Abstract
Streptococcus suis is an important pathogen of pigs and may cause serious disease in humans. Serotyping is one of the important diagnostic tools and is used for the epidemiological study of S. suis. Nontypeable S. suis strains have been reported in many studies; however, the capsular polysaccharide (CPS) synthesis cps loci of nontypeable strains have not been analyzed. In this study, we investigated the genetic characteristics of cps loci in 78 nontypeable strains isolated from healthy pigs. Eight novel cps loci (NCLs) were found, and all of them were located between the orfZ-orfX region and the glf gene. All NCLs possess the wzy and wzx genes, strongly suggesting that the CPSs of these NCLs were synthesized using the Wzx/Wzy-dependent pathway. The cps genes found in the 78 isolates were assigned to 96 homology groups (HGs), 55 of which were NCL specific. The encapsulation of the 78 isolates was also examined using transmission electron microscopy. Fifty-three isolates were found to have a capsule, and these were of varied thicknesses. Our data enhance our understanding of the cps gene cluster diversity of nontypeable S. suis strains and provide insight into the evolution of the S. suis capsular genes.
INTRODUCTION
Streptococcus suis is an important pathogen of pigs (1) and may cause serious disease in humans (2–4). Serotyping is one of the important tools for diagnosis and is used in epidemiological studies of S. suis. A total of 35 S. suis serotypes (serotypes 1 through 34 and 1/2) are known on the basis of the antigenic differences in their capsular polysaccharides (CPSs) (5–8) and the coagglutination test. The S. suis cps gene clusters of the 35 serotypes have been sequenced, and the cps gene clusters were shown to be diverse among different serotypes (9). The predicted products of the cps genes found in the 35 serotypes were assigned to 291 homology groups (HGs). The precise function of many cps genes is still unknown.
The synthesis and export of bacterial polysaccharides are mediated by three pathways known as the Wzx/Wzy-dependent pathway, the synthase-dependent pathway, and the ABC transporter-dependent pathway. The Wzx/Wzy-dependent pathway is most commonly used in Streptococcus pneumoniae capsular biosynthesis (10). Although the precise function of most of the cps genes of S. suis is still unknown, the CPSs of all serotypes of S. suis are also thought to be synthesized by the Wzx/Wzy-dependent pathway. This pathway involves the synthesis of the polysaccharide repeat units, which are initially built on the inner face of the cytoplasmic membrane; transport of the repeat units to the outer face of the membrane by Wzx flippase; and then polymerization of the repeat units by Wzy polymerase. Wzy-dependent polymers usually contain various sugars and glycosidic linkages. The specificity of the Wzy polymerase determines the linkage that it catalyzes between sugars on the growing chain and the next repeat unit. Therefore, the wzy gene is serotype specific. Hence, the serotype-specific wzy gene is an ideal target to discriminate the cps loci for molecular serotyping. Recently, two multiplex PCR (mPCR) assays based on the wzy gene have been developed to identify S. suis serotypes (11, 12). Both methods greatly facilitate the serotype discrimination of S. suis isolates.
Clinically healthy pigs can carry S. suis in their nasal cavities, tonsils, and upper respiratory tract (13, 14). Nontypeable S. suis strains from healthy pigs have been reported in many studies (12, 13, 15–18). The nontypeability of some of these strains is likely due to nonfunctional cps loci which produce an acapsular phenotype. However, nontypeable strains may carry at their cps loci novel cps genes expressing a so far unidentified capsule type. In this study, we obtained the complete sequences of the cps loci from 78 nontypeable isolates through Illumina sequencing and primer walking. Eight novel cps loci (NCLs), designated NCL1 to NCL8, were identified on the basis of the specific polysaccharide polymerase gene wzy. The capsule phenotype of these 78 isolates was also investigated by transmission electron microscopy.
MATERIALS AND METHODS
Bacterial strains and chromosomal DNA preparation.
Of 179 S. suis field isolates recovered from the tonsils of healthy pigs in 2011 and 2012 in a previous study, 96 isolates were unable to be assigned to a known serotype (19). Seventy-eight of these nonserotypeable isolates were investigated in this study (Table 1). Chromosomal DNA was prepared from all isolates using the method described previously (11). Strains were tested as nonserotypeable by both coagglutination and PCR testing (11, 12). Serum for the seroagglutination test was purchased from the Statens Serum Institut (Copenhagen, Denmark).
TABLE 1.
Isolates used in the study
| Isolate | cps locus type | Seroagglutination test result | Capsule thickness (nm) | Yr of isolation | Geographic location | MCG groupa | MLSTb | cps GenBank accession no. |
|---|---|---|---|---|---|---|---|---|
| YS7 | NCL1-6 | Polyagglutinating | No capsule | 2011 | Beijing | 6 | ST305 | KM972281 |
| YS8 | NCL1-6 | Polyagglutinating | No capsule | 2011 | Beijing | 6 | ST305 | KM972290 |
| YS21 | NCL1-3 | Nonagglutinating | 40–60 | 2011 | Sichuan Province | 6 | ST310 | KM972259 |
| YS22 | NCL1-3 | Nonagglutinating | 40–60 | 2011 | Sichuan Province | 6 | ST310 | KM972260 |
| YS23 | NCL2-2 | Polyagglutinating | 30–50 | 2011 | Sichuan Province | 6 | ST328 | KM972261 |
| YS27 | NCL3-1 | Polyagglutinating | 20–40 | 2011 | Beijing | 6 | ST312 | KM972262 |
| YS33 | NCL4 | Autoagglutinating | No capsule | 2011 | Beijing | 7-3 | ST423 | KM972263 |
