated regions on chromosomes III and V. An in-depth, comprehensive analysis of the MG strain in glucose-grown bioreactor batch cultures therefore failed to identify substantial impacts of the minor glycolytic paralogs on fluxes, intracellular metabolite concentrations, or gene expression.
FIG 3.
In vitro enzyme activities and intracellular metabolite concentrations in aerobic batch cultures in bioreactors of the minimal glycolysis (MG) strain and a congenic reference strain. Thirteen minor glycolytic paralogs were deleted in the MG strain. (A) Average values of four independent culture replicates of the in vitro activities for the glycolytic enzymes in the MG strain (white bars) and the prototrophic reference strain CEN.PK113-7D (black bars). HXK, hexokinase/glucokinase; PGI, phosphoglucose isomerase; PFK, phosphofructokinase; FbPA, fructose-bisphosphate aldolase; TPI, triose-phosphate isomerase; GAPdh, glyceraldehyde-3-phosphate dehydrogenase; PGK, phosphoglycerate kinase; GPM, phosphoglycerate mutase; ENO, enolase; PYK, pyruvate kinase; PDC, pyruvate decarboxylase; ADH, alcohol dehydrogenase. The denoted error bars represent standard deviations. (B) Intracellular glycolytic metabolite profiles of the MG strain (open circles) and of CEN.PK113-7D (close circles) from aerobic batch cultures. Average values from two independent culture replicates are shown, and the average deviations of the mean are indicated by error bars. The vertical orange dotted line indicates the time at which glucose was depleted.
Minimal impact of minor glycolytic paralogs under a wide range of conditions.
If during evolution of S. cerevisiae, its glycolytic paralogs have evolved different roles through sub- or neofunctionalization, their deletion may only cause an observable phenotype under specific growth conditions. The growth rates of the MG and reference strains were therefore compared under a wide range of selected growth conditions. During fast growth in shake flasks on complex (yeast extract-peptone-glucose [YPD]) medium (Fig. 4) and during growth at low temperature (12°C) (Fig. 4), the glycolytic pathway operates at rates closer to its maximum capacity than during growth at 30°C on synthetic medium (60, 85). However, no difference in specific growth rates between the MG and reference strains was observed under these conditions. The MG strain also showed the same growth rate as the reference strain at high temperature (37°C) (Fig. 4).
FIG 4.
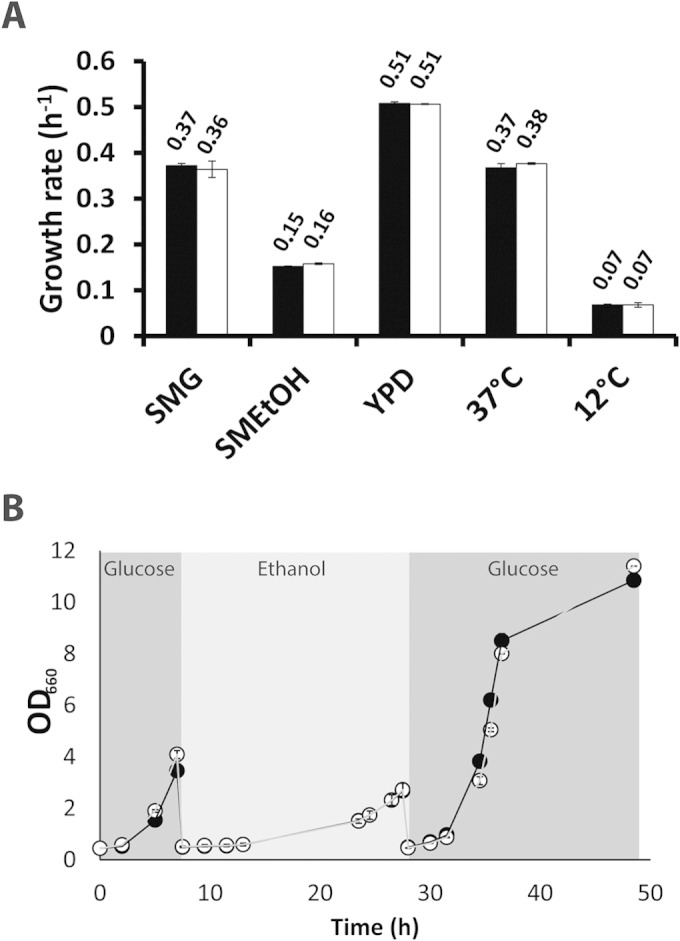
Growth of the minimal glycolysis (MG) strain and a congenic reference strain under different growth conditions in shake flasks. (A) Specific growth rates of S. cerevisiae strains MG (white bars) and CEN.PK113-7D (reference strain, black bars). SMG, synthetic medium with glucose; SMEtOH, synthetic medium with ethanol as the carbon source; YPD, complex medium with glucose as the carbon source. The labels “37°C” and “12°C” indicate growth on SMG at 37° and 12°C, respectively. Average specific growth rates (per hour) are denoted above each bar. (B) Growth, measured as change in the culture's optical density at 660 nm (OD660), of the MG strain (open circles) and reference strain (closed circles) during carbon source switches. Strains were successively grown in glucose, ethanol, and glucose. All data represent the average and average deviation of the mean from two independent culture replicates.
Absence of the minor glycolytic paralogs did not affect growth of the MG strain on ethanol (Fig. 2A and 4A). These results supported the conclusion from the aerobic bioreactor batch cultures that efficient gluconeogenesis does not require any of the minor paralogs (as has previously been proposed for Adh2 [86]). While minor glycolytic paralogs apparently do not contribute to maximum specific growth rates on individual carbon sources, they might be involved in substrate transitions. For instance, Pyk2, the glucose-repressed and fructose-1,6-bisphosphate-insensitive paralog of Pyk1 (21), has been proposed to prevent futile cycling resulting from simultaneous operation of glycolysis and gluconeogenesis during transitions between fermentable and nonfermentable carbon sources (87, 88). However, no significant impact of the deletion of minor glycolytic paralogs was observed during transitions from glucose to ethanol and back to glucose (Fig. 4B).
