Abstract
Timor, an eastern Indonesian island linking mainland Asia with Australia and the Pacific world, had a complex history, including its role as a contact zone between two language families (Austronesian and Trans-New Guinean), as well as preserving elements of a rich Austronesian cultural heritage, such as matrilocal marriage practices. Using an array of biparental (autosomal and X-chromosome single-nucleotide polymorphisms) and uniparental markers (Y chromosome and mitochondrial DNA), we reconstruct a broad genetic profile of Timorese in the Belu regency of West Timor, including the traditional princedom of Wehali, focusing on the effects of cultural practices, such as language and social change, on patterns of genetic diversity. Sex-linked data highlight the different histories and social pressures experienced by women and men. Measures of diversity and population structure show that Timorese men had greater local mobility than women, as expected in matrilocal communities, where women remain in their natal village, whereas men move to the home village of their wife. Reaching further back in time, maternal loci (mitochondrial DNA and the X chromosome) are dominated by lineages with immigrant Asian origins, whereas paternal loci (Y chromosome) tend to exhibit lineages of the earliest settlers in the eastern Indonesian region. The dominance of Asian female lineages is especially apparent in the X chromosome compared with the autosomes, suggesting that women played a paramount role during and after the period of Asian immigration into Timor, perhaps driven by the matrilocal marriage practices of expanding Austronesian communities.
Introduction
Timor, an island three times the size of Hawai'i, is one of the Lesser Sunda Islands, whose western half falls within the Nusa Tenggara Timur province of the Republic of Indonesia. Unlike its more famous eastern cousin (the young country of East Timor), West Timor has not been the subject of extensive genetic study. Nevertheless, as one link in the island chain that connects mainland Asia with Australia and the Pacific, Timor acted as a stepping-stone in the proposed Southern route of early human migration from mainland Asia (Sunda Land) to the landmasses of New Guinea and Australia (Sahul Land). Based on archaeological evidence, the earliest modern human colonization in Timor dates to over 42 000 years ago.1 The island has been radically affected by more recent human migration as well: the Neolithic era saw substantial changes in tools, technology, trade and subsistence. One hypothesis suggests that the spread of farmers, ostensibly from Taiwan, carried the Austronesian languages they spoke through Island Southeast Asia and Melanesia, and later out into Polynesia, in a process known as the Austronesian expansion.2 This movement may have reached eastern Indonesia via the Philippines and Sulawesi rather than through western Indonesia.3, 4 Indeed, Neolithic sites in Timor are extremely rich in shell artifacts and decorative pieces that are rare in Island Southeast Asia outside of Taiwan and the Philippines,5 thus suggesting a close affinity between the eastern series of islands that links Taiwan, the Philippines and Timor. Previous studies of maternal mitochondrial DNA (mtDNA) lineages imply similar patterns of sharing in the biological record.6
Linguistically, Timor hosts two very different language families: Austronesian and Trans-New Guinean (‘Papuan') languages. Two Austronesian languages—Atoni (Dawan or Uab Meto) and Tetun—dominate west Timor, whereas at least 14 distinct languages, both Austronesian and Trans-New Guinean, are spoken in the east. The Austronesian expansion left its mark in other ways as well. Matrilocal residence systems are thought to have dominated ancestral Austronesian societies,7, 8, 9 and despite a widespread regional shift to patrilocal residence, some populations in West Timor still practice matrilocality today.10 Although it is still a matter of contention whether matrilocality in West Timor forms a matrilineal descent system (where clan membership is traced exclusively through female lines to a founding ancestor)7 or is instead simply the practice of matrilocal residence, these communities still clearly exhibit sex-specific dispersal in which populations are regulated by postmarital residence rules.8
The role of Timor as a major Portuguese colonial center simply added to its complex history. Sandalwood, honey, wax and, importantly for their genetic consequences, slaves were among major historical trade commodities.11 The traditional kingdom of Wehali ruled parts of central Timor during the historical period, with Laran as its ritual center. The Wehali kingdom, the religious and political core of the Timor world, was also influential in propagating marriage alliances to outlying regions, thus likely stimulating gene flow to the provinces. Today, people in the Wehali region speak an Austronesian language and still practice matrilocality.
Although an important regional center, few studies have explored the genetic profile of Timor populations. Souto et al.12 studied 15 autosomal short tandem repeats (STRs) in East Timor—once a Portuguese colony and now an independent country. They found high levels of genetic diversity with close affinity to populations along the coasts of New Guinea.13 This result is consistent with recent broad-scale regional studies of Y chromosome and mtDNA diversity across the Indonesian archipelago6, 14 that characterize the islands of Wallacea, including Timor, as having closer links with populations in Papua New Guinea and island Melanesia than mainland Asia.
Yet, Timor is noticeable by its relative absence from these large regional surveys. Its complex history, the presence of multiple language families on a relatively small island and the persistence of ancestral social systems give Timor several unique characteristics. To study the genetic signature of its people, and gain a better understanding of the effects of cultural practices such as language and residence rules on genetic diversity, we present a detailed genetic characterization of the Belu regency of West Timor, focusing on the historical princedom of Wehali.
Materials and methods
Samples and ethics
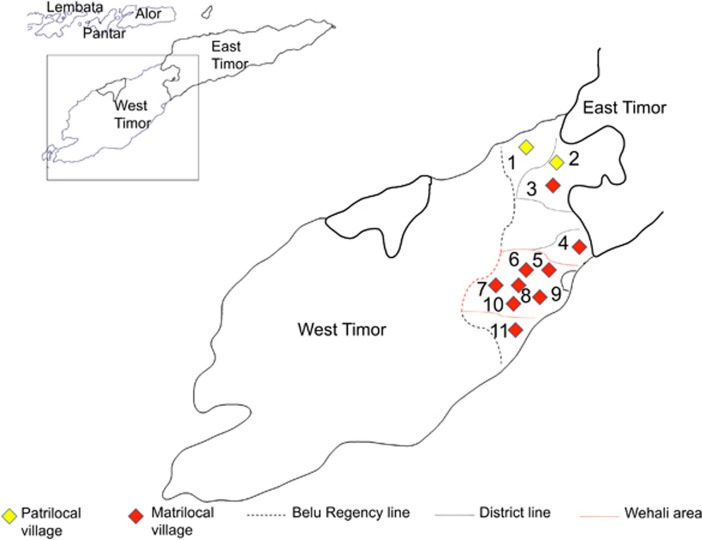
We studied populations from five districts in the Belu regency of West Timor, the Indonesian territory of the island of Timor. Permission to conduct research in Indonesia was granted by the Indonesian Institute of Sciences. Buccal swabs were collected from 529 consenting, closely unrelated and seemingly healthy individuals from 13 villages by JSL, with the assistance of Indonesian Public Health clinic staff. Sample collection followed protocols for the protection of human subjects established by both the Eijkman Institute and the University of Arizona institutional review boards. Participant interviews confirmed ethnic, linguistic and geographic classifications for at least two generations into the past. JSL gathered information on the languages spoken in each community, as well as video-recording 200-word Swadesh lists for later transcription and linguistic analysis. Table 1 lists the populations studied in West Timor, whereas their geographical locations are illustrated in Figure 1.
