Abstract
Cupriavidus pinatubonensis JMP134, like many other environmental bacteria, uses a range of aromatic compounds as carbon sources. Previous reports have shown a preference for benzoate when this bacterium grows on binary mixtures composed of this aromatic compound and 4-hydroxybenzoate or phenol. However, this observation has not been extended to other aromatic mixtures resembling a more archetypal context. We carried out a systematic study on the substrate preference of C. pinatubonensis JMP134 growing on representative aromatic compounds channeled through different catabolic pathways described in aerobic bacteria. Growth tests of nearly the entire set of binary combinations and in mixtures composed of 5 or 6 aromatic components showed that benzoate and phenol were always the preferred and deferred growth substrates, respectively. This pattern was supported by kinetic analyses that showed shorter times to initiate consumption of benzoate in aromatic compound mixtures. Gene expression analysis by real-time reverse transcription-PCR (RT-PCR) showed that, in all mixtures, the repression by benzoate over other catabolic pathways was exerted mainly at the transcriptional level. Additionally, inhibition of benzoate catabolism suggests that its multiple repressive actions are not mediated by a sole mechanism, as suggested by dissimilar requirements of benzoate degradation for effective repression in different aromatic compound mixtures. The hegemonic preference for benzoate over multiple aromatic carbon sources is not explained on the basis of growth rate and/or biomass yield on each single substrate or by obvious chemical or metabolic properties of these aromatic compounds.
INTRODUCTION
Aromatic compounds (AC) are widespread in the environment, displaying a heterogeneous structural diversity. They can be naturally originated by biotic and abiotic processes or released as pollutants into the environment. AC primarily can be found as aromatic amino acids, secondary products abundantly generated by plants, structural components of the very complex lignin heteropolymer in woody plants, and xenobiotic compounds: biocides, industrial by-products, and petroleum derivatives, among others. Microorganisms may degrade hundreds of different AC using specialized biochemical pathways that allow them to grow on these carbon sources (1–3). Typically, bacteria deal with AC as part of complex mixtures in naturally occurring organic compounds, such as those found in plant exudates (4), in soils (5), and even in dissolved organic matter from freshwater and seawater (6). Therefore, microorganisms are concurrently exposed to several structurally heterogeneous AC as potential substrates, which raises the question of whether the components of these mixtures are used simultaneously or in a sequential manner. In the case of the sequential utilization pattern, characterized by diauxic growth, one compound inhibits degradation of the other by exerting metabolite toxicity (7), competitive inhibition of enzymes (8, 9), depletion of electron acceptors (10, 11), or carbon catabolite repression (12, 13). The last phenomenon, which implies that the presence of the preferentially utilized compound represses the expression of genes involved in degradation of the alternative nonpreferred substrate, has been extensively studied using sugars, amino acids, and organic acids as representative of preferred carbon sources in aerobic bacteria (12, 13) and, most recently, has been reported in anaerobic species as well (14, 15). The hierarchical utilization of binary mixtures of AC has also been studied but much less extensively, and these studies focused mostly on substrates that are metabolized by closely related catabolic pathways. The degradation of mixtures of benzoate (Bz) and phenol (Phe), both converted into catechol to be subsequently channeled into the β-ketoadipate pathway by ortho ring cleavage, has been studied in Acinetobacter species (16, 17), pseudomonads (18), and Ralstonia eutropha (19), showing a sharp pattern for the preferential utilization of Bz. The molecular mechanism underlying the inhibition of Phe consumption in this mixture has not been clarified yet. Moreover, molecular studies on the hierarchical utilization of mixtures of AC have been performed mostly on Bz and 4-hydroxybenzoate (4-Hb) mixtures, where different branches of the β-ketoadipate pathway are used to metabolize these single components (20). The inhibition of 4-Hb degradation by Bz has been studied in Pseudomonas putida PRS2000 (21, 22) and Acinetobacter baylyi ADP1 (23), clearly establishing that repression acts at the transcriptional level in these gammaproteobacteria. It has been suggested that in both species, catabolite repression would be mediated by transcriptional regulators of Bz degradation and focused on the pcaK gene, encoding the 4-Hb permease (21, 23). The repression of 4-Hb degradation by Bz in the betaproteobacterium Cupriavidus pinatubonensis JMP134 has also been reported, opening new opportunities to study the use of mixtures of AC in metabolically versatile bacteria (24).
Cupriavidus pinatubonensis JMP134 (25), formerly Alcaligenes eutrophus, Ralstonia eutropha, Wautersia eutropha, and Cupriavidus necator, utilizes roughly 60 different AC as sole energy and carbon sources, and its genome encodes nearly the whole set of degradation pathways of AC reported for the Proteobacteria (26, 27). This is remarkable but not unusual, since several other members of the Proteobacteria and Actinobacteria possess numerous catabolic abilities (3, 28), and in particular, several members of the Burkholderiales group carry an extremely large number of AC catabolism genes (28, 29). C. pinatubonensis and related betaproteobacterial strains have a restricted potential to degrade sugars and small organic acids compared to their ability to degrade AC (27), and therefore, the question of the preference in mixtures of AC is even more relevant with these specialized strains. The study of the preference for Bz over 4-Hb in this strain revealed some interesting features of the catabolite repression phenomenon in mixtures of AC, given that it targets the pobA gene, encoding the first step in 4-Hb degradation, and is mediated by an interaction between Bz and PobR, the transcriptional activator of pobA (24). One of the most striking traits of the reported regulatory circuit is that no Bz turnover is required to exert a strong repression over the target promoter, suggesting that this phenomenon is triggered by a signal unrelated to the metabolism of Bz. This mechanism is also noticeably different from the one mentioned above for the gammaproteobacterial species P. putida and A. baylyi (21, 23).
The unquestionable and marked preponderance of Bz as the preferred substrate in the previously reported systems prompted us to study whether such a prevailing role extends to binary mixtures with AC catabolized by routes other than the β-ketoadipate pathway, in order to establish if such a hierarchy is only a peculiar trait of this route or a global attribute in metabolically versatile bacteria. In addition, studies to clarify if patterns of sequential consumption are maintained in complex mixtures of AC must be addressed, since this is a more realistic approach to understanding bacterial catabolism in nature, where the pool of available substrates is usually highly diverse in composition. We performed a systematic analysis of substrate preferences in C. pinatubonensis with eight AC degraded via the main AC catabolic pathways reported for members of the Proteobacteria (3, 26, 28) (see Fig. 1)—Bz, 4-Hb, Phe, phenylacetate (Pac), 3-hydroxybenzoate (3-Hb), 4-hydroxyphenylacetate (4-Hpa), tyrosine (Tyr), and 2,4-dichlorophenoxyacetate (2,4-D)—establishing the hierarchical relations among them, assessing their transcriptional patterns, and providing a glimpse of the mechanisms supporting these interactions.
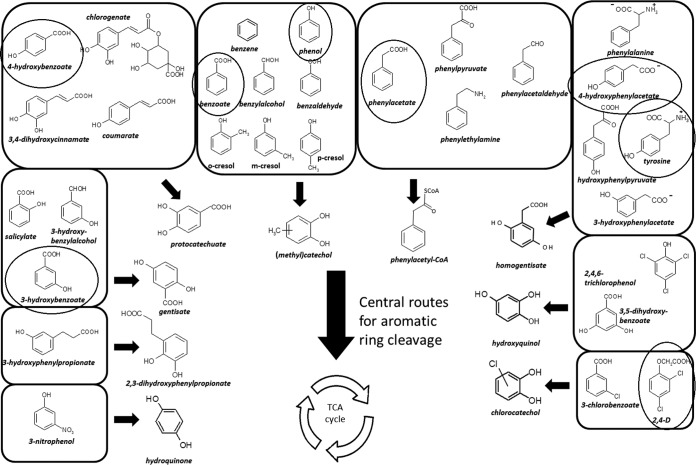
FIG 1.
Main catabolic pathways of aromatic compounds reported for Proteobacteria. A selection of aromatic compounds that are used as carbon and energy sources by the betaproteobacterium Cupriavidus pinatubonensis JMP134 (26) is shown in squares, as well as the corresponding aromatic intermediates that are further metabolized by the respective ring cleavage pathways reported for Proteobacteria (3, 26, 29). Encircled structures correspond to aromatic compounds used in this study. TCA cycle, tricarboxylic acid cycle.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
C. pinatubonensis JMP134 and its ΔbenA mutant (24) were grown at 30°C in mineral salts medium (30) supplemented with 5 mM Bz, 4-Hb, 3-Hb, Pac, 4-Hpa, or Tyr, with 2 mM 2,4-D or Phe, or with 10 mM fructose. These concentrations of AC were used in cultures with single and binary supplements. Some growth tests were performed at other concentrations or with five- or six-member mixtures. Controls without AC were routinely run. E. coli Mach (Invitrogen, Carlsbad, CA) was grown at 37°C in a Luria-Bertani medium. Growth was measured at optical density at 600 nm (OD600). At least three replicates were performed for each growth test.
