Abstract
The endophenotype concept was first proposed as a strategy to use (purportedly) genetically simpler phenotypes in gene identification studies for psychiatric disorders, and is distinct from the closely related concept of intermediate phenotypes. In the area of alcohol use disorder (AUD) research, two candidate endophenotypes have produced replicable genetic associations: level of response to alcohol and neurophysiology markers (e.g., event-related oscillations and event-related potentials). Additional candidate endophenotypes from the cognitive, sensory, and neuroimaging literatures show promise, although more evidence is needed to fully evaluate their potential utility. Translational approaches to AUD endophenotypes have helped characterize the underlying neurobiology and genetics of AUD endophenotypes and identified relevant pharmacological interventions. Future research that capitalizes on the polygenic nature of endophenotypes and emphasizes endophenotypes that may change across development will enhance the usefulness of this concept to understand the genetically-influenced pathways toward AUD.
Keywords: Endophenotypes, alcohol use disorder, addictions, sweet liking, genetics, genomics, delayed reward discounting, brain structure, brain function
Alcohol use disorder (AUD) is genetically complex (i.e., caused by more than one gene) and behaviorally heterogeneous. In view of this heterogeneity, it has been suggested that “alcoholism cannot be reified but reflects a collection of various symptoms and episodic behaviors that collectively make up perhaps as many alcoholisms as there are alcohol abusers” [1]. Against this backdrop, identifying replicable genetic associations for AUD has been challenging.
The concept of using endophenotypes to aid in gene identification was first introduced to the field of psychopathology by Gottesman and Shields [2]. The idea gained widespread attention after Gottesman and Gould [3] reintroduced the concept and argued that psychiatric classification systems by their very nature create heterogeneous groups of affected individuals, and that this heterogeneity hampers our ability to detect susceptibility genes. Further, these broad binary classifications are quite distal from the level of gene action; surely there is no gene “for” AUD, rather, genes affect certain biochemical processes and pathways that alter susceptibility. Since then, there has been a proliferation of interest in endophenotypes across a range of psychiatric disorders [4–6], including AUD [7–11]. A PUBMED search of the terms “endophenotype” plus “alcohol” produces over 150 results (as of November 2014).
Although there are several excellent reviews of specific candidate endophenotypes for AUD, such as subjective responses to alcohol [8] and neurophysiological markers such as brain oscillations [12], as well as novel candidate neurobiological endophenotypes that distinguish between different stages in the development of alcohol dependence [13], there has not been a recent integrative update on where the field stands with respect to using endophenotypes to aid in gene identification for AUD. Our goal here is threefold. First, we evaluate the weight of evidence for various candidate endophenotypes for AUD (including their previously documented genetic associations) and highlight promising candidate endophenotypes from the cognitive/psychological, sensory, and structural neuroimaging domains. Second, we provide illustrative examples of successes and challenges in validating endophenotypes in animal models. Third, we identify themes to guide future research on endophenotypes for AUD.
Evaluating the Weight of Evidence for AUD Candidate Endophenotypes
Gottesman and Gould defined endophenotypes as “measurable components unseen by the unaided eye along the pathways between disease and distal genotype”, and argued that endophenotypes should be “simpler clues to genetic underpinnings than the disease syndrome itself” [2], although it has been more recently recognized that there is likely a gradient of endophenotypes, some of which are closer to gene action and others that are closer to the phenotype [14]. They delineated five criteria: A candidate endophenotype should be (1) associated with illness; (2) heritable; (3) state-independent (present whether or not illness is active); (4) co-segregate with illness within families; and (5) found in a higher rate in the unaffected relatives of affected individuals than in the general population [3]. Others have agreed that endophenotypes should reflect causes rather than effects of disorders, and suggest that endophenotypes should be measured quantitatively [15, 16].
The endophenotype concept is similar to, but distinct from, related concepts such as biomarkers and intermediate phenotypes [17]. Biomarkers refer to measurable indicators of a disease state. As noted by Lenzenweger [17], biomarkers are associated with the disease, but do not necessarily reflect a genetically influenced pathway. For example, in a biomedical context the ratio of aspartate aminotransferase to alanine aminotransferase can be used as a biomarker of alcoholic liver disease. In this case, the biomarker is associated with the disease, but does not reflect a genetically influenced enduring vulnerability to the disease. According to Rasetti and Weinberger, an intermediate phenotype is “a heritable trait that is located in the path of pathogenesis from genetic predisposition to psychopathology” [18]. This concept has been critiqued on account of its ambiguity with respect to where “intermediate” phenotypes lie along the pathway from genes → disorder, which has implications for level of analysis [17]. Thus, although the endophenotype, biomarker, and intermediate phenotype concepts share overlapping goals of clarifying heterogeneity, the terms are not interchangeable. For an extended discussion of these definitional issues, we refer interested readers to Lenzenweger [17].
Flint and Munafo noted that the “endophenotypes” that have been used in studies of different psychiatric diseases typically fell into six categories: anatomical, developmental, electrophysiological, metabolic, sensory, and psychological/cognitive [19], providing a useful framework for classifying potential endophenotypes. We adopted this framework to organize the evidence for a number of AUD candidate endophenotypes according to each of the five Gottesman & Gould [3] criteria (Table 1) as well as genes, regions of interest, and gene sets associated with candidate endophenotypes (summarized in Table 2 and visualized as part of a gene-environment interplay system in Figure 1). We also added a “functional neuroimaging” category in view of the growing number of functional brain candidate endophenotypes for AUD [13]. As Table 1 illustrates, the evidence for many of the strict endophenotype criteria is sparse at present. The two candidate AUD endophenotypes for which there is the greatest evidence, and which have generated the most genetic associations, are neurophysiological phenotypes and level of response to alcohol [for detailed reviews of these as candidate endophenotypes, see 8, 12].
Table 1.
Weight of evidence for candidate endophenotypes for alcohol use disorder, organized by category (anatomical, developmental, electrophysiological, metabolic, sensory, psychological/cognitive, and neuroimaging) and criterion.
| Candidate endophenotype | Endophenotype criterion
|
||||
|---|---|---|---|---|---|
| Associated with AUD | Heritable | State-independent | Co-segregates with illness within families | Presents in nonaffected family members > population | |
| Anatomical | |||||
| Structural brain differences | Boutte et al. [89], Agartz et al. [90], Monnig et al. [137] | Baare et al. [138], Posthuma et al. [139] | Fein, Fein [140], Cheetham et al. [141] | Benegal et al. [91] | |
| Developmental | |||||
| Externalizing behavior | Benegal et al. [91] | Young et al. [67] | Englund et al. [142] | Benegal et al. [91] | |
| Electrophysiological | |||||
| Resting and event-related EEG phenotypes | Ehlers, Phillips [143] Carlson et al. [144]; Roopesh et al. [145], Roopesh et al. [146] | van Beijsterveldt et al. [20] Perlman et al. [147] |
Carlson et al. [144] | c.f. Carlson et al. [144]; c.f. Carlson et al. [32] | Rangaswamy et al. [21] |
| Metabolic | |||||
| HPA axis activity | Wand, Dobs [148], Starcke et al. [149] | Bartels et al. [150] | |||
| Sensory | |||||
| Sweet liking | Kampov-Polevoy et al. [81]; c.f. Kranzler et al. [151] | Keskitalo et al. [152] | Mennella et al. [83], Lange et al. [153] | ||
| Psychological/Cognitive | |||||
| Executive functions | Sjoerds et al. [76] | Friedman et al. [75] | Benzerouk et al. [78], Gierski et al. [77] | ||
| Delayed reward discounting | Mitchell et al. [72] | Anokhin et al. [71] | Petry et al. [73] | ||
| Low subjective response to alcohol (LR) | Schuckit et al. [43], Schuckit et al. [42], Schuckit et al. [44] | Schuckit et al. [154] | Schuckit et al. [43] | Schuckit et al. [43] | |
| Functional Neuroimaging | |||||
| Functional brain differences during spatial working memory tasks | Pfefferbaum et al. [155], Vollstadt-Klein et al. [156], Tapert et al. [157] | Spadoni et al. [133], Spadoni et al. [158] | |||
Notes.
Does not meet endophenotype definition itself, but other candidate endophenotypes (e.g., executive functions, delayed reward discounting) are likely to contribute to individual differences in externalizing across development.
May also be considered a sensory/motor endophenotype. Studies listed should be considered illustrative and not exhaustive.
Table 2.
Genes, regions of interest, and gene sets associated with illustrative candidate endophenotypes.
| Candidate endophenotype | Chromosome | Gene | Potential relevance | References |
|---|---|---|---|---|
| Low subjective response to alcohol | 4 | GABRA2 | Implicated in central nervous system disinhibition | Uhart et al. [48] |
| 6 | OPRM1 | Endogenous opioid system involved in alcohol-induced reward | Ray et al. [50] Ray, Hutchison [51] Ehlers et al. [52] |
|
| 10 | CYP2E1 | Codes CYP2E1 enzyme; involved in ethanol metabolism | Webb et al. [159] | |
| 12 | ALDH2 | Involved in alcohol metabolism; implicated in conversion of acetaldehyde to acetate | Luczak et al. [54] | |
| 15 | CHRNA5-CHRNA3-CHRNB4 genes | Nicotinic acetylcholine receptor genes | Joslyn et al. [55] | |
| 17 | SERT | Involved in serotonin transmission | Hu et al. [49] | |
| multiple | gene sets implicated in neuronal signaling | -- | Joslyn et al. [57] | |
| regions of interest on chromosomes 6, 10, 12, and 17 | -- | -- | Ehlers et al. [53] | |
| EEG variables | 13 | GPC5 | Codes for a cell surface proteoglycan implicated in neural development | Joslyn et al. [101] |
| 2 | GBX2 | Involved in midbrain/hindbrain development and influences the expression of other genes during embryogenesis | Vrieze et al. [28] | |
| 4 | GABRA2 | Implicated in central nervous system disinhibition | Porjesz et al. [22] | |
| 5 | CRH-BP | Codes for a high affinity binding protein for corticotrophin releasing hormone; involved in stress response | Enoch et al. [27] | |
| 7 | CHRM2 | Influences the effects of acetylcholine in the central and peripheral nervous system | Jones et al. [35] | |
| 7 | GRM8 | Involved in glutamatergic system | Chen et al. [36] | |
| 10 | HTR7 | Involved in serotonergic system | Zlojutro et al. [160] | |
| 15 | MYEF2 | Transcriptional repressor of the myelin basic protein gene | Malone et al. [120] | |
| 17 | CRHR1 | Involved in the neuroendocrine stress response | Chen et al. [37] | |
| 21 | KCNJ6 | Codes for the GIRK2 protein involved in dopaminergic signaling | Kang et al. [38] | |
| Executive functions | 11 | BDNF | Codes for BDNF protein involved in brain development and synaptic plasticity | Benzerouk et al. [78] |
| Structural brain differences | 11 | 11q14.2 | -- | Boutte et al. [89] |
| Sweet liking | 1 | TAS1R1, TAS1R2, TAS1R3 | Involved in sweet taste reception | Bachmanov et al. [84] |
| 16 | 16p11.2 | -- | Keskitalo et al. [80] |
Notes. -- indicates not applicable or unknown. Studies listed should be considered illustrative and not exhaustive.
