Abstract
DNA mismatch repair (MMR) acts to repair mispaired bases resulting from misincorporation errors during DNA replication and also recognizes mispaired bases in recombination (HR) intermediates. Exonuclease 1 (Exo1) is a 5′ –3′ exonuclease that participates in a number of DNA repair pathways. Exo1 was identified as an exonuclease that participates in Saccharomyces cerevisiae and human MMR where it functions to excise the daughter strand after mispair recognition, and additionally Exo1 functions in end resection during HR. However, Exo1 is not absolutely required for end resection during HR in vivo. Similarly, while Exo1 is required in MMR reactions that have been reconstituted in vitro, genetics studies have shown that it is not absolutely required for MMR in vivo suggesting the existence of Exo1-independent and Exo1-dependent MMR subpathways. Here, we review what is known about the Exo1-independent and Exo1-dependent subpathways, including studies of mutations in MMR genes that specifically disrupt either subpathway.
Keywords: Msh2–Msh6, Mlh1–Pms1, Mutagenesis, DNA replication fidelity, Mispaired base, Excision, Recombination
1. Exonuclease 1 – redundant roles in multiple DNA metabolism pathways
Exonuclease 1 (Exo1) is a member of the Rad2 family of exonucleases and possesses 5′ –3′ double stranded DNA exonuclease and flap endonuclease activities [1]. Exo1 has an N-terminal catalytic domain that is conserved in other Rad2 family proteins [2], and the C-terminus is predicted to be largely unstructured but is involved in protein–protein interactions (Fig. 1). Exo1 was first identified in Schizosaccharomyces pombe as a meiotic 5′ –3′ exonuclease [3,4]. Exo1 is now known to be involved in multiple pathways for DNA metabolism and repair, including mismatch repair, mitotic and meiotic recombination, Okazaki fragment maturation, response to UV damage, and telomere processing and maintenance.
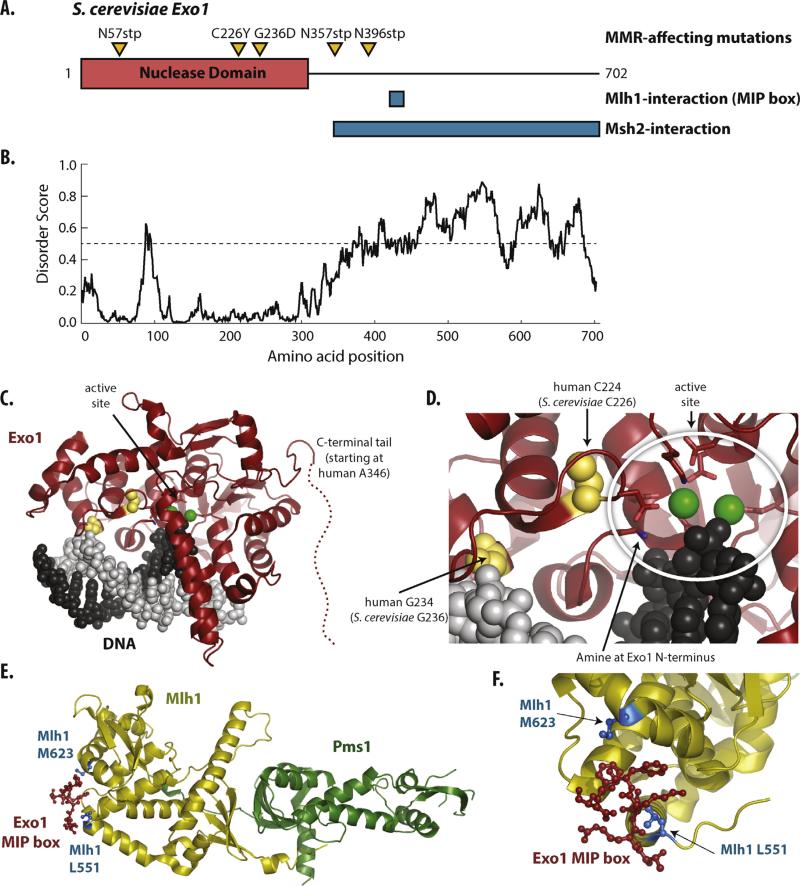
Fig. 1. Exonuclease 1.
(A) Schematic representation of S. cerevisiae Exo1 (702 residues long) depicting the endonuclease domain as a red box and the C-terminal tail as a black line. The positions of amino acids affected by mutations in the EXO1 gene that disrupt Exo1-dependent MMR [13] are shown as orange triangles. Interaction regions with Mlh1 (the MIP box) and Msh2 are shown as blue boxes [5,8]. (B) Predicted long-range disorder for S. cerevisiae Exo1 calculated by IUPRED [113] suggests that the C-terminal tail is largely disordered. (C) Structure of the nuclease domain of human Exo1 in complex with DNA (PDB id 3qea, [2]) is shown with the nuclease domain in red and the DNA strands as light and dark grey. Metal bound to the nuclease active site are displayed as green spheres. (D) Expanded view of the structure with active site residues, including the N-terminal amine, displayed as sticks, and the amino acid substitutions caused by the Exo1-dependent MMR defective mutations shown in yellow. The G236D amino acid substitution likely disrupts an interaction between the Exo1 helix-two-turn-helix (H2TH) motif and the uncleaved strand, and the C226Y amino acid substitution likely has steric interference that disrupts the enzyme active site. (E) Structure of interaction of the S. cerevisiae Exo1 MIP box (red) with the C-terminal domains of S. cerevisiae Mlh1–Pms1 (PDB id 4fmo; [9]). (F) Expanded view of the MIP box–Mlh1 interaction with the amino acid positions that are affected by mutations that disrupt Exo1-dependent MMR (L551 and M623; [13]) shown as blue ball-and-sticks.
Exo1 was identified as a component of Saccharomyces cerevisiae DNA mismatch repair (MMR) due to its physical interaction with the MMR protein Msh2 and the fact that exo1Δ mutations caused a weak mutator phenotype that was epistatic to that caused by an msh2Δ mutation, and human Exo1 was subsequently identified by its homology to S. cerevisiae Exo1 [5–7]. Exo1 binds Msh2 through its C-terminal tail (S. cerevisiae amino acids 368–702) [5] and binds the MMR protein Mlh1 through a conserved Mlh1– interacting protein (MIP) box (S. cerevisiae amino acids 443–448), which is also in the C-terminal tail [8,9] (Fig. 1). Most of the human and S. cerevisiae MMR reactions that have been reconstituted in vitro require Exo1 [10–12]; however, genetics experiments indicate that Exo1 is not absolutely required for MMR in vivo and is at least partially redundant with other proteins that function in MMR [5,13,14].
Exo1 is not strictly required in the other DNA metabolism and repair pathways that it participates in, similar to its redundant role in MMR. The EXO1 gene was discovered to be a high copy suppressor of defects in the Mre11-Rad50-Xrs2 complex [15], which along with other studies have suggested that Exo1 plays a role in recombination [3,4]. Exo1 plays a major, but redundant, role in the resection of double-stranded DNA breaks to generate a 3′ single-stranded DNA tail that is the critical substrate in the initiation of recombination. In the resection of mitotic double-stranded DNA breaks, Exo1 extends the 3′ single-stranded DNA tail at a step downstream of the initial short-range resection by Mre11-Rad50-Xrs2 (MRE11-RAD50-NBS1 in humans) and Sae2 (CtIP in humans) [16]. The role of Exo1 in this long-range resection, however, is redundant with the combined action of the Sgs1 helicase (BLM in humans) and the Dna2 nuclease, as inactivation of both the EXO1 and SGS1 pathways is required to eliminate long-range resection [17–19]. In the resection of meiotic double-strand breaks, loss of Exo1-mediated resection in S. cerevisiae results in a modest loss of spore viability, a modest increase in chromosome nondisjunction during meiosis I, and a decrease in crossing over at some alleles, but little change to heteroduplex formation [15,20–22]. Thus, EXO1 must also be redundant with other resection pathways in meiosis in S. cerevisiae. In contrast, Exo1−/− mice are sterile due to failure of meiosis in spermatocytes and oocytes indicating that Exo1 is most likely a required component for germ cell maturation in mammals [14].
