Abstract
Extra-slow-growing bradyrhizobia from root nodules of field-grown soybeans harbor abundant insertion sequences (ISs) and are termed highly reiterated sequence-possessing (HRS) strains. We analyzed the genome organization of HRS strains with the focus on IS distribution and symbiosis island structure. Using pulsed-field gel electrophoresis, we consistently detected several plasmids (0.07 to 0.4 Mb) in the HRS strains (NK5, NK6, USDA135, 2281, USDA123, and T2), whereas no plasmids were detected in the non-HRS strain USDA110. The chromosomes of the six HRS strains (9.7 to 10.7 Mb) were larger than that of USDA110 (9.1 Mb). Using MiSeq sequences of 6 HRS and 17 non-HRS strains mapped to the USDA110 genome, we found that the copy numbers of ISRj1, ISRj2, ISFK1, IS1632, ISB27, ISBj8, and IS1631 were markedly higher in HRS strains. Whole-genome sequencing showed that the HRS strain NK6 had four small plasmids (136 to 212 kb) and a large chromosome (9,780 kb). Strong colinearity was found between 7.4-Mb core regions of the NK6 and USDA110 chromosomes. USDA110 symbiosis islands corresponded mainly to five small regions (S1 to S5) within two variable regions, V1 (0.8 Mb) and V2 (1.6 Mb), of the NK6 chromosome. The USDA110 nif gene cluster (nifDKENXSBZHQW-fixBCX) was split into two regions, S2 and S3, where ISRj1-mediated rearrangement occurred between nifS and nifB. ISs were also scattered in NK6 core regions, and ISRj1 insertion often disrupted some genes important for survival and environmental responses. These results suggest that HRS strains of soybean bradyrhizobia were subjected to IS-mediated symbiosis island shuffling and core genome degradation.
INTRODUCTION
Soybean bradyrhizobia are soil bacteria that symbiotically fix atmospheric nitrogen in root nodule tissues of soybean plants (1). The first genome sequence of one such bacterium was published in 2002 (2); this bacterium, strain USDA110 of Bradyrhizobium japonicum, has been recently reclassified as Bradyrhizobium diazoefficiens (3). One of the most notable features of this strain is a large symbiosis island (681 kb) on its chromosome; this island contains symbiotic gene clusters and has low GC content (2). Similar symbiosis islands have been detected on the chromosomes of other soybean bradyrhizobial strains, including B. japonicum USDA6T (4) and CPAC 15 (5) and B. diazoefficiens CPAC 7 (5).
Symbiosis islands contain multiple insertion sequences (ISs) and symbiotic gene clusters, including nod, nif, fix, and rhc. The USDA110 genome contains 1 to 15 copies of each of 20 different ISs (2, 4). In the USDA110 genome (9.1 Mb) and USDA6 genome (9.2 Mb), most of these IS copies (78% and 97%, respectively) are within a relatively small region of symbiosis islands (0.68 Mb and 0.86 Mb, respectively) (2, 4). These ISs are not found in the genome of nonsymbiotic Bradyrhizobium sp. strain S23321, which is included in the soybean bradyrhizobial cluster in phylogenetic analysis but lacks a symbiosis island (6), and ISs were probably introduced into soybean bradyrhizobial genomes by horizontal transfer of the symbiosis island (2, 4, 7, 8).
Comparative genomics of soybean bradyrhizobia revealed the peculiar structure of the symbiosis islands: highly conserved backbones that include symbiotic genes are interrupted by strain-specific sequences (4, 5). Strain-specific sequences in symbiosis islands have been also found in the genomes of three strains of Mesorhizobium loti (9). Parker (10, 11) suggested that the evolutionary history of symbiotic genes differs from that of genes outside the symbiosis island. Symbiosis islands are thought to evolve more rapidly than other regions of bradyrhizobial genomes (4, 5).
Extra-slow-growing bradyrhizobia have often been isolated from root nodules of field-grown soybeans in Japan, the United States, and China (12). These strains generally harbor high copy numbers of ISs such as ISRj1 (RSα), ISRj2 (RSβ), and IS1631 (12–14) and are termed highly reiterated sequence-possessing (HRS) strains (12, 14). In other words, the copy numbers of ISRj1, ISRj2, and IS1631 in HRS strains are apparently higher than those in non-HRS strains such as USDA110 (12–14). In particular, IS1631 has been frequently detected in the genomes of HRS strains of soybean bradyrhizobia (12, 14). In the United States, a hyperreiterated IS has been found in B. japonicum serocluster USDA123, a major group indigenous to alkaline soils (15). Because the copy numbers of ISs increase via their transposition (16), we have speculated that the HRS strains originated from non-HRS strains of soybean bradyrhizobia (12–14). No difference between the symbiotic abilities of HRS and non-HRS strains has been observed so far (12–14). The HRS strain NK5 has the potential to transfer the nod genes to nod-minus mutants of Bradyrhizobium elknaii in soil microcosms (17).
The aim of the current study was the genomic characterization of HRS strains in comparison with non-HRS strains to understand the role of symbiosis islands and ISs in the evolution of HRS strains of soybean bradyrhizobia. We attempted to answer two major questions: how does the structure of a symbiosis island change, and how are the ISs distributed inside and outside the symbiosis islands in the HRS strain genome?
MATERIALS AND METHODS
Bacterial strains, growth media, and DNA preparation.
We used 6 well-characterized HRS strains (NK5, NK6, USDA123, T2, USDA135, and 2281) (12) and 17 non-HRS strains collected from many field sites (18) (see Table S1 in the supplemental material). The 23 strains of soybean bradyrhizobia were grown aerobically at 30°C in HEPES- and MES (morpholineethanesulfonic acid)-buffered HM salt medium (1) supplemented with 0.1% (wt/vol) arabinose and 0.025% (wt/vol) yeast extract (Becton, Dickinson and Company, Sparks, MD). Total bacterial DNA was prepared as described earlier (13).
Phylogenetic analysis.
PCR amplification and sequence analysis of 16S rRNA genes and 16S to 23S rRNA gene internal transcribed spacer (ITS) sequences were performed as described previously (1). The accession numbers are listed in Table 1. To construct phylogenetic trees, the neighbor-joining method and Clustal W were used (19).
TABLE 1.
Accession numbers of 16S rRNA genes and internal transcribed spacer sequences of soybean bradyrhizobia used in this study
| Strain | 16S rRNA gene |
ITS |
||
|---|---|---|---|---|
| Accession no. | Reference | Accession no. | Reference | |
| HRS strains | ||||
| NK5 | AB070568 | 12 | AB830105 | This study |
| NK6 | AB070569 | 12 | AB830106 | This study |
| USDA123 | AF208504 | 31 | AB830107 | This study |
| T2 | AB070570 | 12 | AB830108 | This study |
| USDA135 | AB070571 | 12 | AB830109 | This study |
| 2281 | AB029402 | 46 | AB830110 | This study |
| Non-HRS straina | ||||
| USDA110 | AP005940 | 2 | AB100749 | 2 |
Data for other non-HRS strains were previously described (18).
PFGE.
Preparation and restriction digestion of intact genomic DNA in agarose plugs were performed according to the protocol reported previously (20). To detect plasmids, DNA was digested with S1 nuclease (TaKaRa, Tokyo, Japan). To estimate the genome size, DNA was digested with two rare-cutting restriction enzymes, PmeI (NEB, Ipswich, MA) and SmiI (recognition site identical to that of SwaI) (TaKaRa). Pulsed-field gel electrophoresis (PFGE) was carried out in a CHEF-DRII gel electrophoresis system (Bio-Rad, Richmond, CA) in 0.5× Tris-borate-EDTA (TBE) buffer at 14°C in a temperature-controlled cooling unit. To separate DNA fragments of 0.1 to 2 Mb, contour-clamped homogeneous electric field (CHEF) electrophoresis was conducted in a 1% pulsed-field agarose gel (pulse time, 10 to 120 s; field strength, 6 V cm−1; angle, 120°; run time, 24 h). DNA fragments of 1 to 3 Mb were separated in a 0.8% pulsed-field agarose gel (pulse time, 500 s; field strength, 3 V cm−1; angle, 106°; run time, 48 h). DNA fragments of >3 Mb were separated in a 0.8% pulsed-field agarose gel (pulse time, 1,800 s; field strength, 2 V cm−1; angle, 106°; run time, 72 h). Chromosomes of Saccharomyces cerevisiae (0.1 to 2 Mb and 1 to 3 Mb), Schizosaccharomyces pombe (>3 Mb), and Hansenula wingei (>3 Mb) (Bio-Rad) and the lambda DNA ladder (0.1 to 2 Mb) (TaKaRa) were used as size standards.
MiSeq sequencing.