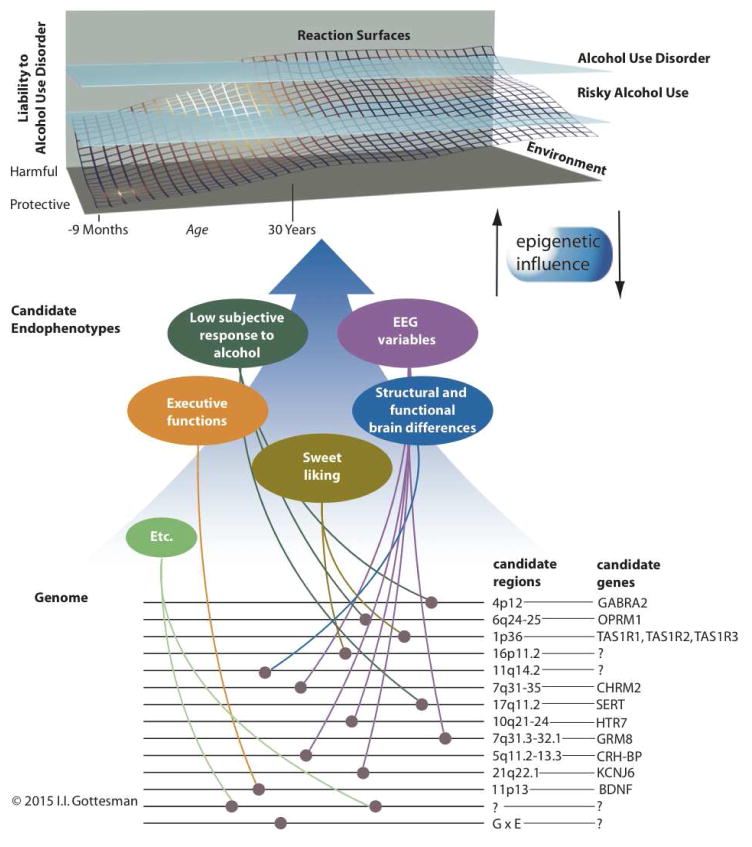
Figure 1.
Depiction of the interplay among genetic and environmental factors that contribute to liability for alcohol use disorder (AUD), and the contribution of candidate endophenotypes and their associated genes and gene regions. Genes and gene regions that are associated with AUD candidate endophenotypes are highlighted here (references are documented in Table 2). These should be considered illustrative and not exhaustive. The “reaction surface” represents the probabilistic interplay among genetic and environmental factors in the development of risky alcohol use (i.e., consuming alcohol in quantities that put individuals at risk for alcohol-related harms) and AUD. Figure adapted from Gottesman & Gould [3] and used with permission.
Neurophysiology
Numerous dimensions of resting and event-related EEG measures (e.g., alpha, theta, and beta oscillations) broadly index information processing and cognitive functioning. Neurophysiological measures are highly heritable [20], and individuals affected with AUD and individuals at high-risk for AUD (offspring of male alcoholics) have elevated resting high-frequency (beta; 12–28 Hz) brain oscillations [21] compared to unaffected and low-risk individuals.
A genome-wide linkage study of EEG beta power in the high-risk Collaborative Study on the Genetics of Alcoholism (COGA) sample found a linkage peak (i.e., a statistical indication that a particular section of a chromosome co-segregates with the trait within families) over a GABAA receptor gene (GABRA2) on chromosome 4 [22]. Subsequent studies across multiple, independent samples have found evidence for association between alcohol dependence and variation in GABRA2 [23–25] (for a recent exception see [26]). A genome-wide linkage study of power for three frequency bands (alpha, theta, and beta) in a sample of Plains American Indians showed evidence for convergent linkage peaks over the corticotropin releasing hormone binding-protein gene (CRH-BP) on chromosome 5 [27]. In the same study, variants in CRH-BP showed association with AUD in a Caucasian replication sample, and anxiety disorders in the Plains Indians, suggesting that CRH-BP may have pleiotropic effects (i.e., associations with multiple disorders). More recently, gene-based tests from a whole-genome sequencing study of EEG beta power identified the gastrulation brain homeobox 2 gene (GBX2) on chromosome 2; however, it is unknown whether this gene is associated with AUD [28].
In another neurophysiology example, the amplitude of the event-related potential P3 wave, which indexes orientation toward novel events and inhibition of ongoing cognitive processing, is reduced in individuals with AUD (and externalizing disorders more generally) and in individuals at familial risk for AUD [29–33], especially in male offspring [34]. Linkage analyses of the frontal theta and parietal-occipital delta event-related oscillations (EROs) underlying the P3 component found a linkage peak over the muscarinic acetylcholine receptor M2 (CHRM2) on chromosome 4 [35]. Subsequent association analyses in the COGA sample found significant association among variants in the glutamate receptor, metabotropic 8 (GRM8) gene and theta EROs [36]; variants in the corticotropin releasing hormone receptor 1 gene (CRHR1) on chromosome 17 and P3 amplitude [37]; and variants in KCNJ6 on chromosome 21 and frontal theta oscillations [38]. Variation in CHRM2, GRM8, CRHR1, and KCNJ6 has also been associated with alcohol and/or drug dependence [39, 40, 37, 36].
Level of response
Level of response (LR) to alcohol is the second candidate endophenotype for AUD to meet many of Gottesman & Gould’s [3] criteria, with the exception that, to our knowledge, there is not evidence that it co-segregates with AUD within families. LR [also known as subjective response to alcohol; 41] is the degree to which a person responds to a specific dose of alcohol or the number of drinks an individual needs to produce specific psychological and motor effects, and is distinct from acquired alcohol tolerance [42]. Low LR is hypothesized to confer risk for AUD because individuals who are less sensitive to alcohol must consume larger quantities of it in order to experience its effects. A program of research led by Schuckit and colleagues demonstrates that low LR is associated with increased alcohol use and problems across multiple samples [43, 42, 44]. As summarized in a recent meta-analysis, populations at risk for AUD, such as individuals with a family history of alcoholism, typically have lower LR compared to other populations [45]. Heritability (h2) estimates for LR are approximately 60% [46, 47].
Variation in a number of genes and gene regions is associated with LR. Variation in GABRA2 [48] and in the 5-HTTLPR polymorphism in the serotonin transporter (SERT) gene on chromosome 17 [49] are associated with subjective responses to alcohol and/or body sway in alcohol challenge studies. Variation in the μ opioid receptor gene (OPRM1) on chromosome 6 is associated with subjective responses to alcohol in alcohol infusion studies of problem and [50] non-problem drinkers [51] and self-rated effects of alcohol in a Native American sample [52]. A systematic genome-wide scan in the same Native American sample identified regions of interest on chromosomes 6, 10, 12, and 17 that were associated with participants’ self-reported subjective LR early in their drinking careers [53]. Variation in the aldehyde dehydrogenase gene (ALDH2) on chromosome 12 was associated with self-reported subjective LR early in the drinking careers of a sample of Chinese- and Korean-American college students [54]. Finally, variants in the cholinergic nicotinic receptor gene cluster (CHRNA5-CHRNA3-CHRNB4) on chromosome 15 [55, 56] and genes sets implicated in neuronal signaling [57] were associated with LR in a sample of young adult offspring of alcoholics.
The evidence for association among these genes and AUD is mixed, with the exception of ALDH2’s well-replicated association [58]. A meta-analysis indicates an association between variation in the 5-HTTLPR polymorphism and AUD; however, there is some indication that this reflects publication bias [59]. A meta-analysis of the commonly studied OPRM1 Asn40Asp (A118G) polymorphism indicated no association with substance dependence (opioid, alcohol, nicotine, or cocaine) [60]. There is some evidence for association among variants in the nicotinic receptor gene cluster on chromosome 15 and AUD [61]; however, other studies have reported null effects [62].
Although LR has received much attention as an endophenotype, it is worth noting that some inconsistencies in the alcohol challenge literature have led others to propose a more nuanced “differentiator model” [63] that takes into account subjective and motor responses across the rise and fall of blood alcohol levels. Under this model, individuals at risk for developing an AUD (by virtue of family history) are hypothesized to show acute sensitization to alcohol as blood/breath alcohol level rises, and acute tolerance as blood/breath alcohol level falls. Thus, these individuals are at risk because of two processes: they experience more pleasurable and excitatory effects of alcohol during initial intoxication, and fewer of the sedative effects of alcohol as blood alcohol level declines. A modified version of the differentiator model [64] suggests that these effects are most pronounced at peak breath alcohol concentration. Recent longitudinal work is consistent with this modified differentiator model; individuals who were more sensitive to the stimulant and rewarding effects of alcohol and who were less sensitive to the sedating effects of alcohol at peak breath alcohol concentration in an alcohol challenge study had the highest number of AUD symptoms six years later [65].
Promising Potential Endophenotypes
Although there are only a small number of examples that come close to meeting the strict definition of an endophenotype, there are many additional candidate endophenotypes for AUD for which some criteria are met. This is especially the case for novel promising endophenotypes for which sufficient data are not yet available. We highlight here four psychological/cognitive, sensory, and neuroimaging candidate endophenotypes that show potential.
Delayed reward discounting
AUD shares genetic influences with several other common externalizing disorders (e.g., illicit drug dependence and antisocial behavior) and measures of impulsivity [66, 67]. Disinhibition, or the inability to control one’s impulses, is the central feature shared among these disorders [68]. Impulsivity is a multifaceted construct that encompasses diverse behaviors that are poorly planned, inappropriate, or unnecessarily risky [69]. Impulsivity as a whole is likely not tractable for study as an endophenotype because it does not represent a unitary construct unseen to the unaided eye, but specific features, such as the ability to delay rewards (i.e., exhibit self-control) have been proposed as a cognitive endophenotype for AUD and substance use disorders more generally [70]. Delayed reward discounting is heritable [30–51% in an adolescent sample; 71]. Abstinent alcoholics are less likely to delay rewards [72] and the unaffected adult daughters (but not adult sons) of alcoholic fathers are less likely to delay reward [73]. To date, however, there have not been systematic gene identification efforts for sub-clinical impulsivity phenotypes such as delayed reward discounting, and so its potential to aid in gene identification for AUD is still relatively unknown.
