ABSTRACT
When expressed alone at high levels, the human adenovirus E4orf4 protein exhibits tumor cell-specific p53-independent toxicity. A major E4orf4 target is the B55 class of PP2A regulatory subunits, and we have shown recently that binding of E4orf4 inhibits PP2AB55 phosphatase activity in a dose-dependent fashion by preventing access of substrates (M. Z. Mui et al., PLoS Pathog 9:e1003742, 2013, http://dx.doi.org/10.1371/journal.ppat.1003742). While interaction with B55 subunits is essential for toxicity, E4orf4 mutants exist that, despite binding B55 at high levels, are defective in cell killing, suggesting that other essential targets exist. In an attempt to identify additional targets, we undertook a proteomics approach to characterize E4orf4-interacting proteins. Our findings indicated that, in addition to PP2AB55 subunits, ASPP-PP1 complex subunits were found among the major E4orf4-binding species. Both the PP2A and ASPP-PP1 phosphatases are known to positively regulate effectors of the Hippo signaling pathway, which controls the expression of cell growth/survival genes by dephosphorylating the YAP transcriptional coactivator. We find here that expression of E4orf4 results in hyperphosphorylation of YAP, suggesting that Hippo signaling is affected by E4orf4 interactions with PP2AB55 and/or ASPP-PP1 phosphatases. Furthermore, knockdown of YAP1 expression was seen to enhance E4orf4 killing, again consistent with a link between E4orf4 toxicity and inhibition of the Hippo pathway. This effect may in fact contribute to the cancer cell specificity of E4orf4 toxicity, as many human cancer cells rely heavily on the Hippo pathway for their enhanced proliferation.
IMPORTANCE The human adenovirus E4orf4 protein has been known for some time to induce tumor cell-specific death when expressed at high levels; thus, knowledge of its mode of action could be of importance for development of new cancer therapies. Although the B55 form of the phosphatase PP2A has long been known as an essential E4orf4 target, genetic analyses indicated that others must exist. To identify additional E4orf4 targets, we performed, for the first time, a large-scale affinity purification/mass spectrometry analysis of E4orf4 binding partners. Several additional candidates were detected, including key regulators of the Hippo signaling pathway, which enhances cell viability in many cancers, and results of preliminary studies suggested a link between inhibition of Hippo signaling and E4orf4 toxicity.
INTRODUCTION
During infection by human adenovirus, the E4orf4 protein product is believed to enhance replication at least in part by introducing novel substrates to protein phosphatase 2A (PP2A) through interactions with one of its major classes of binding partners, the B55 family of PP2A regulatory subunits (1–6). However, when expressed alone at high levels, E4orf4 induces the selective p53-independent death of a variety of human tumor cells (7–21) and is toxic in the yeast Saccharomyces cerevisiae (22–29). Killing of cancer cells by E4orf4 is dose dependent and resembles apoptosis in some cell lines, but it seems to occur by mitotic catastrophe in others (8–13, 15, 30, 31). Recent studies by our group in H1299 human carcinoma cells suggested that E4orf4 expression delays or inhibits transit through mitosis and completion of cytokinesis (32), resulting in an accumulation and death of both G1-arrested diploid and tetraploid cells due to an inability to initiate new rounds of DNA synthesis (21).
This induction of cell death is highly dependent on interactions with the PP2A B55/Cdc55 regulatory subunits (1, 10, 14, 16, 22–28, 31, 33–35). PP2A holoenzymes exist as heterotrimers composed of a catalytic C subunit, an A subunit scaffold, and a B regulatory subunit that determines intracellular localization and substrate specificity (36–39). PP2A functions vary extensively (40–45), due in large part to the 15 or so mammalian B subunits, which have been divided into three structurally divergent classes, designated B/B55, B′/B56, and B″, as well as B‴ striatin/SG2NA (42, 46). Our group found that E4orf4 interacts solely with the B/B55 family (14), and more recent studies showed that E4orf4 associates with the B55α subunit in a region believed to be involved in substrate binding (1, 35). Moreover, we have proposed that E4orf4 acts much like an inhibitory pseudosubstrate for PP2AB55α when expressed at high levels, preventing interactions with and dephosphorylation of substrates such as p107 (1) and inducing cell death by preventing dephosphorylation of key substrates required for cell viability (6, 14). Previous studies by our group and others indicated that such binding to B55 PP2A subunits is essential for E4orf4 toxicity (16, 26, 34, 35), as E4orf4 mutants that are defective in B55α binding (we refer to them as class I mutants) also are defective in cell killing (10). However, members of another class of E4orf4 mutants exist that also are defective for cell killing, even though they bind high levels of B55α (10) (we refer to these as class II mutants), suggesting that B55α binding is necessary but not sufficient for E4orf4 toxicity and that other essential targets exist.
About 90% of all phosphoserine/phosphothreonine phosphatase activity in mammalian cells can be attributed to PP2A and the protein phosphatase 1 (PP1) families of enzymes (38, 46). Unlike heterotrimeric PP2A, the three PP1 catalytic C subunits either can form heterodimeric complexes directly with substrates or employ PP1-interacting proteins to provide additional substrate-binding capacity or to direct the enzyme to unique substrate-containing cell compartments (47). One class of PP1-interacting species is the ASPP (apoptosis-stimulating of p53 protein) family of regulatory proteins (48, 49). ASPP proteins initially were discovered based on their interactions with p53 and with Bcl2 and their stimulation of apoptosis (50–52); however, they also form complexes with PP1, for which they are now considered regulatory subunits (53), and associate with the oncogene YAP (Yes-associated protein), an important transcriptional coactivator (54, 55). Both PP2A and ASPP-PP1 have been implicated in the regulation of several elements of the Hippo signaling pathway that regulates expression of genes involved in proliferation and cell death (55–61).
The Hippo pathway, originally discovered as a signal transduction pathway involved in the regulation of organ size in Drosophila melanogaster, plays an integral role in tissue homeostasis and regeneration, as well as cell differentiation (62, 63). At the heart of the canonical Hippo pathway is an evolutionarily conserved signaling cascade comprised of Mst1 and Mst2 (Mst1/2) kinases (gene names STK4 and STK3), coupled to the WW45 cofactor (gene name SAV1), and downstream kinases Lats1 and Lats2 (Lats1/2), coupled to the Mob1 cofactor (genes MOB1A and MOB1B). Once activated by Mst1/2 phosphorylation, Lats1/2 phosphorylate the homologous transcriptional coactivators YAP and TAZ (transcriptional coactivators with PDZ-binding motif). Phosphorylated forms of YAP/TAZ are sequestered in the cytoplasm through 14-3-3 protein binding and/or proteasomal degradation, keeping these transcriptional coactivators away from nuclear transcription factors which otherwise would result in the transcription of genes involved in cell growth and proliferation (64–68). Interestingly, both PP2A and ASPP-PP1 complexes are known as key regulators of Hippo signaling, in part through dephosphorylation of YAP/TAZ, leading to their translocation to the nucleus and activation of TEAD-dependent transcription of cell growth/survival genes (60, 61, 69).
To date, several E4orf4 interacting partners besides PP2A have been reported and, in some cases, suggested to contribute to E4orf4-induced cell death, including c-Src (12, 18, 31, 70–74), the ATP-dependent chromatin-remodeling factor ACF (3), the NTPDASE4 gene product Golgi UDPase (20, 29), and Nup 205 (2). In an attempt to discover more E4orf4 binding partners, we performed affinity purification/mass spectrometry (AP-MS) using both wild-type E4orf4 and class I and class II E4orf4 mutants. Results from this analysis led to the discovery of many candidates in addition to PP2AB55, most notably all of the ASPP-PP1 subunits. Binding of the ASPP-PP1 subunits was decreased with the class II mutants. Further, we report that expression of E4orf4 reduces dephosphorylation of YAP and knockdown of YAP1 enhances E4orf4 toxicity, potentially implicating effects on Hippo signaling in E4orf4 toxicity
MATERIALS AND METHODS
Affinity purification/mass spectrometry (FLAG AP-MS).
Wild-type (WT), class I (R81A/F84A), and class II (L54A and K88A) (10) coding sequences were cloned in frame in the EcoRI and XhoI sites of pcDNA5/FRT/TO-FLAG (75) and stably expressed in tetracycline-inducible Flp-In T-REx HEK293 cells (Invitrogen) as previously described (76). Briefly, E4orf4 was affinity purified, alongside a set of four negative controls (expressing FLAG only), in at least biological duplicates. The two sets of data from the wild type and R81A/F84A mutants represent experiments for the indicated bait, which were performed roughly 8 months apart (each of which consisted of biological duplicates). Tryptic peptides were analyzed by reversed-phase liquid chromatography coupled to mass spectrometry on an Orbitrap Velos or Orbitrap Elite (Thermo Scientific) mass spectrometer equipped with a nanospray ion source (Proxeon Biosystems). The high-performance liquid chromatography (HPLC) program delivered an acetonitrile gradient over 145 min, and the data were acquired in CID mode (centroid) in a data-dependent manner with MS/MS on the 10 most abundant parent ions (isolated species were put on an exclusion list for 30 s).
RAW mass spectrometry files were converted to mzXML using ProteoWizard (3.0.4468) (77) and analyzed using the iProphet pipeline (78) implemented within the ProHits Laboratory Information Management System (79) as follows. The database consisted of the human and adenovirus complements of the RefSeq protein database (version 57) supplemented with “common contaminants” from the Max Planck Institute (http://maxquant.org/downloads.htm) and the Global Proteome Machine (GPM; http://www.thegpm.org/crap/index.html). The search database consisted of forward and reverse sequences (labeled “DECOY”); in total, 72,226 entries were searched. The search engines used were Mascot (2.3.02; Matrix Science) and Comet (2012.01 rev.3) (80), with trypsin specificity (2 missed cleavages were allowed) and deamidation (NQ) and oxidation (M) as variable modifications. Charges +2, +3, and +4 were allowed, and the parent mass tolerance was set at 12 ppm while the fragment bin tolerance was set at 0.6 amu. The resulting Comet and Mascot search results were processed individually by PeptideProphet, and peptides were assembled into proteins using parsimony rules first described in ProteinProphet into a final iProphet protein output using the Trans-Proteomic Pipeline (TPP; Linux, version 0.0, development trunk rev 0, build 201303061711). TPP options were the following: general options were −p0.05 −x20 −PPM −d“DECOY," the iProphet option was −ipPRIME, and the PeptideProphet option was −OpdP. All proteins with a minimal iProphet protein probability of 0.05 were parsed to the relational module of ProHits. Note that for analysis with SAINT, only proteins with iProphet protein probability of ≥0.95 are considered. This corresponds to an estimated protein-level FDR of ∼0.5%.
For each bait, a cell line was prepared for mass spectrometric analysis as described above. Biological duplicates were grown/treated/processed at different times to maximize the variability and increase the robustness in the detection of true interactors. Negative-control purifications (consisting of cells expressing green fluorescent protein [GFP] and FLAG alone) were processed in parallel. The quality of each sample was assessed manually by aligning the runs for the biological replicates in ProHits. The results were analyzed by SAINTexpress v3.3 (81), a computationally efficient reimplementation of the SAINT (for significance analysis of interactome) method described previously (82). SAINT probabilities computed independently for each bait replicate are averaged, and the average probability (AvgP) is reported as the final SAINT score used to calculate the Bayesian false discovery rate (BFDR). Preys with BFDR of ≤1% were considered true interactors. Visualization of the data was performed using custom-based R scripts (83), with the primary FDR cutoff of 1% and the secondary FDR cutoff of 5%, capping the color intensity of the circle when the spectral counts reach 50.
All RAW mass spectrometry data and downloadable identification and SAINTexpress results tables are deposited in the MassIVE repository housed at the Center for Computational Mass Spectrometry at UCSD (http://proteomics.ucsd.edu/ProteoSAFe/datasets.jsp). The data set has been assigned the MassIVE identifier MSV000079082 and is available for FTP download at http://MSV000079082@massive.ucsd.edu. The data set was assigned the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) identifier PXD001956. Exploration of the data also is available at prohits-web.lunenfeld.ca, a searchable repository of filtered high-confidence interactions (data set name E4orf4).
Quantification of the PP1 catalytic subunit levels.
As PPP1CA, PPP1CB, and PPP1CC share high sequence identity (∼84%), multiple peptides are shared between these paralogs, complicating quantitative analysis solely based on spectral counts. To circumvent this issue, the precursor (MS1) intensity for one unique peptide for each of the PP1 catalytic subunits was extracted. Briefly, RAW files were uploaded into Proteome Discoverer software (V1.3.0.339; Thermo-Fisher Scientific), converted to MGF, and searched using Mascot (Matrix Science, version 2.3) using nearly identical parameters and databases as described above. The only exceptions were that only one missed cleavage site was allowed, and only +2 and +3 spectra were searched. Label-free quantification (with peptide grouping) was enabled in Proteome Discoverer to assess the relative abundance of peptides using parent ion signal intensity in MS1. We also noted that the E4orf4 mutants were expressed at lower levels than wild-type E4orf4, which may affect quantification. Therefore, to compensate for these fluctuations, the MS1 intensity of two unique E4orf4 peptides also was extracted and used for normalization. The selected peptides are PPP1CA (LNLDSIIGR), PPP1CB (IVQMTEAEVR), PPP1CC (NVQLQENEIR), and E4orf4 (AAAHEGVYIEPEAR; EWIYYNYYTER). For all calculations, the MS1 intensity of each unique peptide was averaged between biological duplicates. Additionally, for the E4orf4 bait, the intensity was further averaged between the two selected peptides. Normalization was performed simply by dividing the peptide intensity of the PP1 catalytic subunit by the peptide intensity of the E4orf4 peptides. Note that PPP2CA and PPP2CB are 97% identical, and we have not been able to uniquely assign identification by mass spectrometry; therefore, we merged the two entries and denoted them PPP2CA/PPP2CB in the figures.
