Abstract
The plant hormone abscisic acid (ABA) regulates many key processes involved in plant development and adaptation to biotic and abiotic stresses. Under stress conditions, plants synthesize ABA in various organs and initiate defense mechanisms, such as the regulation of stomatal aperture and expression of defense-related genes conferring resistance to environmental stresses. The regulation of stomatal opening and closure is important to pathogen defense and control of transpirational water loss. Recent studies using a combination of approaches, including genetics, physiology, and molecular biology, have contributed considerably to our understanding of ABA signal transduction. A number of proteins associated with ABA signaling and responses—especially ABA receptors—have been identified. ABA signal transduction initiates signal perception by ABA receptors and transfer via downstream proteins, including protein kinases and phosphatases. In the present review, we focus on the function of ABA in stomatal defense against biotic and abiotic stresses, through analysis of each ABA signal component and the relationships of these components in the complex network of interactions. In particular, two ABA signal pathway models in response to biotic and abiotic stress were proposed, from stress signaling to stomatal closure, involving the pyrabactin resistance (PYR)/PYR-like (PYL) or regulatory component of ABA receptor (RCAR) family proteins, 2C-type protein phosphatases, and SnRK2-type protein kinases.
Keywords: abscisic acid (ABA), ABA receptor, biotic and abiotic stresses, PP2C, SnRK2, stomatal immunity
1. Function of Abscisic Acid (ABA) in Response to Biotic and Abiotic Stresses
Plants are sessile organisms and therefore they constantly encounter diverse biotic and abiotic stresses, including various pathogens, drought, and high salinity. These stresses affect plant growth and development and can severely impair crop production. The plant hormone abscisic acid (ABA) functions as a chemical signal in response to environmental stresses. Stress signals are converted to ABA and this triggers the activation of a number of plant physiological and developmental processes, thereby inducing adaptation to the stress conditions [1,2,3]. Defense responses to biotic and abiotic stress have been extensively investigated [4,5,6,7]. In the present review, we focus on recent research into ABA responses to stomatal defense, and crosstalk of biotic and abiotic responses through the regulation of stomatal movement.
1.1. Function of ABA in the Regulation of Stomatal Movement
Biotic and abiotic stresses adversely affect plant growth and induce severe losses in agricultural crop production. Plants lose water primarily by gaseous exchange through the stomata on their leaves. ABA is a key hormone that regulates water status and stomatal movement. Under drought conditions, plants produce and accumulate increased amounts of ABA in the guard cells, and this induces stomatal closure to conserve water. The cellular and molecular mechanisms underlying ABA-induced stomatal closure have been extensively investigated [4,5,8,9,10,11,12,13]. ABA biosynthesis and catabolism are known to be major determinants of endogenous ABA levels in plant cells [14,15]. The 9-cis-epoxycarotenoid dioxygenase (NCED) genes and cytochrome P450 CYP707A genes encode key enzymes for ABA biosynthesis and ABA catabolism, respectively. The NCED3 gene is induced by drought stress and it upregulates endogenous ABA levels in overexpressed transgenic plants, thereby leading to lower transpiration rates [16,17,18,19,20]. Regarding ABA catabolism, the expression of CYP707A1 to CYP707A4 genes trigger ABA 8′-hydroxylation [21,22]. Among these four members of the CYP707A gene family, the transcripts of CYP707A3 were shown to be the most highly accumulated. Moreover, the results of genetics analysis revealed that the cyp707a3-1 Arabidopsis mutant accumulated higher endogenous ABA levels and exhibited a reduced transpiration rate, thereby resulting in a phenotype exhibiting enhanced tolerance to drought stress [21].
Stomatal control is regulated by a number of environmental factors, including CO2 level, light, and biotic and abiotic stresses. Guard cell turgor pressure is a key parameter regulating stomatal opening and closure, and it in turn is mediated by ionic fluxes via cation and anion channels anchored in the guard cell membrane. Under conditions of biotic and abiotic stresses, ABA functions as a chemical messenger that induces stomatal closure through the activation and inactivation of ion channels by protein kinases and phosphatases [23,24,25,26,27,28,29,30,31,32,33,34,35,36]. The entire sequence of ABA signal transduction, from ABA receptors to stomatal closure, will be discussed later in this review.
1.2. Role of Stomatal Immunity via Restriction of Pathogen Entry
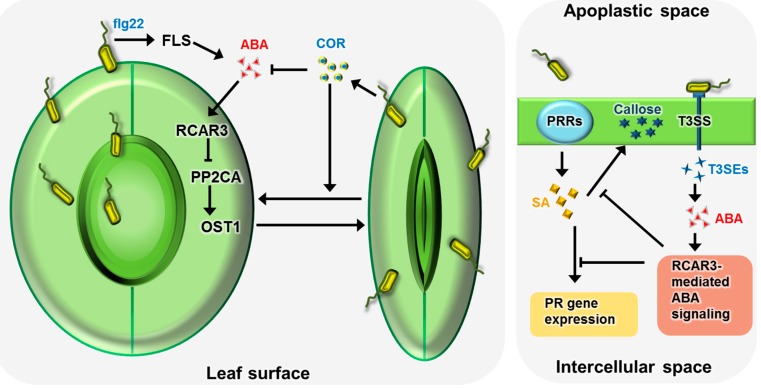
Recently, several studies have demonstrated that ABA plays a crucial role in pathogen response and that ABA signaling overlaps considerably between biotic stress resistance and abiotic stress tolerance. Plants possess physical and biochemical defense barriers that effectively protect them from diverse pathogens. Various foliar pathogens such as bacteria, fungi, and viruses are known to disrupt stomatal movement in order successfully to infect plants [37,38,39]. The first line of defense is the recognition of the evolutionary conserved pathogen materials—termed the pathogen-associated molecular pattern (PAMP)—by plant pattern recognition receptors (PRRS), thereby leading to PAMP-triggered immunity (PTI) [40,41,42]. The second line of defense is the recognition of effectors through plant resistance (R) proteins, thereby leading to effector-triggered immunity (ETI) [40,43]. Melotto et al. [44] showed that stomata constitute not only a path for transpiration, but also a port for pathogen entry. Thus, stomata play a crucial role in the plant immune response, and the regulation of stomatal movement is an early stage of defense mechanism against pathogen infection (Figure 1). Conversely, most pathogens have evolved mechanisms that allow them to overcome or circumvent plant physical barriers, including stomatal closure, thereby enabling them to successfully infect plants [40].
Figure 1.
Schematic representation of a possible mechanism underlying the antagonistic and synergistic role of abscisic acid (ABA) signaling in plant defense response at the pre-invasive and post-invasive stages of Arabidopsis thaliana–Pseudomonas syringae interaction. At the pre-invasive stage, ABA signaling plays a synergistic role in plant resistance to P. syringae attack. Stomata constitute a major route of bacterial entry. Pathogen-associated molecular pattern (PAMP)-induced ABA signaling in the guard cells promotes stomatal closure and actively blocks P. syringae invasion [44,48]. This stomatal immunity involves the ABA receptor–2C-type protein phosphatase–open stomata 1 (RCAR-PP2C-OST1) complex acting as a core component of ABA signaling [12,44]. The phytotoxin coronatine (COR) is a virulence factor produced by P. syringae and it can compromise PAMP-induced stomatal defense by suppressing PAMP-induced ABA signaling and promoting stomatal reopening [12,44,49]. In contrast, ABA signaling plays an antagonistic role in post-invasive defense response. Pseudomonas syringae type III secreted effector (T3SE) proteins upregulate ABA biosynthesis and also the signaling pathways, thereby inhibiting the plant defense response [44,50]. In this process, ABA signaling antagonizes salicylic acid (SA)-mediated pathogenesis-related (PR) gene expression and callose deposition [12,48,50].
1.3. Role of Stomatal Closure in Drought Responses
The primary function of stomatal closure is to prevent water loss, thereby inducing drought tolerance under conditions of osmotic stress. The regulation of stomatal closure is important not only for the defense mechanism to prevent invasion of bacterial pathogens, but also for water conservation. Drought is a major osmotic stress that affects plant growth and development, thereby leading to severe losses in agricultural crop production. Stomatal opening and closure affect various physiological processes and properties, such as photosynthesis and water status. Plants gain water through their roots and lose water primarily via the stomata on their leaves. If the amount of water lost through the leaves exceeds the amount taken up by the roots, plant tissues can be damaged, thereby resulting in cell death. Under drought conditions, ABA is produced or accumulated in the guard cells that surround the stomata, and this induces stomatal closure, thereby conserving water [11,45]. The cellular and molecular regulation of stomatal opening and closure under drought conditions has been extensively investigated and reviewed [4,5,8,9,10,13]. ABA-controlled processes are necessary for plant survival, and ABA-deficient mutants are susceptible to water stress [3,46,47].
