Abstract
We have analyzed patients with previously untreated chronic lymphocytic leukemia with del11q fluorescence in situ hybridization (FISH) abnormality (n = 196) in this study. Detection of the 11q22.3 used a multicolor FISH technique. Patients with del11q fell into two major FISH subsets—sole del11q (n = 64) and del11q with del13q (n = 132). FISH subsets were compared using the median del11q FISH% (>58%, high vs. ≤58%, low). Overall survival (OS) and time to first treatment (TTFT) were estimated using Kaplan–Meier plots (log rank). Multivariate analysis was performed to assess the association between FISH% of del11q and outcomes. Patients with sole del11q were similar to del11q with del13q in terms of TTFT and OS. Patients with high FISH% of del11q had significantly shorter OS and TTFT as compared with patients with low FISH%, particularly in sole del11q; this negative impact of high FISH% of del11q on OS and TTFT was diminished with coexistent del13q. In multivariate analysis, high FISH% of del11q was a significant predictor for shorter OS and TTFT. A comparison of these del11q subsets with a separate cohort of (n = 673) previously untreated patients with sole del13q showed that the high FISH% del11q cohort had a significantly shorter TTFT and OS. In addition, bulky disease by physical examination or computed tomography imaging was infrequent at presentation in patients with del11q. High FISH% of del11q can reliably discriminate higher risk patients with chronic lymphocytic leukemia. Presence of coexistent del13q should be accounted for while prognosticating patients with del11q.
Introduction
The relevance of fluorescence in situ hybridization (FISH) is well established in the prognostication of patients with chronic lymphocytic leukemia (CLL) [1]. Recently, gene expression profiling [2], next generation sequencing [3,4], array comparative genomic hybridization [5], and single nucleotide polymorphism array [6] techniques have been used to interrogate genomic disturbances and mutations in CLL. The relevance of treatment in driving clonal evolution in CLL is also being recognized [3]. FISH-based prognostication is still the most widely used method for cytogenetic risk stratification in patients with newly diagnosed CLL. The clinical significance of chromosomal aberrations detected by FISH in patients with CLL was reported in 2000 [7]. The del11q aberration is regarded as a “high risk” prognostic marker in patients with CLL [8–13]. Chemoimmunotherapy produces high response rates in patients with del11q, but progression-free survival is shorter than that seen in patients without a del11q or del17p [14,15]. In addition, studies have reported that patients with del11q have bulky lymphadenopathy, a factor that could lead to earlier treatment [8,16]. There are various mechanisms by which del11q aberrations may promote CLL cell growth. These include mutations of the ATM gene [17–19], genomic instability due to ATM mutations and disruptions [20–22], modifications in the adhesion molecules and cell signaling receptors [23], increased TCL1 expression [24], insulin receptor overexpression [25,26], spliceosome (SF3B1) mutations [27], other gene expression [28–31], and phosphorylated histones [32].
The current FISH hierarchical model does not account for the prognostic influence of coexistent FISH abnormalities or percentage of positive cells (FISH%). It is unclear whether del13q, which is the most common coexistent FISH abnormality in patients with del11q, has any influence on the prognosis of these patients.
Here, we report on the clinical characteristics (including the incidence of bulky disease at presentation) of patients with del11q, the impact of coexistent del13q, and the prognostic relevance of FISH% of del11q in a cohort of 196 previously untreated patients.
Methods
Data were obtained from a cohort of previously untreated CLL patients (n = 210) with del11q FISH abnormality who presented to our institution between the years 2003 and 2012. All patients provided informed consent as per the declaration of Helsinki and University of Texas, MD Anderson Cancer center (MDACC) institutional review board approved protocol for retrospective chart review. del11q was determined from bone marrow aspirate or peripheral blood by FISH at the time of initial presentation to MDACC. The median time from the outside diagnosis to initial presentation to MDACC was similar across various patient subgroups (Supporting Information Table I). Locus-specific probes for ATM (11q22.3), S13S319 (13q14.3), LAMP1 (13q34), TP53 (17p13.1) as well as the centromeric region of chromosome 12 (12p11.1–q11) were used. Baseline clinical characteristics of all patients with del11q are shown in Table I. Because the majority of patients with del11q have coexistent del13q, we divided the patients into two subsets—sole del11q (n = 64) and del11q with coexistent del13q (n = 132). These two subsets (n = 196) were analyzed for patient outcomes. Clinical features of these two del11q subsets are shown in Supporting Information Table II. Because the number of patients with other FISH aberrations coexistent with del11q was very low, the following patients were excluded from this analysis: eight patients having del11q with +12; three patients having del11q with del17p; and three patients having del11q, del13q with +12. We compared the del11q subsets using the percentage of cells positive on FISH (FISH%). To identify the optimal cutoff of del11q FISH%, we used recursive partitioning (RP). Patients with FISH% >79.7 and >71.2 had a significantly increased risk of early treatment and death (see Supporting Information Figs. 1 and 2). Because the two groups (high vs. low) were found to be highly imbalanced when using the optimal cutoff value by RP analysis and because the results were similar when using median as the cutoff value, we chose the median as the cutoff for del11q FISH% for all analyses (>58%, high vs. ≤58%, low). A separate cohort of previously untreated patients with CLL and a sole del13q (n = 673) was compared with patients with del11q for time to first treatment (TTFT) and overall survival (OS). Similar to del11q FISH%, the median was chosen as the cutoff for sole del13q (>50.5%, high vs. ≤50.5%, low). Both the cohorts (del11q and del13q) presented during the same time period.
TABLE I.