Cell lines.
Human cell lines used include Flp-In 293 T-REx (Invitrogen), H1299 lung carcinoma cells (ATCC CRL-5803) containing a homozygous deletion of the p53 gene, and MDA-MB-231 mammary gland/breast epithelial cells (ATCC HTB-26) that are homozygous mutant for NF2. Cells were cultured at 37°C in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% fetal bovine serum (Wisent).
Generation of YAP1 knockdown cells.
The short hairpin RNA (shRNA)-mediated knockdown (KD) of YAP1 was carried out in H1299 cells based on previously described protocols (84). Briefly, lentiviral supernatants produced by HEK293T cells were used to infect H1299 cells. In parallel, supernatants harboring pLKO were generated and used as controls. Infected cells were selected with puromycin (2 μg/ml; Sigma). The shRNA construct targeting YAP1 (TRCN0000300281) was obtained from the RNAi Consortium (TRC) arrayed human genome-wide shRNA collection (Sigma).
Plasmids and viruses.
For AP-MS studies, wild-type E4orf4 and class I mutant R81A/F84A coding sequences were subcloned into the pcDNA5/FRT/TO-FLAG(N) vector through EcoRI/XhoI restriction enzyme digestion. For coimmunoprecipitation experiments, DNA plasmids for wild-type and mutant pcDNA3-HA-E4orf4 for mammalian cell expression have been described previously (10). Other plasmid DNAs used were pcDNA5/FRT/TO-FLAG(N)-PP1 Cα (EcoRI/XhoI), pDESTpcDNA5/FRT/TO-3FLAG(N)-PP1 Cβ (attB1/attB2), pDESTpcDNA5/FRT/TO-3FLAG(N)-PP1 Cγ (attB1/attB2), and pDESTpcDNA5/FRT/TO-3FLAG(N)-PARD3 (attB1/attB2). The previously described plasmid DNA expressing Myc-tagged RASSF8 was kindly provided by Nicolas Tapon (Cancer Research UK, London Research Institute) (85). Previously described plasmid DNAs expressing V5-tagged ASPP1, ASPP2, iASPP, ASPP2 V922A/F924A, and iASPP F815A were kindly provided by Xin Lu (Ludwig Institute for Cancer Research, University of Oxford) (48). The p2xFLAG CMV2 YAP 2 plasmid was obtained through Addgene (plasmid 19045). AdGFP and AdHA-E4orf4 viruses are adenovirus vectors lacking the viral E1 region that express single gene products under the control of a cytomegalovirus promoter, and they have been described previously (8, 15).
Mammalian cell DNA transfections and adenoviral vector infections.
For transfections, H1299 cells were plated onto 60- or 100-mm-diameter dishes and transfected with plasmid DNAs using either Lipofectamine 2000 (Invitrogen), as specified by the manufacturer, or polyethylenimine (linear PEI; molecular weight, 25,000; Polysciences, Inc.) transfection reagent dissolved in 0.2N HCl, as suggested in reference 86, at 1 μg/μl. Three microliters of Lipofectamine 2000 reagent was used per 1 μg of DNA transfected or 5 μl of PEI reagent per 1 μg of DNA transfected. For adenoviral vector infections, H1299 cells were plated 24 h prior as described above and were infected for 1 h with either AdGFP or AdHA-E4orf4 viral vectors at a multiplicity of infection (MOI) of 35 or 50 PFU per cell in a low volume of infection medium (1× PBS supplemented with 0.2 mM CaCl2, 0.2 mM MgCl2, and 2% serum) before its removal and replacement with the addition of complete medium. For infections and cotransfections, adenoviral vector infections were carried out first, and upon addition of complete medium, transfection mixtures were added.
Cell lysis, coimmunoprecipitations, and Western blotting.
H1299 cells were harvested 24 or 48 h posttransfection and/or infection and lysed for 30 min on ice with various lysis buffers. For experiments requiring only whole-cell extracts (WCE), cells were lysed in radioimmunoprecipitation assay (RIPA) light buffer (50 mM Tris-HCl, pH 8, 150 mM NaCl, 5 mM EDTA, 1% NP-40, 0.1% SDS, 0.1% Triton X-100). For coimmunoprecipitation experiments, cells were lysed in buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 5% glycerol). All lysis buffers were supplemented with the following: 2 mM dithiothreitol (DTT), 4 mM NaF, 2 mM sodium pyrophosphate, 500 μM Na3VO4, 200 μg/ml phenylmethylsulfonyl fluoride (PMSF), 2 μg/ml aprotinin, and 5 μg/ml leupeptin. For coimmunoprecipitation experiments, 300 μg to 1 mg of protein extract was used. Antibodies used in coimmunoprecipitations were mouse monoclonal anti-hemagglutinin (HA.11; Covance), mouse monoclonal anti-FLAG (M2; Sigma-Aldrich), and mouse monoclonal anti-V5 (SV5-Pk1; AbD Serotec). Protein extracts were precleared with a 50% protein G–50% protein A agarose slurry (GenScript). Incubation of lysates with appropriate antibodies was followed by incubation with protein A/G slurry. Immunoprecipitates were washed five times with lysis buffer and heated in sample buffer for 5 min, followed by SDS-PAGE, transfer to polyvinylidene difluoride (PVDF) membranes (Millipore), and Western blotting with appropriate antibodies. WCEs were prepared using 20 to 50 μg of total protein. Antibodies used for Western blotting included anti-FLAG-horseradish peroxidase (Sigma-Aldrich), mouse monoclonal anti-HA (Covance), anti-YAP/TAZ rabbit monoclonal (D24E4; Cell Signaling), rabbit polyclonal anti-phospho-YAP Ser127 (4911; Cell Signaling), goat polyclonal anti-LATS1 (N18; Santa Cruz), rabbit polyclonal anti-phospho LATS Ser909 (9157; Cell Signaling), and mouse monoclonal PP2A B55α (2G9; Upstate). Membranes were incubated with secondary antibody linked to horseradish peroxidase (Jackson ImmunoResearch) followed by ECL detection (PerkinElmer).
Phosphatase assays.
Cells were harvested and lysed for 30 min on ice with λ phosphatase buffer (30 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.5% NP-40) supplemented with 2 mM DTT, 200 μg/ml PMSF, 2 μg/ml aprotinin, and 5 μg/ml leupeptin. Two hundred micrograms of protein extract was treated with 200 U of lambda protein phosphatase (NEB), as specified by the manufacturer, for 30 min at 30°C. Reactions were terminated with the addition of sample buffer, and samples were heated for 5 min prior to SDS-PAGE and transfer to PVDF membranes.
Colony formation assays.
E4orf4 toxicity was measured essentially as done previously (10). Briefly, control and knockdown H1299 cell lines were plated in 6-well dishes in puromycin-containing medium and were transfected with 1 μg of pcDNA3 (containing the neomycin resistance gene) or pcDNA3-HA-E4orf4 for 48 h. Cells then were trypsinized and resuspended in 1 ml medium, and 5 μl was transferred each in three 60-mm dishes. The rest of the cells were pelleted and lysed to check for protein expression. After 10 to 13 days, colonies were stained with Giemsa stain and counted.
RESULTS
Identification of novel E4orf4 binding partners by AP-MS.
Our group previously described two classes of E4orf4 mutants, of which class I mutants fail both to bind B55 and to kill cancer cells, suggesting that interactions with PP2AB55 are essential for toxicity (10). Conversely, class II E4orf4 mutants bind B55 but are defective in cell killing. Although the defects of class II cell killing may be diverse, these findings suggest that one or more additional E4orf4 binding partners also are required for E4orf4-induced toxicity. In an attempt to identify additional E4orf4 targets, we launched a proteomics effort to characterize E4orf4 binding partners. As a starting point, we began by comparing the protein binding patterns obtained between wild-type E4orf4 and a class I E4orf4 mutant, as such a comparison would not only identify additional E4orf4 binding partners but also allow validation of our approach, as the class I mutant would not be expected to associate with PP2AB55 subunits.
E4orf4 wild type and a class I mutant (R81A/F84A) that was shown previously to be unable to bind to B55 were subcloned into the Flp-In T-REx FLAG expression vector and stably expressed in the inducible Flp-In 293 line. E4orf4 species then were affinity purified alongside a set of negative controls in biological replicates, and the interaction partners were determined by AP-MS and statistical analysis. Results from two such analyses (each with two biological replicates) performed 8 months apart are shown in Fig. 1.
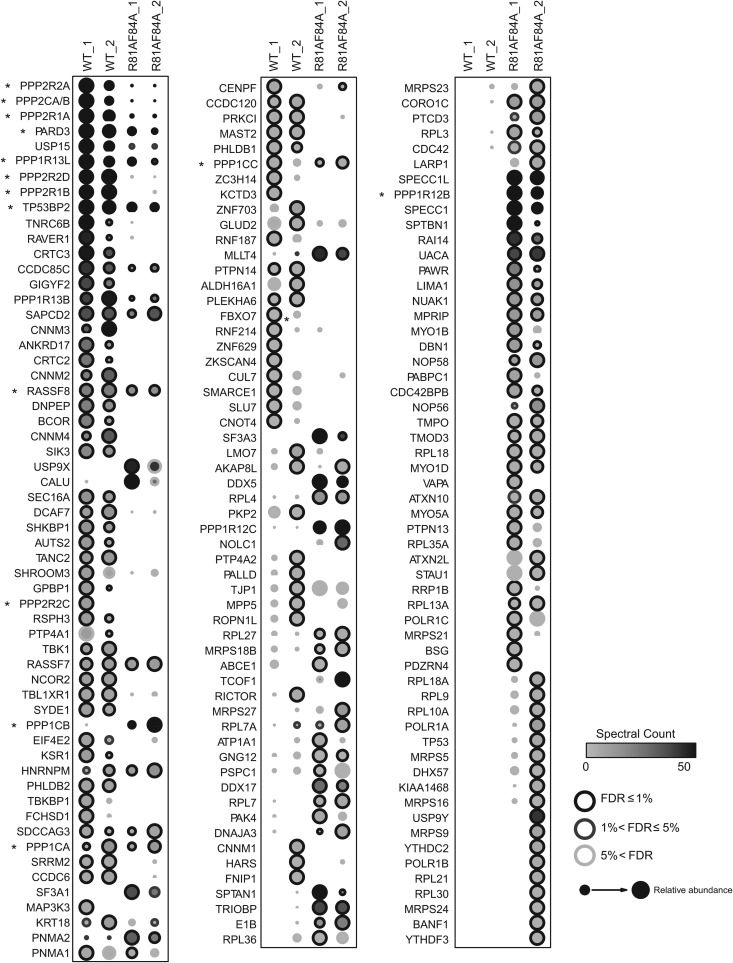
FIG 1.
High-confidence interaction partners for wild-type and class I mutant E4orf4. Comparison of AP-MS results for FLAG-E4orf4 and FLAG-E4orf4-R81A/F84A. Briefly, two biological replicates of human Flp-In T-REx 293 cells stably expressing FLAG-E4orf4 or FLAG-E4orf4-R81A/F84A were harvested in parallel to four negative controls (FLAG alone), and proteins were purified on anti-FLAG beads prior to analysis by liquid chromatography coupled to tandem mass spectrometry. Statistical analysis was employed to score high-confidence interactors, based on a false discovery rate (FDR) estimation, and proteins detected with ≤1% FDR either the wild-type or the mutant purifications are displayed (confidence values are mapped as the circle contour color in bins ≤1%, ≤5%, or >5% across both baits). Circle shading indicates the number of spectra detected (averages) across both biological replicate purifications (this value is capped at 50 spectra; the highest spectral count was 1,086). Circle size is proportional to the relative abundance of each prey across both baits; note, however, that the mutant protein was expressed at much lower levels than the wild-type protein in the stable cell lines used for AP-MS. Results from two studies, each with two biological replicates performed 8 months apart, are shown (denoted “_1” and “_2”). Asterisks denote genes that are discussed in more detail.
As expected, wild-type E4orf4 but not class I mutant R81A/F84A strongly interacted with both isoforms of the PP2A scaffolding A subunit (Aα/PPP2R1A and Aβ/PPP2R1B), as well as the PP2A catalytic subunit (PPP2CA/PPP2CB; these proteins are 97% identical at the amino acid level, and we have not been able to uniquely identify one isoform by mass spectrometry). Moreover, three of the four isoforms of the B55 class of PP2A regulatory subunits (B55α/PPP2R2A, B55γ/PPP2R2C, and B55δ/PPP2R2D) were found to bind significantly to wild-type E4orf4 but not the mutant. The fact that the fourth member of the B55 family was not observed was not surprising, as the B55β subunit is expressed mostly in brain tissues (87, 88). Nonetheless, no PP2A B species other than the B55 class was observed to interact with E4orf4, confirming our earlier findings and validating our AP-MS approach (14).
