Background: The GP12 protein from bacteriophage ϕ29 is a likely wall teichoic acid hydrolase.
Results: GP12 is an efficient catalyst of wall teichoic acid hydrolysis that is influenced by glycosylation of the wall teichioc acid polymer.
Conclusion: GP12 may serve as a tool for the characterization of wall teichoic acid polymers.
Significance: Characterization of a prototypical wall teichoic acid hydrolase using chemically defined substrates.
Keywords: bacteriophage, enzyme, enzyme mechanism, enzyme turnover, phosphodiesterases, Bacillus subtilis, wall teichoic acid
Abstract
The genetics and enzymology of the biosynthesis of wall teichoic acid have been the extensively studied, however, comparatively little is known regarding the enzymatic degradation of this biological polymer. The GP12 protein from the Bacillus subtilis bacteriophage ϕ29 has been implicated as a wall teichoic acid hydrolase. We have studied the wall teichoic acid hydrolase activity of pure, recombinant GP12 using chemically defined wall teichoic acid analogs. The GP12 protein had potent wall teichoic acid hydrolytic activity in vitro and demonstrated ∼13-fold kinetic preference for glycosylated poly(glycerol phosphate) teichoic acid compared with non-glycosylated. Product distribution patterns suggested that the degradation of glycosylated polymers proceeded from the hydroxyl terminus of the polymer, whereas hydrolysis occurred at random sites in the non-glycosylated polymer. In addition, we present evidence that the GP12 protein possesses both phosphodiesterase and phosphomonoesterase activities.
Introduction
Wall teichoic acids (WTAs)2 are polyanionic glycopolymers covalently attached to peptidoglycan in the cell wall of Gram-positive bacteria. Prototypical WTA polymers are repeating units of 1–3-linked glycerol phosphate or 1–5-linked ribitol phosphate and constitute roughly 50% of the wall mass (1). WTAs have been associated with diverse cellular processes including cell division and morphology, peptidoglycan assembly, bacteriophage recognition and adherence, and as a virulence determinant (1). Recently, the disruption of WTA biosynthesis was shown to restore the susceptibility of methicillin-resistant Staphylococcus aureus to certain β-lactam antibiotics (2–4), revealing a surprising role for WTA in β-lactam resistance and highlighting steps in its biosynthetic pathway as attractive antibacterial therapeutic targets.
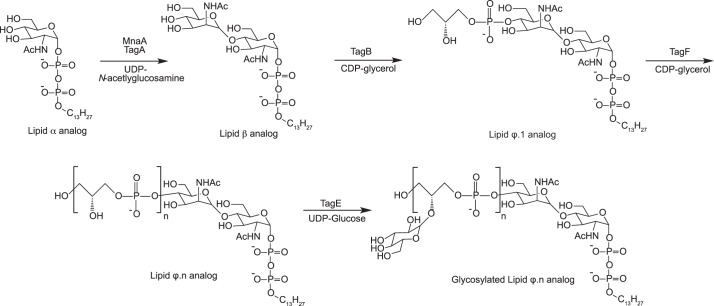
The genetics and biochemistry of WTA biosynthesis have been extensively studied in both the model Gram-positive bacterium Bacillus subtilis and the pathogen S. aureus (1). Biosynthesis of poly(glycerol phosphate) WTA in B. subtilis is illustrative (see Scheme 1). Polymer synthesis begins on the inner cell surface with the addition of N-acetlyglucosamine-1-phosphate to membrane-bound undecaprenyl phosphate, catalyzed by the TagO enzyme and generating lipid α (WTA intermediates are named after the enzymes that act on them (5), summarized in Table 1). Next, TagA adds N-acetlymannosamine to lipid α to form lipid β, the substrate for the primase enzyme TagB that adds a single glycerol phosphate molecule to form lipid ϕ.1 (6). TagF is the polymerase that adds 30 to 50 glycerol phosphate units to this substrate (7), followed by the modification of multiple glycerol units in poly(glycerol phosphate) WTA with α-glucose by TagE (8). The polymer is then transported to the extracellular surface by the TagGH ABC transporter system (9) where the gene products of the dltABCD operon effect d-alalynation (10) and the tagTUV enzymes are believed to catalyze final polymer transfer to N-acetylmuramic acid of peptidoglycan (11).
SCHEME 1.
Synthesis of tridecyl-linked WTA analogs.
TABLE 1.
Nomenclature and chemical composition of analogs for wall teichoic acid precursors and polymers
| Analog | Chemical compositiona |
|---|---|
| Lipid α analog | GlcNAc-1-P-P-tridecane |
| Lipid β analog | ManNAc-β-(1–4)-GlcNAc-1-P-P-tridecane |
| Lipid ϕ.1 analog | GroP-ManNAc-β-(1–4)-GlcNAc-1-P-P-tridecane |
| Lipid ϕ.n analog | (GroP)n-ManNAc-β-(1–4)-GlcNAc-1-P-P-tridecane |
| Glycosylated lipid ϕ.n analog | (α-d-Glu-(1→2)-GroP)n-ManNAc-β-(1–4)-GlcNAc-1-P-P-tridecane |
a P, phosphate; GlcNAc, N-acetyl glucosamine, ManNAc, N-acetyl mannosamine; GroP, glycerol 3-phosphate; Glu, glucose.
In contrast to the thorough understanding of its biosynthesis, the enzymology and biological relevance of WTA degradation have not been well studied. Evidence of such enzymatic activity was first reported more than 40 years ago by Wise and co-workers (12), with the discovery that B. subtilis is capable of utilizing its cell wall as a phosphate source. This was attributed to expression of a so-called teichoicase with glycerophosphodiesterase activity specific for WTA. It was later speculated that the expression of this enzyme may be induced by phosphate starvation (13), enabling the procurement of necessary phosphate from WTA. In line with this, during phosphate-limited growth of B. subtilis, much of the WTA in the cell wall is replaced by the phosphate-free polymer teichuronic acid (14, 15). Apart from the potential importance in the phosphate starvation response, a role for WTA degradation in the regulation of cell wall structure and composition can be envisioned. For instance, WTAs have been shown to direct bacterial peptidoglycan hydrolases (16–19) and consequently, influence wall remodeling. Also, as much as 50% of the wall is recycled during bacterial cell division (20, 21), implying a need perhaps for enzymes capable of processing the very large quantities of WTA in peptidoglycan.
Through activity-guided purification, Kusser and Fielder (22, 23) achieved some enrichment of the wall teichoicase activity from B. subtilis and later from Bacillus pumilus (24), yet these studies were limited in two important aspects. First, the enzyme preparations were never linked to a gene or protein sequence. Second, the substrates used were poorly defined, consisting of crude cell wall preparations or partially purified WTAs. Thus, bacterial teichoicases remain obscure, as are the mechanistic details of their biochemical activity.
Although bacterial enzymes that degrade WTA and their corresponding genes have yet to be identified in any bacterium, the GP12 protein produced by the B. subtilis bacteriophage ϕ29 is thought to possess this activity. GP12 is an appendage protein found in the neck region of ϕ29 that is involved in host cell recognition and entry (25). During phage maturation, GP12 forms homotrimers that undergo autoproteolysis mediated by an intramolecular chaperone domain (26), prior to forming the 12 appendages attached to the neck of phage particles (25, 27). As part of a crystallographic investigation of this large (92 kDa) multifunctional protein, Xiang et al. (27) showed that GP12 could drastically reduce the apparent molecular size of poly(glycerol phosphate) WTA extracted from B. subtilis 168, suggesting that the protein has inherent wall teichoicase activity. The activity was noted to be highly dependent on divalent metal ions, two of which (Ca2+ and Mg2+) were observed bound at the proposed WTA-degrading active site in a pectin lyase domain (28), prompting the suggestion of a two-metal ion mechanism for catalysis of WTA phosphodiester bond hydrolysis (27). Analogous mechanisms for phosphate ester hydrolysis involving a bimetallated active center are known for enzymes from the binuclear metallophosphatase family, which includes glycerophosphodiesterases and purple acid phosphatases (29), and interestingly, GP12 is structurally unrelated to this family.