Table 1. Timor population samples in the present study.
| District | Population | Language spokena | Sample size |
|---|---|---|---|
| Kakuluk Mesak | Fatuketi | Lower Tetun | 35 |
| Kobalima | Raimanawe | Mix of Bunak, Dawan, Ema, Lower and Upper Tetun | 50 |
| Malaka Barat | Besikama | Upper Tetun | 42 |
| Malaka Tengah (Wehali area) | Kakaniuk | Upper Tetun | 49 |
| Kamanasa | Upper Tetun | 67 | |
| Kateri | Upper Tetun | 50 | |
| Kletek Rainan | Upper Tetun | 31 | |
| Kletek Suai | Upper Tetun | 20 | |
| Kletek Wefatuk | Upper Tetun | 20 | |
| Laran | Upper Tetun | 50 | |
| Umanen Lawalu | Upper Tetun | 50 | |
| Tasifeto Timur | Tialai | Mainly Bunak, some Lower and Upper Tetun | 24 |
| Umaklaran | Ema and Lower Tetun | 41 | |
| Total | 529 |
Language affiliations are determined from survey data taken at the time of sample collection.
Figure 1.
Location of populations studied in West Timor: (1) Fatuketi, (2) Umaklaran, (3) Tialai, (4) Raimanawe, (5) Kamanasa, (6) Kateri, (7) Kakaniuk, (8) Laran, (9) Kletek (Kletek Rainan, Kletek Suai and Kletek Wefatuk), (10) Umanen Lawalu and (11) Besikama.
Populations in West Timor mostly speak Austronesian languages as their first language. In the regency of Belu, most speak Tetun (North/Upper Tetun or South/Lower Tetun) (Table 1), although there are small clusters of non-Tetun speakers, including non-Austronesian Bunak-speaking groups.10 These latter communities are found in the area that borders East Timor, where there is more language diversity and a greater number of non-Austronesian-speaking populations (Supplementary Figure 1).
DNA extraction and genetic screening
Full experimental details are provided in Supplementary Text 1. Note that we report three newly discovered Y-chromosome markers that resolve several previously uncharacterized haplogroups in this region.
Mitochondrial DNA hypervariable region I sequences have been deposited in GenBank (accession numbers KJ936094–KJ936619). Y-chromosome STR data are provided as Supplementary Data Set 1.
Statistical and population genetic analyses
Molecular diversity, population structure estimates and genetic distances between populations were calculated using Arlequin v. 3.11 (http://cmpg.unibe.ch/software/arlequin3).15 Pairwise genetic distances between populations were computed as the linearized value FST/(1−FST).16, 17 To evaluate the correlation among linguistic, geographic and genetic distances, Mantel tests were performed in Arlequin.
Median-joining networks were built using Network v. 4.5.1.6 (Fluxus Engineering; http://www.fluxus-engineering.com).18 Haplogroups were tentatively dated with the ρ statistic method19 using a rate of one mutation every 19 171 years.20 Dates are intended only as a rough guide for relative haplogroup ages.21
Differences in mtDNA and Y-chromosome diversity between populations were analyzed using an analysis of molecular variance (AMOVA) implemented in Arlequin. A measure of interlocus differentiation 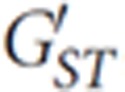 ,22 standardized for different mutation rates, was calculated using code implemented in R (available from the authors on request).23
,22 standardized for different mutation rates, was calculated using code implemented in R (available from the authors on request).23
For autosomal and X-chromosome analyses, we used ancestry informative markers, comprising 37 single-nucleotide polymorphisms (SNPs) selected because of their high information content to discriminate Asian-Melanesian ancestry (for marker details, see Cox et al.9). The two parental populations are Han Chinese and Papua New Guinea highlanders, representing the spectrum of ancestry from Asian to Melanesian. The Bayesian clustering algorithm implemented in STRUCTURE (v. 2.3.4)24 was employed to determine differentiation among populations and compare them with putative parental groups (southern Han Chinese and highland Papua New Guineans) from which our ancestry informative markers were initially chosen.9 We implemented a clustering process as described by Hubisz et al.25 by providing prior information about sampling locations to improve the detection of population structure.
Linguistic analyses
Collection of language data and subsequent linguistic classifications were carried out as described in Lansing et al.26 The ALINE algorithm was employed to obtain quantitative distance metrics between all pairs of languages in the study.27
Results
We screened 529 individuals from 13 communities in West Timor for mtDNA, Y chromosome, X chromosome and autosomal diversity. These communities were drawn from five different districts in the regency of Belu (Table 1 and Figure 1). The Y-chromosome and mtDNA data differ in marked ways: genetic diversity is more uniform for the Y chromosome, ranging from 0.95 to 1.00 (Supplementary Table 1), in contrast to mtDNA diversity that exhibits a much wider range from 0.70 to 0.98 (Table 2) (but see 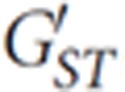 values normalized for the very different mutation rates of these loci below). MtDNA diversity was found to be lowest in the vicinity of Wehali in the district of Malaka Tengah. The six lowest values of mtDNA diversity were observed here, specifically in populations from Kletek (Kletek Rainan, Wefatuk and Suai), Kateri, Umanen Lawalu and Kakaniuk (Table 2). Because of its fast mutation rate, the genetic diversity of mtDNA responds quickly to changes in the size of populations (such as growth and contraction). Summary statistics such as Fu's Fs and Tajima's D show that Timor has experienced at most only weak population growth (Table 2), with statistically significant signals found for only four villages (Raimanawe, Kamanasa, Laran and Besikama). This is consistent with earlier studies that suggest population sizes have been broadly static, with relatively minor increases and declines across Indonesian prehistory, including in Timor.28
values normalized for the very different mutation rates of these loci below). MtDNA diversity was found to be lowest in the vicinity of Wehali in the district of Malaka Tengah. The six lowest values of mtDNA diversity were observed here, specifically in populations from Kletek (Kletek Rainan, Wefatuk and Suai), Kateri, Umanen Lawalu and Kakaniuk (Table 2). Because of its fast mutation rate, the genetic diversity of mtDNA responds quickly to changes in the size of populations (such as growth and contraction). Summary statistics such as Fu's Fs and Tajima's D show that Timor has experienced at most only weak population growth (Table 2), with statistically significant signals found for only four villages (Raimanawe, Kamanasa, Laran and Besikama). This is consistent with earlier studies that suggest population sizes have been broadly static, with relatively minor increases and declines across Indonesian prehistory, including in Timor.28
Table 2. Molecular diversity indices and growth summary statistics for populations in West Timor based on mtDNA.