Executive functions
In recent years there has been a proliferation of interest in executive functions (EF) as they relate to alcohol and other substance use disorders. EF refer to an interrelated set of self-regulatory skills and abilities related to goal-directed behavior [74] including attention, working memory, planning, and cognitive flexibility. EF share a highly heritable (99%) common factor [75] and alcohol dependence severity is associated with impaired EF [76]. These EF impairments among alcohol dependent individuals are to be expected given the toxic effects that alcohol has on the frontal lobes; however, it has been proposed that these EF deficits may predate the onset of disorder and qualify as an endophenotype [77]. For example, unaffected relatives of alcohol or drug dependent probands also have lower EF (as measured in tasks tapping cognitive flexibility and inhibition) compared to healthy controls [78, 77]. In one of these samples, analyses of the brain-derived neurotrophic factor (BDNF) Val66Met polymorphism found that the Met allele was associated with lower EF [78]. However, variation in this same polymorphism is not associated with AUD [79].
Sweet liking
From the sensory domain, sweet liking (i.e., preference for sweet tasting foods) is also a strong yet understudied candidate endophenotype. Perceived pleasantness of sweet foods, the frequency of sweet food consumption, and sweet food cravings are moderately heritable (31–50%) [80]. In a taste preference test, alcoholic men preferred higher concentration sucrose solutions compared to non-alcoholic men [81], and paternal history of alcoholism (an indicator of genetic risk) in an inpatient psychiatric sample predicted sweet liking above and beyond subjects’ own alcohol dependence status [82]. Children with a family history of alcoholism also prefer a higher sucrose concentration compared to children without a family history of alcoholism; however, this was only the case for children who were also experiencing depressive symptomatology [83]. Variation in the taste receptor type one family of genes (TAS1R1, TAS1R2, and TAS1R3) influences sweet liking [84] and there is a linkage peak (LOD = 3.5) on chromosome 16 at 16p11.2 (marker D16S753) for frequency of sweet food consumption [80]. However, whether variation in the TAS1 genes and other sweet-liking genomic regions are associated with AUD has not been systematically examined, representing an important direction for future research.
Brain structure
The rapid advances and growing interest in neuroimaging have begun to identify a number of structural brain features that may qualify as endophenotypes for AUD [13]. Disentangling premorbid differences from the effects of alcohol on the brain is an inherent difficulty of work in this area, as it is known that even subclinical alcohol use disrupts typical neurodevelopment [85] and chronic alcohol abuse accelerates loss of white and gray matter volume [86]. Brain structure is highly heritable (e.g., frontal and language-related structures (h2 > 80%) [87] and white matter volume (h2 = 96%) [88]). Alcohol-dependent individuals have reduced volume in the right hippocampus compared to unaffected controls [89, 90]. Brain volume differences are also observed in alcohol-naïve children, adolescents, and young adults who are at high familial risk for AUD, as evidenced by reduced gray matter volumes across multiple brain regions (superior frontal, cingulate and parahippocampal gyri, amygdala, thalamus, and cerebellum) compared to matched controls at low familial risk for AUD [91]. In this same study, smaller gray matter volumes in many of these regions were associated with elevated externalizing symptoms (attention deficit, hyperactivity, conduct and oppositional defiant disorder symptoms), suggesting that these gray matter differences may predispose individuals to a range of externalizing-spectrum problems, some of which have been previously shown to be genetically correlated with AUD [92, 93]. Thus, some structural brain features appear to meet many of the endophenotype criteria. Systematic gene identification studies for these features may provide evidence for association with AUD and thus become important directions for future research.
AUD Endophenotypes in Animal Models
Translational approaches to endophenotypes that include the development of relevant animal models are important [94] for elucidating the underlying neurobiology and genetics of AUD, which may in turn support the development of pharmacological treatments [95]. As highlighted below, the efforts to develop animal models for AUD endophenotypes have had varying degrees of success with respect to these goals.
Neurophysiology
In findings that mirror the human literature, an alcohol preferring (P) mouse strain had a reduced P300 amplitude compared to a non-alcohol preferring (NP) mouse strain [96]. A follow-up study further indicated that reductions in evoked delta event-related oscillations and decreases in delta and theta phase synchrony contributed to this P300 amplitude reduction [97]. Additional studies are needed to identify the genetic differences between these selectively bred mouse models that are associated with these neurophysiological endophenotypes.
Level of response
Rat models suggest that P animals have lower LR compared to NP animals; for example, in a conditioned place aversion test, P rats avoided an alcohol-paired location less than NP rats [98], suggesting that P rats are less sensitive to the aversive effects of alcohol. here have also been attempts to translate the body sway dimension of LR into rodent models; however, the lack of concordance between the human phenotype and rodent models makes development of novel behavioral assays an important area for continued refinement [for a review see 99]. A series of eleven rodent behavioral assays broadly indexing body sway were tested across eight inbred mouse strains. Interestingly, there was little genetic correlation across the behavioral assays, suggesting that unique sets of genes contribute to foot slippage and wobbly gait, for example [100]. The degree to which unique sets of genes contribute to the component processes of human body sway is unknown. Despite these challenges to phenotype consilience across species, convergent evidence from cross-species (humans, mice, and fruit flies) analyses of locomotor responses to ethanol implicated the glypican gene GPC5 on chromosome 13 [101].
Delayed reward discounting
Delayed reward discounting tasks are very similar in human and non-human animals, making this an ideal translational endophenotype [70]. In findings that mirror the human literature, P mice were more impulsive than NP mice in a delayed reward discounting task [102]. Outbred mice that more steeply discounted delayed rewards also displayed more sensitivity to the stimulant effects of ethanol after repeated exposures [103], suggesting that delayed reward discounting and ethanol sensitization (one component of the differentiator model discussed above) may share underlying predispositions to AUD. We note that this translational literature is not entirely consistent. In one study there were no differences in delayed reward discounting between mice that were bred to either consume high or low amounts of ethanol [104], and there is also some evidence that mice bred to be less sensitive to the reinforcing effects of drugs exhibited greater delay discounting compared to mice that were more sensitive to these reinforcing effects [105]. These differences may be attributable to mouse strain differences, and highlight the difficulties in identifying the appropriate model system for translational studies of candidate endophenotypes.
Sweet liking
Animal models have also provided some validation of sweet liking as a strong candidate endophenotype for AUD and for addiction phenotypes more broadly [106, 84]. Rats selected for high saccharin intake consumed more ethanol relative to rats selected for low saccharin intake [107], and they exhibit greater ethanol withdrawal [108]. Saccharin consumption also appears to offset alcohol consumption. P rats who voluntarily consume saccharin subsequently drink less ethanol compared to alcohol preferring rats who are not given access to saccharin [109]. This suggests that ethanol and saccharin consumption may have overlapping effects on (genetically-influenced) neurobiological systems involved in reward, such as the opioid, serotonin, and dopamine systems [110]. Consistent with this idea, P rats who were administered clonidine (a noradrenergic signaling inhibitor) reduced alcohol consumption and saccharin consumption, but not water consumption [111]. In another example, P rats who were administered TP-10 (a dual-specificity cyclic adenosine monophosphate/cyclic guanosine monophosphate-inhibiting enzyme inhibitor) reduced their alcohol and saccharin self administration [112]. This illustrates the utility of using the endophenotype concept in a translational manner to develop potential therapeutic targets for AUD.
Gene identification for translational endophenotypes
Translational approaches to endophenotypes typically rely on comparisons of mouse strains selectively bred for higher and lower alcohol preference. Mapping these differences to genes and gene networks is critical to fully realizing the promise of a translational approach for endophenotypes. A meta-analysis of three mouse strains that differed in alcohol preference identified 3,800 genes that were differentially expressed in the brains of N and NP mice, and functional gene groups including mitogen-activated protein kinase signaling and transcription regulation pathways were overrepresented among these differentially expressed genes [113]. These results provide numerous candidate genes and pathways to be tested for association with translational candidate endophenotypes and, if merited, further testing in human samples.
The Endophenotype Concept in AUD Research: Redux
The landscape of psychiatric genetics has changed dramatically since the endophenotype concept was most recently reintroduced in the literature. The field has moved towards large, collaborative gene finding research networks for psychiatric outcomes (such as the Psychiatric Genomics Consortium) that have sample sizes in the tens of thousands, moving away from an endophenotype strategy with its emphasis on heritability. However, the trade-off between large-scale genotyping, where amassing very large samples often comes at the cost of less precise measures, and the study of endophenotypes, which may require deeper, more costly measurement on a smaller number of subjects, is currently unknown.
We recognize that an endophenotype approach shares some of the same weaknesses as large-scale genotyping efforts. One is the issue of reliability. As noted by Kendler and Neale [114], although many candidate endophenotypes may seem scientifically “harder” (because they involve, for example, measures of brain structure) than “softer” clinical diagnoses, they do not necessarily have higher reliabilities. This ultimately impacts the power of one’s analyses, and in this respect it does not appear that endophenotypes offer a particular advantage over diagnoses. A second is the issue of whether candidate endophenotypes are truly less heterogeneous than diagnostic categories; for example, a reduction in P3 amplitude can result from several possible differences in underlying event-related oscillations. Thus, whether some endophenotypes for AUD are more homogenous than the diagnostic category for genetic analysis remains an open question. A third is the issue of replication. Only a few of the genes and genetic variants identified in Table 2 have replicable associations with their respective candidate endophenotypes and with AUD, and thus the replicable yield of AUD-associated genes remains small.
Related to the issue of replication, several of the genes summarized in Table 2 were selected based on their prior association with AUD-related phenotypes. This raises the question of whether studies of candidate endophenotypes have identified novel genes for AUD. Let us put the issue of novelty into context. GABRA2 exemplifies the success of an endophenotype approach for AUD in that variation in this gene was initially associated with a neurophysiological endophenotype [23] and numerous subsequent studies have documented its association with alcohol dependence [115]. On the other hand, the largest genome-wide association study of alcohol dependence to date [116] produced associations for genes implicated in alcohol metabolism that were initially identified in the early 1990s [117], as well as a handful of other genetic variants for which the replication results were mixed. Thus, in the absence of other, more successful approaches for AUD gene identification (both in terms of novelty and replicability), it seems reasonable that pursuing both large-scale genotyping strategies and endophenotypes-based approaches may be the most prudent path forward.