In light of the previous qualitative examination, GP12 represents an ideal candidate for the study of enzyme-catalyzed WTA degradation. To that end, we have performed in-depth investigations on this prototypic wall teichoicase (GP12) using chemically defined and soluble WTA substrate analogs. Indeed, the study of WTA biosynthetic enzymes has been greatly enabled by chemoenzymatic syntheses of analogs for the biosynthetic intermediates. In place of the 55-carbon undecaprenyl chain, such analogs feature a more tractable lipid moiety that facilitates manipulation in aqueous systems (8, 30–33). Moreover, soluble poly(glycerol phosphate) polymer is accessible through the polymerization of glycerol phosphate onto CDP-glycerol (34). Equipped with chemically defined substrates, we developed HPLC-based assays for poly(glycerol phosphate) hydrolysis that were used to meticulously characterize the wall teichoicase activity of GP12. We found that polymer glycosylation led to an enhanced rate of polymer degradation, and also appeared to influence the mode of GP12 action. Further interrogation of GP12 catalytic activity suggested that the enzyme utilizes phosphodiesterase and phosphomonoesterase activity in its reaction with WTA polymers.
Experimental Procedures
General Methods
All chemicals were purchased from Sigma with the exception of the following: ammonium bicarbonate was from Thermo Fisher Scientific (Waltham, MA), acetonitrile was from Caledon Laboratory Chemicals (Georgetown, Ontario, Canada), Tris and urea were from BioShop (Burlington, Ontario, Canada), sn-[U-14C]glycerol 3-phosphate, UDP-d-[U-14C]glucose (250 mCi/mmol), and Ultima-Flo M scintillation fluid were from PerkinElmer Life Sciences and N-acetyl-d-[U-14C]glucosamine (300 mCi/mmol) was from American Radiolabeled Chemicals Inc. (St. Louis, MO). The enzymes MnaA, TagA, TagB, TagF, and TagE from B. subtilis and TarD from S. aureus were purified as previously described (6–8, 32, 35). CDP-glycerol and CDP-[U-14C]glycerol (98.7 mCi/mmol) were synthesized using TarD according to established methods (21).
Cloning, Expression, and Purification of GP12
The genetic sequence for a GP12 construct missing the first 88 amino acids was PCR-amplified from pET28b-GP12 (27) using primers ΔN88GP12-Forward and GP12-Reverse (supplemental Table S1). The amplicon was cloned into pET28b plasmid using NcoI and HindIII restriction sites to generate pΔN88GP12. Escherichia coli BL21-CodonPlus (DE3)-RIL transformed with pΔN88GP12 was grown at 25 °C in Luria Bertani (LB) media supplemented with 20 μg/ml of chloramphenicol and 50 μg/ml of kanamycin to an A600 of 0.6 before inducing protein expression with 0.1 mm isopropyl β-d-1-thiogalactopyranoside. Expression was continued for 20 h at 16 °C. Cells were harvested by centrifugation (8500 × g for 15 min at 4 °C) and washed with 0.85% NaCl, prior to resuspension in a buffer containing Tris-HCl (pH 7.5), 300 mm NaCl, 25 mm imidazole. EDTA-free Protease Inhibitor Mixture (Roche Diagnostics, Laval, Québec, Canada) was added to the suspension and the cells were lysed by two consecutive passages through a cell disruptor (Constant Systems, Daventry, Northants, UK) at 30,000 p.s.i. Cell lysates were clarified by centrifugation (40,000 × g for 60 min at 4 °C) prior to loading on a 1-ml HisTrapHP column (GE Life Sciences) pre-equilibrated with resuspension buffer. GP12 was eluted over a 25 to 400 mm imidazole gradient, and fractions containing pure protein were pooled and the buffer exchanged for 50 mm Tris (pH 7.5), 100 mm NaCl, 15% glycerol (v/v), using a HiPrep 26/10 desalting column (GE Life Sciences). Protein concentration was determined according to the method of Gill and von Hippel (36).
Synthesis of WTA Analog Precursors
Lipid α, β, and ϕ.1 analogs were synthesized as described previously (30, 32). [14C]ManNAc-labeled lipid β analog (1 mm stock solution; 20 mCi/mmol) was synthesized by including 20 mCi of UDP-N-acetyl-d-[1-U-14C]glucosamine in the reaction. 1.2 μCi of [14C]ManNAc lipid β was then used to prepare [14C]ManNAc lipid ϕ.1 analog (0.3 mm stock solution; 20 mCi/mmol). Reaction progress was assessed from substrate consumption (monitoring absorbance at 261 nm for uridine containing reagents and at 271 nm for cytidine containing reagents) by reversed phase HPLC using a Waters HPLC system (Waters 1525 HPLC pump and 2998 PDA detector; Waters, Mississauga, Ontario, Canada), or by scintillation counting with an in-line 150TR Flow Scintillation Analyzer (PerkinElmer). Upon reaction completion, enzymes were removed using a centrifugal filter with a 10,000 Da molecular mass cut-off (Pall Life Sciences, Mississauga, Ontario, Canada).
Synthesis of WTA Analogs
The chemoenzymatic approach for the preparation of soluble tricecyl-linked WTA analogs is summarized in Scheme 1. Lipid ϕ.40 analog was synthesized using TagF according to established methods, with the lipid ϕ.1 analog and CDP-glycerol as the glycerol phosphate acceptor and donor, respectively (32, 33). To incorporate 14C into the glycerol phosphate polymer chain, 0.91 μCi of CDP-[U-14C]glycerol (98.7 mCi/mmol) was included in the reaction and the analogswere prepared at 91 mCi/mmol. [14C]ManNAc lipid ϕ.40 analog was similarly prepared at 20 mCi/mmol, by the inclusion of 0.2 μCi of [14C]ManNAc lipid ϕ.1 analog to the reaction. Reaction progress was monitored by anion exchange HPLC using a DNA PAC PA200 anion exchange column (Thermo Fisher Scientific), with visualization by in-line scintillation counting. The solvents used were: A, 20 mm ammonium bicarbonate (pH 8.0) containing 10% acetonitrile (v/v) and B, 20 mm ammonium bicarbonate (pH 8.0) containing 1 m NaCl and 10% acetonitrile (v/v). Substrates and products were separated over the following gradient condition for solvent B: 0–30% (3 min), 30–45% (10 min), 45–100% (5 min), and finally 100% for 5 min. Upon reaction completion, enzymes and low molecular weight by-products were removed using centrifugal filters with molecular mass cut-offs of 30,000 and 3000 Da, respectively (Pall Life Sciences).
Polymer glycosylation reactions were performed as described previously (8), by the incubation of TagE with [14C]glycerol- or [14C]ManNAc lipid ϕ.40 analog and a molar excess of UDP-glucose to glycerol phosphate units, or likewise with non-radioactive lipid ϕ.40 analog and UDP-[14C]glucose. The progress of glycosylation reactions was assessed by anion exchange HPLC using chromatographic conditions identical to those for the non-glycosylated analog, or by UDP-glucose consumption with ion-paired reversed phase HPLC (8). Upon reaction completion, enzymes and reaction by-products were removed by ultrafiltration as above.
Wall Teichoic Acid Degradation Assays
Unless otherwise stated, GP12 reactions with WTA analogs were performed at room temperature in a buffer comprised of 50 mm Tris-HCl (pH 7.5) and 30 mm MgCl2. Reactions were initiated by addition of GP12 and terminated at specified time points by the addition of urea to a final concentration of 6 m. Radioactive reaction products were separated by anion exchange HPLC as described above. Initial rate analyses for GP12 degradation of 14C-labeled WTA analogs were performed with varying concentrations of GP12 and fixed amounts of WTA substrate. The amount of fully polymeric starting substrate remaining after specific time points was quantified by peak integration.
Liquid Chromatography-Mass Spectrometry
Reactions (30 μl) between GP12 and non-radioactive lipid ϕ.40 analogs were quenched with an equal volume of methanol, and product mixtures were analyzed by normal phase liquid chromatography ESI-QTOF mass spectrometry, as described previously (37).
GP12 Hydrolysis of para-Nitrophenyl Phosphates
The catalytic activity of GP12 with pNPP or bis-pNPP was assayed by monitoring the production of para-nitrophenol (pNP). Unless otherwise stated, reactions (100 μl) were performed in buffer containing 50 mm Tris-HCl (pH 7.5), 30 mm MgCl2, and 10 mm CaCl2. After initiation with GP12, the absorbance at 400 nm was measured continuously on a SpectraMax microplate reader (Molecular Devices, Sunnyvale, CA). pNP generated was quantified by interpolation to a standard curve.
