Abstract Abstract
Colombia, one of the world’s megadiverse countries, has a highly diverse mosquito fauna and a high prevalence of mosquito-borne diseases. In order to provide relevant information about the diversity and taxonomy of mosquito species in Colombia and to test the usefulness of DNA barcodes, mosquito species collected at different elevations in the departments of Antioquia and Caldas were identified combining adult morphology and barcode sequences. A total of 22 mosquito species from eight genera were identified using these combined techniques. We generated 77 barcode sequences with 16 species submitted as new country records for public databases. We examined the usefulness of DNA barcodes to discriminate mosquito species from the Neotropics by compiling 1,292 sequences from a total of 133 species and using the tree-based methods of neighbor-joining and maximum likelihood. Both methodologies provided similar results by resolving 105 species of mosquitoes separated into distinct clusters. This study shows the importance of combining classic morphological methodologies with molecular tools to accurately identify mosquitoes from Colombia.
Keywords: Culicomorpha, nematocerous Diptera, Neotropical Region, species identification, combining methodologies
Introduction
Culicidae currently comprises 3,543 formally recognized species distributed throughout most types of habitats and ecosystems of the world (Harbach 2015). Mosquitoes, as a group, occur in a wide array of both aquatic and terrestrial habitats and have correspondingly variable morphological and behavioral adaptations to these (Becker et al. 2010). Due to their hematophagous behavior, mosquitoes are able to transmit many different disease agents such as viruses, bacteria, protozoans, and nematodes from one vertebrate host to another (Becker et al. 2010). The taxonomy of Culicidae has received special attention because of their vector capabilities and medical and veterinary importance (Harbach 2007, Becker et al. 2010). Thus, major mosquito vector species are generally the best studied and understood, both biologically and taxonomically (Harbach 2007).
Mosquito identification is traditionally based on dichotomous keys constructed from morphological features taken for a particular life stage or gender (Munstermann and Conn 1997). The morphological identification of mosquito species is hampered by intraspecific variation, the complexity of some features and the need for specimens in generally excellent condition (Zavortink 1974, Cywinska et al. 2006). The importance of gaining high resolution in mosquito species discrimination requires the implementation of additional tools that can be used for accurate identification (Munstermann and Conn 1997, Cook et al. 2005).
The DNA barcode method was postulated by Hebert et al. (2003a, 2003b) as a DNA sequence-based approach for the accurate identification of specimens and for species discovery. There are three different methods proposed to analyze the efficiency of DNA barcode data to discriminate species: similarity methods based on the match between the query sequence and the reference sequences, tree-based identifications, and the barcode gap (Hebert et al. 2003a, 2004, Ratnasingham and Herbert 2007, Austerlitz et al. 2009). Most published approaches to DNA barcoding use distance-based methods for species designation; however, alternative approaches using character-based phylogenetic analysis have been proposed (DeSalle et al. 2005). Although DNA barcode is now widely use, there are still many open questions about both the advantages and disadvantages of its use, as well as its definition as a methodology (Moritz and Cicero 2004, Brower 2006, Baker et al. 2009, Casiraghi et al. 2010, Jinbo et al. 2011, Wilson et al. 2011, Begsten et al. 2012, Collins and Cruickshank 2013, Stoeckle and Thaler 2014). Despite the known limitations of the method, DNA barcodes have been used to identify specimens and to flag potential new species (e.g. Scheffer et al. 2006, Nagoshi et al. 2011, Tavares et al. 2011, Baldwin and Weigt 2012, Huemer and Mutaten 2015, among others).
In mosquitoes, the effectiveness of the COI barcode marker for specimen identification has been tested in surveys of Canada (Cywinska et al. 2006), India (Kumar et al. 2007), the Iranian islands in the Persian Gulf (Azari-Hamidian et al. 2010), China (Wang et al. 2012), Amazonian Ecuador (Linton et al. 2013), Pakistan (Ashfaq et al. 2014), Singapore (Chan et al. 2014), and Belgium (Versteirt et al. 2015). These studies show correspondence between morphological species and DNA barcode clusters, but also point out the failure of the methodology to distinguish between very similar or cryptic species.
Colombia is located in the northwest of South America. It comprises a variety of biogeographic regions that have contrasting biophysical characteristics and high environmental variability (Etter et al. 2006). Related to this variety of ecosystems, Colombia has high levels of endemism and species richness and has been categorized as a mega-diverse country (Hernandez et al. 1992, Chaves and Arango 1998). Moreover, environmental variability may favor the development and persistence of a great diversity of mosquito species (Foley et al. 2007, Brochero and Quiñones 2008), as well as to favor both the immigration and biological invasion of non-endemic species including vector species (Barreto et al. 1996, Molina et al. 2000, Olano et al. 2001). Furthermore, Colombia is located in a region shown to be a potential hotspot for malaria endemicity and other mosquito borne disease outbreaks (Foley et al. 2008). In Colombia, malaria vector mosquitoes (genus Anopheles Meigen, 1818) have been extensively investigated, the morphological keys for their identification are up-to-date and available, and different genetic techniques have been developed to differentiate cryptic species complexes. Nevertheless, the overall diversity of mosquito fauna in Colombia is understudied and generally poorly known (Olano et al. 2001, Montoya-Lerma et al. 2011).
In order to improve the mosquito knowledge in Colombia, we tested the effectiveness of the barcoding methodology to support the reliable identification of Neotropical species of mosquitoes, previously identified with morphological characters, collected over three different altitudinal ecosystems of the Colombian Andes (Antioquia and Caldas departments).
Materials and methods
Study area
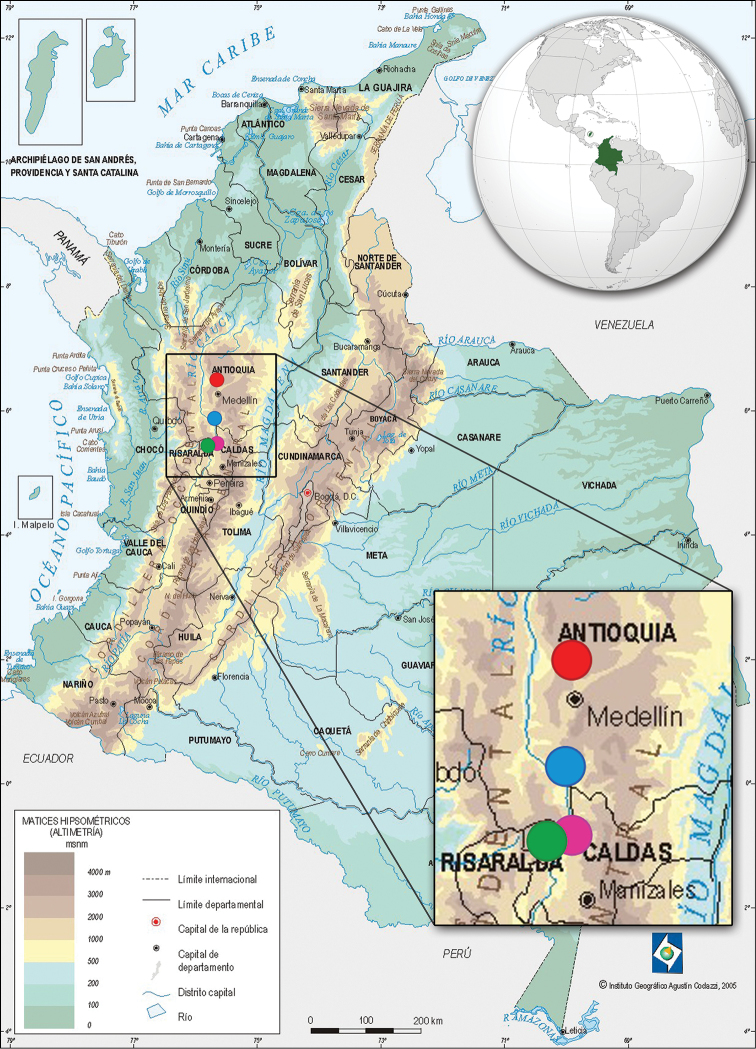
The study area is located in the west central Andean region in Colombia. Fieldwork was conducted during September 2013 in rural areas of Antioquia and Caldas departments (Fig. 1). This study focused on three important biomes: paramo (Belmira, 3,200 masl), cloud forest (Rio Sucio, 1,960 masl), and tropical dry forest (La Pintada, 660 masl). Each sampling location was split into two habitat types: forest (habitat A) and anthropogenic disturbed area (habitat B). Forest sampling in the paramo plots consisted of dense shrubbery and scattered trees associated with sub-paramo zones. Additional collections were made in the rural area of Supia (Caldas) (mountain forest) at an elevation of 1,150 masl.
Figure 1.
Map of Colombia indicating the sampling sites of mosquitoes collected in this study: Belmira, Antioquia, paramo, 3,200 masl (red circle); Rio Sucio, Caldas, cloud forest, 1,960 masl (green circle); La Pintada, Antioquia, tropical dry forest, 660 masl (blue circle); Supia, Caldas, rural area, 1,150 masl (pink circle). Modified from Instituo Geográfico Agustín Codazzi (www.igac.gov.co) and Wikimedia Commons (by Addicted04).