Quantification of transcript levels by quantitative real-time RT-PCR.
Cells of C. pinatubonensis JMP134 were grown overnight in a minimal medium with fructose to inoculate a fresh culture medium, further growing the cells until the OD600 was equal to 0.7. Then they were induced with 5 mM Bz, 3-Hb, 4-Hb, 4-Hpa, Pac, or Tyr, with 2 mM Phe and 2,4-D, or with a mixture of Bz and the other AC and incubated for 1 h. Controls without the addition of AC were also performed. Then, total RNA was obtained from 4-ml cultures of cells, using the bacterial reagent RNAprotect and the RNeasy minikit (Qiagen, Chatsworth, CA, USA). The RNA was quantified using an Eon microplate spectrophotometer (Biotek, Winooski, VT, USA) and treated with the Turbo DNase kit (Ambion, Austin, TX, USA) to remove DNA contamination. Reverse transcription was performed using the ImProm-II reverse transcription system (Promega Corporation, Madison, WI, USA) with 1 μg of RNA in 20-μl reaction mixtures. Real-time reverse transcription-PCR (RT-PCR) was performed using Power SYBR green PCR master mix (Applied Biosystems, Warrington, United Kingdom) and the Eco real-time PCR detection system (Illumina, San Diego, CA, USA). The PCR mixture (15 μl) contained 3.0 μl of template cDNA (diluted 1:10) and a 0.2 μM concentration of each primer. Amplification was performed under the following conditions: 95°C for 10 min, followed by 40 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 40 s, followed by a melting cycle from 55 to 95°C. Relative gene expression calculations were conducted as described in the software manufacturer's instructions: an accurate ratio between the expression of the gene of interest (GOI) and the housekeeping (HK) reference gene was calculated according to the following expression: 2−[ΔCT(GOI) − ΔCT(HK)]. Then, gene expression levels were normalized to the average value of the gene expression determined in the noninduced treatment. The rps7 gene (Reut_A3184) was used as the HK gene. Primer pairs for benA (Reut_B4403), pobA (Reut_B5020), mhbM1 (Reut_B5861), mhbM2 (Reut_B5805), hmgA (Reut_B3923), tfdA (Reut_D6479), tyrB (Reut_B4503), hpaX (Reut_B4218), paaK1 (Reut_B3741), paaA1 (Reut_B3735), paaA2 (Reut_A3206), paaK2 (Reut_A3017), phlK1 (Reut_A1700), and phlK2 (Reut_B5680) are listed in Table 1. All experiments were performed in two biological and two technical replicates.
TABLE 1.
Primer pairs used for real-time RT-PCR
| Forward primer | Sequence (5′→3′) | Reverse primer | Sequence (5′→3′) |
|---|---|---|---|
| tfdAFw | CATCTCGAACGTCAGTCTCG | tfdARv | GGTTGCTGAAAGGAGCTGTC |
| benAFw | GGCTCGTCCACCTACACCTA | benARv | GCTCGTTCTGCTGCTTGC |
| pobAFw | ATCTACGCCAGCCACGAA | pobARv | GCGAGTTCATCCCAGAAGC |
| mhbM1Fw | GCCGTCTTTACCGACTACCT | mhbM1Rv | CGAGAATCGACAGATGGATG |
| mhbM2Fw | AGACATGCCAAAGGACTTGC | mhbM2Rv | TGACGACGAGGTTGTACTGC |
| paaK1Fw | GCCAGACCGAGAAACAGG | paaK1Rv | GCCGAAGATGCCAACCTT |
| paaA1Fw | ACGCACAGTTGGTGGATACC | paaA1Rv | CAGTCCTTCGCTTCGATCTT |
| paaK2Fw | ACTATCCGTTCGGCATGTTC | paaK2Rv | CGATGTCCTTGAGCGTGTAA |
| paaA2Fw | CGGACACGGCCTCTATCTCT | paaA2Rv | CGTCGGGTAGTTGAAGATCG |
| hpaFw | GCTCGCTTATGTGCTGGAC | hpaRv | CTCTTGGGCGTGATGAAATC |
| phlL1Fw | AACCTGGAGTTCGTGGAGAA | phlL1Rv | GGAGCCAAAGCCATACGC |
| phlK2Fw | CGGAGTTGAGCGTAGACCTG | phlK2Rv | TCAGTTTTCCAGCCTTGTCA |
| tyrBFw | CGATCCTTTCGCTCAACG | tyrBRv | CCTTTTCGACAGCTTCCATG |
| hmgAFw | AAGTTCCAGGGCAATCTGTG | hmgARv | TACTTGTACGGCGCGAAGTT |
| rpS7Fw | GAAAAGAAGGCAGGCAAGG | rpS7Rv | TTCGACCGGAACCTGATAGT |
Analytical methods.
AC were detected by high-performance liquid chromatography using cell-free supernatants from cells grown on single AC or mixtures of AC. Samples (20 μl) were obtained at different times of the growth curve and injected into a Hitachi LaChrom Elite chromatograph equipped with an L-2130 pump, an L-2455 diode array detector, an L-2200 autosampler, and a Kromasil 100-3.5 C18 4.6-μm-diameter column. For binary and unitary curves, a methanol-H2O (60:40) mixture (except for the mixtures Bz-Pac, 4-Hb–3-Hb, 3-Hb–4-Hpa [40:60], and 4-Hb–4-Hpa [20:80]) containing 0.1% (vol/vol) phosphoric acid was used as the solvent at a flow rate of 1 ml min−1. The column effluent was monitored at 210 nm for all AC. Retention times for the methanol-H2O (60:40) mixture were as follows: Bz, 3.2 min; 4-Hb, 1.8 min; 3-Hb, 2.0 min; Pac, 3.1 min; 4-Hpa, 1.9 min; 2,4-D, 8.0 min; Phe, 2.6 min; and Tyr, 1.4 min. For the methanol-H2O (40:60) mixture, retention times were as follows: Bz, 3.8 min; 4-Hb, 2.8 min; 3-Hb, 3.4 min; Pac, 3.5 min; and 4-Hpa, 2.7 min. Retention times for the methanol-H2O (20:80) mixture were 8.8 min for 4-Hb and 10.0 min for 4-Hpa. The six- and five-AC mixture samples were eluted with a mobile phase of 15% methanol, 20 mM acetate, and MilliQ water, at pH 3.3.
Chemicals.
AC were purchased from Aldrich Chemical (Milwaukee, WI) except Bz, Tyr, and fructose, which were purchased from Merck (Darmstadt, Germany).
Statistical analysis.
Data were statistically analyzed using one-way analysis of variance. When analysis of variance showed significant effects, Tukey's honestly significant difference (HSD; P < 0.05) test was applied.
RESULTS
Bz and Phe are always the preferred and the deferred growth substrate, respectively, when C. pinatubonensis grows in AC binary mixtures.
To extend the study on the substrate preference of C. pinatubonensis to binary mixtures other than the previously reported Bz–4-Hb combination, catabolized through catechol and protocatechuate ortho ring cleavage pathways, respectively, six additional AC were selected, which are degraded through other key catabolic pathways reported in bacteria (Fig. 1). The substrates were 3-Hb (catabolized through the gentisate pathway), Pac (catabolized via the phenylacetyl coenzyme A [CoA] pathway), 4-Hpa (channeled via 1-hydroxylation into the homogentisate ring cleavage pathway), 2,4-D (degraded via the chlorocatechol ortho ring cleavage pathway), Phe (degraded via the catechol meta and ortho ring cleavage pathways), and Tyr (channeled via 4-hydroxyphenylpyruvate into the homogentisate ring cleavage pathway). The corresponding 28 binary mixtures were scored for bacterial growth and percent substrate removal. Phe and 2,4-D were tested at 2 mM to prevent toxic effects. Phe itself, its catabolic intermediate catechol, and the 2,4-D intermediates 2,4-dichlorophenol and chlorocatechols have been reported to be toxic through mechanisms such as oxidative phosphorylation uncoupling, enhancement of Fenton reaction, and DNA adduct formation, among others (19, 31–34). Controls with the respective single AC were carried out to make proper kinetic comparisons.
Bz was always the preferred substrate in every mixture (Fig. 2A shows results for Bz-Pac as an example; results for all other mixtures containing Bz are shown in Fig. S1 in the supplemental material); i.e., only once was Bz substantially degraded (as judged by percent removal) and the degradation of the other AC started. This lag in onset of degradation by this strain was better observed when the time course of removal in the mixture was compared with those of the respective single cultures with AC (Fig. 2A; also, see Fig. S1). In contrast, Phe was always the deferred growth substrate in these binary mixtures (Fig. 2B shows results for the 4-Hb–Phe combination as an example; results for the other Phe-containing mixtures are shown in Fig. S1). All other combinations, i.e., those where Bz or Phe were absent, showed no clear preference for any member of the binary mixture (Fig. 2C shows results for the 3-Hb–4-Hpa combination as an example; see results for the remaining mixtures in Fig. S1).