| YS34 | NCL4 | Polyagglutinating | No capsule | 2011 | Beijing | 7-3 | ST314 | KM972264 |
| YS35 | NCL5 | Polyagglutinating | 40–60 | 2011 | Beijing | 7-2 | ST315 | KM972265 |
| YS37 | NCL4 | Autoagglutinating | 20–40 | 2011 | Beijing | 7-3 | ST339 | KM972266 |
| YS38 | NCL5 | Polyagglutinating | 30–40 | 2011 | Beijing | 7-2 | ST315 | KM972267 |
| YS41 | NCL1-1 | Polyagglutinating | No capsule | 2011 | Beijing | N | ST340 | KM972268 |
| YS42 | NCL1-4 | Nonagglutinating | No capsule | 2011 | Jiangxi Province | 6 | ST341 | KM972269 |
| YS43 | NCL6 | Nonagglutinating | No capsule | 2011 | Jiangxi Province | 7-3 | ST317 | KM972270 |
| YS46 | NCL7-2 | Polyagglutinating | No capsule | 2011 | Sichuan Province | 6 | NT | KM972271 |
| YS49 | NCL1-3 | Nonagglutinating | No capsule | 2011 | Sichuan Province | 6 | ST329 | KM972272 |
| YS56 | NCL1-1 | Polyagglutinating | 30–50 | 2011 | Sichuan Province | 7-2 | ST331 | KM972273 |
| YS57 | NCL7-1 | Polyagglutinating | 20–40 | 2011 | Sichuan Province | 6 | ST320 | KM972274 |
| YS68 | NCL1-1 | Polyagglutinating | 40–60 | 2011 | Beijing | 6 | ST426 | KM972275 |
| YS71 | NCL1-1 | Polyagglutinating | 40–60 | 2011 | Jiangsu Province | 6 | ST347 | KM972276 |
| YS72 | NCL2-2 | Nonagglutinating | 40–60 | 2011 | Jiangsu Province | 6 | ST332 | KM972277 |
| YS73 | NCL4 | Autoagglutinating | 70–90 | 2011 | Beijing | 7-3 | ST348 | KM972278 |
| YS74 | NCL3-1 | Polyagglutinating | 20–30 | 2011 | Beijing | 6 | ST333 | KM972279 |
| YS75 | NCL3-1 | Polyagglutinating | No capsule | 2011 | Beijing | 6 | ST349 | KM972280 |
| YS80 | NCL1-1 | Polyagglutinating | 40–60 | 2012 | Beijing | 6 | ST429 | KM972282 |
| YS81 | NCL1-2 | Polyagglutinating | 30–50 | 2012 | Jiangsu Province | 6 | ST398 | KM972283 |
| YS82 | NCL1-2 | Polyagglutinating | 30–50 | 2012 | Jiangsu Province | 6 | ST430 | KM972284 |
| YS83 | NCL1-2 | Polyagglutinating | 30–50 | 2012 | Jiangsu Province | 6 | ST430 | KM972285 |
| YS84 | NCL1-2 | Polyagglutinating | 30–50 | 2012 | Jiangsu Province | 6 | ST398 | KM972286 |
| YS85 | NCL8-1 | Nonagglutinating | 50–70 | 2012 | Jiangsu Province | 6 | ST399 | KM972287 |
| YS86 | NCL4 | Autoagglutinating | 20–30 | 2012 | Jiangsu Province | 7-3 | ST400 | KM972288 |
| YS87 | NCL8-1 | Polyagglutinating | No capsule | 2012 | Jiangsu Province | 6 | ST431 | KM972289 |
| YS91 | NCL3-1 | Polyagglutinating | 20–30 | 2012 | Jiangsu Province | 6 | ST451 | KM972291 |
| YS92 | NCL3-1 | Polyagglutinating | 20–30 | 2012 | Jiangsu Province | 6 | ST452 | KM972292 |
| YS93 | NCL3-1 | Polyagglutinating | No capsule | 2012 | Jiangsu Province | 6 | ST452 | KM972293 |
| YS94 | NCL1-1 | Polyagglutinating | 20–30 | 2012 | Jiangsu Province | 6 | ST402 | KM972294 |
| YS95 | NCL2-1 | Polyagglutinating | 20–40 | 2012 | Jiangsu Province | 6 | NT | KM972295 |
| YS96 | NCL2-1 | Nonagglutinating | 20–30 | 2012 | Jiangsu Province | 6 | ST432 | KM972296 |
| YS97 | NCL2-1 | Nonagglutinating | 20–30 | 2012 | Jiangsu Province | 6 | ST432 | KM972297 |
| YS98 | NCL2-1 | Nonagglutinating | 20–30 | 2012 | Jiangsu Province | 6 | ST432 | KM972298 |
| YS99 | NCL2-1 | Nonagglutinating | 20–30 | 2012 | Jiangsu Province | 6 | ST432 | KM972299 |
| YS100 | NCL2-1 | Nonagglutinating | 20–30 | 2012 | Jiangsu Province | 6 | ST432 | KM972222 |
| YS101 | NCL1-7 | Polyagglutinating | No capsule | 2012 | Jiangsu Province | 6 | ST433 | KM972223 |
| YS102 | NCL1-7 | Polyagglutinating | No capsule | 2012 | Jiangsu Province | 6 | ST433 | KM972224 |
| YS103 | NCL2-2 | Polyagglutinating | No capsule | 2012 | Jiangsu Province | 6 | ST403 | KM972225 |
| YS104 | NCL8-1 | Polyagglutinating | No capsule | 2012 | Jiangsu Province | 6 | ST434 | KM972226 |
| YS107 | NCL8-2 | Polyagglutinating | 20–40 | 2012 | Beijing | 6 | ST436 | KM972227 |
| YS109 | NCL8-1 | Polyagglutinating | 20–40 | 2012 | Beijing | 6 | ST453 | KM972228 |
| YS110 | NCL8-1 | Polyagglutinating | 20–30 | 2012 | Beijing | 6 | ST453 | KM972229 |
| YS111 | NCL8-1 | Polyagglutinating | 20–30 | 2012 | Beijing | 6 | ST453 | KM972230 |
| YS113 | NCL3-1 | Polyagglutinating | 35–55 | 2012 | Beijing | 6 | ST437 | KM972231 |
| YS114 | NCL1-2 | Polyagglutinating | 40–60 | 2012 | Beijing | 6 | ST455 | KM972232 |
| YS117 | NCL8-1 | Polyagglutinating | No capsule | 2012 | Beijing | 6 | ST404 | KM972233 |
| YS121 | NCL7-1 | Polyagglutinating | 20–40 | 2012 | Beijing | 6 | ST407 | KM972234 |
| YS122 | NCL8-1 | Polyagglutinating | No capsule | 2012 | Beijing | 6 | ST408 | KM972235 |
| YS123 | NCL8-1 | Nonagglutinating | 30–50 | 2012 | Beijing | 6 | ST453 | KM972236 |
| YS125 | NCL8-1 | Nonagglutinating | 30–50 | 2012 | Beijing | 6 | ST453 | KM972237 |
| YS129 | NCL1-2 | Polyagglutinating | No capsule | 2012 | Beijing | 6 | ST439 | KM972238 |
| YS131 | NCL8-1 | Polyagglutinating | 30–50 | 2012 | Beijing | 6 | ST453 | KM972239 |
| YS132 | NCL8-1 | Nonagglutinating | 20–30 | 2012 | Beijing | 6 | NT | KM972240 |
| YS133 | NCL4 | Polyagglutinating | 30–50 | 2012 | Beijing | 7-3 | ST409 | KM972241 |
| YS140 | NCL7-1 | Polyagglutinating | 20–40 | 2012 | Beijing | 6 | ST442 | KM972242 |
| YS142 | NCL1-2 | Polyagglutinating | 50–70 | 2012 | Beijing | 6 | ST411 | KM972243 |
| YS145 | NCL8-1 | Polyagglutinating | 15–25 | 2012 | Beijing | 6 | ST443 | KM972244 |
| YS146 | NCL8-3 | Polyagglutinating | 70–90 | 2012 | Jiangsu Province | 6 | ST444 | KM972245 |
| YS147 | NCL3-1 | Polyagglutinating | No capsule | 2012 | Sichuan Province | 6 | ST462 | KM972246 |
| YS148 | NCL2-3 | Nonagglutinating | 20–30 | 2012 | Sichuan Province | 7-2 | ST463 | KM972247 |
| YS149 | NCL1-1 | Nonagglutinating | 50–70 | 2012 | Sichuan Province | 6 | ST464 | KM972248 |
| YS150 | NCL3-1 | Polyagglutinating | 20–40 | 2012 | Sichuan Province | 6 | ST465 | KM972249 |
