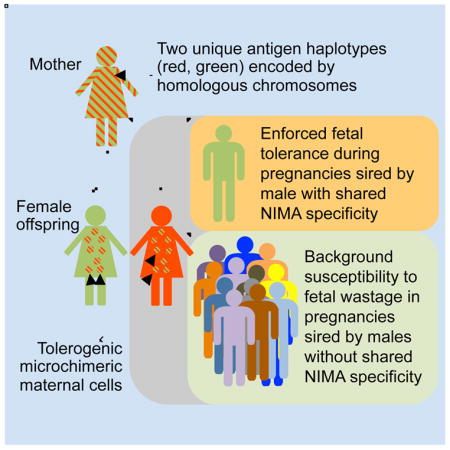
SUMMARY
Exposure to maternal tissue during in utero development imprints tolerance to immunologically foreign non-inherited maternal antigens (NIMA) that persists into adulthood. The biological advantage of this tolerance, conserved across mammalian species, remains unclear. Here we show that maternal cells which establish microchimerism in female offspring during development promote systemic accumulation of immune suppressive regulatory T cells (Tregs) with NIMA specificity. NIMA-specific Tregs expand during pregnancies sired by males expressing alloantigens with overlapping NIMA specificity, thereby averting fetal wastage triggered by prenatal infection and non-infectious disruptions of fetal tolerance. Therefore, exposure to NIMA selectively enhances reproductive success in second-generation females carrying embryos with overlapping paternally inherited antigens. These findings demonstrate that genetic fitness, canonically thought to be restricted to Mendelian inheritance, is enhanced in female placental mammals through vertically transferred maternal cells that promote conservation of NIMA and enforce cross-generational reproductive benefits.
Graphical Abstract
INTRODUCTION
Reproductive health and pregnancy outcomes have traditionally been characterized from the viewpoint of maternal tolerance to immunologically foreign paternal antigens expressed by the fetus (Erlebacher, 2013; Munoz-Suano et al., 2011). However, compulsory fetal exposure to an equally diverse array of discordant non-inherited maternal antigens (NIMA) also occurs during in utero and early postnatal maturation. Maternal antigen stimulation in these developmental contexts imprints remarkably persistent tolerance to NIMA in offspring (Dutta et al., 2009; Hirayama et al., 2012; Mold and McCune, 2012). Pioneering examples of tolerance to NIMA include blunted sensitization to erythrocyte Rh antigen among Rh-negative women born to Rh-positive mothers (Owen et al., 1954), and selective anergy to NIMA-specific HLA haplotypes among transfusion dependent individuals broadly exposed to foreign HLA (Claas et al., 1988). More recently, prolonged survival of NIMA-matched human allografts after solid organ transplantation (Burlingham et al., 1998), and reduced graft versus host disease among NIMA-matched stem cell transplants highlight clinical benefits of NIMA-specific tolerance that persists in individuals through adulthood (Ichinohe et al., 2004; Matsuoka et al., 2006; van Rood et al., 2002).
In human development, tolerance to mother begins in utero with suppressed activation of maturing immune cells with NIMA specificity for infants with a full numerical complement of adaptive immune components at the time of birth (Mold and McCune, 2012; Mold et al., 2008). In this scenario, postnatal persistence of NIMA-specific tolerance represents an expendable developmental remnant of immune suppressive mechanisms essential for in utero survival. However, this reasoning does not explain why tolerance imprinted by exposure to foreign antigens in utero is widely conserved across mammalian species (e.g. non-human primates, ruminants, rodents) regardless of fetal adaptive immune cell maturation relative to parturition (Billingham et al., 1953; Burlingham et al., 1998; Dutta and Burlingham, 2011; Owen, 1945; Picus et al., 1985). For example, prolonged survival of NIMA-matched allografts in humans is consistently reproduced in mice despite the absence of peripheral T cells at the time of birth in this species (Akiyama et al., 2011; Andrassy et al., 2003; Araki et al., 2010; Mold and McCune, 2012). These results illustrating highly engrained phylogenetic roots of NIMA tolerance in mammalian reproduction strongly suggest the existence of universal biological benefits driving conserved tolerance to NIMA that persists through adulthood.
Given the necessity for sustained maternal tolerance to foreign fetal antigens in successful pregnancies across all eutherian placental mammals (Samstein et al., 2012), postnatal NIMA-specific tolerance may be evolutionarily preserved to promote reproductive fitness by reinforcing fetal tolerance in future generation pregnancies. To address this hypothesis, immunological tools that allow precise identification of T cells with NIMA-specificity were uniquely combined with mouse models of allogeneic pregnancy, and pregnancy complications stemming from disruptions in fetal tolerance (Chaturvedi et al., 2015; Rowe et al., 2011; Rowe et al., 2012b). Our data show obligatory developmental exposure to foreign maternal tissue primes expanded accumulation of NIMA-specific immune suppressive regulatory CD4+ T cells (Tregs) that reinforce fetal tolerance during next-generation pregnancies sired by males with overlapping MHC haplotype specificity. Expanded NIMA-specific Treg accumulation requires ongoing postnatal cognate antigen stimulation by maternal cells that establish microchimerism in offspring. In the broader context, cross-generational reproductive benefits conferred by tolerance to NIMA indicates genetic fitness is not restricted only to transmitting homologous chromosomes by Mendelian inheritance, but is enhanced through vertically transferred tolerogenic cells that establish microchimerism in offspring favoring preservation of non-inherited maternal alleles within a population.
RESULTS
Developmental exposure to maternal tissue drives expanded NIMA-specific regulatory T cell accumulation
To investigate the fundamental biology driving conserved persistence of NIMA tolerance across mammalian species, an instructive allogeneic mating strategy that transforms defined model antigens into surrogate NIMA was developed to precisely track T cells with NIMA specificity. Female mice heterozygous for a transgene encoding constitutive expression of a transmembrane recombinant protein containing ovalbumin (OVA) and the 2W1S55-68 variant of I-Eα in all cells (behind the β-actin promoter) (Moon et al., 2011; Rees et al., 1999) were mated with non-transgenic males – thereby transforming 2W1S55-68 and OVA into surrogate NIMA in half the offspring (Figures S1A and S1B). This approach taking advantage of MHC tetramer staining and enrichment techniques for identifying endogenous CD4+ T cells with I-Ab:2W1S55-68 specificity (Moon et al., 2007), combined with tools for manipulating OVA-expressing cells allows NIMA-responsive and NIMA-expressing cells to be simultaneously evaluated. To ensure shifts in I-Ab:2W1S55-68 specific CD4+ T cells reflect developmental exposure to maternal tissue as opposed to 2W1S-OVA+ concepti within the same litter, offspring from reciprocal mating between males heterozygous for the 2W1S-OVA expression transgene and non-transgenic females that transforms 2W1S55-68 peptide and OVA into surrogate non-inherited paternal antigens (NIPA) were used as controls along with genetically identical naive mice without developmental 2W1S exposure (Figures S1C and S1D).