FIG 2.
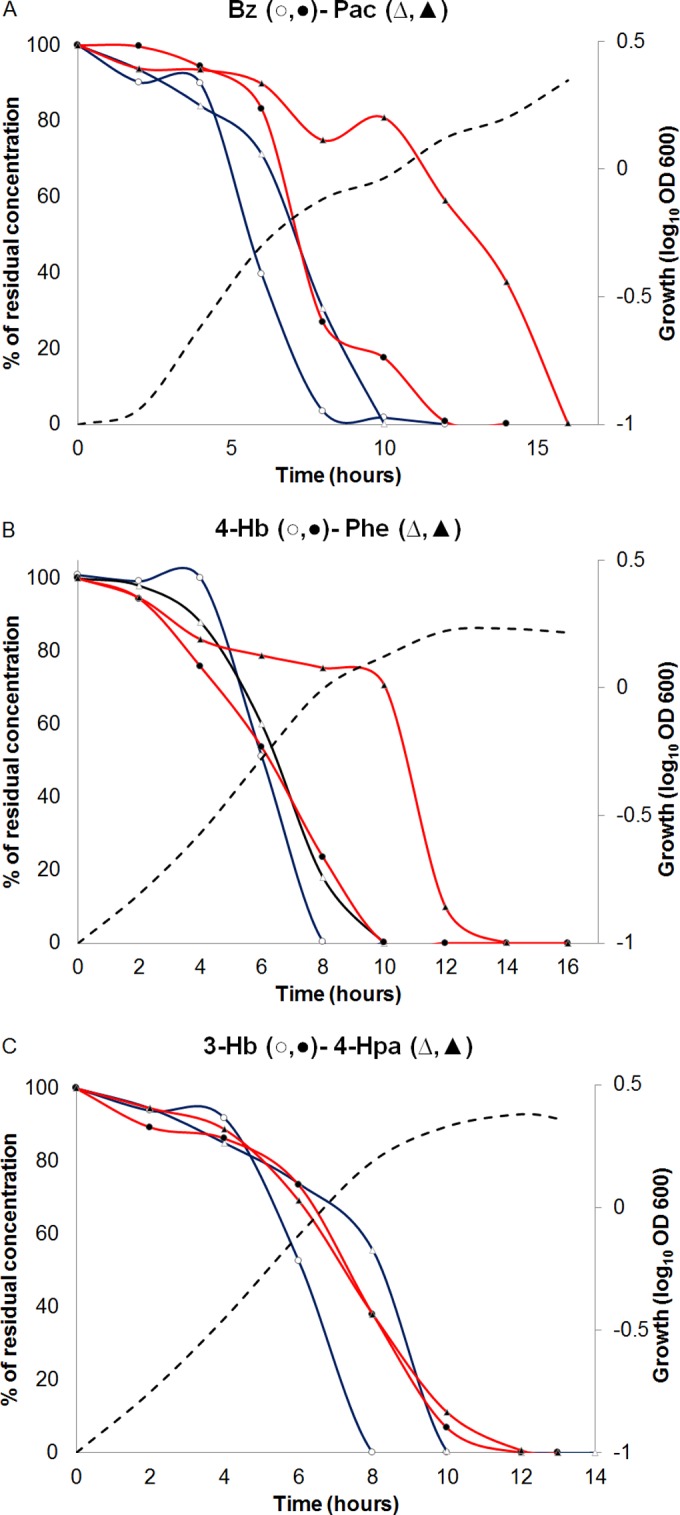
Growth and carbon source degradation curves of Cupriavidus pinatubonensis JMP134 grown on binary mixtures of aromatic compounds and the corresponding carbon source degradation in the respective single-compound cultures. Except for phenol (2 mM), all aromatic compounds were tested at 5 mM in single-compound and binary-mixture cultures. Note that the final growth yields reflect different amounts of added carbon. Open and closed symbols represent substrate removal in cultures grown with single compounds and binary mixtures, respectively. Dashed lines represent growth levels in binary mixtures, determined by OD600 measurements. Plots show representative curves from 4 to 6 biological replicates. Standard deviations of technical replicates were lower than 5% and are not shown for clarity.
To better determine growth substrate removal in mixtures, two parameters were defined and tracked: (i) degradation time overlaps, expressed as the percentage of the total degradation time of the growth substrate whose removal began first, when the catabolism of the other substrate was also occurring, and (ii) degradation start time, corresponding to the time when removal of each compound was first detected. Overlaps in Bz degradation times in binary mixtures with 4-Hb, 3-Hb, Pac, or Phe were near or lower than 10%; i.e., only in the last 10% of the Bz degradation period were the other AC also degraded (see Fig. S2A in the supplemental material). On the hand, in every mixture containing Phe, the degradation of this substrate never started before that of the partner AC (no values in the Phe row in Fig. S2A). Bz exerted the strongest preference over Phe catabolism (12% degradation time overlap) compared with the other six AC that were favored by C. pinatubonensis. In the mixtures of AC where Bz and/or Phe was absent, around 50% or higher overlap values were always observed (see Fig. S2A), indicating that no preference was the more common metabolic trait. Finally, a nonspecific behavior was observed with 2,4-D, given that whether or not its degradation ended before (4-Hb, 3-Hb, Pac, or 4-Hpa) or after (Bz, Phe, or Tyr) that of the other AC (see Fig. S1), >50% overlap values were always found (Fig. S2A).
In addition, the analysis of degradation start times shows that Bz consistently delayed the start time of catabolism of the other AC. Delays ranged from 1.6-fold for Tyr to 3.4-fold for 3-Hb (see Fig. S2B in the supplemental material; compare values in the first column [Bz] with those for the single-AC-supplemented mixtures [gray boxes]). In contrast, the Phe degradation start time (3.2 h for the single compound) was always retarded (from 1.8-fold with Bz to 2.5-fold with Tyr [see Fig. S2B]). Tyr also delayed the degradation start time of other AC, whereas, in general, the other AC did not significantly modify the degradation start time of the other member of the binary mixtures (see Fig. S2B).
The Bz-Phe mixture was further explored, determining the range of Bz concentrations (0.1 to 4 mM) that delayed the use of a fixed Phe concentration (1 mM). The results indicated that as little as 0.25 mM Bz was enough to fully express the preference for Bz, given that only in the 0.1 mM Bz–1.0 mM Phe mixture did Phe degradation start when about half of the initial Bz was still present (data not shown).
Bz exerts repression at different levels on transcription of the genes encoding initial steps of degradation of AC in binary-mixture cultures of C. pinatubonensis.
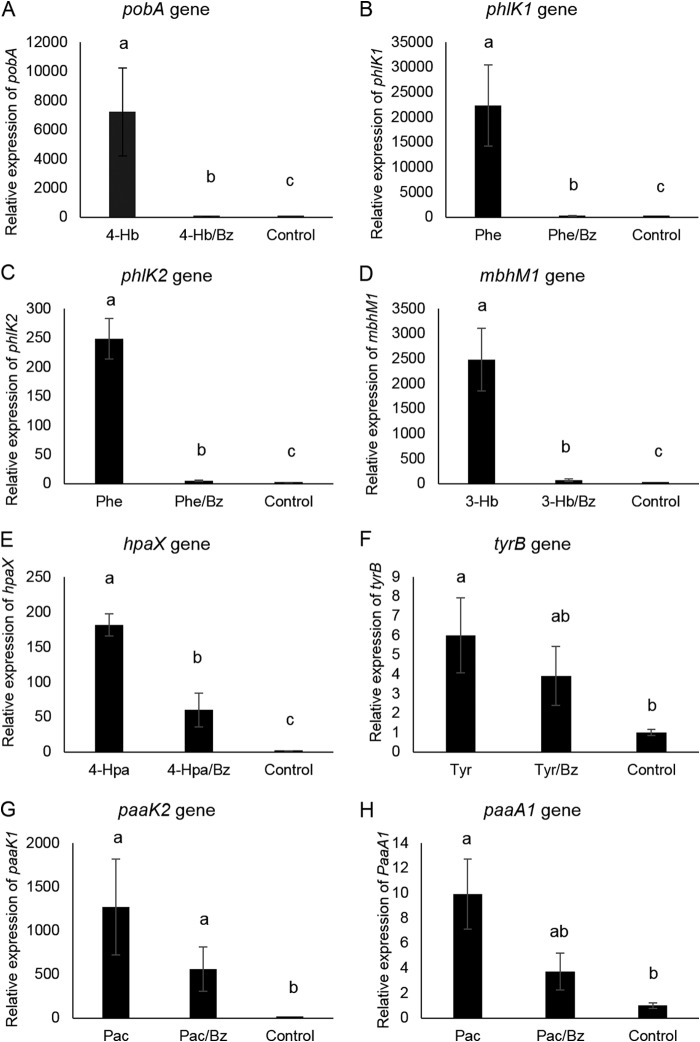
To determine if the mechanism involved in Bz preference was based on transcriptional control, we then carried out a real-time RT-PCR analysis to compare the expression of key degradation genes of AC, in the absence/presence of Bz. The experimental setup consisted of C. pinatubonensis cells previously grown on a nonrepressive carbon source (fructose [data not shown]) and exposed to Bz, to the partner AC, and to the mixture of both. Transcript levels were normalized to that of the rps7 housekeeping reference gene. As previously reported (24), the presence of Bz completely inhibited pobA gene expression (Fig. 3A), encoding the initial monooxygenase converting 4-Hb into protocatechuate (see Fig. S3A in the supplemental material), reflecting that Bz preference is based on transcriptional repression. Bz also completely repressed transcription of phlK1 and phlK2 (Fig. 3B and C), encoding homologous subunits of Phe hydroxylases performing the conversion of Phe to catechol (see Fig. S3B), which is further channeled into ortho and meta ring cleavage pathways, respectively (see Fig. S3B). The promoter activities of both genes, measured in lacZ transcriptional fusions, were determined at two Bz concentrations (0.2 and 2.0 mM), one exerting loose and the other tight Bz preference over Phe (see above). Bz at 2.0 mM significantly prevented expression of both promoters in C. pinatubonensis cells growing on Bz-Phe mixtures, whereas 0.2 mM Bz did so slightly (see Fig. S4 in the supplemental material).