In addition to the redundant roles of Exo1 in MMR and resection of double-stranded DNA breaks, Exo1 has been implicated in other pathways of DNA metabolism as a result of the interaction of exo1 mutations with other defects in DNA metabolism. During Okazaki fragment maturation, flap endonuclease 1 (FEN1, called Rad27 in S. cerevisiae) cleaves off the RNA primer-containing flap; however, Exo1 appears to provide this activity in rad27Δ strains. Deletion of both EXO1 and RAD27 is lethal, whereas overexpression of EXO1 complements the temperature sensitivity and accumulation of Okazaki fragments of a rad27Δ mutant [5,7,23]. In the repair of DNA damaged by ultraviolet (UV) light, most repair is mediated by a nucleotide-excision repair (NER) pathway that involves the Rad2 nuclease; however, deletion of EXO1, which is a UV-inducible gene, results in mild UV sensitivity due to loss of a repair pathway that is distinct from the Rad2-dependent NER pathway [24]. In this repair pathway, Exo1 is thought to extend a subset of repair intermediate gaps generated but not fully repaired by NER leading to DNA damage checkpoint activation [25–27] and repair by recombination [28]. Although Exo1 does not seem to play a pronounced role in telomere maintenance in wild-type cells, during maintenance of telomeres that are uncapped by mutation of YKU70, YKU80, or CDC13, Exo1 resects the exposed chromosomal ends, which leads to shortened telomeres and temperature sensitivity [29,30]. This activity of Exo1 at uncapped telomeres is thought to promote telomere maintenance by recombination in these mutant backgrounds (reviewed in [1]). Thus, most of the characterized roles of Exo1 involve resection that is either redundant with other mechanisms, such as in MMR, or functions as an alternative to pathways when they are compromised by mutations.
2. DNA mismatch repair
MMR promotes genome stability by repairing base–base and insertion/deletion mispairs resulting from DNA replication errors or recombination between DNAs that contain sequence differences. Unrepaired mispairs result in mutations after being replicated in the next cell cycle, and, thus, MMR functions to suppress the accumulation of mutations [31–33]. Inherited MMR defects underlie the human hereditary cancer predisposition syndrome Lynch syndrome (also called hereditary non-polyposis colon cancer or HNPCC; [34–36], and somatic genetic and epigenetic inactivation of MMR genes occurs in many spontaneous tumors [37–40].
MMR is best understood in the bacterium Escherichia coli (reviewed in [41]). The mutS gene was originally identified as a gene that suppresses the accumulation of transition mutations in E. coli [42] and was later found to encode the protein homodimer responsible for the first step of mismatch repair, recognition and binding to the DNA mispair [43]. MutS binding to the mismatch allows recruitment of the MutL protein [44], which can then activate the MutH endonuclease to generate a single-stranded break (e.g., a nick) on the newly synthesized DNA strand at d(GATC) sites [45–48]. These sites can be up to 1–2 kb away from the mispair either 5′ or 3′ of the mispair [48]. Strand discrimination by MutH involves nicking of the unmethylated strand of hemi-methylated d(GATC) sites [45,46]. These sites exist transiently after replication of a fully methylated site and before post-replicative methylation of the newly synthesized strand at d(GATC) sites by the Dam methyltransferase [49–51]. MutL also recruits and activates the UvrD 3′ ≥ 5′ helicase at MutH-generated nicks which displaces the newly synthesized strand [52,53] which is then degraded by one or more of the ExoVII, RecJ, ExoI, and ExoX nucleases depending on the position of the MutH generated nick relative to the mispair in either the 5′ or 3′ direction [54,55]. The resulting gap is then filled by DNA polymerase III and final nick is repaired by DNA ligase [56].
Despite the mechanistic detail known for the E. coli methyl-directed MMR system, E. coli MMR is substantially different from MMR in most other bacteria and eukaryotes. However, most MMR systems share universally conserved steps: (1) mispair recognition by MutS homologs, (2) MutS- and mispair-promoted recruitment of MutL homologs, (3) excision of the newly synthesized strand in a region containing the mispair, and (4) gap filling/repair by DNA synthesis. Use of DNA methylation to identify the newly synthesized strand is restricted to E. coli and closely related gammaproteobacteria [57–59]. How other organisms distinguish the newly synthesized strand from the template strand, which is required to prevent mutation accumulation, is not well understood.
2.1. Eukaryotic MutS homologs
Homodimeric MutS bound to mispairs shows functional asymmetry with one subunit directly recognizing the mispair and one subunit interacting with the DNA backbone [60,61]. In eukaryotes, there are multiple MutS homologs arising from gene duplication and specialization, which interact to form heterodimers. The MutS complexes acting in MMR in fungi and mammals are the Msh2–Msh6 and Msh2–Msh3 heterodimers; plants have an additional heterodimer, Msh2–Msh7, where Msh7 is a Msh6 paralog [32,62–64]. Msh2 is the common subunit, which does not directly interact with mispairs, and deletion of the gene that encodes Msh2 results in a complete loss of MMR [64–67]. In the Msh2–Msh6 heterodimer, Msh6 recognizes base–base mispairs and small insertion/deletions of 1–2 nucleotides, although the C:C mispair is poorly recognized [64,68–70]. In the Msh2–Msh3 heterodimer, Msh3 recognizes some base–base mispairs, specifically A:A, C:C, and to a lesser extent T:G, but primarily recognizes insertion/deletions of 1–14 nucleotides [64,69,71–74]. These differences in recognition stem from changes in the mispair-binding domain of Msh6 and Msh3; a view that is supported by the observation that a chimeric Msh6 with the mispair-binding domain from Msh3 shows Msh3-like mispair recognition specificity [75]. Msh6, like bacterial MutS, extrudes a base at the mispair from the DNA helix and stabilizes it by π-stacking with a conserved phenylalanine [60,61,65]. In contrast, mispair recognition by Msh3 involves using residues as a steric wedge to mediate strand separation, with smaller insertion/deletion loops and base-base mispairs requiring more stabilization by adjacent amino acids than larger insertion deletion loops [76,77]. MutS and MutS homologs also have an ABC ATPase domain. Mispair binding primes Msh2–Msh6 and Msh2–Msh3 to undergo a conformational change upon binding ATP that allows these molecules to recruit MutL homologs and to form clamps that can slide away from the mispair [78,79].
2.2. Eukaryotic MutL homologs
The eukaryotic MutL homologs have also evolved as a result of gene duplication and specialization, and function in MMR as heterodimeric complexes. The primary MutL heterodimeric complex involved in eukaryotic MMR is Mlh1–Pms1 in S. cerevisiae (known as Mlh1–Pms2 in mammals) [80,81], and consistent with this, the genes encoding Mlh1 and Pms1 are required for the bulk of MMR. Eukaryotes also contain additional MutL homologs that play minor roles in MMR, including Mlh1-Mlh3, which also has roles in meiosis [82,83], and Mlh1–Mlh2 in S. cerevisiae (known as Mlh1–Pms1 in mammals) that appears to function as a non-essential accessory factor that helps to promote MMR [84]. Like bacterial MutL, Mlh1–Pms1 is recruited to DNA by both Msh2–Msh6 and Msh2–Msh3 in a mispair- and ATP-promoted reaction [73,79,85,86], and Mlh1–Mlh2 is similarly recruited by both Msh2–Msh6 and Msh2–Msh3 [84]. The Mlh1–Pms1 complex also has an endonuclease activity that is promoted by RFC, PCNA and Msh2–Msh6 [87,88] and is important for MMR, as endonuclease active site mutations inactivate MMR in vivo [88,89]. Similarly, in most organisms, excepting those with methyl-directed MMR, MutL also has an endonuclease activity. In contrast, Mlh1–Mlh2 is not likely to be an endonuclease [84]. Many MMR models have suggested the presence of a stable “ternary” complex between MutS (or MutS homologs) with MutL (or MutL homologs) and DNA [41,79,85,90]; however, recent analyses in vivo discussed below and the fact that stabilization of the interaction in vitro requires crosslinking suggest a transient interaction between MutS homologs and MutL homologs that may be an intermediate in the catalytic loading of MutL (or MutL homologs) onto DNA by MutS (or MutS homologs) in response to mispairs [91,92].
2.3. Strand discrimination
Specific targeting of the newly synthesized DNA strand for excision and resynthesis is necessary to prevent mispairs from leading to mutations. In the methyl-directed MMR system of E. coli, the transient hemi-methylated status of d(GATC) sites, which persist less than 10 min after DNA replication [49,50], serves as the crucial signal. Although the signal for strand discrimination has not been unambiguously identified in organisms without methyl-directed MMR, a number of studies suggest that the mechanism is similarly linked to replication timing and that some features of the replication intermediates themselves may be the strand discrimination signal.