Specimen collection
Mosquito adults were collected using one malaise trap and three Centers for Disease Control-CDC light traps in each habitat type (A or B) of the three biomes, totaling six sites. CDC traps collected for 14 hours, between 5:00 pm and 7:00 am of the next day during two nights in each of our six locations plus the rural area of Supia. Malaise traps were placed and left for 48 hours in each location. Additional specimens were obtained by aspirating mosquitoes attracted to humans during the placement and operation of the traps. All data in the sampled localities were plotted following the protocols of Foley et al. (2010) using a GPS Garmin® Rino 530HCx. Weather data of average wind speed (km/h), temperature (°C), and relative humidity (%) were recorded with a Kestrel® 4000 Weather Tracker.
Wild-caught adults were killed using ethanol (90%) fumes in a lethal chamber to ensure DNA preservation. All mosquitoes were kept dry and individually transferred to labeled 1.5 ml tubes. Each tube was labeled, pierced with a mounted needle (to allow the escape of moisture) and placed in plastic bags containing silica gel. Specimen mounting techniques were conducted using the protocols of Walter Reed Biosystematics Unit-WRBU (Gaffigan and Pecor 1997).
Adult identification using morphology
All the specimens collected were identified by female morphology and male genitalia. Since there is no single morphological key to facilitate the identification of mosquitoes of Colombia, genus level identifications were made with multiple approaches using the dichotomous keys of Lane (1953), Becker et al. (2010), Chaverri (2009), and the multi-entry web-based keys developed by the WRBU for South America (http://wrbu.si.edu/southcom_MQkeys.html). In addition, available dichotomous keys and original species descriptions were compiled through literature freely available on the WRBU website (www.wrbu.org). Important references used for species identifications by taxonomic groups included: Tribe Aedini (Arnell 1976, Reinert 2000), Anopheles (Komp 1942, Cova-Garcia 1961, González and Carrejo 2007), Culex Linnaeus, 1758 (Rozeboom and Komp 1950, Cova-Garcia et al. 1966, Bram 1967, Knight and Haeger 1971, Sirivanakarn 1982, Strickmann and Prat 1989, Pecor et al. 1992), Coquillettidia Dyar, 1905 (Shannon 1934, Cova-Garcia et al. 1966), Haemagogus Williston, 1896 (Kumm et al. 1946, Levi-Castillo 1951, Arnell 1973, Liria and Navarro 2009), Psorophora Robineau-Desvoidy, 1827 (Guedes and Souza 1964, Liria and Navarro 2003), Trichoprosopon Lane & Cerqueira, 1942 (Stone 1944, Zavortink 1979), Wyeomyia Theobald, 1901 (Judd 1998, Motta and de Oliveira 2000). Overall, Lane (1953) was used as a general reference.
Voucher specimens
Voucher specimens and associated genitalia preparations are stored in the entomological collections of the Zoologisches Forschungsmuseum Alexander Koenig (ZFMK), Bonn (Germany). Genomic DNA extracts are stored at –80 °C in the biobank collections of the ZFMK for future reference. Details of reference material are listed by genus in Suppl. material 1, as well as full collection site details including geo-references and environmental conditions. Fully digitized specimen data records are freely available in MosquitoMap (www.mosquitomap.org). COI barcode sequences can be accessed through GenBank (accession numbers KM592986 to KM593062; see Suppl. material 1).
Laboratory protocols, DNA extraction, PCR, and sequencing
DNA was extracted from legs and occasionally abdomens (for specimens without legs or with deficient amplification) following standard protocols of the commercially available DNeasy Blood & Tissue Kit (QIAgen®). The COI barcode region was amplified using the forward primer LCO1490 (5’-GGTCAACAAATCATAAAGATATTGG-3’) and the reverse primer C1N2191 (alias Nancy) (5’CCCGGTAAAATTAAAATATAAACTTC-3’) (Simon et al. 1994). When those primers failed to amplify full-length sequences, the set LCO1490 and HCO2198 (5’-TAAACTTCA GGGTGACCAAAAAATCA-3’) (Folmer et al. 1994) were alternatively used. PCR amplification followed the protocol optimized by the Laboratories of Molecular Biology, Alexander Koenig Museum-ZFMK (Bonn, Germany). Each PCR reaction contained 2.5 μL of DNA, 2.3 μL of ddH20, 2 μL of Q-Solution, 10 μL of Qiagen Multiplex-Mix, and 1.6 μM of each primer. The polymerase chain reaction (PCR) cycle conducted was: 95 °C for 15 min, 25 cycles of 94 °C for 35 s, 55 °C for 90 s and 72 °C for 90 s, 72 °C for 10 min and a 10 °C hold. The PCR product was visualized on 1.5% agarose gels, containing 0.5 mg/ml of GelRed®, followed by 1.75 microliters of the PCR product being removed and mixed with 1.75 μL of loading dye. Gels were run at 120 V for 30 min, prior to ultraviolet visualization. PCR products were cleaned using the commercially available QIAquick PCR Purification Kit (QIAgen®). Bi-directionally sequencing reactions were carried out by Macrogen© Inc. Chromatograms were edited in Geneious 7.0.6 (Biomatters© Ltd). Primer sequences were removed from edited contigs and only high-quality sequences of at least 600 bp were retained for data analysis. Sequences were aligned using Geneious 7.0.6.
Additional sequences
To test variation in the barcoding region across a greater geographical area, barcoding sequences of mosquito species listed for the Neotropics were downloaded from BOLD and GenBank between December 2013 and February 2014 for all identified species with a minimum length of 480 bp of COI barcoding region, no stop codons and alignment without gaps (Suppl. material 2). In order to have a broader representation of mosquito genera, additional sequences of species from other biogeographic areas were downloaded for those Neotropical groups without available sequences. These taxonomic groups include: Coquillettidia (Coquillettidia) Dyar, 1905, Culex (Neoculex) Dyar, 1905, Mansonia (Mansonioides) Theobald, 1907, Ochlerotatus (Protoculex) Felt, 1904, Orthopodomyia Theobald, 1904, Toxorhynchites (Lynchiella) Lahille, 1904, and Toxorhynchites (Toxorhynchites) Theobald, 1901. Moreover, an unpublished data set of 45 sequences of mosquitoes collected in rural areas of Uraba (Antioquia, Colombia) during 2009 was also added to the analysis (Suppl. material 3).
Lutzomyia longipalpis (Lutz & Neiva, 1912) (Diptera: Psychodidae) was constrained as outgroup. We also included several other genera from three different families as outgroups, i.e. Chironomidae [Cricotopus bicinctus (Meigen, 1818), Chironomus decorus Goetghebuer, 1927, Chironomus kiiensis Tokunaga, 1936, Dicrotendipes tritomus (Thienemann & Kieffer, 1916), and Tanytarsus guerlus (Roback, 1957)], Simuliidae [Simulium ochraceum Walker, 1861, Simulium inaequale (Paterson & Shannon, 1927), Gigantodax abalosi Wygodzinsky, 1958, and Gigantodax basinflatus Wygodzinsky & Coscaron, 1989] and Dixidae [Dixella sp.]. A total of 11 outgroup taxa were included. All outgroup taxa sequences were downloaded from BOLD (Suppl. material 4).
Barcoding methodologies
We used similarity methods based on the match between the query sequence and the reference database [e.g. BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and BOLD Identification System (IDS) (http://www.boldsystems.org/index.php/IDS_OpenIdEngine)] and clustering in Tree-based identifications (using Neighbour-joining and maximum likelihood approaches) in order to analyze the DNA barcode region of the mosquitoes collected in Colombia and assign individuals to a given species.
The Neighbour-Joining (NJ) tree (Saitou and Nei 1987) was based on the Tamura-Nei distance model (TN93) (Tamura and Nei 1993) as recommend by Srivathsan and Meier (2012). Bootstrap support values were calculated with 1,000 replicates. The NJ tree, distance matrices and bootstrap were generated using Geneious 7.0.6.
To perform the maximum likelihood analysis, the data set was divided into three partitions according to the codon positions of COI (first, second, and third positions). We determined the best choice of model for each partition using PartionFinder v1.1.0 (Lanfear 2012) under the Akaike Information Criterion (AIC) as recommended by Posada and Buckley (2004). The model chosen for position 1 was SYM+I+G, GTR+I+G for position 2 and GTR+I+G for position 3 of COI gene. Data were analyzed using Garli 2.0 (Zwickl 2006) based on best choice of the model predicted. Default settings (scorethresh-forterm = 0.05; significanttopochange = 0.0001; searchreps = 1) were used to perform 20 replicates. The tree with the highest likelihood was retained. Bootstrap support values were estimated from 1,000 replicates using the same independent models. Analytical runs of ML were performed at the Hydra cluster (Center for Astrophysics, Harvard University), a Linux based cluster running Grid Engine with more than 3,000 CPUs with AMD 64-bit processors.
Results
A total of 77 mosquito specimens were collected from four sampling sites during our study (Table 1). Sampling success among traps varies greatly across location and habitat types. Belmira, the location with highest wind speed and precipitation, only reported one or no specimens for their two habitats. The specimens were identified to 22 species from eight genera. One species collected during this study in La Pintada forest habitat, Wyeomyia luteoventralis Theobald, 1901, is the first record for Colombia (Rozo-Lopez and Mengual, 2015).
Table 1.