FIG 3.
Relative transcript levels of key genes encoding initial steps in the degradation of aromatic compounds by Cupriavidus pinatubonensis JMP134. Fructose-grown cells (OD600 = 0.7) were exposed to each single compound (5 mM Bz, 3-Hb, 4-Hb, 4-Hpa, Pac, or Tyr and 2 mM phenol or 2,4-D) of the binary mixture and to the corresponding binary mixture (same individual concentrations) for 1 h. Values (from three biological and two technical replicates) are averages and standard errors, normalized with respect to the rps7 housekeeping reference gene. Values from inductions with benzoate were negligible and are not shown for clarity. No aromatic compounds were added in controls. Note that different scales are used. Different letters indicate statistically significant differences between values (one-way analysis of variance [ANOVA] and Tukey's HSD test; P < 0.05).
Bz also repressed expression of the mhbM1 gene (Fig. 3D), encoding the monooxygenase transforming 3-Hb into gentisate (see Fig. S3C in the supplemental material). The paralogous mhbM2 gene (see Fig. S3C) was not induced by 3-HB (data not shown) and was therefore not tested. Partial repression by Bz was also observed for expression of hpaX (Fig. 3E), encoding the channeling of 4-Hpa into the homogentisate pathway (see Fig. S3D). Bz did not decrease the expression of the tyrB gene (statistically significant) (Fig. 3F), encoding the initial transformation of Tyr through the homogentisate pathway (see Fig. S3D); furthermore, Bz did not diminish the expression of the hmgA gene (data not shown), encoding the homogentisate dioxygenase (see Fig. S3D), in Bz–4-Hpa or Bz-Tyr mixtures. Similarly, Bz did not affect the expression of the tfdA gene (data not shown), encoding the initial step in 2,4-D degradation. Finally, there was no decrease in expression levels of the paaK1 gene (Fig. 3G) or the paaA1 gene (Fig. 3H), encoding the first and the second steps of the phenylacetyl-CoA ring cleavage pathway, respectively (see Fig. S3E). The paralogous genes paaK2 and paaA2 (see Fig. S3E) were induced at low levels by Pac (data not shown) and, similar to paaK1 and paaA1, their transcript levels were not affected by Bz (data not shown).
To determine if AC, whose degradations were delayed by Bz, activated the expression of the benA gene to some extent, thus favoring Bz degradation, the corresponding quantitative real-time PCR analysis was performed. As expected, Bz triggered benA gene transcription when present in mixtures with the other AC (Fig. 4A). Apparent increases in benA mRNA levels observed in Pac-Bz and Phe-Bz mixtures were not statistically significant, and the rest of the AC tested did not increase benA mRNA levels to any extent (Fig. 4A; also see the expanded data in Fig. 4B).
FIG 4.
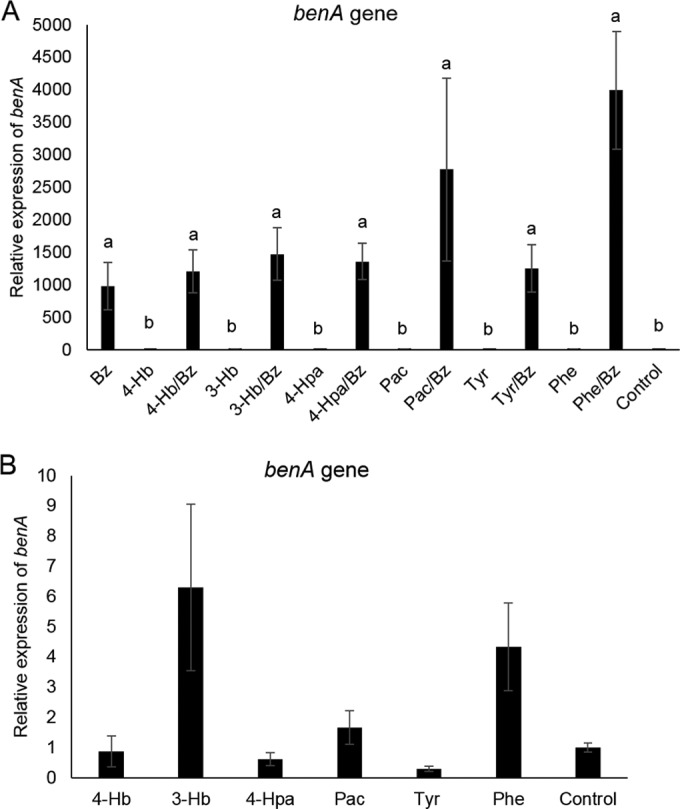
(A) Relative transcript levels of benA, encoding the initial step of benzoate (Bz) degradation by Cupriavidus pinatubonensis JMP134, exposed to binary Bz-containing mixtures of aromatic compounds (AC) or single AC. (B) Expanded data for single AC other than Bz. Fructose-grown cells (OD600 = 0.7) were exposed to the binary mixture (a 5 mM concentration of each AC, except 2 mM phenol) or the single AC (5 mM, except 2 mM phenol) for 1 h. Values (three biological and two technical replicates) correspond to the average and standard error, normalized with respect to the rps7 housekeeping reference gene. Different letters indicate statistically significant differences between values (one-way ANOVA and Tukey's HSD test; P < 0.05).
More than one mechanism probably underlies the preference for Bz of C. pinatubonensis.
As reported previously, inhibition of the activity of the key regulator PobR by Bz molecules per se provokes transcriptional repression of the pobA gene, which controls the first step in 4-Hb degradation by C. pinatubonensis (24). To see if analogous mechanisms also explained the preference for Bz over the other AC, we performed growth tests using a C. pinatubonensis ΔbenA mutant, which is completely unable to grow and to remove Bz due to its lack of the initial dioxygenase involved in Bz degradation. As expected, the nondegraded Bz fully prevented degradation of 4-Hb and, therefore, growth on Bz–4-Hb mixtures (Fig. 5A). Remarkably, the C. pinatubonensis ΔbenA mutant exhibited a significant delay in growth on Bz-Phe mixtures (Fig. 5B), although the final biomass yields for the single-compound and the binary-mixture cultures were essentially similar. It should be noted that toxic effects may explain the delayed growth of this mutant, and final yields in these Bz-Phe mixtures would be recovered by adaptation or mutation. A third behavior was found in that the C. pinatubonensis ΔbenA mutant showed low but noticeable growth delays with Bz–4-Hpa (Fig. 5C), Bz–3-Hb, Bz-Pac, and Bz-Tyr mixtures (data not shown). When cultured in 2,4-D or the Bz–2,4-D mixture, this mutant did not show any difference in lag phase or final biomass yields (data not shown), indicating that different molecular mechanisms may underlie the preference for Bz, because the exclusive presence of nonmetabolized Bz was not always enough for full repression by this compound (with the exception of 4-Hb), and suggesting that the formation of Bz intermediates and/or the operation of downstream enzymes of the catabolic route may be required for this phenomenon.
FIG 5.
