Abstract
Background & Aims
Fibrolamellar hepatocellular carcinoma (FLC) is a rare primary hepatic cancer that develops in children and young adults without cirrhosis. Little is known about its pathogenesis, and it can only be treated with surgery. We performed an integrative genomic analysis of a large series of patients with FLC to identify associated genetic factors.
Methods
Using 78 clinically annotated FLC samples, we performed whole-transcriptome (n=58), single-nucleotide polymorphism array (n=41), and next-generation sequencing (n=48) analyses; we also assessed the prevalence of the DNAJB1–PRKACA fusion transcript associated with this cancer (n=73). We performed class discovery using non-negative matrix factorization, and functional annotation using gene set enrichment analyses, nearest template prediction, ingenuity pathway analyses, and immunohistochemistry. The genomic identification of significant targets in cancer algorithm was used to identify chromosomal aberrations, MuTect and VarScan2 were used to identify somatic mutations, and the random survival forest was used to determine patient prognoses. Findings were validated in an independent cohort.
Results
Unsupervised gene expression clustering revealed 3 robust molecular classes of tumors: the proliferation class (51% of samples) had altered expression of genes that regulate proliferation and mTOR signaling activation; the inflammation class (26% of samples) had altered expression of genes that regulate inflammation and cytokine production; and the unannotated class (23% of samples) had a gene expression signature not previously associated with liver tumors. Expression of genes that regulate neuroendocrine function, as well has histologic markers of cholangiocytes and hepatocytes, were detected in all 3 classes. FLCs had few copy number variations; the most frequent were focal amplification at 8q24.3 (in 12.5% of samples) and deletions at 19p13 (in 28% of samples) and 22q13.32 (in 25% of samples). The DNAJB1–PRKACA fusion transcript was detected in 79% of samples. FLC samples also contained mutations in cancer-related genes such as BRCA2 (in 4.2% of samples), which are uncommon in liver neoplasms. However, FLCs did not contain mutations most commonly detected in liver cancers. We identified an 8-gene signature that predicted survival of patients with FLC.
Conclusions
In a genomic analysis of 78 FLC samples, we identified 3 classes based on gene expression profiles. FLCs contain mutations and chromosomal aberrations not previously associated with liver cancer, and almost 80% contain the DNAJB1–PRKACA fusion transcript. Using this information, we identified a gene signature that is associated with patient survival time.
Keywords: molecular classification, genomic profiling, outcome, targeted therapies
INTRODUCTION
Liver cancer is now the second leading cause of death among cancer patients worldwide1. Fibrolamellar hepatocellular carcinoma (FLC) accounts for 0.85% of all primary hepatic malignancies in the United States2, and its incidence rate is 0.02 cases per 100.000 person-year2, behind hepatocellular carcinoma (HCC)3 and intrahepatic cholangiocarcinoma (ICC)4. Unlike standard HCC, FLC typically arises in non-cirrhotic livers of children and young adults with no specific association with etiologic factors or gender2,5. FLC has a better prognosis than HCC, probably due to the absence of cirrhosis and the earlier age of presentation5,6. Nonetheless, survival is jeopardized by tumor recurrence and metastases6. Treatment options remain limited to surgical resection7, and no effective targeted therapies have been described so far.
Seminal studies have been performed to decipher genomic alterations in FLC, though all of them were performed in small cohorts8–13. The most relevant findings include molecular alterations of the MAPK/ERK9, ErbB10 and Akt-mTOR12 signaling pathways, few chromosomal aberrations based on comparative genomic hybridization data13, and no mutations in EGFR or KRAS genes11. More recently, a DNAJB1-PRKACA fusion protein caused by a deletion at chromosome 19 has been discovered in all 15 FLCs evaluated14.
The scarce understanding of the FLC molecular drivers prevents the initiation of biomarker-driven clinical trials that will allow an improved management of the disease. This prompted us to investigate a large series of 78 FLCs with clinical annotated data [formalin-fixed paraffin-embedded (FFPE, n=54) and fresh-frozen (FF, n=24)] through the application of cutting-edge genomic approaches: whole-genome expression (n=58), single-nucleotide polymorphism (SNP)-array (n=41) and next-generation sequencing (NGS, n=48) [i.e. whole-exome sequencing (WES) and targeted-exome sequencing (TES)]. We also checked for the prevalence of the DNAJB1-PRKACA fusion event (n=73).
Overall, FLC analysis revealed 3 distinct molecular classes, relevant but few chromosomal alterations, a unique mutational portrait compared to HCC and ICC, and a highly frequent fusion transcript. In addition, we generated a prognostic 8-gene expression signature able to accurately predict survival in FLC patients, which was validated in 22 FF FLCs.
MATERIALS and METHODS
Clinicopathological characteristics of FLC patients
A total of 78 FLC patients surgically treated were included in the study (Table 1 and Suppl. Table 1). The training cohort included 42 FFPE FLC and 18 non-tumoral paired samples obtained from patients resected or transplanted during 1987–2009 at 6 academic hospitals: King’s College Hospital (London), Mayo Clinic (Rochester), Icahn School of Medicine at Mount Sinai (New York), IRCCS Istituto Nazionale Tumori (Milan), Hospital Clínic (Barcelona), and Hospital Vall d’Hebron (Barcelona). The study protocol was approved by each center’s Institutional Review Board. FLC diagnosis based on pathology was confirmed by 3 independent pathologists (S.T., S.W. and L.R.). The validation-French cohort included 23 surgically-resected FF FLCs (Suppl. Table 1) obtained from 6 different hospitals in France. We do not obtained non-tumoral samples for this cohort.
Table 1.
Study design
| Study samples | Type of sample | Different cohorts | Applied techniques | ||||
|---|---|---|---|---|---|---|---|
| Molecular profiling | SNP-array | WES | TES | Fusion transcript | |||
| 78 FLCs | 54 FFPE samples | 42 Training cohort | 35 | 32 | 27 | 39 | |
| 12 Validation-Brazilian cohort | 12 | ||||||
| 24 FF samples | 23 Validation-French cohort* | 23 | 9 | 20 | 22 | ||
| 1 additional FF FLC sample | 1 | ||||||
| Total | 58 | 41 | 1 | 47 | 73 | ||
WES = whole-exome sequencing
TES = targeted-exome sequencing
We also obtained 1 additional FF FLC sample that was included in the WES mutational study from a Caucasian male, and 12 additional FFPE FLC and 5 paired non-tumoral samples from a Brazilian cohort (Validation-Brazilian cohort, Suppl. Table 1), where the presence of the DNAJB1-PRKACA fusion transcript was checked.
Gene Expression, SNP-array and NGS
Among the FFPE samples from the training cohort, 35 FLCs were analyzed for molecular characterization, 32 for CNVs, and 27 for mutations. Regarding the FF samples from the validation-French cohort, 23 were profiled at the transcriptional level, 9 by SNP-array and 20 by TES. Whole-genome expression profiling was performed using DASL-array, genome-wide CNVs using HumanOmniExpress FFPE-12 DNA-Analysis BeadChip, WES using HiSeq2000 sequencer and TES using MiSeq Next Generation Sequencer, all platforms from Illumina. For further information see Supplementary Materials and Methods section and Table 1.
Immunohistochemistry, Bioinformatics and Clinical Data Analysis
See Supplementary Materials and Methods section.
