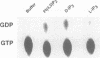
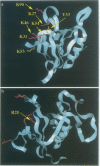
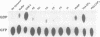Abstract
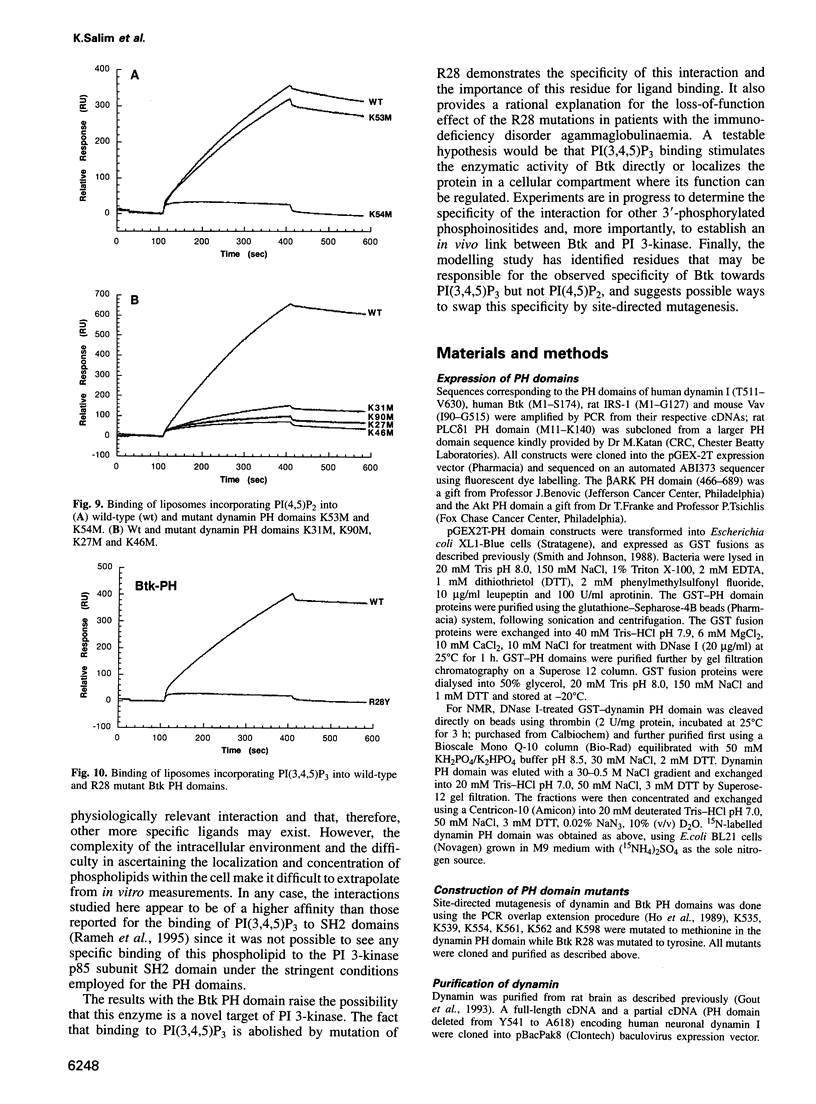
Pleckstrin homology (PH) domains may act as membrane localization modules through specific interactions with phosphoinositide phospholipids. These interactions could represent responses to second messengers, with scope for regulation by soluble inositol polyphosphates. A biosensor-based assay was used here to probe interactions between PH domains and unilamellar liposomes containing different phospholipids and to demonstrate specificity for distinct phosphoinositides. The dynamin PH domain specifically interacted with liposomes containing phosphatidylinositol-4,5-bisphosphate [PI(4,5)P2] and, more weakly, with liposomes containing phosphatidylinositol-4-phosphate [PI(4)P]. This correlates with phosphoinositide activation of the dynamin GTPase. The functional GTPase of a dynamin mutant lacking the PH domain, however, cannot be activated by PI(4,5)P2. The phosphoinositide-PH domain interaction can be abolished selectively by point mutations in the putative binding pocket predicted by molecular modelling and NMR spectroscopy. In contrast, the Bruton's tyrosine kinase (Btk)PH domain specifically bound liposomes containing phosphatidylinositol-3,4,5-trisphosphate [PI(3,4,5)P3]: an interaction requiring Arg28, a residue found to be mutated in some X-linked agammaglobulinaemia patients. A rational explanation for these different specificities is proposed through modelling of candidate binding pockets and is supported by NMR spectroscopy.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abrams C. S., Wu H., Zhao W., Belmonte E., White D., Brass L. F. Pleckstrin inhibits phosphoinositide hydrolysis initiated by G-protein-coupled and growth factor receptors. A role for pleckstrin's PH domains. J Biol Chem. 1995 Jun 16;270(24):14485–14492. doi: 10.1074/jbc.270.24.14485. [DOI] [PubMed] [Google Scholar]
- Belmont J. W. Insights into lymphocyte development from X-linked immune deficiencies. Trends Genet. 1995 Mar;11(3):112–116. doi: 10.1016/S0168-9525(00)89012-5. [DOI] [PubMed] [Google Scholar]
- Burgering B. M., Coffer P. J. Protein kinase B (c-Akt) in phosphatidylinositol-3-OH kinase signal transduction. Nature. 1995 Aug 17;376(6541):599–602. doi: 10.1038/376599a0. [DOI] [PubMed] [Google Scholar]
- Downing A. K., Driscoll P. C., Gout I., Salim K., Zvelebil M. J., Waterfield M. D. Three-dimensional solution structure of the pleckstrin homology domain from dynamin. Curr Biol. 1994 Oct 1;4(10):884–891. doi: 10.1016/s0960-9822(00)00197-4. [DOI] [PubMed] [Google Scholar]