Mosquito species collected. Four collection sites of the Colombian Andes during September 2013. One specimen collected in Rio Sucio (habitat A) still remains undetermined due to damage and lack of conspecific sequences for comparison.
| Mosquito species | Sites | ||||||
|---|---|---|---|---|---|---|---|
| Supia 1,150 masl | Rio Sucio 1,960 masl | La Pintada 660 masl | Belmira 3,200 masl | ||||
| Species | Habitat A | Habitat B | Habitat A | Habitat B | Habitat A | Habitat B | |
| Anopheles neomaculipalpus | 1 | ||||||
| Coquillettidia nigricans | 1 | ||||||
| Culex coniger | 10 | ||||||
| Culex conspirator | 5 | ||||||
| Culex spp. [coronator complex] | 1 | 1 | 3 | ||||
| Culex declarator | 1 | 10 | |||||
| Culex educator | 1 | ||||||
| Culex erraticus | 6 | 2 | |||||
| Culex erythrothorax | 1 | ||||||
| Culex lactator | 3 | ||||||
| Culex lucifugus | 6 | ||||||
| Culex nigripalpus | 1 | 2 | |||||
| Culex spinosus | 1 | ||||||
| Culex spissipes | 1 | ||||||
| Culex theobaldi | 1 | ||||||
| Culex (Culex) sp. | 1 | ||||||
| Culex (Melanoconion) sp. | 4 | 1 | |||||
| Haemagogus janthinomys | 2 | ||||||
| Haemagogus lucifer | 1 | ||||||
| Ochlerotatus angustivittatus | 1 | 1 | |||||
| Ochlerotatus euiris | 1 | ||||||
| Psorophora cingulata | 1 | ||||||
| Psorophora ferox | 2 | 1 | |||||
| Trichoprosopon evansae | 1 | ||||||
| Wyeomyia luteoventralis | 1 | ||||||
| Undetermined | 1 | ||||||
| Total | 4 | 16 | 1 | 45 | 10 | 1 | 0 |
Adult identification using morphology
Twenty one species belonging to seven genera were successfully identified by morphological characteristics of adult females and male genitalia: Coquillettidia nigricans Coquillett, 1904 [n=1 female], Culex conspirator Dyar & Knab, 1906 [n=5 males], Culex corniger Theobald, 1903 [n=8 females], Culex declarator Dyar & Knab, 1906 [n=8; 5 females, 3 males], Culex educator Dyar & Knab, 1906 [n=1 male], Culex erraticus [n=2 males], Culex erythrothorax Dyar, 1907 [n=1 female], Culex lactator Dyar & Knab, 1906 [n=2 females], Culex lucifugus Komp, 1936 [n=3 males], Culex nigripalpus Theobald, 1901 [n=2; 1 female, 1 male], Culex spinosus Lutz, 1905 [n=1 male], Culex spissipes Theobald, 1903 [n=1 female], Culex theobaldi Lutz, 1094 [n=1 male], Haemagogus janthinomys Dyar, 1921 [n=2 females], Haemagogus lucifer Howard, Dyar & Knab, 1913 [n=1 female], Ochlerotatus angustivittatus Dyar & Knab, 1907 [n=2; 1 female, 1 male], Ochlerotatus euiris Dyar, 1922 [n=1 female], Psorophora cingulata Fabricius, 1805 [n=1 female], Psorophora ferox von Humboldt, 1819 [n=3; 2 females, 1 male], Trichoprosopon evansae Antunes, 1942 [n=1 female], and Wyeomyia luteoventralis [n=1 female]. One specimen of the genus Anopheles was identified by morphology to the group: neomaculipalpus / punctimacula. It was only possible to identify the species using its gene sequence.
Mosquito barcoding
All the collected specimens were successfully amplified and sequenced, generating a total of 77 sequences with lengths ranging from 618 to 699 bp. Only high quality sequences were retained. Sequences from 14 species are submitted as new records for public databases: Coquillettidia nigricans, Culex conspirator, Culex educator, Culex lactator, Culex lucifugus, Culex spissipes, Culex theobaldi, Haemagogus janthinomys, Haemagogus lucifer, Ochlerotatus angustivittatus, Ochlerotatus euiris, Psorophora cingulata, Trichoprosopon evansae, Wyeomyia luteoventralis.
All the DNA sequences obtained were compared to those available, by 25 January 2015, in GenBank and BOLD Identification System. At the level of genus, BLAST accurately discriminated 92% of the genera previously identified, whereas BOLD accurately discriminated 70% of genera previously identified. Although BLAST represented a higher percentage of accurate discrimination of genera in our queries, five specimens with a score of more than 89% were wrongly matched: Trichoprosopon was mismatched as Anopheles, Wyeomyia was mismatched as Sabethes Robineau-Desvoidy, 1827, Haemagogus was mismatched as Ochlerotatus (or Aedes), and for the sequence of Haemagogus lucifer mixed results of Spilogona Schnabl, 1911, Haematobia Lepeletier & Serville, 1828 (family Muscidae), and Culex were obtained.
By adding GenBank, BOLD, and unpublished sequences to our data set, it was possible to obtain taxon coverage of 68% of the mosquito genera and 34% of the mosquito species listed for Colombia. Moreover, our data set coverage for Neotropical mosquito species corresponds to 58% of mosquito genera and 12% of mosquito species. The data set of the COI sequences of the Neotropical mosquitoes comprises a total of 1,292 barcode sequences belonging to 133 species and 21 genera (with a minimum length of 640 bp). The alignment was unambiguous: no gaps and amino acid translations without stop codons, indicating that all sequences represented functional protein coding genes, not pseudogenes. The analyzed region starts at the position 45 and stops at position 693 of the COI gene of the mitochondrial genome of Drosophila melanogaster Meigen, 1830 (AJ400907) (Adams et al. 2000) used as the reference genome. The amino acid reading frame starts at the second base of the dataset. The data matrix shows 116 invariant sites and a mean A+T content of 67.4%.
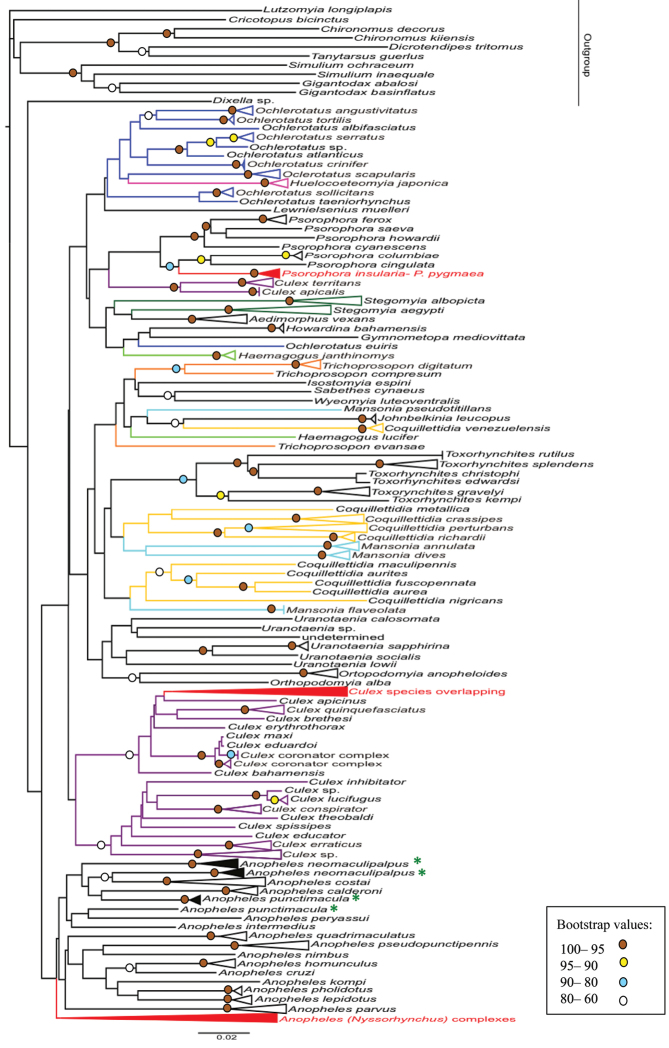
The Neighbour-Joining and Maximum likelihood analyses showed 105 species, from the 133 Neotropical species with available barcodes, separated into distinct clusters. Genera represented by more than one taxon formed cohesive assemblages of five clusters [Anopheles, Orthopodomyia, Psorophora, Toxorhynchites Theobald, 1901, Uranotaenia Lynch Arribalzaga, 1891] in the NJ analysis and eight [Anopheles, Coquillettidia, Culex, Orthopodomyia, Psorophora, Stegomyia Theobald, 1901, Toxorhynchites, Uranotaenia] in the ML analysis.