DNA libraries of the 23 strains of soybean bradyrhizobia were prepared using a Nextera DNA sample preparation kit (Illumina, San Diego, CA). Briefly, tagmentation (i.e., DNA fragmentation and adapter ligation) reactions were performed for 5 min at 55°C. The tagmented DNA was purified with a ZR-96 DNA Clean & Concentrator-5 kit (Zymo Research Corp., Orange, CA) and used as a template for PCR (five cycles; 50-μl reaction mixture). Amplified DNA was purified with AMPure XP DNA purification beads (Beckman Coulter, Inc., Brea, CA). The DNA concentration was measured by using a PicoGreen double-stranded DNA (dsDNA) quantification kit (Life Technologies Corporation, Carlsbad, CA). DNA length was measured by using an Agilent high-sensitivity DNA kit (Agilent Technologies, Santa Clara, CA). Sequencing was performed on a MiSeq next-generation sequencing system (Illumina).
Mapping to the USDA110 genome and IS copy estimation.
Bioinformatic analysis was performed with CLC Genomics Workbench ver. 6.0 (CLC Bio, Aarhus, Denmark). The reads were mapped to a reference genome of USDA110 with the following CLC Genomics Workbench parameters: mismatch cost, 2; insertion cost, 3; deletion cost, 3; length fraction, 0.5; similarity fraction, 0.8; map randomly; no masking. Mapping was also performed with the Burrows-Wheeler alignment (BWA) (21) with default parameters. The copy numbers of these ISs were estimated by their mapping to the USDA110 genome supplemented with one copy of IS1631 at the end of the genome because this IS is present in HRS strains but not in USDA110 (12, 14).
We also selected 10 housekeeping genes (atpD, dnaG, fadD, gltB, lepA, leuS, lysS, recA, tpiA, and trpB) that are present as single copies in the USDA110 genome (2). The bam files from CLC Genomics Workbench or BWA were converted into sam files by SAMtools (http://samtools.sourceforge.net/). Our target sequences were the above-mentioned housekeeping genes and all IS copies. We then summed the numbers of (i) bases in fully mapped reads within each target sequence and (ii) bases within the target sequences that were mapped to their boundaries. The depth of fully and partially mapped reads for the target sequences was normalized by dividing the total base number by the total length of the target sequences. Finally, we calculated the IS copy numbers per genome of each of the 23 strains of soybean bradyrhizobia by dividing the depth of each IS by the average depth of all housekeeping genes. To facilitate this calculation, an in-house program, “IS counter,” was developed by InfoBio Co. Ltd. (Tokyo, Japan).
nifH tree.
Six HRS strain libraries were de novo assembled by using CLC Genomics Workbench ver. 6.0. The generated contigs were subjected to BLASTN searches using the nifH gene of USDA110 as a query. The nifH alignment and tree were constructed with Clustal W (19) and with MEGA5 software (22) using the neighbor-joining method with 1,000 bootstrap replicates. Several reference organisms were also included (6).
Genome analysis of the HRS strain NK6.
The whole genome was sequenced on a PacBio RSII sequencer (Pacific Biosciences, Menlo Park, CA) using 8 cells for single-molecule, real-time (SMRT) DNA sequencing, which was performed by the Dragon Genomics Center, TaKaRa Bio Inc. (Otsu, Japan) (23, 24). The resultant sequences were assembled by using SMRT Analysis version 2.2 (Pacific Biosciences).
The chromosome and plasmid sequences were submitted to the microbial genome annotation pipeline (MiGAP) for automatic annotation (http://www.migap.org/). The chromosome sequence was also submitted to xBASE (25). Genomic comparisons were carried out with GenomeMatcher (26) and BLASTN.
The putative replication origin and terminus were predicted according to the methods of Lee et al. (27) and Kaneko et al. (4) by using GC-skew analysis (28). For terminus prediction, the cumulative distribution of FtsK-orienting polar sequences (KOPS) (29) and dif sequences (30) was also analyzed.
Nodulation and symbiotic nitrogen fixation.
Surface-sterilized soybean seeds (Glycine max cv. Enrei) were inoculated with NK6 and USDA110 as described previously (17). Plants were cultivated in a greenhouse and were repeatedly supplied with nitrogen-free nutrient solution (17). Nitrogen fixation and nodulation were analyzed 50 days after germination. To measure nitrogen fixation, acetylene reduction was assayed by using the nodulated root system over 20 min (17). The nodules excised from the roots were dried, and their weight was measured.
Nucleotide sequence accession numbers.
DNA sequences determined in this study by using PacBio RSII were deposited in the DDBJ database under the accession numbers AP014685 (NK6 chromosome), AP014659 (pNK6a), AP014686 (pNK6b), AP014687 (pNK6c), and AP014688 (pNK6d); accession numbers for 16S rRNA genes and ITS sequences are listed in Table 1. MiSeq sequences were deposited in the DDBJ Sequence Read Archive (DRA001137 [DRX012557 to DRX012562] and DRA002739 [DRX022739 to DRX022754]).
RESULTS
Phylogenetic analysis of 16S rRNA genes, 16S to 23S rRNA gene ITS sequences, and nifH genes.
In the present study, we analyzed six HRS strains, NK5, NK6, USDA123, 2281, USDA135, and T2 (see Table S1 in the supplemental material) (12). Strains USDA123 and USDA135 are indigenous isolates from soybeans grown in soils in the United States (20, 31). Strains NK5 and NK6 were derived from root nodules of soybean grown in paddy fields in Niigata (Japan); T2 originated in the Tokachi upland region (Hokkaido, Japan). Strain 2281 is a type strain of Bradyrhizobium lianoningense isolated from the nodules of soybean grown in alkaline soil in China (46).
In the 16S rRNA gene tree and ITS (16S to 23S rRNA gene internal transcribed spacer) tree (1), the BD (Bradyrhizobium diazoefficiens) and BJ (Bradyrhizobium japonicum) clusters included sequences from HRS strains (shaded in gray in Fig. 1A and B) and non-HRS strains. In both trees, the HRS strains NK5 and NK6 were robustly placed in the BD cluster together with the non-HRS strains USDA110 and USDA122 (Fig. 1A and B). The other four HRS strains belonged to the BJ cluster (Fig. 1A and B).
FIG 1.
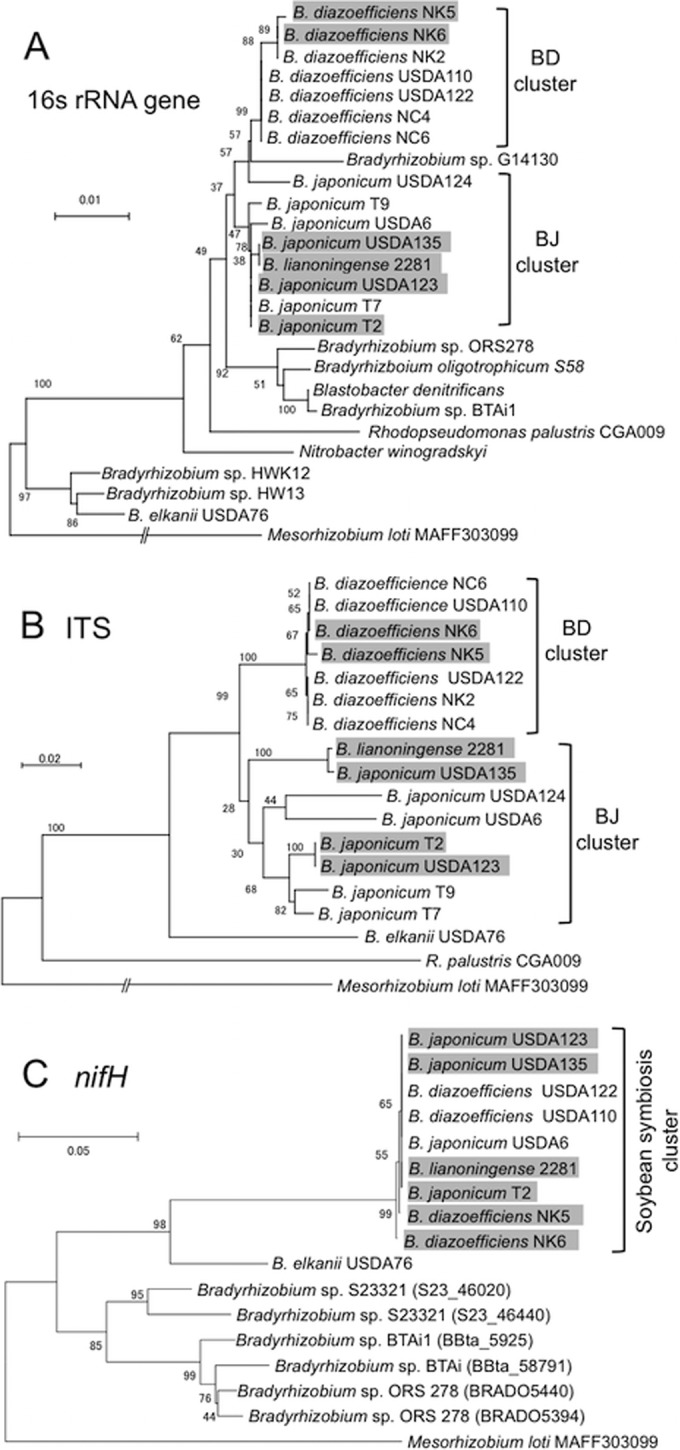
Phylogenetic relationships among HRS and non-HRS strains of soybean bradyrhizobia based on the sequences of 16S rRNA genes (A), internal transcribed spacers (ITS) (B), and nifH genes (C). HRS strains are shaded in gray. For all trees, Mesorhizobium loti MAFF303099 was used as an outgroup (1). Numbers at the nodes are percentages of 1,000 bootstrap replications supporting that partition. Bars show the number of base substitutions per nucleotide. Clusters BD in panels A and B include B. diazoefficiens strains; clusters BJ include B. japonicum and B. lianoningense strains. The soybean symbiosis cluster in panel C includes BD and BJ strains able to nodulate soybean and fix nitrogen. Reference strains of the Bradyrhizobiaceae and their sequences were described previously (6).