Assays for the determination of kinetic parameters contained substrate concentrations ranging between 40.0 and 0.625 mm for pNPP, and from 28.50 to 0.44 mm for bis-pNPP. Initial rates of pNP production (substrate consumption <10%) were plotted as a function of substrate concentration and the data fit by non-linear regression (Prism 6.0; GraphPad, La Jolla, CA) to Equation 1.
 |
Measurement of Inorganic Phosphate
Reactions (50 μl) of GP12 with WTA analogs or bis-pNPP were performed in a buffer containing 50 mm Tris-HCl (pH 7.5), 5 mm MgCl2 and for bis-pNPP hydrolysis, 5 mm CaCl2. At specified time points, reactions were stopped by the addition of EDTA to a final concentration of 10 mm, and then 100 μl of BIOMOL Green reagent (Enzo Life Sciences, Inc., Farmingdale, NY) was added. Pi was detected by measuring absorbance at 640 nm after 15 min, and quantified by interpolation to a standard curve.
Results
Characterization of WTA Analogs
Aiming for a polymer of roughly native length at 40 glycerol phosphate units, we synthesized tridecyl-linked WTA analogs using TagF, and CDP-[14C]glycerol as the glycerol phosphate donor at 39 times the molar concentration of lipid ϕ.1 acceptor. On completion of the reaction, anion exchange HPLC analysis showed a single radioactive product peak, indicating a polyanionic species formed from the polymerization of glycerol phosphate onto the lipid ϕ.1 analog (Fig. 1A). MALDI-TOF analyses of parallel non-radioactive reactions were consistent with the production of lipid ϕ.40 analog, but also showed the polymerization product to comprise a mixture of species differing successively by the mass of the repeating WTA polymer unit (supplemental Fig. S1). This mass distribution coincides with the previous description of non-processive polymerization catalyzed by TagF that precludes the generation of a completely homogenous polymer (33). By controlling the ratio of TagF substrates (e.g. lipid ϕ.1 analog:CDP-glycerol ratio of 1:39), we were able to target the product mixture to a relatively uniform range of species that co-eluted during anion exchange chromatography. The small differences in polymer sizes (1–5 glycerol phosphate units) contrasts favorably with far greater differences for purified WTA or WTA in cell wall extracts (typical polymer sizes of 30–50 glycerol phosphate units) and would not adversely affect subsequent characterizations of the products from degradation reactions.
FIGURE 1.
Characterization of WTA analogs. Analog structures are shown next to radioactive chromatograms for A, [14C]glycerol lipid ϕ.40 analog; B, glycosylated lipid ϕ.40 analog with 14C incorporated into glycerol (i) and glucose (ii) (prepared by the inclusion of 0.42 mCi of UDP-d-[U-14C]glucose (250 mCi/mmol) to a TagE-catalyzed reaction containing non-radiolabeled lipid ϕ.40 analog); and C, CMP-poly(glycerol phosphate) WTA analogs. Polymerization of glycerol phosphate onto CMP-glycerol was accomplished by incubation of CDP-glycerol (3.29 mm) with TagF (34), and glycosylation was performed as with the lipid ϕ.40 analog. 14C was incorporated by the inclusion of either 1 μCi of CDP-[U-14C]glycerol (98.7 mCi/mmol) or 0.45 mCi of UDP-d-[U-14C]glucose (250 mCi/mmol). Chromatograms with glycosylated CMP-poly(glycerol phosphate) show polymer with 14C incorporated as [14C]glycerol (i) or [14C]glucose (ii). Under identical chromatographic conditions, retention times for CMP-poly(glycerol phosphate) analogs were slightly increased compared with tridecyl-linked analogs.
Glycosylation of [14C]glycerol lipid ϕ.40 analog using TagE and UDP-d-glucose likewise yielded a single radioactive product, as judged by anion exchange HPLC (Fig. 1B). A product with an identical retention time was seen on reaction of non-radiolabeled lipid ϕ.40 analog with UDP-d-[14C]glucose, confirming this species as glycosylated polymer (Fig. 1B). MALDI-TOF MS analysis indicated virtually complete modification of all available glycerol phosphate units with glucose (supplemental Fig. S1), also reflected by assessment of UDP-glucose consumed in the reaction (not shown).
TagF most efficiently elongates the primer lipid ϕ.1 (33), but is also capable of using CDP-glycerol as a primer for poly(glycerol phosphate) synthesis, forming CMP-poly(glycerol phosphate) (34). We prepared this nucleotide-linked polymer to test the generality of GP12 activity and verify that the tridecyl tail did not hamper enzymatic degradation. The reaction for [14C]glycerol CMP-poly(glycerol phosphate) WTA showed a single product on anion exchange HPLC analysis (Fig. 1C) and as previously observed, this analog was estimated to exceed the polymer length of the lipid ϕ.40 analog (8, 34). As with the tridecyl-linked analog, glycosylation of CMP-poly(glycerol phosphate) WTA yielded a single product with a shorter retention time than non-glycosylated (Fig. 1C).
GP12 Catalyzes the Degradation of WTA Analogs
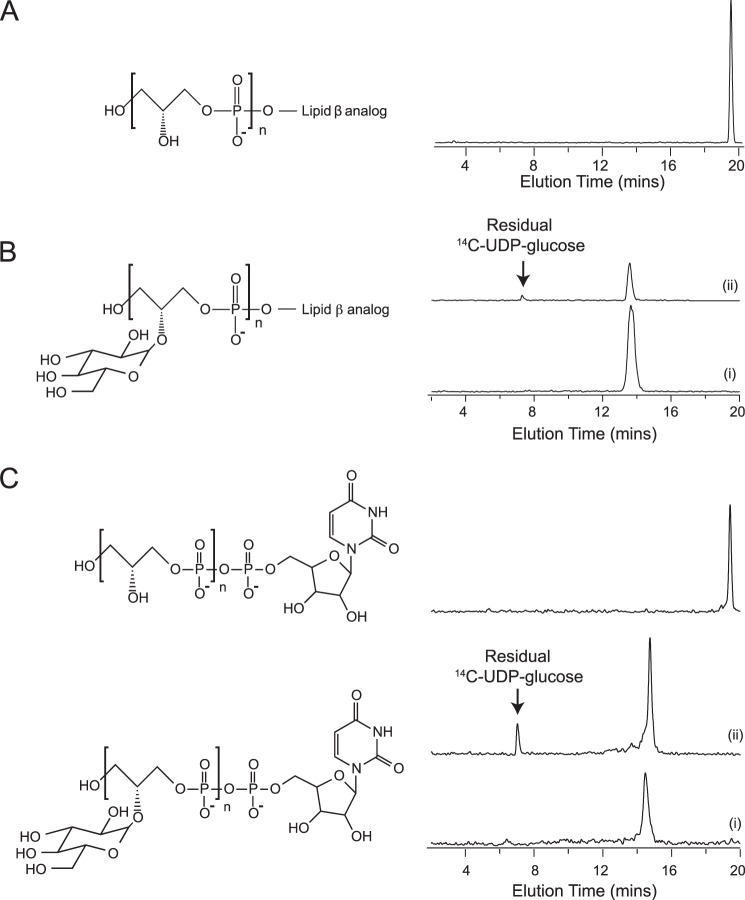
With chemically defined [14C]glycerol-labeled WTA substrates in hand, we examined the activity of GP12 with these analogs by monitoring product formation with anion exchange HPLC. Fig. 2, A and B, show the glycerol containing products generated from lipid ϕ.40 analogs over time, and it was evident that GP12 catalyzed cleavage of the glycerol phosphate polymer chain. Whereas products from the reaction with the non-glycosylated analog spanned a continuum of polymer sizes, a narrow range of products corresponding to species of reduced apparent polymer length was produced from reaction of the glycosylated analog. These emerged after as little as 2 min and were confirmed to contain glucose from a parallel reaction of [14C]glucose-glycosylated lipid ϕ.40 analog (Fig. 2B). As well, degradation occurred to a greater extent and with lower GP12 concentrations for the glycosylated substrate. Under reaction conditions identical to those for WTA analogs, we detected no activity for GP12 with glycerol phosphate (phosphatase), CDP-glycerol (phosphodiesterase), or the lipid ϕ.1 analog (phosphodiesterase).
FIGURE 2.
GP12 catalyzes cleavage of the glycerol phosphate polymer chain in WTA analogs. Reactions contained 100 μm [14C]glycerol-labeled WTA analog and 1 or 0.1 μm GP12, for the non-glycosylated and glycosylated lipid ϕ.40 analog, respectively. After termination of the reactions at specified time points, [14C]glycerol phosphate containing products were separated by anion exchange HPLC. Analog structures, with sites of 14C incorporation marked by asterisks, are shown below radioactive chromatograms for A, products from the degradation of lipid ϕ.40 analog; inset shows the 18-h time point on a magnified scale and B, products from degradation of glycosylated lipid ϕ.40 analog; inset shows the products from reaction of [14C]glucose-glycosylated lipid ϕ.40 analog.