The NJ tree based on Tamura-Nei genetic distances (Fig. 2) revealed most of the species clusters (100 species) with high bootstrap value (97–99). Conversely, 28 species were recovered as non-monophyletic groups during this analysis: Anopheles punctimacula Dyar & Knab, 1906 and Anopheles neomaculipalpus Curry, 1931 as paraphyletic groups, an overlapping of Psorophora insularia (Dyar & Knab, 1906) and Psorophora pygmaea (Theobald, 1903), four species of Culex [Culex declarator, Culex nigripalpus, Culex conspirator, and Culex spinosus], and 20 species of Anopheles (Nyssorynchus) Blanchard, 1902 [myzorynchella section = Anopheles antunesi Galvao & Amarai, 1940, Anopheles lutzii Cruz, 1901, Anopheles pristinus Nagaki & Sallum, 2010; nuneztovari complex = Anopheles goeldi Rozeboom & Gabaldon, 1941, Anopheles dunhami, Anopheles nuneztovari Gabaldon, 1940; oswaldoi complex = Anopheles evansae (Brethes, 1926), Anopheles galvaoi Causey, Deane & Deane, 1943, Anopheles konderi Galvao & Damasceno, 1942, Anopheles oswaldoi (Peryassu, 1922), Anopheles rangeli Gabaldon, Cova-Garcia & Lopez, 1940; strodei complex = Anopheles albertoi Unti, 1941, Anopheles arthuri Unti, 1941, Anopheles rondoni (Neiva & Pinto, 1922), Anopheles strodei Root, 1926; albitarsis complex = Anopheles albitarsis Lynch Arribalzaga, 1878, Anopheles janconnae Wilkerson & Sallum, 2009, Anopheles marajoara Galvao & Damasceno, 1942, Anopheles oryzalimnetes Wilkerson & Motoki, 2009; and Anopheles benarrochi Gabaldon, Cova-Garcia & Lopez, 1941].
Figure 2.
Neighbour-Joining tree of the barcoding sequences of mosquito species listed for Neotropics, based on Tamura-Nei genetic distances. Terminal branches have been collapsed in order to save space (see original tree in Suppl. material 5 and 6). Branches in colors indicate non-monophyletic genera. Red clusters represent groups with problems to discriminate species. Names with green asterisk indicate non- monophyletic species. Bootstrap values above 60 (1,000 replicates) are given at the nodes.
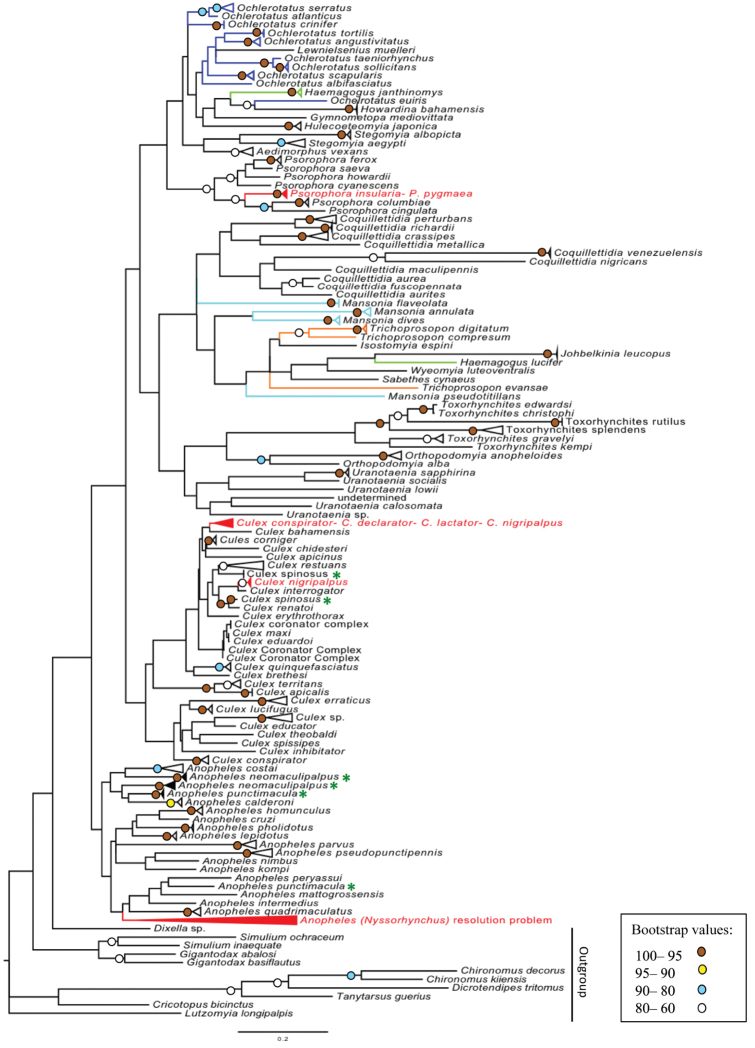
The likelihood score for the best ML tree was –31,760.30891. The overall topologies of the ML (Fig. 3) and NJ trees compared favorably with exception of Coquillettidia, Culex, and Stegomyia, which resolved as monophyletic clusters. The ML tree revealed 92 species clusters with high bootstrap (95–99). The bootstrap on the remaining 13 species ranged from 65 to 93. ML analysis also recovered 28 species as non-monophyletic groups.
Figure 3.
Maximum likelihood tree of the barcoding sequences of mosquito species listed for the Neotropics. Terminal branches have been collapsed in order to save space (see original tree in Suppl. material 7 and 8). Branches in colors indicate non-monophyletic genera. Red clusters represent groups with problems to discriminate species. Names with green asterisk indicate non-monophyletic species. Bootstrap values above 60 (1,000 replicates) are given at the nodes.
Discussion
In this study 22 species belonging to eight genera and 11 subgenera were identified by combining morphological and molecular methodologies. Although our data and sampling size were limited, the combination of methodologies provided better success in species identification. Overall, congruence between morphology and barcode grouping, based on cluster monophyly with high (more than 95%) bootstrap support, was found in 18 of the 22 morphologically defined taxa (82%). Similarity methods based on the match between the query sequence and the reference database (more than 98% identity between BOLD/BLAST) only discriminated three mosquito species of this study. Similarity methods based on the match between the query sequence and the reference databases represent the least suitable method to discriminate mosquito species in this study.
Tree-based diagnostics are graphic criterion for species recognition, which describes genetic similarity in a visually satisfying style (Goldstein and DeSalle 2010). An important advantage of using a tree-based approach is that they present a direct sense of the statistical reliability and do not retrieve a positive result if no matching diagnostics are found (unlike the best match algorithm of BLAST and BOLD which retrieves the closest match but requires the user to interpret its reliability) (Goldstein and DeSalle 2010). Here, we compared two tree-based methods (neighbour-joining and maximum likelihood), which provided similar results. The NJ tree computed was in general agreement with previously published taxonomy based approaches (Hebert et al. 2003a, 2003b, Hebert et al. 2004, Hajibabaei et al. 2007, Cywinska et al. 2006, Kumar et al. 2007). Both tree-based analysis in this study showed 105 species of mosquitoes (from 133 total species) separated into distinct clusters. Since the NJ clustering performed considerably faster than ML approach and indeed has been used in the great majority of published barcoding studies, these results indicate it is an efficient choice for mosquito barcode analyses, similar to what is observed for other organisms (Little and Stevenson 2007, Elias et al. 2007).
The usefulness of DNA barcodes to discriminate mosquito genera of our dataset by using a tree-based approach was poorly supported. Approximately half of currently recognized genera represented by two or more species formed stable clusters in the NJ tree. Similarly, generic monophyly was weakly improved with the ML approach. In other insect genera, the monophyletic clusters based on DNA barcodes varies greatly depending on clustering method: e.g. Lepidoptera: Ithomiinae 50 to 61% recovered generic monophyly (Elias et al. 2007), Diptera: Chironomidae 40% (Ekrem et al. 2007, 2010), Simuliidae 62% (Rivera and Currie 2009) and Muscidae 40% (Renaud et al. 2012), Hymenoptera: Apoidea 100% success (Sheffield et al. 2009). It remains unclear whether this is due to lack of phylogenetic signal in COI at this depth, the type of tree-building method, or to the true lack of monophyly of genera as currently defined (Renaud et al. 2012). Conversely, a high level of correspondence at the species level is observed between morphology and molecular species limits in the tree-based approach in the present study. The performance of DNA-based specimen identification in Diptera using COI differs greatly in the literature, which varies from less than 50% to near 100% congruence levels (Cywinska et al. 2006, Smith et al. 2006, Whitworth et al. 2007, Rivera and Currie 2009, Meiklejohn et al. 2012, Renaud et al. 2012, Smit et al. 2013, Contrearas et al. 2014). In most of the studies, identification success rose upon relaxing the bootstrap requirement.
In tree-based methods, the non-monophyly at the species level represents the greatest challenge for taxon sampling and threshold approach (Meyer and Paulay 2005). Funk and Omland (2003) explained five possible reasons for non-monophyly at the species level, i.e. inadequate phylogenetic information, imperfect taxonomy, interspecific hybridization, incomplete lineage sorting, and unrecognized paralogy. There are cases in which tree-based analyses of DNA barcodes have failed to discriminate species of insects (Wiemers and Fiedler 2007, Whitworth et al. 2007, Foster et al. 2013). The non-monophyletic groups of our study includes thirty species of Anopheles, two species of Psorophora, and four or five, depending on the tree reconstruction method, species of Culex.