RESULTS
Molecular profiling reveals three FLC classes with distinct genomic patterns
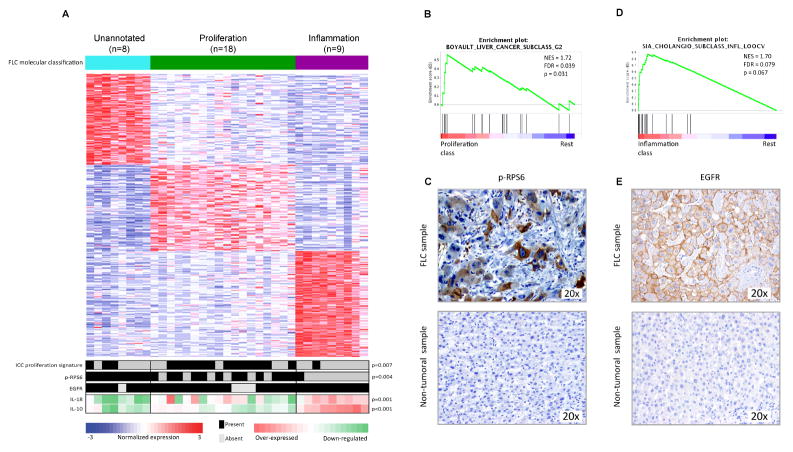
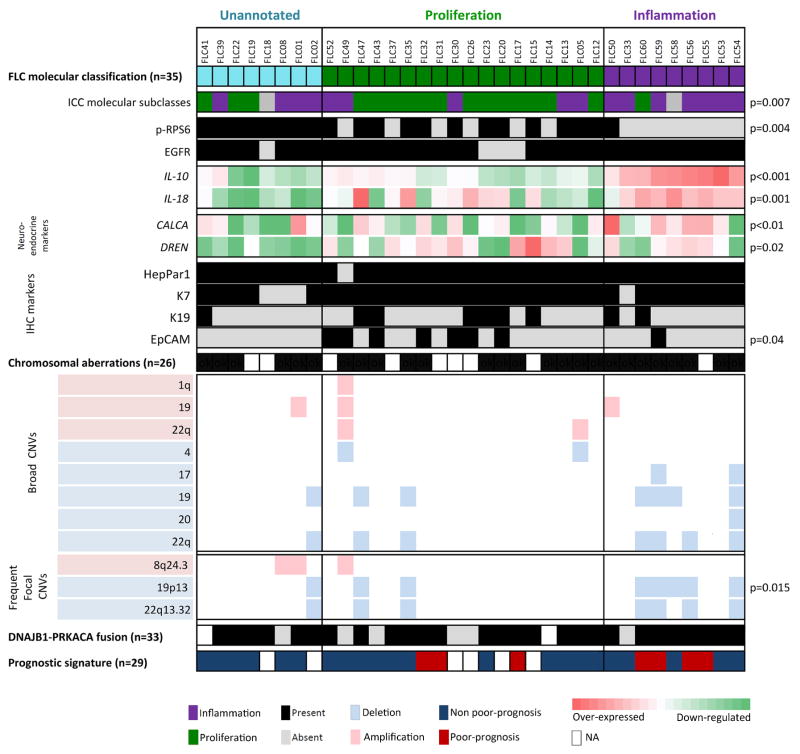
Whole-genome expression profiling was performed on 35 FLCs and 4 paired non-tumoral liver FFPE samples from the training cohort for which good quality RNA was obtained. Unsupervised-clustering of tumoral samples using the Non-negative matrix factorization (NMF) algorithm revealed 3 distinct and robust molecular classes (Figure 1A, Suppl. Figure 1A. We named the classes Proliferation (51% patients, 18/35 cases), Inflammation (26%, 9/35) and Unannotated (23%, 8/35) based on subsequent functional characterization using Gene set enrichment analysis (GSEA) and Nearest template prediction (NTP) -both modules from Gene Pattern15-, Ingenuity pathway analysis (IPA) and immunohistochemistry (IHC). The Proliferation class was enriched in liver cancer proliferation gene signatures such as HCC Boyault-G216, characterized by Akt activation (GSEA: FDR=0.04, Figure 1B), and ICC-Proliferation17 (NTP: p=0.007, Figure 1A). We confirmed Akt-mTOR signaling activation assessing phospho-RPS6 (p-RPS6) levels by IHC (Figure 1C), which was significantly activated in the Proliferation class versus the Inflammation (p<0.004, Figure 1A). The Inflammation class showed enrichment of liver cancer inflammation signatures such as ICC-Inflammation17 (GSEA: FDR=0.079, Figure 1D) and other pathways associated with pro-inflammatory cytokines (IPA: Acute Phase Response, p<0.01). In concordance, a number of interleukins were significantly up-regulated in this class (i.e. IL18, p=0.001 and IL10, p<0.001, Figure 1A). Moreover, the Inflammation class was slightly enriched by patients with liver fibrosis (4/8 vs. 5/23, p=0.18). The Unannotated class presented a distinct molecular profile since it was not enriched by any previously reported liver cancer gene signature, but by a cancer signature of Molecular Mechanisms of Cancer, which contains deregulated MAPKs, TGFB, PIK3, CTNNB1 and RAS genes (IPA: p<0.006). None of the molecular classes was significantly associated with any clinicopathological variable.
Figure 1. Molecular classes of FLC and their distinct genomic profile.
(A) NMF-based algorithm identified 3 robust classes: Proliferation (green), Inflammation (purple) and Unannotated (turquoise). Heat-map shows unsupervised clustering of 35 FLCs based on whole-genome expression showing the top differentially expressed genes for each class. Bottom part of the heat-map shows the overlap of the results from the NTP ICC-Proliferation signature, the IHC results of p-RPS6 and EGFR (panels C and E), and the expression values of IL-10 and 18. (B, D) GSEA plots demonstrated the enrichment in HCC Boyaoult-G2 (B) and ICC-Inflammation (D) gene signatures in FLC-Proliferation and FLC-Inflammation classes, respectively. (C, E) Immunohistochemical pattern of p-RPS6 (C) and EGFR (E) staining in FLC (upper panels) and non-tumoral tissues (lower panels).
The robustness of the NMF-based classification was confirmed by the K-nearest neighbor prediction method through Leave-one-out-cross validation (LOOCV) analysis, which showed 94% of correct prediction (Suppl. Table 2), and by the Affinity propagation clustering, in which the FLC tumors were equally distributed among the 3 classes as in the NMF classification (Suppl. Figure 1B). A gene signature that defined the 3 FLC molecular classes was generated by LOOCV, and included 710 over-expressed genes (Proliferation: 219, Inflammation: 263 and Unannotated: 228, Suppl. Table 3).
We also evaluated the expression levels of several neuroendocrine markers (i.e. NTS, CALCA, PCSK1 and DNER), since its deregulation has been described in FLC10. All the assessed neuroendocrine markers were significantly over-expressed in FLC compared to non-tumoral tissue (i.e. NTS and CALCA: p<0.01, PCSK1 and DNER: p=0.01, Suppl. Figure 2A). The expression pattern of the neuroendocrine genes among the FLC classes was heterogeneous; deregulation of CALCA was significantly enriched in the Inflammation class (p<0.01), and of DNER in the Proliferation and Inflammation classes (p=0.02, Suppl. Figure 2B).