- Eck M. J., Dhe-Paganon S., Trüb T., Nolte R. T., Shoelson S. E. Structure of the IRS-1 PTB domain bound to the juxtamembrane region of the insulin receptor. Cell. 1996 May 31;85(5):695–705. doi: 10.1016/s0092-8674(00)81236-2. [DOI] [PubMed] [Google Scholar]
- Ferguson K. M., Lemmon M. A., Schlessinger J., Sigler P. B. Crystal structure at 2.2 A resolution of the pleckstrin homology domain from human dynamin. Cell. 1994 Oct 21;79(2):199–209. doi: 10.1016/0092-8674(94)90190-2. [DOI] [PubMed] [Google Scholar]
- Ferguson K. M., Lemmon M. A., Schlessinger J., Sigler P. B. Structure of the high affinity complex of inositol trisphosphate with a phospholipase C pleckstrin homology domain. Cell. 1995 Dec 15;83(6):1037–1046. doi: 10.1016/0092-8674(95)90219-8. [DOI] [PubMed] [Google Scholar]
- Franke T. F., Yang S. I., Chan T. O., Datta K., Kazlauskas A., Morrison D. K., Kaplan D. R., Tsichlis P. N. The protein kinase encoded by the Akt proto-oncogene is a target of the PDGF-activated phosphatidylinositol 3-kinase. Cell. 1995 Jun 2;81(5):727–736. doi: 10.1016/0092-8674(95)90534-0. [DOI] [PubMed] [Google Scholar]
- Fushman D., Cahill S., Lemmon M. A., Schlessinger J., Cowburn D. Solution structure of pleckstrin homology domain of dynamin by heteronuclear NMR spectroscopy. Proc Natl Acad Sci U S A. 1995 Jan 31;92(3):816–820. doi: 10.1073/pnas.92.3.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gout I., Dhand R., Hiles I. D., Fry M. J., Panayotou G., Das P., Truong O., Totty N. F., Hsuan J., Booker G. W. The GTPase dynamin binds to and is activated by a subset of SH3 domains. Cell. 1993 Oct 8;75(1):25–36. [PubMed] [Google Scholar]
- Harlan J. E., Hajduk P. J., Yoon H. S., Fesik S. W. Pleckstrin homology domains bind to phosphatidylinositol-4,5-bisphosphate. Nature. 1994 Sep 8;371(6493):168–170. doi: 10.1038/371168a0. [DOI] [PubMed] [Google Scholar]
- Hinshaw J. E., Schmid S. L. Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding. Nature. 1995 Mar 9;374(6518):190–192. doi: 10.1038/374190a0. [DOI] [PubMed] [Google Scholar]
- Hirata M., Kanematsu T., Sakuma K., Koga T., Watanabe Y., Ozaki S., Yagisawa H. D-myo-inositol 1,4,5-trisphosphate binding domain of phospholipase C-delta 1. Biochem Biophys Res Commun. 1994 Dec 30;205(3):1563–1571. doi: 10.1006/bbrc.1994.2845. [DOI] [PubMed] [Google Scholar]
- Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989 Apr 15;77(1):51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Hyvönen M., Macias M. J., Nilges M., Oschkinat H., Saraste M., Wilmanns M. Structure of the binding site for inositol phosphates in a PH domain. EMBO J. 1995 Oct 2;14(19):4676–4685. doi: 10.1002/j.1460-2075.1995.tb00149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James S. R., Downes C. P., Gigg R., Grove S. J., Holmes A. B., Alessi