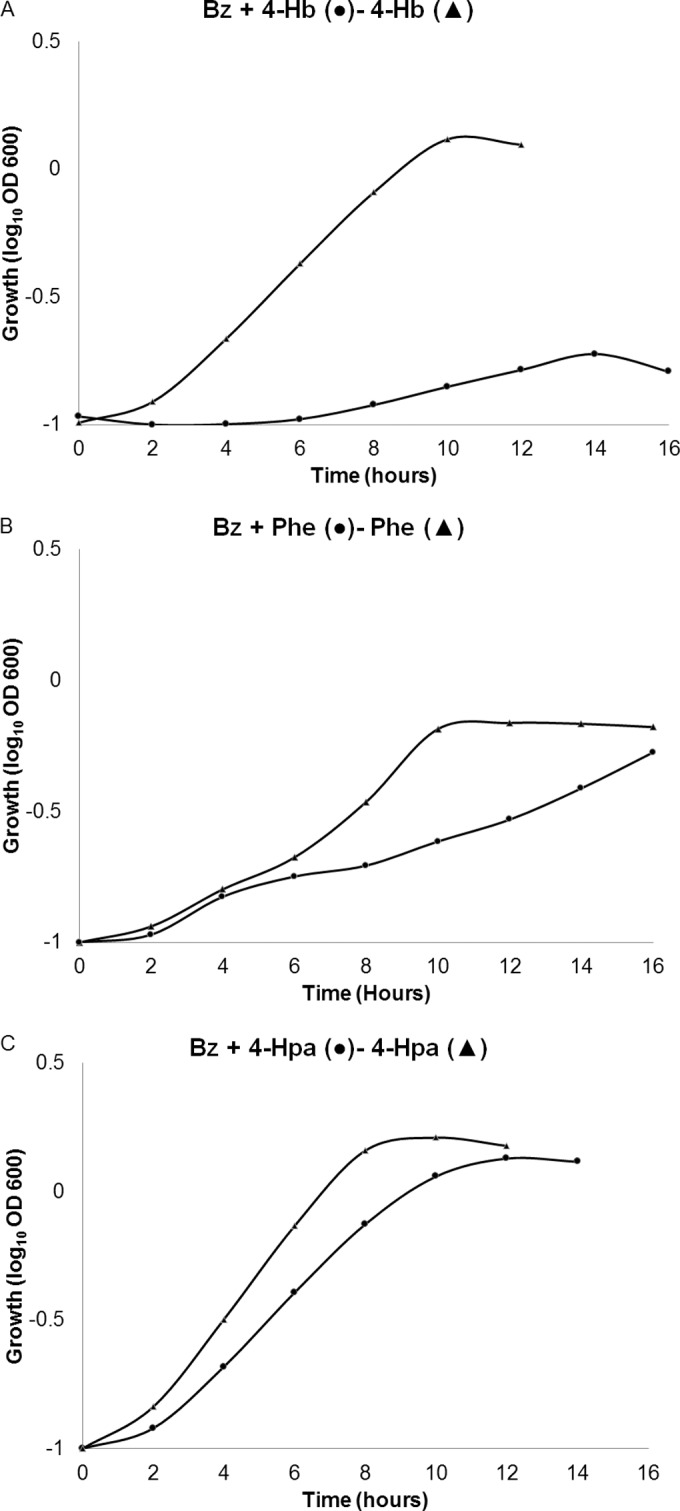
Growth curves of the Cupriavidus pinatubonensis JMP134 ΔbenA mutant grown on benzoate containing binary mixtures of aromatic compounds (2 mM for phenol and 5 mM for 4-hydroxybenzoate and 4-hydroxyphenylacetate) (circles) and the corresponding other single compound (same concentrations) (triangles). Note that final growth yields reflect different amounts of added carbon. Plots show representative curves from at least three biological replicates.
C. pinatubonensis also prefers Bz in multiple-member, low-concentration mixtures.
To better characterize the preference for Bz by C. pinatubonensis, six-member mixtures of AC (Bz, 4-Hb, 3-Hb, Pac, 2,4-D, and Tyr) were used to test growth and degradation. Three low concentrations (50, 100, and 250 μM [each mixture component]) were used to prevent toxicity of summed individual concentrations of AC and, more importantly, to test if Bz preference was also observed at lower concentrations. At these concentrations, cultures with single AC showed low but measurable growth; bacterial cell yields were essentially proportional to the amount of the carbon source added, and complete removals of AC were recorded (data not shown). The low-concentration binary mixtures containing Bz still showed a preference for this compound, but this behavior was more clearly observed in 250 μM than in 100 μM mixtures and practically absent in 50 μM mixtures (data not shown). Notably, the preference for Bz was also clearly observed in six-member-mixture cultures at 100 μM (Fig. 6A) and 250 μM (data not shown), but no clear trend was observed with 50 μM mixtures (data not shown). Five-member-mixture cultures lacking Bz showed no preference for any AC at 250 μM (Fig. 6B), 100 μM, or 50 μM mixture concentrations (data not shown).
FIG 6.
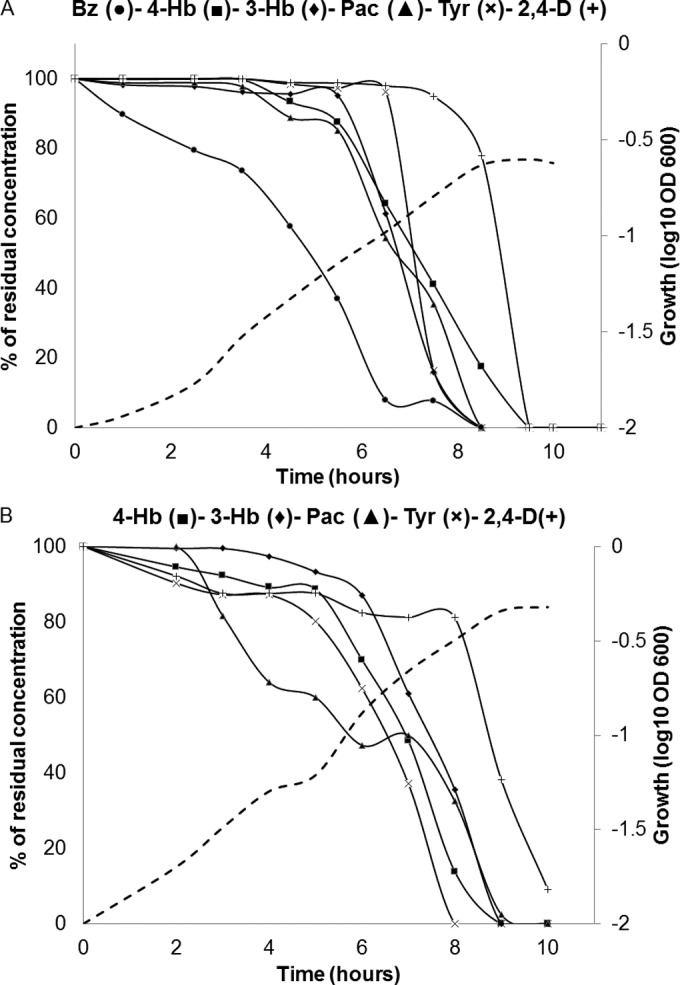
Growth and carbon source degradation by Cupriavidus pinatubonensis JMP134 grown on five- or six-member mixtures of aromatic compounds. The six-member mixture (A) consisted of benzoate, 3- and 4-hydroxybenzoate, phenylacetate, tyrosine, and 2,4-dichlorophenoxyacetate (100 μM each), whereas the five-member mixture (B) (250 μM each) lacked benzoate. Continuous lines represent substrate removal of each aromatic compound. Dashed line represent growth levels determined by OD600 measurements. The curve is representative of 3 to 5 different experiments. Standard deviations for technical replicates were lower than 5% and are not shown for clarity.
It is worth noting that, compared with results obtained in cultures with single AC, the half-lives of Bz, 4-Hb, and 3-Hb decreased in low-concentration six-member mixtures containing Bz (although such an effect was not observed at 250 μM), whereas those of Pac, Tyr, and 2,4-D increased (see Table S1 in the supplemental material). This suggests that Bz negatively affects the degradation of the other AC at lower concentrations, especially at 250 μM. In agreement with this, five-member-mixture cultures lacking Bz showed that the five AC exhibited quite similar and constant half-lives (see Table S1). Cultures of the C. pinatubonensis ΔbenA mutant in low-concentration six-member mixtures showed that the continuous presence of Bz again increased their half-lives while significantly decreasing those of Tyr at 50 μM (see Table S1, gray rows). The absence of Bz in five-member-mixture cultures of this mutant tends to homogenize half-lives in a manner similar to that observed for the five-member-mixture cultures of the wild-type strain (see Table S1), suggesting a role for Bz itself in these effects. In addition, chemostat cultures fed with this six-member mixture (200 μM each) showed essentially complete and similar degradation levels at different dilution rates (see Table S2 in the supplemental material), except for the highest dilution rate (condition 3), which was close to system washout (condition 4), where only Bz and Tyr were fully degraded.