Guard cell turgor pressure is a key parameter regulating stomatal control, and this in turn is mediated by ionic fluxes across the cell membranes through K+ and anion channels. Consequently, these ion channels constitute the major target for regulation by a number of environmental factors, including light, dark, drought, CO2 levels, etc. Under drought conditions, ABA serves as a chemical messenger that induces stomatal closure through second messengers, such as ROS, nitric oxide, Ca2+, and protein kinases; these messengers further target the ion channels [23,24,25,26,27,28,29,30,31,32,33,34,35,36]. The entire sequence of ABA signal transduction, from ABA receptors to stomatal closure, will be discussed later in this review.
2. ABA Signal Transduction Pathway
2.1. Perception and Transfer of the ABA Signal
Previous studies have identified positive and negative regulators of ABA signaling, from ABA receptors to ion channels [8,11,24,26,51,52,53,54,55,56,57,58]. To initiate ABA signal transduction, ABA receptors must be present and must deliver the ABA signal to the downstream pathway in plant cells. Several types of ABA receptor have been identified [9,51,57,59]. ABA-binding activities are known to be present in several locations, including the cell membrane and cytoplasm, indicating that more than one ABA receptor may exist within a single cell [60,61,62]. However, with the exception of the pyrabactin resistance (PYR)/PYR-like (PYL) or regulatory component of ABA receptor (RCAR) family proteins, few studies of ABA receptor function have been conducted. Therefore, here we will focus on the function of PYR/PYL/RCAR proteins in relation to other positive and negative regulators of ABA signaling, including 2C-type protein phosphatases (PP2Cs) and SnRK2 (SNF1-related kinase 2)-type protein kinases.
The PYR/PYL/RCAR protein family comprises 14 members, all of which function in ABA perception and signaling [9,51,57,63,64]. The results of genetic analysis using triple (pyr1:pyl1:pyl4), quadruple (pyr1:pyl1:pyl2:pyl4), and sextuple (pyr1:pyl1:pyl2:pyl4:pyl5:pyl8) mutants revealed that these mutants displayed ABA-insensitive phenotypes during the germinative, seedling, and adult stages [51,64]. In contrast, the single mutant did not display any altered phenotype to ABA [57,65]. Moreover, the results of structural analyses showed that the PYR/PYL/RCAR proteins harbor a ligand-binding pocket, which may function as an ABA- and group A PP2C-binding site [66,67,68,69,70]. Several previous studies have demonstrated that group A PP2Cs function as negative regulators of ABA signaling [13,71,72,73]; moreover, PYR/PYL/RCAR proteins directly inhibit phosphatase activity of PP2Cs in vitro [51,74]. Thus, these PP2Cs constitute direct targets of PYR/PYL/RCAR proteins in the ABA signaling pathway. The results of in vitro and in vivo interaction assays between PYR/PYL/RCAR proteins and their target PP2Cs showed that ABA enhances these interactions; in contrast, other interactions are independent of ABA [12,51,58,63]. These findings imply that ABA binding induces structural changes in PYR/PYL/RCAR proteins and that the ABA-bound PYR/PYL/RCAR proteins can interact tightly with PP2Cs, or that ABA does not change the protein structures and that PP2Cs bind only to the PYR/PYL/RCAR-ABA complex. Whichever is the case, the interactions between PYR/PYL/RCAR proteins and their target PP2Cs induce the activation of downstream targets of the group A PP2Cs, including SnRK2 protein kinases (SnRK2.2, SnRK2.3, SnRK2.6) and the S-type anion channel (SLAC1); these in turn play key roles in the regulation of transcriptional response and stomatal closure [26,56,58]. Several studies have shown that these ABA receptors have a functional redundancy in ABA signaling. In addition, with the exception of PYL13, all the PYR/PYL/RCAR protein family members are able to induce ABA-responsive genes [52]. However, the expression patterns differ from each other, indicating that the functions of PYR/PYL/RCAR proteins and their in vivo downstream target specificity may vary markedly.
The ubiquitination system via the 26S proteasome pathway plays a crucial role in hormone signaling, including the perception of auxin [75,76,77], jasmonate [78], gibberellins [79], and ABA [80,81,82,83,84]. Ubiquitination is a key post-translational modification performed by the sequential action of three enzymes—ubiquitin activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin ligase (E3). In this process, E3 ligase determines and recruits substrate proteins. In the ABA signaling pathway, several E3 ligases induce changes in plant responses to ABA, by degradation of positive and negative regulators of ABA signaling [85]. Many E3 ligases related to ABA signaling have been identified in Arabidopsis and rice [81,83,84,86,87,88,89,90,91,92]; however, only a few substrate proteins have been identified using interaction assays. In addition, most target proteins are restricted to the final stage of ABA signal transduction; these include transcription factors, which regulate the expression of ABA-responsive genes, for example, ABF2 (ABRE-binding bZIP transcription factor 2) (ubiquitinated by ARIA (arm repeat protein interacting with ABF2)), ABI3 (abscisic acid-insensitive 3) (ubiquitinated by AIP2 (ABI3-interacting protein 2)), ABI5 (ABA insensitive 5) (ubiquitinated by KEG (KEEP ON GOING), DWA1 (DWD hypersensitive to ABA1), DWA2, and ABD1 (ABA-hypersensitive DCAF1)), and ERF53 (ethylene response factor 53) (ubiquitinated by RGLG1 (RING domain ligase 1) and RGLG2) [83,91,92,93,94,95]. However, recent studies have demonstrated that ABA receptors (PYR1, PYL4, PYL8, and PYL9) are ubiquitinated by CRL4 (Cullin4-RING E3 ligase) and RSL1, thereby leading to proteasomal degradation and subsequent attenuation of ABA signaling [82,84].
In response to biotic and abiotic stresses, reactive oxygen species (ROS), such as superoxide radical, hydrogen peroxide (H2O2), and nitric oxide (NO) are produced and involved in defense mechanisms [40,96,97]. The ROS function as second messengers of ABA. NADPH (nicotinamide adenine dinucleotide phosphate) oxidases, which referred to as respiratory burst oxidases (RBOH), are induced by biotic and abiotic stresses [96,98,99]. The AtrbohD and AtrbohF are associated with production of ROS in response to ABA [100] and pathogen infection, such as Pseudomonas syringae and Hyaloperonospora Arabidopsis [98]. The atrbohD/atbohF double mutant exhibits impaired ABA signaling phenotypes including stomatal closure, ROS production and cytosolic Ca2+ induction [100]. The atrbohD, atbohF and atrbohD/atbohF mutants also show reduced ROS production and reduced cell deateh in response to pathogen infection. Moreover, these mutants are more susceptible to bacterial pathogen P. syringae pv. tomato DC3000 (avrRpm1) and the oomycete parasite H. Arabidopsis [98].
2.2. Phosphorylation and Dephosphorylation Events in ABA Signaling Transduction
Several protein kinases and phosphatases are involved in protein phosphorylation and dephosphorylation events in ABA signaling in plants [26,56,58,101,102]. The functions of the SnRK2-type protein kinases in relation to ABA were first elucidated using PKABA1 (ABA-induced Ser/Thr protein kinase 1) in wheat [103]. The kinase activity of PKABA1 is induced by ABA and is involved in the phosphorylation and activation of the transcription factor TaABF1 (Triticum aestivum ABRE-binding bZIP transcription factor 1), which induces ABA-responsive gene expression in wheat and barley [103,104,105,106]. Subsequent genetic studies identified open stomata 1 (OST1)/SnRK2.6, which is a Ser/Thr protein kinase, in Arabidopsis [107]. ABA did not induce OST1 expression but did promote kinase activity of this protein [107,108]. The ost1 mutant exhibits an ABA-insensitive and drought-sensitive phenotype, which is characterized by a high level of leaf water loss due to permanently open stomata and a minimal ABA-responsive gene expression [107,108]. OST1 is able to interact with and phosphorylate ABF2 and ABF3, which are transcription factors that bind to ABA-responsive elements (ABRE) and control ABA-responsive gene expression [7,52,109,110]. Furthermore, phosphorylation of ABF3 via OST1 creates the 14-3-3 protein binding motif and this stabilizes ABF3. ABFs, including ABF3 and ABF4, are also involved in drought stress tolerance through the regulation of stomatal closure, thereby leading to a decreased transpirational rate in Arabidopsis [66]. Two other SnRK2-type kinases—SnRK2.2 and SnRK2.3—are induced by ABA, and the snrk2.2/snrk2.3 double mutant exhibits an ABA-insensitive phenotype [53,111]. A triple mutant (snrk2.2/snrk2.3/ost1) displays strongly ABA-insensitive and drought-sensitive phenotypes, implying that these kinases have functional redundancy in ABA and drought signaling [54,109,112]. Moreover, OST1 functions as a positive regulator of freezing tolerance by regulating CBF transcription factors via phosphorylation of ICE1 (inducer of CBF expression 1) [102].