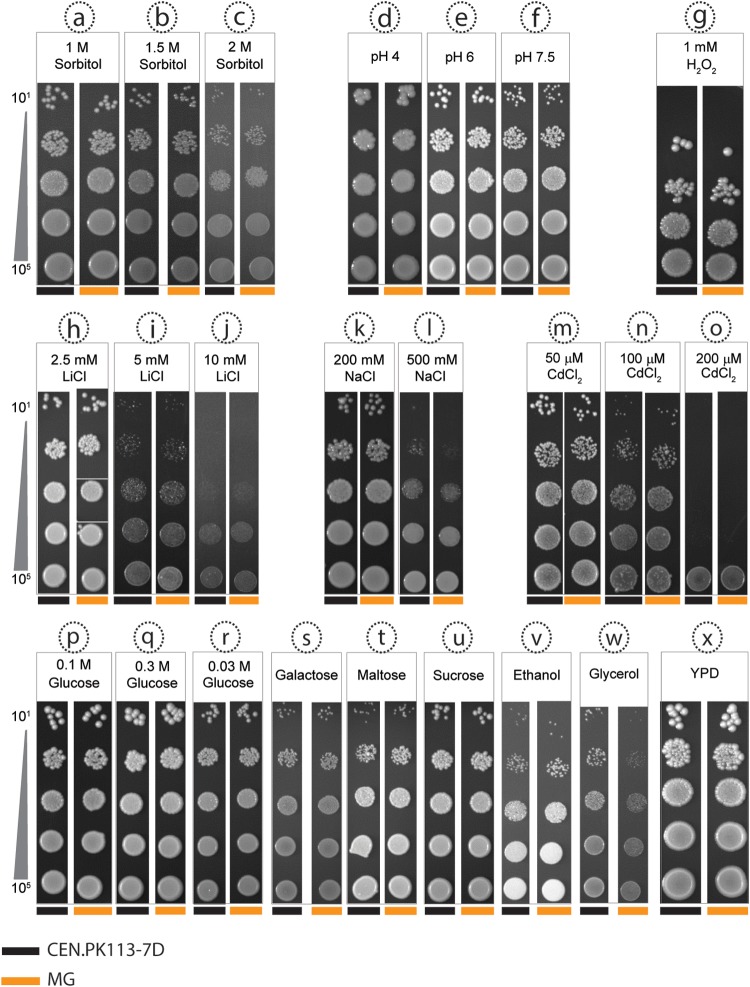
To investigate possible phenotypes of the MG strain under a wider range of environmental conditions, growth on solid medium was investigated under 24 experimental conditions. Some of these were chosen without a specific focus on individual glycolytic paralogs. For example, growth was analyzed at high osmotic pressure (Fig. 5a to c), on different carbon sources (Fig. 5p to w), and at high concentrations of lithium and sodium ions, to which strains from the CEN.PK lineage are hypersensitive (89) (Fig. 5h to l). Only growth with glycerol as a carbon source indicated a slightly reduced growth rate of the MG strain. However, this phenotype was not observed during growth on liquid medium with glycerol (see Fig. S3 in the supplemental material) and may therefore have been caused by a contaminating substrate in the agar used for the plate experiments.
FIG 5.
Growth of the minimal glycolysis (MG) strain and a congenic reference strain on different solid media. Serial dilutions of cell suspensions of the MG strain and of the reference strain CEN.PK113-7D were plated on agar media with 24 different compositions. The conditions included osmotic stress (a to c), different pHs (d to f), oxidative stress (g), three different salts (h to o), six different carbon sources (p to w), and complex medium (x). With the exception of panel x (YPD), all plates contained synthetic medium (SM). All plates contained 2% glucose, with the exception of plates with different carbon sources, which contained galactose (2%), maltose (2%), sucrose (2%), ethanol (3%, vol/vol), or glycerol (6%) and plates with low (0.03 M [5%]) and high (0.3 M [50%]) concentrations of glucose. Colonies of reference and MG strains are denoted by black bands and orange bands, respectively, at the bottom of each panel.
Additional growth conditions tested on solid media were chosen based on information from the literature (74, 90) on sub- or neofunctionalization of specific deleted glycolytic paralogs. In addition to their enolase activity, Eno1 and Eno2 have both been implicated in vacuolar assembly, but in the absence of Eno1, Eno2 is able to support both functions (76). In the vacuole and the ATPase associated with it, disruption causes a deleterious phenotype in S. cerevisiae when cultivated under alkaline conditions (91). As expected, the MG strain grew normally at pH 7.5, thereby indicating the absence of major vacuolar malfunction (Fig. 5f; see Fig. S4 in the supplemental material). Tdh3 and Tdh2, the two minor isoenzymes of glyceraldehyde-3-phosphate dehydrogenase, have been proposed to protect S. cerevisiae against oxidative stress because of their different levels of thiolation (92). However, exposure to oxidative stress by growth on hydrogen peroxide-containing plates (Fig. 5g) showed that the absence of Tdh2 did not affect the oxidative stress resistance of the MG strain. Sub- and neofunctionalization have resulted in a different glucose-dependent regulation of GLK1/HXK1 and PYK2 and in different regulatory properties of the proteins that they encode from those encoded by HXK2 and PYK1, respectively (21, 93). However, growth of the MG strain at low, intermediate, and high glucose concentrations was not impaired relative to that of the reference strain (Fig. 5p to r). Finally, PDC6, which encodes a pyruvate decarboxylase isoenzyme with a low cysteine and methionine content, is specifically induced under sulfur limitation (94). Cultivation in the presence of cadmium increases abundance of Pdc6 (94–96); since cells require a high level of glutathione production for detoxification, sulfur amino acids are then used for this process and are less available for protein synthesis. Despite the absence of PDC6, the MG strain grew as well as the reference strain in the presence of cadmium.
DISCUSSION
Absence of phenotypes upon deletion of 13 glycolytic genes.