DISCUSSION
This work consistently showed that Bz is always degraded preferentially and that, in contrast, Phe is permanently the deferred substrate in mixtures that include AC representing catabolic pathways other than that of β-ketoadipate (3, 28, 29). No clear preference in substrate utilization was found in combinations of AC lacking Bz or Phe, indicating that simultaneous catabolism is the prevailing trait when C. pinatubonensis is exposed to mixtures of AC. A key factor to be considered in the study of sequential aromatic catabolism is the component amounts in the mixtures of AC, since the utilization profile would be substrate concentration dependent. In this bacterium, the identified preferences were observed in a range of concentrations higher than those that can be found in most natural, nonpolluted environments (35–38). However, it is still possible that preference also takes place at lower concentrations and that the utilized growth and degradation tests do not have enough resolution to detect it. In any case, the results reported here indicate a key nutritional and/or regulatory role of Bz for microbial communities in natural environments.
Bz preference over other AC has been shown in several cases (16, 17, 21–24, 39, 40). In addition, Bz is also degraded first by C. pinatubonensis in mixtures with acetate (41) and is preferred over succinate in “Aromatoleum aromaticum” EbN1 (42) and over 4-methylbenzoate and succinate in Magnetospirillum sp. strain pMbN1 (43). Succinate catabolism repression in strain EbN1 was specific for Bz and not a general feature of AC, since Pac and 4-Hb were simultaneously degraded in mixtures with this C4 compound (42). Altogether, it seems clear that Bz is a distinctive preferred substrate for several soil bacteria. There are, of course, examples of simultaneous degradation in Bz-containing mixtures (14, 44), including synergic interactions such as those described in Sagittula stellata E-37, a member of the Roseobacter lineage, where increased growth rates are found for Bz–4-Hb mixtures (45).
There are several possible explanations for the preference for Bz over AC, none of them fully satisfactory. It can be proposed that Bz is more toxic than the other AC, so cells need to degrade it faster for detoxification purposes. However, a comparison of toxicity data for the AC used here indicates that Bz has toxic levels quite similar to those of all other AC except 2,4-D and Phe (see Table S3 in the supplemental material). Even if toxic catabolic intermediates are taken into consideration, such a possibility does not seem to be the case, since catechol formed during Bz catabolism (Fig. 1; also, see Fig. S3A in the supplemental material)—the most toxic of the intermediates generated (31, 33, 34)—is also produced from Phe (Fig. 1; also, see Fig. S3B).
Another possibility is that Bz interferes with the inward transport of the other AC. Such a mechanism of degradation repression is operative in P. putida and A. baylyi (21, 23). However, this is not probable, because uptake of AC takes place without the need for specific transporters at the concentrations used here (20, 46, 47). Bz control by activation of a putative outward transport system (e.g., an efflux pump) is also a possibility, but the little available evidence seems to indicate a role in antibiotic resistance, at least for a salicylic acid efflux pump (48). Permeability comparisons (XlogP3 values [see Table S3 in the supplemental material]) do not provide any additional clues as to differences that might explain the substrate preference through a transport mechanism.
Our results do not show a direct relationship among growth parameters and substrate preferences (compare half-lives for single compounds in Table S1 in the supplemental material). Growth yields, determined as milligrams of cells per millimole of added carbon in 0.25 to 8 mM cultures, did not show any significant difference for Bz, as its growth yield averages were essentially the same as those of Tyr and Pac and somewhat higher (15 to 35%) than those of the other AC except 2,4-D (see Table S3). Consistently, the formation of energy-yielding aerobic metabolism intermediates, assuming channeling through the tricarboxylic acid cycle, did not provide any favorable condition for Bz (see Table S3), given that it produces the same intermediates, such as 4-Hb and 2,4-D, and has even less potential to obtain NADH and ATP than 3-Hb, Pac, 4-Hpa, and Tyr. Consistently, the formation of energy-yielding intermediates (see Table S3) provides only a slightly favorable condition for Bz over 4-Hb because the metabolism of the latter consumes an extra reducing equivalent at the PobA step compared to the BenABC/BenD steps (see Fig. S3A), but compared with 3-Hb, Pac, 4-Hpa, and Tyr, Bz has even less potential to obtain NADH and ATP.
Another explanation for Bz preference would be based on ecological niche. If some AC are more available than others, it would make sense that bacteria exposed to those AC evolve more proficient metabolic routes (increased uptake, sensitive gene induction, robust and efficient enzymes, better detoxification mechanisms, and fine-tuned metabolic fluxes, among others) for the more abundant than for the less abundant AC, thus improving their competitiveness in such habitats. It should be noted that in seawater, a natural environment not expected to be rich in AC, bacteria would degrade Bz and 4-Hb simultaneously, as reported for Sagittula stellata E-37, a member of the very abundant marine Roseobacter lineage (45). There are contrasting details concerning a putative, relatively high natural abundance of Bz in terrestrial environments. Bz may be more chemically stable in oxygenated habitats, because it does not possess reactive hydroxyl groups, in contrast to Phe, 3-Hb, 4-Hb, 4-Hpa, and Tyr, which facilitates the formation of polymeric, less available structures. On the other hand, the Bz unit seems not to be the more abundant plant-related aromatic residue, as compounds with substituted rings and C-6–C-3- or C-6–C-2-structured aromatics are more frequently generated as monomers during lignin decay (49, 50).
No matter what the reason(s) for Bz preference, the results reported here suggest that more than one molecular mechanism underlies this preference in C. pinatubonensis. The first key observation to be considered is that most biodegradation inhibition takes place at the transcriptional level, as revealed by gene expression studies, discarding more simplistic scenarios such as the competitive inhibition of enzymes (8, 9) or depletion of electron acceptors (10, 11). Transcriptional repression of the catabolic pathway for the deferred substrate has been previously demonstrated for the Bz–4-Hb mixture (24), where Bz is the main, if not the only, compound required to block induction of the target pobA gene through a possible interaction with the regulatory protein PobR. As a result, the C. pinatubonensis ΔbenA mutant cannot grow on the Bz–4-Hb mixture, since Bz itself completely represses the 4-Hb catabolism (24; also this work) (Fig. 5A). In contrast, all other Bz-containing mixtures of AC allowed growth of this mutant, but with different delays (Fig. 5), indicating that Bz preference in combinations of AC other than Bz–4-Hb would require some Bz catabolic intermediate, as has been reported for P. putida and A. baylyi (21, 23), and/or an unknown metabolic signal, such as operation of downstream enzymes, a particular redox status, or tricarboxylic acid cycle fluxes. It should be noted, however, that these differences might be also explained if Bz itself (assuming essentially the same repression mechanism as for 4-Hb) has lower affinities for the corresponding target regulators than for PobR, or if mutants arise that allow delayed growth in the presence of Bz. The possible occurrence of more than one mechanism involved in Bz preference is further supported by the varied extents of repression (Fig. 3), since full, partial, and no reduction of the expression of AC degradation genes was detected. It is necessary to point out, however, that no or partial repression may be explained by the wrong choice of target genes. In principle, tested genes encode initial, presumably regulated catabolic steps (26), but the possibility that the expression of other catabolic genes and/or uptake genes is the target of a putative transcriptional repression system cannot be ruled out.
Although four ways to degrade Bz have been reported for bacteria—one under aerobic conditions and the other three under oxygen-limiting conditions (1)—only two of them (encoded by the ben/cat pathway and the box pathway genes) are present in the C. pinatubonensis genome (26). Only the ben/cat pathway is active under the conditions used in this work, since the C. pinatubonensis ΔbenA mutant does not grow on Bz and accumulates Bz (24). This dismisses possibilities such as benzoyl-CoA-type intermediates (generated during Box pathway operation) repressing the expression of genes involved in the degradation of some of the other AC tested here, which has been demonstrated for Box intermediates controlling the degradation of gentisate in Comamonas testosteroni (51).
Contrasting with the hegemonic role of Bz, Phe is consistently the deferred substrate in binary mixtures. Phe has also been reported as the nonpreferred substrate in other species (16, 17, 52). The elucidation of the molecular basis for this is clearly required, and some hints from this report can help future work. Possible explanations for the lack of preference are its toxicity, since along with 2,4-D, Phe is the only AC with lower yields at higher concentrations (see Table S3 in the supplemental material), which may reflect its scored toxicity levels (see Table S3) and the potentially lower energy yielding intermediates when meta ring cleavage is used (see Table S3). Concerning possible molecular mechanisms, Bz clearly represses expression of phlK1 and phlK2 by diminishing their promoter activities (Fig. 3B and C; also see Fig. S4 in the supplemental material), but additional work is required to determine if these genes are also affected by the other AC and if other genes are targets for repression by AC, such as promoters of phlB and catA2 genes encoding the initial steps of meta and ortho ring cleavage pathways, respectively (see Fig. S3B). Interestingly, Phe is the only AC tested here whose degradation is controlled by the sigma factor σ54, which has been frequently reported as the transcription initiation factor involved in expression of catabolic operons that are highly regulated in response to environmental cues (53).
