Abstract
Mec1 (ATR in humans) is the principal kinase responsible for checkpoint activation in response to replication stress and DNA damage in Saccharomyces cerevisiae. Checkpoint initiation requires stimulation of Mec1 kinase activity by specific activators. The complexity of checkpoint initiation in yeast increases with the complexity of chromosomal states during the different phases of the cell cycle. In G1 phase, the checkpoint clamp 9-1-1 is both necessary and sufficient for full activation of Mec1 kinase whereas in G2/M, robust checkpoint function requires both 9-1-1 and the replisome assembly protein Dpb11 (human TopBP1). A third activator, Dna2, is employed specifically during S phase to stimulate Mec1 kinase and to initiate the replication checkpoint. Dna2 is an essential nuclease-helicase that is required for proper Okazaki fragment maturation, for double-strand break repair, and for protecting stalled replication forks. Remarkably, all three Mec1 activators use an unstructured region of the protein, containing two critically important aromatic residues, in order to activate Mec1. A role for these checkpoint activators in channeling aberrant replication structures into checkpoint complexes is discussed.
Keywords: Dna2, nuclease, replication checkpoint, Mec1, ATR
1. Introduction
Dna2 is an essential nuclease-helicase that was initially identified in Saccharomyces cerevisiae in a screen for DNA replication mutants [1,2]. Due to the presence of helicase motifs in its C-terminus as well as the DNA replication defect observed in dna2-1 mutants, Dna2 was originally suggested to be the replicative helicase [3]. However, it soon became clear that the protein also contained vigorous nuclease activity, which was identified as the essential function of Dna2 [4–6]. Based on biochemical interactions and genetic evidence it was shown that Dna2 and Rad27, the yeast homolog of Flap endonuclease 1 (FEN1), work together in the process of lagging strand maturation to cleave the single-stranded flaps formed at the 5′ ends of Okazaki fragments during displacement synthesis by Pol δ [7–9]. In addition to its role in Okazaki fragment maturation, Dna2 has since been implicated in a number of other key processes that are of vital importance for genome maintenance. Here, we will first give a brief overview of the various functions of Dna2 and then focus on its recently reported role in checkpoint activation [10]. Our emphasis will be on the yeast protein, with some reference to the mammalian homologs where relevant.
2. Domain structure and activities of Dna2
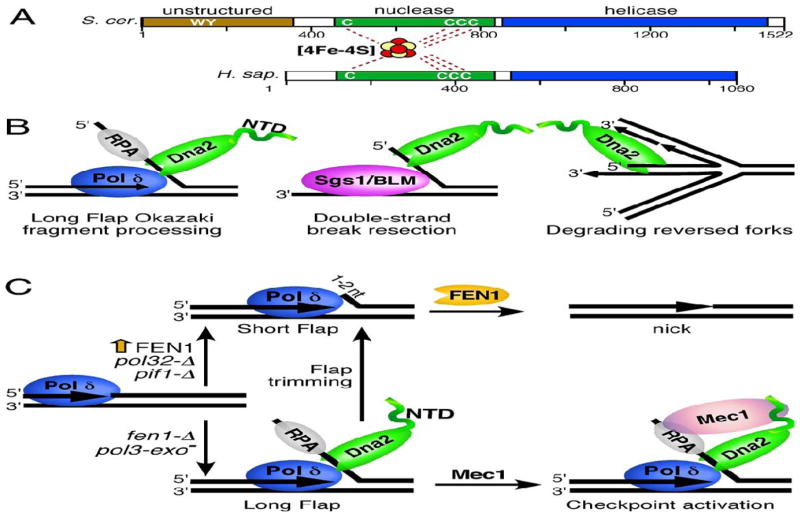
Yeast Dna2 is a 170 kDa protein that contains a C-terminal superfamily I helicase domain, a central RecB family nuclease domain and an unstructured N-terminal tail (Figure 1A). It is found in eukaryotes from yeast to human, but amino acid identity is generally limited to the nuclease and helicase domains while the N-terminus is very poorly conserved [11–13].
Figure 1. The structure and functions of Dna2.
(A) A schematic representation of the domain structure of S. cerevisiae and human Dna2 nuclease/helicase. The nuclease and helicase domains are depicted in green and blue, respectively. The N-terminal unstructured domain of yeast Dna2 is in light brown. The N-terminal domain of human Dna2 is shorter than that of its yeast counterpart and does not contain an appreciable unstructured region. The cysteines that coordinate the iron-sulfur domain as well as the Trp and Tyr that mediate Mec1ATR activation are indicated. (B) Some of the functions of Dna2 in maintenance of nuclear genome stability. Left panel, Dna2 mediates processing of long flaps during Okazaki fragment maturation. Middle panel, Dna2 works in conjunction with Sgs1/BLM helicase in a pathway of DSB end resection. Right panel, Dna2 prevents reversal of replication forks. Additional details are found in the text. (C) The role of Dna2 during Okazaki fragment maturation. On the lagging strand, displacement synthesis by Pol δ generates flaps of 1–2nt that are cleaved by FEN1 to create a ligatable nick (short flap pathway, top of figure). Some flaps escape FEN1 cleavage and grow long enough to bind RPA (long flap pathway, bottom of figure). These long flaps require initial cleavage by Dna2 to allow final processing by FEN1 (flap trimming). Conditions or mutations that promote generation of long flaps (Pol δ exonuclease deficiency or fen1-Δ) are dependent on a functional Dna2, whereas FEN1 overexpression, deletion of the POL32 subunit of Pol δ or deletion of PIF1 promote the short flap pathway and show less dependence on Dna2 [27]. Dna2 bound to long flaps may trigger the checkpoint by activating Mec1ATR.
Dna2 contains ssDNA-specific endonuclease, 5′-3′ helicase and DNA-dependent ATPase activities and is an essential enzyme [3,4,6]. Studies of separation-of-function mutants have established that the nuclease activity of the enzyme is essential for cell survival, whereas helicase-deficient variants are viable but show growth defects [5,6]. The nuclease activity can be either 5′-3′ or 3′-5′ oriented, although RPA stimulates the former and inhibits the latter, making the 5′-3′ polarity the in vivo relevant activity [14,15]. Dna2 preferentially binds to and acts on 5′-flap substrates and, moreover, requires a free ssDNA end for loading. It binds to the 5′-flap, threads over its 5′ end and tracks down the flap in order to cleave its substrate endonucleolytically until it is 5–6 nt from the base of the flap [8,9,16–18].