Baseline Characteristics of Patients With del11q Aberration (n = 196)
| Variable | Median (range) | n (%) |
|---|---|---|
| Age (yrs) | 60 (26–86) | 196 (100) |
| WBC (K/µL) | 36 (3–786) | 196 (100) |
| % Lymphocytes | 85 (23–99) | 196 (100) |
| ALC (K/µL) | 30 (0.8–732) | 196 (100) |
| Gender | ||
| Female | 42 (21) | |
| Male | 154 (78) | |
| CD38 (%) | ||
| Missing | 2 | |
| CD38 low (<30%) | 90 (46) | |
| CD38 high (≥30%) | 104 (54) | |
| Zap-70 (%) | ||
| Missing | 42 | |
| Low (<20%) | 40 (26) | |
| High (≥20%) | 115 (74) | |
| β2 microglobulin (mg/L) | ||
| Missing | 7 | |
| Low (<4) | 130 (69) | |
| High (≥4) | 59 (31) | |
| Rai stage | ||
| Missing | 4 | |
| 0 | 29 (15) | |
| 1 | 102 (53) | |
| 2 | 28 (14) | |
| 3 | 16 (8) | |
| 4 | 17 (9) | |
| Bulky disease by PEa | ||
| Negative | 176 (90) | |
| Positive | 20 (9) | |
| Bulky disease by CTb | ||
| Missing | 94 | |
| Negative | 83 (81) | |
| Positive | 19 (19) | |
| IGHV mutation | ||
| Missing | 52 | |
| Mutated (>2%) | 15 (10) | |
| Unmutated (<2%) | 129 (89) | |
| FISH categoriesc | ||
| Sole del11q | 64 (30) | |
| del11q with del13q | 132 (63) | |
| Other cancers | 42 (21) | |
| No. of subsequently treated patients | 158 (81) |
Bulky disease by physical examination (PE) = any node/nodal mass ≥5 cm and/or spleen ≥6 cm.
Bulky disease by CT scan = any node/nodal mass ≥5 cm and/or spleen ≥10 cm in the longest axis.
Conventional karyotype data included (diploid, n = 85; complex, n = 38; del11q, n = 30; with del17p, n = 1; and not done or inadequate sample, n = 42).
Bulky disease at initial presentation was assessed by physical examination (PE) (defined as any node/nodal mass ≥ 5 cm and/or spleen ≥ 6 cm) and/or baseline computed tomography (CT) scans (any node/nodal mass ≥ 5 cm and/or spleen ≥ 10 cm in the longest axis) among evaluable patients.
For outcomes, TTFT was calculated from the date of initial presentation to MDACC to date of starting therapy or last follow-up date, whichever occurred first. OS was assessed from the initial presentation to the date of death or date of last follow-up, whichever occurred first. Progression-free survival was not evaluated in this study, as we focused only on previously untreated patients and FISH data obtained from the time of initial presentation, because chemotherapy can promote subclonal evolution. Moreover, majority of the patients have received fludarabine, cyclophosphamide, and rituximab (FCR)-based therapy (71%; Supporting Information Table III). The Kaplan–Meier method was used to estimate the probabilities of TTFT and OS, and subgroup comparisons were made via the log-rank test. Univariate Cox proportional hazards regression models were fit for OS and TTFT. A stepwise model selection procedure was applied, and a final multivariable Cox model was determined for OS and TTFT, respectively. The P-value was considered significant when ≤0.05. All statistical analyses were conducted using SAS 9.3 or Splus 8.2.
Results
Clinical characteristics of patients with del11q
A total of (n = 196) untreated patients with CLL and del11q FISH abnormality at presentation were evaluated (see Table I). The median age was 60 yrs, and the majority was men (78%). Median white cell count was 36 K/µL; median absolute lymphocyte count (ALC) was 30 K/µL. Only 17% (n = 33) of patients presented with advanced stage (Rai stage 3–4) disease. High β2 microglobulin (≥4 mg/L) was seen in 31% of patients (n = 59). The majority of patients had an unmutated IGHV (immunoglobulin heavy chain variable region) (n = 129, 89%). High expression of CD38 and Zap-70 was observed in 54% (n = 104) and 74% (n = 115) patients, respectively. Data on VH gene families were available in 89 (45%) patients, and an inclination toward a specific VH gene family was not observed (data not shown). Data on stereotypy status and NOTCH1, TP53, ATM, SF3B1, XPO-1, and BIRC3 mutations were not available. One hundred and fifty-eight (81%) patients eventually received therapy. Most (n = 112, 71%) were treated with the FCR regimen, whereas 29% (n = 46) received non–FCR-based therapies (Supporting Information Table III).
Characteristics of subsets of patients with del11q
The majority (63%) of patients with del11q had a coexistent del13q abnormality at presentation; therefore, to assess the influence of coexistent del13q, we compared del11q subsets according to coexistent del13q. Clinical characteristics of patients with sole del11q (n = 64) vs. those with coexistent del13q (n = 132) are summarized in Supporting Information Table II. Characteristics that were significantly different among patients with sole del11q as compared with patients with del11q with del13q were as follows: younger age (median age, 58 vs. 61 yrs), lower median white blood cell count (29 vs. 41 K/µL), and lower median ALC (24 vs. 34 K/µL) (P ≤ 0.05).
To assess the impact of FISH%, each del11q FISH subset was subdivided into high and low using the median value of FISH% as a cutoff (>58% vs. ≤58%) (see Table II). Patients with low FISH% were more likely to have a normal diploid karyotype compared with patients with high FISH% (73% vs. 42% for sole del11q and 67% vs. 37% for with coexistent del13q).
TABLE II.