Eukaryotic MMR is coupled to the DNA replication machinery. Msh6 and Msh3 bind to the PCNA (proliferating cell nuclear antigen) [93–95], which is a processivity factor for DNA polymerase δ [96,97] suggesting that the Msh2–Msh6 and Msh2–Msh3 complexes interact with PCNA located at replication forks or left on the DNA after replication. More recently, live cell imaging in S. cerevisiae has shown that functional Msh2–Msh6 forms foci that colocalize with components of the replication fork in essentially 100% of S-phase cells in a mispair-independent, PCNA-Msh6 interaction-dependent fashion [92]. In contrast, live cell imaging in S. cerevisiae has shown that Mlh1–Pms1, as well as Mlh1–Mlh2, form foci, which do not colocalize with Msh2–Msh6 foci or replication factories [84,92]. Mlh1–Pms1 foci were present in ~10% of asynchronous wild-type cells and the formation of these foci required Msh2–Msh6. Furthermore, mutations that increased the production of mispairs or disrupted MMR downstream of mispair recognition and Mlh1–Pms1 recruitment, such as deletion of EXO1, led to an increase in the percentage of cells with Mlh1–Pms1 foci [92]. Together these results suggest that Msh2–Msh6 is a constitutive component of replication factories where it acts to detect mispairs formed during replication and that the Mlh1–Pms1 foci are MMR intermediates that appear to be formed by multiple rounds of loading by one or more MutS homolog complexes in response to mispaired bases. The ability to use tagged functional MMR proteins to observe a repair intermediate in vivo opens new exciting experimental possibilities, as mutator assays only monitor the complete MMR reaction.
By temporally restricting the availability of Msh6 to different phases of the cell cycle by fusing it to cell-cycle specific cyclins in an msh3Δ S. cerevisiae strain, MMR in eukaryotes was also shown to only act in a narrow post-replicative window of opportunity similar to E. coli MMR [98]. The accumulation of mutations at a frameshift reversion assay placed at a region of the genome replicated in mid S-phase could be prevented when Msh6 was expressed in S-phase, but not when Msh6 was expressed in G2/M. In contrast, Msh6 expression in G2/M, but not S-phase, prevented the accumulation of mutations in the frameshift reversion assay when the assay was placed at a very late replicating region of the genome. From these data, the window of time for MMR proficiency can be estimated to be on the order of 15 min after the region of the genome was replicated, although the temporal resolution was limited by expression profiles of the Msh6-cyclin constructs [98]. Intriguingly, heteroduplex rejection during recombination between divergent sequences, another Msh2–Msh6–dependent reaction, was not restricted to the same temporal window [98]. Together these observations are consistent with replication-generated single-stranded DNA nicks, gaps, or other DNA structures as possible strand discrimination signals.
Multiple studies have proposed that the signal for strand discrimination may be replication-generated nicks in the genome, consistent with the requirement for a pre-existing nick for reconstitution of eukaryotic MMR in vitro [12,99–101]. Proposed sources of nicks include Okazaki fragments on the lagging strand and RNase H2-mediated cleavage of misincorporated ribonucleotides on the leading strand [102,103]. While nicks generated from ribonucleotides may be the signal for some repair reactions, the mutation rates caused by deletion of the genes encoding RNase H2 are orders of magnitude lower than those expected to be caused by a universal strand discrimination signal defect. In addition, an absolute requirement for pre-existing nicks seems at odds with the absolute requirement for the nick-generating endonuclease function of MutL homologs in eukaryotic MMR [87–89]. PCNA, whether left on the substrate or loaded at nicks, has also been suggested to be a strand discrimination signal [95,101]; however, for PCNA to be a signal, it must loaded onto the DNA with a fixed polarity with regards to the nicked strand and interact with Mlh1–Pms1 in such a way that Mlh1–Pms1 can only nick the already nicked strand; while this is what appears to happen in vitro, it is not clear how the Mlh1–Pms1 endonuclease is targeted to cleave only the pre-nicked strand. The difficulties in finding the strand discrimination signal, particularly on the leading strand, indicate that the reconstituted MMR reactions in vitro do not fully recapitulate MMR in vivo. Other aspects of MMR reactions reconstituted in vitro that appear different from MMR in vivo, include the ability to perform MutL homolog-independent MMR in vitro in contrast to an absolute requirement for MutL homologs in vivo [12,99,100], and the absolute requirement for Exo1 in most MMR reactions in vitro in contrast to the lack of an absolute requirement in vivo [5,13]. In aggregate, these results suggest the existence of other strand discrimination signals and potentially as yet unknown MMR proteins or mechanisms.
3. Exo1-independent and Exo1-dependent MMR subpathways
Despite the requirement of Exo1 in almost all MMR reactions reconstituted in vitro, Exo1 is dispensable for MMR in vivo. In S. cerevisiae, deletion of EXO1 causes only a 2–4 fold increase in mutation rate [5,13], and exo1−/− mouse models do not show the same magnitude or spectrum of mutations or the cancer spectrum associated with mutations in the genes that are absolutely required for MMR [14,104]. Exo1 was, however, identified and implicated in MMR based on the ability to bind Msh2 [5]. Subsequent genetic analyses in S. cerevisiae have revealed the existence of Exo1-dependent and Exo1-independent MMR sub-pathways and identified mutations in different genes, including a number of MMR genes, that appear to specifically inactivate either the Exo1-independent or the Exo1-dependent MMR subpathways [13,75,89,92,105].
3.1. Mutations affecting Exo1-independent MMR
One class of mutations identified that specifically disrupts Exo1-independent MMR are mutations in MLH1 and PMS1 identified from a genome wide screen for exo1 Δ-dependent mutator mutations; however, little is yet known about the biochemical defects caused by these mutations [13]. Mutations modeled to affect the Mlh1–Pms1 ATPase motif also synergize with exo1Δ, although the mechanistic role of the Mlh1–Pms1 ATPase is unclear [106]. In subsequent studies, dominant mutations were identified in MLH1 and PMS1 that when expressed on low copy number plasmids had little effect on MMR in wild-type strains but strongly inactivated MMR in exo1Δ strains [89]. In contrast, the same mutations when present at the MLH1 or PMS1 chromosomal loci caused a complete loss of MMR regardless of whether or not Exo1 was present. Biochemical experiments indicated that all of these novel alleles inactivated the Mlh1–Pms1 endonuclease [89]. These results demonstrate that the Exo1-independent MMR subpathway is hypersensitive to defects in Mlh1–Pms1 and specifically defects affecting its endonuclease activity [13,89]. Based on these results, the original exo1 Δ-dependent mutations [13] and ATPase mutations in MLH1 and PMS1 [106] might be expected to result in a partial endonuclease defect.
PCNA and its loading factor, replication factor C (RFC), are required for the activation of the Mlh1–Pms1 endonuclease activity in vitro, and intriguingly, overexpression of the POL30 gene encoding PCNA in S. cerevisiae suppresses a number of exo1Δ-dependent mutator mutations in several MMR genes [13,87–89]. Consistent with a role for Mlh1–Pms1 endonuclease activity in Exo1-independent MMR, multiple mutations in the POL30 gene have been isolated that cause synergistically increased mutation rates when combined with an exo1Δ mutation [13,92,105,107]. These mutations include pol30-E143K (mislabeled pol30-E143S in [13]), pol30-C22Y (called pol30-201 in [107]), pol30-C81R (called pol30-204 in [107]), and pol30-K217E. Analysis of these and other PCNA mutations isolated in genetic screens has revealed that most fall into two categories based on their distribution on the PCNA structure and the biochemical defects they cause [105]. The first category of pol30 mutations, which affect amino acids near to Cys22 in the structure, result in normal PCNA trimers that appropriately interact with Msh2–Msh6 but have a reduced ability to activate the Mlh1–Pms1 endonuclease in a mispair-independent plasmid nicking assay. The second category of pol30 mutations, which affect amino acids primarily at the interfaces between monomers in the PCNA trimer, result in reduced PCNA trimer stability, reduced or altered interactions with Msh2–Msh6, but normal activation of the Mlh1–Pms1 endonuclease in the mispair-independent plasmid nicking assay. In addition to synergistic interactions with an exo1Δ mutation, both groups of mutations also caused increases in the percentage of cells with Mlh1–Pms1 foci in strains with wild-type EXO1 [92,105], which could indicate a role in affecting the kinetics of Exo1-dependent pathway as well or that the Exo1-independent pathway is involved in the repair of at least one mispair every cell cycle.