| YS155 | NCL2-1 | Polyagglutinating | 20–40 | 2012 | Guizhou Province | 6 | ST446 | KM972250 |
| YS159 | NCL8-1 | Polyagglutinating | No capsule | 2012 | Guizhou Province | 6 | ST448 | KM972251 |
| YS160 | NCL1-4 | Nonagglutinating | No capsule | 2012 | Guizhou Province | 6 | ST467 | KM972252 |
| YS166 | NCL3-1 | Nonagglutinating | No capsule | 2012 | Beijing | 6 | ST469 | KM972253 |
| YS167 | NCL1-5 | Nonagglutinating | No capsule | 2012 | Beijing | 6 | ST470 | KM972254 |
| YS168 | NCL1-5 | Nonagglutinating | No capsule | 2012 | Beijing | 6 | ST470 | KM972255 |
| YS170 | NCL3-1 | Nonagglutinating | 20–30 | 2012 | Beijing | 6 | ST469 | KM972256 |
| YS171 | NCL3-2 | Polyagglutinating | 20–30 | 2012 | Beijing | 6 | ST472 | KM972257 |
| YS173 | NCL8-1 | Polyagglutinating | 40–60 | 2012 | Beijing | 6 | NT | KM972258 |
N, nongroupable.
NT, nontypeable.
Species identification and molecular typing of nontypeable strains.
We used 16S rRNA primers (20) to amplify a nearly complete 16S rRNA gene from the 78 isolates. The recN and gdh genes were also analyzed for taxonomic assignment, as was done previously (21–23). The API 20 Strep biochemical identification system (bioMérieux, Hazelwood, MO) was also used to identify the strains. Multilocus sequence typing (MLST) and minimum core genome sequence (MCG) typing were performed according to the methods described previously (19, 24).
Sequencing of cps loci.
Thirty-six isolates were sequenced by Illumina sequencing: (i) 13 isolates were sequenced in a previous study BioProject accession numbers (PRJNA171455, PRJNA171456, PRJNA171458, PRJNA171460, PRJNA171461, PRJNA171464, PRJNA171466, PRJNA171467, PRJNA171472, PRJNA171473, PRJNA171480, PRJNA171481, and PRJNA171483) (25), and (ii) 23 additional bacterial strains were sequenced by Illumina sequencing in this study. Each cps locus sequence was extracted from the draft genome sequence, and the open reading frames (ORFs) were identified and annotated on the basis of the methods previously reported (11). The TMHMM (v2.0) analysis program (http://www.cbs.dtu.dk/services/TMHMM/) was used to identify putative wzy genes. The distribution of novel wzy genes in an additional 42 isolates was investigated by multiplex PCR (26) for NCL1 to NCL7 and a single PCR for NCL8 (upstream primer, 5′-AAAATTTTCACTTCACCTCGAC; downstream primer, 5′-AATCTTCCAATCAATGCTACGA; annealing temperature, 58°C; product size, 390 bp). The isolates harboring the same wzy gene were clustered into the same NCL. Finally, the cps clusters of 42 isolates were sequenced by directed PCR and primer walking on the basis of the known sequence of each NCL.
Bioinformatics analysis.
The BLASTN and PSI-BLAST software programs (http://blast.ncbi.nlm.nih.gov/Blast.cgi) were used to search several databases, including GenBank (www.ncbi.nlm.nih.gov/GenBank), and the genome sequences of 85 strains sequenced previously (25). Loci were annotated with the Artemis program. The Clusters of Orthologous Groups (COG) and Pfam protein motif databases were used to search for conserved protein domains. cps genes were named according to the nomenclature for the S. suis serotype 2 cps locus (27). The genes from a locus are numbered by a letter from A to Z, in order. We defined the 5′ and 3′ sides of the cps clusters and clustered the proteins of the cps genes into HGs by the methods described in a previous study (9). The genes having a global match region at <50% of the amino acid sequence and with an identity of <50% were identified to be novel HGs compared to the HGs (HG1 to HG291) of the 35 reference serotype strains. Novel HGs were assigned numerically from HG292 onwards, continuing on from the previous HG assignment. Novel HGs present only in all isolates of a single NCL were identified as NCL-specific HGs. The Artemis comparison tool (ACT) (28) was used to visualize the data.
Phylogenetic analysis.
The cpsA to cpsD sequences from 35 serotype reference strains and 78 nontypeable isolates were used to generate a phylogenetic tree. We used the Recombination Detection Program (RDP; v3.0) to remove the recombinational single nucleotide polymorphisms in the cpsA to cpsD sequences. An alignment of the cpsA to cpsD sequences was generated using the CLUSTAL W program (29). A phylogenetic tree was constructed using the neighbor-joining method, and a distance measure was obtained using the Tamura and Nei model implemented in the program MEGA (v5) (30).
Identification of capsule.
The presence or absence of a capsule was confirmed using electron microscopy (31, 32). Briefly, bacteria were grown overnight in Todd-Hewitt broth medium, harvested by centrifugation, and washed once in phosphate-buffered saline. The cells were fixed for 2 h at room temperature in 0.1 M cacodylate buffer (pH 7) containing 5% glutaraldehyde and 0.15% ruthenium red; then, polycationic ferritin (1 mg/ml) was applied for 30 min at 20°C. The cells were then harvested using centrifugation and washed 3 times in 0.1 M cacodylate buffer. Then, the bacterial cells were immobilized in 4% agar, washed five times in cacodylate buffer, and postfixed with 2% osmium tetroxide for 2 h. The samples were dehydrated in a graded series of acetone concentrations (30%, 50%, 70%, 90%, 100%). Samples were then dehydrated in propylene oxide for 10 min and embedded in Spurr low-viscosity resin. Thin sections were cut and stained with uranyl acetate and lead citrate. The sections were examined with a transmission electron microscope (TH7700; Hitachi) at an accelerating voltage of 80 kV.
The capsule thickness between the inner edge and the external edge of the capsular layer was measured (33). Each value is based on 25 to 30 measurements per experiment. Each experiment was done twice independently.