Characteristics of Patients With Sole del11q or del11q With del13q Based on FISH% of del11q
| Variable | Low 11q% (≤58%) (n = 31, sole del11q) |
High 11q% (>58%) (n = 33, sole del11q) |
P-value | Low 11q% (≤58%) (n = 67, with del13q) |
High 11q% (>58%) (n = 65, with del13q) |
P-value |
|---|---|---|---|---|---|---|
| Age (yrs) | 59 (32–78) | 55 (26–78) | 0.21 | 60 (39–86) | 63 (40–80) | 0.20 |
| WBC (K/µL) | 22 (3–127) | 35 (6–346) | 0.09 | 31 (7–345) | 54 (6–786) | 0.08 |
| ALC (K/µL) | 19.4 (0.8–116) | 30 (2.3–308) | 0.12 | 27 (2–732) | 46 (3–707) | 0.53 |
| Gender | 0.48 | 0.84 | ||||
| Female | 3 (10%) | 6 (18%) | 16 (24%) | 17 (26%) | ||
| Male | 28 (90%) | 27 (82%) | 51 (76%) | 48 (74%) | ||
| CD38 (%) | 0.08 | 0.08 | ||||
| CD38 Low (<30%) | 19 (61%) | 12 (36%) | 35 (53%) | 24 (37%) | ||
| CD38 High (≥30%) | 12 (39%) | 21 (64%) | 31 (47%) | 40 (63%) | ||
| Zap-70 (%) | 1.00 | 1.00 | ||||
| Low (<20%) | 4 (19%) | 6 (20%) | 15 (28%) | 15 (30%) | ||
| High (≥20%) | 17 (81%) | 24 (80%) | 38 (72%) | 36 (71%) | ||
| β2 microglobulin (mg/L) | 0.07 | 0.85 | ||||
| Low (<4) | 26 (87%) | 20 (65%) | 42 (65%) | 42 (67%) | ||
| High (≥4) | 4 (13%) | 11 (35%) | 23 (35%) | 21 (33%) | ||
| Rai stage | 0.17 | 0.41 | ||||
| 0 | 7 (23%) | 3 (9%) | 11 (16%) | 8 (13%) | ||
| 1 | 15 (50%) | 18 (55%) | 35 (52%) | 34 (55%) | ||
| 2 | 4 (13%) | 1 (3%) | 11 (16%) | 12 (19%) | ||
| 3 | 2 (7%) | 5 (15%) | 7 (10%) | 2 (3%) | ||
| 4 | 2 (7%) | 6 (18%) | 3 (4%) | 6 (10%) | ||
| Bulky disease by PEa | 0.26 | 0.03 | ||||
| No | 29 (93%) | 27 (82%) | 57 (85%) | 63 (97%) | ||
| Yes | 2 (6%) | 6 (18%) | 10 (15%) | 2 (3%) | ||
| Bulky disease by CTb | 0.66 | 0.77 | ||||
| No | 16 (89%) | 14 (82%) | 27 (77%) | 26 (81%) | ||
| Yes | 2 (11%) | 3 (18%) | 8 (23%) | 6 (19%) | ||
| IGHV Mutation | 1.00 | 0.14 | ||||
| Mutated (>2%) | 1 (5%) | 1 (4%) | 9 (19%) | 4 (8%) | ||
| Unmutated (<2%) | 18 (95%) | 24 (96%) | 39 (81%) | 48 (92%) | ||
| Diploid karyotype | 19 (73%) | 10 (42%) | 0.04 | 39 (67%) | 17 (37%) | 0.003 |
| No. of subsequently treated patients | 25 (81%) | 29 (88%) | 0.50 | 48 (72%) | 56 (86%) | 0.06 |
Bulky disease by physical examination (PE) = any node/nodal mass ≥5 cm and/or spleen ≥6 cm.
Bulky disease by CT scan = any node/nodal mass ≥5 cm and/or spleen ≥10 cm in the longest axis.
Bulky disease at initial presentation (by PE or imaging by CT scan)
Based on two reports with limited numbers of patients with del11q (43 and seven patients) [8,16] and ill-defined criteria for bulky disease, it is generally thought that patients with del11q present with bulky disease. We analyzed data obtained from PE findings or CT scan imaging (neck, thorax, and abdomen) at the time of presentation. Moreover, we used specific criteria for defining bulky disease (see Methods section). Bulky disease by PE (n = 210) or by CT scan imaging (n = 111) was infrequent across all del11q FISH subsets. A higher proportion of patients with sole del11q and high FISH% had bulky disease (18%) as compared with low FISH% (6%). Bulky disease by PE was infrequent even when previously used criteria [10] (>3 cm) were applied.
Analysis of time to event variables among patients with del11q (overall and subsets)
Among all 196 patients (sole del11q and del11q with del13q), 30 patients have died. Seven patients died in the watch-and-wait phase (three other cancers, two unknown, one respiratory failure, and one due to sepsis), whereas 23 patients died after treatment: four with Richter’s transformation, six of unknown cause, three with secondary acute myeloid leukemia, two due to post-transplantation complications, three due to progressive disease, and five from other comorbidities. The median OS has not been reached; median follow-up time is 62 months (range, 0.7–122 months). Overall, 158 (81%) patients have received treatment (71% with an FCR-based regimen and 29% with non–FCR-based regimen). The median TTFT was 7.8 months (95% CI: 2.5–12.2 months).