In addition to PP2A components, multiple other proteins were detected with wild-type E4orf4, while their levels were decreased with the mutant. A list of the major binding species with their respective functions is presented in Table 1. For example, the E4orf4 wild type was found to interact with members of the PRL phosphotyrosine phosphatase family (PTP4A1 and PTP4A2) as well as all four CNNM transmembrane proteins that are known to bind to PRLs and regulate cellular magnesium uptake (CNNM1, CNNM2, CNNM3, and CNNM4), and, as a result, ATP levels to affect cell viability and proliferation (89, 90). The wild-type protein also recovered multiple peptides for the ubiquitin-specific protease USP15, which has been associated with tumor cell apoptosis and immune response via regulation of MDM2 levels (91). Similarly, E4orf4 recovered high peptide counts for the polarity protein Par3 (gene PARD3) (see more below), which has been linked to oncogenic suppression of apoptosis (92, 93). These species, as well as other interacting proteins, may be of interest and will be evaluated in future studies. We noticed that many proteins interacted solely with the class I mutant. These may represent PP2A substrates brought to PP2A through E4orf4 binding or proteins that normally bind weakly to E4orf4 but bind more stably with the loss of PP2A binding; however, it also is possible that such species resulted from a gain of function introduced by the mutation and are of questionable importance in terms of E4orf4 function. Therefore, we have not included these mutant-specific binding proteins in Table 1.
TABLE 1.
Summary of AP-MS analysis from Fig. 1 of proteins found to bind wild-type E4orf4
| Protein found to bind E4orf4 WT through AP-MS | Description |
|---|---|
| PP2A Bα/PPP2R2A | Protein phosphatase 2A 55-kDa regulatory subunit B α isoform; modulates PP2A substrate selectivity and catalytic activity; directs the localization of the enzyme to particular subcellular compartments |
| PP2A Cα/PPP2CA | Protein phosphatase 2A catalytic subunit α and ß isoforms |
| PP2A Cß/PPP2CB | Not distinguishable from each other |
| PP2A Aα/PPP2R1A | Protein phosphatase 2A 65-kDa regulatory subunit A α isoform; scaffolding subunit to coordinate the assembly of the catalytic subunit and the regulatory B subunit |
| PARD3 | Par-3 family cell polarity regulator; involved in directing polarized cell growth |
| USP15 | Ubiquitin specific peptidase 15; part of a family of deubiquitinating enzymes |
| iASPP/PPP1R13L | Protein phosphatase 1, regulatory subunit 13 like; inhibitor of p53-family proteins |
| PP2A Bδ/PPP2R2D | Protein phosphatase 2A 55-kDa regulatory subunit B δ isoform; modulates PP2A substrate selectivity and catalytic activity; directs the localization of the enzyme to particular subcellular compartments |
| PP2A Aβ/PPP2R1B | Protein phosphatase 2A 65-kDa regulatory subunit A β isoform; scaffolding subunit to coordinate the assembly of the catalytic subunit and the regulatory B subunit |
| ASPP2/TP53BP2 | Tumor protein p53 binding protein, 2; required for the induction of apoptosis by p53-family proteins; involved in Hippo signaling pathway |
| TNRC6B | Trinucleotide repeat containing 6B; involved in miRNA-guided gene silencing |
| RAVER1 | Ribonucleoprotein, PTB-binding 1; cooperates with PTBP1 to modulate regulated alternative splicing events |
| CRTC3 | CREB regulated transcription coactivator 3; involved in CREB-dependent gene transcription |
| CCDC85C | Coiled-coil domain containing 85C; may play an important role in cortical development |
| GIGYF2 | GRB10 interacting GYF protein 2; may be involved in the regulation of tyrosine kinase receptor signaling |
| ASPP1/PPP1R13B | Protein phosphatase 1, regulatory subunit 13B; required for the induction of apoptosis by p53-family proteins; involved in Hippo signaling pathway |
| SAPCD2 | Suppressor APC domain containing 2 |
| CNNM3 | CyclinM3; may play an important role in magnesium homeostasis |
| ANKRD17 | Ankyrin repeat domain-containing protein 17; may be involved in liver development |
| CRTC2 | CREB regulated transcription coactivator 2; involved in CREB-dependent gene transcription |
| CNNM2 | CyclinM2; may play an important role in magnesium homeostasis |
| RASSF8 | Ras association (RalGDS/AF-6) domain family (N-terminal) member 8; essential for maintaining adherens junction function in epithelial cells and has a role in epithelial cell migration |
| DNPEP | Aspartyl aminopeptidase |
| BCOR | BCL6 corepressor; transcriptional corepressor |
| CNNM4 | CyclinM4; may play an important role in magnesium homeostasis |
| SIK3 | Salt-inducible kinase 3; energy regulator |
| Sec16A | Homolog A of S. cerevisiae SEC16; may play a role in membrane traffic from the endoplasmic reticulum |
| DCAF7 | DDB1- and CUL4-associated factor 7; may function as a substrate receptor for CUL4-DDB1 E3 ubiquitin-protein ligase complex |
| SHKBP1 | SH3KBP1 binding protein 1 |
| AUTS2 | Autism susceptibility candidate 2; may play a role in neuronal development |
| TANC2 | Tetratricopeptide repeat, ankyrin repeat, and coiled-coil containing 2 |
| SHROOM3 | Shroom family member 3; may be involved in regulating cell shape in certain tissues |
| GPBP1 | GC-rich promoter binding protein 1; functions as a GC-rich promoter-specific transactivating transcription factor |
| PP2A Bγ/PPP2R2C | Protein phosphatase 2A 55-kDa regulatory subunit B γ isoform; modulates PP2A substrate selectivity and catalytic activity; directs the localization of the enzyme to particular subcellular compartments |
| RSPH3 | Radial spoke 3 homolog |
| PRL1/PTP4A1 | Protein tyrosine phosphatase type IVA, member 1; plays regulatory roles in cell proliferation and migration and may be involved in cancer development and metastasis |
| TBK1 | TANK-binding kinase 1; plays an essential role in regulating inflammatory responses to foreign agents |
| RASSF7 | Ras association (RalGDS/AF-6) domain family (N-terminal) member 7; negatively regulates stress-induced JNK activation and apoptosis; required for chromosomal congression by stimulating microtubule polymerization |
| NCOR2 | Nuclear receptor corepressor 2; mediates the transcriptional repression activity of some nuclear receptors by promoting chromatin condensation |
| SYDE1 | Synapse defective 1, Rho GTPase, homolog 1; GTPase activator for the Rho-type GTPases by converting them to an inactive GDP-bound state |
| TBL1XR1 | Transducin (beta)-like 1 X-linked receptor 1; may have transcription corepressor activity |
| EIF4E2 | Eukaryotic translation initiation factor 4E family member 2 |
| KSR1 | Kinase suppressor of ras1; promotes MEK and RAF phosphorylation and activity through assembly of an activated signaling complex |
| PHLDB2 | Pleckstrin homology-like domain, family B, member 2; may be involved in the assembly of the postsynaptic apparatus |
| TBKBP1 | TBK1 binding protein 1/SINTBAD; adaptor protein to TBK1; may be involved in TNF/NFKB pathway |
| FCHSO1 | FCH and double SH3 domain 1 |
| SDCCAG3 | Serologically defined colon cancer antigen 3; may be involved in modulation of TNF response |
| PP1 Cα/PPP1CA | Protein phosphatase PP1-α catalytic subunit; essential for cell division, and participates in the regulation of glycogen metabolism, muscle contractility and protein synthesis |
| SRRM2 | Serine/arginine repetitive matrix 2; involve in pre-mRNA splicing |
| CCDC6 | Coiled-coil domain-containing protein 6; may function as a tumor suppressor |
| MAP3K3 | Mitogen-activated protein kinase kinase kinase 3; regulates the stress-activated protein kinase (SAPK) and extracellular signal-regulated protein kinase (ERK) pathways by activating SEK and MEK1/2 respectively |
| KRT18 | Keratin 18 |
| PNMA1 | Paraneoplastic Ma antigen 1; antineuronal antibody |
| CENPF | Centromere protein F, 350/400ka (mitosin); associates with the centromere-kinetochore complex |
| CCDC120 | Coiled-coil domain containing 120 |
| PRKCI | Protein kinase C, iota |
| MAST2 | Microtubule associated serine/threonine kinase 2; controls TRAF6 and NF-κB activity |
| PHLDB1 | Pleckstrin homology-like domain, family B, member 1 |
| PP1 Cγ/PPP1CC | Protein phosphatase PP1-γ catalytic subunit; essential for cell division, and participates in the regulation of glycogen metabolism, muscle contractility and protein synthesis |
| ZC3H14 | Zinc finger CCCH-type containing 14; belongs to a family of poly(A)-binding proteins that influence gene expression by regulating mRNA stability, nuclear export, and translation |
| KCTD3 | Potassium channel tetramerization domain containing 3 |
| ZNF703 | Zinc finger protein 703; linked with the development of breast cancers |
| GLUD2 | Glutamate dehydrogenase 2 |
| RNF187 | Ring finger protein 187; coactivator of JUN-mediated gene activation |
| PP1 Cβ/PPP1CB | Protein phosphatase PP1-β catalytic subunit; essential for cell division, and participates in the regulation of glycogen metabolism, muscle contractility and protein synthesis |
| MYPT2/PPP1R12B | Protein phosphatase 1, regulatory subunit 12B; the myosin-binding subunit of myosin phosphatase; specific to heart, skeletal, muscle, and brain |
| PPP1R12C | Protein phosphatase 1, regulatory subunit 12C; subunit of myosin phosphatase that regulates the catalytic activity of PP1δ and assembly of the actin cytoskeleton |
A significant new finding was the presence of high levels of all three highly related PP1 catalytic subunits (PP1 Cα/PPP1CA, PP1 Cβ/PPP1CB, and PP1 Cγ/PPP1CC) with both wild-type and mutant E4orf4, although much higher levels of binding of PP1 Cβ/PPP1CB were observed consistently with the mutant (see Fig. 2). In addition to PP1 catalytic subunits, members of the highly related ASPP family of PP1 regulators (ASPP1/PPP1R13B, ASPP2/TP53BP2, and iASPP/PPP1R13L) also were found to associate with wild-type E4orf4 and, to some extent, the class I mutant. Importantly, essentially no other PP1 regulatory subunits were detected with the E4orf4 wild type, suggesting that E4orf4 associates primarily with ASPP-PP1 and not other forms of PP1 (see Fig. 4). Alongside these components, known interaction partners for PP1-ASPP and PARD3, namely, RASSF7, RASSF8, and CCDC85C (57), also were detected, all of which previously were linked to Hippo pathway signaling (Table 1 and see below). It should be noted that the mutant associated strongly with two paralogs of the myosin phosphatase subunits (PPP1R12B and PPP1R12C, which may interact preferentially with PP1 Cβ/PPP1CB), consistent with the detection of actin regulators (SPECC1, SPECC1L, and TRIOBP) with this mutant (see Fig. 3). Thus, these results suggested that E4orf4 can associate with the two major classes of cellular Ser/Thr phosphatases, PP2A and PP1.
FIG 2.
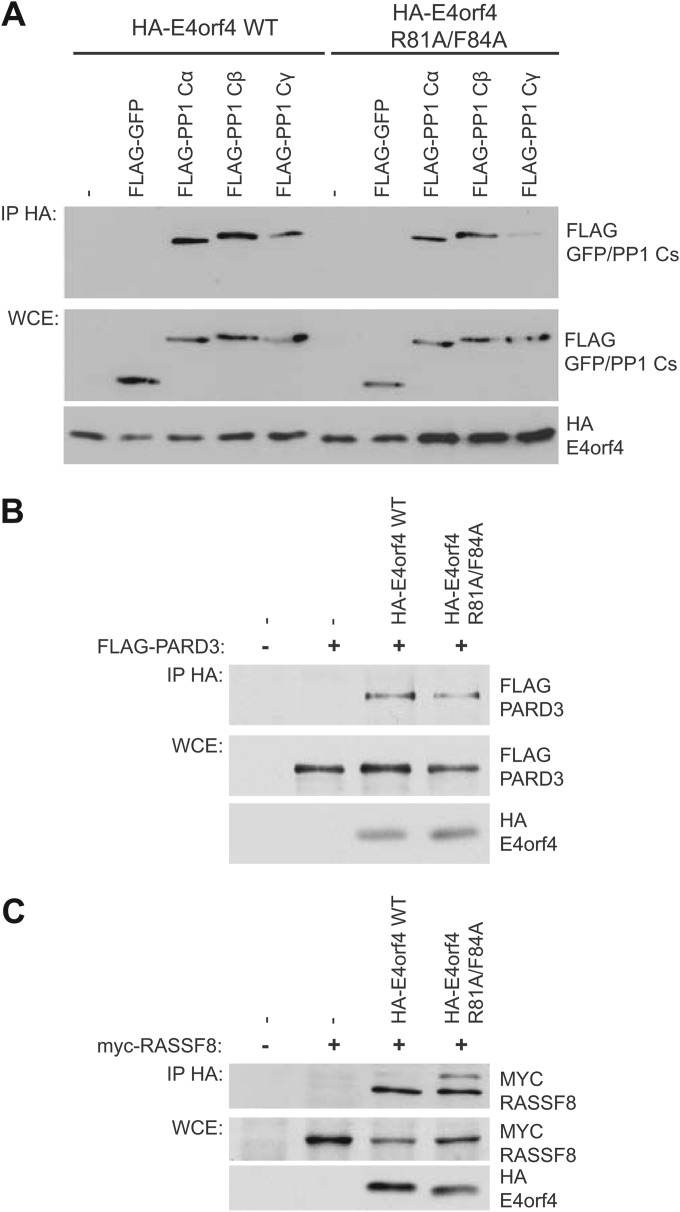
Validation of PP1 catalytic subunits PARD3 and RASSF8 as E4orf4 binding partners. HA-E4orf4 species were immunoprecipitated (IP) using anti-HA antibodies, and Western blotting was performed using appropriate anti-FLAG or anti-MYC antibodies. (A) Binding of FLAG-PP1 catalytic subunits (PP1 Cα, PP1 Cβ, and PP1 Cγ) or FLAG-GFP as a negative control. Whole-cell extracts (WCE) are shown for FLAG-GFP/PP1 catalytic species (PP1 Cα, PP1 Cβ, and PP1 Cγ) and HA-E4orf4 species (E4orf4 wild type and class I mutant R81A/F84A). (B) PARD3. (C) RASSF8.