Nucleotide sequence accession numbers.
The cps sequences of the 78 isolates were deposited in the GenBank database under accession numbers KM972222 to KM972299.
RESULTS
Species identification and molecular typing of the nontypeable isolates.
All 78 nontypeable isolates were unambiguously identified to be S. suis by biochemical tests and comparative sequence analyses of the 16S rRNA, recN, and gdh genes (data not shown).
We further typed the 78 isolates by MLST, and 56 sequence types (STs) were found. The STs of four isolates could not be obtained due to PCR failure of one of the seven housekeeping genes (Table 1) (19). Using genome sequences, they could also be assigned to the MCG groups previously defined (19, 25). The majority of the isolates were from the most ancestral MCG group, group 6 (84.6%, 66/78), while a small proportion of the isolates belonged to group 7 (14.1%, 11/78 isolates). The MCG of one isolate was nongroupable (Table 1).
Molecular grouping of the 78 isolates on the basis of the wzy gene.
The wzy gene has been used as a marker for molecular serotyping of S. suis (11, 12, 26, 34). When the 78 wzy sequences were compared with reference wzy sequences, they were clustered away from the reference sequences as 8 novel clusters. On the basis of these eight novel wzy genes, the cps gene clusters from these nontypeable isolates could be classified into eight different NCLs (Table 1). The number of isolates in each NCL varied: NCL1 had 25 isolates, NCL2 had 11 isolates, NCL3 had 12 isolates, NCL4 had 6 isolates, NCL5 had 2 isolates, NCL6 had 1 isolate, NCL7 had 4 isolates, and NCL8 had 17 isolates.
General features of the NCLs.
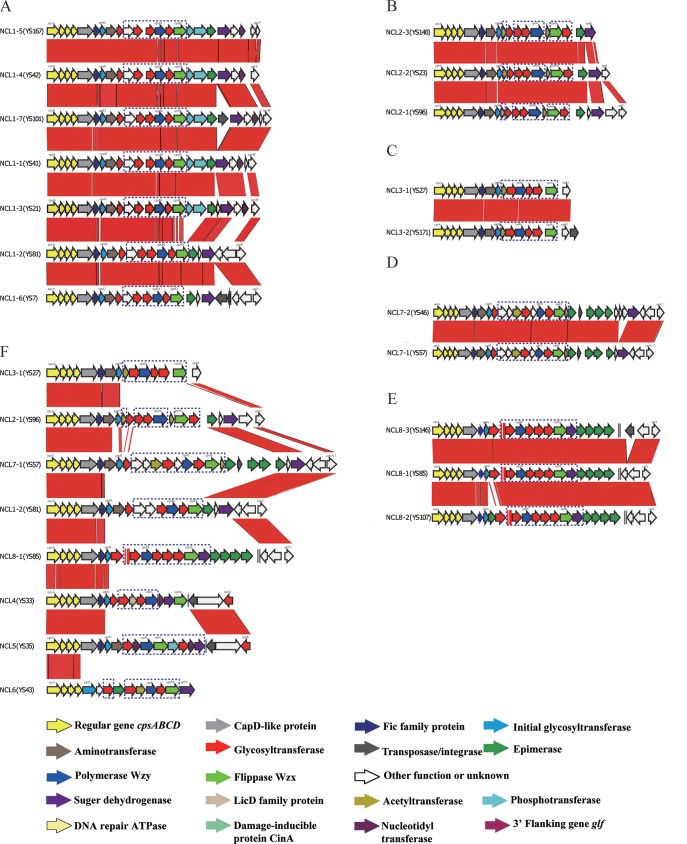
Previous studies showed that not all S. suis cps gene clusters were located at the same chromosomal location, and 5 insertion sites were found (9). We found that all cps loci in these nontypeable strains were located between the orfZ-orfX region and the glf (UDP-galactopyranose mutase) gene, which was previously found to be the cps gene cluster insertion site for reference serotypes 9, 13, 21, 24, 29, 31, and 33 and was defined as location pattern I-b.
The sizes of the NCLs ranged from 15.3 kb to 32.8 kb, and the G+C content of the NCLs varied from 33.4 to 36.8%. The 5′ region of all eight NCLs was conserved, whereas the 3′ region was variable, with the 3′ regions of NCL1, NCL7, and NCL8 sharing high similarity and the 3′ region of NCL4 being nearly identical to that of NCL5 but differing from the 3′ regions of NCL1, NCL7, and NCL8. The central regions of the eight NCLs were highly variable (Fig. 1F). The polysaccharide polymerase (wzy) and flippase (wzx) genes were always present downstream together with various sets of genes coding for glycosyltransferase, acetyltransferase, and modifying enzymes. Sialic acid synthesis genes were not found in any of the NCLs.
FIG 1.
Comparison of the cps loci within NCL1 (A), NCL2 (B), NCL3 (C), NCL7 (D), NCL8 (E), and 8 NCLs (F). Each colored arrow represents the gene whose predicted function is identified at the bottom. The names of the first and last genes, initial transferase genes, and the wzx and wzy genes of each NCL are appended to the corresponding arrows. NCL-specific genes are indicated by dotted blue lines. The red bars in each panel indicate regions conserved among strains. The glf gene is located on the 3′ side of each locus and is not marked.
The cpsA, cpsB, cpsC, and cpsD genes were present and located at the 5′ side of the NCLs. The nucleotide sequences of cpsA to cpsD were conserved across both NCLs and known reference cps loci. In the phylogenetic tree based on the nucleotide sequence alignment of the region from cpsA to cpsD, all our nontypeable strains were clustered together with reference strains with a cps locus of pattern I-a (reference serotypes 1, 1/2, 2, 3, 4, 5, 7, 8, 10, 11, 12, 14, 15, 17, 18, 19, 23, 25, 28, and 30) or pattern I-b (reference serotypes 9, 13, 21, 24, 29, and 31) and were distant from the strains with a cps locus of pattern I-a (reference serotypes 6 and 16), pattern I-b (serotype 33), pattern II (serotype 27), pattern III (serotypes 20, 22, and 26), and pattern IV (serotypes 32 and 34) (see Fig. S1 in the supplemental material).
Comparison of HG contents of NCLs with those of reference cps gene clusters.
A previous study classified the S. suis cps genes in the reference serotypes into HGs, with 291 HGs being named (9). Using the same criteria used previously, the predicted coding sequences in the eight NCLs could be divided into 96 HGs (see Table S1 in the supplemental material). Twenty-six of these 96 HGs were present in the cps loci of the serotype reference strains, while 70 were novel HGs not found in the serotype reference strains and were named HG292 to HG361. Fifty-five novel HGs were NCL specific. Each NCL contained 4 to 10 NCL-specific HGs (see Table S1 in the supplemental material). Eight HGs (HG312, HG313, HG314, HG315, HG329, HG332, HG354, and HG355) were variably present within an NCL. Seven HGs (HG292, HG293, HG294, HG295, HG316, HG330, and HG331) were present in at least two NCLs. Some of the specific enzymes are described below.