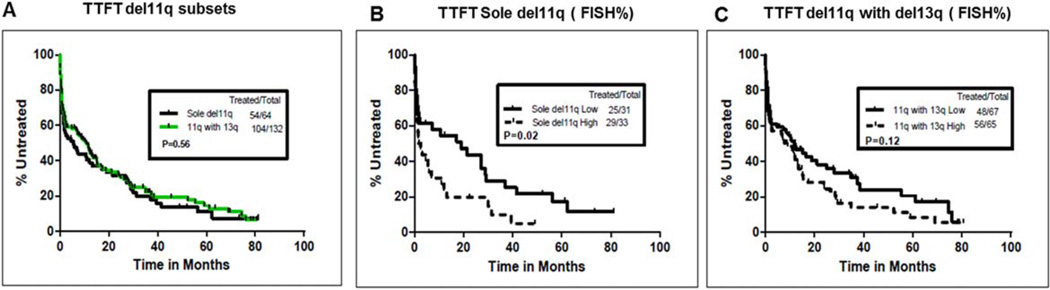
We then analyzed TTFT among the del11q FISH subsets. The median TTFT was not different between those with sole del11q and del11q with del13q (4.5 vs. 10.3 months, P = 0.56) (Fig. 1A). When TTFT was compared based on median FISH% (high vs. low), patients with sole del11q and high FISH% had significantly shorter TTFT (P = 0.02; Fig. 1B). The median TTFT values were 1.7 months (95% CI: 0.67–10.8 months) vs. 19.10 months (95% CI: 1.7–37 months) in high vs. low FISH%, respectively (summarized in Supporting Information Table IV). Patients with del11q and del13q were not significantly different based on median FISH% (Fig. 1C). Table III shows the univariate and multivariate analysis of factors predicting TTFT. In the multivariable Cox model, predictors of shorter TTFT included higher WBC count, bulky disease by PE, Rai stage 3 or 4 disease, and high FISH% of del11q. High FISH% del11q retained statistical significance when an optimal cutoff value of 71.25 was used (Supporting Information Fig. 1 and multivariate analysis (not shown)).
Figure 1.
(A–C) Analysis of time to first treatment (TTFT) in previously untreated CLL patients with del11q FISH subsets (sole del11q and del11q with del13q, n = 64 and n = 132, respectively) and median FISH% del11q. (A) Median TTFT was 4.5 and 10.3 months in sole del11q and del11q with del13q, respectively, and was not significantly different (P = 0.56). (B) Median TTFT was significantly shorter in patients with high FISH% (>58%) as compared with those with low FISH% (≤58%) in sole del11q: 1.7 vs. 19.1 months (P = 0.02). (C) Based on FISH%, TTFT was not significantly different among patients with del11q with del13q (P = 0.12). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
TABLE III.
Univariate and Multivariate Cox Proportional Hazard Model for Time to First Treatment (TTFT) for Patients With del11q FISH Abnormality (n = 196); del11q FISH% Based on Median
| Variable | HR (95% CI) | P-value |
|---|---|---|
| Univariate | ||
| Age (yrs) | 0.99 (0.98–1.01) | 0.28 |
| Log(WBC) | 1.68 (1.45–1.96) | <0.0001 |
| Lymphocyte (%) | 1.03 (1.01–1.04) | <0.0001 |
| Log(ALC) | 1.51 (1.32–1.71) | <0.0001 |
| β2 microglobulin | 1.19 (1.11–1.28) | <0.0001 |
| Male vs. female | 1.00 (0.68–1.46) | 1.00 |
| CD38, high vs. low | 1.48 (1.09–2.01) | 0.01 |
| Zap-70, high vs. low | 1.18 (0.79–1.75) | 0.42 |
| Bulky disease (PE), yes vs. no | 4.21 (2.54–7.00) | <0.0001 |
| Bulky disease (CT), yes vs. no | 2.55 (1.51–4.33) | 0.0005 |
| Rai stage, 3, 4 vs. 0, 1, and 2 | 2.02 (1.40–2.92) | 0.0002 |
| FISH subset, sole del11q vs. del11q with del 13q | 1.10 (0.79–1.53) | 0.56 |
| FISH 11q%, >58% vs. ≤58% | 1.58 (1.16–2.17) | 0.004 |
| Multivariate | ||
| Log(WBC) | 1.63 (1.37–1.93) | <0.0001 |
| Bulky disease (PE), yes vs. no | 3.56 (2.03–6.27) | <0.0001 |
| Rai stage, 3, 4 vs. 0, 1, and 2 | 1.77 (1.17–2.66) | 0.006 |
| FISH 11q%, >58% vs. ≤58% | 1.67 (1.20–2.32) | 0.002 |
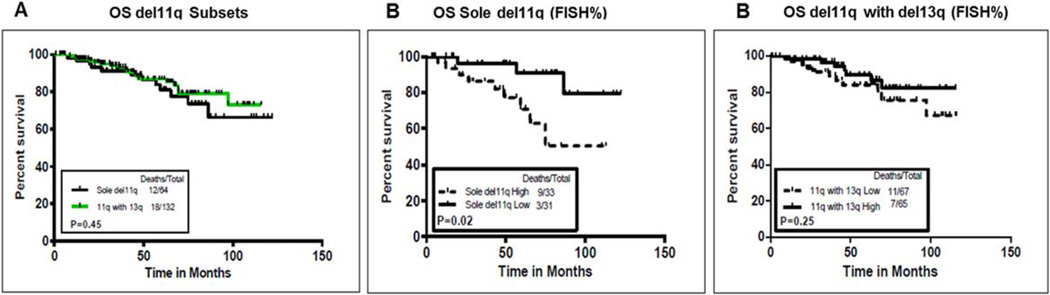
We next assessed whether coexistent deletion 13q had any influence on OS. Figure 2A shows that there was no significant difference between the OS of the two FISH subsets (sole del11q, n = 64 vs. del11q with del13q, n = 132). Median OS was not reached in either group (summarized in Supporting Information Table IV). When patients with sole del11q were compared based on FISH% (high vs. low), patients with high FISH% (>58%) had a significantly inferior OS (P = 0.02; Fig. 2B). The median OS was 75 months, with a 95% CI of 65.2—not estimable in the high FISH% group; the median OS was not reached in the low FISH% group. Patients with del11q and del13q were not significantly different in OS, based on median FISH% (Fig. 1C). Patients with sole del11q with high FISH% had significantly inferior OS as compared with patients with del11q with del13q with high FISH% (P = 0.02) (not shown). Table IV shows the univariate and multivariate analyses of factors predicting OS. In multivariate analysis, shorter survival was associated with older age, higher expression of CD38, and high FISH% (sole del11q). Furthermore, high FISH% del11q retained statistical significance after using an optimal cutoff value of 79.75 (Supporting Information Fig. 2 and multivariate analysis (not shown)).