There is also a growing recognition that endophenotypes may not be genetically simpler than the psychiatric phenotypes with which they are associated [118], and in fact may reflect the contribution of many genetic variants of small effect from across the genome [119]. A recent special section in the journal Psychophysiology (December 2014) devoted to studies of physiological candidate endophenotypes for addiction and schizophrenia in the Minnesota Twin and Family Study reiterates this point. These studies were able to use the same sample of parent and twin pair offspring to conduct biometric modeling (i.e., decomposing variation for a measure into latent genetic and environmental influences based on the pattern of correlations among different degrees of relatives) and genetic association analyses. In one example, biometric modeling indicated that genetic factors accounted for 65% of the variance in P3 amplitude; however, a genome-wide association study in the same sample did not identify any genome-wide significant variants, although a gene-based test did identify the myelin expression factor 2 (MYEF2) gene on chromosome 15 [120]. The absence of genome-wide significant effects suggests that P3 amplitude is likely polygenic. We return to the implications of a polygenic architecture for endophenotypes shortly.
Nonetheless, even if these candidate endophenotypes are not simpler clues to the genetic underpinnings than AUD itself [19], they could still be very useful in terms of delineating underlying mechanisms [118]. And, as illustrated in the neurophysiology, level of response, delayed reward discounting, and sweet liking literatures, endophenotypes can also help to begin to bridge human and non-human animal alcohol research and (in the case of sweet liking) identify possible drug targets, both of which are distinct advantages of the endophenotype concept that Gould and Gottesman [94] emphasized.
Future Directions
Polygenic and network approaches to endophenotypes
AUD has a polygenic architecture, meaning that it includes the effects of many variants of small magnitude across the genome [121, 122]. The advent of low-cost genome-wide genotyping has made it possible to measure polygenic risk for psychiatric disorders such as schizophrenia, and polygenic approaches have shown predictive power in instances where no single marker meets the stringent genome-wide significance threshold [123]. Polygenic approaches can be easily applied to studies of endophenotypes to test whether polygenic risk scores for candidate endophenotypes also show association with AUD [124].
Polygenic effects can be further interrogated using gene network analyses and bioinformatic data to evaluate biological plausibility and relevance (e.g., is the gene expressed in the brain or liver?). Gene network analyses permit examination of whether variants included in polygenic scores are located in functionally related networks of genes [125]. This approach can thus identify the different pathways involved in genetic vulnerability, and the routes by which a set of genes may influence pathways of risk. Knowledge of such networks can be capitalized on to develop novel drug targets. Work in model organisms has begun to identify gene networks associated with initial sensitivity to ethanol, a measure closely associated to level of response [126], and there is some preliminary evidence suggesting that variation in gene networks related to neuronal signaling is associated with level of response in humans [57].
Developmental approaches to endophenotypes
Attention to endophenotypes as developmental phenomena may also provide additional insight into the pathways between genetic predispositions and eventual disorder. To date, the criterion for an endophenotype to be associated with the illness has typically been met using cross-sectional studies that compare individuals with AUD to healthy controls. However, there are also likely to be endophenotypes that emerge or maximally differentiate between those who will and will not go on to have AUD at points earlier in development. In a conceptual example of this, variation in the AUD-associated gene GABRA2 is also associated with childhood conduct disorder symptoms [127] and increased risk (odds ratios ranging from 2.1 to 2.7) of exhibiting an elevated persistent trajectory of externalizing behavior across adolescence and early adulthood [128]. Neither conduct disorder nor externalizing behavior trajectories meet the endophenotype criteria; however, what this example illustrates is the possibility that there may be endophenotypes earlier in development that predict adult AUD, and these can be capitalized on in gene identification efforts.
A corollary of this point, which is particularly relevant for substance use disorder candidate endophenotypes, is that there is likely to be a dynamic relationship between the genes associated with AUD and a necessary environmental exposure (i.e., alcohol). For example, ethanol exposure induces modest differential gene expression in lymphoblastoid cell lines from alcoholics and non-alcoholics [129]. The possibility of identifying AUD genetic predispositions that interact with environmental factors (e.g., adolescent alcohol exposure) to produce variation in a candidate endophenotype is particularly promising. For example, adolescent alcohol exposure may initiate a cascade of biological changes (e.g., gene expression [130]) that contribute to variation in AUD candidate endophenotypes and eventual disorder.
Developmental considerations have been nearly absent in the literature on endophenotypes [131], although it has been noted that one of the more prominent candidate endophenotypes for AUD—P3 amplitude reduction—is more pronounced in adolescence compared to young adulthood in males with a high-risk paternal history of externalizing disorders [132]. Another promising example of a developmental candidate endophenotype comes from a recent fMRI study of spatial working memory. It found that the pattern of functional brain connectivity in early adolescents (12–14 years) with a family history of AUD was less similar to that of older adolescents/young adults (16–20 years) compared to a control sample of early adolescents without a family history [133]. The pattern of findings suggests that neural connectivity is less mature in adolescents with a family history of AUD. Interestingly, this may represent a neuromaturational lag that can only be detected in adolescence. Additional data are needed to determine whether this neurodevelopmental lag in adolescence is indeed associated with subsequent AUD, but this example illustrates that novel candidate endophenotypes that have “sleeper effects” for AUD may be used in gene finding studies.
Attention to developmental changes in and/or the developmental salience of particular genes for candidate endophenotypes may be important for identifying the relevant genes and gene networks implicated in AUD. Fully 95% of genes are expressed in the developing fetal brain [134], in contrast to the 84% of genes expressed in the adult human brain [135]. The structure and function of the brain is largely determined during prenatal development, with a second wave of development in adolescence [136]. Thus, the genes and gene networks that influence neuronal development, cellular migration, and brain anatomy and function may be predominantly expressed quite early in development, but their consequences for cognitive, neurophysiological, and functional brain candidate endophenotypes may only emerge later in development.
Conclusions
The endophenotype concept was initially proposed as a strategy for improving gene identification in view of the complex and heterogeneous nature of psychiatric disorders. In the area of alcohol research, level of response to alcohol and resting and event-related neurophysiological measures have received considerable attention as candidate endophenotypes, and have also led to replicable genetic associations (e.g., CHRM2 [7] and GABRA2 [115]) for AUD. As these examples illustrate, the past successes of endophenotype strategies for AUD gene identification suggest that the concept will continue to remain relevant to AUD research today, even as gene identification efforts move towards large-scale phenotyping at the diagnostic level. A number of other candidate endophenotypes show promise, including delayed reward discounting, executive functions, sweet liking, and structural brain features. Systematic efforts to continue to refine and validate these as elected endophenotypes, and to identify the genes, polygenes, and gene networks that may influence variation in these traits/behaviors (and in turn AUD) by themselves and in the context of environmental exposures (particularly alcohol) represent important directions for future research.
Acknowledgments
Funding. J.E.S. was supported by F32AA022269; I.I.G. was supported by the Grawemeyer Award in Psychology and the Lieber Prize for Schizophrenia Research; and D.M.D was supported by K02AA018755, U10AA008401, P20AA017828, R01AA018333, R01AA015416. The contents of the paper are solely the responsibility of the authors and do not necessarily represent the official views of the funders.
Footnotes
Conflicts of Interest. Jessica E. Salvatore, Irving I. Gottesman, and Danielle M. Dick declare that they have no conflict of interest.
Compliance with Ethical Standards
Human and Animal Rights and Informed Consent. This article does not contain any new data from studies involving human participants or animal subjects.
References
Papers of particular interest, published recently, have been highlighted as:
* Of importance
** Of major importance
- 1.Vaillant GE. The natural history of alcoholism revisited. Cambridge, MA: Harvard University Press; 1995. [Google Scholar]
- 2.Gottesman II, Shields J. Schizophrenia and genetics: A twin study vantage point. Oxford, England: Academic Press; 1972. [Google Scholar]