The pattern of catabolic pathways influenced by preferential utilization of Bz would reflect the evolutive track of C. pinatubonensis. As pointed out by Cases and de Lorenzo (54), the phenomenon of catabolite repression is the result of adaptive regulatory mechanisms specifically evolved during the natural history of each bacterium, considering habitat lifestyle and degree of specialization. It can be hypothesized that the current genome of the metabolically versatile C. pinatubonensis has been shaped as a result of its being a copiotroph (lifestyle) living in soils (habitat) containing Bz as one of the more abundant carbon sources but also including several other additional substrates (generalist, low degree of specialization). During its evolutive track, C. pinatubonensis has gained novel catabolic functions, such as 2,4-D biodegradation, a strain-specific trait (25, 55), but the regulatory mechanisms may not yet be fully adapted to this new capacity, allowing a less constrained expression even in the presence of Bz, as reported here.
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by FONDECYT grant 1110850, CONICYT grant FB 0002-2014, and Millennium Nuclei in “Plant Functional Genomics” grant P10-062F.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.04207-14.
REFERENCES
- 1.Fuchs G, Boll M, Heider J. 2011. Microbial degradation of aromatic compounds—from one strategy to four. Nat Rev Microbiol 9:803–816. doi: 10.1038/nrmicro2652. [DOI] [PubMed] [Google Scholar]
- 2.Jiménez JI, Miñambres B, García JL, Díaz E. 2002. Genomic analysis of the aromatic catabolic pathways from Pseudomonas putida KT2440. Environ Microbiol 4:824–841. doi: 10.1046/j.1462-2920.2002.00370.x. [DOI] [PubMed] [Google Scholar]
- 3.Pérez-Pantoja D, González B, Pieper DH. 2010. Aerobic degradation of aromatic hydrocarbons, p 800–837. In Timmis KN. (ed), Handbook of hydrocarbon and lipid microbiology, vol 2 Springer-Verlag, Berlin, Germany. [Google Scholar]
- 4.Narasimhan K, Basheer C, Bajic VB, Swarup S. 2003. Enhancement of plant-microbe interactions using a rhizosphere metabolomics-driven approach and its application in the removal of polychlorinated biphenyls. Plant Physiol 132:146–153. doi: 10.1104/pp.102.016295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohno T, Parr TB, Gruselle M-CI, Fernández IJ, Sleighter RL, Hatcher PG. 2014. Molecular composition and biodegradability of soil organic matter: a case study comparing two New England forest types. Environ Sci Technol 48:7229–7236. doi: 10.1021/es405570c. [DOI] [PubMed] [Google Scholar]
- 6.Landry C, Tremblay C. 2012. Compositional differences between size classes of dissolved organic matter from freshwater and seawater revealed by an HPLC-FTIR system. Environ Sci Technol 46:1700–1707. doi: 10.1021/es203711v. [DOI] [PubMed] [Google Scholar]
- 7.Reineke W, Jeenes DJ, Williams PA, Knackmuss H-J. 1982. TOL plasmid pWW0 in constructed halobenzoate-degrading Pseudomonas strains: prevention of meta pathway. J Bacteriol 150:195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charng MK, Voice TC, Criddle CS. 1993. Kinetics of competitive inhibition and cometabolism in the biodegradation of benzene, toluene, and p-xylene by two Pseudomonas isolates. Biotechnol Bioeng 41:1057–1065. doi: 10.1002/bit.260411108. [DOI] [PubMed] [Google Scholar]
- 9.Stringfellow WT, Aitken MD. 1995. Competitive metabolism of naphthalene, methylnaphthalenes, and fluorene by phenanthrene-degrading pseudomonads. Appl Environ Microbiol 61:357–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corseuil HX, Hunt CS, Santos RCFD, Alvarez RJJ. 1998. The influence of the gasoline oxygenate ethanol on aerobic and anaerobic BTX biodegradation. Water Res 32:2065–2072. doi: 10.1016/S0043-1354(97)00438-7. [DOI] [Google Scholar]
- 11.Lovanh N, Hunt CS, Alvarez PJ. 2002. Effect of ethanol on BTEX biodegradation kinetics: aerobic continuous culture experiments. Water Res 36:3739–3746. doi: 10.1016/S0043-1354(02)00090-8. [DOI] [PubMed] [Google Scholar]
- 12.Görke B, Stülke J. 2008. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol 6:613–624. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- 13.Rojo F. 2010. Carbon catabolite repression in Pseudomonas: optimizing metabolic versatility and interactions with the environment. FEMS Microbiol Rev 34:658–684. doi: 10.1111/j.1574-6976.2010.00218.x. [DOI] [PubMed] [Google Scholar]
- 14.Marozawa S, Röling WFM, Seifert J, Küffner R, von Bergen M, Meckenstock RU. 2014. Physiology of Geobacter metallireducens under excess and limitation of electron donors. Part I. Batch cultivation with excess of carbon sources. Syst Appl Microbiol 37:277–286. doi: 10.1016/j.syapm.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 15.Valderrama JA, Shingler V, Carmona M, Díaz E. 2014. AccR is a master regulator involved in carbon catabolite repression of the anaerobic catabolism of aromatic compounds in Azoarcus sp. CIB. J Biol Chem 289:1892–1904. doi: 10.1074/jbc.M113.517714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mazzoli R, Pessione E, Giuffrida MG, Fattori P, Barello C, Giunta C, Lindley ND. 2007. Degradation of aromatic compounds by Acinetobacter radioresistens S13: growth characteristics on single substrates and mixtures. Arch Microbiol 188:55–68. doi: 10.1007/s00203-007-0223-z. [DOI] [PubMed] [Google Scholar]
- 17.Zhan Y, Yu H, Yan Y, Ping S, Lu W, Zhang W, Chen M, Lin M. 2009. Benzoate catabolite repression of the phenol degradation in Acinetobacter calcoaceticus PHEA-2. Curr Microbiol 59:368–373. doi: 10.1007/s00284-009-9446-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heinaru E, Viggor S, Vedler E, Truu J, Merimaa M, Heinaru A. 2001. Reversible accumulation of p-hydroxybenzoate and catechol determines the sequential decomposition of phenolic compounds in substrate cultivations in pseudomonads. FEMS Microbiol Ecol 37:79–89. doi: 10.1111/j.1574-6941.2001.tb00855.x. [DOI] [Google Scholar]
- 19.Ampe F, Léonard D, Lindley ND. 1998. Repression of phenol catabolism by organic acids in Ralstonia eutropha. Appl Environ Microbiol 64:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harwood CS, Parales RE. 1996. The beta-ketoadipate pathway and the biology of self-identity. Annu Rev Microbiol 50:553–590. doi: 10.1146/annurev.micro.50.1.553. [DOI] [PubMed] [Google Scholar]
- 21.Cowles CE, Nichols NN, Harwood CS. 2000. BenR, a XylS homologue, regulates three different pathways of aromatic acid degradation in Pseudomonas putida. J Bacteriol 182:6339–6346. doi: 10.1128/JB.182.22.6339-6346.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nichols NN, Harwood CS. 1995. Repression of 4-hydroxybenzoate transport and degradation by benzoate: a new layer of regulatory control in the Pseudomonas putida beta-ketoadipate pathway. J Bacteriol 177:7033–7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brzostowicz PC, Reams AB, Clark TJ, Neidle EL. 2003. Transcriptional cross-regulation of the catechol and protocatechuate branches of the beta-ketoadipate pathway contributes to carbon source-dependent expression of the Acinetobacter sp. strain ADP1 pobA gene. Appl Environ Microbiol 69:1598–1606. doi: 10.1128/AEM.69.3.1598-1606.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Donoso RA, Pérez-Pantoja D, González B. 2011. Strict and direct transcriptional repression of the pobA gene by benzoate avoids 4-hydroxybenzoate degradation in the pollutant degrader Cupriavidus necator JMP134. Environ Microbiol 13:1590–1600. doi: 10.1111/j.1462-2920.2011.02470.x. [DOI] [PubMed] [Google Scholar]
- 25.Don RH, Pemberton JM. 1981. Properties of six pesticide degradation plasmids isolated from Alcaligenes paradoxus and Alcaligenes eutrophus. J Bacteriol 145:681–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pérez-Pantoja D, De la Iglesia R, Pieper DH, González B. 2008. Metabolic reconstruction of aromatic compounds degradation from the genome of the amazing pollutant-degrading bacterium Cupriavidus necator JMP134. FEMS Microbiol Rev 32:736–794. doi: 10.1111/j.1574-6976.2008.00122.x. [DOI] [PubMed] [Google Scholar]