|
Diversity indices |
Growth statistics |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Population | N | h | S | Haplotype diversity | s.d. | MNPD | s.d. | Nucleotide diversity | s.d. | Tajima's D | P | Fu's Fs | P |
| Umaklaran | 41 | 14 | 32 | 0.90 | 0.026 | 7.2 | 3.5 | 0.014 | 0.007 | −0.03 | 0.56 | 0.36 | 0.63 |
| Fatuketi | 35 | 17 | 36 | 0.91 | 0.033 | 8.3 | 4.0 | 0.016 | 0.008 | −0.17 | 0.50 | −1.49 | 0.34 |
| Tialai | 24 | 12 | 29 | 0.93 | 0.027 | 9.4 | 4.5 | 0.018 | 0.009 | 0.77 | 0.84 | 0.54 | 0.62 |
| Raimanawe | 50 | 27 | 46 | 0.95 | 0.015 | 8.8 | 4.1 | 0.017 | 0.009 | −0.50 | 0.36 | −6.61 | 0.042 |
| Kamanasa | 67 | 39 | 58 | 0.98 | 0.007 | 9.0 | 4.2 | 0.017 | 0.009 | −0.87 | 0.17 | −16.56 | <0.001 |
| Kletek Rainan | 30 | 13 | 33 | 0.83 | 0.063 | 8.3 | 4.0 | 0.016 | 0.008 | 0.00 | 0.55 | 0.38 | 0.62 |
| Kletek Suai | 20 | 10 | 29 | 0.86 | 0.055 | 7.8 | 3.8 | 0.015 | 0.008 | −0.20 | 0.48 | 0.59 | 0.60 |
| Kletek Wefatuk | 19 | 10 | 27 | 0.88 | 0.056 | 7.5 | 3.6 | 0.014 | 0.008 | −0.13 | 0.50 | 0.25 | 0.58 |
| Kateri | 50 | 20 | 33 | 0.83 | 0.043 | 6.8 | 3.2 | 0.013 | 0.007 | −0.28 | 0.44 | −2.82 | 0.19 |
| Laran | 50 | 29 | 43 | 0.97 | 0.011 | 8.8 | 4.1 | 0.017 | 0.009 | −0.27 | 0.45 | −8.73 | 0.008 |
| Kakaniuk | 49 | 12 | 31 | 0.70 | 0.067 | 6.7 | 3.2 | 0.013 | 0.007 | −0.11 | 0.49 | 1.88 | 0.77 |
| Umanen Lawalu | 49 | 17 | 38 | 0.77 | 0.062 | 7.1 | 3.4 | 0.014 | 0.007 | −0.55 | 0.34 | −0.77 | 0.42 |
| Besikama | 42 | 25 | 45 | 0.97 | 0.011 | 9.1 | 4.3 | 0.017 | 0.009 | −0.46 | 0.36 | −6.20 | 0.036 |
Abbreviations: h, number of haplotypes; MNPD, mean number of pairwise differences; N, number of sequences; P, probability value; S, number of polymorphic sites.
Significant growth summary statistics are in bold and italicized.
In our samples, 24 Y-chromosome haplogroups (shown in Supplementary Figure 2) were observed (Supplementary Table 2). The C-RPS4Y paragroup has been associated with very early population movements into the Indonesian archipelago.14 It has a patchy distribution throughout Southeast Asia and Indonesia, and is absent or present at low frequency further east in Melanesia and Polynesia (Supplementary Table 3).14 In Timor, this lineage is most frequent in Umaklaran (12.2%) and Kletek Rainan (11.8%) (Supplementary Table 2).
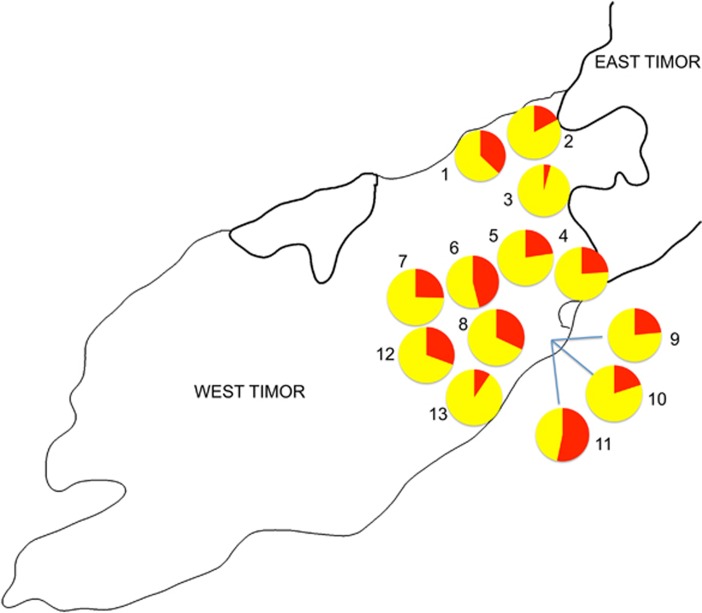
Y-chromosome haplogroups with putative Melanesian origins29, 30 —C-M38, M-P34 and S-M254—account for nearly 40% of Y-chromosome lineages in Timor (38.8%). Based on the distribution of haplotypes, Y- STR diversity and coalescent time estimates, it has been proposed that haplogroup C-M38 arose in Melanesia.29 C-M38 alone accounts for almost one-third of Y chromosomes in Timor (26.6%) and reaches highest frequency in eastern Indonesia rather than further east (Supplementary Table 3). Interestingly, C-M38 is the most common haplogroup in Kletek Wefatuk (53.3%, Figure 2), a relatively new village in Wehali. Its inhabitants only moved to this area from East Timor around 100 years ago (JSL, unpublished survey data). C-M208, a subgroup of C-M38, is the ancestor of the P33 lineage found in Polynesians, and was previously thought to be limited to coastal New Guinea, island Melanesia and the Pacific islands.14 However, we detected C-M208 at low frequency in Timor (0.4%).
Figure 2.
Frequencies of Y-chromosome haplogroup C-M38. Populations: (1) Fatuketi, (2) Umaklaran, (3) Tialai, (4) Raimanawe, (5) Kamanasa, (6) Kateri, (7) Kakaniuk, (8) Laran, (9) Kletek Rainan, (10) Kletek Suai, (11) Kletek Wefatuk, (12) Umanen Lawalu and (13) Besikama.
Surprisingly, the Y-chromosome paragroup O-M122 was not found in Timor, even though it is often associated with the Austronesian migration. However, we did observe several of its derived forms: O-M134, O-JST002611 and O-P201 (Supplementary Table 2). Lineage O-P201 has a wide geographic distribution, with close connections to Indonesia, the Philippines, Taiwan and Oceania.14 Another Asian Y-chromosome lineage, O-M119, dominates indigenous Taiwanese populations29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39 that typically carry the sub-haplogroups O-P203 and O-M110 rather than paragroup O-M119. Although these lineages are infrequent in Timor (O-M119 at 2.4% and O-P203 at 1.2%), O-M110—a haplogroup with putative Taiwanese origins—was found at 8%.
We detected two recently reported K lineage markers in Timor:40 K-P397 and K-P336. Three new haplogroups were also screened: C-P355, C-P343 and S-P377. These markers substantially improve the resolution of the C and K lineages in Island Southeast Asia, and haplogroups K-P397 and C-P355 alone account for almost a quarter of Y chromosomes in West Timor (22.4%). These lineages are shared with other islands in eastern Indonesia, as is P-P295 (10.9% in Timor) that has only been detected in the north–south chain of islands in the east of Island Southeast Asia (Timor, Sumba, Sulawesi and the Philippines; Supplementary Table 3).
A small number of Timorese Y chromosomes belong to haplogroups other than lineages C, K, M, S and O. A total of ∼0.6% belong to haplogroups D-M116, E-P1 and Q-P36, with the D group possibly reflecting wartime connections with Japan. The Timorese also share haplogroup F-P14 (1.8%) with other Lesser Sunda Island populations and Sulawesi. Haplogroup J-M172, potentially a signal of Arab trader contact, is found at very low frequency (0.4%) in Timor and is also shared with Sulawesi (0.6%).
A total of 31 mtDNA haplogroups were identified in Timor, with all lineages falling into macrohaplogroups M (45.4%) and N (54.6%) (Supplementary Figure 3). The predominant haplogroups are F1a4 (19.3%) and various Q lineages (14.9% Supplementary Table 4). F1a4 is common in eastern Indonesia, but nearly absent in the west,6 and connects Indonesia with the Philippines.6, 41 Q lineages are found predominantly in New Guinea and Island Melanesia (Supplementary Table 5).30, 42
Interestingly, the high frequency of haplogroup Q in Timor is linked with a correspondingly low frequency of F1a4 and vice versa (Supplementary Figure 4). Haplogroup F1a4 is found at highest frequency in the Wehali area, where we also observed low levels of haplogroup Q.
