Summary
Mitochondrial respiration is important for cell proliferation, however the specific metabolic requirements fulfilled by respiration to support proliferation have not been defined. Here we show that a major role of respiration in proliferating cells is to provide electron acceptors for aspartate synthesis. This finding is consistent with the observation that cells lacking a functional respiratory chain are auxotrophic for pyruvate, which serves as an exogenous electron acceptor. Further, the pyruvate requirement can be fulfilled with an alternative electron acceptor, alpha-ketobutyrate, which provides cells neither carbon nor ATP. Alpha-ketobutyrate restores proliferation when respiration is inhibited, suggesting that an alternative electron acceptor can substitute for respiration to support proliferation. We find that electron acceptors are limiting for producing aspartate, and supplying aspartate enables proliferation of respiration deficient cells in the absence of exogenous electron acceptors. Together, these data argue a major function of respiration in proliferating cells is to support aspartate synthesis.
Introduction
In mammalian cells, mitochondrial respiration allows coupling of nutrient oxidation to ATP production. Respiration involves a series of redox reactions, where electrons from a reduced substrate are ultimately transferred to molecular oxygen as the final electron acceptor. This results in oxidation of consumed nutrients and reduction of molecular oxygen to water. The free energy released from this series of oxidation-reduction reactions is coupled to production of an electrochemical gradient that can be used to drive ATP synthesis, membrane transport, and thermogenesis (Harms and Seale, 2013; Mitchell, 1961; Schleyer et al., 1982).
While supporting bioenergetics is a critical function of respiration in mammalian cells, many proliferating cells display increased fermentation, which alone can be sufficient to supply ATP (Gottlieb and Tomlinson, 2005). In contrast to most normal tissues, cancer cells consume increased amounts of glucose and metabolize much of this glucose to lactate even in the presence of ample oxygen (Koppenol et al., 2011; Warburg et al., 1924). This phenotype, termed aerobic glycolysis or the Warburg effect, was initially hypothesized to result from diminished mitochondrial function (Warburg, 1956). However, despite utilizing aerobic glycolysis, most cancer cells also consume oxygen (Weinhouse, 1956; Zu and Guppy, 2004). Notably, in cancer cell lines the primary substrate for oxidation is often not glucose but rather glutamine, one of the most heavily consumed nutrients by cells in culture (Fan et al., 2013; Kovacevic, 1971; Zielke et al., 1984). Thus, aerobic glycolysis likely does not replace mitochondrial respiration, but rather, in proliferating cells these processes occur in parallel.
Most cells that engage in aerobic glycolysis are not only capable of respiration but also require respiration for proliferation. Exposure of cancer cells in culture to respiration inhibitors blocks proliferation (Harris, 1980; Howell and Sager, 1979; Kroll et al., 1983; Loffer and Schneider, 1982). In vivo, maintenance of mitochondrial DNA (mtDNA) is required for autochthonous tumor formation (Weinberg et al., 2010), and inhibition of respiration suppresses tumor growth in xenografts (Wheaton et al., 2014; Zhang et al., 2014). These findings argue that mitochondrial respiration is essential for rapid proliferation, but whether respiration is advantageous for proliferation beyond producing ATP is less clear.
Despite the importance of respiration in mammalian cell proliferation, under specific culture conditions proliferation is possible even in the absence of respiration. Serial passage in low dose ethidium bromide produces cells devoid of mtDNA (ρ0 cells) (King and Attardi, 1989, 1996). ρ0 cells lack a functional mitochondrial electron transport chain (ETC), and these respiration-incompetent cells fail to proliferate unless supra-physiological levels of uridine and pyruvate are present in the culture media (King and Attardi, 1989). Uridine auxotrophy is explained by the fact that de novo pyrimidine biosynthesis enzyme dihydroorotate dehydrogenase (DHODH) transfers electrons directly to the ETC to convert dihydroorotate to orotate. Thus, loss of electron transport to O2 prevents this reaction, and exogenous uridine is needed to produce pyrimidines (Gregoire et al., 1984). The requirement for pyruvate, however, was initially unexpected because cells deficient in mtDNA are highly glycolytic and capable of generating large amounts of pyruvate (King and Attardi, 1989).
The fact that adding specific nutrients can substitute for respiration suggests respiration fulfills specific metabolic requirements for proliferating cells. While ATP synthesis via oxidative phosphorylation is often assumed to be the critical output of respiration, neither exogenous uridine nor pyruvate can be oxidized to supply ATP in the absence of respiration. However, other than dihyroorotate to orotate conversion, the metabolic function(s) that become limiting for proliferation in the absence of respiration are unknown.
Here we show that loss of mitochondrial respiration causes proliferating cells to become functionally limited for electron acceptors. This lack of electron acceptors impairs de novo aspartate synthesis and inhibits proliferation. Strikingly, this proliferation block can be overcome by supplementing cells with exogenous electron acceptors or by high levels of aspartate. Taken together our data argue that the most essential metabolic function for proliferation provided by mitochondrial respiration is to provide access to electron acceptors to support aspartate biosynthesis.
Results
Alpha-ketobutyrate can substitute for pyruvate to support proliferation in respiration-incompetent cells
Cells lacking a functional mitochondrial ETC require pyruvate for proliferation (King and Attardi, 1989). This suggests that pyruvate substitutes for an essential metabolic function of respiration. We reasoned that better understanding the role of pyruvate in these cells would allow us to gain insight into how respiration supports the metabolic needs of proliferating cells. To avoid respiration-independent effects of mtDNA depletion, we used 143B ρ0 cells repopulated with mtDNA containing a frame shift deletion in cytochrome B (143B CytB) (Rana et al., 2000). 143B CytB cells have otherwise wild-type mitochondria, but are respiration-incompetent due to lack of cytochrome B in complex III and thus lack a functional ETC. As a control we utilized 143B ρ0 cells which were repopulated with wild-type mtDNA (143B WT cybrid) and are respiration-competent with functional ETC. 143B CytB cells are auxotrophic for pyruvate as they fail to proliferate in the absence of pyruvate (Fig 1A). Conversely, 143B WT cybrid cells cultured with or without pyruvate divide at a similar rate (Fig S1A), confirming that pyruvate auxotrophy accompanies loss of mitochondrial respiration.
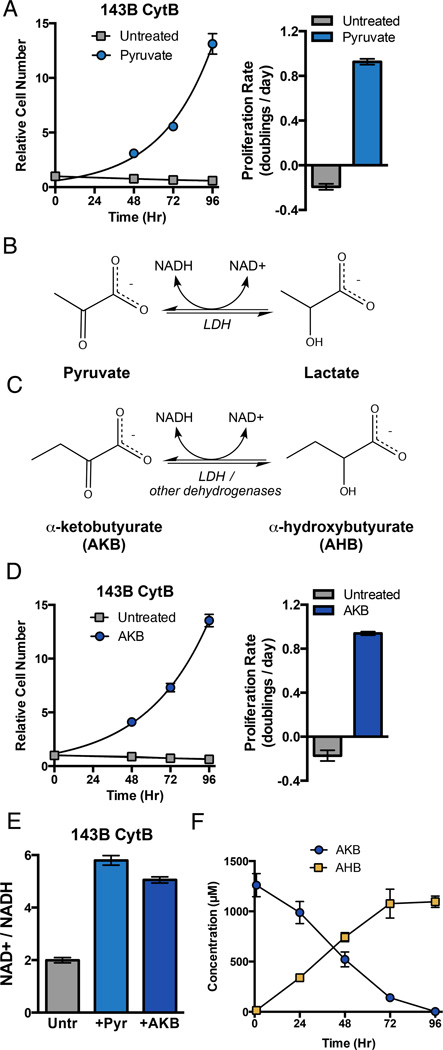
Figure 1. Cytochrome B Mutant 143B Cybrid Cells are Auxotrophic for Electron Acceptors that can Regenerate NAD+.
(A) Proliferation rate of 143B CytB cells was determined in the presence or absence of pyruvate. (left) Cell counts, normalized to cell number at t=0 when media conditions were applied, were assessed over time and used to calculate proliferation rate (right). (B) Pyruvate is a substrate of lactate dehydrogenase (LDH), accepting electrons from NADH to produce NAD+ and lactate. (C) Alpha-ketobutyrate (AKB) can also act as an electron acceptor from NADH and yield NAD+ and alpha-hydroxybutyrate (AHB). (D) Proliferation rate for 143B CytB cells in the presence or absence of AKB was determined as in (A). (E) Intracellular ratio of NAD+/NADH was determined in 143B CytB cells in untreated media or in the presence of pyruvate (Pyr) or AKB. (F) The concentration of AKB and AHB in the media of 143B CytB cells cultured in the presence of AKB was determined over time by GCMS analysis. Values in all figure panels denote mean ± standard error of the mean (SEM), n=3. See also Figure S1.