(i) Initial sugar transferases.
The initial glycosyl phosphotransferase is responsible for linkage of an activated glycosyl phosphate to the lipid carrier (35). Similar to the serotype reference strains, the initial sugar transferase genes were located in the 5′ region in all NCLs and were classified into 4 HGs: HG6 (NCL6), HG8 (NCL4 and NCL8), HG21 (NCL1 and NCL5), and HG295 (NCL2, NCL3, and NCL7) (see Table S1 in the supplemental material).
(ii) Glycosyltransferases.
The glycosyltransferase catalyzes the attachment of sugars to an aglycone in CPS synthesis. Except for the initial glycosyl phosphotransferase, the other glycosyltransferases in all strains tested fell into 31 HGs, and only three of the glycosyltransferases (HG45, HG81, and HG99) were present in serotype reference strains.
(iii) Other transferases.
Acetyltransferase plays an important role in CPS structure determination (36). Acetyltransferases in all tested strains fell into six HGs, only one of which (HG104) existed in a serotype reference strain. The others were NCL-specific novel HGs. Aminotransferase is identified to be an enzyme which can transfer amino groups to sugars (37). Two aminotransferases (HG22 and HG41) were present in the NCLs. Cytidylyltransferases (HG66 and HG347), nucleotidyltransferases (HG313 and HG352), a hexosyltransferase (HG357), and a LicD-family phosphotransferase (HG334) were also present in the NCLs.
(iv) Wzy polymerase and Wzx flippase.
The Wzy polymerase was quite different in each of the eight NCLs and belonged to NCL-specific HGs. The sequences of wzy were highly conserved within the same group.
Eight Wzx flippases were present in NCLs. Except for NCL4, the Wzx flippases of the other seven NCLs also belonged to NCL-specific HGs. The NCL4 Wzx flippase was homologous to the Wzx flippases of reference serotypes 8, 9, and 33 (HG43).
Structural variation within NCLs.
We found genetic structural variations in five of the eight NCLs with the insertion, deletion, or truncation of genes. No genetic heterogeneity was found within NCL4, NCL5, or NCL6, and these NCLs contained few isolates.
(i) NCL1.
Seven subtypes were found in NCL1 (Fig. 1A; see also Table S1 in the supplemental material): NCL1-1 (7 strains), NCL1-2 (7 strains), NCL1-3 (3 strains), NCL1-4 (2 strains), NCL1-5 (2 strains), NCL1-6 (2 strains), and NCL1-7 (2 strains). The subtypes varied due to the variable presence of eight HGs (HG55, HG293, HG294, HG312, HG313, HG314, HG315, and HG329) and transposase genes. Subtype NCL1-7 differed from NCL1-1 by the insertion of a transposase gene. Similarly, differences between NCL1-4 and NCL1-5 and between NCL1-2 and NCL1-6 were also due to transposase gene insertions. NCL1-7 differed from NCL1-4 by the insertion of a transposase gene and the deletion of HG329. NCL1-1 differed from NCL1-3 by the insertion of HG329 and the deletion of HG55. NCL1-2 differed from NCL1-3 by the insertion of HG312, HG313, HG314, and HG315 and the deletion of HG293 and HG294.
(ii) NCL2.
Three types of genetic organizations were found in NCL2 (Fig. 1B; Table 1): NCL2-1 (7 strains), NCL2-2 (3 strains), and NCL2-3 (1 strain). In comparison with NCL2-1, two genes (HG55 and HG332) were deleted in NCL2-2, whereas three genes (HG55, HG292, and HG332) were deleted in NCL2-3.
(iii) NCL3.
Two types of genetic organizations were found in NCL3 (Fig. 1C; Table 1): NCL3-1 (11 strains) differed from NCL3-2 (1 strain) by a transposase gene inserted in the 3′ region in NCL3-2.
(iv) NCL7.
Two types of genetic organizations were found in NCL7 (Fig. 1D, Table 1): NCL7-1 (3 strains) differed from NCL7-2 (1 strain) by two genes (HG354 and HG355) which were inserted in the 3′ region in NCL7-2.
(v) NCL8.
Three types of genetic organizations were found in NCL8 (Fig. 1E, Table 1): NCL8-1 (15 strains), NCL8-2 (1 strain), and NCL8-3 (1 strain). Compared to the sequence of NCL8-1, NCL8-2 had an extra gene (HG23) inserted in the 5′ region. A truncation by the insertion of a transposase gene was found in HG293 (cpsW) of NCL8-3.
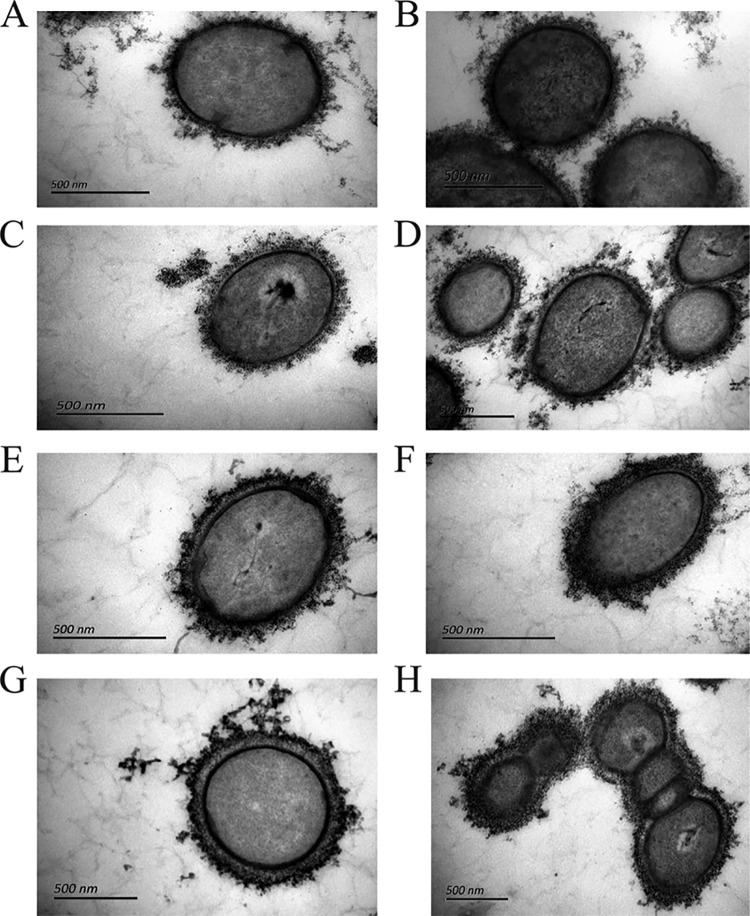
Capsule analysis by transmission electron microscopy.