The rhizobial nif gene cluster, located in the symbiosis island in the non-HRS strains, is required for symbiotic N2 fixation in legume root nodules (2). In the nifH tree (Fig. 1C), all HRS strains analyzed were grouped together with the non-HRS strains USDA6, USDA110, and USDA122. The difference between the nifH tree and the two other trees is in line with observations that the phylogeny of symbiotic genes differs from that of regions outside the symbiosis island (10, 11) and suggests that the nifH gene in the HRS strains was derived from a symbiosis island of a non-HRS strain of soybean bradyrhizobia (10, 11). These data prompted us to undertake a detailed comparison of the genomes of HRS and non-HRS strains.
Plasmid and genome size analyses.
An early report showed that the HRS strain USDA135 carried a plasmid (33); therefore, we analyzed the plasmid profiles of the six HRS strains by PFGE using cell plugs without restriction enzyme treatment. Several plasmids (70 to 400 kb) were consistently detected (Table 2), whereas no plasmids were detected in the non-HRS strain USDA110.
TABLE 2.
Replicon structures and sizes of HRS strains estimated by pulsed-field gel electrophoresis
| Parameter | Treatmenta | Value for strain |
||||||
|---|---|---|---|---|---|---|---|---|
| HRSb |
Non-HRS USDA110 | |||||||
| Group A |
Group B1 |
Group B2 |
||||||
| NK5 | NK6 | USDA135 | 2281 | USDA123 | T2 | |||
| Size (Mb) | ||||||||
| Plasmid | None | 0.36 | 0.20 | 0.20 | 0.25 | 0.40 | 0.40 | |
| 0.20 | 0.15 | 0.22 | 0.28 | 0.28 | ||||
| 0.07 | 0.17 | 0.20 | 0.20 | |||||
| Chromosome | SmiI | 9.9 | 9.5 | 10.7 | 10.2 | 10.6 | 10.4 | 9.1 |
| PmeI | 9.4 | 10 | 10.7 | 10 | ||||
| Avg | 9.65 | 9.75 | 10.7 | 10.2 | 10.65 | 10.2 | 9.1 | |
| Chromosome size relative to that of USDA110 (%) | 106 | 107 | 118 | 112 | 117 | 112 | 100 | |
Plasmid size was estimated by PFGE analysis without digestion, whereas chromosome size was estimated by averaging the total sizes of fragments obtained by digestion with rare-cutting restriction enzymes SmiI and PmeI (see Table S2 in the supplemental material).
HRS strains were previously classified into groups A, B1, and B2 according to estimated copy numbers of ISRj1 and ISRj2 (12).
Genome sizes of the HRS strains were estimated based on the sums of the fragment sizes of DNA digested with PmeI or SmiI (see Table S2 in the supplemental material). The genomes of the six HRS strains (9.4 to 10.7 Mb) were larger than that of USDA110 (9.1 Mb) (Table 2).
IS copy numbers determined by genome mapping.
The copy numbers of two ISs (ISRj1 and ISRj2) have been previously estimated by using Southern hybridization (12). Because of recent advances in sequencing technology and bioinformatics tools, we adopted a genome mapping method. DNAs of 6 HRS strains and 17 non-HRS strains were sequenced on a MiSeq sequencer (see Table S3 in the supplemental material) and mapped. Similar results were obtained by using two mapping methods, BWA and CLC Genomics Workbench. The copy numbers of ISs in HRS strains and the average of those in non-HRS strains are shown in Table 3, whereas detailed data on the non-HRS strains are shown in Table S4 in the supplemental material. Among 21 ISs detected, 7 ISs (ISRj1, ISRj2, ISFK1, IS1632, ISB27, ISBj8, and IS1631) had higher copy numbers in the genomes of the HRS strains than in those of the non-HRS strains (Table 3).
TABLE 3.
Copy numbers of insertion sequences in HRS and non-HRS strains of soybean bradyrhizobia determined by genome mapping
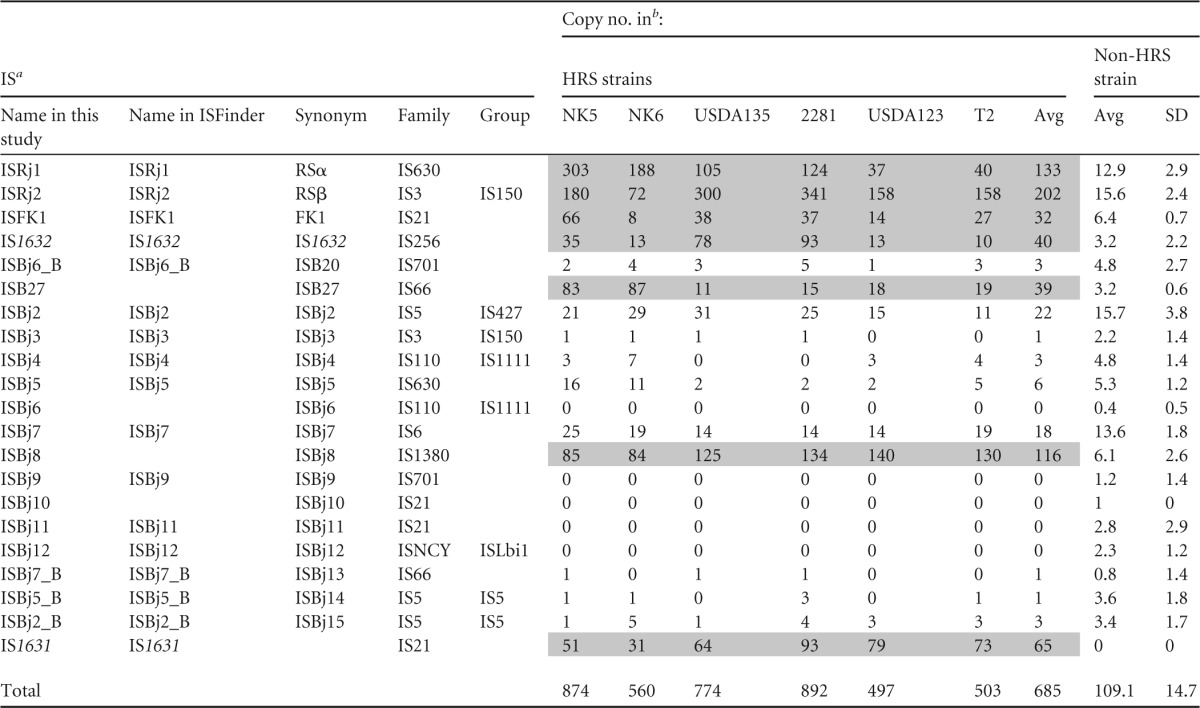
Name in ISFinder indicates the approved IS name in the ISFinder database (https://www-is.biotoul.fr). The IS families and groups are indicated according to ISFinder. Synonyms are those used in the paper on the genome of B. diazoefficiens USDA110 (12).
Copy numbers of ISs were estimated by BWA mapping to USDA110 (see the text). The copy numbers of six IS elements (shaded in gray) were higher in the HRS strains than in non-HRS strains. The detailed data on non-HRS strains are shown in Table S4 in the supplemental material. Strains NK5 and NK6 belong to HRS group A, USDA135 and 2281 to group B1, and USDA123 and T2 to group B2 (12).
Sequencing of the HRS strain genome.