Regionally, haplogroup P is often found associated with haplogroup Q, but P is relatively infrequent in Timor (0.9%). Two patrilocal populations have the highest Q frequencies in Timor: Umaklaran at 31.7%, followed by Fatuketi at 25.7%. Tialai, a matrilocal population located close to Umaklaran and Fatuketi, also carries a high frequency of haplogroup Q (25.0%). This latter case is perhaps less surprising as Tialai is inhabited by people who speak Bunak, a Trans-New Guinea language, whereas the other two populations primarily speak an Austronesian language as their mother tongue.
The Asian mtDNA lineage known as the Polynesian Motif is also found at moderate frequency in West Timor (7.4%), consistent with its potential origin in eastern Indonesia43 and high frequencies on neighboring islands (but see Cox44).
Finally, despite its geographical location as a stepping-stone to Australia, Timorese show little genetic affinity with Australian aborigines (Supplementary Tables 3 and 5). The possible exception is mtDNA haplogroup Q2 that Hudjashov et al.45 suggest may reflect a secondary expansion into Australia, although one that still occurred ∼30 000 years ago. We have not identified more recent connections.
To identify patterns of variation among Timorese populations, we performed an analysis of molecular variance. The variance of mtDNA hypervariable region I (92.0%) and mtDNA SNPs (89.8%) are weakly, but consistently, lower than those of Y-chromosome STRs (97.5%) and SNPs (95.9%). This suggests that Timorese men may have dispersed more widely than women, as expected in communities that practice matrilocality (that is, women stay in their natal village, whereas men are given or sent away to surrounding communities). Moreover, when standardized for the ∼400-fold higher mutation rate of the Y chromosome (in the order of 10−5 mutation events/STR/year)46, 47, 48, 49 relative to mtDNA (in the order of 10−7 mutation events/base pair/year),20 Y-chromosome SNPs 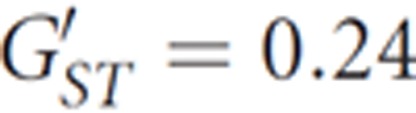 show notably lower population structure than mtDNA SNPs
show notably lower population structure than mtDNA SNPs 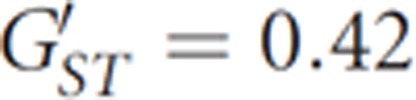 (Supplementary Table 6), thus further suggesting that men have moved more widely than women. Consistent with this finding, when the 13 populations were divided into two groups (patrilocal and matrilocal), the variance among groups was higher for mtDNA markers than for the Y chromosome (Table 3). Again, different social behaviors for men and women—in this case, the practice of matrilocality—appear to have affected patterns of genetic variation differently in males and females.
(Supplementary Table 6), thus further suggesting that men have moved more widely than women. Consistent with this finding, when the 13 populations were divided into two groups (patrilocal and matrilocal), the variance among groups was higher for mtDNA markers than for the Y chromosome (Table 3). Again, different social behaviors for men and women—in this case, the practice of matrilocality—appear to have affected patterns of genetic variation differently in males and females.
Table 3. Analysis of molecular variance (AMOVA) for subsets of Timor populations.
|
Within populations |
Among populations within groups |
Among groups |
|||||
|---|---|---|---|---|---|---|---|
| FST | Variance (%) | FSC | Variance (%) | FCT | Variance (%) | ||
| Y SNPs | |||||||
| All Timor populations | 13 Populations | 0.041 | 95.9 | ||||
| Matrilocal vs patrilocal | Two groups | 0.032 | 96.8 | 0.045 | 4.5 | −0.013 | −1.3 |
| Y STRs | |||||||
| All Timor populations | 13 Populations | 0.025 | 97.5 | ||||
| Matrilocal vs patrilocal | Two groups | 0.018 | 98.2 | 0.028 | 2.8 | −0.010 | −1.0 |
| mtDNA SNPs | |||||||
| All Timor populations | 13 Populations | 0.10 | 89.8 | ||||
| Matrilocal vs patrilocal | Two groups | 0.14 | 85.8 | 0.086 | 8.1 | 0.060 | 6.1 |
| mtDNA HVSI | |||||||
| All Timor populations | 13 Populations | 0.080 | 92.0 | ||||
| Matrilocal vs patrilocal | Two groups | 0.098 | 90.2 | 0.073 | 7.1 | 0.027 | 2.7 |
Abbreviations: HVSI, hypervariable segment I; mtDNA, mitochondrial DNA; SNP, single-nucleotide polymorphism; STR, short tandem repeat.
All values are statistically significant (P≪0.0001).
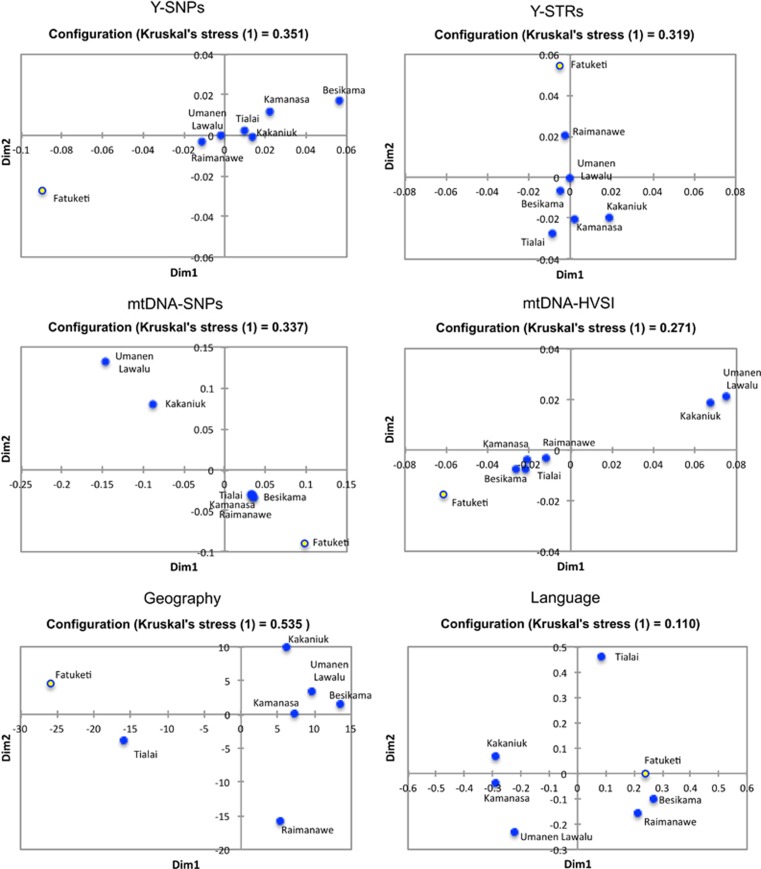
To explore wider regional relationships, multidimensional scaling was performed on seven Timor populations that are predominantly monolingual (Figure 3; note that some populations are strongly bi- or multilingual, thus precluding many of the following language-paired analyses; see Table 1 for details). Maternal lineages consistently show that Umanen Lawalu and Kakaniuk (both from the Wehali region) cluster together, separated from the other five populations: Kamanasa, Fatuketi, Raimanawe, Besikama, and Tialai. Umanen Lawalu and Kakaniuk even vary from their close geographical neighbor in the Wehali area, the village of Kamanasa. However, Umanen Lawalu and Kakaniuk are both older communities in the region, whereas Kamanasa is a new village whose inhabitants arrived from East Timor in 1911 following a period of civil war.