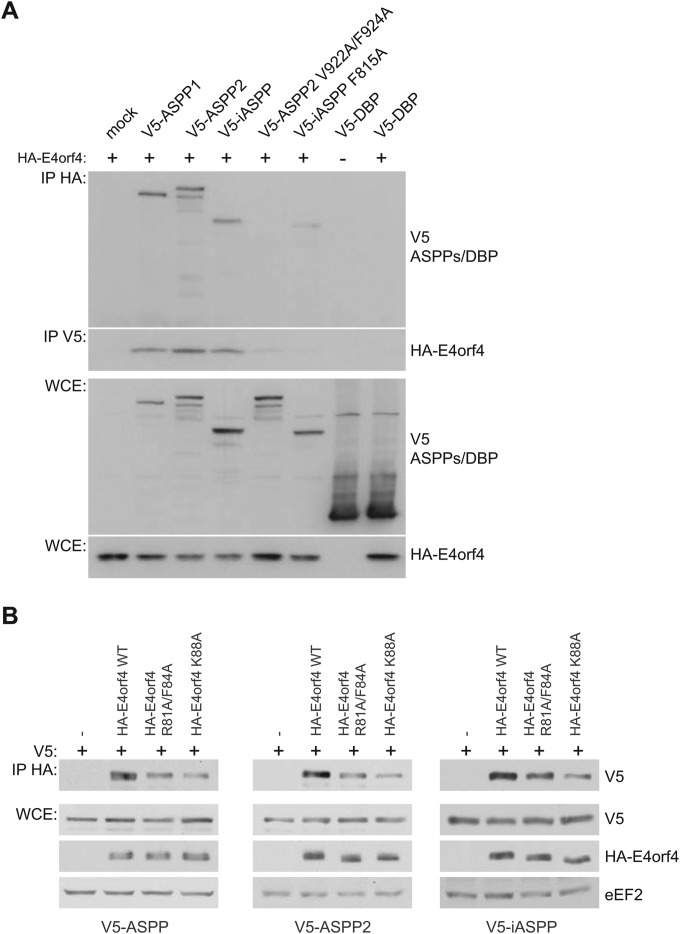
FIG 4.
E4orf4 interacts with all members of the ASPP family of proteins in a PP1 catalytic subunit-dependent manner. (A) HA-E4orf4 was immunoprecipitated using anti-HA antibodies, and Western blotting was performed using anti-V5 antibodies to detect binding of wild-type V5-ASPP subunits (ASPP1, ASPP2, and iASPP), V5-ASPP2 subunit mutated in the RVxF motif that no longer associates with PP1 catalytic subunits (ASPP2 V922A/F924A) (48), V5-iASPP subunit mutated in the RNYF motif that no longer associates with PP1 catalytic subunits (iASPP F815A) (48), or V5-DBP (adenovirus DNA binding protein) as a negative control. Reciprocal coimmunoprecipitation experiments were performed in a similar fashion. Whole-cell extracts (WCE) are shown for V5-ASPP/DBP species and HA-E4orf4. (B) H1299 cells were transfected with the indicated plasmids, and HA-E4orf4 was immunoprecipitated using anti-HA antibodies. Western blotting was performed using anti-V5 antibodies to detect binding of wild-type V5-ASPP subunits (ASPP1, ASPP2, and iASPP). Whole-cell extracts (WCE) are shown for V5-ASPP species and HA-E4orf4 and the loading control eEF2.
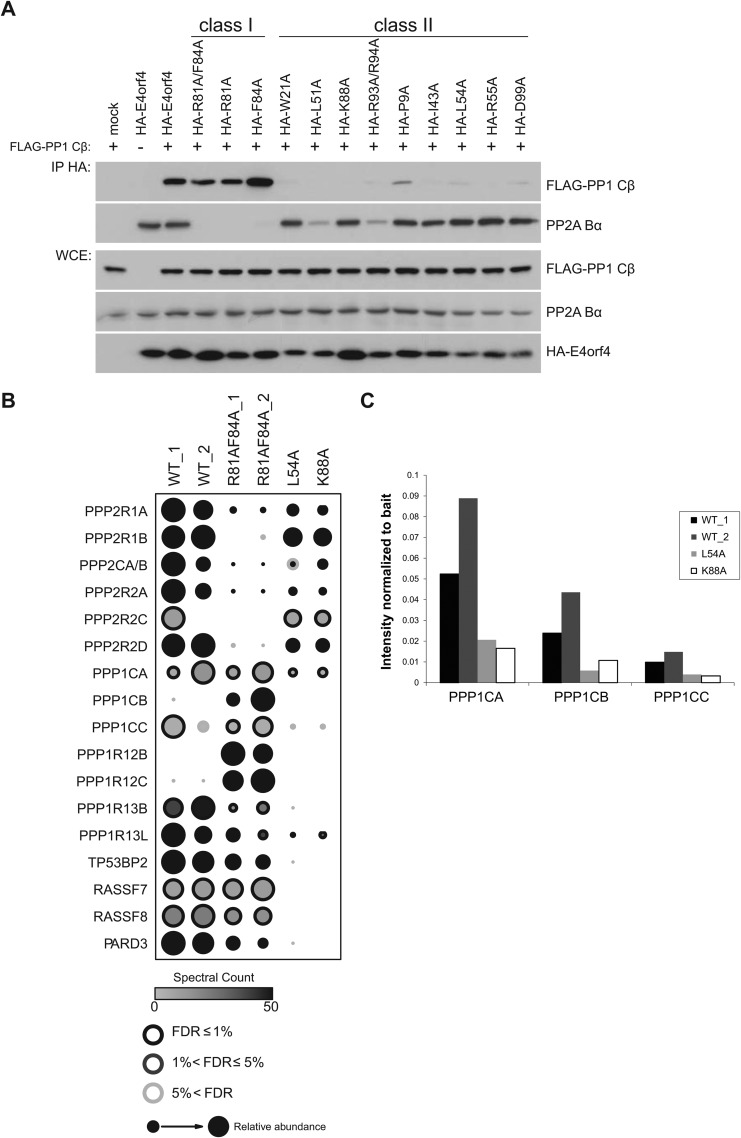
FIG 3.
Binding of E4orf4 mutants to phosphatases and selected components of the Hippo pathway. (A) Binding of E4orf4 mutants to PP1 Cβ. Anti-HA coimmunoprecipitation of HA-E4orf4 wild type, class I mutants (R81A/F84A, R81A, and F84A), or class II mutants (W21A, L51A, K88A, R93A/R94A, P9A, I43A, L54A, R55A, and D99A) was monitored by detection using anti-FLAG (FLAG-PP1 Cβ) or anti-PP2A B55α antibodies. Whole-cell extracts (WCE) are shown for FLAG-PP1 Cβ, PP2A B55α, and HA-E4orf4 species. (B) Analysis by AP-MS. The binding patterns of selected proteins obtained in two AP-MS analyses involving wild-type E4orf4, class I mutant R81A/F84A, and class II mutants L54A and K88A are presented. The identity of E4orf4 binding partners is shown at the left. (C) Binding of individual PP1 C subunits to wild-type E4orf4 and class II mutants derived from AP-MS analyses. Binding to PP1 C subunits was measured by determining the presence of C subunit-specific peptides in AP-MS analyses, as described in Materials and Methods.
E4orf4 interacts with all three isoforms of PP1 catalytic subunit and two components of the Hippo pathway.
To confirm the interaction of PP1 catalytic subunits with E4orf4, coimmunoprecipitation experiments were performed. Plasmid DNAs expressing FLAG-tagged PP1 catalytic subunits were transfected into H1299 cells along with HA-E4orf4-expressing plasmid DNAs to test binding to both wild-type E4orf4 and class I mutant R81A/F84A. Results from coimmunoprecipitation experiments with protein extracts from these cells are shown in Fig. 2A, where HA-E4orf4 species were immunoprecipitated using anti-HA antibodies and Western blotting was performed using anti-FLAG antibodies to detect binding to FLAG-PP1 catalytic species. Both wild-type and mutant E4orf4 were seen to bind to PP1 catalytic subunits with various degrees of efficiency, confirming the AP-MS results, as neither bound to the FLAG-GFP control. These findings further validated the data obtained from the AP-MS analysis and suggested that E4orf4 can interact with all three isoforms of PP1 catalytic subunits. Similar experiments were performed to validate interactions with PARD3 (Fig. 2B) and RASSF8 (Fig. 2C). FLAG-tagged PARD3 or MYC-tagged RASSF8 were cotransfected with HA-tagged wild-type E4orf4 or class I mutant R81A/F84A, and immunoprecipitates obtained using anti-HA antibodies were probed for FLAG or MYC. Both proteins were shown to bind wild-type and mutant HA-E4orf4.
Class II E4orf4 mutants are defective for PP1 binding.
It is believed that much of the toxicity associated with E4orf4 is due to its association with PP2A (1, 10, 14, 16, 34); however, E4orf4 mutational analyses by our group led to the classification of two categories of mutants. While class I E4orf4 mutants are unable to bind to B55α and induce significantly reduced toxicity, class II mutants differ in that they bind B55α yet have reduced toxicity, suggesting that such binding is not sufficient for E4orf4 cell killing (10). Results from a recent study by our group suggested that binding of class II E4orf4 mutants to PP2AB55α holoenzymes differs slightly from that of the wild type; thus, it may not elicit the appropriate effects required for toxicity (1). However, it is also possible that class II mutants are deficient in binding additional essential targets required to induce cell death. Thus, it was of importance to examine directly the interactions of newly detected E4orf4 binding partners with both class I and class II mutants. In the context of the present studies, we were specifically interested in monitoring interactions of these mutants with ASPP-PP1 subunit components of the Hippo pathway.
Using these two classes of E4orf4 mutants, we began by characterizing by coimmunoprecipitation the association with the PP1 catalytic subunit as well as PP2A B55α. HA-E4orf4 species were immunoprecipitated using anti-HA antibodies, and Western blotting was performed using appropriate antibodies. As previously found (10), class I E4orf4 mutants were defective for binding to PP2A B55α, while class II mutants behaved mostly like the wild type in terms of the level of B55α binding (Fig. 3A). However, when anti-FLAG antibodies were used to detect FLAG-PP1 Cβ binding, it was apparent that all class II E4orf4 mutants tested were highly defective in associating with the PP1 Cβ catalytic subunit compared to wild-type E4orf4 and class I mutants.
To examine interactions of the mutants further, we performed an additional round of proteomics analysis, using two class II mutants (the entire data set is deposited in the MassIVE public repository, as described in Materials and Methods, and is available in a user-friendly format at http://prohits-web.lunenfeld.ca). Figure 3B displays the interactions for the PP1 and PP2A proteins and members of the Hippo signaling pathway. As expected, while interaction with all PP2A subunits was nearly abolished in the class I mutant (R81A/F84A), their interactions were largely maintained with the L54A and K88A class II mutants. In contrast, all three members of the ASPP family of PP1 regulators (ASPP1/PPP1R13B, ASPP2/TP53BP2, and iASPP/PPP1R13L) and the Hippo signaling pathway components RASSF7, RASSF8, CCDC85C, PTPN14 (94), and PARD3 were detected with the wild type and the class I mutant but not with the class II E4orf4 mutants. To quantify the interaction of each of the highly homologous PP1 catalytic subunit proteins with E4orf4 class II mutants, the precursor (MS1) intensity for one unique peptide for each of the PP1 catalytic subunits was extracted as described in Materials and Methods. Figure 3C shows that all three interacted with the wild type but did so much more poorly with both class II mutants, indicating that class II mutants indeed were defective in such associations. In total, the results shown in Fig. 3 suggested that the inability to associate with ASPP-PP1 and components of the Hippo pathway contribute to the loss of toxicity of class II E4orf4 mutants.
E4orf4 binds to the ASPP-PP1 complex.
