Abstract
The growing epidemic of obesity and diabetes, the aging population as well as prevalence of drug abuse has led to significant increases in the rates of the closely associated acute and chronic kidney diseases, including diabetic nephropathy. Furthermore, evidence shows that parental behavior and diet can affect the phenotype of subsequent generations via epigenetic transmission mechanisms. These data suggest a strong influence of the environment on disease susceptibility and that, apart from genetic susceptibility, epigenetic mechanisms need to be evaluated to gain critical new information about kidney diseases. Epigenetics is the study of processes that control gene expression and phenotype without alterations in the underlying DNA sequence. Epigenetic modifications, including cytosine DNA methylation and covalent post translational modifications of histones in chromatin are part of the epigenome, the interface between the stable genome and the variable environment. This dynamic epigenetic layer responds to external environmental cues to influence the expression of genes associated with disease states. The field of epigenetics has seen remarkable growth in the past few years with significant advances in basic biology, contributions to human disease, as well as epigenomics technologies. Further understanding of how the renal cell epigenome is altered by metabolic and other stimuli can yield novel new insights into the pathogenesis of kidney diseases. In this review, we have discussed the current knowledge on the role of epigenetic mechanisms (primarily DNA me and histone modifications) in acute and chronic kidney diseases, and their translational potential to identify much needed new therapies.
Keywords: AKI, CKD, diabetic nephropathy, inflammation, fibrosis, transcription regulation, epigenetics, metabolic memory
Introduction
The rates of acute and chronic kidney diseases (CKDs) are growing worldwide and in the United States which in turn has increased the incidence of end stage renal disease (ESRD) requiring painful dialysis. This is projected to escalate further due to increased longevity, and the growing epidemic of diabetes and obesity which can lead to diabetic nephropathy (DN), a widespread CKD that can progress to ESRD. Chemical pollutants, aging, over and under nutrition, sedentary lifestyles, and other environmental stressors are implicated in many diseases, including CKD, a common complex gene-environmental disease1-4. Thus environmental factors can affect phenotypes via “epigenetic” transmission mechanisms to alter disease susceptibility, suggesting that, apart from genetic traits, epigenetic variations might yield valuable information about the initiation and progression of various kidney diseases5-7. Several genetic studies including Genome-wide Association studies (GWAS) have identified susceptibility loci and candidate genes for renal impairment and CKDs, including DN, albeit with relatively small effects that explain only a small proportion of heritability8. These observations, along with studies on disease-discordant monozygotic twins, provide a strong rationale to examine how epigenetic modifications may orchestrate the pathology of various kidney diseases.
Epigenetics refers to the study of heritable changes in gene expression and phenotype that are not mediated by alterations in the underlying DNA sequence of the genome. Although heritable traits refer to transmission from parent to offspring, in the context of the cell, it can refer to memories and patterns of gene expression and cellular states being passed on dynamically during replication to daughter cells9, 10. Epigenetic modifications include cytosine methylation of DNA (DNA methylation, DNAme), Histone post translational modifications (PTMs), and non-coding RNAs11-14. Together, they form an epigenetic layer over the genetic layer that can respond to environmental cues and external stimuli to alter the expression of genes associated with normal or disease phenotypes. Epigenetics plays critical roles in cell identity, development, genomic imprinting, X-inactivation and differential disease susceptibility between monozygotic twins10, 11, 13.
Recently, there has been remarkable progress in epigenetics research due in part to technological breakthroughs in next generation sequencing (NGS) and Epigenomics (genomewide study of epigenetics)15-17. The importance of epigenetic alterations in fibrosis, inflammation and immunity associated with various renal disorders, as well as in kidney development is increasingly being appreciated5, 18-20. This review summarizes the current knowledge on the role of epigenetics (primarily DNAme and histone PTMs) in acute and CKDs, and its translational potential to identify much needed new therapies.
Epigenetic Modifications and the Epigenome: Rationale for studies in the kidney
Epigenetic marks including DNAme, chromatin histone tail PTMs and non-coding RNAs collectively form the “epigenome”. Nucleosomes, the building blocks of chromatin, are made up of chromosomal DNA wrapped around core Histones (H2A, H2B, H3 and H4)11, 21. The epigenome is the interface between the stable genome and the variable environment and dictates cell-type-specific gene expression despite similarities in genetic DNA sequence5, 6, 12, 13. Perturbations in the epigenome have been implicated in various pathological conditions including cancer and diabetes6, 22, 23.
Methylation at the fifth position of cytosine (5mC) in DNA (DNAme) is a well established epigenetic mark13, 24. It is distributed throughout the genome generally at CpG dinucleotides which can occur in clusters known as CpG islands (CGIs). DNAme patterns are established during development by de novo DNA methyl transferases (DNMT) 3A and DNMT3B, while DNMT1 acts as a maintenance MT in later stages13. Promoter DNAme can repress transcription via interference with transcription factor binding or recruiting repressor complexes consisting of methyl-DNA binding proteins25. However, the regulatory effects of DNAme can vary from gene repression to activation depending on genomic contexts such as promoters, gene bodies, enhancers and repeat sequences13. DNAme is a relatively stable epigenetic modification, and DNA de-methylation was thought to occur mainly by passive mechanisms during development and cell division. But more recently, enzymes of Ten eleven translocation (TET) family (TET1/TET2/TET3) and thymine DNA glycosylases (TDG) have been shown to mediate active DNA de-methylation through oxidation of 5mc to 5-hydroxymethyl cytosine (5hmc) which is involved in diverse biological processes26, 27. Overall, the role of DNAme in gene regulation appears to be more complex than previously thought.
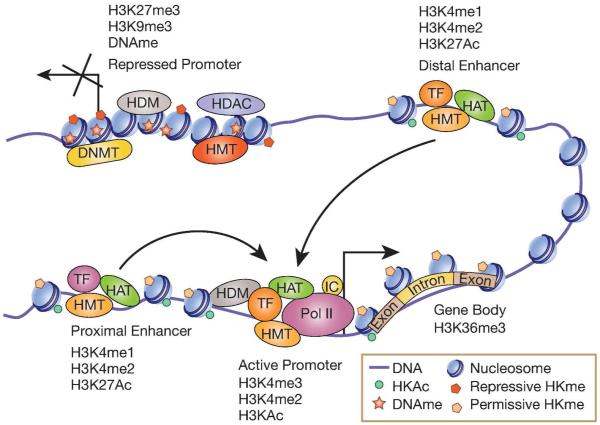
Covalent PTMs of core histones in chromatin, including histone lysine acetylation (HKAc) and methylation (HKme) are also epigenetic marks that regulate chromatin structure and gene expression11, 21. H3KAc is generally associated with relaxed chromatin and active gene expression. On the other hand, HKme can serve as active or repressive mark depending on the lysine residue modified and extent of methylation (mono-, di- or tri-). HKAc is catalyzed by histone acetyl transferases (HATs) and HKMe by histone methyltransferases (HMTs). In contrast, histone deacetylases (HDACs) and histone demethylases (HDMs) erase HKAc and HKme respectively. Histone modifications serve as docking sites for co-activators, co-repressors, chromatin remodeling proteins, and proteins that bind to modified histones11. Combinatorial effects of various histone PTMs form a ‘Histone code’ that dictates transcriptional outcomes by controlling compact (heterochromatin) or relaxed (euchromatin) states of chromatin28. Epigenomic profiling revealed that specific histone PTMs can mark and define distinct regulatory regions of the genome15, 21, 29. Histone H3 lysine 4 trimethylation (H3K4me3) is mostly associated with transcription start sites at promoters, and H3K36me3 with transcribed regions/gene bodies. H3/H4KAc and H3K4me2 are generally associated with active promoters, whereas, promoters of repressed genes are enriched with H3K9me3, H3K27me3 and H4K20me3. Enhancers are associated with H3K4me1 and H3K27Ac (Fig. 1).
Figure 1. Schematic of epigenetic modifications associated with chromatin function.
Active promoters are enriched with permissive histone modifications (H3KAc and H3K4me), while transcribed gene bodies are enriched with H3K36me3. In contrast, inactive promoters are associated with repressive epigenetic modifications including DNA methylation (DNAme) and histone modifications (H3K9me3 and H3K27me3) and reduced H3KAc due to actions of histone deacetylases (HDACs). In addition, inactive promoters could be enriched with specific histone demethylases (HDM) that erase permissive marks, while active promoters could be enriched with specific with HDMs that erase repressive marks. Enhancers are enriched with H3K4me1 and H3K4me2, and active enhancers are marked with H3K27Ac and histone acetyl transferases (HATs) such as P300. Enhancers can regulate the transcription of genes located several kilobases away by promoting chromatin conformation changes (bending) to promote long range interactions between enhancer bound transcription factors (TF) and co-activators with target gene promoters. Post translational modifications of other core histones, including histone H4, such as H4KAc and H4K20me3, as well as on other amino acids like arginine, are also important for chromatin function (not shown). HMT-histone methyl transferase; DNMT-DNA methyl transferase; IC-Transcription initiation complexes; Pol II-RNA Polymerase II.
Non coding (nc) RNAs such as short microRNAs (about 22 nucleotides in length) and long non coding RNAs (lncRNAs, >200 nucleotides long) are also part of the epigenetic layer and demonstrated to work via epigenetic mechanisms30-33. The roles of several miRNAs and a few lncRNAs in renal disorders including DN has recently been studied5, 34 and will not be discussed in this review.
Because epigenetic modifications are reversible, the epigenome is dynamically regulated in response to changing environment. In addition, the epigenome can exhibit a memory of prior exposure to environmental cues and disease conditions, with consequent long-lasting effects even after the initial trigger has been removed. Epigenetic changes can be inherited at cellular and tissue levels, and also transmitted to the offspring.35-37 Compelling evidence suggests that adverse intrauterine environment and maternal/paternal lifestyles or dietary habits could place offspring at risk for metabolic disorders, including obesity, through epigenetic transmission35, 38-40 which, in turn could lead to various renal complications. Interestingly, changes in metabolism can regulate epigenetic modifications due to alteration in metabolites such as Acetyl-coA, S-adenosyl methionine and α-ketoglutarate because they are utilized as co-factors by histone and DNA modifying enzymes13, 41, 42. Sedentary life style implicated in metabolic disorders could be associated with epigenetic abnormalities43.