To understand the structure of symbiosis islands and IS distribution in the genomes of the HRS strains, we tried to sequence their whole genomes. However, this was difficult even by using a conventional 454 sequencer and paired-end library (8 kb) because of long repeated DNA sequences containing many IS copies. Thus, we adopted a new PacBio sequencing strategy for longer reads with hybrid error correction (23, 24). We focused on NK6, which is phylogenetically very close to USDA110 (Fig. 1B), and we obtained five contigs (Table 4) tentatively designated the NK6 chromosome (9.78 Mb), pNK6a (0.21 Mb), pNK6b (0.21 Mb), pNK6c (0.14 Mb), and pNK6d (0.14 Mb) (Fig. 2). The size of the NK6 chromosome (9.78 Mb) estimated by PacBio sequencing was almost identical to that estimated by PFGE (9.75 Mb) (Table 4). Plasmids pNK6a and pNK6b corresponded to the 0.20-Mb band, whereas pNK6c and pNK6d corresponded to the 0.15-Mb band. Thus, the validity of the NK6 genome sequence was supported by the results of PFGE analyses (Table 4).
TABLE 4.
General features of the genomes of strains NK6, USDA6, and USDA110
| Parameter | Value for: |
||||||
|---|---|---|---|---|---|---|---|
| HRS strain NK6 |
Non-HRS strains |
||||||
| Chromosome | pNK6a | pNK6b | pNK6c | pNK6d | USDA6 | USDA110 | |
| Size determined by: | |||||||
| PacBio sequencing (bp) | 9,780,023 | 212,255 | 210,438 | 136,288 | 136,153 | 9,207,384 | 9,105,828 |
| PFGE (Mb)a | 9.75 | 0.20 | 0.20 | 0.15 | 0.15 | 9.1 | |
| G+C content (%) | 63.7 | 61.1 | 61.2 | 61.1 | 61.7 | 63.7 | 64.1 |
| No. of: | |||||||
| tRNA-coding genes | 55 | 1 | 0 | 2 | 0 | 51 | 50 |
| rRNA gene clusters | 3 | 0 | 0 | 0 | 0 | 2 | 1 |
| Protein-coding genes | 9,814 | 218 | 215 | 152 | 149 | 8,829 | 8,317 |
| Genomic islands | 12 | 0 | 0 | 0 | 0 | 15 | 14 |
| Insertion sequences | 306 | 41 | 27 | 8 | 22 | 69 | 104 |
The sizes of the respective replicons were determined by pulsed-field gel electrophoresis (see Table S2 in the supplemental material).
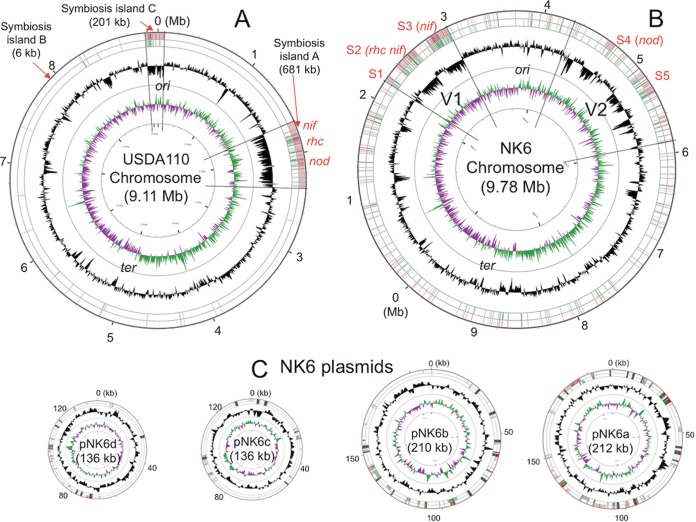
FIG 2.
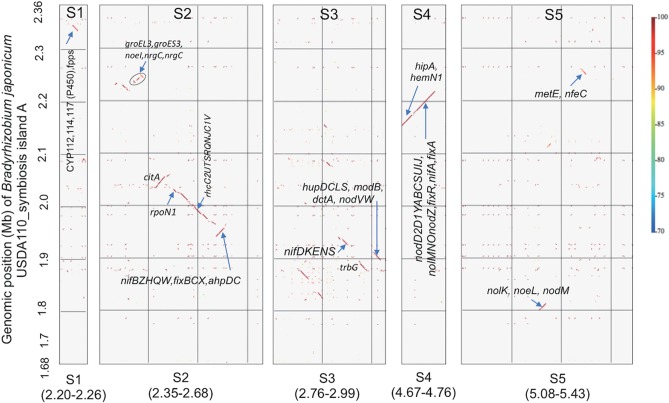
Schematic presentations of the USDA110 genome (A), NK6 chromosome (B), and NK6 plasmids (C). The HRS strain NK6 has one chromosome (B) and four plasmids (C), whereas the non-HRS strain USDA110 has a chromosome (A) but no plasmids (2). The outermost circles show the DNA regions homologous to symbiosis islands A and C of USDA110 (brown). The second circles show the distribution of 20 different insertion sequences (green). The third circles show the G+C content (black). The innermost circles show the GC skew. Two variable regions, V1 and V2, show no colinearity with the USDA110 genome (Fig. 3). Regions S1 to S5 in the NK6 chromosome correspond to the symbiosis islands A and C of the USDA110 genome. Note that the insertion sequences are more abundant on the NK6 chromosome than on the USDA110 chromosome.
Analysis of the replicon regions in the NK6 genome.
The putative chromosomal origin of replication was predicted to be located at 3.77 Mb (bp 3765634 to 3766154) based on GC-skew analysis (28) and the similarity of the gene clusters associated with origins of replication among Bradyrhizobiaceae (4, 27) (Fig. 2). The putative terminus (ter) was predicted to be located at 8.94 Mb based on GC-skew analysis (Fig. 2), cumulative distribution of the KOPS (29), and the position of the dif sequence (bp 8944569 to 8944542 bp) (30). Plasmids pNK6a to pNK6d possess one or two sets of the repABC operon (see Table S5 in the supplemental material), which is responsible for replication and segregation of alphaproteobacterial low-copy-number plasmids, because their putative origins of replication are located within repC (32).
The GC content of the largest replicon of the NK6 chromosome (63.7%) was similar to those of the USDA6 and USDA110 genomes (63.7% to 64.1%) but was higher than those of the four small replicons (pNK6a to pNK6d) (61.1% to 61.7%), suggesting an external origin of the small replicons (Table 4). These analyses strongly suggest that the NK6 genome is composed of a single chromosome and four plasmids (Fig. 2).
Genome comparison between the HRS strain NK6 and non-HRS strain USDA110.
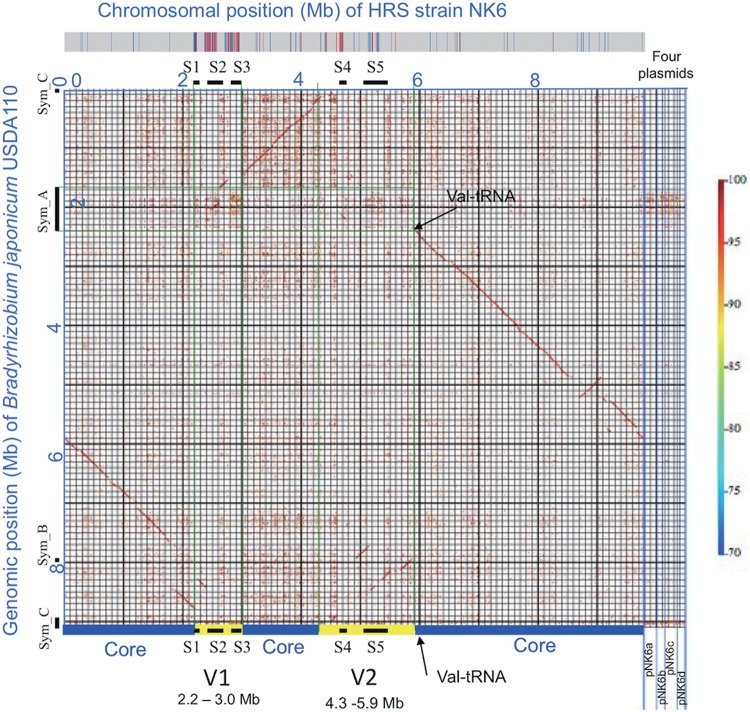
Using GenomeMatcher (26), we found strong and extended colinearity between the chromosomes of NK6 and USDA110 (Fig. 3). However, two variable genomic regions, V1 (0.8 Mb) and V2 (1.6 Mb), were detected on the NK6 chromosome (Fig. 3). None of the NK6 plasmids showed colinearity with the USDA110 genome (Fig. 3). To identify the genomic regions of the HRS strains corresponding to the symbiosis islands of the non-HRS strains, we compared the DNA sequences of major symbiosis islands A and C of the USDA110 genome (Fig. 2A) with the NK6 genome by BLASTN (Fig. 2). The sequences of the NK6 chromosome homologous to symbiosis islands A and C of USDA110 were concentrated in the regions S1 to S3 in V1 and S4 and S5 in V2 (brown in the outermost circle in Fig. 2B and C). The regions S1 to S5 had lower GC contents (black in the third-outermost circle in Fig. 2B) in the NK6 chromosome, similarly to symbiosis islands A and C in USDA110 (Fig. 2A).