- 3.Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160(4):636–45. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 4.Courtet P, Gottesman II, Jollant F, Gould TD. The neuroscience of suicidal behaviors: what can we expect from endophenotype strategies? Translational Psychiatry. 2011;1(5):e7. doi: 10.1038/tp.2011.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wood AC, Neale MC. Twin studies and their implications for molecular genetic studies: endophenotypes integrate quantitative and molecular genetics in ADHD research. J Am Acad Child Adolesc Psychiatry. 2010;49(9):874–83. doi: 10.1016/j.jaac.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hasler G, Drevets WC, Gould TD, Gottesman II, Manji HK. Toward constructing an endophenotype strategy for bipolar disorders. Biol Psychiatry. 2006;60(2):93–105. doi: 10.1016/j.biopsych.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 7.Dick DM, Jones K, Saccone N, Hinrichs A, Wang JC, Goate A, et al. Endophenotypes successfully lead to gene identification: results from the collaborative study on the genetics of alcoholism. Behav Genet. 2006;36(1):112–26. doi: 10.1007/s10519-005-9001-3. [DOI] [PubMed] [Google Scholar]
- 8.Ray LA, Mackillop J, Monti PM. Subjective responses to alcohol consumption as endophenotypes: advancing behavioral genetics in etiological and treatment models of alcoholism. Subst Use Misuse. 2010;45(11):1742–65. doi: 10.3109/10826084.2010.482427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hines LM, Ray L, Hutchison K, Tabakoff B. Alcoholism: the dissection for endophenotypes. Dialogues Clin Neurosci. 2005;7(2):153–63. doi: 10.31887/DCNS.2005.7.2/lhines. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rangaswamy M, Porjesz B. Uncovering genes for cognitive (dys)function and predisposition for alcoholism spectrum disorders: A review of human brain oscillations as effective endophenotypes. Brain Res. 2008;1235:153–71. doi: 10.1016/j.brainres.2008.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ooteman W. Psychobiological pathways to alcohol craving and their relevance for patient-treatment matching. University of Amsterdam; 2006. [Google Scholar]
- 12.Porjesz B, Rangaswamy M. Neurophysiological endophenotypes, CNS disinhibition, and risk for alcohol dependence and related disorders. Scientific World Journal. 2007;7:131–41. doi: 10.1100/tsw.2007.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karoly HC, Hagerty SL, Hutchison KE. Developing neurobiological endophenotypes that reflect failure to control alcohol consumption and dependence. Current Addiction Reports. 2014;1:10–8. doi: 10.1007/s40429-013-0007-2. [DOI] [Google Scholar]
- 14.Gottesman II, McGue M. Endophenotype. In: Cautin RL, Lilienfeld SO, editors. Encyclopedia of Clinical Psychology. Wiley-Blackwell; in press. [Google Scholar]
- 15.Almasy L, Blangero J. Endophenotypes as quantitative risk factors for psychiatric disease: rationale and study design. Am J Med Genet. 2001;105(1):42–4. doi: 10.1002/1096-8628(20010108)105:1<42::AID-AJMG1055>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 16.Cannon TD, Keller MC. Endophenotypes in the genetic analyses of mental disorders. Annu Rev Clin Psychol. 2006;2:267–90. doi: 10.1146/annurev.clinpsy.2.022305.095232. [DOI] [PubMed] [Google Scholar]
- **17.Lenzenweger MF. Thinking clearly about the endophenotype-intermediate phenotype-biomarker distinctions in developmental psychopathology research. Dev Psychopathol. 2013;25(4 Pt 2):1347–57. doi: 10.1017/s0954579413000655. A careful delineation of the differences and similarities among the endophenotype, biomarker, and intermediate phenotype concepts. [DOI] [PubMed] [Google Scholar]
- 18.Rasetti R, Weinberger DR. Intermediate phenotypes in psychiatric disorders. Curr Opin Genet Dev. 2011;21(3):340–8. doi: 10.1016/j.gde.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flint J, Munafo MR. The endophenotype concept in psychiatric genetics. Psychol Med. 2007;37(2):163–80. doi: 10.1017/s0033291706008750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Beijsterveldt CE, Molenaar PC, de Geus EJ, Boomsma DI. Heritability of human brain functioning as assessed by electroencephalography. Am J Hum Genet. 1996;58:562–573. [PMC free article] [PubMed] [Google Scholar]
- 21.Rangaswamy M, Porjesz B, Chorlian DB, Wang K, Jones KA, Kuperman S, et al. Resting EEG in offspring of male alcoholics: beta frequencies. Int J Psychophysiol. 2004;51:239–251. doi: 10.1016/j.ijpsycho.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 22.Porjesz B, Almasy L, Edenberg HJ, Wang K, Chorlian DB, Foroud T, et al. Linkage disequilibrium between the beta frequency of the human EEG and a GABAA receptor gene locus. Proc Natl Acad Sci U S A. 2002;99:3729–33. doi: 10.1073/pnas.052716399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edenberg HJ, Dick DM, Xuei X, Tian H, Almasy L, Bauer LO, et al. Variations in GABRA2, encoding the α2 subunit of the GABA-A receptor are associated with alcohol dependence and with brain oscillations. Am J Hum Genet. 2004;74(4):705–14. doi: 10.1086/383283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Covault J, Gelernter J, Hesselbrock V, Nellissery M, Kranzler HR. Allelic and haplotypic association of GABRA2 with alcohol dependence. Am J Med Genet B Neuropsychiatr Genet. 2004;129B(1):104–9. doi: 10.1002/ajmg.b.30091. [DOI] [PubMed] [Google Scholar]
- 25.Enoch MA, Hodgkinson CA, Yuan Q, Albaugh B, Virkkunen M, Goldman D. GABRG1 and GABRA2 as independent predictors for alcoholism in two populations. Neuropsychopharmacol. 2009;34:1245–54. doi: 10.1038/npp.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Irons DE, Iacono WG, Oetting WS, Kirkpatrick RM, Vrieze SI, Miller MB, et al. Gamma-aminobutyric Acid system genes-no evidence for a role in alcohol use and abuse in a community-based sample. Alcohol Clin Exp Res. 2014;38(4):938–47. doi: 10.1111/acer.12352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Enoch MA, Shen PH, Ducci F, Yuan Q, Liu J, White KV, et al. Common genetic origins for EEG, alcoholism and anxiety: the role of CRH-BP. PloS one. 2008;3(10):e3620. doi: 10.1371/journal.pone.0003620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vrieze SI, Malone SM, Vaidyanathan U, Kwong A, Kang HM, Zhan X, et al. In search of rare variants: Preliminary results from whole genome sequencing of 1,325 individuals with psychophysiological endophenotypes. Psychophysiology. 2014;51(12):1309–20. doi: 10.1111/psyp.12350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Porjesz B, Rangaswamy M, Kamarajan C, Jones KA, Padmanabhapillai A, Begleiter H. The utility of neurophysiological markers in the study of alcoholism. Clin Neurophysiol. 2005;116(5):993–1018. doi: 10.1016/j.clinph.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 30.Begleiter H, Porjesz B, Bihari B, Kissin B. Event-related brain potentials in boys at risk for alcoholism. Science. 1984;225:1493–6. doi: 10.1126/science.6474187. [DOI] [PubMed] [Google Scholar]
- 31.Campanella S, Noel X, Tomberg C. Cognitive event-related potentials and alcoholism. J Psychophysiol. 2010;24(4):231–9. doi: 10.1027/0269-8803/a000036. [DOI] [Google Scholar]
- 32.Carlson SR, Iacono WG, McGue M. P300 amplitude in adolescent twins discordant and concordant for alcohol use disorders. Biol Psychol. 2002;61(1–2):203–27. doi: 10.1016/s0301-0511(02)00059-5. [DOI] [PubMed] [Google Scholar]
- 33.Hesselbrock V, Begleiter H, Porjesz B, O’Connor S, Bauer L. P300 event-related potential amplitude as an endophenotype of alcoholism--evidence from the collaborative study on the genetics of alcoholism. J Biomed Sci. 2001;8(1):77–82. doi: 10.1007/bf02255974. [DOI] [PubMed] [Google Scholar]
- 34.Euser AS, Arends LR, Evans BE, Greaves-Lord K, Huizink AC, Franken IHA. The P300 event-related brain potential as a neurobiological endophenotype for substance use disorders: A meta-analytic investigation. Neurosci Biobehav R. 2012;36(1):572–603. doi: 10.1016/j.neubiorev.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 35.Jones KA, Porjesz B, Almasy L, Bierut L, Goate A, Wang JC, et al. Linkage and linkage disequilibrium of evoked EEG oscillations with CHRM2 receptor gene polymorphisms: implications for human brain dynamics and cognition. Int J Psychophysiol. 2004;53(2):75–90. doi: 10.1016/j.ijpsycho.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 36.Chen AC, Tang Y, Rangaswamy M, Wang JC, Almasy L, Foroud T, et al. Association of single nucleotide polymorphisms in a glutamate receptor gene (GRM8) with theta power of event-related oscillations and alcohol dependence. Am J Med Genet B Neuropsychiatr Genet. 2009;150B(3):359–68. doi: 10.1002/ajmg.b.30818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen AC, Manz N, Tang Y, Rangaswamy M, Almasy L, Kuperman S, et al. Single-nucleotide polymorphisms in corticotropin releasing hormone receptor 1 gene (CRHR1) are associated with quantitative trait of event-related potential and alcohol dependence. Alcohol Clin Exp Res. 2010;34(6):988–96. doi: 10.1111/j.1530-0277.2010.01173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *38.Kang SJ, Rangaswamy M, Manz N, Wang JC, Wetherill L, Hinrichs T, et al. Family-based genome-wide association study of frontal theta oscillations identifies potassium channel gene KCNJ6. Genes Brain Behav. 2012;11(6):712–9. doi: 10.1111/j.1601-183X.2012.00803.x. A genome-wide association study of frontal theta oscillations that identified KCNJ6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo XG, Kranzler HR, Zuo LJ, Wang S, Blumberg HP, Gelernter J. CHRM2 gene predisposes to alcohol dependence, drug dependence and affective disorders: results from an extended case-control structured association study. Hum Mol Genet. 2005;14(16):2421–34. doi: 10.1093/hmg/ddi244. [DOI] [PubMed] [Google Scholar]
- 40.Clarke TK, Laucht M, Ridinger M, Wodarz N, Rietschel M, Maier W, et al. KCNJ6 is associated with adult alcohol dependence and involved in gene x early life stress interactions in adolescent alcohol drinking. Neuropsychopharmacol. 2011;36(6):1142–8. doi: 10.1038/npp.2010.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morean ME, Corbin WR. Subjective response to alcohol: a critical review of the literature. Alcohol Clin Exp Res. 2010;34(3):385–95. doi: 10.1111/j.1530-0277.2009.01103.x. [DOI] [PubMed] [Google Scholar]
- 42.Schuckit MA, Smith TL, Danko GP, Trim R, Bucholz KK, Edenberg HJ, et al. An evaluation of the full level of response to alcohol model of heavy drinking and problems in COGA offspring. J Stud Alcohol Drugs. 2009;70(3):436–45. doi: 10.15288/jsad.2009.70.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schuckit MA, Smith TL, Anderson KG, Brown SA. Testing the level of response to alcohol: social information processing model of alcoholism risk--a 20-year prospective study. Alcohol Clin Exp Res. 2004;28(12):1881–9. doi: 10.1097/01.alc.0000148111.43332.a5. [DOI] [PubMed] [Google Scholar]