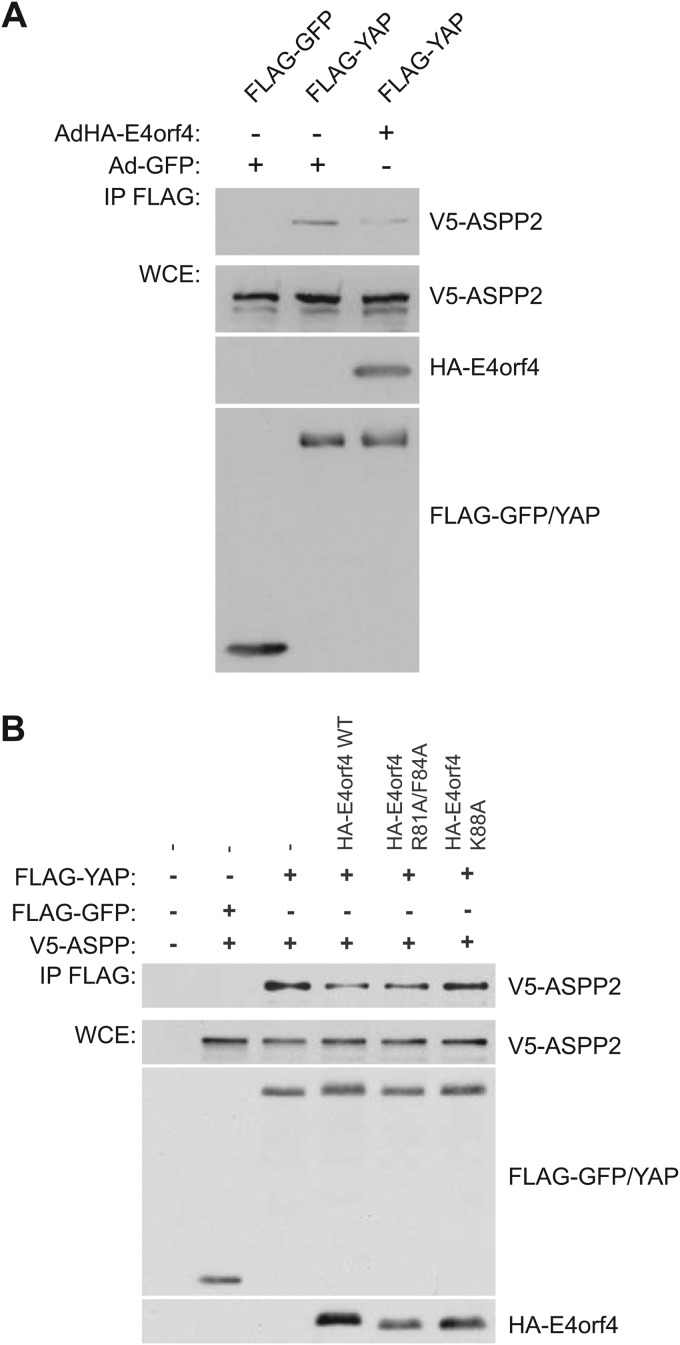
The interaction of all ASPP proteins with wild-type E4orf4 was validated in coimmunoprecipitation experiments. As shown in Fig. 4A, HA-E4orf4 species were immunoprecipitated using anti-HA antibodies, and Western blotting was performed using anti-V5 antibodies to detect binding to V5-ASPP species. Reciprocal coimmunoprecipitation experiments showed that E4orf4 bound to all members of the ASPP family, namely, the functionally similar ASPP1 and ASPP2 species and the divergent iASPP. Interestingly, the mutants ASPP2 V922A/F924A and iASPP F815A, which are unable to associate with PP1 catalytic subunits (48), displayed a reduced ability to interact with E4orf4. These results again confirmed the detection of ASPP proteins as E4orf4 partners and also suggested that E4orf4 associates preferentially with PP1-bound forms of ASPP proteins (see Discussion). We also examined the ability of class I (R81A/F84A) and class II (K88A) E4orf4 mutants to bind to all three ASPP proteins. As shown in Fig. 4B, the class I mutant showed a reduced binding to all three ASPP proteins, but a further decrease was observed with the class II mutant. These data corresponded very well with the mass spectrometry data shown in Fig. 3B.
Effects of E4orf4 on the YAP component of the Hippo signaling pathway.
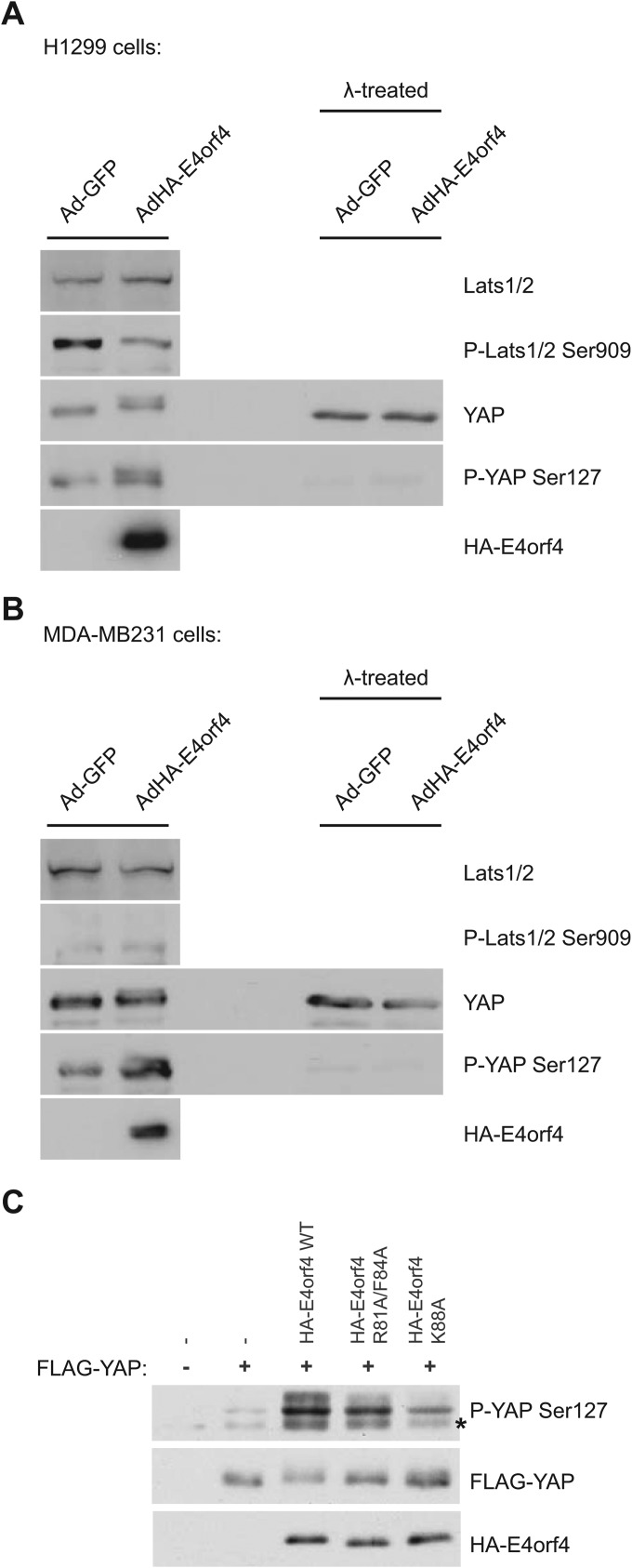
As noted above, both PP2A and ASPP-PP1 complexes are key regulators of Hippo signaling, in part through their dephosphorylation of YAP/TAZ (60, 61, 69). Thus, the mechanism of E4orf4 killing may involve effects on the Hippo signaling pathway. Additionally, other members of the Hippo pathway also were detected as E4orf4 binding partners, including RASSF7, RASSF8, CCDC85C, and PARD3 (Fig. 1, 2, and 3B) (56–58). To explore the possibility of a link between E4orf4 and this critical signaling pathway, we examined the effects of E4orf4 on the phosphorylation state of key Hippo signaling proteins, as the pathway is highly regulated by phosphorylation. Figure 5A shows that H1299 human lung carcinoma cells infected with adenoviral vectors expressing HA-E4orf4 accumulated hyperphosphorylated YAP, as observed using anti-YAP antibodies and antibodies against the phosphoserine 127 residue of YAP, the major phosphorylation site on YAP for the Hippo Lats kinases (95). This effect was evident by the presence of a slower-migrating species that was not detected in the absence of E4orf4. Figure 5A also shows that the doublet of YAP species detected with both antibodies in E4orf4-expressing cells was converted into a single faster-migrating species when samples were treated with λ phosphatase or disappeared completely in Western blots using total and phospho-YAP antibodies, respectively. Moreover, this hyperphosphorylation of YAP in E4orf4-expressing cells was not due to an activation of the Lats1/2 kinases, as Lats activation was not induced under these conditions and was, if anything, reduced at Ser909, an autophosphorylation site that acts as an indicator of Lats activity (96). Comparable results were observed in MDA-MB-231 breast cancer cells, which are deficient in an upstream component of the Hippo cascade, NF2/Merlin (97) (Fig. 5B). To examine the effect of class I and class II E4orf4 mutants on YAP phosphorylation, the experiment was repeated, but in this case in cells expressing E4orf4 proteins following cDNA transfection. Figure 5C shows that expression of wild-type E4orf4 greatly increased the phosphorylation of YAP at Ser127. The increase of phosphorylation was only slightly reduced by the class I mutant (R81A/F84A) but was significantly reduced with the class II K88A mutant (see further comments in Discussion).
FIG 5.
E4orf4 induces Lats kinase-independent hyperphosphorylation of YAP. (A) H1299 lung carcinoma cells were infected with adenoviral vectors expressing either GFP (AdGFP) or HA-E4orf4 (AdHA-E4orf4) at a multiplicity of infection (MOI) of 35 and harvested, and whole-cell extracts (WCE) were analyzed by SDS-PAGE and Western blotting using the indicated antibodies. WCEs also were treated with λ phosphatase as described in Materials and Methods. (B) Analysis similar to that described for panel A, with MDA-MB-231 breast cancer cells infected with adenoviral vectors expressing either GFP (AdGFP) or HA-E4orf4 (AdHA-E4orf4) at an MOI of 50. (C) Analysis of YAP phosphorylation was determined in cells expressing wild-type HA-E4orf4, class I mutant HA-R81A/F84A, or class II mutant K88A expressed from transfected plasmid cDNAs. Immunoprecipitates obtained using anti-HA antibodies were blotted using anti-YAP phosphoserine-127, anti-FLAG, or anti-HA antibodies as indicated. An asterisk indicates an unspecific band.
E4orf4 reduces the association of ASPP2 and YAP.
Our results suggested that YAP hyperphosphorylation in cells expressing E4orf4 was not due to higher Lats kinase activity; thus, another explanation for this effect is that E4orf4 inhibits YAP dephosphorylation. ASPP-PP1 complexes previously have been shown to be involved in the dephosphorylation of YAP (and TAZ), with which they physically associate (56, 57, 59–61); thus, we examined the association of YAP with ASPP2 in the presence and absence of E4orf4 in coimmunoprecipitation experiments. Figure 6A shows that the expression of wild-type E4orf4 significantly reduced YAP interactions with ASPP2 to only 36.6% of the signal detected without E4orf4 (as quantified by ImageJ), suggesting that the binding of E4orf4 to ASPP-PP1 complexes prevents the association of the phosphatase complex with YAP and other substrates, leading to their hyperphosphorylation. Figure 6B shows, in cells expressing E4orf4 proteins from plasmid cDNAs, that both wild-type E4orf4 and class I mutant R81A/F84A reduced interactions between ASPP2 and YAP; however, little effect was seen with the class II mutant K88A that fails to interact with ASPP-PP1 complexes (see further comments in Discussion). Interestingly, despite the use of a very different E4orf4 expression system, the E4orf4 protein induced a very similar decrease in YAP binding, as seen in Fig. 6A, at 32.9% of signal detected without E4orf4 (as quantified by ImageJ).
FIG 6.
E4orf4 reduces the binding of YAP to ASPP2. (A) FLAG-YAP was immunoprecipitated using anti-FLAG antibodies, and Western blotting was performed using anti-V5 antibodies to detect binding of V5-ASPP2. Whole-cell extracts (WCE) are shown for FLAG-YAP, V5-ASPP2, and HA-E4orf4 species in the presence or absence of wild-type E4orf4 expressed from the AdHA-E4orf4 vector. (B) Analysis similar to that described for panel A, with wild-type E4orf4 or class I or II mutants (as indicated) expressed from transfected plasmid cDNAs. Band intensities were quantified using Image J software.
E4orf4 toxicity is enhanced by knocking down YAP1 expression.