In Arabidopsis, 76 out of 112 phosphatase genes have been identified as PP2Cs [71,113]. These include nine group A PP2Cs, of which six are involved in the regulation of ABA signaling [71,114]. The results of genetic analysis revealed that group A PP2Cs—ABI1 (ABA-insensitive 1), ABI2, HAB1 (hypersensitive to ABA 1), HAB2, AHG1 (ABA-hypersensitive germination 1), and PP2CA (protein phosphatase 2CA)—are negative regulators of ABA signaling and that loss-of-function mutants in Arabidopsis exhibit an ABA-hypersensitive phenotype during seed germination [13,72,73,115,116,117,118,119,120,121]. The PP2C function related to ABA was first demonstrated using the dominant mutants abi1-1 and abi2-1, which display an ABA-insensitive phenotype during seed germination, seedling growth, and stomatal closure [122,123,124,125].
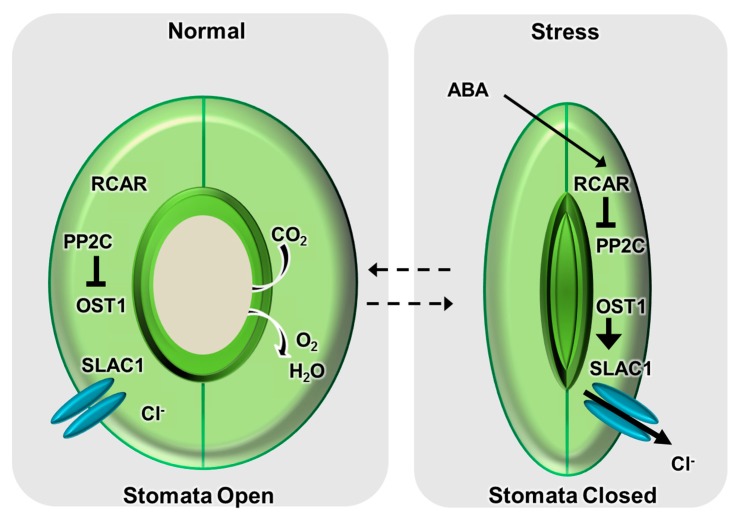
Several studies have evaluated the functional and physical interactions between PYR/PYL/RCAR, PP2C, and SnRK2 [26,51,56,57,58,126,127]. These interactions were initially suggested to occur via physical interactions between OST1 and ABI1/ABI2 [128]. More recently, studies have identified several PYR/PYL/RCAR-PP2C-SnRK2 interactions that clearly function in ABA and stress signaling [12,64,102]. For example, the physical interaction between RCAR2 and PP2CA induces inactivation of OST1 kinase, thereby leading to downregulation of SLAC1 activity in guard cells [58]. These PP2Cs have a functional redundancy in ABA signaling; hence, double or triple mutants have strongly ABA-insensitive phenotypes [53,115,126]. The triple mutant abi1-2/hab1-1/pp2ca-1 displays constitutive activation of SnRK2-type kinases [52], suggesting that the ABA-dependent activation of SnRK2-type kinases is derived from removal of the inhibitory effect of PP2Cs. The ABA-dependent physical interactions of PYR/PYL/RCAR induce the activation of SnRKs, thereby promoting transfer of the ABA signal to the downstream pathway (Figure 2) [51,52,57,127].
Figure 2.
Simplified overview of stomatal movement via the abscisic acid (ABA) signaling pathway. Under normal conditions, 2C-type protein phosphatase (PP2C) family members, which are negative regulators of ABA signaling, suppress open stomata 1 (OST1) kinase activity via physical interaction, leaving the S-type anion channel (SLAC1) with basal activity. Under conditions of stress, including drought, the ABA concentration in leaves increases rapidly [9,10]. ABA perception occurs via regulatory component of ABA receptor (RCAR) family members functioning as ABA receptors in combination with PP2Cs [9,135]. The formation of the RCAR–PP2C complex breaks the PP2C–OST1 complex, thereby releasing active OST1 kinase from inhibition. In turn, OST1-mediated phosphorylation induces activation of the SLAC1 channel, thereby releasing anions and depolarizing the membrane [26]. This depolarization induces a further drop in turgor and closure of the stomatal pores.
2.3. ABA Signal Transduction in Biotic Stress
The regulation of stomatal closure constitutes an important layer of the PAMP-triggered defense mechanism at the pre-invasive level. ABA-mediated stomatal closure inhibits pathogen entry to the apoplastic space; hence, ABA functions as a positive regulator of the defense response (Figure 1). Plant recognition of PAMPs via PRRs leads to stomatal closure, thereby restricting pathogen entry [11,44,129,130,131,132]. Several studies have suggested that ABA is associated with PAMP-triggered stomatal closure. Melotto et al. [44] used the ost1 (snrk2.6) mutant, which is a positive regulator of ABA [107], and the ABA-deficient aba3-1 mutant [133] to show that stomatal closure is not induced by PAMPs. Conversely, overexpression of ABA receptors, such as RCAR3, RCAR4, and RCAR5, which function as positive regulators of ABA, induced the maintenance of stomatal closure during Pseudomonas syringae pv. tomato (Pst) DC3000 inoculation and PAMP treatment, thereby leading to enhanced resistance to Pst DC3000 [12]. Moreover, pp2ca and hab1, the group A 2C type protein phosphatases, mutants were positively correlated with stomatal immunity, similar to overexpression of RCAR3 and RCAR4, respectively [12,134].
After pathogen entry to the apoplastic space, ABA affects pathogen response by interacting with defense-related hormones such as salicylic acid (SA), jasmonic acid (JA), and ethylene, and by acting antagonistically towards these hormones [38,44,65,136,137]. Typically, SA is associated withresistance to biotrophic and hemibiotrophic pathogens and expression of acidic pathogenesis-related (PR) genes; whereas JA and ethylene are associated with resistance to necrotrophic pathogens and expression of basic PR genes [138,139,140]. Previous studies have suggested that ABA signaling transduction negatively regulates SA-mediated pathogen responses. For instance, overexpression of RCAR3, which functions as a positive regulator of ABA, enhanced susceptibility to virulent bacterial pathogens, by suppressing SA-mediated defense responses such as callose deposition and gene expression at the post-invasive level (Figure 1) [12,135]. In addition, overexpression and mutation of the NCED and ABA3 genes, which are involved in ABA biosynthesis, conferred susceptible and resistant phenotypes, respectively, to Pseudomonas syringae pv. tomato (Pst) DC3000 [67]. Previous genetic studies have investigated the negative effect of ABA on JA- and ethylene-dependent pathogen resistance. The OsMPK5 (Oryza sativa mitogen-activated protein kinase 5) is induced by ABA and by biotic and abiotic stresses [141]. Overexpression of OsMPK5 enhances the ABA level, but reduces ethylene accumulation, thereby resulting in a phenotype exhibiting enhanced susceptibility to the bacterial and fungal pathogens Burkholderia glumae and Magnaporthe oryzae, respectively [137,141]. However, it has been shown that ethylene functions as both positive and negative regulators of stomatal opening and closing, depending on the tissue and environmental conditions [96,142,143,144,145]. Thus, cooperated regulation of ABA and ethylene signaling in response to the prevailing environmental conditions is necessary for fine-tuning of stomatal opening and closure, thereby optimizing gaseous exchange for photosynthesis while limiting transpirational water loss and/or pathogen invasion [146,147]. Based on the above findings, we propose that PAMP-induced stomatal closure requires the activation of the ABA signal transduction pathway in guard cells and that ABA functions as a positive regulator of disease resistance at the pre-invasive level. In contrast, defense hormone-triggered resistance inhibited by ABA in the apoplastic space and ABA has a negative effect on disease resistance at the post-invasive level.