The kidney is a complex multicellular organ that can be differentially affected by diverse stimuli. Although the component cells have basically the same genetic sequence, they have distinct epigenomes and elegant studies have demonstrated epigenetic cues in renal development and nephrogenesis19, 44, 45. Further understanding of how the renal cell epigenome is altered by metabolic and other stimuli can yield novel new insights into the pathogenesis of AKI and CKDs that can be translated to new therapies.
Tools to Study Epigenetic Modifications and the Epigenome
Epigenetics research has been spurred by the development of highly sensitive techniques and advances in genome sequencing that have aided in detecting epigenetic modifications like histone modifications and DNAme, chromatin structure (open or condensed), as well as long range interaction of enhancers in transcription regulation. Utility of these approaches to detect epigenetic changes at candidate genes has been extended to unbiased genome-wide location analyses by using microarrays and next generation sequencing (NGS)15, 16, 46-48. Whole transcriptome profiling by NGS (for coding and noncoding genes) and epigenome-wide association studies (EWAS), which combine immunoisolation of epigenetic marks and NGS, can yield information on genome-scale dynamic changes (Table 1), while sophisticated bioinformatic tools can analyze the emanating terabytes of data15, 46, 49-51. Such approaches have made it possible to predict different regulatory elements in chromatin, identify lncRNAs and alternate splicing based on specific genome locations of epigenetic modifications 21, 30, 46, 52. Notably, epigenomic profiling led to the characterization of enhancers in mammalian cells15, 29, 50, 51, including ‘super enhancers’53, enhancer derived RNAs54 and stimulus specific ‘latent enhancers’55. Chromatin conformation capture (3C) assays followed by sequencing (4C-seq) demonstrate long range enhancer interactions and regulation of promoters several hundred kilobases away56. EWAS also revealed that SNPs in non-coding regions may regulate chromatin structure and possibly enhancer functions in disease conditions57-60. Each of the methods used for epigenetic profiling has strengths and weaknesses. Major advantage of NGS based studies such as RNA-seq, ChIP-seq, bisulfite-Seq and others (Table 1) is that these unbiased approaches provide genome-wide and quantitative information unlike microarrays. However, nucleotide resolution and differences in costs as well as the complexity of data analyses are also key factors to be considered. In particular, there is a lot of interest in examining DNAme in clinical cohorts and choice of the specific technique among those available for the analysis of DNAme patterns (Table 1) would weigh in these specific advantages and disadvantages16, 61. Genome-wide quantification of sodium bisulfite-conversion based cytosine-DNAme is the most commonly used method which can be performed by either NGS platforms or the Infinium® Human Methylation 450K Bead-chips. 450K is widely used, especially for large scale clinical projects, as an easy and relatively affordable platform with base resolution and coverage of most CpG loci and islands. Bisulfite-Seq provides better resolution and genome-wide coverage compared with affinity based methods such as Methylated DNA immunoprecipitation-seq (MeDIP-seq) or MBD-seq, but is more expensive and involves more complex bioinformatics analysis62. Heterogeneity of cell types in renal tissues also poses a challenge in EWAS studies, since epigenomes and transcriptomes are cell-type specific. With the rapid advent in better technologies and reduced costs, EWAS are increasingly being performed in wide ranging experimental and clinical studies. Advances in epigenomics platforms and data analysis tools as well as publicly available datasets can therefore be exploited to gain new insights into the pathologies of common kidney diseases16, 61 and uncover targets for novel epigenetic therapies.
Table 1. Techniques used in Epigenome Profiling studies.
RNA-seq, RNA expression (transcriptome) analysis by sequencing166; GRO-seq, global nuclear run-on coupled with massively parallel sequencing167; smRNA-seq, isolation of small RNA fraction followed by sequencing for miRNAs168; DNase-seq, DNase I digestion followed by sequencing for chromatin accessibility50; MNase-seq, micrococcal nuclease digestion followed by sequencing169; FAIRE-seq, formaldehyde-assisted isolation of regulatory elements followed by sequencing for evaluating chromatin accessibility170; ATAC-seq, assay for transposase-accessible chromatin using sequencing171; MeDIP-seq, Methylated DNA immunoprecipitation followed by sequencing for DNA methylation (DNAme profiling)172; MBD-seq, methyl CpG binding domain (MBD) precipitation of genomic DNA followed by sequencing for DNAme173; Pyrosequencing, used to quantify DNA methylation at specific CpG sites174; Infinium® Human Methylation 450K BeadChip, microarray based methods widely used to detect changes in DNAme at over 485,000 loci covering 99% of Refseq genes and 96% of CpG sites175; ChIP-seq, chromatin immunoprecipitation followed by sequencing for detecting enrichment of chromatin binding factors genome-wide46; NOMe-seq, nucleosome occupancy and methylation sequencing176; X-ChIP–seq, crosslinking ChIP followed by sequencing177, 4C, chromatin confirmation capture (3C) followed by sequencing to study genome-wide interaction profile for a single locus178; Hi-C, 3C followed by enrichment of ligated products with biotin labeling and sequencing179.
| Chromatin Features | Techniques used | References |
|---|---|---|
| Transcriptome | RNA-seq | 166 |
| Gro-seq | 167 | |
| smRNA-seq | 168 | |
| Chromatin Structure, Chromatin Accessibilty | DNAse-seq | 50 |
| Mnase-seq | 169 | |
| FAIRE-seq | 170 | |
| ATAC-seq | 171 | |
| DNA methylation | Bisulfite-seq | 62 |
| MeDIP-seq | 172 | |
| MBD-seq | 173 | |
| Pyrosequencing | 174 | |
| 450 k bead chip | 175 | |
| Histone Modifications, Nucleosome Positioning |
ChIP-seq | 46 |
| NOME-seq | 176 | |
| X-ChIP-Seq | 177 | |
| Chromatin interacting Factors | ChIP-seq | 46 |
| Long rage interactions of Enhancers | 4C-seq | 178 |
| Hi-C | 179 |
Acute Kidney Injury (AKI) – Contribution of epigenetic mechanisms
AKI, a serious kidney disorder due to various causes, presents as rapid decline in renal function along with significant renal pathology. AKI is usually associated with high rates of mortality, and also increased the risk for CKD and ESRD63, 64. Several cellular and molecular mechanisms have been described for AKI65. AKI can affect renal vascular endothelial cells and tubular epithelial cells, with tubular necrosis apoptosis and tubular obstruction. Inflammation is major consequence of AKI with increased release of proinflammatory cytokines/chemokines, such as TNF-α, IL-6, IL-10, and MCP-1, by tubular and endothelial cells, as well as dendritic cells and infiltrating leukocytes/monocytes66. The kidney can repair/regenerate after AKI via various mechanisms. In recent years, epigenetic mechanisms have been shown to play a role in AKI which suggest potential therapeutical implications.
Chromatin remodeling from compact to open states allows transcription factor binding, and Pol II mediated transcription. This is facilitated by several mechanisms, including Adenosine triphosphate (ATPase)-catalyzed nucleosome remodeling67 mediated by numerous chromatin remodeling complexes , including the SWI/SNF complex which contains a key protein called Brahma-related gene-1 (BRG1). In early studies, nucleosome remodeling was demonstrated in AKI. Several inflammatory genes including Tnf and Ccl2 were transcriptionally upregulated in a renal ischemia reperfusion (I/R) model of AKI, with increased promoter Pol II and BRG1 recruitment along with enrichment of active chromatin histone marks, demonstrating an important role for BRG1 in the induction of these genes in tubular cells in AKI68-71.
A number of studies have evaluated the role of chromatin histone PTMs, especially HKAc, in the expression of genes involved in AKI66 . In one study, 70 increases in HKAc was associated with transcription of High mobility group (HMG) CoA reductase (Hmgcr) which contributes to cholesterol synthesis and cytoprotection during I/R in a model of AKI70. Another report examined the transcriptional repressor, ATF3 which is downregulated during AKI and, based on several lines of data, implicated ATF3 as a protective factor72. The results demonstrated that ATF3-mediated recruitment of HDAC1 at inflammatory gene promoters is protective during renal I/R injury72. Another study showed that I/R transiently reduced H3KAc in renal proximal tubules. But, during the recovery phase H3KAc was restored via HDAC5 downregulation and it was also associated with increased expression of the protective factor BMP7. These results suggest that HDAC5 could be potentially targeted to augment BMP7 in I/R73. In a mouse model of unilateral ureteral obstruction (UUO), H3KAc was decreased in the injured kidney with parallel increase in HDAC1/2 74. In a mouse model of AKI, there was a steady increase in H3K14Ac levels, but decreases in H4K5Ac and H3K12Ac levels75 further demonstrating that, in AKI there are dynamic changes in HKAc, a labile mark. Interestingly, in another study, the increases in renal HKAc after I/R evident one day after injury persisted even after three weeks76, suggesting that this memory of epigenetic chromatin modifications seen long after the initial AKI may possibly lead to CKD. I/R injury in rodents was also shown to up-regulate histone modifying systems in vivo, along with enrichment of histone modifications at promoters of inflammatory and fibrotic genes like Ccl2, Tgfb1 and collagens71.
The key role of HKAc in AKI and subsequent CKD is further supported by data showing protective effects of certain HKAc modifying drugs66. Such epigenetic modulators could be novel new therapeutic modalities for human AKI. The HDAC inhibitor Vorinostat had protective effects in some experimental models of renal injury77, 78, while another HDAC inhibitor conferred reno-protection in the UUO model79. The deactylase Sirt1 which targets both histone and non-histone proteins, displayed protective effects in several models by suppressing stress and aging related genes. Sirt1 knockdown augmented renal apoptosis and fibrosis in the UUO model80 while, conversely chemical activators of Sirt1 improved cell survival. Sirt1 activators could protect tubular functions after I/R in relatively older mice81. On the contrary, the deacetylase Sirt2 was found to have a proinflammatory role in the kidney during lipopolysaccharide (LPS) induced acute injury by mechanisms that included NF-κB activation and chemokine induction in proximal tubular epithelial cells. Sirt2 deficiency in mice was protective against LPS-induced infiltration of neutrophils and macrophages, acute tubular injury, and renal function decline82.
Certain natural products that modulate chromatin factors have also been evaluated. These include curcumin (the active component of turmeric) which is known to target the HAT, p300/CBP83. In animal studies, curcumin treatment had renoprotective effects in cisplatin induced AKI, along with decreased production of inflammatory cytokines such as TNF83, 84. Thus specific HATs, HDACs, SIRT isoforms and their modulators regulate renal inflammation in AKI by distinct mechanisms.