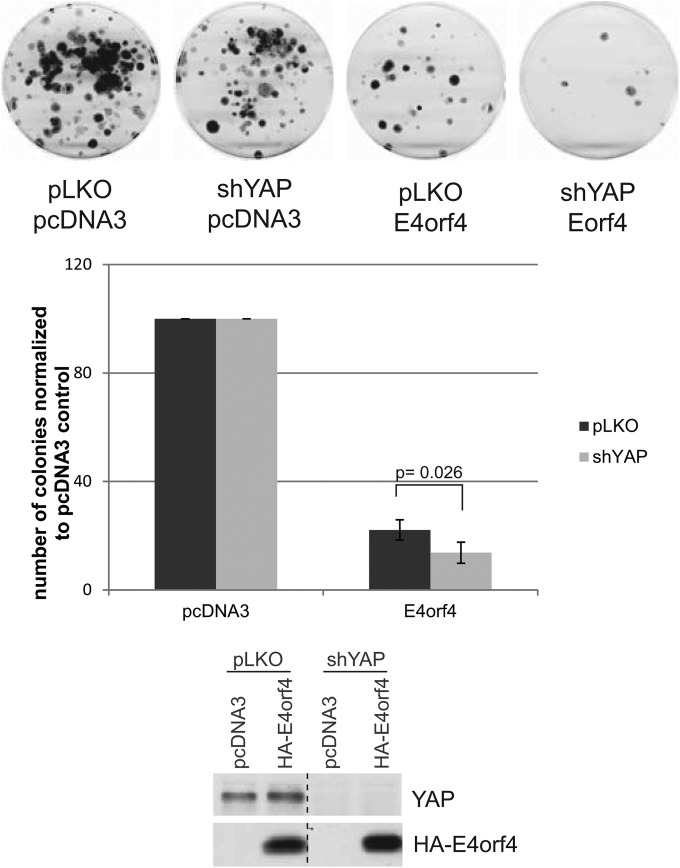
To obtain more direct biological evidence linking effects of YAP phosphorylation and Hippo signaling with E4orf4 toxicity, H1299 cells were generated in which YAP1 expression was reduced by expression of an appropriate shRNA. If inhibition of Hippo signaling plays a role in E4orf4 toxicity, knockdown of YAP1 should enhance cell killing by E4orf4. Figure 7 (bottom) shows that YAP1 expression was greatly reduced in the shYAP-H1299 cells relative to that of control H1299 cells selected using a control pLKO shRNA (pLKO-H1299). These cells then were tested for their ability to be killed by the expression of E4orf4 using our standard colony formation assay in which cells selected for expression of either plasmid DNA pcDNA3 or pcDNA3-E4orf4 were plated and colony formation measured. Figure 7 shows plates from a single experiment (top) as well as the average results from four different experiments (middle). Whereas expression of E4orf4 in control pLKO-H1299 cells resulted in a reduction in colony formation of about 75%, E4orf4 induced a significant decrease in colony formation in shYAP-H1299 cells of around 40% (from 22.1% to 13.7%). Thus, these results are consistent with the possibility that E4orf4 toxicity involves interference of Hippo signaling through effects on YAP, resulting in cell death due to the failure to induce prosurvival gene expression.
FIG 7.
E4orf4 cell killing in control and YAP1 knockdown cells. Cell killing was measured by colony formation assays in control H1299 cells and those in which YAP1 expression was reduced by the expression of an appropriate shRNA following expression of wild-type E4orf4, as described in Materials and Methods. (Top and middle) Effect on colony formation. (Bottom) YAP1 and HA-E4orf4 expression as measured by Western blotting. Error bars represent standard deviations, and the P value was calculated from a t test with two paired samples for means.
DISCUSSION
Although several E4orf4 binding partners have been discovered thus far and characterized in the context of viral replication or E4orf4-induced cell death, our current findings with AP-MS technology have not only confirmed an association with PP2AB55 holoenzymes but also revealed new associated proteins involved in pathways that may be affected by E4orf4 expression, potentially providing new insights into some of these pathways.
In addition to PP2A, E4orf4 was found to interact with another important class of phosphatases, PP1 (PP1 Cα, PP1 Cβ, and PP1 Cγ), an association that was validated in coimmunoprecipitation experiments. Furthermore, PP1-interacting proteins from the ASPP family (ASPP1, ASPP2, and iASPP) also were found in both the AP-MS analysis and coimmunoprecipitation experiments and shown to be dependent on the interaction between ASPP species and PP1 catalytic subunits. Taken together, these results suggested that E4orf4 preferentially binds to ASPP-PP1 heterodimers. It is possible that E4orf4 binds directly to the three highly related PP1 catalytic subunits and blocks association with all PP1-interacting proteins other than the ASPP species. More likely, however, E4orf4 may bind ASPP-PP1 complexes like a pseudosubstrate, as is the case with PP2AB55.
Although additional studies clearly will be required, the current results suggest that the association of E4orf4 with PP1 is important for E4orf4-induced cell death. We previously reported that class II E4orf4 mutants, which bind to PP2AB55α but display reduced toxicity, may be deficient in exerting a biological effect because they bound to B55α inappropriately (1); however, it now seems possible that they are defective because they fail to interact with additional essential E4orf4 targets, one of which may be ASPP-PP1 enzymes. While there is no reason to assume that all class II mutants have the same defect, our current results demonstrated that all of the class II mutants tested, despite being able to bind to B55α, were defective in their associations with all ASPP-PP1 complexes. Although the precise mechanism by which PP1 contributes to E4orf4-induced toxicity is unclear, an attractive hypothesis is that both PP1 and PP2A must be targeted by E4orf4 to control the phosphorylation of common pathways and substrates implicated in the regulation of cell death, one of which is the Hippo signaling target, YAP. Consistent with the potential biological links of E4orf4 to Hippo is the fact that several other components of Hippo signaling, including PTPN14, RASSF7, RASSF8, CCDC85C, and PARD3, are among the E4orf4-interacting species detected in our AP-MS analysis. It is not completely clear yet at what step(s) all of these proteins interface with the Hippo pathway, although they all are found associated with the transcriptional activator YAP1 (56, 57).
The revelation that regulators of Hippo signaling are involved in E4orf4-induced toxicity and, perhaps, adenovirus replication suggests a new common target for some viruses. It was shown very recently that polyomavirus small T antigen (PyST) binds to YAP1 and YAP2, resulting in decreased YAP phosphorylation and subsequent degradation from enhanced YAP association with PP2A (98). Additionally, both SV40 small T (ST) and Merkel cell polyomavirus small T proteins have been shown to activate YAP (99). Unlike PyST, adenoviral E4orf4 does not appear to interact with YAP (M. Z. Mui and P. E. Branton, unpublished results), and unlike other small DNA tumor viruses, adenovirus E4orf4, at least when expressed alone at high levels, appears to induce opposite effects on YAP, leading to Lats-independent hyperphosphorylation of YAP. It should be remembered that during normal adenovirus infection, the levels of E4orf4 expression are much lower than those required for toxicity, and at least in the case of PP2A, we believe that the role of E4orf4 binding is to introduce new substrates for dephosphorylation by this enzyme to further the infectious process (1). This also may be the case with ASPP-PP1. Nevertheless, at high levels in H1299 lung cancer cell lines, E4orf4 appeared to downregulate Lats kinase activity, further strengthening a link between E4orf4 and the Hippo signaling pathway. In MDA-MB-231 cells that exhibit downregulated Hippo signaling, Lats activity was unaffected by E4orf4 (Fig. 5B), yet YAP hyperphosphorylation still was apparent. Taken together, these results suggested that another mechanism exists by which YAP is hyperphosphorylated in the presence of E4orf4. PP1 and PP2A have been linked to the dephosphorylation of both YAP and TAZ (59–61, 69); thus, it is not surprising that, when expressed at high levels, E4orf4, through interactions with both PP2A and PP1, could affect YAP phosphorylation through inhibition of one or both enzymes. We have shown previously that E4orf4 binds to PP2AB55α like a pseudosubstrate and prevents the binding of normal substrates, including p107 (1). Thus, it is possible that E4orf4 functions in a similar way with ASPP-PP1; in fact, we demonstrated that E4orf4 reduced interactions with YAP. Finally, although the Striatin/B‴ form of PP2A was found to be involved in regulating the Hippo signaling pathway (56, 100), a recent study reported B55 and A subunit interactions with LATS2 (56). Our current results suggested that in terms of both effects on YAP hyperphosphorylation and reduction of the association of YAP with ASPP-PP1, class II mutants were more defective than class I, suggesting that interactions with ASPP-PP1 are more significant than those with PP2AB55. Clearly, further studies will be required to resolve this issue.
The biological significance of the effect E4orf4 has on YAP is not yet clear and will be investigated in future studies; however, from the perspective of E4orf4-induced cell death, the hyperphosphorylation and inhibition of YAP would be consistent with the attenuation of transcription of genes required for proliferation. Interestingly, cytoskeleton reorganization and cell detachment was found to activate Lats1/2, leading to YAP phosphorylation and inhibition, events which are required for anoikis in nontransformed cells, while in cancer cells with a deregulated Hippo pathway, the knockdown of YAP and TAZ restored anoikis (101). E4orf4 may induce these events in a manner independent from that of the canonical Hippo kinases through inhibition of ASPP-PP1 and possibly PP2A. Further evidence supporting a link between E4orf4 toxicity and the Hippo pathway was evident in our finding that knockdown of YAP1, which should reduce Hippo signaling, enhanced E4orf4 killing.
Although E4orf4 protein is localized largely in the nucleus, low levels also are present in the cytoplasm (12, 18, 70, 71, 73). It is believed that these two subpopulations are responsible for the distinct cytoplasmic and nuclear death activities attributed to E4orf4, where the former involves mostly c-Src kinases and the latter PP2A (3, 12, 21, 70–73). The implication of Hippo signaling regulators as being involved in the mechanism by which E4orf4 induces cell death revealed a possible link between the cytoplasmic and nuclear pathways. Cytoplasmic events such as cell polarity, cell-cell contacts, the actin cytoskeleton, and G protein-coupled receptors have been known to influence the Hippo signaling pathway, so that the effectors of the pathway, YAP and TAZ, are properly regulated in the nucleus to ultimately control cell proliferation and fate (65, 67, 102).
We believe that our study offers important new avenues of research toward an understanding of the mechanism by which E4orf4 induces cell death by implicating the Hippo signaling pathway. Furthermore, this implication may even explain the tumor cell specificity of E4orf4-induced toxicity, as the Hippo signaling pathway often is deregulated in cancer cells. Clearly further studies will be required to identify the role of the highly complex Hippo pathway in E4orf4-induced cell killing.
ACKNOWLEDGMENTS
We thank X. Lu and N. Tapon for generously supplying reagents.
This work was supported by grants from the Canadian Institutes of Health Research (CIHR), MOP-93753 (P.E.B.), MOP-84314 (A.-C.G.), MOP-130540 (S. H.), and MOP-106635 (M.P.), and from the Canadian Cancer Society Research Institute and the Cancer Research Society (A.-C.G.).
A.-C.G. is the Canada Research Chair in Functional Proteomics and the Lea Reichmann Chair in Cancer Proteomics, S.H. is a recipient of a Canada Research Chair (CRC) in Functional Genomics, and A.I.P. is a recipient of a CIHR-funded McGill Chemical Biology Postdoctoral Fellowship.