3. Conclusions and Perspective
The plant hormone ABA is involved in several biotic and abiotic responses and is associated with the regulation of complex signal transductions including seed dormancy, growth and development. Although a diversity of studies have been performed on ABA and its role in defense responses to biotic and abiotic stresses, no clear model of ABA on these stresses has been proposed, nor have its relation between biotic and abiotic stress been adequately determined. Several key components compose the ABA signaling pathway and each component plays a role as a positive and negative regulator in each step. Several members of the group A PP2Cs and SnRK2s ABA receptors are known to exist. Thus, many possible interactions can occur and each interaction has the potential to regulate downstream targets. Consequently, regulatory fine-tuning is extremely complex. Further detailed studies are required to clarify the complex network of interactions occurring in biotic and abiotic signaling pathways. The results of these studies will facilitate the development of genetically engineering agricultural crops with strong resistance to environmental stresses.
Acknowledgments
This work was supported by a grant from “the Next-Generation BioGreen 21 Program for Agriculture & Technology Development (Project No. PJ01100501)” Rural Development Administration, Republic of Korea and the Chung-Ang University Research Scholarship Grants in 2014.
Abbreviations
- ABA
Abscisic acid
- ABRE
ABA-responsive elements
- CRL4
Cullin4-RING E3 ligase
- E1
Ubiquitin-activating enzyme
- E2
Ubiquitin-conjugating enzyme
- E3
Ubiquitin ligase
- ETI
Effector-triggered immunity
- JA
Jasmonic acid
- NCED
9-cis-epoxycarotenoid dioxygenase
- NO
Nitric oxide
- OST1
Open Stomata 1
- PAMP
Pathogen-associated molecular pattern
- PP2C
2C-type protein phosphatases
- PR
Pathogenesis related
- PRR
Pattern recognition receptor
- PTI
PAMP triggered immunity
- PYR
Pyrabactin resistance
- RCAR
Regulatory component of ABA receptor
- ROS
Reactive oxygen species
- SA
Salicylic acid
- SLAC
S-type anion channel
- SnRK
SNF related kinase
Author Contributions
Chae Woo Lim, Woonhee Baek and Jangho Jung performed the major work on literature search and drafting the text; Jung-Hyun Kim and Sung Chul Lee designed the structures of the manuscript; Sung Chul Lee was responsible for preparation of the manuscript, its submission and revision. All authors have read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Ton J., Flors V., Mauch-Mani B. The multifaceted role of ABA in disease resistance. Trends Plant Sci. 2009;14:310–317. doi: 10.1016/j.tplants.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 2.Robert-Seilaniantz A., Navarro L., Bari R., Jones J.D. Pathological hormone imbalances. Curr. Opin. Plant Biol. 2007;10:372–379. doi: 10.1016/j.pbi.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 3.Finkelstein R.R., Gampala S.S., Rock C.D. Abscisic acid signaling in seeds and seedlings. Plant Cell. 2002;14:S15–S45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Popko J., Hansch R., Mendel R.R., Polle A., Teichmann T. The role of abscisic acid and auxin in the response of poplar to abiotic stress. Plant Biol. (Stuttg.) 2010;12:242–258. doi: 10.1111/j.1438-8677.2009.00305.x. [DOI] [PubMed] [Google Scholar]
- 5.Wilkinson S., Davies W.J. Drought, ozone, ABA and ethylene: New insights from cell to plant to community. Plant Cell Environ. 2010;33:510–525. doi: 10.1111/j.1365-3040.2009.02052.x. [DOI] [PubMed] [Google Scholar]
- 6.Chinnusamy V., Gong Z., Zhu J.K. Abscisic acid-mediated epigenetic processes in plant development and stress responses. J. Integr. Plant Biol. 2008;50:1187–1195. doi: 10.1111/j.1744-7909.2008.00727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sirichandra C., Davanture M., Turk B.E., Zivy M., Valot B., Leung J., Merlot S. The Arabidopsis ABA-activated kinase OST1 phosphorylates the bZIP transcription factor ABF3 and creates a 14-3-3 binding site involved in its turnover. PLoS ONE. 2010;5:e13935. doi: 10.1371/journal.pone.0013935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Assmann S.M. OPEN STOMATA1 opens the door to ABA signaling in Arabidopsis guard cells. Trends Plant Sci. 2003;8:151–153. doi: 10.1016/S1360-1385(03)00052-9. [DOI] [PubMed] [Google Scholar]
- 9.Cutler S.R., Rodriguez P.L., Finkelstein R.R., Abrams S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 10.Hubbard K.E., Nishimura N., Hitomi K., Getzoff E.D., Schroeder J.I. Early abscisic acid signal transduction mechanisms: Newly discovered components and newly emerging questions. Genes Dev. 2010;24:1695–1708. doi: 10.1101/gad.1953910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee S.C., Luan S. ABA signal transduction at the crossroad of biotic and abiotic stress responses. Plant Cell Environ. 2012;35:53–60. doi: 10.1111/j.1365-3040.2011.02426.x. [DOI] [PubMed] [Google Scholar]
- 12.Lim C.W., Luan S., Lee S.C. A prominent role for RCAR3-mediated ABA signaling in response to Pseudomonas syringae pv. tomato DC3000 infection in Arabidopsis. Plant Cell Physiol. 2014;55:1691–1703. doi: 10.1093/pcp/pcu100. [DOI] [PubMed] [Google Scholar]
- 13.Wasilewska A., Vlad F., Sirichandra C., Redko Y., Jammes F., Valon C., dit Frey N.F., Leung J. An update on abscisic acid signaling in plants and more. Mol. Plant. 2008;1:198–217. doi: 10.1093/mp/ssm022. [DOI] [PubMed] [Google Scholar]
- 14.Nambara E., Marion-Poll A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 2005;56:165–185. doi: 10.1146/annurev.arplant.56.032604.144046. [DOI] [PubMed] [Google Scholar]
- 15.Nilson S.E., Assmann S.M. The control of transpiration. Insights from Arabidopsis. Plant Physiol. 2007;143:19–27. doi: 10.1104/pp.106.093161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iuchi S., Kobayashi M., Taji T., Naramoto M., Seki M., Kato T., Tabata S., Kakubari Y., Yamaguchi-Shinozaki K., Shinozaki K. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 2001;27:325–333. doi: 10.1046/j.1365-313x.2001.01096.x. [DOI] [PubMed] [Google Scholar]
- 17.Schwartz S.H., Qin X., Zeevaart J.A. Elucidation of the indirect pathway of abscisic acid biosynthesis by mutants, genes, and enzymes. Plant Physiol. 2003;131:1591–1601. doi: 10.1104/pp.102.017921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwartz S.H., Tan B.C., Gage D.A., Zeevaart J.A., McCarty D.R. Specific oxidative cleavage of carotenoids by VP14 of maize. Science. 1997;276:1872–1874. doi: 10.1126/science.276.5320.1872. [DOI] [PubMed] [Google Scholar]
- 19.Tan B.C., Schwartz S.H., Zeevaart J.A., McCarty D.R. Genetic control of abscisic acid biosynthesis in maize. Proc. Natl. Acad. Sci. USA. 1997;94:12235–12240. doi: 10.1073/pnas.94.22.12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson A.J., Jackson A.C., Symonds R.C., Mulholland B.J., Dadswell A.R., Blake P.S., Burbidge A., Taylor I.B. Ectopic expression of a tomato 9-cis-epoxycarotenoid dioxygenase gene causes over-production of abscisic acid. Plant J. 2000;23:363–374. doi: 10.1046/j.1365-313x.2000.00789.x. [DOI] [PubMed] [Google Scholar]
- 21.Umezawa T., Okamoto M., Kushiro T., Nambara E., Oono Y., Seki M., Kobayashi M., Koshiba T., Kamiya Y., Shinozaki K. CYP707A3, a major ABA 8′-hydroxylase involved in dehydration and rehydration response in Arabidopsis thaliana. Plant J. 2006;46:171–182. doi: 10.1111/j.1365-313X.2006.02683.x. [DOI] [PubMed] [Google Scholar]
- 22.Kushiro T., Okamoto M., Nakabayashi K., Yamagishi K., Kitamura S., Asami T., Hirai N., Koshiba T., Kamiya Y., Nambara E. The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: Key enzymes in ABA catabolism. EMBO J. 2004;23:1647–1656. doi: 10.1038/sj.emboj.7600121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schroeder J.I., Hagiwara S. Cytosolic calcium regulates ion channels in the plasma membrane of Vicia faba guard cells. Nature. 1989;338:427–430. doi: 10.1038/338427a0. [DOI] [Google Scholar]