Culex was the most diverse and species rich genus in the sampled mosquitoes. Culex is a cosmopolitan genus and one of the largest groups of the family Culicidae (768 species divided among 26 subgenera). There are many areas of uncertainty regarding phylogenetic relationships within the genus, as well as some problems with the identification of some species. In our study, many difficulties arose attempting to identify specimens of Culex species. In many species, female identification is very problematic due to polymorphisms and ambiguities of the morphological characters (Forattini 2002, Laurito et al. 2013). Morphological identification of Culex species is based primarily on differences in male genitalia (Harbach 2011). Moreover, the presence of unknown species complexes within Culex makes species identification challenging (Harbach 2011, Laurito et al. 2013). As research previously suggested, the non-monophyly of given species of Culex approached during this study is not a surprise. Furthermore, in earlier studies only 42% of the Culex (Culex) previously identified morphologically to species were clustered with their conspecifics in the NJ tree (Laurito et al. 2013). Although our data are limited, present results point in the same direction and more taxonomic work is needed to assess the monophyly of Culex and the phylogenetic relationships within the genus.
Classification of the species within the genus Anopheles has always been challenging. The current system of subgeneric classification is based primarily on characteristics of the male genitalia (Harbach 2013). Many Anopheles species are morphologically indistinguishable and form cryptic species complexes, which require a molecular approach as the only effective tool for resolving their identification (Foster et al. 2013). The DNA barcode approach has been successfully used in corroborating lineages within the Anopheles albitarsis complex (Ruiz-Lopez et al. 2012). In the present study, difficulties in resolving species among the sequences of the subgenus Anopheles (Nyssorhynchus) were expected. Most of the overlapping of species was obtained among one section (myzorynchella section) and four of the five complexes comprising this group (nuneztovari, oswaldoi, strodei, albitarsis). Even if species level could not be reached for this difficult subgenus, barcodes allowed us to clearly identify the different species complexes present. Single genes, including the COI barcode region, are poor at confirming morphologically defined species and to estimate phylogenetic relationships within the subgenus Nyssorhynchus (Foster et al. 2013). However, a multi-locus approach (COI barcode region, nuclear white and CAD genes) was able to discriminate Nyssorhynchus species with greater accuracy (Foster et al. 2013).
The most important factor affecting the accuracy of species identification through public databases is the coverage and reliability of available sequences (Ekrem et al. 2007, Wiemers and Fiedler 2007, Virgilio et al. 2010). Unfortunately, the GenBank and BOLD databases have many records believed to be from misidentified specimens (Harris 2003, Meier and Dikow 2004, Meier et al. 2008), which are only obvious by incorporating accurately identified specimens into databases (Meyer and Paulay 2005, Meier et al. 2006). The importance of voucher specimens which can be reexamined is paramount. Furthermore, erroneous sequences may lead to an underestimation of the potential for species discrimination by DNA barcodes (Pape et al. 2009, Collins and Cruickshank 2013). In total, sequences for 133 Neotropical species were included in the tree-based analysis, which displayed species discrimination among most of the taxa, but not for all. We expect that as more studies provide sampling data, the number of entries will increase in databases, which should greatly improve the accuracy of search queries. Nonetheless, results of best match retrieved from any of the databases should not be used indiscriminately without considering the reliability of the comparison. Although BLAST and BOLD Identification System retrieved incongruent identifications for some of our sequences, these search tools are useful to check for sequence contaminations, especially when a species is sequenced for first time.
It is clear that reference libraries with properly identified sequences will facilitate the association of conspecific specimens and the detection of identification errors. The DNA barcodes produced in this work allowed for the identification of females and damaged specimens, which could not be done using morphological characteristics alone. This study highlights the potential of barcoding methodology to resolve taxonomic problems associated with limitations in morphological identification, but also draws attention to its limitations to discriminate species in some Neotropical mosquito genera, especially in Anopheles (Nyssorhynchus) species complexes and some Culex species. A larger taxon barcode library with correctly identified vouchers will help future studies. Despite the limitations in our survey, the DNA barcodes produced in this work are an important contribution to increase the scope of reference libraries with properly identified sequences and to assist in the identification of mosquito species from Colombia.
Acknowledgments
Many thanks to James Pecor (WRBU, USA) for confirming the identification of some specimens and for sharing useful taxonomic literature. Also, thanks to Claudia Etzbauer (ZFMK, Germany) for her support while obtaining molecular data and to two anonymous reviewers for improving our manuscript with helpful comments. Special thanks to David Fowler (UTK, USA) for logistical support during fieldwork.
Citation
Rozo-Lopez P, Mengual X (2015) Mosquito species (Diptera, Culicidae) in three ecosystems from the Colombian Andes: identification through DNA barcoding and adult morphology. ZooKeys 513: 39–64. doi: 10.3897/zookeys.513.9561
Supplementary materials
Mosquito specimens collected in the present study
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Paula Rozo-Lopez, Ximo Mengual
Data type: occurrence
Explanation note: Full collection site details, including geo-references and environmental conditions, of the mosquito taxa identified in this study.
COI mosquito sequences downloaded from NCBI and BOLD
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Paula Rozo-Lopez, Ximo Mengual
Data type: Text
Explanation note: Sequences from NCBI and BOLD. Sequences downloaded between December 2013 and February 2014 for all identified species with a minimum length of 480 bp of COI barcoding region, no stop codons and possible alignment among the majority of the sequences. Total of 1,159 sequences. In blue: Additional species sequences from other geographic areas for those Neotropical groups without available sequences from the Neotropics.
Mosquito specimens collected in Uraba during 2009 (unpublish data)
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Paula Rozo-Lopez, Ximo Mengual
Data type: occurrence
Explanation note: Additional sequences of mosquitoes collected in rural areas of Uraba (Antioquia, Colombia) during 2009, including sampling information.
Outgroup taxa used in the present study
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Paula Rozo-Lopez, Ximo Mengual
Data type: Text
Explanation note: Outgroup taxa used in the present study. Sequences downloaded from BOLD between December 2013 and February 2014.
NJ tree based on 1,292 sequences of Neotropical mosquitoes
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Paula Rozo-Lopez, Ximo Mengual
Data type: picture
Explanation note: Original Neighbour-Joining tree of the barcoding sequences of mosquito species listed for the Neotropics, based on Tamura-Nei genetic distances.
NJ tree based on 1,292 sequences of Neotropical mosquitoes
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Paula Rozo-Lopez, Ximo Mengual
Data type: multimedia
Explanation note: Original Neighbour-Joining tree of the barcoding sequences of mosquito species listed for the Neotropics, based on Tamura-Nei genetic distances (.tre).
ML tree based on 1,292 sequences of Neotropical mosquitoes
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Paula Rozo-Lopez, Ximo Mengual
Data type: picture
Explanation note: Original Maximum likelihood tree of the barcoding sequences of mosquito species listed for the Neotropics.
ML tree based on 1,292 sequences of Neotropical mosquitoes
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Paula Rozo-Lopez, Ximo Mengual
Data type: multimedia
Explanation note: Original Maximum likelihood tree of the barcoding sequences of mosquito species listed for the Neotropics (.tre).