Deletion of 13 of the 27 glycolytic paralogs in S. cerevisiae yielded a “minimal glycolysis” (MG) yeast strain whose most spectacular characteristic was the absence of any pronounced phenotype under a wide range of laboratory growth conditions. One of the hypotheses that has been proposed to explain retention of functionally redundant paralogs during evolution is a contribution to gene dosage and, thereby, to the capacity of the pathway or process in which they operate (97, 98). The high glycolytic rates in anaerobic cultures of the MG strain (18 mmol glucose per g of dry biomass per h) did not significantly differ from those of wild-type strains (99) (Fig. 2). This result argues against gene dosage effects as a means for fixing minor glycolytic paralogs in the yeast genome. Instead, our observations indicate that duplication of glycolytic genes is not a prerequisite for achieving the high glycolytic fluxes and fermentative capacities that are characteristic of S. cerevisiae and essential for many of its industrial applications (100, 101). It might be argued that the gene dosage hypothesis does apply to phosphofructokinase, as deletion of either PFK1 or PFK2 substantially reduces enzyme activity and fitness (79–81) (see Fig. S1 in the supplemental material). However, the presence of Pfk1 and Pfk2 in a hetero-octameric complex can also be seen as a case of neofunctionalization, in which two paralogs have been fixed by an acquired mutual dependency. The near-wild-type growth kinetics of the MG strain in aerobic and anaerobic cultures is difficult to reconcile with the hypothesis that duplication of glycolytic genes during the WGD event played a major role in increasing its glycolytic capacity or in causing the phenomenon of aerobic fermentation (the Crabtree effect) (20).
Our inability to identify a phenotype after deletion of all minor paralogs of glycolytic genes in S. cerevisiae does not imply that such a phenotype does not exist. The range of conditions tested represents an infinitesimal fraction of the environmental conditions to which S. cerevisiae may have been exposed to in its evolutionary history. The absence of a clear phenotype under standard laboratory conditions makes the MG strain an even more interesting platform for future high-throughput studies to investigate its phenotype under more conditions. Other characteristic features of S. cerevisiae like tolerance to high ethanol concentrations or growth at high gravity are interesting conditions to test, as well as the robustness of the MG strain under nonstandard conditions, such as sporulation, starvation, severe calorie restriction, and dynamic nutrient supply regimens. However, guessing the environmental factors that have conferred an evolutionary advantage to strains carrying gene duplications presents a formidable challenge, more particularly because the real niche for S. cerevisiae is still a matter of debate, to the extent that S. cerevisiae may be considered a generalist that does not necessarily favor a specific type of environment (102). For further functional analysis studies of the minor glycolytic paralogs, the set of intermediate strains used for construction of the MG strains (see Table S3 in the supplemental material) can be used to rapidly identify which minor paralog or paralogs contribute to any newly identified phenotypes. Moreover, the sensitivity of fitness analyses can be improved—for example, by competitive cultivation of wild-type and MG strains in mixed cultures.
In S. cerevisiae, overrepresentation of paralogous gene sets is not unique for glycolysis (23 of 27 glycolytic genes are paralogs, corresponding to 85%) but also occurs for metabolic genes in general. Of all metabolic genes, 44% are paralogs (98), while this percentage is only 5% for the entire yeast genome (5, 17). Currently, a large research effort is under way to enable design and assembly of entirely synthetic yeast genomes (36). For the design of compact, functional synthetic genomes, it will be highly interesting to investigate whether our results on yeast glycolysis can be extrapolated to minor paralogs of genes encoding key enzymes in other central metabolic pathways. At present, predictions of the outcome of such experiments can only be highly speculative. The clear impact of “minor paralogs” of transaldolase and transketolase on pentose fermentation kinetics by engineered S. cerevisiae (103) is a clear example that, at least under some conditions, minor paralogs can make a clear contribution to metabolic flux.
The deletion of all minor glycolytic paralogs had a surprisingly small effect on the yeast transcriptome in aerobic, glucose-grown batch cultures. This result is in marked contrast with several studies in which deletion of individual major glycolytic paralogs led to transcriptional upregulation of its minor paralogs. An example of such a “compensatory” upregulation of minor paralogs is the upregulation of PDC5 upon deletion of PDC1 (104). Such an asymmetric cross-regulation is consistent with the backup theory for fixation of duplicated genes (17), which postulates that (minor) paralogs provide a buffer against deleterious mutations. Although upregulation of minor paralogs and even repair of a null mutation in a major glycolytic paralog by recombination with a minor paralog have been demonstrated (pdc1/PDC5) (104), the general evolutionary significance of the backup theory is still a matter of debate (98, 105). The selective advantages responsible for the overrepresentation of paralogous genes among the structural genes of yeast glycolysis therefore remain an intriguing conundrum.
Minimal glycolysis yeast: a versatile new platform for quantitative research.
As the MG strain was constructed before the advent of CRISPR-Cas9 technology (106), its construction involved 16 consecutive rounds of transformation and 11 marker recycling steps. A previous study (107) in which all 20 hexose transporter (HXT) genes in S. cerevisiae were deleted by repeated rounds of transformation and marker recycling resulted in massive genomic rearrangements, which were recently explained from repeated use of the LoxP system for marker recovery (120). Repeated use of the LoxP system enables recombination across LoxP sites that are left in the genome after marker removal (108). The “HXT-null” strain is mainly used as an excellent platform for functional analysis of individual transporter strains (107, 109–117), in which role its chromosomal rearrangements are hardly relevant. While the MG strain similarly offers an interesting platform for functional analysis of (heterologous) glycolytic genes by one-step gene replacement, we designed and constructed it with the specific aim of building a robust platform for quantitative studies in systems biology. By using URA3 and amdSYM (53) as counterselectable, recyclable marker genes, major chromosomal rearrangements could be avoided. Furthermore, interference of auxotrophies in the interpretation of quantitative growth studies (57) was avoided by making the MG strain prototrophic.
The availability of a well-defined yeast platform with a minimal complement of glycolytic enzymes should provide clear advantages for quantitative modeling of the kinetics and regulation of glycolysis, as it eliminates the intrinsic uncertainties caused by the simultaneous, context-dependent expression of different isoenzymes. In view of the important role of glucose transport in the kinetics of yeast glycolysis, we are currently constructing MG variants with a single hexose transporter. In addition to providing a relevant test bed for mathematical modeling of glycolysis, the MG strain provides an interesting, simplified starting point for laboratory studies on yeast glycolysis. Previous studies have indicated that long-term laboratory evolution can have a major impact on glycolytic genes and their expression (59, 118). Comparison of evolution of the MG strain with that of strains that carry a full complement of glycolytic genes provides an interesting starting point for studies on the impact of gene duplication on evolutionary flexibility of a ubiquitous central metabolic pathway.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Technology Foundation STW (Vidi grant 10776).
We thank Walter van Gulik for expert support for metabolome analysis and Lizanne Bosman for experimental contribution to the construction of the MG strain.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00064-15.