- 24.Negi J., Matsuda O., Nagasawa T., Oba Y., Takahashi H., Kawai-Yamada M., Uchimiya H., Hashimoto M., Iba K. CO2 regulator SLAC1 and its homologues are essential for anion homeostasis in plant cells. Nature. 2008;452:483–486. doi: 10.1038/nature06720. [DOI] [PubMed] [Google Scholar]
- 25.Pei Z.M., Murata Y., Benning G., Thomine S., Klusener B., Allen G.J., Grill E., Schroeder J.I. Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature. 2000;406:731–734. doi: 10.1038/35021067. [DOI] [PubMed] [Google Scholar]
- 26.Lee S.C., Lan W., Buchanan B.B., Luan S. A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc. Natl. Acad. Sci. USA. 2009;106:21419–21424. doi: 10.1073/pnas.0910601106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vahisalu T., Kollist H., Wang Y.F., Nishimura N., Chan W.Y., Valerio G., Lamminmaki A., Brosche M., Moldau H., Desikan R., et al. SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature. 2008;452:487–491. doi: 10.1038/nature06608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ward J.M., Maser P., Schroeder J.I. Plant ion channels: Gene families, physiology, and functional genomics analysis. Annu. Rev. Physiol. 2009;71:59–82. doi: 10.1146/annurev.physiol.010908.163204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vahisalu T., Puzorjova I., Brosche M., Valk E., Lepiku M., Moldau H., Pechter P., Wang Y.S., Lindgren O., Salojarvi J., et al. Ozone-triggered rapid stomatal response involves the production of reactive oxygen species, and is controlled by SLAC1 and OST1. Plant J. 2010;62:442–453. doi: 10.1111/j.1365-313X.2010.04159.x. [DOI] [PubMed] [Google Scholar]
- 30.Grabov A., Blatt M.R. Membrane voltage initiates Ca2+ waves and potentiates Ca2+ increases with abscisic acid in stomatal guard cells. Proc. Natl. Acad. Sci. USA. 1998;95:4778–4783. doi: 10.1073/pnas.95.8.4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grabov A., Blatt M.R. A steep dependence of inward-rectifying potassium channels on cytosolic free calcium concentration increase evoked by hyperpolarization in guard cells. Plant Physiol. 1999;119:277–288. doi: 10.1104/pp.119.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamilton D.W., Hills A., Kohler B., Blatt M.R. Ca2+ channels at the plasma membrane of stomatal guard cells are activated by hyperpolarization and abscisic acid. Proc. Natl. Acad. Sci. USA. 2000;97:4967–4972. doi: 10.1073/pnas.080068897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Z.H., Hills A., Lim C.K., Blatt M.R. Dynamic regulation of guard cell anion channels by cytosolic free Ca2+ concentration and protein phosphorylation. Plant J. 2010;61:816–825. doi: 10.1111/j.1365-313X.2009.04108.x. [DOI] [PubMed] [Google Scholar]
- 34.Garcia-Mata C., Lamattina L. Nitric oxide induces stomatal closure and enhances the adaptive plant responses against drought stress. Plant Physiol. 2001;126:1196–1204. doi: 10.1104/pp.126.3.1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia-Mata C., Gay R., Sokolovski S., Hills A., Lamattina L., Blatt M.R. Nitric oxide regulates K+ and Cl− channels in guard cells through a subset of abscisic acid-evoked signaling pathways. Proc. Natl. Acad. Sci. USA. 2003;100:11116–11121. doi: 10.1073/pnas.1434381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sokolovski S., Hills A., Gay R., Garcia-Mata C., Lamattina L., Blatt M.R. Protein phosphorylation is a prerequisite for intracellular Ca2+ release and ion channel control by nitric oxide and abscisic acid in guard cells. Plant J. 2005;43:520–529. doi: 10.1111/j.1365-313X.2005.02471.x. [DOI] [PubMed] [Google Scholar]
- 37.Arnaud D., Hwang I. A Sophisticated network of signaling pathways regulates stomatal defenses to bacterial pathogens. Mol. Plant. 2015;8:566–581. doi: 10.1016/j.molp.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 38.Melotto M., Underwood W., He S.Y. Role of stomata in plant innate immunity and foliar bacterial diseases. Annu. Rev. Phytopathol. 2008;46:101–122. doi: 10.1146/annurev.phyto.121107.104959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McLachlan D.H., Kopischke M., Robatzek S. Gate control: Guard cell regulation by microbial stress. New Phytol. 2014;203:1049–1063. doi: 10.1111/nph.12916. [DOI] [PubMed] [Google Scholar]
- 40.Jones J.D., Dangl J.L. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 41.Monaghan J., Zipfel C. Plant pattern recognition receptor complexes at the plasma membrane. Curr. Opin. Plant Biol. 2012;15:349–357. doi: 10.1016/j.pbi.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 42.Schwessinger B., Zipfel C. News from the frontline: Recent insights into PAMP-triggered immunity in plants. Curr. Opin. Plant Biol. 2008;11:389–395. doi: 10.1016/j.pbi.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 43.Chisholm S.T., Coaker G., Day B., Staskawicz B.J. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 44.Melotto M., Underwood W., Koczan J., Nomura K., He S.Y. Plant stomata function in innate immunity against bacterial invasion. Cell. 2006;126:969–980. doi: 10.1016/j.cell.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 45.Bauer H., Ache P., Lautner S., Fromm J., Hartung W., Al-Rasheid K.A., Sonnewald S., Sonnewald U., Kneitz S., Lachmann N., et al. The stomatal response to reduced relative humidity requires guard cell-autonomous ABA synthesis. Curr. Biol. 2013;23:53–57. doi: 10.1016/j.cub.2012.11.022. [DOI] [PubMed] [Google Scholar]
- 46.Kang J.Y., Choi H.I., Im M.Y., Kim S.Y. Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell. 2002;14:343–357. doi: 10.1105/tpc.010362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frey A., Effroy D., Lefebvre V., Seo M., Perreau F., Berger A., Sechet J., To A., North H.M., Marion-Poll A. Epoxycarotenoid cleavage by NCED5 fine-tunes ABA accumulation and affects seed dormancy and drought tolerance with other NCED family members. Plant J. 2012;70:501–512. doi: 10.1111/j.1365-313X.2011.04887.x. [DOI] [PubMed] [Google Scholar]
- 48.Cao F.Y., Yoshioka K., Desveaux D. The roles of ABA in plant–pathogen interactions. J. Plant Res. 2011;124:489–499. doi: 10.1007/s10265-011-0409-y. [DOI] [PubMed] [Google Scholar]
- 49.Brooks D.M., Bender C.L., Kunkel B.N. The Pseudomonas syringae phytotoxin coronatine promotes virulence by overcoming salicylic acid-dependent defences in Arabidopsis thaliana. Mol. Plant Pathol. 2005;6:629–639. doi: 10.1111/j.1364-3703.2005.00311.x. [DOI] [PubMed] [Google Scholar]
- 50.De Torres-Zabala M., Truman W., Bennett M.H., Lafforgue G., Mansfield J.W., Egea P.R., Bogre L., Grant M. Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. EMBO J. 2007;26:1434–1443. doi: 10.1038/sj.emboj.7601575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., Grill E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 52.Fujii H., Chinnusamy V., Rodrigues A., Rubio S., Antoni R., Park S.Y., Cutler S.R., Sheen J., Rodriguez P.L., Zhu J.K. In vitro reconstitution of an abscisic acid signalling pathway. Nature. 2009;462:660–664. doi: 10.1038/nature08599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fujii H., Verslues P.E., Zhu J.K. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell. 2007;19:485–494. doi: 10.1105/tpc.106.048538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fujii H., Zhu J.K. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. USA. 2009;106:8380–8385. doi: 10.1073/pnas.0903144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Geiger D., Scherzer S., Mumm P., Marten I., Ache P., Matschi S., Liese A., Wellmann C., Al-Rasheid K.A., Grill E., et al. Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc. Natl. Acad. Sci. USA. 2010;107:8023–8028. doi: 10.1073/pnas.0912030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Geiger D., Scherzer S., Mumm P., Stange A., Marten I., Bauer H., Ache P., Matschi S., Liese A., Al-Rasheid K.A., et al. Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc. Natl. Acad. Sci. USA. 2009;106:21425–21430. doi: 10.1073/pnas.0912021106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park S.Y., Fung P., Nishimura N., Jensen D.R., Fujii H., Zhao Y., Lumba S., Santiago J., Rodrigues A., Chow T.F., et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee S.C., Lim C.W., Lan W., He K., Luan S. ABA signaling in guard cells entails a dynamic protein-protein interaction relay from the PYL-RCAR family receptors to ion channels. Mol. Plant. 2013;6:528–538. doi: 10.1093/mp/sss078. [DOI] [PubMed] [Google Scholar]