Furthermore, we assessed EGFR protein levels as they were previously reported to be altered in FLC8. Most of the FLCs showed diffuse and strong membranous EGFR IHC staining in comparison to the non-tumoral tissues (88%, 37/42 vs. 18%, 3/17: p<0.0001, Suppl. Table 4, Figure 1A and 1E). Increased EGFR protein expression was observed across the different molecular classes.
In parallel we tested our gene-signature in the validation-French cohort, also profiled at the transcriptional level. Results showed the following distribution in the 3 FLC molecular classes (FDR=0.25): Proliferation - 41.2%, Inflammation - 41.2% and Unannotated – 17.64% (Suppl. Figure 1C).
In summary, molecular profiling of FLC reveals 3 distinct molecular classes, 2 of them associated with proliferative or inflammatory molecular traits of liver cancers -mainly of ICC-, and an overall enrichment of neuroendocrine markers and EGFR protein over-expression.
FLC shows a dual, cholangiocyte and hepatocyte, differentiation pattern
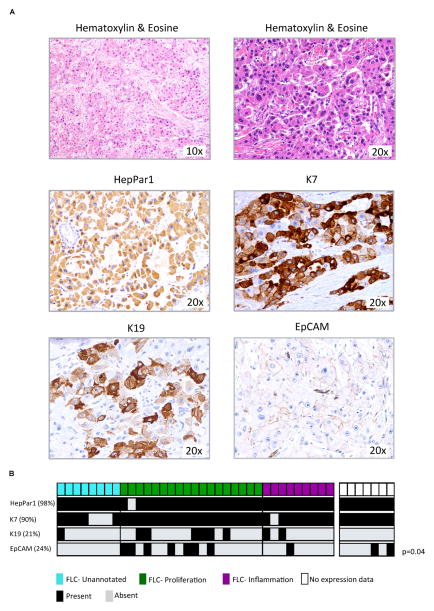
We characterized the cellular differentiation pattern of FLC by IHC and investigated the observed molecular association of FLC with ICC. As shown in Figure 2A, our FLCs had the characteristic pathological features of this tumor: large polygonal tumor cells with abundant, granular and eosinophilic cytoplasm with occasional pale bodies, large vesiculated nuclei with prominent nucleoli, and bands of fibrous stroma5. To study the cellular differentiation pattern of FLC, a panel of known markers associated with ICC (K7, K19), HCC (HepPar1) and the hepatic progenitor lineage (EpCAM and K19) were evaluated in 42 FLC and their paired non-tumoral liver when available. Most of the cohort was HepPar1 positive (98%, 41/42) and strongly stained for K7 (90%, 38/42, Figure 2A, Suppl. Table 3). Additionally, 38% (16/42) of tumors stained for progenitor-like markers, including 21% (9/42) of the FLCs with focal but intense staining for K19, and 24% (10/42) with EpCAM positive membrane staining. Integration of these results with our molecular classification (Figure 2B) showed a significant enrichment of EpCAM positive staining in the Proliferation class (39%, 7/18: p=0.04), and a paucity of progenitor cell markers in the Unannotated class (EpCAM: 0/8 vs. 8/27 and K19: 1/8 vs. 9/27).
Figure 2. Immunohistochemical characterization of FLC.
(A) FLC pathological characterization by hematoxylin and eosin staining (upper panels), and HepPar1, K7, K19 and EpCAM immunostaining (median and lower panels). (B) Distribution of the IHC results within the molecular classes.
These results support the previously suggested18 dual -cholangiocyte and hepatocyte-differentiation pattern of FLC, and validate the uniform positivity of HepPar1 and K7 markers in FLC18. In addition, we find an enrichment of progenitor cell features (EpCAM) in the Proliferation class.
FLC is characterized by a low number of chromosomal aberrations
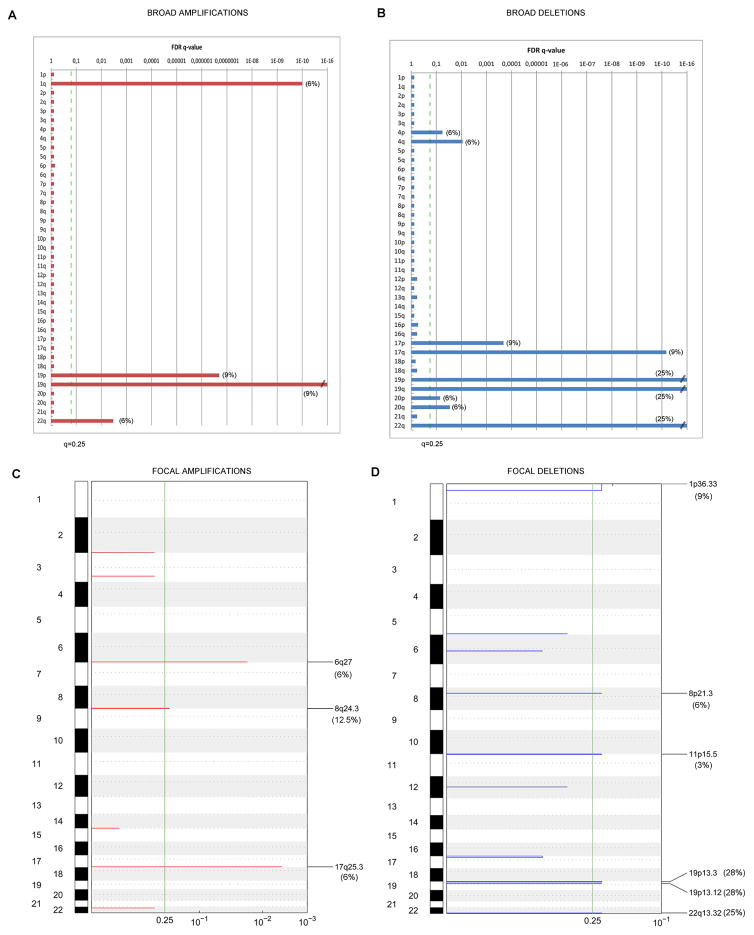
Overall, the FLC cohort showed a low number of CNVs (Suppl. Figure 3) compared with HCC and ICC. The key significant broad and focal aberrations are summarized in Figure 3. Main broad chromosomal gains were found at chromosome 19 (9%), and broad losses at chromosome 19 (25%) and 22q (25%), (see details in Figure 3A–B and Suppl. Table 5). These aberrations were equally distributed among the 3 molecular classes (Suppl. Table 6).
Figure 3. Significant broad and focal chromosomal alterations in FLC.
GISTIC algorithm identified significant CNVs in 32 FLC samples. Chromosomes are displayed in descending order along the vertical axis. GISTIC q-values (x-axis) for amplifications (left, red) and deletions (right, blue) corresponding to the FDR q-value obtained from GISTIC are plotted across the genome (y-axis). Vertical green line stands for the significance threshold of q < 0.25. A–B) represent graphically the significance of broad arm level CNVs, and C–D) the significance of focal CNVs.
Significant focal amplifications were identified at 3 different chromosomal regions affecting 22% of cases (Figure 3C, Suppl. Table 6). The most frequent focally amplified locus was at 8q24.3 in 4/32 patients (12.5%) spanning several genes as RPL8 and COMMD5, followed by 6q27 (6%) covering KIF25, FRMD and MLLT4, and 17q25.3 (6%) which includes a single gene, RBFOX3 (Table 2, Suppl. Table 7).