Several hypotheses have been proposed to explain pyruvate auxotrophy (Howell and Sager, 1979; King and Attardi, 1996; Morais et al., 1994; van den Bogert et al., 1992). Pyruvate carbon has many metabolic fates including conversion to oxaloacetate via pyruvate carboxylase, malate via malic enzyme, and acetyl-CoA via the pyruvate dehydrogenase complex. Thus, one hypothesis is that pyruvate acts as a carbon substrate for synthesis of biosynthetic intermediates that are normally dependent on respiration. An alternative hypothesis is that in the absence of functional ETC, cells cannot adequately oxidize cellular NADH. Hence, pyruvate is required as an exogenous electron acceptor to regenerate NAD+ via the lactate dehydrogenase (LDH) reaction (Fig 1B). Importantly, the continued production of pyruvate from glycolysis requires NAD+, and therefore, the use of glucose-derived pyruvate as a biosynthetic intermediate requires an exogenous source of NAD+ regeneration to maintain redox balance.
In order to decouple the potential roles of pyruvate, we sought alternative substrates that could support 143B CytB cell proliferation in the absence of pyruvate. One candidate we identified is the four-carbon metabolite alpha-ketobutyrate (AKB), which can act as a substrate for LDH or other intracellular dehydrogenases (Fig 1C). While not the preferred substrate for LDH, we confirmed that LDH can utilize AKB to regenerate NAD+ from NADH with reasonable kinetics (Fig S1B). We tested whether AKB is sufficient to support proliferation of 143B CytB cells in the absence of pyruvate. Indeed, AKB restores proliferation of 143B CytB cells to similar levels as cells cultured with pyruvate (Fig 1D). AKB addition does not restore oxygen consumption (Fig S1C), alleviate uridine auxotrophy (Fig S1D), or impact proliferation of 143B WT cybrids (Fig S1E), demonstrating that AKB acts similarly to pyruvate in supporting proliferation of respiration-deficient 143B CytB cells.
As exogenous electron acceptors, AKB and pyruvate are both expected to regenerate NAD+. We measured the NAD+/NADH ratios in 143B CytB cells cultured in the absence or presence of either pyruvate or AKB. Addition of either pyruvate or AKB is sufficient to increase the NAD+/NADH ratio, consistent with these molecules serving as electron acceptors (Fig 1E). Given that AKB and pyruvate have different metabolic carbon-fates, we hypothesized that AKB is used primarily as an electron acceptor and not as a carbon substrate in other metabolic pathways. When AKB is metabolized by a dehydrogenase that uses NADH to reduce the alpha-ketone to a hydroxyl and regenerate NAD+, the expected product is alpha-hydroxybutyrate (AHB) (Fig 1C). To test whether this is the fate of exogenous AKB in 143B CytB cells, we quantitatively measured the consumption of AKB and excretion of AHB in the media of 143B CytB cells cultured in the absence of pyruvate. We found that AKB consumption matched AHB excretion (Fig 1F). AKB and AHB are both four-carbon metabolites that differ only by oxidation state, and levels of these metabolites in the culture media are negligible when AKB is not added. Thus, this result strongly suggests that AKB acts only as an electron acceptor and does not directly contribute carbon to metabolism. To further confirm that pyruvate and AKB carbon are metabolized differently, we find that pyruvate, but not AKB, maintains viability of 143B WT cybrid and HeLa cells deprived of glucose and glutamine (Fig S1F). Since AKB is sufficient to replace pyruvate in supporting 143B CytB cell proliferation, the pyruvate auxotrophy of these respiration incompetent cells is best explained as a requirement for exogenous electron acceptors. Taken together, these data suggest that access to exogenous electron acceptors provided by respiration (O2), pyruvate, or AKB are required to support proliferation.
Inhibition of respiration causes auxotrophy for electron acceptors
Generation of 143B CytB cells involves a selection process that could result in uncharacterized changes that alter the metabolic requirements provided by respiration. To test whether parental cells with functional ETC have the same requirements for respiration, we treated wild-type 143B cells with an array of respiration inhibitors. Given the known DHODH requirement for respiration and the finding that AKB can substitute for pyruvate as an electron acceptor, this and all subsequent experiments were performed in the presence of uridine and in the absence of pyruvate. Inhibitors of mitochondrial complex I, complex III, complex IV, and ATP synthase were all sufficient to suppress proliferation and most resulted in decreased cell count over time (Fig 2A). Exposure to ETC inhibitors at these doses decreased oxygen consumption (Fig S2) and thus decreased access to electron acceptors. To determine if respiration is required for proliferation because it provides access to electron acceptors, we tested if AKB supplementation could restore proliferation. In all cases, AKB addition rescued proliferation to rates comparable to that of untreated cells (Fig 2A). To confirm that AKB also acts as an electron acceptor in this context, we measured the NAD+/NADH ratio in cells treated with ETC inhibitors in the absence or presence of AKB. All ETC inhibitors decreased cellular NAD+/NADH ratio in the absence of AKB (Fig 2B). AKB addition restored NAD+/NADH, while having no effect on O2 consumption, confirming that AKB is sufficient to increase oxidized cofactor pools (Fig 2B, S2).
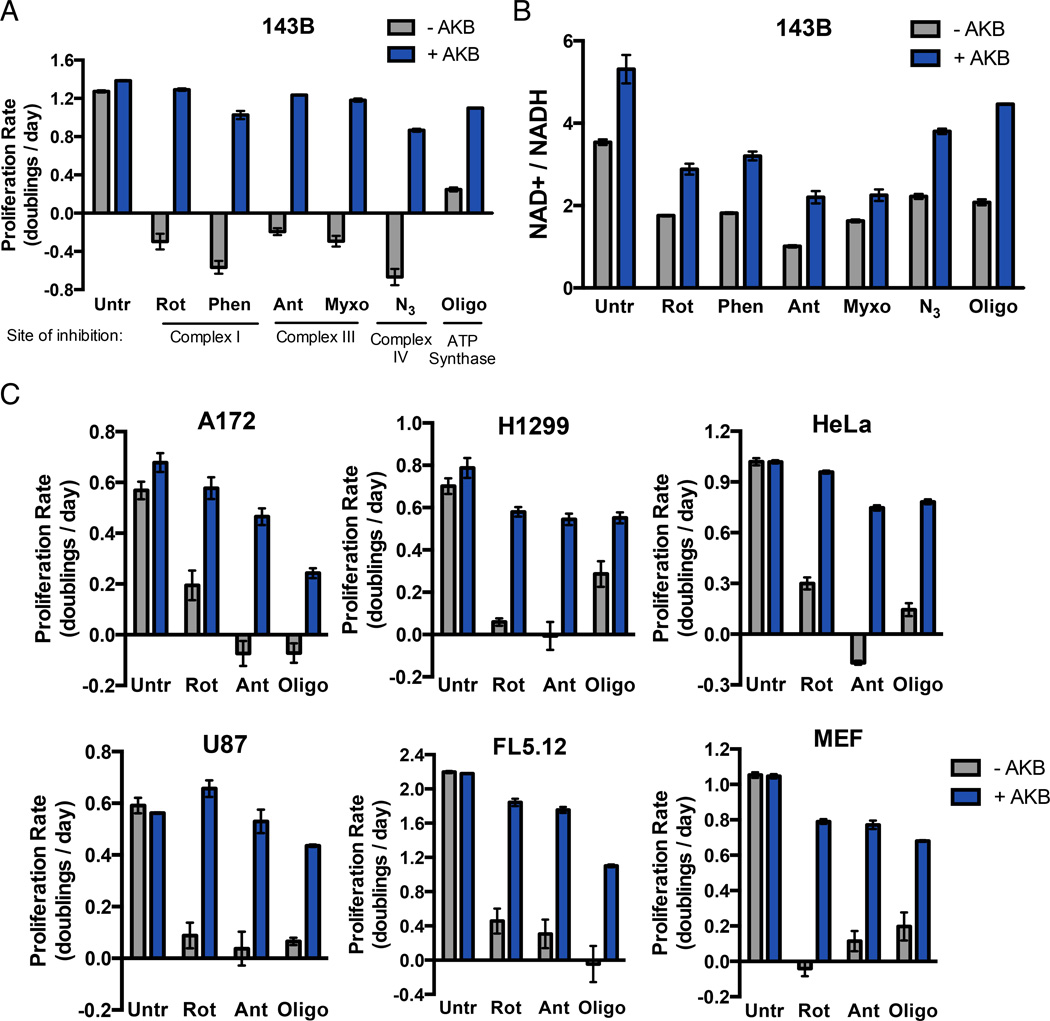
Figure 2. Proliferation of Respiration-Inhibited Cells is Restored by Exogenous Electron Acceptors.