With regards to HKme, changes in the active mark, H3K4me3 and H3KAc, were observed at inflammatory and other genes in various models of AKI68-70. The expression of the H3K4 MT- Set1 was also enhanced71. Overall, several active chromatin marks (but not repressive) depict dynamic changes in AKI. Together these studies suggest the importance of chromatin histone PTMs in the pathogenesis of AKI (Fig. 2). This is an active area of research that is expected to soon reveal additional histone PTMs altered in AKI.
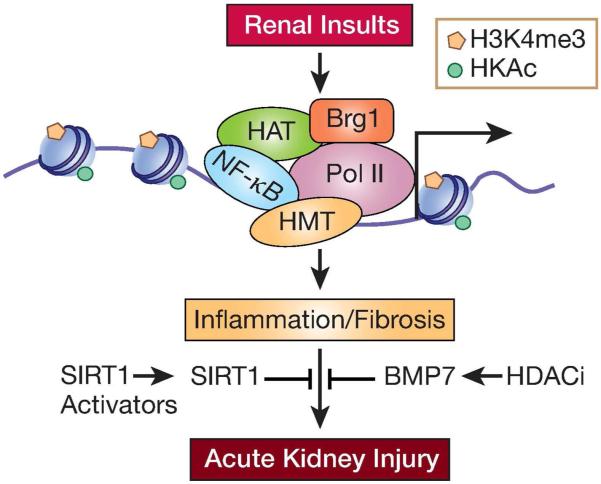
Figure 2. Histone modifications in AKI.
Epigenetic mechanisms have been implicated in the expression of inflammatory and fibrotic genes during AKI. AKI activates transcription factors such as NF-κB and increases permissive histone modifications (H3KAc and H3K4me) via increased recruitment of histone acetyl transferases (HATs) and relevant histone methyl transferases (HMTs), which promote chromatin remodeling by Brg1 and increased transcription by RNA Polymerase II. AKI has also been shown to promote DNA-demethylation to de-repress certain genes (not shown). Reversal of these epigenetic events (for example by SIRT1 activators) could inhibit pathological gene expression associated with AKI. TF-transcription factor; HAT-histone acetyl transferase; HMT-histone methyl transferase. Pol II-RNA Polymerase II.
A few studies have reported that AKI can cause DNAme (5mC) alterations in AKI 66. The first report showed de-methylation of a cytosine residue within the complement C3 promoter along with upregulation of C3 by renal I/R injury85. More recently, in a mouse model of AKI, global levels of 5hmC (but not 5mC) were decreased86. Interestingly, this was associated with changes in levels of TET1 and TET2 which are involved in 5mC to 5hmC conversion and DNA de-methylation. Thus it appears that DNA de-methylation and dynamic changes in 5hmc are more likely to be involved in AKI. This is quite logical since, DNA-de-methylation is quite dynamic and amenable to alterations during a rapidly developing disorder like AKI to affect the expression of pathological or protective genes.
Epigenetic Modifications and CKD, including Diabetic Nephropathy
CKD is a major global health problem affecting nearly 20% of adults. It is characterized by proteinuria and reduced glomerular filtration rate, and can lead to ESRD requiring dialysis. Diabetes and obesity are the leading cause of CKD. Features of CKD and DN include glomerular hypertrophy, mesangial expansion, tubulointerstial fibrosis and excess accumulation of extracellular matrix proteins, basement membrane thickening and podocyte loss87-89. High glucose (HG) is a major pathological mediator that drives DN via multiple biochemical and signaling mechanisms in kidney cells including, tubular epithelial cells, fibroblasts, endothelial cells, mesangial cells and podocytes87-89. This leads to production of related diabetogenic agents such as AGEs, growth factors such as TGF-β and Angiotensin II, and inflammatory cytokines87, 88. TGF-β is a major downstream effector of HG induced renal damage and is also induced by the other diabetogenic factors, which further amplifies HG mediated pathological events87, 90, 91. Numerous studies have identified various mechanisms, effector transcription factors, such as Smads and NF-κB, and gene regulation mechanisms in the pathogenesis of DN that will not be detailed here. Despite this extensive knowledge, further investigation is needed, because currently used treatments cannot effectively prevent progression of DN in many participants92. Recently, miRNAs have been demonstrated to be important players in DN by regulating the expression of fibrotic, inflammatory and other genes5, 34, 93, 94. Notably, epigenetic mechanisms such as DNAme and histone PTMs may be key missing links worth investigating, since pro-inflammatory and pro-fibrotic genes can be regulated by HG via epigenetic mechanisms in vascular cells, monocytes and mesangial cells5, 95-97.
Compelling evidence comes from clinical studies demonstrating the role of ‘metabolic memory’, described as the long term protective effects of prior good glycemic control on diabetic complications development, or persistence of the deleterious effects of prior hyperglycemic exposure long after glucose normalization. The Diabetes Control and Complications Trial (DCCT) showed that glucose normalization with intensive insulin treatment reduced microvascular complications including DN relative to conventional therapy. In the follow up Epidemiology of Diabetes Intervention and Complications (EDIC) study, all DCCT participants were placed on intensive therapy, which led to similar HbA1c levels in both treatment groups98. However, participants who were on conventional therapy in DCCT still continue to develop vascular complications, including DN, at higher rates during EDIC relative to the original intensive treatment group, implicating ‘metabolic memory’98, 99. Beneficial effects of early glycemic control termed ‘legacy effect’ have also been observed in clinical trials with Type 2 diabetes participants100. In addition, metabolic memory of vascular inflammation, fibrosis and related pathological gene expression has been demonstrated in several experimental models of diabetic complications using cell culture and animal models, though CKD was not evaluated6, 101-107. These studies revealed persistent changes in epigenetic modifications and related histone modifying enzymes at pathological gene promoters along with corresponding gene expression even after removal from diabetic conditions, thus supporting a role for epigenetics in metabolic memory. This notion is further supported by the recent epigenomics study in human diabetic complications and metabolic memory that examined variations in key histone PTMs in white blood cells from two groups of patients the DCCT/EDIC cohort, namely a control group (from the former intensive treated) and a case group (former conventional treated), with the cases experiencing greater complications (including DN) than controls. The results showed significant enrichment of promoter H3K9Ac in monocytes of cases versus controls at a set of genes with functions related to inflammation and diabetic complications. Monocyte H3KAc also showed strong association with mean HbA1c over DCCT and EDIC time periods108.
CKD and DN are common diseases affected by genetic and environment factors. Therefore, more in-depth studies of epigenetic mechanisms related to CKD, DN and metabolic memory that can take advantage of advances in epigenomics tools and publicly available databases can yield novel new information that could be translated to better treatment options.
DNAme in CKD and DN
Increasing evidence is implicating DNAme variations in CKDs including DN6. Changes in DNAme were observed at diabetes susceptibility genes including UNC13B in whole blood from diabetes subjects with versus without DN 109. Saliva from patients with ESRD participants also showed changes in DNAme versus those with CKD alone110. Because the epigenome is cell-type specific, the heterogeneity of blood cells and renal tissues poses a challenge while analyzing epigenetic data from such material. Refined laser capture microdissection techniques can aid in procuring enriched glomeruli and tubuli. from limited amount of renal biopsies to gain valuable insights into the epigenome in CKD versus normal states. In one such study, DNAme profiling of tubuli from subjects with CKD demonstrated significant changes in DNAme versus normal controls, with predominant variations occurring at enhancers that were also correlated with the increased expression of key fibrotic genes111. These results provide the first evidence of association between DNAme variations and fibrosis in human CKD using relevant kidney samples and represent an important step in CKD epigenome research. Another study using genomic DNA from participants in the Chronic Renal Insufficiency (CRIC) cohort found correlations between the rate of loss of renal function and changes in DNAme at genes involved in epithelial to mesenchymal transition, inflammation and fibrosis112. In a recent study that analyzed methylome-wide loci in participants with CKD found significant changes at CGIs of 23 genes including ELMO1 and PRKAG2113, further supporting the role of DNAme in CKD.
Involvement of DNAme in kidney disease is also supported by experimental studies. Profibrotic TGF-β inhibited the expression of Rasal1, a negative regulator of Ras signaling, by promoting DNAme at its promoter via Dnmt1, leading to Ras activation and increased fibrosis in fibroblasts114. This was reversed by BMP-7, which upregulated Rasal1 by inducing Tet3-mediated promoter DNA de-methylation 115. Furthermore, HG-induced DNA hypomethylation and concomitant increases in H3K9Ac were involved in the upregulation of p66Shc which promotes mitochondrial oxidant stress in DN116. Communication between renal cells like tubular cells and podocytes plays important roles in kidney function and a recent extensive study showed the involvement of epigenetic mechanisms117. Using proximal tubule specific Sirt1 deficient and transgenic mice, it was shown that tubular Sirt1 attenuates albuminuria by epigenetic mechanisms including DNAme to suppress podocyte expression of Claudin-1, a tight junction protein. Sirt1 was downregulated in diabetes and this could promote albuminuria and DN117. Clinical samples and experimental animal models of nephropathy depicted downregulation of the transcription factor KLF4 in podocytes, which was associated with increased DNAme at the nephrin (Nphs1) promoter and downregulation of nephrin, thus leading to podocyte apoptosis and proteinuria. KLF4 overexpresion had renoprotective effects, whereas, in contrast its knockout enhanced proteinuria further supporting a role for KLF4 in podocyte survival118. Recently, aberrant DNAme was reported at several gene promoters including hypomethylation of Angiotensinogen gene (Agt) in renal proximal tubules from diabetic db/db mice. Treatment of cultured proximal tubular cells with inhibitors of DNMT and HDAC increased Agt expression, supporting an epigenetic switch in the expression of key genes involved in DN. Some of these changes in db/db mice were resistant to pioglitazone treatment, suggesting that anti-diabetic therapy alone is not effective in reversing epigenetic changes119.Together, these emerging findings clearly demonstrate that DNAme variations are evident in CKD (Fig. 3). Further studies are likely to shed light on when and how DNAme is altered during the course of CKD, whether reversal of DNAme variations seen in the early stages of the disease can ameliorate renal dysfunction and/or prevent further progression to ESRD. Examination of DNAme changes at key regulatory regions including enhancers, and near SNPs reported to be associated with renal function decline, can illuminate functional relationships since such changes can affect the expression of genes directly related to CKD.