FIG 3.
Comparison between the NK6 and USDA110 genomes. The color scale on the right indicates the percent nucleotide identity in the BLASTN alignment output. Blue and yellow in the lowermost color bar indicate core and variable regions, respectively, of the NK6 chromosome. Variable regions V1 and V2 show no colinearity with the USDA110 genome. Regions S1 to S5 within V1 and V2 correspond to the symbiosis islands in the USDA110 genome (see also Fig. 2). Blue and red in the uppermost color bar show regions of the NK6 chromosome with higher identity to IS transposase and other genes, respectively, in USDA110 symbiosis island A.
IS distribution.
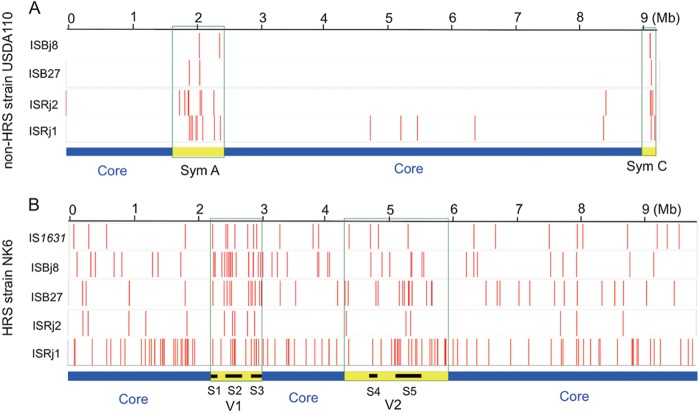
In NK6, insertion sequences (ISs) were more abundant even in the core regions (i.e., sequences outside the variable regions) and in the plasmids than in the USDA110 regions outside the symbiosis islands (green in the second-outermost circles in Fig. 2). Using BLASTN, we found high copy numbers of several ISs in the NK6 chromosome and plasmids, i.e., ISRj1 (114 copies in the NK6 chromosome and 11 copies in plasmids), ISRj2 (16 and 2 copies), ISB27 (49 and 33 copies), ISBj8 (52 and 12 copies), and IS1631 (27 and 4 copies) (see Table S6 in the supplemental material), which was similar to the results of mapping analysis (Table 3). A substantial number of copies (45% to 67%) were in the core regions of the NK6 chromosome, whereas most of these ISs were confined within symbiosis islands A and C in the USDA110 genome (Fig. 4).
FIG 4.
Distribution of insertion sequences (ISs) in the genomes of the non-HRS strain USDA110 and HRS strain NK6. Blue bars, core regions shared between USDA110 and NK6 genomes; yellow bars, symbiosis islands in the USDA110 genome and variable regions of the NK6 chromosome (Fig. 3). Red lines indicate the positions of ISs (listed on the left) whose copy numbers were higher in the NK6 genome than in the USDA110 genome (Table 3).
Among ISs with higher copy number on NK6 genome, the sequences of ISRj1 and IS1631 copies were better conserved than those of other ISs in terms of length and homology, suggesting that ISRj1 and IS1631 mediated ancestral transposition events.
Organization of symbiotic genes on the NK6 chromosome.
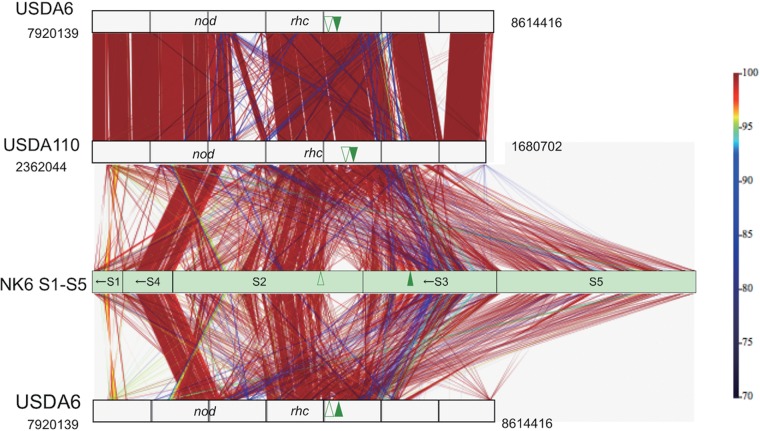
In symbiosis islands of non-HRS strains, highly conserved backbones that include symbiotic genes are interrupted by strain-specific sequences (4, 5) (see, for example, USDA6 versus USDA110 in Fig. 5). However, the NK6 regions homologous to symbiosis island A of USDA110 were shuffled considerably even when we tried to align parts of regions S1 to S5 (Fig. 5) in the best possible way. The most conserved region was S4, which contained the nod gene cluster, whereas the nif clusters were separated into at least two regions (S2 and S3; green triangles in Fig. 5). The center of region S2, including the rhc gene cluster (type III protein secretion system), showed partial colinearity with symbiosis island A of USDA110 but contained small inversions and NK6-specific spacer sequences (Fig. 5).
FIG 5.
Multiple comparison of the symbiosis islands of non-HRS strains USDA6 and USDA110 and HRS strain NK6. The color scale indicates the percent nucleotide identity in the alignment output by BLASTN. The direction and order of fragmented regions (S1 to S5) homologous to symbiosis island A in the USDA110 genome were optimized to show their colinearity with the symbiosis islands of USDA110 and USDA6. Arrows indicate the reverse directions of the S1, S4, and S3 regions in NK6. Green arrowheads indicate the positions of separate nif clusters nifBZHQW (open reading frame arrowheads) and nifDKENS (filled arrowheads) (Fig. 1B and 6). The numbers indicate the positions in the USDA6 and USDA110 genomes.
To identify symbiotic gene clusters within the S1 to S5 regions, these regions were precisely compared with USDA110 symbiosis island A (Fig. 6). Small and fragmented clusters of symbiotic genes were scattered in regions S1 to S5. The major clusters were P450 genes in S1, rhcC2UTSRQNJC1V-nifBZHQW-fixBCX in S2, nifDKENS in S3, nodD2D1YABCSUIJZ-nolMNO-fixRA-nifA in S4, and nolK-noeE-nodM in S5 (Fig. 6).
FIG 6.
Comparison between the USDA110 genome and five fragmented regions of the NK6 genome (S1 to S5) homologous to the USDA110 symbiosis islands. The color scale indicates the percent nucleotide identity in the BLASTN alignment output.
These results indicate that the basic structures of symbiosis islands, clearly present in USDA6 and USDA110, are disrupted in NK6 and their remnants are found mainly within the V1 and V2 regions.
Role of ISRj1 in nif gene cluster rearrangement in NK6.
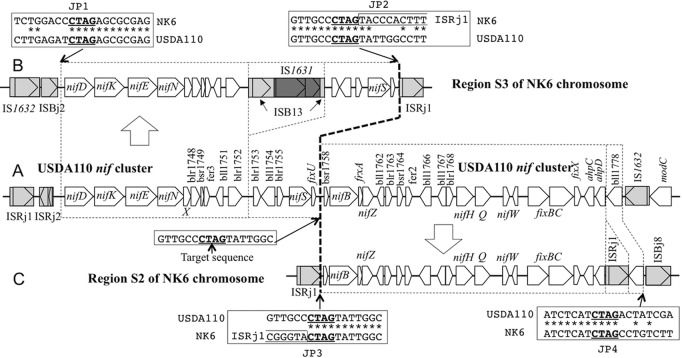
To examine whether ISs mediate shuffling of symbiotic regions, we compared the nif gene clusters on the chromosomes of USDA110 (Fig. 7A) and NK6 (S3 [Fig. 7B] and S2 [Fig. 7C]). At a 4-bp sequence (5′-CTAG, underlined), the major target sequence of ISRj1 transposase (2), between nifS and nifB in the USDA110 genome, we found a trace of the ISRj1-mediated division of the USDA110 nif cluster into two nif gene clusters in regions S3 and S2 of the NK6 chromosome. Comparison of the DNA sequences at junction positions JP2 and JP3 clearly indicates that ISRj1 was inserted into the target sequence of an ancestral non-HRS strain similar to USDA110. The presence of 5′-CTAG at the left (JP1) and right (JP4) ends of the nif cluster shared between USDA110 and NK6 suggests that ISRj1 mediated nif cluster rearrangement in the NK6 chromosome. In addition, IS1631-nested ISB13 and ISRj1 were inserted into the fragmented nif clusters in S3 and S2, respectively. These events occurred consistently at intergenic regions of the nif gene cluster, suggesting that nif gene cluster-dependent symbiotic nitrogen fixation was preserved.
FIG 7.