Figure 2.
(A–C) Analysis of impact of coexistent del13q on overall survival (OS) in previously untreated CLL patients with del11q FISH subsets (sole del11q and del11q with del13q, n = 64 and n = 132, respectively). (A) Median OS was not reached (NR) in either group (P = 0.45). (B) Median OS was significantly shorter in patients with high FISH% (>58%) as compared with those with low FISH% (≤58%) in sole del11q: NR months vs. NR (P = 0.02). (C) Based on FISH%, OS was not significantly different among patients with del11q with del13q (P = 0.25). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
TABLE IV.
Univariate and Multivariate Cox Proportional Hazard Model for Overall Survival (OS) for Patients With del11q FISH Abnormality (n = 196); del11q FISH% Based on Median
| Analysis | HR (95% CI) | P-value |
|---|---|---|
| Univariate | ||
| Age (yrs) | 1.05 (1.00–1.09) | 0.01 |
| Log(WBC) | 0.86 (0.61–1.21) | 0.39 |
| Lymphocyte (%) | 1.00 (0.98–1.02) | 0.94 |
| Log(ALC) | 0.89 (0.68–1.18) | 0.43 |
| β2 microglobulin | 1.27 (1.05–1.53) | 0.01 |
| Male vs. female | 1.11 (0.43–2.90) | 0.83 |
| CD38, high vs. low | 2.56 (1.18–5.56) | 0.02 |
| Zap-70, high vs. low | 0.75 (0.33–1.71) | 0.49 |
| Bulky disease (PE), yes vs. no | 1.82 (0.70–4.73) | 0.22 |
| Bulky disease (CT), yes vs. no | 1.21 (0.41–3.63) | 0.73 |
| Rai stage, 3, 4 vs. 0, 1, and 2 | 2.38 (1.09–5.20) | 0.03 |
| FISH subset, sole del11q vs. del11q with del 13q | 1.33 (0.64–2.76) | 0.45 |
| FISH 11q%, >58% vs. ≤58% | 1.19 (0.58–2.44) | 0.64 |
| Multivariate | ||
| Age (yrs) | 1.05 (1.02–1.09) | 0.006 |
| CD38, high vs. low | 3.12 (1.35–7.20) | 0.008 |
| aFISH 11q%, >58% vs. ≤58% | 4.29 (1.12–16.36) | 0.03 |
In FISH subset (sole del11q).
Comparison of time to event variables among patients with del11q subsets and sole del13q aberration
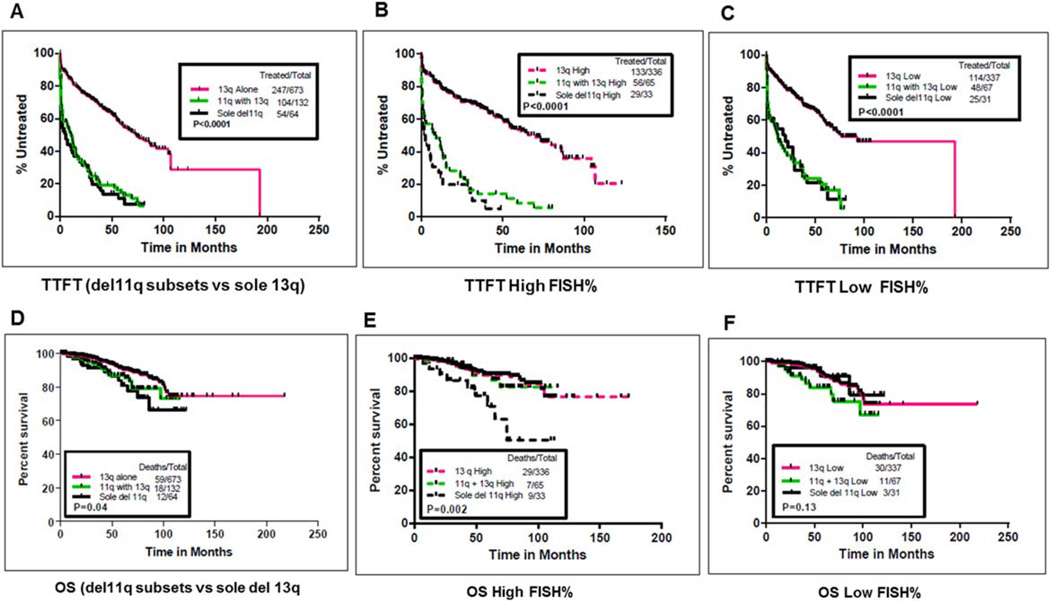
After comparing the TTFT and OS between the two del11q FISH subsets (sole del11q and coexistent del13q) and impact of del11q FISH%, we compared TTFT and OS of del11q FISH subsets with a separate cohort of (n = 673) previously untreated patients presenting with sole del13q (Fig. 3A–F), and the results are summarized in Supporting Information Tables III and IV. Median follow-up time for patients with sole del13q was 52.3 months (range, 0–218 months). TTFT was significantly shorter in patients with del11q (all FISH subsets) as compared with patients with sole del13q (P < 0.0001; Fig. 3A). Within each subgroup of patients with high or low median FISH%, respectively, the TTFT was significantly shorter in del11q FISH subsets as compared with sole del13q (P < 0.0001; Fig. 3B,C). When comparisons were made to OS, del11q FISH subsets (sole del11q and del11q with del13q) had a shorter OS compared with sole del13q (P = 0.04; Fig. 3D). OS in patients with high FISH% in sole del11q was significantly inferior as compared with high FISH% in coexistent del13q and sole del13q (P = 0.002; Fig. 3E). Among patients with low FISH%, OS was not significantly different among the three groups, but very few events had occurred in any group (Fig. 3F). These results were more significant when the three groups (sole del11q, with coexistent del13q, and sole del13q) were compared based on optimal cutoff values of FISH% obtained from RP method (see Supporting Information Fig. 3).