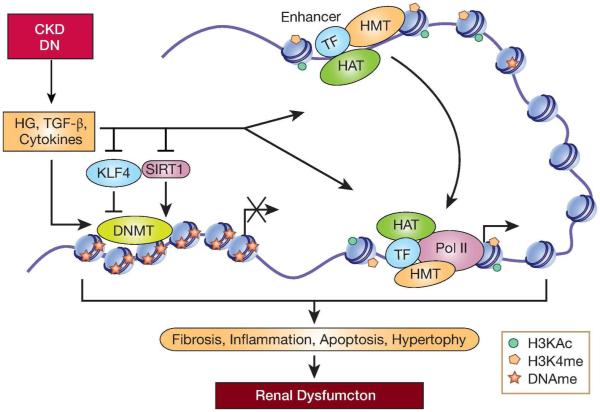
Figure 3. Role of DNA Methylation in CKD.
DNAme can regulate genes associated with CKD in various renal cells. In fibroblasts, the pro-fibrotic TGF-β can promote fibrosis by inhibiting RASAL1 expression through promoter hypermethylation leading to activation of Ras signaling and fibrosis. In the normal kidney KLF4 regulates DNAme to increase nephrin expression in podocytes. But inhibition of KLF4 expression in disease conditions increases DNAme and inhibits nephrin expression leading to podocyte apoptosis. In contrast, metabolites generated by SIRT1 in normal tubular epithelial cells repress Claudin-1 expression in glomerular podocytes via promoter hypermethylation.SIRT1 downregulation under diabetic conditions relieves this repression leading to Claudin-1 expression and glomerular dysfunction. Furthermore, EWAS studies using tubular biopsies of patients with CKD showed changes in DNAme at enhancers that potentially regulate fibrotic genes. TF-transcription factor; HAT-histone acety transferase; HMT-histone methyl transferase. Pol II-RNA Polymerase II. DNMT, DNA methyl transferases.
Histone PTMs and CKD
Several lines of evidence also support the role of histone PTM variations in CKD. Key events related to CKD such as dyregulated expression of fibrotic, cell cycle and inflammatory genes have recently been associated with changes in histone PTMs, particularly, in cell culture models treated with HG and TGF-β. These studies showed TGF-β induced enrichment of active epigenetic marks H3K9/14Ac, H3K4me1 and H3K4me3, and decreased levels of repressive marks like H3K9me3 at pro-fibrotic gene promoters. Furthermore, TGF-β upregulated the H3K4-methyltransferase SET7, which enhanced H3K4me1 at TGF-β induced genes, and SET7 knockdown inhibited TGF-β-induced fibrotic gene expression, implicating SET7 as a key player in DN97. Interestingly, HG also triggered similar epigenetic events, which were inhibited by a TGF-β neutralizing antibody, implicating TGF-β in HG-induced effects97. Furthermore, TGF-β also promoted p300 recruitment, with concomitant increase in H3K9/14Ac and chromatin relaxation at fibrotic gene promoters and Smad2/3 acetylation/activation, leading to increased fibrosis120. Thus, TGF-β uses both epigenetic histone PTMs and non-epigenetic mechanisms to regulate the expression of genes relevant to DN pathogenesis.
Endoplasmic reticulum (ER) stress plays a key role in DN by regulating inflammatory and apoptotic pathways121, 122 and one report examined the role of epigenetic mechanisms. ER-stress related genes including Xbp1 were up-regulated in the kidneys from diabetic db/db mice. ER-stress increased SET7 levels through XBP-1, which led to increased H3K4me1 at the Ccl2 promoter and its upregulation. These events were blocked by treatment of db/db mice with the ER-stress inhibitor betaine. Furthermore, SET7 siRNAs also blocked Ccl2 expression123. In another study, myocardin-related transcription factor A (MRTF-A) was induced by HG in tubular epithelial cells, which in turn promoted recruitment of HAT p300 and WD repeat-containing protein 5 (WDR5), a key component of the active H3K4 methyltransferase complex, at the Collagen 1 promoter and increased H3K18/K27Ac as well as H3K4me3, to up-regulate its expression. In Mrtf-A knockout mice, fibrotic gene expression and DN development were attenuated, further implicating Mrtf-A in fibrosis124. Promoter HKAc mediated chromatin relaxation were also demonstrated in TGF-β induced sustained up-regulation of miR-192, a key pro-fibrotic miRNA in MC. This study demonstrated a novel interplay between Smads and Ets-1 transcription factors, HATs and HKAc as well as Akt kinase activation in miR-192 expression125.The mediatory role of H3KAc was also demonstrated in HG induced downregulation of miR-29 via HDAC4 induced de-acetylation of miR-29 promoter in podocytes, leading to up-regulation of miR-29 targets including HDAC4 and fibrotic genes126. Thus, there is substantial evidence to demonstrate the pivotal role of both histone and non-histone lysine-Ac in regulating genes associated with DN pathogenesis (Fig. 4). Results from diabetic mice models also revealed similar changes in H3K9Ac at fibrotic and inflammatory gene promoters120, 125, 127-129. One study found increased expression of HDAC-2, -4 and -5 in kidneys of type 1 diabetic rats and biopsies from diabetic patients. In vitro experiments showed HDAC4 was upregulated by HG, TGF-β and AGEs, and that HDAC4-STAT1 signaling promotes podocyte injury. Furthermore, HDAC4 knockdown using intrarenal delivery of lentivruses ameliorated renal injury in diabetic rats128.
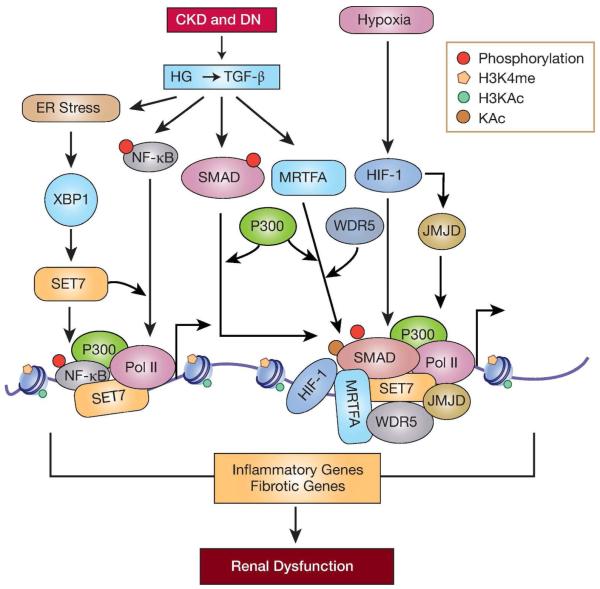
Figure 4. Fibrotic and inflammatory gene regulation by histone modifications in CKD.
In diabetic nephropathy and other CKDs signal transduction events triggered by pathological factors such as high glucose (HG) and downstream effectors including TGF-β induce the expression and activation of key transcription factors such as SMAD, NF-κB, XBP1 and MRTFA as well as histone modifying enzymes such as SET7 in glomerular and tubular cells. These signaling events also lead to recruitment of HMTs (such as SET7), HATs (such as P300) and chromatin proteins such as (WDR5) at pro-fibrotic and pro-inflammatory gene promoters leading to increases in permissive histone modifications (H3KAc and H3K4me) and chromatin relaxation. This enhances chromatin access to transcription factors and RNA Polymerase II (Pol II), which increases expression of pro-fibrotic and pro-inflammatory genes implicated in renal dysfunction. . Renal hypoxia also promotes tubular fibrosis in CKD. Hypoxia-inducible factor -1 (HIF-1) co-operates with histone modifying enzymes (JMJD) to regulate target genes. Persistence of such changes in epigenetic modifications (Fig 2-4) could be the underlying mechanisms involved in transcription memory (in AKI) or ‘metabolic memory’ (in diabetic nephropathy) associated with sustained increased risk for CKD and ESRD; JMJD-JMJD1A and JMJD2B. Pol II-RNA polymerase II.
The in vivo role of HKme in DN has been reported in a few studies. Kidneys from diabetic rats and mice showed increased H3K4me2 and RNA Polymerase II, but decreased H3K27me3 levels, at fibrotic genes. However, the corresponding histone modifying enzymes showed different expression patterns in rats versus mice kidneys130, suggesting species-specific differences (possibly related to severity of DN). Another in vivo study examined multiple histone PTMs in glomeruli from diabetic db/db mice and control db/+ mice129 to obtain a comprehensive picture of histone code alterations and cross talk amongst histone PTMs during DN. Results showed alteration in some of the histone PTMs examined, including H3K9/14Ac, at genes encoding PAI-1 and RAGE in db/db mice relative to db/+, which could facilitate open chromatin states as indicated by enhanced enrichment of RNA polymerase II, and increased gene expression. Several HATs including Tip60, and H3K4-MTs including SET7 were also upregulated in db/db mice129.
Notably, the protective effects of HDAC inhibitors have been demonstrated in animal models of DN78, 131-134, further supporting the role of HKAc and epigenetic mechanisms in renal dysfunction. However, non-histone acetylation mediated by HATs cannot be ignored. This was highlighted in a recent study demonstrating a critical role for STAT3 acetylation in DN. It was found that diabetes increases NF-κB and STAT3 acetylation via downregulation of Sirt1 in podocytes to promote pathologic gene expression and this effect can be mimicked by Sirt1 deletion in db/db mice135. Thus, a complex network of epigenetic factors and transcription factors regulate pathologic gene expression in CKD. Cell necrosis and apoptosis in later stages of CKD might significantly affect epigenetic profiles. Further understanding of these mechanisms and determination of time course of changes during CKD are needed which will be of utmost importance when designing epigenetic therapies.
Chronic hypoxia in renal tubular interstitial compartments can lead to increased fibrosis in CKD, and other kidney disorders136-140. Gene expression under hypoxic conditions is controlled by a master regulatory transcription factor, hypoxia-inducible factor-1 (HIF1) which binds to the promoter of target genes to induce their expression141. Under normoxic conditions, HIF-1 degradation is promoted by prolyl hydroxylase, but under hypoxic conditions, prolyl hydroxylase activity is lost leading to stabilization of HIF-1141. Interestingly several epigenetic changes were reported to be associated with hypoxia142, 143. Moreover, HIF-1 was shown to induce the expression of several epigenetic regulators such as histone demethylases in mammalian cells144, 145. HIF-1 can assist in promoting dynamic changes in chromatin conformation146. The epigenetic modifying properties of HIF-1 could be exploited for better approaches to curb chronic hypoxia induced tubulointerstitial fibrosis in various kidney disorders140.