D. R. Specific binding of the Akt-1 protein kinase to phosphatidylinositol 3,4,5-trisphosphate without subsequent activation. Biochem J. 1996 May 1;315(Pt 3):709–713. doi: 10.1042/bj3150709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. B., Rhee S. G. Significance of PIP2 hydrolysis and regulation of phospholipase C isozymes. Curr Opin Cell Biol. 1995 Apr;7(2):183–189. doi: 10.1016/0955-0674(95)80026-3. [DOI] [PubMed] [Google Scholar]
- Lemmon M. A., Ferguson K. M., O'Brien R., Sigler P. B., Schlessinger J. Specific and high-affinity binding of inositol phosphates to an isolated pleckstrin homology domain. Proc Natl Acad Sci U S A. 1995 Nov 7;92(23):10472–10476. doi: 10.1073/pnas.92.23.10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon M. A., Ferguson K. M., Schlessinger J. PH domains: diverse sequences with a common fold recruit signaling molecules to the cell surface. Cell. 1996 May 31;85(5):621–624. doi: 10.1016/s0092-8674(00)81022-3. [DOI] [PubMed] [Google Scholar]
- Liu J. P., Robinson P. J. Dynamin and endocytosis. Endocr Rev. 1995 Oct;16(5):590–607. doi: 10.1210/edrv-16-5-590. [DOI] [PubMed] [Google Scholar]
- Macias M. J., Musacchio A., Ponstingl H., Nilges M., Saraste M., Oschkinat H. Structure of the pleckstrin homology domain from beta-spectrin. Nature. 1994 Jun 23;369(6482):675–677. doi: 10.1038/369675a0. [DOI] [PubMed] [Google Scholar]
- Myers M. G., Jr, Grammer T. C., Brooks J., Glasheen E. M., Wang L. M., Sun X. J., Blenis J., Pierce J. H., White M. F. The pleckstrin homology domain in insulin receptor substrate-1 sensitizes insulin signaling. J Biol Chem. 1995 May 19;270(20):11715–11718. doi: 10.1074/jbc.270.20.11715. [DOI] [PubMed] [Google Scholar]
- Nolde D. E., Sobol A. G., Pluzhnikov K. A., Grishin E. V., Arseniev A. S. Three-dimensional structure of ectatomin from Ectatomma tuberculatum ant venom. J Biomol NMR. 1995 Jan;5(1):1–13. doi: 10.1007/BF00227465. [DOI] [PubMed] [Google Scholar]
- Pawson T. Protein modules and signalling networks. Nature. 1995 Feb 16;373(6515):573–580. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- Pitcher J. A., Touhara K., Payne E. S., Lefkowitz R. J. Pleckstrin homology domain-mediated membrane association and activation of the beta-adrenergic receptor kinase requires coordinate interaction with G beta gamma subunits and lipid. J Biol Chem. 1995 May 19;270(20):11707–11710. doi: 10.1074/jbc.270.20.11707. [DOI] [PubMed] [Google Scholar]
- Rameh L. E., Chen C. S., Cantley L. C. Phosphatidylinositol (3,4,5)P3 interacts with SH2 domains and modulates PI 3-kinase association with tyrosine-phosphorylated proteins. Cell. 1995 Dec 1;83(5):821–830. doi: 10.1016/0092-8674(95)90195-7. [DOI] [PubMed] [Google Scholar]