(A) The proliferation rate of 143B cells was determined in media with or without AKB supplementation in the absence (Untr) or in the presence of the mitochondrial respiration inhibitors rotenone (Rot), phenformin (Phen), antimycin (Ant), myxothiazol (Myxo), azide (N3), or oligomycin (Oligo). (B) The ratio of NAD+/NADH was determined in 143B cells with or without AKB supplementation in the absence or presence of respiration inhibitors as in (A). (C) The proliferation rate of A172, H1299, HeLa, U87, FL5.12, and MEF cell lines was determined with or without AKB supplementation in the absence or presence of rotenone, antimycin, or oligomycin. Values in all figure panels denote mean ± SEM, n=3. See also Figure S2.
To determine if a similar dependence on respiration exists in other proliferating cells, we treated a panel of genetically diverse cell lines including both transformed and non-transformed cells with three representative respiration inhibitors: rotenone (complex I inhibitor), antimycin (complex III inhibitor), and oligomycin (ATP synthase inhibitor). For all cell lines, treatment with any of the inhibitors blocked proliferation and AKB was sufficient to restore proliferation (Fig 2C).
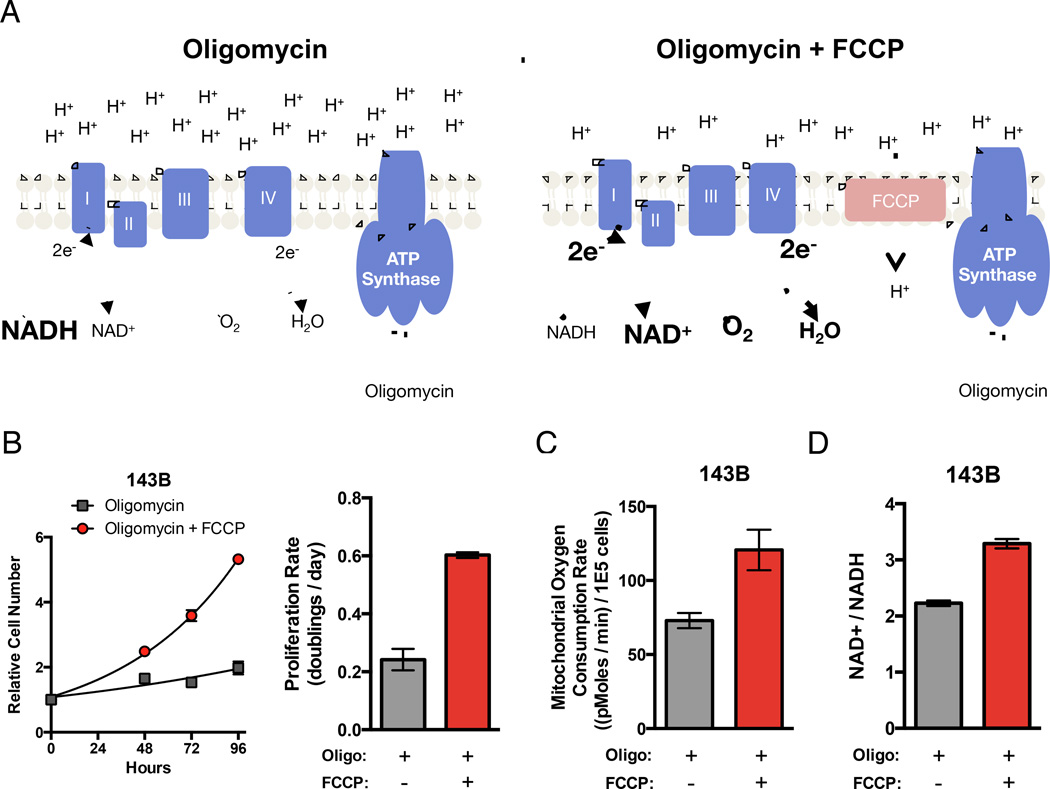
The use of oxygen as a terminal electron acceptor by mitochondrial respiration is well described. However, this role for oxygen is classically considered in the context of enabling NADH oxidation by the ETC to produce a membrane potential across the mitochondrial inner membrane and support ATP synthesis. Notably, reduction of AKB allows regeneration of NAD+ but does not support ATP production, arguing that mitochondrial ATP production is not required for proliferation. To further test this idea, we utilized oligomycin, a specific ATP synthase inhibitor, and the ionophore carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP), an uncoupler of the mitochondrial membrane potential. In normal respiration, electrons from NADH are transferred to the ETC and then ultimately to oxygen to generate an electrochemical proton gradient across the mitochondrial inner membrane, which can drive ATP synthesis. Oligomycin does not directly inhibit components of the ETC, but rather slows ETC electron transfer to oxygen by hyperpolarizing the mitochondrial membrane potential (Fig 3A) (Brand and Nicholls, 2011). Thus, treatment with oligomycin alone decreases oxygen consumption (Fig S2C). However, addition of the uncoupling agent FCCP can restore the ability to transfer electrons from the ETC to oxygen in the presence of oligomycin by relieving membrane hyperpolarization without reversing inhibition of ATP synthase function (Fig 3A). Thus, if providing access to electron acceptors is the essential function of respiration independent of ATP production, restoring oxygen consumption with a low dose of FCCP that does not completely dissipate all intracellular proton membrane potentials should restore proliferation of oligomycin treated cells. We compared the proliferation rate of 143B cells cultured in oligomycin in the absence or presence of low dose FCCP. Consistent with the hypothesis, FCCP addition increased proliferation of oligomycin treated cells (Fig 3B). Also in agreement with the hypothesis, FCCP increased both oxygen consumption and the NAD+/NADH ratio (Fig 3C, D). Since FCCP and oligomycin act independently, FCCP addition does not restore mitochondrial ATP production. These data argue that providing access to electron acceptors, rather than supporting mitochondrial ATP production, is a limiting function of mitochondrial respiration for supporting proliferation.
Figure 3. Oxygen Utilization in the Absence of Mitochondrial ATP Production is Sufficient for Cell Proliferation.
(A) Schematic illustrating the effects of oligomycin and FCCP on mitochondrial membrane potential and ATP synthesis. Oligomycin treatment inhibits ATP synthase, resulting in a hyperpolarized mitochondrial membrane. This hyperpolarization inhibits proton pumping and thereby inhibits ETC activity resulting in decreased NADH oxidation and O2 consumption (left). Treatment with FCCP in addition to oligomycin relieves the hyperpolarization of the mitochondrial membrane allowing restoration of NADH oxidation and mitochondrial O2 consumption without restoring ATP production (right). (B) Proliferation rate of 143B cells treated with oligomycin in the presence or absence of FCCP treatment. (C) Mitochondrial oxygen consumption rate of 143B cells treated with oligomycin with or without FCCP. (D) Intracellular NAD+/NADH ratio in 143B cells treated with oligomycin in the presence or absence of FCCP. Values in all figure panels denote mean ± SEM, n=3 (B, D), n=5 (C).