The C-terminal half of Dna2 contains its ATPase/helicase domain. The helicase activity is ATP-dependent, and mutation of the Walker A domain abolishes not only the ATPase activity but also the helicase function of Dna2 [3]. Dna2 cannot unwind fully duplex DNA but requires a ssDNA region and, similarly to the nuclease activity, has an absolute requirement for a free 5′ end in order to load [16]. The helicase activity of Dna2 has been considered weak; in fact, its presence in the human homolog has been questioned [19–21]. Although the helicase function of Dna2 is not absolutely essential for viability, helicase-deficient variants exhibit growth defects and sensitivity to DNA damaging agents [5], indicating that the helicase activity nonetheless contributes to Dna2 function in vivo. Furthermore, recent work on a nuclease-dead variant of Dna2 reported strong helicase activity comparable to that of the vigorous Sgs1 helicase [22]. It was therefore suggested that the robust nuclease function of the enzyme masks the helicase activity, possibly by cleaving the 5′ overhangs that are required for helicase binding and initiation of unwinding.
The N-terminal domain (NTD) of Dna2 shows no strict conservation between species, and displays large variation in size (Figure 1A). The NTD of yeast Dna2 is predicted to be unstructured [10], while human Dna2 shows no unstructured N-terminal domain. However, the exact translation start site of human Dna2 is uncertain, and longer variants have been proposed (one 1146 aa in length) that would have a significant unstructured NTD [21,23]. Despite being redundant for nuclease and helicase activity, the NTD of yeast Dna2 is required for normal growth, as deletion of the first 405 amino acids of Dna2 leads to a temperature-sensitive phenotype [24]. This may be attributable to the ability of the N-terminal domain to mediate Dna2 binding to secondary structure DNA [25]. It is conceivable that cells expressing an N-terminal deletion mutant of Dna2 may be deficient in processing of flaps with secondary structure, causing the accumulation of these harmful intermediates. Consistent with this model, overexpression of RAD27, the gene for yeast FEN1, suppresses the temperature sensitivity of the dna2-Δ405 mutant [24]. Likely, the increased efficiency of the FEN1-dependent pathway reduces the dependence on Dna2, and its sub-optimal functionality is therefore tolerated.
More recently, the N-terminal domain has been found to mediate checkpoint activation by activating the essential Mec1ATR checkpoint kinase [10]. This new function requires Trp128 and Tyr130 of yeast Dna2 and will be discussed in further detail in section 6. It should be noted that the temperature-sensitive phenotype that was caused by deletion of the entire 405 aa Dna2 NTD [24] is much more severe than that caused by the targeted point mutations that inactivate the checkpoint function of Dna2 [10]. Therefore, the 405 aa NTD contains at least two activities, checkpoint activation and hairpin DNA binding, and whether and to what extent these activities overlap has not been determined.
Dna2 belongs to the growing list of proteins that has been found to contain an iron-sulfur domain. Conserved cysteines 519, 768, 771 and 777 contribute to an Fe-S cluster that flanks the nuclease active site (Figure 1A). Interestingly, mutants that lack the intact Fe-S domain have reduced nuclease as well as reduced helicase activity, suggesting that the Fe-S cluster is not only important for nuclease activity, but has a structural role and thus affects the stability of the entire protein [26].
3. Functions of Dna2
3.1. Role of Dna2 in Okazaki fragment maturation
The best-studied role for Dna2 is in the process of Okazaki fragment processing (Figure 1C). During DNA replication, the lagging strand is synthesized in discontinuous fragments that are primed by short RNA primers. When Pol δ lays down an Okazaki fragment, it displaces the 5′-end of the previous fragment, giving rise to a 5′-flap structure. The structure-specific Flap endonuclease 1 (FEN1) cuts at the base of these flaps, removing the RNA/DNA primer and giving rise to a ligatable end that is joined to the flanking Okazaki fragment by ligase 1. Generally, the length of the 5′-flap is restricted to only 1–2 nt by the action of FEN1, which efficiently cuts the emerging flap, and the action of the 3′-exonuclease activity of Pol δ, which degrades the replicated DNA back to the nick position [27] (Figure 1C). However, a subset of flaps escapes these controls and grows to a length exceeding 25 nt. These long flaps become coated with RPA and are thus refractory to FEN1 cleavage [28]. Dna2, however, can process these RPA-coated long flaps. It acts by digesting them close to their base to produce a short flap, which no longer binds RPA and can be further processed by FEN1 [7,9,29,30] (Figure 1C). Mutations or conditions that promote the generation of long flaps through increased strand displacement synthesis by Pol δ, e.g. Pol δ exonuclease deficiency, are very sensitive to Dna2 dysfunction [31]. Conversely, mutations or conditions that limit strand displacement synthesis and generate shorter flaps, e.g. overexpression of FEN1, deletion of the PIF1 helicase, or deletion of the POL32 subunit of Pol δ, suppress the phenotype of certain DNA2 mutants, and can even suppress the lethality of DNA2 deletion [27,29,32].
Because Dna2 is critically required for the processing of long flaps during Okazaki fragment maturation, this process is considered the most important physiological function of Dna2. In wild-type cells, deletion of DNA2 leads to the persistent accumulation of long flaps that become RPA-coated and activate the checkpoint, leading to permanent cell cycle arrest [33].
3.2. Role of Dna2 in DNA end resection
In addition to its well-established role in Okazaki fragment maturation, Dna2 has an important function in DNA end resection during homologous recombination. DNA double-strand breaks are one of the most cytotoxic forms of DNA damage in the cell and they can be repaired by homologous recombination (HR). HR requires resection of the 5′ DNA end in order to produce a 3′ single-stranded overhang that becomes coated with Rad51 to form a nucleoprotein filament that catalyzes homologous pairing and strand invasion. One of the complexes mediating end resection consists of Dna2 working together with the Sgs1-Top3-Rmi1 complex [14,15,34,35]. Interestingly, the helicase activity of Dna2 is dispensable for end resection, suggesting that Sgs1 provides the helicase activity while Dna2 acts as nuclease to degrade the 5′ strand [34]. RPA directs the nuclease activity of Dna2 to the 5′ DNA end and prevents degradation of the 3′ strand [14,15]. For a more detailed description of the role of different nucleases in end resection, the reader is directed to excellent recent reviews [36,37].