The synergistic interaction between pol30 mutations resulting in mutant PCNA trimers that cannot efficiently activate the Mlh1–Pms1 endonuclease with loss of EXO1 is consistent with a role for high levels of Mlh1–Pms1 endonuclease activity in the Exo1-independent MMR subpathway. In contrast, the PCNA mutants that can activate the Mlh1–Pms1 endonuclease in vitro but have reduced or altered interactions with Msh2–Msh6 are more puzzling. In eukaryotes, Msh6 and Msh3 interact with PCNA using a PCNA interaction peptide (PIP-box) at the very N-terminus of the proteins [93–95]. Similar to the PCNA defects causing altered or loss of the PCNAMsh6 interaction, mutations inactivating the Msh6 PIP-box cause weak MMR defects (Shell 2007) and show strong interactions with loss of EXO1 [92]. Although disruption of the PCNA–Msh6 interaction causes loss of Msh2–Msh6 from replication forks [92], it is not clear how features of this upstream MMR step of mispair recognition could alter the requirement for Exo1 in the downstream steps of MMR. In contrast, if an additional role of the PCNA–Msh6 interaction is to recruit or retain PCNA for Mlh1–Pms1 endonuclease activation, the effect of the PIP-box mutations would be consistent with other mutations that affect activation of the Mlh1–Pms1 endonuclease [105]. A role for PCNA recruitment and/or retention by Msh2–Msh6 would be consistent with the fact that that deletion of the Msh6 PIP-box synergizes with mutations that partially disrupt Mlh1–Pms1 endonuclease activity [105] and that the Msh6 N-terminal region containing the PIP box is entirely unstructured and in S. cerevisiae is ~1000 Å long [75]. Importantly, small angle X-ray scattering reveals that PCNA does not stably associate with other regions of Msh2–Msh6 but rather the PCNA–Msh2–Msh6 complex contains two domains joined but separated by the Msh6 NTR, and consistent with this the NTRs of Msh6 and Msh3 are interchangeable and can also be fused to the N-terminus of Msh2 and retain function [75]. In the evolution of the deuterostome lineage leading to humans, the N-terminal region of Msh6 was additionally modified to include a methyl histone-binding PWWP domain that promotes recruitment of Msh2–Msh6 to chromatin, possibly supplementing the function of PCNA in recruiting MMR proteins to replicating DNA [108].
Overall, the available results indicate that the majority of mutations identified as affecting the Exo1-independent MMR sub-pathway cause defects in the efficient activation of the Mlh1–Pms1 endonuclease. A small number of additional mutations that interact with a deletion of EXO1 have not yet been directly linked to the activation of the Mlh1–Pms1 endonuclease and may affect other steps in the pathway. These mutations affect genes encoding a ribonucleotide reductase subunit (RNR1), a subunit of DNA polymerase δ and the Rev3-translesion polymerase (POL32), the DNA polymerase δ mutator allele pol3-L612M, and the DNA polymerase α mutator allele pol1-L868M [13,92,109]. Additional studies will be required to understand how these latter defects specifically affect Exo1-independent MMR.
3.2. Mutations affecting Exo1-dependent MMR
In comparison to extensive genetics studies described above, the Exo1-dependent MMR subpathway has been less well characterized in vivo. A number of mutations affecting this subpathway were identified by screening for mutations that enhanced the S. cerevisiae pms1-A99V mutation (annotated as pms1-A130V in [13] due to a wrongly annotated start codon), which inactivates the Exo1-independent MMR subpathway [13]. Consistent with the requirement of EXO1, one class of these mutations alter EXO1, including mutations that affect amino acid residues near the catalytic and DNA binding sites (exo1-C226Y and exo1-G236D) [2,110], truncate the nuclease domain (exo1-N57*), or truncate the unstructured C-terminal tail of Exo1 (exo1-N357* and exo1-N396*) (Fig. 1A,C and D). Mutations that truncate or affect amino acids in the Exo1 MIP-box have little effect on mutation rates unless combined with mutations disrupting the Exo1-independent MMR subpathway [13,111]. Other mutations likely affecting the Exo1–Mlh1 interaction were also found in the screen for enhancers of pms1-A99V, including mlh1-L511F and mlh1-M623I, which affect amino acids at the Mlh1 MIP-box binding site (Fig. 1E and F) [9,13]. Biochemical studies of the related Mlh1-L511A and Mlh1-M623A variants, designed based on structural analysis of the Mlh1–MIP box interaction, revealed that Mlh1-L511A disrupts the interaction with the Exo1 MIP box [9].
Together, the available data indicate that the Exo1–Mlh1 interaction is required in the Exo1-dependent pathway. This interaction may represent the recruitment of Exo1 in MMR, although Exo1 also binds Msh2, which could also potentially recruit Exo1 to act in MMR [5]. The role of Mlh1–Pms1 in the Exo1-dependent subpathway must extend beyond Exo1 recruitment, however, as Mlh1–Pms1 endonuclease mutations disrupt both MMR subpathways [87–89]. In addition to mutations affecting both Exo1 and the Exo1–Mlh1 interaction, other enhancers of pms1-A99V were isolated [13]; however, it is not yet known if these enhancer mutations disrupt the Exo1-dependent MMR subpathway or if they are specific to the pms1-A99V allele.
3.3. Eukaryotic MMR model
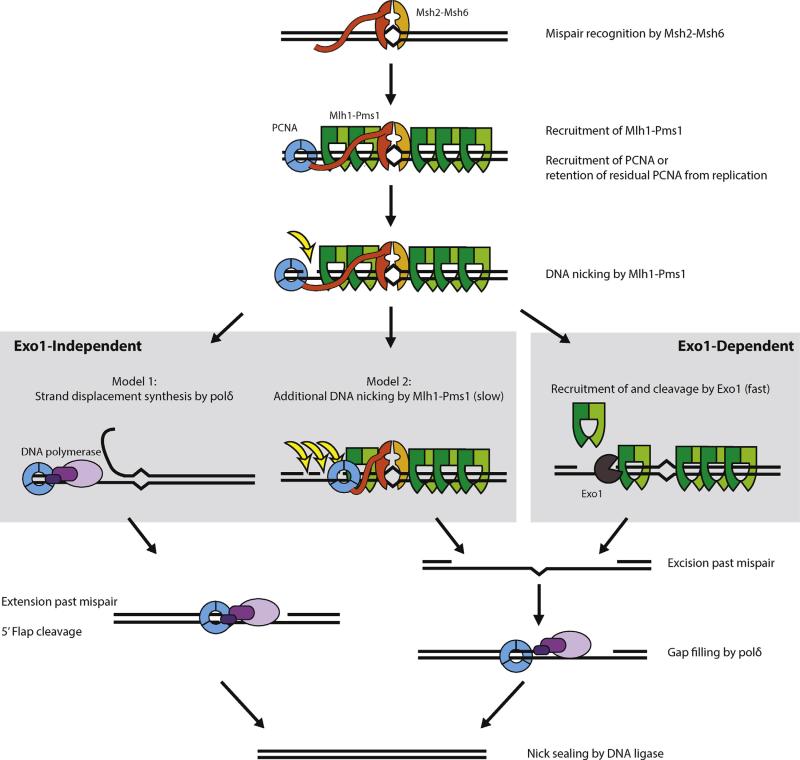
The delineation of the Exo1-dependent and Exo1-independent MMR subpathways allows a new model for eukaryotic MMR to be proposed. Here, we have limited the discussion to primarily the Msh2–Msh6 and Mlh1–Pms1 heterodimers, although analogous pathways in which Msh2–Msh6 is replaced with Msh2–Msh3 and/or Mlh1–Pms1 is replaced with Mlh1–Mlh3 are likely similar (Fig. 2). The initial steps of the MMR reaction including replication coupling are identical in both Exo1-independent and Exo1-dependent MMR. A mispair is recognized by Msh2–Msh6 when it is in an ADP-bound or nucleotide-free state. The Msh2–Msh6 heterodimer that recognizes the mispair can either be associated with or independent of a replication fork. After binding ATP, Msh2–Msh6 forms sliding clamps and becomes proficient for Mlh1–Pms1 (human Mlh1–Pms2) recruitment. Multiple Mlh1–Pms1 heterodimers are recruited in response to each mispair and are visible as foci, which are either completely or substantially devoid of Msh2–Msh6 [92]. PCNA, either recruited by Msh2–Msh6 and loaded by RFC at nicks in the vicinity of the mispair or retained by Msh2–Msh6 from PCNA available during ongoing replication, activates Mlh1–Pms1 endonuclease activity on the newly synthesized strand. Strand discrimination by Mlh1–Pms1 likely involves recognition of DNA replication intermediates such as nicks or asymmetrically loaded components such as PCNA involving mechanisms that are not well understood. In the Exo1-dependent subpathway, Exo1 is recruited by Mlh1–Pms1 at the Mlh1–Pms1 generated nick to excise DNA in a 5′ ≥ 3′ direction until the mispair is converted to a gap. Termination of excision could be mediated by weak processivity of Exo1 in the absence of tethering to mispair-recruited Mlh1–Pms1 or Msh2–Msh6 (e.g., [12]). In the Exo1-independent subpathway, multiple nicking events by Mlh1–Pms1 appear to be required. Processive nicking could lead to excision of the newly synthesized strand in the vicinity of the mispair [105] or could generate 3′ ends that could initiate strand displacement synthesis by DNA polymerase δ [112]. Resynthesis of DNA gaps or strand displacement synthesis by DNA polymerase δ in combination with PCNA would then lead to products with nicks that are then repaired by ligation.