References
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, et al. (2000) The genome sequence of Drosophila melanogaster. Science 287(5461): 2185–2195. doi: 10.1126/science.287.5461.2185 [DOI] [PubMed] [Google Scholar]
- Arnell JH. (1973) Mosquito studies (Diptera, Culicidae) XXXII. A revision of the genus Haemagogus. Contributions of the American Entomological Institute 10: 1–174. [Google Scholar]
- Arnell JH. (1976) Mosquito studies (Diptera, Culicidae) XXXIII. A revision of the Scapularis group of Aedes (Ochlerotatus). Contributions of the American Entomological Institute 13(3): 1–144. [Google Scholar]
- Ashfaq M, Hebert PDN, Mirza JH, Khan AM, Zafar Y, Mirza MS. (2014) Analyzing Mosquito (Diptera: Culicidae) Diversity in Pakistan by DNA Barcoding. PLoS ONE 9(5): . doi: 10.1371/journal.pone.0097268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austerlitz F, David O, Schaeffer B, Bleakley K, Olteanu M, Leblois R, Veuille M, Laredo C. (2009) DNA barcode analysis: a comparison of phylogenetic and statistical classification methods. BMC Bioinformatics 10 (Suppl 14): . doi: 10.1186/1471-2105-10-S14-S10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azari-Hamidian S, Linton YM, Abai MR, Ladonni H, Oshaghi MA, Hanafi-Bojd AA, Moosa-Kazemi SH, Shabkhiz H, Pakari A, Harbach RE. (2010) Mosquito (Diptera: Culicidae) fauna of the Iranian islands in the Persian Gulf. Journal of Natural History 44: 913–925. doi: 10.1080/00222930903437358 [Google Scholar]
- Baker AJ, Tavares ES, Elbourne RF. (2009) Countering criticisms of single mitochondrial DNA gene barcoding in birds. Molecular Ecology Resources 9(1): 257–268. doi: 10.1111/j.1755-0998.2009.02650.x [DOI] [PubMed] [Google Scholar]
- Baldwin CC, Weigt LA. (2012) A new species of Soapfish (Teleostei: Serranidae: Rypticus), with redescription of R. subbifrenatus and comments on the use of DNA barcoding in systematic studies. Copeia 1: 23–36. doi: 10.1643/CG-11-035 [Google Scholar]
- Barreto M, Burbano ME, Suárez M, Barreto P. (1996) Psorophora ciliata y otros mosquitos (Diptera: Culicidae) en Yolombó. Colombia Medica 27(2): 62–65. [Google Scholar]
- Becker N, Petric D, Zgomba M, Boase C, Madon M, Dahl C, Kaiser A. (2010) Mosquitoes and their control. Springer, Heidelbeg, 577 pp. doi: 10.1007/978-3-540-92874-4 [Google Scholar]
- Bergsten J, Bilton DT, Fujisawa T, Elliott M, Monaghan MT, Balke M, Hendrich L, Geijer J, Herrmann J, Foster GN, Ribera I, Nilsson AN, Barraclough TG, Vogler AP. (2012) The effect of geographical scale of sampling on DNA barcoding. Systematic Biology 61(5): 851–869. doi: 10.1093/sysbio/sys037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bram RA. (1967) The classification of the Culex subgenus Culex in the New World (Diptera, Culicidae). Proceedings of the US Natural History Museum 120: 1–122. doi: 10.5479/si.00963801.120-3557.1 [Google Scholar]
- Brochero H, Quiñones ML. (2008) Retos de la entomología médica para la vigilancia en salud pública en Colombia: Reflexión para el caso de malaria. Biomedica 28: 18–24. doi: 10.7705/biomedica.v28i1.105 [PubMed] [Google Scholar]
- Brower AVZ. (2006) Problems with DNA barcodes for species delimitation: ‘ten species’ of Astraptes fulgerator reassessed (Lepidoptera: Hesperiidae). Systematics and Biodiversity 4: 127–132. doi: 10.1017/S147720000500191X [Google Scholar]
- Casiraghi M, Labra M, Ferri E, Galimberti A, De Mattia F. (2010) DNA barcoding: a six-question tour to improve users’ awareness about the method. Briefing in Bioinformatics 11: 440–453. doi: 10.1093/bib/bbq003 [DOI] [PubMed] [Google Scholar]
- Chan A, Chiang LP, Hapuarachchi HC, Tan CH, Pang SC, Lee R, Lee KS, Ng LC, Lam-Phua SG. (2014) DNA barcoding: complementing morphological identification of mosquito species in Singapore. Parasites and Vectors 7: 569. doi: 10.1186/s13071-014-0569-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaverri LG. (2009) Culicidae (mosquitos, zancudos). In: Brown BV, Borkent A, Cumming JM, Wood DM, Woodley NE, Zumbado MA. (Eds) Manual of Central American Diptera Vol. 1 NRC-CNRC Press, Ontario, 369–388. [Google Scholar]
- Chaves ME, Arango N. (1998) Informe Nacional Sobre el Estado de la Biodiversidad en Colombia-1997. I.A. von Humboldt, Bogota. [Google Scholar]
- Collins RA, Cruickshank RH. (2013) The seven deadly sins of DNA barcoding. Molecular Ecology Resources 13(6): 969–975. doi: 10.1111/1755-0998.12046 [DOI] [PubMed] [Google Scholar]
- Contreras MA, Vivero RJ, Velez ID, Porter CH, Uribe S. (2014) DNA Barcoding for the identification of sand fly species (Diptera, Psychodidae, Phlebotominae) in Colombia. PLoS ONE 9(1): . doi: 10.1371/journal.pone.0085496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook S, Diallo M, Sall AA, Cooper A, Holmes EC. (2005) Mitochondrial markers for molecular identification of Aedes mosquitoes (Diptera: Culicidae) involved in transmission of arboviral disease in West Africa. Morphology, Sistematics and Evolution 42(1): 19–28. doi: 10.1603/0022-2585(2005)042[0019:MMFMIO]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Cova-Garcia P. (1961) Notas sobre los Anofelinos de Venezuela y su identificación. Grafos, Caracas, 212 pp. [Google Scholar]
- Cova-Garcia P, Sutil E, Rausseo JA. (1966) Mosquitos (culicinos) de Venezuela. Ministerio de Sanidad y Asistencia Social, Caracas, 410 pp. [Google Scholar]
- Cywinska A, Hunter FF, Hebert PDN. (2006) Identifying Canadian mosquito species through DNA barcodes. Medical and Veterinary Entomology 20(4): 413–424. doi: 10.1371/journal.pone.0047051 [DOI] [PubMed] [Google Scholar]
- DeSalle R, Egan MG, Siddall M. (2005) The unholy trinity: taxonomy, species delimitation and DNA barcoding. Philosophical Transactions of Royal Society of London B Biological Science 360(1462): 1905–1916. doi: 10.1098/rstb.2005.1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekrem T, Willassen E, Stur E. (2007) A comprehensive DNA sequence library is essential for identification with DNA barcodes. Molecular Phylogenetics and Evolution 43: 530–542. doi: 10.1016/j.ympev.2006.11.021 [DOI] [PubMed] [Google Scholar]
- Ekrem T, Willassen E, Stur E. (2010) Phylogenetic utility of five genes for dipteran phylogeny: A test case in the Chironomidae leads to generic synonymies. Molecular Phylogenetics and Evolution 57: 561–571. doi: 10.1016/j.ympev.2010.06.006 [DOI] [PubMed] [Google Scholar]
- Elias M, Hill RI, Willmott KR, Dasmahapatra KK, Brower AVZ, Mallet J, Jiggins CD. (2007) Limited performance of DNA barcoding in a diverse community of tropical butterflies. Proceedings of the Royal Society B 274: 2881–2889. doi: 10.1098/rspb.2007.1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etter A, McAlpine C, Wilson K, Phinn S, Possingham H. (2006) Regional patterns of agricultural land use and deforestation in Colombia. Agriculture, Ecosystems & Environment 114(2): 369–386. doi: 10.1016/j.agee.2005.11.013 [Google Scholar]
- Foley DH, Rueda LM, Wilkerson RC. (2007) Insight into global mosquito biogeography from country species records. Journal of Medical Entomology 44(4): 554–567. doi: 10.1603/0022-2585(2007)44[554:IIGMBF]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Foley DH, Weitzman AL, Miller SE, Faran ME, Rueda LM, Wilkerson RC. (2008) The value of georeferenced collection records for predicting patterns of mosquito species richness and endemism in the Neotropics. Ecological Entomology 33: 12–23. doi: 10.1111/j.1365-2311.2007.00927.x [Google Scholar]
- Foley DH, Wilkerson RC, Birney I, Harrison S, Christensen J, Rueda LM. (2010) MosquitoMap and the Mal-area calculator: New web tools to relate mosquito species distribution with vector borne disease. International Journal of Health Geographics 9(11): 1–8. doi: 10.1186/1476-072X-9-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology 3(5): 294–299. [PubMed] [Google Scholar]
- Forattini OP. (2002) Culicidologia Médica. Vol.2. Identificação, Biologia, Epidemiologia. Edusp, São Paulo, 860 pp. [Google Scholar]
- Foster PG, Bergo ES, Bourke BP, Oliveira TMP, Nagaki SS, Sant’Ana DC, Sallum MA. (2013) Phylogenetic analysis and DNA-based species confirmation in Anopheles (Nyssorhynchus). PLoS ONE 8(2): . doi: 10.1371/journal.pone.0054063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk DJ, Omland KE. (2003) Species-level paraphyly and polyphyly: Frequency, causes, and consequences, with insights from animal mitochondrial DNA. Annual Review of Ecology, Evolution, and Systematics 34: 397–423. doi: 10.1146/annurev.ecolsys.34.011802.132421 [Google Scholar]
- Gaffigan T, Pecor J. (1997) WRBU: Collecting, Rearing, Mounting and Shipping Mosquitoes. http://wrbu.si.edu/Techniques.html [accessed 23 February 2015]
- Goldstein PZ, DeSalle R. (2011) Integrating DNA barcode data and taxonomic practice: Determination, discovery, and description. Bioessays 33(2): 135–47. doi: 10.1002/bies.201000036 [DOI] [PubMed] [Google Scholar]
- González R, Carrejo N. (2007) Introducción al estudio taxonómico de Anopheles de Colombia: Claves y notas de distribución. Universidad del Valle, Cali, 237 pp. [Google Scholar]
- Guedes AS, Souza MA. (1964) Sobre Psorophora (Janthinosoma) albigenu Lutz, 1908 e Psorophora (Janthinosoma) albipes (Theobald, 1907) (Diptera, Culicidae). Revista Brasileira de Malaiologia e Doencas Tropicais 16: 471–486. [Google Scholar]