Table 2.
Significant Focal Copy Number Alterations in the 26 molecularly profiled FLC samples.
| Cytoband | Total number affected genes | Gene name | Wide peak boundaries | Alteration q value# | Proliferation Class (n = 12)
|
Inflammation Class (n = 8)
|
Unnanotated Class (n = 6)
|
All molecularly profiled samples (n = 26)
|
|---|---|---|---|---|---|---|---|---|
| Patients harboring alteration, n (%) | Patients harboring alteration, n (%) | Patients harboring alteration, n (%) | Total frequency n (%) | |||||
| Focal amplifications
| ||||||||
| 6q27 | 4 | KIF25 MLLT4 FRMD1 |
chr6:168352106-168552064 | 0.01897 | 2 (16) | 0 (0) | 0 (0) | 2 (10) |
| 8q24.3 | 12 | RPL8 COMMD5 ZNF7 ZNF250 |
chr8:145919127-146364022 | 0.22437 | 1 (8) | 0 (0) | 2 (33) | 3 (12) |
| 17q25.3 | 1 | RBFOX3 | chr17:77364582-77395624 | 0.00402 | 1 (8) | 1 (12.5) | 0 (0) | 2 (10) |
| Focal deletions
| ||||||||
| 1p36.33 | 56 | MIR429 MIR200A MIR200B |
chr1:1-1388267 | 0.22421 | 1 (8) | 0 (0) | 2 (33) | 3 (12) |
| 8p21.3 | 10 | EPB49 SFTPC |
chr8:21888571-22058121 | 0.22421 | 0 (0) | 1 (8) | 1 (17) | 2 (10) |
| 11p15.5 | 104 | CDKN1C | chr11:1-3009437 | 0.22421 | 1 (8) | 0 (0) | 0 (0) | 1 (5) |
| 19p13.3 | 150 | STK11 MIR637 |
chr19:14276712-14623757 | 0.22421 | 2 (16) | 5 (62.5)* | 1 (17) | 8 (31) |
| 19p13.12 | 7 | LPHN1 | chr19:1-4110491 | 0.22421 | 2 (16) | 5 (62.5)* | 1 (17) | 8 (31) |
| 22q13.32 | 2 | FAM19A5 | chr22:48929172-49004394 | 0.22421 | 2 (16) | 4 (50) | 1 (17) | 7 (27) |
Significant q value < 0.25
Significant enrichment p=0.0151
Six focal deletions were found in 31% of the cohort (Figure 3D, Suppl. Table 6) affecting protein coding genes and relevant miRNAs (Table 2, Suppl. Table 8). The most frequent focal deletions were at 19p13.12 and 19p13.3, simultaneously observed in 8 patients (28%), 5 of which belonged to the Inflammation class (p=0.015, Table 2). The focal deletion at 19p13.12 involved several G-protein coupled-receptors such as LPHN1, and is responsible for generating the predominant fusion protein recently reported DNAJB1-PRKACA14. Remarkably, 79% (58/73) of our FLCs [training cohort: 85% (33/39), validation-French cohort: 73% (16/22) and the validation-Brazilian cohort: 75% (9/12)] had this chimeric transcript originated from the fusion of DNAJB1 exon 1, with exons 2 to 10 from the PRKACA gene (Suppl. Figure 3B). None of the 5 paired non-tumoral samples showed this fusion event. The 19p13.3 focal deletion affected the tumor-suppressor gene (TSG) STK1, while deletion at 22q13.32 (25%) included a candidate TSG locus described in several tumors19, and at 11p15.5 (3%) covered the TSG CDKN1C (see details in Figure 3C–D, Table 2, Suppl. Table 8).
In parallel, we studied the CNVs in 9 FF FLCs from the validation-French cohort (Suppl. Table 9) where most of the frequent CNVs (i.e. 8q gains, and 18p, 18q, 21q and 22q losses) were also found in the FFPE results (Suppl. Table 5). Interestingly, the CNVs of 2 additional samples from separate parts of the same tumor (92T, 93T) showed the same background CNVs, while one had additional alterations, suggesting a sub-clone expansion event (Suppl. Table 9).
Herein, we demonstrate that FLC does not have a highly altered chromosomal profile even though a number of key loci are affected. The focal deletions at chromosome 19 are the most frequent chromosomal aberrations, being the one at 19p13.12 responsible for the occurrence of the highly prevalent DNAJB1-PRKACA fusion protein.
NGS analysis reveals a unique mutational portrait of FLC
To evaluate the mutational landscape of FLC, WES was run in 1 FLC-normal liver pair from FF tissue. In order to address the low purity of this tumor (around 50% of cells were tumoral), a pipeline specifically designed for low purity/heterogeneous tumors20 was applied. A total of 276 somatic single nucleotide variants (SNVs) were identified (Suppl. Figure 4A). One-third of them (90/276) were located in coding regions, of which 68 (76%) were non-synonymous (Suppl. Figure 4A). Almost all of them (98.5%, 67/68) were missense and just one nonsense. Thirty-four percent (23/67) of the missense non-synonymous mutations had a damaging role (Suppl. Figure 4A & Suppl. Table 12). Among the damagingly mutated genes identified, 4 out of 5 mutations (i.e. BRCA2-Y2789C, CSMD2-G1055E, ARMCX1-L246F and COL6A6-L929I) were technically validated by TES (Suppl. Figure 4B), and BRCA2-Y2789C also by Sanger (Suppl. Figure 4C). Due to the biological relevance of BRCA2 as TSG, we sequenced the entire gene by TES in 47 patients (training and validation-French cohorts) and results showed another mutation in BRCA2, P2612S. Then, we can confirm that BRCA2 is mutated in 4.2% of FLCs. In parallel, none of the most prevalent mutations in HCC (i.e. TP53, CTNNB1, ARID1A, TTN, NFE2L2 and AXIN1)21 or in other liver cancers (i.e. EGFR, BRAF, KRAS, NRAS, IDH1 and IDH2)21 were found mutated in FLC.
Taken together, FLC presents a unique liver cancer mutational portrait which does not include any of the most prevalent liver cancer mutated genes21. We identified BRCA2 damaging mutations in 4.2% of the FLC cases (2/47).
8-gene expression signature predicts survival in FLC
Overall, clinicopathological features of our cohorts resemble those previously reported2,5,6. Complete clinical follow-up data for the training cohort was available for 38 patients (Suppl. Table 1), which were mostly young (median age of 25.5 years-old, range 11–65) and Caucasian (61%, 23/38). The cohort showed a balanced gender distribution (58% females, 22/38) and non-significant underlying liver disease (cirrhosis in 3%, 1/38), with 95% of the patients non-viral infected (1/38 HCV and 1/38 HBV). All patients were surgically treated [resected (89%, 34/38) or transplanted (11%, 4/38)]. Median tumor size was 11cm (25–75% quartile: 7–13cm), 18% (7/38) of the tumors were multinodular, 16% (6/38) had satellite lesions, and 13% (5/38) had macrovascular invasion. Median survival was 58 months, with a median follow-up of 48 months. Twenty-one patients recurred (54%) during follow-up, with a median time to recurrence of 19 months. Complete clinical data was available for 22 FLCs from the validation-French cohort, which were mostly females (72% females, 16/22), with a median tumor size of 10cm (25–75% quartile: 2.5–17cm), and a median age of 26 years (range 17–65). One patient had alcohol-related liver damage (5%, 1/22). Median survival for this cohort was of 68 months, with a median follow-up of 58 months. The validation-Brazilian cohort included resected patients, mostly females (67% females, 8/22), with a median tumor size of 13cm (25–75% quartile: 10–17cm), 23 years-old (range 18–33) of median age, and none of them presented underlying liver disease (Suppl. Table 1). The median survival for these patients was 48 months, and 39 months of median follow-up.