3.3. Dna2 in telomere maintenance
Dna2 also appears to play a role in telomere maintenance. One clue pointing at this function came from localization studies, where Dna2 was shown to localize to telomeres during G1, redistribute throughout the genome in S-phase, and return to telomeres in late S-phase or G2 [38]. Also mammalian Dna2 is localized to telomeres and has been found to be essential for proper telomere maintenance. In fact, Dna2 haploinsufficiency leads to an increase in fragile telomeres and other telomere defects in mouse mouse embryonic fibroblasts [39].
There is mounting evidence that telomeres, as well as other internal G-rich sequences, fold into stable G-quadruplex structures in vivo [40,41]. These secondary structures are problematic for DNA replication because they may block the progression of the replication fork if not resolved by ancillary proteins. Interestingly, both human and yeast Dna2 have been reported to bind and act on G-quadruplexes by unwinding and/or cleaving them [39,42]. The role of Dna2 at telomeres may therefore involve removal of G-quadruplexes in order to permit continuing replication. Its activity on G-quadruplexes may also be of use in resolving these structures on the lagging strand during Okazaki fragment maturation.
3.4. Dna2 in mitochondrial DNA maintenance
In 2006, a study of the suppression of Dna2 lethality by pif1Δ implicated Dna2 in mitochondrial DNA maintenance in yeast [32]. Pif1 exists in both a nuclear and mitochondrial form, but the pif1-m2 variant localizes only to mitochondria and shows full mitochondrial function [43]. Deletion of PIF1 suppresses the lethality of dna2Δ cells. However, although the pif1-m2 mutant grew normally on a non-fermentable carbon source because it still produced the mitochondrial form of Pif1, pif1-m2 dna2Δ double mutants failed to show growth on glycerol media, suggestive of a role for Dna2 in mitochondrial DNA maintenance [32]. Two years later, human Dna2 was reported to be partially targeted to mitochondria [23,44]. A fraction of it colocalizes with mitochondrial DNA nucleoids and with the mitochondrial helicase Twinkle. Furthermore, Dna2 was recruited to mitochondrial DNA nucleoids upon replication stalling and its depletion hindered repair of damaged mtDNA [44]. Finally, the discovery of Dna2 mutations in patients suffering from progressive myopathy and mitochondrial DNA deletions leaves little doubt that this multifaceted protein is involved in the maintenance of not only the nuclear, but also the mitochondrial genome [45].
3.5. Additional roles for Dna2 in maintenance of genetic stability
As is evident from the summary above, Dna2 is involved in a plethora of processes that all contribute in one way or another to the maintenance of genetic stability. The list above is not complete, however, and additional functions may be added in the future. Recently, Hu et al. presented appealing evidence for Dna2’s nuclease involvement in preventing fork reversal into so-called chicken foot structures in Schizosaccharomyces pombe (Figure 1B). Checkpoint-dependent phosphorylation of Dna2 was required for its continued association with stalled replication forks and prevention of fork reversal upon replication stress, induced by hydroxyurea or MMS treatment [46]. Importantly, checkpoint signaling was not compromised in the dna2− spores, indicating that the increased fork reversal in dna2− or dna2ts cells was not due to a defect in checkpoint activation but rather to a role of Dna2 that lies downstream of checkpoint initiation. Overexpression of FEN1 did not suppress the increased fork reversal phenotype of dna2− cells, suggesting that this function of Dna2 is unrelated to that of Okazaki fragment maturation.
Human Dna2 has also been suggested to have a role in genome stability that appears to be distinct from its role in Okazaki fragment maturation or telomere maintenance. Cells depleted of hDna2 accumulate in S/G2 phase of the cell cycle and accumulate internuclear chromatin bridges, a defect that was not suppressed by FEN1 overexpression [44,47]. Furthermore, a recent elegant study implicates even human Dna2 in processing and restart of reversed replication forks [48], suggesting that the reported function of S. pombe Dna2 in replication restart may be conserved [46]. As in S. pombe, the nuclease, but not the helicase, activity of Dna2 was required for the processing of reversed forks in a process that also involves the WRN helicase [48].
4. DNA damage and replication checkpoint pathways
Genomic stability is of vital importance for all cells and the cell cycle checkpoint response is one of the fundamental mechanisms guarding it in eukaryotes. These signal transduction pathways recognize and respond to DNA damage or replication stress and act to slow down or stop cell cycle progression until the DNA damage or aberrant DNA structures have been cleared. Triggering of the checkpoint leads to stabilization of stalled replication forks, increased repair and dNTP synthesis, inhibition of late origin firing and changes in gene expression that allow the cell to overcome the DNA damage or, if the damage is too severe, initiates apoptosis (reviewed in [49]). In this way, the accurate transmission of genomic information to daughter cells is guaranteed. The importance of functional checkpoints is underscored by their involvement in human disease: defects in the different checkpoint proteins lead to conditions such as Ataxia telangiectasia, the rare Seckel syndrome and increased cancer disposition as a consequence of the increased genome instability [50,51].
In S. cerevisiae the checkpoints rely on two apical protein kinases of the phosphatidylinositol 3-kinase-related protein kinase (PIKK) family, namely Mec1 and Tel1. Classically, Mec1 (the homolog of human ATR) is considered to respond to stretches of RPA-coated ssDNA [52], while Tel1 (ATM in human) becomes activated in response to DNA double-strand breaks (DSBs) [53]. However, it is clear that partial redundancy between the Mec1ATR and Tel1ATM pathways exists [54]. Tel1ATM and Mec1ATR can also be sequentially activated within one process, as is the case with DSB repair [55]. DSBs initially activate Tel1ATM, but resection of the 5′-strand results in exposure of ssDNA regions and acts as an efficient switch from Tel1ATM to Mec1ATR signaling [56].
Tel1ATM exists as a homodimer that dissociates into an active monomer in response to DSBs [57,58]. In contrast, Mec1ATR is always found tightly associated with Ddc2 (the ortholog of human ATRIP) and there is no evidence of it acting as a monomer [59]. Mec1ATR-Ddc2ATRIP forms a heterodimeric complex, and there is evidence for the existence of higher-ordered complexes [60,61].
Mec1ATR and Tel1ATM are the critical upstream regulators of the checkpoint but they rely on interaction with other proteins in order to initiate checkpoint signaling. Upon recognition of DNA damage or extensive lengths of RPA-coated ssDNA, the PIKKs are recruited to the site of damage, where they are activated by binding of sensor proteins that have been independently recruited. This “two-man rule” of checkpoint activation prevents aberrant triggering of the signaling pathway. Once activated, the checkpoint kinases phosphorylate downstream mediator and effector proteins, triggering a cascade of phosphorylation events that regulate numerous target proteins and cellular processes (reviewed in [49]).