REFERENCES
- 1.Innan H, Kondrashov F. 2010. The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet 11:97–108. doi: 10.1038/nrg2689. [DOI] [PubMed] [Google Scholar]
- 2.Ohno S. 1970. Evolution by gene duplication. Springer-Verlag, New York, NY. [Google Scholar]
- 3.Zhang JZ. 2003. Evolution by gene duplication: an update. Trends Ecol Evol 18:292–298. doi: 10.1016/S0169-5347(03)00033-8. [DOI] [Google Scholar]
- 4.Wolfe KH, Shields DC. 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 5.Kellis M, Birren BW, Lander ES. 2004. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- 6.Dean EJ, Davis JC, Davis RW, Petrov DA. 2008. Pervasive and persistent redundancy among duplicated genes in yeast. PLoS Genet 4:e1000113. doi: 10.1371/journal.pgen.1000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeLuna A, Vetsigian K, Shoresh N, Hegreness M, Colon-Gonzalez M, Chao S, Kishony R. 2008. Exposing the fitness contribution of duplicated genes. Nat Genet 40:676–681. doi: 10.1038/ng.123. [DOI] [PubMed] [Google Scholar]
- 8.Gu X, Zhang ZQ, Huang W. 2005. Rapid evolution of expression and regulatory divergences after yeast gene duplication. Proc Natl Acad Sci U S A 102:707–712. doi: 10.1073/pnas.0409186102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Musso G, Costanzo M, Huangfu MQ, Smith AM, Paw J, Luis BJS, Boone C, Giaever G, Nislow C, Emili A, Zhang ZL. 2008. The extensive and condition-dependent nature of epistasis among whole-genome duplicates in yeast. Genome Res 18:1092–1099. doi: 10.1101/gr.076174.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conant GC, Wolfe KH. 2008. Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet 9:938–950. doi: 10.1038/nrg2482. [DOI] [PubMed] [Google Scholar]
- 11.Romano AH, and Conway T. 1996. Evolution of carbohydrate metabolic pathways. Res Microbiol 147:448–455. doi: 10.1016/0923-2508(96)83998-2. [DOI] [PubMed] [Google Scholar]
- 12.Fothergill-Gilmore LA, Michels PAM. 1993. Evolution of glycolysis. Prog Biophys Mol Biol 59:105–235. doi: 10.1016/0079-6107(93)90001-Z. [DOI] [PubMed] [Google Scholar]
- 13.Dandekar T, Schuster S, Snel B, Huynen M, Bork P. 1999. Pathway alignment: application to the comparative analysis of glycolytic enzymes. Biochem J 343:115–124. doi: 10.1042/0264-6021:3430115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steinke D, Hoegg S, Brinkmann H, Meyer A. 2006. Three rounds (1R/2R/3R) of genome duplications and the evolution of the glycolytic pathway in vertebrates. BMC Biol 4:16. doi: 10.1186/1741-7007-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dashko S, Zhou N, Compagno C, Piskur J. 2014. Why, when, and how did yeast evolve alcoholic fermentation? FEMS Yeast Res 14:826–832. doi: 10.1111/1567-1364.12161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Dijken JP, Weusthuis RA, Pronk JT. 1993. Kinetics of growth and sugar consumption in yeasts. Antonie Van Leeuwenhoek 63:343–352. doi: 10.1007/BF00871229. [DOI] [PubMed] [Google Scholar]
- 17.Gu ZL, Steinmetz LM, Gu X, Scharfe C, Davis RW, Li WH. 2003. Role of duplicate genes in genetic robustness against null mutations. Nature 421:63–66. doi: 10.1038/nature01198. [DOI] [PubMed] [Google Scholar]
- 18.Gordon JL, Byrne KP, Wolfe KH. 2009. Additions, losses, and rearrangements on the evolutionary route from a reconstructed ancestor to the modern Saccharomyces cerevisiae genome. PLoS Genet 5:e1000485. doi: 10.1371/journal.pgen.1000485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merico A, Sulo P, Piskur J, Compagno C. 2007. Fermentative lifestyle in yeasts belonging to the Saccharomyces complex. FEBS J 274:976–989. doi: 10.1111/j.1742-4658.2007.05645.x. [DOI] [PubMed] [Google Scholar]
- 20.Conant GC, Wolfe KH. 2007. Increased glycolytic flux as an outcome of whole-genome duplication in yeast. Mol Syst Biol 3:129. doi: 10.1038/msb4100170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boles E, Schulte F, Miosga T, Freidel K, Schluter E, Zimmermann FK, Hollenberg CP, Heinisch JJ. 1997. Characterization of a glucose-repressed pyruvate kinase (Pyk2p) in Saccharomyces cerevisiae that is catalytically insensitive to fructose-1,6-bisphosphate. J Bacteriol 179:2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heinisch JJ, Muller S, Schluter E, Jacoby J, Rodicio R. 1998. Investigation of two yeast genes encoding putative isoenzymes of phosphoglycerate mutase. Yeast 14:203–213. [DOI] [PubMed] [Google Scholar]
- 23.Hohmann S. 1991. Characterization of PDC6, a third structural gene for pyruvate decarboxylase in Saccharomyces cerevisiae. J Bacteriol 173:7963–7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McAlister L, Holland MJ. 1985. Differential expression of the three yeast glyceraldehyde-3-phosphate dehydrogenase genes. J Biol Chem 260:15019–15027. [PubMed] [Google Scholar]
- 25.Walsh RB, Clifton D, Horak J, Fraenkel DG. 1991. Saccharomyces cerevisiae null mutants in glucose phosphorylation: metabolism and invertase expression. Genetics 128:521–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conant GC, Wagner A. 2004. Duplicate genes and robustness to transient gene knock-downs in Caenorhabditis elegans. Proc Biol Sci 271:89–96. doi: 10.1098/rspb.2003.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Smet R, Van de Peer Y. 2012. Redundancy and rewiring of genetic networks following genome-wide duplication events. Curr Opin Plant Biol 15:168–176. doi: 10.1016/j.pbi.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 28.Hsiao TL, Vitkup D. 2008. Role of duplicate genes in robustness against deleterious human mutations. PLoS Genet 4:e1000014. doi: 10.1371/journal.pgen.1000014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kafri R, Springer M, Pilpel Y. 2009. Genetic redundancy: new tricks for old genes. Cell 136:389–392. doi: 10.1016/j.cell.2009.01.027. [DOI] [PubMed] [Google Scholar]