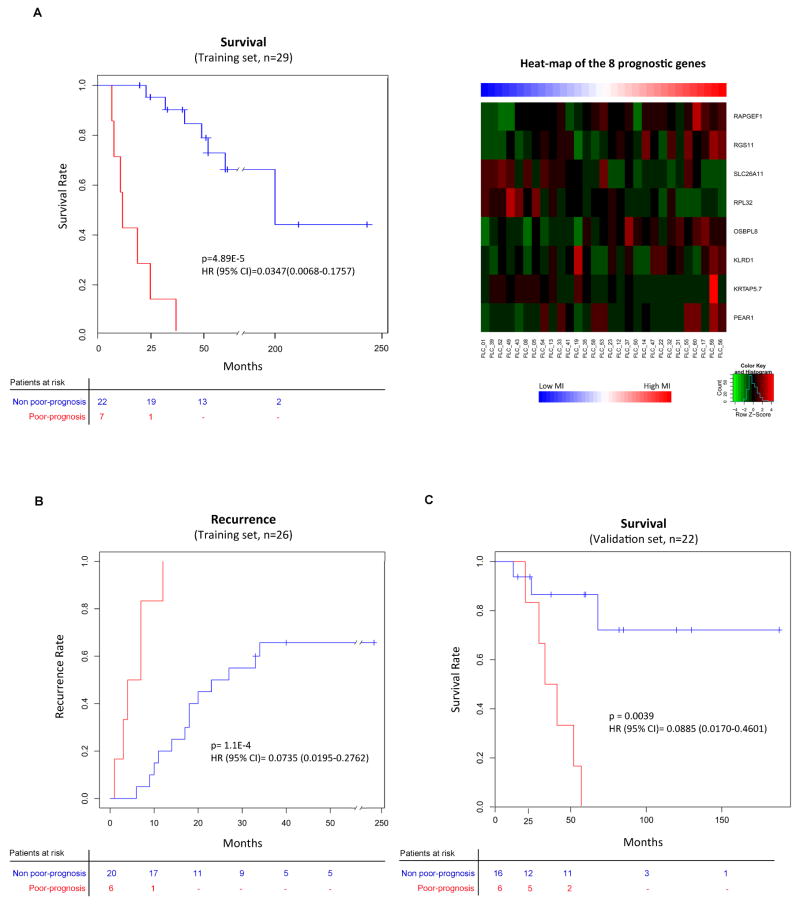
Since the molecular classes of FLC did not show a clear association with survival, we generated a gene expression-based prognostic signature that discriminated patients according to their survival using the Random survival forest analysis in the training set (n=29). This signature was composed of 8 genes: PEAR1, KRTAP, KLRD1, OSBPL8, RPL32, SLC26A11, RGS11 and RAPGEF1 (Figure 4A, right panel). The prognostic signature allowed us to develop a predictive quantitative risk score for mortality based on the weighted hazard of death for each resected individual (i.e. mortality index (MI), range 6.7–42, Suppl. Table 11). Patients with higher MI (upper quartile, MI≥21) had a worse outcome (Poor-prognosis) with a median survival of 12 months, compared to 197 months for those patients with lower MI (Non poor-prognosis, MI<21). Thus, based on the upper quartile division of the MI values, FLC patients can be accurately stratified for their survival times following surgical resection as Poor-prognosis and Non poor-prognosis (Figure 4A, left panel). This is evident in the Kaplan-Meier analysis (HR: 0.035, 95% CI: 0.007–0.176, p<0.001). Interestingly, the same prognostic signature was able to predict tumor recurrence in the same cohort (training set: n=26, HR: 0.074, 95% CI: 0.02–0.28, p<0.001, Figure 4B). In this case the upper MI quartile threshold was set at ≥25, since the number of patients was reduced due to the lack of recurrence information in 3 patients. When integrating these data into our molecular classification, we identified that patients in the Unannotated class showed a trend towards a more indolent outcome since none of the patients with the Poor-prognosis signature clustered in this class (0/6 vs. 7/23, p=0.32).
Figure 4. Gene expression-based prognostic signature and its validation.
(A, B & C) Kaplan-Meier plots estimating overall survival in training (n=29, A, left panel) and validation-French (n=22, C) cohort, and overall recurrence in those patients from the training set for whom data on recurrence was available (n=26, B). Heat-map shows expression values of the 8 genes that constitute the prognostic signature (A, right panel). Patients in the Poor-prognosis class (red) showed shorter survival and earlier recurrence.
Next, we validated the 8-gene prognostic signature in an independent cohort (validation-French set: n=22, Figure 4C). We found that patients with the Poor-prognosis signature (upper quartile, MI≥8), had significantly worse survival outcome (33 months median survival vs. not reached; HR: 0.089, 95% CI: 0.017–0.46, p=0.0039) than those with the Non poor-prognosis signature (MI<8).
Overall, we generate and validate a signature that can stratify resected FLC patients based on their prognosis by the interaction in the expression of 8 genes.
DISCUSSION
There is little known about the molecular pathogenesis of FLC. Previous publications provided some insights into the molecular characteristics of this rare tumor, but are limited in terms of number of patients and technology used8–13. Herein, by using NGS, SNP-array and whole-transcriptome analysis we provide a comprehensive genomic study of a large FLC cohort. This demonstrates a unique portrait of genomic aberrations distinct from the most prevalent primary liver neoplasms, HCC and ICC. In addition, we identify key pathways and candidate genes as targets for molecular therapies, and describe a gene-based prognostic signature.
In this study we identified 3 different molecular classes of FLC named Proliferation, Inflammation and Unannotated. Tumors in the most common class, Proliferation (51% of patients), were enriched in liver cancer proliferative gene signatures (ICC and HCC), markers of progenitor cell origin (EpCAM) and mTOR signaling activation (p-RPS6) (Figure 5). The Inflammation class was characterized by up-regulation of liver inflammatory gene signatures and interleukins (i.e. IL-8 and IL-10), and was slightly enriched in patients with fibrosis, which could explain its inflammatory profile. In addition, DNA deletions in chromosome 19 were particularly enriched in this class. Finally, the Unannotated class was found to be up-regulated by non-liver-related cancer signatures, it lacked progenitor traits and showed a trend for better outcome (Figure 5). These molecular classes were reproduced in an independent cohort, which showed a more prevalent inflammatory profile than in the training cohort (41% vs. 26% in training) at the expense of patients with proliferative features (41% vs. 51%). In order to compare the expression pattern of our samples to a recently reported FLC classification10, we confirmed over-expression of neuroendocrine markers, which were similarly distributed among all the 3 classes
Figure 5. Integrative genomic analysis.