The thickness of the capsule of the 78 isolates was measured by transmission electron microscopy (Table 1; representative strains are shown in Fig. 2). Capsule analysis was performed twice for each strain, and the capsule thickness was consistent between the experiments. Strain SC84 (serotype 2) was used as a control and was well encapsulated, and its capsule had a thickness of from 110 nm to 130 nm. Fifty-three nontypeable strains were most likely encapsulated with varied capsule thicknesses (Table 1). The capsule thickness also showed wide variation between strains within an NCL or NCL subtypes. The remaining 25 nontypeable strains are likely to be nonencapsulated (Table 1). The nonencapsulated strains were distributed among the different NCLs and subtypes.
FIG 2.
Visualization of the S. suis capsule of representative strains using transmission electron microscopy following ferritin stabilization. (A) YS74 (NCL3-1); (B) YS121 (NCL7-1); (C) YS35 (NCL5); (D) YS72 (NCL2-2); (E) YS133 (NCL4); (F) YS149 (NCL1-1); (G) YS146 (NCL8-3); (H) S. suis serotype 2 control strain SC84.
DISCUSSION
In this study, we analyzed the cps loci of 78 nontypeable strains isolated from healthy pigs and found eight NCLs with 70 novel HGs. The isolates that harbored identical novel cps gene clusters were from diverse geographic locations and belonged to different MLST types, suggesting that these strains have been widely circulating in the pig population. They shared highly conserved cpsA, cpsB, cpsC, and cpsD genes with known pattern I-a and pattern I-b serotype reference strains, indicating that they probably evolved from a common ancestor. The possession of wzy and wzx genes in NCLs strongly suggests that their CPSs were synthesized by the Wzx/Wzy-dependent pathway. The sequences of NCL-specific wzy genes were quite variable in different types of cps loci but were highly conserved within the same group. Consistent with previous reports, wzy is an ideal target with which to discriminate the cps loci in nontypeable S. suis strains.
By comparison with known cps loci of serotype reference strains, it was found that genes responsible for the synthesis and polymerization of the repeat unit were located in the central region of the gene cluster and were highly variable in the NCLs. The 70 novel HGs are likely to have been acquired by lateral gene transfer from other species. The source of these novel genes is unclear, but they are likely to have originated from the microbiota in the nasopharynx, which provides a large gene pool for S. suis cps loci. The exchange of these genes may be facilitated by the transposase-like regions which were found in the NCLs. Further, as cps gene reservoirs, the NCLs may play an important role in generating the cps gene cluster variation in S. suis CPS by intraspecies gene transfer. The presence of many different cps loci in S. suis suggests a continuous and ongoing evolution of cps gene clusters in this species.
Among the 55 NCL-specific novel HGs, 25 genes encoded glycosyltransferases and 9 genes encoded additional transferases. The presence of these unique transferases implies that the oligosaccharide structure repeat units that they transfer are unique. Most of the NCL-specific novel HGs showed weak similarity to the products of genes of other species. However, NCL-specific novel HGs of NCL8 (from HG305 to HG311) showed high similarity to S. pneumoniae serotype 12A cps genes (from SPC12A_0012 to SPC12A_0018). These genes are also conserved in S. pneumoniae serotypes 12B, 12F, 44, and 46. Homologs of the l-FucpNAc pathway and d-MnapNAcA pathway genes present in S. pneumoniae serotypes 12A and 12F were also found in NCL8. Thus, the oligosaccharide structure repeat unit in NCL8 may be similar to the oligosaccharide structure repeat units of S. pneumoniae serotypes 12, 44, and 46 and different from those of S. suis serotypes 2 and 14 (38, 39). A similar observation was also made for NCL6. Homologs of some S. pneumoniae serotype 9N cps genes (10) were found in NCL6 (HG357, HG359, HG360, and HG361).
It is interesting to note that encapsulated strains predominantly distributed in NCL1-1, NCL1-2, NCL2-1, NCL3, NCL5, NCL7-1, and NCL8 in this study. Pan et al. also reported on the isolation of a novel S. suis serotype from pigs with meningitis (34). The cps locus of a novel variant serotype was similar to NCL1 on the basis of sequence data, whereas they shared wzx (HG301) and six other novel HGs (HG292, HG300, HG312, HG313, HG314, and HG315). We also found strains that harbor NCL1, NCL3, and NCL7 from the lungs of diseased pigs (our unpublished data). CPS plays important roles in S. suis colonization of the host, resistance to phagocytosis, and persistence in the blood (40–42). Therefore, further characterization of strains isolated from diseased pigs is needed to determine the importance of these strains harboring NCL1, NCL2, NCL3, NCL5, NCL7, and NCL8 as disease-causing serotypes.
A total of 25 isolates were nonencapsulated. These strains were mainly distributed in two NCLs (NCL4 and NCL6) and seven subtypes (NCL1-4 to NCL1-7, NCL3-1, NCL7-2, and NCL8-1). Since these strains, as well as capsulated S. suis, can colonize pig nasopharynges, it is possible that some of these strains may produce a thin capsule that was present at levels below the detection limit of the method that we used or the capsule is produced in vivo. In this study, we used polycationic ferritin as a substitute for homologous antiserum to stabilize the capsule during the dehydration steps (33, 43, 44). It is possible that polycationic ferritin incompletely stabilized the capsular material, leading to the collapse of the thin capsule during the dehydration process. It is also possible that some of these strains were defective in CPS production because of a defective cps gene cluster. HG34 in strains YS160, YS167, and YS168 was truncated by a nonsense mutation. The glycosyltransferase genes, HG297 in strain YS160 and HG300 in YS103, were disrupted by a single base deletion, resulting in a frameshift mutation. The gene HG292 in YS167 and YS168 was disrupted by the insertion of a transposase gene. These truncation mutations, along with the other nonsynonymous changes, may account for the defective cps gene cluster. In addition, it is known that certain genes outside the cps locus are involved in the modification or transcriptional regulation of capsular polysaccharides in other species (45), and thus, the lack of production of a capsule may not be due to a defect in the cps gene cluster.
In conclusion, we determined the genetic characteristics of the cps loci from nontypeable S. suis isolates, and the novel genes identified facilitated their discrimination. Our data also contribute to the understanding of the genetic diversity of the cps loci in nontypeable S. suis isolates and provide insight into the evolution of capsular genes in S. suis.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants (2013ZX10004221 and 2013ZX10004216-001-002) from the Ministry of Science and Technology, People’s Republic of China, and grants (81290340, 81290345, and 81261120559) from the National Natural Science Foundation of China.