Similar results were obtained when CMP-poly(glycerol phosphate) WTAs were used as substrates, suggesting no significant influence of the tridecyl tail on activity (Figs. 3, A and B). Subsequent investigations were focused on the tridecyl-linked analogs, which could be quantified based on definitive knowledge of the starting concentration of glycerol phosphate acceptor and the polymer size.
FIGURE 3.
Reactions of GP12 with CMP-poly(glycerol phosphate) WTA analogs. Radioactive chromatograms from the anion exchange HPLC separation of the products are shown for A, products from [14C]glycerol CMP-poly(glycerol phosphate) degradation; the broad product peak with a slightly reduced retention time indicates co-elution of multiple species of similar lengths; and B, products from glycosylated CMP-poly(glycerol phosphate) WTA containing [14C]glycerol (i) and [14C]glucose (ii).
GP12 More Efficiently Degrades Glycosylated Lipid ϕ.40 Analog
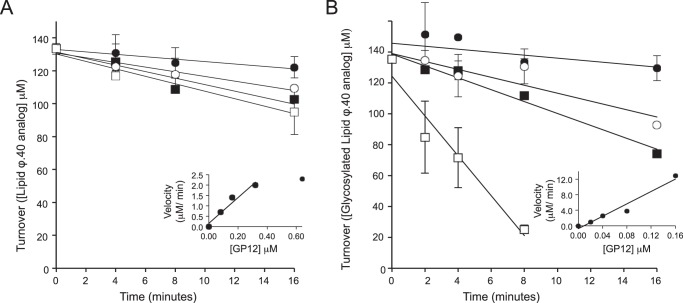
The above results implied GP12 was more active toward glycosylated WTA analogs. To explore this apparent preference, we examined activity as a function of time and GP12 concentration, monitoring substrate consumption with anion exchange HPLC. We note here that substrate concentrations were in large excess of GP12 and substrate consumption was limited to 15% to ensure measurement of initial rates and to favor turnover of the fully polymeric, time 0 substrate. The assay showed a linear response with respect to time and enzyme concentration and under these conditions, apparent turnover by GP12 was estimated at 6.2 and 80 min−1 for non-glycosylated and glycosylated lipid ϕ.40 analog, respectively (Fig. 4, A and B).
FIGURE 4.
GP12 exhibits a preference for glycosylated WTA analogs. Activity was assayed by monitoring the disappearance of full-length [14C]glycerol-labeled WTA analogs by anion exchange HPLC. Data points are the mean from three independent experiments and error bars are the S.D. Insets show plots of the initial rates as a function of GP12 concentration and enzyme turnover, calculated as the slope of the linear regression line, was 6.2 and 80 μm min−1, for non-glycosylated lipid ϕ.40 and glycosylated lipid ϕ.40, respectively. A, reactions contained lipid ϕ.40 analog at 135 μm and GP12 at 0.08 (●), 0.16 (○), 0.32(■), and 0.64 (□) μm. B, reactions contained glycosylated lipid ϕ.40 analog at 138 μm and GP12 at 0.02 (●), 0.04 (○), 0.08 (■), and 0.16 (□) μm.
Product Distribution Analyses
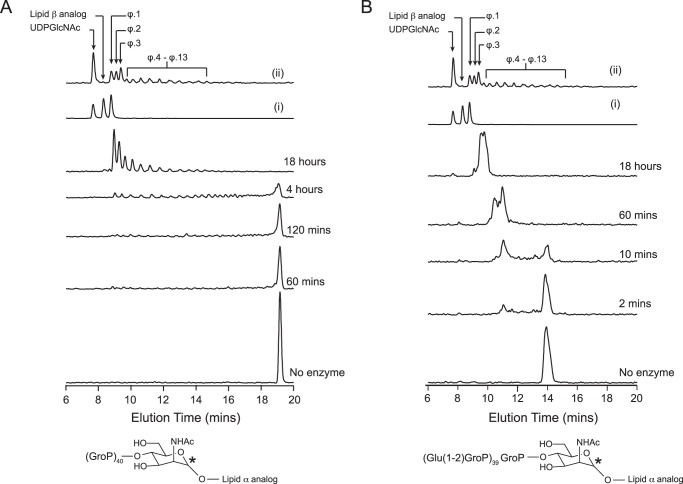
The degradation of non-glycosylated and glycosylated analogs seemed to produce different outcomes with respect to the size and distribution of products. We hypothesized that this was indicative of different modes of degradation for the two substrates. However, because the starting material in either case was labeled throughout the polymer chain, products without the tridecyl moiety were indistinguishable from those having this appendage. To differentiate between these and thereby gain insight into the mode of polymer degradation, we synthesized WTA analogs labeled with 14C exclusively at ManNAc to ensure the detection of only tridecyl-containing products, as GP12 was not active on the lipid ϕ.1 moiety.
Fig. 5A shows HPLC chromatograms of products from the reaction of GP12 with lipid ϕ.40 analog radioactively labeled as described above. Between 0 and 4 h, the product distribution resembled that from reactions with the [14C]glycerol-labeled polymer. Further reaction yielded a product distribution skewed toward products with smaller apparent polymer sizes, as opposed to the relatively uniform distribution observed for [14C]glycerol-labeled products. Hence, the broad variability in product composition from the lipid ϕ.40 analog reaction includes both tridecyl-linked and tridecyl-free species. In contrast, tridecyl-linked products, significantly reduced in apparent polymer size, were detected after 2 min of reaction with [14C]ManNAc-glycosylated lipid ϕ.40 (Fig. 5B) and the product distribution remained skewed toward smaller species. Consideration of these findings alongside observations with the equivalent [14C]glycerol-labeled analog suggests that these truncated, tridecyl-linked species were co-produced with 2–5-mers of (glucose) glycerol phosphate in the early stages of glycosylated lipid ϕ.40 analog degradation. Collectively, these results portray distinct modes of GP12 action against glycosylated and non-glycosylated substrates; the former is preferentially hydrolyzed at regular intervals, whereas hydrolysis occurs at random sites throughout the latter. Notably, degradation reactions ultimately generated tridecyl-linked products containing 2–5 monomer units, irrespective of polymer glycosylation. It follows that poly(glycerol phosphate) substrates of GP12 contain at least 3 monomer units.
FIGURE 5.
Degradation of non-glycosylated versus glycosylated lipid ϕ.40 analogs produces different distributions of tridecyl-linked products. Reactions were performed with 200 μm [14C]ManNAc lipid ϕ.40 analogs. The top two traces in both panels show the elution profiles for standard mixtures containing (14C-labeled) UDP-GlcNAc (i), lipid β analog and lipid ϕ.1 analog and UDP-GlcNAc (ii) and the product mixture from TagF-catalyzed polymerization reaction containing a 3:1 ratio of CDP-glycerol to the lipid ϕ.1 analog. Analog structures with sites of 14C incorporation marked by asterisks are shown below radioactive chromatograms for A, products from reactions with lipid ϕ.40 analog and 1 μm GP12; and B, glycosylated lipid ϕ.40 analog and 0.1 μm GP12.
LC-MS analyses of WTA degradation products offered additional support for the inferred modes of polymer degradation (Table 2, supplemental Fig. S2). First, this analysis confirmed that cleavage of the polymer occurred at phosphate esters. Furthermore, various glycerol phosphate-containing species were identified from a reaction with the non-glycosylated analog, whereas species containing one or two (glycosylated) glycerol units were detected from a reaction with glycosylated WTA. Surprisingly, phosphomonoester products anticipated from phosphodiester hydrolysis were largely absent from the species detected, which was suggestive of monoesterase activity by GP12.
TABLE 2.