5. Mec1ATR activation
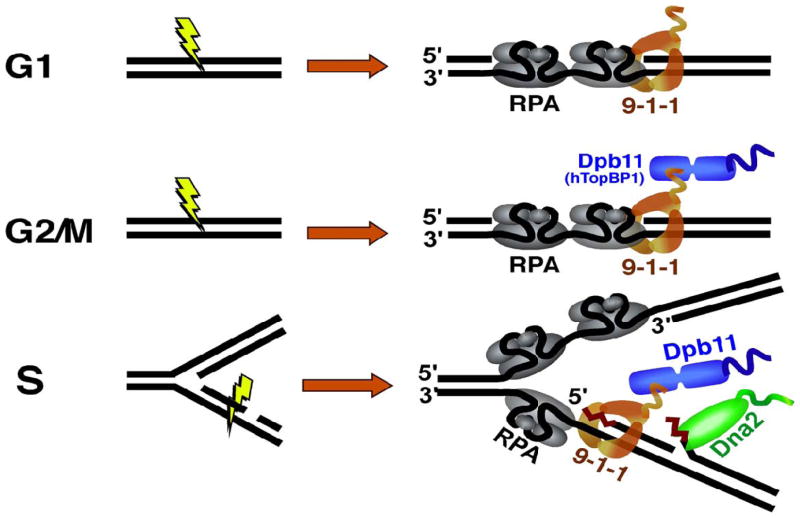
Mec1ATR-Ddc2ATRIP is recruited to stretches of ssDNA through the interaction of the N-terminus of Ddc2ATRIP with the RPA70 subunit [52,62,63]. However, stimulation of Mec1ATR kinase activity requires its interaction with a sensor protein, three of which have been discovered in S. cerevisiae. These three activators show partial redundancy for checkpoint activation in a manner that is dependent on the cell cycle phase (Figure 2). The first activator is the Ddc1-Rad17-Mec3 (human Rad9-Rad1-Hus1; hence the designation 9-1-1) checkpoint clamp that is essential for checkpoint activation in G1- and G2- cell cycle phases but dispensable in S-phase [64–67]. The 9-1-1 complex shows structural similarity with the replication clamp PCNA (proliferating-cell nuclear antigen) [68,69]. Unlike PCNA, which is loaded onto 3′-ds-ssDNA junctions, 9-1-1 is loaded by the Rad24-RFC (replication factor C) clamp loader with an opposite polarity onto 5′-ds-ssDNA junctions [70–72]. In S. cerevisiae, the 9-1-1 complex can directly stimulate Mec1ATR kinase [60], but this activity has not been reported in S. pombe or in metazoans [67]. Consistent with Ddc1 mediating the stimulation by 9-1-1, mutation of two critical Trp residues in the C-terminal tail of Ddc1 completely abrogates the ability of 9-1-1 to trigger the checkpoint in G1 [67]. Therefore, the 9-1-1 complex is the only activator of Mec1 in the G1 phase of the cell cycle [73].
Figure 2. Mec1ATR activation during different cell cycle phases in S. cerevisiae.
Top panel, 9-1-1 (orange) is the sole activator of Mec1 in G1 phase. Middle panel, In G2/M, Mec1 can be activated through two redundant pathways that are separable. The first involves direct activation by 9-1-1 and depends on two aromatic residues in the unstructured C-terminus of the Ddc1 subunit of 9-1-1. The second pathway relies on activation by Dpb11 (in blue), although 9-1-1 is still required for Dpb11 recruitment. Bottom panel, Dna2 (green), 9-1-1 (orange) and Dpb11 (blue) act in a redundant fashion to stimulate Mec1 upon replication stalling induced by hydroxyurea. Dna2 is depicted as bound to long flaps, while 9-1-1 and thereby Dpb11 bind 5′-ssDNA-dsDNA junctions.
In G2, 9-1-1 plays a dual role in checkpoint activation: in addition to directly activating Mec1ATR, it is also required for recruitment of the replication initiation protein Dpb11TopBP1, which in turn binds Mec1ATR and activates its kinase [67]. The C-terminus of Ddc1Rad9 contains a conserved serine/threonine phosphorylation site that is involved in Dpb11TopBP1 recruitment [74]. Indeed, phosphorylation of yeast Ddc1Rad9 on Thr-602 is essential for recruitment of Dpb11TopBP1 to the vicinity of Mec1 and the consequent stimulation of Mec1ATR by Dpb11TopBP1 [74,75]. The direct activation of Mec1ATR by Ddc1Rad9 and the indirect activation by Ddc1Rad9 via Dpb11TopBP1 constitute two separable and partially redundant activation mechanisms; the G2 checkpoint in response to treatment with the UV-mimetic chemical 4-nitroquinoline-oxide is only abolished if both mechanisms are disabled. Conversely, full G2 checkpoint signal requires that both stimulatory pathways are intact [67]. As mentioned above, the direct stimulation of Mec1ATR by Ddc1Rad9 has only been established for S. cerevisiae. In all other systems studied, Dpb11TopBP1 is the only factor that has been reported to directly activate Mec1ATR, although 9-1-1 is required for Dpb11TopBP1 recruitment [74,76,77].
6. Dna2 as a Mec1ATR activator
While the checkpoint is mediated by 9-1-1 in G1 and by both Dpb11TopBP1 and 9-1-1 in G2, the S-phase checkpoint shows even more complexity. The existence of an additional S-phase activator(s) was indicated by the remaining activity of the replication checkpoint even in the absence of the checkpoint functions of Ddc1Rad9 and Dpb11TopBP1 [78]. A biochemical screen for Mec1ATR activators identified Dna2 as an S-phase-specific activator of the Mec1ATR kinase. The stimulatory activity of Dna2 was independent of its nuclease and helicase activities, and was determined to derive from its N-terminal domain, anchored by the two aromatic residues Trp128 and Tyr130 [10]. Replacement of these two residues with alanines resulted in a mutant (Dna2-WY-AA) that lacked Mec1ATR stimulatory activity both in vitro and in vivo when replication forks were stalled by hydroxyurea-induced dNTP depletion. The checkpoint function of all three proteins, Dna2, Ddc1Rad9 and Dpb11TopBP1 had to be inactivated in order to completely abrogate Mec1 function in S-phase, consistent with the contribution of all three activators to Mec1 stimulation at stalled replication forks. Dna2 does not significantly contribute to the checkpoints in G1 or G2.