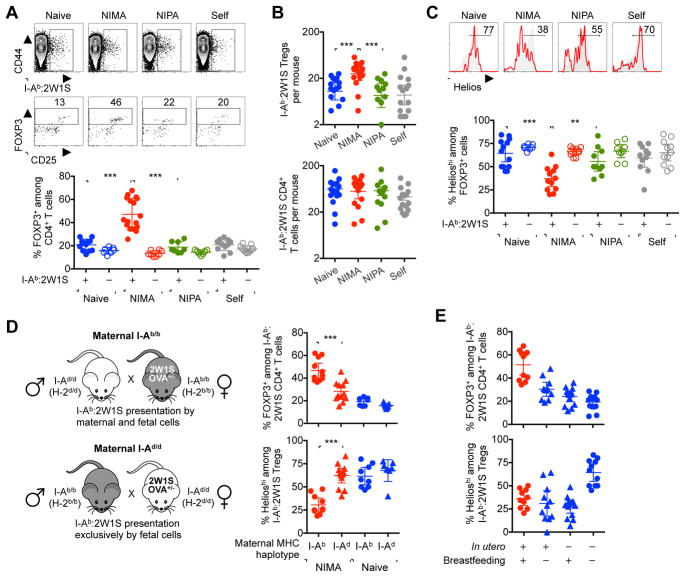
Sharply increased proportions of immune suppressive regulatory T cells (Tregs) identified by expression of the lineage defining FOXP3 transcriptional regulator (Fontenot et al., 2003; Hori et al., 2003) were found among CD4+ T cells with I-Ab:2W1S55-68 specificity in adult NIMA-2W1S mice compared with age matched naive mice, and additional control mice exposed to the identical 2W1S-OVA recombinant protein as a surrogate NIPA or ubiquitous ‘self’ antigen (Figures 1A and S1). Interestingly, while the percentage and number of FOXP3+ Tregs with I-Ab:2W1S55-68 specificity were significantly increased in the spleen and peripheral lymph nodes for NIMA-2W1S offspring, the total number of I-Ab:2W1S55-68-specific CD4+ T cells remained similar regardless of developmental 2W1S stimulation (Figure 1B). Along with sharply reduced expression of Helios that marks thymus derived Tregs among cells with I-Ab:2W1S55-68 specificity in NIMA-2W1S compared with each group of control mice (Thornton et al., 2010) (Figure 1C), these results strongly suggest developmental exposure to immunologically discordant maternal tissue primes induced FOXP3 expression among NIMA-specific CD4+ T cells. Importantly, these shifts were restricted to CD4+ T cells with NIMA-specificity since expanded Tregs and diminished Helios expression were eliminated among bulk CD4+ T cells in each group of mice regardless of developmental 2W1S stimulation (Figures 1A and 1C).
Figure 1. Developmental exposure to maternal tissue primes expanded NIMA-specific FOXP3+ Tregs.
(A) Representative plots showing the gating strategy used to identify I-Ab:2W1S specific among CD4+ T cells (top), FOXP3+ Tregs among I-Ab:2W1S specific CD4+ T cells (middle), and composite data (bottom) for percent FOXP3+ among CD4+ T cells with I-Ab:2W1S specificity (filled) compared with bulk CD4+ T cells (open) in the spleen plus peripheral lymph nodes of naive (blue), NIMA-2W1S (red), NIPA-2W1S (green), or 2W1S-self (gray) 8 week old adult mice.
(B) Total number of I-Ab:2W1S specific FOXP3+ Tregs (top) and CD4+ T cells (bottom) for each group of mice described in panel A.
(C) Percent Helioshi among I-Ab:2W1S specific (red line) or bulk (gray shaded) FOXP3+ CD4+ T cells for each group of mice described in panel A.
(D) Mating strategy for generating genetically identical NIMA-2W1S offspring born to either MHC class II I-Ab/b (I-Ab:2W1S55-68 peptide presented by cells of both maternal and fetal origin) or I-Ad/d (I-Ab:2W1S55-68 peptide only presented by cells of fetal origin) haplotype mothers, and composite data for percent FOXP3+ among I-Ab:2W1S specific CD4+ T cells and Helioshi among I-Ab:2W1S specific FOXP3+ cells for each group of NIMA-2W1S (red) compared with naive (blue) mice.
(E) Percent FOXP3+ among I-Ab:2W1S specific CD4+ T cells, and Helioshi among I-Ab:2W1S specific FOXP3+ cells for each group of cross-fostered offspring exposed to 2W1S-OVA in utero and/or postnatally through breastfeeding by 2W1S-OVA+ mothers. Each point represents the result from an individual female mouse, and these data are representative of at least three separate experiments each with similar results. Bars, mean ± 95% confidence interval. ** P < 0.01, *** P < 0.001. See also Figure S1.
Considering exposure to maternal tissue begins in utero when fetal immune components are undergoing maturation, related experiments addressed whether expanded NIMA-specific Tregs require antigen presentation by maternal cells. Here, the I-Ab restricted nature of 2W1S55-68 peptide presentation was exploited to compare NIMA-specific Tregs in genetically identical NIMA-2W1S offspring born to I-Ab/b or I-Ad/d mothers (Rees et al., 1999) (Figure 1D). Interestingly, expanded proportions of FOXP3+ Tregs and diminished cell-intrinsic Helios expression were each significantly reduced for I-Ab:2W1S55-68 specific CD4+ T cells in NIMA-2W1S offspring born to I-Ad/d mothers to levels comparable to naive control mice (Figure 1D). The results highlighting tolerogenic properties of maternal cells in mouse offspring parallel the presence of maternal hematopoietic cells in human fetal lymph nodes during in utero development (Mold et al., 2008), and suggest compulsory developmental exposure to immunologically foreign maternal tissue actively primes expansion of immune suppressive Tregs with NIMA specificity.
Expanded NIMA-specific Treg accumulation may reflect in utero and/or postnatal exposure to immunologically foreign maternal antigen (e.g. soluble maternal HLA alloantigens, intact maternal cells in breast milk) (Molitor et al., 2004; Zhou et al., 2000). To dissociate the contributions of maternal antigen stimulation during each developmental context, the individual impacts of in utero and early postnatal NIMA exposure through breastfeeding were evaluated by cross-fostering offspring after birth with naive or 2W1S-OVA+ nursing mothers. In agreement with improved survival of NIMA-matched allografts in transplant recipients exposed to maternal antigen both in utero and through breastfeeding (Andrassy et al., 2003; Campbell et al., 1984), maximal NIMA-specific Treg expansion required in utero plus postnatal maternal antigen stimulation (Figure 1E). Comparatively, Helios expression remained at diminished levels with maternal tissue exposure in utero or through breastfeeding suggesting maternal antigen stimulation in either developmental context primes enriched proportions of NIMA-specific CD4+ T cells poised for induced FOXP3 expression (Figure 1E). Taken together, these findings indicate immunologically foreign maternal antigen stimulation in utero and through breastfeeding work synergistically to promote expanded peripheral accumulation of NIMA-specific Tregs.
Microchimeric maternal cells maintain expanded NIMA-specific Tregs
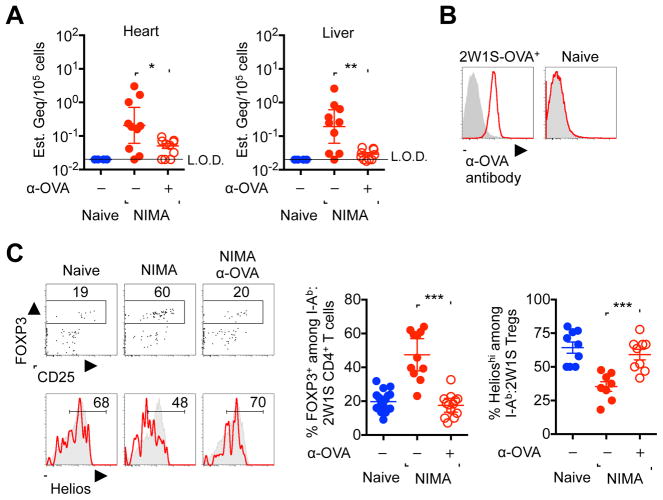
Persistence of NIMA-specific tolerance coincides with postnatal retention of microchimeric maternal cells in adult human and rodent offspring (Dutta et al., 2009; Loubiere et al., 2006; Maloney et al., 1999; Mold et al., 2008). Nonetheless, the immunological cause and effect relationship between these two interrelated phenomena engrained in mammalian reproduction remain undefined. One possibility is that postnatal maintenance of NIMA-specific tolerance actively prevents rejection of antigenically discordant maternal cells fostering their long-term survival in offspring. Alternatively, retained microchimeric maternal cells may provide an essential postnatal source of cognate antigen required for maintaining expanded Tregs with NIMA-specificity. Having established early developmental exposure to 2W1S-OVA+ maternal tissues imprints persistent accumulation of NIMA-2W1S-specific Tregs, analysis of cells expressing these model antigens was extended to investigate the necessity for postnatal stimulation by microchimeric 2W1S-OVA+ maternal cells. We found OVA-encoding DNA, reflective of 2W1S-OVA+ maternal cells in systemic organs (e.g. heart, liver), at levels ranging from 1 in 105 to 106 cells in NIMA offspring consistent with quantities of microchimeric maternal cells identified using PCR for MHC haplotype alleles (Bakkour et al., 2014; Dutta et al., 2009; Mold et al., 2008) (Figures 2A and S2A).