- 27.Lykidis A, Pérez-Pantoja D, Ledger T, Mavromatis K, Anderson IJ, Ivanova NN, Hooper S, Lapidus A, González B, Kyrpides N. 2010. The complete multipartite genome sequence of Cupriavidus necator JMP134, a versatile pollutant degrader. PLoS One 5:e9729. doi: 10.1371/journal.pone.0009729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pérez-Pantoja D, Donoso R, Junca H, González B, Pieper DH. 2010. Phylogenomics of aerobic bacterial degradation of aromatics, p 1355–1397. In Timmis KN. (ed), Handbook of hydrocarbon and lipid microbiology, vol 2 Springer-Verlag, Berlin, Germany. [Google Scholar]
- 29.Pérez-Pantoja D, Donoso R, Agulló L, Córdova M, Seeger M, Pieper DH, González B. 2012. Genomic analysis of aromatic compounds biodegradation in Burkholderiales. Environ Microbiol 14:1091–1117. doi: 10.1111/j.1462-2920.2011.02613.x. [DOI] [PubMed] [Google Scholar]
- 30.Dorn E, Hellwig M, Reineke W, Knackmuss HJ. 1974. Isolation and characterization of a 3-chlorobenzoate degrading pseudomonad. Arch Microbiol 99:61–70. doi: 10.1007/BF00696222. [DOI] [PubMed] [Google Scholar]
- 31.Jiménez JI, Pérez-Pantoja D, Chavarría M, Díaz E, de Lorenzo V. 2014. A second chromosomal copy of the catA gene endows Pseudomonas putida mt-2 with an enzymatic safety valve for excess of catechol. Environ Microbiol 16:1767–1778. doi: 10.1111/1462-2920.12361. [DOI] [PubMed] [Google Scholar]
- 32.Ledger T, Pieper DH, González B. 2006. Chlorophenol hydroxylases encoded by pJP4 plasmid differentially contribute to chlorophenoxyacetic acid degradation. Appl Environ Microbiol 72:2783–2792. doi: 10.1128/AEM.72.4.2783-2792.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pérez-Pantoja D, Ledger T, Pieper DH, González B. 2003. Efficient turnover of chlorocatechols is essential for growth of Ralstonia eutropha JMP134(pJP4) in 3-chlorobenzoic acid. J Bacteriol 185:1534–1542. doi: 10.1128/JB.185.5.1534-1542.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schweigert N, Zehnder AJB, Eggen RIL. 2001. Chemical properties of catechols and their molecular modes of toxic action in cells, from microorganisms to mammals. Environ Microbiol 3:81–91. doi: 10.1046/j.1462-2920.2001.00176.x. [DOI] [PubMed] [Google Scholar]
- 35.Iannucci A, Fragasso M, Platani C, Papa R. 2013. Plant growth and phenolic compounds in the rhizosphere soil of wild oat (Avena fatua L.). Front Plant Sci 4:509. doi: 10.3389/fpls.2013.00509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones DL, Simfukwe P, Hill PW, Mills RT, Emmett BA. 2014. Evaluation of dissolved organic carbon as a soil quality indicator in national monitoring schemes. PLoS One 9:e90882. doi: 10.1371/journal.pone.0090882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Padilla-Sánchez JA, Plaza-Bolaños P, Romero-González R, Garrido-Frenich A, Martínez-Vidal JL. 2010. Application of a quick, easy, cheap, effective, rugged and safe-based method for the simultaneous extraction of chlorophenols, alkylphenols, nitrophenols and cresols in agricultural soils, analysed by using gas chromatography-triple quadrupole-mass spectrometry/mass spectrometry. J Chromatogr A 1217:5724–5731. doi: 10.1016/j.chroma.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 38.Sène M, Gallet C, Doré T. 2001. Phenolic compounds in a Saheliam sorghum (Sorghum bicolor) genotype (CE145-66) and associated soils. J Chem Ecol 27:81–92. doi: 10.1023/A:1005620000835. [DOI] [PubMed] [Google Scholar]
- 39.Choi KY, Zylstra GJ, Kim E. 2007. Benzoate catabolite repression of the phthalate degradation pathway in Rhodococcus sp. strain DK17. Appl Environ Microbiol 73:1370–1374. doi: 10.1128/AEM.02379-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Higgins SJ, Mandelstam J. 1972. Regulation of pathways degrading aromatic substrates in Pseudomonas putida. Enzymic response to binary mixtures of substrates. Biochem J 126:901–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ampe F, Lindley ND. 1995. Acetate utilization is inhibited by benzoate in Alcaligenes eutrophus: evidence for transcriptional control of the expression of acoE coding for acetyl coenzyme A synthetase. J Bacteriol 177:5826–5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trautwein K, Grundmann O, Wöhlbrand L, Eberlein C, Boll M, Rabs R. 2012. Benzoate mediates repression of C4-dicarboxylate utilization in “Aromatoleum aromaticum” EbN1. J Bacteriol 194:518–528. doi: 10.1128/JB.05072-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lahme S, Trautwein K, Strijkstra A, Dörries M, Wöhlbrand L, Rabus R. 2014. Benzoate mediates the simultaneous repression of anaerobic 4-methylbenzoate and succinate utilization in Magnetospirillum sp. strain pMbN1. BMC Microbiol 14:269. doi: 10.1186/s12866-014-0269-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang C, Anderson AJ. 2013. Utilization of pyrene and benzoate in Mycobacterium isolate KMS is regulated differentially by catabolic repression. J Basic Microbiol 53:81–92. doi: 10.1002/jobm.201100480. [DOI] [PubMed] [Google Scholar]
- 45.Gulvik CA, Buchan A. 2013. Simultaneous catabolism of plant-derived aromatic compounds results in enhanced growth for members of the Roseobacter lineage. Appl Environ Microbiol 79:3716–3723. doi: 10.1128/AEM.00405-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ledger T, Flores-Aceituno F, González B. 2009. 3-Chlorobenzoate is taken up by a chromosomally encoded transport system in Cupriavidus necator JMP134. Microbiology 155:2757–2765. doi: 10.1099/mic.0.029207-0. [DOI] [PubMed] [Google Scholar]
- 47.Leveau JH, Zehnder AJ, van der Meer JR. 1998. The tfdK gene product facilitates uptake of 2,4-dichlorophenoxyacetate by Ralstonia eutropha JMP134(pJP4). J Bacteriol 180:2237–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen Z, Pu X-Y, Zhang Q. 2011. Salicylate functions as an efflux pump inducer and promote the emergence of fluoroquinolone-resistant Campylobacter jejuni mutants. Appl Environ Microbiol 77:7128–7133. doi: 10.1128/AEM.00763-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bugg TDH, Ahmad M, Hardiman EM, Rahmanpour R. 2011. Pathways for degradation of lignin in bacteria and fungi. Nat Prod Rep 28:1883–1896. doi: 10.1039/c1np00042j. [DOI] [PubMed] [Google Scholar]
- 50.Ruíz-Dueñas FJR, Martínez AT. 2009. Microbial degradation of lignin: how a bulky recalcitrant polymer is efficiently recycled in nature and how we can take advantage of this. Microb Biotechnol 2:164–177. doi: 10.1111/j.1751-7915.2008.00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen DW, Zhang Y, Jiang CY, Liu SJ. 2014. Benzoate metabolism intermediate benzoyl coenzyme A affects gentisate pathway regulation in Comamonas testosteroni. Appl Environ Microbiol 80:4051–4062. doi: 10.1128/AEM.01146-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Szököl J, Rucká L, Simciková M, Halada P, Nesvera J, Pátek M. 2014. Induction and carbon catabolite repression of phenol degradation genes in Rhodococcus erythropolis and Rhodococcus jostii. Appl Microbiol Biotechnol 98:8267–8279. doi: 10.1007/s00253-014-5881-6. [DOI] [PubMed] [Google Scholar]
- 53.Shingler V. 2011. Signal sensory systems that impact σ54-dependent transcription. FEMS Microbiol Rev 35:425–440. doi: 10.1111/j.1574-6976.2010.00255.x. [DOI] [PubMed] [Google Scholar]
- 54.Cases I, de Lorenzo V. 2005. Promoters in the environment: transcriptional regulation in its natural context. Nat Rev Microbiol 3:105–118. doi: 10.1038/nrmicro1084. [DOI] [PubMed] [Google Scholar]
- 55.Trefault N, De la Iglesia R, Molina AM, Manzano M, Ledger T, Pérez-Pantoja D, Sánchez MA, Stuardo M, González B. 2004. Genetic organization of the catabolic plasmid pJP4 from Ralstonia eutropha JMP134 (pJP4) reveals mechanisms of adaptation to chloroaromatic pollutants and evolution of specialized chloroaromatic degradation pathways. Environ Microbiol 6:655–668. doi: 10.1111/j.1462-2920.2004.00596.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.