Figure 3.
Multidimensional scaling (MDS) plot of seven populations in West Timor based on Y-chromosome single-nucleotide polymorphisms (SNPs), Y-chromosome short tandem repeats (STRs), mitochondrial DNA (mtDNA) SNPs, mtDNA hypervariable region I sequences, geography and language distances (a patrilocal village, Fatuketi, is shown as an open circle; the other villages are matrilocal).
Conversely, Y-chromosome plots show that Fatuketi is the only village that clusters far away from other populations (Figure 3). The fact that this village is patrilocal, with men remaining in their home village, might contribute to this outlier pattern. The language data present a different pattern again (Figure 3), with Tialai, the village whose inhabitants speak Bunak, a Trans-New Guinea language, unsurprisingly separated from the remaining Austronesian-speaking communities. Nevertheless, no statistically significant correlations were observed between genetic diversity, language or geography (Supplementary Table 7).
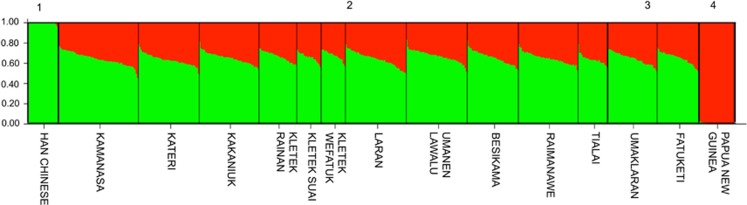
Asian admixture rates across the genome were determined using a suite of ancestry informative markers:9 18 from the autosomes and 19 from the X chromosome. Timor (64% Asian ancestry) fits with regional expectations (Supplementary Table 8), falling both geographically and in terms of Asian ancestry between Flores/Lembata (66%) and Alor (51%).9 Within Timor, Asian ancestry exhibits a surprisingly large range among populations (61–72%) (Supplementary Table 9), but there is no evidence of subdivision by language or social system (Figure 4). Nevertheless, consistent with previous research on Asian-Melanesian ancestry across the Indo-Pacific region,9, 50 we found that Asian admixture is biased toward women (that is, Asian ancestry is higher in the X chromosome (70%) relative to the autosomes (58%); Supplementary Table 9). Interestingly, such a large difference in admixture rates between the X chromosome and autosomes (11%) is only observed elsewhere on Sumba and in Vanuatu (Supplementary Table 8), thus suggesting that differences in male/female dynamics were amplified in Timor during, and likely continuing after, the initial admixture event.51
Figure 4.
Admixture plot produced by STRUCTURE. Putative parental populations, (1) Southern Han Chinese (green) and (4) Papua New Guinea Highlanders (red), are clearly shown as well differentiated ancestral groups. Populations in West Timor are presented as sets of (2) matrilocal and (3) patrilocal villages. Despite variation in ancestry components among individuals, little difference in admixture proportions is observed between communities.
Tialai, the only population in our study whose inhabitants speak a non-Austronesian language (Bunak), had the lowest Asian admixture rate in the X chromosome (65%). Curiously, the bias in Asian admixture rates between the X chromosome and the autosomes is also lowest in Tialai. Conversely, the highest bias is found in Umanen Lawalu, in the Wehali region (Supplementary Table 9). This finding further suggests that matrilocality, which persists to the present in Wehali, may have been a driving force behind this admixture bias in the X chromosome and autosomes.
Discussion
Comparison of uniparental and biparental genetic markers reveals the sheer complexity of prehistoric Timor, including periods of population isolation, long-distance contact and the effect of social systems. Mitochondrial DNA, Y chromosome, autosomal and X-linked lineages reflect different aspects of this history, but all emphasize a substantial contribution from the first settlers to reach Timor. Mitochondrial lineages P and Q, and Y-chromosome lineages C, M and S, are all associated with the first colonization of the Indonesian archipelago by modern humans ∼50 000 years ago14 (Supplementary Tables 10 and 11). In the autosomes and X chromosome, a little over a third of the average Timorese genome (34%) traces back to these first settlers too. Traces of the first settlers are also found in the languages spoken in Timor, where hints of ancestral languages (unrelated to either Austronesian or the Trans New Guinean language group) are preserved through loan words borrowed in the modern languages.52
And yet the primary story told by a range of genetic loci is one of more recent contact with settlers ultimately having Asian origins. Mitochondrial lineages B and F, and Y-chromosome lineage O, attest to considerable mixing with more recent Asian immigrants. This contact was neither minor nor insubstantial. Two-thirds of the average Timorese genome today (66%) has an ultimate, and relatively recent, Asian origin. Despite its current perception as an isolated outpost, Timor was once a major contact zone in eastern Island Southeast Asia.
Exactly how and when this contact occurred remains unclear. Certainly, some connection with the spread of Austronesian languages remains a major contender. Analysis of complete mtDNA genomes suggests that the Austronesian expansion is responsible for much of the dominance of Asian maternal ancestry in Oceania, whereas contact with earlier groups is demonstrated by the ongoing presence of more locally ancient lineages.53 A recent analysis of genome-wide SNPs revealed that admixture between Asian and Melanesian sources began in eastern Indonesia ∼4000 years ago, consistent with a mid-Holocene period of expansion in Neolithic lifeways and the spread of Austronesian languages.50 Yet, the process of transitioning to a farming lifestyle seems to have been a complex and lengthy one. Timor's neighbor, New Guinea, has a long agricultural tradition based on root crops and, at lower altitudes, bananas.54 Bananas are still one of the main agricultural products, and an important source of income, for people living in the fertile plain of Wehali Wewiku in West Timor,10 and the dispersal of banana cultivars west from New Guinea may well have predated other elements of the Neolithic package that are presumed to have been introduced through later Asian contact.55, 56 Indeed, this westward dispersal of bananas into eastern Indonesia may be associated with the spread of Trans-New Guinea languages into Timor, a Papuan language family that is thought to be a relatively recent translocation from western New Guinea.57 Proposed Y-chromosome markers putatively associated with this expansion (M-P34 and S-M254) occur at moderate frequency in Timor, and have been dated to 6000–10 000 years ago,34, 58 thus likely pre-dating the Austronesian expansion.