Figure 2. NIMA-specific Treg expansion requires persistent postnatal exposure to microchimeric maternal cells.
(A) Maternal 2W1S-OVA+ microchimeric cell encoding DNA levels in each tissue of naive (blue filled), NIMA-2W1S (red filled), or NIMA-2W1S mice treated with anti-OVA depleting antibody (red open). (B) Cell surface OVA expression levels among splenocytes from 2W1S-OVA+ compared with naive control mice after staining with anti-OVA (red line) or rabbit IgG isotype (gray shaded) antibodies. (C) Representative plots and composite data for FOXP3+ Tregs among I-Ab:2W1S specific CD4+ T cells, and Helios expression among I-Ab:2W1S specific FOXP3+ cells for each group of mice described in panel A. Each point represents the result from an individual female mouse at 8 weeks of age, and these data are representative of at least three separate experiments each with similar results. Bars, mean ± 95% confidence interval. * P < 0.05, ** P < 0.01, *** P < 0.001. See also Figure S2.
To definitively address the cause and effect relationship between NIMA-specific tolerance and microchimeric maternal cells, anti-OVA antibody that uniformly binds 2W1S-OVA+ cells was used to deplete microchimeric 2W1S-OVA+ maternal cells (Figure 2B). In line with the efficiency whereby anti-OVA antibody depletes congenically marked 2W1S-OVA+ cells after adoptive transfer into non-transgenic recipients (Figure S2B), OVA encoding DNA representative of microchimeric 2W1S-OVA+ cells in each organ of NIMA offspring declined sharply within 12 days following in vivo anti-OVA antibody administration (Figure 2A). Remarkably, NIMA-2W1S-specific Treg accumulation and cell-intrinsic Helios down-regulation both returned to background levels found in naive control mice after elimination of microchimeric 2W1S-OVA+ cells within this time frame (Figure 2C). Thus, microchimeric maternal cells provide an essential source of cognate maternal antigen required for sustaining postnatal NIMA-specific tolerance.
Selectively enriched NIMA-specific Treg expansion in female offspring accentuated during pregnancy with NIMA-matched fetal antigen stimulation
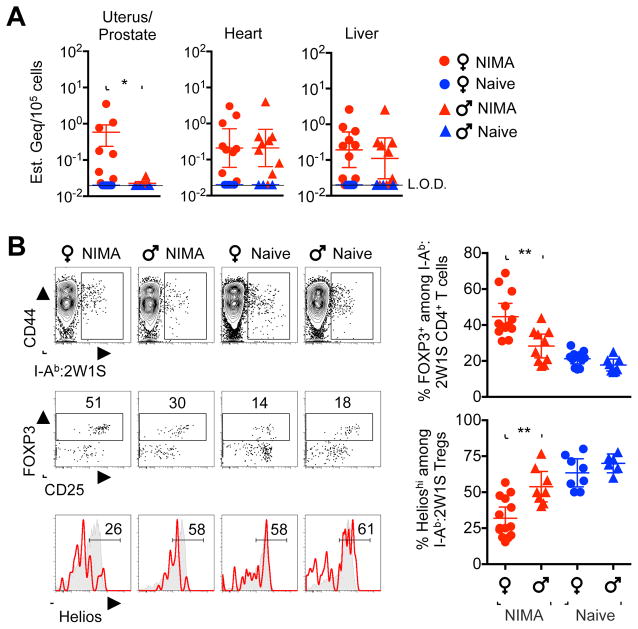
Given the necessity for expanded maternal tolerance that encompasses immunologically foreign paternal-fetal antigens in successful pregnancy shared by all eutherian placental mammals (Erlebacher, 2013; Munoz-Suano et al., 2011; Samstein et al., 2012), we reasoned reinforced fetal tolerance that promotes reproductive fitness may represent a more universal evolutionary driver for conserved NIMA-specific tolerance. This notion is supported by highly enriched microchimeric 2W1S-OVA+ maternal cells in female reproductive tissue (uterus) of NIMA offspring, and their conspicuous absence in analogous male reproductive tissue (prostate) (Figure 3A). In turn, NIMA-specific Treg expansion and reduced Helios expression were markedly more pronounced in female compared with male NIMA-2W1S littermate offspring (Figure 3B). Thus, gender-specific differences favoring more robust NIMA-specific Treg expansion in females parallel the selective accumulation of microchimeric maternal cell in female reproductive tissue.
Figure 3. Expanded NIMA-specific Treg accumulation in female offspring parallels discordant maternal cell microchimerism in gender-specific reproductive tissue.
(A) 2W1S-OVA+ encoding DNA levels in each tissue among NIMA-2W1S female (red circle), littermate 2W1S-NIMA male (red triangle), naive female (blue circle) and naive male (blue triangle) mice. (B) Representative plots and composite data showing I-Ab:2W1S specific CD4+ T cells (top), FOXP3+ Tregs among I-Ab:2W1S specific CD4+ T cells (middle), and Helios expression among Tregs with I-Ab:2W1S specificity (red line) or bulk specificity (gray shaded) among NIMA-2W1S female compared with NIMA-2W1S littermate male mice. Each point represents the result from an individual mouse at 8 weeks of age, and these data are representative of at least three separate experiments each with similar results. Bars, mean ± 95% confidence interval. * P < 0.05, ** P < 0.01
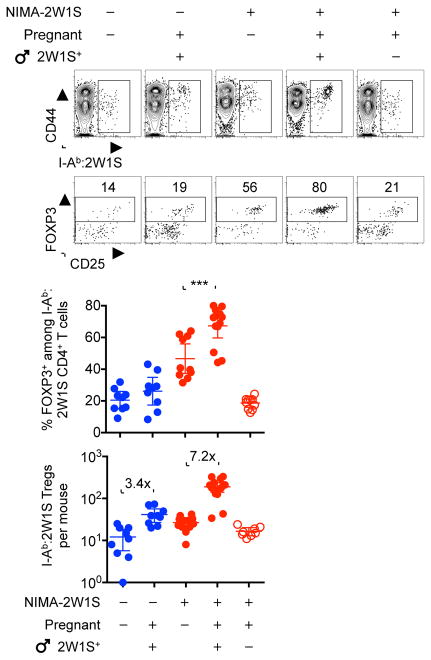
To further investigate the reproductive significance for gender-specific differences in postnatal persistence of NIMA-specific tolerance, shifts in NIMA-specific CD4+ T cells were evaluated in females during pregnancy after cognate fetal antigen stimulation. During allogeneic pregnancies sired by 2W1S-OVA+ transgenic males, sharply accelerated expansion tempo occurred among 2W1S-specific Tregs in NIMA-2W1S female mice compared with naive control mice (7.2-fold compared with 3.4-fold expansion by midgestation in NIMA and naive mice, respectively [P = 0.004]) (Figure 4). Accelerated NIMA-specific Treg expansion tempo during pregnancy represents a targeted response to cognate 2W1S stimulation by shared fetal-expressed antigen because NIMA-2W1S-specific Tregs did not expand during pregnancies sired by non-transgenic male mice (Figure 4). Thus, mammalian females contain an enriched pool of NIMA-specific Tregs poised for accelerated re-expansion upon encounter with paternal-fetal antigen of overlapping specificity during pregnancy.
Figure 4. NIMA-specific Treg expansion accelerated during pregnancy with NIMA-matched fetal antigen stimulation.