Evidently, there is a high level of redundancy in activation of the S-phase checkpoint. In agreement with earlier reports [79,80], Kumar and Burgers found that Tel1ATM could partially mediate the S-phase checkpoint signal in response to 4-NQO treatment or hydroxyurea-induced replication stalling. Therefore, full abrogation of the S-phase checkpoint required inactivation of all three Mec1ATR activators (or Mec1ATR itself) and Tel1ATM [10]. It should be noted here that MEC1 deficiency confers lethality in yeast, but this can be suppressed by deletion of SML1, an inhibitor of ribonucleotide reductase. Therefore, mec1Δ sml1Δ strains are viable [81,82].
The reason for the extensive redundancy in activation of the replication checkpoint is somewhat unclear, but may relate to the critical importance of the S-phase checkpoint. Indeed, ddc1Δ cells that lack a functional G1 and G2 checkpoint do not show a significant growth defect [67], but cells that are completely deficient in S-phase checkpoint signaling are very sick and grow poorly even in the absence of DNA damaging agents [10,83]. This suggests that some checkpoint function is required during normal DNA replication. In support of this hypothesis, checkpoint-defective cells fail to complete DNA replication efficiently even in the absence of induced DNA damage. However, normal S-phase progression and cell growth is restored when any single activation mechanism (either by DNA2, DPB11, DDC1 or TEL1) is restored.
7. Unique and common features of Mec1ATR activators
One possible explanation for the presence of multiple Mec1ATR activators is that they recognize and react to different DNA structures or different types of DNA damage. The specificity of 9-1-1 loading onto 5′-ds-ssDNA junctions provides a unique targeting mechanism at the 5′-ends of Okazaki fragments, at resected double-strand DNA breaks, and at ssDNA gaps that have been generated during the process of nucleotide excision repair. Since Dpb11TopBP1 is recruited by Ddc1Rad9 [75], it is expected to be found at the same DNA structures as 9-1-1. Conversely, Dna2 prefers 5′-flap structures [18,72]. Both 5′-flaps and 5′-dsDNA junctions are natural intermediates during lagging strand replication (Figure 1C), and therefore, the major replication checkpoint targeting mode likely proceeds through the lagging strand. However, if re-priming on the leading strand occurs as a consequence of replication fork stalling, 5′-junctions should also be available for 9-1-1 loading at gaps on the leading strand [84]. Furthermore, in light of the reported ability of Dna2 to bind secondary structures and G4 DNA [25,39,42], it could also mediate checkpoint activation in response to replication fork stalling at such structures. Since Dna2 travels with the replication fork [46], its recruitment to stalled forks would not be required, while 9-1-1 and Dpb11TopBP1are only recruited once forks stall.
Although the three Mec1ATR activators are structurally unrelated and have different overall functions in the cell, their similarity with regard to Mec1ATR-activating features is remarkable. All three proteins contain both structured domain(s) with highly specific functions as well as an unstructured region. Our current understanding of these activators is that the structured domain has both checkpoint-independent and checkpoint-dependent functions. As part of checkpoint initiation the structured domain binds certain forms of DNA damage or aberrant DNA structures that may occur during different phases of the cell cycle. The DNA-bound activator will then signal checkpoint initiation via its unstructured activation tail provided that it is localized near sufficient RPA-coated ssDNA in order to recruit Mec1ATR through its Ddc2ATRIP subunit. As an example of this two-component activator model, the G1 DNA damage checkpoint is absolutely dependent on 9-1-1, which localizes to DNA repair gaps and activates Mec1ATR through the C-terminal tail of the Ddc1 subunit. Removal of this tail abrogates the G1 checkpoint. However, it can be restored by fusing the N-terminal tail of Dna2 to the truncated C-terminus of Ddc1 [10]. Even though the NTD of Dna2 normally only functions in S phase, it now has gained G1 function by virtue of being linked to 9-1-1.
All three unstructured tails contain two critical aromatic residues that are essential for stimulation of Mec1ATR kinase activity. However, these two aromatic residues can be separated by as little as one amino acid in Dna2 and by as many as 192 amino acid residues in Ddc1 [10,67,78]. Furthermore, no significant sequence similarity is evident even within the immediate surroundings of the activating aromatic residues. The specificity of activation may instead stem from the aromatic residues being presented within a certain secondary structure context. The activation motif of Ddc1Rad9 forms a potential β strand-loop-β strand motif [68,69] and mutations that abolish either β strand eliminate the ability of the protein to activate Mec1ATR [67]. The key aromatic residues are located at the β strand -loop boundary, and this property appears to be conserved for Dna2 and Dpb11TopBP1 (P. Wanrooij, unpublished observations). However, further studies are required in order for us to gain full understanding of the specificity and mechanism of Mec1ATR activation.
Understanding the distinctive roles of the three different Mec1ATR activators when and where chromosomes are challenged, along with mechanistic insight of their interaction with this essential kinase, are intriguing directions for future investigation.
Acknowledgments
Work in the laboratory of P.B. is supported by grants GM032431 and GM083970 from the National Institutes of Health. P.W. is funded by the Emil Aaltonen Foundation and the Swedish Cultural Foundation in Finland.