FLC gene expression classification and its overlap with the previously published consensus ICC classification17 and the FLC results of the copy number alterations, immunohistochemical stainings and gene expression of interleukines and neuroendocrine markers, together with the presence of the fusion transcript and the enrichment results of the prognostic signature. The integrative analysis revealed an indolent 20 profile together with a paucity of progenitor markers for the Unannotated class, a highly enrichment of progenitor cells traits for the Proliferation class, and an enrichment of focal deletions and interleukines at the Inflammation class. Moreover, these analysis demonstrate the highly prevalence of the DNAJB1-PRKACA fusion transcript.
Once we had defined the molecular classes, we aimed to explore unanswered questions regarding the pathogenesis of this neoplasm: a) cell of origin, b) potential drivers amenable for targeted therapies and c) prognostic stratification. Regarding the immunohistochemical pattern of FLC, our results demonstrated a common and universal dual pattern of morphological characteristics and markers of hepatocytes and HCC (HepPar1), which are not usually found in ICC22, along with specific markers of bile duct cells usually negative in HCC (K7). All these information points towards a dual bile duct and hepatocyte differentiation pattern of the FLC, previously proposed by a tissue microarray study of 26 FLCs18. In fact, our data suggest that FLC could be derived from a hepatic precursor cell with the ability to differentiate into hepatocytes (HepPar1) and bile duct cells (K7) that maintain both markers once the cell become mature. In parallel, around 40% of FLCs show progenitor cell markers (K19 or EpCAM) in contrast to the 15% in conventional HCC cases18. Furthermore, the uniform and strong positivity of K7 observed in our cohort, supports the use of this marker as an additional tool during the diagnostic workup of FLC, as previously suggested18.
In order to identify key drivers of tumor progression we performed an integrative analysis by using whole-transcriptome, SNP-array and NGS data. In contrast to other cancers17,23, FLC shows an overall flat profile of chromosomal alterations with some focal and broad amplifications and deletions pointing towards candidate regions harboring oncogenes and TSGs. Three focal amplifications were identified at 6q27, 8q24.3 and 17q25.3 loci, which involved 22% of the cases. The most critical genes affected by these amplifications are RPL8, reportedly up-regulated in different cancers (e.g. HCC, melanoma, glioma and breast carcinoma24,25) and MLLT4, described to generate a fusion oncogene together with AF6 inducing leukemic transformation26. In terms of focal deletions, these ones affect 31% of cases, being the most common at 19p13.2 locus (containing STK11, a TSG serine/threonine-protein kinase that regulates mTOR signaling), at 19p13.12 (covering LPHN, with a protective role in non-small cell lung cancer27) and at 22q13.32, described as a candidate TSG locus in other tumors19. Moreover, it was striking to observe that the 19p13.12 focal deletions has been described to be the cause of the DNAJB1-PRKACA fusion protein14, which was identified in 79% (58/73) of our FLCs (Suppl. Figure 3B). Despite this prevalence is very high for a potentially druggable target, it was inferior to the 100% previously reported14, probably reflecting both a larger size of our cohort and eventually a less aggressive phenotype. In both studies none of the adjacent tissues presented the fusion event.
Next, we looked for structural somatic mutations in FLC using NGS. Despite previous attempts to find frequently mutated cancer genes in FLC (i.e. KRAS and EGFR11), none had been reported. By WES, FLC showed 23 damaging mutations, which included the well-known TSG BRCA2 implicated in DNA repair. This mutation was confirmed by Sanger sequencing and TES, and this last technique revealed another BRCA2 mutation. Then, BRCA2 mutations are present in FLC but showed a low prevalence of 4.2%. (2/47). In addition, we confirmed that none of the most prevalent mutations in HCC or ICC cancer types (i.e. TP53, CTNNB1, ARID1A, TTN, NFE2L2, AXIN1, EGFR, BRAF, KRAS, NRAS, IDH1 and IDH2)21 were altered in FLC. Hence, from the mutational standpoint, FLC presents a unique mutational profile different from other primary liver cancers, with potential candidates as therapeutic targets.
Thus, after exploring NGS and SNP-array, along with signaling cascades and the fusion protein, we identified a number of potential candidates for targeted therapy in FLC. First, we confirmed the high prevalence of the recently described predominant fusion protein DNAJB1-PRKACA in our cohort, which can be a therapeutic target, since it has been proved to retain the kinase activity of PRKACA, the catalytic subunit of Protein Kinase A14. Secondly, structural alterations highlight BRCA2 mutations (4.2%), which are potentially druggable by PARP inhibitors, and MLLT4 (6%), as novel targets. Finally, mTOR signaling, activated in more than 70% of the FLC cases, and aberrant EGFR protein expression in 88% of the tumors, are both candidate targetable pathways that have been reported to be effectively blockade (mTOR: everolimus and temsirolimus28, EGF: gefitinib and cetuximab29) in other epithelial neoplasms (e.g. colon, lung)29. However, the effectiveness of erlotinib/gefitinib in lung and other cancers is usually associated with the occurrence of mutations or amplifications, which is not the case in FLC. Exploring all these potential targets in the setting of early clinical trials or marker-driven proof-of-concept trials is recommended.
None of the molecular classes was linked to clinical outcomes. Certainly, the small sample size precludes general conclusion, and only the Unannotated class, which lacks progenitor-cell features, showed a non-significant trend to better prognosis. In an effort to elucidate prognostic markers beyond the biological classes, as has been the case in HCC30,31, we generated a gene signature able to stratify FLC patients according to their prognosis. The prognostic signature was successfully validated in an independent cohort of 22 FF FLCs. This signature included genes involved in PI3K/PTEN signaling such as PEAR132, or KLRD1, natural killer cell inhibitory receptor33, and RAPGEF1, regulator of the GTPases/Ras protein family34. Overall, the prognostic signature composed of relevant cancer genes will allow prognostic stratification of FLC patients, and could also improve the design of clinical trials. At the same time, the molecular classification will help to understand the distinct response of FLC patients to certain molecular targeted therapies, and it could be an alternative tool for stratification of patients prior randomization in clinical trials based on their molecular profile.
Our genomic study characterizes FLC as a singular molecular entity, with certainly distinct traits compared with other liver tumors. Structural alterations are unique compared with those of classical HCC or ICC. FLC presents high prevalence of the DNAJB1-PRKACA fusion protein that contains a kinase, chromosomal focal amplifications of MLL4, and few BRCA2 mutations. Conversely, prevalent mutations or chromosomal aberrations in HCC or ICC are lacking in FLC. Our results indicate that FLC has a potential origin in a common precursor cell, since it retains molecular markers of hepatocyte and cholangiocyte lineages, and progenitor cell markers (EpCAM and K19:38%). Finally, FLC gene signatures herein reported are more frequently enriched in ICC17 (80%) than in HCC35 (28%, Suppl. Table 12). Nonetheless, gene signatures of poor-prognosis and invasiveness generated in several cancer types (e.g. breast, melanoma)16,36–39 are not enriched in FLC samples, as opposed to other liver cancers (Suppl. Table 12).