Fig. 2. Eukaryotic MMR.
Eukaryotic mismatch repair downstream of mispair recognition by a replication-coupled or replication-uncoupled Msh2–Msh6 (or Msh2–Msh3) heterodimer (not shown) involves common and subpathway-specific steps. Mispair- and ATP-dependent recruitment of Mlh1–Pms1 by Msh2–Msh6, recruitment and/or retention of PCNA, and at least one endonucleolytic cleavage of the newly synthesized strand by Mlh1–Pms1 are common upstream steps in eukaryotic MMR. Mlh1–Pms1 foci are repair intermediates that contain either substoichiometric amounts of Msh2–Msh6 or no Msh2–Msh6 [92]. After at least one initial cleavage event by PCNA-activated Mlh1–Pms1, eukaryotic MMR follows either an Exo1-independent or Exo1-dependent subpathway. In Exo1-dependent MMR, a nick 5′ to the mispair allows Exo1 to be recruited and excise the newly synthesized strand to a position past the mispair. Exo1-independent MMR has been proposed to follow one of two models. In the first model, an Mlh1-Pms1 dependent nick 5′ to the mispair initiates strand displacement synthesis by DNA polymerase δ to a position past the mispair. The 5′ flap is then cleaved and the resulting nick is sealed by DNA ligase [112]. In the second model, Mlh1-Pms1 stimulated by PCNA performs multiple rounds of endonuclease cleavage leading to DNA degradation past the mispair, generating a product similar to that generated by Exo1 [105]. The two Exo1-independent models are not necessarily exclusive. Multiple rounds of cleavage by Mlh1–Pms1 could precede a mixture of gap filling and strand displacement synthesis by DNA polymerase δ, and the balance of the two mechanisms could vary in vivo from mispair to mispair.
Genetics studies demonstrate that both the Exo1-dependent and Exo1-independent MMR pathways act in mutation avoidance [5,13,75,89,92,105], but do not reveal the relative importance of each pathway in a wild-type cell. Monitoring Mlh1–Pms1 foci, which are MMR intermediates, gives some insight into the relative kinetics. Mutations eliminating the Mlh1–Pms1 endonuclease activity, which cause a slight dominant mutator phenotype in the presence of wild type Mlh1–Pms1, show a dramatic increase in the percentage of cells with Mlh1–Pms1 foci from ~10% to ~90%[89]. Similarly mutations affecting the ability of PCNA to activate the Mlh1–Pms1 endonuclease also shows an increased percentage of cells with foci, even in the presence of wild-type EXO1 [105], indicating that these mutations probably slow both the Exo1-dependent and Exo1-independent MMR subpathways. In addition, deletion of EXO1 increases the percentage of cells with Mlh1–Pms1 foci from ~10% to 50% [98]. This increase in MMR intermediates suggests that the Exo1-dependent MMR subpathway is likely faster in vivo, and thus, more frequently used.
Acknowledgements
This work was supported by NIH Grant R01GM50006 to R.D.K., NIH NSRA F32GM106598 to E.M.G., and the Ludwig Institute for Cancer Research to R.D.K. and C.D.P.
References
- 1.Tran PT, Erdeniz N, Symington LS, Liskay RM. EXO1–a multi-tasking eukaryotic nuclease. DNA Repair. 2004;3:1549–1559. doi: 10.1016/j.dnarep.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 2.Orans J, McSweeney EA, Iyer RR, Hast MA, Hellinga HW, Modrich P, Beese LS. Structures of human exonuclease 1 DNA complexes suggest a unified mechanism for nuclease family. Cell. 2011;145:212–223. doi: 10.1016/j.cell.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Szankasi P, Smith GR. A role for exonuclease I from S. pombe in mutation avoidance and mismatch correction. Science (New York, N.Y.) 1995;267:1166–1169. doi: 10.1126/science.7855597. [DOI] [PubMed] [Google Scholar]
- 4.Szankasi P, Smith GR. A DNA exonuclease induced during meiosis of Schizosaccharomyces pombe. J. Biol. Chem. 1992;267:3014–3023. [PubMed] [Google Scholar]
- 5.Tishkoff DX, Boerger AL, Bertrand P, Filosi N, Gaida GM, Kane MF, Kolodner RD. Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc. Natl. Acad. Sci. U. S. A. 1997;94:7487–7492. doi: 10.1073/pnas.94.14.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmutte C, Marinescu RC, Sadoff MM, Guerrette S, Overhauser J, Fishel R. Human exonuclease I interacts with the mismatch repair protein hMSH2. Cancer Res. 1998;58:4537–4542. [PubMed] [Google Scholar]
- 7.Tishkoff DX, Amin NS, Viars CS, Arden KC, Kolodner RD. Identification of a human gene encoding a homologue of Saccharomyces cerevisiae EXO1, an exonuclease implicated in mismatch repair and recombination. Cancer Res. 1998;58:5027–5031. [PubMed] [Google Scholar]
- 8.Dherin C, Gueneau E, Francin M, Nunez M, Miron S, Liberti SE, Rasmussen LJ, Zinn-Justin S, Gilquin B, Charbonnier JB, Boiteux S. Characterization of a highly conserved binding site of Mlh1 required for exonuclease I-dependent mismatch repair. Mol. Cell. Biol. 2009;29:907–918. doi: 10.1128/MCB.00945-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gueneau E, Dherin C, Legrand P, Tellier-Lebegue C, Gilquin B, Bonnesoeur P, Londino F, Quemener C, Le Du MH, Marquez JA, Moutiez M, Gondry M, Boiteux S, Charbonnier JB. Structure of the MutLalpha C-terminal domain reveals how Mlh1 contributes to Pms1 endonuclease site. Nat. Struct. Mol. Biol. 2013;20:461–468. doi: 10.1038/nsmb.2511. [DOI] [PubMed] [Google Scholar]
- 10.Genschel J, Bazemore LR, Modrich P. Human exonuclease I is required for 5′ and 3′ mismatch repair. J. Biol. Chem. 2002;277:13302–13311. doi: 10.1074/jbc.M111854200. [DOI] [PubMed] [Google Scholar]
- 11.Genschel J, Modrich P. Mechanism of 5′-directed excision in human mismatch repair. Mol. Cell. 2003;12:1077–1086. doi: 10.1016/s1097-2765(03)00428-3. [DOI] [PubMed] [Google Scholar]
- 12.Bowen N, Smith CE, Srivatsan A, Willcox S, Griffith JD, Kolodner RD. Reconstitution of long and short patch mismatch repair reactions using Saccharomyces cerevisiae proteins. Proc. Natl. Acad. Sci. U. S. A. 2013;110:18472–18477. doi: 10.1073/pnas.1318971110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amin NS, Nguyen MN, Oh S, Kolodner RD. exo1-Dependent mutator mutations: model system for studying functional interactions in mismatch repair. Mol. Cell. Biol. 2001;21:5142–5155. doi: 10.1128/MCB.21.15.5142-5155.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei K, Clark AB, Wong E, Kane MF, Mazur DJ, Parris T, Kolas NK, Russell R, Hou H, Jr., Kneitz B, Yang G, Kunkel TA, Kolodner RD, Cohen PE, Edelmann W. Inactivation of Exonuclease 1 in mice results in DNA mismatch repair defects, increased cancer susceptibility, and male and female sterility. Genes Dev. 2003;17:603–614. doi: 10.1101/gad.1060603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsubouchi H, Ogawa H. Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae. Mol. Biol. Cell. 2000;11:2221–2233. doi: 10.1091/mbc.11.7.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mimitou EP, Symington LS. DNA end resection – unraveling the tail. DNA Repair. 2011;10:344–348. doi: 10.1016/j.dnarep.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fiorentini P, Huang KN, Tishkoff DX, Kolodner RD, Symington LS. Exonuclease I of Saccharomyces cerevisiae functions in mitotic recombination in vivo and in vitro. Mol. Cell. Biol. 1997;17:2764–2773. doi: 10.1128/mcb.17.5.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khazanehdari KA, Borts RH. EXO1 and MSH4 differentially affect crossing-over and segregation. Chromosoma. 2000;109:94–102. doi: 10.1007/s004120050416. [DOI] [PubMed] [Google Scholar]