Abbreviations
- 9-1-1
human Rad9, Rad1, Hus1 (yeast Ddc1, Rad17, Mec3) checkpoint clamp
- ATM
ataxia-telangiectasia mutated
- ATR
ATM and Rad3 –related
- ATRIP
ATR interacting protein
- DSB
double-strand break
- dsDNA
double-stranded DNA
- ssDNA
single-stranded DNA
- FEN1
flap endonuclease 1
- NTD
N-terminal domain
- PIKK
phosphatidylinositol 3-kinase-related protein kinase
- RPA
replication protein A
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Budd ME, Campbell JL. A yeast gene required for DNA replication encodes a protein with homology to DNA helicases. Proc Natl Acad Sci USa. 1995;92:7642–7646. doi: 10.1073/pnas.92.17.7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuo C, Nuang H, Campbell JL. Isolation of yeast DNA replication mutants in permeabilized cells. Proc Natl Acad Sci USa. 1983;80:6465–6469. doi: 10.1073/pnas.80.21.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Budd ME, Choe WC, Campbell JL. DNA2 encodes a DNA helicase essential for replication of eukaryotic chromosomes. J Biol Chem. 1995;270:26766–26769. doi: 10.1074/jbc.270.45.26766. [DOI] [PubMed] [Google Scholar]
- 4.Bae SH, Choi E, Lee KH, Park JS, Lee SH, Seo YS. Dna2 of Saccharomyces cerevisiae possesses a single-stranded DNA-specific endonuclease activity that is able to act on double-stranded DNA in the presence of ATP. J Biol Chem. 1998;273:26880–26890. doi: 10.1074/jbc.273.41.26880. [DOI] [PubMed] [Google Scholar]
- 5.Formosa T, Nittis T. Dna2 mutants reveal interactions with Dna polymerase alpha and Ctf4, a Pol alpha accessory factor, and show that full Dna2 helicase activity is not essential for growth. Genetics. 1999;151:1459–1470. doi: 10.1093/genetics/151.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Budd ME, Choe WC, Campbell JL. The nuclease activity of the yeast DNA2 protein, which is related to the RecB-like nucleases, is essential in vivo. J Biol Chem. 2000;275:16518–16529. doi: 10.1074/jbc.M909511199. [DOI] [PubMed] [Google Scholar]
- 7.Ayyagari R, Gomes XV, Gordenin DA, Burgers PMJ. Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 AND DNA2. J Biol Chem. 2003;278:1618–1625. doi: 10.1074/jbc.M209801200. [DOI] [PubMed] [Google Scholar]
- 8.Bae SH, Seo YS. Characterization of the enzymatic properties of the yeast dna2 Helicase/endonuclease suggests a new model for Okazaki fragment processing. J Biol Chem. 2000;275:38022–38031. doi: 10.1074/jbc.M006513200. [DOI] [PubMed] [Google Scholar]
- 9.Bae SH, Bae KH, Kim JA, Seo YS. RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature. 2001;412:456–461. doi: 10.1038/35086609. [DOI] [PubMed] [Google Scholar]
- 10.Kumar S, Burgers PM. Lagging strand maturation factor Dna2 is a component of the replication checkpoint initiation machinery. Genes Dev. 2013;27:313–321. doi: 10.1101/gad.204750.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Q, Choe W, Campbell JL. Identification of the Xenopus laevis homolog of Saccharomyces cerevisiae DNA2 and its role in DNA replication. J Biol Chem. 2000;275:1615–1624. doi: 10.1074/jbc.275.3.1615. [DOI] [PubMed] [Google Scholar]
- 12.Eki T, Okumura K, Shiratori A, Abe M, Nogami M, Taguchi H, et al. Assignment of the closest human homologue (DNA2L:KIAA0083) of the yeast Dna2 helicase gene to chromosome band 10q21.3-q22.1. Genomics. 1996;37:408–410. doi: 10.1006/geno.1996.0581. [DOI] [PubMed] [Google Scholar]
- 13.Gould KL, Burns CG, Feoktistova A, Hu CP, Pasion SG, Forsburg SL. Fission yeast cdc24(+) encodes a novel replication factor required for chromosome integrity. Genetics. 1998;149:1221–1233. doi: 10.1093/genetics/149.3.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cejka P, Cannavo E, Polaczek P, Masuda-Sasa T, Pokharel S, Campbell JL, et al. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature. 2010;467:112–116. doi: 10.1038/nature09355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niu H, Chung W-H, Zhu Z, Kwon Y, Zhao W, Chi P, et al. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature. 2010;467:108–111. doi: 10.1038/nature09318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balakrishnan L, Polaczek P, Pokharel S, Campbell JL, Bambara RA. Dna2 exhibits a unique strand end-dependent helicase function. J Biol Chem. 2010;285:38861–38868. doi: 10.1074/jbc.M110.165191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kao H-I, Campbell JL, Bambara RA. Dna2p helicase/nuclease is a tracking protein, like FEN1, for flap cleavage during Okazaki fragment maturation. J Biol Chem. 2004;279:50840–50849. doi: 10.1074/jbc.M409231200. [DOI] [PubMed] [Google Scholar]
- 18.Stewart JA, Campbell JL, Bambara RA. Dna2 is a structure-specific nuclease, with affinity for 5′-flap intermediates. Nucleic Acids Research. 2010;38:920–930. doi: 10.1093/nar/gkp1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masuda-Sasa T, Imamura O, Campbell JL. Biochemical analysis of human Dna2. Nucleic Acids Research. 2006;34:1865–1875. doi: 10.1093/nar/gkl070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masuda-Sasa T, Polaczek P, Campbell JL. Single strand annealing and ATP-independent strand exchange activities of yeast and human DNA2: possible role in Okazaki fragment maturation. J Biol Chem. 2006;281:38555–38564. doi: 10.1074/jbc.M604925200. [DOI] [PubMed] [Google Scholar]
- 21.Kim J-H, Kim H-D, Ryu G-H, Kim D-H, Hurwitz J, Seo Y-S. Isolation of human Dna2 endonuclease and characterization of its enzymatic properties. Nucleic Acids Research. 2006;34:1854–1864. doi: 10.1093/nar/gkl102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levikova M, Klaue D, Seidel R, Cejka P. Nuclease activity of Saccharomyces cerevisiae Dna2 inhibits its potent DNA helicase activity. Proc Natl Acad Sci USA. 2013 doi: 10.1073/pnas.1300390110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng L, Zhou M, Guo Z, Lu H, Qian L, Dai H, et al. Human DNA2 Is a Mitochondrial Nuclease/Helicase for Efficient Processing of DNA Replication and Repair Intermediates. Mol Cell. 2008;32:325–336. doi: 10.1016/j.molcel.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bae SH, Kim JA, Choi E, Lee KH, Kang HY, Kim HD, et al. Tripartite structure of Saccharomyces cerevisiae Dna2 helicase/endonuclease. Nucleic Acids Research. 2001;29:3069–3079. doi: 10.1093/nar/29.14.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee C-H, Lee M, Kang H-J, Kim D-H, Kang Y-H, Bae S-H, et al. The N-terminal 45-kDa Domain of Dna2 Endonuclease/Helicase Targets the Enzyme to Secondary Structure DNA. Journal of Biological Chemistry. 2013;288:9468–9481. doi: 10.1074/jbc.M112.418715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pokharel S, Campbell JL. Cross talk between the nuclease and helicase activities of Dna2: role of an essential iron-sulfur cluster domain. Nucleic Acids Research. 2012;40:7821–7830. doi: 10.1093/nar/gks534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stith CM, Sterling J, Resnick MA, Gordenin DA, Burgers PM. Flexibility of eukaryotic Okazaki fragment maturation through regulated strand displacement synthesis. J Biol Chem. 2008;283:34129–34140. doi: 10.1074/jbc.M806668200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murante RS, Rust L, Bambara RA. Calf 5′ to 3′ exo/endonuclease must slide from a 5′ end of the substrate to perform structure-specific cleavage. J Biol Chem. 1995;270:30377–30383. doi: 10.1074/jbc.270.51.30377. [DOI] [PubMed] [Google Scholar]