- Rawlings D. J., Saffran D. C., Tsukada S., Largaespada D. A., Grimaldi J. C., Cohen L., Mohr R. N., Bazan J. F., Howard M., Copeland N. G. Mutation of unique region of Bruton's tyrosine kinase in immunodeficient XID mice. Science. 1993 Jul 16;261(5119):358–361. doi: 10.1126/science.8332901. [DOI] [PubMed] [Google Scholar]
- Shaw G. The pleckstrin homology domain: an intriguing multifunctional protein module. Bioessays. 1996 Jan;18(1):35–46. doi: 10.1002/bies.950180109. [DOI] [PubMed] [Google Scholar]
- Smith D. B., Johnson K. S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988 Jul 15;67(1):31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Sognier M. A., Neft R. E., Roe A. L., Eberle R. L., Belli J. A. Dot-blot hybridization: quantitative analysis with direct beta counting. Biotechniques. 1991 Oct;11(4):520–525. [PubMed] [Google Scholar]
- Takei K., McPherson P. S., Schmid S. L., De Camilli P. Tubular membrane invaginations coated by dynamin rings are induced by GTP-gamma S in nerve terminals. Nature. 1995 Mar 9;374(6518):186–190. doi: 10.1038/374186a0. [DOI] [PubMed] [Google Scholar]
- Thomas J. D., Sideras P., Smith C. I., Vorechovský I., Chapman V., Paul W. E. Colocalization of X-linked agammaglobulinemia and X-linked immunodeficiency genes. Science. 1993 Jul 16;261(5119):355–358. doi: 10.1126/science.8332900. [DOI] [PubMed] [Google Scholar]
- Timm D., Salim K., Gout I., Guruprasad L., Waterfield M., Blundell T. Crystal structure of the pleckstrin homology domain from dynamin. Nat Struct Biol. 1994 Nov;1(11):782–788. doi: 10.1038/nsb1194-782. [DOI] [PubMed] [Google Scholar]
- Touhara K., Inglese J., Pitcher J. A., Shaw G., Lefkowitz R. J. Binding of G protein beta gamma-subunits to pleckstrin homology domains. J Biol Chem. 1994 Apr 8;269(14):10217–10220. [PubMed] [Google Scholar]
- Tsukada S., Simon M. I., Witte O. N., Katz A. Binding of beta gamma subunits of heterotrimeric G proteins to the PH domain of Bruton tyrosine kinase. Proc Natl Acad Sci U S A. 1994 Nov 8;91(23):11256–11260. doi: 10.1073/pnas.91.23.11256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuma P. L., Stachniak M. C., Collins C. A. Activation of dynamin GTPase by acidic phospholipids and endogenous rat brain vesicles. J Biol Chem. 1993 Aug 15;268(23):17240–17246. [PubMed] [Google Scholar]
- Vihinen M., Iwata T., Kinnon C., Kwan S. P., Ochs H. D., Vorechovský I., Smith C. I. BTKbase, mutation database for X-linked agammaglobulinemia (XLA). Nucleic Acids Res. 1996 Jan 1;24(1):160–165. doi: 10.1093/nar/24.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihinen M., Zvelebil M. J., Zhu Q., Brooimans R. A., Ochs H. D., Zegers B. J., Nilsson L., Waterfield M. D., Smith C. I. Structural basis for pleckstrin homology domain mutations in X-linked agammaglobulinemia. Biochemistry. 1995 Feb 7;34(5):1475–1481. doi: 10.1021/bi00005a002. [DOI] [PubMed] [Google Scholar]