Figure 3.
(A–F) Comparison of TTFT and OS among del11q FISH subsets (sole del11q and del11q with del13q) with a cohort of untreated CLL patients with sole del13q (n = 673). Time to first treatment (TTFT) is compared among sole del11q (n = 64), del11q with del13q (n = 132), and sole del13q (n = 673). (A) Median TTFT was significantly shorter in del11q FISH subsets compared with sole del13q cohort: 4.5, 10.3, and 73.5 months, respectively (P < 0.0001). (B, C) Median TTFT was significantly shorter in del11q FISH subsets as compared with sole del13q, with respect to FISH%. Subgroup of patients with high FISH% (>58% for del11q subsets and >50.5% for sole del13q) and low FISH% (≤58% for del11q categories and ≤50.5% for sole del13q) are shown (P < 0.0001 for high and low FISH% subgroups). (D) OS was significantly different among the three groups (sole del11q, del11q with del13q, and sole del13q). Median OS was not reached (NR) in all the three groups (P = 0.04). (E, F) Median OS was significantly shorter in sole del11q with high FISH% when compared with coexistent del13q and sole del13q with high FISH% (P = 0.002). Within low FISH% patients, OS was not significantly different among the three groups (P = 0.13). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Discussion
Among the various molecular and cytogenetic techniques, FISH-based prognostication is still the most widely utilized for initial risk stratification in patients with CLL. The FISH hierarchical model was initially proposed in 2000 [7]. Among the 325 (treated and untreated) patients with CLL studied, 268 (82%) exhibited a FISH abnormality. del11q was detectable in 58 (18%) patients, second in frequency only to del13q. Sole del11q was seen in 19 of 58 (33%) patients. Information from this model and studies reported before [8,9,12,20] and after 2000 [10,16] indicated that the del11q FISH category identifies patients with high-risk CLL and that they often have bulky disease. However, it is of note that most of these studies were limited by small numbers of patients with sole del11q, a mixed patient population (untreated and treated patients), lack of del11q subset analysis, and demonstration of bulky disease by ill-defined criteria [8–12]. In this study, we have focused on analysis of 196 previously untreated patients with CLL and del11q at initial presentation.
Contrary to previous reports, we found that bulky disease (PE and/or CT scans) was infrequent in patients with del11q at initial presentation. Although no uniform guidelines exist to define “bulky disease” in CLL, the criteria used in our analysis for defining bulky disease by PE (splenomegaly > 6 cm) are similar to those included under the indications for treatment in the 2008 International Working Group of CLL guidelines [33] (except that lymph node > 5 cm is considered as bulky disease in this analysis). This criterion for bulky disease (lymph node > 5 cm) was used in another study [34] and in our previous report on 66 patients with del11q [14]. Another study reported bulky disease (lymph node > 7 cm) by CT scan in 11 of 77 patients with CLL [35].
Clinical features of patients with del11q reported in our study were similar to those in previous reports [10,14]. Patients with del11q have high frequencies of associated del13q (63%), unmutated IGHV, and overexpression of CD38 and Zap-70. Patients were analyzed based on del11q FISH categories: sole del11q vs. del11q with del13q. Other FISH abnormalities such as del17p were uncommon with del11q at the time of initial presentation. These results are similar to those of another analysis, where among the 2,184 patients only 1% of patients showed a coexistent del11q with del17p [36]. We further divided these del11q FISH categories based on median FISH% of del11q (high and low). Of note, patients with sole del11q and high FISH% (>58%) had a higher incidence of complex karyotype as compared with those with low FISH%.
We then looked at the impact of coexistent del13q on TTFT and OS. Both TTFT and OS were not significantly different between the two FISH subsets (Figs. 1A and 2A). Nonetheless, patients with sole del11q and high FISH% were distinct in having shorter TTFT and OS as compared with those with low FISH% (Figs. 1B and 2B). Marasca et al. [10] reported that FISH% (>25%) predicted for significantly inferior TTFT but not worse OS. In their study, patients with sole del11q vs. those with del11q and del13q were not compared; in addition the reason for a cutoff of 25% was unclear (the median FISH% reported was 52%). Similarly, another analysis (abstract form) showed that patients with a higher copy number of del11q ≥ 40% had a significantly shorter TTFT and OS [37]. The subdivision of the FISH subsets was important, as we have shown that the negative effect of high FISH% on OS was absent in the presence of coexistent del13q.
Moreover, high FISH% was a predictor for shorter TTFT and OS in multivariate analysis (Tables III and III). Of note, by RP, we identified an optimal cutoff for FISH% for TTFT and OS (71.75% and 79.75%, respectively) dividing the patients into high vs. low FISH% (Supporting Information Figs. 1–3). The reasons for choosing median FISH% as a cutoff were as follows: using optimal cutoffs provided an imbalanced population and results using median FISH% were similar to those obtained with optimal cutoffs.
Our findings suggest that there is an interaction between del13q and del11q. Although these results are clinically relevant, questions with regard to the molecular biology, i.e., interactive mechanisms of Rb gene, length of 13q deletion and ATM deletions, and novel mutations such as those in SF3B1 and BIRC3, are beyond the scope of our study.