Representative plots and composite data showing I-Ab:2W1S specific CD4+ T cells (top), FOXP3+ Tregs among I-Ab:2W1S specific CD4+ T cells (middle), and composite data for percent and number of FOXP3+ CD4+ T cells with I-Ab:2W1S specificity in virgin and midgestation (E11.5) naive female (blue) compared with NIMA-2W1S (red) female mice after mating with 2W1S-OVA+ transgenic male mice or non-transgenic controls. Each point represents the result from an individual mouse, these data are representative of at least three separate experiments each with similar results. Bars, mean ± 95% confidence interval. *** P < 0.001
Microchimeric maternal cells enforce cross-generational protection against fetal wastage
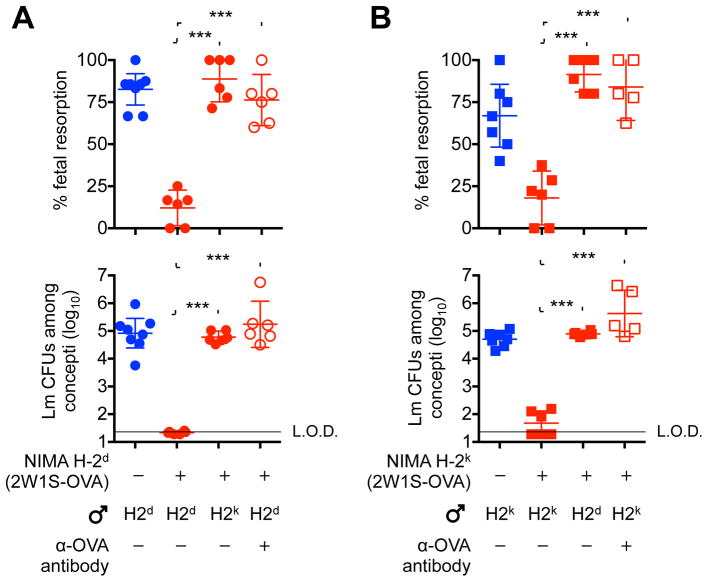
Since the immunological identity of individuals is primarily defined by unique expression of MHC haplotype alleles (Zinkernagel and Doherty, 1979), MHC haplotype alleles (e.g. H-2d, H-2k –along with 2W1S and OVA antigens), were transformed into surrogate NIMA to investigate functional properties of tolerance in the setting of broader NIMA overlap (Figure S3). In turn, the protective properties of NIMA-specific tolerance were probed by infection with the prenatal bacterial pathogen, Listeria monocytogenes, which disrupts fetal tolerance with ensuing fetal wastage (Chaturvedi et al., 2015; Mylonakis et al., 2002; Rowe et al., 2012a). Remarkably, fetal resorption and in utero L. monocytogenes invasion after prenatal infection in naive mice bearing allogeneic pregnancy were eliminated by overlap between NIMA and paternal-fetal MHC haplotype antigens (NIMA H-2d females mated with H-2d Balb/c male mice) (Figures 5A and S3). Protection against fetal wastage occurred in an antigen-specific fashion requiring commonality between NIMA and paternal-fetal antigens since fetal resorption and in utero bacterial invasion each rebounded when third-party males bearing irrelevant MHC haplotype alleles (e.g. H-2k CBA/J mice) were used to sire allogeneic pregnancy in NIMA H-2d female mice (Figures 5A and S3). Thus, protection against prenatal infection conferred by non-inherited antigenic overlap between maternal grandmother and the developing fetus and highlight profound cross-generational benefits of persistent NIMA-specific tolerance.
Figure 5. Overlap between NIMA and paternal-fetal alloantigen protects against fetal resorption and in utero bacterial invasion following prenatal infection.
(A) Percent fetal resorption (top) and average recoverable bacterial CFUs from each concepti per litter (bottom) five days following L. monocytogenes intravenous maternal infection initiated midgestation (E11.5) for naive female (blue) compared with NIMA-H-2d-(2W1S-OVA) (red) female mice during allogeneic pregnancy sired by H-2d or third party H-2k males, or depletion of microchimeric 2W1S-OVA+ maternal cells with anti-OVA antibody prior to mating.
(B) Percent fetal resorption (top) and average recoverable bacterial CFUs from each concepti per litter (bottom) five days following L. monocytogenes intravenous maternal infection initiated midgestation (E11.5) for naive female (blue) compared with NIMA-H-2k-(2W1S-OVA) (red) female mice during allogeneic pregnancy sired by H-2k or third party H-2d males, or depletion of microchimeric 2W1S-OVA+ maternal cells with anti-OVA antibody prior to mating. Each point represents the result from an individual mouse, and these data are representative of at least three separate experiments each with similar results. Bars, mean ± 95% confidence interval. *** P < 0.001. See also Figure S3
Given the requirement for postnatal maternal microchimerism to maintain expanded NIMA-specific tolerance (Figure 2), we next addressed the necessity for microchimeric maternal cells to protect against fetal wastage after prenatal L. monocytogenes infection. Here, cross-reactivity between anti-OVA antibody used to deplete H-2d-2W1S-OVA+ microchimeric maternal cells and fetal-expressed OVA antigen was avoided by exclusively using non-transgenic H-2d Balb/c male mice to sire allogeneic pregnancy in H-2d-2W1S-OVA NIMA female mice (Figure S3). This analysis showed depletion of microchimeric maternal cells prior to mating efficiently overturns protection against fetal resorption and in utero L. monocytogenes invasion in NIMA female mice (Figure 5A). Importantly, these reproductive benefits were not restricted to NIMA H-2d haplotype alleles since fetal resorption and in utero L. monocytogenes invasion were each similarly averted among NIMA H-2k female mice during allogeneic pregnancy sired by H-2k CBA/J male mice (Figure 5B). Conversely, protection against fetal wastage was lost in NIMA H-2k female mice if NIMA mismatched H-2d males were used to sire allogeneic pregnancy, or if H-2k-2W1S-OVA+ microchimeric maternal cells were depleted using anti-OVA antibody prior to mating with non-transgenic H-2k CBA/J male mice (Figure 5B). Thus, persistent postnatal tolerance to NIMA protects against fetal wastage triggered by prenatal infection.
To further extend this analysis to non-infectious disruptions in fetal tolerance stemming from blunted expansion of maternal Tregs (e.g. spontaneous abortion, preeclampsia) (Jiang et al., 2014; Santner-Nanan et al., 2009; Sasaki et al., 2004), the protective benefits of NIMA-specific tolerance on fetal wastage triggered by partial depletion of bulk maternal FOXP3+ Tregs during allogeneic pregnancy were investigated. Diphtheria toxin administration to female mice heterozygous for co-expression of the high-affinity human diphtheria toxin receptor (DTR) with FOXP3 during pregnancy causes partial transient depletion of bulk maternal Tregs to levels comparable to virgin control mice with disruptions in fetal tolerance and ensuing fetal wastage (Kim et al., 2007; Rowe et al., 2011; Rowe et al., 2012b). Therefore, our breeding strategy was modified to transform MHC haplotype alleles (e.g. H-2d, H-2k) along with 2W1S-OVA into surrogate NIMA in genetically identical H-2b FOXP3DTR/WT female mice (Figure S4). Remarkably, fetal resorption triggered by partial depletion of bulk maternal Tregs during allogeneic pregnancy in naive control female mice was reversed to near completion by overlap between NIMA and fetal expressed MHC haplotype antigens (NIMA H-2d females mated with H-2d Balb/c male mice, or NIMA H-2k females mated with H-2k CBA/J male mice) (Figures 6A and 6B). Protection against fetal wastage induced by partial maternal Treg depletion was paternal-antigen specific and required ongoing stimulation by microchimeric maternal cells since fetal resorption rebounded during pregnancies sired by third-party males that express irrelevant MHC haplotype alleles or if 2W1S-OVA+ microchimeric maternal cells were depleted prior to mating (Figures 6A and 6B). Taken together, these findings demonstrate resiliency against fetal wastage conferred by persistent postnatal NIMA-specific tolerance encompasses both infectious and non-infectious perturbations in fetal tolerance during next generation pregnancies.
Figure 6. Overlap between NIMA and paternal-fetal antigen protects against fetal wastage triggered by partial depletion of maternal FOXP3+ regulatory T cells.