Epigenetic Mechanisms in Hypertension
Hypertension is a major risk factor for cardiovascular disease and ESRD147. Disorders of Na+-transport, re-absorption and excretion in the renal collecting duct contribute to abnormal blood pressure in humans. This process is mostly regulated by the mineralocorticoid aldosterone via upregulation of the tubular epithelial sodium channel (ENaC). ENaC is composed of three sub units (α, β and γ), of which ENACα (ENaCa) regulates ion-transport. Recent studies have demonstrated epigenetic control mechanisms for basal and aldosterone induced ENaCa transcription148. Under basal conditions, the H3K79-methyltransferase Disrupter of Telomere Silencing (Dot)1a and corresponding H3K79me were enriched at the ENaCa promoter keeping it constrained but poised for activation by aldosterone and other stimuli. This repression was mediated by Dot1a complex formation with Sirt1 and other key proteins at the ENaCa promoter. A series of studies suggested that EnaCa repression can be relieved by multiple mechanisms,149-154 in all of which, levels of H3K79me were reduced at the ENaCa promoter supporting a critical role for Dot1a activity. However, several questions remain unanswered such as how H3K79me, which is normally associated with transcription activation, mediates repression. Furthermore, deacetylase activity of Sirt1 was not required for this repression suggesting interaction of the Dot1a -Sirt1 complex with additional repressors is critical. Angiotensin 1converting enzyme (ACE1) also plays important role in hypertension by activating the renin-angiotensin system. Hypertension induced by high salt diet is associated with upregulation of ACE1 (Ace) via increases in activation marks (H3KAc and H3K4me) and decreases in repressive mark (H3K9me2) at the Ace promoter155. Epigenetic mechanisms have also been implicated in Ace upregulation in hypertensive offspring of mothers exposed to low protein diet156. Study of epigenetics in hypertension and its potential heritability is clearly warranted and more investigations are anticipated157.
Perspectives
Increasing evidence suggests a critical role for epigenetic modifications in AKI and CKD. Importantly, the reversibility of epigenetic marks yields a window of opportunity for the development of novel epigenetic drugs. Several studies have explored the potential beneficial effects of HDAC inhibitors in animal models of AKI and CKD78, 127, 131-133, 158-160. However, since the specificity and the mechanism of action of these inhibitors are not fully clear, more work is needed before they can be evaluated in humans. Regulators of other epigenetic modifications including histone KMe and DNAme are also worth investigating, suggesting the need for novel screening approaches to discover additional epigenetic modulators specific to renal diseases. Recently, a zebra fish model was used to screen novel HDAC inhibitors that could accelerate recovery in AKI models of mice161. Several epigenetic drugs targeting histone modifying enzymes that are currently being used in cancer treatment13, 162, 163 could be tested for CKDs. Currently available drugs such as Losartan cannot prevent progression to CKD in many patients164. Experimental evidence shows that these drugs could not completely inhibit diabetes induced epigenetic mechanisms in the kidney129, suggesting that a combination of epigenetic drugs with conventional therapies might be more effective for certain kidney diseases. This was tested in a mouse model of HIV-induced nephropathy. Results showed that a combination of benazepril (ACE inhibitor) and vorinostat (HDAC inhibitor) significantly reduced nephropathy compared with each drug alone.165 Epigenetic therapy is clearly an exciting emerging field that is expected to gain momentum in the upcoming years as more epigenomic data emanates from ongoing basic and clinical studies with renal disease models. Several roadblocks and challenges should also be overcome, including low specificity/selectivity and unwanted side effects.
Acknowledgements
We gratefully acknowledge funding from the National Institute of Health (grants R01 DK081705, R01 DK058191, R01 HL106089-01 and R01 DK065073), and from the Juvenile Diabetes Research Foundation (17-2012-480)
Footnotes
Disclosures
None
References
- 1.Jain N, Reilly RF. Effects of dietary interventions on incidence and progression of CKD. Nat Rev Nephrol. 2014;10:712–724. doi: 10.1038/nrneph.2014.192. [DOI] [PubMed] [Google Scholar]
- 2.Menon V, Wang X, Sarnak MJ, et al. Long-term outcomes in nondiabetic chronic kidney disease. Kidney Int. 2008;73:1310–1315. doi: 10.1038/ki.2008.67. [DOI] [PubMed] [Google Scholar]
- 3.Leung KC, Tonelli M, James MT. Chronic kidney disease following acute kidney injury-risk and outcomes. Nat Rev Nephrol. 2013;9:77–85. doi: 10.1038/nrneph.2012.280. [DOI] [PubMed] [Google Scholar]
- 4.Jha V, Garcia-Garcia G, Iseki K, et al. Chronic kidney disease: global dimension and perspectives. Lancet. 2013;382:260–272. doi: 10.1016/S0140-6736(13)60687-X. [DOI] [PubMed] [Google Scholar]
- 5.Kato M, Natarajan R. Diabetic nephropathy-emerging epigenetic mechanisms. Nat Rev Nephrol. 2014;10:517–530. doi: 10.1038/nrneph.2014.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reddy MA, Tak Park J, Natarajan R. Epigenetic modifications in the pathogenesis of diabetic nephropathy. Semin Nephrol. 2013;33:341–353. doi: 10.1016/j.semnephrol.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tonna S, El-Osta A, Cooper ME, et al. Metabolic memory and diabetic nephropathy: potential role for epigenetic mechanisms. Nat Rev Nephrol. 2010;6:332–341. doi: 10.1038/nrneph.2010.55. [DOI] [PubMed] [Google Scholar]
- 8.McKnight AJ, McKay GJ, Maxwell AP. Genetic and epigenetic risk factors for diabetic kidney disease. Adv Chronic Kidney Dis. 2014;21:287–296. doi: 10.1053/j.ackd.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 9.Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 10.Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science. 2010;330:612–616. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 12.Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28:1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 13.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 14.Peschansky VJ, Wahlestedt C. Non-coding RNAs as direct and indirect modulators of epigenetic regulation. Epigenetics. 2014;9:3–12. doi: 10.4161/epi.27473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunham I, Kundaje A, Aldred SF, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mimura I, Kanki Y, Kodama T, et al. Revolution of nephrology research by deep sequencing: ChIP-seq and RNA-seq. Kidney Int. 2014;85:31–38. doi: 10.1038/ki.2013.321. [DOI] [PubMed] [Google Scholar]
- 17.Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 18.Natarajan R. Introduction: epigenetics and the kidney. Semin Nephrol. 2013;33:311–313. doi: 10.1016/j.semnephrol.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 19.Patel SR, Dressler GR. The genetics and epigenetics of kidney development. Semin Nephrol. 2013;33:314–326. doi: 10.1016/j.semnephrol.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beckerman P, Ko YA, Susztak K. Epigenetics: a new way to look at kidney diseases. Nephrol Dial Transplant. 2014;29:1821–1827. doi: 10.1093/ndt/gfu026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 22.You JS, Jones PA. Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell. 2012;22:9–20. doi: 10.1016/j.ccr.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ling C, Groop L. Epigenetics: a molecular link between environmental factors and type 2 diabetes. Diabetes. 2009;58:2718–2725. doi: 10.2337/db09-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen ZX, Riggs AD. DNA methylation and demethylation in mammals. J Biol Chem. 2011;286:18347–18353. doi: 10.1074/jbc.R110.205286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bird AP, Wolffe AP. Methylation-induced repression--belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 26.Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502:472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song CX, Yi C, He C. Mapping recently identified nucleotide variants in the genome and transcriptome. Nat Biotechnol. 2012;30:1107–1116. doi: 10.1038/nbt.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 29.Heintzman ND, Stuart RK, Hon G, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 30.Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20:300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 32.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kato M, Natarajan R. MicroRNA circuits in transforming growth factor-beta actions and diabetic nephropathy. Semin Nephrol. 2012;32:253–260. doi: 10.1016/j.semnephrol.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szyf M. Nongenetic inheritance and transgenerational epigenetics. Trends Mol Med. 2015;21:134–144. doi: 10.1016/j.molmed.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 37.Hackett JA, Sengupta R, Zylicz JJ, et al. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science. 2013;339:448–452. doi: 10.1126/science.1229277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei Y, Yang CR, Wei YP, et al. Paternally induced transgenerational inheritance of susceptibility to diabetes in mammals. Proc Natl Acad Sci U S A. 2014;111:1873–1878. doi: 10.1073/pnas.1321195111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simmons R. Epigenetics and maternal nutrition: nature v. nurture. Proc Nutr Soc. 2011;70:73–81. doi: 10.1017/S0029665110003988. [DOI] [PubMed] [Google Scholar]