Finally, we compared patients with del11q and a separate cohort of previously untreated patients with CLL and sole del13q (n = 673). The TTFT was significantly inferior in all del11q categories (Fig. 3A–C). The OS was shorter in patients with del11q (Fig. 3D). When the OS was compared based on FISH%, patients with sole del11q with high FISH% had shorter OS, whereas this difference was not observed in patients with low FISH% (Fig. 3E,F). However, few deaths have occurred in those arms; so, the follow-up may be too short to detect differences. A limitation of this study is the median follow-up of 62 months and fewer events, which may limit a robust analysis of OS. The relevance of the median FISH% should be validated at other centers.
In summary, our data provide novel insights on the clinical relevance of the del11q FISH category. The majority of previously untreated patients with del11q do not have bulky disease at the time of presentation. We have shown that patients with sole del11q and high FISH% have a shorter TTFT and OS. We propose that coexistent del13q and FISH% should be accounted for at the time of initial presentation when analyzing patients with del11q. We conclude that the highest risk status for del11q FISH category may be restricted to patients with sole del11q and high FISH%.
Supplementary Material
Footnotes
Additional Supporting Information may be found in the online version of this article.
Conflict of interest: None of the authors report any competing conflicts of interest.
Author Contributions
P.J., S.O.B., and M.K. designed the study. P.J., X.W., M.K., P.T., and S.O.B., analyzed the results. P.J., L.T., X.W., P.T., L.A., M.K., and S.O.B. wrote the article. P.J., L.T., X.W., M.K., and S.O.B did the clinical correlation. M.K., S.O.B., H.K., W.W., A.F., Z.E., and J.B. contributed to patient samples. All authors have reviewed and contributed to the manuscript.
References
- 1.Cramer P, Hallek M. Prognostic factors in chronic lymphocytic leukemia—What do we need to know? Nat Rev Clin Oncol. 2011;8:38–47. doi: 10.1038/nrclinonc.2010.167. [DOI] [PubMed] [Google Scholar]
- 2.Friedman DR, Lucas JE, Weinberg JB. Clinical and biological relevance of genomic heterogeneity in chronic lymphocytic leukemia. PLoS One. 2013;8:e57356. doi: 10.1371/journal.pone.0057356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Landau DA, Carter SL, Stojanov P, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152:714–726. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Puente XS, Pinyol M, Quesada V, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gunn SR, Mohammed MS, Gorre ME, et al. Whole-genome scanning by array comparative genomic hybridization as a clinical tool for risk assessment in chronic lymphocytic leukemia. J Mol Diagn. 2008;10:442–451. doi: 10.2353/jmoldx.2008.080033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mian M, Rinaldi A, Mensah AA, et al. Large genomic aberrations detected by SNP array are independent prognosticators of a shorter time to first treatment in chronic lymphocytic leukemia patients with normal FISH. Ann Oncol. 2013;24:1378–1384. doi: 10.1093/annonc/mds646. [DOI] [PubMed] [Google Scholar]
- 7.Dohner H, Stilgenbauer S, Benner A, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 8.Dohner H, Stilgenbauer S, James MR, et al. 11q deletions identify a new subset of B-cell chronic lymphocytic leukemia characterized by extensive nodal involvement and inferior prognosis. Blood. 1997;89:2516–2522. [PubMed] [Google Scholar]
- 9.Neilson JR, Auer R, White D, et al. Deletions at 11q identify a subset of patients with typical CLL who show consistent disease progression and reduced survival. Leukemia. 1997;11:1929–1932. doi: 10.1038/sj.leu.2400819. [DOI] [PubMed] [Google Scholar]
- 10.Marasca R, Maffei R, Martinelli S, et al. Clinical heterogeneity of de novo 11q deletion chronic lymphocytic leukaemia: Prognostic relevance of extent of 11q deleted nuclei inside leukemic clone. Hematol Oncol. 2013;31:88–95. doi: 10.1002/hon.2028. [DOI] [PubMed] [Google Scholar]
- 11.Zenz T, Gribben JG, Hallek M, et al. Risk categories and refractory CLL in the era of chemo-immunotherapy. Blood. 2012;119:4101–4107. doi: 10.1182/blood-2011-11-312421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fegan C, Robinson H, Thompson P, et al. Karyotypic evolution in CLL: Identification of a new sub-group of patients with deletions of 11q and advanced or progressive disease. Leukemia. 1995;9:2003–2008. [PubMed] [Google Scholar]
- 13.Dewald GW, Brockman SR, Paternoster SF, et al. Chromosome anomalies detected by interphase fluorescence in situ hybridization: Correlation with significant biological features of B-cell chronic lymphocytic leukaemia. Br J Haematol. 2003;121:287–295. doi: 10.1046/j.1365-2141.2003.04265.x. [DOI] [PubMed] [Google Scholar]
- 14.Tsimberidou AM, Tam C, Abruzzo LV, et al. Chemoimmunotherapy may overcome the adverse prognostic significance of 11q deletion in previously untreated patients with chronic lymphocytic leukemia. Cancer. 2009;115:373–380. doi: 10.1002/cncr.23993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hallek M, Fischer K, Fingerle-Rowson G, et al. Addition of rituximab to fludarabine and cyclophosphamide in patients with chronic lymphocytic leukaemia: A randomised, open-label, phase 3 trial. Lancet. 2010;376:1164–1174. doi: 10.1016/S0140-6736(10)61381-5. [DOI] [PubMed] [Google Scholar]
- 16.Dickinson JD, Gilmore J, Iqbal J, et al. 11q22.3 deletion in B-chronic lymphocytic leukemia is specifically associated with bulky lymphadenopathy and ZAP-70 expression but not reduced expression of adhesion/cell surface receptor molecules. Leuk Lymphoma. 2006;47:231–244. doi: 10.1080/10428190500254141. [DOI] [PubMed] [Google Scholar]