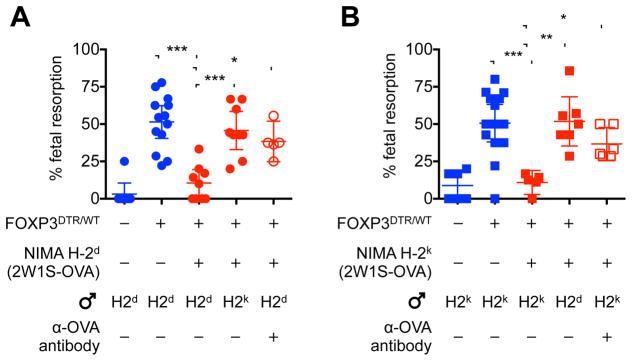
(A) Percent fetal resorption for naive (blue) FOXP3WT/WT and FOXP3DTR/WT female mice compared with each group of NIMA-H-2d FOXP3DTR/WT (red) female mice five days after initiating diphtheria toxin during allogeneic pregnancy sired by H-2d or third party H-2k males, or depletion of microchimeric 2W1S-OVA+ maternal cells with anti-OVA antibody prior to mating.
(B) Percent fetal resorption for naive (blue) FOXP3WT/WT and FOXP3DTR/WT female mice compared with each group of NIMA-H-2k FOXP3DTR/WT (red) female mice five days after initiating diphtheria toxin during allogeneic pregnancy sired by H-2k or third party H-2d males, or depletion of microchimeric 2W1S-OVA+ maternal cells with anti-OVA antibody prior to mating. Each point represents the result from an individual mouse, these data are representative of at least three separate experiments each with similar results. Bars, mean ± 95% confidence interval. * P < 0.05, ** P < 0.01, *** P < 0.001. See also Figure S4.
DISCUSSION
Reproductive success in female placental mammals requires sustained maternal tolerance to immunologically foreign paternal antigens expressed by the developing fetus (Erlebacher, 2013; Munoz-Suano et al., 2011; Samstein et al., 2012). Reciprocally, disruptions in fetal tolerance are increasingly recognized in human cases of spontaneous abortion, preeclampsia and prematurity (Jiang et al., 2014; Santner-Nanan et al., 2009; Sasaki et al., 2007; Sasaki et al., 2004). In humans, over 10% of pregnancies are afflicted by these complications linked with disrupted fetal tolerance (Blencowe et al., 2012; Duley, 2009). Given this sustained backdrop of refining selection that likely occurs across all outbred mammalian species, conservation of phenotypic traits that improve reproductive fitness is a biological imperative.
Herein, this reasoning was applied to investigate the ontological conservation of NIMA-specific tolerance and maternal cell microchimerism across placental mammalian species (Andrassy et al., 2003; Bakkour et al., 2014; Dutta and Burlingham, 2011; Gammill et al., 2015). Using mice with defined MHC haplotype alleles in multi-generational breeding that transforms MHC haplotype alleles into surrogate NIMA, we show sharply increased resiliency against fetal wastage in the presence of overlap between NIMA and paternal-fetal antigen encountered during next generation pregnancies. Cross-generational reproductive benefits conferred by NIMA-specific tolerance shown here for mice are consistent with pioneering observations of reduced erythrocyte Rh antigen sensitization among Rh-negative women born to Rh-positive mothers (Owen et al., 1954). However, while developmental exposure to this single minor alloantigen does not prevent hemolytic disease of the newborn (Booth et al., 1953; Owen et al., 1954), we find broader non-inherited antigenic overlap between maternal grandmother and offspring that encompasses MHC haplotype alleles efficiently protects against fetal wastage (Figure 7). By establishing clear reproductive benefits for NIMA-specific tolerance applicable to all placental mammalian species, these results highlight broad evolutionary advantages for persistent postnatal NIMA-specific tolerance beyond averting anti-maternal immunity for human and other species with comparatively more developed fetal adaptive immune components at the time of birth (Mold and McCune, 2012; Mold et al., 2008).
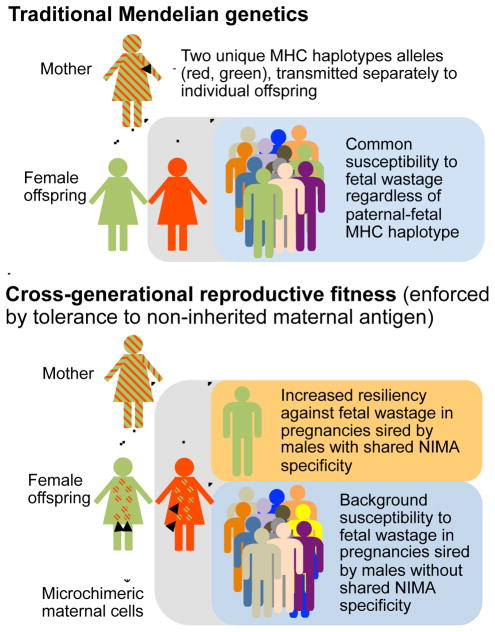
Figure 7. Cross-generational reproductive fitness enforced by vertically transferred microchimeric maternal cells in eutherian placental mammals.
In traditional Mendelian genetics (top), pregnancies among female offspring are equally susceptible to fetal wastage or other complications stemming from disruptions in fetal tolerance regardless of paternal MHC haplotype specificity. Comparatively, persistent postnatal maintenance of tolerogenic microchimeric maternal cells in female offspring promotes cross-generational reproductive fitness (bottom) by selectively protecting against fetal wastage during next generation pregnancies sired by males with shared overlapping NIMA specificity.
Dissecting the mechanistic relationship between NIMA-specific tolerance and microchimeric maternal cells that both persist in offspring through adulthood requires strategies for precisely identifying NIMA-specific immune components along with manipulation of microchimeric maternal cells. Prior limitations restricting these analyses were simultaneously bypassed by transforming defined model antigens into surrogate NIMA using female mice heterozygous for a transgene encoding constitutive expression of model antigens for breeding with non-transgenic males (Figure S1). Using antigen-specific tools, endogenous CD4+ T cells with surrogate NIMA specificity were shown to be highly enriched for expression of the Treg lineage defining transcriptional regulator, FOXP3 (Fontenot et al., 2003; Hori et al., 2003). By establishing NIMA specificity for this essential immune regulatory CD4+ T cell subset, these results extend previously described reversal of NIMA-specific tolerance by depleting of bulk CD4+ T cells or Tregs (Akiyama et al., 2011; Matsuoka et al., 2006; Mold et al., 2008; Molitor-Dart et al., 2007). In turn, protection against fetal wastage conferred by expanded Tregs with shared NIMA plus fetal specificity also reinforce beneficial properties of expanded maternal Tregs with pre-exiting fetal specificity retained after prior pregnancy in partner specific protection against complications in subsequent pregnancy (Campbell et al., 1985; Rowe et al., 2012b; Trupin et al., 1996).
More importantly, the concurrent ability to deplete maternal cells retained in offspring allowed us to definitively establish the causative relationship between microchimeric maternal cells and postnatal persistence of NIMA-specific tolerance. Similar to the necessity of low-level exposure to cognate antigen in numerical maintenance of ‘memory’ effector CD4+ T cells with foreign microbial specificity (Belkaid et al., 2002; Nelson et al., 2013; Uzonna et al., 2001), sustained expansion of NIMA-specific FOXP3+ CD4+ T cells also requires postnatal exposure to cognate maternal antigen expressed by microchimeric maternal cells. Reciprocally, in vivo depletion of microchimeric maternal cells efficiently overturned both expanded NIMA-specific Treg accumulation and protection against fetal wastage. Thus, vertically transferred maternal cells that establish microchimerism in offspring promote cross-generational reproductive fitness by preserving tolerance to NIMA along with non-inherited genetic alleles within a population (Figure 7).
In the broader context, these results indicate genetic fitness, canonically thought to be restricted to transmitting only half of homologous chromosomes through Mendelian inheritance, is enhanced in female placental mammals to also promote conservation of non-inherited antigens by vertical transmission of tolerogenic maternal cells that establish microchimerism in offspring. However in nature, this engrained drive for genetic fitness in each individual is likely counterbalanced by pathogen-mediated selection for MHC diversity across the entire population (Spurgin and Richardson, 2010). Nonetheless, our findings suggest more extended cross-generational analysis will illuminate the ongoing controversy regarding how MHC haplotype similarity impacts mate selection and pregnancy outcomes (Chaix et al., 2008; Israeli et al., 2014; Ober et al., 1997). Finally, reproductive advantages actively maintained by tolerogenic microchimeric maternal cells underscore the need for renewed consideration of immune tolerance from the intriguing perspective of constitutive chimerism beyond engrained pillars of binary “self” versus “non-self” antigen distinction defined using genetically homogenous inbred mice that artificially eliminates cross-generational tolerance (Jenkins et al., 2010; Nelson, 2012).