- Hajibabaei M, Singer GAC, Hebert PDN, Hickey DA. (2007) DNA barcoding: How it complements taxonomy, molecular phylogenetics, and population genetics. Trends in Genetics 23(4): 167–72. doi: 10.1016/j.tig.2007.02.001 [DOI] [PubMed] [Google Scholar]
- Harbach RE. (2007) The Culicidae (Diptera): A review of taxonomy, classification and Phylogeny. Zootaxa 638: 591–638. doi: 10.1186/1471-2148-9-298 [Google Scholar]
- Harbach RE. (2011) Classification within the cosmopolitan genus Culex (Diptera: Culicidae): The foundation for molecular systematics and phylogenetic research. Acta Tropica 120: 1–14. doi: 10.1016/j.actatropica.2011.06.005 [DOI] [PubMed] [Google Scholar]
- Harbach RE. (2013) Genus Anopheles Meigen, 1818. http://mosquito-taxonomic-inventory.info/genus-emanophelesem-meigen-1818-0 [accessed 23 February 2015]
- Harbach RE. (2015) Mosquito Taxonomic Inventory. Valid Species List. http://mosquito-taxonomic-inventory.info/valid-species-list [accessed 12 February 2015]
- Harris DJ. (2003) Can you bank on GenBank? Trends in Ecology and Evolution 18(7): 317–319. doi: 10.1016/S0169-5347(03)00150-2 [Google Scholar]
- Hebert PDN, Cywinska A, Ball SL, deWaard JR. (2003a) Biological identifications through DNA barcodes. Proceedings of the Royal Society of London B 270: 313–321. doi: 10.1098/rspb.2002.2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert PDN, Ratnasingham S, DeWaard JR. (2003b) Barcoding animal life: Cytochrome C oxidase subunit 1 divergences among closely related species. Proceedings of the Royal Society of London B 270: S96–S99. doi: 10.1098/rsbl.2003.0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert PDN, Stoeckle MY, Zemlak TS, Francis CM. (2004) Identification of birds through DNA barcodes. PLoS Biology 2: 1657–1663. doi: 10.1371/journal.pbio.0020312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández J, Ortiz R, Walschburger T, Hurtado A. (1992) Estado de la Biodiversidad en Colombia. La Diversidad Biológica de Iberoamérica. I CYTED, Mexico, DF, 146 pp. [Google Scholar]
- Huemer P, Mutaten M. (2015) Alpha taxonomy of the genus Kessleria Nowicki, 1864, revisited in light of DNA-barcoding (Lepidoptera, Yponomeutidae). ZooKeys 503: 89–133. doi: 10.3897/zookeys.503.9590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinbo U, Kato T, Ito M. (2011) Current progress in DNA barcoding and future implications for entomology. Entomological Science 14: 107–124. doi: 10.1111/j.1479-8298.2011.00449.x [Google Scholar]
- Judd DD. (1998) Review of a bromeliad-ovipositing lineage in Wyeomyia and the resurrection of Hystatomyia (Diptera: Culicidae). Annals of the Entomological Society of America 91: 572–589. doi: 10.1093/aesa/91.5.572 [Google Scholar]
- Knight JW, Haeger JS. (1971) Key to adults of the Culex subgenera Melanoconion and Mochlostyrax of eastern North America. Journal of Medical Entomology 8(5): 551–555. doi: 10.1093/jmedent/8.5.551 [DOI] [PubMed] [Google Scholar]
- Komp WHW. (1942) The Anopheline mosquitoes of the Caribbean region. Bulletin 179 U.S.PHS. National Institute of Health, Washington D.C., 200 pp. [Google Scholar]
- Kumar NP, Rajavel AR, Natarajan R, Jambulingam P. (2007) DNA barcodes can distinguish species of Indian mosquitoes (Diptera: Culicidae). Journal of Medical Entomology 44(1): 1–7. doi: 10.1603/0022-2585(2007)44[1:DBCDSO]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Kumm HW, Osorno-Mesa E, Boshell-Manrique J. (1946) Studies on mosquitoes of the genus Haemagogus in Colombia (Diptera, Culicidae). American Journal of Hygiene 43(1): 13–28. [DOI] [PubMed] [Google Scholar]
- Lane J. (1953) Neotropical Culicidae Vol. 1 & 2 University of São Paulo, São Paulo, 1112 pp. [Google Scholar]
- Lanfear R, Calcott B, Ho SYW, Guindon S. (2012) PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Molecular Biology and Evolution 29(6): 1695–1701. doi: 10.1093/molbev/mss020 [DOI] [PubMed] [Google Scholar]
- Laurito M, de Oliveira TM, Almirón WR, Sallum MA. (2013) COI barcode versus morphological identification of Culex (Culex) (Diptera: Culicidae) species: A case study using samples from Argentina and Brazil. Memórias Do Instituto Oswaldo 108(I): 110–122. doi: 10.1590/0074-0276130457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi-Castillo R. (1951) Los mosquitos del género de Haemagogus Williston, 1896 en America del Sur. Don Bosco, Guayaquil, 74 pp. [Google Scholar]
- Linton YM, Pecor JE, Porter CH, Mitchell L, Garzón-Moreno A, Foley DH, Brooks D, Wilkerson RC. (2013) Mosquitoes of eastern Amazonian Ecuador: biodiversity, bionomics and barcodes. Memórias do Instituto Oswaldo Cruz 108(1): 100–109. doi: 10.1590/0074-0276130440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liria J, Navarro JC. (2003) Psorophora (Psorophora) lineata (Humboldt, 1819) y Psorophora (Pso.) saeva Dyar & Knab, 1906 (Diptera: Culicidae). Correcciones en su identificacion. Entomotropica 18(2): 113–119. [Google Scholar]
- Liria J, Navarro JC. (2009) Clave fotográfica para hembras de Haemagogus Williston, 1896 (Diptera: Culicidae) de Venezuela, con un nuevo registro para el país. Boletin de Malariologia y Salud Ambiental 2: 283–282. [Google Scholar]
- Little DP, Stevenson DW. (2007) A comparison of algorithms for the identification of specimens using DNA barcodes: Examples from gymnosperms. Cladistics 23(1): 1–21. doi: 10.1111/j.1096-0031.2006.00126.x [DOI] [PubMed] [Google Scholar]
- Meier R, Dikow T. (2004) Significance of specimen databases from taxonomic revisions for estimating and mapping the global species diversity of invertebrates and repatriating reliable and complete specimen data. Conservation Biology 18: 478–488. doi: 10.1111/j.1523-1739.2004.00233.x [Google Scholar]
- Meier R, Shiyang K, Vaidya G, Ng PKL. (2006) DNA barcoding and taxonomy in Diptera: A tale of high intraspecific variability and low identification success. Systematic Biology 55(5): 715–728. doi: 10.1080/10635150600969864 [DOI] [PubMed] [Google Scholar]
- Meier R, Zhang GY, Ali F. (2008) The use of mean instead of smallest interspecific distances exaggerates the size of the “barcoding gap” and leads to misidentification. Systematic Biology 57(5): 809–813. doi: 10.1080/10635150802406343 [DOI] [PubMed] [Google Scholar]
- Meiklejohn KA, Wallman JF, Cameron SL, Dowton M. (2012) Comprehensive evaluation of DNA barcoding for the molecular species identification of forensically important Australian Sarcophagidae (Diptera). Invertebrate Systematics 26(6): 515–525. doi: 10.1071/IS12008 [Google Scholar]
- Meyer CP, Paulay G. (2005) DNA barcoding: Error rates based on comprehensive sampling. PLoS Biology 3: 2229–2238. doi: 10.1371/journal.pbio.0030422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina JA, Hildebrand P, Olano VA, Muñoz P, Barreto M, Guhl F. (2000) Fauna de insectos hematófagos del sur del parque natural nacional Chiribiquete, Caquetá, Colombia. Biomedica 20: 314–326. doi: 10.7705/biomedica.v20i4.1075 [Google Scholar]
- Montoya-Lerma J, Solarte YA, Giraldo-Calderón GI, Quiñones ML, Ruiz-López F, Wilkerson RC, González R. (2011) Malaria vector species in Colombia: A review. Memórias do Instituto Oswaldo Cruz 106: 223–238. doi: 10.1590/S0074-02762011000900028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz C, Cicero C. (2004) DNA Barcoding: promise and pitfalls. PLoS Biology 2: . doi: 10.1371/journal.pbio.0020354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta MA, de Oliveira RL. (2000) The subgenus Dendromyia Theobald: A review with redescriptions of four species (Diptera: Culicidae). Memorias do Instituto Oswaldo Cruz 95(5): 649–683. [DOI] [PubMed] [Google Scholar]
- Munstermann LE, Conn JE. (1997) Systematics of mosquito disease vectors (Diptera, Culicidae): Impact of molecular biology and cladistic analysis. Annual Review of Entomology 42: 351–369. doi: 10.1146/annurev.ento.42.1.351 [DOI] [PubMed] [Google Scholar]
- Nagoshi RN, Brambila J, Meagher RL. (2011) Use of DNA barcodes to identify invasive armyworm Spodoptera species in Florida. Journal of Insect Science 11: . doi: 10.1673/031.011.15401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olano VA, Brochero HL, Sáenz R, Quiñones ML, Molina JA. (2001) Mapas preliminares de la distribución de especies de Anopheles vectores de malaria en Colombia. Biomedica 21: 402–408. [Google Scholar]
- Pape T, Bickel D, Meier R. (2009) Diptera Diversity: Status, Challenges and Tools. Brill Academic Publishers, Leiden, 459 pp. [Google Scholar]
- Pecor JE, Mallampalli VL, Harbach RE, Peyton EL. (1992) Catalog and illustrated review of the subgenus Melanoconion of Culex (Diptera: Culicidae). Contributions of the American Entomological Institute 27: 1–228. [Google Scholar]
- Posada D, Buckley TR. (2004) Model selection and model averaging in phylogenetics: Advantages of Akaike information criterion and bayesian approaches over likelihood ratio tests. Systematic Biology 53(5): 793–808. doi: 10.1080/10635150490522304 [DOI] [PubMed] [Google Scholar]
- Ratnasingham S, Herbert PDN. (2007) BOLD: The Barcode of Life Data System BOLD. Molecular Ecology Notes 7(3): 355–364. doi: 10.1111/j.1471-8286.2007.01678.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinert JF. (2000) Comparative anatomy of the female genitalia of genera and subgenera in tribe Aedini (Diptera: Culicidae). Genus Aedes Meigen. Contributions of the American Entomological Institute 32(3): 1–102. [Google Scholar]