- Warnock D. E., Terlecky L. J., Schmid S. L. Dynamin GTPase is stimulated by crosslinking through the C-terminal proline-rich domain. EMBO J. 1995 Apr 3;14(7):1322–1328. doi: 10.1002/j.1460-2075.1995.tb07118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendoloski J. J., Shen J., Oliva M. T., Weber P. C. Biophysical tools for structure-based drug design. Pharmacol Ther. 1993 Nov;60(2):169–183. doi: 10.1016/0163-7258(93)90005-x. [DOI] [PubMed] [Google Scholar]
- Yoon H. S., Hajduk P. J., Petros A. M., Olejniczak E. T., Meadows R. P., Fesik S. W. Solution structure of a pleckstrin-homology domain. Nature. 1994 Jun 23;369(6482):672–675. doi: 10.1038/369672a0. [DOI] [PubMed] [Google Scholar]
- Zhang O., Kay L. E., Olivier J. P., Forman-Kay J. D. Backbone 1H and 15N resonance assignments of the N-terminal SH3 domain of drk in folded and unfolded states using enhanced-sensitivity pulsed field gradient NMR techniques. J Biomol NMR. 1994 Nov;4(6):845–858. doi: 10.1007/BF00398413. [DOI] [PubMed] [Google Scholar]
- Zhang P., Talluri S., Deng H., Branton D., Wagner G. Solution structure of the pleckstrin homology domain of Drosophila beta-spectrin. Structure. 1995 Nov 15;3(11):1185–1195. doi: 10.1016/s0969-2126(01)00254-4. [DOI] [PubMed] [Google Scholar]
- Zheng J., Cahill S. M., Lemmon M. A., Fushman D., Schlessinger J., Cowburn D. Identification of the binding site for acidic phospholipids on the pH domain of dynamin: implications for stimulation of GTPase activity. J Mol Biol. 1996 Jan 12;255(1):14–21. doi: 10.1006/jmbi.1996.0002. [DOI] [PubMed] [Google Scholar]
- Zhou M. M., Ravichandran K. S., Olejniczak E. F., Petros A. M., Meadows R. P., Sattler M., Harlan J. E., Wade W. S., Burakoff S. J., Fesik S. W. Structure and ligand recognition of the phosphotyrosine binding domain of Shc. Nature. 1995 Dec 7;378(6557):584–592. doi: 10.1038/378584a0. [DOI] [PubMed] [Google Scholar]
- de Weers M., Mensink R. G., Kraakman M. E., Schuurman R. K., Hendriks R. W. Mutation analysis of the Bruton's tyrosine kinase gene in X-linked agammaglobulinemia: identification of a mutation which affects the same codon as is altered in immunodeficient xid mice. Hum Mol Genet. 1994 Jan;3(1):161–166. doi: 10.1093/hmg/3.1.161. [DOI] [PubMed] [Google Scholar]
- van Nuland N. A., Kroon G. J., Dijkstra K., Wolters G. K., Scheek R. M., Robillard G. T. The NMR determination of the IIA(mtl) binding site on HPr of the Escherichia coli phosphoenol pyruvate-dependent phosphotransferase system. FEBS Lett. 1993 Jan 2;315(1):11–15. doi: 10.1016/0014-5793(93)81122-g. [DOI] [PubMed] [Google Scholar]