REFERENCES
- 1.Mui MZ, Kucharski M, Miron MJ, Hur WS, Berghuis AM, Blanchette P, Branton PE. 2013. Identification of the adenovirus E4orf4 protein binding site on the B55alpha and Cdc55 regulatory subunits of PP2A: implications for PP2A function, tumor cell killing and viral replication. PLoS Pathog 9:e1003742. doi: 10.1371/journal.ppat.1003742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu Y, Kucharski TJ, Gamache I, Blanchette P, Branton PE, Teodoro JG. 2014. Interaction of adenovirus type 5 e4orf4 with the nuclear pore subunit nup205 is required for proper viral gene expression. J Virol 88:13249–13259. doi: 10.1128/JVI.00933-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brestovitsky A, Sharf R, Mittelman K, Kleinberger T. 2011. The adenovirus E4orf4 protein targets PP2A to the ACF chromatin-remodeling factor and induces cell death through regulation of SNF2h-containing complexes. Nucleic Acids Res 39:6414–6427. doi: 10.1093/nar/gkr231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Estmer Nilsson C, Petersen-Mahrt S, Durot C, Shtrichman R, Krainer AR, Kleinberger T, Akusjarvi G. 2001. The adenovirus E4-ORF4 splicing enhancer protein interacts with a subset of phosphorylated SR proteins. EMBO J 20:864–871. doi: 10.1093/emboj/20.4.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kanopka A, Muhlemann O, Petersen-Mahrt S, Estmer C, Ohrmalm C, Akusjarvi G. 1998. Regulation of adenovirus alternative RNA splicing by dephosphorylation of SR proteins. Nature 393:185–187. doi: 10.1038/30277. [DOI] [PubMed] [Google Scholar]
- 6.O'Shea C, Klupsch K, Choi S, Bagus B, Soria C, Shen J, McCormick F, Stokoe D. 2005. Adenoviral proteins mimic nutrient/growth signals to activate the mTOR pathway for viral replication. EMBO J 24:1211–1221. doi: 10.1038/sj.emboj.7600597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marcellus RC, Teodoro JG, Wu T, Brough DE, Ketner G, Shore GC, Branton PE. 1996. Adenovirus type 5 early region 4 is responsible for E1A-induced p53-independent apoptosis. J Virol 70:6207–6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marcellus RC, Lavoie JN, Boivin D, Shore GC, Ketner G, Branton PE. 1998. The early region 4 orf4 protein of human adenovirus type 5 induces p53-independent cell death by apoptosis. J Virol 72:7144–7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shtrichman R, Kleinberger T. 1998. Adenovirus type 5 E4 open reading frame 4 protein induces apoptosis in transformed cells. J Virol 72:2975–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marcellus RC, Chan H, Paquette D, Thirlwell S, Boivin D, Branton PE. 2000. Induction of p53-independent apoptosis by the adenovirus E4orf4 protein requires binding to the Balpha subunit of protein phosphatase 2A. J Virol 74:7869–7877. doi: 10.1128/JVI.74.17.7869-7877.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lavoie JN, Nguyen M, Marcellus RC, Branton PE, Shore GC. 1998. E4orf4, a novel adenovirus death factor that induces p53-independent apoptosis by a pathway that is not inhibited by zVAD-fmk. J Cell Biol 140:637–645. doi: 10.1083/jcb.140.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robert A, Miron MJ, Champagne C, Gingras MC, Branton PE, Lavoie JN. 2002. Distinct cell death pathways triggered by the adenovirus early region 4 ORF 4 protein. J Cell Biol 158:519–528. doi: 10.1083/jcb.200201106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livne A, Shtrichman R, Kleinberger T. 2001. Caspase activation by adenovirus e4orf4 protein is cell line specific and is mediated by the death receptor pathway. J Virol 75:789–798. doi: 10.1128/JVI.75.2.789-798.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li S, Brignole C, Marcellus R, Thirlwell S, Binda O, McQuoid MJ, Ashby D, Chan H, Zhang Z, Miron MJ, Pallas DC, Branton PE. 2009. The adenovirus E4orf4 protein induces G2/M arrest and cell death by blocking protein phosphatase 2A activity regulated by the B55 subunit. J Virol 83:8340–8352. doi: 10.1128/JVI.00711-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li S, Szymborski A, Miron MJ, Marcellus R, Binda O, Lavoie JN, Branton PE. 2009. The adenovirus E4orf4 protein induces growth arrest and mitotic catastrophe in H1299 human lung carcinoma cells. Oncogene 28:390–400. doi: 10.1038/onc.2008.393. [DOI] [PubMed] [Google Scholar]
- 16.Shtrichman R, Sharf R, Barr H, Dobner T, Kleinberger T. 1999. Induction of apoptosis by adenovirus E4orf4 protein is specific to transformed cells and requires an interaction with protein phosphatase 2A. Proc Natl Acad Sci U S A 96:10080–10085. doi: 10.1073/pnas.96.18.10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miron MJ, Gallouzi IE, Lavoie JN, Branton PE. 2004. Nuclear localization of the adenovirus E4orf4 protein is mediated through an arginine-rich motif and correlates with cell death. Oncogene 23:7458–7468. doi: 10.1038/sj.onc.1207919. [DOI] [PubMed] [Google Scholar]
- 18.Lavoie JN, Champagne C, Gingras MC, Robert A. 2000. Adenovirus E4 open reading frame 4-induced apoptosis involves dysregulation of Src family kinases. J Cell Biol 150:1037–1056. doi: 10.1083/jcb.150.5.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kleinberger T. 2014. Induction of cancer-specific cell death by the adenovirus E4orf4 protein. Adv Exp Med Biol 818:61–97. [DOI] [PubMed] [Google Scholar]
- 20.Avital-Shacham M, Sharf R, Kleinberger T. 2014. NTPDASE4 gene products cooperate with the adenovirus E4orf4 protein through PP2A-dependent and -independent mechanisms and contribute to induction of cell death. J Virol 88:6318–6328. doi: 10.1128/JVI.00381-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cabon L, Sriskandarajah N, Mui MZ, Teodoro JG, Blanchette P, Branton PE. 2013. Adenovirus E4orf4 protein-induced death of p53−/− H1299 human cancer cells follows a G1 arrest of both tetraploid and diploid cells due to a failure to initiate DNA synthesis. J Virol 87:13168–13178. doi: 10.1128/JVI.01242-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Afifi R, Sharf R, Shtrichman R, Kleinberger T. 2001. Selection of apoptosis-deficient adenovirus E4orf4 mutants in Saccharomyces cerevisiae. J Virol 75:4444–4447. doi: 10.1128/JVI.75.9.4444-4447.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kornitzer D, Sharf R, Kleinberger T. 2001. Adenovirus E4orf4 protein induces PP2A-dependent growth arrest in Saccharomyces cerevisiae and interacts with the anaphase-promoting complex/cyclosome. J Cell Biol 154:331–344. doi: 10.1083/jcb.200104104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roopchand DE, Lee JM, Shahinian S, Paquette D, Bussey H, Branton PE. 2001. Toxicity of human adenovirus E4orf4 protein in Saccharomyces cerevisiae results from interactions with the Cdc55 regulatory B subunit of PP2A. Oncogene 20:5279–5290. doi: 10.1038/sj.onc.1204693. [DOI] [PubMed] [Google Scholar]
- 25.Mui MZ, Roopchand DE, Gentry MS, Hallberg RL, Vogel J, Branton PE. 2010. Adenovirus protein E4orf4 induces premature APCCdc20 activation in Saccharomyces cerevisiae by a protein phosphatase 2A-dependent mechanism. J Virol 84:4798–4809. doi: 10.1128/JVI.02434-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Z, Mui MZ, Chan F, Roopchand DE, Marcellus RC, Blanchette P, Li S, Berghuis AM, Branton PE. 2011. Genetic analysis of B55alpha/Cdc55 protein phosphatase 2A subunits: association with the adenovirus E4orf4 protein. J Virol 85:286–295. doi: 10.1128/JVI.01381-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koren R, Rainis L, Kleinberger T. 2004. The scaffolding A/Tpd3 subunit and high phosphatase activity are dispensable for Cdc55 function in the Saccharomyces cerevisiae spindle checkpoint and in cytokinesis. J Biol Chem 279:48598–48606. doi: 10.1074/jbc.M409359200. [DOI] [PubMed] [Google Scholar]
- 28.Li Y, Wei H, Hsieh TC, Pallas DC. 2008. Cdc55p-mediated E4orf4 growth inhibition in Saccharomyces cerevisiae is mediated only in part via the catalytic subunit of protein phosphatase 2A. J Virol 82:3612–3623. doi: 10.1128/JVI.02435-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mittelman K, Ziv K, Maoz T, Kleinberger T. 2010. The cytosolic tail of the Golgi apyrase Ynd1 mediates E4orf4-induced toxicity in Saccharomyces cerevisiae. PLoS One 5:e15539. doi: 10.1371/journal.pone.0015539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pechkovsky A, Salzberg A, Kleinberger T. 2013. The adenovirus E4orf4 protein induces a unique mode of cell death while inhibiting classical apoptosis. Cell Cycle 12:2343–2344. doi: 10.4161/cc.25707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pechkovsky A, Lahav M, Bitman E, Salzberg A, Kleinberger T. 2013. E4orf4 induces PP2A- and Src-dependent cell death in Drosophila melanogaster and at the same time inhibits classic apoptosis pathways. Proc Natl Acad Sci U S A 110:E1724–E1733. doi: 10.1073/pnas.1220282110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sriskandarajah N, Blanchette P, Kucharski TJ, Teodoro J, Branton PE. 2015. Analysis by live imaging of effects of the adenovirus E4orf4 protein on passage through mitosis of H1299 tumor cells. J Virol 89:4685–4689. doi: 10.1128/JVI.03437-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kleinberger T, Shenk T. 1993. Adenovirus E4orf4 protein binds to protein phosphatase 2A, and the complex down regulates E1A-enhanced junB transcription. J Virol 67:7556–7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shtrichman R, Sharf R, Kleinberger T. 2000. Adenovirus E4orf4 protein interacts with both Balpha and B′ subunits of protein phosphatase 2A, but E4orf4-induced apoptosis is mediated only by the interaction with Balpha. Oncogene 19:3757–3765. doi: 10.1038/sj.onc.1203705. [DOI] [PubMed] [Google Scholar]
- 35.Horowitz B, Sharf R, Avital-Shacham M, Pechkovsky A, Kleinberger T. 2013. Structure- and modeling-based identification of the adenovirus E4orf4 binding site in the protein phosphatase 2A B55alpha subunit. J Biol Chem 288:13718–13727. doi: 10.1074/jbc.M112.343756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sents W, Ivanova E, Lambrecht C, Haesen D, Janssens V. 2013. The biogenesis of active protein phosphatase 2A holoenzymes: a tightly regulated process creating phosphatase specificity. FEBS J 280:644–661. doi: 10.1111/j.1742-4658.2012.08579.x. [DOI] [PubMed] [Google Scholar]
- 37.Janssens V, Longin S, Goris J. 2008. PP2A holoenzyme assembly: in cauda venenum (the sting is in the tail). Trends Biochem Sci 33:113–121. doi: 10.1016/j.tibs.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 38.Ferrigno P, Langan TA, Cohen P. 1993. Protein phosphatase 2A1 is the major enzyme in vertebrate cell extracts that dephosphorylates several physiological substrates for cyclin-dependent protein kinases. Mol Biol Cell 4:669–677. doi: 10.1091/mbc.4.7.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kamibayashi C, Estes R, Lickteig RL, Yang SI, Craft C, Mumby MC. 1994. Comparison of heterotrimeric protein phosphatase 2A containing different B subunits. J Biol Chem 269:20139–20148. [PubMed] [Google Scholar]
- 40.Hunt T. 2013. On the regulation of protein phosphatase 2A and its role in controlling entry into and exit from mitosis. Adv Biol Regul 53:173–178. doi: 10.1016/j.jbior.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 41.Mumby MC, Walter G. 1993. Protein serine/threonine phosphatases: structure, regulation, and functions in cell growth. Physiol Rev 73:673–699. [DOI] [PubMed] [Google Scholar]
- 42.Janssens V, Goris J. 2001. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem J 353:417–439. doi: 10.1042/0264-6021:3530417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kurimchak A, Grana X. 2012. PP2A holoenzymes negatively and positively regulate cell cycle progression by dephosphorylating pocket proteins and multiple CDK substrates. Gene 499:1–7. doi: 10.1016/j.gene.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sablina AA, Hector M, Colpaert N, Hahn WC. 2010. Identification of PP2A complexes and pathways involved in cell transformation. Cancer Res 70:10474–10484. doi: 10.1158/0008-5472.CAN-10-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Westermarck J, Hahn WC. 2008. Multiple pathways regulated by the tumor suppressor PP2A in transformation. Trends Mol Med 14:152–160. doi: 10.1016/j.molmed.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 46.Virshup DM, Shenolikar S. 2009. From promiscuity to precision: protein phosphatases get a makeover. Mol Cell 33:537–545. doi: 10.1016/j.molcel.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 47.Bollen M, Peti W, Ragusa MJ, Beullens M. 2010. The extended PP1 toolkit: designed to create specificity. Trends Biochem Sci 35:450–458. doi: 10.1016/j.tibs.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Llanos S, Royer C, Lu M, Bergamaschi D, Lee WH, Lu X. 2011. Inhibitory member of the apoptosis-stimulating proteins of the p53 family (iASPP) interacts with protein phosphatase 1 via a noncanonical binding motif. J Biol Chem 286:43039–43044. doi: 10.1074/jbc.M111.270751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Skene-Arnold TD, Luu HA, Uhrig RG, De Wever V, Nimick M, Maynes J, Fong A, James MN, Trinkle-Mulcahy L, Moorhead GB, Holmes CF. 2013. Molecular mechanisms underlying the interaction of protein phosphatase-1c with ASPP proteins. Biochem J 449:649–659. doi: 10.1042/BJ20120506. [DOI] [PubMed] [Google Scholar]