- 44.Schuckit MA, Smith TL, Trim R, Kreikebaum S, Hinga B, Allen R. Testing the level of response to alcohol-based model of heavy drinking and alcohol problems in offspring from the San Diego Prospective Study. J Stud Alcohol Drugs. 2008;69(4):571–9. doi: 10.15288/jsad.2008.69.571. [DOI] [PubMed] [Google Scholar]
- 45.Quinn PD, Fromme K. Subjective response to alcohol challenge: a quantitative review. Alcohol Clin Exp Res. 2011;35(10):1759–70. doi: 10.1111/j.1530-0277.2011.01521.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heath AC, Madden PAF, Bucholz KK, Dinwiddie SH, Slutske WS, Bierut LJ, et al. Genetic differences in alcohol sensitivity and the inheritance of alcoholism risk. Psychol Med. 1999;29:1069–81. doi: 10.1017/s0033291799008909. [DOI] [PubMed] [Google Scholar]
- 47.Kalu N, Ramchandani VA, Marshall V, Scott D, Ferguson C, Cain G, et al. Heritability of level of response and association with recent drinking history in nonalcohol-dependent drinkers. Alcohol Clin Exp Res. 2012;36(6):1034–41. doi: 10.1111/j.1530-0277.2011.01699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uhart M, Weerts EM, McCaul ME, Guo X, Yan X, Kranzler HR, et al. GABRA2 markers moderate the subjective effects of alcohol. Addict Biol. 2013;18(2):357–69. doi: 10.1111/j.1369-1600.2012.00457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu X, Oroszi G, Chun J, Smith TL, Goldman D, Schuckit MA. An expanded evaluation of the relationship of four alleles to the level of response to alcohol and the alcoholism risk. Alcohol Clin Exp Res. 2005;29(1):8–16. doi: 10.1097/01.alc.0000150008.68473.62. [DOI] [PubMed] [Google Scholar]
- 50.Ray LA, Bujarski S, MacKillop J, Courtney KE, Monti PM, Miotto K. Subjective response to alcohol among alcohol-dependent individuals: Effects of the mu-opioid receptor (OPRM1) gene and alcoholism severity. Alcohol Clin Exp Res. 2013;37:E116–E24. doi: 10.1111/j.1530-0277.2012.01916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ray LA, Hutchison KE. A polymorphism of the u-opioid receptor gene (OPRM1) and sensitivty to the effects of alcohol in humans. Alcohol Clin Exp Res. 2004;28(12):1780–95. doi: 10.1097/01.alc.0000148114.34000.b9. [DOI] [PubMed] [Google Scholar]
- 52.Ehlers CL, Lind PA, Wilhelmsen KC. Association between single nucleotide polymorphisms in the mu opioid receptor gene (OPRM1) and self-reported responses to alcohol in American Indians. BMC Med Genet. 2008;9:35. doi: 10.1186/1471-2350-9-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ehlers CL, Gizer IR, Schuckit MA, Wilhelmsen KC. Genome-wide scan for self-rating of the effects of alcohol in American Indians. Psychiatr Genet. 2010;20(5):221–8. doi: 10.1097/YPG.0b013e32833add87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luczak SE, Pandika D, Shea SH, Eng MY, Liang T, Wall TL. ALDH2 and ADH1B interactions in retrospective reports of low-dose reactions and initial sensitivity to alcohol in Asian American college students. Alcohol Clin Exp Res. 2011;35(7):1238–45. doi: 10.1111/j.1530-0277.2011.01458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Joslyn G, Brush G, Robertson M, Smith TL, Kalmijn J, Schuckit M, et al. Chromosome 15q25.1 genetic markers associated with level of response to alcohol in humans. Proc Natl Acad Sci U S A. 2008;105(51):20368–73. doi: 10.1073/pnas.0810970105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choquet H, Joslyn G, Lee A, Kasberger J, Robertson M, Brush G, et al. Examination of rare missense variants in the CHRNA5-A3-B4 gene cluster to level of response to alcohol in the San Diego Sibling Pair study. Alcohol Clin Exp Res. 2013;37(8):1311–6. doi: 10.1111/acer.12099. [DOI] [PubMed] [Google Scholar]
- 57.Joslyn G, Ravindranathan A, Brush G, Schuckit M, White RL. Human variation in alcohol response is influenced by variation in neuronal signaling genes. Alcohol Clin Exp Res. 2010;34(5):800–12. doi: 10.1111/j.1530-0277.2010.01152.x. [DOI] [PubMed] [Google Scholar]
- 58.Luczak SE, Glatt SJ, Wall TL. Meta-analyses of ALDH2 and ADH1B with alcohol dependence in Asians. Psychol Bull. 2006;132(4):607–21. doi: 10.1037/0033-2909.132.4.607. [DOI] [PubMed] [Google Scholar]
- 59.McHugh RK, Hofmann SG, Asnaani A, Sawyer AT, Otto MW. The serotonin transporter gene and risk for alcohol dependence: a meta-analytic review. Drug Alcohol Depend. 2010;108(1–2):1–6. doi: 10.1016/j.drugalcdep.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arias A, Feinn R, Kranzler HR. Association of an Asn40Asp (A118G) polymorphism in the mu-opioid receptor gene with substance dependence: A meta-analysis. Drug Alcohol Depend. 2006;83(3):262–8. doi: 10.1016/j.drugalcdep.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 61.Wang JC, Grucza R, Cruchaga C, Hinrichs AL, Bertelsen S, Budde JP, et al. Genetic variation in the CHRNA5 gene affects mRNA levels and is associated with risk for alcohol dependence. Mol Psychiatry. 2009;14(5):501–10. doi: 10.1038/mp.2008.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen LS, Xian H, Grucza RA, Saccone NL, Wang JC, Johnson EO, et al. Nicotine dependence and comorbid psychiatric disorders: examination of specific genetic variants in the CHRNA5-A3-B4 nicotinic receptor genes. Drug Alcohol Depend. 2012;123 (Suppl 1):S42–51. doi: 10.1016/j.drugalcdep.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Newlin DB, Thomas JB. Alcohol challenge with sons of alcoholics: A critical review and analysis. Psychol Bull. 1990;108(3):383–402. doi: 10.1037/0033-2909.108.3.383. [DOI] [PubMed] [Google Scholar]
- 64.King AC, de Wit H, McNamara PJ, Cao D. Rewarding, stimulant, and sedative alcohol response and relationship to future binge drinking. Arch Gen Psychiat. 2011;68(4):389–99. doi: 10.1001/archgenpsychiatry.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.King AC, McNamara PJ, Hasin DS, Cao D. Alcohol challenge responses predict future alcohol use disorder symptoms: a 6-year prospective study. Biol Psychiatry. 2014;75(10):798–806. doi: 10.1016/j.biopsych.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kendler KS, Prescott CA, Myers J, Neale MC. The structure of genetic and environmental risk factors for common psychiatric and substance use disorders in men and women. Arch Gen Psychiat. 2003;60:929–37. doi: 10.1001/archpsyc.60.9.929. [DOI] [PubMed] [Google Scholar]
- 67.Young SE, Stallings MC, Corley RP, Krauter KS, Hewitt JK. Genetic and environmental influences on behavioral disinhibition. Am J Med Genet. 2000;96(5):684–95. doi: 10.1002/1096-8628(20001009)96:5<684::aid-ajmg16>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 68.Krueger RF, Hicks BM, Patrick CJ, Carlson SR, Iacono WG, McGue M. Etiological connections among substance dependence, antisocial behavior, and personality: Modeling the externalizing spectrum. J Abnorm Psychol. 2002;111(3):411–24. doi: 10.1037//0021-843X.111.3.411. [DOI] [PubMed] [Google Scholar]
- 69.Evenden JL. Varieties of impulsivity. Psychopharmacology. 1999;146(4):348–61. doi: 10.1007/pl00005481. [DOI] [PubMed] [Google Scholar]
- 70.MacKillop J. Integrating behavioral economics and genetics: Delayed reward discounting as an endophenoype for addictive disorders. J Exp Anal Behav. 2013;99(1):14–31. doi: 10.1002/jeab.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Anokhin AP, Golosheykin S, Grant JD, Heath AC. Heritability of delay discounting in adolescence: a longitudinal twin study. Behav Genet. 2011;41(2):175–83. doi: 10.1007/s10519-010-9384-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mitchell JM, Fields HL, D’Esposito M, Boettiger CA. Impulsive responding in alcoholics. Alcohol Clin Exp Res. 2005;29(12):2158–69. doi: 10.1097/01.alc.0000191755.63639.4a. [DOI] [PubMed] [Google Scholar]
- 73.Petry NM, Kirby KN, Kranzler HR. Effects of gender and family history of alcohol dependence on a behavioral task of impulsivity in healthy subjects. J Stud Alcohol. 2002;63:83–90. [PubMed] [Google Scholar]
- 74.Barkley RA. Behavioral inhibition, sustained attention, and executive functions: Constructing a unifying theory of ADHD. Psychol Bull. 1997;121(1):65–94. doi: 10.1037//0033-2909.121.1.65. [DOI] [PubMed] [Google Scholar]
- 75.Friedman NP, Miyake A, Young SE, Defries JC, Corley RP, Hewitt JK. Individual differences in executive functions are almost entirely genetic in origin. J Exp Psychol Gen. 2008;137(2):201–25. doi: 10.1037/0096-3445.137.2.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sjoerds Z, van den Brink W, Beekman AT, Penninx BW, Veltman DJ. Response inhibition in alcohol-dependent patients and patients with depression/anxiety: a functional magnetic resonance imaging study. Psychol Med. 2013:1–13. doi: 10.1017/s0033291713002274. [DOI] [PubMed] [Google Scholar]
- 77.Gierski F, Hubsch B, Stefaniak N, Benzerouk F, Cuervo-Lombard C, Bera-Potelle C, et al. Executive functions in adult offspring of alcohol-dependent probands: toward a cognitive endophenotype? Alcohol Clin Exp Res. 2013;37 (Suppl 1):E356–63. doi: 10.1111/j.1530-0277.2012.01903.x. [DOI] [PubMed] [Google Scholar]
- 78.Benzerouk F, Gierski F, Gorwood P, Ramoz N, Stefaniak N, Hubsch B, et al. Brain-derived neurotrophic factor (BDNF) Val66Met polymorphism and its implication in executive functions in adult offspring of alcohol-dependent probands. Alcohol. 2013;47(4):271–4. doi: 10.1016/j.alcohol.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 79.Nedic G, Perkovic MN, Sviglin KN, Muck-Seler D, Borovecki F, Pivac N. Brain-derived neurotrophic factor Val66Met polymorphism and alcohol-related phenotypes. Prog Neuropsychopharmacol Biol Psychiatry. 2013;40:193–8. doi: 10.1016/j.pnpbp.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 80.Keskitalo K, Knaapila A, Kallela M, Palotie A, Wessman M, Sammalisto S, et al. Sweet taste preferences are partly genetically determined: Identification of a trait locus on chromosome 16. Am J Clin Nutr. 2007;86:55–63. doi: 10.1093/ajcn/86.1.55. [DOI] [PubMed] [Google Scholar]
- 81.Kampov-Polevoy A, Garbutt JC, Janowsky D. Evidence of preference for a high-concentration sucrose solution in alcoholic men. Am J Psychiat. 1997;154(2):269–70. doi: 10.1176/ajp.154.2.269. [DOI] [PubMed] [Google Scholar]