- 40.Park JH, Stoffers DA, Nicholls RD, et al. Development of type 2 diabetes following intrauterine growth retardation in rats is associated with progressive epigenetic silencing of Pdx1. J Clin Invest. 2008;118:2316–2324. doi: 10.1172/JCI33655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen L, Song CX, He C, et al. Mechanism and function of oxidative reversal of DNA and RNA methylation. Annu Rev Biochem. 2014;83:585–614. doi: 10.1146/annurev-biochem-060713-035513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaelin WG, Jr., McKnight SL. Influence of metabolism on epigenetics and disease. Cell. 2013;153:56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ronn T, Volkov P, Davegardh C, et al. A six months exercise intervention influences the genome-wide DNA methylation pattern in human adipose tissue. PLoS Genet. 2013;9:e1003572. doi: 10.1371/journal.pgen.1003572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen S, Bellew C, Yao X, et al. Histone deacetylase (HDAC) activity is critical for embryonic kidney gene expression, growth, and differentiation. J Biol Chem. 2011;286:32775–32789. doi: 10.1074/jbc.M111.248278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adli M, Parlak M, Li Y, et al. Epigenetic States of Nephron Progenitors and Epithelial Differentiation. J Cell Biochem. 2015 Jan 5; doi: 10.1002/jcb.25048. doi: 10.1002/jcb.25048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hawkins RD, Hon GC, Ren B. Next-generation genomics: an integrative approach. Nat Rev Genet. 2010;11:476–486. doi: 10.1038/nrg2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Laird PW. Principles and challenges of genomewide DNA methylation analysis. Nat Rev Genet. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 48.Pepke S, Wold B, Mortazavi A. Computation for ChIP-seq and RNA-seq studies. Nat Methods. 2009;6:S22–32. doi: 10.1038/nmeth.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Michels KB, Binder AM, Dedeurwaerder S, et al. Recommendations for the design and analysis of epigenome-wide association studies. Nat Methods. 2013;10:949–955. doi: 10.1038/nmeth.2632. [DOI] [PubMed] [Google Scholar]
- 50.Neph S, Vierstra J, Stergachis AB, et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489:83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mouse EC, Stamatoyannopoulos JA, Snyder M, et al. An encyclopedia of mouse DNA elements (Mouse ENCODE) Genome Biol. 2012;13:418. doi: 10.1186/gb-2012-13-8-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hon GC, Hawkins RD, Ren B. Predictive chromatin signatures in the mammalian genome. Hum Mol Genet. 2009;18:R195–201. doi: 10.1093/hmg/ddp409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hnisz D, Abraham BJ, Lee TI, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lam MT, Cho H, Lesch HP, et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013;498:511–515. doi: 10.1038/nature12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ostuni R, Piccolo V, Barozzi I, et al. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152:157–171. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 56.de Wit E, de Laat W. A decade of 3C technologies: insights into nuclear organization. Genes Dev. 2012;26:11–24. doi: 10.1101/gad.179804.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grundberg E, Meduri E, Sandling JK, et al. Global analysis of DNA methylation variation in adipose tissue from twins reveals links to disease-associated variants in distal regulatory elements. Am J Hum Genet. 2013;93:876–890. doi: 10.1016/j.ajhg.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gaulton KJ, Nammo T, Pasquali L, et al. A map of open chromatin in human pancreatic islets. Nat Genet. 2010;42:255–259. doi: 10.1038/ng.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leung A, Parks BW, Du J, et al. Open chromatin profiling in mice livers reveals unique chromatin variations induced by high fat diet. J Biol Chem. 2014;289:23557–23567. doi: 10.1074/jbc.M114.581439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leung A, Schones DE, Natarajan R. Using epigenetic mechanisms to understand the impact of common disease causing alleles. Curr Opin Immunol. 2012;24:558–563. doi: 10.1016/j.coi.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ko YA, Susztak K. Epigenomics: the science of no-longer-junk DNA. Why study it in chronic kidney disease? Semin Nephrol. 2013;33:354–362. doi: 10.1016/j.semnephrol.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harris RA, Wang T, Coarfa C, et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotechnol. 2010;28:1097–1105. doi: 10.1038/nbt.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chertow GM, Burdick E, Honour M, et al. Acute kidney injury, mortality, length of stay, and costs in hospitalized patients. J Am Soc Nephrol. 2005;16:3365–3370. doi: 10.1681/ASN.2004090740. [DOI] [PubMed] [Google Scholar]
- 64.Coca SG. Long-term outcomes of acute kidney injury. Curr Opin Nephrol Hypertens. 2010;19:266–272. doi: 10.1097/MNH.0b013e3283375538. [DOI] [PubMed] [Google Scholar]
- 65.Bonventre JV, Yang L. Cellular pathophysiology of ischemic acute kidney injury. J Clin Invest. 2011;121:4210–4221. doi: 10.1172/JCI45161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bomsztyk K, Denisenko O. Epigenetic alterations in acute kidney injury. Semin Nephrol. 2013;33:327–340. doi: 10.1016/j.semnephrol.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mellor J. Dynamic nucleosomes and gene transcription. Trends Genet. 2006;22:320–329. doi: 10.1016/j.tig.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 68.Naito M, Zager RA, Bomsztyk K. BRG1 increases transcription of proinflammatory genes in renal ischemia. J Am Soc Nephrol. 2009;20:1787–1796. doi: 10.1681/ASN.2009010118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Naito M, Bomsztyk K, Zager RA. Endotoxin mediates recruitment of RNA polymerase II to target genes in acute renal failure. J Am Soc Nephrol. 2008;19:1321–1330. doi: 10.1681/ASN.2007121368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Naito M, Bomsztyk K, Zager RA. Renal ischemia-induced cholesterol loading: transcription factor recruitment and chromatin remodeling along the HMG CoA reductase gene. Am J Pathol. 2009;174:54–62. doi: 10.2353/ajpath.2009.080602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zager RA, Johnson AC. Renal ischemia-reperfusion injury upregulates histone-modifying enzyme systems and alters histone expression at proinflammatory/profibrotic genes. Am J Physiol Renal Physiol. 2009;296:F1032–1041. doi: 10.1152/ajprenal.00061.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li HF, Cheng CF, Liao WJ, et al. ATF3-mediated epigenetic regulation protects against acute kidney injury. J Am Soc Nephrol. 2010;21:1003–1013. doi: 10.1681/ASN.2009070690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marumo T, Hishikawa K, Yoshikawa M, et al. Epigenetic regulation of BMP7 in the regenerative response to ischemia. J Am Soc Nephrol. 2008;19:1311–1320. doi: 10.1681/ASN.2007091040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marumo T, Hishikawa K, Yoshikawa M, et al. Histone deacetylase modulates the proinflammatory and -fibrotic changes in tubulointerstitial injury. Am J Physiol Renal Physiol. 2010;298:F133–141. doi: 10.1152/ajprenal.00400.2009. [DOI] [PubMed] [Google Scholar]
- 75.Havasi A, Haegele JA, Gall JM, et al. Histone acetyl transferase (HAT) HBO1 and JADE1 in epithelial cell regeneration. Am J Pathol. 2013;182:152–162. doi: 10.1016/j.ajpath.2012.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zager RA, Johnson AC, Becker K. Acute unilateral ischemic renal injury induces progressive renal inflammation, lipid accumulation, histone modification, and “end-stage” kidney disease. Am J Physiol Renal Physiol. 2011;301:F1334–1345. doi: 10.1152/ajprenal.00431.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zacharias N, Sailhamer EA, Li Y, et al. Histone deacetylase inhibitors prevent apoptosis following lethal hemorrhagic shock in rodent kidney cells. Resuscitation. 2011;82:105–109. doi: 10.1016/j.resuscitation.2010.09.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Van Beneden K, Geers C, Pauwels M, et al. Valproic acid attenuates proteinuria and kidney injury. J Am Soc Nephrol. 2011;22:1863–1875. doi: 10.1681/ASN.2010111196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu N, He S, Ma L, et al. Blocking the class I histone deacetylase ameliorates renal fibrosis and inhibits renal fibroblast activation via modulating TGF-beta and EGFR signaling. PLoS One. 2013;8:e54001. doi: 10.1371/journal.pone.0054001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.He W, Wang Y, Zhang MZ, et al. Sirt1 activation protects the mouse renal medulla from oxidative injury. J Clin Invest. 2010;120:1056–1068. doi: 10.1172/JCI41563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fan H, Yang HC, You L, et al. The histone deacetylase, SIRT1, contributes to the resistance of young mice to ischemia/reperfusion-induced acute kidney injury. Kidney Int. 2013;83:404–413. doi: 10.1038/ki.2012.394. [DOI] [PubMed] [Google Scholar]
- 82.Jung YJ, Lee AS, Nguyen-Thanh T, et al. SIRT2 Regulates LPS-Induced Renal Tubular CXCL2 and CCL2 Expression. J Am Soc Nephrol. 2014 Oct 27; doi: 10.1681/ASN.2014030226. ASN.2014030226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhou H, Beevers CS, Huang S. The targets of curcumin. Curr Drug Targets. 2011;12:332–347. doi: 10.2174/138945011794815356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kuhad A, Pilkhwal S, Sharma S, et al. Effect of curcumin on inflammation and oxidative stress in cisplatin-induced experimental nephrotoxicity. J Agric Food Chem. 2007;55:10150–10155. doi: 10.1021/jf0723965. [DOI] [PubMed] [Google Scholar]
- 85.Pratt JR, Parker MD, Affleck LJ, et al. Ischemic epigenetics and the transplanted kidney. Transplant Proc. 2006;38:3344–3346. doi: 10.1016/j.transproceed.2006.10.112. [DOI] [PubMed] [Google Scholar]
- 86.Huang N, Tan L, Xue Z, et al. Reduction of DNA hydroxymethylation in the mouse kidney insulted by ischemia reperfusion. Biochem Biophys Res Commun. 2012;422:697–702. doi: 10.1016/j.bbrc.2012.05.061. [DOI] [PubMed] [Google Scholar]
- 87.Kanwar YS, Sun L, Xie P, et al. A glimpse of various pathogenetic mechanisms of diabetic nephropathy. Annu Rev Pathol. 2011;6:395–423. doi: 10.1146/annurev.pathol.4.110807.092150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Forbes JM, Cooper ME. Mechanisms of diabetic complications. Physiol Rev. 2013;93:137–188. doi: 10.1152/physrev.00045.2011. [DOI] [PubMed] [Google Scholar]
- 89.Reidy K, Kang HM, Hostetter T, et al. Molecular mechanisms of diabetic kidney disease. J Clin Invest. 2014;124:2333–2340. doi: 10.1172/JCI72271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sanchez AP, Sharma K. Transcription factors in the pathogenesis of diabetic nephropathy. Expert Rev Mol Med. 2009;11:e13. doi: 10.1017/S1462399409001057. [DOI] [PubMed] [Google Scholar]