- 17.Schaffner C, Stilgenbauer S, Rappold GA, et al. Somatic ATM mutations indicate a pathogenic role of ATM in B-cell chronic lymphocytic leukemia. Blood. 1999;94:748–753. [PubMed] [Google Scholar]
- 18.Skowronska A, Parker A, Ahmed G, et al. Biallelic ATM inactivation significantly reduces survival in patients treated on the United Kingdom Leukemia Research Fund Chronic Lymphocytic Leukemia 4 Trial. J Clin Oncol. 2012;30:4524–4532. doi: 10.1200/JCO.2011.41.0852. [DOI] [PubMed] [Google Scholar]
- 19.Rose-Zerilli MJ, Forster J, Parker H, et al. ATM mutation rather than BIRC3 deletion and/or mutation predicts reduced survival in 11q-deleted chronic lymphocytic leukemia: Data from the UK LRF CLL4 trial. Haematologica. 2014;99:736–742. doi: 10.3324/haematol.2013.098574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Starostik P, Manshouri T, O’Brien S, et al. Deficiency of the ATM protein expression defines an aggressive subgroup of B-cell chronic lymphocytic leukemia. Cancer Res. 1998;58:4552–4557. [PubMed] [Google Scholar]
- 21.Rossi D, Gaidano G. ATM and chronic lymphocytic leukemia: Mutations, and not only deletions, matter. Haematologica. 2012;97:5–8. doi: 10.3324/haematol.2011.057109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guarini A, Marinelli M, Tavolaro S, et al. ATM gene alterations in chronic lymphocytic leukemia patients induce a distinct gene expression profile and predict disease progression. Haematologica. 2012;97:47–55. doi: 10.3324/haematol.2011.049270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sembries S, Pahl H, Stilgenbauer S, et al. Reduced expression of adhesion molecules and cell signaling receptors by chronic lymphocytic leukemia cells with 11q deletion. Blood. 1999;93:624–631. [PubMed] [Google Scholar]
- 24.Herling M, Patel KA, Weit N, et al. High TCL1 levels are a marker of B-cell receptor pathway responsiveness and adverse outcome in chronic lymphocytic leukemia. Blood. 2009;114:4675–4686. doi: 10.1182/blood-2009-03-208256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saiya-Cork K, Collins R, Parkin B, et al. A pathobiological role of the insulin receptor in chronic lymphocytic leukemia. Clin Cancer Res. 2011;17:2679–2692. doi: 10.1158/1078-0432.CCR-10-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown JR. Insulin receptor activation in deletion 11q chronic lymphocytic leukemia. Clin Cancer Res. 2011;17:2605–2607. doi: 10.1158/1078-0432.CCR-11-0295. [DOI] [PubMed] [Google Scholar]
- 27.Wang L, Lawrence MS, Wan Y, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med. 2011;365:2497–2506. doi: 10.1056/NEJMoa1109016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Visone R, Rassenti LZ, Veronese A, et al. Karyo-type-specific microRNA signature in chronic lymphocytic leukemia. Blood. 2009;114:3872–3879. doi: 10.1182/blood-2009-06-229211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kalla C, Scheuermann MO, Kube I, et al. Analysis of 11q22–q23 deletion target genes in B-cell chronic lymphocytic leukaemia: Evidence for a pathogenic role of NPAT, CUL5, and PPP2R1B. Eur J Cancer. 2007;43:1328–1335. doi: 10.1016/j.ejca.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 30.Haslinger C, Schweifer N, Stilgenbauer S, et al. Microarray gene expression profiling of B-cell chronic lymphocytic leukemia subgroups defined by genomic aberrations and VH mutation status. J Clin Oncol. 2004;22:3937–3949. doi: 10.1200/JCO.2004.12.133. [DOI] [PubMed] [Google Scholar]
- 31.Aalto Y, El-Rifa W, Vilpo L, et al. Distinct gene expression profiling in chronic lymphocytic leukemia with 11q23 deletion. Leukemia. 2001;15:1721–1728. doi: 10.1038/sj.leu.2402282. [DOI] [PubMed] [Google Scholar]
- 32.Ouillette P, Fossum S, Parkin B, et al. Aggressive chronic lymphocytic leukemia with elevated genomic complexity is associated with multiple gene defects in the response to DNA double-strand breaks. Clin Cancer Res. 2010;16:835–847. doi: 10.1158/1078-0432.CCR-09-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hallek M, Cheson BD, Catovsky D, et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: A report from the international workshop on chronic lymphocytic leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood. 2008;111:5446–5456. doi: 10.1182/blood-2007-06-093906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eichhorst BF, Fischer K, Fink AM, et al. Limited clinical relevance of imaging techniques in the follow-up of patients with advanced chronic lymphocytic leukemia: Results of a meta-analysis. Blood. 2011;117:1817–1821. doi: 10.1182/blood-2010-04-282228. [DOI] [PubMed] [Google Scholar]
- 35.Norin S, Kimby E, Lundin J. Tumor burden status evaluated by computed tomography scan is of prognostic importance in patients with chronic lymphocytic leukemia. Med Oncol. 2010;27:820–825. doi: 10.1007/s12032-009-9292-y. [DOI] [PubMed] [Google Scholar]
- 36.Greipp PT, Smoley SA, Viswanatha DS, et al. Patients with chronic lymphocytic leukaemia and clonal deletion of both 17p13.1 and 11q22.3 have a very poor prognosis. Br J Haematol. 2013;163:326–333. doi: 10.1111/bjh.12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hernandez JA, Rodriguez AE, Santacruz R, et al. A high number of losses in 11q chromosome is associated with short time to first treatment (TFT) and overall survival (OS) in patients with chronic lymphocytic leukemia (CLL); Paper presented at the proceedings of the European Hematology Association (EHA); 2013. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.