- 82.Kampov-Polevoy AB, Ziedonis D, Steinberg ML, Pinsky I, Krejci J, Eick C, et al. Association between sweet preference and paternal history of alcoholism in psychiatric and substance abuse patients. Alcohol Clin Exp Res. 2003;27(12):1929–36. doi: 10.1097/01.ALC.0000099265.60216.23. [DOI] [PubMed] [Google Scholar]
- 83.Mennella JA, Pepino MY, Lehmann-Castor SM, Yourshaw LM. Sweet preferences and analgesia during childhood: effects of family history of alcoholism and depression. Addiction. 2010;105(4):666–75. doi: 10.1111/j.1360-0443.2009.02865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bachmanov AA, Bosak NP, Floriano WB, Inoue M, Li X, Lin C, et al. Genetics of sweet taste preferences. Flavour Fragr J. 2011;26(4):286–94. doi: 10.1002/ffj.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Luciana M, Collins PF, Muetzel RL, Lim KO. Effects of alcohol use initiation on brain structure in typically developing adolescents. Am J Drug Alcohol Abuse. 2013;39(6):345–55. doi: 10.3109/00952990.2013.837057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pfefferbaum A, Lim KO, Zipursky RB, Mathalon DH, Rosenbloom MJ, Lane B, et al. Brain gray and white matter volume loss accelerates with aging in chronic alcoholics: A quantitative MRI study. Alcohol Clin Exp Res. 1992;16(6):1078–89. doi: 10.1111/j.1530-0277.1992.tb00702.x. [DOI] [PubMed] [Google Scholar]
- 87.Thompson PM, Cannon TD, Narr KL, van Erp T, Poutanen VP, Huttunen M, et al. Genetic influences on brain structure. Nat Neurosci. 2001;4(12):1253–8. doi: 10.1038/nn758. [DOI] [PubMed] [Google Scholar]
- 88.Bohlken MM, Mandl RC, Brouwer RM, van den Heuvel MP, Hedman AM, Kahn RS, et al. Heritability of structural brain network topology: A DTI study of 156 twins. Hum Brain Mapp. 2014;35(10):5295–305. doi: 10.1002/hbm.22550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Boutte D, Calhoun VD, Chen J, Sabbineni A, Hutchison K, Liu J. Association of genetic copy number variations at 11 q14.2 with brain regional volume differences in an alcohol use disorder population. Alcohol. 2012;46(6):519–27. doi: 10.1016/j.alcohol.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Agartz I, Momenan R, Rawlings RR, Kerich MJ, Hommer DW. Hippocampal volume in patients with alcohol dependence. Arch Gen Psychiatry. 1999;56(4):356–63. doi: 10.1001/archpsyc.56.4.356. [DOI] [PubMed] [Google Scholar]
- 91.Benegal V, Antony G, Venkatasubramanian G, Jayakumar PN. Gray matter volume abnormalities and externalizing symptoms in subjects at high risk for alcohol dependence. Addict Biol. 2007;12(1):122–32. doi: 10.1111/j.1369-1600.2006.00043.x. [DOI] [PubMed] [Google Scholar]
- 92.Slutske WS, Heath AC, Dinwiddie SH, Madden PAF, Bucholz KK, Dunne MP, et al. Common genetic risk factors for conduct disorder and alcohol dependence. J Abnorm Psychol. 1998;107(3):363–74. doi: 10.1037/0021-843X.107.3.363. [DOI] [PubMed] [Google Scholar]
- 93.Edwards AC, Kendler KS. Twin study of the relationship between adolescent attention-deficit/hyperactivity disorder and adult alcohol dependence. J Stud Alcohol Drugs. 2012;73(2):185–94. doi: 10.15288/jsad.2012.73.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gould TD, Gottesman II. Psychiatric endophenotypes and the development of valid animal models. Genes Brain Behav. 2006;5(2):113–9. doi: 10.1111/j.1601-183X.2005.00186.x. [DOI] [PubMed] [Google Scholar]
- 95.Fernando AB, Robbins TW. Animal models of neuropsychiatric disorders. Annu Rev Clin Psychol. 2011;7:39–61. doi: 10.1146/annurev-clinpsy-032210-104454. [DOI] [PubMed] [Google Scholar]
- 96.Ehlers CL, Somes C. Long latency event-related potentials in mice: Effects of stimulus characteristics and strain. Brain Res. 2002;957:117–28. doi: 10.1016/s0006-8993(02)03612-0. [DOI] [PubMed] [Google Scholar]
- 97.Criado JR, Ehlers CL. Event-related oscillations as risk markers in genetic mouse models of high alcohol preference. Neuroscience. 2009;163(2):506–23. doi: 10.1016/j.neuroscience.2009.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stewart RB, Murphy JM, McBride WJ, Lumeng L, Li T-K. Place conditioning with alcohol in alcohol-preferring and -nonpreferring rats. Pharmacol Biochem Behav. 1996;53(3):487–91. doi: 10.1016/0091-3057(95)02102-7. [DOI] [PubMed] [Google Scholar]
- 99.Crabbe JC, Bell RL, Ehlers CL. Human and laboratory rodent low response to alcohol: is better consilience possible? Addict Biol. 2010;15(2):125–44. doi: 10.1111/j.1369-1600.2009.00191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Crabbe JC, Metten P, Cameron AJ, Wahlsten D. An analysis of the genetics of alcohol intoxication in inbred mice. Neurosci Biobehav Rev. 2005;28(8):785–802. doi: 10.1016/j.neubiorev.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 101.Joslyn G, Wolf FW, Brush G, Wu LQ, Schuckit M, White RL. Glypican gene GPC5 participates in the behavioral response to ethanol: evidence from humans, mice, and fruit flies. G3-Genes Genomes Genetics. 2011;1(7):627–35. doi: 10.1534/g3.111.000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Oberlin BG, Grahame NJ. High-alcohol preferring mice are more impulsive than low-alcohol preferring mice as measured in the delay discounting task. Alcohol Clin Exp Res. 2009;33(7):1294–303. doi: 10.1111/j.1530-0277.2009.00955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mitchell SH, Reeves JM, Li N, Phillips TJ. Delay discounting predicts behavioral sensitization to ethanol in outbred WSC mice. Alcohol Clin Exp Res. 2006;30(3):429–37. doi: 10.1111/j.1530-0277.2006.00047.x. [DOI] [PubMed] [Google Scholar]
- 104.Wilhelm CJ, Reeves JM, Phillips TJ, Mitchell SH. Mouse lines selected for alcohol consumption differ on certain measures of impulsivity. Alcohol Clin Exp Res. 2007;31(11):1839–45. doi: 10.1111/j.1530-0277.2007.00508.x. [DOI] [PubMed] [Google Scholar]
- 105.Helms CM, Reeves JM, Mitchell SH. Impact of strain and D-amphetamine on impulsivity (delay discounting) in inbred mice. Psychopharmacology (Berl) 2006;188(2):144–51. doi: 10.1007/s00213-006-0478-0. [DOI] [PubMed] [Google Scholar]
- 106.Carroll ME, Morgan AD, Anker JJ, Perry JL, Dess NK. Selective breeding for differential saccharin intake as an animal model of drug abuse. Behav Pharmacol. 2008;19(5–6):435–60. doi: 10.1097/FBP.0b013e32830c3632. [DOI] [PubMed] [Google Scholar]
- 107.Gahtan E, Labounty LP, Wyvell C, Carroll ME. The relationships among saccharin consumption, oral ethanol, and IV cocaine self-administration. Pharmacol Biochem Behav. 1995;53(4):919–25. doi: 10.1016/0091-3057(95)02148-5. [DOI] [PubMed] [Google Scholar]
- 108.Dess NK, O’Neill P, Chapman CD. Ethanol withdrawal and proclivity are inversely related in rats selectively bred for differential saccharin intake. Alcohol. 2005;37(1):9–22. doi: 10.1016/j.alcohol.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 109.Kampov-Polevoy AB, Overstreet DH, Rezvani AH, Janowsky DS. Suppression of ethanol intake in alcohol-preferring rats by prior voluntary saccharin consumption. Pharmacol Biochem Behav. 1995;52(1):59–64. doi: 10.1016/0091-3057(94)00430-q. [DOI] [PubMed] [Google Scholar]
- 110.Fortuna JL. Sweet preference, sugar addiction and the familial history of alcohol dependence: Shared neural pathways and genes. J Psychoactive Drugs. 2010;42(2):147–51. doi: 10.1080/02791072.2010.10400687. [DOI] [PubMed] [Google Scholar]
- **111.Rasmussen DD, Alexander L, Malone J, Federoff D, Froehlich JC. The alpha-adrenergic receptor agonist, clonidine, reduces alcohol drinking in alcohol-preferring (P) rats. Alcohol. 2014 doi: 10.1016/j.alcohol.2014.07.002. Illustrates the translational potential of endophenotypes to develop therapeutic drug targets. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Logrip ML, Vendruscolo LF, Schlosburg JE, Koob GF, Zorrilla EP. Phosphodiesterase 10A regulates alcohol and saccharin self-administration in rats. Neuropsychopharmacol. 2014;39(7):1722–31. doi: 10.1038/npp.2014.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mulligan MK, Ponomarev I, Hitzemann RJ, Belknap JK, Tabakoff B, Harris RA, et al. Toward understanding the genetics of alcohol drinking through transcriptome meta-analysis. Proc Natl Acad Sci U S A. 2006;103(16):6368–73. doi: 10.1073/pnas.0510188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kendler KS, Neale MC. Endophenotype: a conceptual analysis. Mol Psychiatry. 2010;15(8):789–97. doi: 10.1038/mp.2010.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zintzaras E. Gamma-aminobutyric acid A receptor, alpha-2 (GABRA2) variants as individual markers for alcoholism: a meta-analysis. Psychiatr Genet. 2012;22(4):189–96. doi: 10.1097/YPG.0b013e328353ae53. [DOI] [PubMed] [Google Scholar]
- **116.Gelernter J, Kranzler HR, Sherva R, Almasy L, Koesterer R, Smith AH, et al. Genome-wide association study of alcohol dependence: Significant findings in African- and European-Americans including novel risk loci. Mol Psychiatr. 2013;19:41–9. doi: 10.1038/mp.2013.145. The largest genome-wide association study of alcohol dependence to date. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Thomasson HR, Edenberg HJ, Crabb DW, Mai XL, Jerome RE, Li TK, et al. Alcohol and aldehyde dehydrogenase genotypes and alcoholism in Chinese men. Am J Hum Genet. 1991;48:667–81. [PMC free article] [PubMed] [Google Scholar]
- *118.Flint J, Timpson N, Munafo M. Assessing the utility of intermediate phenotypes for genetic mapping of psychiatric disease. Trends Neurosci. 2014 doi: 10.1016/j.tins.2014.08.007. Discusses the utility of endophenotypes in psychiatry to understand mechanism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **119.Iacono WG, Vaidyanathan U, Vrieze SI, Malone SM. Knowns and unknowns for psychophysiological endophenotypes: Integration and response to commentaries. Psychophysiology. 2014;51(12):1339–47. doi: 10.1111/psyp.12358. Integrative summary of genetic association studies of 17 psychophysiological endophenotypes in the Minnesota Twin Family Study. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Malone SM, Vaidyanathan U, Basu S, Miller MB, McGue M, Iacono WG. Heritability and molecular-genetic basis of the P3 event-related brain potential: A genome-wide association study. Psychophysiology. 2014;51(12):1246–58. doi: 10.1111/psyp.12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Plomin R, Haworth CMA, Davis OSP. Common disorders as quantitative traits. Nat Rev Genet. 2009;10:872–8. doi: 10.1038/nrg2670. [DOI] [PubMed] [Google Scholar]