Nevertheless, Asian ancestry is a dominant feature of modern Timorese. Although questions have been raised about the provenance of the language family, the spread of Austronesian languages must still have been a defining moment in Island Southeast Asian prehistory. With its deepest branches in Taiwan, Austronesian languages are spoken without exception across most of modern Island Southeast Asia. All but one of our West Timor populations speak Austronesian languages, although Trans-New Guinea languages are more prevalent in East Timor. These linguistic hints are reinforced by genetic signals. The Timorese carry mtDNA lineages (such as B4a, B4b, B4c, B4c1b3, B5a, B5b, B5b1, D and E) that are distributed widely across Mainland and Island Southeast Asia,59, 60 and are thought to reflect multiple population movements from mainland Asia.6 Other maternal lineages connect Timor with the Philippines and Taiwan (F1a4, E1a1a, M7c3c and Y2). Haplogroup F1a4 is particularly noteworthy: it accounts for almost 20% of some populations and shows an almost exclusive connection with the Philippines.6, 41 Curiously, F1a4 has a higher diversity in Indonesian populations compared with the Philippines (Supplementary Figure 4), and may have greater antiquity in the southern part of this range rather than being part of any dispersal from the north (Supplementary Table 10). This north–south connection is also observed in Y-chromosome lineages (for example, P-P295, O-M110 and O-P201), and is consistent with linguistic affinity between Timor and the northern island chain, including Sulawesi and the Maluku Islands, that is perhaps explained by an eastern route of Austronesian language dispersal from the Philippines.52 Furthermore, a network of Y-chromosome haplogroup O-M110 lineages (Supplementary Figure 5) shows that Timor shares an ancestral haplotype with indigenous Taiwanese. Descendant lineages exhibit a star-like pattern indicative of population growth and/or geographical expansion. Although the network is largely uninformative about the direction of migration, a recent admixture study using genome-wide data infers that gene flow from Taiwan to Island Southeast Asia best explains admixture patterns in Austronesian-speaking populations.61
One curious disconnect is the predominance of Asian genetic lineages and languages in Timor, coupled with notable aspects of Papuan cultural traits. The rice cultivation that underpins western Indonesian society is largely absent, perhaps because of climatic conditions that make this region unsuitable for sustained rice agriculture.51, 62 Such apparent inconsistencies may also partly reflect the passage of time. Today, languages and genetics are mostly unlinked across large parts of the Austronesian contact zone. Although associations remain in Sumba,26 they are not apparent in Timor (Supplementary Table 7 and Figure 4). Similarly, no association between language and genetics is observed in the neighboring Maluku islands, where—like in Timor—there is still a clear linguistic contact zone between Austronesian and Papuan language speakers.63 Lansing et al.51 propose a process of extensive Asian admixture, emphasizing ongoing heterogeneous language and culture replacement, in which speakers of the two language families continue to influence each other.64 The history of Timor does not seem to comprise merely an ancestral Melanesian substratum with a single expansion of Asian Austronesian speakers, but is instead compounded by the bidirectional ebb and flow of later populations from western Indonesia and New Guinea and, more recently, from Arab traders and colonists venturing from much further afield. Since Indonesian independence, movements linked to economic and political concerns have also contributed to population dynamics across the Indonesian archipelago,65, 66 although our sampling scheme was designed to avoid such individuals.
The genetic patterns are also affected in other ways. Mitochondrial DNA and Y-chromosome data show that women and men experienced different histories and were subjected to different social pressures. In general, maternal loci are dominated by lineages with immigrant Asian origins, whereas paternal loci are dominated by lineages with local Melanesian origins. This female/male division is not simply a result of genetic drift in the haploid genetic loci, but also stands out strongly in biparental and sex-linked nuclear markers. The female dominance of Asian lineages is clear in the X chromosome compared with the autosomes (Supplementary Table 9 and Figure 4), suggesting that female migrants played a leading role during the period of Asian immigration into eastern Indonesia, including Timor. This finding provides support for an Austronesian ‘house society' model51 in which the Austronesian expansion led to the dispersal of matrilocal societies with small numbers of neighboring non-Austronesian males marrying into Austronesian matrilocal, matrilineal houses.51 These admixture rates seem to capture not only the initial contact event during the mid-Holocene, but also ongoing interactions over the subsequent ∼4000 years.
Sex-specific differences in social practices continued to affect the genetic diversity of Timorese. A recent broad-scale regional study of both mtDNA and Y chromosomes in Indonesian populations showed that women have moved widely between communities, whereas men historically remained at home.6, 14 This pattern is typical of patrilocal societies, but ancestral Austronesian societies are thought to have been characterized by matrilocal systems7, 8, 9 that still dominate many communities in West Timor today (JSL, unpublished survey data).10 Consistent with this social relict, the genetic profile of Timorese shows that men moved widely between communities, whereas women stayed local (Table 3). Similarly, Lansing et al.51 showed that the effective population size of Timor women is reduced compared with the island's men. This lower female than male effective population size is also consistent with male-biased migration in a matrilocal society.
The persistence of these Austronesian social behaviors has shaped recent population structure across the island. This is seen most clearly in the Malaka Tengah district, the site of the historical Wehali kingdom and ritual center, and still a stronghold of matrilineal, matrilocal communities.67 Wehali is considered ‘female land', a matrilineal area where all the land, houses and property belong to women. Wehali's politico-ideological structures extended beyond the limited area of the princedom, eventually comprising 17 domains in both the west and east of the island. The spread of Wehali's power was primarily achieved through marriage alliances.10 Although surrounding Tetun and Atoni populations transferred wealth to Wehali, indigenous concepts of power gave Wehali a notable role as the ‘husband-giver' to more peripheral areas in Timor. Consistent with this tradition, putative Austronesian mtDNA lineages like B4a1a1a (that is, the Polynesian motif) reach their highest frequencies in the inner Wehali villages of Kateri (36%) and Laran (18%). This process perhaps also explains the higher mobility of male lineages across West Timor compared with female lineages.
This study thus illustrates the influential roles of isolation, contact and social behavior in producing the genetic profile of modern West Timorese. Isolation led to the persistence of genetic lineages from the very first settlers in Timor; contact created genomes characterized by rampant Asian and Melanesian admixture; and social behavior, particularly a tenacious holding to the Austronesian practice of matrilocality while surrounding populations transitioned to patrilocality, created patterns of male and female dispersal that differ from neighboring regions. Far from being an island outlier, Timor stands firmly at the heart of the Austronesian world.