Identification of products from GP12-catalyzed degradation of tridecyl WTA analogs by LC-MS
Reactions containing lipid ϕ.40 and glycosylated lipid ϕ.40 were quenched after 60 and 5 min, respectively. Extracted ion chromatograms and associated mass spectra are provided in supplemental Fig. S2.
| Ion observed (m/z) | Species |
|---|---|
| Lipid ϕ.40 analog | |
| 171.02 | [GroP]− |
| 245.06 | [Gro-P-Gro]− |
| 399.07 | [Gro-P-Gro-P-Gro]− |
| 553.07 | [Gro-P-Gro-P-Gro-P-Gro]− |
| 707.07 | [Gro-P-Gro-P-Gro-P-Gro-P-Gro]− |
| 701.16 | [Gro-P-Gro-P-Gro-P-Gro-P-Gro-P-Gro-P-Gro-P-Gro-P-Gro]−2 |
| 536.17 | [(GroP)2-ManNAc-β-(1–4)-GlcNAc-1-P-P-tridecane]−2 |
| 613.17 | [(GroP)3-ManNAc-β-(1–4)-GlcNAc-1-P-P-tridecane]−2 |
| 690.27 | [(GroP)4-ManNAc-β-(1–4)-GlcNAc-1-P-P-tridecane]−2 |
| 767.18 | [(GroP)5-ManNAc-β-(1–4)-GlcNAc-1-P-P-tridecane]−2 |
| Glycosylated lipid ϕ.40 analog | |
| 333.04 | [Gro(Glu)-P]− |
| 569.18 | [Gro(Glu)-P-Gro(Glu)]− |
GP12 Phosphomonoesterase Activity and the Mechanism of Hydrolysis
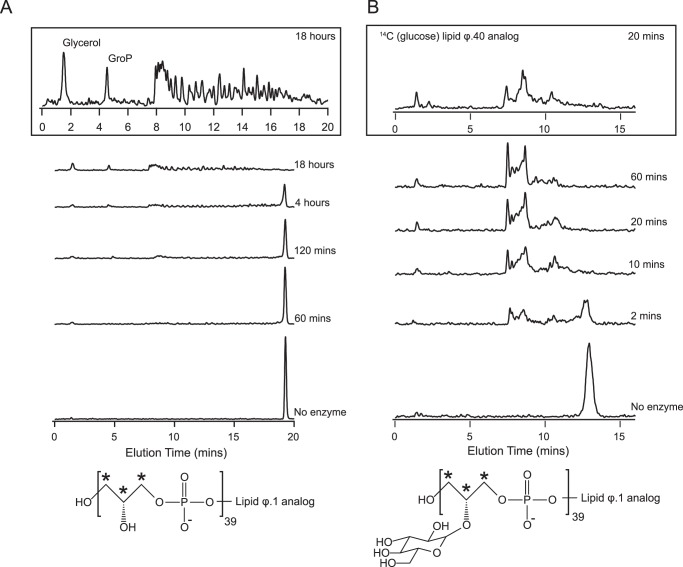
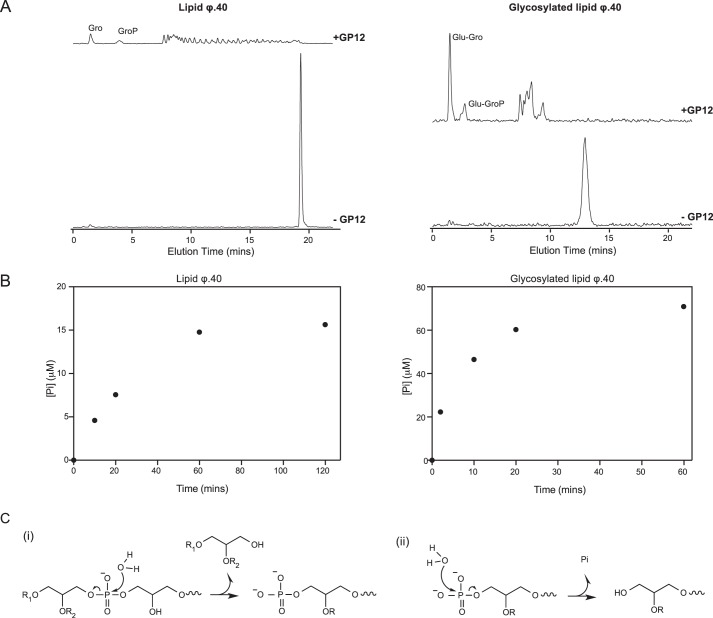
The findings from LC-MS analyses were not fully representative of WTA degradation products apparent from the HPLC studies, likely because of low ion abundance and/or ion suppression in these experiments. We therefore sought to establish whether GP12 was additionally capable of catalyzing monoester hydrolysis by initially examining the products from extensive degradation of [14C]glycerol WTA analogs. Dephosphorylated WTA monomers (glycerol, glucose-glycerol) were observed as major products from these reactions (Fig. 6A), and could have possibly arisen from hydrolysis of the corresponding phosphomonoesters generated in preceding phosphodiester hydrolysis. Alternatively, dephosohorylated WTA monomers might be the product of diester hydrolysis, especially as glycerol phosphate was found not to be a substrate for GP12. To clarify, we queried whether Pi was generated during GP12 reactions with WTA analogs under conditions commensurate with significant polymer degradation (see Figs. 2 and 5). The results of these experiments, presented in Fig. 6B, show that Pi release was concomitant with polymer degradation, but was not stoichiometric relative to the input of phosphate in the form of phosphodiester linkages. This was consistent with the modes of action deduced for GP12, whereby not all available phosphodiester linkages are subject to catalytic hydrolysis. Moreover, higher Pi output for the reaction of the glycosylated analog indicated the presence of increased amounts of hydrolyzable monoesters, as could be expected from the greater extent of degradation observed for this analog. We conclude from these findings that GP12 exercises phosphomonoesterase activity in situ, on products formed during ongoing WTA hydrolysis.
FIGURE 6.
Phosphomonoesterase activity of GP12 during WTA analog degradation. A, radioactive chromatograms for products from overnight (18 h) reactions of GP12 with [14C]glycerol lipid ϕ.40 analogs, after separation by anion exchange HPLC. Control reactions lacking enzyme showed no evidence of non-enzymatic degradation. B, time course for Pi generation from degradation of lipid ϕ.40 analogs. Reaction mixtures contained 70 μm lipid ϕ.40 analog and 1 μm GP12 or 70 μm glycosylated lipid ϕ.40 analog and 0.25 μm GP12. Data points are the mean from two independent experiments. C, summary of the reactions catalyzed by GP12 with WTA substrates. (i) a glycerophosphoglycerol-type product is released as the alcohol leaving group from GP12 phosphodiester hydrolysis. R1 = H or [glycerol 3-phosphate]n; R2 = H or glucose. (ii) GP12 hydrolyzes the phosphomonoester product from preceding phosphodiester hydrolysis to release Pi.
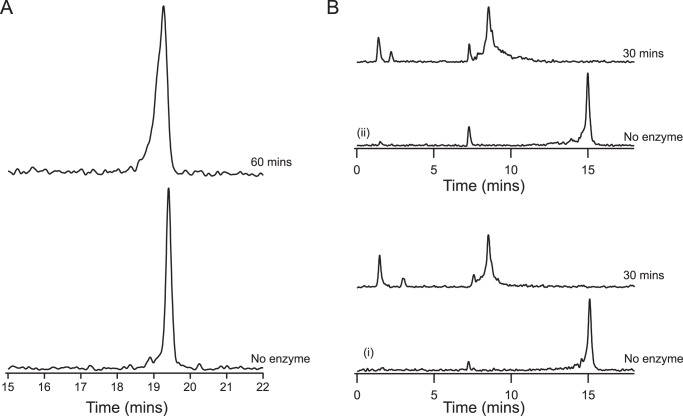
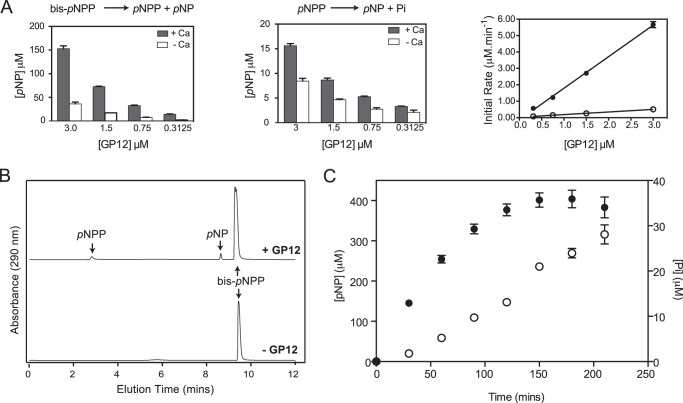
The activity of GP12 against WTA substrates therefore appeared to involve two hydrolytic reactions, illustrated in Fig. 6C. The relative abundance of the products of phosphodiester cleavage (i.e. alcohol and phosphate monoester) was not clear from these reactions with polyphosphodiester WTA substrates, presenting a challenge to ascertaining the extent of phosphate ester hydrolysis. As a result, we employed bis-pNPP and pNPP as simple substrates to model the reactions catalyzed by GP12. On the basis of pNP liberation, GP12 was active on both molecules in an enzyme-concentration dependent manner that was stimulated by Ca2+ (Fig. 7A). Enzyme turnover was greater with the diester-containing substrate and indeed, markedly higher kcat and kcat/Km were observed for bis-pNPP (Table 3). In general, the kinetic parameters reflected poor binding and modest turnover for either substrate, but were nonetheless illustrative of preferential diesterase activity by GP12. Hence, the hydrolysis of bis-pNPP by GP12 was predicted to yield equimolar amounts of pNPP and pNP in the initial stages of the reaction when [bis-pNPP] ≫ [pNPP]. To determine the end result of pNPP generated during bis-pNPP hydrolysis, we analyzed the reaction products by HPLC and found a greater abundance of pNP, indicating substantial monoesterase activity, i.e. pNPP hydrolysis (Fig. 7B). The ratio of pNPP:pNP implied that GP12 did not catalyze diester and monoester hydrolysis consecutively, which would have resulted in immediate conversion of the pNPP produced. This was verified by monitoring the course of Pi release during bis-pNPP turnover. Pi generation was observed at a significantly lower rate than pNP (Fig. 7C), providing strong evidence for catalysis of non-consecutive hydrolytic reactions by GP12 with phosphodiester-containing substrates.