Electron acceptor insufficiency causes inhibition of nucleotide biosynthesis
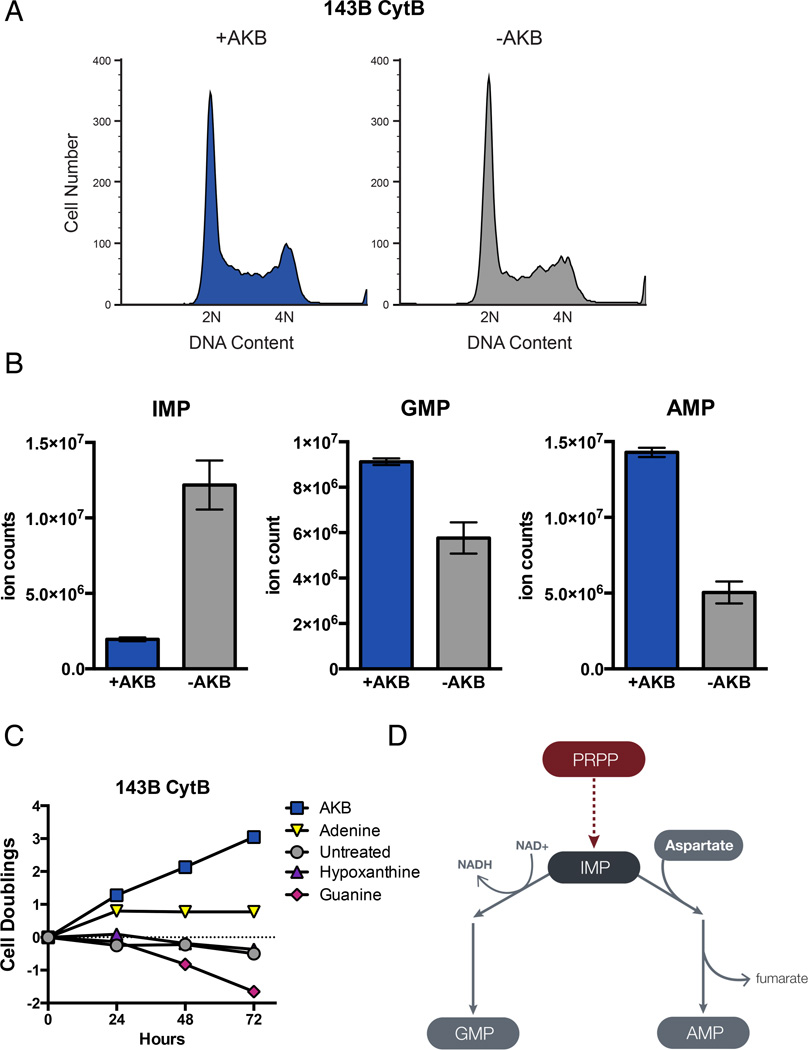
To gain mechanistic insight into why electron acceptors are required for proliferation, we characterized the phenotype of respiration incompetent cells under conditions where access to electron acceptors is limiting. Analysis of DNA content in non-proliferating 143B CytB cells without AKB suggested that these cells do not arrest at a specific stage of the cell cycle (Fig 4A). Surprisingly, despite complete cessation of proliferation for over 72 hours these cells remained viable, and a substantial fraction of the cells had a DNA content between 2N and 4N suggesting that some cells were unable to progress through S-phase. Furthermore, compared to cells proliferating in the presence of AKB, the non-proliferating population showed a subtle accumulation of cells with a DNA content less than 4N. An inability to generate sufficient nucleotides to support DNA synthesis can prevent progression through S-phase (Lunt et al., 2015). To determine if a deficiency in nucleotide synthesis contributes to the inability to proliferate in the absence of AKB, we quantified nucleotide pools in 143B CytB cells in the presence and absence of AKB using liquid chromatography mass spectrometry (LCMS). Because pyrimidine nucleotide pools are confounded by excess uridine supplemented in the media, we focused analysis on purine nucleotides. Compared to AKB replete cells, cells cultured without AKB had increased levels of the purine nucleotide inosine-5’-monophosphate (IMP) (Fig 4B). IMP is a precursor of both guanine-5’-monophosphate (GMP) and adenine-5’-monophosphate (AMP). However, despite an increase in IMP, both GMP and AMP levels were decreased in the absence of AKB (Fig 4B). To determine the functional significance of the purine nucleotide deficiency, we supplemented 143B CytB cells with exogenous adenine, guanine, and hypoxanthine. Using nucleotide salvage pathways, these nucleobases are converted to AMP, GMP, and IMP, respectively. Whereas hypoxanthine had no effect on proliferation and guanine supplementation was slightly toxic, adenine supplementation restored proliferation for one doubling, and alleviated the accumulation of cells with DNA content less than 4N (Fig 4C, S3A). These data suggest that loss of respiration impairs adenine nucleotide synthesis and that adenine nucleotide deficiency partially explains the inability of cells to proliferate in the absence of electron acceptors.
Figure 4. Electron Acceptor Insufficiency Affects Purine Nucleotide Levels.
(A) Analysis of DNA content by propidium iodide staining and flow cytometry of 143B CytB cells cultured with our without AKB supplementation. (B) LCMS quantification of purine nucleotide levels in 143B CytB cells cultured with or without AKB supplementation. (C) Cell doublings were measured over time of 143B CytB cells cultured in unsupplemented media or media supplemented with AKB, adenine, hypoxanthine, or guanine. (D) Schematic illustrating the use of IMP for GMP and AMP synthesis. Synthesis of GMP uses NAD+, whereas synthesis of AMP requires aspartate. Values in (B) and (C) denote mean ± SEM, n=3. See also Figure S3.
Both the AMP/IMP ratio and the GMP/IMP ratio dramatically decrease when AKB is withdrawn from cells cultured in the presence of this exogenous electron acceptor (Fig S3B). This decrease in the AMP/IMP and GMP/IMP ratios suggests an inability to convert IMP to either AMP or GMP. Conversion of IMP to GMP consumes an NAD+ by inosine 5’monophosphate dehydrogenase (IMPDH); thus, this reaction may be inhibited by decreased NAD+ availability (Fig 4D). In contrast, conversion of IMP to AMP does not directly require oxidation but instead consumes aspartate (Fig 4D). This suggests the possibility that decreased AMP production could be downstream of an aspartate deficiency. Consistent with this, SAICAR, a precursor to IMP that also requires aspartate for its synthesis is also depleted when AKB is withdrawn (Fig S3C). Importantly, an underlying aspartate deficiency could explain the transitory nature of adenine rescue; adenine can only restore proliferation to those cells in a phase of cell cycle where adenine is limiting and fails to compensate for other roles that aspartate might play in cell growth and division.
Aspartate synthesis is inhibited by electron acceptor insufficiency
For most cells in culture, carbon for de novo aspartate synthesis is supplied by anaplerotic glutamine (DeBerardinis et al., 2007; Hensley et al., 2013). Glutamine is converted to glutamate, which enters the TCA cycle upon conversion to alpha-ketoglutarate (AKG) by glutamate dehydrogenase (GDH) or transamination (Fig 5A). While GDH uses NAD+ as a co-substrate, the glutamate transaminases utilize alpha-ketoacids, such as pyruvate, as co-substrates. Regardless of which pathway produces AKG from glutamate, conversion of a carbon-nitrogen single bond to a carbon-oxygen double bond necessitates that an electron pair is transferred to an electron acceptor (Fig S4A).
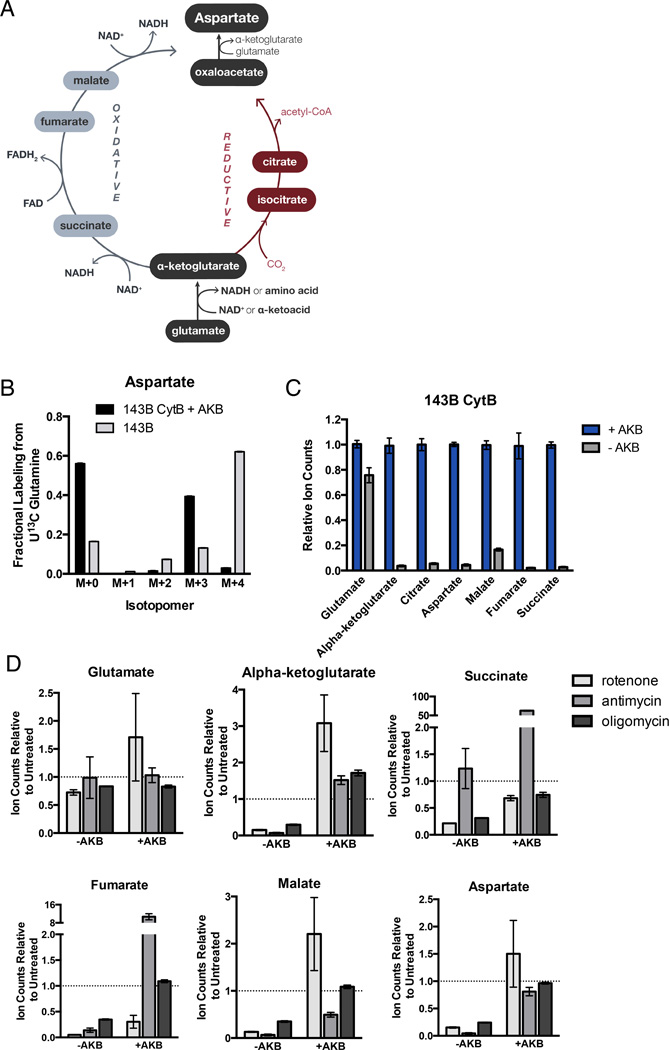
Figure 5. Electron Acceptor Insufficiency Suppresses TCA Metabolite and Aspartate Levels.