- 30.Krakauer DC, Plotkin JB. 2002. Redundancy, antiredundancy, and the robustness of genomes. Proc Natl Acad Sci U S A 99:1405–1409. doi: 10.1073/pnas.032668599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nowak MA, Boerlijst MC, Cooke J, Smith JM. 1997. Evolution of genetic redundancy. Nature 388:167–171. doi: 10.1038/40618. [DOI] [PubMed] [Google Scholar]
- 32.Thatcher JW, Shaw JM, Dickinson WJ. 1998. Marginal fitness contributions of nonessential genes in yeast. Proc Natl Acad Sci U S A 95:253–257. doi: 10.1073/pnas.95.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeLuna A, Springer M, Kirschner MW, Kishony R. 2010. Need-based up-regulation of protein levels in response to deletion of their duplicate genes. PLoS Biol 8:e1000347. doi: 10.1371/journal.pbio.1000347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fell DA, Thomas S. 1995. Physiological control of metabolic flux: the requirement for multisite modulation. Biochem J 311:35–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kacser H, Burns JA, Fell DA. 1995. The control of flux. Biochem Soc Trans 23:341–366. [DOI] [PubMed] [Google Scholar]
- 36.Dymond JS, Richardson SM, Coombes CE, Babatz T, Muller H, Annaluru N, Blake WJ, Schwerzmann JW, Dai JB, Lindstrom DL, Boeke AC, Gottschling DE, Chandrasegaran S, Bader JS, Boeke JD. 2011. Synthetic chromosome arms function in yeast and generate phenotypic diversity by design. Nature 477:471–476. doi: 10.1038/nature10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teusink B, Passarge J, Reijenga CA, Esgalhado E, van der Weijden CC, Schepper M, Walsh MC, Bakker BM, van Dam K, Westerhoff HV, Snoep JL. 2000. Can yeast glycolysis be understood in terms of in vitro kinetics of the constituent enzymes? Testing biochemistry. Eur J Biochem 267:5313–5329. doi: 10.1046/j.1432-1327.2000.01527.x. [DOI] [PubMed] [Google Scholar]
- 38.Smallbone K, Messiha HL, Carroll KM, Winder CL, Malys N, Dunn WB, Murabito E, Swainston N, Dada JO, Khan F, Pir P, Simeonidis E, Spasic I, Wishart J, Weichart D, Hayes NW, Jameson D, Broomhead DS, Oliver SG, Gaskell SJ, McCarthy JEG, Paton NW, Westerhoff HV, Kell DB, Mendes P. 2013. A model of yeast glycolysis based on a consistent kinetic characterisation of all its enzymes. FEBS Lett 587:2832–2841. doi: 10.1016/j.febslet.2013.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bertani G. 1951. Studies on lysogenesis. 1. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol 62:293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bertani G. 2004. Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J Bacteriol 186:595–600. doi: 10.1128/JB.186.3.595-600.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Entian KD, Kötter P. 2007. 25 yeast genetic strain and plasmid collections, p 629–666. In Stansfield I, Stark J (ed), Methods in microbiology. Academic Press, Amsterdam, The Netherlands. [Google Scholar]
- 42.Nijkamp JF, van den Broek M, Datema E, de Kok S, Bosman L, Luttik MA, Daran-Lapujade P, Vongsangnak W, Nielsen J, Heijne WHM, Klaassen P, Paddon CJ, Platt D, Kotter P, van Ham RC, Reinders MJT, Pronk JT, de Ridder D, Daran JM. 2012. De novo sequencing, assembly and analysis of the genome of the laboratory strain Saccharomyces cerevisiae CEN.PK113-7D, a model for modern industrial biotechnology. Microb Cell Fact 11:36. doi: 10.1186/1475-2859-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Dijken JP, Bauer J, Brambilla L, Duboc P, Francois JM, Gancedo C, Giuseppin MLF, Heijnen JJ, Hoare M, Lange HC, Madden EA, Niederberger P, Nielsen J, Parrou JL, Petit T, Porro D, Reuss M, van Riel N, Rizzi M, Steensma HY, Verrips CT, Vindelov J, Pronk JT. 2000. An interlaboratory comparison of physiological and genetic properties of four Saccharomyces cerevisiae strains. Enzyme Microb Technol 26:706–714. doi: 10.1016/S0141-0229(00)00162-9. [DOI] [PubMed] [Google Scholar]
- 44.Wach A, Brachat A, Alberti-Segui C, Rebischung C, Philippsen P. 1997. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast 13:1065–1075. [DOI] [PubMed] [Google Scholar]
- 45.Gueldener U, Heinisch J, Koehler GJ, Voss D, Hegemann JH. 2002. A second set of LoxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res 30:e23. doi: 10.1093/nar/30.6.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shuster JR, Moyer D, Irvine B. 1987. Sequence of the Kluyveromyces lactis URA3 gene. Nucleic Acids Res 15:8573–8573. doi: 10.1093/nar/15.20.8573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wach A, Brachat A, Pohlmann R, Philippsen P. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 48.Guldener U, Heck S, Fiedler T, Beinhauer J, Hegemann JH. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res 24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Kok S, Yilmaz D, Suir E, Pronk JT, Daran JM, van Maris AJA. 2011. Increasing free-energy (ATP) conservation in maltose-grown Saccharomyces cerevisiae by expression of a heterologous maltose phosphorylase. Metab Eng 13:518–526. doi: 10.1016/j.ymben.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 50.Gritz L, and Davies J. 1983. Plasmid encoded hygromycin-B resistance: the sequence of hygromycin-B phosphotransferase gene and its expression in Escherichia coli and Saccharomyces cerevisiae. Gene 25:179–188. doi: 10.1016/0378-1119(83)90223-8. [DOI] [PubMed] [Google Scholar]