Comparison of the nif loci on the USDA110 and NK6 chromosomes. At a 4-bp sequence (5′-CTAG, underlined) between nifS and nifB in the USDA110 genome (A), a trace of ISRj1-mediated major division of the USDA110 nif cluster into two nif gene clusters in regions S3 (B) and S2 (C) on the NK6 chromosome was found. The bold dashed line shows the major nif division boundary. Dotted boxes show regions conserved between the two strains. DNA sequences at junction positions JP1 to JP4 contain 5′-CTAG (underlined), the preferred target sequence of ISRj1 transposase. Asterisks indicate nucleotides identical in the two strains.
Symbiotic phenotypes of NK6 and USDA110.
To confirm that the ability of the HRS strain NK6 to fix nitrogen is compatible with that of the non-HRS strain USDA110, we compared the symbiotic phenotypes of the two strains. Inoculation of soybean plants showed that nodulation (179 ± 51 mg dry nodule weight per plant) and N2 fixation (15.1 ± 3.2 μmol acetylene reduced h−1 g−1 fresh nodule weight) after inoculation with NK6 were similar to nodulation (114 ± 17 mg dry nodule weight per plant) and N2 fixation (12.1 ± 2.7 μmol acetylene reduced h−1 g−1 fresh nodule weight) after inoculation with USDA110. Therefore, NK6 has preserved the symbiotic capability despite the apparent ISRj1-mediated division and IS insertions of the nif gene cluster on its genome.
Gene disruption in the NK6 chromosome by ISRj1 insertion.
Although disruption of nif genes and loss of function were not observed in the NK6 genome, we suspected that this example might be a consequence of selection pressure because the strain was isolated from soybean nodules. Therefore, we surveyed other genes on the NK6 chromosome that may have been disrupted by ISRj1 insertion; we chose ISRj1 because it has the highest copy number (114 copies) and is widely distributed on the NK6 chromosome (Fig. 4). We compared the border sequences around all ISRj1 copies of the NK6 chromosome with the USDA110 genome by BLASTN and found that 11 genes were structurally disrupted (Table 5). For example, three copies of ISRj1 (NK6α4, NK6α19, and NK6α31) were inserted into genes encoding the flagellar hook protein (flgE), glutathione S-transferase, and adenylate cyclase, respectively (Table 5; see Fig. S1 in the supplemental material). The accumulation of genes disrupted by ISRj1 and other ISs may cause the phenotype of extra-slow growth.
TABLE 5.
Gene disruption in NK6 by ISRj1 insertionsa
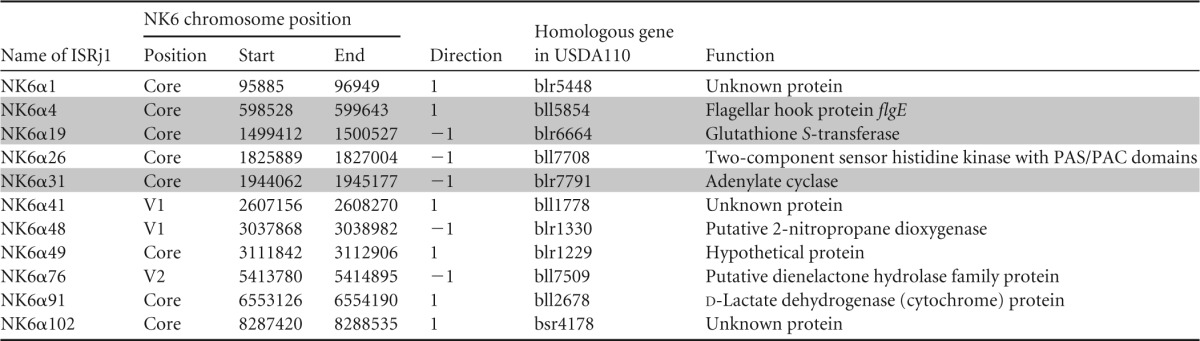
Genes relevant to survival and environmental responses are shaded in gray.
DISCUSSION
Bradyrhizobia prefer legumes (34, 35) and nonlegumes (36–38) that tend to form symbiosis (39). The present study advances genomic characterization of HRS strains of soybean bradyrhizobia with abundant ISs. Phylogenetic (Fig. 1) and comparative genome (Fig. 3) analyses indicated that both the HRS strain NK6 and non-HRS strain USDA110 are derived from a common ancestor. As depicted in Fig. 8 (upper part), the ISs of soybean bradyrhizobia were probably introduced into ancestral nonsymbiotic bradyrhizobial genomes by horizontal transfer of a symbiosis island (2, 4, 7, 8), because ISs are not found in the bradyrhizobial strains ORS278, BTAi1, and S58, which lack symbiosis islands and nod genes (40, 41), or in the nonsymbiotic strain S23321 (6). The HRS strains accumulated abundant ISs (Table 3), and at least in NK6 the symbiosis island was rearranged (Fig. 2) into symbiotic gene clusters (Fig. 5 and 6) within the variable regions V1 and V2, which can be partially explained by IS-mediated DNA rearrangements (Fig. 7). Although the core chromosome structures outside variable regions were well conserved in NK6 (Fig. 3), IS transposition (Fig. 4) and IS-mediated gene disruption (Table 5) were observed in these regions.
FIG 8.
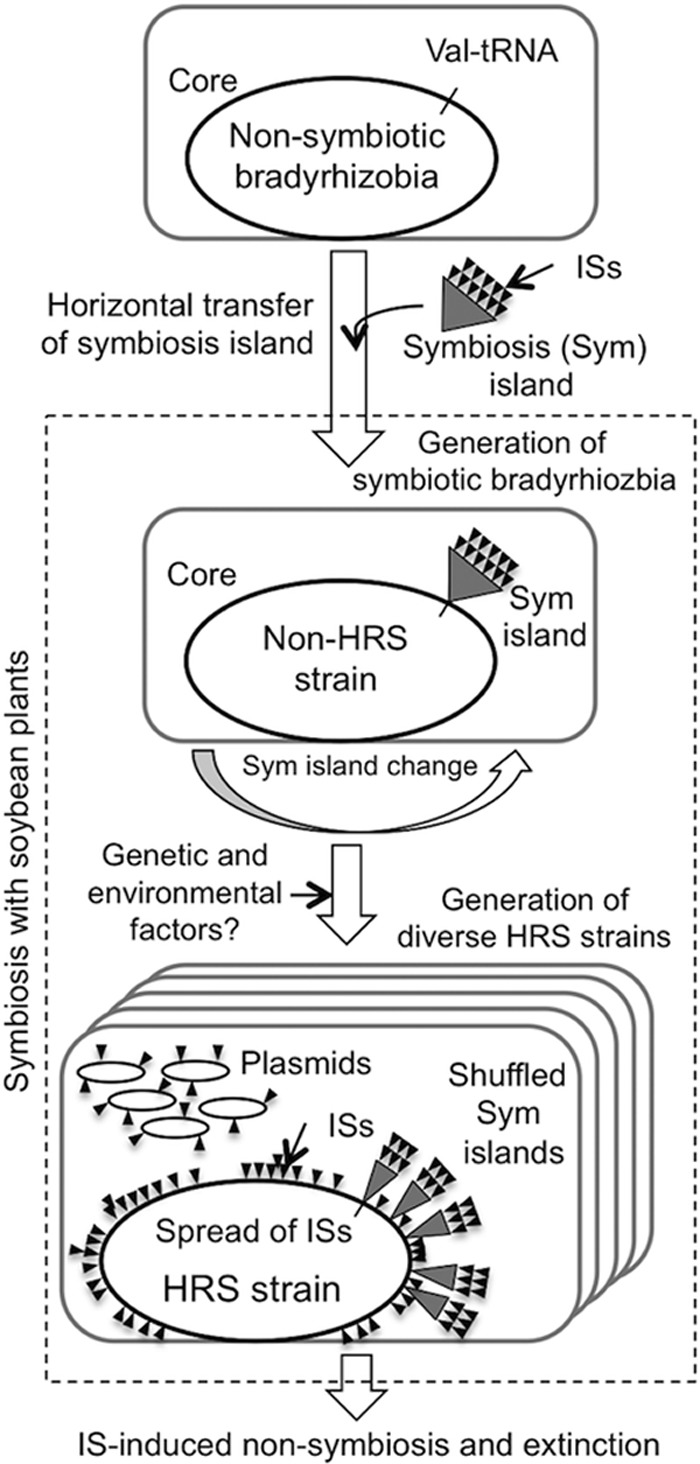
Possible role of symbiosis islands and ISs in the evolution of HRS strains of soybean bradyrhizobia.
The genetic and environmental factors that induced generation of HRS strains remain to be identified. However, plasmid acquisition may be an important cue (Fig. 8, lower part), because all HRS strains tested carried plasmids (Table 2) and NK6 plasmids pNK6a and pNK6d had a high density of ISs, including HRS strain-specific IS1631 (see Fig. S2 in the supplemental material).