- 91.Sharma K, Ziyadeh FN. Hyperglycemia and diabetic kidney disease. The case for transforming growth factor-beta as a key mediator. Diabetes. 1995;44:1139–1146. doi: 10.2337/diab.44.10.1139. [DOI] [PubMed] [Google Scholar]
- 92.Perico N, Benigni A, Remuzzi G. Present and future drug treatments for chronic kidney diseases: evolving targets in renoprotection. Nat Rev Drug Discov. 2008;7:936–953. doi: 10.1038/nrd2685. [DOI] [PubMed] [Google Scholar]
- 93.Kato M, Castro NE, Natarajan R. MicroRNAs: potential mediators and biomarkers of diabetic complications. Free Radic Biol Med. 2013;64:85–94. doi: 10.1016/j.freeradbiomed.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kantharidis P, Wang B, Carew RM, et al. Diabetes complications: the microRNA perspective. Diabetes. 2011;60:1832–1837. doi: 10.2337/db11-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Reddy MA, Natarajan R. Epigenetic mechanisms in diabetic vascular complications. Cardiovasc Res. 2011;90:421–429. doi: 10.1093/cvr/cvr024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pirola L, Balcerczyk A, Okabe J, et al. Epigenetic phenomena linked to diabetic complications. Nat Rev Endocrinol. 2010;6:665–675. doi: 10.1038/nrendo.2010.188. [DOI] [PubMed] [Google Scholar]
- 97.Sun G, Reddy MA, Yuan H, et al. Epigenetic histone methylation modulates fibrotic gene expression. J Am Soc Nephrol. 2010;21:2069–2080. doi: 10.1681/ASN.2010060633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gubitosi-Klug RA, Group DER The diabetes control and complications trial/epidemiology of diabetes interventions and complications study at 30 years: summary and future directions. Diabetes Care. 2014;37:44–49. doi: 10.2337/dc13-2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sustained effect of intensive treatment of type 1 diabetes mellitus on development and progression of diabetic nephropathy: the Epidemiology of Diabetes Interventions and Complications (EDIC) study. JAMA. 2003;290:2159–2167. doi: 10.1001/jama.290.16.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Colagiuri S, Cull CA, Holman RR. Are lower fasting plasma glucose levels at diagnosis of type 2 diabetes associated with improved outcomes?: U.K. prospective diabetes study 61. Diabetes Care. 2002;25:1410–1417. doi: 10.2337/diacare.25.8.1410. [DOI] [PubMed] [Google Scholar]
- 101.Roy S, Sala R, Cagliero E, et al. Overexpression of fibronectin induced by diabetes or high glucose: phenomenon with a memory. Proc Natl Acad Sci U S A. 1990;87:404–408. doi: 10.1073/pnas.87.1.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li SL, Reddy MA, Cai Q, et al. Enhanced proatherogenic responses in macrophages and vascular smooth muscle cells derived from diabetic db/db mice. Diabetes. 2006;55:2611–2619. doi: 10.2337/db06-0164. [DOI] [PubMed] [Google Scholar]
- 103.Villeneuve LM, Reddy MA, Lanting LL, et al. Epigenetic histone H3 lysine 9 methylation in metabolic memory and inflammatory phenotype of vascular smooth muscle cells in diabetes. Proc Natl Acad Sci U S A. 2008;105:9047–9052. doi: 10.1073/pnas.0803623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.El-Osta A, Brasacchio D, Yao D, et al. Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. J Exp Med. 2008;205:2409–2417. doi: 10.1084/jem.20081188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhong Q, Kowluru RA. Epigenetic changes in mitochondrial superoxide dismutase in the retina and the development of diabetic retinopathy. Diabetes. 2011;60:1304–1313. doi: 10.2337/db10-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kowluru RA, Santos JM, Mishra M. Epigenetic modifications and diabetic retinopathy. Biomed Res Int. 2013:635284. doi: 10.1155/2013/635284. doi: 10.1155/2013/635284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Keating ST, El-Osta A. Glycemic memories and the epigenetic component of diabetic nephropathy. Curr Diab Rep. 2013;13:574–581. doi: 10.1007/s11892-013-0383-y. [DOI] [PubMed] [Google Scholar]
- 108.Miao F, Chen Z, Genuth S, et al. Evaluating the role of epigenetic histone modifications in the metabolic memory of type 1 diabetes. Diabetes. 2014;63:1748–1762. doi: 10.2337/db13-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bell CG, Teschendorff AE, Rakyan VK, et al. Genome-wide DNA methylation analysis for diabetic nephropathy in type 1 diabetes mellitus. BMC Med Genomics. 2010;3:33–42. doi: 10.1186/1755-8794-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sapienza C, Lee J, Powell J, et al. DNA methylation profiling identifies epigenetic differences between diabetes patients with ESRD and diabetes patients without nephropathy. Epigenetics. 2011;6:20–28. doi: 10.4161/epi.6.1.13362. [DOI] [PubMed] [Google Scholar]
- 111.Ko YA, Mohtat D, Suzuki M, et al. Cytosine methylation changes in enhancer regions of core pro-fibrotic genes characterize kidney fibrosis development. Genome Biol. 2013;14:R108. doi: 10.1186/gb-2013-14-10-r108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wing MR, Devaney JM, Joffe MM, et al. DNA methylation profile associated with rapid decline in kidney function: findings from the CRIC study. Nephrol Dial Transplant. 2014;29:864–872. doi: 10.1093/ndt/gft537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Smyth LJ, McKay GJ, Maxwell AP, et al. DNA hypermethylation and DNA hypomethylation is present at different loci in chronic kidney disease. Epigenetics. 2014;9:366–376. doi: 10.4161/epi.27161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bechtel W, McGoohan S, Zeisberg EM, et al. Methylation determines fibroblast activation and fibrogenesis in the kidney. Nat Med. 2010;16:544–550. doi: 10.1038/nm.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tampe B, Tampe D, Muller CA, et al. Tet3-mediated hydroxymethylation of epigenetically silenced genes contributes to bone morphogenic protein 7-induced reversal of kidney fibrosis. J Am Soc Nephrol. 2014;25:905–912. doi: 10.1681/ASN.2013070723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bock F, Shahzad K, Wang H, et al. Activated protein C ameliorates diabetic nephropathy by epigenetically inhibiting the redox enzyme p66Shc. Proc Natl Acad Sci U S A. 2013;110:648–653. doi: 10.1073/pnas.1218667110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hasegawa K, Wakino S, Simic P, et al. Renal tubular Sirt1 attenuates diabetic albuminuria by epigenetically suppressing Claudin-1 overexpression in podocytes. Nat Med. 2013;19:1496–1504. doi: 10.1038/nm.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hayashi K, Sasamura H, Nakamura M, et al. KLF4-dependent epigenetic remodeling modulates podocyte phenotypes and attenuates proteinuria. J Clin Invest. 2014;124:2523–2537. doi: 10.1172/JCI69557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Marumo T, Yagi S, Kawarazaki W, et al. Diabetes Induces Aberrant DNA Methylation in the Proximal Tubules of the Kidney. J Am Soc Nephrol. 2015 Feb 4; doi: 10.1681/ASN.2014070665. 10.1681/ASN.2014070665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yuan H, Reddy MA, Sun G, et al. Involvement of p300/CBP and epigenetic histone acetylation in TGF-beta1-mediated gene transcription in mesangial cells. Am J Physiol Renal Physiol. 2013;304:F601–613. doi: 10.1152/ajprenal.00523.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lindenmeyer MT, Rastaldi MP, Ikehata M, et al. Proteinuria and hyperglycemia induce endoplasmic reticulum stress. J Am Soc Nephrol. 2008;19:2225–2236. doi: 10.1681/ASN.2007121313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kitamura M. Endoplasmic reticulum stress and unfolded protein response in renal pathophysiology: Janus faces. Am J Physiol Renal Physiol. 2008;295:F323–334. doi: 10.1152/ajprenal.00050.2008. [DOI] [PubMed] [Google Scholar]
- 123.Chen J, Guo Y, Zeng W, et al. ER stress triggers MCP-1 expression through SET7/9-induced histone methylation in the kidneys of db/db mice. Am J Physiol Renal Physiol. 2014;306:F916–925. doi: 10.1152/ajprenal.00697.2012. [DOI] [PubMed] [Google Scholar]
- 124.Xu H, Wu X, Qin H, et al. Myocardin-Related Transcription Factor A Epigenetically Regulates Renal Fibrosis in Diabetic Nephropathy. J Am Soc Nephrol. 2014 Oct 27; doi: 10.1681/ASN.2014070678. 10.1681/ASN.2014070678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kato M, Dang V, Wang M, et al. TGF-beta induces acetylation of chromatin and of Ets-1 to alleviate repression of miR-192 in diabetic nephropathy. Sci Signal. 2013;6:ra43. doi: 10.1126/scisignal.2003389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Du B, Ma LM, Huang MB, et al. High glucose down-regulates miR-29a to increase collagen IV production in HK-2 cells. FEBS Lett. 2010;584:811–816. doi: 10.1016/j.febslet.2009.12.053. [DOI] [PubMed] [Google Scholar]
- 127.Lin CL, Lee PH, Hsu YC, et al. MicroRNA-29a promotion of nephrin acetylation ameliorates hyperglycemia-induced podocyte dysfunction. J Am Soc Nephrol. 2014;25:1698–1709. doi: 10.1681/ASN.2013050527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wang X, Liu J, Zhen J, et al. Histone deacetylase 4 selectively contributes to podocyte injury in diabetic nephropathy. Kidney Int. 2014;86:712–725. doi: 10.1038/ki.2014.111. [DOI] [PubMed] [Google Scholar]
- 129.Reddy MA, Sumanth P, Lanting L, et al. Losartan reverses permissive epigenetic changes in renal glomeruli of diabetic db/db mice. Kidney Int. 2014;85:362–373. doi: 10.1038/ki.2013.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Komers R, Mar D, Denisenko O, et al. Epigenetic changes in renal genes dysregulated in mouse and rat models of type 1 diabetes. Lab Invest. 2013;93:543–552. doi: 10.1038/labinvest.2013.47. [DOI] [PubMed] [Google Scholar]
- 131.Van Beneden K, Geers C, Pauwels M, et al. Comparison of trichostatin A and valproic acid treatment regimens in a mouse model of kidney fibrosis. Toxicol Appl Pharmacol. 2013;271:276–284. doi: 10.1016/j.taap.2013.05.013. [DOI] [PubMed] [Google Scholar]
- 132.Gilbert RE, Huang Q, Thai K, et al. Histone deacetylase inhibition attenuates diabetes-associated kidney growth: potential role for epigenetic modification of the epidermal growth factor receptor. Kidney Int. 2011;79:1312–1321. doi: 10.1038/ki.2011.39. [DOI] [PubMed] [Google Scholar]