FIGURE 7.
Catalytic activity of GP12 with bis-pNPP and pNPP. A, reactions (100 μl) were performed with 5 mm bis-pNPP or pNPP and GP12 at the concentrations indicated. Left and middle panels show the pNP produced after 30 min. Data points are the mean from three independent experiments; errors bars are the S.D. Right panel shows that the initial rate of pNP production from bis-pNPP (black circles) and pNPP (open circles) was linear with GP12 concentration. B, HPLC analysis of the products from GP12 hydrolysis of bis-pNPP. After 30 min, reaction mixtures from A were injected onto a C18 reversed phase column. Separation was achieved by a 10-min gradient of 2–95% CH3CN + 0.1% TFA in ddH2O + 0.1% TFA. The product mixture was determined to contain 12.1 nmol of pNPP and 15.4 nmol of pNP, based on interpolation to standard curves. C, reactions of GP12 with bis-pNPP were assayed for pNP (black circles) and Pi (open circles), at the time points indicated. Data points are the mean from three independent experiments; error bars are the S.D. The rates of pNP and Pi production, determined from the slope of the linear regression line between 30 and 120 min, were 2.57 and 0.13 μm/min, respectively.
TABLE 3.
Kinetic parameters for GP12 hydrolysis of bis-(para-nitrophenyl) phosphate and para-nitrophenyl phosphate

Kinetic parameters were derived by fitting the average initial rates from three independent experiments to Eqn. 1. The S.E. of the fit is reported.
Discussion
WTAs make up a substantial proportion of the cell wall in Gram-positive bacteria, where they adopt diverse functional roles. In B. subtilis, WTA degradation is thought to be part of the phosphate starvation response, but may also be important for cell wall recycling. An understanding of this process has been hampered by the absence of known enzymes possessing this activity, and limited, in previous investigations, to impure enzyme preparations and/or uncharacterized WTA substrates. Here, we have initiated the clarification of this facet of WTA biochemistry by using synthetic WTA analogs to investigate teichoicase activity by the GP12 protein from bacteriophage ϕ29. To our knowledge, this represents the first detailed study of such activity for a known enzyme, with chemically defined WTA substrates.
Given the large amount of WTA in the B. subtilis cell wall, it may not be surprising to find teichoicase activity of bacteriophage origin. An ability to efficiently degrade the glycosylated WTA encountered at the extracellular surface undoubtedly aids in overcoming the impediment to penetration posed by the cell wall. Still, such activity has only been reported for ϕ29, although WTAs are well known bacteriophage cell-surface receptors (38). It is conceivable that other bacteriophages harbor teichoicase activity and indeed, a BLAST query on GP12 returns a number of putative homologs from Bacillus and Staphylococcus bacteriophages sharing the intramolecular chaperone and pectin lyase domains in common with GP12. Interestingly, these domains are also putatively identified in the product from the B. subtilis gene of unknown function, yobO. This gene product lacks significant sequence similarity to hypothetical B. subtilis teichoicases such as the phosphate starvation proteins PhoD and GlpQ (39), which invokes the possibility of multiple teichoicases encoded by the B. subtilis genome. Whether these proteins exhibit teichoicase activity is the subject of ongoing work in our laboratory.
That WTA glycosylation was shown to promote more efficient polymer degradation by GP12 also seems to coincide with the presumed biological function of this protein. In all likelihood, this was primarily due to the different modes that were discerned for the action of GP12 against non-glycosylated and glycosylated WTA. Glycosylation unexpectedly appeared to exert an effect on the charge properties of WTA, evidenced by chromatographic profiles (anion exchange) showing the elution of glycosylated analogs in much lower NaCl concentrations than non-glycosylated analogs with equivalent phosphate content. Comparable observations have been reported from polyacrylamide gel electrophoresis of biological WTAs, where d-alanyl modifications can be ruled out as the cause as they are known to be lost during the WTA extraction process (40). Therefore, glycosylation may counteract potentially unfavorable interactions with the highly negatively charged WTA polymer that could hinder productive associations with GP12. As well, degradation of non-glycosylated WTA gave rise to diverse products that supplied multiple additional substrates for GP12. Hence, as the reaction progresses, there may be increasing competition for the formation of enzyme complexes with the full-length (non-glycosylated) substrate, further contributing to a reduced rate of degradation. One can imagine testing this with WTA substrates of different polymer lengths and glycosylation states, although such substrates are difficult to create with current methods.
Arguably, the product distribution resulting from degradation of the glycosylated analog matches the expectations for an enzyme that acts processively on a polymeric substrate (41–44). Moreover, the GP12 trimer has the capacity to accommodate multiple polymeric substrates (27), a property associated with processively acting enzymes (45). Even so, taking into account that GP12 catalyzes two distinct hydrolytic reactions, it would seem that non-processive polymer degradation is a likely mode of action for this enzyme.
The catalysis of two hydrolytic reactions by GP12 is reminiscent of certain binuclear metallophosphatases that utilize a bimetallated active site to catalyze the hydrolysis of both phosphate ester bonds in a diester substrate. Non-consecutive catalysis of these hydrolytic reactions has been shown for the promiscuous glycerophosphodiesterase GdpQ from Enterobacter aerogenes (46–48), bacteriophage λ phosphatase (49), and a cyclic phosphatase dihydrolase from Eggerthella lenta (50). Alternatively, purple acid phosphatases from pig and red kidney bean (51), as well as CthPnkp from Clostridium thermocellum (52), are binuclear metallophosphatases that reportedly catalyze consecutive phosphate ester hydrolyzes; the second hydrolytic reaction appears to occur without prior enzyme release of the monoester product from the first reaction. The hallmark of this mechanism is the production of Pi at roughly half the rate of the alcohol products from hydrolysis. Accordingly, the rates of Pi and pNP production during the reaction of GP12 with bis-pNPP reflect discontinuous release of Pi. The distinction between consecutive versus non-consecutive phosphate ester hydrolysis with WTA substrates could not be easily gleaned due to the difficulty in resolving monoester and alcohol products from complex degradation product mixtures. However, considering the starting concentrations of diester linkages and extent of GP12-catalyzed depolymerization under the experimental conditions, diester hydrolysis appeared to greatly exceed Pi release. We surmise that GP12 catalyzes non-consecutive hydrolytic reactions with WTA substrates.