- 29.Pike JE, Burgers PMJ, Campbell JL, Bambara RA. Pif1 helicase lengthens some Okazaki fragment flaps necessitating Dna2 nuclease/helicase action in the two-nuclease processing pathway. Journal of Biological Chemistry. 2009;284:25170–25180. doi: 10.1074/jbc.M109.023325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burgers PMJ. Polymerase Dynamics at the Eukaryotic DNA Replication Fork. Journal of Biological Chemistry. 2008;284:4041–4045. doi: 10.1074/jbc.R800062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Budd ME, Tong AHY, Polaczek P, Peng X, Boone C, Campbell JL. A Network of Multi-Tasking Proteins at the DNA Replication Fork Preserves Genome Stability. PLoS Genet. 2005;1:e61. doi: 10.1371/journal.pgen.0010061.st004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Budd ME, Reis CC, Smith S, Myung K, Campbell JL. Evidence suggesting that Pif1 helicase functions in DNA replication with the Dna2 helicase/nuclease and DNA polymerase delta. Molecular and Cellular Biology. 2006;26:2490–2500. doi: 10.1128/MCB.26.7.2490-2500.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Budd ME, Antoshechkin IA, Reis C, Wold BJ, Campbell JL. Inviability of a DNA2 deletion mutant is due to the DNA damage checkpoint. Cell Cycle. 2011;10:1690–1698. doi: 10.4161/cc.10.10.15643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu Z, Chung W-H, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, et al. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011;25:350–362. doi: 10.1101/gad.2003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Symington LS, Gautier J. Double-Strand Break End Resection and Repair Pathway Choice. Annu Rev Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- 37.Mimitou EP, Symington LS. DNA end resection: many nucleases make light work. DNA Repair (Amst) 2009;8:983–995. doi: 10.1016/j.dnarep.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choe W, Budd M, Imamura O, Hoopes L, Campbell JL. Dynamic localization of an Okazaki fragment processing protein suggests a novel role in telomere replication. Molecular and Cellular Biology. 2002;22:4202–4217. doi: 10.1128/MCB.22.12.4202-4217.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin W, Sampathi S, Dai H, Liu C, Zhou M, Hu J, et al. Mammalian DNA2 helicase/nuclease cleaves G-quadruplex DNA and is required for telomere integrity. The EMBO Journal. 2013;32:1425–1439. doi: 10.1038/emboj.2013.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps H. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat Struct Mol Biol. 2005;12:847–854. doi: 10.1038/nsmb982. nsmb982 [pii] [DOI] [PubMed] [Google Scholar]
- 41.Paeschke K, Capra JA, Zakian VA. DNA Replication through G-Quadruplex Motifs Is Promoted by the Saccharomyces cerevisiae Pif1 DNA Helicase. Cell. 2011;145:678–691. doi: 10.1016/j.cell.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Masuda-Sasa T, Polaczek P, Peng X, Chen L, Campbell J. Processing of G4 DNA by Dna2 helicase/nuclease and replication protein A (RPA) provides insights into the mechanism of Dna2/RPA substrate recognition. J Biol Chem. 2008;283:24359–24373. doi: 10.1074/jbc.M802244200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schulz VP, Zakian VA. The saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell. 1994;76:145–155. doi: 10.1016/0092-8674(94)90179-1. [DOI] [PubMed] [Google Scholar]
- 44.Duxin J, Dao B, Martinsson P, Rajala N, Guittat L, Campbell J, et al. Human Dna2 is a nuclear and mitochondrial DNA maintenance protein. Mol Cell Biol. 2009;29:4274–4282. doi: 10.1128/MCB.01834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ronchi D, Di Fonzo A, Lin W, Bordoni A, Liu C, Fassone E, et al. Mutations in DNA2 Link Progressive Myopathy to Mitochondrial DNA Instability. The American Journal of Human Genetics. 2013 doi: 10.1016/j.ajhg.2012.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu J, Sun L, Shen F, Chen Y, Hua Y, Liu Y, et al. The Intra-S Phase Checkpoint Targets Dna2 to Prevent Stalled Replication Forks from Reversing. Cell. 2012;149:1221–1232. doi: 10.1016/j.cell.2012.04.030. [DOI] [PubMed] [Google Scholar]
- 47.Duxin JP, Moore HR, Sidorova J, Karanja K, Honaker Y, Dao B, et al. Okazaki Fragment Processing-independent Role for Human Dna2 Enzyme during DNA Replication. Journal of Biological Chemistry. 2012;287:21980–21991. doi: 10.1074/jbc.M112.359018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thangavel S, Berti M, Levikova M, Pinto C, Gomathinayagam S, Vujanovic M, et al. DNA2 drives processing and restart of reversed replication forks in human cells. J Cell Biol. 2015;208:545–562. doi: 10.1083/jcb.201406100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harrison JC, Haber JE. Surviving the breakup: the DNA damage checkpoint. Annu Rev Genet. 2006;40:209–235. doi: 10.1146/annurev.genet.40.051206.105231. [DOI] [PubMed] [Google Scholar]
- 50.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 51.O’Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat Genet. 2003;33:497–501. doi: 10.1038/ng1129. [DOI] [PubMed] [Google Scholar]