(A) Schematic detailing the TCA cycle reaction routes for the biosynthesis of aspartate from glutamine. (B) Isotopomer distribution of aspartate from 143B CytB cells supplemented with AKB and 143B cells cultured in the presence of U-13C Glutamine for 8 hours. (C) GCMS quantification of TCA cycle metabolites, glutamate, and aspartate from 143B CytB cells cultured with or without AKB. (D) GCMS quantification of TCA metabolites, glutamate, and aspartate from 143B cells cultured with or without AKB in the presence or absence of the indicated mitochondrial inhibitors. Ion counts are relative to untreated 143B cells, which is denoted by the dashed grey line in each panel. Values denote mean ± SEM, n=3. See also Figure S4.
Aspartate can be produced from AKG via both reductive and oxidative pathways. Reductive synthesis of aspartate by reductive carboxylation of AKG requires high levels of AKG (Fendt et al., 2013), and electron acceptors are needed to maintain increased AKG pools. In addition, NAD+ dependent oxidation of AKG is required to support reductive carboxylation of AKG, providing another mechanism by which electron acceptors are required for reductive aspartate synthesis (Mullen et al., 2014). Oxidative synthesis of aspartate from AKG requires three additional oxidation reactions requiring net transfer of two electron pairs, thus imposing a further requirement for additional electron acceptors to produce aspartate oxidatively from AKG (Fig 5A). Given that both aspartate synthesis pathways utilize electron acceptors, we reasoned that electron acceptor deficiency in respiration-inhibited cells could limit aspartate production and result in aspartate deficiency.
We used gas chromatography mass spectrometry (GCMS) to measure the electron acceptor dependency of glutamate, aspartate, and TCA cycle metabolite levels in cells. In 143B CytB cells, which metabolize glutamine by reductive carboxylation (Fig 5B, S4B) (Mullen et al., 2012) glutamate levels are only modestly lower upon AKB withdrawal, whereas pool sizes of TCA cycle metabolites are drastically decreased (Fig 5C). Aspartate was also dramatically decreased in these cells, supporting the hypothesis that aspartate limitation may explain the observed AMP/IMP imbalance in cells with electron acceptor insufficiency. In wild-type 143B cells, which normally produce aspartate by oxidative TCA cycle metabolism (Fig 5B, S4B), treatment with the respiration inhibitors rotenone, antimycin, and oligomycin yielded similar results, with decreases in both TCA metabolites and aspartate but not glutamate (Fig 5D). In all cases, treatment with AKB restored aspartate to levels comparable to or higher than untreated cells. These data demonstrate that lack of available electron acceptors restricts aspartate biosynthesis.
Aspartate restores proliferation in cells with electron acceptor insufficiency
To determine if electron acceptor deficiency inhibits proliferation because of an effect on aspartate biosynthesis, we tested whether exogenous aspartate could replace the requirement for exogenous electron acceptors. Strikingly, in the absence of exogenous electron acceptors, supplementation with supra-physiological levels of aspartate was capable of supporting exponential growth of 143B CytB cells (Fig 6A). Given the decrease in other TCA cycle intermediates observed following electron acceptor withdrawal, we also tested whether other TCA cycle intermediates or their cell-permeable derivatives could restore proliferation of these cells. None were able to restore proliferation to a similar degree as aspartate (Fig S5A, B). Respiration-competent 143B WT cybrid cells proliferated at similar rates in the presence or absence of aspartate (Fig S5C). Importantly, aspartate does not function as an exogenous electron acceptor as it neither increases mitochondrial oxygen consumption, nor does it alter the NAD+/NADH ratio in 143B CytB cells (Fig S5D, E). These data imply that aspartate is not an electron acceptor but rather is itself the biosynthetic demand that becomes limiting for proliferation in electron acceptor deficient cells.
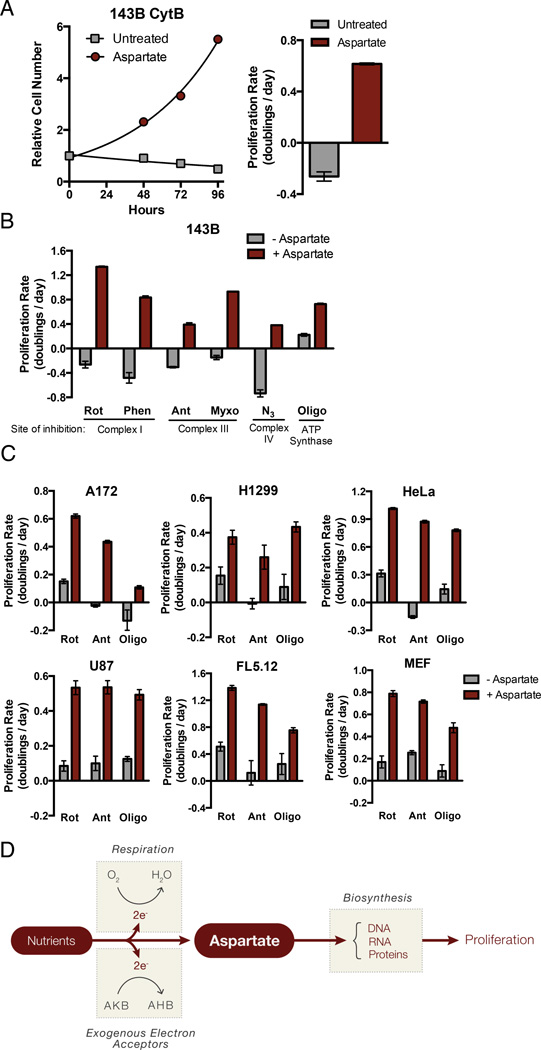
Figure 6. Aspartate is the Key Biosynthetic Precursor Provided by Respiration.
(A) Proliferation rate of 143B CytB cells determined in the presence or absence of aspartate. (B) The proliferation rate of 143B cells cultured with or without aspartate in the presence of the mitochondrial respiration inhibitors rotenone (Rot), phenformin (Phen), antimycin (Ant), myxothiazol (Myxo), azide (N3), or oligomycin (Oligo). (C) The proliferation rate of A172, H1299, HeLa, U87, FL5.12, and MEF cells cultured with or without aspartate in the presence of rotenone, antimycin, or oligomycin. (D) Schematic detailing the role of respiration and exogenous electron acceptors in aspartate biosynthesis. The conversion of nutrients into aspartate requires the removal of electrons and therefore requires access to electron acceptors, which can be supplied by respiration (O2) or exogenous electron acceptors such as AKB. Maintenance of aspartate pools supports nucleotide and protein biosynthesis. Values in all figure panels denote mean ± SEM, n=3. See also Figure S5.
Measurement of purine nucleotides shows that in 143B CytB cells aspartate addition relieves IMP accumulation and restores AMP pools (Fig S5F). This is consistent with the hypothesis that under electron acceptor deficiency, aspartate is limiting for adenine nucleotide synthesis. Aspartate treatment also restored SAICAR levels, another intermediate limited by aspartate availability (Fig S5G). Interestingly, GMP levels are not restored by aspartate treatment, consistent with the conversion of IMP to GMP being dependent on NAD+, which is not restored by aspartate (Fig S5E, F). Indeed, GMP levels were lower in aspartate treated cells, likely a result of decreased IMP availability.