EXPERIMENTAL PROCEDURES
Mice
C57BL/6 (H-2b; CD45.2+), Balb/c (H-2d), CBA/J (H-2k), B6.C-H2d/bByJ (H-2d), B6.Ak-H2k/J (H-2k), and B6.SJL-PtprcaPepcb/BoyJ (H-2b; CD45.1+) mice were purchased from The Jackson Laboratory. 2W1S-OVA+ transgenic mice that constitutively express recombinant 2W1S55-68-OVA protein behind the β-actin promoter, and FOXP3DTR/DTR mice where FOXP3+ cells are susceptible to diphtheria toxin induced ablation have each been described (Kim et al., 2007; Moon et al., 2011). 2W1S-OVA+ mice were maintained on either the C57BL/6 or Balb/c strain backgrounds after backcrossing for >10 generations. For cross-fostering, pregnant mice were checked twice daily for birth timing, and newborn offspring introduced to lactating foster mothers within 12 hours after birth, with weaning 21 days thereafter and analysis at 8 weeks of age. For partial transient maternal Treg depletion, FOXP3DTR/WT pregnant females were administered purified diphtheria toxin daily (Sigma-Aldrich, USA) (0.5 μg first dose, followed by 0.1 μg/dose) beginning midgestation (E11.5) for 5 consecutive days, and the frequency of fetal resorption evaluated E16.5. All experiments were performed using sex and aged matched controls under Cincinnati Children’s Hospital IACUC approved protocols.
Tetramer enrichment and flow cytometry
Cell surface staining with phycoerythrin (PE)-conjugated MHC class II I-Ab:2W1S55-68tetramer followed by enrichment using anti-PE-conjugated magnetic beads (Miltenyi Biotec) have been described (Moon et al., 2007; Moon et al., 2009; Rowe et al., 2012b). To identify CD4+ T cells with I-Ab:2W1S specificity, cells in secondary lymphoid tissue (spleen plus axillary, brachial, cervical, inguinal, mesenteric, pancreatic, para-aortic/uterine lymph nodes) of each mouse were combined, enriched with PE conjugated I-Ab:2W1S55-68 tetramer, and stained for cell-surface CD4 (GK1.5), CD8α (53-7.3), CD25 (PC61), CD44 (IM7), CD11b (M1/70), CD11c (N418), B220 (RA3-B62), F4/80 (BM8), along with intranuclear FOXP3 (FJK-16s) or Helios (22F6) expression using commercially available antibodies and cell permeabilization reagents (BD PharMingen or eBioscience). For cell surface ovalbumin expression, cells were stained initially with polyclonal rabbit α-OVA (EMP Millipore) or IgG isotype antibodies followed by secondary staining with PE conjugated anti-rabbit IgG (eBioscience) antibody. Cells stained with fluorochrome-conjugated tetramer and/or antibody were acquired using a FACSCanto cytometer (Becton Dickinson), and analyzed using FlowJo (TreeStar) software.
Bacteria
For infection, Listeria monocytogenes (wildtype strain 10403s) was grown to early log phase (OD600 0.1) in brain heart infusion media at 37°C, washed and diluted with sterile saline, and inoculated intravenously via the lateral tail vein (104 CFUs) at midgestation (E11.5) as described (Chaturvedi et al., 2015; Rowe et al., 2011). The inoculum for each experiment was confirmed by spreading diluted aliquots onto agar plates. Five days thereafter, fetal resorption and in utero bacteria invasion was evaluated by sterilely dissecting each concepti, homogenization in sterile saline containing 0.05% Triton X-100 to release intracellular bacteria, plating serial dilutions of each concepti homogenate onto agar plates, and enumeration after incubation at 37°C for 24 hours.
DNA extraction and quantitative PCR
The heart, liver, uterus or prostate was sterilely dissected, and DNA extracted from each tissue using the QIAamp DNA extraction kit (Qiagen). Thereafter, PCR for enumerating 2W1S-OVA+ DNA was performed in 20 separate wells per tissue each containing 333 ng genomic DNA (~3.33 × 105 cells) in 20 μl total volume supplemented with 10 μl Taqman Gene Expression Master Mix and 1 μl ovalbumin Taqman assay (Applied Biosystems), for a detection limit of ~1 in 6.66 × 106 cells per tissue. Amplification was performed using the 7500 Fast Real-Time PCR System (Life Technologies) under the following program: 95°C for 10 minutes, followed by forty cycles of 95°C for 15 seconds and 60°C for 1 minute. For generating standard curve for 2W1S-OVA+ DNA, DNA from 2W1S-OVA+ splenocytes or C57BL/6 control mice were isolated, and combined with six serial 10-fold dilutions (10−1 to 10−6) of 2W1S-OVA+ DNA into C57BL/6 control DNA so that the DNA concentration remained identical in each well (333 ng total DNA in 20 μl). The resulting linear regression equation y = −1.137ln(x) + 38.443 (R2 = 0.986) was used to calculate the amount of 2W1S-OVA+ DNA in each tissue sample.
Depletion of microchimeric 2W1S-OVA+ maternal cells
To deplete 2W1S-OVA+ cells, 2W1S-NIMA mice were administered 650 μg purified rabbit α-OVA antibody (EMP Millipore) or IgG isotype antibody (Sigma-Aldrich) by intraperitoneal injection, followed 10 days later by a second treatment with 325 μg of the same antibody. Two days after the second antibody inoculation, the level of 2W1S-OVA+ cells in each tissue was analyzed by quantitative real-time PCR, antigen-specific CD4+ T cells investigated using I-Ab:2W1S55-68 tetramer staining, or used for mating with H-2d Balb/c or H-2k CBA/J males to investigate pregnancy outcomes.
Statistical analysis
Where applicable, NIMA mice in each group were randomized for either administration of anti-OVA or isotype antibody, or for breeding with either NIMA-matched or NIMA-discordant MHC haplotype males. Considering data sets did not consistently show a normal distribution, differences between groups were analyzed using the Mann-Whitney non-parametric test (Prism, GraphPad); and P < 0.05 was taken as statistical significance.
Supplementary Material
Acknowledgments
We thank Dr. Marc Jenkins for providing 2W1S-OVA transgenic mice; and Drs. James Moon, Louis Muglia, Joseph Qualls, Anne Stevens, Kevin Urdahl for helpful discussions. This work as supported by the NIH-NIAID through awards R01-AI100934 and R21-AI112186 (to SSW), and NIH-NHLBI through award R01-HL103745 (to AFS). SSW holds an Investigator in the Pathogenesis of Infectious Disease award from the Burroughs Wellcome Fund.