- 51.Alani E, Cao L, Kleckner N. 1987. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics 116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Akada R, Kitagawa T, Kaneko S, Toyonaga D, Ito S, Kakihara Y, Hoshida H, Morimura S, Kondo A, Kida K. 2006. PCR-mediated seamless gene deletion and marker recycling in Saccharomyces cerevisiae. Yeast 23:399–405. doi: 10.1002/yea.1365. [DOI] [PubMed] [Google Scholar]
- 53.Solis-Escalante D, Kuijpers NG, Bongaerts N, Bolat I, Bosman L, Pronk JT, Daran JM, Daran-Lapujade P. 2013. amdSYM, a new dominant recyclable marker cassette for Saccharomyces cerevisiae. FEMS Yeast Res 13:126–139. doi: 10.1111/1567-1364.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Solis-Escalante D, Kuijpers NG, van der Linden FH, Pronk JT, Daran JM, Daran-Lapujade P. 2014. Efficient simultaneous excision of multiple selectable marker cassettes using I-SceI-induced double-strand DNA breaks in Saccharomyces cerevisiae. FEMS Yeast Res 14:741–754. doi: 10.1111/1567-1364.12162. [DOI] [PubMed] [Google Scholar]
- 55.Gietz RD, Woods RA. 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol 350:87–96. [DOI] [PubMed] [Google Scholar]
- 56.Verduyn C, Postma E, Scheffers WA, Vandijken JP. 1992. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous culture study on the regulation of respiration and alcoholic fermentation. Yeast 8:501–517. doi: 10.1002/yea.320080703. [DOI] [PubMed] [Google Scholar]
- 57.Pronk JT. 2002. Auxotrophic yeast strains in fundamental and applied research. Appl Environ Microbiol 68:2095–2100. doi: 10.1128/AEM.68.5.2095-2100.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Postma E, Verduyn C, Scheffers WA, Vandijken JP. 1989. Enzymic analysis of the Crabtree effect in glucose-limited chemostat cultures of Saccharomyces cerevisiae. Appl Environ Microbiol 55:468–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jansen MLA, Diderich JA, Mashego M, Hassane A, de Winde JH, Daran-Lapujade P, Pronk JT. 2005. Prolonged selection in aerobic, glucose-limited chemostat cultures of Saccharomyces cerevisiae causes a partial loss of glycolytic capacity. Microbiology 151:1657–1669. doi: 10.1099/mic.0.27577-0. [DOI] [PubMed] [Google Scholar]
- 60.Cruz ALB, Hebly M, Duong GH, Wahl SA, Pronk JT, Heijnen JJ, Daran-Lapujade P, van Gulik WM. 2012. Similar temperature dependencies of glycolytic enzymes: an evolutionary adaptation to temperature dynamics? BMC Syst Biol 6:151. doi: 10.1186/1752-0509-6-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. 1951. Protein measurement with the Folin phenol reagent. J Biol Chem 193:265–275. [PubMed] [Google Scholar]
- 62.Lange HC, Heijnen JJ. 2001. Statistical reconciliation of the elemental and molecular biomass composition of Saccharomyces cerevisiae. Biotechnol Bioeng 75:334–344. doi: 10.1002/bit.10054. [DOI] [PubMed] [Google Scholar]
- 63.Canelas AB, Ras C, ten Pierick A, van Dam JC, Heijnen JJ, van Gulik WM. 2008. Leakage-free rapid quenching technique for yeast metabolomics. Metabolomics 4:226–239. doi: 10.1007/s11306-008-0116-4. [DOI] [Google Scholar]
- 64.Wu L, Mashego MR, van Dam JC, Proell AM, Vinke JL, Ras C, van Winden WA, van Gulik WM, Heijnen JJ. 2005. Quantitative analysis of the microbial metabolome by isotope dilution mass spectrometry using uniformly 13C-labeled cell extracts as internal standards. Anal Biochem 336:164–171. doi: 10.1016/j.ab.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 65.Cipollina C, ten Pierick A, Canelas AB, Seifar RM, van Maris AJA, van Dam JC, Heijnen JJ. 2009. A comprehensive method for the quantification of the non-oxidative pentose phosphate pathway intermediates in Saccharomyces cerevisiae by GC-IDMS. J Chromatogr B Analyt Technol Biomed Life Sci 877:3231–3236. doi: 10.1016/j.jchromb.2009.07.019. [DOI] [PubMed] [Google Scholar]
- 66.van Dam JC, Eman MR, Frank J, Lange HC, van Dedem GWK, Heijnen SJ. 2002. Analysis of glycolytic intermediates in Saccharomyces cerevisiae using anion exchange chromatography and electrospray ionization with tandem mass spectrometric detection. Anal Chim Acta 460:209–218. doi: 10.1016/S0003-2670(02)00240-4. [DOI] [Google Scholar]
- 67.Kozak BU, van Rossum HM, Benjamin KR, Wu L, Daran JMG, Pronk JT, van Maris AIJA. 2014. Replacement of the Saccharomyces cerevisiae acetyl-CoA synthetases by alternative pathways for cytosolic acetyl-CoA synthesis. Metab Eng 21:46–59. doi: 10.1016/j.ymben.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 68.Piper MDW, Daran-Lapujade P, Bro C, Regenberg B, Knudsen S, Nielsen J, Pronk JT. 2002. Reproducibility of oligonucleotide microarray transcriptome analyses: an interlaboratory comparison using chemostat cultures of Saccharomyces cerevisiae. J Biol Chem 277:37001–37008. doi: 10.1074/jbc.M204490200. [DOI] [PubMed] [Google Scholar]
- 69.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ewing B, Hillier L, Wendl MC, Green P. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 8:175–185. [DOI] [PubMed] [Google Scholar]
- 73.Ewing B, Green P. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8:186–194. [PubMed] [Google Scholar]
- 74.Gancedo C, Flores CL. 2008. Moonlighting proteins in yeasts. Microbiol Mol Biol Rev 72:197–210. doi: 10.1128/MMBR.00036-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Knijnenburg TA, Daran JMG, van den Broek MA, Daran-Lapujade PAS, de Winde JH, Pronk JT, Reinders MJT, Wessels LFA. 2009. Combinatorial effects of environmental parameters on transcriptional regulation in Saccharomyces cerevisiae: a quantitative analysis of a compendium of chemostat-based transcriptome data. BMC Genomics 10:53. doi: 10.1186/1471-2164-10-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Decker BL, Wickner WT. 2006. Enolase activates homotypic vacuole fusion and protein transport to the vacuole in yeast. J Biol Chem 281:14523–14528. doi: 10.1074/jbc.M600911200. [DOI] [PubMed] [Google Scholar]