To determine whether aspartate is sufficient to restore proliferation in wild-type cells where respiration is inhibited, wild-type 143B cells were treated with various respiration inhibitors with or without aspartate supplementation. Aspartate restored proliferation of cells treated with all ETC inhibitors (Fig 6B). Aspartate addition also rescued ETC inhibitor-induced proliferation decreases in all other cell lines tested (Fig 6C).
Taken together, these data support a model where a major metabolic requirement for proliferating cells fulfilled by respiration is providing access to electron acceptors in the form of oxygen. This access to electron acceptors is required to support de novo aspartate biosynthesis and in the absence of mitochondrial respiration, the demand for oxygen can be met through supplementation of other electron acceptors such as pyruvate or AKB. Alternatively, if the demand for aspartate can be met exogenously, electron acceptors become dispensable for proliferation. Surprisingly, this implies that the major function of respiration in proliferating cells is to support aspartate synthesis (Fig 6D).
Discussion
In this study we examine the role of mitochondrial respiration in proliferative metabolism. Whereas respiration is primarily an ATP-producing catabolic process in non-proliferating cells, in proliferating cells respiration serves a crucial anabolic role by providing access to electron acceptors to support aspartate synthesis. Furthermore, mitochondrial ATP production appears dispensable in proliferating cells with access to sufficient glucose. Proliferating cells have different metabolic requirements than non-proliferating cells (Lunt and Vander Heiden, 2011); yet, in both cases the components of the metabolic network are largely the same (Hu et al., 2013). During proliferation these same network components must take on distinct roles to balance the contrasting anabolic and catabolic needs of the cell. While respiration likely supports ATP production in addition to aspartate biosynthesis in many contexts, our finding that respiration is specifically required for aspartate biosynthesis in proliferating cells highlights a distinct anabolic role for respiration.
All cells must perform thermodynamically unfavorable processes. One way to accomplish this is to harness the free-energy of nutrient oxidation. The source of electron donors for oxidation varies widely across species from inorganic material for chemolithotrophic bacteria to reduced carbon for most heterotrophs including mammalian cells. In all cases, nutrient oxidation requires net transfer of electrons to a terminal electron acceptor. In mammalian cells, substrates for nutrient oxidation include carbohydrates, lipids, and amino acids. Various metabolic pathways, including the TCA cycle, are employed for nutrient oxidation with electrons transferred initially to electron accepting cofactors such as NAD+ or FAD. These reactions yield NADH or FADH2, respectively, and result in production of intermediates with more oxidized carbon. The electrons from the reduced cofactors are transferred to the ETC, which uses O2 as a terminal electron acceptor to produce water and facilitate ATP production. Importantly, ATP production through this process intimately couples the oxidation of carbon substrates to O2 consumption.
In contexts where mammalian cells cannot utilize O2 as a terminal electron acceptor, cells are unable to regenerate oxidized cofactors via the ETC. Cells can ferment pyruvate to lactate to regenerate NAD+; however, because the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) step of glycolysis consumes NAD+, use of glucose-derived pyruvate for lactate fermentation does not net yield NAD+. Thus, in the absence of access to exogenous electron acceptors, in order to maintain redox balance and sustain glycolytic ATP production most glucose carbon metabolized via glycolysis past the GAPDH step must be excreted as lactate. While fermentation can produce sufficient ATP for proliferation, it is a redox neutral pathway. In the absence of exogenous electron acceptors cells cannot net synthesize molecules that are more oxidized than the nutrients consumed. Thus, losing oxygen as an electron acceptor results in the inability to net remove electrons from consumed carbon substrates.
Proliferating cells must duplicate all cell components, some of which are more oxidized than the nutrients consumed. This suggests that net removal of electrons from carbon is an intrinsic anabolic requirement to support production of oxidized molecules, such as nucleotides, for biomass accumulation. The need to oxidize nutrients to generate biomass may result in electron acceptor insufficiency and limit proliferation even in the presence of oxygen. Notably, the NAD+/NADH ratio in normoxic cancer cells is more reduced than previously assumed (Hung et al., 2011; Zhao et al., 2015), consistent with oxidation capacity being constrained. Because the NAD+/NADH ratio and the pyruvate/lactate ratio are tightly coupled in cells (Williamson et al., 1967), a low NAD+/NADH ratio is expected to drive increased net pyruvate to lactate conversion. This raises the possibility that the Warburg effect is a reflection of electron acceptor insufficiency.
Here we find that electron acceptors are most limiting for de novo aspartate synthesis. While cells in culture are exposed to atmospheric oxygen, oxygen availability in animal tissues is much lower (Bertout et al., 2008). Therefore, it may be advantageous for cells to utilize less oxidative pathways when electron acceptors become more limiting. The need for electron acceptors is decreased when metabolic pathways that fix CO2 are used for de novo aspartate synthesis. For instance, pyruvate carboxylation produces oxaloacetate that can be transaminated to form aspartate. Alternatively, reductive AKG carboxylation can generate isocitrate, which can be isomerized to citrate, cleaved to form oxaloacetate, and transaminated to produce aspartate. In both cases, fully oxidized carbon is incorporated to produce aspartate without directly requiring net removal of electrons. However, pyruvate and AKG are themselves oxidized relative to the major nutrients glucose and glutamine, imposing a need for electron acceptors to produce these carboxylation pathway substrates. Furthermore, the ability to maintain increased levels of pyruvate or AKG to support pyruvate carboxylation or reductive isocitrate dehydrogenase metabolism is compromised in electron acceptor deficient cells as illustrated by the dramatic fall in AKG levels when respiration is inhibited (Fig 5B, C). Therefore, regardless of the biosynthesis pathway used, electron acceptor insufficiency will limit aspartate synthesis.
Beyond its role as an amino acid in proteins, aspartate is required for conversion of IMP to AMP in de novo purine synthesis and provides the carbon backbone for de novo pyrimidines synthesis. Thus, aspartate deficiency will impair protein, purine nucleotide, and pyrimidine nucleotide synthesis. Notably, aspartate is inefficiently transported into most mammalian cells (Birsoy, et al., co-submitted), with supra-physiological concentrations required to restore growth in electron acceptor deficient cells. The concentration of aspartate in blood is around 10 µM, among the lowest levels of all circulating amino acids (Mayers and Vander Heiden, 2015). Additionally, aspartate aminotransferase is among the most abundant enzymes in the liver, suggesting that circulating aspartate may be actively maintained at low levels. Given the critical roles of aspartate in biosynthesis and the challenges associated with obtaining aspartate from the blood, access to electron acceptors for aspartate synthesis may be a requirement for proliferation in many tumors. Consistent with this idea, pharmacologic inhibition of ETC activity inhibits tumor growth in several cancer models (Shackelford et al., 2013; Wheaton et al., 2014; Zhang et al., 2014). Additionally, while ρ0 cells are unable to form xenografts, reconstituting mtDNA in these cells restores tumorigenicity (Hayashi et al., 1992). This requirement for mtDNA appears so stringent that, in one study, injected ρ0 cells formed tumors only after acquiring mtDNA from the host (Tan et al., 2015). While ρ0 cells are also expected to be pyrimidine-limited due to loss of DHODH activity, genetic loss of complex I, which blocks respiration but not DHODH, also impairs tumorigenesis (Park et al., 2009).
Beyond loss of aspartate biosynthesis, we find loss of ETC activity affects the NAD+/NADH ratio. This redox change will also influence other pathways. For example, NAD+ is integral to sirtuin and PARP activity, and thus, loss of ETC activity may perturb signaling pathways (Canto and Auwerx, 2011). Additionally, ETC activity impacts apoptosis (Newmeyer and Ferguson-Miller, 2003; Wallace, 2012), processes that depend on the mitochondrial membrane potential (Chen et al., 2014; Geissler et al., 2000), and reactive oxygen species (ROS) levels in cells (Schieber and Chandel, 2014). Mutations in ETC components resulting in partial inhibition of respiration may even be advantageous for proliferation in certain contexts by increasing ROS production (Petros et al., 2005). Nevertheless, exogenous aspartate addition is sufficient to restore proliferation of cells that otherwise stop proliferating or die when ETC activity is impaired. Thus, a primary role for mitochondrial respiration in cell proliferation must be to provide access to electron acceptors in support of aspartate synthesis.