- 52.Zou L, Elledge SJ. Sensing DNA Damage Through ATRIP Recognition of RPA-ssDNA Complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 53.Maser RS, Monsen KJ, Nelms BE, Petrini JH. hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Molecular and Cellular Biology. 1997;17:6087–6096. doi: 10.1128/mcb.17.10.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Craven RJ, Greenwell PW, Dominska M, Petes TD. Regulation of genome stability by TEL1 and MEC1, yeast homologs of the mammalian ATM and ATR genes. Genetics. 2002;161:493–507. doi: 10.1093/genetics/161.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jazayeri A, Falck J, Lukas C, Bartek J, Smith GCM, Lukas J, et al. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- 56.Shiotani B, Zou L. Single-stranded DNA orchestrates an ATM-to-ATR switch at DNA breaks. Mol Cell. 2009;33:547–558. doi: 10.1016/j.molcel.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 58.Lee J-H, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 59.Paciotti V, Clerici M, Lucchini G, Longhese MP. The checkpoint protein Ddc2, functionally related to S. pombe Rad26, interacts with Mec1 and is regulated by Mec1-dependent phosphorylation in budding yeast. Genes Dev. 2000;14:2046–2059. [PMC free article] [PubMed] [Google Scholar]
- 60.Majka J, Niedziela-Majka A, Burgers PMJ. The checkpoint clamp activates Mec1 kinase during initiation of the DNA damage checkpoint. Mol Cell. 2006;24:891–901. doi: 10.1016/j.molcel.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ball HL, Cortez D. ATRIP oligomerization is required for ATR-dependent checkpoint signaling. J Biol Chem. 2005;280:31390–31396. doi: 10.1074/jbc.M504961200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rouse J, Jackson SP. Lcd1p recruits Mec1p to DNA lesions in vitro and in vivo. Mol Cell. 2002;9:857–869. doi: 10.1016/s1097-2765(02)00507-5. [DOI] [PubMed] [Google Scholar]
- 63.Ball HL, Ehrhardt MR, Mordes DA, Glick GG, Chazin WJ, Cortez D. Function of a conserved checkpoint recruitment domain in ATRIP proteins. Molecular and Cellular Biology. 2007;27:3367–3377. doi: 10.1128/MCB.02238-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang H, Elledge SJ. Genetic and physical interactions between DPB11 and DDC1 in the yeast DNA damage response pathway. Genetics. 2002;160:1295–1304. doi: 10.1093/genetics/160.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Longhese MP, Paciotti V, Fraschini R, Zaccarini R, Plevani P, Lucchini G. The novel DNA damage checkpoint protein ddc1p is phosphorylated periodically during the cell cycle and in response to DNA damage in budding yeast. Embo J. 1997;16:5216–5226. doi: 10.1093/emboj/16.17.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pellicioli A, Lucca C, Liberi G, Marini F, Lopes M, Plevani P, et al. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. Embo J. 1999;18:6561–6572. doi: 10.1093/emboj/18.22.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Navadgi Patil VM, Burgers PM. The unstructured C-terminal tail of the 9-1-1 clamp subunit Ddc1 activates Mec1/ATR via two distinct mechanisms. Mol Cell. 2009;36:743–753. doi: 10.1016/j.molcel.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Doré AS, Kilkenny ML, Rzechorzek NJ, Pearl LH. Crystal structure of the rad9-rad1-hus1 DNA damage checkpoint complex--implications for clamp loading and regulation. Mol Cell. 2009;34:735–745. doi: 10.1016/j.molcel.2009.04.027. [DOI] [PubMed] [Google Scholar]
- 69.Sohn SY, Cho Y. Crystal structure of the human rad9-hus1-rad1 clamp. J Mol Biol. 2009;390:490–502. doi: 10.1016/j.jmb.2009.05.028. [DOI] [PubMed] [Google Scholar]
- 70.Majka J, Burgers PMJ. Yeast Rad17/Mec3/Ddc1: a sliding clamp for the DNA damage checkpoint. Proc Natl Acad Sci USa. 2003;100:2249–2254. doi: 10.1073/pnas.0437148100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ellison V, Stillman B. Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5′ recessed DNA. PLoS Biol. 2003;1:E33. doi: 10.1371/journal.pbio.0000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Majka J, Binz SK, Wold MS, Burgers PMJ. Replication protein A directs loading of the DNA damage checkpoint clamp to 5′-DNA junctions. J Biol Chem. 2006;281:27855–27861. doi: 10.1074/jbc.M605176200. [DOI] [PubMed] [Google Scholar]
- 73.Pfander B, Diffley JFX. Dpb11 coordinates Mec1 kinase activation with cell cycle-regulated Rad9 recruitment. The EMBO Journal. 2011;30:4897–4907. doi: 10.1038/emboj.2011.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Furuya K, Poitelea M, Guo L, Caspari T, Carr AM. Chk1 activation requires Rad9 S/TQ-site phosphorylation to promote association with C-terminal BRCT domains of Rad4TOPBP1. Genes Dev. 2004;18:1154–1164. doi: 10.1101/gad.291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Puddu F, Granata M, Di Nola L, Balestrini A, Piergiovanni G, Lazzaro F, et al. Phosphorylation of the budding yeast 9-1-1 complex is required for Dpb11 function in the full activation of the UV-induced DNA damage checkpoint. Mol Cell Biol. 2008;28:4782–4793. doi: 10.1128/MCB.00330-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Delacroix S, Wagner JM, Kobayashi M, Yamamoto K-I, Karnitz LM. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 2007;21:1472–1477. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee J, Kumagai A, Dunphy WG. The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. J Biol Chem. 2007;282:28036–28044. doi: 10.1074/jbc.M704635200. [DOI] [PubMed] [Google Scholar]
- 78.Navadgi Patil VM, Kumar S, Burgers PM. The unstructured C-terminal tail of yeast Dpb11 (human TopBP1) protein is dispensable for DNA replication and the S phase checkpoint but required for the G2/M checkpoint. Journal of Biological Chemistry. 2011;286:40999–41007. doi: 10.1074/jbc.M111.283994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Morrow DM, Tagle DA, Shiloh Y, Collins FS, Hieter P. TEL1, an S. cerevisiae homolog of the human gene mutated in ataxia telangiectasia, is functionally related to the yeast checkpoint gene MEC1. Cell. 1995;82:831–840. doi: 10.1016/0092-8674(95)90480-8. [DOI] [PubMed] [Google Scholar]
- 80.Myung K, Kolodner RD. Suppression of genome instability by redundant S-phase checkpoint pathways in Saccharomyces cerevisiae. Proc Natl Acad Sci USa. 2002;99:4500–4507. doi: 10.1073/pnas.062702199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhao X, Muller EG, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
- 82.Chabes A, Georgieva B, Domkin V, Zhao X, Rothstein R, Thelander L. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell. 2003;112:391–401. doi: 10.1016/s0092-8674(03)00075-8. [DOI] [PubMed] [Google Scholar]
- 83.Mallory JC, Petes TD. Protein kinase activity of Tel1p and Mec1p, two Saccharomyces cerevisiae proteins related to the human ATM protein kinase. Proc Natl Acad Sci USa. 2000;97:13749–13754. doi: 10.1073/pnas.250475697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lopes M, Foiani M, Sogo JM. multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]