Footnotes
AUTHOR CONTRIBUTIONS
J.K., T.J., J.E., L.X., and B.S. performed the experiments. All authors participated in the experimental design and data analysis. J.K. and S.S.W. wrote the manuscript with editorial input from all the authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akiyama Y, Caucheteux SM, Vernochet C, Iwamoto Y, Tanaka K, Kanellopoulos-Langevin C, Benichou G. Transplantation tolerance to a single noninherited MHC class I maternal alloantigen studied in a TCR-transgenic mouse model. Journal of immunology. 2011;186:1442–1449. doi: 10.4049/jimmunol.1003023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrassy J, Kusaka S, Jankowska-Gan E, Torrealba JR, Haynes LD, Marthaler BR, Tam RC, Illigens BM, Anosova N, Benichou G, et al. Tolerance to noninherited maternal MHC antigens in mice. Journal of immunology. 2003;171:5554–5561. doi: 10.4049/jimmunol.171.10.5554. [DOI] [PubMed] [Google Scholar]
- Araki M, Hirayama M, Azuma E, Kumamoto T, Iwamoto S, Toyoda H, Ito M, Amano K, Komada Y. Prediction of reactivity to noninherited maternal antigen in MHC-mismatched, minor histocompatibility antigen-matched stem cell transplantation in a mouse model. Journal of immunology. 2010;185:7739–7745. doi: 10.4049/jimmunol.1001226. [DOI] [PubMed] [Google Scholar]
- Bakkour S, Baker CA, Tarantal AF, Wen L, Busch MP, Lee TH, McCune JM. Analysis of maternal microchimerism in rhesus monkeys (Macaca mulatta) using real-time quantitative PCR amplification of MHC polymorphisms. Chimerism. 2014;5:6–15. doi: 10.4161/chim.27778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y, Piccirillo CA, Mendez S, Shevach EM, Sacks DL. CD4+CD25+ regulatory T cells control Leishmania major persistence and immunity. Nature. 2002;420:502–507. doi: 10.1038/nature01152. [DOI] [PubMed] [Google Scholar]
- Billingham RE, Brent L, Medawar PB. Actively acquired tolerance of foreign cells. Nature. 1953;172:603–606. doi: 10.1038/172603a0. [DOI] [PubMed] [Google Scholar]
- Blencowe H, Cousens S, Oestergaard MZ, Chou D, Moller AB, Narwal R, Adler A, Vera Garcia C, Rohde S, Say L, et al. National, regional, and worldwide estimates of preterm birth rates in the year 2010 with time trends since 1990 for selected countries: a systematic analysis and implications. Lancet. 2012;379:2162–2172. doi: 10.1016/S0140-6736(12)60820-4. [DOI] [PubMed] [Google Scholar]
- Booth PB, Dunsford I, Grant J, Murray S. Haemolytic disease in first-born infants. British Medical Journal. 1953;2:41–42. [Google Scholar]
- Burlingham WJ, Grailer AP, Heisey DM, Claas FH, Norman D, Mohanakumar T, Brennan DC, de Fijter H, van Gelder T, Pirsch JD, et al. The effect of tolerance to noninherited maternal HLA antigens on the survival of renal transplants from sibling donors. N Engl J Med. 1998;339:1657–1664. doi: 10.1056/NEJM199812033392302. [DOI] [PubMed] [Google Scholar]
- Campbell D, MacGillivray I, Carr-Hill R. Pre-eclampsia in second pregnancy. Br J Obstet Gynaecol. 1985;92:131–140. doi: 10.1111/j.1471-0528.1985.tb01064.x. [DOI] [PubMed] [Google Scholar]
- Campbell DA, Jr, Lorber MI, Sweeton JC, Turcotte JG, Niederhuber JE, Beer AE. Breast feeding and maternal-donor renal allografts. Possibly the original donor-specific transfusion. Transplantation. 1984;37:340–344. doi: 10.1097/00007890-198404000-00004. [DOI] [PubMed] [Google Scholar]
- Chaix R, Cao C, Donnelly P. Is mate choice in humans MHC-dependent? PLoS Genet. 2008;4:e1000184. doi: 10.1371/journal.pgen.1000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi V, Ertelt JM, Jiang TT, Kinder JM, Xin L, Owens KJ, Jones HN, Way SS. CXCR3 blockade protects against Listeria monocytogenes infection induced fetal wastage. Journal of Clinical Investigation. 2015 doi: 10.1172/JCI78578. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claas FH, Gijbels Y, van der Velden-de Munck J, van Rood JJ. Induction of B cell unresponsiveness to noninherited maternal HLA antigens during fetal life. Science. 1988;241:1815–1817. doi: 10.1126/science.3051377. [DOI] [PubMed] [Google Scholar]
- Duley L. The global impact of pre-eclampsia and eclampsia. Semin Perinatol. 2009;33:130–137. doi: 10.1053/j.semperi.2009.02.010. [DOI] [PubMed] [Google Scholar]
- Dutta P, Burlingham WJ. Microchimerism: tolerance vs. sensitization. Curr Opin Organ Transplant. 2011;16:359–365. doi: 10.1097/MOT.0b013e3283484b57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta P, Molitor-Dart M, Bobadilla JL, Roenneburg DA, Yan Z, Torrealba JR, Burlingham WJ. Microchimerism is strongly correlated with tolerance to noninherited maternal antigens in mice. Blood. 2009;114:3578–3587. doi: 10.1182/blood-2009-03-213561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlebacher A. Mechanisms of T cell tolerance towards the allogeneic fetus. Nat Rev Immunol. 2013;13:23–33. doi: 10.1038/nri3361. [DOI] [PubMed] [Google Scholar]
- Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- Gammill HS, Stephenson MD, Aydelotte TM, Nelson JL. Microchimerism in women with recurrent miscarriage. Chimerism. 2015 doi: 10.1080/19381956.2015.1017241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama M, Azuma E, Komada Y. Tolerogenic effect of non-inherited maternal antigens in hematopoietic stem cell transplantation. Front Immunol. 2012;3:135. doi: 10.3389/fimmu.2012.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor FoxP3. Science. 2003;299:1057–1061. doi: 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- Ichinohe T, Uchiyama T, Shimazaki C, Matsuo K, Tamaki S, Hino M, Watanabe A, Hamaguchi M, Adachi S, Gondo H, et al. Feasibility of HLA-haploidentical hematopoietic stem cell transplantation between noninherited maternal antigen (NIMA)-mismatched family members linked with long-term fetomaternal microchimerism. Blood. 2004;104:3821–3828. doi: 10.1182/blood-2004-03-1212. [DOI] [PubMed] [Google Scholar]
- Israeli M, Kristt D, Nardi Y, Klein T. Genetic Considerations in Human Sex-Mate Selection: Partners Share Human Leukocyte Antigen but not Short-Tandem-Repeat Identity Markers. Am J Reprod Immunol. 2014;71:467–471. doi: 10.1111/aji.12213. [DOI] [PubMed] [Google Scholar]
- Jenkins MK, Chu HH, McLachlan JB, Moon JJ. On the composition of the preimmune repertoire of T cells specific for Peptide-major histocompatibility complex ligands. Annu Rev Immunol. 2010;28:275–294. doi: 10.1146/annurev-immunol-030409-101253. [DOI] [PubMed] [Google Scholar]
- Jiang TT, Chaturvedi V, Ertelt JM, Kinder JM, Clark DR, Valent AM, Xin L, Way SS. Regulatory T cells: new keys for further unlocking the enigma of fetal tolerance and pregnancy complications. Journal of immunology. 2014;192:4949–4956. doi: 10.4049/jimmunol.1400498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Rasmussen JP, Rudensky AY. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat Immunol. 2007;8:191–197. doi: 10.1038/ni1428. [DOI] [PubMed] [Google Scholar]
- Loubiere LS, Lambert NC, Flinn LJ, Erickson TD, Yan Z, Guthrie KA, Vickers KT, Nelson JL. Maternal microchimerism in healthy adults in lymphocytes, monocyte/macrophages and NK cells. Lab Invest. 2006;86:1185–1192. doi: 10.1038/labinvest.3700471. [DOI] [PubMed] [Google Scholar]
- Maloney S, Smith A, Furst DE, Myerson D, Rupert K, Evans PC, Nelson JL. Microchimerism of maternal origin persists into adult life. J Clin Invest. 1999;104:41–47. doi: 10.1172/JCI6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka K, Ichinohe T, Hashimoto D, Asakura S, Tanimoto M, Teshima T. Fetal tolerance to maternal antigens improves the outcome of allogeneic bone marrow transplantation by a CD4+ CD25+ T-cell-dependent mechanism. Blood. 2006;107:404–409. doi: 10.1182/blood-2005-07-3045. [DOI] [PubMed] [Google Scholar]