- 59.McCourt P., Creelman R. The ABA receptors—We report you decide. Curr. Opin. Plant Biol. 2008;11:474–478. doi: 10.1016/j.pbi.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 60.Kitahata N., Nakano T., Kuchitsu K., Yoshida S., Asami T. Biotin-labeled abscisic acid as a probe for investigating abscisic acid binding sites on plasma membranes of barley aleurone protoplasts. Bioorg. Med. Chem. 2005;13:3351–3358. doi: 10.1016/j.bmc.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 61.Pedron J., Brault M., Nake C., Miginiac E. Detection of abscisic-acid-binding proteins in the microsomal protein fraction of Arabidopsis thaliana with abscisic-acid-protein conjugates used as affinity probes. Eur. J. Biochem. 1998;252:385–390. doi: 10.1046/j.1432-1327.1998.2520385.x. [DOI] [PubMed] [Google Scholar]
- 62.Zhang D.P., Chen S.W., Peng Y.B., Shen Y.Y. Abscisic acid-specific binding sites in the flesh of developing apple fruit. J. Exp. Bot. 2001;52:2097–2103. doi: 10.1093/jexbot/52.364.2097. [DOI] [PubMed] [Google Scholar]
- 63.Santiago J., Rodrigues A., Saez A., Rubio S., Antoni R., Dupeux F., Park S.Y., Marquez J.A., Cutler S.R., Rodriguez P.L. Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J. 2009;60:575–588. doi: 10.1111/j.1365-313X.2009.03981.x. [DOI] [PubMed] [Google Scholar]
- 64.Gonzalez-Guzman M., Pizzio G.A., Antoni R., Vera-Sirera F., Merilo E., Bassel G.W., Fernandez M.A., Holdsworth M.J., Perez-Amador M.A., Kollist H., et al. Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell. 2012;24:2483–2496. doi: 10.1105/tpc.112.098574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mosher S., Moeder W., Nishimura N., Jikumaru Y., Joo S.H., Urquhart W., Klessig D.F., Kim S.K., Nambara E., Yoshioka K. The lesion-mimic mutant cpr22 shows alterations in abscisic acid signaling and abscisic acid insensitivity in a salicylic acid-dependent manner. Plant Physiol. 2010;152:1901–1913. doi: 10.1104/pp.109.152603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nishimura N., Hitomi K., Arvai A.S., Rambo R.P., Hitomi C., Cutler S.R., Schroeder J.I., Getzoff E.D. Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science. 2009;326:1373–1379. doi: 10.1126/science.1181829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yin P., Fan H., Hao Q., Yuan X., Wu D., Pang Y., Yan C., Li W., Wang J., Yan N. Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nat. Struct. Mol. Biol. 2009;16:1230–1236. doi: 10.1038/nsmb.1730. [DOI] [PubMed] [Google Scholar]
- 68.Santiago J., Dupeux F., Round A., Antoni R., Park S.Y., Jamin M., Cutler S.R., Rodriguez P.L., Marquez J.A. The abscisic acid receptor PYR1 in complex with abscisic acid. Nature. 2009;462:665–668. doi: 10.1038/nature08591. [DOI] [PubMed] [Google Scholar]
- 69.Miyazono K., Miyakawa T., Sawano Y., Kubota K., Kang H.J., Asano A., Miyauchi Y., Takahashi M., Zhi Y., Fujita Y., et al. Structural basis of abscisic acid signalling. Nature. 2009;462:609–614. doi: 10.1038/nature08583. [DOI] [PubMed] [Google Scholar]
- 70.Melcher K., Ng L.M., Zhou X.E., Soon F.F., Xu Y., Suino-Powell K.M., Park S.Y., Weiner J.J., Fujii H., Chinnusamy V., et al. A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature. 2009;462:602–608. doi: 10.1038/nature08613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schweighofer A., Hirt H., Meskiene I. Plant PP2C phosphatases: Emerging functions in stress signaling. Trends Plant Sci. 2004;9:236–243. doi: 10.1016/j.tplants.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 72.Gosti F., Beaudoin N., Serizet C., Webb A.A., Vartanian N., Giraudat J. ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell. 1999;11:1897–1910. doi: 10.1105/tpc.11.10.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Merlot S., Gosti F., Guerrier D., Vavasseur A., Giraudat J. The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 2001;25:295–303. doi: 10.1046/j.1365-313x.2001.00965.x. [DOI] [PubMed] [Google Scholar]
- 74.Szostkiewicz I., Richter K., Kepka M., Demmel S., Ma Y., Korte A., Assaad F.F., Christmann A., Grill E. Closely related receptor complexes differ in their ABA selectivity and sensitivity. Plant J. 2010;61:25–35. doi: 10.1111/j.1365-313X.2009.04025.x. [DOI] [PubMed] [Google Scholar]
- 75.Dharmasiri N., Dharmasiri S., Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 76.Kepinski S., Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435:446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- 77.Kepinski S., Leyser O. Plant development: Auxin in loops. Curr. Biol. 2005;15:R208–R210. doi: 10.1016/j.cub.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 78.Sheard L.B., Tan X., Mao H., Withers J., Ben-Nissan G., Hinds T.R., Kobayashi Y., Hsu F.F., Sharon M., Browse J., et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature. 2010;468:400–405. doi: 10.1038/nature09430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Murase K., Hirano Y., Sun T.P., Hakoshima T. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature. 2008;456:459–463. doi: 10.1038/nature07519. [DOI] [PubMed] [Google Scholar]
- 80.Santner A., Estelle M. Recent advances and emerging trends in plant hormone signalling. Nature. 2009;459:1071–1078. doi: 10.1038/nature08122. [DOI] [PubMed] [Google Scholar]
- 81.Zhang H., Cui F., Wu Y., Lou L., Liu L., Tian M., Ning Y., Shu K., Tang S., Xie Q. The RING finger Ubiquitin E3 ligase SDIR1 targets SDIR1-INTERACTING PROTEIN1 for degradation to modulate the salt stress response and ABA signaling in Arabidopsis. Plant Cell. 2015;27:214–227. doi: 10.1105/tpc.114.134163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Irigoyen M.L., Iniesto E., Rodriguez L., Puga M.I., Yanagawa Y., Pick E., Strickland E., Paz-Ares J., Wei N., de Jaeger G., et al. Targeted degradation of abscisic acid receptors is mediated by the ubiquitin ligase substrate adaptor DDA1 in Arabidopsis. Plant Cell. 2014;26:712–728. doi: 10.1105/tpc.113.122234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Seo K.I., Lee J.H., Nezames C.D., Zhong S., Song E., Byun M.O., Deng X.W. ABD1 is an Arabidopsis DCAF substrate receptor for CUL4-DDB1-based E3 ligases that acts as a negative regulator of abscisic acid signaling. Plant Cell. 2014;26:695–711. doi: 10.1105/tpc.113.119974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bueso E., Rodriguez L., Lorenzo-Orts L., Gonzalez-Guzman M., Sayas E., Munoz-Bertomeu J., Ibanez C., Serrano R., Rodriguez P.L. The single-subunit RING-type E3 ubiquitin ligase RSL1 targets PYL4 and PYR1 ABA receptors in plasma membrane to modulate abscisic acid signaling. Plant J. 2014;80:1057–1071. doi: 10.1111/tpj.12708. [DOI] [PubMed] [Google Scholar]
- 85.Lyzenga W.J., Stone S.L. Abiotic stress tolerance mediated by protein ubiquitination. J. Exp. Bot. 2012;63:599–616. doi: 10.1093/jxb/err310. [DOI] [PubMed] [Google Scholar]
- 86.Ryu M.Y., Cho S.K., Kim W.T. The Arabidopsis C3H2C3-type RING E3 ubiquitin ligase AtAIRP1 is a positive regulator of an abscisic acid-dependent response to drought stress. Plant Physiol. 2010;154:1983–1997. doi: 10.1104/pp.110.164749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bu Q., Li H., Zhao Q., Jiang H., Zhai Q., Zhang J., Wu X., Sun J., Xie Q., Wang D., et al. The Arabidopsis RING finger E3 ligase RHA2a is a novel positive regulator of abscisic acid signaling during seed germination and early seedling development. Plant Physiol. 2009;150:463–481. doi: 10.1104/pp.109.135269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ko J.H., Yang S.H., Han K.H. Upregulation of an Arabidopsis RING-H2 gene, XERICO, confers drought tolerance through increased abscisic acid biosynthesis. Plant J. 2006;47:343–355. doi: 10.1111/j.1365-313X.2006.02782.x. [DOI] [PubMed] [Google Scholar]