We thank Wen Zhang for technical assistance.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00315-15.
REFERENCES
- 1.Gottschalk M, Xu J, Calzas C, Segura M. 2010. Streptococcus suis: a new emerging or an old neglected zoonotic pathogen? Future Microbiol 5:371–391. doi: 10.2217/fmb.10.2. [DOI] [PubMed] [Google Scholar]
- 2.Gottschalk M, Segura M, Xu J. 2007. Streptococcus suis infections in humans: the Chinese experience and the situation in North America. Anim Health Res Rev 8:29–45. doi: 10.1017/S1466252307001247. [DOI] [PubMed] [Google Scholar]
- 3.Ye C, Zhu X, Jing H, Du H, Segura M, Zheng H, Kan B, Wang L, Bai X, Zhou Y, Cui Z, Zhang S, Jin D, Sun N, Luo X, Zhang J, Gong Z, Wang X, Wang L, Sun H, Li Z, Sun Q, Liu H, Dong B, Ke C, Yuan H, Wang H, Tian K, Wang Y, Gottschalk M, Xu J. 2006. Streptococcus suis sequence type 7 outbreak, Sichuan, China. Emerg Infect Dis 12:1203–1208. doi: 10.3201/eid1708.060232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu H, Jing H, Chen Z, Zheng H, Zhu X, Wang H, Wang S, Liu L, Zu R, Luo L, Xiang N, Liu H, Liu X, Shu Y, Lee SS, Chuang SK, Wang Y, Xu J, Yang W, Streptococcus suis Study Groups. 2006. Human Streptococcus suis outbreak, Sichuan, China. Emerg Infect Dis 12:914–920. doi: 10.3201/eid1206.051194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gottschalk M, Higgins R, Jacques M, Beaudoin M, Henrichsen J. 1991. Characterization of six new capsular types (23 through 28) of Streptococcus suis. J Clin Microbiol 29:2590–2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gottschalk M, Higgins R, Jacques M, Mittal KR, Henrichsen J. 1989. Description of 14 new capsular types of Streptococcus suis. J Clin Microbiol 27:2633–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Higgins R, Gottschalk M, Boudreau M, Lebrun A, Henrichsen J. 1995. Description of six new capsular types (29 to 34) of Streptococcus suis. J Vet Diagn Invest 7:405–406. doi: 10.1177/104063879500700322. [DOI] [PubMed] [Google Scholar]
- 8.Perch B, Pedersen KB, Henrichsen J. 1983. Serology of capsulated streptococci pathogenic for pigs: six new serotypes of Streptococcus suis. J Clin Microbiol 17:993–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okura M, Takamatsu D, Maruyama F, Nozawa T, Nakagawa I, Osaki M, Sekizaki T, Gottschalk M, Kumagai Y, Hamada S. 2013. Genetic analysis of capsular polysaccharide synthesis gene clusters from all serotypes of Streptococcus suis: potential mechanisms for generation of capsular variation. Appl Environ Microbiol 79:2796–2806. doi: 10.1128/AEM.03742-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bentley SD, Aanensen DM, Mavroidi A, Saunders D, Rabbinowitsch E, Collins M, Donohoe K, Harris D, Murphy L, Quail MA, Samuel G, Skovsted IC, Kaltoft MS, Barrell B, Reeves PR, Parkhill J, Spratt BG. 2006. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet 2:e31. doi: 10.1371/journal.pgen.0020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Z, Zheng H, Gottschalk M, Bai X, Lan R, Ji S, Liu H, Xu J. 2013. Development of multiplex PCR assays for the identification of the 33 serotypes of Streptococcus suis. PLoS One 8:e72070. doi: 10.1371/journal.pone.0072070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okura M, Lachance C, Osaki M, Sekizaki T, Maruyama F, Nozawa T, Nakagawa I, Hamada S, Rossignol C, Gottschalk M, Takamatsu D. 2014. Development of a two-step multiplex PCR assay for typing of capsular polysaccharide synthesis gene clusters of Streptococcus suis. J Clin Microbiol 52:1714–1719. doi: 10.1128/JCM.03411-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marois C, Le Devendec L, Gottschalk M, Kobisch M. 2007. Detection and molecular typing of Streptococcus suis in tonsils from live pigs in France. Can J Vet Res 71:14–22. [PMC free article] [PubMed] [Google Scholar]
- 14.Mwaniki CG, Robertson ID, Trott DJ, Atyeo RF, Lee BJ, Hampson DJ. 1994. Clonal analysis and virulence of Australian isolates of Streptococcus suis type 2. Epidemiol Infect 113:321–334. doi: 10.1017/S095026880005175X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottschalk M, Lacouture S, Bonifait L, Roy D, Fittipaldi N, Grenier D. 2013. Characterization of Streptococcus suis isolates recovered between 2008 and 2011 from diseased pigs in Quebec, Canada. Vet Microbiol 162:819–825. doi: 10.1016/j.vetmic.2012.10.028. [DOI] [PubMed] [Google Scholar]
- 16.Han DU, Choi C, Ham HJ, Jung JH, Cho WS, Kim J, Higgins R, Chae C. 2001. Prevalence, capsular type and antimicrobial susceptibility of Streptococcus suis isolated from slaughter pigs in Korea. Can J Vet Res 65:151–155. [PMC free article] [PubMed] [Google Scholar]
- 17.Sanchez del Rey V, Fernandez-Garayzabal JF, Briones V, Iriso A, Dominguez L, Gottschalk M, Vela AI. 2013. Genetic analysis of Streptococcus suis isolates from wild rabbits. Vet Microbiol 165:483–486. doi: 10.1016/j.vetmic.2013.04.025. [DOI] [PubMed] [Google Scholar]
- 18.Wang S, Liu P, Li C, Tan Y, Cai X, Zhou D, Jiang Y. 2012. Isolation and characterization of 89K pathogenicity island-positive ST-7 strains of Streptococcus suis serotype 2 from healthy pigs, northeast China. Sci World J 2012:302386. doi: 10.1100/2012/302386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng H, Ji S, Lan R, Liu Z, Bai X, Zhang W, Gottschalk M, Xu J. 2014. Population analysis of Streptococcus suis isolates from slaughtered swine by use of minimum core genome sequence typing. J Clin Microbiol 52:3568–3572. doi: 10.1128/JCM.00536-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baker GC, Smith JJ, Cowan DA. 2003. Review and re-analysis of domain-specific 16S primers. J Microbiol Methods 55:541–555. doi: 10.1016/j.mimet.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 21.Okwumabua O, O'Connor M, Shull E. 2003. A polymerase chain reaction (PCR) assay specific for Streptococcus suis based on the gene encoding the glutamate dehydrogenase. FEMS Microbiol Lett 218:79–84. doi: 10.1111/j.1574-6968.2003.tb11501.x. [DOI] [PubMed] [Google Scholar]