- 77.Pelaez R, Herrero P, Moreno F. 2010. Functional domains of yeast hexokinase 2. Biochem J 432:181–190. doi: 10.1042/BJ20100663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kopperschlager G, Bar J, Nissler K, Hofmann E. 1977. Physicochemical parameters and subunit composition of yeast phosphofructokinase. Eur J Biochem 81:317–325. doi: 10.1111/j.1432-1033.1977.tb11954.x. [DOI] [PubMed] [Google Scholar]
- 79.Breitenbach-Schmitt I, Heinisch J, Schmitt HD, Zimmermann FK. 1984. Yeast mutants without phosphofructokinase activity can still perform glycolysis and alcoholic fermentation. Mol Gen Genet 195:530–535. doi: 10.1007/BF00341458. [DOI] [Google Scholar]
- 80.Clifton D, Fraenkel DG. 1982. Mutant studies of yeast phosphofructokinase. Biochemistry 21:1935–1942. doi: 10.1021/bi00537a037. [DOI] [PubMed] [Google Scholar]
- 81.Lobo Z, Maitra PK. 1983. Phosphofructokinase mutants of yeast: biochemistry and genetics. J Biol Chem 258:1444–1449. [PubMed] [Google Scholar]
- 82.Bakker BM, Bro C, Kotter P, Luttik MAH, van Dijken JP, Pronk JT. 2000. The mitochondrial alcohol dehydrogenase adh3p is involved in a redox shuttle in Saccharomyces cerevisiae. J Bacteriol 182:4730–4737. doi: 10.1128/JB.182.17.4730-4737.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cakar ZP, Turanli-Yildiz B, Alkim C, Yilmaz U. 2012. Evolutionary engineering of Saccharomyces cerevisiae for improved industrially important properties. FEMS Yeast Res 12:171–182. doi: 10.1111/j.1567-1364.2011.00775.x. [DOI] [PubMed] [Google Scholar]
- 84.Teng X, Dayhoff-Brannigan M, Cheng WC, Gilbert CE, Sing CN, Diny NL, Wheelan SJ, Dunham MJ, Boeke JD, Pineda FJ, Hardwick JM. 2013. Genome-wide consequences of deleting any single gene. Mol Cell 52:485–494. doi: 10.1016/j.molcel.2013.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tai SL, Daran-Lapujade P, Luttik MAH, Walsh MC, Diderich JA, Krijger GC, van Gulik WM, Pronk JT, Daran JM. 2007. Control of the glycolytic flux in Saccharomyces cerevisiae grown at low temperature: a multi-level analysis in anaerobic chemostat cultures. J Biol Chem 282:10243–10251. doi: 10.1074/jbc.M610845200. [DOI] [PubMed] [Google Scholar]
- 86.de Smidt O, du Preez JC, Albertyn J. 2012. Molecular and physiological aspects of alcohol dehydrogenases in the ethanol metabolism of Saccharomyces cerevisiae. FEMS Yeast Res 12:33–47. doi: 10.1111/j.1567-1364.2011.00760.x. [DOI] [PubMed] [Google Scholar]
- 87.Gruning NM, Rinnerthaler M, Bluemlein K, Mulleder M, Wamelink MMC, Lehrach H, Jakobs C, Breitenbach M, Ralser M. 2011. Pyruvate kinase triggers a metabolic feedback loop that controls redox metabolism in respiring cells. Cell Metab 14:415–427. doi: 10.1016/j.cmet.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ring J, Sommer C, Carrnona-Gutierrez D, Ruckenstuhl C, Eisenberg T, Madeo F. 2012. The metabolism beyond programmed cell death in yeast. Exp Cell Res 318:1193–1200. doi: 10.1016/j.yexcr.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Daran-Lapujade P, Daran JM, Luttik MAH, Almering MJH, Pronk JT, Kötter P. 2009. An atypical PMR2 locus is responsible for hypersensitivity to sodium and lithium cations in the laboratory strain Saccharomyces cerevisiae CEN.PK113-7D. FEMS Yeast Res 9:789–792. doi: 10.1111/j.1567-1364.2009.00530.x. [DOI] [PubMed] [Google Scholar]
- 90.Kim JW, Dang CV. 2005. Multifaceted roles of glycolytic enzymes. Trends Biochem Sci 30:142–150. doi: 10.1016/j.tibs.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 91.Kane PA. 2006. The where, when, and how of organelle acidification by the yeast vacuolar H+-ATPase. Microbiol Mol Biol Rev 70:177–191. doi: 10.1128/MMBR.70.1.177-191.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Grant CM, Quinn KA, Dawes IW. 1999. Differential protein S-thiolation of glyceraldehyde-3-phosphate dehydrogenase isoenzymes influences sensitivity to oxidative stress. Mol Cell Biol 19:2650–2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rodriguez A, de la Cera T, Herrero P, Moreno F. 2001. The hexokinase 2 protein regulates the expression of the GLK1, HXK1 and HXK2 genes of Saccharomyces cerevisiae. Biochem J 355:625–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Boer VM, de Winde JH, Pronk JT, Piper MDW. 2003. The genome-wide transcriptional responses of Saccharomyces cerevisiae grown on glucose in aerobic chemostat cultures limited for carbon, nitrogen, phosphorus, or sulfur. J Biol Chem 278:3265–3274. doi: 10.1074/jbc.M209759200. [DOI] [PubMed] [Google Scholar]
- 95.Cormier L, Barbey R, Kuras L. 2010. Transcriptional plasticity through differential assembly of a multiprotein activation complex. Nucleic Acids Res 38:4998–5014. doi: 10.1093/nar/gkq257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fauchon M, Lagniel G, Aude JC, Lombardia L, Soularue P, Petat C, Marguerie G, Sentenac A, Werner M, Labarre J. 2002. Sulfur sparing in the yeast proteome in response to sulfur demand. Mol Cell 9:713–723. doi: 10.1016/S1097-2765(02)00500-2. [DOI] [PubMed] [Google Scholar]
- 97.Qian WF, Liao BY, Chang AYF, Zhang JZ. 2010. Maintenance of duplicate genes and their functional redundancy by reduced expression. Trends Genet 26:425–430. doi: 10.1016/j.tig.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kuepfer L, Sauer U, Blank LM. 2005. Metabolic functions of duplicate genes in Saccharomyces cerevisiae. Genome Res 15:1421–1430. doi: 10.1101/gr.3992505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Verduyn C, Postma E, Scheffers WA, Vandijken JP. 1990. Physiology of Saccharomyces cerevisiae in anaerobic glucose-limited chemostat cultures. J Gen Microbiol 136:395–403. doi: 10.1099/00221287-136-3-395. [DOI] [PubMed] [Google Scholar]