- 89.Li H., Jiang H., Bu Q., Zhao Q., Sun J., Xie Q., Li C. The Arabidopsis RING finger E3 ligase RHA2b acts additively with RHA2a in regulating abscisic acid signaling and drought response. Plant Physiol. 2011;156:550–563. doi: 10.1104/pp.111.176214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Park G.G., Park J.J., Yoon J., Yu S.N., An G. A RING finger E3 ligase gene, Oryza sativa Delayed Seed Germination 1 (OsDSG1), controls seed germination and stress responses in rice. Plant Mol. Biol. 2010;74:467–478. doi: 10.1007/s11103-010-9687-3. [DOI] [PubMed] [Google Scholar]
- 91.Lee J.H., Yoon H.J., Terzaghi W., Martinez C., Dai M., Li J., Byun M.O., Deng X.W. DWA1 and DWA2, two Arabidopsis DWD protein components of CUL4-based E3 ligases, act together as negative regulators in ABA signal transduction. Plant Cell. 2010;22:1716–1732. doi: 10.1105/tpc.109.073783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cheng M.C., Hsieh E.J., Chen J.H., Chen H.Y., Lin T.P. Arabidopsis RGLG2, functioning as a RING E3 ligase, interacts with AtERF53 and negatively regulates the plant drought stress response. Plant Physiol. 2012;158:363–375. doi: 10.1104/pp.111.189738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang X., Garreton V., Chua N.H. The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev. 2005;19:1532–1543. doi: 10.1101/gad.1318705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu H., Stone S.L. Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. Plant Cell. 2010;22:2630–2641. doi: 10.1105/tpc.110.076075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim S., Choi H.I., Ryu H.J., Park J.H., Kim M.D., Kim S.Y. ARIA, an Arabidopsis arm repeat protein interacting with a transcriptional regulator of abscisic acid-responsive gene expression, is a novel abscisic acid signaling component. Plant Physiol. 2004;136:3639–3648. doi: 10.1104/pp.104.049189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Daszkowska-Golec A., Szarejko I. Open or close the gate—Stomata action under the control of phytohormones in drought stress conditions. Front. Plant Sci. 2013;4:138. doi: 10.3389/fpls.2013.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.He P., Shan L., Sheen J. Elicitation and suppression of microbe-associated molecular pattern-triggered immunity in plant–microbe interactions. Cell. Microbiol. 2007;9:1385–1396. doi: 10.1111/j.1462-5822.2007.00944.x. [DOI] [PubMed] [Google Scholar]
- 98.Torres M.A., Dangl J.L., Jones J.D. Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA. 2002;99:517–522. doi: 10.1073/pnas.012452499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Groom Q.J., Torres M.A., Fordham-Skelton A.P., Hammond-Kosack K.E., Robinson N.J., Jones J.D. rbohA, a rice homologue of the mammalian gp91phox respiratory burst oxidase gene. Plant J. 1996;10:515–522. doi: 10.1046/j.1365-313X.1996.10030515.x. [DOI] [PubMed] [Google Scholar]
- 100.Kwak J.M., Mori I.C., Pei Z.M., Leonhardt N., Torres M.A., Dangl J.L., Bloom R.E., Bodde S., Jones J.D., Schroeder J.I. NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 2003;22:2623–2633. doi: 10.1093/emboj/cdg277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vlad F., Rubio S., Rodrigues A., Sirichandra C., Belin C., Robert N., Leung J., Rodriguez P.L., Lauriere C., Merlot S. Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell. 2009;21:3170–3184. doi: 10.1105/tpc.109.069179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ding Y., Li H., Zhang X., Xie Q., Gong Z., Yang S. OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis. Dev. Cell. 2015;32:278–289. doi: 10.1016/j.devcel.2014.12.023. [DOI] [PubMed] [Google Scholar]
- 103.Anderberg R.J., Walker-Simmons M.K. Isolation of a wheat cDNA clone for an abscisic acid-inducible transcript with homology to protein kinases. Proc. Natl. Acad. Sci. USA. 1992;89:10183–10187. doi: 10.1073/pnas.89.21.10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Johnson R.R., Wagner R.L., Verhey S.D., Walker-Simmons M.K. The abscisic acid-responsive kinase PKABA1 interacts with a seed-specific abscisic acid response element-binding factor, TaABF, and phosphorylates TaABF peptide sequences. Plant Physiol. 2002;130:837–846. doi: 10.1104/pp.001354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gomez-Cadenas A., Verhey S.D., Holappa L.D., Shen Q., Ho T.H., Walker-Simmons M.K. An abscisic acid-induced protein kinase, PKABA1, mediates abscisic acid-suppressed gene expression in barley aleurone layers. Proc. Natl. Acad. Sci. USA. 1999;96:1767–1772. doi: 10.1073/pnas.96.4.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Johnson R.R., Shin M., Shen J.Q. The wheat PKABA1-interacting factor TaABF1 mediates both abscisic acid-suppressed and abscisic acid-induced gene expression in bombarded aleurone cells. Plant Mol. Biol. 2008;68:93–103. doi: 10.1007/s11103-008-9354-0. [DOI] [PubMed] [Google Scholar]
- 107.Mustilli A.C., Merlot S., Vavasseur A., Fenzi F., Giraudat J. Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell. 2002;14:3089–3099. doi: 10.1105/tpc.007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yoshida R., Hobo T., Ichimura K., Mizoguchi T., Takahashi F., Aronso J., Ecker J.R., Shinozaki K. ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol. 2002;43:1473–1483. doi: 10.1093/pcp/pcf188. [DOI] [PubMed] [Google Scholar]
- 109.Fujita Y., Nakashima K., Yoshida T., Katagiri T., Kidokoro S., Kanamori N., Umezawa T., Fujita M., Maruyama K., Ishiyama K., et al. Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 2009;50:2123–2132. doi: 10.1093/pcp/pcp147. [DOI] [PubMed] [Google Scholar]
- 110.Furihata T., Maruyama K., Fujita Y., Umezawa T., Yoshida R., Shinozaki K., Yamaguchi-Shinozaki K. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. USA. 2006;103:1988–1993. doi: 10.1073/pnas.0505667103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Boudsocq M., Barbier-Brygoo H., Lauriere C. Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J. Biol. Chem. 2004;279:41758–41766. doi: 10.1074/jbc.M405259200. [DOI] [PubMed] [Google Scholar]
- 112.Nakashima K., Fujita Y., Kanamori N., Katagiri T., Umezawa T., Kidokoro S., Maruyama K., Yoshida T., Ishiyama K., Kobayashi M., et al. Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 2009;50:1345–1363. doi: 10.1093/pcp/pcp083. [DOI] [PubMed] [Google Scholar]
- 113.Kerk D., Bulgrien J., Smith D.W., Barsam B., Veretnik S., Gribskov M. The complement of protein phosphatase catalytic subunits encoded in the genome of Arabidopsis. Plant Physiol. 2002;129:908–925. doi: 10.1104/pp.004002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xue T., Wang D., Zhang S., Ehlting J., Ni F., Jakab S., Zheng C., Zhong Y. Genome-wide and expression analysis of protein phosphatase 2C in rice and Arabidopsis. BMC Genomics. 2008;9:550. doi: 10.1186/1471-2164-9-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rubio S., Rodrigues A., Saez A., Dizon M.B., Galle A., Kim T.H., Santiago J., Flexas J., Schroeder J.I., Rodriguez P.L. Triple loss of function of protein phosphatases type 2C leads to partial constitutive response to endogenous abscisic acid. Plant Physiol. 2009;150:1345–1355. doi: 10.1104/pp.109.137174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Robert N., Merlot S., N'Guyen V., Boisson-Dernier A., Schroeder J.I. A hypermorphic mutation in the protein phosphatase 2C HAB1 strongly affects ABA signaling in Arabidopsis. FEBS Lett. 2006;580:4691–4696. doi: 10.1016/j.febslet.2006.07.047. [DOI] [PubMed] [Google Scholar]