- 22.Kirkpatrick DT, Ferguson JR, Petes TD, Symington LS. Decreased meiotic intergenic recombination and increased meiosis I nondisjunction in exo1 mutants of Saccharomyces cerevisiae. Genetics. 2000;156:1549–1557. doi: 10.1093/genetics/156.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun X, Thrower D, Qiu J, Wu P, Zheng L, Zhou M, Bachant J, Wilson DM, 3rd, Shen B. Complementary functions of the Saccharomyces cerevisiae Rad2 family nucleases in Okazaki fragment maturation, mutation avoidance, and chromosome stability. DNA Repair. 2003;2:925–940. doi: 10.1016/s1568-7864(03)00093-4. [DOI] [PubMed] [Google Scholar]
- 24.Qiu J, Guan MX, Bailis AM, Shen B. Saccharomyces cerevisiae exonuclease-1 plays a role in UV resistance that is distinct from nucleotide excision repair. Nucleic Acids Res. 1998;26:3077–3083. doi: 10.1093/nar/26.13.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giannattasio M, Follonier C, Tourriere H, Puddu F, Lazzaro F, Pasero P, Lopes M, Plevani P, Muzi-Falconi M. Exo1 competes with repair synthesis, converts NER intermediates to long ssDNA gaps, and promotes checkpoint activation. Mol. Cell. 2010;40:50–62. doi: 10.1016/j.molcel.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 26.Sertic S, Pizzi S, Cloney R, Lehmann AR, Marini F, Plevani P, Muzi-Falconi M. Human exonuclease 1 connects nucleotide excision repair (NER) processing with checkpoint activation in response to UV irradiation. Proc. Natl. Acad. Sci. U. S. A. 2011;108:13647–13652. doi: 10.1073/pnas.1108547108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindsey-Boltz LA, Kemp MG, Reardon JT, DeRocco V, Iyer RR, Modrich P, Sancar A. Coupling of human DNA excision repair and the DNA damage checkpoint in a defined in vitro system. J. Biol. Chem. 2014;289:5074–5082. doi: 10.1074/jbc.M113.542787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yin Y, Petes TD. The role of Exo1p Exonuclease in DNA end resection to generate gene conversion tracts in Saccharomyces cerevisiae. Genetics. 2014;197:1097–1109. doi: 10.1534/genetics.114.164517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maringele L, Lydall D. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Delta mutants. Genes Dev. 2002;16:1919–1933. doi: 10.1101/gad.225102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bertuch AA, Lundblad V. EXO1 contributes to telomere maintenance in both telomerase-proficient and telomerase-deficient Saccharomyces cerevisiae. Genetics. 2004;166:1651–1659. doi: 10.1534/genetics.166.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18:85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- 32.Kolodner RD, Marsischky GT. Eukaryotic DNA mismatch repair. Curr. Opin. Genet. Dev. 1999;9:89–96. doi: 10.1016/s0959-437x(99)80013-6. [DOI] [PubMed] [Google Scholar]
- 33.Jiricny J. Postreplicative mismatch repair. Cold Spring Harbor perspect. Biol. 2013;5:a012633. doi: 10.1101/cshperspect.a012633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fishel R, Lescoe MK, Rao MR, Copeland NG, Jenkins NA, Garber J, Kane M, Kolodner R. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 35.Parsons R, Li GM, Longley MJ, Fang WH, Papadopoulos N, Jen J, de la Chapelle A, Kinzler KW, Vogelstein B, Modrich P. Hypermutability and mismatch repair deficiency in RER+ tumor cells. Cell. 1993;75:1227–1236. doi: 10.1016/0092-8674(93)90331-j. [DOI] [PubMed] [Google Scholar]
- 36.de la Chapelle A. Genetic predisposition to colorectal cancer. Nat. Rev. Cancer. 2004;4:769–780. doi: 10.1038/nrc1453. [DOI] [PubMed] [Google Scholar]
- 37.Borresen AL, Lothe RA, Meling GI, Lystad S, Morrison P, Lipford J, Kane MF, Rognum TO, Kolodner RD. Somatic mutations in the hMSH2 gene in microsatellite unstable colorectal carcinomas. Hum. Mol. Genet. 1995;4:2065–2072. doi: 10.1093/hmg/4.11.2065. [DOI] [PubMed] [Google Scholar]
- 38.Kane MF, Loda M, Gaida GM, Lipman J, Mishra R, Goldman H, Jessup JM, Kolodner R. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997;57:808–811. [PubMed] [Google Scholar]
- 39.Peltomaki P. Role of DNA mismatch repair defects in the pathogenesis of human cancer. J. Clin. Oncol. 2003;21:1174–1179. doi: 10.1200/JCO.2003.04.060. [DOI] [PubMed] [Google Scholar]
- 40.The Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chem. Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 42.Cox EC, Degnen GE, Scheppe ML. Mutator gene studies in Escherichia coli: the mutS gene. Genetics. 1972;72:551–567. doi: 10.1093/genetics/72.4.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Su SS, Modrich P. Escherichia coli mutS-encoded protein binds to mismatched DNA base pairs. Proc. Natl. Acad. Sci. U. S. A. 1986;83:5057–5061. doi: 10.1073/pnas.83.14.5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grilley M, Welsh KM, Su SS, Modrich P. Isolation and characterization of the Escherichia coli mutL gene product. J. Biol. Chem. 1989;264:1000–1004. [PubMed] [Google Scholar]
- 45.Langle-Rouault F, Maenhaut-Michel G, Radman M. GATC sequences, DNA nicks and the MutH function in Escherichia coli mismatch repair. EMBO J. 1987;6:1121–1127. doi: 10.1002/j.1460-2075.1987.tb04867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lahue RS, Su SS, Modrich P. Requirement for d(GATC) sequences in Escherichia coli mutHLS mismatch correction. Proc. Natl. Acad. Sci. U. S. A. 1987;84:1482–1486. doi: 10.1073/pnas.84.6.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Welsh KM, Lu AL, Clark S, Modrich P. Isolation and characterization of the Escherichia coli mutH gene product. J. Biol. Chem. 1987;262:15624–15629. [PubMed] [Google Scholar]
- 48.Au KG, Welsh K, Modrich P. Initiation of methyl-directed mismatch repair. J. Biol. Chem. 1992;267:12142–12148. [PubMed] [Google Scholar]
- 49.Campbell JL, Kleckner N. E. coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell. 1990;62:967–979. doi: 10.1016/0092-8674(90)90271-f. [DOI] [PubMed] [Google Scholar]
- 50.Ogden GB, Pratt MJ, Schaechter M. The replicative origin of the E. coli chromosome binds to cell membranes only when hemimethylated. Cell. 1988;54:127–135. doi: 10.1016/0092-8674(88)90186-9. [DOI] [PubMed] [Google Scholar]
- 51.Schlagman SL, Hattman S, Marinus MG. Direct role of the Escherichia coli Dam DNA methyltransferase in methylation-directed mismatch repair. J. Bacteriol. 1986;165:896–900. doi: 10.1128/jb.165.3.896-900.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hall MC, Jordan JR, Matson SW. Evidence for a physical interaction between the Escherichia coli methyl-directed mismatch repair proteins MutL and UvrD. EMBO J. 1998;17:1535–1541. doi: 10.1093/emboj/17.5.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamaguchi M, Dao V, Modrich P. MutS and MutL activate DNA helicase II in a mismatch-dependent manner. J. Biol. Chem. 1998;273:9197–9201. doi: 10.1074/jbc.273.15.9197. [DOI] [PubMed] [Google Scholar]
- 54.Cooper DL, Lahue RS, Modrich P. Methyl-directed mismatch repair is bidirectional. J. Biol. Chem. 1993;268:11823–11829. [PubMed] [Google Scholar]