- 100.Hong K, Nielsen J. 2012. Metabolic engineering of Saccharomyces cerevisiae: a key cell factory platform for future biorefineries. Cell Mol Life Sci 69:2671–2690. doi: 10.1007/s00018-012-0945-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nevoigt E. 2008. Progress in metabolic engineering of Saccharomyces cerevisiae. Microbiol Mol Biol Rev 72:379–412. doi: 10.1128/MMBR.00025-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Goddard M, Greig D. 2015. Saccharomyces cerevisiae: a nomadic yeast with no niche? FEMS Yeast Res 15:fov009. doi: 10.1093/femsyr/fov009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wisselink HW, Cipollina C, Oud B, Crimi B, Heijnen JJ, Pronk JT, van Maris AJA. 2010. Metabolome, transcriptome and metabolic flux analysis of arabinose fermentation by engineered Saccharomyces cerevisiae. Metab Eng 12:537–551. doi: 10.1016/j.ymben.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 104.Hohmann S, Cederberg H. 1990. Autoregulation may control the expression of yeast pyruvate decarboxylase structural genes PDC1 and PDC5. Eur J Biochem 188:615–621. doi: 10.1111/j.1432-1033.1990.tb15442.x. [DOI] [PubMed] [Google Scholar]
- 105.Zhang JZ. 2012. Advances in experimental medicine and biology, p 279–300. Springer, New York, NY. [Google Scholar]
- 106.DiCarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM. 2013. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res 41:4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wieczorke R, Krampe S, Weierstall T, Freidel K, Hollenberg CP, Boles E. 1999. Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett 464:123–128. doi: 10.1016/S0014-5793(99)01698-1. [DOI] [PubMed] [Google Scholar]
- 108.Delneri D, Tomlin GC, Wixon JL, Hutter A, Sefton M, Louis EJ, Oliver SG. 2000. Exploring redundancy in the yeast genome: an improved strategy for use of the Cre-LoxP system. Gene 252:127–135. doi: 10.1016/S0378-1119(00)00217-1. [DOI] [PubMed] [Google Scholar]
- 109.Coelho MA, Goncalves C, Sampaio JP, Goncalves P. 2013. Extensive intra-kingdom horizontal gene transfer converging on a fungal fructose transporter gene. PLoS Genet 9:e1003587. doi: 10.1371/journal.pgen.1003587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dae-Hee L, Soo-Jung K, Jin-Ho S. 2014. Molecular cloning and characterization of two novel fructose-specific transporters from the osmotolerant and fructophilic yeast Candida magnoliae JH110. Appl Microbiol Biotechnol 98:3569–3578. doi: 10.1007/s00253-013-5225-y. [DOI] [PubMed] [Google Scholar]
- 111.Price DRG, Tibbles K, Shigenobu S, Smertenko A, Russell CW, Douglas AE, Fitches E, Gatehouse AMR, Gatehouse JA. 2010. Sugar transporters of the major facilitator superfamily in aphids; from gene prediction to functional characterization. Insect Mol Biol 19:97–112. doi: 10.1111/j.1365-2583.2009.00918.x. [DOI] [PubMed] [Google Scholar]
- 112.Schneidereit A, Scholz-Starke J, Buttner M. 2003. Functional characterization and expression analyses of the glucose-specific AtSTP9 monosaccharide transporter in pollen of Arabidopsis. Plant Physiol 133:182–190. doi: 10.1104/pp.103.026674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Schussler A, Martin H, Cohen D, Fitz M, Wipf D. 2006. Characterization of a carbohydrate transporter from symbiotic glomeromycotan fungi. Nature 444:933–936. doi: 10.1038/nature05364. [DOI] [PubMed] [Google Scholar]
- 114.Stasyk OG, Maidan MM, Stasyk OV, Van Dijck P, Thevelein JM, Sibirny AA. 2008. Identification of hexose transporter-like sensor HXS1 and functional hexose transporter HXT1 in the methylotrophic yeast Hansenula polymorpha. Eukaryot Cell 7:735–746. doi: 10.1128/EC.00028-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Vignault C, Vachaud M, Cakir B, Glissant D, Dedaldechamp F, Buttner M, Atanassova R, Fleurat-Lessard P, Lemoine R, Delrot S. 2005. VvHT1 encodes a monosaccharide transporter expressed in the conducting complex of the grape berry phloem. J Exp Bot 56:1409–1418. doi: 10.1093/jxb/eri142. [DOI] [PubMed] [Google Scholar]
- 116.Xuan YH, Hu YB, Chen LQ, Sosso D, Ducat DC, Hou BH, Frommer WB. 2013. Functional role of oligomerization for bacterial and plant SWEET sugar transporter family. Proc Natl Acad Sci U S A 110:E3685–E3694. doi: 10.1073/pnas.1311244110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Young E, Poucher A, Comer A, Bailey A, Alper H. 2011. Functional survey for heterologous sugar transport proteins, using Saccharomyces cerevisiae as a host. Appl Environ Microbiol 77:3311–3319. doi: 10.1128/AEM.02651-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mashego MR, Jansen MLA, Vinke JL, van Gulik WM, Heijnen JJ. 2005. Changes in the metabolome of Saccharomyces cerevisiae associated with evolution in aerobic glucose-limited chemostats. FEMS Yeast Res 5:419–430. doi: 10.1016/j.femsyr.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 119.Byrne KP, Wolfe KH. 2005. The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res 15:1456–1461. doi: 10.1101/gr.3672305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Solis-Escalante D, van den Broek M, Kuijpers NG, Pronk JT, Boles E, Daran JM, Daran-Lapujade P. 2015. The genome sequence of the popular hexose-transport-deficient Saccharomyces cerevisiae strain EBY.VW4000 reveals LoxP/Cre-induced translocations and gene loss. FEMS Yeast Res 15:fou004. doi: 10.1093/femsyr/fou004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.