- 122.Mather K, Jinks JL. Biometrical genetics: The study of continuous variation. London: Chapman & Hall; 1982. [Google Scholar]
- 123.The International Schizophrenia Consortium. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460(7256):748–52. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *124.Evans DM, Brion MJ, Paternoster L, Kemp JP, McMahon G, Munafo M, et al. Mining the human phenome using allelic scores that index biological intermediates. PLoS Genet. 2013;9(10):e1003919. doi: 10.1371/journal.pgen.1003919. Uses a polygenic scoring procedure to screen for genetic associations for complex disorders. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wang L, Jia P, Wolfinger RD, Chen X, Zhao Z. Gene set analysis of genome-wide association studies: Methodological issues and perspectives. Genomics. 2011;98:1–8. doi: 10.1016/j.ygeno.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wolen AR, Phillips CA, Langston MA, Putman AH, Vorster PJ, Bruce NA, et al. Genetic dissection of acute ethanol responsive gene networks in prefrontal cortex: functional and mechanistic implications. PloS one. 2012;7(4):e33575. doi: 10.1371/journal.pone.0033575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dick DM, Bierut L, Hinrichs A, Fox L, Bucholz KK, Kramer J, et al. The role of GABRA2 in risk for conduct disorder and alcohol and drug dependence across developmental stages. Behav Genet. 2006;36(4):577–90. doi: 10.1007/s10519-005-9041-8. [DOI] [PubMed] [Google Scholar]
- 128.Dick DM, Latendresse SJ, Lansford JE, Budde JP, Goate A, Dodge KA, et al. Role of GABRA2 in trajectories of externalizing behavior across development and evidence of moderation by parental monitoring. Arch Gen Psychiat. 2009;66(6):649–57. doi: 10.1001/archgenpsychiatry.2009.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.McClintick JN, Brooks AI, Deng L, Liang L, Wang JC, Kapoor M, et al. Ethanol treatment of lymphoblastoid cell lines from alcoholics and non-alcoholics causes many subtle changes in gene expression. Alcohol. doi: 10.1016/j.alcohol.2014.07.004. online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wang K, Song H, Jin M, Xiao H, Zhao G, Zou H, et al. Chronic alcohol consumption from adolescence to adulthood in mice--hypothalamic gene expression changes in insulin-signaling pathway. Alcohol. 2014;48(6):571–8. doi: 10.1016/j.alcohol.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 131.Iacono WG, Malone SM. Developmental endophenotypes: indexing genetic risk for substance abuse with the P300 brain event-related potential. Child Development Perspectives. 2011;5(4):239–47. doi: 10.1111/j.1750-8606.2011.00205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Carlson SR, Iacono WG. Deviant P300 amplitude development in males is associated with paternal externalizing psychopathology. J Abnorm Psychol. 2008;117(4):910–23. doi: 10.1037/a0013443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *133.Spadoni AD, Simmons AN, Yang TT, Tapert SF. Family history of alcohol use disorders and neuromaturation: a functional connectivity study with adolescents. Am J Drug Alcohol Abuse. 2013;39(6):356–64. doi: 10.3109/00952990.2013.818680. Takes a developmental perspective on endophenotype research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Miller JA, Ding SL, Sunkin SM, Smith KA, Ng L, Szafer A, et al. Transcriptional landscape of the prenatal human brain. Nature. 2014;508(7495):199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489(7416):391–9. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Spear LP. The adolescent brain and age-related behavioral manifestations. Neurosci Biobehav Rev. 2000;24:417–463. doi: 10.1016/s0149-7634(00)00014-2. [DOI] [PubMed] [Google Scholar]
- 137.Monnig MA, Tonigan JS, Yeo RA, Thoma RJ, McCrady BS. White matter volume in alcohol use disorders: a meta-analysis. Addict Biol. 2013;18(3):581–92. doi: 10.1111/j.1369-1600.2012.00441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Baare WF, Hulshoff Pol HE, Boomsma DI, Posthuma D, de Geus EJ, Schnack HG, et al. Quantitative genetic modeling of variation in human brain morphology. Cereb Cortex. 2001;11(9):816–24. doi: 10.1093/cercor/11.9.816. [DOI] [PubMed] [Google Scholar]
- 139.Posthuma D, de Geus EJ, Neale MC, Hulshoff Pol HE, Baare WEC, Kahn RS, et al. Multivariate genetic analysis of brain structure in an extended twin design. Behav Genet. 2000;30(4):311–9. doi: 10.1023/a:1026501501434. [DOI] [PubMed] [Google Scholar]
- 140.Fein G, Fein D. Subcortical volumes are reduced in short-term and long-term abstinent alcoholics but not those with a comorbid stimulant disorder. NeuroImage Clinical. 2013;3:47–53. doi: 10.1016/j.nicl.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cheetham A, Allen NB, Whittle S, Simmons J, Yucel M, Lubman DI. Volumetric differences in the anterior cingulate cortex prospectively predict alcohol-related problems in adolescence. Psychopharmacology. 2014;231(8):1731–42. doi: 10.1007/s00213-014-3483-8. [DOI] [PubMed] [Google Scholar]
- 142.Englund MM, Egeland B, Oliva EM, Collins WA. Childhood and adolescent predictors of heavy drinking and alcohol use disorders in early adulthood: a longitudinal developmental analysis. Addiction. 2008;103:23–35. doi: 10.1111/j.1360-0443.2008.02174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ehlers CL, Phillips E. Association of EEG alpha variants and alpha power with alcohol dependence in Mexican American young adults. Alcohol. 2007;41(1):13–20. doi: 10.1016/j.alcohol.2007.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Carlson SR, Iacono WG, McGue M. P300 amplitude in nonalcoholic adolescent twin pairs who become discordant for alcoholism as adults. Psychophysiology. 2004;41(6):841–4. doi: 10.1111/j.0048-5772.2004.00238.x. [DOI] [PubMed] [Google Scholar]
- 145.Roopesh BN, Rangaswamy M, Kamarajan C, Chorlian DB, Stimus A, Bauer LO, et al. Priming deficiency in male subjects at risk for alcoholism: the N4 during a lexical decision task. Alcohol Clin Exp Res. 2009;33(12):2027–36. doi: 10.1111/j.1530-0277.2009.01042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Roopesh BN, Rangaswamy M, Kamarajan C, Chorlian DB, Pandey AK, Porjesz B. Reduced resource optimization in male alcoholics: N400 in a lexical decision paradigm. Alcohol Clin Exp Res. 2010;34(11):1905–14. doi: 10.1111/j.1530-0277.2010.01279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Perlman G, Johnson W, Iacono WG. The heritability of P300 amplitude in 18-year-olds is robust to adolescent alcohol use. Psychophysiology. 2009;46(5):962–9. doi: 10.1111/j.1469-8986.2009.00850.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wand GS, Dobs AS. Alterations in the hypothalamic-pituitary-adrenal axis in actively drinking alcoholics. J Clin Endocrinol Metab. 1991;72(6):1290–5. doi: 10.1210/jcem-72-6-1290. [DOI] [PubMed] [Google Scholar]
- 149.Starcke K, van Holst RJ, van den Brink W, Veltman DJ, Goudriaan AE. Physiological and endocrine reactions to psychosocial stress in alcohol use disorders: Duration of abstinence matters. Alcohol Clin Exp Res. 2013;37(8):1343–50. doi: 10.1111/acer.12103. [DOI] [PubMed] [Google Scholar]
- 150.Bartels M, Van den Berg M, Sluyter F, Boomsma DI, de Geus EJ. Heritability of cortisol levels: review and simultaneous analysis of twin studies. Psychoneuroendocrinology. 2003;28(2):121–37. doi: 10.1016/s0306-4530(02)00003-3. [DOI] [PubMed] [Google Scholar]
- 151.Kranzler HR, Sandstrom KA, Van Kirk J. Sweet taste preference as a risk factor for alcohol dependence. Am J Psychiat. 2001;158(5):813–5. doi: 10.1176/appi.ajp.158.5.813. [DOI] [PubMed] [Google Scholar]
- 152.Keskitalo K, Tuorila H, Spector TD, Cherkas LF, Knaapila A, Silventoinen K, et al. Same genetic components underlie different measures of sweet taste preference. Am J Clin Nutr. 2007;86:1663–9. doi: 10.1093/ajcn/86.5.1663. [DOI] [PubMed] [Google Scholar]
- 153.Lange LA, Kampov-Polevoy AB, Garbutt JC. Sweet liking and high novelty seeking: independent phenotypes associated with alcohol-related problems. Alcohol Alcoholism. 2010;45(5):431–6. doi: 10.1093/alcalc/agq040. [DOI] [PubMed] [Google Scholar]
- 154.Schuckit MA, Smith TL, Danko G, Kuperman S, Bierut LJ, Hesselbrock V. Correlations among first-degree relatives for responses on the Self-Rating of the Effects of Alcohol Questionnaire in teenagers. J Stud Alcohol. 2005;66(1):62–5. doi: 10.15288/jsa.2005.66.62. [DOI] [PubMed] [Google Scholar]
- 155.Pfefferbaum A, Desmond JE, Galloway C, Menon V, Glover GH, Sullivan EV. Reorganization of frontal systems used by alcoholics for spatial working memory: an fMRI study. Neuroimage. 2001;14(1 Pt 1):7–20. doi: 10.1006/nimg.2001.0785. [DOI] [PubMed] [Google Scholar]
- 156.Vollstadt-Klein S, Hermann D, Rabinstein J, Wichert S, Klein O, Ende G, et al. Increased activation of the ACC during a spatial working memory task in alcohol-dependence versus heavy social drinking. Alcohol Clin Exp Res. 2010;34(5):771–6. doi: 10.1111/j.1530-0277.2010.01149.x. [DOI] [PubMed] [Google Scholar]
- 157.Tapert SF, Brown GG, Kindermann SS, Cheung EH, Frank LR, Brown SA. fMRI measurement of brain dysfunction in alcohol-dependent young women. Alcohol Clin Exp Res. 2001;25(2):236–45. doi: 10.1111/j.1530-0277.2001.tb02204.x. [DOI] [PubMed] [Google Scholar]
- 158.Spadoni AD, Norman AL, Schweinsburg AD, Tapert SF. Effects of family history of alcohol use disorders on spatial working memory BOLD response in adolescents. Alcohol Clin Exp Res. 2008;32(7):1135–45. doi: 10.1111/j.1530-0277.2008.00694.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Webb A, Lind PA, Kalmijn J, Feiler HS, Smith TL, Schuckit MA, et al. The investigation into CYP2E1 in relation to the level of response to alcohol through a combination of linkage and association analysis. Alcohol Clin Exp Res. 2011;35(1):10–8. doi: 10.1111/j.1530-0277.2010.01317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zlojutro M, Manz N, Rangaswamy M, Xuei X, Flury-Wetherill L, Koller D, et al. Genome-wide association study of theta band event-related oscillations identifies serotonin receptor gene HTR7 influencing risk of alcohol dependence. Am J Med Genet B Neuropsychiatr Genet. 2011;156B(1):44–58. doi: 10.1002/ajmg.b.31136. [DOI] [PMC free article] [PubMed] [Google Scholar]