- 117.Saez A., Apostolova N., Gonzalez-Guzman M., Gonzalez-Garcia M.P., Nicolas C., Lorenzo O., Rodriguez P.L. Gain-of-function and loss-of-function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signalling. Plant J. 2004;37:354–369. doi: 10.1046/j.1365-313X.2003.01966.x. [DOI] [PubMed] [Google Scholar]
- 118.Kuhn J.M., Boisson-Dernier A., Dizon M.B., Maktabi M.H., Schroeder J.I. The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA. Plant Physiol. 2006;140:127–139. doi: 10.1104/pp.105.070318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nishimura N., Yoshida T., Murayama M., Asami T., Shinozaki K., Hirayama T. Isolation and characterization of novel mutants affecting the abscisic acid sensitivity of Arabidopsis germination and seedling growth. Plant Cell Physiol. 2004;45:1485–1499. doi: 10.1093/pcp/pch171. [DOI] [PubMed] [Google Scholar]
- 120.Saez A., Robert N., Maktabi M.H., Schroeder J.I., Serrano R., Rodriguez P.L. Enhancement of abscisic acid sensitivity and reduction of water consumption in Arabidopsis by combined inactivation of the protein phosphatases type 2C ABI1 and HAB1. Plant Physiol. 2006;141:1389–1399. doi: 10.1104/pp.106.081018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yoshida T., Nishimura N., Kitahata N., Kuromori T., Ito T., Asami T., Shinozaki K., Hirayama T. ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol. 2006;140:115–126. doi: 10.1104/pp.105.070128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Koornneef M., Reuling G., Karssen C.M. The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol. Plant. 1984;61:377–383. doi: 10.1111/j.1399-3054.1984.tb06343.x. [DOI] [Google Scholar]
- 123.Allen G.J., Kuchitsu K., Chu S.P., Murata Y., Schroeder J.I. Arabidopsis abi1-1 and abi2-1 phosphatase mutations reduce abscisic acid-induced cytoplasmic calcium rises in guard cells. Plant Cell. 1999;11:1785–1798. doi: 10.1105/tpc.11.9.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Finkelstein R.R., Somerville C.R. Three classes of abscisic acid (ABA)-insensitive mutations of Arabidopsis define genes that control overlapping subsets of ABA responses. Plant Physiol. 1990;94:1172–1179. doi: 10.1104/pp.94.3.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Leung J., Merlot S., Giraudat J. The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell. 1997;9:759–771. doi: 10.1105/tpc.9.5.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bhaskara G.B., Nguyen T.T., Verslues P.E. Unique drought resistance functions of the highly ABA-induced clade A protein phosphatase 2Cs. Plant Physiol. 2012;160:379–395. doi: 10.1104/pp.112.202408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Umezawa T., Sugiyama N., Mizoguchi M., Hayashi S., Myouga F., Yamaguchi-Shinozaki K., Ishihama Y., Hirayama T., Shinozaki K. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2009;106:17588–17593. doi: 10.1073/pnas.0907095106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yoshida R., Umezawa T., Mizoguchi T., Takahashi S., Takahashi F., Shinozaki K. The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J. Biol. Chem. 2006;281:5310–5318. doi: 10.1074/jbc.M509820200. [DOI] [PubMed] [Google Scholar]
- 129.Montillet J.L., Leonhardt N., Mondy S., Tranchimand S., Rumeau D., Boudsocq M., Garcia A.V., Douki T., Bigeard J., Lauriere C., et al. An abscisic acid-independent oxylipin pathway controls stomatal closure and immune defense in Arabidopsis. PLoS Biol. 2013;11:e1001513. doi: 10.1371/journal.pbio.1001513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cho S.M., Kang B.R., Han S.H., Anderson A.J., Park J.Y., Lee Y.H., Cho B.H., Yang K.Y., Ryu C.M., Kim Y.C. 2R,3R-butanediol, a bacterial volatile produced by Pseudomonas chlororaphis O6, is involved in induction of systemic tolerance to drought in Arabidopsis thaliana. Mol. Plant Microbe Interact. 2008;21:1067–1075. doi: 10.1094/MPMI-21-8-1067. [DOI] [PubMed] [Google Scholar]
- 131.Zeng W., He S.Y. A prominent role of the flagellin receptor FLAGELLIN-SENSING2 in mediating stomatal response to Pseudomonas syringae pv tomato DC3000 in Arabidopsis. Plant Physiol. 2010;153:1188–1198. doi: 10.1104/pp.110.157016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zeng W., Melotto M., He S.Y. Plant stomata: A checkpoint of host immunity and pathogen virulence. Curr. Opin. Biotechnol. 2010;21:599–603. doi: 10.1016/j.copbio.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Leon-Kloosterziel K.M., Gil M.A., Ruijs G.J., Jacobsen S.E., Olszewski N.E., Schwartz S.H., Zeevaart J.A., Koornneef M. Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J. 1996;10:655–661. doi: 10.1046/j.1365-313X.1996.10040655.x. [DOI] [PubMed] [Google Scholar]
- 134.Lim C.W., Lee S.C. Arabidopsis abscisic acid receptors play an important role in disease resistance. Plant Mol. Biol. 2015;88:313–324. doi: 10.1007/s11103-015-0330-1. [DOI] [PubMed] [Google Scholar]
- 135.Miyakawa T., Fujita Y., Yamaguchi-Shinozaki K., Tanokura M. Structure and function of abscisic acid receptors. Trends Plant Sci. 2013;18:259–266. doi: 10.1016/j.tplants.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 136.Anderson J.P., Badruzsaufari E., Schenk P.M., Manners J.M., Desmond O.J., Ehlert C., Maclean D.J., Ebert P.R., Kazan K. Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell. 2004;16:3460–3479. doi: 10.1105/tpc.104.025833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Asselbergh B., de Vleesschauwer D., Hofte M. Global switches and fine-tuning-ABA modulates plant pathogen defense. Mol. Plant Microbe Interact. 2008;21:709–719. doi: 10.1094/MPMI-21-6-0709. [DOI] [PubMed] [Google Scholar]
- 138.Pieterse C.M., Leon-Reyes A., van der Ent S., van Wees S.C. Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 2009;5:308–316. doi: 10.1038/nchembio.164. [DOI] [PubMed] [Google Scholar]
- 139.Alvarez M.E., Pennell R.I., Meijer P.J., Ishikawa A., Dixon R.A., Lamb C. Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell. 1998;92:773–784. doi: 10.1016/S0092-8674(00)81405-1. [DOI] [PubMed] [Google Scholar]
- 140.Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005;43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- 141.Xiong L., Yang Y. Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid-inducible mitogen-activated protein kinase. Plant Cell. 2003;15:745–759. doi: 10.1105/tpc.008714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Tanaka Y., Sano T., Tamaoki M., Nakajima N., Kondo N., Hasezawa S. Ethylene inhibits abscisic acid-induced stomatal closure in Arabidopsis. Plant Physiol. 2005;138:2337–2343. doi: 10.1104/pp.105.063503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Merritt F., Kemper A., Tallman G. Inhibitors of ethylene synthesis inhibit auxin-induced stomatal opening in epidermis detached from leaves of Vicia faba L. Plant Cell Physiol. 2001;42:223–230. doi: 10.1093/pcp/pce030. [DOI] [PubMed] [Google Scholar]
- 144.Huang D., Wu W., Abrams S.R., Cutler A.J. The relationship of drought-related gene expression in Arabidopsis thaliana to hormonal and environmental factors. J. Exp. Bot. 2008;59:2991–3007. doi: 10.1093/jxb/ern155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Nemhauser J.L., Hong F., Chory J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell. 2006;126:467–475. doi: 10.1016/j.cell.2006.05.050. [DOI] [PubMed] [Google Scholar]
- 146.Zheng Z., Xu X., Crosley R.A., Greenwalt S.A., Sun Y., Blakeslee B., Wang L., Ni W., Sopko M.S., Yao C., et al. The protein kinase SnRK2.6 mediates the regulation of sucrose metabolism and plant growth in Arabidopsis. Plant Physiol. 2010;153:99–113. doi: 10.1104/pp.109.150789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wilson I.D., Ribeiro D.M., Bright J., Confraria A., Harrison J., Barros R.S., Desikan R., Neill S.J., Hancock J.T. Role of nitric oxide in regulating stomatal apertures. Plant Signal. Behav. 2009;4:467–469. doi: 10.4161/psb.4.5.8545. [DOI] [PMC free article] [PubMed] [Google Scholar]