In conclusion, we present a comprehensive genomic characterization of FLC at multiple levels. We identified a robust 3 class molecular classification highlighting key biological traits of FLC with potential implications for future biomarker-based clinical trials, and provided a clear picture of the chromosomal and mutational profile revealing potential molecular targets. Moreover, the prognostic gene-signature we generated might aid in the stratification of patients for trials beyond the clinicopathological variables, and allow for inclusion in surveillance programs for the FLC patients with worst prognosis. On the other hand, our results further support the feasibility of high-throughput profiling in FFPE tissues that will facilitate the knowledge of numerous malignancies and different diseases. Hence, the FLC, a rare primary hepatic malignancy which was first described in 1956 by Edmondson40 as a variant of HCC, is now considered to be a distinct form of primary liver cancer with respect to its epidemiology, etiology and prognosis, and herein we clearly demonstrate its distinctive genomic profile.
Supplementary Material
Acknowledgments
Grant support: JML is supported by grants from the U.S. National Institute of Diabetes and Digestive and Kidney Diseases (J.M.L: 1R01DK076986), the Samuel Waxman Cancer Research Foundation, the Spanish National Health Institute (J.M.L: SAF-2013-41027), the Asociación Española Contra el Cáncer (AECC) and the European Commission Framework Programme 7 (Heptromic, proposal number 259744). HC is funded by a fellowship from Instituto de Salud Carlos III (ISCIII/FIS FI10/00143), YH by the U.S. National Institute of Diabetes and Digestive and Kidney Diseases (R01 DK099558), MVC by the Spanish Ministry of Education (FPU12/05138), SCW by the Innate/Adaptive Immune Interactions in Gut Inflammation (NIH/NIDDK P01/DK072201), ME by Cellex Foundation, Botin Foundation, Health and Science Departments of the Catalan Government (Generalitat de Catalunya), LRR by a grant from the U.S. National Cancer Institute and support from the Mayo Clinic Center for Cell Signaling in Gastroenterology (L.R.R.: R01CA165076 and NIDDK P30DK084567), and JZR by a grant from INCa (PAIR-CHC, NoFLIC project). VM is partially supported by the AIRC (Italian Association for Cancer Research) and the INT-Milano Institutional Funds (5×1000 on Hepato-Oncology) and FC by a grant from the Alves de Queiroz Family Fund for Research from Brazil.
The authors would like to thank Monica Higuera, Roser Pinyol, Sara Torrecilla, Yumi Kasai, Judit Peix, Juan Joseé Lozano, Nasra H. Giama and Poh Seng Tan for their generous help and technical support. This work was supported in part through the computational resources and staff expertise provided by the Department of Scientific Computing at the Icahn School of Medicine at Mount Sinai. We also thank the clinicians and pathologists that collected the samples and clinical data in Brazil, Venancio Alves, and in France: Jeanne Tran Van Nhieu (AP-HP, Henri Mondor Hospital), Daniel Cherqui (AP-HP, Henri Mondor Hospital), Jacques Belghiti (AP-HP, Beaujon Hospital), Geneviève Monges (IPC, Marseille), Dominique Franco (AP-HP, Antoine Béclère hospital), Charles Balabaud (Bordeaux hospital) and Jean Yves Scoazec (Lyon Hospital).
Abbreviations
- CI
confidence interval
- CNV
copy number variations
- FDR
false discovery rate
- FF
fresh-frozen
- FFPE
formalin-fixed paraffin-embedded
- FLC
fibrolamellar hepatocellular carcinoma
- HCC
hepatocellular carcinoma
- HR
hazard ratio
- ICC
intrahepatic cholangiocarcinoma
- LOOCV
leave-one-out cross validation
- NGS
next-generation sequencing
- NMF
non-negative matrix factorization
- SNP
single-nucleotide polymorphism
- TES
targeted-exome sequencing
- TSG
tumor-suppressor gene
- WES
whole-exome sequencing
Footnotes
Conflict of interests: Authors declare no conflicts of interest related to this manuscript.
Data repository: Submitted microarray data GEO accession numbers: GSE57727 and GSE59443. Sequence data have been deposited in the European Nucleotide Archive (ENA), under the accession number PRJEB7902.
Author contribution:
Conception and design: HC, AL, JML.
Financial support: JML.
Provision of study materials or patients: AQ, DMN, BM, FC, LRR, VM, MS, NH, JZ, JML.
Data analysis and interpretation: HC, CA, SS, ZZ, KH, YH, MVC, AV, ST, SCW, LR, JML.
Manuscript writing: HC, CA, JML.
Final approval of manuscript: HC, CA, SS, ZZ, KH, LC, YH, MVC, AV, ST, SCW, LR, SI, AL, AQ, DMN, BM, ME, FC, LRR, SW, VM, MS, NH, JZ, JML.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lozano R, Naghavi M, Foreman K, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095–128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.El-Serag HB, Davila JA. Is fibrolamellar carcinoma different from hepatocellular carcinoma? A US population-based study. Hepatology. 2004;39:798–803. doi: 10.1002/hep.20096. [DOI] [PubMed] [Google Scholar]
- 3.EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2012;56:908–43. doi: 10.1016/j.jhep.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 4.Shaib Y, El-Serag HB. The epidemiology of cholangiocarcinoma. Semin Liver Dis. 2004;24:115–25. doi: 10.1055/s-2004-828889. [DOI] [PubMed] [Google Scholar]
- 5.Torbenson M. Review of the clinicopathologic features of fibrolamellar carcinoma. Adv Anat Pathol. 2007;14:217–23. doi: 10.1097/PAP.0b013e3180504913. [DOI] [PubMed] [Google Scholar]
- 6.Kakar S, Burgart LJ, Batts KP, et al. Clinicopathologic features and survival in fibrolamellar carcinoma: comparison with conventional hepatocellular carcinoma with and without cirrhosis. Mod Pathol. 2005;18:1417–23. doi: 10.1038/modpathol.3800449. [DOI] [PubMed] [Google Scholar]
- 7.Mavros MN, Mayo SC, Hyder O, et al. A systematic review: treatment and prognosis of patients with fibrolamellar hepatocellular carcinoma. J Am Coll Surg. 2012;215:820–30. doi: 10.1016/j.jamcollsurg.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 8.Buckley AF, Burgart LJ, Kakar S. Epidermal growth factor receptor expression and gene copy number in fibrolamellar hepatocellular carcinoma. Hum Pathol. 2006;37:410–4. doi: 10.1016/j.humpath.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 9.Kannangai R, Vivekanandan P, Martinez-Murillo F, et al. Fibrolamellar carcinomas show overexpression of genes in the RAS, MAPK, PIK3, and xenobiotic degradation pathways. Hum Pathol. 2007;38:639–44. doi: 10.1016/j.humpath.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 10.Malouf GG, Job S, Paradis V, et al. Transcriptional profiling of pure fibrolamellar hepatocellular carcinoma reveals an endocrine signature. Hepatology. 2014 doi: 10.1002/hep.27018. [DOI] [PubMed] [Google Scholar]