Acknowledgments
This research was supported by a US National Science Foundation grant (SES 0725470) to JSL, MFH, TMK and Joe C Watkins, which funded the doctoral research of MKT. The Royal Society of New Zealand provided support through a Rutherford Fellowship (RDF-10-MAU-001) and Marsden Grant (11-MAU-007) to MPC. We gratefully acknowledge the assistance with sample collection of Agustini Leonita and Alida Harahap of the Eijkman Institute for Molecular Biology, Jakarta, Indonesia.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Journal of Human Genetics website (http://www.nature.com/jhg)
Supplementary Material
References
- O'Connor S. New evidence from East Timor contributes to our understanding of earliest modern human colonization east of the Sunda Shelf. Antiquity. 2007;81:523–535. [Google Scholar]
- Bellwood P. First Migrants: Ancient Migration in Global Perspective. Wiley Blackwell, Chichester, West Sussex, UK; 2013. [Google Scholar]
- Pawley A.Selected Papers from the Eighth International Conference on Austronesian Linguisticseds Zeitoun, E. & Li, P. J.-K.) 95–138.Institute of Linguistics, Academia Sinica, Taipei; 1999 [Google Scholar]
- Spriggs M. Chronology of the Neolithic transition in Island Southeast Asia and the western Pacific: a view from 2003. Rev. Archaeol. 2003;24:57–80. [Google Scholar]
- Bellwood P.Prehistory of the Indo-Malaysian ArchipelagoRevised edn (University of Hawaii Press, Honolulu; 2007 [Google Scholar]
- Tumonggor M. K., Karafet T. M., Hallmark B., Lansing J. S., Sudoyo H., Hammer M. F., et al. The Indonesian archipelago: an ancient genetic highway linking Asia and the Pacific. J. Hum. Genet. 2013;58:165–173. doi: 10.1038/jhg.2012.154. [DOI] [PubMed] [Google Scholar]
- Hage P., Marck J. Matrilineality and the Melanesian origin of Polynesian Y chromosomes. Curr. Anthropol. 2003;44:S121–S127. [Google Scholar]
- Jordan F. M., Gray R. D., Greenhill S. J., Mace R. Matrilocal residence is ancestral in Austronesian societies. Proc. R. Soc. B. 2009;276:1957–1964. doi: 10.1098/rspb.2009.0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox M. P., Karafet T. M., Lansing J. S., Sudoyo H., Hammer M. F. Autosomal and X-linked single nucleotide polymorphisms reveal a steep Asian-Melanesian ancestry cline in eastern Indonesia and a sex bias in admixture rates. Proc. R. Soc. B. 2010;277:1589–1596. doi: 10.1098/rspb.2009.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therik T. Wehali: The Female Land. Traditions of a Timorese Ritual Centre. Pandanus Books, in association with the Australian National University, Department of Anthropology, Research School of Pacific and Asian Studies, Canberra; 2004. [Google Scholar]
- Kammen D. Master-slave, traitor-nationalist, opportunist-oppressed: political metaphors in East Timor, Indonesia. Indonesia. 2003;76:70–85. [Google Scholar]
- Souto L., Alves C., Gusmão L., Ferreira E., Amorim A., Côrte-Real F., et al. Population data on 15 autosomal STRs in a sample from East Timor. Forensic. Sci. Int. 2005;155:77–80. doi: 10.1016/j.forsciint.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Souto L., Gusmão L., Amorim A., Côrte-Real F., Vieira D. N. Y-STR haplotype diversity in distinct linguistic groups from East Timor. Am. J. Hum. Biol. 2006;18:691–701. doi: 10.1002/ajhb.20553. [DOI] [PubMed] [Google Scholar]
- Karafet T. M., Hallmark B., Cox M. P., Sudoyo H., Downey S. S., Lansing J. S., et al. Major east-west division underlies Y chromosome stratification across Indonesia. Mol. Biol. Evol. 2010;27:1833–1844. doi: 10.1093/molbev/msq063. [DOI] [PubMed] [Google Scholar]
- Excoffier L., Lischer H. E. L. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Res. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Slatkin M. Inbreeding coefficients and coalescence times. Genet. Res. Camb. 1991;58:167–175. doi: 10.1017/s0016672300029827. [DOI] [PubMed] [Google Scholar]
- Slatkin M. A measure of population subdivision based on microsatellite allele frequencies. Genetics. 1995;139:457–462. doi: 10.1093/genetics/139.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt H. J., Forster P., Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Forster P., Harding R., Torroni A., Bandelt H.-J. Origin and evolution of Native American mtDNA variation: a reappraisal. Am. J. Hum. Genet. 1996;59:935–945. [PMC free article] [PubMed] [Google Scholar]
- Soares P., Ermini L., Thomson N., Mormina M., Rito T., Röhl A., et al. Correcting for purifying selection: an improved human mitochondrial molecular clock. Am. J. Hum. Genet. 2009;84:740–759. doi: 10.1016/j.ajhg.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox M. P. Accuracy of molecular dating with the rho statistic: deviations from coalescent expectations under a range of demographic models. Hum. Biol. 2008;80:335–357. doi: 10.3378/1534-6617-80.4.335. [DOI] [PubMed] [Google Scholar]
- Hedrick P. W. A standardized genetic differentiation measure. Evolution. 2005;59:1633–1638. [PubMed] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing (2014 ) http://www.r-project.org .
- Pritchard J. K., Stephens M., Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubisz M., Falush D., Stephens M., Pritchard J. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Res. 2009;9:1322–1332. doi: 10.1111/j.1755-0998.2009.02591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansing J. S., Cox M. P., Downey S. S., Gabler B. M., Hallmark B., Karafet T. M., et al. Coevolution of languages and genes on the island of Sumba, Eastern Indonesia. Proc. Natl Acad. Sci. USA. 2007;104:16022–16026. doi: 10.1073/pnas.0704451104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downey S. S., Hallmark B., Cox M. P., Norquest P., Lansing J. S. Computational feature-sensitive reconstruction of language relationships: developing the ALINE distance for comparative historical linguistic reconstruction. J. Quant. Ling. 2008;15:340–369. [Google Scholar]
- Guillot E. G., Tumonggor M. K., Lansing J. S., Sudoyo H., Cox M. P. Climate change influenced female population sizes through time across the Indonesian archipelago. Hum. Biol. 2013;85:135–152. doi: 10.3378/027.085.0306. [DOI] [PubMed] [Google Scholar]
- Kayser M., Brauer S., Weiss G., Schiefenhovel W., Underhill P., Shen P., et al. Reduced Y chromosome, but not mitochondrial DNA, diversity in human populations from West New Guinea. Am. J. Hum. Genet. 2003;72:281–302. doi: 10.1086/346065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser M., Brauer S., Cordaux R., Casto A., Lao O., Zhivotovsky L. A., et al. Melanesian and Asian origins of Polynesians: mtDNA and Y chromosome gradients across the Pacific. Mol. Biol. Evol. 2006;23:2234–2244. doi: 10.1093/molbev/msl093. [DOI] [PubMed] [Google Scholar]
- Su B., Xiao J., Underhill P., Deka R., Zhang W., Akey J., et al. Y chromosome evidence for a northward migration of modern humans into Eastern Asia during the last Ice Age. Am. J. Hum. Genet. 1999;65:1718–1724. doi: 10.1086/302680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su B., Jin L., Underhill P., Martinson J., Saha N., McGarvey S. T., et al. Polynesian origins: insights from the Y chromosome. Proc. Natl Acad. Sci. USA. 2000;97:8225–8228. doi: 10.1073/pnas.97.15.8225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser M., Brauer S., Weiss G., Underhill P. A., Roewer L., Schiefenhövel W., et al. Melanesian origin of Polynesian Y chromosomes. Curr. Biol. 2000;10:1237–1246. doi: 10.1016/s0960-9822(00)00734-x. [DOI] [PubMed] [Google Scholar]
- Kayser M., Brauer S., Weiss G., Schiefenhövel W., Underhill P. A., Stoneking M. Independent histories of human Y chromosomes from Melanesia and Australia. Am. J. Hum. Genet. 2001;68:173–190. doi: 10.1086/316949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser M., Choi Y., van Oven M., Mona S., Brauer S., Trent R. J., et al. The impact of the Austronesian expansion: evidence from mtDNA and Y chromosome diversity in the Admiralty Islands of Melanesia. Mol. Biol. Evol. 2008;25:1362–1374. doi: 10.1093/molbev/msn078. [DOI] [PubMed] [Google Scholar]