It is implicit from the GP12 crystal structure that the two hydrolytic reactions occur at the same catalytic center (27), although different amino acid residues may be important for diesterase and monoesterase activities (52, 53). In support of this, it was observed that as diester hydrolysis slowed during the bis-pNPP reaction (due to product inhibition by pNP; not shown), the rate of Pi release was unaffected (see Fig. 7C). Interestingly, GP12 did not catalyze Pi release from glycerol phosphate, but was active on pNPP, a molecule that bears some structural resemblance to the ligand co-crystallized with GP12 (N-cyclohexyl-2-aminoethanesulfonic acid) (27). In view of this, the regions flanking the phosphate group appear relevant to the specificity of GP12 hydrolysis. This notion is exemplified by the aforementioned purple acid phosphatases, for which hydrolytic activity was shown to be affected by such factors as the steric accessibility of the substrate and the pKa of the potential leaving group (51, 54). Our findings suggest that GP12 has an increased propensity toward the hydrolysis of phosphodiester bonds flanked by bulky substituents (e.g. glucose-glycerol, nitrophenol), and these considerations could have implications for the activity of GP12 in a biological setting, where WTA substrates possess an unspecified organization of α-glucose and d-alanine modifications.
In summary, we have conducted the first detailed biochemical investigation of a purified WTA degrading enzyme, demonstrating GP12 to be a glycerophosphodiesterase that exercises diesterase and monoesterase activities in its degradation of WTA polymers. This dual capability makes GP12 exceptional among glycerophosphodiesterases and offers an intriguing potential paradigm for WTA degrading enzymes by highlighting a mechanism for phosphate liberation from WTA. The influence of glycosylation on the efficiency, specificity, and modality of polymer degradation was an important discovery because the dispersal of glycosylation sites and other modifications throughout the biological polymer remains unclear. GP12 and related enzymes may therefore find utility as tools to investigate structural modifications to WTAs. We hope that the insight into the enzymology of WTA degradation provided by this current work will aid the characterization of novel teichoicases, and find further application in elucidating the physiological relevance of this activity.
Author Contributions
E. D. B. conceived the study and designed the experiments with C. L. M. R. G. I. purified recombinant GP12, synthesized WTA analog precursors and performed preliminary biochemical characterizations. T. A. G. performed LC-MS analyses. C. L. M. performed all other experiments and wrote the manuscript with E. D. B.
Supplementary Material
Acknowledgments
pET28b-GP12 plasmid was the kind gift of Michael Rossman. We thank the Biointerfaces Institute at McMaster University for performing MALDI-TOF MS experiments.
This work was supported by a Canada Research Chair award (to E. D. B.) and Canadian Institutes for Health Research Grant MOP-15496. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Figs. S1 and S2 and Table S2.
- WTA
- wall teichoic acid
- bis-pNPP
- bis-para-nitrophenyl phosphate
- pNP
- para-nitrophenol.
References
- 1. Sewell E. W., Brown E. D. (2014) Taking aim at wall teichoic acid synthesis: new biology and new leads for antibiotics. J. Antibiot. 67, 43–51 [DOI] [PubMed] [Google Scholar]
- 2. Brown S., Xia G., Luhachack L. G., Campbell J., Meredith T. C., Chen C., Winstel V., Gekeler C., Irazoqui J. E., Peschel A., Walker S. (2012) Methicillin resistance in Staphylococcus aureus requires glycosylated wall teichoic acids. Proc. Natl. Acad. Sci. U.S.A. 109, 18909–18914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Farha M. A., Leung A., Sewell E. W., D'Elia M. A., Allison S. E., Ejim L., Pereira P. M., Pinho M. G., Wright G. D., Brown E. D. (2013) Inhibition of WTA synthesis blocks the cooperative action of PBPs and sensitizes MRSA to β-lactams. ACS Chem. Biol. 8, 226–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang H., Gill C. J., Lee S. H., Mann P., Zuck P., Meredith T. C., Murgolo N., She X., Kales S., Liang L., Liu J., Wu J., Santa Maria J., Su J., Pan J., Hailey J., Mcguinness D., Tan C. M., Flattery A., Walker S., Black T., Roemer T. (2013) Discovery of wall teichoic acid inhibitors as potential anti-MRSA β-lactam combination agents. Chem. Biol. 20, 272–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pereira M. P., Brown E. D. (2009) in Microbial Glycobiology (Moran A. P., Holst O., Brennan P., Itzstein, von M., eds) 1st Ed., pp. 337–350, Academic Press/Elsevier, New York [Google Scholar]
- 6. Bhavsar A. P., Truant R., Brown E. D. (2005) The TagB protein in Bacillus subtilis 168 is an intracellular peripheral membrane protein that can incorporate glycerol phosphate onto a membrane-bound acceptor in vitro. J. Biol. Chem. 280, 36691–36700 [DOI] [PubMed] [Google Scholar]
- 7. Schertzer J. W., Brown E. D. (2003) Purified, recombinant Tagf protein from Bacillus subtilis 168 catalyzes the polymerization of glycerol phosphate onto a membrane acceptor in vitro. J. Biol. Chem. 278, 18002–18007 [DOI] [PubMed] [Google Scholar]
- 8. Allison S. E., D'Elia M. A., Arar S., Monteiro M. A., Brown E. D. (2011) Studies of the genetics, function, and kinetic mechanism of TagE, the wall teichoic acid glycosyltransferase in Bacillus subtilis 168. J. Biol. Chem. 286, 23708–23716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lazarevic V., Karamata D. (1995) The tagGH operon of Bacillus subtilis 168 encodes a two-component ABC transporter involved in the metabolism of two wall teichoic acids. Mol. Microbiol. 16, 345–355 [DOI] [PubMed] [Google Scholar]
- 10. Reichmann N. T., Cassona C. P., Gründling A. (2013) Revised mechanism of d-alanine incorporation into cell wall polymers in Gram-positive bacteria. Microbiology 159, 1868–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kawai Y., Marles-Wright J., Cleverley R. M., Emmins R., Ishikawa S., Kuwano M., Heinz N., Bui N. K., Hoyland C. N., Ogasawara N., Lewis R. J., Vollmer W., Daniel R. A., Errington J. (2011) A widespread family of bacterial cell wall assembly proteins. EMBO J. 30, 4931–4941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wise E. M., Jr., Glickman R. S., Teimer E. (1972) Teichoic acid hydrolase activity in soil bacteria. Proc. Natl. Acad. Sci. U.S.A. 69, 233–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grant W. D. (1979) Cell wall teichoic acid as a reserve phosphate source in Bacillus subtilis. J. Bacteriol. 137, 35–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lang W. K., Glassey K., Archibald A. R. (1982) Influence of phosphate supply on teichoic acid and teichuronic acid content of Bacillus subtilis cell walls. J. Bacteriol. 151, 367–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Soldo B., Lazarevic V., Pagni M., Karamata D. (1999) Teichuronic acid operon of Bacillus subtilis 168. Mol. Microbiol. 31, 795–805 [DOI] [PubMed] [Google Scholar]
- 16. Park J. T., Uehara T. (2008) How bacteria consume their own exoskeletons (turnover and recycling of cell wall peptidoglycan). Microbiol. Mol. Biol. Rev. 72, 211–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vollmer W., Joris B., Charlier P., Foster S. (2008) Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol. Rev. 32, 259–286 [DOI] [PubMed] [Google Scholar]
- 18. Schlag M., Biswas R., Krismer B., Kohler T., Zoll S., Yu W., Schwarz H., Peschel A., Götz F. (2010) Role of staphylococcal wall teichoic acid in targeting the major autolysin Atl. Mol. Microbiol. 75, 864–873 [DOI] [PubMed] [Google Scholar]
- 19. Biswas R., Martinez R. E., Göhring N., Schlag M., Josten M., Xia G., Hegler F., Gekeler C., Gleske A.-K., Götz F., Sahl H.-G., Kappler A., Peschel A. (2012) Proton-binding capacity of Staphylococcus aureus wall teichoic acid and its role in controlling autolysin activity. PLoS ONE 7, e41415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mauck J., Chan L., Glaser L. (1971) Turnover of the cell wall of Gram-positive bacteria. J. Biol. Chem. 246, 1820–1827 [PubMed] [Google Scholar]