In the NK6 genome, the nod gene cluster was preserved better than other symbiotic genes (Fig. 5 and 6), probably because NK6 was isolated from root nodules under selection pressure of symbiosis with soybeans (13, 14). Although the nif gene clusters of NK6 were partially disrupted by IS-mediated rearrangements, the functional units of nif gene clusters were conserved (Fig. 6 and 7), in line with the fact that HRS strain NK6 was collected from healthy grown soybean plants with active symbiotic nitrogen fixation. In a sense, it was a limitation in the present study not to access nonnodulating HRS strains of soybean bradyrhizobia (Fig. 8).
We found a disruption of three potentially important genes by ISRj1. The mutation in the flagellar hook protein, a component of flagellar assembly (42), may reduce NK6 motility. The disruption of glutathione S-transferase may compromise cellular metabolism and protection against chemical and oxidative stresses (43). The disruption of adenylate cyclase, which catalyzes conversion of ATP into cAMP and regulates signal transduction systems that perceive environmental stimuli may disrupt corresponding signaling pathways (44). Although these genes are often redundant in soybean bradyrhizobia (2), their disruption may have reduced the above functions in NK6 and induced extra-slow growth.
Previous studies have indicated that HRS isolates from soybean nodules are diverse in terms of nifDK polymorphisms, IS number profile, and growth rate (12, 13). It is thus likely that diverse IS-mediated rearrangements and IS insertion occasionally disrupt symbiotic genes, including nod and nif and genes essential for survival. If so, these HRS strains undergo nonnodulation, non-nitrogen fixation, and extinction in soil-soybean systems, respectively (Fig. 8). This idea is supported by a recent phylogenetic analysis indicating that loss of nodulation capability had occurred in a natural population of bradyrhizobia isolated from nodules and root surfaces of Lotus strigosus (45).
Genome reduction of endosymbionts occurs in obligate intercellular lifestyles in insects and animals through IS expansion and elimination and genome streamlining (47, 48). On the other hand, genome expansion is common in plant symbionts due to their facultative intercellular lifestyles in soil and plant environments (47). In addition, plant symbiosis is supported by symbiosis islands or plasmids with numerous ISs in (brady)rhizobia (12, 47). Thus, the existence of HRS strains may have unique evolutionary implications relevant to IS expansion in plant-associated bacteria.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by grants from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Genomics-Based Technology for Agricultural Improvement SFC-2003 and the Bio-Oriented Technology Research Advancement Institution [BRAIN]) and by Grants-in-Aid for Scientific Research (A) 23248052 and 26252065 from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00741-15.
REFERENCES
- 1.Itakura M, Saeki K, Omori H, Yokoyama T, Kaneko T, Tabata S, Ohwada T, Tajima S, Uchiumi T, Honnma K, Fujita K, Iwata H, Saeki Y, Hara Y, Ikeda S, Eda S, Mitsui H, Minamisawa K. 2009. Genomic comparison of Bradyrhizobium japonicum strains with different symbiotic nitrogen-fixing capabilities and other Bradyrhizobiaceae members. ISME J 3:326–339. doi: 10.1038/ismej.2008.88. [DOI] [PubMed] [Google Scholar]
- 2.Kaneko T, Nakamura Y, Sato S, Minamisawa K, Uchiumi T, Sasamoto S, Watanabe A, Idesawa K, Iriguchi M, Kawashima K, Kohara M, Matsumoto M, Shimpo S, Tsuruoka H, Wada T, Yamada M, Tabata S. 2002. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res 9:189–197. doi: 10.1093/dnares/9.6.189. [DOI] [PubMed] [Google Scholar]
- 3.Delamuta JR, Ribeiro RA, Ormeño-Orrillo E, Melo IS, Martínez-Romero E, Hungria M. 2013. Polyphasic evidence supporting the reclassification of Bradyrhizobium japonicum group Ia strains as Bradyrhizobium diazoefficiens sp. nov. Int J Sys Evol Microbiol 63:3342–3351. doi: 10.1099/ijs.0.049130-0. [DOI] [PubMed] [Google Scholar]
- 4.Kaneko T, Maita H, Hirakawa H, Uchiike N, Minamisawa K, Watanabe A, Sato S. 2011. Complete genome sequence of the soybean symbiont Bradyrhizobium japonicum strain USDA6T. Genes 2:763–787. doi: 10.3390/genes2040763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siqueira AF, Ormeño-Orrillo E, Souza RC, Rodrigues EP, Almeida LG, Barcellos FG, Batista JS, Nakatani AS, Martínez-Romero E, Vasconcelos AT, Hungria M. 2014. Comparative genomics of Bradyrhizobium japonicum CPAC 15 and Bradyrhizobium diazoefficiens CPAC 7: elite model strains for understanding symbiotic performance with soybean. BMC Genomics 15:420. doi: 10.1186/1471-2164-15-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okubo T, Tsukui T, Maita H, Okamoto S, Oshima K, Fujisawa T, Saito A, Futamata H, Hattori R, Shimomura Y, Haruta S, Morimoto S, Wang Y, Sakai Y, Hattori M, Aizawa S, Nagashima KV, Masuda S, Hattori T, Yamashita A, Bao Z, Hayatsu M, Kajiya-Kanegae H, Yoshinaga I, Sakamoto K, Toyota K, Nakao M, Kohara M, Anda M, Niwa R, Jung-Hwan P, Sameshima-Saito R, Tokuda S, Yamamoto S, Yamamoto S, Yokoyama T, Akutsu T, Nakamura Y, Nakahira-Yanaka Y, Takada-Hoshino Y, Hirakawa H, Mitsui H, Terasawa K, Itakura M, Sato S, Ikeda-Ohtsubo W, Sakakura N, Kaminuma E, Minamisawa K. 2012. Complete genome sequence of Bradyrhizobium sp. S23321: insights into symbiosis evolution in soil oligotrophs. Microbes Environ 27:306–315. doi: 10.1264/jsme2.ME11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lozano L, Hernández-González I, Bustos P, Santamaría RI, Souza V, Young JPW, Dávila G, González V. 2010. Evolutionary dynamics of insertion sequences in relation to the evolutionary histories of the chromosome and symbiotic plasmid genes of Rhizobium etli populations. Appl Environ Microbiol 76:6504–6513. doi: 10.1128/AEM.01001-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Menna P, Hungria M. 2011. Phylogeny of nodulation and nitrogen-fixation genes in Bradyrhizobium: supporting evidence for the theory of monophyletic origin, and spread and maintenance by both horizontal and vertical transfer. Int J Syst Evol Microbiol 61:3052–3067. doi: 10.1099/ijs.0.028803-0. [DOI] [PubMed] [Google Scholar]
- 9.Kasai-Maita H, Hirakawa H, Nakamura Y, Kaneko T, Miki K, Maruya J, Okazaki S, Tabata S, Saeki K, Sato S. 2013. Commonalities and differences among symbiosis islands of three Mesorhizobium loti strains. Microbes Environ 28:275–278. doi: 10.1264/jsme2.ME12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parker MS. 2012. Legume select symbiosis island sequence variants in Bradyrhizobium. Mol Ecol 21:1769–1778. doi: 10.1111/j.1365-294X.2012.05497.x. [DOI] [PubMed] [Google Scholar]
- 11.Parker MA. 2015. The spread of Bradyrhizobium lineages across host legume clades: from Abarema to Zygia. Microb Ecol 69:630–640. doi: 10.1007/s00248-014-0503-5. [DOI] [PubMed] [Google Scholar]
- 12.Sameshima R, Isawa T, Sadowsky MJ, Hamada T, Kasai H, Shutsrirung A, Mitsui H, Minamisawa K. 2003. Phylogeny and distribution of extra-slow-growing Bradyrhizobium japonicum harboring high copy numbers of RSα, RSβ and IS1631. FEMS Microbiol Ecol 44:191–202. doi: 10.1016/S0168-6496(03)00009-6. [DOI] [PubMed] [Google Scholar]
- 13.Minamisawa K, Isawa T, Nakatsuka Y, Ichikawa N. 1998. New Bradyrhizobium japonicum strains that possess high copy numbers of the repeated sequence RSα. Appl Environ Microbiol 65:1845–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Isawa T, Sameshima R, Mitsui H, Minamisawa K. 1999. IS1631 occurrence in Bradyrhizobium japonicum highly reiterated sequence-possessing strains with high copy numbers of repeated sequences RSα and RSβ. Appl Environ Microbiol 65:3493–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodriguez-Quinones F, Judd AK, Sadowsky MJ, Liu RL, Cregan PB. 1992. Hyperreiterated DNA regions are conserved among Bradyrhizobium japonicum serocluster 123 strains. Appl Environ Microbiol 58:1878–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahillon J, Chandler M. 1998. Insertion sequences. Microbiol Mol Biol Rev 62:725–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minamisawa K, Itakura M, Suzuki M, Chige K, Isawa T, Yuhashi K, Mitsui H. 2002. Horizontal transfer of nodulation genes in soil and microocosms from Bradyrhizobium japonicum to B. elkanii. Microbes Environ 17:82–90. doi: 10.1264/jsme2.2002.82. [DOI] [Google Scholar]