- Capelli C., Wilson J. F., Richards M., Stumpf M. P., Gratrix F., Oppenheimer S., et al. A predominantly indigenous paternal heritage for the Austronesian-speaking peoples of insular Southeast Asia and Oceania. Am. J. Hum. Genet. 2001;68:432–443. doi: 10.1086/318205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurles M. E., Nicholson J., Bosch E., Renfrew C., Sykes B. C., Jobling M. A. Y chromosomal evidence for the origins of oceanic-speaking peoples. Genetics. 2002;160:289–303. doi: 10.1093/genetics/160.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karafet T. M., Lansing J. S., Redd A. J., Watkins J. C., Surata S. P. K., Arthawiguna W. A., et al. Balinese Y-chromosome perspective on the peopling of Indonesia: genetic contributions from pre-Neolithic hunter-gatherers, Austronesian farmers, and Indian traders. Hum. Biol. 2005;77:93–114. doi: 10.1353/hub.2005.0030. [DOI] [PubMed] [Google Scholar]
- Li H., Wen B., Chen S. J., Su B., Pramoonjago P., Liu Y., et al. Paternal genetic affinity between Western Austronesians and Daic populations. BMC Evol. Biol. 2008;8:146. doi: 10.1186/1471-2148-8-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karafet T. M., Mendez F. L., Sudoyo H., Lansing J. S., Hammer M. F.Improved phylogenetic resolution and rapid diversification of Y chromosome haplogroup K-M256 in Southeast Asia Eur. J. Hum. Genet.(epub ahead of print 4 June 2014; doi: 10.1038/ejhg.2014.104 [DOI] [PMC free article] [PubMed]
- Tabbada K. A., Trejaut J., Loo J. H., Chen Y. M., Lin M., Mirazón-Lahr M., et al. Philippine mitochondrial DNA diversity: A populated viaduct between Taiwan and Indonesia. Mol. Biol. Evol. 2010;27:21–31. doi: 10.1093/molbev/msp215. [DOI] [PubMed] [Google Scholar]
- Friedlaender J. S., Friedlaender F. R., Hodgson J. A., Stoltz M., Koki G., Horvat G., et al. Melanesian mtDNA Complexity. PLoS ONE. 2007;2:e248. doi: 10.1371/journal.pone.0000248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redd A. J., Takesaki N., Sherry S. T., McGarvey S. T., Sofro A. S., Stoneking M. Evolutionary history of the COII/tRNALys intergenic 9-bp deletion in human mitochondrial DNAs from the Pacific. Mol. Biol. Evol. 1995;12:604–615. doi: 10.1093/oxfordjournals.molbev.a040240. [DOI] [PubMed] [Google Scholar]
- Cox M. P. Indonesian mitochondrial DNA and its opposition to a pleistocene era origin of Proto-Polynesians in Island Southeast Asia. Hum. Biol. 2005;77:179–188. doi: 10.1353/hub.2005.0037. [DOI] [PubMed] [Google Scholar]
- Hudjashov G., Kivisild T., Underhill P. A., Endicott P., Sanchez J. J., Lin A. A., et al. Revealing the prehistoric settlement of Australia by Y chromosome and mtDNA analysis. Proc. Natl Acad. Sci. USA. 2007;104:8726–8730. doi: 10.1073/pnas.0702928104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuy B. M., Stenersen M., Egeland T., Olaisen B. Y-chromosomal microsatellite mutation rates: differences in mutation rate between and within loci. Hum. Mutat. 2004;23:117–124. doi: 10.1002/humu.10294. [DOI] [PubMed] [Google Scholar]
- Fenner J. N. Cross-cultural estimation of the human generation interval for use in genetics-based population divergence studies. Am. J. Phys. Anthropol. 2005;128:415–423. doi: 10.1002/ajpa.20188. [DOI] [PubMed] [Google Scholar]
- Gusmão L., Sánchez-Diz P., Calafell F., Martín P., Alonso C. A., Álvarez-Fernández F., et al. Mutation rates at Y chromosome specific microsatellites. Hum. Mutat. 2005;26:520–528. doi: 10.1002/humu.20254. [DOI] [PubMed] [Google Scholar]
- Zhivotovsky L. A., Underhill P. A., Cinnioǧlu C., Kayser M., Morar B., Kivisild T., et al. The effective mutation rate at Y chromosome short tandem repeats, with application to human population-divergence time. Am. J. Hum. Genet. 2004;74:50–61. doi: 10.1086/380911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., Pugach I., Stoneking M., Kayser M., Jin L., the HUGO Pan-Asian SNP Consortium Genetic dating indicates that the Asian-Papuan admixture through Eastern Indonesia corresponds to the Austronesian expansion. Proc. Natl Acad. Sci. USA. 2012;109:4574–4579. doi: 10.1073/pnas.1118892109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansing S. J., Cox M. P., de Vet T. A., Downey S. S., Hallmark B., Sudoyo H. An ongoing Austronesian expansion in Island Southeast Asia. J. Anthropol. Archaeol. 2011;30:262–272. [Google Scholar]
- Hull G. Studies in Languages and Cultures of East Timor. University of Western Sydney; 2002. pp. 1–38. [Google Scholar]
- Duggan A. T., Evans B., Friedlaender F. R., Friedlaender J. S., Koki G., Merriwether D. A., et al. Maternal history of Oceania from complete mtDNA genomes: contrasting ancient diversity with recent homogenization due to the Austronesian expansion. Am. J. Hum. Genet. 2014;94:721–733. doi: 10.1016/j.ajhg.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor S.Unpacking the Island Southeast Asian Neolithic cultural package, and finding local complexity. Selected Papers from the 10th International Conference of the European Association of Southeast Asian Archaeologists: the British Museum, London1474–87.2006
- Denham T., Donohue M. Pre-Austronesian dispersal of banana cultivars West from New Guinea: linguistic relics from Eastern Indonesia. Archaeol. Oceania. 2009;44:18–28. [Google Scholar]
- De Langhe E. Relevance of banana seeds in archaeology. Ethnobot. Res. Appl. 2009;7:271–281. [Google Scholar]
- Pawley A.Examining the Farming/Language Dispersal Hypothesis(eds Bellwood P. & Renfrew, C.) 251–273.McDonald Institute for Archaeological Research, Cambridge; 2002 [Google Scholar]
- Mona S., Tommaseo-Ponzetta M., Brauer S., Sudoyo H., Marzuki S., Kayser M. Patterns of Y chromosome diversity intersect with the Trans-New Guinea hypothesis. Mol. Bio. Evol. 2007;24:2546–2555. doi: 10.1093/molbev/msm187. [DOI] [PubMed] [Google Scholar]
- Hill C., Soares P., Mormina M., Macaulay V., Clarke D., Blumbach P. B., et al. A mitochondrial stratigraphy for island southeast Asia. Am. J. Hum. Genet. 2007;80:29–43. doi: 10.1086/510412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinam T. A., Hong L. C., Phipps M. E., Stoneking M., Ameen M., Edo J., et al. Evolutionary history of continental southeast Asians: ‘Early train' hypothesis based on genetic analysis of mitochondrial and autosomal DNA data. Mol. Biol. Evol. 2012;29:3513–3527. doi: 10.1093/molbev/mss169. [DOI] [PubMed] [Google Scholar]
- Lipson M., Loh P. R., Patterson N., Moorjani P., Ko Y. C., Stoneking M., et al. Reconstructing Austronesian population history in Island Southeast Asia. bioRxivdoi:http://dx.doi.org/10.1101/005603 ( 2014 [DOI] [PMC free article] [PubMed]
- Cox M. P.Population Genetics Research Progress(ed Koven, V. T. )Ch. 245–83.Nova Science Publishers, New York; 2008 [Google Scholar]
- Wilder J. A., Cox M. P., Paquette A. M., Alford R., Satyagraha A. W., Harahap A., et al. Genetic continuity across a deeply divergent linguistic contact zone in North Maluku, Indonesia. BMC Genet. 2011;12:100. doi: 10.1186/1471-2156-12-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J. J., Soares D. B. Out of the Ashes. Australian National University Press, Canberra; 2003. [Google Scholar]
- Hägerdal H. The slaves of Timor: life and death on the fringes of early colonial society. Itinerario. 2010;34:19–44. [Google Scholar]
- Ahmad J. Out of the Ashes. Australian National University Press, Canberra, Australia; 2003. [Google Scholar]
- Hägerdal H. Lords of the Land, Lords of the Sea: Conflict and Adaptation in Early Colonial Timor. KITLV Press, Leiden; 2012. pp. 1600–1800. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.