- 133.Advani A, Huang Q, Thai K, et al. Long-term administration of the histone deacetylase inhibitor vorinostat attenuates renal injury in experimental diabetes through an endothelial nitric oxide synthase-dependent mechanism. Am J Pathol. 2011;178:2205–2214. doi: 10.1016/j.ajpath.2011.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Noh H, Oh EY, Seo JY, et al. Histone deacetylase-2 is a key regulator of diabetes- and transforming growth factor-beta1-induced renal injury. Am J Physiol Renal Physiol. 2009;297:F729–739. doi: 10.1152/ajprenal.00086.2009. [DOI] [PubMed] [Google Scholar]
- 135.Liu R, Zhong Y, Li X, et al. Role of transcription factor acetylation in diabetic kidney disease. Diabetes. 2014;63:2440–2453. doi: 10.2337/db13-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mimura I, Nangaku M. The suffocating kidney: tubulointerstitial hypoxia in end-stage renal disease. Nat Rev Nephrol. 2010;6:667–678. doi: 10.1038/nrneph.2010.124. [DOI] [PubMed] [Google Scholar]
- 137.Nangaku M. Chronic hypoxia and tubulointerstitial injury: a final common pathway to end-stage renal failure. J Am Soc Nephrol. 2006;17:17–25. doi: 10.1681/ASN.2005070757. [DOI] [PubMed] [Google Scholar]
- 138.Tanaka T, Kojima I, Ohse T, et al. Cobalt promotes angiogenesis via hypoxia-inducible factor and protects tubulointerstitium in the remnant kidney model. Lab Invest. 2005;85:1292–1307. doi: 10.1038/labinvest.3700328. [DOI] [PubMed] [Google Scholar]
- 139.Tanaka T, Kato H, Kojima I, et al. Hypoxia and expression of hypoxia-inducible factor in the aging kidney. J Gerontol A Biol Sci Med Sci. 2006;61:795–805. doi: 10.1093/gerona/61.8.795. [DOI] [PubMed] [Google Scholar]
- 140.Mimura I, Tanaka T, Nangaku M. Novel therapeutic strategy with hypoxia-inducible factors via reversible epigenetic regulation mechanisms in progressive tubulointerstitial fibrosis. Semin Nephrol. 2013;33:375–382. doi: 10.1016/j.semnephrol.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 141.Kaelin WG, Jr., Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 142.Mimura I, Tanaka T, Wada Y, et al. Pathophysiological response to hypoxia - from the molecular mechanisms of malady to drug discovery: epigenetic regulation of the hypoxic response via hypoxia-inducible factor and histone modifying enzymes. J Pharmacol Sci. 2011;115:453–458. doi: 10.1254/jphs.10r19fm. [DOI] [PubMed] [Google Scholar]
- 143.Perez-Perri JI, Acevedo JM, Wappner P. Epigenetics: new questions on the response to hypoxia. Int J Mol Sci. 2011;12:4705–4721. doi: 10.3390/ijms12074705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Beyer S, Kristensen MM, Jensen KS, et al. The histone demethylases JMJD1A and JMJD2B are transcriptional targets of hypoxia-inducible factor HIF. J Biol Chem. 2008;283:36542–36552. doi: 10.1074/jbc.M804578200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Krieg AJ, Rankin EB, Chan D, et al. Regulation of the histone demethylase JMJD1A by hypoxia-inducible factor 1 alpha enhances hypoxic gene expression and tumor growth. Mol Cell Biol. 2010;30:344–353. doi: 10.1128/MCB.00444-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mimura I, Nangaku M, Kanki Y, et al. Dynamic change of chromatin conformation in response to hypoxia enhances the expression of GLUT3 (SLC2A3) by cooperative interaction of hypoxia-inducible factor 1 and KDM3A. Mol Cell Biol. 2012;32:3018–3032. doi: 10.1128/MCB.06643-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Coffman TM. Under pressure: the search for the essential mechanisms of hypertension. Nat Med. 2011;17:1402–1409. doi: 10.1038/nm.2541. [DOI] [PubMed] [Google Scholar]
- 148.Kone BC. Epigenetics and the control of the collecting duct epithelial sodium channel. Semin Nephrol. 2013;33:383–391. doi: 10.1016/j.semnephrol.2013.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhang W, Yu Z, Wu H, et al. An Af9 cis-element directly targets Dot1a to mediate transcriptional repression of the alphaENaC gene. Am J Physiol Renal Physiol. 2013;304:F367–375. doi: 10.1152/ajprenal.00537.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zhang W, Xia X, Reisenauer MR, et al. Aldosterone-induced Sgk1 relieves Dot1a-Af9-mediated transcriptional repression of epithelial Na+ channel alpha. J Clin Invest. 2007;117:773–783. doi: 10.1172/JCI29850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zhang W, Xia X, Reisenauer MR, et al. Dot1a-AF9 complex mediates histone H3 Lys-79 hypermethylation and repression of ENaCalpha in an aldosterone-sensitive manner. J Biol Chem. 2006;281:18059–18068. doi: 10.1074/jbc.M601903200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhang D, Yu ZY, Cruz P, et al. Epigenetics and the control of epithelial sodium channel expression in collecting duct. Kidney Int. 2009;75:260–267. doi: 10.1038/ki.2008.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zhang D, Li S, Cruz P, et al. Sirtuin 1 functionally and physically interacts with disruptor of telomeric silencing-1 to regulate alpha-ENaC transcription in collecting duct. J Biol Chem. 2009;284:20917–20926. doi: 10.1074/jbc.M109.020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yu Z, Kong Q, Kone BC. Aldosterone reprograms promoter methylation to regulate alphaENaC transcription in the collecting duct. Am J Physiol Renal Physiol. 2013;305:F1006–1013. doi: 10.1152/ajprenal.00407.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lee HA, Cho HM, Lee DY, et al. Tissue-specific upregulation of angiotensin-converting enzyme 1 in spontaneously hypertensive rats through histone code modifications. Hypertension. 2012;59:621–626. doi: 10.1161/HYPERTENSIONAHA.111.182428. [DOI] [PubMed] [Google Scholar]
- 156.Bogdarina I, Welham S, King PJ, et al. Epigenetic modification of the renin-angiotensin system in the fetal programming of hypertension. Circ Res. 2007;100:520–526. doi: 10.1161/01.RES.0000258855.60637.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Liang M, Cowley AW, Jr., Mattson DL, et al. Epigenomics of hypertension. Semin Nephrol. 2013;33:392–399. doi: 10.1016/j.semnephrol.2013.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Van Beneden K, Mannaerts I, Pauwels M, et al. HDAC inhibitors in experimental liver and kidney fibrosis. Fibrogenesis Tissue Repair. 2013;6:1. doi: 10.1186/1755-1536-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kosanam H, Thai K, Zhang Y, et al. Diabetes induces lysine acetylation of intermediary metabolism enzymes in the kidney. Diabetes. 2014;63:2432–2439. doi: 10.2337/db12-1770. [DOI] [PubMed] [Google Scholar]
- 160.Yoshikawa M, Hishikawa K, Marumo T, et al. Inhibition of histone deacetylase activity suppresses epithelial-to-mesenchymal transition induced by TGF-beta1 in human renal epithelial cells. J Am Soc Nephrol. 2007;18:58–65. doi: 10.1681/ASN.2005111187. [DOI] [PubMed] [Google Scholar]
- 161.Cianciolo Cosentino C, Skrypnyk NI, Brilli LL, et al. Histone deacetylase inhibitor enhances recovery after AKI. J Am Soc Nephrol. 2013;24:943–953. doi: 10.1681/ASN.2012111055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 163.Fenaux P, Mufti GJ, Hellstrom-Lindberg E, et al. Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: a randomised, open-label, phase III study. Lancet Oncol. 2009;10:223–232. doi: 10.1016/S1470-2045(09)70003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ruggenenti P, Cravedi P, Remuzzi G. The RAAS in the pathogenesis and treatment of diabetic nephropathy. Nat Rev Nephrol. 2010;6:319–330. doi: 10.1038/nrneph.2010.58. [DOI] [PubMed] [Google Scholar]
- 165.Zhong Y, Chen EY, Liu R, et al. Renoprotective effect of combined inhibition of angiotensin-converting enzyme and histone deacetylase. J Am Soc Nephrol. 2013;24:801–811. doi: 10.1681/ASN.2012060590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Mortazavi A, Williams BA, McCue K, et al. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 167.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zhou L, Li X, Liu Q, et al. Small RNA transcriptome investigation based on next-generation sequencing technology. J Genet Genomics. 2011;38:505–513. doi: 10.1016/j.jgg.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 169.West JA, Cook A, Alver BH, et al. Nucleosomal occupancy changes locally over key regulatory regions during cell differentiation and reprogramming. Nat Commun. 2014;5:4719. doi: 10.1038/ncomms5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Simon JM, Giresi PG, Davis IJ, et al. Using formaldehyde-assisted isolation of regulatory elements (FAIRE) to isolate active regulatory DNA. Nat Protoc. 2012;7:256–267. doi: 10.1038/nprot.2011.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Buenrostro JD, Giresi PG, Zaba LC, et al. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Taiwo O, Wilson GA, Morris T, et al. Methylome analysis using MeDIP-seq with low DNA concentrations. Nat Protoc. 2012;7:617–636. doi: 10.1038/nprot.2012.012. [DOI] [PubMed] [Google Scholar]
- 173.Serre D, Lee BH, Ting AH. MBD-isolated Genome Sequencing provides a high-throughput and comprehensive survey of DNA methylation in the human genome. Nucleic Acids Res. 2010;38:391–399. doi: 10.1093/nar/gkp992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Colyer HA, Armstrong RN, Sharpe DJ, et al. Detection and analysis of DNA methylation by pyrosequencing. Methods Mol Biol. 2012;863:281–292. doi: 10.1007/978-1-61779-612-8_17. [DOI] [PubMed] [Google Scholar]
- 175.Touleimat N, Tost J. Complete pipeline for Infinium((R)) Human Methylation 450K BeadChip data processing using subset quantile normalization for accurate DNA methylation estimation. Epigenomics. 2012;4:325–341. doi: 10.2217/epi.12.21. [DOI] [PubMed] [Google Scholar]
- 176.Kelly TK, Liu Y, Lay FD, et al. Genome-wide mapping of nucleosome positioning and DNA methylation within individual DNA molecules. Genome Res. 2012;22:2497–2506. doi: 10.1101/gr.143008.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kasinathan S, Orsi GA, Zentner GE, et al. High-resolution mapping of transcription factor binding sites on native chromatin. Nat Methods. 2014;11:203–209. doi: 10.1038/nmeth.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.van de Werken HJ, de Vree PJ, Splinter E, et al. 4C technology: protocols and data analysis. Methods Enzymol. 2012;513:89–112. doi: 10.1016/B978-0-12-391938-0.00004-5. [DOI] [PubMed] [Google Scholar]
- 179.Belton JM, McCord RP, Gibcus JH, et al. Hi-C: a comprehensive technique to capture the conformation of genomes. Methods. 2012;58:268–276. doi: 10.1016/j.ymeth.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]