- 55.Burdett V, Baitinger C, Viswanathan M, Lovett ST, Modrich P. In vivo requirement for RecJ, ExoVII, ExoI, and ExoX in methyl-directed mismatch repair. Proc. Natl. Acad. Sci. U. S. A. 2001;98:6765–6770. doi: 10.1073/pnas.121183298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lahue RS, Au KG, Modrich P. DNA mismatch correction in a defined system. Science (New York, N.Y.) 1989;245:160–164. doi: 10.1126/science.2665076. [DOI] [PubMed] [Google Scholar]
- 57.Lobner-Olesen A, Skovgaard O, Marinus MG. Dam methylation: coordinating cellular processes. Curr. Opin. Microbiol. 2005;8:154–160. doi: 10.1016/j.mib.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 58.Hiraga S, Ichinose C, Onogi T, Niki H, Yamazoe M. Bidirectional migration of SeqA-bound hemimethylated DNA clusters and pairing of oriC copies in Escherichia coli. Genes Cells. 2000;5:327–341. doi: 10.1046/j.1365-2443.2000.00334.x. [DOI] [PubMed] [Google Scholar]
- 59.Eisen JA, Hanawalt PC. A phylogenomic study of DNA repair genes, proteins, and processes. Mutat. Res. 1999;435:171–213. doi: 10.1016/s0921-8777(99)00050-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lamers MH, Perrakis A, Enzlin JH, Winterwerp HH, de Wind N, Sixma TK. The crystal structure of DNA mismatch repair protein MutS binding to a G × T mismatch. Nature. 2000;407:711–717. doi: 10.1038/35037523. [DOI] [PubMed] [Google Scholar]
- 61.Obmolova G, Ban C, Hsieh P, Yang W. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature. 2000;407:703–710. doi: 10.1038/35037509. [DOI] [PubMed] [Google Scholar]
- 62.Culligan KM, Hays JB. Arabidopsis MutS homologs-AtMSH2, AtMSH3, AtMSH6, and a novel AtMSH7-form three distinct protein heterodimers with different specificities for mismatched DNA. Plant Cell. 2000;12:991–1002. doi: 10.1105/tpc.12.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu SY, Culligan K, Lamers M, Hays J. Dissimilar mispair-recognition spectra of Arabidopsis DNA-mismatch-repair proteins MSH2 × MSH6 (MutSalpha) and MSH2 × MSH7 (MutSgamma) Nucleic Acids Res. 2003;31:6027–6034. doi: 10.1093/nar/gkg780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marsischky GT, Filosi N, Kane MF, Kolodner R. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 1996;10:407–420. doi: 10.1101/gad.10.4.407. [DOI] [PubMed] [Google Scholar]
- 65.Warren JJ, Pohlhaus TJ, Changela A, Iyer RR, Modrich PL, Beese LS. Structure of the human MutSalpha DNA lesion recognition complex. Mol. Cell. 2007;26:579–592. doi: 10.1016/j.molcel.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 66.Reenan RA, Kolodner RD. Isolation and characterization of two Saccharomyces cerevisiae genes encoding homologs of the bacterial HexA and MutS mismatch repair proteins. Genetics. 1992;132:963–973. doi: 10.1093/genetics/132.4.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reenan RA, Kolodner RD. Characterization of insertion mutations in the Saccharomyces cerevisiae MSH1 and MSH2 genes: evidence for separate mitochondrial and nuclear functions. Genetics. 1992;132:975–985. doi: 10.1093/genetics/132.4.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Acharya S, Wilson T, Gradia S, Kane MF, Guerrette S, Marsischky GT, Kolodner R, Fishel R. hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc. Natl. Acad. Sci. U. S. A. 1996;93:13629–13634. doi: 10.1073/pnas.93.24.13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marsischky GT, Kolodner RD. Biochemical characterization of the interaction between the Saccharomyces cerevisiae MSH2–MSH6 complex and mispaired bases in DNA. J. Biol. Chem. 1999;274:26668–26682. doi: 10.1074/jbc.274.38.26668. [DOI] [PubMed] [Google Scholar]
- 70.Miret JJ, Milla MG, Lahue RS. Characterization of a DNA mismatch-binding activity in yeast extracts. J. Biol. Chem. 1993;268:3507–3513. [PubMed] [Google Scholar]
- 71.Habraken Y, Sung P, Prakash L, Prakash S. Binding of insertion/deletion DNA mismatches by the heterodimer of yeast mismatch repair proteins MSH2 and MSH3. Curr. Biol. 1996;6:1185–1187. doi: 10.1016/s0960-9822(02)70686-6. [DOI] [PubMed] [Google Scholar]
- 72.Palombo F, Iaccarino I, Nakajima E, Ikejima M, Shimada T, Jiricny J. hMutSbeta, a heterodimer of hMSH2 and hMSH3, binds to insertion/deletion loops in DNA. Curr. Biol. 1996;6:1181–1184. doi: 10.1016/s0960-9822(02)70685-4. [DOI] [PubMed] [Google Scholar]
- 73.Srivatsan A, Bowen N, Kolodner RD. Mispair-specific recruitment of the Mlh1–Pms1 complex identifies repair substrates of the Saccharomyces cerevisiae Msh2–Msh3 complex. J. Biol. Chem. 2014;289:9352–9364. doi: 10.1074/jbc.M114.552190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Harrington JM, Kolodner RD. Saccharomyces cerevisiae Msh2–Msh3 acts in repair of base–base mispairs. Mol. Cell. Biol. 2007;27:6546–6554. doi: 10.1128/MCB.00855-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shell SS, Putnam CD, Kolodner RD. The N terminus of Saccharomyces cerevisiae Msh6 is an unstructured tether to PCNA. Mol. Cell. 2007;26:565–578. doi: 10.1016/j.molcel.2007.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dowen JM, Putnam CD, Kolodner RD. Functional studies and homology modeling of Msh2–Msh3 predict that mispair recognition involves DNA bending and strand separation. Mol. Cell. Biol. 2010;30:3321–3328. doi: 10.1128/MCB.01558-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gupta S, Gellert M, Yang W. Mechanism of mismatch recognition revealed by human MutSbeta bound to unpaired DNA loops. Nat. Struct. Mol. Biol. 2012;19:72–78. doi: 10.1038/nsmb.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gradia S, Subramanian D, Wilson T, Acharya S, Makhov A, Griffith J, Fishel R. hMSH2–hMSH6 forms a hydrolysis-independent sliding clamp on mismatched DNA. Mol. Cell. 1999;3:255–261. doi: 10.1016/s1097-2765(00)80316-0. [DOI] [PubMed] [Google Scholar]
- 79.Mendillo ML, Mazur DJ, Kolodner RD. Analysis of the interaction between the Saccharomyces cerevisiae MSH2–MSH6 and MLH1–PMS1 complexes with DNA using a reversible DNA end-blocking system. J. Biol. Chem. 2005;280:22245–22257. doi: 10.1074/jbc.M407545200. [DOI] [PubMed] [Google Scholar]
- 80.Prolla TA, Pang Q, Alani E, Kolodner RD, Liskay RM. MLH1, PMS1, and MSH2 interactions during the initiation of DNA mismatch repair in yeast. Science (New York, N.Y.) 1994;265:1091–1093. doi: 10.1126/science.8066446. [DOI] [PubMed] [Google Scholar]
- 81.Li GM, Modrich P. Restoration of mismatch repair to nuclear extracts of H6 colorectal tumor cells by a heterodimer of human MutL homologs. Proc. Natl. Acad. Sci. U. S. A. 1995;92:1950–1954. doi: 10.1073/pnas.92.6.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang TF, Kleckner N, Hunter N. Functional specificity of MutL homologs in yeast: evidence for three Mlh1-based heterocomplexes with distinct roles during meiosis in recombination and mismatch correction. Proc. Natl. Acad. Sci. U. S. A. 1999;96:13914–13919. doi: 10.1073/pnas.96.24.13914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Flores-Rozas H, Kolodner RD. The Saccharomyces cerevisiae MLH3 gene functions in MSH3-dependent suppression of frameshift mutations. Proc. Natl. Acad. Sci. U. S. A. 1998;95:12404–12409. doi: 10.1073/pnas.95.21.12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Campbell CS, Hombauer H, Srivatsan A, Bowen N, Gries K, Desai A, Putnam CD, Kolodner RD. Mlh2 is an accessory factor for DNA mismatch repair in Saccharomyces cerevisiae. PLoS Genet. 2014;10:e1004327. doi: 10.1371/journal.pgen.1004327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Blackwell LJ, Wang S, Modrich P. DNA chain length dependence of formation and dynamics of hMutSalpha. hMutLalpha. heteroduplex complexes. J. Biol. Chem. 2001;276:33233–33240. doi: 10.1074/jbc.M105076200. [DOI] [PubMed] [Google Scholar]