- 18.Shiina Y, Itakura M, Choi H, Saeki Y, Hayatsu M, Minamisawa K. 2014. Correlation between soil type and N2O reductase genotype (nosZ) of indigenous soybean bradyrhizobia: nosZ-minus populations are dominant in Andosols. Microbes Environ 29:420–426. doi: 10.1264/jsme2.ME14130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sobral BW, Sadowsky MJ, Atherly AG. 1990. Genome analysis of Bradyrhizobium japonicum serocluster 123 field isolates by using field inversion gel electrophoresis. Appl Environ Microbiol 56:1949–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koren S, Schatz MC, Walenz BP, Martin J, Howard JT, Ganapathy G, Wang Z, Rasko DA, McCombie WR, Jarvis ED, Phillippy AM. 2012. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat Biotechnol 30:693–700. doi: 10.1038/nbt.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 25.Chaudhuri RR, Pallen MJ. 2006. xBASE, a collection of online databases for bacterial comparative genomics. Nucleic Acids Res 34:D335–D337. doi: 10.1093/nar/gkj140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohtsubo Y, Ikeda-Ohtsubo W, Nagata Y, Tsuda M. 2008. GenomeMatcher: a graphical user interface for DNA sequence comparison. BMC Bioinformatics 9:376. doi: 10.1186/1471-2105-9-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee KB, De Backer P, Aono T, Liu CT, Suzuki S, Suzuki T, Kaneko T, Yamada M, Tabata S, Kupfer DM, Najar FZ, Wiley GB, Roe B, Binnewies TT, Ussery DW, D'Haeze W, Herder JD, Gevers D, Vereecke D, Holsters M, Oyaizu H. 2008. The genome of the versatile nitrogen fixer Azorhizobium caulinodans ORS571. BMC Genomics 4:271–285. doi: 10.1186/1471-2164-9-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lobry JR. 1996. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol Biol Evol 13:660–665. doi: 10.1093/oxfordjournals.molbev.a025626. [DOI] [PubMed] [Google Scholar]
- 29.Bigot S, Saleh OA, Lesterlin C, Pages C, El Karoui M, Dennis C, Grigoriev M, Allemand JF, Barre FX, Cornet F. 2005. KOPS: DNA motifs that control E. coli chromosome segregation by orienting the FtsK translocase. EMBO J 24:3770–3780. doi: 10.1038/sj.emboj.7600835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blakely GW, Davidson AO, Sherratt DJ. 2000. Sequential strand exchange by XerC and XerD during site-specific recombination at dif. J Biol Chem 275:9930–9936. doi: 10.1074/jbc.275.14.9930. [DOI] [PubMed] [Google Scholar]
- 31.van Berkum P, Fuhrmann JF. 2000. Evolutionary relationships among the soybean bradyrhizobia reconstructed from 16S rRNA gene and internal transcribed spacer region sequence divergence. Int J Sys Evol Microbiol 50:2165–2172. doi: 10.1099/00207713-50-6-2165. [DOI] [PubMed] [Google Scholar]
- 32.Cevallos MA, Cervantes-Rivera R, Gutiérrez-Ríos RM. 2008. The repABC plasmid family. Plasmid 60:19–37. doi: 10.1016/j.plasmid.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 33.Cytryn E, Jitacksorn S, Giraudo E, Sadowsky MJ. 2008. Insights learned from pBTAi1, a 229-kb accessory plasmid from Bradyrhizobium sp. strain BTAi1 and prevalence of accessory plasmids in other Bradyrhizobium sp. strains. ISME J 2:158–170. doi: 10.1038/ismej.2007.105. [DOI] [PubMed] [Google Scholar]
- 34.Sugawara M, Sadowsky MJ. 2013. Influence of elevated atmospheric carbon dioxide on transcriptional responses of Bradyrhizobium japonicum in the soybean rhizoplane. Microbes Environ 28:217–227. doi: 10.1264/jsme2.ME12190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saeki Y, Shiro S, Tajima T, Yamamoto A, Sameshima-Saito R, Sato T, Yamakawa T. 2013. Mathematical ecology analysis of geographical distribution of soybean-nodulating bradyrhizobia in Japan. Microbes Environ 28:470–478. doi: 10.1264/jsme2.ME13079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okubo T, Tokida T, Ikeda S, Bao Z, Tago K, Hayatsu M, Nakamura H, Sakai H, Usui Y, Hayashi K, Hasegawa T, Minamisawa K. 2014. Effects of elevated carbon dioxide, elevated temperature, and rice growth stage on the community structure of rice root–associated bacteria. Microbes Environ 29:184–190. doi: 10.1264/jsme2.ME14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okazaki K, Iino T, Kuroda Y, Taguchi K, Takahashi H, Ohwada T, Tsurumaru H, Okubo T, Minamisawa K, Ikeda S. 2014. An assessment of the diversity of culturable bacteria from main root of sugar beet. Microbes Environ 29:220–223. doi: 10.1264/jsme2.ME13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ikeda S, Sasaki K, Okubo T, Yamashita A, Terasawa K, Bao Z, Liu D, Watanabe T, Murase J, Asakawa S, Eda S, Mitsui H, Sato T, Minamisawa K. 2014. Low nitrogen fertilization adapts rice root microbiome to low nutrient environment by changing biogeochemical functions. Microbes Environ 29:50–59. doi: 10.1264/jsme2.ME13110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takeshima K, Hidaka T, Wei M, Yokoyama T, Minamisawa K, Mitsui H, Itakura M, Kaneko T, Tabata S, Saeki K, Oomori H, Tajima S, Uchiumi T, Abe M, Tokuji Y, Ohwada T. 2013. Involvement of a novel genistein-inducible multidrug efflux pump of Bradyrhizobium japonicum early in the interaction with Glycine max (L.) merr. Microbes Environ 28:414–421. doi: 10.1264/jsme2.ME13057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giraud E, Moulin L, Vallenet D, Barbe V, Cytryn E, Avarre JC, Jaubert M, Simon D, Cartieaux F, Prin Y, Bena G, Hannibal L, Fardoux J, Kojadinovic M, Vuillet L, Lajus A, Cruveiller S, Rouy Z, Mangenot S, Segurens B, Dossat C, Franck WL, Chang WS, Saunders E, Bruce D, Richardson P, Normand P, Dreyfus B, Pignol D, Stacey G, Emerich D, Verméglio A, Médigue C, Sadowsky MJ. 2007. Legume symbioses: absence of Nod genes in photosynthetic bradyrhizobia. Science 316:1307–1312. doi: 10.1126/science.1139548. [DOI] [PubMed] [Google Scholar]
- 41.Okubo T, Fukushima S, Itakura M, Oshima K, Longtonglang A, Teaumroong N, Mitsui H, Hattori M, Hattori R, Hattori T, Minamisawa K. 2013. Genome anlaysis suggests that the soil oligotrophic bacterium Agromonas oligotrophica (Bradyrhizobium oligotrophicum) is a nitrogen-fixing symbiont of Aeschynomene indica. Appl Environ Microbiol 79:2542–2551. doi: 10.1128/AEM.00009-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanbe M, Yagasaki J, Zehner S, Göttfert M, Aizawa S. 2007. Characterization of two sets of subpolar flagella in Bradyrhizobium japonicum. J Bacteriol 189:1083–1089. doi: 10.1128/JB.01405-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Allocati N, Federici L, Masulli M, Di Ilio C. 2009. Glutathione transferases in bacteria. FEBS J 276:58–75. doi: 10.1111/j.1742-4658.2008.06743.x. [DOI] [PubMed] [Google Scholar]
- 44.Catanese CA, Emerich DW, Zahler WL. 1989. Adenylate cyclase and cyclic AMP phosphodiesterase in Bradyrhizobium japonicum bacteroids. J Bacteriol 171:4531–4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sachs JL, Russell JE, Hollowell AC. 2011. Evolutionary instability of symbiotic function in Bradryhizobium japonicum. PLoS One, 6:e26370. doi: 10.1371/journal.pone.0026370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu LM, Ge C, Cui Z, Li J, Fan H. 1995. Bradyrhizobium liaoningense sp. nov., isolated from the root nodules of soybeans. Int J Syst Bacteriol 45:706–711. doi: 10.1099/00207713-45-4-706. [DOI] [PubMed] [Google Scholar]
- 47.Batut J, Anderson SGE, O'Callaghan D. 2004. The evolution of chronic infection strategies in the α-Proteobacteria. Nat Rev Microbiol 2:933–945. doi: 10.1038/nrmicro1044. [DOI] [PubMed] [Google Scholar]
- 48.Siguier P, Goubeyre E, Chandler M. 2014. Bacterial insertion sequences: their genomic impact and diversity. FEMS Microbiol Rev 38:865–891. doi: 10.1111/1574-6976.12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.