- 50.Iwabuchi K, Bartel PL, Li B, Marraccino R, Fields S. 1994. Two cellular proteins that bind to wild-type but not mutant p53. Proc Natl Acad Sci U S A 91:6098–6102. doi: 10.1073/pnas.91.13.6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Naumovski L, Cleary ML. 1996. The p53-binding protein 53BP2 also interacts with Bc12 and impedes cell cycle progression at G2/M. Mol Cell Biol 16:3884–3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Samuels-Lev Y, O'Connor DJ, Bergamaschi D, Trigiante G, Hsieh JK, Zhong S, Campargue I, Naumovski L, Crook T, Lu X. 2001. ASPP proteins specifically stimulate the apoptotic function of p53. Mol Cell 8:781–794. doi: 10.1016/S1097-2765(01)00367-7. [DOI] [PubMed] [Google Scholar]
- 53.Helps NR, Barker HM, Elledge SJ, Cohen PT. 1995. Protein phosphatase 1 interacts with p53BP2, a protein which binds to the tumour suppressor p53. FEBS Lett 377:295–300. doi: 10.1016/0014-5793(95)01347-4. [DOI] [PubMed] [Google Scholar]
- 54.Espanel X, Sudol M. 2001. Yes-associated protein and p53-binding protein-2 interact through their WW and SH3 domains. J Biol Chem 276:14514–14523. [DOI] [PubMed] [Google Scholar]
- 55.Vigneron AM, Ludwig RL, Vousden KH. 2010. Cytoplasmic ASPP1 inhibits apoptosis through the control of YAP. Genes Dev 24:2430–2439. doi: 10.1101/gad.1954310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Couzens AL, Knight JD, Kean MJ, Teo G, Weiss A, Dunham WH, Lin ZY, Bagshaw RD, Sicheri F, Pawson T, Wrana JL, Choi H, Gingras AC. 2013. Protein interaction network of the mammalian Hippo pathway reveals mechanisms of kinase-phosphatase interactions. Sci Signal 6:rs15. doi: 10.1126/scisignal.2004712. [DOI] [PubMed] [Google Scholar]
- 57.Hauri S, Wepf A, van Drogen A, Varjosalo M, Tapon N, Aebersold R, Gstaiger M. 2013. Interaction proteome of human Hippo signaling: modular control of the co-activator YAP1. Mol Syst Biol 9:713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang W, Li X, Huang J, Feng L, Dolinta KG, Chen J. 2014. Defining the protein-protein interaction network of the human hippo pathway. Mol Cell Proteomics 13:119–131. doi: 10.1074/mcp.M113.030049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Royer C, Koch S, Qin X, Zak J, Buti L, Dudziec E, Zhong S, Ratnayaka I, Srinivas S, Lu X. 2014. ASPP2 links the apical lateral polarity complex to the regulation of YAP activity in epithelial cells. PLoS One 9:e111384. doi: 10.1371/journal.pone.0111384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu CY, Lv X, Li T, Xu Y, Zhou X, Zhao S, Xiong Y, Lei QY, Guan KL. 2011. PP1 cooperates with ASPP2 to dephosphorylate and activate TAZ. J Biol Chem 286:5558–5566. doi: 10.1074/jbc.M110.194019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang P, Bai Y, Song B, Wang Y, Liu D, Lai Y, Bi X, Yuan Z. 2011. PP1A-mediated dephosphorylation positively regulates YAP2 activity. PLoS One 6:e24288. doi: 10.1371/journal.pone.0024288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Low BC, Pan CQ, Shivashankar GV, Bershadsky A, Sudol M, Sheetz M. 2014. YAP/TAZ as mechanosensors and mechanotransducers in regulating organ size and tumor growth. FEBS Lett 588:2663–2670. doi: 10.1016/j.febslet.2014.04.012. [DOI] [PubMed] [Google Scholar]
- 63.Varelas X. 2014. The Hippo pathway effectors TAZ and YAP in development, homeostasis and disease. Development 141:1614–1626. doi: 10.1242/dev.102376. [DOI] [PubMed] [Google Scholar]
- 64.Hao J, Zhang Y, Wang Y, Ye R, Qiu J, Zhao Z, Li J. 2014. Role of extracellular matrix and YAP/TAZ in cell fate determination. Cell Signal 26:186–191. doi: 10.1016/j.cellsig.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 65.Piccolo S, Dupont S, Cordenonsi M. 2014. The biology of YAP/TAZ: Hippo signaling and beyond. Physiol Rev 94:1287–1312. doi: 10.1152/physrev.00005.2014. [DOI] [PubMed] [Google Scholar]
- 66.Harvey KF, Zhang X, Thomas DM. 2013. The Hippo pathway and human cancer. Nat Rev Cancer 13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 67.Yu FX, Guan KL. 2013. The Hippo pathway: regulators and regulations. Genes Dev 27:355–371. doi: 10.1101/gad.210773.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hong W, Guan KL. 2012. The YAP and TAZ transcription co-activators: key downstream effectors of the mammalian Hippo pathway. Semin Cell Dev Biol 23:785–793. doi: 10.1016/j.semcdb.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schlegelmilch K, Mohseni M, Kirak O, Pruszak J, Rodriguez JR, Zhou D, Kreger BT, Vasioukhin V, Avruch J, Brummelkamp TR, Camargo FD. 2011. Yap1 acts downstream of alpha-catenin to control epidermal proliferation. Cell 144:782–795. doi: 10.1016/j.cell.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Champagne C, Landry MC, Gingras MC, Lavoie JN. 2004. Activation of adenovirus type 2 early region 4 ORF4 cytoplasmic death function by direct binding to Src kinase domain. J Biol Chem 279:25905–25915. doi: 10.1074/jbc.M400933200. [DOI] [PubMed] [Google Scholar]
- 71.Robert A, Smadja-Lamere N, Landry MC, Champagne C, Petrie R, Lamarche-Vane N, Hosoya H, Lavoie JN. 2006. Adenovirus E4orf4 hijacks rho GTPase-dependent actin dynamics to kill cells: a role for endosome-associated actin assembly. Mol Biol Cell 17:3329–3344. doi: 10.1091/mbc.E05-12-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Landry MC, Sicotte A, Champagne C, Lavoie JN. 2009. Regulation of cell death by recycling endosomes and Golgi membrane dynamics via a pathway involving Src-family kinases, Cdc42 and Rab11a. Mol Biol Cell 20:4091–4106. doi: 10.1091/mbc.E09-01-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gingras MC, Champagne C, Roy M, Lavoie JN. 2002. Cytoplasmic death signal triggered by SRC-mediated phosphorylation of the adenovirus E4orf4 protein. Mol Cell Biol 22:41–56. doi: 10.1128/MCB.22.1.41-56.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Landry MC, Champagne C, Boulanger MC, Jette A, Fuchs M, Dziengelewski C, Lavoie JN. 2014. A functional interplay between the small GTPase Rab11a and mitochondria-shaping proteins regulates mitochondrial positioning and polarization of the actin cytoskeleton downstream of Src family kinases. J Biol Chem 289:2230–2249. doi: 10.1074/jbc.M113.516351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kean MJ, Ceccarelli DF, Goudreault M, Sanches M, Tate S, Larsen B, Gibson LC, Derry WB, Scott IC, Pelletier L, Baillie GS, Sicheri F, Gingras AC. 2011. Structure-function analysis of core STRIPAK proteins: a signaling complex implicated in Golgi polarization. J Biol Chem 286:25065–25075. doi: 10.1074/jbc.M110.214486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kean MJ, Couzens AL, Gingras AC. 2012. Mass spectrometry approaches to study mammalian kinase and phosphatase associated proteins. Methods 57:400–408. doi: 10.1016/j.ymeth.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 77.Kessner D, Chambers M, Burke R, Agus D, Mallick P. 2008. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics 24:2534–2536. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shteynberg D, Deutsch EW, Lam H, Eng Sun JKZ, Tasman N, Mendoza L, Moritz RL, Aebersold R, Nesvizhskii AI. 2011. iProphet: multi-level integrative analysis of shotgun proteomic data improves peptide and protein identification rates and error estimates. Mol Cell Proteomics 10:M111.007690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu G, Zhang J, Larsen B, Stark C, Breitkreutz A, Lin ZY, Breitkreutz BJ, Ding Y, Colwill K, Pasculescu A, Pawson T, Wrana JL, Nesvizhskii AI, Raught B, Tyers M, Gingras AC. 2010. ProHits: integrated software for mass spectrometry-based interaction proteomics. Nat Biotechnol 28:1015–1017. doi: 10.1038/nbt1010-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Eng JK, Jahan TA, Hoopmann MR. 2013. Comet: an open-source MS/MS sequence database search tool. Proteomics 13:22–24. doi: 10.1002/pmic.201200439. [DOI] [PubMed] [Google Scholar]
- 81.Teo G, Liu G, Zhang J, Nesvizhskii AI, Gingras AC, Choi H. 2014. SAINTexpress: improvements and additional features in significance analysis of INTeractome software. J Proteomics 100:37–43. doi: 10.1016/j.jprot.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Choi H, Larsen B, Lin ZY, Breitkreutz A, Mellacheruvu D, Fermin D, Qin ZS, Tyers M, Gingras AC, Nesvizhskii AI. 2011. SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nat Methods 8:70–73. doi: 10.1038/nmeth.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Knight JD, Liu G, Zhang JP, Pasculescu A, Choi H, Gingras AC. 2014. A web-tool for visualizing quantitative protein-protein interaction data. Proteomics 15:1432–1436. doi: 10.1002/pmic.201400429. [DOI] [PubMed] [Google Scholar]
- 84.Papadakis AI, Sun C, Knijnenburg TA, Xue Y, Grernrum W, Holzel M, Nijkamp W, Wessels LF, Beijersbergen RL, Bernards R, Huang S. 2015. SMARCE1 suppresses EGFR expression and controls responses to MET and ALK inhibitors in lung cancer. Cell Res 25:445–458. doi: 10.1038/cr.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Langton PF, Colombani J, Chan EH, Wepf A, Gstaiger M, Tapon N. 2009. The dASPP-dRASSF8 complex regulates cell-cell adhesion during Drosophila retinal morphogenesis. Curr Biol 19:1969–1978. doi: 10.1016/j.cub.2009.10.027. [DOI] [PubMed] [Google Scholar]
- 86.Fukumoto Y, Obata Y, Ishibashi K, Tamura N, Kikuchi I, Aoyama K, Hattori Y, Tsuda K, Nakayama Y, Yamaguchi N. 2010. Cost-effective gene transfection by DNA compaction at pH 4.0 using acidified, long shelf-life polyethylenimine. Cytotechnology 62:73–82. doi: 10.1007/s10616-010-9259-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mayer RE, Hendrix P, Cron P, Matthies R, Stone SR, Goris J, Merlevede W, Hofsteenge J, Hemmings BA. 1991. Structure of the 55-kDa regulatory subunit of protein phosphatase 2A: evidence for a neuronal-specific isoform. Biochemistry 30:3589–3597. doi: 10.1021/bi00229a001. [DOI] [PubMed] [Google Scholar]
- 88.Zolnierowicz S, Csortos C, Bondor J, Verin A, Mumby MC, DePaoli-Roach AA. 1994. Diversity in the regulatory B-subunits of protein phosphatase 2A: identification of a novel isoform highly expressed in brain. Biochemistry 33:11858–11867. doi: 10.1021/bi00205a023. [DOI] [PubMed] [Google Scholar]
- 89.Hardy S, Uetani N, Wong N, Kostantin E, Labbe DP, Begin LR, Mes-Masson A, Miranda-Saavedra D, Tremblay ML. 2014. The protein tyrosine phosphatase PRL-2 interacts with the magnesium transporter CNNM3 to promote oncogenesis. Oncogene 34:986–995. doi: 10.1038/onc.2014.33. [DOI] [PubMed] [Google Scholar]
- 90.Stuiver M, Lainez S, Will C, Terryn S, Gunzel D, Debaix H, Sommer K, Kopplin K, Thumfart J, Kampik NB, Querfeld U, Willnow TE, Nemec V, Wagner CA, Hoenderop JG, Devuyst O, Knoers NV, Bindels RJ, Meij IC, Muller D. 2011. CNNM2, encoding a basolateral protein required for renal Mg2+ handling, is mutated in dominant hypomagnesemia. Am J Hum Genet 88:333–343. doi: 10.1016/j.ajhg.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zou Q, Jin J, Hu H, Li HS, Romano S, Xiao Y, Nakaya M, Zhou X, Cheng X, Yang P, Lozano G, Zhu C, Watowich SS, Ullrich SE, Sun SC. 2014. USP15 stabilizes MDM2 to mediate cancer-cell survival and inhibit antitumor T cell responses. Nat Immunol 15:562–570. doi: 10.1038/ni.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Archibald A, Mihai C, Macara IG, McCaffrey L. 2014. Oncogenic suppression of apoptosis uncovers a Rac1/JNK proliferation pathway activated by loss of Par3. Oncogene 34:3199–3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Iden S, van Riel WE, Schafer R, Song JY, Hirose T, Ohno S, Collard JG. 2012. Tumor type-dependent function of the par3 polarity protein in skin tumorigenesis. Cancer Cell 22:389–403. doi: 10.1016/j.ccr.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 94.Wang W, Huang J, Wang X, Yuan J, Li X, Feng L, Park JI, Chen J. 2012. PTPN14 is required for the density-dependent control of YAP1. Genes Dev 26:1959–1971. doi: 10.1101/gad.192955.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, Zheng P, Ye K, Chinnaiyan A, Halder G, Lai ZC, Guan KL. 2007. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev 21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xiao L, Chen Y, Ji M, Dong J. 2011. KIBRA regulates Hippo signaling activity via interactions with large tumor suppressor kinases. J Biol Chem 286:7788–7796. doi: 10.1074/jbc.M110.173468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, Elvassore N, Piccolo S. 2011. Role of YAP/TAZ in mechanotransduction. Nature 474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 98.Hwang JH, Pores Fernando AT, Faure N, Andrabi S, Hahn WC, Schaffhausen BS, Roberts TM. 2014. Polyomavirus small T antigen interacts with yes-associated protein to regulate cell survival and differentiation. J Virol 88:12055–12064. doi: 10.1128/JVI.01399-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nguyen HT, Hong X, Tan S, Chen Q, Chan L, Fivaz M, Cohen SM, Voorhoeve PM. 2014. Viral small T oncoproteins transform cells by alleviating hippo-pathway-mediated inhibition of the YAP proto-oncogene. Cell Rep 8:707–713. doi: 10.1016/j.celrep.2014.06.062. [DOI] [PubMed] [Google Scholar]
- 100.Ribeiro PS, Josue F, Wepf A, Wehr MC, Rinner O, Kelly G, Tapon N, Gstaiger M. 2010. Combined functional genomic and proteomic approaches identify a PP2A complex as a negative regulator of Hippo signaling. Mol Cell 39:521–534. doi: 10.1016/j.molcel.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 101.Zhao B, Li L, Wang L, Wang CY, Yu J, Guan KL. 2012. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev 26:54–68. doi: 10.1101/gad.173435.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gaspar P, Tapon N. 2014. Sensing the local environment: actin architecture and Hippo signalling. Curr Opin Cell Biol 31C:74–83. [DOI] [PubMed] [Google Scholar]