- 11.Patonai A, Erdelyi-Belle B, Korompay A, et al. Molecular characteristics of fibrolamellar hepatocellular carcinoma. Pathol Oncol Res. 2012;19:63–70. doi: 10.1007/s12253-012-9558-0. [DOI] [PubMed] [Google Scholar]
- 12.Sahin F, Kannangai R, Adegbola O, et al. mTOR and P70 S6 kinase expression in primary liver neoplasms. Clin Cancer Res. 2004;10:8421–5. doi: 10.1158/1078-0432.CCR-04-0941. [DOI] [PubMed] [Google Scholar]
- 13.Ward SC, Waxman S. Fibrolamellar carcinoma: a review with focus on genetics and comparison to other malignant primary liver tumors. Semin Liver Dis. 2011;31:61–70. doi: 10.1055/s-0031-1272835. [DOI] [PubMed] [Google Scholar]
- 14.Honeyman JN, Simon EP, Robine N, et al. Detection of a recurrent DNAJB1-PRKACA chimeric transcript in fibrolamellar hepatocellular carcinoma. Science. 2014;343:1010–4. doi: 10.1126/science.1249484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reich M, Liefeld T, Gould J, et al. GenePattern 2. 0. Nat Genet. 2006;38:500–1. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 16.Boyault S, Rickman DS, de Reynies A, et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45:42–52. doi: 10.1002/hep.21467. [DOI] [PubMed] [Google Scholar]
- 17.Sia D, Hoshida Y, Villanueva A, et al. Integrative molecular analysis of intrahepatic cholangiocarcinoma reveals 2 classes that have different outcomes. Gastroenterology. 2013;144:829–40. doi: 10.1053/j.gastro.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ward SC, Huang J, Tickoo SK, et al. Fibrolamellar carcinoma of the liver exhibits immunohistochemical evidence of both hepatocyte and bile duct differentiation. Mod Pathol. 2010;23:1180–90. doi: 10.1038/modpathol.2010.105. [DOI] [PubMed] [Google Scholar]
- 19.Diaz de Stahl T, Hartmann C, de Bustos C, et al. Chromosome 22 tiling-path array-CGH analysis identifies germ-line- and tumor-specific aberrations in patients with glioblastoma multiforme. Genes Chromosomes Cancer. 2005;44:161–9. doi: 10.1002/gcc.20226. [DOI] [PubMed] [Google Scholar]
- 20.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–9. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Villanueva A, Llovet JM. Liver cancer in 2013: Mutational landscape of HCC--the end of the beginning. Nat Rev Clin Oncol. 2014;11:73–4. doi: 10.1038/nrclinonc.2013.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wennerberg AE, Nalesnik MA, Coleman WB. Hepatocyte paraffin 1: a monoclonal antibody that reacts with hepatocytes and can be used for differential diagnosis of hepatic tumors. Am J Pathol. 1993;143:1050–4. [PMC free article] [PubMed] [Google Scholar]
- 23.Chiang DY, Villanueva A, Hoshida Y, et al. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer Res. 2008;68:6779–88. doi: 10.1158/0008-5472.CAN-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swoboda RK, Somasundaram R, Caputo L, et al. Shared MHC class II-dependent melanoma ribosomal protein L8 identified by phage display. Cancer Res. 2007;67:3555–9. doi: 10.1158/0008-5472.CAN-06-2763. [DOI] [PubMed] [Google Scholar]
- 25.Liu Y, Zhu X, Zhu J, et al. Identification of differential expression of genes in hepatocellular carcinoma by suppression subtractive hybridization combined cDNA microarray. Oncol Rep. 2007;18:943–51. [PubMed] [Google Scholar]
- 26.Deshpande AJ, Chen L, Fazio M, et al. Leukemic transformation by the MLL-AF6 fusion oncogene requires the H3K79 methyltransferase Dot1l. Blood. 2013;121:2533–41. doi: 10.1182/blood-2012-11-465120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsu YC, Yuan S, Chen HY, et al. A four-gene signature from NCI-60 cell line for survival prediction in non-small cell lung cancer. Clin Cancer Res. 2009;15:7309–15. doi: 10.1158/1078-0432.CCR-09-1572. [DOI] [PubMed] [Google Scholar]
- 28.Bhat M, Sonenberg N, Gores GJ. The mTOR pathway in hepatic malignancies. Hepatology. 2013;58:810–8. doi: 10.1002/hep.26323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wheeler DL, Dunn EF, Harari PM. Understanding resistance to EGFR inhibitors-impact on future treatment strategies. Nat Rev Clin Oncol. 2010;7:493–507. doi: 10.1038/nrclinonc.2010.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoshida Y, Villanueva A, Sangiovanni A, et al. Prognostic gene expression signature for patients with hepatitis C-related early-stage cirrhosis. Gastroenterology. 2013;144:1024–30. doi: 10.1053/j.gastro.2013.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nault JC, De Reynies A, Villanueva A, et al. A hepatocellular carcinoma 5-gene score associated with survival of patients after liver resection. Gastroenterology. 2013;145:176–87. doi: 10.1053/j.gastro.2013.03.051. [DOI] [PubMed] [Google Scholar]
- 32.Kauskot A, Vandenbriele C, Louwette S, et al. PEAR1 attenuates megakaryopoiesis via control of the PI3K/PTEN pathway. Blood. 2013;121:5208–17. doi: 10.1182/blood-2012-10-462887. [DOI] [PubMed] [Google Scholar]
- 33.Moser JM, Gibbs J, Jensen PE, et al. CD94-NKG2A receptors regulate antiviral CD8(+) T cell responses. Nat Immunol. 2002;3:189–95. doi: 10.1038/ni757. [DOI] [PubMed] [Google Scholar]
- 34.Schonherr C, Yang HL, Vigny M, et al. Anaplastic lymphoma kinase activates the small GTPase Rap1 via the Rap1-specific GEF C3G in both neuroblastoma and PC12 cells. Oncogene. 2010;29:2817–30. doi: 10.1038/onc.2010.27. [DOI] [PubMed] [Google Scholar]
- 35.Villanueva APA, Sayols S, Battiston, Hoshida Y, Méndez-González J, Letouzé E, Hernandez-Gea V, Cornella H, Pinyol R, Solé M, Fuster J, Zucman-Rossi J, Mazzaferro V, Esteller M, Llovet JM. DNA Methylation-based prognosis and epidrivers in hepatocellular carcinoma. Hepatology. 2015 doi: 10.1002/hep.27732. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coulouarn C, Factor VM, Thorgeirsson SS. Transforming growth factor-beta gene expression signature in mouse hepatocytes predicts clinical outcome in human cancer. Hepatology. 2008;47:2059–67. doi: 10.1002/hep.22283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winnepenninckx V, Lazar V, Michiels S, et al. Gene expression profiling of primary cutaneous melanoma and clinical outcome. J Natl Cancer Inst. 2006;98:472–82. doi: 10.1093/jnci/djj103. [DOI] [PubMed] [Google Scholar]
- 38.Montero-Conde C, Martin-Campos JM, Lerma E, et al. Molecular profiling related to poor prognosis in thyroid carcinoma. Combining gene expression data and biological information. Oncogene. 2008;27:1554–61. doi: 10.1038/sj.onc.1210792. [DOI] [PubMed] [Google Scholar]
- 39.Sarrio D, Rodriguez-Pinilla SM, Hardisson D, et al. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res. 2008;68:989–97. doi: 10.1158/0008-5472.CAN-07-2017. [DOI] [PubMed] [Google Scholar]
- 40.Edmondson HA. Differential diagnosis of tumors and tumor-like lesions of liver in infancy and childhood. AMA J Dis Child. 1956;91:168–86. doi: 10.1001/archpedi.1956.02060020170015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.