- 21. Glaser L., Lindsay B. (1977) Relation between cell wall turnover and cell growth in Bacillus subtilis. J. Bacteriol. 130, 610–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kusser W., Fiedler F. (1982) Purification, Mr-value and subunit structure of a teichoic acid hydrolase from Bacillus subtilis. FEBS Lett. 149, 67–70 [DOI] [PubMed] [Google Scholar]
- 23. Kusser W., Fiedler F. (1983) Teichoicase from Bacillus subtilis Marburg. J. Bacteriol. 155, 302–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kusser W., Fiedler F. (1984) A novel glycerophosphodiesterase from Bacillus pumilus. FEBS Lett. 166, 301–306 [DOI] [PubMed] [Google Scholar]
- 25. Xiang Y., Morais M. C., Battisti A. J., Grimes S., Jardine P. J., Anderson D. L., Rossmann M. G. (2006) Structural changes of bacteriophage ϕ29 upon DNA packaging and release. EMBO J. 25, 5229–5239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schulz E. C., Ficner R. (2011) Knitting and snipping: chaperones in β-helix folding. Curr. Opin. Struct. Biol. 21, 232–239 [DOI] [PubMed] [Google Scholar]
- 27. Xiang Y., Leiman P. G., Li L., Grimes S., Anderson D. L., Rossmann M. G. (2009) Crystallographic insights into the autocatalytic assembly mechanism of a bacteriophage tail spike. Mol. Cell 34, 375–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jones P., Binns D., Chang H.-Y., Fraser M., Li W., McAnulla C., McWilliam H., Maslen J., Mitchell A., Nuka G., Pesseat S., Quinn A. F., Sangrador-Vegas A., Scheremetjew M., Yong S.-Y., Lopez R., Hunter S. (2014) InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schenk G., Mitić N., Gahan L. R., Ollis D. L., McGeary R. P., Guddat L. W. (2012) Binuclear metallohydrolases: complex mechanistic strategies for a simple chemical reaction. Acc. Chem. Res. 45, 1593–1603 [DOI] [PubMed] [Google Scholar]
- 30. Ginsberg C., Zhang Y.-H., Yuan Y., Walker S. (2006) In vitro reconstitution of two essential steps in wall teichoic acid biosynthesis. ACS Chem. Biol. 1, 25–28 [DOI] [PubMed] [Google Scholar]
- 31. Brown S., Zhang Y.-H., Walker S. (2008) A revised pathway proposed for Staphylococcus aureus wall teichoic acid biosynthesis based on in vitro reconstitution of the intracellular steps. Chem. Biol. 15, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pereira M. P., Schertzer J. W., D'Elia M. A., Koteva K. P., Hughes D. W., Wright G. D., Brown E. D. (2008) The wall teichoic acid polymerase TagF efficiently synthesizes poly(glycerol phosphate) on the TagB product lipid III. ChemBioChem 9, 1385–1390 [DOI] [PubMed] [Google Scholar]
- 33. Sewell E. W., Pereira M. P., Brown E. D. (2009) The wall teichoic acid polymerase TagF is non-processive in vitro and amenable to study using steady state kinetic analysis. J. Biol. Chem. 284, 21132–21138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schertzer J. W., Brown E. D. (2008) Use of CDP-glycerol as an alternate acceptor for the teichoic acid polymerase reveals that membrane association regulates polymer length. J. Bacteriol. 190, 6940–6947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Badurina D. S., Zolli-Juran M., Brown E. D. (2003) CTP:glycerol 3-phosphate cytidylyltransferase (TarD) from Staphylococcus aureus catalyzes the cytidylyl transfer via an ordered Bi–Bi reaction mechanism with micromolar Km values. Biochim. Biophys. Acta 1646, 196–206 [DOI] [PubMed] [Google Scholar]
- 36. Gill S. C., von Hippel P. H. (1989) Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182, 319–326 [DOI] [PubMed] [Google Scholar]
- 37. Bulat E., Garrett T. A. (2011) Putative N-acylphosphatidylethanolamine synthase from Arabidopsis thaliana is a lysoglycerophospholipid acyltransferase. J. Biol. Chem. 286, 33819–33831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Winstel V., Xia G., Peschel A. (2014) Pathways and roles of wall teichoic acid glycosylation in Staphylococcus aureus. Int. J. Med. Microbiol. 304, 215–221 [DOI] [PubMed] [Google Scholar]
- 39. Botella E., Hübner S., Hokamp K., Hansen A., Bisicchia P., Noone D., Powell L., Salzberg L. I., Devine K. M. (2011) Cell envelope gene expression in phosphate-limited Bacillus subtilis cells. Microbiology 157, 2470–2484 [DOI] [PubMed] [Google Scholar]
- 40. Xia G., Maier L., Sanchez-Carballo P., Li M., Otto M., Holst O., Peschel A. (2010) Glycosylation of wall teichoic acid in Staphylococcus aureus by TarM. J. Biol. Chem. 285, 13405–13415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Horn S. J., Sørbotten A., Synstad B., Sikorski P., Sørlie M., Vårum K. M., Eijsink V. G. (2006) Endo/exo mechanism and processivity of family 18 chitinases produced by Serratia marcescens. FEBS J. 273, 491–503 [DOI] [PubMed] [Google Scholar]
- 42. Levengood M. R., Splain R. A., Kiessling L. L. (2011) Monitoring processivity and length control of a carbohydrate polymerase. J. Am. Chem. Soc. 133, 12758–12766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Horn S. J., Sørlie M., Vårum K. M., Väljamäe P., Eijsink V. G. (2012) Measuring processivity. Methods Enzymol. 510, 69–95 [DOI] [PubMed] [Google Scholar]
- 44. Beckham G. T., Ståhlberg J., Knott B. C., Himmel M. E., Crowley M. F., Sandgren M., Sørlie M., Payne C. M. (2014) Towards a molecular-level theory of carbohydrate processivity in glycoside hydrolases. Curr. Opin. Biotech. 27, 96–106 [DOI] [PubMed] [Google Scholar]
- 45. Sobhanifar S., Worrall L. J., Gruninger R. J., Wasney G. A., Blaukopf M., Baumann L., Lameignere E., Solomonson M., Brown E. D., Withers S. G., Strynadka N. C. J. (2015) Structure and mechanism of Staphylococcus aureus TarM, the wall teichoic acid α-glycosyltransferase. Proc. Natl. Acad. Sci. U.S.A.112, E576–E585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hadler K. S., Tanifum E. A., Yip S. H.-C., Mitić N., Guddat L. W., Jackson C. J., Gahan L. R., Nguyen K., Carr P. D., Ollis D. L., Hengge A. C., Larrabee J. A., Schenk G. (2008) Substrate-promoted formation of a catalytically competent binuclear center and regulation of reactivity in a glycerophosphodiesterase from Enterobacter aerogenes. J. Am. Chem. Soc. 130, 14129–14138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hadler K. S., Mitić N., Ely F., Hanson G. R., Gahan L. R., Larrabee J. A., Ollis D. L., Schenk G. (2009) Structural flexibility enhances the reactivity of the bioremediator glycerophosphodiesterase by fine-tuning its mechanism of hydrolysis. J. Am. Chem. Soc. 131, 11900–11908 [DOI] [PubMed] [Google Scholar]
- 48. Hadler K. S., Gahan L. R., Ollis D. L., Schenk G. (2010) The bioremediator glycerophosphodiesterase employs a non-processive mechanism for hydrolysis. J. Inorg. Biochem. 104, 211–213 [DOI] [PubMed] [Google Scholar]
- 49. Keppetipola N., Shuman S. (2007) Characterization of the 2′,3′ cyclic phosphodiesterase activities of Clostridium thermocellum polynucleotide kinase-phosphatase and bacteriophage phosphatase. Nucleic Acids Res. 35, 7721–7732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ghodge S. V., Cummings J. A., Williams H. J., Raushel F. M. (2013) Discovery of a cyclic phosphodiesterase that catalyzes the sequential hydrolysis of both ester bonds to phosphorus. J. Am. Chem. Soc. 135, 16360–16363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cox R. S., Schenk G., Mitić N., Gahan L. R., Hengge A. C. (2007) Diesterase activity and substrate binding in purple acid phosphatases. J. Am. Chem. Soc. 129, 9550–9551 [DOI] [PubMed] [Google Scholar]
- 52. Keppetipola N., Shuman S. (2006) Distinct enzymic functional groups are required for the phosphomonoesterase and phosphodiesterase activities of Clostridium thermocellum polynucleotide kinase/phosphatase. J. Biol. Chem. 281, 19251–19259 [DOI] [PubMed] [Google Scholar]
- 53. Wiersma-Koch H., Sunden F., Herschlag D. (2013) Site-directed mutagenesis maps interactions that enhance cognate and limit promiscuous catalysis by an alkaline phosphatase superfamily phosphodiesterase. Biochemistry 52, 9167–9176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schenk G., Gahan L. R., Carrington L. E., Mitić N., Valizadeh M., Hamilton S. E., de Jersey J., Guddat L. W. (2005) Phosphate forms an unusual tripodal complex with the Fe-Mn center of sweet potato purple acid phosphatase. Proc. Natl. Acad. Sci. U.S.A. 102, 273–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.