- 86.Mendillo ML, Hargreaves VV, Jamison JW, Mo AO, Li S, Putnam CD, Woods VL, Jr., Kolodner RD. A conserved MutS homolog connector domain interface interacts with MutL homologs. Proc. Natl. Acad. Sci. U. S. A. 2009;106:22223–22228. doi: 10.1073/pnas.0912250106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006;126:297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 88.Kadyrov FA, Holmes SF, Arana ME, Lukianova OA, O'Donnell M, Kunkel TA, Modrich P. Saccharomyces cerevisiae MutLalpha is a mismatch repair endonuclease. J. Biol. Chem. 2007;282:37181–37190. doi: 10.1074/jbc.M707617200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Smith CE, Mendillo ML, Bowen N, Hombauer H, Campbell CS, Desai A, Putnam CD, Kolodner RD. Dominant mutations in S. cerevisiae PMS1 identify the Mlh1–Pms1 endonuclease active site and an Exonuclease 1-independent mismatch repair pathway. PLoS Genet. 2013;9:e1003869. doi: 10.1371/journal.pgen.1003869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Acharya S, Foster PL, Brooks P, Fishel R. The coordinated functions of the E. coli MutS and MutL proteins in mismatch repair. Mol. Cell. 2003;12:233–246. doi: 10.1016/s1097-2765(03)00219-3. [DOI] [PubMed] [Google Scholar]
- 91.Winkler I, Marx AD, Lariviere D, Heinze RJ, Cristovao M, Reumer A, Curth U, Sixma TK, Friedhoff P. Chemical trapping of the dynamic MutS-MutL complex formed in DNA mismatch repair in Escherichia coli. J. Biol. Chem. 2011;286:17326–17337. doi: 10.1074/jbc.M110.187641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hombauer H, Campbell CS, Smith CE, Desai A, Kolodner RD. Visualization of eukaryotic DNA mismatch repair reveals distinct recognition and repair intermediates. Cell. 2011;147:1040–1053. doi: 10.1016/j.cell.2011.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Clark AB, Valle F, Drotschmann K, Gary RK, Kunkel TA. Functional interaction of proliferating cell nuclear antigen with MSH2–MSH6 and MSH2–MSH3 complexes. J. Biol. Chem. 2000;275:36498–36501. doi: 10.1074/jbc.C000513200. [DOI] [PubMed] [Google Scholar]
- 94.Kleczkowska HE, Marra G, Lettieri T, Jiricny J. hMSH3 and hMSH6 interact with PCNA and colocalize with it to replication foci. Genes Dev. 2001;15:724–736. doi: 10.1101/gad.191201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Flores-Rozas H, Clark D, Kolodner RD. Proliferating cell nuclear antigen and Msh2p–Msh6p interact to form an active mispair recognition complex. Nat. Genet. 2000;26:375–378. doi: 10.1038/81708. [DOI] [PubMed] [Google Scholar]
- 96.Prelich G, Tan CK, Kostura M, Mathews MB, So AG, Downey KM, Stillman B. Functional identity of proliferating cell nuclear antigen and a DNA polymerase-delta auxiliary protein. Nature. 1987;326:517–520. doi: 10.1038/326517a0. [DOI] [PubMed] [Google Scholar]
- 97.Krishna TS, Kong XP, Gary S, Burgers PM, Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 98.Hombauer H, Srivatsan A, Putnam CD, Kolodner RD. Mismatch repair, but not heteroduplex rejection, is temporally coupled to DNA replication. Science (Newyork, N.Y.) 2011;334:1713–1716. doi: 10.1126/science.1210770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang Y, Yuan F, Presnell SR, Tian K, Gao Y, Tomkinson AE, Gu L, Li GM. Reconstitution of 5′-directed human mismatch repair in a purified system. Cell. 2005;122:693–705. doi: 10.1016/j.cell.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 100.Constantin N, Dzantiev L, Kadyrov FA, Modrich P. Human mismatch repair: reconstitution of a nick-directed bidirectional reaction. J. Biol. Chem. 2005;280:39752–39761. doi: 10.1074/jbc.M509701200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pluciennik A, Dzantiev L, Iyer RR, Constantin N, Kadyrov FA, Modrich P. PCNA function in the activation and strand direction of MutLalpha endonuclease in mismatch repair. Proc. Natl. Acad. Sci. U. S. A. 2010;107:16066–16071. doi: 10.1073/pnas.1010662107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lujan SA, Williams JS, Clausen AR, Clark AB, Kunkel TA. Ribonucleotides are signals for mismatch repair of leading-strand replication errors. Mol. Cell. 2013;50:437–443. doi: 10.1016/j.molcel.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ghodgaonkar MM, Lazzaro F, Olivera-Pimentel M, Artola-Boran M, Cejka P, Reijns MA, Jackson AP, Plevani P, Muzi-Falconi M, Jiricny J. Ribonucleotides misincorporated into DNA act as strand-discrimination signals in eukaryotic mismatch repair. Mol. Cell. 2013;50:323–332. doi: 10.1016/j.molcel.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Edelmann L, Edelmann W. Loss of DNA mismatch repair function and cancer predisposition in the mouse: animal models for human hereditary nonpolyposis colorectal cancer. Am. J. Med. Genet. Part C Semin. Med. Genet. 2004;129C:91–99. doi: 10.1002/ajmg.c.30021. [DOI] [PubMed] [Google Scholar]
- 105.Goellner EM, Smith CE, Campbell CS, Hombauer H, Desai A, Putnam CD, Kolodner RD. PCNA and Msh2–Msh6 activate an Mlh1–Pms1 endonuclease pathway required for Exo1-independent mismatch repair. Mol. Cell. 2014;55:291–304. doi: 10.1016/j.molcel.2014.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tran PT, Simon JA, Liskay RM. Interactions of Exo1p with components of MutLalpha in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 2001;98:9760–9765. doi: 10.1073/pnas.161175998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lau PJ, Flores-Rozas H, Kolodner RD. Isolation and characterization of new proliferating cell nuclear antigen (POL30) mutator mutants that are defective in DNA mismatch repair. Mol. Cell. Biol. 2002;22:6669–6680. doi: 10.1128/MCB.22.19.6669-6680.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Li F, Mao G, Tong D, Huang J, Gu L, Yang W, Li GM. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSalpha. Cell. 2013;153:590–600. doi: 10.1016/j.cell.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liberti SE, Larrea AA, Kunkel TA. Exonuclease 1 preferentially repairs mismatches generated by DNA polymerase alpha. DNA Repair. 2013;12:92–96. doi: 10.1016/j.dnarep.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lee Bi BI, Nguyen LH, Barsky D, Fernandes M, Wilson DM., 3rd Molecular interactions of human Exo1 with DNA. Nucleic Acids Res. 2002;30:942–949. doi: 10.1093/nar/30.4.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tran PT, Fey JP, Erdeniz N, Gellon L, Boiteux S, Liskay RM. A mutation in EXO1 defines separable roles in DNA mismatch repair and post-replication repair. DNA Repair. 2007;6:1572–1583. doi: 10.1016/j.dnarep.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kadyrov FA, Genschel J, Fang Y, Penland E, Edelmann W, Modrich P. A possible mechanism for exonuclease 1-independent eukaryotic mismatch repair. Proc. Natl. Acad. Sci. U. S. A. 2009;106:8495–8500. doi: 10.1073/pnas.0903654106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dosztanyi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]