Experimental Procedures
Cell Culture
143B, A172, H1299, HeLa, U87, 143B cybrid cell lines (WT Cybrid and CytB), FL5.12, and immortalized MEF cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) (Gibco) supplemented with 10% fetal bovine serum and penicillin-streptomycin. FL5.12 cells were supplemented with 5 µg/mL recombinant mouse IL-3 (R&D Systems), and 143B cybrid cells were supplemented 0.1 mg/mL uridine. All cells were incubated at 37°C with 5% CO2.
Proliferation Rates
Cell were plated onto replicate 6 well dishes (Corning), with initial seeding density of 20,000 cells per well for 143B, A172, H1299, HeLa, 143B WT Cybrid, FL5.12, and MEFs and 30,000 cells for 143B CytB and U87 cells. After overnight incubation for cells to adhere, 6 wells were counted to determine initial count at time of treatment. Cells were washed twice in phosphate buffered saline (PBS) and 4 mL of treatment media was added. Final cell counts were measured 4 days after treatment, and proliferation rate was calculated. See Supplemental Experimental procedures for detailed protocol. Concentrations of all metabolites and compounds added to culture media for each cell line are included as a table in supplemental methods (Table S1).
Mitochondrial Oxygen Consumption
Oxygen consumption rates were determined using a Seahorse Bioscience Extracellular Flux Analyzer (XF24). See Supplemental Experimental procedures for detailed protocol.
Purine Nucleotide Metabolite Extraction and LCMS Analysis
Cells were treated as indicated for 15 hours and polar metabolites were extracted. A Dionex UltiMate 3000 ultra-high performance liquid chromatography system connected to a Q Exactive benchtop Orbitrap mass spectrometer, equipped with an Ion Max source and a HESI II probe (Thermo Fisher Scientific) was used to quantify metabolites. See Supplemental Experimental Procedures for detailed extraction and LCMS conditions.
Amino Acid and TCA cycle Metabolite Extraction and GCMS Analysis
Cells were seeded at 400,000 cells/well in 6 well dishes overnight. The following day cells were washed twice in PBS and media was changed to media containing the indicated treatments. After 8 hours, polar metabolites were extracted using 80% methanol in water with 1 µg norvaline standard added per sample. Soluble content was dried under nitrogen gas. Polar samples were derivatized and measured as previously detailed (Lewis et al., 2014). Relative metabolite abundances were determined by integrating ion peak area (Metran) and normalized to norvaline internal extraction standard.
Cell Cycle Distribution Measurements
Cells were incubated with or without AKB or 100 µM adenine as indicated for 78 hours before being washed with PBS, trypsinized, pelleted, and resuspended in 500 µL PBS. Cells were fixed by adding 4.5 mL 70% ethanol and incubated at 4°C overnight. Cells were then pelleted and resuspe nded in 1 mL PBS + 0.1% (v/v) Triton X-100. RNase A and propidium iodide (PI) were added to 0.2 mg/mL and 20 µg/mL, respectively. Samples were incubated at 37°C for 15 minutes and filtered into a flow cytometry tube. DNA content was measured by flow cytometry (BD FACS Canto II) and analyzed (FACS Diva Software).
Measurement of NAD+/NADH
Cells were plated in the same manner as proliferation assays and treated as indicated prior to preparation of cell extracts 6 hours after treatment. NAD+/NADH was measured using a modified version of the manufacturer instructions supplied with the NAD/NADH Glo Assay (Promega). See Supplemental Experimental Procedures for a detailed protocol.
Supplementary Material
Acknowledgements
This work was supported by the Burroughs Wellcome Fund and the NIH (P30CA1405141, GG006413, and R01 CA168653) to M.V.H., a postdoctoral fellowship, PF-15-096-01-TBE from the American Cancer Society to L.S., NIH (T32 GM007753) to D.G., HHMI International Student Research fellowship and the Vertex Scholars Program to A.H., Alex’s Lemonade Stand Undergraduate Research Fellowship to L.B., We thank Alena Heath for graphic design and members of the Vander Heiden lab for thoughtful discussion. We also thank C. Moraes, I.F.M. de Coo, and Navdeep Chandel for the 143B CytB and 143B WT cybrid cell lines.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions L.S. and D.G. performed experiments to determine proliferation rates, NAD+/NADH, metabolite quantifications, and oxygen consumption rates. A.H. performed cell cycle analysis experiments. L.B. performed proliferation rate experiments. E.F. performed LCMS quantification. L.S., D.G., and M.V.H. designed the study and wrote the manuscript.
References
- Bertout JA, Patel SA, Simon MC. The impact of O2 availability on human cancer. Nature reviews Cancer. 2008;8:967–975. doi: 10.1038/nrc2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand MD, Nicholls DG. Assessing mitochondrial dysfunction in cells. The Biochemical journal. 2011;435:297–312. doi: 10.1042/BJ20110162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Auwerx J. NAD+ as a signaling molecule modulating metabolism. Cold Spring Harbor symposia on quantitative biology. 2011;76:291–298. doi: 10.1101/sqb.2012.76.010439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WW, Birsoy K, Mihaylova MM, Snitkin H, Stasinski I, Yucel B, Bayraktar EC, Carette JE, Clish CB, Brummelkamp TR, et al. Inhibition of ATPIF1 ameliorates severe mitochondrial respiratory chain dysfunction in mammalian cells. Cell reports. 2014;7:27–34. doi: 10.1016/j.celrep.2014.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Kamphorst JJ, Mathew R, Chung MK, White E, Shlomi T, Rabinowitz JD. Glutamine-driven oxidative phosphorylation is a major ATP source in transformed mammalian cells in both normoxia and hypoxia. Molecular systems biology. 2013;9:712. doi: 10.1038/msb.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendt SM, Bell EL, Keibler MA, Olenchock BA, Mayers JR, Wasylenko TM, Vokes NI, Guarente L, Vander Heiden MG, Stephanopoulos G. Reductive glutamine metabolism is a function of the alpha-ketoglutarate to citrate ratio in cells. Nature communications. 2013;4:2236. doi: 10.1038/ncomms3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler A, Krimmer T, Bomer U, Guiard B, Rassow J, Pfanner N. Membrane potential-driven protein import into mitochondria. The sorting sequence of cytochrome b(2) modulates the deltapsi-dependence of translocation of the matrix-targeting sequence. Molecular biology of the cell. 2000;11:3977–3991. doi: 10.1091/mbc.11.11.3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb E, Tomlinson IP. Mitochondrial tumour suppressors: a genetic and biochemical update. Nature reviews Cancer. 2005;5:857–866. doi: 10.1038/nrc1737. [DOI] [PubMed] [Google Scholar]
- Gregoire M, Morais R, Quilliam MA, Gravel D. On auxotrophy for pyrimidines of respiration-deficient chick embryo cells. European journal of biochemistry / FEBS. 1984;142:49–55. doi: 10.1111/j.1432-1033.1984.tb08249.x. [DOI] [PubMed] [Google Scholar]
- Harms M, Seale P. Brown and beige fat: development, function and therapeutic potential. Nature medicine. 2013;19:1252–1263. doi: 10.1038/nm.3361. [DOI] [PubMed] [Google Scholar]
- Harris M. Pyruvate blocks expression of sensitivity to antimycin A and chloramphenicol. Somatic cell genetics. 1980;6:699–708. doi: 10.1007/BF01538969. [DOI] [PubMed] [Google Scholar]
- Hayashi J, Takemitsu M, Nonaka I. Recovery of the missing tumorigenicity in mitochondrial DNA-less HeLa cells by introduction of mitochondrial DNA from normal human cells. Somatic cell and molecular genetics. 1992;18:123–129. doi: 10.1007/BF01233159. [DOI] [PubMed] [Google Scholar]
- Hensley CT, Wasti AT, DeBerardinis RJ. Glutamine and cancer: cell biology, physiology, and clinical opportunities. The Journal of clinical investigation. 2013;123:3678–3684. doi: 10.1172/JCI69600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell N, Sager R. Cytoplasmic genetics of mammalian cells: conditional sensitivity to mitochondrial inhibitors and isolation of new mutant phenotypes. Somatic cell genetics. 1979;5:833–845. doi: 10.1007/BF01542645. [DOI] [PubMed] [Google Scholar]