- Renaud AK, Savage J, Adamowicz SJ. (2012) DNA barcoding of Northern Nearctic Muscidae (Diptera) reveals high correspondence between morphological and molecular species limits. BMC Ecology 12(24): 1–15. doi: 10.1186/1472-6785-12-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera J, Currie D. (2009) Identification of Nearctic black flies using DNA barcodes (Diptera: Simuliidae). Molecular Ecology Resources 9: 224–236. doi: 10.1111/j.1755-0998.2009.02648.x [DOI] [PubMed] [Google Scholar]
- Rozeboom LE, Komp WHW. (1950) A review of the species of Culex of the subgenus Melanoconion (Diptera, Culicidae). Annals of the Entomological Society of America 43: 75–114. doi: 10.1093/aesa/43.1.75 [Google Scholar]
- Rozo-Lopez P, Mengual X. (2015) Updated list of the mosquitoes of Colombia (Diptera: Culicidae). Biodiversity Data Journal 3: . doi: 10.3897/BDJ.3.e4567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Lopez F, Wilkerson RC, Conn JE, McKeon SN, Levin DM, Quiñones ML, Povoda MM, Linton YM. (2012) DNA barcoding reveals both known and novel taxa in the albitarsis group (Anopheles: Nyssorhynchus) of Neotropical malaria vectors. Parasites and Vectors 5(44): 1–12. doi: 10.1186/1756-3305-5-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. (1987) The neighbour-joining method: A new method for reconstructing phylogenetic tress. Molecular Biology and Evolution 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Scheffer SJ, Lewis ML, Joshi RC. (2006) DNA barcoding applied to invasive leafminers (Diptera: Agromyzidae) in the Philippines. Annals of the Entomological Society of America 99(2): 204–210. doi: 10.1603/0013-8746(2006)099[0204:DBATIL]2.0.CO;2 [Google Scholar]
- Shannon RC. (1934) The genus Mansonia (Culicidae) in the Amazon valley. Proceeding of the Entomological Society of Washington 36(5): 99–110. [Google Scholar]
- Sheffield CS, Hebert PDN, Kevan PG, Packer L. (2009) DNA barcoding a regional bee (Hymenoptera: Apoidea) fauna and its potential for ecological studies. Molecular Ecology Resources 9: 196–207. doi: 10.1111/j.1755-0998.2009.02645.x [DOI] [PubMed] [Google Scholar]
- Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook P. (1994) Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Annals of the Entomological Society of America 87(6): 651–701. doi: 10.1146/annurev.ecolsys.37.091305.110018 [Google Scholar]
- Sirivanakarn S. (1982) A review of the systematics and a proposed scheme of internal classification of the New World subgenus Melanoconion of Culex (Diptera, Culicidae). Mosquito Systematics 14: 265–338. [Google Scholar]
- Smit J, Reijnen B, Stokvis F. (2013) Half of the European fruit fly species barcoded (Diptera, Tephritidae); a feasibility test for molecular identification. ZooKeys 365: 279–305. doi: 10.3897/zookeys.365.5819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MA, Woodley NE, Janzen DH, Hallwachs W, Hebert PDN. (2006) DNA barcodes reveal cryptic host-specificity within the presumed polyphagous members of a genus of parasitoid flies (Diptera: Tachinidae). PNAS 103: 3657–3662. doi: 10.1073/pnas.0511318103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivathsan A, Meier R. (2012) On the inappropriate use of the Kimura-2- parameter (K2P) divergences in the barcoding literature. Cladistics 28: 190–194. doi: 10.1111/j.1096-0031.2011.00370.x [DOI] [PubMed] [Google Scholar]
- Stoeckle MY, Thaler DS. (2014) DNA Barcoding Works in Practice but Not in (Neutral) Theory. PLoS ONE 9(7): . doi: 10.1371/journal.pone.0100755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone A. (1944) Notes on the genus Trichoprosopon (Diptera, Culicidae). Revista de Entomologia 15: 335–341. [Google Scholar]
- Strickman D, Pratt J. (1989) Redescription of Culex corniger and elevation of Culex (Culex) lactator Dyar and Knab from synonymy based on specimens from Central America (Diptera: Culicidae). Proceedings of Entomological Society 91: 551–574. [Google Scholar]
- Tamura K, Nei M. (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molecular Biology and Evolution 10(3): 512–526. [DOI] [PubMed] [Google Scholar]
- Tavares ES, Gonçalves P, Miyaki CY, Baker AJ. (2011) DNA barcode detects high genetic structure within Neotropical bird species. PLoS ONE 6(12): . doi: 10.1371/journal.pone.0028543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteirt V, Nagy ZT, Roelants P, Denis L, Breman FC, Damiens D, Dekoninck W, Backeljau T, Coosemans M, Van Bortel W. (2015) Identification of Belgian mosquito species (Diptera: Culicidae) by DNA barcoding. Molecular Ecology Recourses (2): 449–457. doi: 10.1111/1755-0998.12318 [DOI] [PubMed]
- Virgilio M, Backeljau T, Nevado B, Meyer MD. (2010) Comparative performances of DNA barcoding across insect orders. BMC Bioinformatics 11: . doi: 10.1186/1471-2105-11-206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Li C, Guo X, Xing D, Dong Y, Wang Z, Zhang Y, Liu M, Zheng Z, Zhang H, Zhu X, Wu Z, Zhao T. (2012) Identifying the main mosquito species in China based on DNA barcoding. PLoS ONE 7(10): . doi: 10.1371/journal.pone.0047051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitworth T, Dawson R, Magalon H, Baudry E. (2007) DNA barcoding cannot reliably identify species of the blowfly genus Protocalliphora (Diptera: Calliphoridae). Proceedings of the Royal B 274: 1731–1739. doi: 10.1098/rspb.2007.0062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemers M, Fiedler K. (2007) Does the DNA barcoding gap exist? A case study in blue butterflies (Lepidoptera: Lycaenidae). Frontiers in Zoology 4: . doi: 10.1186/1742-9994-4-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JJ, Rougerie R, Schonfeld J, Janzen DH, Hallwachs W, Hajibabaei W, Kitching IJ, Haxaire J, Hebert PDN. (2011) When species matches are unavailable are DNA barcodes correctly assigned to higher taxa? An assessment using sphingid moths. BMC Ecology 11: . doi: 10.1186/1472-6785-11-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavortink TJ. (1974) The status of taxonomy of mosquitoes by the use of morphological characters. Mosquito Systematics 6(2): 130–133. [Google Scholar]
- Zavortink TJ. (1979) The new Sabethine genus Johnbelkinia and a preliminary reclassification of the composite genus Trichoprosopon. Contributions of the American Entomological Institute 17: 1–61. [Google Scholar]
- Zwickl DJ. (2006) Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. PhD thesis, University of Texas at Austin, Austin, USA.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mosquito specimens collected in the present study
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Paula Rozo-Lopez, Ximo Mengual
Data type: occurrence
Explanation note: Full collection site details, including geo-references and environmental conditions, of the mosquito taxa identified in this study.
COI mosquito sequences downloaded from NCBI and BOLD
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Paula Rozo-Lopez, Ximo Mengual
Data type: Text
Explanation note: Sequences from NCBI and BOLD. Sequences downloaded between December 2013 and February 2014 for all identified species with a minimum length of 480 bp of COI barcoding region, no stop codons and possible alignment among the majority of the sequences. Total of 1,159 sequences. In blue: Additional species sequences from other geographic areas for those Neotropical groups without available sequences from the Neotropics.
Mosquito specimens collected in Uraba during 2009 (unpublish data)
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Paula Rozo-Lopez, Ximo Mengual
Data type: occurrence
Explanation note: Additional sequences of mosquitoes collected in rural areas of Uraba (Antioquia, Colombia) during 2009, including sampling information.
Outgroup taxa used in the present study
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Paula Rozo-Lopez, Ximo Mengual
Data type: Text
Explanation note: Outgroup taxa used in the present study. Sequences downloaded from BOLD between December 2013 and February 2014.
NJ tree based on 1,292 sequences of Neotropical mosquitoes
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Paula Rozo-Lopez, Ximo Mengual
Data type: picture
Explanation note: Original Neighbour-Joining tree of the barcoding sequences of mosquito species listed for the Neotropics, based on Tamura-Nei genetic distances.
NJ tree based on 1,292 sequences of Neotropical mosquitoes
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Paula Rozo-Lopez, Ximo Mengual
Data type: multimedia
Explanation note: Original Neighbour-Joining tree of the barcoding sequences of mosquito species listed for the Neotropics, based on Tamura-Nei genetic distances (.tre).
ML tree based on 1,292 sequences of Neotropical mosquitoes
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Paula Rozo-Lopez, Ximo Mengual
Data type: picture
Explanation note: Original Maximum likelihood tree of the barcoding sequences of mosquito species listed for the Neotropics.
ML tree based on 1,292 sequences of Neotropical mosquitoes
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Paula Rozo-Lopez, Ximo Mengual
Data type: multimedia
Explanation note: Original Maximum likelihood tree of the barcoding sequences of mosquito species listed for the Neotropics (.tre).