- 22.Tien LHT, Nishibori T, Nishitani Y, Nomoto R, Osawa R. 2013. Reappraisal of the taxonomy of Streptococcus suis serotypes 20, 22, 26, and 33 based on DNA-DNA homology and sodA and recN phylogenies. Vet Microbiol 162:842–849. doi: 10.1016/j.vetmic.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 23.Ishida S, Tien LHT, Osawa R, Tohya M, Nomoto R, Kawamura Y, Takahashi T, Kikuchi N, Kikuchi K, Sekizaki T. 2014. Development of an appropriate PCR system for the reclassification of Streptococcus suis. J Microbiol Methods 107:66–70. doi: 10.1016/j.mimet.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 24.King SJ, Leigh JA, Heath PJ, Luque I, Tarradas C, Dowson CG, Whatmore AM. 2002. Development of a multilocus sequence typing scheme for the pig pathogen Streptococcus suis: identification of virulent clones and potential capsular serotype exchange. J Clin Microbiol 40:3671–3680. doi: 10.1128/JCM.40.10.3671-3680.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen C, Zhang W, Zheng H, Lan R, Wang H, Du P, Bai X, Ji S, Meng Q, Jin D, Liu K, Jing H, Ye C, Gao GF, Wang L, Gottschalk M, Xu J. 2013. Minimum core genome sequence typing of bacterial pathogens: a unified approach for clinical and public health microbiology. J Clin Microbiol 51:2582–2591. doi: 10.1128/JCM.00535-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Z, Bai X, Ji S, Xu J, Zheng H. 2014. Development of a multiplex PCR assay to identify seven new capsular gene loci of Streptococcus suis. Chin J Zoonoses 30:337–346. [Google Scholar]
- 27.Smith HE, de Vries R, van't Slot R, Smits MA. 2000. The cps locus of Streptococcus suis serotype 2: genetic determinant for the synthesis of sialic acid. Microb Pathog 29:127–134. doi: 10.1006/mpat.2000.0372. [DOI] [PubMed] [Google Scholar]
- 28.Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, Parkhill J. 2005. ACT: the Artemis comparison tool. Bioinformatics 21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- 29.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar S, Tamura K, Nei M. 1994. MEGA: molecular evolutionary genetics analysis software for microcomputers. Comput Appl Biosci 10:189–191. [DOI] [PubMed] [Google Scholar]
- 31.Bonifait L, Gottschalk M, Grenier D. 2010. Cell surface characteristics of nontypeable isolates of Streptococcus suis. FEMS Microbiol Lett 311:160–166. doi: 10.1111/j.1574-6968.2010.02086.x. [DOI] [PubMed] [Google Scholar]
- 32.Vanrobaeys M, De Herdt P, Charlier G, Ducatelle R, Haesebrouck F. 1999. Ultrastructure of surface components of Streptococcus gallolyticus (S. bovis) strains of differing virulence isolated from pigeons. Microbiology 145(Pt 2):335–342. doi: 10.1099/13500872-145-2-335. [DOI] [PubMed] [Google Scholar]
- 33.Jacques M, Gottschalk M, Foiry B, Higgins R. 1990. Ultrastructural study of surface components of Streptococcus suis. J Bacteriol 172:2833–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pan Z, Ma J, Dong W, Song W, Wang K, Lu C, Yao H. 2015. Novel variant serotype of Streptococcus suis isolated from piglets with meningitis. Appl Environ Microbiol 81:976–985. doi: 10.1128/AEM.02962-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pelosi L, Boumedienne M, Saksouk N, Geiselmann J, Geremia RA. 2005. The glucosyl-1-phosphate transferase WchA (Cap8E) primes the capsular polysaccharide repeat unit biosynthesis of Streptococcus pneumoniae serotype 8. Biochem Biophys Res Commun 327:857–865. doi: 10.1016/j.bbrc.2004.12.082. [DOI] [PubMed] [Google Scholar]
- 36.Calix JJ, Nahm MH. 2010. A new pneumococcal serotype, 11E, has a variably inactivated wcjE gene. J Infect Dis 202:29–38. doi: 10.1086/653123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beynon LM, Dutton GG, Richards JC. 1990. Structure of the amino acid-containing capsular polysaccharide from Escherichia coli O8:K49:H21. Carbohydr Res 205:347–359. doi: 10.1016/0008-6215(90)80152-S. [DOI] [PubMed] [Google Scholar]
- 38.Van Calsteren MR, Gagnon F, Calzas C, Goyette-Desjardins G, Okura M, Takamatsu D, Gottschalk M, Segura M. 2013. Structure determination of Streptococcus suis serotype 14 capsular polysaccharide. Biochem Cell Biol 91:49–58. doi: 10.1139/bcb-2012-0036. [DOI] [PubMed] [Google Scholar]
- 39.Van Calsteren MR, Gagnon F, Lacouture S, Fittipaldi N, Gottschalk M. 2010. Structure determination of Streptococcus suis serotype 2 capsular polysaccharide. Biochem Cell Biol 88:513–525. doi: 10.1139/O09-170. [DOI] [PubMed] [Google Scholar]
- 40.Chabot-Roy G, Willson P, Segura M, Lacouture S, Gottschalk M. 2006. Phagocytosis and killing of Streptococcus suis by porcine neutrophils. Microb Pathog 41:21–32. doi: 10.1016/j.micpath.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 41.Gottschalk M, Segura M. 2000. The pathogenesis of the meningitis caused by Streptococcus suis: the unresolved questions. Vet Microbiol 76:259–272. doi: 10.1016/S0378-1135(00)00250-9. [DOI] [PubMed] [Google Scholar]
- 42.Smith HE, Damman M, van der Velde J, Wagenaar F, Wisselink HJ, Stockhofe-Zurwieden N, Smits MA. 1999. Identification and characterization of the cps locus of Streptococcus suis serotype 2: the capsule protects against phagocytosis and is an important virulence factor. Infect Immun 67:1750–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jacques M, Foiry B. 1987. Electron microscopic visualization of capsular material of Pasteurella multocida types A and D labeled with polycationic ferritin. J Bacteriol 169:3470–3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jacques M, Foiry B, Higgins R, Mittal KR. 1988. Electron microscopic examination of capsular material from various serotypes of Actinobacillus pleuropneumoniae. J Bacteriol 170:3314–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Knirel YA, Wang J, Luo X, Senchenkova SN, Lan R, Shpirt AM, Du P, Shashkov AS, Zhang N, Xu J, Sun Q. 2014. Genetic and structural identification of an O-acyltransferase gene (oacC) responsible for the 3/4-O-acetylation on rhamnose III in Shigella flexneri serotype 6. BMC Microbiol 14:266. doi: 10.1186/s12866-014-0266-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.