- Hu J, Locasale JW, Bielas JH, O'Sullivan J, Sheahan K, Cantley LC, Vander Heiden MG, Vitkup D. Heterogeneity of tumor-induced gene expression changes in the human metabolic network. Nature biotechnology. 2013;31:522–529. doi: 10.1038/nbt.2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung YP, Albeck JG, Tantama M, Yellen G. Imaging cytosolic NADH-NAD(+) redox state with a genetically encoded fluorescent biosensor. Cell metabolism. 2011;14:545–554. doi: 10.1016/j.cmet.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MP, Attardi G. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- King MP, Attardi G. Isolation of human cell lines lacking mitochondrial DNA. Methods in enzymology. 1996;264:304–313. doi: 10.1016/s0076-6879(96)64029-4. [DOI] [PubMed] [Google Scholar]
- Koppenol WH, Bounds PL, Dang CV. Otto Warburg's contributions to current concepts of cancer metabolism. Nature reviews Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- Kovacevic Z. The pathway of glutamine and glutamate oxidation in isolated mitochondria from mammalian cells. The Biochemical journal. 1971;125:757–763. doi: 10.1042/bj1250757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll W, Loffler M, Schneider F. Energy parameters, macromolecular synthesis and cell cycle progression of in vitro grown Ehrlich ascites tumor cells after inhibition of oxidative ATP synthesis by oligomycin. Zeitschrift fur Naturforschung Section C: Biosciences. 1983;38:604–612. [PubMed] [Google Scholar]
- Lewis CA, Parker SJ, Fiske BP, McCloskey D, Gui DY, Green CR, Vokes NI, Feist AM, Vander Heiden MG, Metallo CM. Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Molecular cell. 2014;55:253–263. doi: 10.1016/j.molcel.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffer M, Schneider F. Further characterization of the growth inhibitory effect of rotenone on in vitro cultured Ehrlich ascites tumour cells. Molecular and cellular biochemistry. 1982;48:77–90. doi: 10.1007/BF00227608. [DOI] [PubMed] [Google Scholar]
- Lunt SY, Muralidhar V, Hosios AM, Israelsen WJ, Gui DY, Newhouse L, Ogrodzinski M, Hecht V, Xu K, Acevedo PN, et al. Pyruvate kinase isoform expression alters nucleotide synthesis to impact cell proliferation. Molecular cell. 2015;57:95–107. doi: 10.1016/j.molcel.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annual review of cell and developmental biology. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- Mayers JR, Vander Heiden MG. Famine versus feast: understanding the metabolism of tumors in vivo. Trends in biochemical sciences. 2015;40:130–140. doi: 10.1016/j.tibs.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature. 1961;191:144–148. doi: 10.1038/191144a0. [DOI] [PubMed] [Google Scholar]
- Morais R, Zinkewich-Peotti K, Parent M, Wang H, Babai F, Zollinger M. Tumor-forming ability in athymic nude mice of human cell lines devoid of mitochondrial DNA. Cancer research. 1994;54:3889–3896. [PubMed] [Google Scholar]
- Mullen AR, Hu Z, Shi X, Jiang L, Boroughs LK, Kovacs Z, Boriack R, Rakheja D, Sullivan LB, Linehan WM, et al. Oxidation of alpha-ketoglutarate is required for reductive carboxylation in cancer cells with mitochondrial defects. Cell reports. 2014;7:1679–1690. doi: 10.1016/j.celrep.2014.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen AR, Wheaton WW, Jin ES, Chen PH, Sullivan LB, Cheng T, Yang Y, Linehan WM, Chandel NS, DeBerardinis RJ. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481:385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newmeyer DD, Ferguson-Miller S. Mitochondria: releasing power for life and unleashing the machineries of death. Cell. 2003;112:481–490. doi: 10.1016/s0092-8674(03)00116-8. [DOI] [PubMed] [Google Scholar]
- Park JS, Sharma LK, Li H, Xiang R, Holstein D, Wu J, Lechleiter J, Naylor SL, Deng JJ, Lu J, et al. A heteroplasmic, not homoplasmic, mitochondrial DNA mutation promotes tumorigenesis via alteration in reactive oxygen species generation and apoptosis. Human molecular genetics. 2009;18:1578–1589. doi: 10.1093/hmg/ddp069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petros JA, Baumann AK, Ruiz-Pesini E, Amin MB, Sun CQ, Hall J, Lim S, Issa MM, Flanders WD, Hosseini SH, et al. mtDNA mutations increase tumorigenicity in prostate cancer. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:719–724. doi: 10.1073/pnas.0408894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana M, de Coo I, Diaz F, Smeets H, Moraes CT. An out-of-frame cytochrome b gene deletion from a patient with parkinsonism is associated with impaired complex III assembly and an increase in free radical production. Annals of neurology. 2000;48:774–781. [PubMed] [Google Scholar]
- Schieber M, Chandel NS. ROS function in redox signaling and oxidative stress. Current biology : CB. 2014;24:R453–R462. doi: 10.1016/j.cub.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleyer M, Schmidt B, Neupert W. Requirement of a membrane potential for the posttranslational transfer of proteins into mitochondria. European journal of biochemistry / FEBS. 1982;125:109–116. doi: 10.1111/j.1432-1033.1982.tb06657.x. [DOI] [PubMed] [Google Scholar]
- Shackelford DB, Abt E, Gerken L, Vasquez DS, Seki A, Leblanc M, Wei L, Fishbein MC, Czernin J, Mischel PS, et al. LKB1 inactivation dictates therapeutic response of non-small cell lung cancer to the metabolism drug phenformin. Cancer cell. 2013;23:143–158. doi: 10.1016/j.ccr.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan AS, Baty JW, Dong LF, Bezawork-Geleta A, Endaya B, Goodwin J, Bajzikova M, Kovarova J, Peterka M, Yan B, et al. Mitochondrial genome acquisition restores respiratory function and tumorigenic potential of cancer cells without mitochondrial DNA. Cell metabolism. 2015;21:81–94. doi: 10.1016/j.cmet.2014.12.003. [DOI] [PubMed] [Google Scholar]
- van den Bogert C, Spelbrink JN, Dekker HL. Relationship between culture conditions and the dependency on mitochondrial function of mammalian cell proliferation. Journal of cellular physiology. 1992;152:632–638. doi: 10.1002/jcp.1041520323. [DOI] [PubMed] [Google Scholar]
- Wallace DC. Mitochondria and cancer. Nature reviews Cancer. 2012;12:685–698. doi: 10.1038/nrc3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Warburg O, Posener K, Negelein E. On the metabolism of carcinoma cells. Biochem Z. 1924;152:309–344. [Google Scholar]
- Weinberg F, Hamanaka R, Wheaton WW, Weinberg S, Joseph J, Lopez M, Kalyanaraman B, Mutlu GM, Budinger GR, Chandel NS. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhouse S. On respiratory impairment in cancer cells. Science. 1956;124:267–269. doi: 10.1126/science.124.3215.267. [DOI] [PubMed] [Google Scholar]
- Wheaton WW, Weinberg SE, Hamanaka RB, Soberanes S, Sullivan LB, Anso E, Glasauer A, Dufour E, Mutlu GM, Budigner GS, et al. Metformin inhibits mitochondrial complex I of cancer cells to reduce tumorigenesis. Elife. 2014;3:e02242. doi: 10.7554/eLife.02242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson DH, Lund P, Krebs HA. The redox state of free nicotinamide-adenine dinucleotide in the cytoplasm and mitochondria of rat liver. The Biochemical journal. 1967;103:514–527. doi: 10.1042/bj1030514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Fryknas M, Hernlund E, Fayad W, De Milito A, Olofsson MH, Gogvadze V, Dang L, Pahlman S, Schughart LA, et al. Induction of mitochondrial dysfunction as a strategy for targeting tumour cells in metabolically compromised microenvironments. Nature communications. 2014;5:3295. doi: 10.1038/ncomms4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Hu Q, Cheng F, Su N, Wang A, Zou Y, Hu H, Chen X, Zhou HM, Huang X, et al. SoNar, a Highly Responsive NAD(+)/NADH Sensor, Allows High-Throughput Metabolic Screening of Anti-tumor Agents. Cell metabolism. 2015;21:777–789. doi: 10.1016/j.cmet.2015.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielke HR, Zielke CL, Ozand PT. Glutamine: a major energy source for cultured mammalian cells. Federation proceedings. 1984;43:121–125. [PubMed] [Google Scholar]
- Zu XL, Guppy M. Cancer metabolism: facts, fantasy, and fiction. Biochemical and biophysical research communications. 2004;313:459–465. doi: 10.1016/j.bbrc.2003.11.136. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.