- Mold JE, McCune JM. Immunological tolerance during fetal development: from mouse to man. Adv Immunol. 2012;115:73–111. doi: 10.1016/B978-0-12-394299-9.00003-5. [DOI] [PubMed] [Google Scholar]
- Mold JE, Michaelsson J, Burt TD, Muench MO, Beckerman KP, Busch MP, Lee TH, Nixon DF, McCune JM. Maternal alloantigens promote the development of tolerogenic fetal regulatory T cells in utero. Science. 2008;322:1562–1565. doi: 10.1126/science.1164511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molitor ML, Haynes LD, Jankowska-Gan E, Mulder A, Burlingham WJ. HLA class I noninherited maternal antigens in cord blood and breast milk. Hum Immunol. 2004;65:231–239. doi: 10.1016/j.humimm.2003.12.006. [DOI] [PubMed] [Google Scholar]
- Molitor-Dart ML, Andrassy J, Kwun J, Kayaoglu HA, Roenneburg DA, Haynes LD, Torrealba JR, Bobadilla JL, Sollinger HW, Knechtle SJ, et al. Developmental exposure to noninherited maternal antigens induces CD4+ T regulatory cells: relevance to mechanism of heart allograft tolerance. Journal of immunology. 2007;179:6749–6761. doi: 10.4049/jimmunol.179.10.6749. [DOI] [PubMed] [Google Scholar]
- Moon J, Chu H, Pepper M, McSorley S, Jameson S, Kedl R, Jenkins M. Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity. 2007;27:203–213. doi: 10.1016/j.immuni.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon JJ, Chu HH, Hataye J, Pagan AJ, Pepper M, McLachlan JB, Zell T, Jenkins MK. Tracking epitope-specific T cells. Nat Protoc. 2009;4:565–581. doi: 10.1038/nprot.2009.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon JJ, Dash P, Oguin TH, 3rd, McClaren JL, Chu HH, Thomas PG, Jenkins MK. Quantitative impact of thymic selection on Foxp3+ and Foxp3− subsets of self-peptide/MHC class II-specific CD4+ T cells. Proc Natl Acad Sci USA. 2011;108:14602–14607. doi: 10.1073/pnas.1109806108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Suano A, Hamilton AB, Betz AG. Gimme shelter: the immune system during pregnancy. Immunol Rev. 2011;241:20–38. doi: 10.1111/j.1600-065X.2011.01002.x. [DOI] [PubMed] [Google Scholar]
- Mylonakis E, Paliou M, Hohmann EL, Calderwood SB, Wing EJ. Listeriosis during pregnancy: a case series and review of 222 cases. Medicine (Baltimore) 2002;81:260–269. doi: 10.1097/00005792-200207000-00002. [DOI] [PubMed] [Google Scholar]
- Nelson JL. The otherness of self: microchimerism in health and disease. Trends Immunol. 2012;33:421–427. doi: 10.1016/j.it.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson RW, McLachlan JB, Kurtz JR, Jenkins MK. CD4+ T cell persistence and function after infection are maintained by low-level peptide:MHC class II presentation. Journal of immunology. 2013;190:2828–2834. doi: 10.4049/jimmunol.1202183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober C, Weitkamp LR, Cox N, Dytch H, Kostyu D, Elias S. HLA and mate choice in humans. Am J Hum Genet. 1997;61:497–504. doi: 10.1086/515511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen RD. Immunogenetic Consequences of Vascular Anastomoses between Bovine Twins. Science. 1945;102:400–401. doi: 10.1126/science.102.2651.400. [DOI] [PubMed] [Google Scholar]
- Owen RD, Wood HR, Foord AG, Sturgeon P, Baldwin LG. Evidence for actively acquired tolerance to Rh antigens. Proc Natl Acad Sci USA. 1954;40:420–424. doi: 10.1073/pnas.40.6.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picus J, Aldrich WR, Letvin NL. A naturally occurring bone-marrow-chimeric primate. I. Integrity of its immune system. Transplantation. 1985;39:297–303. doi: 10.1097/00007890-198503000-00018. [DOI] [PubMed] [Google Scholar]
- Rees W, Bender J, Teague TK, Kedl RM, Crawford F, Marrack P, Kappler J. An inverse relationship between T cell receptor affinity and antigen dose during CD4(+) T cell responses in vivo and in vitro. Proc Natl Acad Sci USA. 1999;96:9781–9786. doi: 10.1073/pnas.96.17.9781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe JH, Ertelt JM, Aguilera MN, Farrar MA, Way SS. Foxp3(+) regulatory T cell expansion required for sustaining pregnancy compromises host defense against prenatal bacterial pathogens. Cell Host Microbe. 2011;10:54–64. doi: 10.1016/j.chom.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe JH, Ertelt JM, Xin L, Way SS. Listeria monocytogenes cytoplasmic entry induces fetal wastage by disrupting maternal FoxP3+ regulatory cell-sustained fetal tolerance. PLoS Pathogens. 2012a;8 doi: 10.1371/journal.ppat.1002873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe JH, Ertelt JM, Xin L, Way SS. Pregnancy imprints regulatory memory that sustains anergy to fetal antigen. Nature. 2012b;490:102–106. doi: 10.1038/nature11462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samstein RM, Josefowicz SZ, Arvey A, Treuting PM, Rudensky AY. Extrathymic generation of regulatory T cells in placental mammals mitigates maternal-fetal conflict. Cell. 2012;150:29–38. doi: 10.1016/j.cell.2012.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santner-Nanan B, Peek MJ, Khanam R, Richarts L, Zhu E, Fazekas de St Groth B, Nanan R. Systemic increase in the ratio between Foxp3+ and IL-17-producing CD4+ T cells in healthy pregnancy but not in preeclampsia. Journal of immunology. 2009;183:7023–7030. doi: 10.4049/jimmunol.0901154. [DOI] [PubMed] [Google Scholar]
- Sasaki Y, Darmochwal-Kolarz D, Suzuki D, Sakai M, Ito M, Shima T, Shiozaki A, Rolinksi J, Saito S. Proportion of peripheral blood and decidual CD4+ CD25(bright) regulatory T cells in pre-eclampsia. Clin Exp Immunol. 2007;149:139–145. doi: 10.1111/j.1365-2249.2007.03397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Sakai M, Miyazaki S, Higuma S, Shiozaki A, Saito S. Decidual and peripheral blood CD4+CD25+ regulatory T cells in early pregnancy subjects and spontaneous abortion cases. Mol Hu Reprod. 2004;10:347–353. doi: 10.1093/molehr/gah044. [DOI] [PubMed] [Google Scholar]
- Spurgin LG, Richardson DS. How pathogens drive genetic diversity: MHC, mechanisms and misunderstandings. Proc Biol Sci. 2010;277:979–988. doi: 10.1098/rspb.2009.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton AM, Korty PE, Tran DQ, Wohlfert EA, Murray PE, Belkaid Y, Shevach EM. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. Journal of immunology. 2010;184:3433–3441. doi: 10.4049/jimmunol.0904028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trupin L, Simon L, Eskenazi B. Change in paternity: a risk factor for preeclampsia in multiparas. Epidemiology. 1996;7:240–244. doi: 10.1097/00001648-199605000-00004. [DOI] [PubMed] [Google Scholar]
- Uzonna JE, Wei G, Yurkowski D, Bretscher P. Immune elimination of Leishmania major in mice: implications for immune memory, vaccination, and reactivation disease. Journal of immunology. 2001;167:6967–6974. doi: 10.4049/jimmunol.167.12.6967. [DOI] [PubMed] [Google Scholar]
- van Rood JJ, Loberiza FR, Jr, Zhang MJ, Oudshoorn M, Claas F, Cairo MS, Champlin RE, Gale RP, Ringden O, Hows JM, et al. Effect of tolerance to noninherited maternal antigens on the occurrence of graft-versus-host disease after bone marrow transplantation from a parent or an HLA-haploidentical sibling. Blood. 2002;99:1572–1577. doi: 10.1182/blood.v99.5.1572. [DOI] [PubMed] [Google Scholar]
- Zhou L, Yoshimura Y, Huang Y, Suzuki R, Yokoyama M, Okabe M, Shimamura M. Two independent pathways of maternal cell transmission to offspring: through placenta during pregnancy and by breast-feeding after birth. Immunology. 2000;101:570–580. doi: 10.1046/j.1365-2567.2000.00144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkernagel RM, Doherty PC. MHC-restricted cytotoxic T cells: studies on the biological role of polymorphic major transplantation antigens determining T-cell restriction-specificity, function, and responsiveness. Adv Immunol. 1979;27:51–177. doi: 10.1016/s0065-2776(08)60262-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.